- 1Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai)/School of Marine Sciences, Sun Yat-sen University, Guangzhou, China
- 2State Key Laboratory of Biocontrol/School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 3Guangxi Key Laboratory of Beibu Gulf Marine Biodiversity Conservation, Beibu Gluf University, Qinzhou, China
Simple sequence repeats (SSRs) of short nucleotide motifs occur very frequently in the 5′ untranslated coding region (5′-UTR) of genes and have been implicated in the regulation of gene expression. In this study, we identified an SSR with a variable number of CT repeats in the 5′-UTR of the Litopenaeus vannamei IRF (LvIRF) gene that has been shown to mediate antiviral responses by inducing the expression of Vago, a functional homolog of mammalian IFN. We then explored the effects of varying the number of (CT)n repeats on the expression of LvIRF using both dual-luciferase reporter assays and Western blots. Our results demonstrate that the length of the (CT)n-SSR in this gene can influence the expressional level of LvIRF, in that a shorter (CT)n repeat had a stronger ability to induce the expression of LvIRF. Moreover, we found that the (CT)n repeat in LvIRF was associated with viral resistance in shrimp. Individual shrimps with shorter (CT)n repeats in the 5′-UTR of LvIRF exhibited high tolerance to white spot syndrome virus (WSSV), and this trait was inherited in offspring. Taken together, these results indicated that this (CT)n-SSR could be used as a molecular marker for shrimp breeding for WSSV resistance.
Introduction
Simple sequence repeats (SSRs), also known as microsatellites or short tandem repeats (STRs), are tandem repeats of short sequence motifs occurring ubiquitously in eukaryotic genomes. They exhibit extensive polymorphism due to variations in the copy number of each specific repeat motif, and are considered useful as genetic markers for genetic diversity analysis, DNA fingerprinting, and linkage mapping. Several studies have suggested that SSRs are non-randomly distributed in the genome, as untranslated coding regions (UTRs), and have more SSRs than coding regions, as well as exhibit a strong bias towards di- and tri-nucleotides repeats (Metzgar et al., 2000; Wren et al., 2000; Li et al., 2002; Morgante et al., 2002; Fujimori et al., 2003). SSRs in UTRs have been found to be associated with gene expression by affecting transcription factor binding, methylation of CpG and/or DNA structure modification (Li et al., 2004).
Some of the best-known examples of SSRs affecting gene expression come from human genetic disorders. For example, fragile X syndrome (FXS) has been attributed to the absence of fragile X mental retardation 1 (FMR1) gene expression due to a long CGG tri-nucleotide repeat in the FMR1 gene 5′-UTR adjacent to the promoter that results in the FMR1 gene’s epigenetic silencing (Colak et al., 2014). In another study, a polymorphic (CA)n microsatellite identified in the 5′-UTR of the prolactin 1 (prl 1) gene of tilapia has been associated with differences in prl 1 gene expression and the growth responses of salt-challenged tilapia (Streelman and Kocher, 2002). In Drosophila, a series of polymorphic (GA)n microsatellite sequences in promoter regions have been shown to bind to a protein family called GAGA factors, which are involved in gene expression (Tsukiyama et al., 1994; Berger and Dubreucq, 2012). In mammalian cells, three novel downstream elements, which show homology to GAGA factor binding sequences, can regulate promoter activity and preferentially affect transcription start site (TSS) selection at the 5′-UTR end of promoters (Lee et al., 2010).
Disease resistance is a low heritability trait that is easily influenced by external environments. Designer breeding using molecular markers is considered to be an effective method for the cultivation of disease resistance and other complex traits in crops and livestock (Xue et al., 2013). Whole genome resources and transcriptome resources provide opportunities to dissect the genes controlling complex traits and can be used to reveal the coupling mechanisms of different genes, which in turn can contribute to the use of functional genes as molecular markers for designer breeding projects (Xue et al., 2015). In order to do this, however, a thorough understanding of the functional genes related to disease resistance is needed.
The shrimp species Litopenaeus vannamei is a worldwide aquaculture species that was first introduced to China in the 1980s, and its production has increased rapidly in the 21st century. However, various shrimp diseases are responsible for huge losses, especially the white spot syndrome virus (WSSV) (Flegel, 2012; Li et al., 2019b). To date, there is no effective method to prevent WSSV infection in shrimp, but selective breeding of WSSV-resistant species should be an effective way to solve this problem.
In shrimp, innate immunity plays a key role in the defense against a wide variety of invading microbes such as bacteria, fungi, and viruses. Several signaling pathways are essential components of innate immunity, including the Toll, IMD, and JAK/STAT pathways (Li et al., 2019a; Li et al., 2019b). In recent studies, the shrimp IRF-Vago-JAK/STAT pathway, which is similar to the IRF-IFN-JAK/STAT pathway in vertebrates, has been functionally identified to play a significant role in defense against viruses including WSSV (Li et al., 2015). The L. vannamei IRF (LvIRF) is the first interferon regulatory factor (IRF) identified in crustaceans, and has a similar protein nature as mammalian IRFs. LvIRF can be activated during viral infection, and then translocates to the nucleus to initiate the expression of the L. vannamei Vago4 (LvVago4) gene. This is effectively an arthropod cytokine encoding a viral-activated secreted peptide through activating the JAK-STAT pathway to restrict viral infection, similar to mammalian IFNs (Chen et al., 2011; Paradkar et al., 2012; Wang and He, 2019).
The key constituents of innate immunity were regarded as molecular markers for breeding disease resistance in shrimp. In this study, we found a (CT)n microsatellite with a variable number of CT motifs present in the 5′-UTR of the LvIRF gene. We demonstrated that the number of (CT)n repeats modulates the promoter activity of the LvIRF gene in a length dependent manner, and observed that shrimp with different numbers of CT repeats showed distinct tolerances to WSSV. Furthermore, we demonstrate the use of the number of (CT)n repeats at this locus as a molecular marker to selectively breed a new generation of shrimp resistant to WSSV. In F2 offspring, the populations with smaller numbers of (CT)n repeats were more resistant to WSSV.
Materials and Methods
Experimental Animals and Pathogens
In order to investigate the relationship between (CT)n repeats and the antiviral traits of L. vannamei, three different populations of 100 healthy shrimp (TH01, TH03 and SP07) were collected from the Hengxing shrimp farm in Zhanjiang city, China. TH01 was collected from Vietnam, TH03 was collected from China, and SP07 was collected from Saipan. The shrimp body weight was 5.0 ± 1.0 g each, and each population of shrimp was cultured in filtered sea water with a 2.5% salinity at 26°C in a recirculating water tank and fed with fodder at rate of 5% of body weight per day. WSSV was prepared from shrimp muscle tissue previously infected with WSSV and stored at −80°C. The muscle tissue was homogenized to prepare it as a WSSV inoculum at a final concentration of 106 virions/50 μl, which was injected into each shrimp (Li et al., 2014). All tanks were checked every 2 h and dead shrimp were collected and marked. On the 10th day (240 h) after infection, all shrimp were collected and marked. In a study of the WSSV resistance of L. vannamei, the survival rate curve showed a normal distribution using this injection infection method (Huang et al., 2011). Therefore, we used mortality peaks as the cut-off point for determining resistance, and each population was divided into two groups: WSSV-susceptible and WSSV-resistant. All experimental materials were stored at −20°C with DNA holder and RNA holder (TakaRa, Dalian, China) for downstream DNA and RNA extractions.
Analysis of Allelic Polymorphism, Promoter Sequences and Preparation of Expression Vectors
Genomic DNA was extracted from muscle tissue using the E.Z.N.A. Tissue DNA Kit (Omega Bio-tek, Doraville, GA, USA). The LvIRF cDNA had previously been deposited in the NCBI GenBank (KM277954), and the L. vannamei IRF-5′-UTR was used as a DNA template for PCR amplification (Table 1) (Li et al., 2015). PCR products were electrophoresed on 20% polyacrylamide gels at 80 V for 8 h in 1 × TBE running buffer (89 mM Tris-boric acid, 2 mM EDTA, pH 8.0). The polymorphic bands were purified using the E.Z.N.A. Gel Extraction Kit (Omega Bio-tek, Doraville, GA, USA), followed by cloning into the pMD-19T vector (TakaRa, Dalian, China) and sequence confirmation. The sequences were analyzed and compared using BioEdit (Hall, 1999), and those with different in length were chosen for polymorphism analysis by sequencing.
