- Girona Neuroimmunology and Multiple Sclerosis Unit (UNIEM), Dr. Josep Trueta University Hospital, Girona Biomedical Research Institute (IDIBGI), Girona, Spain
Multiple sclerosis (MS) is an autoimmune disease of the central nervous system characterized by loss of coordination, weakness, dysfunctions in bladder capacity, bowel movement, and cognitive impairment. Thus, the disease leads to a significant socioeconomic burden. In the pathophysiology of the disease, both genetic and environmental risk factors are involved. Gene x environment interaction is modulated by epigenetic mechanisms. Epigenetics refers to a sophisticated system that regulates gene expression with no changes in the DNA sequence. The most studied epigenetic mechanism is the DNA methylation. In this review, we summarize the data available from the current literature by grouping sets of differentially methylated genes in distinct biological categories: the immune system including innate and adaptive response, the DNA damage, and the central nervous system.
Introduction
Multiple sclerosis (MS) is a potentially disabling central nervous system (CNS) disease characterized by inflammation, demyelination, and axonal degeneration. The pathophysiological mechanisms involved in this autoimmune disease differ between patients. In MS pathogenesis, antigen-presenting cells (APCs) stimulate CD4+ T cells in the periphery, favoring their differentiation into CD4+ T helper (Th) cells. Upon their activation, these immune cells cross the blood–brain barrier (BBB) and migrate into the brain, where they are reactivated by resident APCs. Proinflammatory cytokines and chemokines are released by reactivated CD4+ T cells, while infiltrated cytotoxic CD8+ T cells impair neuronal integrity. In parallel, plasma B cells release antibodies against self myelin epitopes, contributing to demyelination (Ghasemi et al., 2017) (Figure S1).
An individual’s genetic background and exposure to environmental factors confer risk of developing MS through epigenetic modifications (Figure 1). DNA methylation (DNAme) is the most common epigenetic mechanism in somatic cells. This process occurs mainly in regulatory and promoter regions, where cytosine-guanine dinucleotides are markedly present (Weber et al., 2007). Although the precise mechanism of action of DNAme in MS remains to be fully elucidated, several studies have reported differentially methylated regions in either lymphocytes or peripheral blood mononuclear cells (PBMCs) and in post-mortem brain tissue (Table S1).
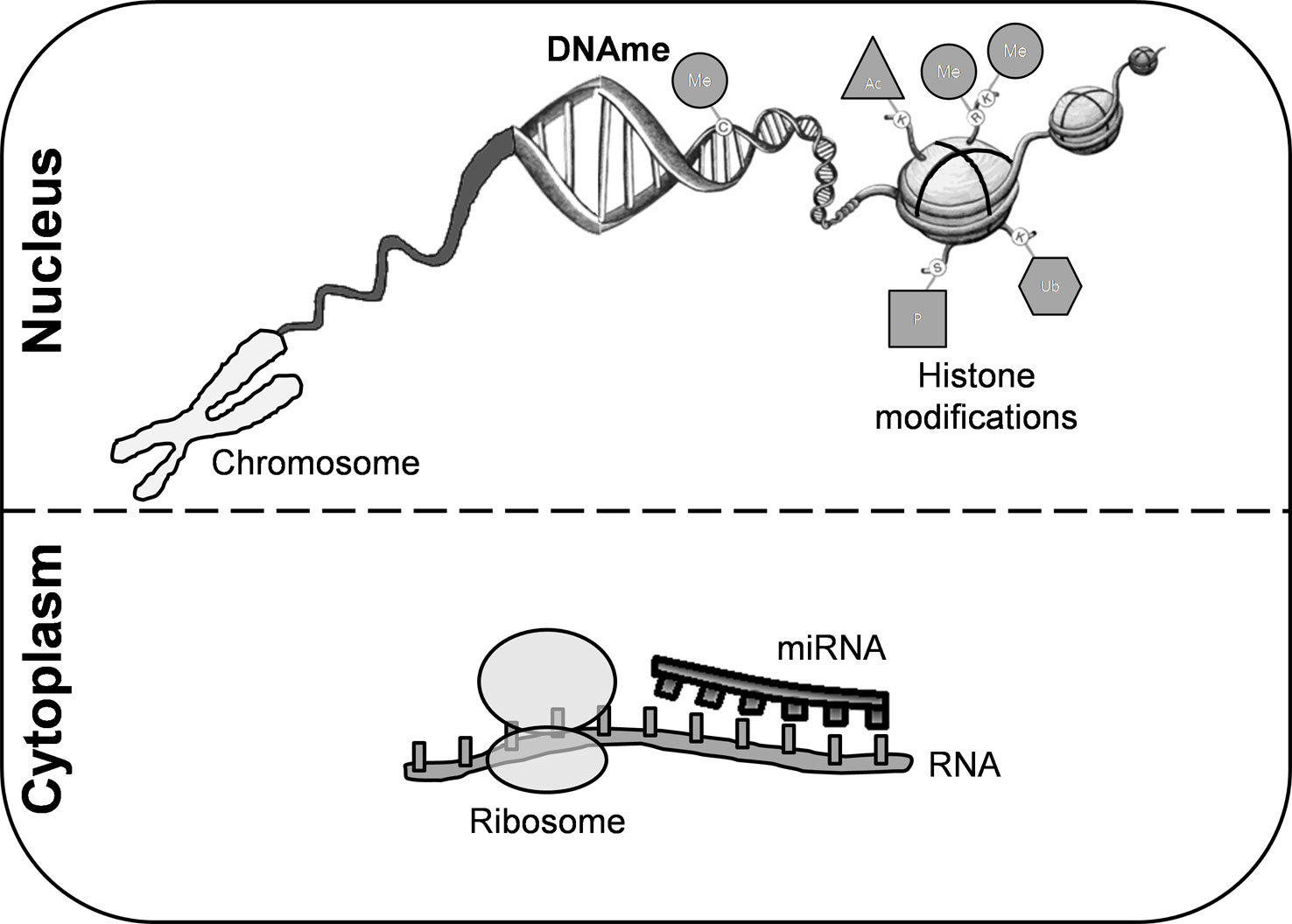
Figure 1 Epigenetic modifications. These mechanisms are crucial for regulating gene transcription and chromatin architecture. Among them, we can highlight histone modifications, DNA methylation, and microRNAs. Covalent modifications of histones include acetylation, phosphorylation, sumoylation, ubiquitination, and methylation. DNA methylation is the most common epigenetic mechanism that occurs mainly in enriched CG dinucleotides regions in somatic cells. miRNAs are small non-coding RNA molecules that participate in RNA silencing. DNAme, DNA methylation; miRNA, microRNA; Me, methylation; Ub, ubiquitination; Ac, acetylation; P, phosphorylation.
