
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 20 December 2019
Sec. Epigenomics and Epigenetics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.01285
This article is part of the Research Topic Mitochondrial Genetics and Epigenetics View all 13 articles
The higher order organization of eukaryotic and prokaryotic genomes is pivotal in the regulation of gene expression. Specifically, chromatin accessibility in eukaryotes and nucleoid accessibility in bacteria are regulated by a cohort of proteins to alter gene expression in response to diverse physiological conditions. By contrast, prior studies have suggested that the mitochondrial genome (mtDNA) is coated solely by mitochondrial transcription factor A (TFAM), whose increased cellular concentration was proposed to be the major determinant of mtDNA packaging in the mitochondrial nucleoid. Nevertheless, recent analysis of DNase-seq and ATAC-seq experiments from multiple human and mouse samples suggest gradual increase in mtDNA occupancy during the course of embryonic development to generate a conserved footprinting pattern which correlate with sites that have low TFAM occupancy in vivo (ChIP-seq) and tend to adopt G-quadruplex structures. These findings, along with recent identification of mtDNA binding by known modulators of chromatin accessibility such as MOF, suggest that mtDNA higher order organization is generated by cross talk with the nuclear regulatory system, may have a role in mtDNA regulation, and is more complex than once thought.
The genome of all organisms undergoes concerted cycles of packaging to reduce its volume and to control access to the regulatory mechanisms of transcription and replication. In the eukaryotic nucleus, DNA is compacted into chromatin, which provides differential accessibility in response to a variety of histone modifications (Zhu and Li, 2016). The bacterial genomes, which lack histones, are folded into nucleoids using a set of dedicated proteins, entitled Nucleoid-Associated Proteins (NAPs), such as HU, Histone-like Nucleoid Structuring protein (H-NS) and Structural Maintenance of Chromosomes proteins (SMC). Alongside their architectural role in DNA packaging, these proteins also play a role in other processes, such as replication and chromosome segregation (Badrinarayanan et al., 2015; Dame and Tark-Dame, 2016). Notably, the most commonly used models for investigation of nucleoid organization are Escherichia coli, Bacillus subtilis and Caulobacter crescentus (Dame and Tark-Dame, 2016); the latter is an alphaproteobacterium, which belongs to the same phylogenetic branch from which the mitochondria originated (Wang and Wu, 2015).
The circular genomes of C. crescentus are organized in ellipsoidal and helical structures between two opposite poles of the cell, creating two ‘arms’ that are folded around each other (Le et al., 2013). While analyzing interactions between different regions within the C. crescentus genome by genome-wide chromatin conformational capture (Hi-C) (Le et al., 2013), 23 preferential Chromosomal Interaction Domains (CID) were identified. CID boundaries seem to closely associate with transcription and replication units. The boundaries tend to reestablish shortly after, or even during DNA replication, possibly to disentangle the newly formed DNA molecules. Additionally, the CID boundaries can be disrupted by transcription inhibition (Le et al., 2013). Novel CID boundaries can be created by artificially moving loci of highly expressed genes into inherently low expressed regions (Le et al., 2013). These findings, strongly suggest that the bacterial nucleoid, including that of alphaproteobacteria, is a highly regulated structure with great importance to DNA replication and transcription.
In addition to their nuclear genome, all eukaryotic cells contain a much smaller cytoplasmic genome—the mitochondrial DNA (mtDNA). This genome originated ~2.5 billion years ago from an ancient endosymbiosis between a former free-living alphaproteobacterium and the progenitor of all eukaryotic cells (Sagan, 1967; Pittis and Gabaldon, 2016). Although during the course of evolution the ancient bacterium lost most of its inherent genetic material either due to transfer to the nucleus, or due to natural selection, the mitochondria in the vast majority of eukaryotes still harbor their own genomes. Despite its modest size, the mammalian mtDNA encodes 13 critical subunits of the oxidative phosphorylation system (OXPHOS), two ribosomal RNA genes and 22 tRNAs that are required for cellular energy production. Mammalian mtDNA forms a protein-DNA structure that was termed ‘nucleoid’, to highlight its ancient bacterial heritage (see below). The animal mtDNA is four orders of magnitude smaller than the nuclear genome, and has been long thought to be separately regulated from the nuclear genome (Gustafsson et al., 2016). Accordingly, mitochondrial transcription factor A (TFAM) is believed to be sufficient for mitochondrial nucleoid formation (Kaufman et al., 2007) and the primary driver of mtDNA packaging (Gustafsson et al., 2016; Farge and Falkenberg, 2019). The role of TFAM in mtDNA packaging and higher order organization has been recently thoroughly reviewed, and therefore will be mentioned here only briefly (Farge and Falkenberg, 2019). TFAM is highly conserved across species, and despite the apparently linear mtDNA organization in yeast (Gerhold et al., 2010), the yeast orthologue (Abf2p) of TFAM packs this genome as well (Farge and Falkenberg, 2019). mtDNA condensation positively correlates with the cellular concentration of TFAM so that increased TFAM concentration leads to higher degrees of mtDNA compaction (Kukat et al., 2015).
Thus, our current view of mtDNA regulation suggests that a nuclear-encoded yet mitochondrially restricted set of proteins modulates mtDNA transcription, replication and packaging (Gustafsson et al., 2016). For example, mtDNA genes are transcribed by POLRMT, and not RNA Polymerase II which transcribes nuclear mRNAs, and the mtDNA is replicated by DNA polymerase gamma (POLG), which has no accepted role in replication of the nuclear DNA. However, it would be surprising from an evolutionary point of view if the past 2.5 billion years since mitochondrial endosymbiosis had not led to significant adaptation of the regulation of the mitochondrial and nuclear DNA. Is it plausible that the longtime co-existence of the mitochondrion and its host have been accompanied by adaptation of mtDNA to the host regulatory and packaging systems? Co-adaptation of the nuclear and mitochondrial genomes had been demonstrated in the context of the OXPHOS and in the mitoribosomes, which use nuclear DNA-encoded proteins, and either exclusively mtDNA-encoded proteins (in OXPHOS) or mtDNA-encoded rRNA and tRNA transcripts (in the mitoribosome) (Levin et al., 2014). However, the discovery of transcription factors that directly regulate transcription in both the nucleus and in the mtDNA has suggested that the control of gene expression is coordinated not only by signals, but by dual localization of transcription factors (Barshad et al., 2018). Hence, adaptation of mtDNA regulation to the nuclear regulatory system is plausible.