Total RNA was extracted using the RNeasy Mine Kit (Qiagen, Hilden, Germany) and reverse transcribed into cDNA using the PrimeScript RT reagent kit (TakaRa, Dalian, China). The ORF of LvIRF without a stop codon was used as a cDNA template for PCR amplification (Table 1). PCR products were then cloned into the pAc5.1-His/V5 A vector (Invitrogen, Carlsbad, CA, USA) to generate pAc-IRF-V5 and the sequence was confirmed. Different length LvIRF-5′-UTR sequences were then amplified from pMD-19T vectors and cloned into the pAc-IRF-V5 vector to generate pAc-IRF-(CT)n-V5. These were sequenced, and a pair wise alignment was then performed. The promoter regions of LvVago4 were cloned into the PGL-3 basic vector (Promega, Madison, WI, USA) to generate a luciferase reporter gene plasmid (Chen et al., 2011).
Dual-Luciferase Reporter Assays
To detect the effects of LvIRF on the promoters of LvVago4 genes, dual-luciferase reporter assays were performed using Drosophila Schneider 2 (S2) cells with pAc-IRF-(CT)n-V5 that contained different numbers of CT repeats as IRF-expressing vectors. Cells in each well of a 96-well plate (TPP, Switzerland) were transfected with 0.05 μg PGL-LvVago4 vector as a reporter plasmid, 0.005 μg pRL-TK renilla luciferase (Promega, Madison, WI, USA), and 0.05 μg of expression plasmid, LvIRF vector or empty expression vector as controls. The pRL-TK renilla luciferase plasmid was used as an internal control. 48 h after transfection the firefly and renilla luciferase activities were measured, according to the manufacturer’s instructions. Each experiment was performed in triplicate.
Western Blot and Quantitative RT-PCR
The pAc-IRF-(CT)n-V5 vector was transfected into S2 cells. After 72 h, cells were harvested and lysed in NP-40 lysis buffer with protease inhibitor cocktail (Sigma, St. Louis, MO, USA). Western blot was performed with a rabbit anti-V5 antibody (Sigma, St. Louis, MO, USA) as a primary antibody, and alkaline phosphatase-conjugated goat anti-rabbit as a secondary antibody (Sigma, St. Louis, MO, USA). ImageJ was then used to measure the gray-level values and calculate the ratio of LvIRF and β-actin. Quantitative RT-PCR was performed for the analysis of IRF gene expression, and the EF-1α gene was detected as an internal control (Table 1).
Microsatellite Genotyping
The CT microsatellite from the LvIRF 5′-UTR region was genotyped using FAM fluorogenic probes followed by capillary electrophoresis to discriminate allele size. Primers (Table 1) were designed to amplify a 202 bp fragment containing 18 CT repeats that was used as a reference, and deviation from this size allowed us to deduce the number of CT repeats for different alleles. Samples were sequenced and analyzed using an ABI Genetic Analyzer 3730 XL. We classified the alleles into two groups: short (S) alleles were ≤ 18 repeats, and long (L) alleles were > 18 repeats, based on the median (the (CT)n repeat numbers ranged from 13 to 24) that was adopted by referring to other reports (Gregorek et al., 2013; Dias et al., 2017).
Establishment of HX-CTS and HX-CTL Generations
The L. vannamei breeding program was carried out at the Hengxing shrimp farm in Zhanjiang city, China. Choosing the HX150301 and HX1403 populations as founder stocks, two ponds were stocked with spawners, with one pond named the HX-CTS population where the parents contained short CT repeats in the LvIRF 5′-UTR, while the other one pond only contained long CT repeats, which we named the HX-CTL population (Figure 5A). When the shrimp from each pond had a body weight of 5 g, we used the artificial WSSV challenge experiment mentioned above, to draw survival rate curves.
Statistical Analysis
Allelic frequency calculations were performed using PopGen32 software (Yeh and Boyle, 1997). We used 2 × 2 contingency tables and Fisher’s exact test to obtain P-values with odds ratios (ORs) and 95% confidence intervals (95% CIs) to calculate significance, as well as two-tailed unpaired t-tests and ANOVA. Bonferroni corrections were applied to account for the multiple testing of the CT genotype frequency [*p corr < 0.0042 (0.05/12)] and genotype groups [*p corr < 0.0167 (0.05/3)], following Fisher exact tests (Lewitter et al., 2012). ANOVA tests were followed by Tukey post hoc tests (*p < 0.05).
Results
Analysis of the (CT)n Variation in the 5′-UTR of the LvIRF Gene
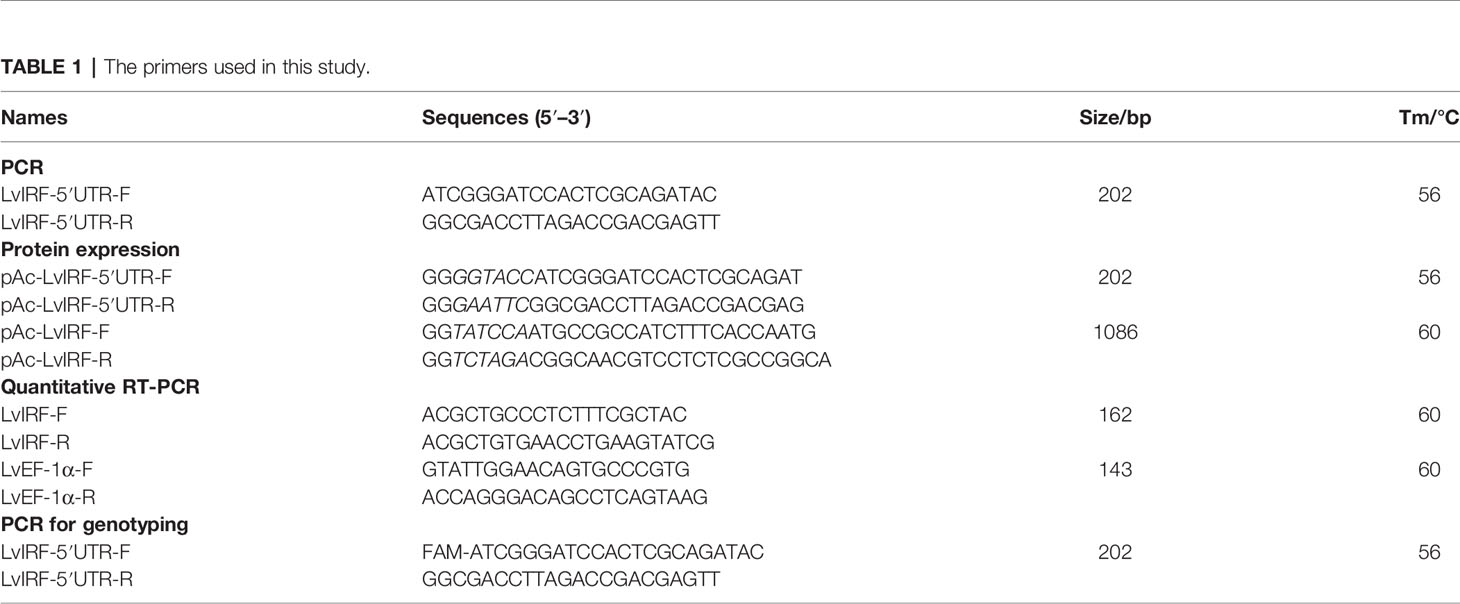
LvIRF has been shown to bind the promoter of LvVago4 to regulate its transcription, which plays a role in the defense against WSSV infection (Li et al., 2015). In the present study, we found a (CT)n repeat in the 5′-UTR region of the LvIRF gene and designed primers to amplify the 5′-UTR including the (CT)n motif. PCR amplification of genomic DNA from 20 shrimp collected from diverse genealogies was performed and the amplified products had various lengths, ranging in size from 190 to 230 bp (Figure 1A). Sequencing results showed that the (CT)n microsatellite was polymorphic in this region, and could be further divided into a long (CT)n repeat and a short (CT)n repeat. Fourteen sequences, different in (CT)n motif number only, were chosen for further study. A pair wise alignment was then made using part of these sequences, such as (CT)10+5, (CT)14+5, and (CT)18+7 (Figure 1B).