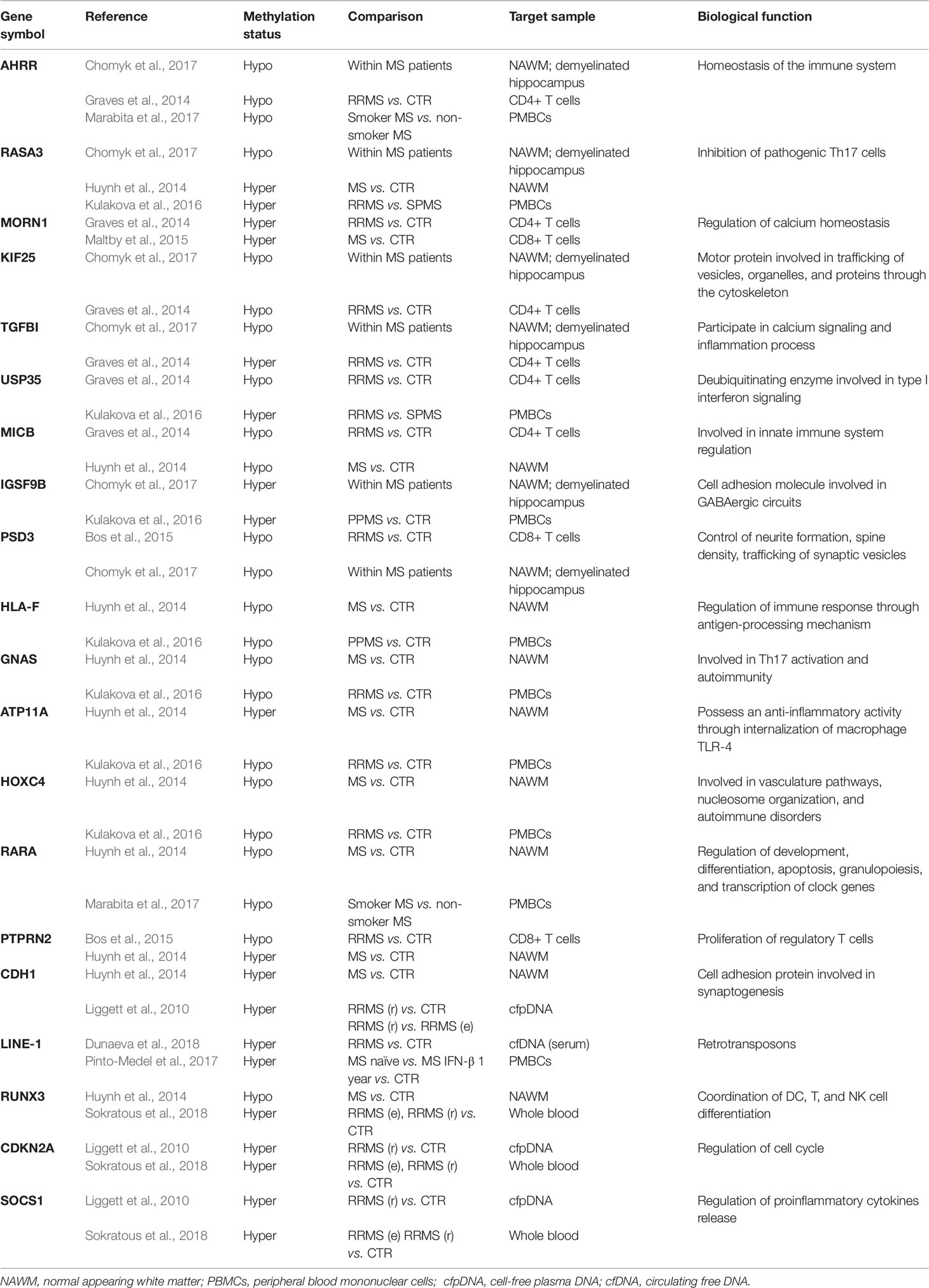
In this study, sets of differentially methylated genes described in the relevant literature are compared using Venn diagrams in order to determine the common, overlapping genes (Table 1). Although the studies included in this review have used a variety of methods, target samples, and subjects at different stages of the disease with distinct demographic characteristics that may contribute to DNAme heterogeneity, we assume that the common results reported at cell type level by different case studies could potentially explain in part MS pathophysiology. These results are summarized below.
