- 1Department of Biochemistry and Molecular Biology, Miller School of Medicine, University of Miami, Miami, FL, United States
- 2Sylvester Comprehensive Cancer Center, Miller School of Medicine, University of Miami, Miami, FL, United States
- 3Department of Urology, Miller School of Medicine, University of Miami, FL, United States
This year marks the 20th anniversary of the discovery that the nucleolus can temporarily immobilize proteins, a process known as nucleolar sequestration. This review reflects on the progress made to understand the physiological roles of nucleolar sequestration and the mechanisms involved in the immobilization of proteins. We discuss how protein immobilization can occur through a highly choreographed amyloidogenic program that converts the nucleolus into a large fibrous organelle with amyloid-like characteristics called the amyloid body (A-body). We propose a working model of A-body biogenesis that includes a role for low-complexity ribosomal intergenic spacer RNA (rIGSRNA) and a discrete peptide sequence, the amyloid-converting motif (ACM), found in many proteins that undergo immobilization. Amyloid bodies provide a unique model to study the multistep assembly of a membraneless compartment and may provide alternative insights into the pathological amyloidogenesis involved in neurological disorders.
Nucleolar Sequestration: Visitors to the Nucleolus
The role of the nucleolus as the site of ribosome biosynthesis has been established since the mid-1960s (Perry, 1960; Perry, 1962; Miller and Beatty, 1969). Nucleoli are built around tandem repeats of ribosomal DNA (rDNA) in nucleolar organizing regions (NORs) and structurally dependent on active transcription of rDNA (Hernandez-Verdun, 2006; Raska et al., 2006). Each nucleolus consists of a tripartite organization which is classically defined by their different appearances under electron microscopy (EM) (Pederson, 2011): the fibrillar center (FC), where the RNA polymerase I machinery is active; the dense fibrillar component (DFC) that is enriched in fibrillarin (FIB1); and the granular component (GC) that harbors B23. RNA polymerase I activity is believed to occur at the interface between the FC and the DFC, while processing of newly synthesized ribosomal RNA (rRNA) and assembly with ribosomal proteins occur within the GC (Scheer and Hock, 1999; Thiry and Lafontaine, 2005). The traditional role of the nucleolus as a hub of rRNA synthesis and ribosome assembly has been the subject of many excellent literature surveys (Boisvert, 2007; Pederson, 2011; Nemeth and Grummt, 2018). This review focuses on a lesser known phenomenon originally coined “nucleolar sequestration,” which describes the ability of the nucleolus to sequester regulatory proteins in response to cellular cues (Emmott and Hiscox, 2009; Pederson and Tsai, 2009; Boulon, 2010).
2019 marks 20 years since nucleolar sequestration was first hypothesized (Bachant and Elledge, 1999) following the discoveries that cell cycle regulator Cdc14 phosphatase and E3 ubiquitin ligase MDM2 could be temporarily localized in nucleoli to affect cell cycle progression (Shou et al., 1999; Visintin et al., 1999; Weber, 1999; Lohrum, 2000; Bernardi et al., 2004). In Saccharomyces cerevisiae, Cdc14 is sequestered in the nucleolus by Cfi1/Net1 until anaphase onset when it is released to promote exit from mitosis (Shou et al., 1999; Visintin et al., 1999). In mammalian cells, nucleolar sequestration of MDM2 prevents it from binding and exporting p53 into the cytoplasm for degradation (Weber, 1999; Lohrum, 2000; Bernardi et al., 2004). This stabilizes p53 in the nucleus, where it acts as a transcription factor that promotes growth arrest or apoptosis. The E3 ubiquitin ligase von Hippel–Lindau protein (VHL) is another example of nucleolar sequestration (Mekhail, 2004). VHL promotes the degradation of the transcription factor hypoxia-inducible factor (HIF) under normal oxygen conditions. Low extracellular pH triggers nucleolar sequestration of VHL, enabling HIF to evade degradation and promote transcription of target genes involved in oxygen homeostasis. Table 1 summarizes the reports of nucleolar sequestration that have been observed for many other proteins under various cellular conditions. Taken together, these underscore a fundamental cellular strategy that uses the nucleolus to regulate protein dynamics in response to cellular cues.
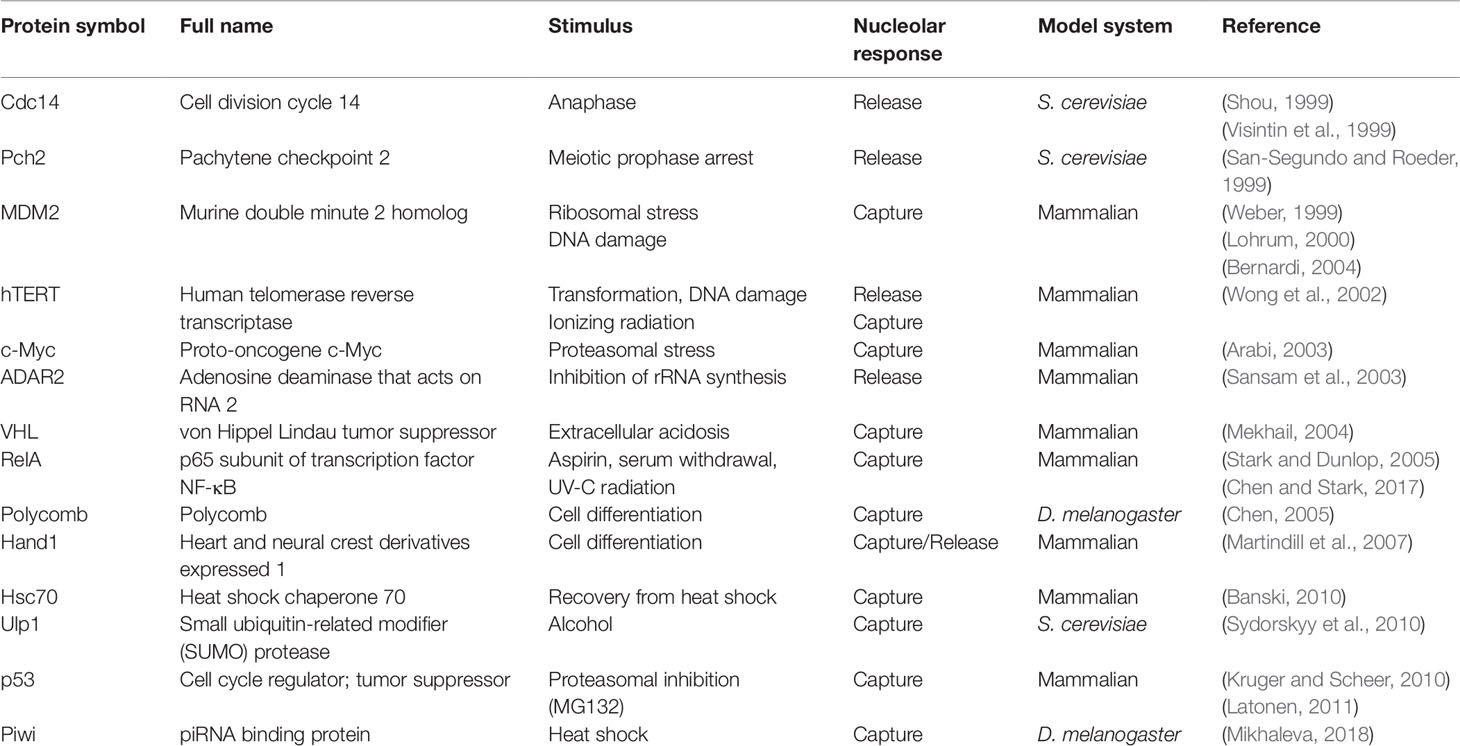
Table 1 List of the proteins whose activities have been reported to be regulated by nucleolar sequestration.
Nucleolar Sequestration: A Case of Protein Immobilization
The Nucleolar Detention Centers
Pioneering work by Phair and Misteli (2000) showed that proteins are highly mobile and rapidly exchange between affinity interactions and the cellular milieu. The nucleolus is considered a dynamic droplet, assembled by demixing of its three sub-structural liquid phases (i.e., FC, DFC, and GC), which are composed of highly mobile proteins (Brangwynne et al., 2011; Feric et al., 2016; Hult et al., 2017; Sawyer et al., 2019). It is remarkable, then, that proteins undergoing nucleolar sequestration are non-dynamic or immobile (Mekhail, 2004; Mekhail, 2005; Audas et al., 2012; Jacob, 2013). This has been demonstrated for Cdc14 (Tomson et al., 2009), MDM2 (Audas et al., 2012), VHL (Mekhail, 2005), RNF8 (Mekhail, 2007), DNMT1 (Audas et al., 2012), and Piwi (Mikhaleva, 2018), amongst other proteins. Even resident nucleolar proteins such as RNA polymerase I subunit RPA16, Pescadillo, and SENP3 can undergo cycles of mobility/immobility (Jacob, 2013). For example, VHL switches from a highly dynamic, uniform distribution under standard growth conditions (21% O2, pH 7.4) to an immobilized state in the nucleolus on exposure to extracellular acidosis (1% O2, pH 6.0) (Mekhail, 2005). Only upon neutralization of extracellular pH is VHL released from the nucleolus to return to its original distribution and mobility (Mekhail, 2005). These nuclear foci that contain sequestered/immobile proteins were originally called “nucleolar detention centers,” as targets are both localized and detained within the nucleolus, unable to freely diffuse elsewhere (Mekhail, 2005; Audas et al., 2012b; Jacob, 2013). From this perspective, the function of nucleolar detention or immobilization is to temporarily inactivate relevant proteins, inhibiting their access to downstream effectors. Just as possible, though, is that the clustering of detained proteins may render an enzymatic reaction more efficient. For example, immobilized nucleolar Cdc14 maintains Spo12 dephosphorylation to regulate cell cycle progression (Tomson et al., 2009). In addition, Piwi switches from its canonical role as a non-nucleolar transposable element repressor to a rDNA-specific repressor when it is sequestered in the nucleolus (Mikhaleva, 2018). From these studies, it is clear that cells have evolved a strategy to regulate molecular networks by reversibly switching proteins between a mobile and an immobile state. Whether nucleolar sequestration represents a loss- or gain-of-function might depend on the proteins undergoing immobilization.