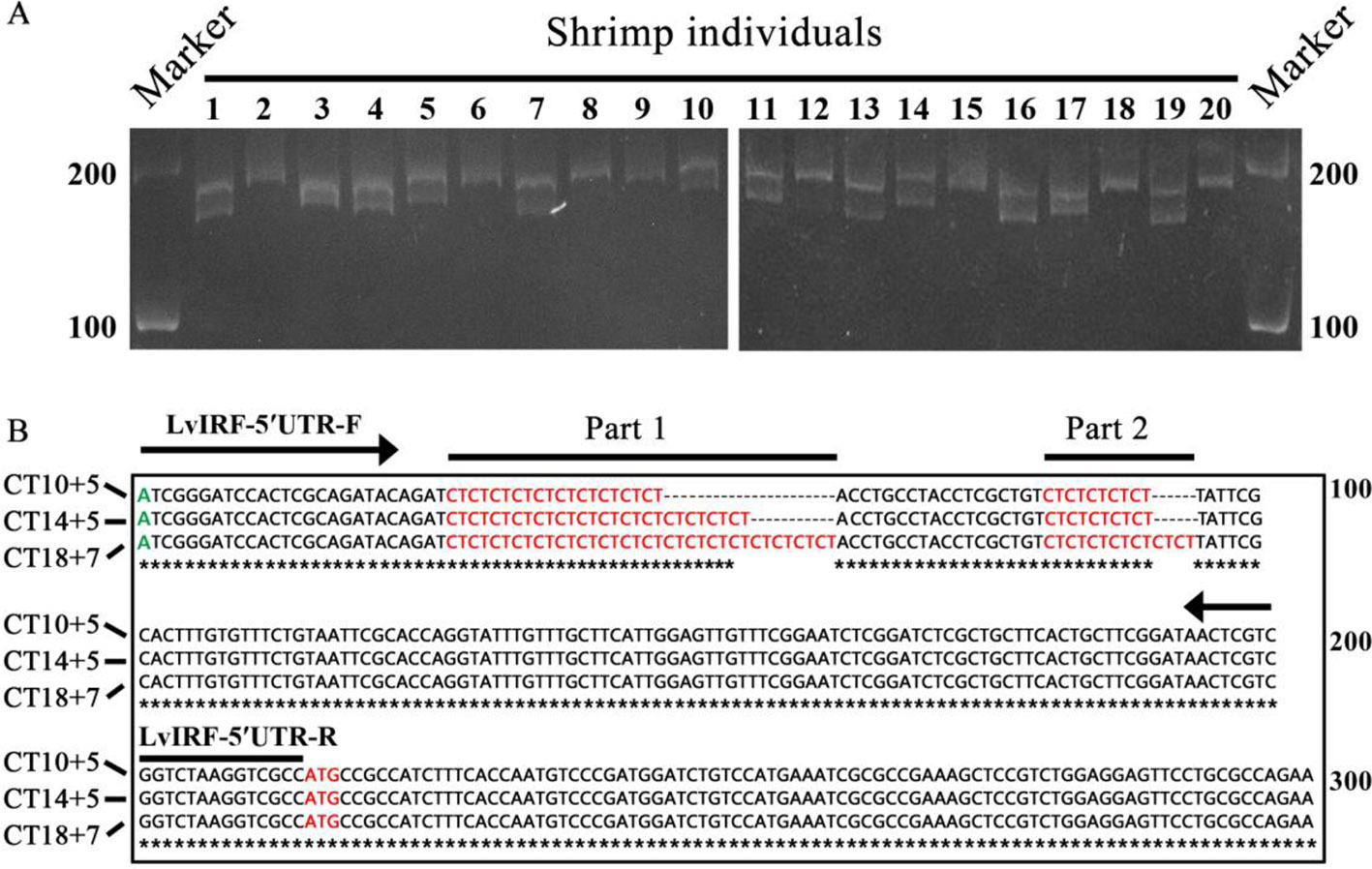
Figure 1 Polymorphisms in the 5′-UTR of LvIRF. (A) PCR amplified 5′-UTRs from 20 L. vannamei individuals were analyzed by agarose gel. Length polymorphisms between individuals was observed. The (CT)n repeat motifs was marked in red, transcription start site (TSS) was marked in green, and translation start site (ATG) was marked in red. The locations of primers LvIRF-5′UTR-F/R (expected size 202 bp) was shown to analysis the polymorphism of (CT)n repeat in shrimp. (B) Multiple sequence alignment of LvIRF 5’-UTRs. Variation in the numbers of (CT)n motifs, with a long (CT)n repeat (Part 1) and a short (CT)n repeat (Part 2), were distinctly observed.
Estimating the Effect of (CT)n Motif Number Variation on LvIRF Expression
To investigate whether the variable numbers of (CT)n motif in the 5′-UTR of LvIRF are implicated with its expression, dual-luciferase reporter analysis in Drosophila S2 cells was carried out. The 5′-UTRs of the LvIRF gene harboring various (CT)n motifs were cloned into pAc-5.1-His/V5 A to generate pAc-IRF-(CT)n-V5 vectors for protein expression (Figure 2A). Three vectors, including 15, 19, and 25 (CT)n repeats, were constructed and transfected into S2 cells. The pAc-IRF-V5 and pAc-5.1-His/V5 A empty vector were also transfected into S2 cells as controls. To estimate the effect of (CT)n motif number variation on LvIRF expression, Western blotting was then performed. The results showed that the ratio of LvIRF and β-actin with (CT)15 was twice as large as that with (CT)25 in terms of gray-level values (Figure 2B). Previous reports have shown that the promoter of LvVago4 contains a conserved IRF binding motif that was confirmed to be targeted by LvIRF (Li et al., 2015). To further confirm the above results (Figure 2B), dual luciferase reporter assays were performed using luciferase-expressing vectors containing the LvVago4 promoter (Figure 2C). Maximal expression of LvIRF was observed in cells transfected with pAc-IRF-(CT)15, which was 1.5-fold higher than in cells with pAc-IRF-(CT)25 (p < 0.001), and 2.3-fold higher than in cells with pAc-IRF-V5 (p < 0.001). Considering that the (CT)n repeat is composed of two parts, we explored whether the different numbers of the two (CT)n repeats were implicated in LvIRF expression. We observed that the pAc-IRF-(CT)11+5 and pAc-IRF-(CT)10+6 showed no significant differences on the promoter activities of LvVago4. Similar results were observed in three other groups that contained a total of 17, 18, and 19 repeats, respectively (Figure 2D). Taken together, these results revealed that the expression of LvIRF was only affected by the total number of (CT)n copies.
