- College of Agronomy, Anhui Agricultural University, Key Laboratory of Wheat Biology and Genetic Improvement on Southern Yellow & Huai River Valley, Ministry of Agriculture and Rural Affairs, Hefei, China
Seed dormancy and germination are important agronomic traits in wheat (Triticum aestivum L.) because they determine pre-harvest sprouting (PHS) resistance and thus affect grain production. These processes are regulated by Gibberellic Acid-Stimulated Regulator (GASR) genes. In this study, we identified 37 GASR genes in common wheat, which were designated TaGASR1-37. Moreover, we identified 40 pairs of paralogous genes, of which only one had a Ka/Ks value greater than 1, indicating that most TaGASR genes have undergone negative selection. Chromosomal location and duplication analysis revealed 25 pairs of segmentally duplicated genes and seven pairs of tandemly duplicated genes, suggesting that large-scale duplication events may have contributed to the expansion of TaGASR gene family. Microarray analysis of the expression of 18 TaGASR genes indicated that these genes play diverse roles in different biological processes. Using wheat varieties with contrasting seed dormancy phenotypes, we investigated the expression patterns of TaGASR genes and the corresponding seed germination index phenotypes in response to water imbibition, exogenous ABA and GA treatment, and low- and high-temperature treatment. Based on these data, we identified the TaGASR34 gene as potentially associated with seed dormancy and germination. Further, we used a SNP mutation of the TaGASR34 promoter (-16) to develop the CAPS marker GS34-7B, which was then used to validate the association of TaGASR34 with seed dormancy and germination by evaluating two natural populations across environments. Notably, the frequency of the high-dormancy GS34-7Bb allele was significantly lower than that of the low-dormancy GS34-7Ba allele, implying that the favorable GS34-7Bb allele has not previously been used in wheat breeding. These results provide valuable information for further functional analysis of TaGASR genes and present a useful gene and marker combination for future improvement of PHS resistance in wheat.
Introduction
Common wheat (Triticum aestivum L.) is an important food crop grown throughout the world. One of the most important agronomic traits for wheat production is seed dormancy, which is defined as the prevention of germination of an intact viable seed under favorable conditions (Bewley, 1997). In modern varieties of domesticated wheat, low levels of dormancy (or lack of dormancy) have been selected to achieve higher yield by fast and uniform germination of seeds. However, this strategy has undesirable side effects as, under conditions of excess rainfall or humidity during harvest, low dormancy may promote germination of mature seeds while they remain within the spike of the mother plant (a phenomenon known as pre-harvest sprouting, PHS) (Clerkx et al., 2003; Finkelstein et al., 2008). It is estimated that global direct losses caused by PHS amount to one billion USD annually (Brown et al., 2018). Therefore, improving our understanding of the molecular mechanisms involved in seed dormancy and germination may be helpful for the improvement of PHS resistance in cultivated wheat.
Abscisic acid (ABA) and gibberellic acid (GA, also known as gibberellin) are two plant hormones that have decisive roles in regulating seed dormancy and germination. ABA is involved in the induction and maintenance of dormancy, whereas GA regulates the breaking of seed dormancy and thereby promotes germination (Kucera et al., 2005; Finkelstein et al., 2008). The roles of ABA and GA in dormancy and germination have been confirmed by physiological, biochemical, and genetic evidence in diverse plant species (Koornneef and van der Veen, 1980; Koornneef et al., 1984; Jacobsen and Olszewski, 1993; Finkelstein et al., 2002; Lee et al., 2002; Nambara and Marion-Poll, 2003; Kushiro et al., 2004; Appleford et al., 2007; Yamauchi et al., 2007; Shu et al., 2013; Ibrahim, 2016; Huang et al., 2016; Shu et al., 2017). For example, the tobacco ABA biosynthesis gene encoding 9-cis-epoxycarotenoid dioxygenase (LeNCED1) has been shown to enhance seed dormancy when overexpressed (Thompson et al., 2000). In Arabidopsis thaliana, three mutations in ABA-insensitive 1, -2, and -3 genes, known as abi1, abi2, and abi3, respectively, are associated with reduced seed dormancy (Koornneef et al., 1984). In addition, overexpression of the runner bean GA catabolism gene GA2-oxidase 1 (PcGA2ox1) has been shown to be associated with increased seed dormancy in transgenic wheat (Appleford et al., 2007). GA-deficient mutants (including ga1 and ga2) have been found to show strong seed dormancy, since seeds of these lines did not germinate without the addition of exogenous GA (Koornneef and van der Veen, 1980; Lee et al., 2002; Yamauchi et al., 2007; Shu et al., 2013). Finally, mutations in DELLA genes such as RGL2 (RGA-LIKE2) and SPY (SPINDLY), both of which are negative regulators of GA signaling, can rescue the ga1 non-germinating seed phenotype (Jacobsen and Olszewski, 1993; Lee et al., 2002). Taken together, these findings indicate that GA and ABA synthesis and signaling are necessary to control seed dormancy and germination. However, to date the detailed mechanisms responsible for these processes, especially in hexaploid wheat, remain poorly understood.
Temperature has been shown to be an important environmental factor influencing seed dormancy. Low temperatures during seed development enhance dormancy (Rodriguez et al., 2001; Chiang et al., 2011; Kendall et al., 2011; Nakamura et al., 2011; He et al., 2014), whereas dormancy of imbibed seeds can be lost after a short exposure to low temperature (Finch-Savage and Leubner-Metzger, 2006). By contrast, incubation at high temperatures can increase the level of dormancy by affecting GA synthesis and response pathways as well as responsiveness to ABA (Walker-Simmons, 1987; Corbineau et al., 1991; Yamauchi et al., 2004; Benech-Arnold et al., 2006; Leymarie et al., 2008). These results imply the presence of crosstalk between GA and ABA synthesis and response and temperature in controlling seed dormancy and germination.
Gibberellic Acid-Stimulated Regulator (GASR, also known as GASA and GAST) genes are a family of GA-responsive genes that play important roles in regulating seed germination. GASR proteins encoded by GASR genes are composed of a spliceable hydrophobic signal peptide at the N-terminal, a hydrophilic region of different lengths in the middle (usually consisting of polar amino acid residues), and a C-terminal containing 12 conserved cysteines (i.e. the GASA domain) (Herzog et al., 1995; Ben-Nissan et al., 2004; De la Fuente et al., 2006; Tomoyuki et al., 2006; Zimmermann et al., 2010; Ling et al., 2013). Bioinformatic analysis has identified a GA response element (GARE), an ABA response element (ABRE), and other GA- and ABA-related cis-elements in the GASA promoter (Zhang and Wang, 2008), indicating a relationship between GASA genes and these two plant hormones.
Since Shi et al. (1992) first identified a GAST gene (GAST1) in tomato (Solanum lycopersicon), numerous GAST1 homologues have subsequently been identified in other plant species, including AtGASA in Arabidopsis (Herzog et al., 1995; Aubert et al., 1998), OsGASR in rice (Oryza sativa) (Furukawa et al., 2006), StSN in potato (Solanum tuberosum) (Segura et al., 1999; Berrocal-Lobo et al., 2002), ZmGSL in maize (Zea mays) (Zimmermann et al., 2010), GEG in gerbera (Gerbera hybrida) (Kotilainen, 1999), FaGAST in strawberry (Fragaria vesca) (De la Fuente et al., 2006; Moyano-Cañete et al., 2013), MdGASA in apple (Malus domestica) (Fan et al., 2017), and GIP in Petunia (Petunia hybrida) (Ben-Nissan and Weiss, 1996; Ben-Nissan et al., 2004).
Members of the GASR family are involved in diverse plant growth, development, and biotic/abiotic stress response functions, including shoot and petal growth (Shi et al., 1992; Ben-Nissan and Weiss, 1996), stem growth (Ben-Nissan et al., 2004; Wigoda et al., 2006; Zhang et al., 2009), leaf expansion (Sun et al., 2013), root formation (Taylor and Scheuring, 1994; Zimmermann et al., 2010), flowering time regulation (Herzog et al., 1995; Zhang et al., 2009), seed growth and maturation (Roxrud et al., 2007; Dong et al., 2014; Zhang et al., 2016; Li et al., 2017), seed germination (Rubinovich and Weiss, 2010), fruit development and ripening (De la Fuente et al., 2006; Moyano-Cañete et al., 2013), fiber development (Liu et al., 2013), and heat tolerance (Ko et al., 2007; Zhang and Wang, 2011), as well as plant responses to saline (Alonso-Ramirez et al., 2009), oxidative (Wigoda et al., 2006; Alonso-Ramirez et al., 2009), wounding, and pathogen infection stresses (Segura et al., 1999; Berrocal-Lobo et al., 2002). In addition, Rubinovich and Weiss (2010) reported that seeds overexpressing GASA4 showed partial resistance to paclobutrazol (an inhibitor of GA biosynthesis) and a higher germination percentage than wild-type Arabidopsis seeds. The same study also reported higher rates of germination in seeds containing artificial miRGASA RNA to suppress GASA5, a repressor of the GA response. Similarly, Alonso-Ramirez et al. (2009) reported that overexpressing FsGASA4, a GASA-family gene found in Fagus sylvatica, increased the seed germination rates of transgenic Arabidopsis exposed to saline, oxidative, and heat stress. Moreover, Zhang and Wang (2008) reported that GASA4 expression was induced by GA3 and inhibited by ABA, whereas GASA5 expression showed the opposite trend. With respect to GASA6, Zhong et al. (2015) reported that AtGASA6-overexpressing seeds displayed early germination, whereas reduced AtGASA6 expression in transfer DNA (T-DNA) insertion and RNA interference (RNAi) knockout/knockdown mutants resulted in delayed seed germination in response to ABA, paclobutrazol, and glucose (Glc) stress treatments. These results suggest that AtGASA6 integrates GA, ABA, and Glc signaling in the regulation of seed germination. Taken together, GASA4, GASA5, and GASA6 likely play an important role in controlling dormancy and germination by modulating plant responses to GA and ABA. However, the roles that these GASA homologs play in common wheat remain unclear.
The objectives of this study were to identify GASR genes in wheat (TaGASR genes) and perform bioinformatic analyses, including the generation of a phylogenetic tree and the examination of gene structure, conserved domains, chromosomal location, expression patterns, duplication events, and promoter sequences; clone TaGASR genes associated with seed dormancy and germination and introduce these TaGASR genes into wheat varieties with contrasting seed dormancy phenotypes; and validate the association of TaGASR genes with seed dormancy and germination in different natural populations.
Materials and Methods
Plant Materials and Field Trials
Six wheat varieties with contrasting dormancy levels were selected to examine the expression of TaGASR genes. These included: Zhongmai 895 (ZM895, average germination index (GI): 0.92), Jing 411 (J411, average GI: 0.96), Zhongyou 9507 (ZY9507, average GI: 0.97), Yangxiaomai (YXM, average GI: 0.07), Suiningtuotuo (SNTT, Caverage GI: 0.15), Hongmangchun 21 (HMC21, average GI: 0.09). Varieties J411 and HMC21, which have especially low and especially high levels of seed dormancy, respectively, were used for cloning the TaGASR34 gene.
We validated the association of TaGASR34 with seed dormancy and germination using the Chinese wheat mini-core collection (CMCC), a small core collection consisting of 260 Chinese wheat varieties (Table S1), as well as a natural population (NP) consisting of 260 Chinese wheat varieties (including 179 from the Yellow and Huai Valleys Winter Wheat region, 30 from the Southwest Winter Wheat region, 22 from the Middle and Lower Yangtze River Valley Winter Wheat region, 21 from the Northern Winter Wheat region, and 8 from outside China) (Table S2). The CMCC and NP were planted at the Dayangdian experimental station of Anhui Agricultural University in Hefei, China (31° 58′ N, 117° 240′ E). CMCC plants were grown during the 2014–2015 and 2015–2016 growing seasons, and NP plants were grown during the 2013–2014 and 2014–2015 growing seasons. Field trials were conducted in plots containing two 2 m rows 25 cm apart. Forty seeds were planted in each row. All experiments were performed in randomized complete blocks with two independent replicates. Field management followed local agricultural practices.
