- 1Hebei Key Laboratory of Environment and Human Health, Department of Epidemiology and Statistics, School of Public Health, Hebei Medical University, Shijiazhuang, China
- 2The Hebei Key Laboratory of Gastroenterology, Division of Gastroenterology, The Second Hospital of Hebei Medical University, Shijiazhuang, China
- 3Antenatal Diagnosis Center, The Fourth Hospital of Shijiazhuang, Shijiazhuang, China
- 4Hebei Key Laboratory of Environment and Human Health, Department of Social Medicine and Health Care Management, School of Public Health, Hebei Medical University, Shijiazhuang, China
- 5School of Basic Medicine, Hebei Medical University, Shijiazhuang, China
Oxidative stress is closely related to the occurrence and development of various diseases such as cancer, diabetes, and cardiovascular and infectious diseases. We identified six critical genetic variants related to oxidative stress, and evaluated their main effects and their interaction effects on hepatitis B virus (HBV)-induced liver diseases. We enrolled 3,128 Han Chinese subjects into five groups: healthy controls, chronic hepatitis B (CHB), liver cirrhosis (LC), hepatocellular carcinoma (HCC), and natural clearance. We then determined the genotypes in each group for CYBA-rs4673, NCF4-rs1883112, NOX4-rs1836882, rs3017887, SOD2-rs4880, and GCLM-rs41303970, and evaluated the association between these variants and HBV-induced liver diseases. Gene-gene interactions were evaluated using generalized multifactor dimensionality reduction, logistic regression, and four-by-two tables. Significant associations were observed between healthy controls and the CIB group (CHB+LC+HCC). The CYBA-rs4673AG genotype was associated with a 1.356 rate of susceptibility of HBV-induced liver disease compared to the wild type GG genotype. The NCF4-rs1883112G allele occurred more frequently in healthy controls than in the CIB group in all three models (dominant, codominant, and recessive). Nox4-rs1836882 TC showed a protective association, being more frequent in healthy controls compared to the wild type TT genotype. GCLM-rs41303970A was associated with HBV-induced liver disease. The overall best model by multifactor dimensionality reduction was a five factor interaction model that had the highest cross validation consistency (10/10) and test accuracy (0.5669), P = 0.001. Oxidative stress-related gene polymorphisms are likely to be associated with HBV-induced liver disease, suggesting that information on these variations is useful for risk assessment of HBV-induced liver disease.
Introduction
Oxidative stress (OS) is the process by which the body is stimulated by harmful exogenous or endogenous factors (Patterson et al., 2019). Reactive oxygen species (ROS)-induced oxidative DNA damage can also increase chromosomal aberrations that are closely related to the occurrence and development of infectious diseases, cancer, diabetes, and cardiovascular diseases (Fetoni et al., 2019).
There is evidence that hepatitis B virus (HBV) can induce a pro-oxidative state. Sulfhydryl and lipid peroxidation are generated in CHB patients by increasing the OS level (Duygu et al., 2012). In addition, ROS products are increased in carriers with high HBV DNA titers (Fujita et al., 2008). In vitro and in vivo experiments confirm that HBV infection can induce OS occurrence. A similar increase is also found in chronic hepatitis B (CHB) patients who have significantly higher total peroxide levels, which is considered a parameter of OS, compared to asymptomatic carriers indicate that OS plays a critical role in hepatic injury (Ha et al., 2010). These findings indicate that OS, which is provoked in patients with viral risk factors including HBV, plays critical roles in the pathogenesis of HBV-induced liver diseases such as CHB, liver cirrhosis (LC), and even hepatocellular carcinoma (HCC) (Xianyu et al., 2018). Susceptibility to OS is partly determined by genetic background, also twin studies and segregation analysis support the role of genetic polymorphisms in impacting the host response to HBV infection. Therefore it may be beneficial to detect whether the following gene polymorphisms that regulate key oxidation-reduction enzymes affect the occurrence of HBV-related liver disease.
Nicotinamide adenine dinucleotide phosphate (NADPH) oxidases, also known as nitrogen oxides (Nox), consist of Nox1, Nox2, Nox3, Nox4, Nox5, dual oxidase 1 (Duox1), and Duox2; these oxides transfer electrons through membranes and catalyze the NADPH-dependent reduction of O2 to O2− to produce ROS (Choi et al., 2014). NADPH oxidase consists of six different subunits, a catalytic core formed by p22phox (encoded by the CYBA gene) and Nox2, and four cytoplasmic subunits responsible for activation: p40phox (encoded by the NCF4 gene), p47phox, p67phox, and rac2 (encoded by RAC2 gene). Of the catalytic NADPH oxidase subunits, Nox4 (Entrez Gene: 50,507) is the most widely distributed isoform. Together with Nox1 and Nox2, Nox4 is expressed in the liver, and the biological functions of all three Nox subunits need to be combined with another membrane-associated protein subunit, p22phox (Altenhofer et al., 2012). The C242T transformation (rs4673) in the CYBA gene encoding the p22phox protein can regulate NADPH oxidase activity. The C allele is related to a significant increase in enzyme activity and OS (Tang et al., 2017). Recently, rs1836882 in the Nox4 gene promoter region was found to modulate the association between dietary caloric intake and ROS levels in peripheral blood mononuclear cells (PBMCs) (Liu et al., 2013). Another single-nucleotide polymorphism (SNP), rs3017887 in the Nox4 gene 5′ UTR region, was chosen as it is the most informative SNP (can capture 12 of 124 SNPs with a minor allelic frequency of at least 0.05) in a haplotype block composed of four SNPs possibly related to oxidative burden (Lim et al., 2009). Another NADPH oxidase polymorphism is 212A>G (rs1883112), located in the NCF4 promoter (encoding p40phox), and is involved in NADPH oxidase down-regulation (Lopes et al., 2004).
ROS clearance in tissues depends on manganese-dependent superoxide dismutase (MnSod), which can catalyze the transformation of active superoxide radicals to hydrogen peroxide and plays an important role in anti-OS response. This mitochondrial enzyme is encoded by the nuclear SOD2 gene. The substitution of Ala for Val on the 16th amino acid in the signal sequence (Val16Ala, rs4880) leads to a decrease in the efficiency of MnSOD entering the mitochondrial matrix and weakens the ability of cells to neutralize superoxide radicals (Wang et al., 2018).
Glutathione (GSH) is the most effective and abundant low molecular weight thiol antioxidant. As a key enzyme in GSH metabolism, the higher expression of glutamate-cysteine ligase (GCL) can significantly reduce the ROS content in tumor cells, and reduce the proliferation rate of tumor cells. GCL is composed of a catalytic subunit (GCLC) and a modifier subunit (GCLM) (Rains and Jain, 2011). The C588T polymorphism (rs41303970) in the GCLM gene promoter suppresses GCLM gene expression, whereas low GCLM gene expression is associated with lower plasma GSH levels and enhances OS (Nakamura et al., 2002). In the current study, we tried to clarify whether this polymorphism was involved in HBV predisposition.
Considering the role of OS in the formation and progression of HBV-induced liver diseases, it is important to look for genetic markers capable of identifying individuals at risk for disease progression. Moore (2003) suggests that gene-gene interactions are more important than the independent main effects of any single susceptibility gene, and may synergistically or antagonistically contribute to the increased risk of disease. Therefore, in addition to studying the main effect of these six gene loci, we also verified the interaction effects between them using three different modeling methods.