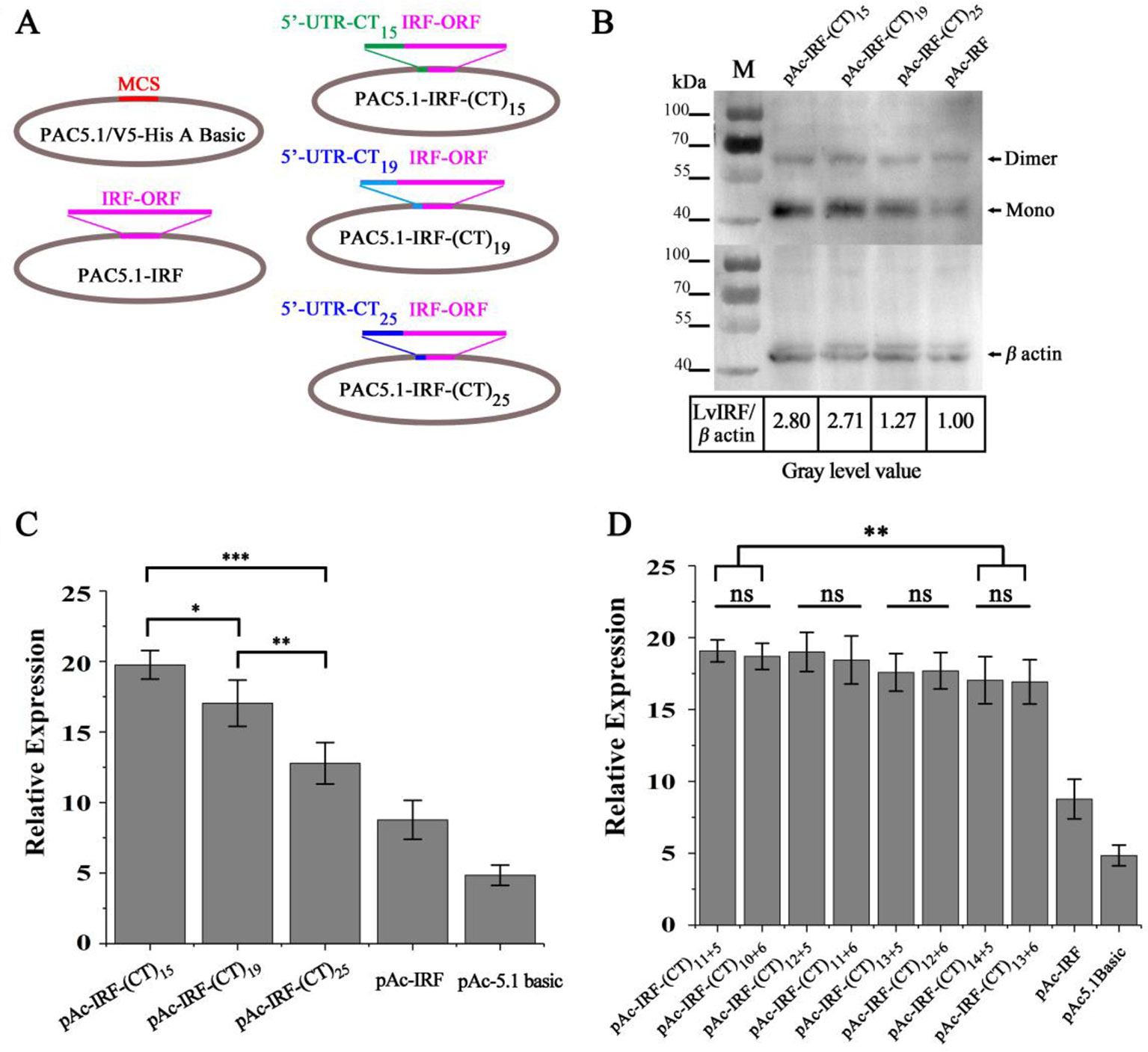
Figure 2 (CT)n repeats implicated in the expression of LvIRF. (A) Structures of 5′-UTRs harboring different (CT)n repeats and IRF coding sequence in pAc-5.1/His A vectors. (B) Western blots of LvIRF expression driven by different (CT)n repeat numbers. Gray level value ratios of LvIRF and β-actin in table, and the expression level from cells with pAc-IRF was set as the baseline (1.0). The ratio of LvIRF and β-actin with (CT)15 was twice as much as that with (CT)25. (C-D) Dual-luciferase reporter assays of LvIRF expression, (C) LvIRF expression with different total numbers of (CT)n repeats, (D) LvIRF expression with same total numbers of (CT)n repeats but different numbers of the two distinct (CT)n repeats. LvIRF expression with (CT)15 was 1.2-fold higher than that with pAc-IRF-(CT)19 (p < 0.05) and 1.5-fold higher than that with pAc-IRF-(CT)25 (p < 0.001). The pAc-IRF-(CT)11+5 and pAc-IRF-(CT)10+6 transfected cells showed no significant differences on the promoter activities of LvVago4, similar to the other three groups. *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant.
Specific (CT)n Microsatellite Genotypes Were Associated With WSSV-Resistance
According to the survival rate curve, we chose the mortality peaks (Figure 3, arrow) as the cut-off points for dividing each population of shrimp into two groups: WSSV-susceptible (Sus., died before mortality peaks) and WSSV-resistant (Res., died after mortality peaks) (Table 2). This phenotype was then genotyped in all three shrimp populations (Figure 4). The allelic distribution profile for each population was different. The number of (CT)n repeats in TH01 were skewed towards 18, 19, or 20 (Figure 4A), while TH03 was rich in 17, 18, or 20 (Figure 4B), and SP07 had an isolated peak at 20 (Figure 4C). The large difference in this allelic distribution indicated that the three populations used in this study had different genetic backgrounds, and showed a large degree of genetic stratification.
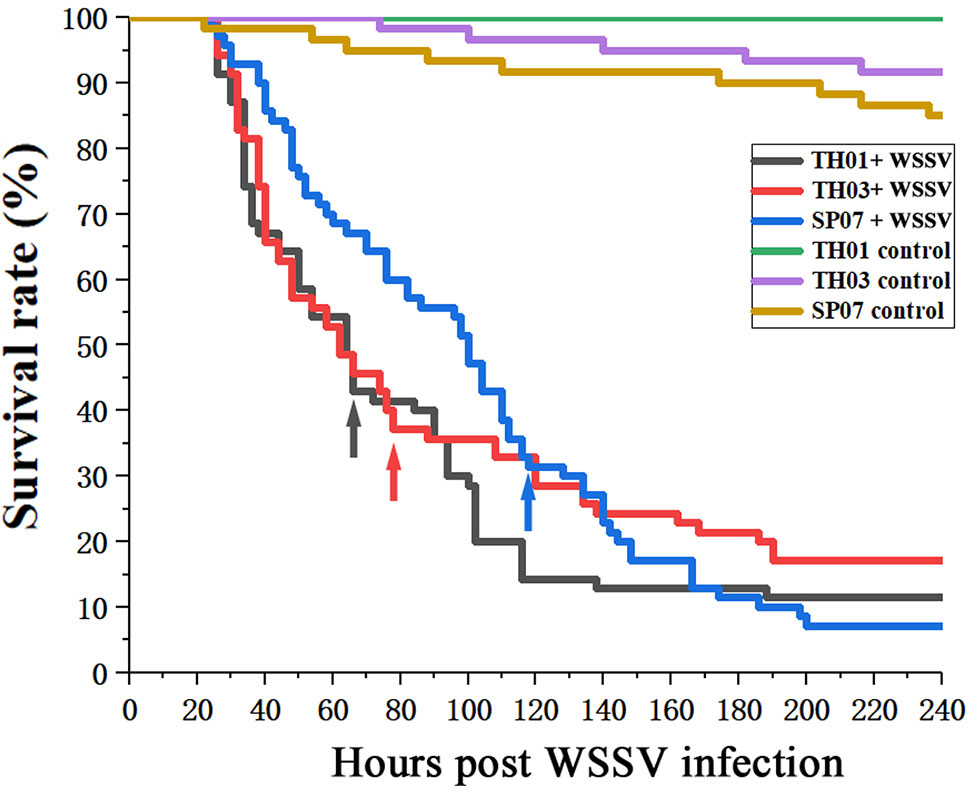
Figure 3 Survival rate curves for TH01, TH03 and SP07 shrimp. Three shrimp populations from different sources; TH01 from Vietnam, TH03 from China, and SP07 from Saipan. Arrows used to indicate the mortality peak times for the Sus. group (died before mortality peaks) and Res. group (died after mortality peaks).