Flowering time was scored when 50% of florets were open in a plot. Sixty spikes of each plot were collected at physiological maturity (i.e. after loss of chlorophyll from the spike, leaf and peduncle) (Trethowan, 1995), naturally air dried for 3 days avoiding direct sunlight and high temperature, hand-threshed to minimize damage to embryos and seed coat, then stored at -20°C until all were harvested. After all varieties were threshed, they were used for subsequent seed germination index (GI) assay.
Germination Index Assays
Fifty seeds from each genotype were placed in Φ 90 Petri dishes on filter paper with 9 ml distilled water, and then grown in a 20°C greenhouse with a 14 h day/10 h night photoperiod cycle at 80% humidity. The number of germinated seeds in each culture dish was counted at the same time every day and removed. The GI values were calculated after 7 days. Germination was defined as visible rupture of the pericarp and testa (Mares, 1983; Chang et al., 2010).
All GI tests were conducted twice at 5 and 15 days after harvest. For CMCC plants, GI measurements made 5 and 15 days after harvest in 2014, 2015, and 2016 were designated 14GI5-CMCC, 14GI15-CMCC, 15GI5-CMCC, 15GI15-CMCC, 16GI5-CMCC, and 16GI15-CMCC, respectively. For NP plants, GI measurements made 5 and 15 after harvest in 2013, 2014, and 2015 were designated 13GI5-NP, 13GI15-NP, 14GI5-NP, 14GI15-NP, 15GI5-NP, and 15GI15-NP, respectively.
Identification of TaGASR Genes in Common Wheat
We obtained the full sequence of the wheat genome from the Ensembl database (http://plants.ensembl.org/index.html). All candidate TaGASR gene sequences were obtained by BLAST search using a hidden Markov model (HMM) of the Pfam database. Sequences of candidate genes were confirmed by querying the Pfam, SMART, and NCBI databases (Chen et al., 2015). Bioinformatics analysis of the TaGASR genes were performed, including the determination of ORFs and the calculation of pI values, Mw values, and nucleic acid lengths of all genes, using the ExPASy website (www.expasy.org).
Phylogenetic Tree, Multiple Alignment and Gene Structure Analysis
Phylogenetic trees were constructed using the NJ method as implemented by MEGA version 7 with the number of bootstraps set to 1,000 (Chu et al., 2016; Cheng et al., 2018). In addition, the CDS and gene sequences of TaGASR genes were analyzed using Gene Structure Display Server (GSDS) version 2.0 (gsds.cbi.pku.edu.cn) to determine the structure of their exons/introns (Wang et al., 2017; Cheng et al., 2018). Multiple sequence alignments of the 37 TaGASR full-length protein sequences were performed using ClustalX 2.11 (Gao et al., 2017; Liu et al., 2017).
Conserved Domain and Promoter Analysis
We used MEME Suite version 5.0.5 to identify conservative motifs, and performed all searches using the default parameter settings (Wu et al., 2016). We also used the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) to analyze the regions 1,500 bp up- and downstream of TaGASR family genes in order to identify the type and number of cis-acting elements in the promoters of these genes (Zhao et al., 2018).
Microarray Analysis
We obtained microarray data for three biological replicates of 13 different tissue samples from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) database of the National Center for Biotechnology Information (NCBI) (Barrett and Edgar, 2006) using the login number GSE12508. The online probe matching tool provided by the NetAffx Analysis Center (Wilkins et al., 2009) (https://www.affymetrix.com/analysis/index.affx) was used to identify the probes corresponding to the putative TaGASR genes. When the gene had more than one probe group, the probe with the highest matching value was used. All data was normalized, logarithmized, averaged, and saved as tab-delimited files before importing into Cluster (version 3.0) (Sturn et al., 2002) to generate heat maps. Finally, heat maps were obtained using Heat-mapper Plus (www.heatmapper.ca) (Kiana et al., 2005).
Identifying Homologous Pairs and Calculating Ka/Ks Values
Paralogous pairs (gene pairs originating from duplication events within genome of a single species) and orthologous pairs (gene pairs in different genomes that have diverged by speciation) were identified according to the method described in Altschul et al. (1997). We identified paralogous pairs as aligned sequences longer than 300 bp with identity ≥ 40%, and identified orthologous pairs as aligned sequences longer than 300 bp (Blanc and Wolfe, 2004).
Ka and Ks were calculated according to the method described in Wang et al. (2015). Sequence alignment was performed using MEGA 7.0, and Ka/Ks values were calculated using DnaSP version 5 (Wang et al., 2017).
Chromosomal Location and Duplication Analysis
The physical locations of TaGASR genes were obtained from the Ensembl database and constructed chromosomal maps using MapGene2Chromosome version 2.0 (http://mg2c.iask.in/mg2c_v2.0/) (Voorrips, 2002). To classify the expansion of TaGASR genes, putative tandem duplications of gene family members were examined in the same gene region and in adjacent gene regions (Cannon et al., 2004). All GASR genes were analyzed and compared using pairwise BLASTP with E-values < 10-10. The coordinates of segmental duplications of target genes were searched by querying the Vista Synteny browser (pipeline.lbl.gov/cgibin/gateway2). If genes of interest were located in duplicated chromosomal blocks, these paralogs were deemed to be generated by segmental duplication. Two genes found within a 100-kb region that were separated by five or fewer genes were deemed to be tandemly duplicated. Using the Smith-Waterman algorithm (http://www.ebi.ac.uk/Tools/psa/) we calculated the local alignment between the two protein sequences of duplicated genes. Finally, we generated synchronized maps using Circos version 0.69 (Jorge et al., 2000); putative duplicated genes are connected by colored lines.
ABA, GA, Low and High Temperature Treatments
Seeds of two wheat varieties (J411 and HMC21) were treated with 50 μM GA3, 50 μM ABA, low temperature (4°C), or high temperature (36°C) treatments. Distilled water was used as a control. Seed samples were collected at 48 h after the start of the treatment. Collected seeds were immediately frozen in liquid nitrogen and stored at -80°C for RNA isolation.
Determination and Analysis of Endogenous Hormones ABA and GA
Collected seeds treated with GA3, ABA, low temperature, or high temperature treatments were immediately frozen in liquid nitrogen, ground into a powder, and 0.1 g of the sample was mixed with a methanol–water (80:20 V/V) solution. The standard compounds within the mixture were separated by electrospray ionization liquid chromatography tandem mass spectrometry (LC-ESI-MS/MS), as described by Yoshimoto et al. (2009). Hormones were extracted from at least three independent samples harvested.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from seeds by the Trizol method. cDNA was synthesized using a Primer Script RT Master Mix (Takara, Tokyo, Japan) according to the manufacturer’s instructions. Specific primers for 37 TaGASR genes were designed using Primer Premier 5.0 (Table S3), and TaActin was used as a reference gene (Sun et al., 2015).
The total volume of PCR reactions used for qRT-PCR analysis was 20 μl. Each reaction included 10 μl TransStart Tip Green qPCR SuperMix, 0.4 μl Passive Reference Dye, 0.4 μl each of forward and reverse primers, and 8.8 μl ddH2O. The reaction procedure was as follows: an initial denaturation at 94°C for 30 s, followed by 40–45 cycles of 94°C for 5 s and 50–60°C for 15 s and a final extension step at 72°C for 10 s. We performed three biological replicates for each sample. Finally, we used GraphPad version 5 to process data and generate charts (Bryfczynski, 2009).
DNA Extraction and Cloning of TaGASR34
Genomic DNA was isolated from undamaged dry kernels of the J411 and HMC21 varieties using a modified phenol–chloroform method (Hu et al., 2016; Jiang et al., 2018). The full-length sequence of the TaGASR34 gene was obtained by querying the Chinese spring wheat genome. Gene-specific primers were designed to selectively amplify the GASR34 gene (Table S4). We then isolated the GASR gene sequence from both the J411 and HMC21 varieties. These amplicons were then cloned and sequenced (Table S5).
The total reaction volume used for the cloning PCR was 20 μl, including 4.0 μl TransStart® FastPfu buffer, 1.6 μl 2.5 mmol/L dNTPs, 0.4 μl 2.5 U/μl TransStart® FastPfu DNA polymerase, 0.4 μl each of 10 μmol/L forward and reverse primer, 2.0 μl of (50–60 ng/μl) template DNA, and 10.4 μl ddH2O. The cloning PCR reaction procedure was as follows: an initial denaturation at 94°C for 5 min, 37 cycles of 95°C for 30 s and 60°C for 30 s, and a final elongation at 72°C for 2 min. PCR products were then separated in 1.5% agarose gels, and the target fragment was recovered from the gel matrix. The recovered product was introduced into Trans1-T1 competent cells, gently mixed, and cultured 8 h. Liquid samples containing positive clones were identified by sequencing (Sangon Biotech, Shanghai, China).
DNAMAN version 7.0 was used to compare sequencing results to identify different allelic variations. Known sequence information from the TaGASR34 CDS was used to analyze gene structure (e.g. promoter, exon, intron, and 3’UTR regions) and SNP variation (Table S5).
Development of Gene-Specific Markers for TaGASR34
One gene-specific primer pair (designated GS34-7B) was designed based on a SNP mutation of the TaGASR34 promoter (-16) using Primer Premier version 5.0 (Table S4). The resulting amplification product was digested by BsaI at 37°C to introduce one SNP mutation (C/G) in the TaGASR34 promoter. We amplified the GS34-7B marker using the cloning PCR reaction conditions described above. The resulting PCR product was digested with BsaI for 6 h. Digested fragments were separated on 2% agarose gels.
Validation of Gene-Specific Markers for TaGASR34
The gene-specific marker GS34-7B for TaGASR34 was validated in the CMCC (Table S1) and NP groups of wheat varieties (Table S2). Descriptive statistics and Mann-Whitney U-tests were performed to analyze significant differences in GI values between varieties with the two alleles of GS34-7B. Our genotyping results found that the GS34-7B marker identified two alleles, including the allele GS34-7Ba, which was associated with higher GI values, and GS34-7Bb, which was associated with lower GI values.
Statistical Analysis
Excel and SPSS version 18.0 were used for data analysis. We calculated mean values and standard deviation (SD) from three technical replicates each of three biological replicates. Student t-tests were used to determine whether there were significant differences between the mean values of treatment and control plants. The significance threshold used was *P < 0.05.