Mitochondrial DNA is compacted through its interactions with TFAM (Kukat et al., 2015), but there is growing evidence for the involvement of additional nuclear-encoded proteins that also regulate nuclear chromatin. This includes MOF (Chatterjee et al., 2016), members of the AP1 family (c-Jun and JunD) as well as CEBPB (Blumberg et al., 2014) and MEF2D (She et al., 2011). The discovery of mtDNA binding and mitochondrial transcriptional regulation by MOF, a histone lysine acetyltransferase that remodels chromatin, was particularly surprising, as it raises questions about its acetyltransferase target in the mitochondria, and its possible role in mtDNA organization. Secondly, c-Jun and JunD, which were recently shown to bind negatively selected sites in the mtDNA (Blumberg et al., 2014), tend to bind nuclear DNA enhancer regions and affect nuclear DNA gene regulation (Phanstiel et al., 2017). Third, CEBPB, a known chromatin remodeler (Bornstein et al., 2014), not only binds the mtDNA in vivo, but also serves as a candidate repressor of human mtDNA gene expression (Barshad et al., 2018). Fourth, DNase-seq and ATAC-seq analysis in multiple human and mouse cells revealed a conserved footprinting pattern, which overlapped known mtDNA regulatory elements, yet correlated with low TFAM occupancy in HeLa cells (Blumberg et al., 2018). This ATAC-seq mtDNA footprinting pattern was gradually formed during the course of embryogenesis in both mouse and humans, as reflected by gradually increasing mtDNA occupancy (Marom et al., 2019). Hence, it is possible that there are mtDNA sites which are consistently occupied, and sites that are consistently under-occupied across the mtDNA, and that the mtDNA is bound not only by TFAM but rather by other additional proteins in an organized manner. This reflects the existence of an organized protein–DNA structure in the mitochondrial genome, thus providing first clues for the existence of a structured higher order organization of the mitochondrial genome.
We would argue that the investigation of the regulation of the mitochondrial nucleoid in the frame of protein-DNA patterns of interactions and their impact on regulation of mtDNA gene expression and replication is of equivalent importance to our understanding of the organization and compaction of the nuclear chromosome, but that it is markedly less well studied and understood. In this essay we will discuss current knowledge regarding the nature of the higher order organization of the mitochondrial genome (Figure 1), and assess its functional potential from an evolutionary perspective.
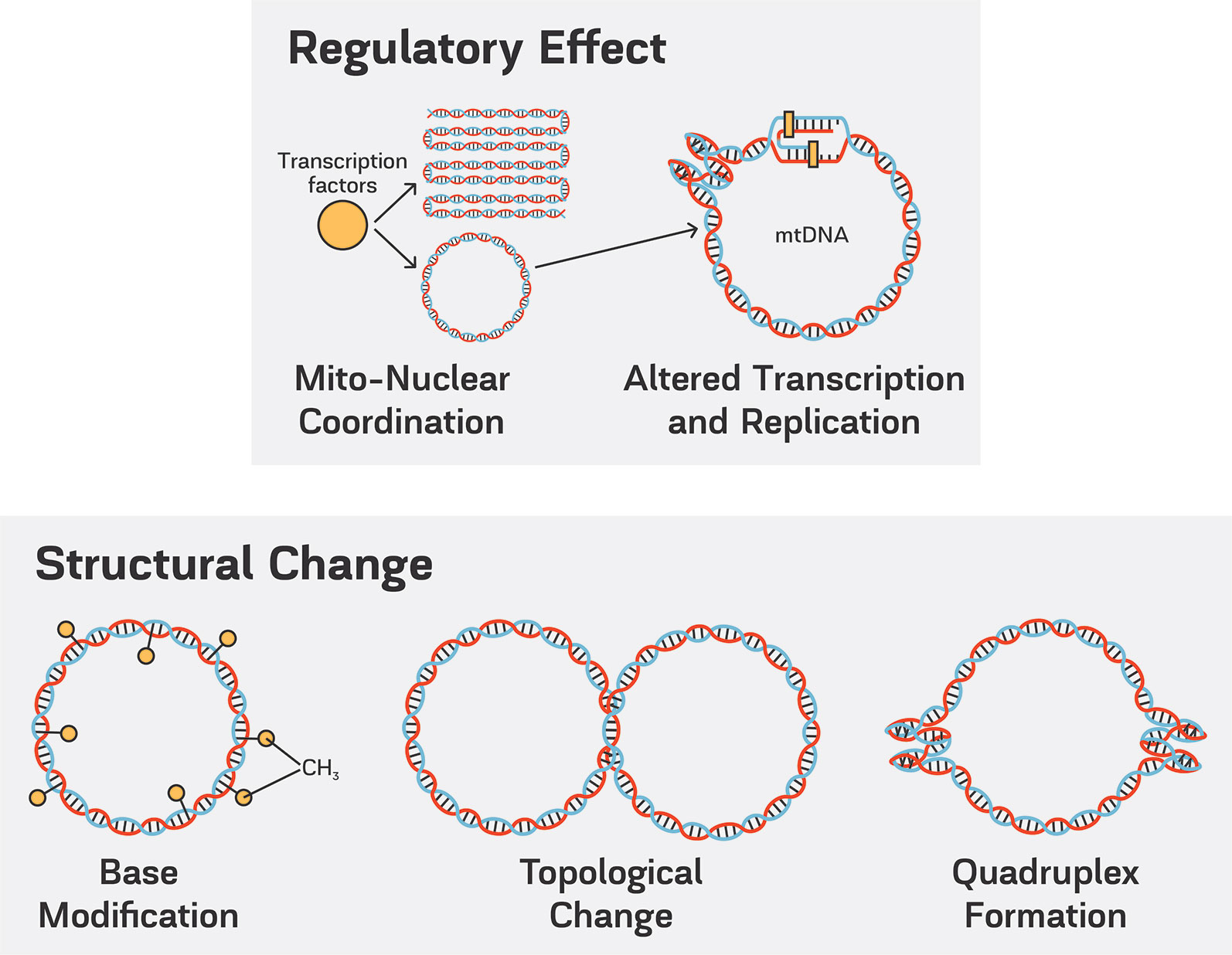
Figure 1 The possible structural changes in the mtDNA and the cross-talk between mtDNA higher order organization and regulation.
The only known structural unit of the mitochondrial genome is the nucleoid, which contains mtDNA and closely interacting proteins (Hensen et al., 2014). The nucleoids are vital for mitochondrial function as they coordinate transcription (Rebelo et al., 2011), translation (He et al., 2012) and interact with enzymatic activities of the mitochondrial inner membrane (Wang and Bogenhagen, 2006). There are a correspondingly large number of proteins that can be pulled down by crosslinking to nucleoids, thus reflecting these diverse activities (Bogenhagen et al., 2008). High-resolution microscopy techniques were used to show that nucleoids are compact and ellipsoidal, suggesting the exclusion of non-nucleoid proteins, and that nucleoids are associated with the mitochondrial inner membrane (Brown et al., 2011; Kukat et al., 2011; Kukat et al., 2015). The number of mtDNA molecules in each nucleoid has been a matter of considerable debate. Logically, the nucleoids must at least transiently contain multiple mitochondrial genomes after the completion of replication. Most studies have observed that the copy number is stably higher with estimates ranging from 1.4 to 7.5 genomes per nucleoid [(reviewed in (Lee and Han, 2017)].