Mechanisms of Nucleolar Sequestration
The ability of the nucleolus to sequester a wide variety of proteins in various cellular contexts suggests multiple mechanisms for nucleolar sequestration. Pathway analysis of interactions between resident nucleolar proteins and visitor proteins indicates that sequestered proteins interact either directly or indirectly, with the same small subset of “hub” nucleolar proteins, which primarily includes B23/NPM and Nucleolin (Emmott and Hiscox, 2009). Nucleolar retention of highly dynamic proteins through interactions with less mobile “hub” nucleolar partner(s) anchored by multivalent protein and RNA interactions contribute to nucleolar plasticity (Mitrea et al., 2016). Cdc14 is anchored in the nucleolus for most of the cell cycle through its association with Cfi1/Net1 (Shou et al., 1999; Visintin et al., 1999). During anaphase onset, a signaling cascade of phosphorylation inhibits the interaction of Cdc14 with Net1, releasing it to act as a mitotic exit activator (Azzam et al., 2004). ARF binding to MDM2 unmasks a cryptic nucleolar localization signal (NoLS) within its C-terminal RING domain, which is essential for MDM2 nucleolar sequestration (Tao and Levine, 1999; Weber, 1999; Lohrum, 2000; Weber et al., 2000). During DNA damage and acidosis, MDM2 is shuttled into the nucleolus by direct binding to PML (promyelocytic leukemia protein) (Bernardi et al., 2004), itself a target of nucleolar sequestration (Mattsson, 2001). MDM2 can also be sequestered in nucleoli by binding to ATP, independently of ARF (Poyurovsky et al., 2003). Alternatively, MDM2 binds ribosomal proteins (e.g., RPL5 and RPL11) that are released into the nucleoplasm during ribosomal stress, which stabilizes p53 (Boulon, 2010; Liu et al., 2016). Mapping analysis of VHL identified an approximately 30-amino acid fragment referred to as a nucleolar detention signal (NoDS) that is necessary and sufficient to immobilize proteins in nucleoli (Mekhail, 2007). The NoDS is composed of an arginine/histidine-rich sequence followed by two or more hydrophobic LXV motifs where X can be any hydrophobic residue (e.g., LWL, LLV, LFV, and LQV). A survey of proteins containing NoDS identified many candidates harboring this motif, including DNA methyltransferase I DNMT1, PML, and DNA polymerase delta catalytic subunit POLD1, all shown to be immobilized in nucleoli under extracellular acidosis (Mekhail, 2007; Audas et al., 2012a). The NoDS interacts with inducible long noncoding RNA derived from the ribosomal intergenic spacer (rIGSRNA) (Audas et al., 2012a). Silencing of rIGSRNA is sufficient to prevent both the formation of nucleolar detention centers and immobilization of NoDS-containing proteins (Audas et al., 2012a). The identification of the NoDS in many proteins provided evidence that nucleolar sequestration is a common cellular strategy to regulate protein function (Mekhail, 2007).
Biochemical Properties of Nucleolar Detention Centers
Nucleolar Detention Centers Display Amyloidogenic-Like Characteristics
In various physiological settings, proteins with limited mobility often display amyloid-like properties (Kayatekin et al., 2014; Berchowitz et al., 2015). Amyloidogenesis is the process whereby soluble proteins assemble into aggregates known as amyloid fibrils (Knowles et al., 2014). Because of their association to neurodegenerative disorders, including Alzheimer’s and Parkinson’s disease, amyloids were classically perceived as exclusively toxic protein aggregates (Schnabel, 2010; Knowles et al., 2014). The discovery of functional amyloid Pmel17 in 2006 challenged the notion that the amyloid state is merely pathological (Fowler, 2006). Since then, several groups have reported the existence of functional/physiological amyloids in different organisms (Chiti and Dobson, 2006; Fowler, 2007; Maji et al., 2009; Falabella et al., 2012; Fowler and Kelly, 2012; Li et al., 2012; Kayatekin et al., 2014; Berchowitz et al., 2015; Tang et al., 2015; Saad, 2017; Cereghetti, 2018). Nucleolar detention centers composed of proteins such as VHL, MDM2, POLD1, etc., share many properties beyond immobility that are associated with the amyloid state. First, these nuclear foci stained positive with amyloidophilic dyes such as Congo red, Thioflavin S/T, and Amylo-Glo, all of which recognize different biochemical features of amyloids (Audas et al., 2016). Second, the nucleolar detention centers stain positive for the OC fibril antibody that specifically targets the amyloid fibril conformation (Kayed et al., 2007). Third, immobilized proteins found in these nuclear foci displayed biochemical properties associated with amyloids, including resistance to proteinase K, insolubility in common detergents, and can only be dissociated into monomers by SDS/high temperature (Audas et al., 2016). Arguably the utmost unique feature of amyloid bodies is their electron-dense fibrillar organization, which would be as predicted for a condensate enriched in amylogenic proteins (Jacob, 2013; Audas et al., 2016). Consequently, the terms “nucleolar detention centers” and NoDS were replaced with “amyloid bodies” (A-bodies) and “amyloid-converting motif” (ACM), respectively, to reflect the transformation of the nucleolus into an A-body, a molecular prison of proteins in their amyloid-like state. Figure 1A shows an example of this dramatic and reversible transformation of the tripartite nucleolus into the fibrillar A-body in cells responding to stimuli.
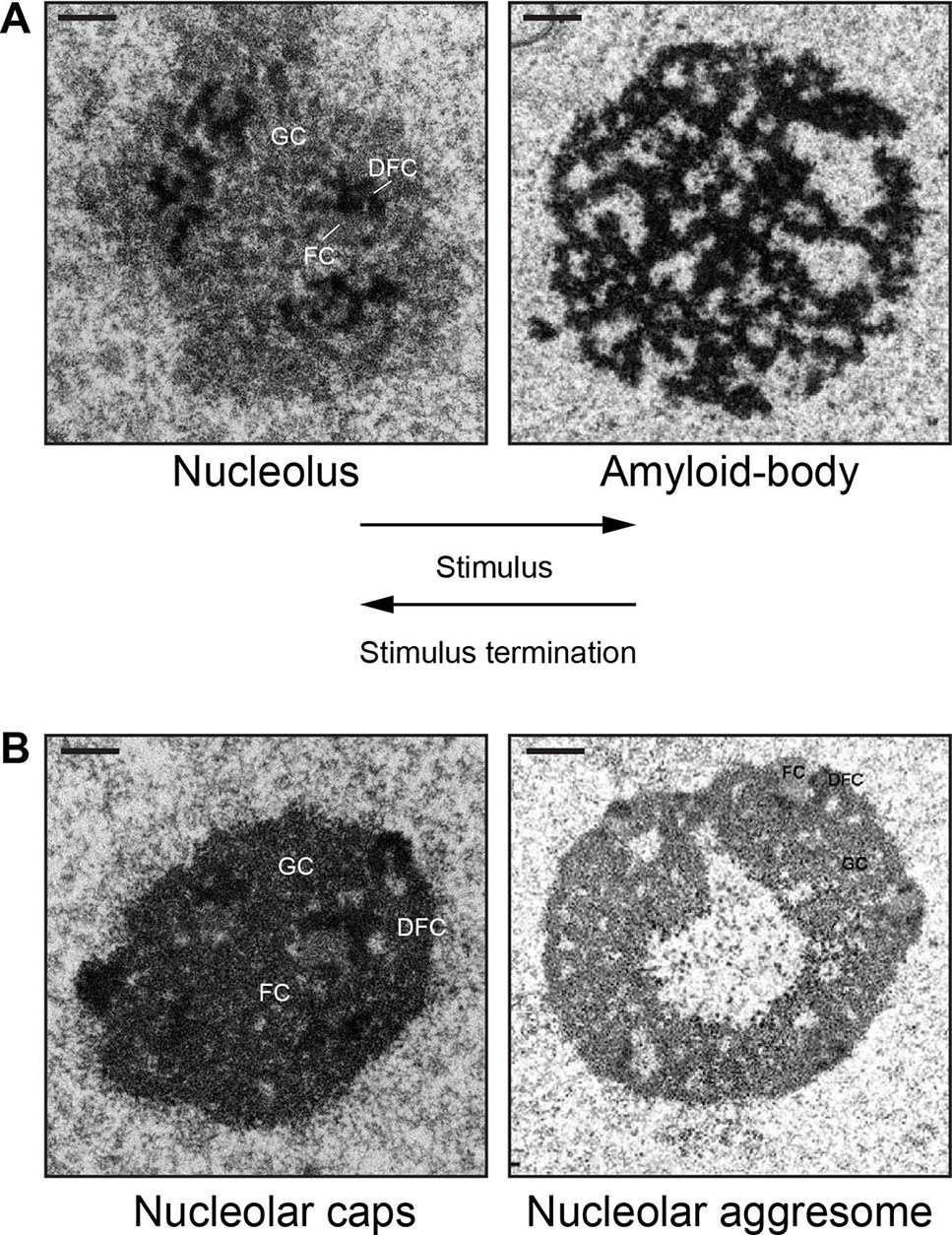
Figure 1 Nucleolar sequestration: the reversible remodeling of the nucleolus into an amyloid body. (A) During stimuli (heat shock or extracellular acidosis), the tripartite nucleolus undergoes a dramatic transformation into electron-dense fibrillar organization that characterizes an amyloid body. The fibers contain immobilized proteins in an amyloid-like state. After stimuli termination, an amyloid body is disaggregated and transforms back into the tripartite nucleolus. (B) The fibrillar amyloid bodies are distinct from the amorphous, electron-dense nucleolar caps (16 h cisplatin) or the electron-light nucleolar aggresomes (16 h MG132). FC, fibrillar component; DFC, dense fibrillar component; GC, granular component. Scale represents 1 µm. Amyloid body and nucleolar aggresome taken from (Audas, 2016) and (Kruger and Scheer, 2010), with permission.