Materials and Methods
Participants
A total of 3,128 unrelated individuals of Han Chinese were recruited from the First, Second, and Fourth Affiliated Hospital of Hebei Medical University and Infectious Disease Hospital of Shijiazhuang City between 2009 and July 2016. The samples included 840 healthy individuals, 691 cases of CHB, 680 cases of LC, 421 HBV-related HCC patients, and 496 HBV natural clearances. The CIB group consisted of the combined values from the CHB, LC, and HCC groups. All participants self-reported as living in the Hebe province of China for 6 months or more.
Healthy individuals were free of hepatitis B surface antigen (HBsAg), anti-HBs, antibodies to hepatitis B core (anti-HBc), and other HBV biomarkers, and normal biochemical parameters as validated by routine blood examinations. They possessed no history of cardiovascular events, diabetes, liver- and kidney-associated diseases, or other autoimmune diseases. HBV natural clearances were defined as being negative for HBsAg plus anti-HBs positive, or positive for both anti-HBs and anti-HBc based on normal liver function. Patients (including CHB, LC, and HCC) were diagnosed by being HBsAg positive, anti-HBc positive, and anti-HBe or HBeAg positive for 6 months or more. The diagnostic criteria for CHB was positivity for HBV-DNA and elevation of alanine aminotransfease (ALT) or aspartate aminotransferase (AST), a histopathological diagnosis of liver injury either by ultrasonography or laboratory testing. LC diagnosis depended on clinical, histological, biochemical, or radiological findings (Levesque et al., 2019). HCC was diagnosed based on magnetic resonance imaging of the liver and/or computed tomography according to published guidelines for HCC diagnosis and management (Sastre et al., 2015). Patients with hepatitis C virus (HCV), alcoholic liver disease, human immunodeficiency virus, or suspected autoimmune diseases were excluded from this study.
The following information was collected for all subjects: basal information (age, gender, history of smoking or drinking, and history of HBV vaccination), disease background (course of the disease, presence or absence of other complications), and biochemistry examination results (five hepatitis B markers, quantity of HBV-DNA, ALT, AST, and AFP concentrations).
The research protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki and has been approved by the Ethics Committee of Hebei Medical University. All subjects signed an informed consent before being included in the study.
Selection of Gene Polymorphisms
Candidate SNPs were screened by searching the Ensembl genome browser (http://asia.ensembl.org/index.html), UCSC Genome Browser Home (http://genome.ucsc.edu/), NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP). Criteria for selecting SNPs included: i) SNPs have not been shown to be associated with any HBV-induced disease in Chinese individuals; ii) the minor allele frequency (MAF) of SNPs was greater than 5% in the northern China population; iii) functional SNPs of exons, promoters, or the 3' UTR and 5' UTR regions of genes were preferred; iv) upstream gene variants and splice region variants were also defined as functional SNPs; and v) SNPinfo Web Server was used to predict the function of multiple sites. A total of six SNPs met these criteria (Table 1).
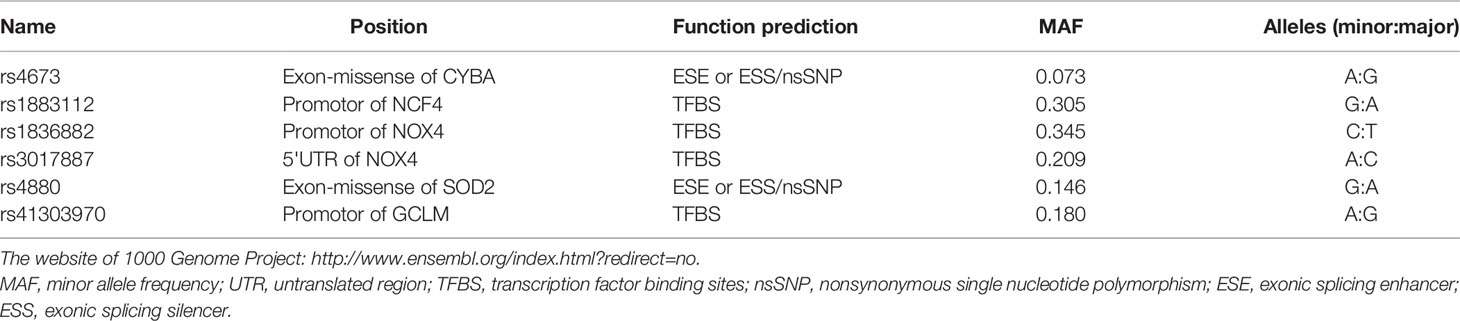
Table 1 Background information on selected single-nucleotide polymorphisms in Han Chinese people downloaded from 1000 Genome.
Genotyping
Two milliliter venous blood samples from all subjects were collected under ethylene diamine tetraacetic acid (EDTA) anticoagulation conditions. Genomic DNA Purification Kits from Promega (San Luis Obispo, CA, USA) were used for genomic DNA extraction, and time of flight mass spectrometry technology (MALDI-TOF MS) from Sequenom Inc. (San Diego, CA, USA) was used for SNP genotyping. Primers for the six SNP alleles were designed by the Bio Miao Biological Company (Beijing, China). Experimental data were processed using MassARRAY Typer 4.0.5 Software (Sequenom). The software determined that more than 97% of the genotyping results of the six SNPs were of conservative (high quality). The genotyping call rate of rs4673, rs1883112, rs1836882, rs3017887, rs4880, and rs41303970 were 99.46, 97.35, 98.95, 99.23, 97.92, and 97.92%, respectively. The genotyping call cluster plots of six SNPs are shown in Supplementary Figure 1. For quality control, 1% of each plate was reserved for template positive and negative controls, and repetitive genotyping was randomly selected for 10% of samples. The consistency of two genotyping results reached 100%.
Statistical Analysis
Statistical analyses were performed using SPSS version 21.0 for windows (SPSS Inc., Chicago, IL, USA). A two-sided P value of 0.05 was considered statistically significant in all analyses. Continuous variables of the normal distribution are shown as the mean ± SD. Categorical variables are presented as frequencies. Continuous data were compared using Student's t-test and Wilcoxon test. Univariate and multivariate logistic regression analyses were performed to determine the differences in categorical variables between groups. In addition, multivariate analysis was used to correct for confounding factors. To evaluate the influence of SNPs, we set up three models (codominant, dominant, and recessive). Considering the wild type of a SNP as (w) and the variant as (v), then in the codominant model, genotypes w/w, w/v, and v/v were coded as 0, 1, and 2, respectively. Genotype w/w was coded as 0, and w/v and v/v were both coded as 1 in a dominant model. Similarly, when a recessive effect was assumed, w/w and w/v were both scored as 0, and v/v was scored as 1.
Hardy-Weinberg expectations (HWE) were analyzed with SNPStats software (Sole et al., 2006) using the codominant model. Haplotype analysis was conducted by Haploview v4.2 (Barrett et al., 2005). Gene-gene interactions were initially analyzed by the multifactor dimensionality reduction (MDR) algorithm, which was applied using the generalized MDR (GMDR) software package (v0.9). The statistical significance of the MDR models was evaluated by permutation testing. Multiplicative interaction effects were examined with multiple logistic regression analysis. The model included two polymorphisms (two-way) and their interactive term to calculate the P-value and odds ratio (OR) for each gene-gene interaction. For better interpretation of the results, we defined each genotype as a binary variable (risk genotype versus non-risk genotype). Additivity interaction was evaluated using a four-by-two table. Like the variable assignment method of the multiplicative model, risk genotypes and non-risk genotypes were classified into two categories. Three effect indices were used to evaluate the additive model: Synergy index (S), attributable proportion of interaction (AP), and relative excess risk of interaction (RERI). Detailed description of the calculation methods for the three indices are described by Andersson et al. (2005). In the absence of interaction, 95% CI of RERI and AP include 0, 95% CI of S include 1.