The Immune System
The homeostasis of the immune system is modulated by the aryl hydrocarbon receptor (AHR). AHR activity is negatively regulated by the encoded protein for the aryl hydrocarbon receptor repressor (AHRR). MS patients showed lower expression levels of circulating AHR than their matched controls (Neavin et al., 2018). In line with these findings, lower DNAme levels for AHRR have been measured in demyelinated hippocampi (Chomyk et al., 2017), CD4+ T cells (Graves et al., 2014), and PBMCs (Marabita et al., 2017) of MS patients. This suggests that immune differentiation as well as the clinical course are compromised in MS (Neavin et al., 2018). Furthermore, it is widely accepted that the major histocompatibility complex (MHC) plays a key role in the genetic susceptibility to MS. Two polymorphic genes, termed MHC class I chain-related gene A (MICA) and MHC class I chain-related gene B (MICB), are located within the MHC class I region. These molecules interact with specific receptors constitutively expressed in natural killer (NK) and T cells. The expression of MICB proteins in circulating PBMCs stimulates autoreactive T cells and favors MS progression (Abediankenari et al., 2011). Similarly, Fernandez-Morera et al. (2008) found that the MICB*004 allele was significantly higher in MS patients than their matched controls. MS patients displayed lower DNAme levels for MICB compared to controls (Graves et al., 2014; Huynh et al., 2014), in agreement with Abediankenari et al. (2011) and Fernandez-Morera et al. (2008). Runt-related transcription factor 3 (RUNX3) is expressed in dendritic cells (DCs), as well as T and NK cells, regulating their differentiation. In contrast, cyclin-dependent kinase inhibitor 2A (CDKN2A) controls the cell cycle, and it is abundantly expressed in oligodendrocytes and CD4+ and CD8+ T cells. In PBMCs collected from MS patients, both genes were underexpressed, which indicates a misbalance in CD4/CD8 T cell differentiation (Parnell et al., 2014). In line with these findings, elevated levels of DNAme for RUNX3 and CDKN2A were found in MS patients compared to controls (Liggett et al., 2010; Sokratous et al., 2018).
Innate Immune Response
In DCs, high levels of cyclic adenosine monophosphate (cAMP) activate Th17 response through the stimulation of guanine nucleotide-binding protein, alpha stimulating (GNAS). High levels of activated Th17 cells are associated with autoimmunity (Lee et al., 2015). In line with these findings, other studies have revealed a lower number of DNAme groups to GNAS (Huynh et al., 2014; Kulakova et al., 2016) and consequently, an overactivation of Th17 cells. It is well-known that the recognition of CNS self-epitopes by monocytes facilitates type I interferon (IFN) release and thus, autoimmunity. IFN signaling is regulated by ubiquitination mechanisms. Ubiquitin-specific peptidase 35 (USP35), a member of the deubiquitinating enzyme family, reverses the process of ubiquitination and confers neuroprotection. During an inflammatory response, USP35 is underexpressed in monocytes. The expression of this deubiquitinating enzyme is higher when the inflammation is mitigated (Liu et al., 2018). Thus, we can speculate that a high number of DNAme groups to USP35 might occur in parallel with a relapsing MS course (Kulakova et al., 2016), while a hypomethylated pattern could favor a remitting course for the disease (Graves et al., 2014). In macrophages, the group of phosphatidylserines, a canonical substrate for ATPase phospholipid transporting 11A (ATP11A), is charged negatively and participates in the internalization of toll-like receptor 4 (TLR4). The internalization of TLR4 is essential to restricting long-term inflammatory responses. Deletion of ATP11A in humans causes an exacerbated inflammatory response (van der Mark et al., 2017). The addition of DNAme groups to ATP11A, which Huynh et al. (2014) described in the CNS of MS patients, might result in lower protein content and, consequently, chronic inflammation. However, Kulakova et al. (2016) found an opposite effect, indicating that the expression of ATP11A was presumably higher in MS patients than controls. This discrepancy might suggest that the study of DNAme derived from PBMCs (Kulakova et al., 2016) was conducted during the remitting course of MS, when no signs of inflammation were present.