Results
Identification and Evolutionary Analysis of TaGASR Genes
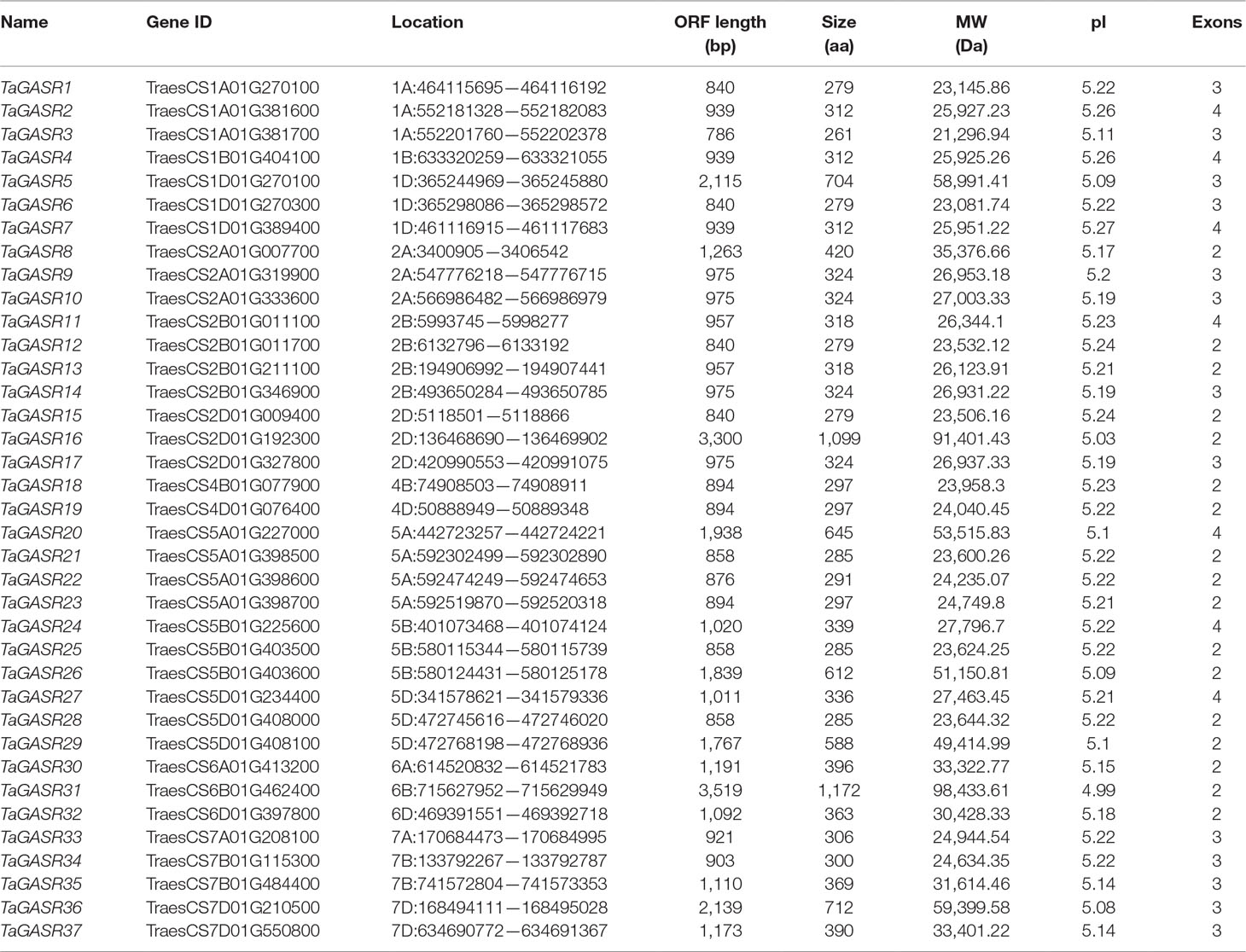
We identified 37 GASR genes in common wheat based on a typical GASR motif (PF02704, one HMM model). These were designated TaGASR1-37 according to the name of the species and their chromosomal location (Table 1). The amino acid (aa) lengths of all 37 TaGASR genes ranged from 261 to 1,172 aa. The longest gene was TaGASR31, the shortest was TaGASR3, and the lengths of their open reading frames (ORFs) ranged from 786–3,519 bp. The predicted protein molecular weights (MW) of TaGASR proteins ranged from 21,296.94 to 98,433.61 Da, and their theoretical isoelectric points (pI) varied between 4.99 and 5.27.
Next, we constructed a phylogenetic tree of all GASR family genes. Based on the classification of GASR genes in rice and Arabidopsis (Table S6), members of the GASR gene family in the phylogenetic tree were divided into three subfamilies (G1, G2, and G3) (Figure 1). Of these, subfamily G3 contained the most members (20), while subfamily G1 had the fewest members (6).
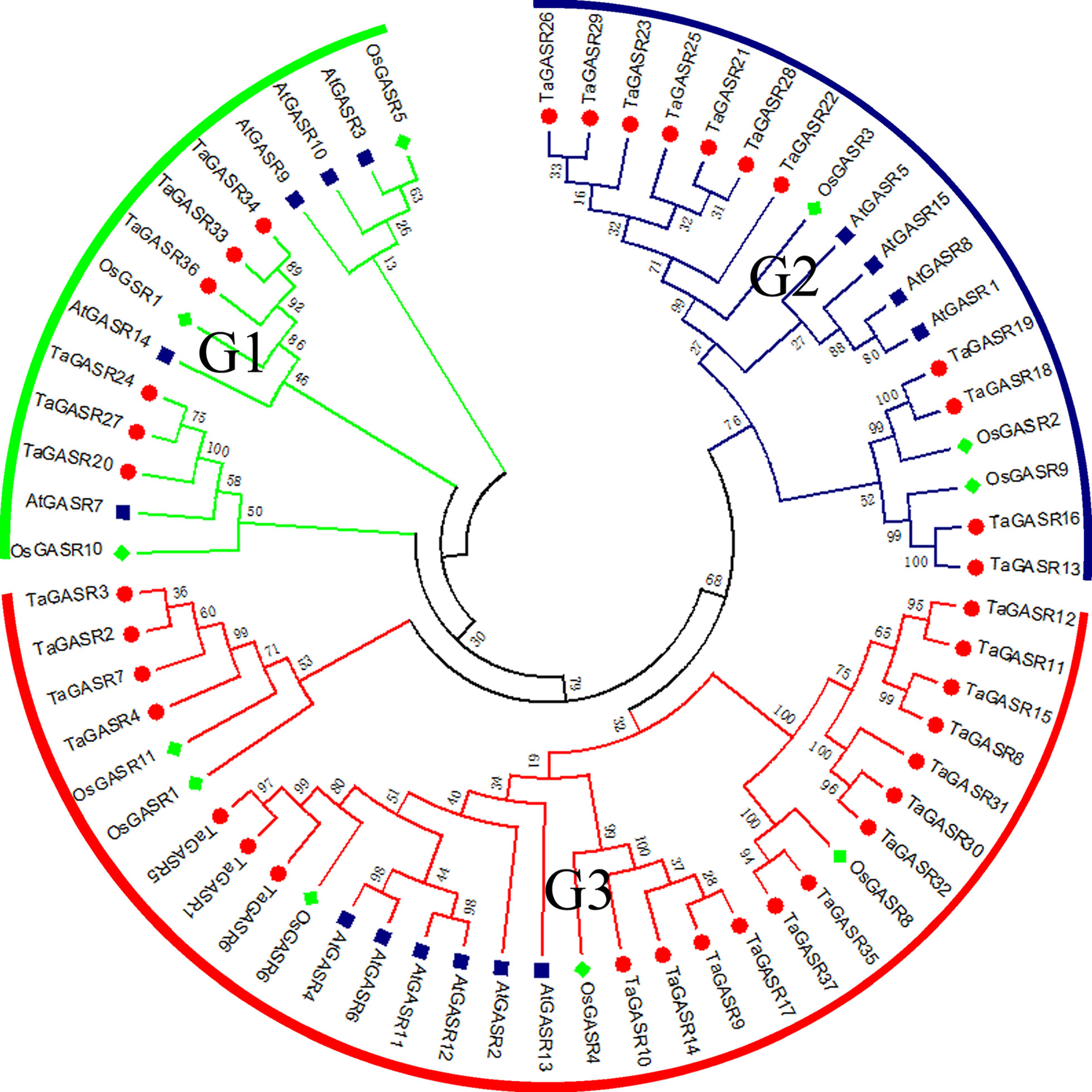
Figure 1 Phylogeny of GASRs from wheat, rice and Arabidopsis. The 37 TaGASR genes, 11 OsGASR genes, and 15 AtGASR genes are clustered into three subfamilies. Details of GASR genes from Arabidopsis and rice are listed in Table S6. The tree was generated using ClustalX version 2.11 using the neighbor-joining (NJ) method.
Most TaGASR genes had 2–4 exons (Figure 2A). In addition, 37 (92.5%) of the 40 paralogous pairs had the same number of exons and similar gene structures (Figure 2A). Twenty motifs were detected in the 37 TaGASR gene family members using MEME. Among these, motif 2 (a variable region) and motif 5 (a GASR domain) were identified in all TaGASR genes, and motif 3 (a putative signal peptide) was found in 36 TaGASR genes (all except TaGASR8; Figure 2B). In addition, multiple alignment analysis of GASR protein sequences of rice, Arabidopsis thaliana, and wheat showed that all putative TaGASR proteins had a conserved GASA domain (Figure 2C).
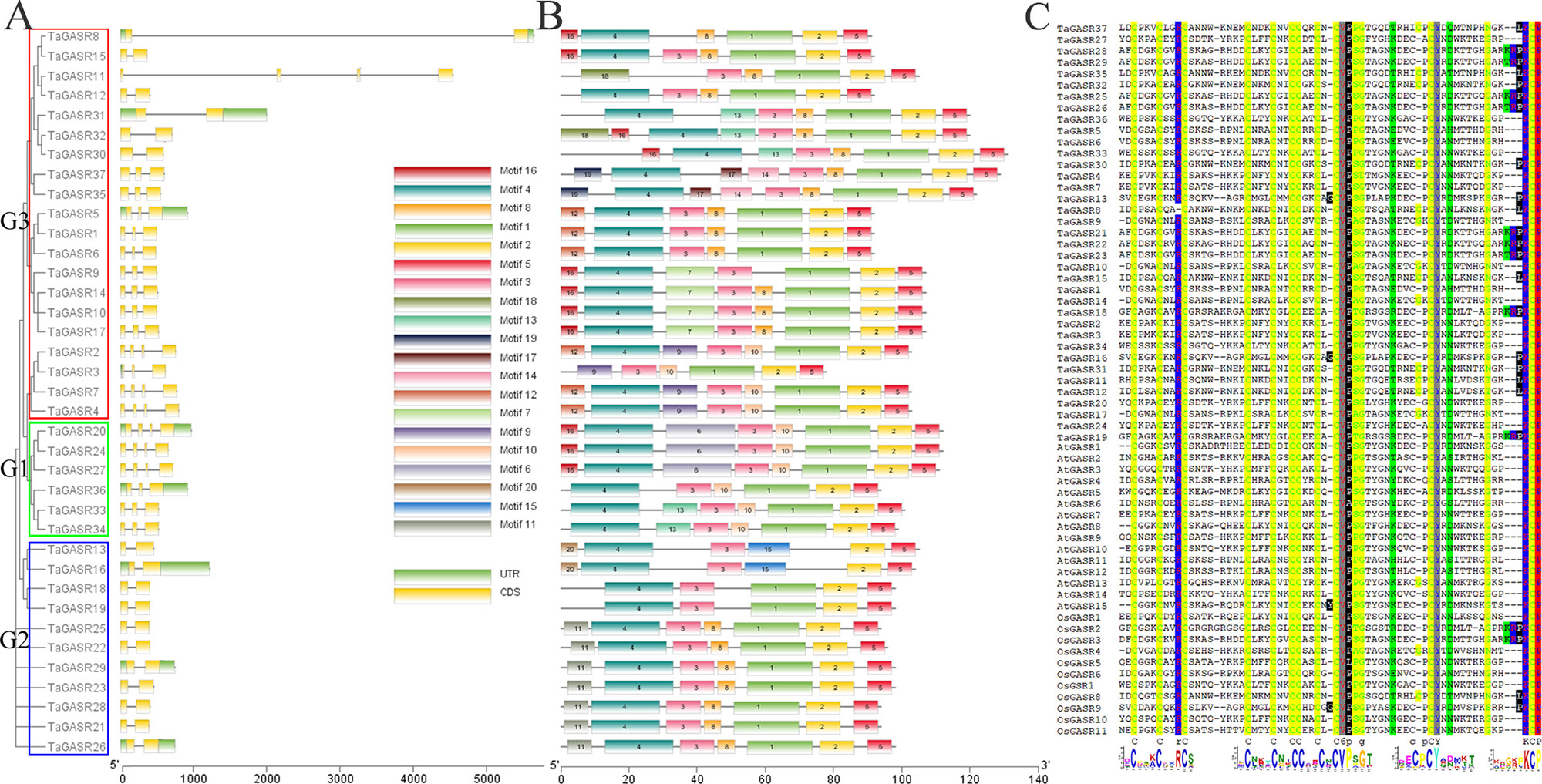
Figure 2 Evolutionary and gene structure analysis of TaGASR genes. (A) Phylogenetic relationships and gene structures of TaGASR genes. Exons, introns and untranslated regions (UTRs) are indicated by yellow rectangles, gray lines, and green rectangles, respectively. Colored boxes indicate the subfamily based on the phylogenetic analysis. (B) Schematic representation of 20 conserved motifs in TaGASR genes. Conserved motifs in TaGASR genes were identified using MEME. Different colored boxes represent different motifs. Box lengths in the figure do not represent actual relative motif sizes. (C) Multiple sequence alignment of TaGASR proteins. Sequences were aligned using DNAMAN software. The GASA motif is clearly highly conserved.