Higher order organization of both the eukaryotic nuclear genome and the bacterial nucleoid involve regulated steps of protein binding followed by bending and folding to allow the interaction of sequences that are distant in the primary DNA sequence. In a study of the mtDNA binding pattern of the mitochondrial transcription termination factor MTERF1 in mammalian cells, simultaneous binding of MTERF1 was observed at the proximal heavy strand promoter (HSP1) and within the MT-TL1 sequence (Martin et al., 2005). This interaction increased the expression of genes regulated by HSP1, and a model was proposed that would allow the direct recycling of transcription complexes from the termination point of HSP1 transcription back to its origin. While attractive, the phenotype of an Mterf1 deficient mouse cast doubt upon this elegant concept, as the predicted loss in HSP1 activity was not observed (Terzioglu et al., 2013). Other cis interactions along mtDNA are yet to be discovered. Interactions between distant nuclear genomic regions are currently being investigated using sequencing-based techniques such as the Chromosome conformation capture (3C) and subsequent derivative of this methodology (i.e., 4C, 5C and HiC) (Oluwadare et al., 2019), yet all of these techniques are currently designed to identify interactions between regions that are megabases apart, which limit their utility in the study of the human mtDNA. Although a recent study of HiC data claimed to observe direct interactions between the mitochondrial and nuclear genomes (Doynova et al., 2016), no independent study supported such findings. Given the above, there is a need for the development of techniques that will allow mapping of interactions between mtDNA sequences, while taking into account the small size of this genome and its circularity.
Mitochondrial transcription uses unique features to allow the differential expression of a very tightly packed genome with a limited number of primary transcripts. Since mitochondrial transcription has recently been reviewed (Gustafsson et al., 2016), here we will only consider the role of the physical structure of mtDNA in the initiation and termination of transcription.
One key challenge for mitochondrial transcription is the use of oppositely oriented promoters that transcribe the same regions in both directions—with strand specific promoters in mammals but bidirectional promoters in birds (L’Abbe et al., 1991; Randi and Lucchini, 1998) and amphibians (Bogenhagen et al., 1986; Bogenhagen and Romanelli, 1988). The human mtDNA harbors a single light-stranded promoter, which is responsible for expression of the OXPHOS complex I subunit ND6 as well as eight tRNA. The activation of this promoter requires the binding of TFAM, which creates a pronounced U-turn bending in mtDNA, proximal to the site of transcription initiation (Ngo et al., 2011; Rubio-Cosials et al., 2011). The two heavy-strand promoters are closely adjacent to each other, with HSP1 principally driving expression of the two rRNA genes (i.e. the 12S and 16S rRNAs), and HSP2 driving the expression of the remaining twelve protein-coding genes and the distal tRNA genes along the heavy strand (Montoya et al., 1983). Like LSP, TFAM activates HSP1, although studies have come to different conclusions as to the topology of TFAM’s interaction at HSP1 (Ngo et al., 2014; Morozov and Temiakov, 2016; Hillen et al., 2017; Uchida et al., 2017).
The balancing of expression of HSP1 and HSP2 has been a matter of some debate. It seems reasonable that some mechanism must exist, since HSP1 is primarily devoted to rRNA and HSP2 to the expression of protein coding genes, which was also shown in living cells (Blumberg et al., 2017). Our group and others have shown that HSP2 is distinct in that TFAM is not only dispensable for activation, but actively inhibits it (Lodeiro et al., 2012; Zollo et al., 2012). We have further shown that the topological state of mtDNA may be important for HSP2 activation. Unique among the promoters, HSP2 is activated by negative supercoiling in a fashion reminiscent of bacterial systems, but no similar effect is seen at LSP and HSP1 (Zollo and Sondheimer, 2017).
The termination of mitochondrial transcription is also regulated by the physical state of mtDNA. Because the molecule is circular and has oppositely oriented promoters, processive transcriptional complexes are at risk of collision. Because of the positioning of genes, the termination of LSP and HSP1 at a point between mt.3229 (the end of the 16S ribosomal RNA as transcribed by HSP1) and mt.4329 (the end of MT-TQ as transcribed by LSP) would allow the simultaneous utilization of LSP and HSP1 without promoter collision. Considerable evidence has been provided for the role of MTERF1 in interaction with mt.3232–3253 (within the coding sequence of MT-TL1), including the crystal structure of the interaction of MTERF1 with its mtDNA target sequences (Yakubovskaya et al., 2010). As noted above, evidence from mouse knockout studies of Mterf1 agreed only partially with this concept, and suggested that the insulation of the LSP against transcription proceeding back through the promoter might also be important (Terzioglu et al., 2013). It is important to recognize that the regulation of both transcription and mtDNA physical structure in mouse and human may not be identical, but the organization of mtDNA regulatory elements clearly influences interactions with transcription factors to exert control over gene expression. The means of controlling interactions between transcriptional complexes arising from LSP and HSP2 remains undiscovered.
G-Quadruplexes (GQs) are non-canonical nucleic acid secondary structures that use Hoogsteen hydrogen bonding between guanines on the same strand (Rhodes and Lipps, 2015). The occurrence of GQs within the DNA is not random, and is notably conserved across species, thus supporting selective constraints and hence potential functional importance (Murat and Balasubramanian, 2014). Moreover, whereas transient GQs correlate with binding sites of chromatin remodeling-related transcription factors, genome-wide sites with more stable GQs have been implicated in replication stalling and inhibition of chromatin remodeling (Varizhuk et al., 2019), which support their involvement in regulation of higher order DNA organization. For example, GQ-ChIP-seq experiments revealed that most GQs tend to form within nucleosome-depleted regions with increased transcription activity (Hansel-Hertsch et al., 2016). As GQ structures are mostly resolved by RecQ helicases (Mendoza et al., 2016; Sauer and Paeschke, 2017; Varizhuk et al., 2019), it is noteworthy that one such helicase, RecQ4, is transported into the mitochondria, interacts with DNA POLG and promotes mtDNA replication (Ding and Liu, 2015). Indeed, due to the asymmetric composition of nucleotides in the heavy (more guanine-rich) and light (more cytosine-rich) strands of mtDNA, the heavy mtDNA strand is prone to GQ formation. Previously, in silico analysis suggested the existence of G-quadruplex-forming motifs throughout the human mtDNA (Falabella et al., 2019). Imaging of mtDNA using GQ binding dyes showed that they are widely present (Huang et al., 2015), and the application of compounds that bind to GQ impact mtDNA transcription and replication (Falabella et al., 2019). We have recently demonstrated that GQ formation can even selectively bias the replication of a mixed mtDNA population (heteroplasmy) (Naeem et al., 2019). Hence, similar to the nuclear genome, GQ formation in the human mtDNA affects the regulation of this genome.