Amyloid Bodies Are Distinct From Liquid-Like Biomolecular Condensates and Other Nucleolar Organizations
Purification and mass spectrometry analysis have identified hundreds of cellular proteins that are captured in A-bodies, many of which were shown to undergo immobilization by photobleaching experiments (Mekhail, 2007; Audas et al., 2012a; Audas et al., 2016). Because A-bodies contain an array of immobilized proteins in an amyloid-like state, we suggested the term “systemic physiological amyloidogenesis” to describe A-body biogenesis, in keeping with the original terminology to describe functional amyloids (Chiti and Dobson, 2006; Fowler, 2007; Maji et al., 2009). Other laboratories have also proposed that A-body formation represents an amyloidogenic liquid-to-solid phase transition (Lyons and Anderson, 2016; Latonen, 2019; Hall et al., 2019) and used the terms “solid,” “solid-like,” or “non-dynamic” biomolecular condensates to describe this membraneless organelle (Lyons and Anderson, 2016; Weber, 2017; Cereghetti, 2018; Holehouse and Pappu, 2018; Woodruff et al., 2018; Itakura et al., 2018; Fay and Anderson, 2018; Latonen, 2019). Prior to the discovery of A-bodies, the terms “liquid-to-solid phase transition” and “solid” had been reserved to describe the formation of pathological aggregates (Patel et al., 2015; Murakami et al., 2015; Jain and Vale, 2017; Mateju et al., 2017; Peskett, 2018; Posey et al., 2018). Other physiological examples of “solid-like” structures include Balbiani bodies observed in Xenopus (Boke et al, 2016) and pH-regulated fluid-to-solid transition of the cytoplasm in yeast (Munder et al, 2016). The fibrous, amyloid-like characteristic of the A-body differentiates it from other biomolecular condensates that display liquid-like properties (Brangwynne et al., 2009; Brangwynne et al., 2011; Weber and Brangwynne, 2012; Zhu and Brangwynne, 2015; Banani, 2017; Shin and Brangwynne, 2017), such as stress granules, nucleoli, and paraspeckles, amongst others (Hodges, 1998; Gall, 2000; Lamond and Spector, 2003; Rizzi et al., 2004; Dellaire and Bazett-Jones, 2004; Valgardsdottir, 2005; Parker and Sheth, 2007; Sasaki and Hirose, 2009; Bond and Fox, 2009; Machyna et al., 2013; Pederson, 2011; Protter and Parker, 2016). Liquid-like biomolecular condensates are dynamic, their constituents are mobile, they do not form fibers detectable by EM nor do they typically stain with amyloidophilic dyes (Phair and Misteli, 2000; Shin and Brangwynne, 2017; Weber, 2017). The biochemical and biophysical differences between dynamic, liquid-like, and non-dynamic or solid-like condensates are summarized within Table 2.

Table 2 Biochemical, biophysical, and dynamic properties of liquid-like condensates or solid-like condensates with amyloid characteristics.
A-bodies may also be contrasted from other stress-induced nucleolar structures, namely, nucleolar caps (Shav-Tal et al., 2005) and nucleolar aggresomes (Latonen, 2011; Latonen, 2011; Latonen, 2019) (Figure 1B). Transcriptional inhibition of Pol I results in nucleolar segregation, in which the FC and GC phases separate, perhaps as a consequence of changes in surface tension (Shin and Brangwynne, 2017) resulting in the formation of perinucleolar caps that surround the segregated nucleolus (Shav-Tal et al., 2005). Proteasomal inhibition induces nucleolar inclusions called nucleolar aggresomes that contain proteins marked for degradation, and various RNAs (Latonen, 2011). By EM, A-bodies have a unique fibrillar organization characteristic of amyloids, whereas nucleolar caps appear as electron-dense amorphous structures and nucleolar aggresomes are cavernous, occupying a large electron-light central space of the nucleolus (Kruger and Scheer, 2010) (Figure 1B). Interestingly, while the formation of nucleolar caps and A-bodies is accompanied by a redistribution of nucleolar components and subsequent arrest of ribosomal biogenesis (Shav-Tal et al., 2005; Mekhail, 2006; Jacob, 2013), nucleolar aggresomes require transcriptionally active nucleoli to form (Latonen, 2011; Latonen, 2011), with all nucleolar components intact and visible by EM. Additionally, proteins are dynamically sorted into nucleolar caps and remain mobile (Shav-Tal et al., 2005), while several proteins in A-bodies and nucleolar aggresomes are immobile (Mekhail, 2005; Audas et al., 2012a; Jacob, 2013; Audas et al., 2016). Another recently identified nucleolar stress body, nucleolar amyloid bodies (NoABs), is induced by prematurely terminated peptides that diffuse through the nuclear pores and aggregate within the nucleolus (Mediani et al., 2019). EM has not been reported for NoABs. It remains to be tested if NoABs are rIGSRNA-dependent, as are A-bodies, and if prematurely terminated peptides are involved in nucleolar sequestration of full-length proteins. While there are definitive similarities between the different stress-induced nucleolar organizations and that proteins can undergo sequestration in nucleolar aggresomes (both are discussed in (Latonen, 2019), A-bodies represent a unique nucleolar structure based on their fibrous properties and dependency on rIGSRNA.
Protein Immobilization Into A-Bodies: A Choreographed Multistep Pathway
How membraneless subcellular condensates maintain their unique identities and how proteins and/or RNA are sorted into these condensates remain subjects of active research. New insights into the biophysical properties of biomolecular condensates have demonstrated unexpected links between the sequence-encoded information in protein and RNA components of compartments and the material properties they impart (Kato et al., 2012; Altmeyer et al., 2015; Brangwynne et al., 2015; Elbaum-Garfinkle et al., 2015; Molliex et al., 2015; Nott et al., 2015; Pak et al., 2016; Shin and Brangwynne, 2017; Weber, 2017; Langdon, 2018). Recent studies demonstrated the importance of RNA-binding proteins with prion-like, low-complexity domains in forming biomolecular condensates. Equally as exciting has been the work done to uncover the specific sequences or structural elements embedded in architectural RNA that dictate the biophysical properties of biomolecular condensates (Chujo, 2017; Chujo and Hirose, 2017; Yamazaki et al., 2018; Hirose et al., 2019). With these new tools, we have begun to understand how specific elements within RNA and proteins are involved in condensate biogenesis.
A Working Model of A-Body Biogenesis
Figure 2 proposes a stepwise working model of A-body biogenesis that integrates different opinions in the literature, showcasing this process as a precisely choreographed multistep routine rather than simply a random aggregation of misfolded proteins. The first step in the formation of A-bodies is the appearance of several transient foci that are distinct from known nucleolar layers (Wang et al., 2018). These foci are spherical, contain mobile proteins, and can undergo fusion, thereby displaying some of the properties associated with dynamic condensates. An interesting feature of the inducible nucleolar foci is that they stain positive for 1-anilino-8-naphthalenesulfonate (ANS), a dye specific for hydrophobic regions of proteins and used in vitro to detect the molten globule state, a precursor of amyloid fibrils (Booth, 1997). EM revealed that ANS-positive foci correspond to electron-dense, amorphous aggregates concomitant with loss of the typical tripartite organization of transcriptionally active nucleoli. In the second step, the stimulus-induced foci rapidly mature into Congo red-positive aggregates that contain immobilized proteins, limiting their ability to diffuse within the nucleolus. We coined these maturing foci “nascent A-bodies.” Photobleaching analysis showed that, once formed, nascent A-bodies expand by directly capturing and immobilizing free proteins (step 3) (Wang et al, 2018). Maturation of A-bodies terminates once the pools of cellular mobile proteins have been depleted, culminating in a distinct fibrous organization. Disassembly of A-bodies occurs within 1–2 h after stimulus termination, a process that requires heat shock proteins hsp70 and hsp90 (step 4) (Audas et al., 2016).
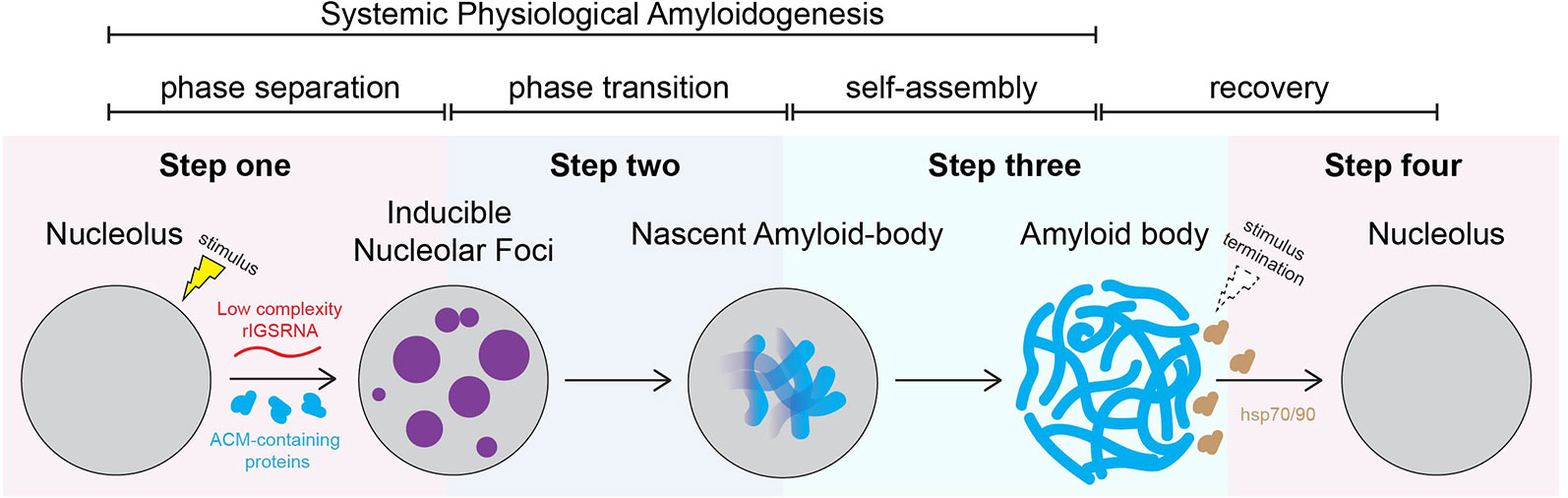
Figure 2 Working model: amyloid body biogenesis is a precisely choreographed routine. We propose that, on stimulus, low-complexity ribosomal intergenic spacer RNA (rIGSRNA) derived from the rDNA intergenic spacer accumulate in the nucleolus. Step 1: Low-complexity rIGSRNA interact with short cationic peptides, such as the R/H-rich sequence of the ACM (formally NoDS), to form nucleolar liquid-like foci. Step 2: Local concentration of proteins with amyloidogenic propensity in the foci triggers physiological amyloidogenesis and generates nascent amyloid bodies (A-bodies). Step 3: Once seeded, nascent A-bodies self-assemble into fibrillar, solid-like A-bodies. A-bodies enable cells to rapidly and reversibly store a large array of proteins and enter cellular dormancy in response to stress. Step 4: Upon recovery/stimulus termination, A-body disaggregation is mediated by heat shock protein (hsp) chaperones 70 and 90. Through these steps, A-body biogenesis may represent a physiological liquid-to-solid phase transition.
