- 1Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences, Novosibirsk, Russia
- 2Natural Science Department, Novosibirsk State University, Novosibirsk, Russia
Aggressiveness is a hereditary behavioral pattern that forms a social hierarchy and affects the individual social rank and accordingly quality and duration of life. Thus, genome-wide studies of human aggressiveness are important. Nonetheless, the aggressiveness-related genome-wide studies have been conducted on animals rather than humans. Recently, in our genome-wide study, we uncovered natural selection against underexpression of human aggressiveness-related genes and proved it using F1 hybrid mice. Simultaneously, this natural selection equally supports two opposing traits in humans (dominance and subordination) as if self-domestication could have happened with its disruptive natural selection. Because there is still not enough scientific evidence that this could happen, here, we verified this natural selection pattern using quantitative PCR and two outbred rat lines (70 generations of artificial selection for aggressiveness or tameness, hereinafter: domestication). We chose seven genes—Cacna2d3, Gad2, Gria2, Mapk1, Nos1, Pomc, and Syn1—over- or underexpression of which corresponds to aggressive or domesticated behavior (in humans or mice) that has the same direction as natural selection. Comparing aggressive male rats with domesticated ones, we found that these genes are overexpressed statistically significantly in the hypothalamus (as a universal behavior regulator), not in the periaqueductal gray, where there was no aggressiveness-related expression of the genes in males. Database STRING showed statistically significant associations of the human genes homologous to these rat genes with long-term depression, circadian entrainment, Alzheimer’s disease, and the central nervous system disorders during chronic IL-6 overexpression. This finding more likely supports positive perspectives of further studies on self-domestication syndromes.
Introduction
Ethologists define aggressiveness as a hereditary behavioral pattern important for preservation of the species. It is associated with the establishment of a social hierarchy in society and contributes to an increase in the individual reproductive potential (Lorenz, 2002). Because the individual social rank affects both quality and duration of life (Michopoulos et al., 2012), the genome-wide studies on human aggressiveness are vitally important. Yet, this research is still performed on laboratory animals: mice (Kudryavtseva et al., 2017), rats (Alttoa et al., 2010), rabbits (Albert et al., 2012), pigs (Zhou et al., 2014), dogs (Zapata et al., 2016), foxes (Kukekova et al., 2018), chickens (Natt et al., 2012), and trout (Christie et al., 2016), without the reference human genome (Telenti et al., 2016).
The reference human genome is represented by the databases Ensembl (Zerbino et al., 2015) [as a summary of over 104 individual genomes (Telenti et al., 2016)], dbSNP (Sherry et al., 2001) [as over 108 single-nucleotide polymorphisms (SNPs)], and dbWGFP (Wu et al., 2016) as data on all the 1010 possible SNPs intended for personalized medicine (Trovato, 2014). The basic concept of personalized medicine is a clinical SNP marker of human diseases identified as the statistically significant difference in the occurrence of its alleles within the representative cohorts of patients versus relatively healthy volunteers [see, e.g. (Varzari et al., 2018)]. Because these markers affect diagnosis and treatment of humans, they need to be studied, but the conventional way to identify and validate each of the 1010 possible human SNPs will be prohibitively expensive. Because of neutrality of the majority of human SNPs (Haldane, 1957; Kimura, 1968), the genome-wide search for and discarding neutral SNPs before identification of the clinical SNP markers can reduce the cost of this research.
Earlier, we created Web service SNP_TATA_Comparator1 for genome-wide SNP analysis (Ponomarenko et al., 2015). Using it, we predicted candidate SNP markers of human aggressiveness (Chadaeva et al., 2016) and natural selection against underexpression of human aggressiveness-related genes (Chadaeva et al., 2017). Recently, we proved this natural selection pattern using F1 hybrid male mice as an animal model of human heredity (Chadaeva et al., 2019). Simultaneously, this natural selection equally supports two opposing traits in humans (i.e., dominance and subordination) as if self-domestication could have happened with its characteristic disruptive natural selection as Belyaev (1979) discovered in wild and domesticated foxes half a century ago.
Because there is still not enough scientific evidence that this human self-domestication could happen, here, we verified this natural selection pattern using quantitative PCR (qPCR) and two outbred rat lines selected artificially for aggressiveness or tameness (hereinafter: domestication). We discuss the results as evidence either in favor or against recently discovered self-domestication syndrome (Theofanopoulou et al., 2017).
Materials and Methods
Study Design
According to many studies, various phenomena can affect gene expression during artificial selection, e.g., pleiotropic and epistatic effects, co-expression of the genes affecting target traits and the genes neutral toward them as well as stress-induced epigenetic reprogramming of genes that is inheritable for the next several generations as an expression modulator of these genes. Thus, nothing can be claimed about changes in the expression of any given gene during artificial selection, with the only exception when both artificial and natural selection change the expression of this gene in the same known direction, which becomes the actual direction of its expression change. Thus, taking advantage of the natural selection against underexpression of aggressiveness-related genes, using PubMed (Lu, 2011), we chose the genes to be analyzed whose over- or underexpression corresponds to aggressive or nonaggressive (hereinafter: domesticated) behavior in various animal models of human behavior.
Additionally, for a comparison between aggressive and domesticated male rats in the expression of these genes, we used only the hypothalamus–periaqueductal gray (PAG) pair as described elsewhere (Gorinski et al., 2019). We chose the hypothalamus as a universal brain region most often used in the studies on aggressiveness of female and male animals of all ages [see, e.g. (Walker et al., 2016)], while PAG is specific for both postpartum sexual and maternal aggressiveness (Lonstein and Stern, 1998), which are both absent in the male rats being analyzed here.