Results
Demographic Characteristics of the Samples and Candidate Single-Nucleotide Polymorphism Information
Supplementary Table 1 describes the demographic characteristics and laboratory parameters of the patients and control subjects. There were significant differences in age, sex, smoking habits, and alcohol consumption among the groups. There were also significant differences in the course of the disease, and the HbeAg positive rate between the three patient groups (P < 0.001). Table 1 lists the background information of the candidate SNPs in our study, including the loci position, function prediction, MAF, and mutation base. According to the HWE test (Table 2), there was no difference between the observed genotype distribution and the expected distribution of the six SNPs. The MAF values of these six SNPs were consistent with the frequencies listed in the Ensembl database.
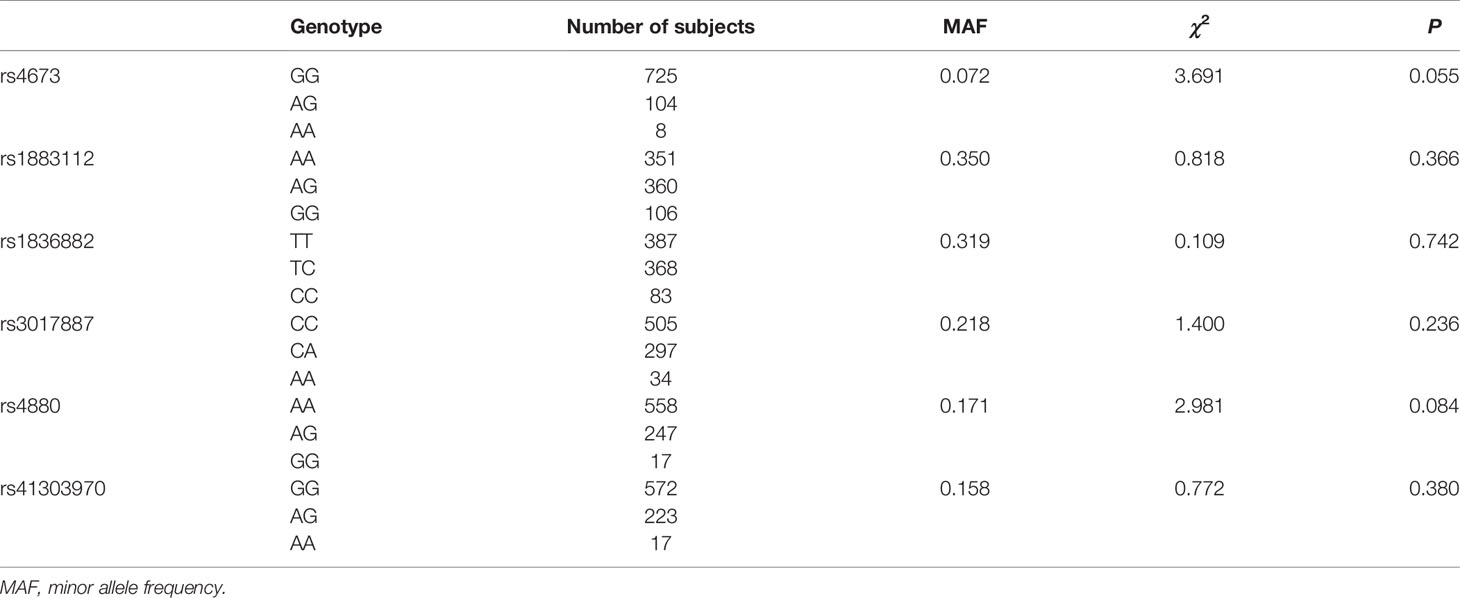
Table 2 Test of Hardy-Weinberg Equilibrium for the six single-nucleotide polymorphisms in healthy individuals.
Association of Susceptibility of Hepatitis B Virus-Induced Liver Disease and Candidate Single-Nucleotide Polymorphisms
Table 3 lists the distribution of candidate SNPs genotypes in the CIB and healthy groups. Adjusted logistic regression analysis of factors including age, gender, smoking habits, and alcohol consumption suggested that rs4673 in CYBA was correlated with susceptibility to HBV-induced liver disease. In the codominant genetic model, AG genotype carriers showed a higher risk of HBV-induced liver disease than GG genotype carriers (OR: 1.356, P = 0.016), with the same relationship (OR: 1.315, P = 0.026) in the dominant genetic model. Using multivariable logistic analysis, the protective association of the NCF4-rs1883112G allele was more frequent in healthy controls than in the CIB group in all three models (adjusted OR = 0.815, P = 0.027 for AG, OR = 0.707, P = 0.015 for GG in the codominant model; corrected OR = 0.790, P = 0.007 in the dominant model; adjusted OR = 0.733, P = 0.029 in the recessive model). The same beneficial action of rs1836882TC on HBV-induced liver disease was shown in the codominant genetic model (adjusted OR = 0.798, P = 0.014), and in the dominant genetic model (adjusted OR = 0.805, P = 0.013). SOD2-rs4880G was significantly associated with susceptibility of HBV-induced liver disease under univariable analysis in the dominant genetic model, but after adjusting for confounders, the loci’s protective effect was not significant (P = 0.080). With regards to the GCLM gene, compared to reference subjects with the rs41303970G allele, carriage of the A allele was associated with a higher risk of HBV-induced liver disease susceptibility (adjusted OR = 1.916, P = 0.022 in the codominant model, and adjusted OR = 1.976, P = 0.016 in the recessive model). There was no association detected between Nox4-rs3017887 and HBV-induced liver disease in any models.