Low-Complexity rIGSRNA Drive Formation of Inducible Nucleolar Foci
In mammals, the nucleolus is organized around a scaffold of ∼400 rDNA tandem repeats of 43 kb, of which approximately half are transcriptionally active (Nemeth and Grummt, 2018; Sharifi and Bierhoff, 2018). Each repeat consists of an rDNA enhancer/promoter located directly upstream of rRNA genes separated by a ribosomal intergenic spacer (rIGS) of variable length and organization (Gonzalez and Sylvester, 1995; Smirnov, 2016) (Figure 3A). The rIGS is an enigmatic region of the human genome historically, and erroneously, called the “non-transcribed region” (Smirnov, 2016). Interestingly, in recent years, species conservation (Agrawal and Ganley, 2018) and functional studies have demonstrated that these regions of the genome are transcriptionally active, generating various non-coding RNA (ncRNA) (Jacob, 2012; Audas and Lee, 2016). These ncRNA from the rIGS are involved in regulating rRNA expression (Mayer, 2006; Mayer et al., 2008; Schmitz, 2010; Zhao, 2016; Zhao, 2016; Zhao, 2018) and thereby responsible for maintaining a significant fraction of the rDNA cassettes in a heterochromatic, transcriptionally silent chromatin structure (Grummt and Pikaard, 2003; Santoro, 2005), controlling PTBP1-regulated alternative splicing (Yap, 2018), and assembling A-bodies (Audas et al., 2012a; Jacob, 2013; Audas et al., 2016; Wang et al., 2018) (Figure 3A). The various rIGS ncRNA appear to be products of RNA polymerase I (Mayer, 2006; Audas et al., 2012a), except for the antisense PAPAS, which is transcribed by RNA polymerase II (Zhao, 2016).
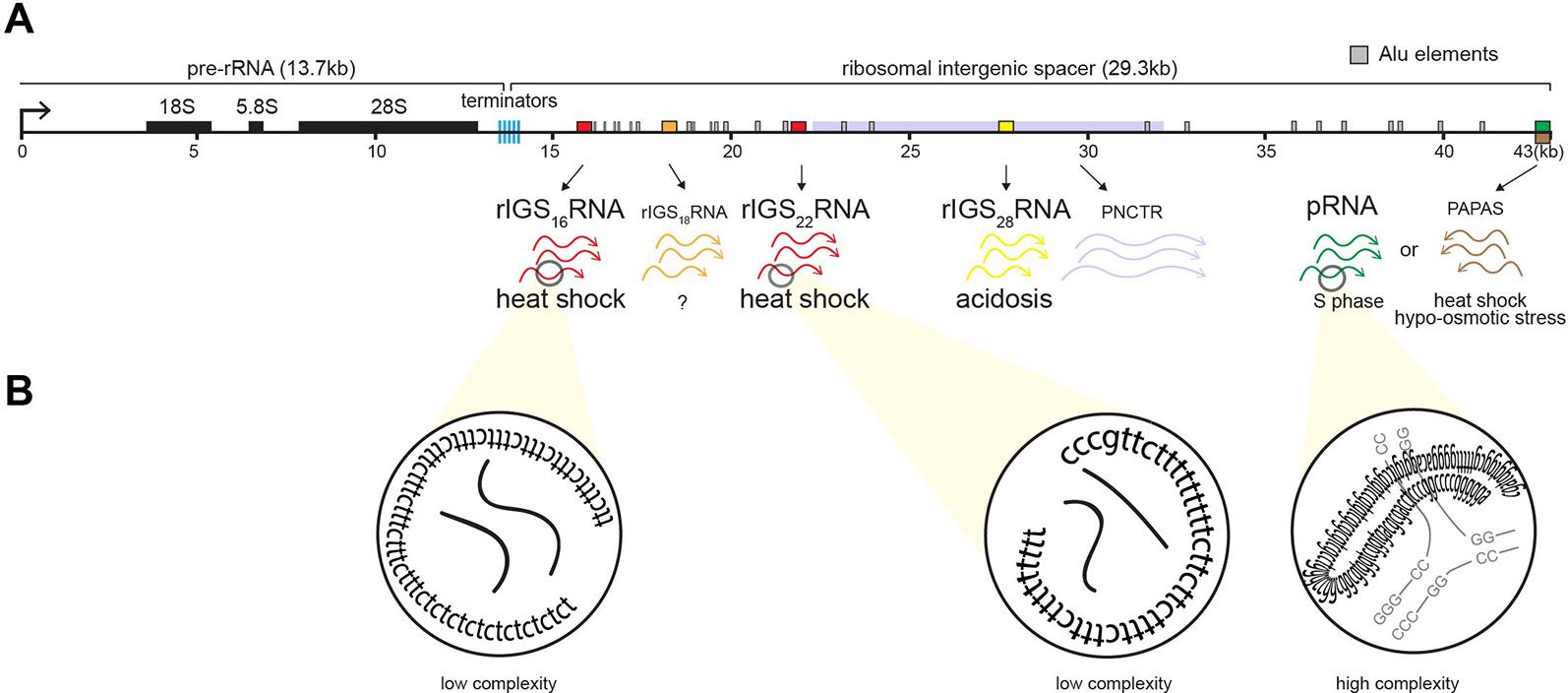
Figure 3 Induction of non-coding RNA (ncRNA) from the ribosomal cassette. (A) Schematic of a single human rDNA repeat unit, which is composed of a ∼13-kb pre-rRNA coding region flanked by a ∼30-kb intergenic spacer (rIGS). The rIGS transcribes several functional non-coding RNA. Stimuli-specific loci of rIGS produce ribosomal intergenic spacer RNA (rIGSRNA) required for A-body formation. rIGS28RNA and rIGS16RNA/rIGS22RNA are produced under acidotic (yellow) and heat shock (red) conditions, respectively. No function has been ascribed to rIGS18RNA yet. Other ncRNA found in the rIGS include a >10-kb transcript called PNCTR (pyrimidine-rich non-coding transcript) involved in PTBP1 binding (purple), pRNA (green), and antisense PAPAS (brown) involved in rRNA regulation, as well as Alu element-derived (gray boxes) RNA involved in nucleolar architecture. (B) rIGSRNA contain low-complexity sequences comprising of long dinucleotide repeats, as determined by RNA sequencing, RT cloning, and RNA-FISH. This is in contrast to other ncRNA that display high complexity, i.e., possess secondary structure, such as pRNA.
The appearance of nucleolar foci under stress coincides with the induction of rIGS28RNA in acidosis, and rIGS16RNA and rIGS22RNA in heat shock (Figure 3). Recent work suggests that low-complexity dinucleotide repetitive sequences operate as the architectural determinants of rIGSRNA to recruit proteins to A-bodies (Wang et al., 2018) (Figure 3B). Live cell imaging and in vitro assays indicated that positively charged R/H-rich peptide domains of the ACM (Figure 4) co-assemble more efficiently with the negatively charged low-complexity sequences of rIGSRNA than with flanking high-complexity RNA sequences to form the inducible nucleolar foci (Wang et al., 2018). The rIGSRNA/ACM interactions in vitro and in vivo are particularly sensitive to even the slightest increase in salt concentration. This contrasts other phase-separated compartments of the nucleolus, which are disrupted at considerably higher salt concentrations. Thus, the interactions between low-complexity RNA and the R/H-rich region of the ACM characteristic of step 1 in A-body biogenesis (Figure 2) appear to be electrostatic in nature and are likely driven by complex coacervation, a charge-based form of liquid–liquid phase separation that has been observed in other cellular settings in vivo (Altmeyer et al., 2015; Pak et al., 2016).
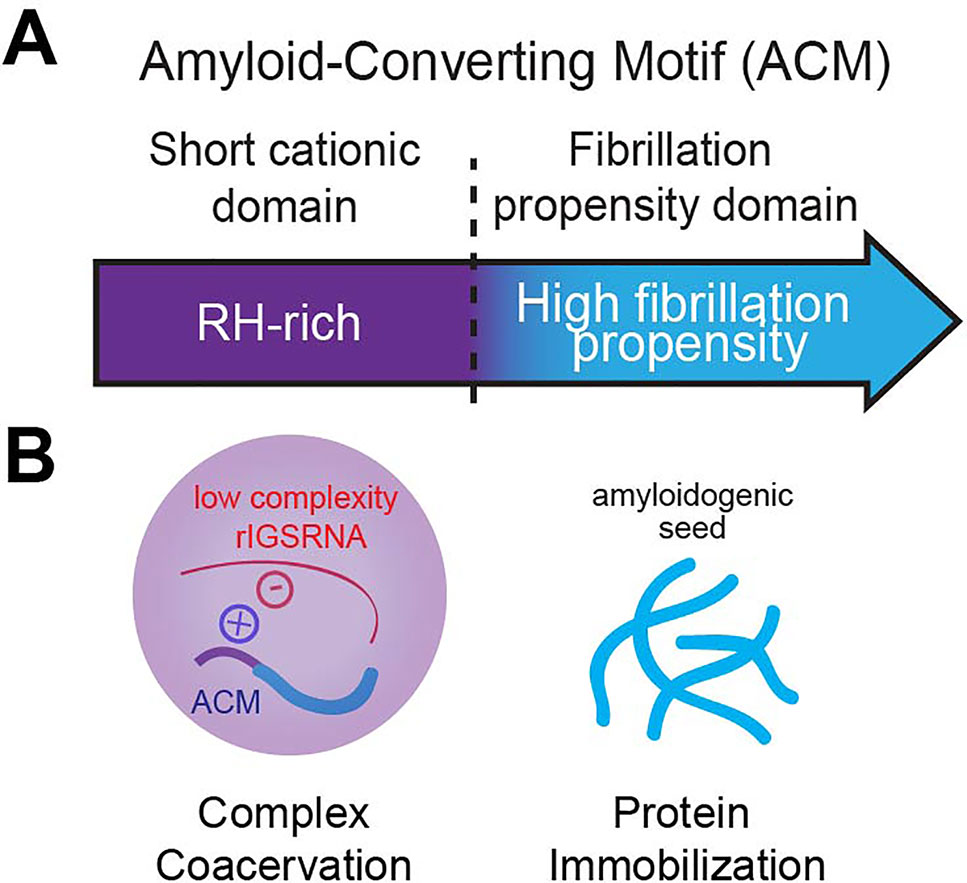
Figure 4 The amyloid-converting motif (ACM). (A) The ACM is necessary and sufficient to target and immobilize proteins in amyloid bodies (A-bodies). It consists of a R/H-rich short cationic domain flanking a high fibrillation propensity domain (determined by Rosetta energy of less than −23 kcal/mol). (B) We propose that it is its bipartite nature that allows the ACM to traverse phase boundaries. Complex coacervation of short cationic domains and low-complexity rIGSRNA (liquid–liquid phase separation) concentrates fibrillation propensity domains to activate a liquid-to-solid phase transition to form A-bodies.