Promoter and Microarray Analysis of the TaGASR Gene Family
Thirty-six TaGASR genes (i.e. all putative genes except TaGASR13) were found to contain two components. One is a cis-acting regulatory element that responsive to biological stress. These elements, include CGTCA- (present in 23.41% of all putative TaGASR genes) and/or TGACG-motifs (23.41%) involved in plant methyl jasmonic acid (MeJA) response, TCA-elements (9.57%) involved in salicylic acid (SA) response, ABREs (29.79%) involved in ABA response, GARE-motifs (2.48%), and P-boxes (1.77%) involved in GA response, and AuxRR-cores (1.06%) and TGA-elements (8.51%) involved in auxin (IAA) response (Figure 3 and Table S7). The second element was a cis-acting regulatory element involved in abiotic stress responses, including low temperature stress response (LTR), drought inducibility (MBS), and general defense and stress response cis-acting elements (TC-rich repeats) (Figure 3 and Table S7).
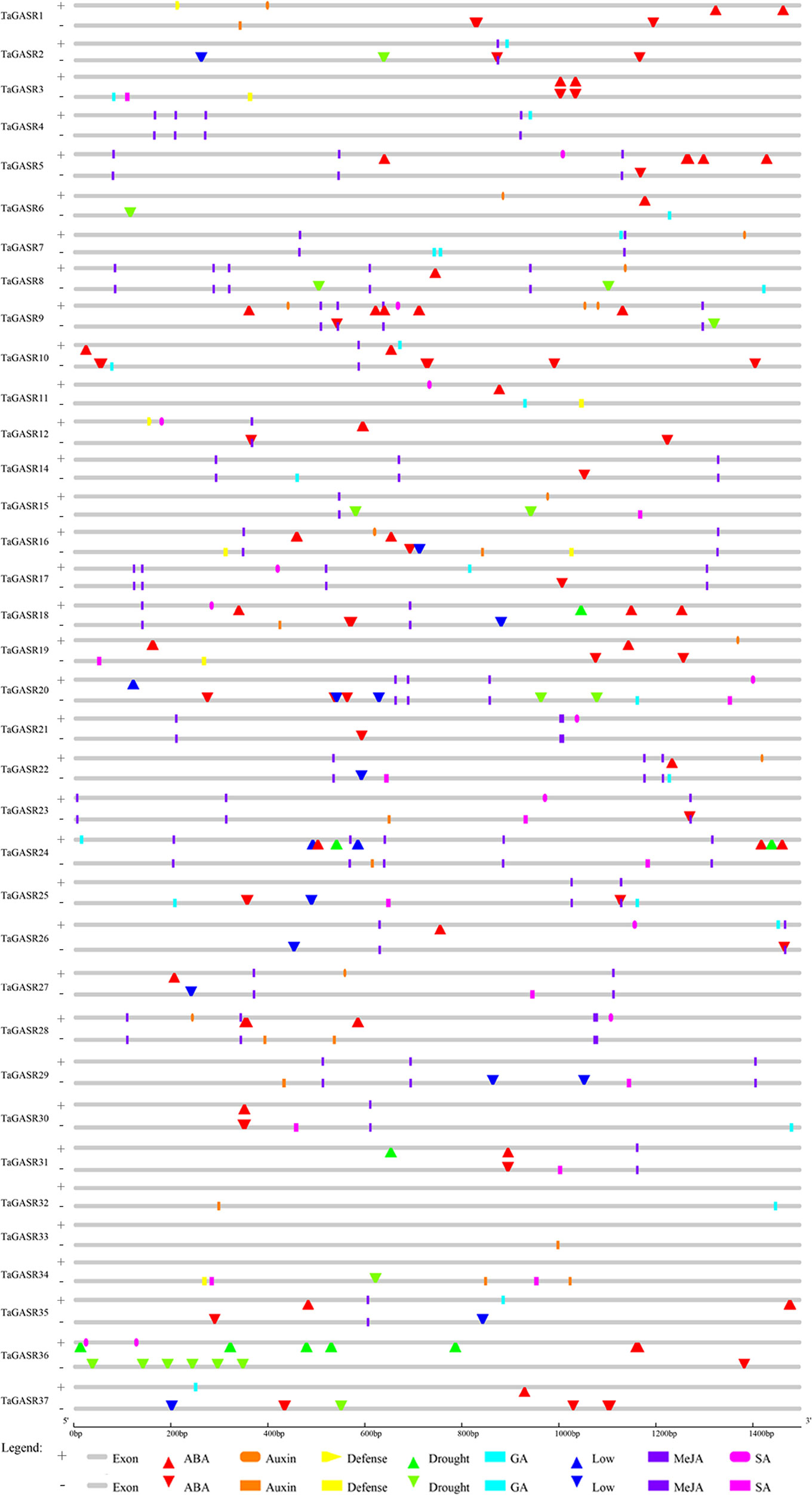
Figure 3 Cis-acting element analysis of the promoter regions of TaGASR genes. Based on functional annotation data, cis-acting elements were classified into two major classes: phytohormone responsive elements (i.e. those responsive to ABA, auxin, GA, MeJA, and/or SA) and abiotic stress response cis-acting elements (e.g. those involved in plant defense, drought stress response, and/or low temperature stress response).
Microarray expression data was obtained for 18 of the TaGASR genes from the NCBI database (accession number GSE12508). Most TaGASR genes showed tissue-specific expression patterns. In particular, TaGASR1, TaGASR5, and TaGASR6 were highly expressed in anthers, both before anthesis (Aba) and 22 days post-anthesis (22.DAP.EM). Moreover, we obtained microarray data showing the relative expression levels of 27 of the 40 paralogous pairs in the TaGASR gene family. Of these, 22 showed similar expression patterns, and five (i.e. TaGASR1/-5, TaGASR5/-6, TaGASR21/-22, TaGASR21/-25, and TaGASR21/-28) showed differential expression patterns (Figure 4 and Table S8).
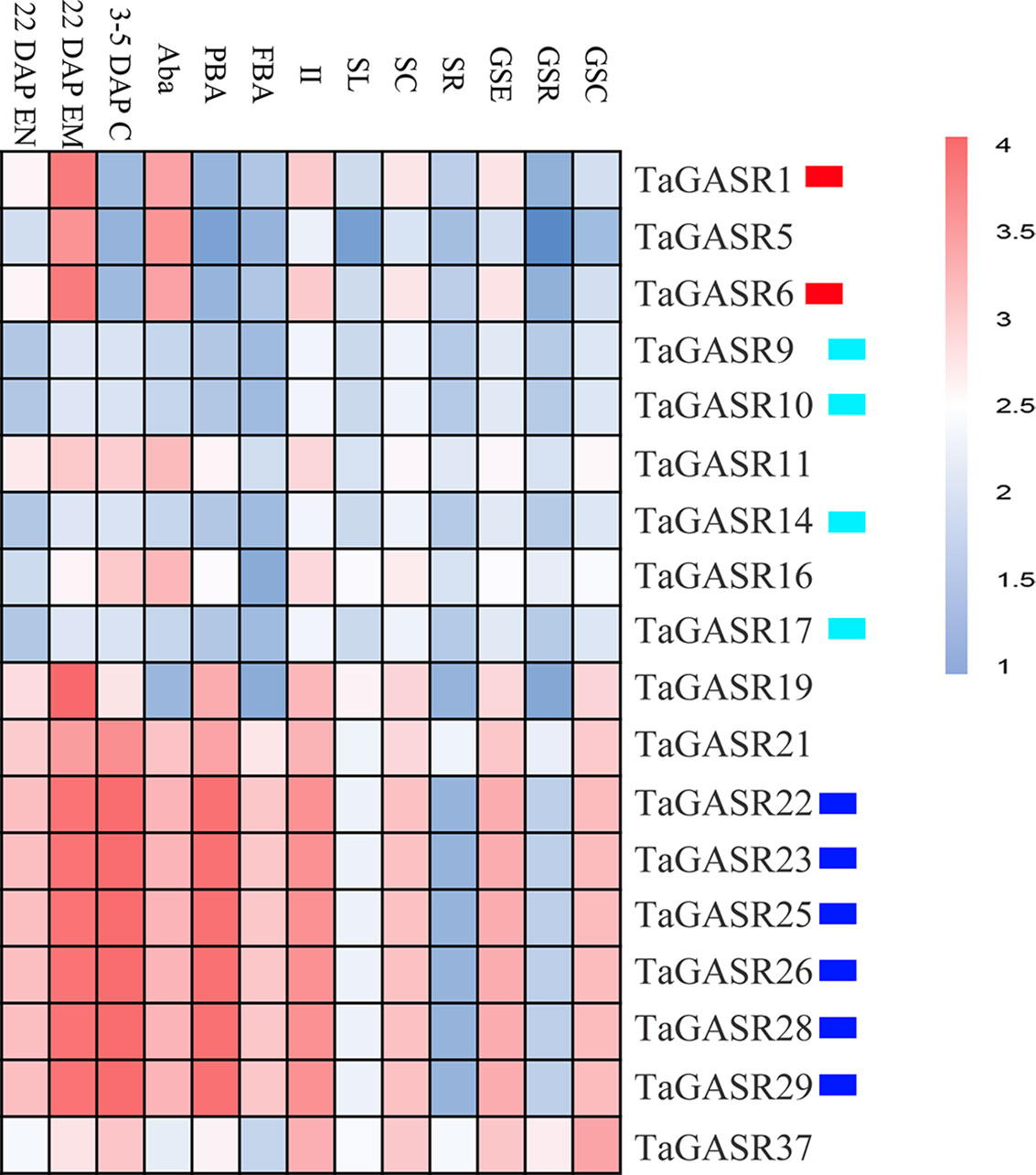
Figure 4 Expression profiles of TaGASR genes in different tissues and at different developmental stages. Heatmap shows hierarchical clustering of the 18 TaGASR genes among different tissues. Abbreviations represent specific developmental stages: GSC, germinating seed, coleoptile; GSR, germinating seed, root; GSE, germinating seed, embryo; SR, seedling, root; SC, seedling, crown; SL, seedling, leaf; II, immature inflorescence; FBA, floral bracts, before anthesis; PBA, pistil, before anthesis; Aba, anthers, before anthesis; 3–5 DAP C, 3–5 DAP caryopsis; 22 DAP EM , 22 DAP embryo; 22 DAP EN, 22 DAP endosperm.