Accordingly, we heuristically predicted significant overexpression of the analyzed genes in the hypothalamus, but not PAG, of the aggressive male rats in comparison with the domesticated ones. Because it is still unknown whether this pattern is true, we tested this prediction by qPCR.
Animals
The study was performed on adult male gray rats (Rattus norvegicus) selectively bred for over 70 generations for aggressive or domesticated behavior (as two outbred lines) under standard conditions of the Conventional Animal Facility of the Institute of Cytology and Genetics (Novosibirsk, Russia) as described elsewhere (Belyaev and Borodin, 1982; Naumenko et al., 1989; Plyusnina and Oskina, 1997).
The total number of rats was 12: six aggressive and six domesticated, each weighing 250–270 g, 4 months old, all from different unrelated litters. All the rats were decapitated. Following a handbook (Paxinos and Watson, 2013), we extracted samples of the hypothalamus and PAG, which were then flash-frozen in liquid nitrogen and stored at −70°C until use. Every effort was made to minimize the number of animals studied and their suffering.
All the procedures were in line with Directive 2010/63/EU of the European Parliament and of the European Council of September 22, 2010. The research protocol was approved by the Interinstitutional Commission on Bioethics at the Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences.
The Supplementary File “Supplementary experiment” describes these procedures in more detail.
RNA Extraction and qPCR
From each above-mentioned sample of either the hypothalamus or PAG from 12 male rats, total RNA was extracted with the TRIzol™ Reagent (Invitrogen, cat. #15596018) and TURBO DNA-free™ Kit (Ambion, #AM1907). Total RNA was quantified on an Invitrogen Qubit™ 2.0 fluorometer (Invitrogen–Life Technologies). Integrity of total RNA was verified by means of Agilent Bioanalyzer 2100 (Agilent). cDNA was synthesized with the Phusion RT-PCR Kit (Thermo Scientific). Using PrimerBLAST, oligonucleotide primers for qPCR were designed (Table 1). qPCR was carried out by means of the EVA Green I Kit (Syntol, #R-441)) at least twice for each sample on a CFX-96 Touch System (Bio-Rad, USA), whose output was mean ± standard error of the mean (SEM) in arbitrary units as a gene expression metric. Rpl30 (ribosomal protein L30) served as a reference gene.
Statistical Analysis
For each gene being analyzed, we converted the qPCR data (mean ± SEM) into two equivalent Dn and Up statistics corresponding the lower and upper boundaries of the [mean ± standard deviation] interval of these data (Table S, hereinafter: see Supplementary file “Supplementary experiment”), which passed both nonparametric and parametric tests, while neither the mean nor the SEM information is lost.
We conducted the Kolmogorov–Smirnov test as well as Fisher’s Z-test and the Mann–Whitney U test combined within the option “Mann–Whitney test” of software package Statistica (Statsoft™, USA).
Results
Seven Aggressiveness-Related Genes Chosen for Analysis in This Work
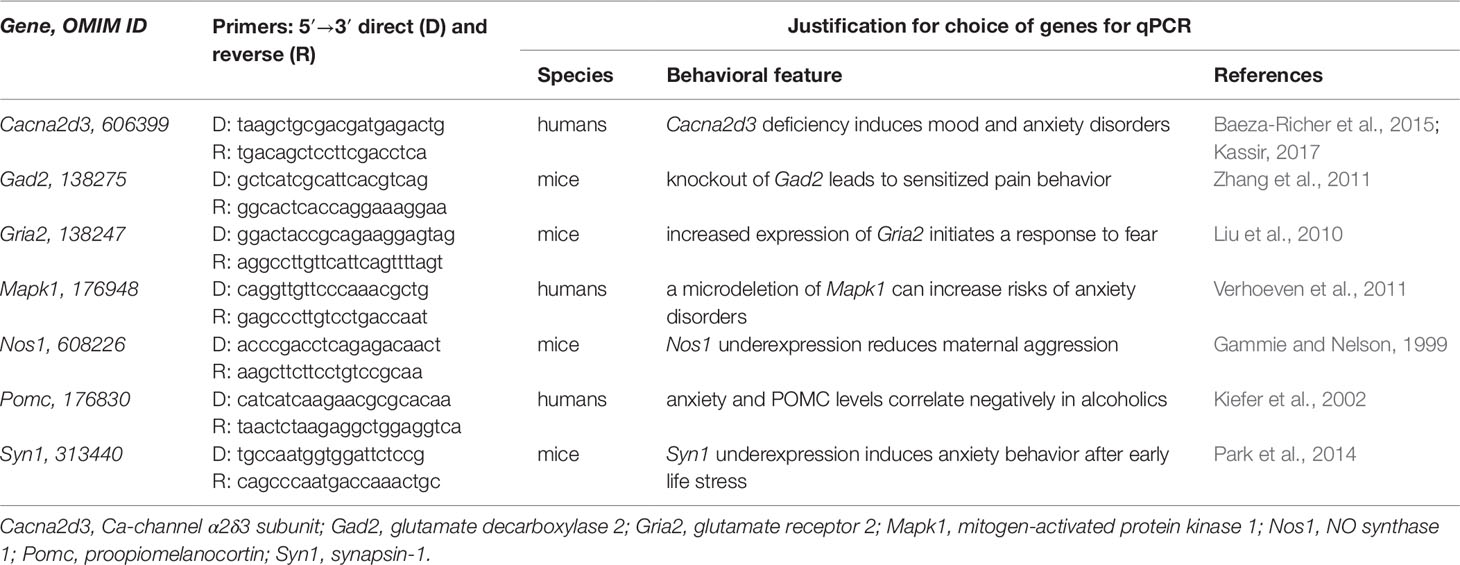