Amyloidogenic Properties of the ACM Trigger Protein Immobilization
In principle, A-bodies and Balbiani bodies should be composed of proteins that possess fibrillation propensity, i.e., increased likelihood of forming fibrils. Bioinformatic analysis of the consensus ACM revealed the hydrophobic LXV motifs make up a region of high fibrillation propensity, as predicted by a Rosetta energy score of less than −23 kcal/mol (Goldschmidt, 2010) (Figure 4A). As a whole, the ACM bears a striking resemblance to that of the prototypical pathological β-amyloid, historically associated with Alzheimer’s disease, in which a R/H-rich sequence is in close proximity to a region of high fibrillation propensity referred to in literature as the P3 fragment (Figure 4A) (Dulin et al., 2008). The ACM exhibits classic amyloidogenic properties previously observed for β-amyloid, namely, the cross-β X-ray diffraction pattern in vivo and the ability to form fibrils in vitro (Audas et al., 2016).
It has been proposed that the function of the initial liquid state is to locally concentrate proteins with fibrillation propensity that would otherwise be at levels below the critical threshold in the cell (Knowles et al., 2014). Concentration-dependent activation of protein fibrillation has been well-documented in vitro (Eisenberg and Jucker, 2012; Knowles et al., 2014). Indeed, markers of early or nascent A-bodies include ANS and A11 staining (Wang et al., 2018) that are typically used in vitro to indicate accumulating pre-amyloidogenic structures (Booth, 1997; Kayed et al., 2007; Hawe et al., 2008; Tang et al., 2015). In addition, photobleaching analyses revealed that the core of nucleolar foci contain immobile proteins. Nascent A-bodies expand by self-assembly, during which soluble proteins are added directly and autonomously to the growing amyloid structure (Figure 2) (Wang et al., 2018).
The ACM Traverses Across Phase Boundaries to Confer A-Body Identity
As described above, low-complexity RNA sequences are important in conferring A-bodies their unique identity (Wang et al., 2018), adding to the list of architectural determinants, which includes sequence-specific RNA (Yamazaki et al., 2018), mRNA secondary structure (Jain and Vale, 2017; Langdon, 2018), and short unstructured RNA in vitro (Nott et al., 2016). The low-complexity rIGSRNA-mediated foci themselves appear immiscible from the three canonical compartments of the nucleolus (Feric et al., 2016; Wang et al., 2018), suggesting they exhibit distinct properties that may exclude mixing. Ultimately, it is the bipartite nature of the ACM that confers A-body identity and differentiates the ACM from other motifs. One possibility is that the R/H-rich “short cationic domain” mediates complex coacervation while the “fibrillation propensity domain” initiates amyloidogenesis to immobilize proteins in A-bodies (Audas et al., 2016; Wang et al., 2018) (Figure 4B). In this model, proteins without a region of high fibrillation propensity are unlikely to be incorporated into A-bodies. In other words, while many proteins containing positively charged domains can form transient rIGSRNA-dependent nucleolar foci, only the ones that harbor high fibrillation propensity sequences will be found immobilized in A-bodies. The balance of positive to negative charge ratio, presence of polar and/or aromatic residues, and modifications (Aumiller and Keating, 2016; Monahan et a.l, 2017) will affect how ACM engage in intermolecular interactions with other proteins and low-complexity RNA. Therefore, the ACM is versatile as it can physiologically transition proteins across phase boundaries.
Physiological Amyloids Promote Cell Dormancy
Endogenous A-bodies have been found in primary cultures and cell lines exposed to various stimuli, the cores of human tumors, and subsets of cells in normal human and mouse tissues (Audas et al., 2016). So, while A-bodies share structural features with pathological amyloids, their ubiquitous and reversible nature is indicative of a physiological function. Pathway enrichment analysis determined that many of the proteins sequestered in A-bodies are involved in cell cycle regulation and DNA synthesis, such as CDK1, POLD1, and DNMT1 (Audas et al., 2016). By temporarily detaining key factors from their sites of activity into A-bodies, major molecular networks are disrupted, suppressing metabolic activity (Mekhail, 2006) and arresting proliferation/DNA synthesis (Audas et al., 2016). This is different from the quiescent state (G0) in the cell cycle where cells arrest proliferation but remain metabolically active. Therefore, the biological role of A-body formation is to promote cellular dormancy as an adaptive response to environmental stressors. These findings are consistent with reports that show Balbiani bodies (Boke et al., 2016) and fluid-to-solid transitions in yeast (Munder et al, 2016) promote dormancy, reinforcing the concept that cells utilize different states of matter to perform various biological functions (Table 2). The material properties of “solid-like” systems—non-dynamic, amyloid-like—result in loss of metabolic activity and promote cellular dormancy, while those of ”liquid-like” systems—dynamic, fluid-like—concentrate and facilitate biochemical reactions.
A-Bodies as Targets for Therapeutic Discovery
Implications for Pathological Amyloids
A-bodies that are physiologically produced remain confined within the nucleus and are considered non-toxic amyloids (Wang et al., 2017). How the nuclear environment alters the interaction properties of aggregation-prone proteins to prevent toxicity (Woerner et al., 2016; Maharana, 2018) compared to the cytoplasm is unclear. These results imply the cell tightly regulates the induction and degradation of low-complexity rIGSRNA within the nucleolus in response to stimuli. It is interesting that the pathological β-amyloid involved in Alzheimer’s disease is an ACM and undergoes immobilization in A-bodies. Mutations that decrease the fibrillation potential of β-amyloid prevent its immobilization in A-bodies, providing a link between β-amyloid fibrillation and A-body biogenesis. It is tempting to speculate that disruptions of A-body biogenesis, for example by cell death, may provide the initial seeds for pathological amyloidogenesis. Whether aberrant production or localization of low-complexity RNA or exposure of A-bodies to the extracellular environment is involved in amyloid pathologies such as Alzheimer’s and Huntington’s diseases will be the subject of future studies.
Implications for Tumor Cell Dormancy and Metastasis
Given the evidence that A-bodies are observed in the cores of tumors and promote cell dormancy (Audas et al., 2016), perhaps there is a link between A-body formation and tumor cell dormancy. This could allow cancer cells to adapt to the harsh hypoxic/acidotic conditions of the tumor microenvironment. Such a connection has pertinent implications for chemotherapeutic resistance and metastasis (Li, 2014; Li, 2015). With major metabolic pathways shut down and key drug targets stored away in A-bodies, a small population of dormant tumor cells may be resistant to treatment, survive for a prolonged period of time, and contribute to disease recurrence or metastasis upon their metabolic reactivation. Preventing cancer cells from forming A-bodies and going dormant or, more realistically, preventing cancer cells from reactivating may become a viable treatment option.
An interesting avenue of research that may offer a more effective cancer treatment strategy involves manipulating the proteins that evade capture into A-bodies. Interestingly, proteins that remain active to sustain basal metabolism and viability under stress tend to be devoid of fibrillation propensity domains and evade capture into A-bodies. These proteins would be more susceptible to manipulation and serve as better chemotherapeutic targets than proteins captured in A-bodies.
Discussion
Twenty years after its discovery, the study of nucleolar sequestration has led to important conceptual and mechanistical advances in our understanding of the role of membraneless bodies and how they are constructed. The ability of cells to reversibly cycle proteins from a mobile to immobile state in A-bodies represents an effective posttranslational mechanism to regulate molecular networks. In this review, we propose a stepwise working model of A-body biogenesis that highlights this process as a precisely choreographed multistep routine rather than a random aggregation of misfolded proteins. This also makes A-bodies unique from the liquid compartments that populate the cell. A-bodies are characterized by electron-dense fibers composed of an array of immobilized proteins that stain positive with various amyloidogenic dyes. Liquid condensates do not display amyloidogenic features and generally contain mobile proteins. Obviously, this raises the key question of why liquid condensates typically do not mature into bodies with amyloidogenic properties. One possibility is that the clusters of low-complexity rIGSRNA are able to recruit sufficient quantity of proteins by simple electrostatic interactions, thereby reaching the critical concentration threshold to trigger amyloidogenesis (Knowles et al., 2014). This model is supported by the observation of electron-dense material early on in A-body biogenesis. The bipartite nature of the ACM, which can both electrostatically interact with low-complexity rIGSRNA and contains a high fibrillation propensity domain necessary for immobilization, may also explain the maturation of A-bodies. Perhaps the architectural determinants of RNA that seed various liquid bodies are too restrictive to certain binding partners to reach a concentration threshold required for amyloidogenesis. Additionally, there is evidence that suggests high nuclear RNA concentration acts as a buffer to prevent condensates from becoming amyloidogenic (Maharana et al., 2018). Indeed, mutated proteins with increased propensity to form amyloids are preferentially formed in the cytoplasm, which has low RNA concentration. While this appears to be the case for cytoplasmic stress granules enriched in amyloidogenic mutant proteins, this likely does not explain A-body maturation as the nucleolus is highly enriched in rRNA. How A-bodies progress from liquid-like condensates to solid-like condensates is a major question in the field of condensate biology, which we are actively pursuing. We are also keen to explore other stimuli, particularly those that alter nucleolar architecture, that may promote the accumulation of rIGSRNA and the formation of A-bodies, including viral infection (Wang, 2010), inflammation, and diurnal cycles of metabolism. Diurnal oscillations in nucleolar size and abundance of nucleolar RNAs have been seen in mammalian systems (Sinturel et al., 2017; Aitken and Semple, 2017), as well as an emerging connection between circadian disturbances and Alzheimer’s disease (Musiek, 2015). Despite most proteins having inherent fibrillation propensity, the dominant view remains that amyloids are inherently toxic, rather than a physiological fold exploited by cells to regulate various peptide functions. The challenge will be to decipher fundamental differences between A-bodies and other physiological or pathological membraneless compartments that could inform our understanding on how to prevent, detect, and treat amyloid-based diseases.
Author Contributions
MW, MB, PT, and SL designed the manuscript, discussed the conceptual implications, wrote the paper, and made the figures.