Although in vitro experiments suggested that TFAM binds to GQ at non-physiological concentrations (Lyonnais et al., 2017), analysis of ChIP-seq TFAM binding experiments in HeLa cells revealed TFAM occupancy throughout the mtDNA (Wang et al., 2013), yet low occupancy of TFAM at GQ-forming regions (Blumberg et al., 2018). Moreover, we showed that G-quadruplex-forming motifs tend to co-localize with conserved DNase-seq footprinting sites in adult cells (Blumberg et al., 2018) and during development (Marom et al., 2019). Other proteins such as the ATP-dependent Lon protease bind GQ sequences in vitro (Lu et al., 2003), and in vivo (Lu et al., 2007). Thus, it is plausible that investigation of the conformation assumed by such motifs in vivo will offer clues for the discovery of novel mtDNA binding proteins that may be involved in the construction and regulation of its higher order organization. Interestingly, nuclear DNA regions that tend to be packed late during the cell cycle, and are prone to breakage, also harbor non-B DNA structures (Dong et al., 2014). Specifically, GQ structures are resolved at the DNA, likely by the Pif1 helicase, to allow maintenance of the mtDNA (Bannwarth et al., 2016). Indeed, double mutant Pif1 mice exhibit elevated levels of mtDNA damage. As in the nuclear genome, hotspots for chromosomal aberrations and fragile sites tend to correlate with the state of chromatin accessibility (Mishmar et al., 1999). Further investigating the patterns of non-canonical DNA structure may offer additional insights to differential accessibility of sites across the mitochondrial genome.
Chromosomal aberrations of various types in the nuclear genome (i.e. inversions, deletions, insertions, duplications and translocations) not only change the location of genes, but also change the location of regulatory elements, thus changing the chromatin structure and regulatory landscape of the modified region. As discussed above, regulatory factors bind the mtDNA not only within the non-coding promoters’ region, but rather throughout the mitochondrial genome [reviewed in: (Barshad et’al., 2018)].
Therefore, it is logical that mtDNA aberrations such as deletions, duplications, inversions and insertions may not only change the coding content, but will change the location of regulatory elements and hence have the potential impact on mtDNA regulation. Consistent with this hypothesis, and because of the high gene density of mtDNA, structural rearrangements and deletions are poorly tolerated. The association between mitochondrial deletions and pathology is robust. The accumulation of deletions during the process of aging was discovered nearly thirty years ago (Cortopassi and Arnheim, 1990) and at nearly the same time it was recognized that the Kearns-Sayre syndrome was also linked to deletions in mtDNA (Shoffner et al., 1989). The phenotypic impact of mtDNA deletions has been largely interpreted as the result of the loss of genetic material. The effect of such mtDNA aberrations on mtDNA regulation in vivo merits further investigation.
Is it possible that structural aberrations are not random, preferentially occurring at positions of special mtDNA organization? Indeed, the 4,977 bp deletion has previously been shown to be flanked by simple repeat sequences with the tendency to form non-B DNA structures (Hou and Wei, 1998). Interestingly, non-B DNA structures tend to co-localize in general with other types of mtDNA deletions that accumulated with aging (Hou and Wei, 1996; Damas et al., 2012). Specifically, as already discussed above, G-quadruplex forming sequences tend to occur at such breakpoints (Dong et al., 2014), and affect mtDNA transcription in vitro (Hillen et al., 2017). Hence, it is logical to suggest the existence of mtDNA hotspots for aberrations. In the nuclear genome hot spots for chromosomal aberrations tend to occur in regions with special chromatin organization (Mishmar et al., 1999; Fungtammasan et al., 2012), which calls for assessing such connection in the mtDNA as well.
mtDNA aberrations do not only associate with human pathologies, but also led to changes in mtDNA gene order and content during the course of evolution. As an example, although the mitochondrial genome remained circular in most studied metazoans, it is linear in Medusozoa (Kayal et al., 2012). Secondly, although most vertebrate mtDNAs contain a non-coding region, which harbors most known regulatory elements, the chordate amphioxus nearly lacks a non-coding region (Spruyt et al., 1998; Boore et al., 1999), which prevents identification of the positions of orthologous regulatory elements. Third, fragmentation of the mtDNA into several co-segregating parts that together comprise the full gene content seen in vertebrates has been described in organisms such as lice (Shao et al., 2012) and certain nematodes (Phillips et al., 2016). Do such mtDNA rearrangements affect mtDNA regulation? A recent study of in vivo mtDNA transcription using the precision global run-on transcription-sequencing (PRO-seq) revealed, that although the mtDNA gene contents in Drosophila and Caenorhabditis elegans are nearly identical to that of humans, the gene order and gene content per mtDNA strand profoundly changed (Blumberg et al., 2017). We recently showed that such changes were accompanied by the emergence of a very different mtDNA transcriptional initiation and termination schemes in vivo (Blumberg et al., 2017). Specifically, we observed that in contrast to human mtDNA which harbors two heavy strand and one light strand transcriptional initiation sites, Drosophila had 5-7 initiation sites, and C elegans had a single transcription initiation site, consistent with their mtDNA strand-gene contents. These phenomena exemplify how changes in mtDNA organization, during the course of evolution and in human diseases, likely lead to changes in mtDNA regulation.
As the recently identified mtDNA DNAse-seq and ATAC-seq footprinting patterns appears to be conserved between human and mouse (Blumberg et al., 2018), it would be of interest to study such in organisms with different mtDNA organization, as well as in human cells with pathological mtDNA deletions. Such study will directly assess the impact of mtDNA aberrations on mtDNA higher order organization and while engaging such study with techniques that assess transcriptional pattern in vivo (such as PRO-seq) one will be able to assess the connection between such changes with alteration in mtDNA regulation.
The structure of mtDNA and its accessibility is also impacted by topoisomerases, single or double-strand DNA-cleavage proteins that are used to alter the topological state of DNA, keeping it available for transcription and replication and preventing the formation of knots or other unusable structures (Vos et al., 2011). The issues faced by mtDNA that must be resolved by topoisomerase are distinct from those seen in linear chromosomes and include the resolution of concatameric structures formed by mtDNA replication (Kolesar et al., 2013).