Table 1 presents the seven analyzed aggressiveness-related genes found in PubMed (Lu, 2011)—Cacna2d3, Gad2, Gria2, Mapk1, Nos1, Pomc, and Syn1—over- or underexpression of which corresponds to aggressive or domesticated behavior that has the same direction as natural selection as described in subsection “Study design.” Cacna2d3 deficiency causes iron anemia (Baeza-Richer et al., 2015), which is associated with depressive behavior (Kassir, 2017). Gad2 knockout mice are characterized by sensitized pain behavior (Zhang et al., 2011). It was shown that exposure to fox urine as a fear inducer causes Gria2 overexpression in mice (Liu et al., 2010). Verhoeven et al. (2011) described the first adolescent patient with a long history of complaints about anxiety and a distal 22q11 microdeletion containing the human Mapk1 gene, deletion of which can increase the neurobiological susceptibility to anxiety disorders as they concluded. Gammie and Nelson (1999) reported reduced maternal aggression in Nos1-deficient female mice. Kiefer et al. (2002) observed statistically significant negative linear correlations between anxiety and POMC levels in alcoholics during the two most adverse days: 1 and 14 days after alcohol withdrawal. A mouse model of human behavior (Park et al., 2014) revealed that early life stress can epigenetically reprogram Syn1 for underexpression, which causes overanxiety in adulthood.
Differential Expression of the Seven Analyzed Genes Within the Two Indicator Brain Regions
Table S presents our qPCR data on the expression levels of the seven analyzed aggressiveness-related genes in both the hypothalamus and PAG of the aggressive and domesticated male rats. These magnitudes vary from 0.12 to 3.41.
First, we conducted the Kolmogorov–Smirnov test for the normal distribution, the results of which are presented in Table S. As one can see, none of the genes being analyzed at the same time follows a normal distribution and shows equality of dispersion of its expression in the aggressive and domesticated male rats being compared. This result justifies the replacement of a pair of traditionally used statistical metrics—mean and SEM (characterizing the assumed normal distribution of experimental values)—by an equivalent pair of statistics, Dn and Up (see the main section “Materials and methods”) free of such an assumption. This assumption turned out to be erroneous in the case of our experiment.
This is why we used the “Mann–Whitney test” option of Statistica (Statsoft™, USA) to test the significance of the overexpression of these genes in the aforementioned brain regions of the aggressive male rats versus domesticated ones. The results are presented in the three rightmost columns of Table S and, additionally, in Figure 1.
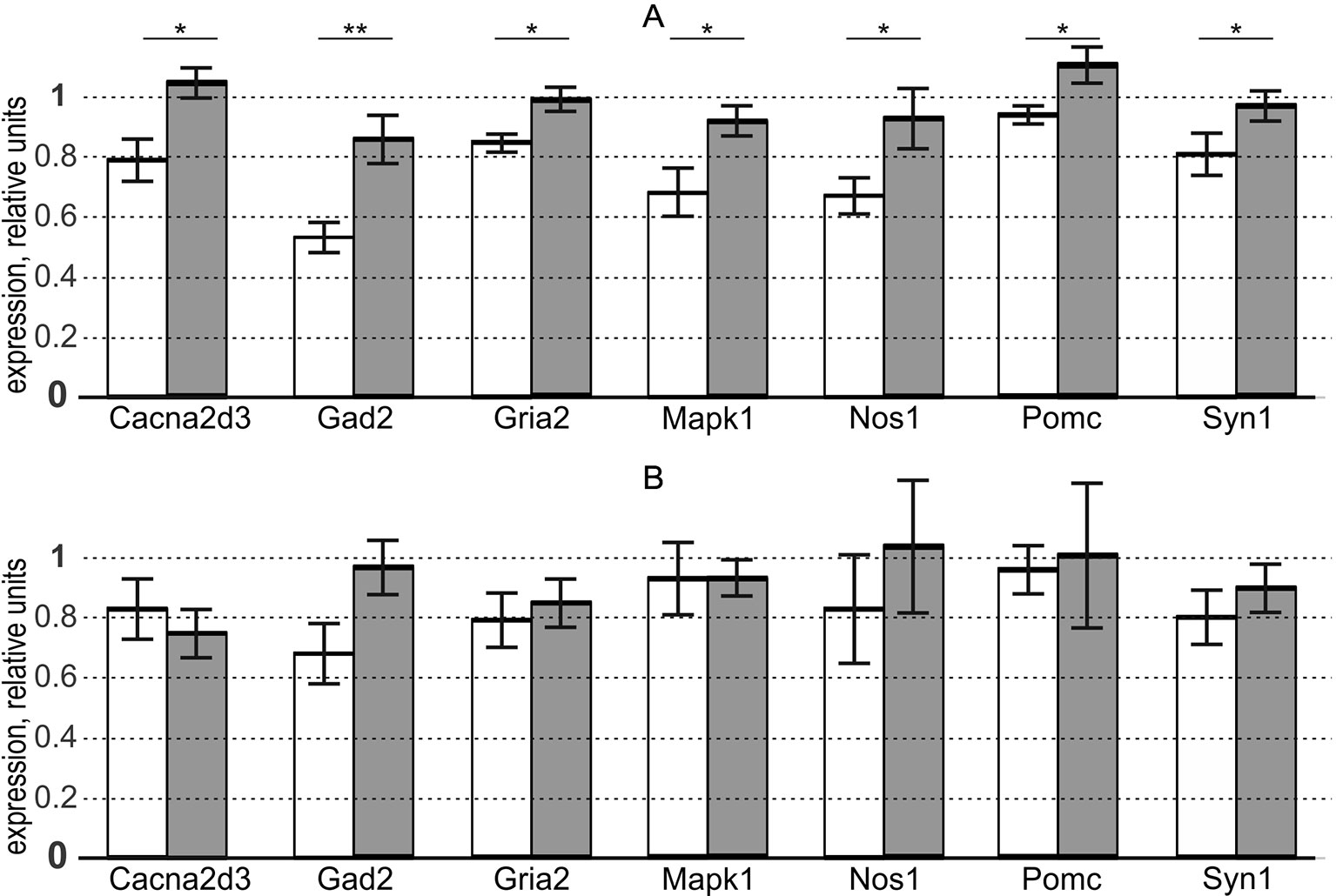
Figure 1 In aggressive male adult rats versus domesticated ones, seven genes selected via the criteria of our study design are statistically significantly overexpressed in the hypothalamus, but not in PAG, where there is no expression of the aggressiveness-related genes in males. white bars, domesticated rats; grey bars, aggressive rats; bar height, mean; error bars, standard error of mean (SEM); Asterisks * and ** denote statistical significance at p < 0.05 and p < 0.01, respectively. (A) hypothalamus (as an indicator of the presence of the aggressiveness-related expression of genes). (B) periaqueductal gray (as an indicator of the absence of the aggressiveness-related expression of genes, as a control within the framework of this study). Table S presents the gene expression levels revealed by qPCR and their statistical analysis performed in this work (see Supplementary file “Supplementary experiment”).
Panel (A) of the figure shows differential expression of the chosen genes in the hypothalamus. As readers can see, we found that the aggressive and domesticated male rats differ in the expression of these genes, in agreement with the rationale for the choice of these genes (Table 1) and widespread use of the hypothalamus as a universal behavioral regulator (Walker et al., 2016).