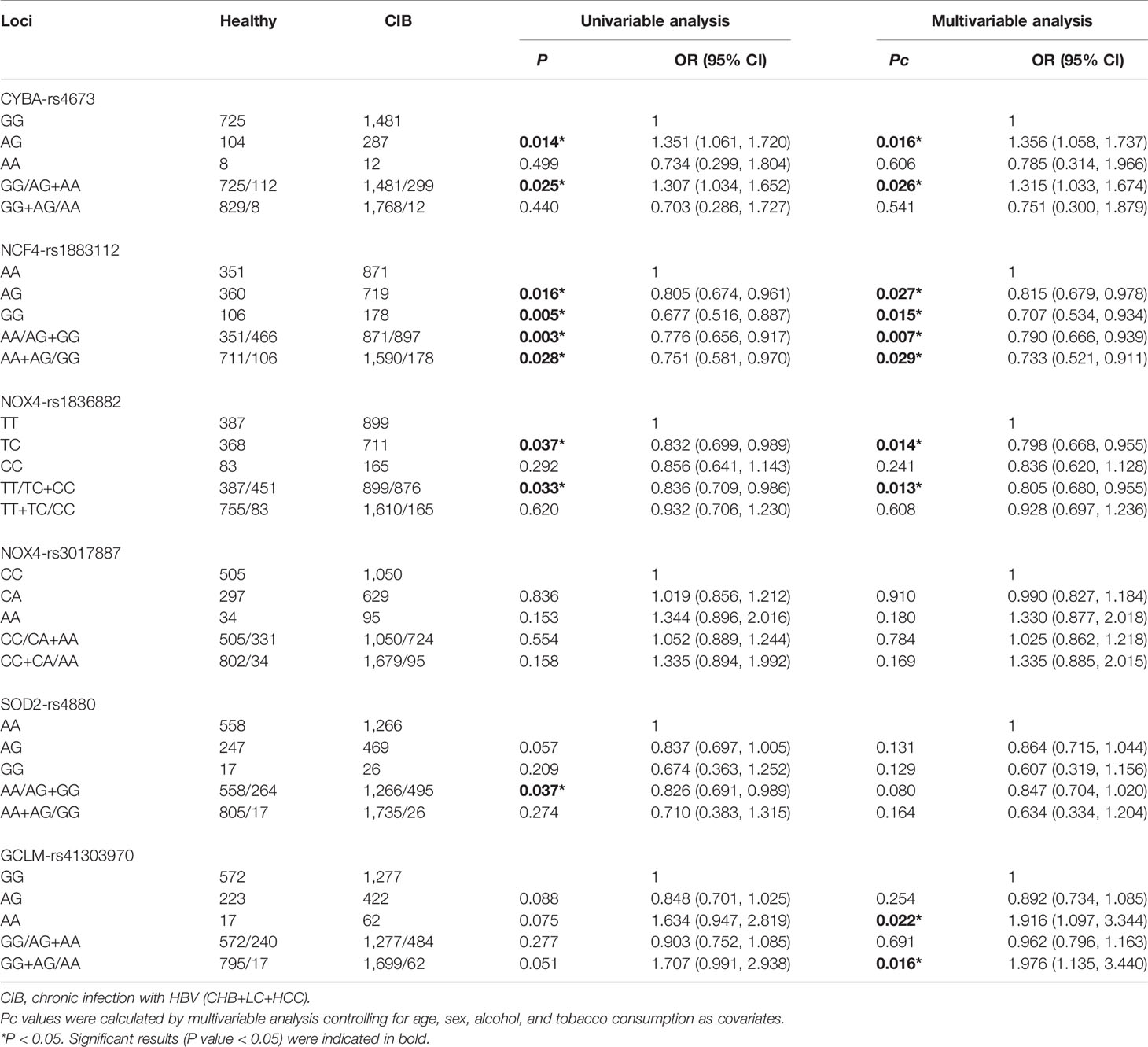
Table 3 Logistic analysis of predictive factors with regard to the risk of hepatitis B virus-induced liver disease between healthy individuals and CIB group.
Rs1836882 and rs3017887 are located on the same chromosome and, therefore, their close physical location resulted in linkage disequilibrium (LD), which required haplotype block analysis to determine their relationship to susceptibility of HBV-induced liver disease. Supplementary Figure 2 illustrates that they were in weakened LD. Three haplotypes, ht1(CT), ht2(CC), ht3(AT), accounted for 100% of the distribution and are shown in Table 4. The protective association of the CT haplotype was more frequent in the control group than the CIB group, but the difference was not significant (P = 0.063).

Table 4 Haplotype analysis for two SNPs within Nox4 gene between CIB and healthy individuals by Haploview.
Association of Hepatitis B Virus Natural Clearance and Candidate Single-Nucleotide Polymorphisms
Supplementary Table 2 lists the distribution of the six SNP genotypes for the CIB and natural clearance groups. With the CIB subjects as cases, and natural clearance individuals as a control group, we found no significant genotypic differences in the six SNP alleles.
Association of Hepatitis B Virus-Induced Liver Disease Progression and Candidate Single-Nucleotide Polymorphism
Supplementary Table 3 lists the genotype distribution of the six SNPs for the CHB and LC groups. No positive results were obtained. Supplementary Table 4 shows the distribution of genotypes for the HCC group and CHB+LC group. These SNPs were ineffective as a tool to differentiate HCC from CHB and LC in the northern Chinese Han population.
Gene-Gene Interaction in Susceptibility of Hepatitis B Virus-Induced Liver Disease
In the above analysis, almost all candidate SNPs were associated with susceptibility of HBV-induced liver disease, but not with HBV clearance and disease progression. Therefore, interaction analysis was performed only in the CIB and healthy control groups. In the GMDR analysis, all three genotypes of the six SNPs were assigned to 0, 1, 2, according to their pathogenic risk. Table 5 shows the results of GMDR analysis: the overall best model by MDR was the five-factor interaction model that had the highest cross validation consistency (10/10) and test accuracy (0.5669), P = 0.001. The risk of HBV-induced liver disease in the “high-risk” population was 2.221 (1.384, 3.341) times that of the “low-risk” population. Supplementary Figure 3 presents the distribution of high-risk and low-risk genotypes in the best five-locus model. Supplementary Table 5 lists the results of the multiplicative interaction analysis of SNPs with HBV-induced liver disease by logistic regression. We found no interaction between the 15 pairs of loci. However, a trend of multiplicative interaction was detected between rs1883112 and rs1836882 (P = 0.069) that was consistent with the results of the GMDR analysis. Similarly, the additive model did not find any significant interaction terms (Supplementary Table 6: 95% CI of RERI and AP included 0, 95% CI of S included 1).
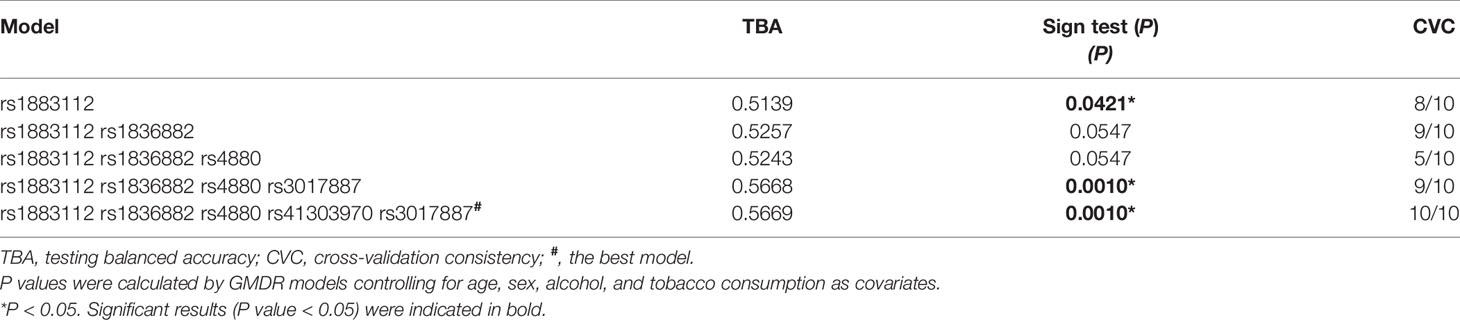
Table 5 Generalized multifactor dimensionality reduction models of multi-genes of hepatitis B virus-induced liver disease.
Discussion
Although OS susceptibility is partly determined by genetic background, few studies have been performed on the relationship between functional gene polymorphisms of key enzymes regulated by redox and the risk of HBV-induced liver disease. Consequently, identifying the missing heritability of key enzymes regulated by redox has become a challenging task for HBV-induced liver disease research. In addition, genetic interaction is another significant source of missing heredity. Based on the above hypothesis, we initially assessed the relationship between six genetic polymorphisms related to OS and HBV-induced liver disease in 3,128 samples through four case-control studies.