There is a single known topoisomerase that is specific for the mitochondrion, TOP1MT (Zhang et al., 2001). This is a type IB topoisomerase, capable of relaxing supercoiling by single-strand cleavage and strand passage. Surprisingly, mice deleted for the homologous Top1mt, are viable, although they do show evidence of increased supercoiling of their mtDNA (Zhang et al., 2014). Instead, Top1mt−/− animals had increased activity of type IIA topoisomerases, suggesting the capacity for compensation for the loss of Top1mt activity.
The presence of type IIA topoisomerases is probably required in mitochondria, since these proteins fulfill the requirement for the de-catenation of linked molecules of mtDNA. Top2β has dual localization to the mitochondria and nucleus, with a shortened isoform present in the mitochondrion (Low et al., 2003). Top2β is canonically responsible for type IIA activity in non-proliferating cells. Although Top2α was not initially identified in the mitochondrion, recent studies have confirmed that it does locate within the organelle (Zhang et al., 2014).
The topoisomerases collectively appear to play important roles in regulating the supercoiling and also the transcription of mtDNA (Sobek et al., 2013). This provides a striking echo of our growing understanding of the role of topoisomerases in regulating nuclear transcription (McKinnon, 2016). The continuing studies, particularly of the bigenomic type IIA topoisomerases, may increase our understanding of how nuclear and mitochondrial transcription are coordinately regulated using template topology, a mechanism of control that is strikingly conserved from bacteria to man.
Nuclear chromatin is regulated by DNA and protein modifications including the methylation of cytosines and the acetylation of specific lysine residues in histones. Such changes directly correlate with chromatin accessibility and have antagonistic impact on gene expression: whereas H3K27 tri-methylation correlates with gene silencing, K27 acetylation correlates with gene activation (Rada-Iglesias et al., 2011). As histones are not imported into the mitochondria, there is considerable interest in the possibility that equivalent modifications occur in the mtDNA or proteins that bind to it. Recent work reported that acetylation and phosphorylation of TFAM can fine-tune TFAM-DNA binding affinity (King et al., 2018). However, as such results were obtained while testing the binding capacity of TFAM (modified and unmodified) to non-specific DNA, it still remains to be assessed whether such modifications affect TFAM binding to mtDNA in living cells. More intriguing is the discovery of several types of mtDNA methylation in different porcine tissues, which correlated with different patterns of mtDNA transcription and mtDNA copy numbers (Liu et al., 2019). The extent to which mtDNA CpG and GpC methylation affect mitochondrial function in cells and in the entire organism remains still in open discussion (Mposhi et’al., 2017), and its very existence has been questioned (Matsuda et al., 2018). Nevertheless, there are reports of association between altered levels of mtDNA methylation and Alzheimer’s disease (Stoccoro et al., 2017), suggesting physiological relevance. Taken together, it seems that similar to the nuclear genome, the mtDNA might be ‘epigenetically’ modified, which correlates with downstream activity. However, the connection between such modifications and mitochondrial higher order organization, and with mitochondrial activities, still remains to be tested.
The higher order organization of the bacterial nucleoid and the nuclear chromatin are tightly regulated, and the impact of such structures on regulation has been widely studied. In the current essay we discussed current knowledge of the higher order organization of the mitochondrial genome in light of evolution and of the growing usage of functional genomics techniques (Figure 1). Recent analysis of DNase-seq and ATAC-seq suggest a conserved mtDNA footprinting pattern between tissues, which does not correlate with the binding sites pattern of the only known mtDNA coating protein—TFAM. As such pattern is conserved between man and mouse, the time is ripe to hypothesize that mtDNA–protein interactions, and hence mtDNA higher order organization, are more complex, and more regulated, than once thought. As functional genomics techniques that determine interactions between genomic regions (such as HiC) grow gradually more sensitive, they could shed light on the packaging of this small genome, and its impact on regulation.
DM and NS conceived the idea and wrote the manuscript. RL and MN participated in critical reading. RL most contributed to writing the text discussing the bacterial nucleoid and nuclear chromatin. MN participated in writing the text discussing G-quadruplex.
This study was funded by the Israeli Science Foundation grant 372/17, and by the US Army Life Sciences Division LS67993 grant, both awarded to DM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Ms Neta Kasher for the artwork.
Badrinarayanan, A., Le, T. B. K., Laub, M. T. (2015). Bacterial chromosome organization and segregation. Annu. Rev. Cell Dev. Bi. 31, 171–199. doi: 10.1146/annurev-cellbio-100814-125211
Bannwarth, S., Berg-Alonso, L., Auge, G., Fragaki, K., Kolesar, J. E., Lespinasse, F., et al. (2016). Inactivation of Pif1 helicase causes a mitochondrial myopathy in mice. Mitochondrion 30, 126–137. doi: 10.1016/j.mito.2016.02.005
Barshad, G., Marom, S., Cohen, T., Mishmar, D. (2018). Mitochondrial DNA transcription and its regulation: an evolutionary perspective. Trends In Genet.: T. I. G. 34 (9), 682–692. doi: 10.1016/j.tig.2018.05.009
Barshad, G., Blumberg, A., Cohen, T., Mishmar, D. (2018). Human primitive brain displays negative mitochondrial-nuclear expression correlation of respiratory genes. Genome Res. 28, 952–967. doi: 10.1101/gr.226324.117
Blumberg, A., Sailaja, B. S., Kundaje, A., Levin, L., Dadon, S., Shmorak, S., et al. (2014). Transcription factors bind negatively-selected sites within human mtDNA genes. Genome Biol. Evol. 6, 2634–2646. doi: 10.1093/gbe/evu210
Blumberg, A., Rice, E. J., Kundaje, A., Danko, C. G., Mishmar, D. (2017). Initiation of mtDNA transcription is followed by pausing, and diverges across human cell types and during evolution. Genome Res. 27, 362–373. doi: 10.1101/gr.209924.116
Blumberg, A., Danko, C. G., Kundaje, A., Mishmar, D. (2018). A common pattern of DNase I footprinting throughout the human mtDNA unveils clues for a chromatin-like organization. Genome Res. 28, 1158–1168. doi: 10.1101/gr.230409.117
Bogenhagen, D. F., Romanelli, M. F. (1988). Template sequences required for transcription of Xenopus laevis mitochondrial DNA from two bidirectional promoters. Mol. Cell. Biol. 8, 2917–2924. doi: 10.1128/mcb.8.7.2917
Bogenhagen, D. F., Yoza, B. K., Cairns, S. S. (1986). Identification of initiation sites for transcription of Xenopus laevis mitochondrial DNA. J. Biol. Chem. 261, 8488–8494.