Chromosomal Location and Duplication Analysis of the TaGASR Gene Family
Thirty-seven TaGASR genes were distributed on wheat chromosome groups 1–7, except none were found on groups 3 and 4A (Figure 5). More than three genes each were found on chromosomes 1A, 1D, 2A, 2B, 2D, 5A, 5B, and 5D, and four were present on chromosomes 2B and 5A. Other chromosomes contained fewer than three TaGASR genes. According to Sturn et al. (2002), chromosomal regions smaller than 200 Kb containing two or more genes can be defined as a single gene cluster. In this study, we identified six gene clusters containing a total of thirteen genes of the TaGASR gene family. These were evenly distributed on chromosomes 1D, 2A, 2B, 5A, 5B, and 5D (Figure 5).
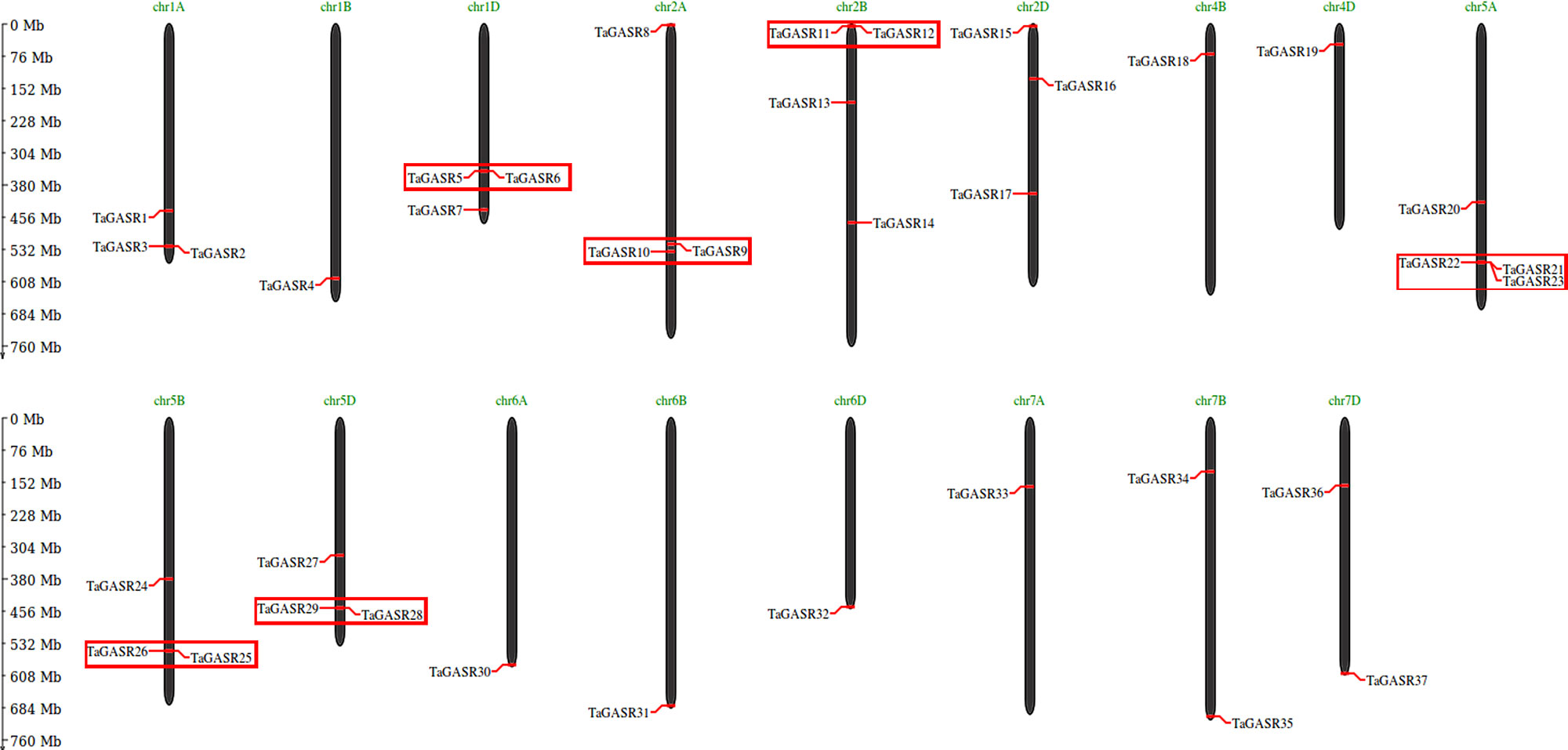
Figure 5 Chromosomal localization and gene duplication events of TaGASR genes. Respective chromosome numbers are indicated above each bar. Duplicated paralogous pairs of GASR genes in tandem duplication blocks are indicated by small boxes of the same color.
In addition, we identified 25 TaGASR genes unevenly distributed on 21 wheat linkage groups (LGs), although no genes were found on LGs 3A, 3B, 3D, and 4A. The most TaGASR genes were found in LGs 2B and 2D (3), and some LGs have only one gene (e.g. LG 1A). We also found no significant positive correlation between LG length and the number of TaGASR genes (Figure 6). Furthermore, we detected 25 pairs of segmentally duplicated genes and seven pairs of tandemly duplicated genes in the 37 genes of the TaGASR gene family. These were found to be unevenly distributed on chromosomes 1D, 2A, 2B, 5A, 5B, and 5D (Table 5).
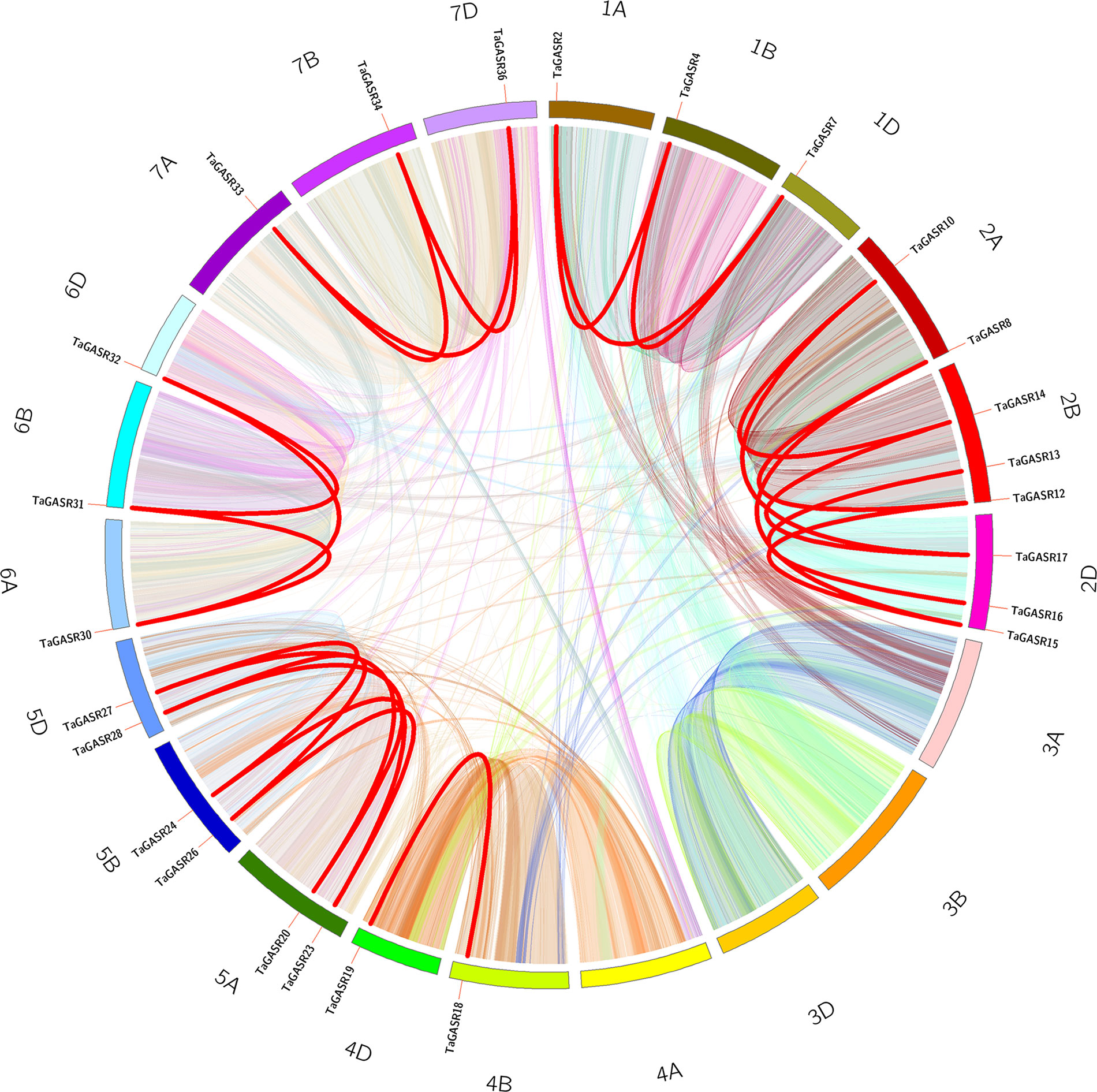
Figure 6 Microsynteny related to TaGASR family in wheat. Wheat chromosomes are shown in different colors. Each chromosome box indicates sequence length in megabases. Different color lines represent syntenic relationships between TaGASR regions, whereas thick red lines represent paralogous TaGASR genes.
Expression of TaGASR Genes During Seed Imbibition
The expression patterns of the 37 TaGASR genes were investigated at 0 h and 10 h after seed imbibition in six wheat varieties with contrasting seed dormancy phenotypes. After 10 h of imbibition, seeds from three varieties (HMC21, YXM, and SNTT) with high levels of seed dormancy showed no seed germination, whereas seeds from three different varieties (J411, ZY9507, and ZM895) with low levels of seed dormancy showed obvious germination (average GI: 0.97, 0.91, and 0.93, respectively; Table S9). Relative to that unimbibed seeds, most of the 37 TaGASR genes were up-regulated in response to imbibition, whereas a few were down-regulated or showed no significant differences in gene expression (e.g. TaGASR21). For each TaGASR gene, we also found obvious differences in relative transcript levels among the six compared wheat varieties. In particular, five specific TaGASR genes (TaGASR15/-24/-25/-34/-35) were more highly transcribed in the three varieties with low levels of seed dormancy than in the three varieties with high levels of seed dormancy. In contrast, we found the opposite trend among the transcription levels of seven TaGASR genes (e.g. TaGASR10/-14/-20/-27/-29/-30/-33) (Figure 7).