The exon-missense polymorphism of CYBA rs4673 results in a substitution of histidine for tyrosine and an alteration of NADPH activity by destroying the heme binding site (Mata-Balaguer et al., 2004). Rs4673 has been widely demonstrated to be related to coronary artery disease (CAD) (Mazaheri et al., 2017), metabolic disorders (Snahnicanova et al., 2018), and cancer (Megias-Vericat et al., 2018). The association between this locus and HBV-induced liver disease has been studied here for the first time. We therefore inferred the soundness of our results from the function of this locus. Bioinformatics analysis (Mazaheri et al., 2017) indicates a significant impact of rs4673 on the structure and function of the p22phox protein. It was demonstrated that substitution of tyrosine by histidine on codon 72 of p22phox protein results in a functional mutated protein. Hydrophobicity analysis shows that the hydrophobicity of codon 72 decreases from −0.311 to −0.522 after Y72H substitution. The proportion of sheet structures decreases from 57.9 to 55.4% in the mutated protein. However, the rs4673 polymorphism has no effect on the structure of the messenger (mRNA). Several teams have researched the biological function of this locus. A study found that carriers of the T(A) allele have reduced enzymatic activity both in stimulated human blood vessel cells and basal cells (Guzik et al., 2000). However, another in vitro test showed that the mutant T(A) allele produces higher levels of ROS (Shimo-Nakanishi et al., 2004). Schirmer et al. (2008) found no changes in enzymatic activity owing to mutations at these loci. As the functional study results are highly controversial, the relationship between the mutation site and disease is also controversial. Our findings suggest that the AG (TC) genotype carriers had a higher risk of HBV-induced liver disease than GG (CC) genotype carriers, but there was no significant difference between the AA (TT) and GG (CC) genotypes in the two groups. Overall comparison of the A(T) allele with the G(C) allele produced a non-significant risk estimate for HBV-induced liver disease. These results should be verified with a larger sample size in other ethnic groups.
Another NADPH oxidase subunit analyzed in this study was p40phox, which is involved in the down-regulation of NADPH oxidase (Lopes et al., 2004) and is coded by the NCF4 gene. Rs1883112 is located in the promoter of NCF4 and potentially affects gene expression and ROS generation. Schirmer et al. (2008) found that this SNP does not affect NCF4 mRNA expression in stimulated granulocytes and basal cells. According to generalized estimating equations (GEE) analysis, NCF4 rs1883112 is an SNP in the p40phox gene regulating NAD(P)H oxidase (Quinn and Gauss, 2004; Olsson et al., 2007). It is an independent host factor and can protect the body from infection, blood toxicity, and cardiotoxicity. Gustafson came to a similar conclusion. He proved that rs1883112 is the representative of nearby SNP, which has a functional effect on NADPH oxidase activity (Gustafson et al., 2014). As with rs4673, the effect of this polymorphism in HBV-induced liver disease was unknown prior to the current study. We found that the rs1883112G allele was more frequent in healthy controls than the CIB group and thus had a protective effect. The protective effect of the variant G allele has been demonstrated in other diseases, and is related to higher rate of disease-free survival in diffuse large B-cell lymphoma (DLBCL) (Rossi et al., 2009) and aggressive B-cell non-Hodgkin's lymphoma (NHL) (Gustafson et al., 2014). For acute myeloid leukemia (AML) patients, the rs1883112 variant G allele is related to higher frequency of complete remission (Megias-Vericat et al., 2018). Because the variant allele has been shown to be protective in different diseases, functional studies on this locus are strongly encouraged.
As the most widely distributed isoform of NADPH oxidase subunits, the Nox4 gene is reported to be associated with a variety of deleterious effects, including cell senescence and apoptosis, angiogenesis, vascular aging, atherosclerosis (Chen et al., 2012), and other malignancies (Fitzgerald et al., 2012). Jiao et al. (2012) found that Nox4 increases ROS generation, reduces insulin gene expression, and increases β-cell apoptosis both in mice and in vitro. The relationship between certain liver diseases and Nox4 has also been confirmed (de Mochel et al., 2010; Choi et al., 2014; Rabelo et al., 2018). Rs1836882 and rs3017887 are located in the Nox4 promoter and the 5' UTR region, respectively, and may alter the transcriptional activity of the Nox4 gene, thus being included in our research. Liu et al. (2013) suggested that the Nox4 promoter region carrying rs1836882-TT genotype has a higher binding affinity for HNF3γ protein and a lower transcriptional activity than with the other two genotypes. By silencing the Nox4 gene or over-expressing HNF3γ, compared with the CT or CC genotypes, the level of ROS is more reduced in the PBMC of high calorie dieters with the TT genotype. In our population study, we found that the TC genotype was significantly more frequent in the healthy group than the CIB group compared to the TT genotype. However, there was no statistical difference in the distribution of CC and TT genotypes between the two groups. Overall comparison of the C allele with the T allele produced a non-significant risk estimate for HBV-induced liver disease. These results are inconsistent with Liu's proposed mechanism. The reason may be that the serum samples in Liu's study were all taken from people on high-calorie diets and experiments were not conducted in a large unbiased sample set. McCaughan et al. (2014) found that this locus is associated with new-onset diabetes after renal transplantation. No other research has been performed on this locus. We did not find that rs3017887 was related to HBV-induced liver disease. Siqueira et al. (2015) reached a similar conclusion, and no association between the locus and fibrosis in HCV patients was found in his study. Rabelo et al. (2018) confirmed that the rs3017887 locus was not associated with nonalcoholic steato-hepatitis (NASH). Because the samples of these three studies belong to different liver diseases, repetitive experiments using a large number of samples must be carried out in different races.
Superoxide dismutase (SOD) is one of the most important antioxidants in human beings. SOD2 is the only form that has been certified to be critical for the survival of aerobic life. Genetic mutation of SOD2 was reported to affect SOD activity, of which the most important was rs4880. Rs4880 has been researched as a possible predisposing factor in several diseases where OS is deemed to play a key role in pathogenesis, including drug induced liver injury (Lucena et al., 2010), non-alcoholic fatty liver disease, cancers, and alcoholic liver diseases (Huang et al., 2016). Several papers report a relationship between this locus and HCV-related cirrhosis and HCC (Nahon et al., 2007; Jin et al., 2011). To our knowledge, this is the first report to study the effect of SOD2 polymorphism on HBV-related disease. We found that the AA genotype confers a susceptibility to HBV-induced liver disease compared to the AG and GG genotypes in univariable analysis, but there was no difference between the two groups in genotype distribution when assessed using multivariable analysis. The A to G base substitution at rs4880 results in the conversion of alanine to valine in the leader amino acid sequence responsible for mitochondrial enzyme localization (Crawford et al., 2012). Xu (Xu et al., 2017) found that the A allele is related to a decrease in SOD2 mRNA and protein levels. Wang et al. (2012) confirmed the down-regulation of SOD2 and GSH peroxidase expression in HBV transgenic mice. Namiduru et al. (2010) also found a decrease in SOD2 activity in patients with chronic hepatitis B and C. The results of these functional studies are consistent with those of our population study. In our next phase of work, bioinformatics analysis will be performed to evaluate the impact of the polymorphism on structure and function of the corresponding protein using PolyPhen-2 and SNAP software. In addition, the impact of the Val16Ala-SOD2 polymorphism on protein hydrophobicity will be evaluated through the Kyte-Doolittle scale. RNAsnp web server will be used to evaluate the impact of rs4880 transitions on mRNA structure.