Panel (B) of Figure 1 reveals the absence of differences between the two groups of male rats (aggressive and domesticated) in the expression of these genes in PAG. This finding is consistent with the specificity of this indicator brain region for both postpartum sexual aggression and maternal aggression (Lonstein and Stern, 1998), which are both absent in the male rats used here (as a control within the framework of this study).
Given that the “Mann–Whitney test” option of Statistica (Statsoft™, USA) additionally carries out Fisher’s Z-test, Table S contains its results too. Thus, both the nonparametric Mann–Whitney U test and parametric Fisher’s Z-test independently confirmed our prediction of the overexpression of the analyzed genes in the hypothalamus, but not PAG, of the aggressive male rats versus domesticated ones (see subsection “Study design”). This means robustness of the match between our prediction and qPCR data with respect to the variation of the statistical tests.
Using Bonferroni’s correction for multiple tests, we evaluated the statistical significance of the overall result of our study as a whole as follows. Within the approximation of equiprobable positive and negative outcomes (p0 = 1/2) of each of the 14 comparisons between our prediction and qPCR data (Table S), the probability of the observed or better final result is pΣ = p014 = (1/2)14 = 2−14. Because our final result generalizes N = 56 partial statistical tests (i.e., 28 Kolmogorov–Smirnov tests, 14 Mann–Whitney U tests, and 14 Fisher’s Z-tests, as shown in Table S), Bonferroni’s correction of the statistical significance is pBonferroni = NpΣ = 56 × 2-14 < 100/10000 = 0.01. This result means that the overall findings of our study as a whole are statistically significant too (i.e., we analyzed sufficient and balanced numbers and sufficient diversity of statistical criteria, animals, brain regions, and genes within this project).
Discussion
On the basis of our prediction of natural selection against underexpression of aggressiveness-related genes (Chadaeva et al., 2017), which was already proved on F1 hybrid mice (Chadaeva et al., 2019), here we predicted and then demonstrated overexpression of Cacna2d3, Gad2, Gria2, Mapk1, Nos1, Pomc, and Syn1 in the hypothalamus, but not PAG, of aggressive male rats versus domesticated ones. This result means predictability of this phenomenon only within the framework of the basic principle of breeding genetics taken into account together with the natural selection against underexpression of aggressiveness-related genes rather than exceptional importance of these genes for aggressive behavior. For example, many studies show the key roles of genes 5HTT, Comp, Maoa, and Drd4 for manifestation of aggressive behavior. By contrast, overexpression of Comp reduces aggressiveness (Wilhelm et al., 2013), whereas either overexpression or underexpression of 5HTT, depending on the brain region, can either weaken or enhance this trait (Kudryavtseva et al., 2017). Underexpression of genes Maoa (Bortolato et al., 2018) and Drd4 (Keck et al., 2013) can increase it. Consequently, these genes well known for their association with aggressiveness cannot be in our set of analyzed genes (see “Study Design” subsection).
According to Bonferroni’s correction for multiple tests, our study as a whole is statistically significant and involves both sufficient and balanced diversity of animals, brain regions, and genes. Indeed, Cushing (2016) exemplified such a qPCR study using the same number of animals as in our study, as did Ilchibaeva et al. (2018) in terms of the total number of the genes being analyzed.