Bogenhagen, D. F., Rousseau, D., Burke, S. (2008). The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283, 3665–3675. doi: 10.1074/jbc.M708444200
Boore, J. L., Daehler, L. L., Brown, W. M. (1999). Complete sequence, gene arrangement, and genetic code of mitochondrial DNA of the cephalochordate Branchiostoma floridae (Amphioxus). Mol. Biol. Evol. 16, 410–418. doi: 10.1093/oxfordjournals.molbev.a026122
Bornstein, C., Winter, D., Barnett-Itzhaki, Z., David, E., Kadri, S., Garber, M., et al. (2014). A negative feedback loop of transcription factors specifies alternative dendritic cell chromatin States. Mol. Cell 56, 749–762. doi: 10.1016/j.molcel.2014.10.014
Brown, T. A., Tkachuk, A. N., Shtengel, G., Kopek, B. G., Bogenhagen, D. F., Hess, H. F., et al. (2011). Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol. 31, 4994–5010. doi: 10.1128/MCB.05694-11
Chatterjee, A., Seyfferth, J., Lucci, J., Gilsbach, R., Preissl, S., Bottinger, L., et al. (2016). Acetyl transferase regulates transcription and respiration in mitochondria. Cell 167, 722–738 e23. doi: 10.1016/j.cell.2016.09.052
Cortopassi, G. A., Arnheim, N. (1990). Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 18, 6927–6933. doi: 10.1093/nar/18.23.6927
Damas, J., Carneiro, J., Goncalves, J., Stewart, J. B., Samuels, D. C., Amorim, A., et al. (2012). Mitochondrial DNA deletions are associated with non-B DNA conformations. Nucleic Acids Res. 40, 7606–7621. doi: 10.1093/nar/gks500
Dame, R. T., Tark-Dame, M. (2016). Bacterial chromatin: converging views at different scales. Curr. Opin. Cell Biol. 40, 60–65. doi: 10.1016/j.ceb.2016.02.015
Ding, L., Liu, Y. (2015). Borrowing nuclear DNA helicases to protect mitochondrial DNA. Int. J. Mol. Sci. 16, 10870–10887. doi: 10.3390/ijms160510870
Dong, D. W., Pereira, F., Barrett, S. P., Kolesar, J. E., Cao, K., Damas, J., et al. (2014). Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genomics 15, 677. doi: 10.1186/1471-2164-15-677
Doynova, M. D., Berretta, A., Jones, M. B., Jasoni, C. L., Vickers, M. H., O’Sullivan, J. M. (2016). Interactions between mitochondrial and nuclear DNA in mammalian cells are non-random. Mitochondrion 30, 187–196. doi: 10.1016/j.mito.2016.08.003
Falabella, M., Kolesar, J. E., Wallace, C., de Jesus, D., Sun, L., Taguchi, Y. V., et al. (2019). G-quadruplex dynamics contribute to regulation of mitochondrial gene expression. Sci. Rep. 9, 5605. doi: 10.1038/s41598-019-41464-y
Farge, G., Falkenberg, M. (2019). Organization of DNA in Mammalian Mitochondria. Int. J. Mol. Sci. 20 (11), E2770. doi: 10.3390/ijms20112770
Fungtammasan, A., Walsh, E., Chiaromonte, F., Eckert, K. A., Makova, K. D. (2012). A genome-wide analysis of common fragile sites: what features determine chromosomal instability in the human genome? Genome Res. 22, 993–1005. doi: 10.1101/gr.134395.111
Gerhold, J. M., Aun, A., Sedman, T., Joers, P., Sedman, J. (2010). Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol. Cell 39, 851–861. doi: 10.1016/j.molcel.2010.09.002
Gustafsson, C. M., Falkenberg, M., Larsson, N. G. (2016). Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 85, 133–160. doi: 10.1146/annurev-biochem-060815-014402
Hansel-Hertsch, R., Beraldi, D., Lensing, S. V., Marsico, G., Zyner, K., Parry, A., et al. (2016). G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 48, 1267–1272. doi: 10.1038/ng.3662
He, J., Cooper, H. M., Reyes, A., Di Re, M., Sembongi, H., Litwin, T. R., et al. (2012). Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 40, 6109–6121. doi: 10.1093/nar/gks266
Hensen, F., Cansiz, S., Gerhold, J. M., Spelbrink, J. N. (2014). To be or not to be a nucleoid protein: a comparison of mass-spectrometry based approaches in the identification of potential mtDNA-nucleoid associated proteins. Biochimie 100, 219–226. doi: 10.1016/j.biochi.2013.09.017
Hillen, H. S., Morozov, Y. I., Sarfallah, A., Temiakov, D., Cramer, P. (2017). Structural basis of mitochondrial transcription initiation. Cell 171, 1072–1081. doi: 10.1016/j.cell.2017.10.036
Hillen, H. S., Parshin, A. V., Agaronyan, K., Morozov, Y. I., Graber, J. J., Chernev, A., et al. (2017). Mechanism of transcription anti-termination in human mitochondria. Cell 171, 1082–1093. doi: 10.1016/j.cell.2017.09.035
Hou, J. H., Wei, Y. H. (1996). The unusual structures of the hot-regions flanking large-scale deletions in human mitochondrial DNA. Biochem. J. 318 (Pt 3), 1065–1070. doi: 10.1042/bj3181065
Hou, J. H., Wei, Y. H. (1998). AT-rich sequences flanking the 5’-end breakpoint of the 4977-bp deletion of human mitochondrial DNA are located between two bent-inducing DNA sequences at assume distorted structure in organello. Mutat. Res. 403, 75–84. doi: 10.1016/s0027-5107(98)00054-2
Huang, W. C., Tseng, T. Y., Chen, Y. T., Chang, C. C., Wang, Z. F., Wang, C. L., et al. (2015). Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 43, 10102–10113. doi: 10.1093/nar/gkv1061
Kaufman, B. A., Durisic, N., Mativetsky, J. M., Costantino, S., Hancock, M. A., Grutter, P., et al. (2007). The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell 18, 3225–3236. doi: 10.1091/mbc.e07-05-0404
Kayal, E., Bentlage, B., Collins, A. G., Kayal, M., Pirro, S., Lavrov, D. V. (2012). Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol. Evol. 4, 1–12. doi: 10.1093/gbe/evr123
King, G. A., Hashemi Shabestari, M., Taris, K. H., Pandey, A. K., Venkatesh, S., Thilagavathi, J., et al. (2018). Acetylation and phosphorylation of human TFAM regulate TFAM-DNA interactions via contrasting mechanisms. Nucleic Acids Res. 46, 3633–3642. doi: 10.1093/nar/gky204
Kolesar, J. E., Wang, C. Y., Taguchi, Y. V., Chou, S. H., Kaufman, B. A. (2013). Two-dimensional intact mitochondrial DNA agarose electrophoresis reveals the structural complexity of the mammalian mitochondrial genome. Nucleic Acids Res. 41, e58. doi: 10.1093/nar/gks1324
Kukat, C., Wurm, C. A., Spahr, H., Falkenberg, M., Larsson, N. G., Jakobs, S. (2011). Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U.S.A. 108, 13534–13539. doi: 10.1073/pnas.1109263108
Kukat, C., Davies, K. M., Wurm, C. A., Spahr, H., Bonekamp, N. A., Kuhl, I., et al. (2015). Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 112, 11288–11293. doi: 10.1073/pnas.1512131112
L’Abbe, D., Duhaime, J. F., Lang, B. F., Morais, R. (1991). The transcription of DNA in chicken mitochondria initiates from one major bidirectional promoter. J. Biol. Chem. 266, 10844–10850.