Funding
This work was supported by grants from the National Institute of General Medical Sciences (R01GM115342) (SL), the National Cancer Institute (R01CA200676) of the NIH, and the Sylvester Comprehensive Cancer Center (SL).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agrawal, S., Ganley, A. R. D. (2018). The conservation landscape of the human ribosomal RNA gene repeats. PloS One 13 (12), e0207531. doi: 10.1371/journal.pone.0207531
Aitken, S., Semple, C. A. (2017). The circadian dynamics of small nucleolar RNA in the mouse liver. J. R. Soc. Interface 14 (130). doi: 10.1098/rsif.2017.0034
Altmeyer, M., Neelsen, K. J., Teloni, F., Pozdnyakova, I., Pellegrino, S., Grofte, M., et al. (2015). Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088. doi: 10.1038/ncomms9088
Arabi, A. (2003). Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J. Cell Sci. 116 (9), 1707–1717. doi: 10.1242/jcs.00370
Audas, T. E., Audas, D. E., Jacob, M. D., Ho, J. J., Khacho, M., Wang, M., et al. (2016). Stressing out over long noncoding RNA. Biochim. Biophys. Acta 1859 (1), 184–191. doi: 10.1016/j.bbagrm.2015.06.010
Audas, T. E., Jacob, M. D., Lee, S. (2012a). Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 45 (2), 147–157. doi: 10.1016/j.molcel.2011.12.012
Audas, T. E., Jacob, M. D., Lee, S. (2012b). The nucleolar detention pathway: a cellular strategy for regulating molecular networks. Cell Cycle 11 (11), 2059–2062. doi: 10.4161/cc.20140
Audas, T. E. (2016). Adaptation to stressors by systemic protein Amyloidogenesis. Dev. Cell 39 (2), 155–168. doi: 10.1016/j.devcel.2016.09.002
Audas, T.E., Lee, S. (2016). Stressing out over long noncoding rna. Biochim. Biophys. Acta 1859, 184–191.
Aumiller, W. M., Jr., Keating, C. D. (2016). Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat. Chem. 8 (2), 129–137. doi: 10.1038/nchem.2414
Azzam, R., Chen, S. L., Shou, W., Mah, A. S., Alexandru, G., Nasmyth, K., et al. (2004). Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 305 (5683), 516–519. doi: 10.1126/science.1099402
Bachant, J. B., Elledge, S. J. (1999). Mitotic treasures in the nucleolus. Nature 398 (6730), 757–758. doi: 10.1038/19641
Banani, S. F., Lee, H. O., Hyman, A. A., Rosen, M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18 (5), 285–298. doi: 10.1038/nrm.2017.7
Banski, P., Mahboubi, H., Kodiha, M., Shrivastava, S., Kanagaratham, C., Stochaj, U. (2010). Nucleolar targeting of the chaperone hsc70 is regulated by stress, cell signaling, and a composite targeting signal which is controlled by autoinhibition. J. Biol. Chem. 285 (28), 21858–21867. doi: 10.1074/jbc.M110.117291
Berchowitz, L. E., Kabachinski, G., Walker, M. R., Carlile, T. M., Gilbert, W. V., Schwartz, T. U., et al. (2015). Regulated formation of an Amyloid-like translational repressor governs gametogenesis. Cell 163 (2), 406–418. doi: 10.1016/j.cell.2015.08.060
Bernardi, R., Scaglioni, P. P., Bergmann, S., Horn, H. F., Vousden, K. H., Pandolfi, P. P. (2004). PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 6 (7), 665–672. doi: 10.1038/ncb1147
Boisvert, F. M., van Koningsbruggen, S., Navascues, J., Lamond, A. I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8 (7), 574–585. doi: 10.1038/nrm2184
Boke, E., Ruer, M., Wuhr, M., Coughlin, M., Lemaitre, R., Gygi, S. P., et al. (2016). Amyloid-like self-assembly of a cellular compartment. Cell 166 (3), 637–650. doi: 10.1016/j.cell.2016.06.051
Bond, C. S., Fox, A. H. (2009). Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186 (5), 637–644. doi: 10.1083/jcb.200906113
Booth, D. R., Sunde, M., Bellotti, V., Robinson, C. V., Hutchinson, W. L., Fraser, P. E., et al. (1997). Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature 385 (6619), 787–793. doi: 10.1038/385787a0
Boulon, S., Westman, B. J., Hutten, S., Boisvert, F. M., Lamond, A. I. (2010). The nucleolus under stress. Mol. Cell 40 (2), 216–227. doi: 10.1016/j.molcel.2010.09.024
Brangwynne, C. P., Eckmann, C. R., Courson, D. S., Rybarska, A., Hoege, C., Gharakhani, J., et al. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324 (5935), 1729–1732. doi: 10.1126/science.1172046
Brangwynne, C. P., Mitchison, T. J., Hyman, A. A. (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 108 (11), 4334–4339. doi: 10.1073/pnas.1017150108
Brangwynne, C. P., Tompa, P., Pappu, Rohit V. (2015). Polymer physics of intracellular phase transitions. Nat. Phys. 11 (11), 899–904. doi: 10.1038/nphys3532
Cereghetti, G., Saad, S., Dechant, R., Peter, M. (2018). Reversible, functional amyloids: towards an understanding of their regulation in yeast and humans. Cell Cycle 17 (13), 1545–1558. doi: 10.1080/15384101.2018.1480220
Chen, J., Stark, L. A. (2017). Aspirin prevention of colorectal cancer: focus on NF-kappaB signalling and the nucleolus. Biomedicines 5 (3). doi: 10.3390/biomedicines5030043
Chen, X., Hiller, M., Sancak, Y., Fuller, M. T. (2005). Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science 310 (5749), 869–872. doi: 10.1126/science.1118101
Chiti, F., Dobson, C. M. (2006). Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366. doi: 10.1146/annurev.biochem.75.101304.123901
Chujo, T., Hirose, T. (2017). Nuclear bodies built on architectural long noncoding RNAs: unifying principles of their construction and function. Mol. Cells 40 (12), 889–896. doi: 10.14348/molcells.2017.0263
Chujo, T., Yamazaki, T., Kawaguchi, T., Kurosaka, S., Takumi, T., Nakagawa, S., et al. (2017). Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J. 36 (10), 1447–1462. doi: 10.15252/embj.201695848
Dellaire, G., Bazett-Jones, D. P. (2004). PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26 (9), 963–977. doi: 10.1002/bies.20089
Dulin, F., Leveille, F., Ortega, J. B., Mornon, J. P., Buisson, A., Callebaut, I., et al. (2008). P3 peptide, a truncated form of A beta devoid of synaptotoxic effect, does not assemble into soluble oligomers. FEBS Lett. 582 (13), 1865–1870. doi: 10.1016/j.febslet.2008.05.002
Eisenberg, D., Jucker, M. (2012). The amyloid state of proteins in human diseases. Cell 148 (6), 1188–1203. doi: 10.1016/j.cell.2012.02.022
Elbaum-Garfinkle, S., Kim, Y., Szczepaniak, K., Chen, C. C., Eckmann, C. R., Myong, S., et al. (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112 (23), 7189–7194. doi: 10.1073/pnas.1504822112
Emmott, E., Hiscox, J. A. (2009). Nucleolar targeting: the hub of the matter. EMBO Rep. 10 (3), 231–238. doi: 10.1038/embor.2009.14
Falabella, P., Riviello, L., Pascale, M., Lelio, I. D., Tettamanti, G., Grimaldi, A., et al. (2012). Functional amyloids in insect immune response. Insect Biochem. Mol. Biol. 42 (3), 203–211. doi: 10.1016/j.ibmb.2011.11.011
Fay, M. M., Anderson, P. J. (2018). The role of RNA in biological phase separations. J. Mol. Biol. 430 (23), 4685–4701. doi: 10.1016/j.jmb.2018.05.003
Feric, M., Vaidya, N., Harmon, T. S., Mitrea, D. M., Zhu, L., Richardson, T. M., et al. (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell 165 (7), 1686–1697. doi: 10.1016/j.cell.2016.04.047
Fowler, D. M., Kelly, J. W. (2012). Functional amyloidogenesis and cytotoxicity-insights into biology and pathology. PloS Biol. 10 (12), e1001459. doi: 10.1371/journal.pbio.1001459
Fowler, D. M., Koulov, A. V., Alory-Jost, C., Marks, M. S., Balch, W. E., Kelly, J. W. (2006). Functional amyloid formation within mammalian tissue. PloS Biol. 4 (1), e6. doi: 10.1371/journal.pbio.0040006
Fowler, D. M., Koulov, A. V., Balch, W. E., Kelly, J. W. (2007). Functional amyloid–from bacteria to humans. Trends Biochem. Sci. 32 (5), 217–224. doi: 10.1016/j.tibs.2007.03.003
Gall, J. G. (2000). Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16, 273–300. doi: 10.1146/annurev.cellbio.16.1.273
Goldschmidt, L., Teng, P. K., Riek, R., Eisenberg, D. (2010). Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. U. S. A. 107 (8), 3487–3492. doi: 10.1073/pnas.0915166107
Gonzalez, I. L., Sylvester, J. E. (1995). Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 27 (2), 320–328. doi: 10.1006/geno.1995.1049
Grummt, I., Pikaard, C. S. (2003). Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4 (8), 641–649. doi: 10.1038/nrm1171
Hall, A. C., Ostrowski, L. A., Mekhail, K. (2019). Phase separation as a melting pot for DNA repeats. Trends Genet. doi: 10.1016/j.tig.2019.05.001
Hawe, A., Sutter, M., Jiskoot, W. (2008). Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 25 (7), 1487–1499. doi: 10.1007/s11095-007-9516-9
Hernandez-Verdun, D. (2006). Nucleolus: from structure to dynamics. Histochem. Cell Biol. 125 (1–2), 127–137. doi: 10.1007/s00418-005-0046-4
Hirose, T., Yamazaki, T., Nakagawa, S. (2019). Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdiscip. Rev. RNA, e1545. doi: 10.1002/wrna.1545
Hodges, M., Tissot, C., Howe, K., Grimwade, D., Freemont, P. S. (1998). Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am. J. Hum. Genet. 63 (2), 297–304. doi: 10.1086/301991
Holehouse, A. S., Pappu, R. V. (2018). Functional Implications of Intracellular Phase Transitions. Biochemistry 57 (17), 2415–2423. doi: 10.1021/acs.biochem.7b01136
Hult, C., Adalsteinsson, D., Vasquez, P. A., Lawrimore, J., Bennett, M., York, A., et al. (2017). Enrichment of dynamic chromosomal crosslinks drive phase separation of the nucleolus. Nucleic Acids Res. 45 (19), 11159–11173. doi: 10.1093/nar/gkx741