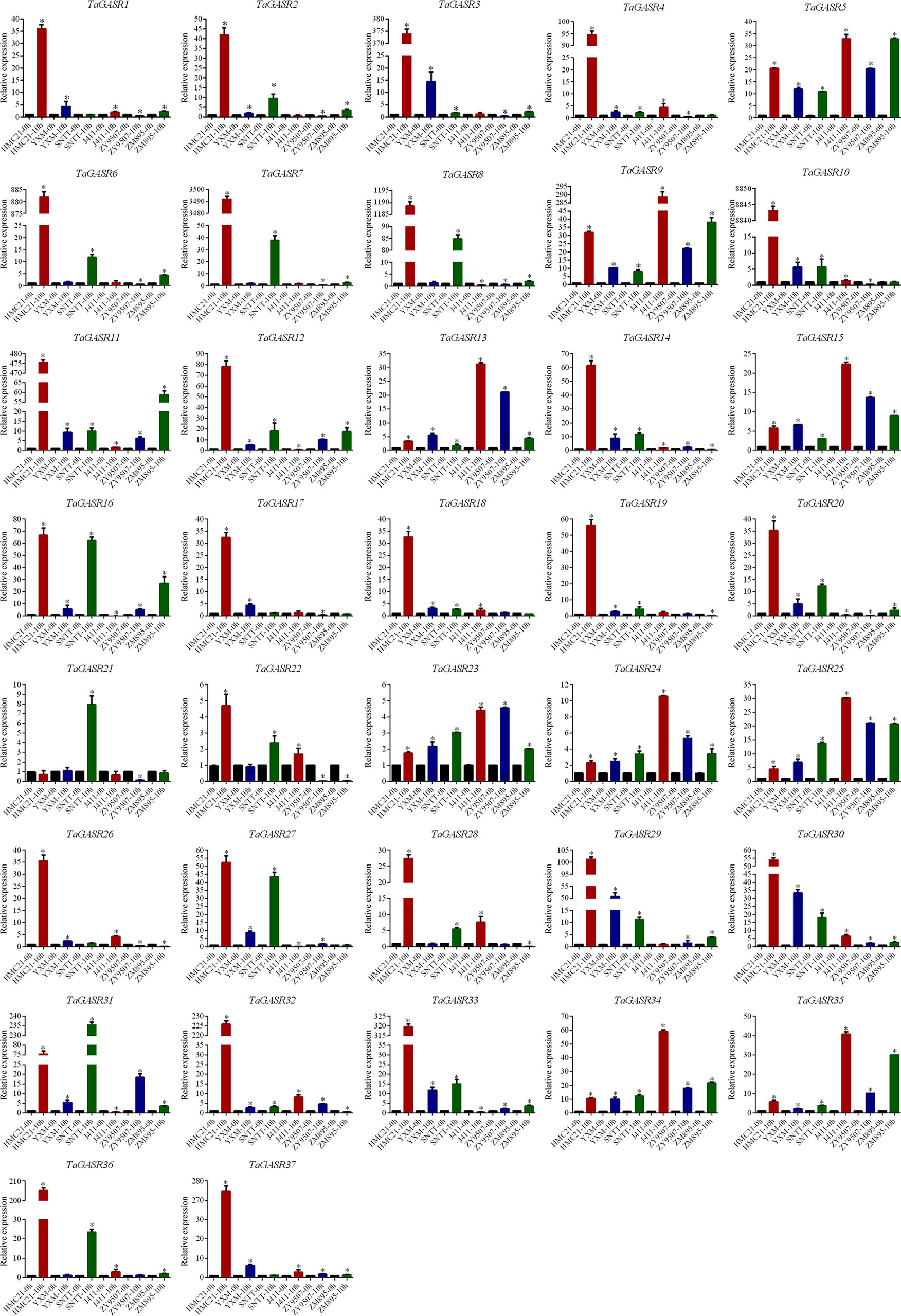
Figure 7 Expression patterns of 37 TaGASR genes during seed imbibition in six wheat varieties. Y-axis: relative expression; Error bars indicate 6 ± SE. *Statistical analysis of materials and methods has been labeled.
Expression Patterns of TaGASR Genes in Response to Exogenous GA, ABA, Low and High Temperature Treatments
We further investigated the expression patterns of five TaGASR genes (TaGASR15/-24/-25/-34/-35) in response to exogenous GA, ABA, low temperature (LT), and high temperature (HT) treatments in varieties HMC21 and J411, which show very high and very low levels of seed dormancy, respectively. Moreover, we assessed the GI values of the two varieties. After 50 µM GA treatment, HMC21 (high dormancy) seeds showed no sensitivity to GA and remained dormant (average GI: 0.00). In contrast, J411 (low dormancy) seeds showed strong sensitivity to GA resulting in high levels of germination (average GI: 0.92; Table S9). In addition, in HMC21 and J411 we found different levels of transcription for all five of the TaGASR genes examined. Both TaGASR15 and TaGASR34 were up-regulated in J411 seeds, but down-regulated in HMC21 seeds. After 50 µM ABA treatment, HMC21 seeds retained strong dormancy (average GI: 0.00), but J411 seeds showed little sensitivity to ABA (average GI: 0.77) (Table S9). Moreover, all five genes tested were up-regulated in J411 seeds but were down-regulated in HMC21 seeds. Similarly, after HT (36°C) treatment, HMC21 seeds showed high levels of dormancy (average GI: 0.00), whereas J411 seeds showed low levels of dormancy (average GI: 0.71; Table S9). All five genes were also up-regulated in J411 seeds, but TaGASR15 and TaGASR34 were down-regulated in HMC21 seeds. After LT (4°C) treatment, HMC21 seeds showed no sensitivity to LT and remained dormant (average GI: 0.00), whereas J411 seeds showed strong sensitivity to LT with high-level germination (average GI: 0.89; Table S9). Each of the five genes showed different expression patterns in HMC21 and J411, but only TaGASR34 was down-regulated in HMC21 seeds yet up-regulated in J411 seeds (Figure 8A).
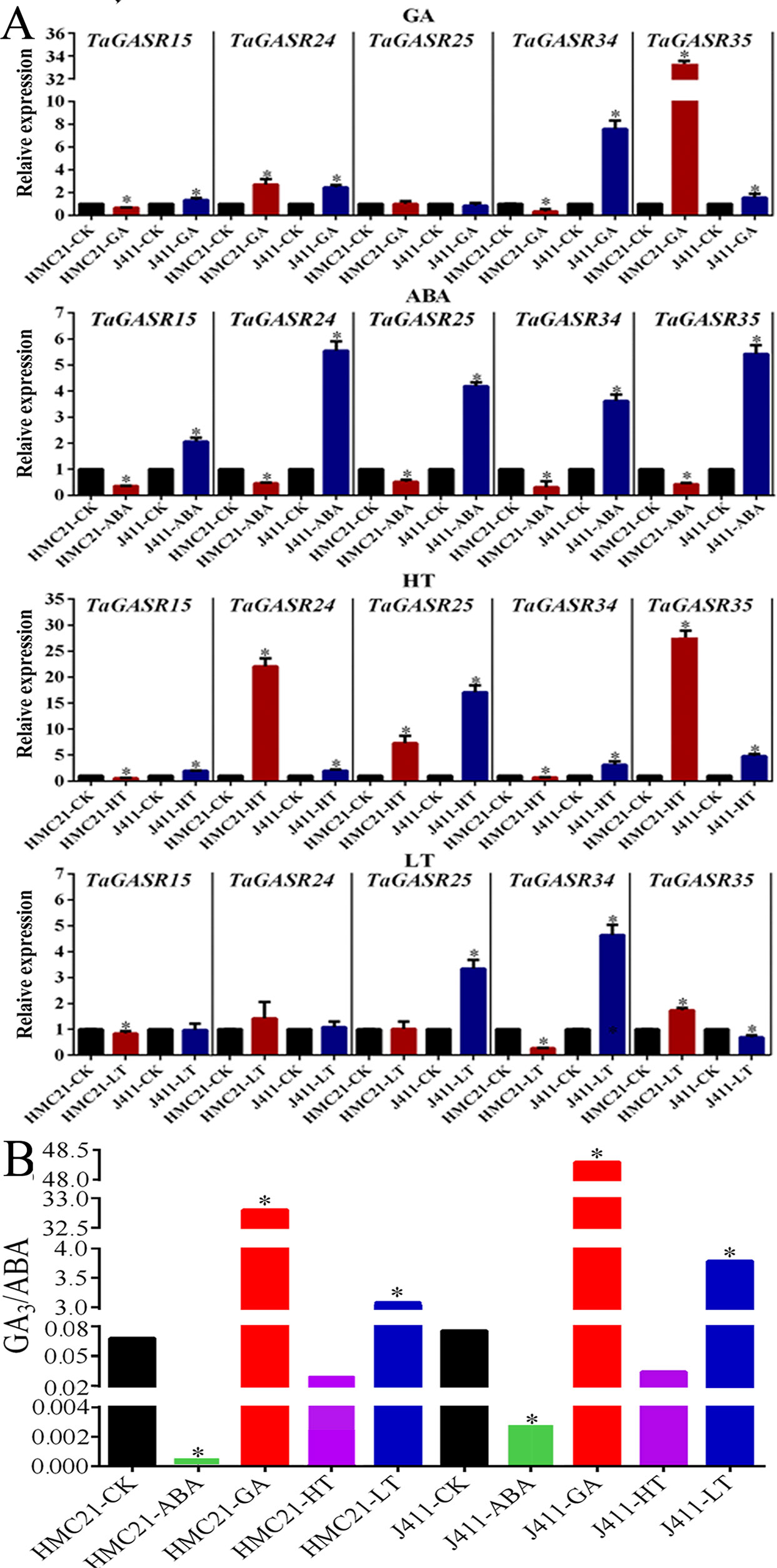
Figure 8 Expression in response to ABA, GA, High Temperature (HT), and Low Temperature (LT) treatments in wheat varieties Jing411 (J411) and Hongmangchun 21 (HMC21). (A) The levels of endogenous GA3/ABA in J411 and HMC21 (B) Expression patterns of five TaGASR genes in response to ABA, GA, High Temperature (HT), and Low Temperature (LT) treatments in wheat varieties Jing411 (J411) and Hongmangchun 21 (HMC21). Y-axis: relative expression; Error bars, 6 ± SE.
Simultaneously, we also examined the levels of endogenous ABA and GA3 in J411 and HMC21 seeds after ABA, GA, HT, and LT treatments, with deionized water as a control. In both J411 and HMC21 seeds, after ABA and HT treatments, the ratios of endogenous GA3:ABA were lower compared to control; nevertheless, after GA3 and LT treatments, the ratios of endogenous GA3:ABA were significantly higher than control. Notably, the ratios of endogenous GA3:ABA were consistently lower in HMC21 seeds than in J411 seeds after above four treatments (Figure 8B).
Based on the consistent trends between gene expression patterns and corresponding GI phenotypes, we speculated that TaGASR34 was a candidate gene strongly associated with seed dormancy and germination.
Cloning and Sequence Analysis of TaGASR34
A primer pair (GASR34-7B; Table S4) was designed to isolate the TaGASR34 gene in the J411 and HMC21 varieties. The TaGASR34 gene was 1,974 bp in length, including a 995 bp promoter sequence, a 458 bp 3′UTR, 3 exons, and 2 introns. Sequence alignment analysis revealed 6 SNP mutations in the TaGASR34 promoter, and no variation was detected in the TaGASR34 coding region (Figure S1).
In addition, 12 cis-acting elements were identified in the promoter of TaGASR34, including one TC-rich repeat element, five MBS (MYB transcription factor binding site) elements, one CE3 element (related to ABA and VP1 response), two Skn-1 elements (related to endosperm expression), two ARE elements, and one box E element. Notably, the replacement of the G/A base at the -16 position resulted in the absence of a box E element (Figure S2).