Although many brain regions are studied regarding aggressiveness [see, e.g. Ilchibaeva et al. (2018)], in this work, we chose the hypothalamus–PAG pair as two indicators (“presence” and “absence,” respectively) of the aggressiveness-related expression of genes, to compare their gene expression patterns as did Gorinski et al. (2019) using prefrontal cortex and cerebellum in the case of depression.
Besides, according to the recommendations of Bustin et al. (2009), several reference genes should be used for any quantitative analysis of any gene expression changes during embryogenesis, postnatal ontogenesis, puberty, pregnancy, aging, pathogenesis, and recovery as well as for the early diagnosis of pathologies and for monitoring of treatment efficacy. That is why, without quantitative measures, we qualitatively answered the binary question “Do intergroup differences exceed the intergroup similarities statistically significantly or insignificantly in one animal model of human aggressiveness (Figure 1) designed using the fact that this difference is already proven in another model of human aggressiveness (Table 1)?” To give an example, wild and domesticated foxes differ in colors (a qualitative trait, not a quantitative trait): red (main) and white (albino) versus red (main) and white (albino) as well as “Black crystal,” “Ermine-like,” “Silver sable-like,” “Amber-gold pastel,” “Steel-blue,” “Platinum,” “Cobalt,” “Purple,” “Pearl,” “Ashen,” “Beige,” “Peach,” “Straw,” and many other commercial fur colors rarely or never seen in the wild (Ponomarenko et al., 2017).
As for the amount of research data on the human genes homologous to the seven rat genes studied here, using these genes and search option “Human,” public database STRING2 (von Mering et al., 2005) associated them statistically significantly with long-term depression (p < 0.001), circadian entrainment (p < 0.001), Alzheimer’s disease (p < 0.025), Cushing’s syndrome (p < 0.025), and other health problems. In addition, it associated these genes with the research articles on disorders of the central nervous system during chronic IL-6 overexpression at p < 0.00001 (Gruol et al., 2011) and with articles on stress-caused transcriptome changes at p < 0.001 (Liu et al., 2008), among other research papers. In particular, more than a half (four of seven: Cacna2d3, Mapk1, Pomc, and Syn1) of underexpression cases found here associate domestication with anxiety, which is the key trait for mutual trust within a human–pet pair, as demonstrated for dogs (Zapata et al., 2016), sheep (Coulon et al., 2014), and guinea pigs (Kaiser et al., 2015). Notably, the original research on sports medicine (Tiric-Campara et al., 2012) related to healthy young boxers, kick boxers, and karate fighters, whose combat intensity increases with their increasing anxiety, indicates that this phenomenon prevents injuries under extreme conditions in the arena until the end of a fight or sparring. Finally, Nobel laureate Daniel Kahneman and his colleagues (Keinan et al., 1999) have shown the importance of anxiety within the framework of Stroop-like interference effects (Stroop, 1935) during human economic decision making under chronic psychological or social stress.
Conclusion
Altogether, our findings seem to support positive perspectives of further studies on recently discovered domestication syndromes (Theofanopoulou et al., 2017). Consequently, because the current study considered human aggression under the influence of a heuristically limited number of genes that have many additional functions in many other traits, it would be relevant and worthwhile to extend uniformly this gene set to differentially expressed genes (DEGs) in genome-wide transcriptomes for aggressive and domesticated animals (e.g., rats as in this study) compared with each other. In this case, next, the search for the statistically significant similarities between such DEGs of a given aggressive animal versus domesticated animal (in comparison with the DEGs seen in patients with a certain disease versus relatively healthy volunteers) may point to target genes as an answer to the question “Which domesticated animals (and how) can serve as models for which human diseases?” Finally, on this basis, using our public Web service SNP_TATA_Comparator (Ponomarenko et al., 2015), it may be possible to predict candidate SNP markers within these target genes for the above-mentioned diseases in humans as self-domestication syndromes. These computer-based predictions could accelerate the clinical arbitrary search for such genetic markers in cohorts of patients as compared to volunteers. Accordingly, Table 1 and Figure 1 suggest that the domesticated male rats studied here represent a candidate animal model of either CACNA2D3-, MAPK1, POMC-, or SYN1-deficient chronic anxiety disorders as human self-domestication syndromes. In this regard, Theodore Dobzhansky (1963) generalized self-domestication–related traits as follows: “man is genetically specialized to be unspecialized.”
Ethics Statement
The research protocol was approved by the Interinstitutional Commission on Bioethics at the Institute of Cytology and Genetics of the Siberian Branch of the Russian Academy of Sciences (Novosibirsk, Russia).
Author Contributions
MP wrote the manuscript. SS and RK performed the experiment on rats. NK and IC performed the qPCR experiment. AB contributed to data analysis. ES contributed to corrections in the revised manuscript. DO contributed to the keyword search in Web databases.