Rs41303970-T is located in the promoter region of the GCLM gene, suppresses up-regulation of GCLM gene expression, and is related to lower plasma GSH levels (Nakamura et al., 2002). Patients with HBV infection showed significantly lower serum GSH levels than healthy individuals (P < 0.01). Nakamura confirmed lower promoter activities in cells for GCLM-rs41303970T allele carriers. Therefore, the functional consequence predicted from the SNP would be lower GCLM protein level in carriers of the variant allele, which we hypothesized may lead to a decrease in GSH synthesis. There is evidence for tissue-specific regulation of rs41303970, where the variant allele shows lower expression in all tissues (data was obtained from the GTEx Portal data base). Samples suitable for gene expression analysis will be necessary for further investigation of this hypothesis. Previous studies show that this mutation is associated with the development of a variety of diseases, such as ischemic heart disease, type 2 diabetes (Katakami et al., 2010), and asthma (Polonikov et al., 2007). Although the disease types differ, their conclusions are consistent with ours: rs41303970T is the susceptible base for these diseases, and is consistent with the functional studies of this locus.
The MDR method simplifies high-dimensional genetic data into one-dimension, making gene-gene and gene-environmental interactions detection possible in relatively small sample size with high statistical power. The GMDR method used in our study has the advantage of controlling confounding factors compared to the conventional MDR method. The interaction of testing SNPs using GMDR showed a five factor model with a cross validation consistency of 10 as the strongest risk predictor to HBV-induced liver disease. Six SNPs of the five genes we selected were all OS-related variants, the functional significance of these polymorphisms was confirmed by experimental, case-control studies, or cohort studies. In addition, there is a close connection between these genes; for example, the biological functions of Nox4 need to be combined with another membrane-associated protein subunit, p22phox. Therefore, they may influence each other in terms of pathogenicity. We further tested two-way interaction by multiplicative interaction and additive interaction analysis. Tendency toward multiplicative interaction were detected between NCF4-rs1883112 and Nox4-rs1836882 (P = 0.069), which was consistent with the two-factor MDR method model (P = 0.0547). Researchers are displaying a growing interest in detecting the genetic interactions that may affect HBV infection susceptibility (Bose et al., 2013; He et al., 2015). However, there are still several factors that limit the analysis of genetic interaction. First, at present there is no optimal analysis method, and different analytical models may lead to conflicting results. Secondly, as statistical interactions are not equivalent to biological interactions, genetic interactions are difficult to explain, which may be the biggest limiting factor hindering this series of studies. However, detection of genetic epistasis may provide new insights into the biological and biochemical pathways that illuminate disease mechanisms, and aid in generating new pathogenesis hypotheses.
This is the first study to examine OS-related gene polymorphisms implicated in susceptibility of HBV-induced liver disease, clearance, and progression in the Han Chinese population, and found that CYBA-rs4673, NCF4-rs1883112, Nox4-rs1836882, and GCLM-rs41303970 are associated with HBV-induced liver disease. It is worth noting that the data obtained from this study should be carefully interpreted and we must note several limitations. To verify these relationships and interactions, larger sample sizes in different ethnic groups are needed for further research. In addition, functional validation of statistically significant SNPs is needed to determine their biological significance. Last but not least, we do not know the significance and biological mechanism underlying the interaction with susceptibility of HBV-induced liver disease. It will be interesting to estimate the functional relationship between SNPs identified in the interaction model and to explore their possible effects on HBV-induced liver disease. However, current biological function trials and biochemical information are still limited. We can only conclude that Nox4-rs1836882, rs3017887, NCF4-rs1883112, SOD2-rs4880, and GCLM-rs41303970 have statistically significant interactive functions in the development of HBV-induced liver disease. Despite the above limitations and concerns, the analysis of gene interactions can provide important avenues of research for discovering biological pathways and mechanisms of disease occurrence, thus forming scientific hypotheses (Cordell, 2009).
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Hebei Medical University. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
Concept and design: NM, DL, and LY. Writing of article: NM. Statistical analysis: NM and LZ. Specimen collection and questionnaire investigation: XG, FY, WG, PG, JZ, MY, YiM, SQ, and LL. Inputing data: NM, WL, and XZ. SNP sample management: LW, YaM, and QS. Experiments and procedures: NM, WL, and XZ. Financial support: DL and LY.
Funding
This work was supported by the Project of Science and Technology Department of Hebei Province (172777123), Key Project of Natural Science Foundation of Hebei Province (H2016206576), National Natural Science Foundation of China (81601876), Science and Technology Research Project of Colleges and Universities in Hebei Province (QN2015006), Innovative experiment program for college students of Hebei Medical University (USIP2019211); National Natural Science Foundation of China (No. 81400322).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01290/full#supplementary-material
References
Altenhofer, S., Kleikers, P. W., Radermacher, K. A., Scheurer, P., Rob Hermans, J. J., Schiffers, P., et al. (2012). The NOX toolbox: validating the role of NADPH oxidases in physiology and disease. Cell Mol. Life Sci. 69, 2327–2343. doi: 10.1007/s00018-012-1010-9
Andersson, T., Alfredsson, L., Kallberg, H., Zdravkovic, S., Ahlbom, A. (2005). Calculating measures of biological interaction. Eur. J. Epidemiol. 20, 575–579. doi: 10.1007/s10654-005-7835-x
Barrett, J. C., Fry, B., Maller, J., Daly, M. J. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265. doi: 10.1093/bioinformatics/bth457