Itakura, A. K., Futia, R. A., Jarosz, D. F. (2018). It pays to be in phase. Biochemistry 57 (17), 2520–2529. doi: 10.1021/acs.biochem.8b00205
Jacob, M. D., Audas, T. E., Mullineux, S. T., Lee, S. (2012). Where no RNA polymerase has gone before: novel functional transcripts derived from the ribosomal intergenic spacer. Nucleus 3 (4), 315–319. doi: 10.4161/nucl.20585
Jacob, M. D., Audas, T. E., Uniacke, J., Trinkle-Mulcahy, L., Lee, S. (2013). Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus. Mol. Biol. Cell 24 (18), 2943–2953. doi: 10.1091/mbc.e13-04-0223
Jain, A., Vale, R. D. (2017). RNA phase transitions in repeat expansion disorders. Nature 546 (7657), 243–247. doi: 10.1038/nature22386
Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149 (4), 753–767. doi: 10.1016/j.cell.2012.04.017
Kayatekin, C., Matlack, K. E., Hesse, W. R., Guan, Y., Chakrabortee, S., Russ, J., et al. (2014). Prion-like proteins sequester and suppress the toxicity of huntingtin exon 1. Proc. Natl. Acad. Sci. U. S. A. 111 (33), 12085–12090. doi: 10.1073/pnas.1412504111
Kayed, R., Head, E., Sarsoza, F., Saing, T., Cotman, C. W., Necula, M., et al. (2007). Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2, 18. doi: 10.1186/1750-1326-2-18
Knowles, T. P., Vendruscolo, M., Dobson, C. M. (2014). The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15 (6), 384–396. doi: 10.1038/nrm3810
Kruger, T., Scheer, U. (2010). p53 localizes to intranucleolar regions distinct from the ribosome production compartments. J. Cell Sci. 123 (Pt 8), 1203–1208. doi: 10.1242/jcs.062398
Lamond, A. I., Spector, D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4 (8), 605–612. doi: 10.1038/nrm1172
Langdon, E. M., Qiu, Y., Ghanbari Niaki, A., McLaughlin, G. A., Weidmann, C. A., Gerbich, T. M. (2018). mRNA structure determines specificity of a polyQ-driven phase separation. Science 360 (6391), 922–927. doi: 10.1126/science.aar7432
Latonen, L. (2011). Nucleolar aggresomes as counterparts of cytoplasmic aggresomes in proteotoxic stress. Proteasome inhibitors induce nuclear ribonucleoprotein inclusions that accumulate several key factors of neurodegenerative diseases and cancer. Bioessays 33 (5), 386–395. doi: 10.1002/bies.201100008
Latonen, L. (2019). Phase-to-phase with nucleoli - stress responses, protein aggregation and novel roles of RNA. Front. Cell Neurosci. 13, 151. doi: 10.3389/fncel.2019.00151
Latonen, L.Moore, H. M., Bai, B., Jaamaa, S., Laiho, M. (2011). Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene 30 (7), 790–805. doi: 10.1038/onc.2010.469
Li, J., McQuade, T., Siemer, A. B., Napetschnig, J., Moriwaki, K., Hsiao, Y. S., et al. (2012). The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150 (2), 339–350. doi: 10.1016/j.cell.2012.06.019
Li, J., Jiang, E., Wang, X., Shangguan, A. J., Zhang, L., Yu, Z. (2015). Dormant Cells: the original cause of tumor recurrence and metastasis. Cell Biochem. Biophys. 72 (2), 317–320. doi: 10.1007/s12013-014-0477-4
Li, S., Kennedy, M., Payne, S., Kennedy, K., Seewaldt, V. L., Pizzo, S. V., et al. (2014). Model of tumor dormancy/recurrence after short-term chemotherapy. PloS One 9 (5), e98021. doi: 10.1371/journal.pone.0098021
Liu, Y., Deisenroth, C., Zhang, Y. (2016). RP-MDM2-p53 pathway: linking ribosomal biogenesis and tumor surveillance. Trends Cancer 2 (4), 191–204. doi: 10.1016/j.trecan.2016.03.002
Lohrum, M. A., Ashcroft, M., Kubbutat, M. H., Vousden, K. H. (2000). Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2 (3), 179–181. doi: 10.1038/35004057
Lyons, S. M., Anderson, P. (2016). RNA-Seeded functional amyloids balance growth and survival. Dev. Cell 39 (2), 131–132. doi: 10.1016/j.devcel.2016.10.005
Machyna, M., Heyn, P., Neugebauer, K. M. (2013). Cajal bodies: where form meets function. Wiley Interdiscip. Rev. RNA 4 (1), 17–34. doi: 10.1002/wrna.1139
Maharana, S., Wang, J., Papadopoulos, D. K., Richter, D., Pozniakovsky, A., Poser, I., et al. (2018). RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360 (6391), 918–921. doi: 10.1126/science.aar7366
Maji, S. K., Perrin, M. H., Sawaya, M. R., Jessberger, S., Vadodaria, K., Rissman, R. A., et al. (2009). Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325 (5938), 328–332. doi: 10.1126/science.1173155
Martindill, D. M., Risebro, C. A., Smart, N., Franco-Viseras Mdel, M., Rosario, C. O., Swallow, C. J., et al. (2007). Nucleolar release of Hand1 acts as a molecular switch to determine cell fate. Nat. Cell Biol. 9 (10), 1131–1141. doi: 10.1038/ncb1633
Mateju, D., Franzmann, T. M., Patel, A., Kopach, A., Boczek, E. E., Maharana, S., et al. (2017). An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 36 (12), 1669–1687. doi: 10.15252/embj.201695957
Mattsson, K., Pokrovskaja, K., Kiss, C., Klein, G., Szekely, L. (2001). Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. Proc. Natl. Acad. Sci. U.S.A. 98 (3), 1012–1017. doi: 10.1073/pnas.98.3.1012
Mayer, C., Neubert, M., Grummt, I. (2008). The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 9 (8), 774–780. doi: 10.1038/embor.2008.109
Mayer, C., Schmitz, K. M., Li, J., Grummt, I., Santoro, R. (2006). Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 22 (3), 351–361. doi: 10.1016/j.molcel.2006.03.028
Mediani, L., Guillen-Boixet, J., Vinet, J., Franzmann, T. M., Bigi, I., Mateju, D., et al. (2019). Defective ribosomal products challenge nuclear function by impairing nuclear condensate dynamics and immobilizing ubiquitin. EMBO J., e101341. doi: 10.15252/embj.2018101341
Mekhail, K., Gunaratnam, L., Bonicalzi, M. E., Lee, S. (2004). HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 6 (7), 642–647. doi: 10.1038/ncb1144
Mekhail, K.Khacho, M., Carrigan, A., Hache, R. R., Gunaratnam, L., Lee, S. (2005). Regulation of ubiquitin ligase dynamics by the nucleolus. J. Cell Biol. 170 (5), 733–744. doi: 10.1083/jcb.200506030
Mekhail, K.Rivero-Lopez, L., Khacho, M., Lee, S. (2006). Restriction of rRNA synthesis by VHL maintains energy equilibrium under hypoxia. Cell Cycle 5 (20), 2401–2413. doi: 10.4161/cc.5.20.3387
Mekhail, K.Rivero-Lopez, L., Al-Masri, A., Brandon, C., Khacho, M., Lee, S. (2007). Identification of a common subnuclear localization signal. Mol. Biol. Cell 18 (10), 3966–3977. doi: 10.1091/mbc.e07-03-0295
Mikhaleva, E. A., Leinsoo, T. A., Ishizu, H., Gvozdev, V. A., Klenov, M. S. (2018). The nucleolar transcriptome regulates Piwi shuttling between the nucleolus and the nucleoplasm. Chromosome Res. doi: 10.1007/s10577-018-9595-y
Miller, O. L., Jr., Beatty, B. R. (1969). Visualization of nucleolar genes. Science 164 (3882), 955–957. doi: 10.1126/science.164.3882.955
Mitrea, D. M., Cika, J. A., Guy, C. S., Ban, D., Banerjee, P. R., Stanley, C. B., et al. (2016). Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 5. doi: 10.7554/eLife.13571
Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., et al. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163 (1), 123–133. doi: 10.1016/j.cell.2015.09.015
Monahan, Z., Ryan, V. H., Janke, A. M., Burke, K. A., Rhoads, S. N., Zerze, G. H., et al. (2017). Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36 (20), 2951–2967. doi: 10.15252/embj.201696394
Munder, M. C.Midtvedt, D., Franzmann, T., Nuske, E., Otto, O., Herbig, M., et al. (2016). A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife 5. doi: 10.7554/eLife.09347
Murakami, T., Qamar, S., Lin, J. Q., Schierle, G. S., Rees, E., Miyashita, A., et al (2015). ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88 (4), 678–690. doi: 10.1016/j.neuron.2015.10.030
Musiek, E. S. (2015). Circadian clock disruption in neurodegenerative diseases: cause and effect? Front. Pharmacol. 6, 29. doi: 10.3389/fphar.2015.00029
Nemeth, A., Grummt, I. (2018). Dynamic regulation of nucleolar architecture. Curr. Opin. Cell Biol. 52, 105–111. doi: 10.1016/j.ceb.2018.02.013
Nott, T. J., Craggs, T. D., Baldwin, A. J. (2016). Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem. 8 (6), 569–575. doi: 10.1038/nchem.2519
Nott, T. J., Petsalaki, E., Farber, P., Jervis, D., Fussner, E., Plochowietz, A., et al. (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57 (5), 936–947. doi: 10.1016/j.molcel.2015.01.013
Pak, C. W., Kosno, M., Holehouse, A. S., Padrick, S. B., Mittal, A., Ali, R., et al. (2016). Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63 (1), 72–85. doi: 10.1016/j.molcel.2016.05.042
Parker, R., Sheth, U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25 (5), 635–646. doi: 10.1016/j.molcel.2007.02.011
Patel, A., Lee, H.O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162 (5), 1066–1077. doi: 10.1016/j.cell.2015.07.047
Pederson, T., Tsai, R. Y. (2009). In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 184 (6), 771–776. doi: 10.1083/jcb.200812014
Pederson, T. (2011). The nucleolus. Cold Spring Harb Perspect. Biol. 3 (3). doi: 10.1101/cshperspect.a000638
Perry, R. P. (1960). On the nucleolar and nuclear dependence of cytoplasmic RNA synthesis in HeLa cells. Exp. Cell Res. 20, 216–220. doi: 10.1016/0014-4827(60)90240-8