Adaptive Immune Response
CD4+ T cells proliferate into effector T cells in order to provide the most effective response to maintain immune homeostasis. Among CD4+ T cells, the Th17 subset regulates the immune response against autoimmunity. The generation of pathogenic Th17 (pTh17) cells is associated with an upregulation of RAS p21 protein activator 3 (RASA3). In contrast, reduced expression of RASA3 suppresses pTh17 cell generation via enhanced interleukin 4 synthesis (Wu et al., 2018). An elevated number of methyl groups for RASA3 was measured in normal-appearing white matter (NAWM) (Huynh et al., 2014) and PBMCs (Kulakova et al., 2016), presumably due to the fact that MS patients displayed no radiological disease activity and absence of pTh17 cells. In contrast, Chomyk et al. (2017) demonstrated a hypomethylated RASA3 pattern in the hippocampus during ongoing demyelination. Therefore, the migration of pathogenic lymphocytes into the MS brain originates in the destruction of myelin sheath and axonal degeneration through elevated levels of RASA3 (Wu et al., 2018). The activation of homeobox C4 (HOXC4) promotes the proliferation and differentiation of B cells (Park et al., 2013). High mRNA transcription rates for HOXC4 are associated with dysfunctions in vasculature pathways and nucleosome organization (Marchetti et al., 2018) and have also been observed in autoimmune disorders (Park et al., 2013). A lower number of methylated groups to HOXC4 would result in elevated protein levels, contributing to autoimmunity (Huynh et al., 2014; Kulakova et al., 2016). The proliferation of regulatory T cells is achieved through the interaction of protein tyrosine phosphatase receptor type N2 (PTPRN2) with SMAD family member 3 (SMAD3) (Lee et al., 2016). A hypomethylated pattern of PTPRN2 has been described in T cells collected from MS patients (Bos et al., 2015). Thus, high levels of PTPRN2 might potentiate the proliferation of regulatory T cells. Interestingly, transforming growth factor, beta-induced (TGFBI) prevents autoimmunity by promoting T cell activation through Ca2+-calcineurin signaling (Graca, 2007). The addition of methyl groups to TGFBI in CD4+ T cells derived from MS patients (Graves et al., 2014) might prevent autoimmune reactivity favoring Ca2+-calcineurin signaling and T cell activation (Graca, 2007). Cytokines participate in the differentiation, maturation, and survival of immune cells. Suppressor of cytokine signaling 1 (SOCS1) regulates specifically the release of proinflammatory cytokines in MS. Under pathophysiological conditions, a significant reduction of SOCS1 has been measured in parallel with the synthesis of proinflammatory cytokines in MS (Toghi et al., 2017). In agreement with Toghi et al. (2017), a substantial number of DNAme groups to SOCS1 might result in an acute inflammatory response, as has been reported in the context of MS (Liggett et al., 2010; Sokratous et al., 2018).
Deoxyribonucleic Acid Damage
The endoplasmic reticulum (ER), along with the plasma membrane (PM) junctions, is essential for Ca2+ homeostasis. ER Ca2+ depletion potentiates the inflammatory response, ER stress and, lastly, DNA damage (Woo et al., 2016). Upon activation of T-cell receptors (TCRs), protein junctions containing membrane occupation and recognition nexus (MORN) motifs stimulate a Ca2+ influx in T cells. Deficiency of protein junctions containing MORN motifs aggravates the store-operated Ca2+ entry in T cells and causes DNA damage (Woo et al., 2016). An elevated number of DNAme groups to MORN repeat containing 1 (MORN1) has been reported in both CD4+ and CD8+ T cells collected from MS patients (Graves et al., 2014; Maltby et al., 2015) and might affect T cell viability (Woo et al., 2016). On the other hand, pleckstrin and Sec7 domain-containing 3 (PSD3) are predominantly expressed in the CNS (Schraut et al., 2014), as well as in T cells, macrophages, and neutrophils (Okada et al., 2012). The Sec7 domain of PSD3 protein is a guanine nucleotide exchange factor for small GTPases that contributes to neurite formation, spine density, trafficking of synaptic vesicles, and receptor internalization (Okada et al., 2012). High levels of PSD3 activate Fas-induced apoptosis, while its depletion disrupts cell shape and polarity (Okada et al., 2012). In MS, a hypomethylation of PSD3 has been reported in both CD8+ T cells (Bos et al., 2015) and demyelinated hippocampi (Chomyk et al., 2017). This might suggest that immune homeostasis is unbalanced under these conditions (Okada et al., 2012).
De novo telomere addition of long interspersed nuclear elements (LINE-1) into genomic DNA causes genetic defects, alters the regulatory mechanisms and the structural properties of the genome at their insertion place and might lead to genomic remodeling (Furano, 2000). Changes in LINE-1 methylation status have been described in MS patients. Indeed, LINE-1 CpG sites were more frequently methylated in MS patients than in controls (Pinto-Medel et al., 2017; Dunaeva et al., 2018). A hypermethylated DNA pattern for LINE-1 in MS might be related to chromosomal abnormalities and DNA damage (Pinto-Medel et al., 2017).