Bose, S., Tripathi, D. M., Sukriti, Sakhuja, P., Kazim, S. N., Sarin, S. K. (2013). Genetic polymorphisms of CYP2E1 and DNA repair genes HOGG1 and XRCC1: association with hepatitis B related advanced liver disease and cancer. Gene 519, 231–237. doi: 10.1016/j.gene.2013.02.025
Chen, F., Haigh, S., Barman, S., Fulton, D. J. (2012). From form to function: the role of Nox4 in the cardiovascular system. Front. Physiol. 3, 412. doi: 10.3389/fphys.2012.00412
Choi, J., Corder, N. L., Koduru, B., Wang, Y. (2014). Oxidative stress and hepatic Nox proteins in chronic hepatitis C and hepatocellular carcinoma. Free Radic. Biol. Med. 72, 267–284. doi: 10.1016/j.freeradbiomed.2014.04.020
Cordell, H. J. (2009). Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 10, 392–404. doi: 10.1038/nrg2579
Crawford, A., Fassett, R. G., Geraghty, D. P., Kunde, D. A., Ball, M. J., Robertson, I. K., et al. (2012). Relationships between single nucleotide polymorphisms of antioxidant enzymes and disease. Gene 501, 89–103. doi: 10.1016/j.gene.2012.04.011
de Mochel, N. S., Seronello, S., Wang, S. H., Ito, C., Zheng, J. X., Liang, T. J., et al. (2010). Hepatocyte NAD(P)H oxidases as an endogenous source of reactive oxygen species during hepatitis C virus infection. Hepatology 52, 47–59. doi: 10.1002/hep.23671
Duygu, F., Karsen, H., Aksoy, N., Taskin, A. (2012). Relationship of oxidative stress in hepatitis B infection activity with HBV DNA and fibrosis. Ann. Lab. Med. 32, 113–118. doi: 10.3343/alm.2012.32.2.113
Fetoni, A. R., Paciello, F., Rolesi, R., Paludetti, G., Troiani, D. (2019). Targeting dysregulation of redox homeostasis in noise-induced hearing loss: Oxidative stress and ROS signaling. Free Radic. Biol. Med. 135, 46–59. doi: 10.1016/j.freeradbiomed.2019.02.022
Fitzgerald, J. P., Nayak, B., Shanmugasundaram, K., Friedrichs, W., Sudarshan, S., Eid, A. A., et al. (2012). Nox4 mediates renal cell carcinoma cell invasion through hypoxia-induced interleukin 6- and 8- production. PloS One 7, e30712. doi: 10.1371/journal.pone.0030712
Fujita, N., Sugimoto, R., Ma, N., Tanaka, H., Iwasa, M., Kobayashi, Y., et al. (2008). Comparison of hepatic oxidative DNA damage in patients with chronic hepatitis B, and C. J. Viral Hepat 15, 498–507. doi: 10.1111/j.1365-2893.2008.00972.x
Gustafson, H. L., Yao, S., Goldman, B. H., Lee, K., Spier, C. M., LeBlanc, M. L., et al. (2014). Genetic polymorphisms in oxidative stress-related genes are associated with outcomes following treatment for aggressive B-cell non-Hodgkin lymphoma. Am. J. Hematol. 89, 639–645. doi: 10.1002/ajh.23709
Guzik, T. J., West, N. E., Black, E., McDonald, D., Ratnatunga, C., Pillai, R., et al. (2000). Functional effect of the C242T polymorphism in the NAD(P)H oxidase p22phox gene on vascular superoxide production in atherosclerosis. Circulation 102, 1744–1747. doi: 10.1161/01.CIR.102.15.1744
Ha, H. L., Shin, H. J., Feitelson, M. A., Yu, D. Y. (2010). Oxidative stress and antioxidants in hepatic pathogenesis. World J. Gastroenterol. 16, 6035–6043. doi: 10.3748/wjg.v16.i48.6035
He, D., Tao, S., Guo, S., Li, M., Wu, J., Huang, H., et al. (2015). Interaction of TLR-IFN and HLA polymorphisms on susceptibility of chronic HBV infection in Southwest Han Chinese. Liver Int. 35, 1941–1949. doi: 10.1111/liv.12756
Huang, Y. S., Wang, L. Y., Chang, C. H., Perng, C. L., Lin, H. C. (2016). Superoxide dismutase 2 genetic variation as a susceptibility risk factor for alcoholic cirrhosis. Alcohol Alcohol 51, 633–637. doi: 10.1093/alcalc/agw004
Jiao, J., Dou, L., Li, M., Lu, Y., Guo, H. B., Man, Y., et al. (2012). NADPH oxidase 2 plays a critical role in dysfunction and apoptosis of pancreatic beta-cells induced by very low-density lipoprotein. Mol. Cell Biochem. 370, 103–113. doi: 10.1007/s11010-012-1402-z
Jin, F., Xiong, W. J., Jing, J. C., Feng, Z., Qu, L. S., Shen, X. Z. (2011). Evaluation of the association studies of single nucleotide polymorphisms and hepatocellular carcinoma: a systematic review. J. Cancer Res. Clin. Oncol. 137, 1095–1104. doi: 10.1007/s00432-010-0970-0
Katakami, N., Kaneto, H., Matsuoka, T. A., Takahara, M., Imamura, K., Ishibashi, F., et al. (2010). Accumulation of gene polymorphisms related to oxidative stress is associated with myocardial infarction in Japanese type 2 diabetic patients. Atherosclerosis 212, 534–538. doi: 10.1016/j.atherosclerosis.2010.06.010
Levesque, E., Ait-Ammar, N., Dudau, D., Clavieras, N., Feray, C., Foulet, F., et al. (2019). Invasive pulmonary aspergillosis in cirrhotic patients: analysis of a 10-year clinical experience. Ann. Intensive Care 9, 31. doi: 10.1186/s13613-019-0502-2
Lim, S. C., Liu, J. J., Low, H. Q., Morgenthaler, N. G., Li, Y., Yeoh, L. Y., et al. (2009). Microarray analysis of multiple candidate genes and associated plasma proteins for nephropathy secondary to type 2 diabetes among Chinese individuals. Diabetologia 52, 1343–1351. doi: 10.1007/s00125-009-1368-x
Liu, Q., Li, H., Wang, N., Chen, H., Jin, Q., Zhang, R., et al. (2013). Polymorphism of rs1836882 in NOX4 gene modifies associations between dietary caloric intake and ROS levels in peripheral blood mononuclear cells. PloS One 8, e85660. doi: 10.1371/journal.pone.0085660
Lopes, L. R., Dagher, M. C., Gutierrez, A., Young, B., Bouin, A. P., Fuchs, A., et al. (2004). Phosphorylated p40PHOX as a negative regulator of NADPH oxidase. Biochemistry 43, 3723–3730. doi: 10.1021/bi035636s
Lucena, M. I., Garcia-Martin, E., Andrade, R. J., Martinez, C., Stephens, C., Ruiz, J. D., et al. (2010). Mitochondrial superoxide dismutase and glutathione peroxidase in idiosyncratic drug-induced liver injury. Hepatology 52, 303–312. doi: 10.1002/hep.23668
Mata-Balaguer, T., de la Herran, R., Ruiz-Rejon, C., Ruiz-Rejon, M., Garrido-Ramos, M. A., Ruiz-Rejon, F. (2004). Angiotensin-converting enzyme and p22(phox) polymorphisms and the risk of coronary heart disease in a low-risk Spanish population. Int. J. Cardiol. 95, 145–151. doi: 10.1016/j.ijcard.2003.05.017
Mazaheri, M., Karimian, M., Behjati, M., Raygan, F., Hosseinzadeh Colagar, A. (2017). Association analysis of rs1049255 and rs4673 transitions in p22phox gene with coronary artery disease: A case-control study and a computational analysis. Ir J. Med. Sci. 186, 921–928. doi: 10.1007/s11845-017-1601-4
McCaughan, J. A., McKnight, A. J., Maxwell, A. P. (2014). Genetics of new-onset diabetes after transplantation. J. Am. Soc. Nephrol. ,25, 1037–1049. doi: 10.1681/ASN.2013040383