Validation of the Relationship Between TaGASR34, Seed Dormancy, and Seed Germination
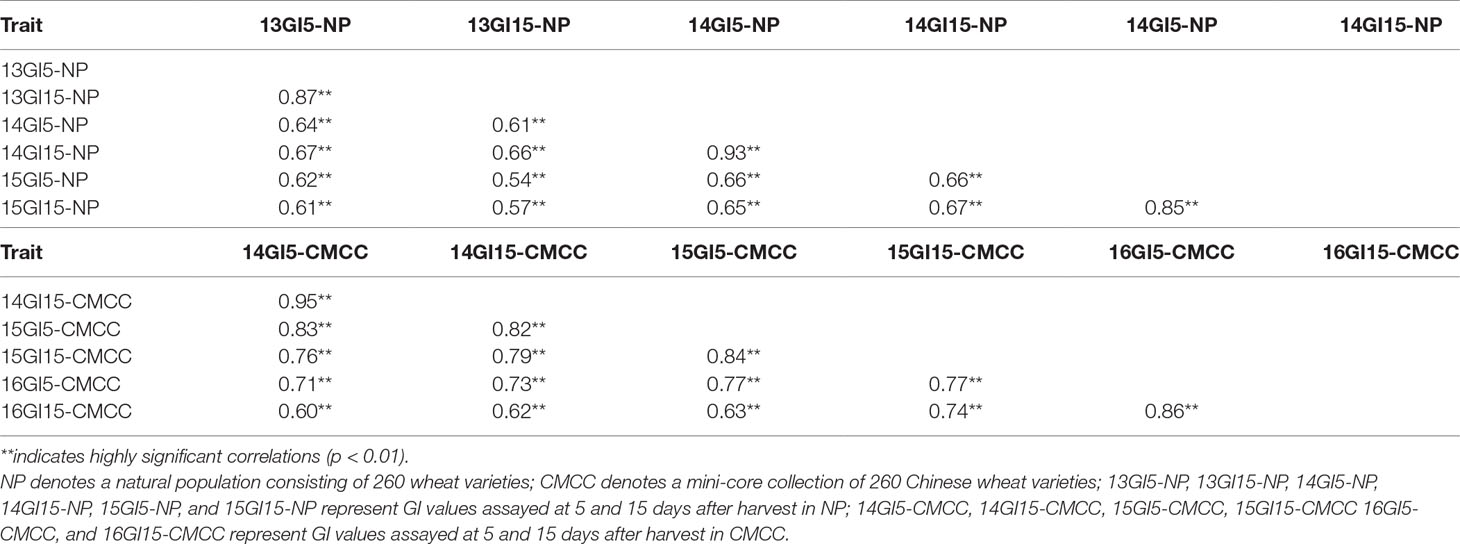
All GI phenotypic data showed wide variations within both the NP and CMCC populations across environments, with coefficients of variance of 25.46–55.21% and 38.76–85.79%, respectively (Table S10). In NP plants, the average GI value of 13GI15-NP plants was the highest (mean GI = 0.72), ranging from 0.07 to 0.98, followed by the 15GI15-NP (mean GI: 0.64, range: 0.02–0.98) and 13GI5-NP (mean GI: 0.56, range: 0.04–0.91). In CMCC plants, the mean GI values of both the 15GI15-CMCC and 16GI15-CMCC were the highest (mean GI: 0.56), ranging from 0.04 to 0.99 and 0.05 to 0.96, respectively. Significant correlations were detected in GI values assayed under different environments in both NP and CMCC plants, with correlation coefficients of 0.54–0.93 and 0.60–0.95 (P < 0.01), respectively (Table 2).
Based on the SNP mutation in the TaGASR34 promoter listed above, the cleaved amplified polymorphic site (CAPS) marker GS34-7B was developed and used to validate the association between TaGASR34 and seed dormancy and germination in both CMCC and NP plants. Two allelic variations were identified. These were designated GS34-7Ba, which was associated with increasing GI and could be digested into 900-bp and 410-bp fragments, and GS34-7Bb, which was associated with decreasing GI and was present as a single undigested 1310-bp fragment (Figure 9). In CMCC plants, 224 (86.15% of the total) were found to contain the GS34-7Ba allele, whereas 36 (13.85%) carried GS34-7Bb. In NP plants, 165 (63.46%) contained the GS34-7Ba allele, whereas 95 (36.54%) harbored GS34-7Bb. We detected significant differences (P < 0.01 or 0.05) in mean GI values between varieties with the two alleles of TaGASR34 in both populations across environments (Table 3). Notably, in both CMCC and NP, the frequency distribution of GS34-7Bb (13.85% and 36.54%, respectively) was consistently lower than that of GS34-7Ba (86.15% and 63.46%, respectively).
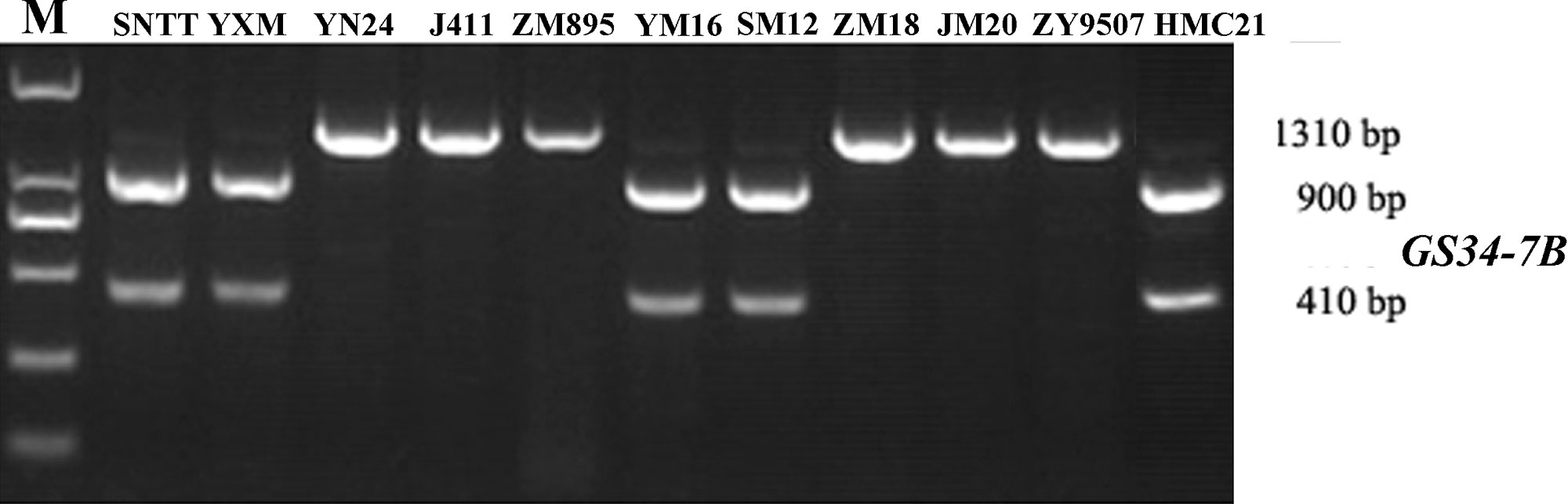
Figure 9 Different genotypes identified by the functional marker GS34-7B in different wheat varieties. Shown are: Suiningtuotuo (SNTT), Yangxiaomai (YXM), Yangnong 24 (YN24), Jing 411 (J411), Zhongmai 895 (ZM895), Yangmai 16 (YM16), Shimai12 (SM12), Zhongmai 18 (ZM18), Jimai 20 (JM20), Zhongyou 9507 (ZY9507), and Hongmangchun 21 (HMC21).
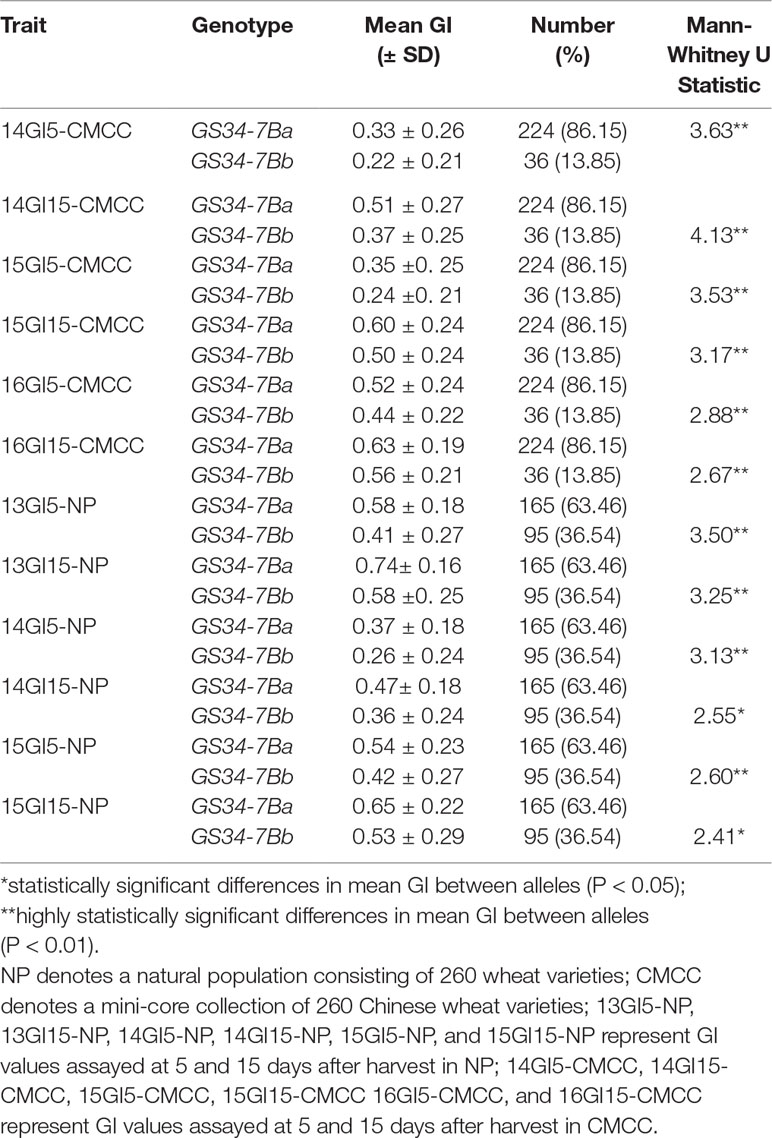
Table 3 Descriptive statistics and Mann-Whitney U test results for seed germination index (GI) values between the two alleles of TaGASR34 in NP and CMCC.
Frequency Distribution of TaGASR34 in Non-Chinese Wheat Germplasms
We also investigated the frequency distribution of TaGASR34 alleles in 580 wheat germplasms from 23 countries in four continents, including Europe (312), Asia (191), Africa (50), and Oceania (27; Table S11). The frequency of GS34-7Bb (present in 95 varieties, 16.38% of the total) was significantly lower than the frequency of GS34-7Ba (485, 83.62%). On each continent, GS34-7Bb also had a consistently lower frequency than GS34-7Ba (Figure S3 and Table S11).
Discussion
Genome-Wide Identification of TaGASR Genes in Common Wheat
Many GASA homologs have been studied extensively in a variety of plant species because of their roles in plant development and biotic/abiotic stresses (Taylor and Scheuring, 1994; Ben-Nissan and Weiss, 1996; Aubert et al., 1998; Shi and Olszewski, 1998; Tomoyuki et al., 2006; Zhang and Wang, 2008; Alonso-Ramírez et al., 2009; Zhang et al., 2009; Rubinovich and Weiss, 2010; Zimmermann et al., 2010; Sun et al., 2013; Zhong et al., 2015; Fan et al., 2017). However, little is known regarding GASR homologs in common wheat. Only seven GASR homologs have yet been identified and characterized. These include TaGASR7-A1, which is associated with grain length and weight, and TaGASR7-B1, TaGASR7-D1 (Dong et al., 2014), TaGAST1, TaGAST2, TaGAST3, and TaGAST4, all of which are involved in spike/seedling development (Kim et al., 2016).