Le, T. B. K., Imakaev, M. V., Mirny, L. A., Laub, M. T. (2013). High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734. doi: 10.1126/science.1242059
Lee, S. R., Han, J. (2017). Mitochondrial nucleoid: shield and switch of the mitochondrial genome. Oxid. Med. Cell Longev. 8060949. doi: 10.1155/2017/8060949
Levin, L., Blumberg, A., Barshad, G., Mishmar, D. (2014). Mito-nuclear co-evolution: the positive and negative sides of functional ancient mutations. Front. In Genet. 5, 448. doi: 10.3389/fgene.2014.00448
Liu, H., Yin, T., Zhang, X., Zhao, X. (2019). Identifying pig mitochondrial TSS: structure and functional features. Mitochondrion 49, 19–24. doi: 10.1016/j.mito.2019.07.001
Lodeiro, M. F., Uchida, A., Bestwick, M., Moustafa, I. M., Arnold, J. J., Shadel, G.’S., et al. (2012). Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro. Proc. Natl. Acad. Sci. U.S.A. 109, 6513–6518. doi: 10.1073/pnas.1118710109
Low, R. L., Orton, S., Friedman, D. B. (2003). A truncated form of DNA topoisomerase IIbeta associates with the mtDNA genome in mammalian mitochondria. Eur. J. Biochem. 270, 4173–4186. doi: 10.1046/j.1432-1033.2003.03814.x
Lu, B., Liu, T., Crosby, J. A., Thomas-Wohlever, J., Lee, I., Suzuki, C. K. (2003). The ATP-dependent Lon protease of Mus musculus is a DNA-binding protein that is functionally conserved between yeast and mammals. Gene 306, 45–55. doi: 10.1016/s0378-1119(03)00403-7
Lu, B., Yadav, S., Shah, P. G., Liu, T., Tian, B., Pukszta, S., et al. (2007). Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. J. Biol. Chem. 282, 17363–17374. doi: 10.1074/jbc.M611540200
Lyonnais, S., Tarres-Sole, A., Rubio-Cosials, A., Cuppari, A., Brito, R., Jaumot, J., et al. (2017). The human mitochondrial transcription factor A is a versatile G-quadruplex binding protein. Sci. Rep. 7, 43992. doi: 10.1038/srep43992
Marom, S., Blumberg, A., Kundaje, A., Mishmar, D. (2019). mtDNA Chromatin-like organization is gradually established during Mammalian Embryogenesis. iScience 12, 141–151. doi: 10.1016/j.isci.2018.12.032
Martin, M., Cho, J., Cesare, A. J., Griffith, J. D., Attardi, G. (2005). Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell 123, 1227–1240. doi: 10.1016/j.cell.2005.09.040
Matsuda, S., Yasukawa, T., Sakaguchi, Y., Ichiyanagi, K., Unoki, M., Gotoh, K., et al. (2018). Accurate estimation of 5-methylcytosine in mammalian mitochondrial DNA. Sci. Rep. 8, 5801. doi: 10.1038/s41598-018-24251-z
McKinnon, P. J. (2016). Topoisomerases and the regulation of neural function. Nat. Rev. Neurosci. 17, 673–679. doi: 10.1038/nrn.2016.101
Mendoza, O., Bourdoncle, A., Boule, J. B., Brosh, R. M., Jr., Mergny, J. L. (2016). G-quadruplexes and helicases. Nucleic Acids Res. 44, 1989–2006. doi: 10.1093/nar/gkw079
Mishmar, D., Mandel-Gutfreund, Y., Margalit, H., Rahat, A., Kerem, B. (1999). Common fragile sites: G-band characteristics within an R-band. Am. J. Hum. Genet. 64, 908–910. doi: 10.1086/302299
Montoya, J., Gaines, G. L., Attardi, G. (1983). The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell 34, 151–159. doi: 10.1016/0092-8674(83)90145-9
Morozov, Y. I., Temiakov, D. (2016). Human mitochondrial transcription initiation complexes have similar topology on the light and heavy strand promoters. J. Biol. Chem. 291, 13432–13435. doi: 10.1074/jbc.C116.727966
Mposhi, A., Van der Wijst, M. G., Faber, K. N., Rots, M. G. (2017). Regulation of mitochondrial gene expression, the epigenetic enigma. Front. Biosci. (Landmark Ed) 22, 1099–1113. doi: 10.2741/4535
Murat, P., Balasubramanian, S. (2014). Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 25, 22–29. doi: 10.1016/j.gde.2013.10.012
Naeem, M. M., Maheshan, R., Costford, S. R., Wahedi, A., Trajkovski, M., Plavec, J., et al. (2019). G-Quadruplex mediated reduction of a pathogenic mitochondrial heteroplasmy. Hum. Mol. Genet. 28 (19), 3163–3174. doi: 10.1093/hmg/ddz153
Ngo, H. B., Kaiser, J. T., Chan, D. C. (2011). The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 18, 1290–1296. doi: 10.1038/nsmb.2159
Ngo, H. B., Lovely, G. A., Phillips, R., Chan, D. C. (2014). Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 5, 3077. doi: 10.1038/ncomms4077
Oluwadare, O., Highsmith, M., Cheng, J. (2019). An overview of methods for reconstructing 3-D chromosome and genome structures from Hi-C data. Biol. Proc. Online 21, 7. doi: 10.1186/s12575-019-0094-0
Phanstiel, D. H., Van Bortle, K., Spacek, D., Hess, G. T., Shamim, M. S., Machol, I., et al. (2017). Static and Dynamic DNA Loops form AP-1-Bound activation hubs during macrophage development. Mol. Cell 67, 1037–1048 e6. doi: 10.1016/j.molcel.2017.08.006
Phillips, W. S., Brown, A. M., Howe, D. K., Peetz, A. B., Blok, V. C., Denver, D. R., et al. (2016). The mitochondrial genome of Globodera ellingtonae is composed of two circles with segregated gene content and differential copy numbers. BMC Genomics 17, 706. doi: 10.1186/s12864-016-3047-x
Pittis, A. A., Gabaldon, T. (2016). Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531, 101–104. doi: 10.1038/nature16941
Rada-Iglesias, A., Bajpai, R., Swigut, T., Brugmann, S. A., Flynn, R. A., Wysocka, J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283. doi: 10.1038/nature09692
Randi, E., Lucchini, V. (1998). Organization and evolution of the mitochondrial DNA control region in the avian genus Alectoris. J. Mol. Evol. 47, 449–462. doi: 10.1007/pl00006402