Megias-Vericat, J. E., Montesinos, P., Herrero, M. J., Moscardo, F., Boso, V., Rojas, L., et al. (2018). Impact of NADPH oxidase functional polymorphisms in acute myeloid leukemia induction chemotherapy. Pharmacogenomics J. 18, 301–307. doi: 10.1038/tpj.2017.19
Moore, J. H. (2003). The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum. Hered 56, 73–82. doi: 10.1159/000073735
Nahon, P., Sutton, A., Pessayre, D., Rufat, P., Ziol, M., Ganne-Carrie, N., et al. (2007). Manganese superoxide dismutase dimorphism and iron overload, hepatocellular carcinoma, and death in hepatitis C virus-infected patients. Clin. Gastroenterol. Hepatol 5, 630–635. doi: 10.1016/j.cgh.2006.11.022
Nakamura, S., Kugiyama, K., Sugiyama, S., Miyamoto, S., Koide, S., Fukushima, H., et al. (2002). Polymorphism in the 5'-flanking region of human glutamate-cysteine ligase modifier subunit gene is associated with myocardial infarction. Circulation 105, 2968–2973. doi: 10.1161/01.CIR.0000019739.66514.1E
Namiduru, E. S., Namiduru, M., Tarakcioglu, M., Toy, A. (2010). Antioxidant defense in patients with chronic viral hepatitis B and C type. Clin. Lab. 56, 207–213. doi: 10.1515/CCLM.2010.025
Olsson, L. M., Lindqvist, A. K., Kallberg, H., Padyukov, L., Burkhardt, H., Alfredsson, L., et al. (2007). A case-control study of rheumatoid arthritis identifies an associated single nucleotide polymorphism in the NCF4 gene, supporting a role for the NADPH-oxidase complex in autoimmunity. Arthritis Res. Ther. 9, R98. doi: 10.1186/ar2299
Patterson, J. C., Joughin, B. A., van de Kooij, B., Lim, D. C., Lauffenburger, D. A., Yaffe, M. B. (2019). ROS and oxidative stress are elevated in mitosis during asynchronous cell cycle progression and are exacerbated by mitotic arrest. Cell Syst. 8, 163–167 e162. doi: 10.1016/j.cels.2019.01.005
Polonikov, A. V., Ivanov, V. P., Solodilova, M. A., Khoroshaya, I. V., Kozhuhov, M. A., Panfilov, V. I. (2007). The relationship between polymorphisms in the glutamate cysteine ligase gene and asthma susceptibility. Respir. Med. 101, 2422–2424. doi: 10.1016/j.rmed.2007.06.012
Quinn, M. T., Gauss, K. A. (2004). Structure and regulation of the neutrophil respiratory burst oxidase: comparison with nonphagocyte oxidases. J. Leukoc Biol. 76, 760–781. doi: 10.1189/jlb.0404216
Rabelo, F., Stefano, J. T., Cavaleiro, A. M., Lima, R. V. C., de Campos Mazo, D. F., Carrilho, F. J., et al. (2018). Association between the CYBA and NOX4 genes of NADPH oxidase and its relationship with metabolic syndrome in non-alcoholic fatty liver disease in Brazilian population. Hepatobiliary Pancreat Dis. Int. 17, 330–335. doi: 10.1016/j.hbpd.2018.06.005
Rains, J. L., Jain, S. K. (2011). Oxidative stress, insulin signaling, and diabetes. Free Radic. Biol. Med. 50, 567–575. doi: 10.1016/j.freeradbiomed.2010.12.006
Rossi, D., Rasi, S., Franceschetti, S., Capello, D., Castelli, A., De Paoli, L., et al. (2009). Analysis of the host pharmacogenetic background for prediction of outcome and toxicity in diffuse large B-cell lymphoma treated with R-CHOP21. Leukemia 23, 1118–1126. doi: 10.1038/leu.2008.398
Sastre, J., Diaz-Beveridge, R., Garcia-Foncillas, J., Guardeno, R., Lopez, C., Pazo, R., et al. (2015). Clinical guideline SEOM: hepatocellular carcinoma. Clin. Transl. Oncol. 17, 988–995. doi: 10.1007/s12094-015-1451-3
Schirmer, M., Hoffmann, M., Kaya, E., Tzvetkov, M., Brockmoller, J. (2008). Genetic polymorphisms of NAD(P)H oxidase: variation in subunit expression and enzyme activity. Pharmacogenomics J. 8, 297–304. doi: 10.1038/sj.tpj.6500467
Shimo-Nakanishi, Y., Hasebe, T., Suzuki, A., Mochizuki, H., Nomiyama, T., Tanaka, Y., et al. (2004). Functional effects of NAD(P)H oxidase p22(phox) C242T mutation in human leukocytes and association with thrombotic cerebral infarction. Atherosclerosis 175, 109–115. doi: 10.1016/j.atherosclerosis.2004.01.043
Siqueira, E. R., Pereira, L. B., Stefano, J. T., Patente, T., Cavaleiro, A. M., Silva Vasconcelos, L. R., et al. (2015). Association of a variant in the regulatory region of NADPH oxidase 4 gene and metabolic syndrome in patients with chronic hepatitis C. Eur. J. Med. Res. 20, 45. doi: 10.1186/s40001-015-0136-2
Snahnicanova, Z., Mendelova, A., Grendar, M., Holubekova, V., Kostkova, M., Pozorciakova, K., et al. (2018). Association of polymorphisms in CYBA, SOD1, and CAT genes with Type 1 Diabetes and Diabetic Peripheral Neuropathy in children and adolescents. Genet. Test Mol. Biomarkers 22, 413–419. doi: 10.1089/gtmb.2018.0018
Sole, X., Guino, E., Valls, J., Iniesta, R., Moreno, V. (2006). SNPStats: a web tool for the analysis of association studies. Bioinformatics 22, 1928–1929. doi: 10.1093/bioinformatics/btl268
Tang, R. N., Wu, P., An, L. (2017). NADPH oxidase p22phox C242T polymorphism is associated with macroalbuminuria in diabetic patients: A meta-analysis. J. Diabetes Complications 31, 1207–1211. doi: 10.1016/j.jdiacomp.2017.02.015
Wang, Q., Na, B., Ou, J. H., Pulliam, L., Yen, T. S. (2012). Hepatitis B virus alters the antioxidant system in transgenic mice and sensitizes hepatocytes to Fas signaling. PloS One 7, e36818. doi: 10.1371/journal.pone.0036818
Wang, P., Zhu, Y., Xi, S., Li, S., Zhang, Y. (2018). Association between MnSOD Val16Ala polymorphism and cancer risk: evidence from 33,098 cases and 37,831 controls. Dis. Markers 2018, 3061974. doi: 10.1155/2018/3061974
Xianyu, J., Feng, J., Yang, Y., Tang, J., Xie, G., Fan, L. (2018). Correlation of oxidative stress in patients with HBV-induced liver disease with HBV genotypes and drug resistance mutations. Clin. Biochem. 55, 21–27. doi: 10.1016/j.clinbiochem.2018.03.014
Keywords: gene polymorphisms, hepatitis B virus, oxidative stress, CYBA, NCF4, Nox4, GCLM
Citation: Ma N, Liu W, Zhang X, Gao X, Yu F, Guo W, Meng Y, Gao P, Zhou J, Yuan M, Mi Y, Zhang L, Qi S, Li L, Wang L, Su Q, Yang L and Liu D (2020) Oxidative Stress-Related Gene Polymorphisms Are Associated With Hepatitis B Virus-Induced Liver Disease in the Northern Chinese Han Population. Front. Genet. 10:1290. doi: 10.3389/fgene.2019.01290
Received: 11 June 2019; Accepted: 22 November 2019;
Published: 08 January 2020.
Edited by:
Amelia Marti, University of Navarra, SpainReviewed by:
Tingting Geng, New York Medical College, United StatesShiying Xuan, Qingdao Municipal Hospital, China
Copyright © 2020 Ma, Liu, Zhang, Gao, Yu, Guo, Meng, Gao, Zhou, Yuan, Mi, Zhang, Qi, Li, Wang, Su, Yang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Yang, bGVpbGVpeTEwQDEyNi5jb20=; Dianwu Liu, ZGlhbnd1TDE5NTZAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Ning Ma
Ning Ma Wenxuan Liu1†
Wenxuan Liu1†