Funding
Manuscript writing was supported by SB RAS Integration Project #0324-2018-0021 (for MP). The use of the equipment at the Center for Genetic Resources of Laboratory Animals at ICG SB RAS was supported by project #RFMEFI62117X0015 from the Russian Ministry of Science and Higher Education and by project #0324-2019-0041 from the Russian Government Budget (for SS). The data compilation was supported by the 5-100 Excellence Programme (for RK). The qPCR experiment was performed on the equipment of the Genomic Research Center at ICG SB RAS that was supported by the project #0324-2019-0042 from the Russian Government Budget (for NK). The concept and study design were supported by grant #18-34-00496 from the Russian Foundation for Basic Research (for IC). The data analysis was supported by projects: #0324-2019-0040 from the Russian Government Budget (for AB). Correction of the revised manuscript was supported by the project “Investigation, analysis and complex independent expertise of projects of the National technological initiatives, including the accompanying of projects of ‘road map’ ‘NeuroNet’,” which is executed within the framework of the state assignment #28.12487.2018/12.1 of the Ministry of Science and Higher Education of the Russian Federation (for ES). The Web database search was supported by the Russian Federal Science & Technology Program for the Development of Genetic Technologies (for DO).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Shevchuk Editing 3 (Brooklyn, NY, USA) for English-language editing.
Footnotes
- ^ http://wwwmgs.bionet.nsc.ru/cgi-bin/mgs/tatascan/start.pl
- ^ https://string-db.org/
- ^ http://www.shevchuk-editing.com
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01267/full#supplementary-material
References
Albert, F. W., Somel, M., Carneiro, M., Aximu-Petri, A., Halbwax, M., et al. (2012). A comparison of brain gene expression levels in domesticated and wild animals. PloS Genet. 8, e1002962. doi: 10.1371/journal.pgen.1002962
Alttoa, A., Koiv, K., Hinsley, T. A., Brass, A., Harro, J. (2010). Differential gene expression in a rat model of depression based on persistent differences in exploratory activity. Eur. Neuropsychopharmacol. 20, 288–300. doi: 10.1016/j.euroneuro.2009.09.005
Baeza-Richer, C., Arroyo-Pardo, E., Blanco-Rojo, R., Toxqui, L., Remacha, A., Vaquero, M. P., et al. (2015). Genetic contribution to iron status: SNPs related to iron deficiency anaemia and fine mapping of CACNA2D3 calcium channel subunit. Blood Cells Mol. Dis. 55, 273–280. doi: 10.1016/j.bcmd.2015.07.008
Belyaev, D. K., Borodin, P. M. (1982). The influence of stress on variation and its role in evolution. Biol. Zent. Bl. 100, 705–714.
Belyaev, D. K. (1979). The Wilhelmine E. Key 1978 invitational lecture. Destabilizing selection as a factor in domestication. J. Hered. 70, 301–308. doi: 10.1093/oxfordjournals.jhered.a109263
Bortolato, M., Floris, G., Shih, J. C. (2018). From aggression to autism: new perspectives on the behavioral sequelae of monoamine oxidase deficiency. J. Neural Transm. (Vienna) 125, 1589–1599. doi: 10.1007/s00702-018-1888-y
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Chadaeva, I., Ponomarenko, M., Rasskazov, D., Sharypova, E., Kashina, E., Matveeva, M., et al. (2016). Candidate SNP markers of aggressiveness-related complications and comorbidities of genetic diseases are predicted by a significant change in the affinity of TATA-binding protein for human gene promoters. BMC Genomics 17, 995. doi: 10.1186/s12864-016-3353-3
Chadaeva, I., Rasskazov, D., Sharypova, E., Savinkova, L., Ponomarenko, P., Ponomarenko, M. (2017). Candidate SNP markers of social dominance, which may affect the affinity of the TATA-binding protein for human gene promoters. Russ. J. Genet. Appl. Res. 7, 523–537. doi: 10.1134/S2079059717050045
Chadaeva, I., Ponomarenko, P., Rasskazov, D., Sharypova, E., Kashina, E., Kleshchev, M., et al. (2019). Natural selection equally supports the human tendencies in subordination and domination: a genome-wide study with in silico confirmation and in vivo validation in mice. Front. Genet. 10, 73. doi: 10.3389/fgene.2019.00073
Christie, M. R., Marine, M. L., Fox, S. E., French, R. A., Blouin, M. S. (2016). A single generation of domestication heritably alters the expression of hundreds of genes. Nat. Commun. 7, 10676. doi: 10.1038/ncomms10676
Coulon, M., Levy, F., Ravel, C., Nowak, R., Boissy, A. (2014). Mild effects of gestational stress and social reactivity on the onset of mother-young interactions and bonding in sheep. Stress 17, 460–470. doi: 10.3109/10253890.2014.969238
Cushing, B. S. (2016). Estrogen receptor alpha distribution and expression in the social neural network of Monogamous and Polygynous Peromyscus. PloS One 11, e0150373. doi: 10.1371/journal.pone.0150373
Dobzhansky, T. (1963). “Genetic entities in hominid evolution,” in Classification and human evolution. Ed. Washburn, S. L. (Chicago: Aldine), 347–362.