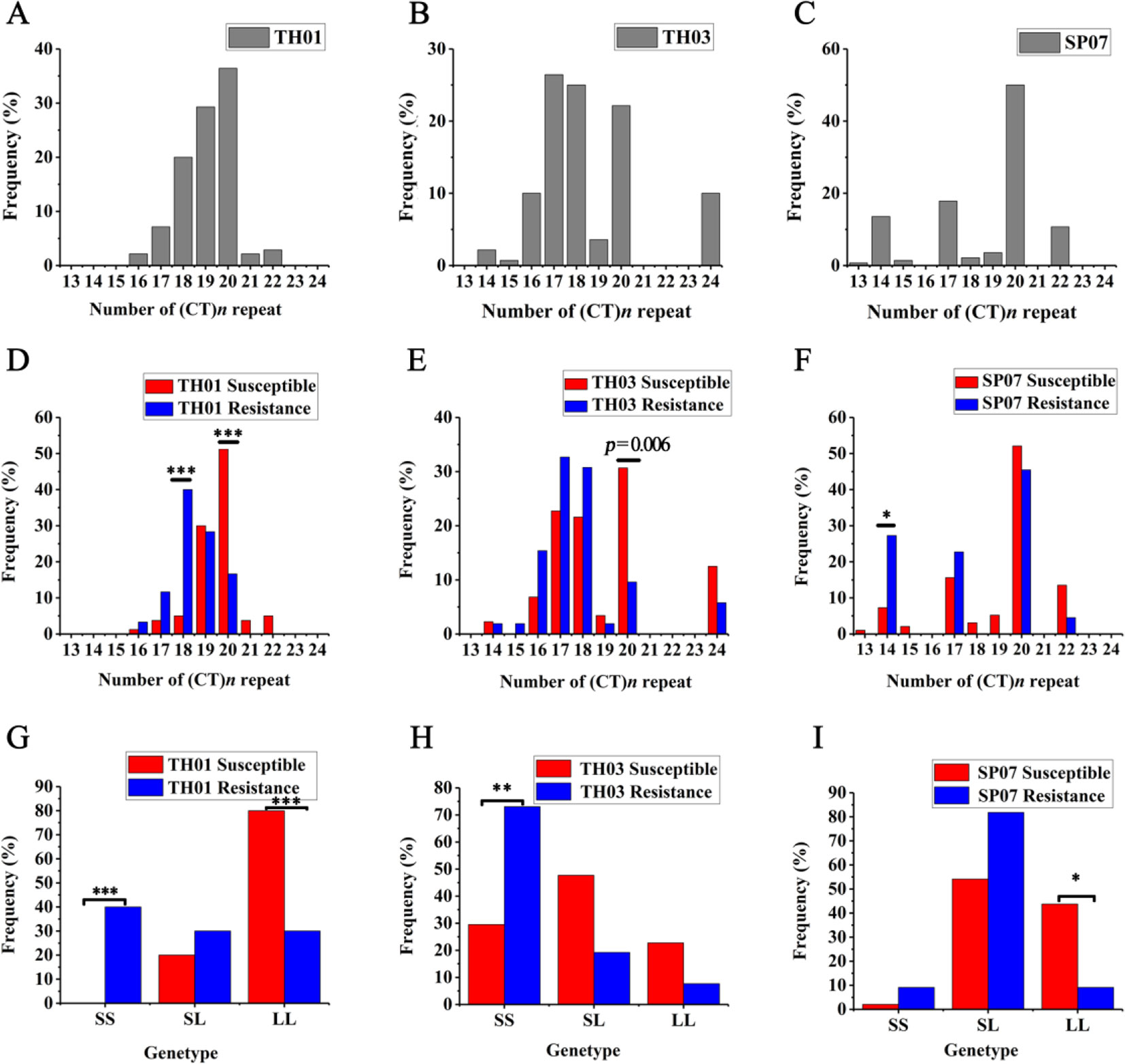
Figure 4 (CT)n microsatellite polymorphism allele distribution and genotype frequency in three different shrimp populations. (A–C) The allelic distribution in three population: TH01 (A), TH03 (B) and SP07 (C). (D–F) The allelic distribution in two groups of the three population (Fisher exact test: *p = 0.0042). (G–I) The genotype frequency in two groups of the three populations (Fisher exact test: *p = 0.0167). Associated with WSSV resistance, the TH01 and TH03 populations showed a significant difference in the S/S genotype, and the L/L genotype show a significant difference and was associated with WSSV susceptible in the SP07 population. *p < 0.05, **p < 0.01, ***p < 0.001.
Moreover, there was a significant difference when comparing the allelic distribution between Sus. group and Res. groups. In the TH01 population, the (CT)18 genotype was enriched in the Res. group associated with WSSV resistance, and showed a very significant difference with the Sus. Group (Fisher exact test: p < 0.00008, OR = 12.667, 95% CI = 4.09 - 39.225) (Figure 4D). On the contrary, the (CT)20 genotype was enriched in the Sus. Group (Fisher exact test: p = 0.00004, OR = 0.19, 95% CI = 0.085 - 0.427) (Figure 4D). In the SP07 population, the (CT)14 genotype was enriched in the Res. group (Fisher exact test: p = 0.0022, OR = 4.768, 95% CI = 1.726 - 13.168) (Figure 4F). In the TH03 population, the (CT)20 genotype was enriched in the Sus. Group (Fisher exact test: p = 0.0061, OR = 0.24, 95% CI = 0.086 - 0.671), although this difference was only nominally significant (Figure 4E).
Since the (CT)n repeat numbers ranged from 13 to 24 in the three populations, the repeat length cut-offs for allelic categorization were defined as ≤18 repeats for short (S) alleles and >18 repeats for long (L) alleles. Following allele classification, we could attribute to each individual one of three bi-allelic genotypes: S/S, S/L, or L/L. Genotype distribution frequencies were then determined and are shown for each population in Figures 4G–I. Associated with WSSV resistance, the TH01 population showed a very significant difference in the S/S genotype (Fisher exact test: p < 0.0003, OR = 28.053, 95% CI = 3.417 - 230.33) and the L/L genotype (Fisher exact test: p < 0.0003, OR = 0.107, 95% CI = 0.036 - 0.322). Similar to the TH01 population, the S/S genotype associated with WSSV resistance showed a significant difference in the TH03 population (Fisher exact test: p = 0.0005, OR = 6.476, 95% CI = 2.194 - 19.095), while the L/L genotype showed a significant difference in the SP07 population (Fisher exact test: p = 0.0054, OR = 0.129, 95% CI = 0.027 - 0.613).
Allelic distribution showed that S alleles tended to be enriched in Res. groups and L alleles tended to Sus. groups. We then compared the bi-allelic genotype distribution between the Sus. and Res. groups, and shrimp with the S/S genotype in the 5′-UTR of the LvIRF gene were resistant to WSSV, while those with L/L were susceptible to WSSV. The above results suggested that the shrimp with a short (CT)n repeat in the 5′-UTR of LvIRF were more resistant to WSSV than those with longer (CT)n repeats.
Short CT Microsatellite Genotypes Improve the Resistance of Shrimp Populations to WSSV
Based on our breeding scheme illustrated in Figure 5A, we obtained two new shrimp populations. HX-CTS, containing short (CT)n repeats in the 5′-UTR of LvIRF, and HX-CTL, which only contained long (CT)n repeats. Figure 5B shows the allelic distribution in these two populations, which were as expected, and the HX-CTS population contained the (CT)14 and (CT)15 genotypes, while the HX-CTL population did not. The survival rate curve after WSSV infection is shown in Figure 5C. The survival rate curves of the two populations could be easily separated, as the HX-CTS mortality rate reached 50% 16 h later than the HX-CTL population. We also measured the LvIRF gene expression in gills by quantitative RT-PCR (Figure 5D). There was no difference at 0 h, but after 12 h of WSSV infection, the expression of LvIRF in the HX-CTS population was twice that of the HX-CTL population, which was a significant difference (p = 0.02). These results suggested that shrimp containing a smaller number of (CT)n repeats in the 5′-UTR of LvIRF were more resistant to WSSV infection, thereby improving their survival rate in this assay.