The Central Nervous System
Observations of human MS plaques have revealed an increase of protein tyrosine phosphatases (PTPs) during ongoing remyelination (Hendriks et al., 2013). In NAWM derived from MS patients, the addition of DNAme to PTPRN2 (Huynh et al., 2014), a member of the PTP family, indicates that the CNS integrity remains unaltered. An upregulation of TGFBI is involved in CNS repair after brain injury through the inhibition of the inflammatory response, mediated by activated microglia. Therefore, TGFBI acts as a suppressor of microgliosis (Kim and Lee, 2011). Consequently, lower DNAme levels to TGFBI in the demyelinated hippocampus of MS patients might occur in order to counteract the deleterious effect associated with the activation of microglia (Kim and Lee, 2011). Kinesins (KIFs), a family of motor proteins mainly expressed in neurons, immune cells, and oligodendrocytes, are involved in the trafficking of vesicles, organelles, and proteins through the cytoskeleton (Hirokawa and Takemura, 2013). An overexpression of KIF improves axonal transport, while its downregulation leads to neurodegeneration and CNS atrophy (Lo Giudice et al., 2006). Dysregulation of certain KIFs has been postulated to aggravate MS disease (Hares et al., 2014). Lower methylated levels for KIF family member 25 (KIF25) have been measured in the hippocampi (Chomyk et al., 2017) and CD4+ T cells (Graves et al., 2014) of MS patients. Thus, it is plausible that lower DNAme levels could increase the production of KIF motor proteins to compensate the increased CNS molecule transport demand in MS patients. Cell adhesion molecules (CAMs) participate in axon guidance, synaptogenesis, and neuronal regeneration (Hansen and Walmod, 2013). Immunoglobulin superfamily member 9B (IGSF9B) is a member of the CAM family highly expressed in GABAergic interneurons, macrophages/microglia, and astrocytes (Gil-Varea et al., 2018). IGSF9B is also constitutively expressed in circulating monocytes (Tserel et al., 2014); however, its biological function in this cell type remains unclear and thus requires further investigation. Elevated expression of IGSF9B favors the maintenance of inhibitory synapses (Gil-Varea et al., 2018), while low levels result in a selective loss of GABAergic interneurons (Mishra et al., 2014). A prominent loss of GABAergic circuits has been described in MS (Gil-Varea et al., 2018). The findings reported by Chomyk et al. (2017) and Kulakova et al. (2016) suggest that a substantial number of DNAme to IGSF9B could be accompanied by lower transcription rates as well as a selective loss of GABAergic interneurons (Gil-Varea et al., 2018). In MS, HLA class I histocompatibility antigen, alpha chain F (HLA-F) cooperates with the antigen-processing peptide-loading complex, regulating the immune response via NK receptors (Alcina et al., 2012). An overexpression of HLA-F protects neurons from astrocyte-mediated neurotoxicity (Song et al., 2016). Thus, it can be hypothesized that a lower number of methyl groups to HLA-F might confer neuroprotection. In this regard, Huynh et al. (2014) found a hypomethylation of HLA-F promoter in NAWM collected from MS patients compared to their controls. It is noteworthy that RUNX3, a transcription factor importantly involved in DC and T and NK cell differentiation, is upregulated in active and chronic MS lesions (Sokratous et al., 2018). Similarly, Huynh et al. (2014) reported a hypomethylated DNA pattern for RUNX3 in NAWM collected from MS patients. This indicates that RUNX3 DNAme status could be used as an inflammatory biomarker even when inflammation is apparently not present in the CNS (Huynh et al., 2014). When retinoic acid (RA) interacts with retinoic acid receptor alpha (RARA), several transcriptional changes affecting the immune system and CNS repair can take place. Indeed, the inflammatory activity observed in MS patients is attenuated upon stimulation of RARA (Kim et al., 2017). In line with these findings, Huynh et al. (2014) reported a hypomethylated DNAme pattern for RARA in MS NAWM, which suggests no inflammatory activity (Huynh et al., 2014). Additionally, Marabita et al. (2017) found a smaller number of DNAme groups for RARA in tobacco smokers with MS compared to non-smokers diagnosed with MS, which is apparently related to tobacco consumption (Su et al., 2016). Cadherins are widespread calcium-dependent cell adhesion proteins involved in cell-cell adhesion, mobility, and proliferation of epithelial cells. In particular, cadherin 1 (CDH1) participates in many aspects of synapse formation and function (Almeida and Lyons, 2014). Recently, James et al. (2018) identified a large percentage of single nucleotide polymorphisms (SNPs) that may confer risk of MS. Among them, rs1886700 is statistically associated with lower expression of CDH1. Therefore, an increase of DNAme to CDH1 (Liggett et al., 2010; Huynh et al., 2014) probably leads to lower mRNA transcription rates and a worse disease course in MS (James et al., 2018).
Conclusions and Perspectives
Multiple sclerosis is a chronic inflammatory CNS disease originating from a complex interaction between genes and the environment. As far as we know, genetics accounts for almost 30% of MS prevalence worldwide, while the remaining percentage corresponds to epigenetic modifications due to exposure to distinct environmental factors (Weber et al., 2007). This review summarized the common differentially methylated genes expressed in distinct cell types and tissues derived from MS patients in order to elucidate their role in the pathophysiology of the disease. We conclude that changes in the methylation status of certain genes cause chromosomal abnormalities, DNA damage, and the generation of pathogenic immune cells resulting in inadequate innate and adaptive immune response. Furthermore, we assume that aberrant epigenetic profile in the context of disease potentiates microgliosis, alterations in synapse formation and function, and a selective loss of GABAergic interneurons which in turn favors neurodegeneration. A deeper understanding of the underlying physiological mechanisms mediated by DNAme will contribute to the development of new strategies in MS prognosis and therapy.