Rebelo, A. P., Dillon, L. M., Moraes, C. T. (2011). Mitochondrial DNA transcription regulation and nucleoid organization. J. Inherited Metab. Dis. 34, 941–951. doi: 10.1007/s10545-011-9330-8
Rhodes, D., Lipps, H. J. (2015). G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 43, 8627–8637. doi: 10.1093/nar/gkv862
Rubio-Cosials, A., Sidow, J. F., Jimenez-Menendez, N., Fernandez-Millan, P., Montoya, J., Jacobs, H. T., et al. (2011). Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 18, 1281–1289. doi: 10.1038/nsmb.2160
Sagan, L. (1967). On the origin of mitosing cells. J. Theor. Biol. 14, 255–274. doi: 10.1016/0022-5193(67)90079-3
Sauer, M., Paeschke, K. (2017). G-quadruplex unwinding helicases and their function in vivo. Biochem. Soc. Trans. 45, 1173–1182. doi: 10.1042/BST20170097
Shao, R., Zhu, X. Q., Barker, S. C., Herd, K. (2012). Evolution of extensively fragmented mitochondrial genomes in the lice of humans. Genome Biol. Evol. 4, 1088–1101. doi: 10.1093/gbe/evs088
She, H., Yang, Q., Shepherd, K., Smith, Y., Miller, G., Testa, C., et al. (2011). Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J. Clin. Invest. 121, 930–940. doi: 10.1172/JCI43871
Shoffner, J. M., Lott, M. T., Voljavec, A. S., Soueidan, S. A., Costigan, D. A., Wallace, D. C. (1989). Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc. Natl. Acad. Sci. United States America 86, 7952–7956. doi: 10.1073/pnas.86.20.7952
Sobek, S., Dalla Rosa, I., Pommier, Y., Bornholz, B., Kalfalah, F., Zhang, H., et al. (2013). Negative regulation of mitochondrial transcription by mitochondrial topoisomerase I. Nucleic Acids Res. 41, 9848–9857. doi: 10.1093/nar/gkt768
Spruyt, N., Delarbre, C., Gachelin, G., Laudet, V. (1998). Complete sequence of the amphioxus (Branchiostoma lanceolatum) mitochondrial genome: relations to vertebrates. Nucleic Acids Res. 26, 3279–3285. doi: 10.1093/nar/26.13.3279
Stoccoro, A., Siciliano, G., Migliore, L., Coppede, F. (2017). Decreased methylation of the mitochondrial D-loop region in late-onset Alzheimer’s disease. J. Alzheimers Dis. 59, 559–564. doi: 10.3233/JAD-170139
Terzioglu, M., Ruzzenente, B., Harmel, J., Mourier, A., Jemt, E., Lopez, M. D., et al. (2013). MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab. 17, 618–626. doi: 10.1016/j.cmet.2013.03.00
Uchida, A., Murugesapillai, D., Kastner, M., Wang, Y., Lodeiro, M. F., Prabhakar, S., et al. (2017). Unexpected sequences and structures of mtDNA required for efficient transcription from the first heavy-strand promoter. Elife 6, e27283. doi: 10.7554/eLife.27283
Varizhuk, A., Isaakova, E., Pozmogova, G. (2019). DNA G-Quadruplexes (G4s) modulate epigenetic (Re)Programming and chromatin remodeling: transient genomic G4s Assist in the establishment and maintenance of epigenetic marks, while persistent G4s may erase epigenetic marks. Bioessays 41, e1900091. doi: 10.1002/bies.201900091
Vos, S. M., Tretter, E. M., Schmidt, B. H., Berger, J. M. (2011). All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 12, 827–841. doi: 10.1038/nrm3228
Wang, Y., Bogenhagen, D. F. (2006). Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 281, 25791–25802. doi: 10.1074/jbc.M604501200
Wang, Z., Wu, M. (2015). An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 5, 7949. doi: 10.1038/srep07949
Wang, Y. E., Marinov, G. K., Wold, B. J., Chan, D. C. (2013). Genome-wide analysis reveals coating of the mitochondrial genome by TFAM. PloS One 8, e74513. doi: 10.1371/journal.pone.0074513
Yakubovskaya, E., Mejia, E., Byrnes, J., Hambardjieva, E., Garcia-Diaz, M. (2010). Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription. Cell 141, 982–993. doi: 10.1016/j.cell.2010.05.018
Zhang, H., Barcelo, J. M., Lee, B., Kohlhagen, G., Zimonjic, D. B., Popescu, N. C., et al. (2001). Human mitochondrial topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 98, 10608–10613. doi: 10.1073/pnas.191321998
Zhang, H., Zhang, Y. W., Yasukawa, T., Dalla Rosa, I., Khiati, S., Pommier, Y. (2014). Increased negative supercoiling of mtDNA in TOP1mt knockout mice and presence of topoisomerases IIalpha and IIbeta in vertebrate mitochondria. Nucleic Acids Res. 42, 7259–7267. doi: 10.1093/nar/gku384
Zhu, P., Li, G. (2016). Structural insights of nucleosome and the 30-nm chromatin fiber. Curr. Opin. Struct. Biol. 36, 106–115. doi: 10.1016/j.sbi.2016.01.013
Zollo, O., Sondheimer, N. (2017). Topological requirements of the mitochondrial heavy-strand promoters. Transcription 8, 307–312. doi: 10.1080/21541264.2017.1331156
Keywords: ATAC-seq, DNase-seq, G-quadruplex, higher order organization, mtDNA, mitochondrial transcription factor A
Citation: Mishmar D, Levin R, Naeem MM and Sondheimer N (2019) Higher Order Organization of the mtDNA: Beyond Mitochondrial Transcription Factor A. Front. Genet. 10:1285. doi: 10.3389/fgene.2019.01285
Received: 14 August 2019; Accepted: 21 November 2019;
Published: 20 December 2019.
Edited by:
Aurora Gomez-Duran, University of Cambridge, United KingdomReviewed by:
Maria Solà, Instituto de Biología Molecular de Barcelona (IBMB), SpainCopyright © 2019 Mishmar, Levin, Naeem and Sondheimer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Mishmar, ZG1pc2htYXJAYmd1LmFjLmls; Neal Sondheimer, bmVhbC5zb25kaGVpbWVyQHNpY2traWRzLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.