Perry, R. P. (1962). The cellular sites of synthesis of Ribosomal and 4s Rna. Proc. Natl. Acad. Sci. U. S. A. 48 (12), 2179–2186. doi: 10.1073/pnas.48.12.2179
Peskett, T. R., Rau, F., O’Driscoll, J., Patani, R., Lowe, A. R., Saibil, H. R. (2018). A liquid to solid phase transition underlying pathological huntingtin Exon1 aggregation. Mol. Cell 70 (4), 588–601, e6. doi: 10.1016/j.molcel.2018.04.007
Phair, R. D., Misteli, T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404 (6778), 604–609. doi: 10.1038/35007077
Posey, A. E., Ruff, K. M., Harmon, T. S., Crick, S. L., Li, A., Diamond, M. I., et al. (2018). Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J. Biol. Chem. 293 (10), 3734–3746. doi: 10.1074/jbc.RA117.000357
Poyurovsky, M. V., Jacq, X., Ma, C., Karni-Schmidt, O., Parker, P. J., Chalfie, M., et al. (2003). Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Mol. Cell 12 (4), 875–887. doi: 10.1016/S1097-2765(03)00400-3
Protter, D. S., Parker, R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26 (9), 668–679. doi: 10.1016/j.tcb.2016.05.004
Raska, I., Shaw, P. J., Cmarko, D. (2006). Structure and function of the nucleolus in the spotlight. Curr. Opin. Cell Biol. 18 (3), 325–334. doi: 10.1016/j.ceb.2006.04.008
Rizzi, N., Denegri, M., Chiodi, I., Corioni, M., Valgardsdottir, R., Cobianchi, F., et al. (2004). Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol. Biol. Cell 15 (2), 543–551. doi: 10.1091/mbc.e03-07-0487
Saad, S., Cereghetti, G., Feng, Y., Picotti, P., Peter, M., Dechant, R. (2017). Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat. Cell Biol. 19 (10), 1202–1213. doi: 10.1038/ncb3600
Sansam, C. L., Wells, K. S., Emeson, R. B. (2003). Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl. Acad. Sci. U. S. A. 100 (24), 14018–14023. doi: 10.1073/pnas.2336131100
San-Segundo, P. A., Roeder, G. S. (1999). Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97 (3), 313–324. doi: 10.1016/S0092-8674(00)80741-2
Santoro, R. (2005). The silence of the ribosomal RNA genes. Cell Mol. Life Sci. 62 (18), 2067–2079. doi: 10.1007/s00018-005-5110-7
Sasaki, Y. T., Hirose, T. (2009). How to build a paraspeckle. Genome Biol. 10 (7), 227. doi: 10.1186/gb-2009-10-7-227
Sawyer, I. A., Sturgill, D., Dundr, M. (2019). Membraneless nuclear organelles and the search for phases within phases. Wiley Interdiscip. Rev. RNA 10 (2), e1514. doi: 10.1002/wrna.1514
Scheer, U., Hock, R. (1999). Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11 (3), 385–390. doi: 10.1016/S0955-0674(99)80054-4
Schmitz, K. M., Mayer, C., Postepska, A., Grummt, I. (2010). Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24 (20), 2264–2269. doi: 10.1101/gad.590910
Schnabel, J. (2010). Protein folding: the dark side of proteins. Nature 464 (7290), 828–829. doi: 10.1038/464828a
Sharifi, S., Bierhoff, H. (2018). Regulation of RNA Polymerase I transcription in development, disease, and aging. Annu. Rev. Biochem. doi: 10.1146/annurev-biochem-062917-012612
Shav-Tal, Y., Blechman, J., Darzacq, X., Montagna, C., Dye, B. T., Patton, J. G., et al. (2005). Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell 16 (5), 2395–2413. doi: 10.1091/mbc.e04-11-0992
Shin, Y., Brangwynne, C. P. (2017). Liquid phase condensation in cell physiology and disease. Science 357 (6357). doi: 10.1126/science.aaf4382
Shou, W., Seol, J. H., Shevchenko, A., Baskerville, C., Moazed, D., Chen, Z. W., et al. (1999). Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 (2), 233–244. doi: 10.1016/S0092-8674(00)80733-3
Sinturel, F., Gerber, A., Mauvoisin, D., Wang, J., Gatfield, D., Stubblefield, J. J., et al. (2017). Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169 (4), 651–663 e14. doi: 10.1016/j.cell.2017.04.015
Smirnov, E., Cmarko, D., Mazel, T., Hornacek, M., Raska, I. (2016). Nucleolar DNA: the host and the guests. Histochem Cell Biol. 145 (4), 359–372. doi: 10.1007/s00418-016-1407-x
Stark, L. A., Dunlop, M. G. (2005). Nucleolar sequestration of RelA (p65) regulates NF-kappaB-driven transcription and apoptosis. Mol. Cell Biol. 25 (14), 5985–6004. doi: 10.1128/MCB.25.14.5985-6004.2005
Sydorskyy, Y., Srikumar, T., Jeram, S. M., Wheaton, S., Vizeacoumar, F. J., Makhnevych, T., et al. (2010). A novel mechanism for SUMO system control: regulated Ulp1 nucleolar sequestration. Mol. Cell Biol. 30 (18), 4452–4462. doi: 10.1128/MCB.00335-10
Tang, Z., Dai, S., He, Y., Doty, R. A., Shultz, L. D., Sampson, S. B., et al. (2015). MEK guards proteome stability and inhibits tumor-suppressive amyloidogenesis via HSF1. Cell 160 (4), 729–744. doi: 10.1016/j.cell.2015.01.028
Tao, W., Levine, A. J. (1999). P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. U.S.A. 96 (12), 6937–6941. doi: 10.1073/pnas.96.12.6937
Thiry, M., Lafontaine, D. L. (2005). Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 15 (4), 194–199. doi: 10.1016/j.tcb.2005.02.007
Tomson, B. N., Rahal, R., Reiser, V., Monje-Casas, F., Mekhail, K., Moazed, D., et al. (2009). Regulation of Spo12 phosphorylation and its essential role in the FEAR network. Curr. Biol. 19 (6), 449–460. doi: 10.1016/j.cub.2009.02.024
Valgardsdottir, R., Chiodi, I., Giordano, M., Cobianchi, F., Riva, S., Biamonti, G. (2005). Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell 16 (6), 2597–2604. doi: 10.1091/mbc.e04-12-1078
Visintin, R., Hwang, E. S., Amon, A. (1999). Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398 (6730), 818–823. doi: 10.1038/19775
Wang, L., Ren, X. M., Xing, J. J., Zheng, A. C. (2010). The nucleolus and viral infection. Virol. Sin. 25 (3), 151–157. doi: 10.1007/s12250-010-3093-5
Wang, M., Audas, T. E., Lee, S. (2017). Disentangling a bad reputation: changing perceptions of amyloids. Trends Cell Biol. 27 (7), 465–467. doi: 10.1016/j.tcb.2017.03.001
Wang, M., Tao, X., Jacob, M. D., Bennett, C. A., Ho, J. J. D., Gonzalgo, M. L., et al. (2018). Stress-induced low complexity rna activates physiological amyloidogenesis. Cell Rep. 241713-241721 (7), e4. doi: 10.1016/j.celrep.2018.07.040
Weber, S. C., Brangwynne, C. P. (2012). Getting RNA and protein in phase. Cell 149 (6), 1188–1191. doi: 10.1016/j.cell.2012.05.022
Weber, J. D., Taylor, L. J., Roussel, M. F., Sherr, C. J., Bar-Sagi, D. (1999). Nucleolar arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1 (1), 20–26. doi: 10.1038/8991
Weber, J. D., Kuo, M. L., Bothner, B., DiGiammarino, E. L., Kriwacki, R. W., Roussel, M. F., et al. (2000). Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol. Cell Biol. 20 (7), 2517–2528. doi: 10.1128/MCB.20.7.2517-2528.2000
Weber, S. C. (2017). Sequence-encoded material properties dictate the structure and function of nuclear bodies. Curr. Opin. Cell Biol. 46, 62–71. doi: 10.1016/j.ceb.2017.03.003
Woerner, A. C., Frottin, F., Hornburg, D., Feng, L. R., Meissner, F., Patra, M., et al. (2016). Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351 (6269), 173–176. doi: 10.1126/science.aad2033
Wong, J. M., Kusdra, L., Collins, K. (2002). Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol. 4 (9), 731–736. doi: 10.1038/ncb846
Woodruff, J. B., Hyman, A. A., Boke, E. (2018). Organization and function of non-dynamic biomolecular condensates. Trends Biochem. Sci. 43 (2), 81–94. doi: 10.1016/j.tibs.2017.11.005
Yamazaki, T., Souquere, S., Chujo, T., Kobelke, S., Chong, Y. S., Fox, A. H., et al. (2018). Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70 (6), 1038–1053 e7. doi: 10.1016/j.molcel.2018.05.019
Yap, K., Mukhina, S., Zhang, G., Tan, J. S. C., Ong, H. S., Makeyev, E. V. (2018). A short tandem repeat-enriched RNA Assembles a nuclear compartment to control alternative splicing and promote cell survival. Mol. Cell 72 (3), 525–540 e13. doi: 10.1016/j.molcel.2018.08.041
Zhao, Z., Dammert, M. A., Grummt, I., Bierhoff, H. (2016a). lncRNA-induced nucleosome repositioning reinforces transcriptional repression of rRNA genes upon hypotonic stress. Cell Rep. 14 (8), 1876–1882. doi: 10.1016/j.celrep.2016.01.073
Zhao, Z., Dammert, M. A., Hoppe, S., Bierhoff, H., Grummt, I. (2016b). Heat shock represses rRNA synthesis by inactivation of TIF-IA and lncRNA-dependent changes in nucleosome positioning. Nucleic Acids Res. 44 (17), 8144–8152. doi: 10.1093/nar/gkw496
Zhao, Z., Senturk, N., Song, C., Grummt, I. (2018). lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 32 (11–12), 836–848. doi: 10.1101/gad.311688.118
Keywords: heat shock (HS), acidosis, architectural RNA (arcRNA), Alzheimer’s disease, cellular dormancy, physiological amyloidogenesis, beta-amyloid protein
Citation: Wang M, Bokros M, Theodoridis PR and Lee S (2019) Nucleolar Sequestration: Remodeling Nucleoli Into Amyloid Bodies. Front. Genet. 10:1179. doi: 10.3389/fgene.2019.01179
Received: 20 July 2019; Accepted: 24 October 2019;
Published: 21 November 2019.
Edited by:
Michael E. Symonds, University of Nottingham, United KingdomReviewed by:
Alexander Kouzmenko, Tokiwa Foundation, JapanJanine M. LaSalle, University of California, Davis, United States
Copyright © 2019 Wang, Bokros, Theodoridis and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen Lee, U3RlcGhlbmxlZUBtZWQubWlhbWkuZWR1
 Miling Wang
Miling Wang Michael Bokros
Michael Bokros Phaedra Rebecca Theodoridis1,2
Phaedra Rebecca Theodoridis1,2 Stephen Lee
Stephen Lee