Author Contributions
NC and JT-R researched the literature and drafted the manuscript. JT-R critically reviewed and edited the work. Both authors read and approved the final manuscript.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This review was funded by the Deutsche Forschungsgemeinschaft to Dr. Jordi Tomas Roig (ref. TO 977/1-1) and the University of Girona to Mrs Naiara Celarain Sanz (ref. IFUdG2017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
MS, multiple sclerosis; CNS, central nervous system; APC, antigen presenting cells; Th, T helper cells; BBB, blood brain barrier; miRNA, microRNA; Me, methylation; Ub, ubiquitination; Ac, acetylation; P, phosphorylation; DNAme, DNA methylation; PBMCs, peripheral blood mononuclear cells; AHR, aryl hydrocarbon receptor; AHRR, aryl-hydrocarbon receptor repressor; MHC, major histocompatibility complex; MICA, MHC class I chain-related gene A; MICB, MHC class I chain-related gene B; NK, natural killer; RUNX3, runt related transcription factor 3; DC, dendritic cells; CDKN2A, cyclin dependent kinase inhibitor 2A; GNAS, guanine nucleotide binding protein alpha stimulating; IFN, type I interferon; USP35, ubiquitin specific peptidase 35; ATP11A, ATPase phospholipid transporting 11A; TLR4, toll-like receptor 4; pTh17, pathogenic Th17 cells; RASA3, RAS p21 protein activator 3; NAWM, normal appearing white matter; HOXC4, homeobox C4; PTPRN2, protein tyrosine phosphatase receptor type N2; SMAD3, SMAD family member 3; TGFBI, transforming growth factor beta induced; SOCS1, suppressor of cytokine signaling 1; ER, endoplasmic reticulum; PM, plasma membrane; TCRs, T-cell receptors; MORN1, MORN repeat containing 1; PSD3, pleckstrin and sec7 domain containing 3; LINE-1, long interspersed nuclear element; PTPs, protein tyrosine phosphatases; KIF25, kinesin family member 25; CAM, cell adhesion molecules; IGSF9B, immunoglobulin superfamily member 9B; HLA-F, HLA class I histocompatibility antigen alpha chain F; RA, retinoic acid; RARA, retinoic acid receptor alpha; CDH1, cadherin 1.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01138/full#supplementary-material
References
Abediankenari, S., Yousefzadeh, Y., Majidi, M., Ghasemi, M., Nasehi, M., Ghaffari, J., et al. (2011). MICB gene expression on peripheral blood mononuclear cells and susceptibility to multiple sclerosis in North of Iran. Iran J. Allergy Asthma Immunol. 10 (4), 261–265. doi: 010.04/ijaai.261265
Alcina, A., de Abad-Grau, M. M., Fedetz, M., Izquierdo, G., Lucas, M., Fernández, Ó, et al. (2012). Multiple sclerosis risk variant HLA-DRB1*1501 associates with high expression of DRB1 gene in different human populations. PloS One 7 (1), e29819. doi: 10.1371/journal.pone.0029819
Almeida, R. G., Lyons, D. A. (2014). On the resemblance of synapse formation and CNS myelination. Neuroscience 276, 98–108. doi: 10.1016/j.neuroscience.2013.08.062
Bos, S. D., Page, C. M., Andreassen, B. K., Elboudwarej, E., Gustavsen, M. W., Briggs, F., et al. (2015). Genome-wide DNA methylation profiles indicate CD8+ T cell hypermethylation in multiple sclerosis. PloS One 10 (3), e0117403. doi: 10.1371/journal.pone.0117403
Chomyk, A. M., Volsko, C., Tripathi, A., Deckard, S. A., Trapp, B. D., Fox, R. J., et al. (2017). DNA methylation in demyelinated multiple sclerosis hippocampus. Sci. Rep. 7, 8696. doi: 10.1038/s41598-017-08623-5
Dunaeva, M., Derksen, M., Pruijn, G. J. M. (2018). LINE-1 hypermethylation in serum cell-free DNA of relapsing remitting multiple sclerosis patients. Mol. Neurobiol. 55 (6), 4681–4688. doi: 10.1007/s12035-017-0679-z
Fernandez-Morera, J. L., Rodriguez-Rodero, S., Tunon, A., Martinez-Borra, J., Vidal-Castineira, J. R., Lopez-Vazquez, A., et al. (2008). Genetic influence of the nonclassical major histocompatibility complex class I molecule MICB in multiple sclerosis susceptibility. Tissue Antigens 72 (1), 54–59. doi: 10.1111/j.1399-0039.2008.01066.x
Furano, A. V. (2000). The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog. Nucleic Acid Res. Mol. Biol. [Internet] 64, 255–294.
Ghasemi, N., Razavi, S., Nikzad, E. (2017). Multiple sclerosis: pathogenesis, symptoms, diagnoses and cell-based therapy. Cell J. 19, 1–10. doi: 10.22074/cellj.2016.4867
Gil-Varea, E., Urcelay, E., Vilariño-Güell, C., Costa, C., Midaglia, L., Matesanz, F., et al. (2018). Exome sequencing study in patients with multiple sclerosis reveals variants associated with disease course. J. Neuroinflamm. 15(1), 265. doi: 10.1186/s12974-018-1307-1
Graca, L. (2007). The Immune Synapse as a Novel Target for Therapy. Berlin, Germany: Springer Science & Business Media;, 192.
Graves, M. C., Benton, M., Lea, R. A., Boyle, M., Tajouri, L., Macartney-Coxson, D., et al. (2014). Methylation differences at the HLA-DRB1 locus in CD4+ T-Cells are associated with multiple sclerosis. Multiple Sclerosis (Houndmills Basingstoke England) 20, 1033–1041. doi: 10.1177/1352458513516529
Hansen, M., Walmod, P. S. (2013). IGSF9 family proteins. Neurochem. Res. 38, 1236–1251. doi: 10.1007/s11064-013-0999-y
Hares, K., Kemp, K., Rice, C., Gray, E., Scolding, N., Wilkins, A. (2014). Reduced axonal motor protein expression in non-Lesional grey matter in multiple sclerosis. Mult. Scler. J. 20 (7), 812–821. doi: 10.1177/1352458513508836
Hendriks, W. J. A. J., Elson, A., Harroch, S., Pulido, R., Stoker, A., Den Hertog, J. (2013). Protein tyrosine phosphatases in health and disease. FEBS J. 280p, 708–730. doi: 10.1111/febs.12000
Hirokawa, N., Takemura, R., (2013). “Kinesin Superfamily Proteins,” in Encyclopedia of Biological Chemistry: (Waltham: Academic Press) Second Edition, 679–687.