Gammie, S. C., Nelson, R. J. (1999). Maternal aggression is reduced in neuronal nitric oxide synthase-deficient mice. J. Neurosci. 19, 8027–8035. doi: 10.1523/jneurosci.19-18-080271999
Gorinski, N., Bijata, M., Prasad, S., Wirth, A., Abdel Galil, D., Zeug, A., et al. (2019). Attenuated palmitoylation of serotonin receptor 5-HT1A affects receptor function and contributes to depression-like behaviors. Nat. Commun. 10, 3924. doi: 10.1038/s41467-019-11876-5
Gruol, D. L., Puro, A., Hao, C., Blakely, P., Janneke, E., Vo, K. (2011). Neuroadaptive changes in cerebellar neurons induced by chronic exposure to IL-6 . J. Neuroimmunol. 239, 28–36. doi: 10.1016/j.jneuroim.2011.08.009
Haldane, J. B. S. (1957). The cost of natural selection. J. Genet. 55, 511–524. doi: 10.1007/bf02984069
Ilchibaeva, T. V., Tsybko, A. S., Kozhemyakina, R. V., Kondaurova, E. M., Popova, N. K., Naumenko, V. S. (2018). Genetically defined fear-induced aggression: focus on BDNF and its receptors. Behav. Brain Res. 343, 102–110. doi: 10.1016/j.bbr.2018.01.034
Kaiser, S., Hennessy, M. B., Sachser, N. (2015). Domestication affects the structure, development and stability of biobehavioural profiles. Front. Zool. 12, S19. doi: 10.1186/1742-9994-12-S1-S19
Kassir, A. (2017). Iron deficiency: a diagnostic and therapeutic perspective in psychiatry. Encephale. 43, 85–89. doi: 10.1016/j.encep.2016.08.002
Keck, T. M., Suchland, K. L., Jimenez, C. C., Grandy, D. K. (2013). Dopamine D4 receptor deficiency in mice alters behavioral responses to anxiogenic stimuli and the psychostimulant methylphenidate. Pharmacol. Biochem. Behav. 103, 831–841. doi: 10.1016/j.pbb.2012.12.006
Keinan, G., Friedland, N., Kahneman, D., Roth, D. (1999). The effect of stress on the suppression of erroneous competing responses. Anxiety Stress Coping. 12, 455–476. doi: 10.1080/10615809908249321
Kiefer, F., Horntrich, M., Jahn, H., Wiedemann, K. (2002). Is withdrawal-induced anxiety in alcoholism based on beta-endorphin deficiency? Psychopharmacol. (Berl). 162, 433–437. doi: 10.1007/s00213-002-1118-y
Kimura, M. (1968). Evolutionary rate at the molecular level. Nature. 217, 624–626. doi: 10.1038/217624a0
Kudryavtseva, N. N., Smagin, D. A., Kovalenko, I. L., Galyamina, A. G., Vishnivetskaya, G. B., Babenko, V. N., et al. (2017). Serotonergic genes in the development of anxiety/depression-like state and pathology of aggressive behavior in male mice: RNA-seq data. Mol. Biol. (Mosk). 51, 288–300. doi: 10.1134/S0026893317020133
Kukekova, A. V., Johnson, J. L., Xiang, X., Feng, S., Liu, S. (2018). Red fox genome assembly identifies genomic regions associated with tame and aggressive behaviors. Nat. Ecol. Evol. 2, 1479–1491. doi: 10.1038/s41559-018-0611-6
Liu, X., Serova, L., Kvetnansky, R., Sabban, E. L. (2008). Identifying the stress transcriptome in the adrenal medulla following acute and repeated immobilization. Ann. N Y Acad. Sci. 1148, 1–28. doi: 10.1196/annals.1410.082
Liu, Y., Formisano, L., Savtchouk, I., Takayasu, Y., Szabo, G., Zukin, R. S., et al. (2010). A single fear-inducing stimulus induces a transcription-dependent switch in synaptic AMPAR phenotype. Nat. Neurosci. 13, 223–231. doi: 10.1038/nn2474
Lonstein, J. S., Stern, J. M. (1998). Site and behavioral specificity of periaqueductal gray lesions on postpartum sexual, maternal, and aggressive behaviors in rats. Brain Res. 804, 21–35. doi: 10.1016/S0006-8993(98)00642-8
Lu, Z. (2011). PubMed and beyond: a survey of web tools for searching biomedical literature. Database (Oxford) 2011, baq036. doi: 10.1093/database/baq036
Michopoulos, V., Higgins, M., Toufexis, D., Wilson, M. E. (2012). Social subordination produces distinct stress-related phenotypes in female rhesus monkeys. Psychoneuroendocrinology 37, 1071–1085. doi: 10.1016/j.psyneuen.2011.12.004
Natt, D., Rubin, C. J., Wright, D., Johnsson, M., Belteky, J., Andersson, L., et al. (2012). Heritable genome-wide variation of gene expression and promoter methylation between wild and domesticated chickens. BMC Genomics 13, 59. doi: 10.1186/1471-2164-13-59
Naumenko, E. V., Popova, N. K., Nikulina, E. M., Dygalo, N. N., Shishkina, G. T., Borodin, P. M., et al. (1989). Behavior, adrenocortical activity, and brain monoamines in Norway rats selected for reduced aggressiveness towards man. Pharmacol. Biochem. Behav. 33, 85–91. doi: 10.1016/0091-3057(89)90434-6
Park, H. J., Kim, S. K., Kang, W. S., Chung, J. H., Kim, J. W. (2014). Increased activation of synapsin 1 and mitogen-activated protein kinases/extracellular signal-regulated kinase in the amygdala of maternal separation rats. CNS Neurosci. Ther. 20, 172–181. doi: 10.1111/cns.12202
Paxinos, G., Watson, C. (2013). The rat brain in stereotaxic coordinates. 7th edition (London: Academic Press, Elsevier Inc.), 472.