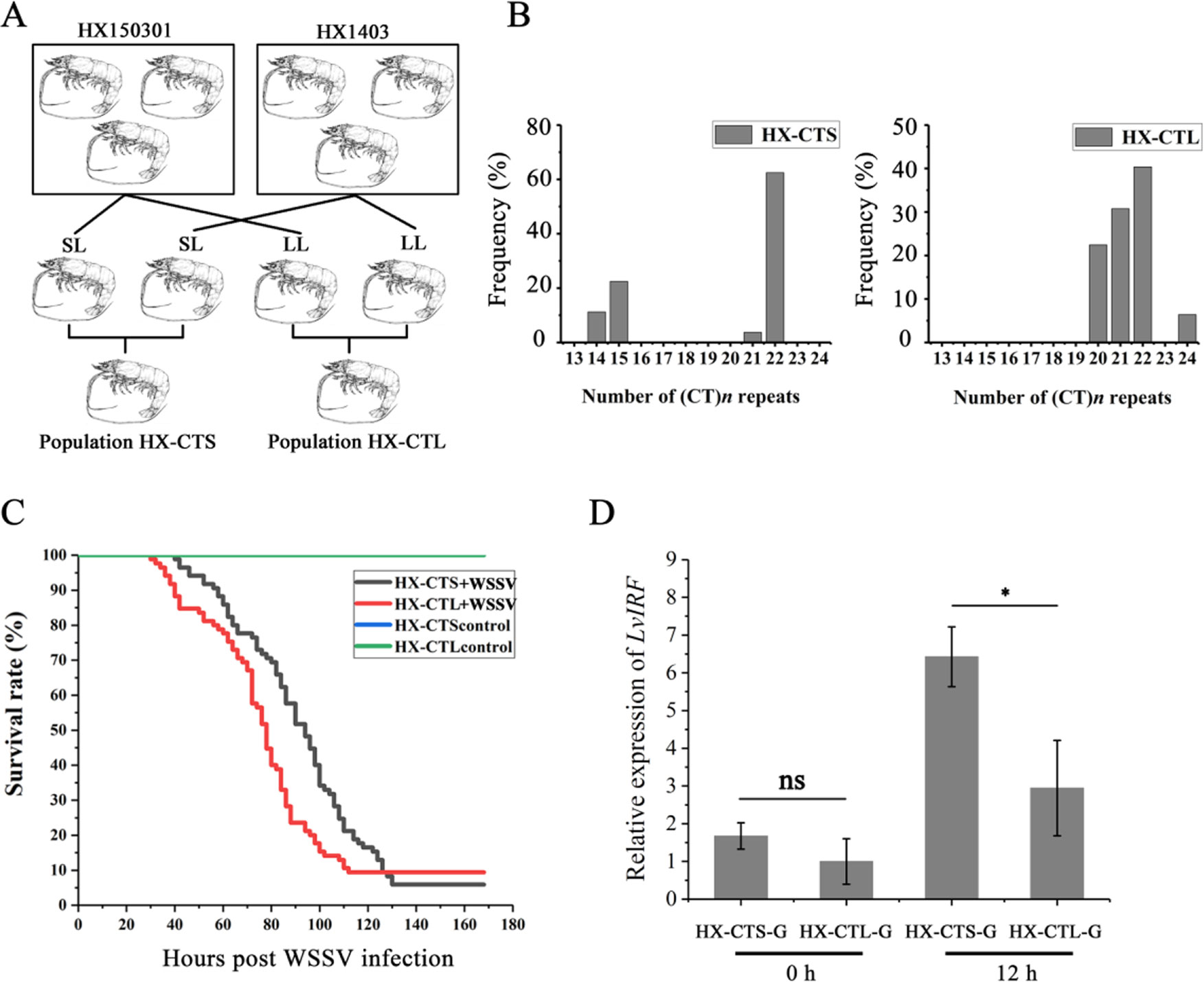
Figure 5 WSSV challenge in selective breeding offspring. (A) Schematic representation of the selective generations. (B) The allelic distribution in two populations, HX-CTS and HX-CTL. (C) The survival rate curves of HX-CTS and HX-CTL. (D) Quantitative RT-PCR was performed in triplicate for each sample using the EF-1α gene as an internal control by the Livak (2−△△CT) method. Expression levels in gill were used to determine the mean fold change (means ± SD, n = 5), and the expression level of the HX-CTL population at 0 h was used as a baseline (1.0). 12 h after infection with WSSV, LvIRF expression in HX-CTS was 2.1-fold higher than that of HX-CTL 12 h after infection with WSSV. *p < 0.05, ns, not significant.
Discussion
Our study examined the role of an SSR with a (CT)n motif in the 5′-UTR, as well as its repeat length variation, in the transcriptional regulation of the IRF gene of L. vannamei and the association of its genetic variations with WSSV resistance. Microsatellite repeats, especially (CT)n motifs, exist in abundance in the 5′-UTRs of several eukaryotic genes (Bhattacharyya et al., 2004; Mohammadparast et al., 2014; Zhao et al., 2014). The microsatellites are present in the vicinity of TSSs, indicating that they are likely involved in regulating target gene transcription (Castillo-Davis, 2005; Simpson and Ayyar, 2008). Our first goal was to investigate the highly polymorphic CT repeat located in the core promoter of the LvIRF-5’-UTR. Genotyping demonstrated a high heterogeneity of alleles, with lengths ranging from 13 to 24 repeats. In order to elucidate the role of (CT)n variation in the 5’-UTR of the LvIRF gene, we selected (CT)n variant alleles from different individuals and found that shrimp with smaller numbers of (CT)n repeats exhibited an enhanced tolerance to WSSV infection.
SSRs in the UTRs sequences or within promoter regions are thought to play a role in the regulation of gene expression. In Catharanthus roseus, Tryptophan decarboxylase (Tdc) gene expression is affected by the length of a (CT)n microsatellite in the 5’-UTR of this gene (Kumar and Bhatia, 2016). The chickpea CaIMP gene, with length variations of (CT)n repeat motifs present in the 5’-UTR, were differentially transcribed, and were shown to be associated with drought tolerance (Joshi-Saha and Reddy, 2015). Recently, a genome-wide survey of the contribution of microsatellites to gene expression in humans identified 2,060 significantly expressed microsatellites that were enriched in conserved regions, colocalize with regulatory elements, and may modulated certain histone modifications (Gymrek et al., 2016; Lieben, 2016). In our study, dual-luciferase reporter assays and Western blots clearly showed that the expression of the LvIRF gene was affected by the total of (CT)n repeat numbers. In particular, shorter repeats [(CT)15] showed higher expression of LvIRF than longer repeats [(CT)25].