Huynh, J. L., Garg, P., Thin, T. H., Yoo, S., Dutta, R., Trapp, B. D., et al. (2014). Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat. Neurosci. 17, 121–130. doi: 10.1038/nn.3588
James, T., Lindén, M., Morikawa, H., Fernandes, S. J., Ruhrmann, S., Huss, M., et al. (2018). Impact of genetic risk loci for multiple sclerosis on expression of proximal genes in patients. Hum. Mol. Genet. 27 (5), 912–928. doi: 10.1093/hmg/ddy001
Kim, M., Lee, E. H. (2011). Molecular mechanisms of microglial deactivation by TGF-β-inducible protein βig-h3. Integr. Biosci. 9 (2), 101–105. doi: 10.1080/17386357.2005.9647258
Kim, S. Y., Kelland, E. E., Kim, J. H., Lund, B. T., Chang, X., Wang, K., et al. (2017). The influence of retinoic acid on the human oligodendrocyte precursor cells by RNA-sequencing. Biochem. Biophys. Rep. 9, 166–172. doi: 10.1016/j.bbrep.2016.12.004
Kulakova, O. G., Kabilov, M. R., Danilova, L. V., Popova, E. V., Baturina, O. A., Tsareva, E. Y., et al. (2016). Whole-Genome DNA methylation analysis of peripheral blood mononuclear cells in multiple sclerosis patients with different disease courses. Acta Naturae 8, 103–110.
Lee, J., Kim, T. H., Murray, F., Li, X., Choi, S. S., Broide, D. H., et al. (2015). Cyclic AMP concentrations in dendritic cells induce and regulate Th2 immunity and allergic asthma. Proc. Natl. Acad. Sci. 112 (5), 1529–1534. doi: 10.1073/pnas.1417972112
Lee, S. A., Mefford, J. A., Huang, Y., Witte, J. S., Martin, J. N., Haas, D. W., et al. (2016). Host genetic predictors of the kynurenine pathway of tryptophan catabolism among treated HIV-infected Ugandans. AIDS 30 (11), 1807–1815. doi: 10.1097/QAD.0000000000001124
Liggett, T., Melnikov, A., Tilwalli, S., Yi, Q., Chen, H., Replogle, C., et al. (2010). Methylation patterns of cell-free plasma DNA in relapsing-remitting multiple sclerosis. J. Neuro. Sci. 290, 16–21. doi: 10.1016/j.jns.2009.12.018
Liu, Q., Wu, Y., Qin, Y., Hu, J., Xie, W., Xiao-Feng Qin, F., et al. (2018). Broad and diverse mechanisms used by deubiquitinase family members in regulating the type i interferon signaling pathway during antiviral responses. Sci. Adv. 4(5), eaar2824. doi: 10.1126/sciadv.aar2824
Lo Giudice, M., Neri, M., Falco, M., Sturnio, M., Calzolari, E., Di Benedetto, D., et al. (2006). A missense mutation in the coiled-coil domain of the KIF5A gene and late-onset hereditary spastic paraplegia. Arch. Neurol. 63 (2), 284–287.
Maltby, V. E., Graves, M. C., Lea, R. A., Benton, M. C., Sanders, K. A., Tajouri, L., et al. (2015). Genome-wide DNA methylation profiling of CD8+ T cells shows a distinct epigenetic signature to CD4+ T cells in multiple sclerosis patients. Clin. Epigenet. 9, 71. doi: 10.1186/s13148-017-0371-1
Marabita, F., Almgren, M., Sjöholm, L. K., Kular, L., Liu, Y., James, T., et al. (2017). Smoking induces DNA methylation changes in Multiple Sclerosis patients with exposure-response relationship. Sci. Rep. 7 (1), 14589. doi: 10.1038/s41598-017-14788-w
Marchetti, G., Ziliotto, N., Meneghetti, S., Baroni, M., Lunghi, B., Menegatti, E., et al. (2018). Changes in expression profiles of internal jugular vein wall and plasma protein levels in multiple sclerosis. Mol. Med. 24(1), 42. doi: 10.1186/s10020-018-0043-4
Mishra, A., Traut, M. H., Becker, L., Klopstock, T., Stein, V., Klein, R. (2014). genetic evidence for the adhesion protein IgSF9/Dasm1 to regulate inhibitory synapse development independent of its intracellular domain. J. Neurosci. 34 (12), 4187–4199. doi: 10.1523/JNEUROSCI.3671-13.2014
Neavin, D. R., Liu, D., Ray, B., Weinshilboum, R. M. (2018). The role of the aryl hydrocarbon receptor (AHR) in immune and inflammatory diseases. Int. J. Mol. Sci. 19(12). pii: E3851. doi: 10.3390/ijms19123851.