Plyusnina, I., Oskina, I. (1997). Behavioral and adrenocortical responses to open-field test in rats selected for reduced aggressiveness toward humans. Physiol. Behav. 61, 381–385. doi: 10.1016/S0031-9384(96)00445-3
Ponomarenko, M., Rasskazov, D., Arkova, O., Ponomarenko, P., Suslov, V., Savinkova, L., et al. (2015). How to use SNP_TATA_Comparator to find a significant change in gene expression caused by the regulatory SNP of this gene’s promoter via a change in affinity of the TATA-binding protein for this promoter. BioMed. Res. Int. 2015, 359835. doi: 10.1155/2015/359835
Ponomarenko, M., Gunbin, K., Doroshkov, A., Suslov, V., Kolchanov, N. (2017). “Cladogenesis,” in Reference Module in Life Sciences (Elsevier Inc.). doi: 10.1016/b978-0-12-809633-8.06219-1
Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M., et al. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311. doi: 10.1093/nar/29.1.308
Stroop, J. R. (1935). Studies of interference in serial verbal reactions. J. Exp. Psychol. 18, 643–662. doi: 10.1037/h0054651
Telenti, A., Pierce, L. C., Biggs, W. H., di Iulio, J., Wong, E. H., Fabani, M. M., et al. (2016). Deep sequencing of 10,000 human genomes. Proc. Natl. Acad. Sci. USA. 113, 11901–11906. doi: 10.1073/pnas.1613365113
Theofanopoulou, C., Gastaldon, S., O’Rourke, T., Samuels, B. D., Martins, P. T., Delogu, F., et al. (2017). Self-domestication in Homo sapiens: insights from comparative genomics. PloS One 12, e0185306. doi: 10.1371/journal.pone.0185306
Tiric-Campara, M., Tupkovic, E., Mazalovic, E., Karalic, E., Biscevic, M., Djelilovic-Vranic, J., et al. (2012). Correlation of aggressiveness and anxiety in fighting sports. Med. Arh. 66, 116–121. doi: 10.5455/medarh.2012.66.116-121
Trovato, G. M. (2014). Sustainable medical research by effective and comprehensive medical skills: overcoming the frontiers by predictive, preventive and personalized medicine. EPMA J. 5, 14. doi: 10.1186/1878-5085-5-14
Varzari, A., Tudor, E., Bodrug, N., Corloteanu, A., Axentii, E., Deyneko, I. V. (2018). Age-specific association of CCL5 gene polymorphism with pulmonary tuberculosis: a case-control study. Genet. Test Mol. Biomarkers 22, 281–287. doi: 10.1089/gtmb.20170250
Verhoeven, W., Egger, J., Brunner, H., de Leeuw, N. (2011). A patient with a de novo distal 22q11.2 microdeletion and anxiety disorder. Am. J. Med. Genet. A. 155A, 392–397. doi: 10.1002/ajmg.a.33802
von Mering, C., Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., et al. (2005). STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33, D433–D437. doi: 10.1093/nar/gki005
Walker, S. E., Papilloud, A., Huzard, D., Sandi, C. (2016). The link between aberrant hypothalamic–pituitary–adrenal axis activity during development and the emergence of aggression - animal studies. Neurosci. Biobehav. Rev. 91, 138–152. doi: 10.1016/j.neubiorev.2016.10.008
Wilhelm, C. J., Choi, D., Huckans, M., Manthe, L., Loftis, J. M. (2013). Adipocytokine signaling is altered in Flinders sensitive line rats, and adiponectin correlates in humans with some symptoms of depression. Pharmacol. Biochem. Behav. 103, 643–651. doi: 10.1016/j.pbb.2012.11.001
Wu, J., Wu, M., Li, L., Liu, Z., Zeng, W., Jiang, R. (2016). dbWGFP: a database and web server of human whole-genome single nucleotide variants and their functional predictions. Database (Oxford). 2016, baw024. doi: 10.1093/database/baw024
Zapata, I., Serpell, J. A., Alvarez, C. E. (2016). Genetic mapping of canine fear and aggression. BMC Genomics 17, 572. doi: 10.1186/s12864-016-2936-3
Zerbino, D. R., Wilder, S. P., Johnson, N., Juettemann, T., Flicek, P. R. (2015). The Ensembl regulatory build. Genome Biol. 16, 56. doi: 10.1186/s13059-015-0621-5
Zhang, Z., Cai, Y. Q., Zou, F., Bie, B., Pan, Z. Z. (2011). Epigenetic suppression of GAD65 expression mediates persistent pain. Nat. Med. 17, 1448–1455. doi: 10.1038/nm2442
Keywords: behavior, rat model, aggressiveness, domestication, differentially expressed gene, hypothalamus, periaqueductal gray
Citation: Oshchepkov D, Ponomarenko M, Klimova N, Chadaeva I, Bragin A, Sharypova E, Shikhevich S and Kozhemyakina R (2019) A Rat Model of Human Behavior Provides Evidence of Natural Selection Against Underexpression of Aggressiveness-Related Genes in Humans. Front. Genet. 10:1267. doi: 10.3389/fgene.2019.01267
Received: 15 October 2018; Accepted: 18 November 2019;
Published: 13 December 2019.
Edited by:
Frank Middleton, Upstate Medical University, United StatesReviewed by:
Elena E. Terenina, Institut National de la Recherche Agronomique de Toulouse, FranceEser Ercil, Independent Researcher, United States
Copyright © 2019 Oshchepkov, Ponomarenko, Klimova, Chadaeva, Bragin, Sharypova, Shikhevich and Kozhemyakina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Irina Chadaeva, aWNoYWRhZXZhQGJpb25ldC5uc2MucnU=
 Dmitry Oshchepkov1
Dmitry Oshchepkov1 Mikhail Ponomarenko
Mikhail Ponomarenko Natalya Klimova
Natalya Klimova Irina Chadaeva
Irina Chadaeva