IFNs are a group of secreted cytokines with activities that inhibit viral replication, and are able to regulate the functions of several types of immune cells. In mammals, type I and III IFNs exhibit significant antiviral activities, and are considered to be central to antiviral innate immunity (Levy et al., 2011). The IRF family is a group of transcription factors that plays critical roles in the activation of IFNs (Ikushima et al., 2013). LvIRF is an IRF in crustaceans, with functions similar to mammalian IRFs. LvIRF mediates the IRF-Vago-JAK/STAT pathway in shrimp, and has been shown to inhibit viral (WSSV) replication, analogous to the IRF-IFN-JAK/STAT pathway in vertebrates (Li et al., 2015). We used shrimp populations with allelic distribution differences to analyze the relationship between LvIRF genotype and WSSV resistance. Our results demonstrated that the LvIRF (CT)n repeat number was associated with WSSV resistance in shrimp. Smaller numbers of repeats showed significant resistance to WSSV, perhaps due to the higher LvIRF expression. We selected three populations of shrimp with different genetic backgrounds and allelic distributions of LvIRF to analyze the association between (CT)n repeats and WSSV resistance, and obtained similar results. This indicated that the regulation of LvIRF expression by an SSR with a (CT)n motif in the 5′-UTR was widespread in L. vannamei. Studies have shown that the IRF-IFN-JAK/STAT pathway is a broad-spectrum antiviral pathway in vertebrates (Thompson et al., 2014; Qin et al., 2016). LvIRF had similar functions to mammalian IRF, indicating that the IRF-Vago-JAK/STAT pathway may induce immune responses to a wide range of viruses, including WSSV. Microsatellite polymorphisms associated with disease and biological traits have also been reported. For example, more than 25 inherited human disorders have been shown to be caused by STRs (Sun et al., 2018), including FXS (Colak et al., 2014), Huntington’s disease (Wheeler et al., 2007), Friedreich’s ataxia (Shishkin et al., 2009), and idiopathic short stature (Dias et al., 2017).
We also used the microsatellite of LvIRF as a molecular marker to breed two populations of offspring, which contained different SSR genotypes and exhibited distinct tolerances to viral infection. The shrimp with small numbers of (CT)n repeats had a stronger antiviral immune response, as manifested by observation of an increased LvIRF gene expression and the prolonged survival time after WSSV infection. Thus, the identified SSR could be candidate marker for shrimp breeding for WSSV resistance. In fact, molecular markers have been widely reported in the breeding of several cash crops, including rice and wheat (Xue et al., 2018). For example, by analyzing the agronomic traits of rice varieties throughout the world, a number of molecular markers have been obtained for marketable traits, such as grain size: GLW7/OsSPL13 (Si et al., 2016), chilling tolerance: COLD1 (Ma et al., 2015), disease resistance: Pigm (Deng et al., 2017), and nitrate-use efficiency: NITR1.1 (Hu et al., 2015). Some successful cases of molecular marker assisted breeding have also been reported in aquatic animals. For instance, using transcriptome sequencing analysis, eight IFN system genes were identified as anti-disease molecular markers for resistance breeding of gibel carp (Mou et al., 2018). The use of molecular markers has made shrimp designer breeding possible in the future.
In summary, we identified an SSR of a (CT)n repeat located in the 5′-UTR of LvIRF and demonstrated that the total length of these (CT)n -SSRs could influence the levels of gene expression. In addition, we also demonstrated that this SSR was associated with WSSV resistance. Our results provide some insights into how this SSR could be used as a molecular marker in breeding of WSSV-resistant shrimp. Further work needs to be done to investigate the feasibility of other molecular markers in shrimp breeding.
Data Availability Statement
All reagents and experimental data are available within the article or supplementary information or from corresponding author upon reasonable request.
Author Contributions
CL and JH conceived and designed the experiments. BY, HW, PZ, and SW performed the experiments and analyzed data. CL and BY wrote the draft manuscript. CL was responsible for forming the hypothesis, project development, data coordination, and writing, finalizing, and submitting the manuscript. All authors discussed the results and approved the final version.
Funding
This research was supported by National Key Research and Development Program of China (2018YFD0900502/5), the Science and Technology Planning Project of Guangdong Province (2018B020204001); National Natural Science Foundation of China (31772883); Guangdong Natural Science Funds for Distinguished Young Scholars (2016A030306041); Tip-Top Scientific and Technical Innovative Youth Talents of Guangdong Special Support Program (No. 2016TQ03N504), and Fundamental Research Funds for the Central Universities (17lgpy62). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JX declared a shared affiliation, with no collaboration, with the authors to the handling editor at time of review.
References
Berger, N., Dubreucq, B. (2012). Evolution goes GAGA: GAGA binding proteins across kingdoms. Biochim. Biophys. Acta 1819 (8), 863–868. doi: 10.3892/or.11.1.231
Bhattacharyya, N., Tao, J. M., Klein, E. A., Banerjee, S. (2004). Alterations of transforming growth factor beta receptor II, insulin growth factor receptor II genes in microsatellite unstable prostate carcinomas. Oncol. Rep. 11 (1), 231–236.
Castillo-Davis, C. I. (2005). The evolution of noncoding DNA: how much junk, how much func? Trends Genet. 21 (10), 533–536. doi: 10.1016/j.tig.2005.08.001
Chen, Y. H., Jia, X. T., Zhao, L., Li, C. Z., Zhang, S., Chen, Y. G., et al. (2011). Identification and functional characterization of Dicer2 and five single VWC domain proteins of Litopenaeus vannamei. Dev. Comp. Immunol. 35 (6), 661–671. doi: 10.1016/j.dci.2011.01.010
Colak, D., Zaninovic, N., Cohen, M. S., Rosenwaks, Z., Yang, W. Y., Gerhardt, J., et al. (2014). Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science 343 (6174), 1002–1005. doi: 10.1126/science.1245831
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355 (6328), 962–965. doi: 10.1126/science.aai8898
Dias, C., Giordano, M., Frechette, R., Bellone, S., Polychronakos, C., Legault, L., et al. (2017). Genetic variations at the human growth hormone receptor (GHR) gene locus are associated with idiopathic short stature. J. Cell Mol. Med. 21 (11), 2985–2999. doi: 10.1111/jcmm.13210
Flegel, T. W. (2012). Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 110 (2), 166–173. doi: 10.1016/j.jip.2012.03.004
Fujimori, S., Washio, T., Higo, K., Ohtomo, Y., Murakami, K., Matsubara, K., et al. (2003). A novel feature of microsatellites in plants: a distribution gradient along the direction of transcription. FEBS Lett. 554 (1-2), 17–22. doi: 10.1016/s0014-5793(03)01041-x
Gregorek, A. C., Gornik, K. C., Polancec, D. S., Dabelic, S. (2013). GT microsatellite repeats in the heme oxygenase-1 gene promoter associated with abdominal aortic aneurysm in Croatian patients. Biochem. Genet. 51 (5-6), 482–492. doi: 10.1007/s10528-013-9579-8
Gymrek, M., Willems, T., Guilmatre, A., Zeng, H., Markus, B., Georgiev, S., et al. (2016). Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet. 48 (1), 22–29. doi: 10.1038/ng3461
Hall, T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Sym. Ser. 41, 95–98.