Okada, S., Suzuki, K., Takaharu, K., Noda, H., Kamiyama, H., Maeda, T., et al. (2012). Aberrant methylation of the Pleckstrin and Sec7 domain-containing gene is implicated in ulcerative colitis-associated carcinogenesis through its inhibitory effect on apoptosis. Int. J. Oncol. 40 (3), 686–694. doi: 10.3892/ijo.2011.1231
Park, S. R., Kim, P. H., Lee, K. S., Lee, S. H., Seo, G. Y., Yoo, Y. C., et al. (2013). APRIL stimulates NF-κB-mediated HoxC4 induction for AID expression in mouse B cells. Cytokine 61 (2), 608–613. doi: 10.1016/j.cyto.2012.10.018
Parnell, G. P., Gatt, P. N., Krupa, M., Nickles, D., McKay, F. C., Schibeci, S. D., et al. (2014). The autoimmune disease-associated transcription factors EOMES and TBX21 are dysregulated in multiple sclerosis and define a molecular subtype of disease. Clin. Immunol. 151 (1), 16–24. doi: 10.1016/j.clim.2014.01.003
Pinto-Medel, M. J., Oliver-Martos, B., Urbaneja-Romero, P., Hurtado-Guerrero, I., Ortega-Pinazo, J., Serrano-Castro, P., et al. (2017). Global methylation correlates with clinical status in multiple sclerosis patients in the first year of IFNbeta treatment. Sci. Rep. 7 (1), 8727. doi: 10.1038/s41598-017-09301-2
Schraut, K. G., Jakob, S. B., Weidner, M. T., Schmitt, A. G., Scholz, C. J., Strekalova, T., et al. (2014). Prenatal stress-induced programming of genome-wide promoter DNA methylation in 5-HTT-deficient mice. Transl. Psychiatry 4, e473. doi: 10.1038/tp.2014.107
Sokratous, M., Dardiotis, E., Bellou, E., Tsouris, Z., Michalopoulou, A., Dardioti, M., et al. (2018). CpG island methylation patterns in relapsing-remitting multiple sclerosis. J. Mol. Neurosci. J. Mol. Neurosci. 64, 478–484. doi: 10.1007/s12031-018-1046-x
Song, S. W., Miranda, C. J., Braun, L., Meyer, K., Frakes, A. E., Ferraiuolo, L., et al. (2016). Major histocompatibility complex class I molecules protect motor neurons from astrocyte-induced toxicity in amyotrophic lateral sclerosis. Nat. Med. 22 (4), 397–403. doi: 10.1038/nm.4052
Su, D., Wang, X., Campbell, M. R., Porter, D. K., Pittman, G. S., Bennett, B. D., et al. (2016). Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. PloS One 11 (12), e0166486. doi: 10.1371/journal.pone.0166486
Toghi, M., Taheri, M., Arsang-Jang, S., Ohadi, M., Mirfakhraie, R., Mazdeh, M., et al. (2017). SOCS gene family expression profile in the blood of multiple sclerosis patients. J. Neurol. Sci. 375, 481–485. doi: 10.1016/j.jns.2017.02.015
Tserel, L., Limbach, M., Saare, M., Kisand, K., Metspalu, A., Milani, L., et al. (2014). CpG sites associated with NRP1, NRXN2 and miR-29b-2 are hypomethylated in monocytes during ageing. Immun. Ageing 11 (1), 1. doi: 10.1186/1742-4933-11-1
van der Mark, V. A., Ghiboub, M., Marsman, C., Zhao, J., van Dijk, R., Hiralall, J. K., et al. (2017). Phospholipid flippases attenuate LPS-induced TLR4 signaling by mediating endocytic retrieval of Toll-like receptor 4. Cell Mol. Life Sci. 74 (4), 715–730. doi: 10.1007/s00018-016-2360-5
Weber, M., Hellmann, I., Stadler, M. B., Ramos, L., Pääbo, S., Rebhan, M., et al. (2007). Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 39, 457–466. doi: 10.1038/ng1990
Woo, J. S., Srikanth, S., Nishi, M., Ping, P., Takeshima, H., Gwack, Y. (2016). Junctophilin-4, a component of the endoplasmic reticulum–plasma membrane junctions, regulates Ca2+ dynamics in T cells. Proc. Natl. Acad. Sci. [Internet] 113 (10), 2762–2767. doi: 10.1073/pnas.1524229113
Keywords: multiple sclerosis, deoxyribonucleic acid methylation, immune cells, central nervous system, deoxyribonucleic acid damage
Citation: Celarain N and Tomas-Roig J (2019) Changes in Deoxyribonucleic Acid Methylation Contribute to the Pathophysiology of Multiple Sclerosis. Front. Genet. 10:1138. doi: 10.3389/fgene.2019.01138
Received: 03 June 2019; Accepted: 21 October 2019;
Published: 12 November 2019.
Edited by:
Trygve Tollefsbol, University of Alabama at Birmingham, United StatesReviewed by:
Richard Koche, Memorial Sloan Kettering Cancer Center, United StatesBruce V Taylor, University of Tasmania, Australia
Copyright © 2019 Celarain and Tomas-Roig. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naiara Celarain, bmNlbGFyYWluQGlkaWJnaS5vcmc=; Jordi Tomas-Roig, anRvbWFzQGlkaWJnaS5vcmc=
 Naiara Celarain*
Naiara Celarain* Jordi Tomas-Roig
Jordi Tomas-Roig