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47 (7), 834–838. doi: 10.1038/ng3337
Huang, Y.-C., Yin, Z.-X., Ai, H.-S., Huang, X.-D., Li, S.-D., Weng, S.-P., et al. (2011). Characterization of WSSV resistance in selected families of Litopenaeus vannamei. Aquaculture 311 (1-4), 54–60. doi: 10.1016/j.aquaculture.2010.11.032
Ikushima, H., Negishi, H., Taniguchi, T. (2013). The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb. Symp. Quant. Biol. 78, 105–116. doi: 10.1101/sqb.2013.78.020321
Joshi-Saha, A., Reddy, K. S. (2015). Repeat length variation in the 5’UTR of myo-inositol monophosphatase gene is related to phytic acid content and contributes to drought tolerance in chickpea (Cicer arietinum L.). J. Exp. Bot. 66 (19), 5683–5690. doi: 10.1093/jxb/erv156
Kumar, S., Bhatia, S. (2016). A polymorphic (GA/CT)n- SSR influences promoter activity of Tryptophan decarboxylase gene in Catharanthus roseus L. Don. Sci. Rep. 6, 33280. doi: 10.1038/srep33280
Lee, N., Iyer, S. S., Mu, J., Weissman, J. D., Ohali, A., Howcroft, T. K., et al. (2010). Three novel downstream promoter elements regulate MHC class I promoter activity in mammalian cells. PloS One 5 (12), e15278. doi: 10.1371/journal.pone.0015278
Levy, D. E., Marie, I. J., Durbin, J. E. (2011). Induction and function of type I and III interferon in response to viral infection. Curr. Opin. Virol. 1 (6), 476–486. doi: 10.1016/j.coviro.2011.11.001
Lewitter, F., Bush, W. S., Moore, J. H., Kann, M. (2012). Chapter 11: genome-wide association studies. PloS Comput. Biol. 8 (12), e1002822. doi: 10.1371/journal.pcbi.1002822
Li, Y. C., Korol, A. B., Fahima, T., Beiles, A., Nevo, E. (2002). Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 11 (12), 2453–2465. doi: 10.1046/j.1365-294X.2002.01643.x
Li, Y. C., Korol, A. B., Fahima, T., Nevo, E. (2004). Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol. 21 (6), 991–1007. doi: 10.1093/molbev/msh073
Li, C. Z., Chen, Y. X., Weng, S. P., Li, S. D., Zuo, H. L., Yu, X. Q., et al. (2014). Presence of tube isoforms in Litopenaeus vannamei suggests various regulatory patterns of signal transduction in invertebrate NF-kappa B pathway. Dev. Comp. Immunol. 42 (2), 174–185. doi: 10.1016/j.dci.2013.08.012
Li, C., Li, H., Chen, Y., Chen, Y., Wang, S., Weng, S. P., et al. (2015). Activation of Vago by interferon regulatory factor (IRF) suggests an interferon system-like antiviral mechanism in shrimp. Sci. Rep. 5, 15078. doi: 10.1038/srep15078
Li, C., Wang, S., He, J. (2019a). The two NF-κB pathways regulating bacterial and WSSV infection of shrimp. Front. Immunol. 10 (1785). doi: 10.3389/fimmu.2019.01785
Li, C., Weng, S., He, J. (2019b). WSSV-host interaction: Host response and immune evasion. Fish Shellfish Immunol. 84, 558–571. doi: 10.1016/j.fsi.2018.10.043
Lieben, L. (2016). Complex traits: repeat, repeat, repeat–gene expression variability explained. Nat. Rev. Genet. 17 (2), 68–69. doi: 10.1038/nrg.2015.27
Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X., Zeng, D., et al. (2015). COLD1 confers chilling tolerance in rice. Cell 160 (6), 1209–1221. doi: 10.1101/gr.10.1.72
Metzgar, D., Bytof, J., Wills, C. (2000). Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 10 (1), 72–80.
Mohammadparast, S., Bayat, H., Biglarian, A., Ohadi, M. (2014). Exceptional expansion and conservation of a CT-repeat complex in the core promoter of PAXBP1 in primates. Am. J. Primatol. 76 (8), 747–756. doi: 10.1002/ajp.22266
Morgante, M., Hanafey, M., Powell, W. (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30 (2), 194–200. doi: 10.1038/ng822
Mou, C. Y., Wang, Y., Zhang, Q. Y., Gao, F. X., Li, Z., Tong, J. F., et al. (2018). Differential interferon system gene expression profiles in susceptible and resistant gynogenetic clones of gibel carp challenged with herpesvirus CaHV. Dev. Comp. Immunol. 86, 52–64. doi: 10.1016/j.dci.2018.04.024
Paradkar, P. N., Trinidad, L., Voysey, R., Duchemin, J. B., Walker, P. J. (2012). Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. U. S. A. 109 (46), 18915–18920. doi: 10.1073/pnas.1205231109
Qin, Y. F., Liu, Q. X., Tian, S., Xie, W. H., Cui, J., Wang, R. F. (2016). TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3 beta to TBK1. Cell Res. 26 (5), 613–628. doi: 10.1038/cr.2016.27
Shishkin, A. A., Voineagu, I., Matera, R., Cherng, N., Chernet, B. T., Krasilnikova, M. M., et al. (2009). Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol. Cell 35 (1), 82–92. doi: 10.1016/j.molcel.2009.06.017
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48 (4), 447–456. doi: 10.1038/ng3518
Simpson, P., Ayyar, S. (2008). Evolution of cis-regulatory sequences in drosophila. Long-Range Control Gene Expression 61, 67–106. doi: 10.1016/S0065-2660(07)00003-X
Streelman, J. T., Kocher, T. D. (2002). Microsatellite variation associated with prolactin expression and growth of salt-challenged tilapia. Physiol. Genomics 9 (1), 1–4. doi: 10.1152/physiolgenomics.001052001
Sun, J. H., Zhou, L. D., Emerson, D. J., Phyo, S. A., Titus, K. R., Gong, W. F., et al. (2018). Disease-associated short tandem repeats co-localize with chromatin domain boundaries. Cell 175 (1), 224–22+. doi: 10.1016/j.cell.2018.08.005
Thompson, M. R., Sharma, S., Atianand, M., Jensen, S. B., Carpenter, S., Knipe, D. M., et al. (2014). Interferon gamma-inducible protein (IFI) 16 transcriptionally regulates type i interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J. Biol. Chem. 289 (34), 23568–23581. doi: 10.1074/jbc.M114.554147
Tsukiyama, T., Becker, P. B., Wu, C. (1994). ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367 (6463), 525–532. doi: 10.1038/367525a0
Wang, P. H., He, J. G. (2019). Nucleic acid sensing in invertebrate antiviral immunity. Int. Rev. Cell Mol. Biol. 345, 287–360. doi: 10.1016/bs.ircmb.2018.11.002
Wheeler, V. C., Persichetti, F., McNeil, S. M., Mysore, J. S., Mysore, S. S., MacDonald, M. E., et al. (2007). Factors associated with HD CAG repeat instability in Huntington disease. J. Med. Genet. 44 (11), 695–701. doi: 10.1136/jmg.2007.050930
Wren, J. D., Forgacs, E., Fondon, J. W., 3rd, Pertsemlidis, A., Cheng, S. Y., Gallardo, T., et al. (2000). Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am. J. Hum. Genet. 67 (2), 345–356. doi: 10.1086/303013
Xue, Y., Duan, Z., Chong, K., Yao, Y. (2013). Next-generation biotechnological breeding techniques for the future——designer breading by molecular modules. Bull. Chin. Acad. Sci. (3), 308–314. doi: 10.3969/j.issn.1000-3045.2013.03.003
Xue, Y., Zhong, K., Han, B., Gui, J., Wang, T., Fu, X., et al. (2015). New chapter of designer breeding in china: update on strategic program of molecular module-based designer breeding systems. Bull. Chin. Acad. Sci. (3), 393–402. doi: 10.16418/j.issn.1000-3045.2015.03.014
Xue, Y., Zhong, K., Han, B., Gui, J., Jing, H. (2018). Innovation and achievements of designer breeding by molecular modules in china. Bull. Chin. Acad. Sci. 33 (9), 893–899. doi: 10.16418/j.issn.1000-3045.2018.09.001
Yeh, F. C., Boyle, T. J. B. (1997). Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian J. Bot. 129 (2), 157.
Keywords: computed tomography (CT) microsatellite, Litopenaeus vannamei IRF, white spot syndrome virus (WSSV) resistance, molecular marker, Litopenaeus vannamei
Citation: Yin B, Wang H, Zhu P, Weng S, He J and Li C (2019) A Polymorphic (CT)n-SSR Influences the Activity of the Litopenaeus vannamei IRF Gene Implicated in Viral Resistance. Front. Genet. 10:1257. doi: 10.3389/fgene.2019.01257
Received: 29 August 2019; Accepted: 14 November 2019;
Published: 06 December 2019.
Edited by:
Shikai Liu, Ocean University of China, ChinaReviewed by:
Jun Hong Xia, Sun Yat-sen University, ChinaJie Mei, Huazhong Agricultural University, China
Copyright © 2019 Yin, Wang, Zhu, Weng, He and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianguo He, bHNzaGpnQG1haWwuc3lzdS5lZHUuY24=; Chaozheng Li, bGljaGFvemhAbWFpbDIuc3lzdS5lZHUuY24=
 Bin Yin1,2
Bin Yin1,2 Haiyang Wang
Haiyang Wang Jianguo He
Jianguo He Chaozheng Li
Chaozheng Li