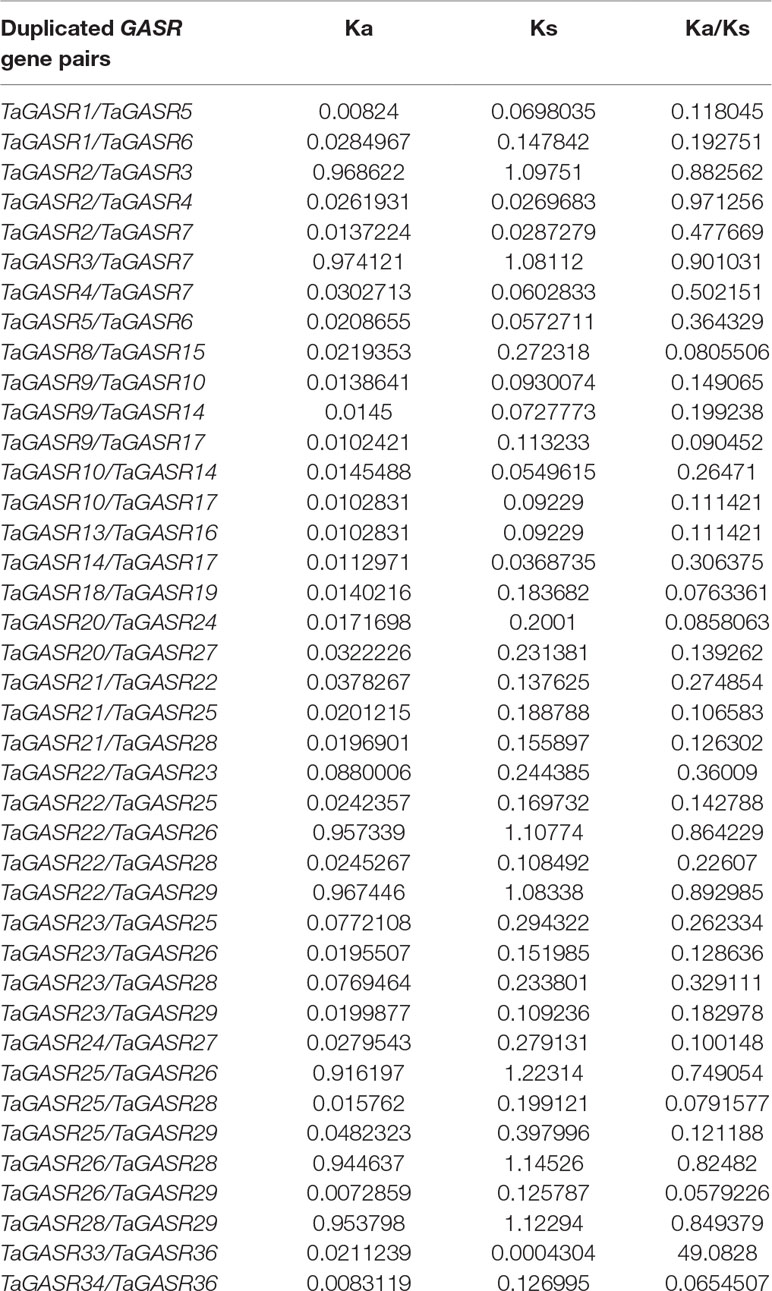
In this study, we identified 37 TaGASR genes and 40/40 paralogous/orthologous pairs in common wheat (Table S12). In general, Ka/Ks ratios > 1 indicates accelerated evolution with positive selection, Ka/Ks ratios approximately equal to 1 indicates neutral selection, whereas Ka/Ks ratios < 1 indicates functional constraint by purifying selection (Cui et al., 2019). Here, we found that the Ka/Ks ratio of only one homologous pair was greater than 1, implying that most TaGASR genes have undergone negative selection in wheat (Table 4). We also identified 27 pairs of Ta/Os orthologous genes as well as 13 pairs of Ta/At orthologous genes, suggesting that the genetic relationship between wheat and rice was closer than that between wheat and Arabidopsis.
Evolutionary and Microanalysis Analysis of GASR Genes
Our structural analysis of the 37 TaGASR genes revealed varying numbers of exons and introns, indicating that the wheat GASR gene family is diverse (Figure 2B). Previous studies have reported the number of exons in GASR genes from different species ranging from 2 to 5, and the number of introns ranging from 1 to 4. For example, the comparative structures of GASR genes in potato and apple suggest stable numbers of introns and exons have been maintained during evolution (Marta et al., 2002; Fan et al., 2017).
During evolution, eukaryotic genomes retain genes and associated regulatory and noncoding sequences on corresponding chromosomes to varying degrees. In the present study, intraspecific microanalysis revealed many collinear genes in wheat (Figure 6 and Table 5), suggesting that the TaGASR gene family may have underwent large-scale duplication (e.g. whole-genome or segmental duplication) or tandem duplication events. Structural analysis revealed that segmental duplication was more frequent than tandem duplication in the TaGASR gene family. During subsequent evolution, duplicated genes generally experience one of three alternative fates: nonfunctionalization, neofunctionalization, and subfunctionalization (Lynch and Conery, 2000). Many previous studies have reported that gene duplication plays an important role in genome rearrangement and expansion as well as an important role in the generation of gene functional diversity (Zhang et al., 2013; Cui et al., 2019). Together, these results provide a new resource to study the evolution of the GASR gene family among different plant species.
TaGASR Gene Expression Profiles and Potential Functions
In this study, we found cis-acting regulatory elements responsive to five important plant hormones (ABA, SA, GA, IAA, and MeJA) among the 36 TaGASR genes (although not in TaGASR13). In addition, we also found three cis-acting regulatory elements that regulate responses to abiotic stress (e.g. drought, low temperature, and defense). In particular, cis-acting regulatory elements associated with drought and low-temperature response were most prevalent among TaGASR genes (Figure 3 and Table S7). Taken together, our results suggest that elements responsive to the five plant hormones and elements associated with abiotic stress responses may play important roles in regulating the growth of wheat.
A total of 18 TaGASR gene expression profiles were obtained using publicly available microarray data (GSE12508) (Sun et al., 2015). Of these, 72% (13/18) were found to be highly expressed in 22 DAP embryos (22 DAP EM), and 67% (12/18) were highly expressed in anthers before anthesis (Aba). These results indicate that many TaGASR genes may play significant roles during wheat growth. We also found that many paralogous gene pairs sharing a high degree of sequence homology had similar patterns of expression (e.g. TaGASR1/-6 and TaGASR9/-10 in 22 DAP EM and Aba plants, as well as TaGASR14/-17, TaGASR22/-23 and TaGASR23/-29 in PBA and II plants) (Figure 4 and Table S8), implying that paralogous genes may have redundant functions during tissue development (Figure 4). These results provide a basis for further investigation of the functions of TaGASR genes in wheat.
Screening of TaGASR Genes Associated With Seed Dormancy and Germination and Its Application in Wheat Breeding
The prevalence of PHS in wheat is predominantly due to insufficient dormancy at harvest when seeds are mature (Mares and Mrva, 2001; Ogbonnaya et al., 2008). It is now recognized that moderate to high levels of seed dormancy are required for protection against PHS. Therefore, identification of genes controlling seed dormancy may help to decrease yield losses in wheat caused by PHS. Previous studies have shown that Arabidopsis AtGASA4, AtGASA5 (Rubinovich and Weiss, 2010), and AtGASA6 (Zhong et al., 2015), as well as Fagus sylvatica FsGASA4 (Alonso-Ramírez et al., 2009) play key roles in controlling seed dormancy and germination in those two species. However, the roles played by GASR homologous genes in wheat are largely unknown.
In this study, we investigated the expression patterns of 37 TaGASR genes during seed imbibition in six wheat varieties with contrasting patterns of seed dormancy, and found that the transcript levels of five specific TaGASR genes (TaGASR15/-24/-25/-34/-35) were consistently higher in the three varieties with low dormancy levels than that in the three varieties with high dormancy levels. This suggests that these five TaGASR genes may be involved in regulating seed dormancy and germination. In many plant species, seed dormancy and germination are controlled by two major plant hormones (ABA and GA) and temperature (Graeber et al., 2012; Shu et al., 2013; He et al., 2014). Subsequently, we analyzed differences in expression of these genes in varieties J411 and HMC21 following GA3, ABA, HT, and LT treatments. We found that only TaGASR34 was consistently down-regulated in dormant seeds and up-regulated in non-dormant seeds (Figure 8A). Also, we analyzed the two endogenous ABA and GA3 contents after GA3, ABA, HT, and LT treatments, and found that the ratios of endogenous GA3:ABA after the above four treatments was consistently lower in HMC21 seeds compared to J411 seeds, which is consistent with the differences in sensitivity of J411 and HMC21 seeds to the above four treatments and their GI phenotypes. These findings indicate that four stress treatments could affect the endogenous hormone levels of the two varieties and thus modulate seed dormancy and germination, which is in accordance with the previous results reported by Yamauchi et al. (2004) (Figure 8B). Taken together, this result in combination with GI phenotypic data from different treatments led to speculation that TaGASR34 may be a candidate gene for the regulation of seed dormancy and germination.
We further isolated the TaGASR34 gene and found that the G/A replacement of its promoter at the -16 position resulted in the deletion of a box E component. Next, we developed a CAPS marker (GS34-7B) based on the SNP variation. This marker was used to further validate the association of TaGASR34 with seed dormancy and germination using two natural populations in different environments, suggesting that the allelic version of TaGASR34 may underlie phenotypic differences in seed dormancy and germination. However, the specific functions of the box E component have not yet been determined, and the detailed regulatory mechanism by which TaGASR34 is associated with differences in seed dormancy and germination should be explored in future studies.
It is noteworthy that in both Chinese and foreign wheat germplasms, the frequency distribution of the TaGASR34 allele GS34-7Bb, which was associated with higher dormancy levels, was found to be significantly lower than GS34-7Ba, which was associated with lower dormancy levels. This result suggests that the favored GS34-7Bb allele is not frequently used in wheat breeding.
Previously, Dong et al. (2014) found that a C/G SNP variation at the -3 bp position upstream of the start codon of TaGASR7-A1 (corresponding to TaGASR33 identified in this study) affected grain length in common wheat. However, no variation was detected for TaGASR7-B1 (corresponding to TaGASR34 identified in this study) or TaGASR7-D1 (corresponding to TaGASR36 identified in this study). Zhang et al. (2016) reported that TaGASR7 was associated with significantly elevated thousand kernel weight (TKW) in aabbdd mutant plants with frameshift mutations in all six alleles. Interestingly, our present results indicate that the SNP variation (G/A) at the -16 position of the TaGASR34 promoter had a significant effect on seed dormancy and germination, however, no effect was observed on thousand grain weight (TGW), grain length (GL) and width (GW) (Table S13 and Table S14). In addition, the presence of different TaGASR33 alleles had little effect on seed dormancy and germination (data not shown). Therefore, pyramiding the two preferred allelic variants of TaGASR33 and TaGASR34 in a single variety may help achieve simultaneous improvement of both grain yield and dormancy.
According to a phylogenetic tree of GASR family members, we found that TaGASR34 was most closely related to the rice homolog OsGASR7 and the Arabidopsis homolog GASA14, implying that they may have similar functions. Wang et al. (2009) showed that OsGSR1 was a positive regulator of GA signaling. Similarly, here we found that TaGASR34 was up-regulated after GA treatment and showed increased sensitivity to GA, supporting that TaGASR34 is also involved in GA signaling. However, the role played by OsGSR1 in regulating seed dormancy and germination is unknown and should be further investigated in future studies. Sun et al. (2013) reported that Arabidopsis GASA14 expression was up-regulated by GA and down-regulated by transcriptional regulators that repress GA responses, including the DELLA proteins GAI and RGA. The same study also reported that germination rate of the gasa14-1 GASA14 null mutant was lower than those of Col wild-type plants, thereby further supporting the hypothesis that TaGASR34 plays a role in regulating seed dormancy and germination.
Conclusion
In this study, we performed a basic bioinformatics analysis of TaGASR gene family in common wheat, and cloned TaGASR34 as a likely candidate gene involved in the regulation of seed dormancy and germination. Further, we validated the association of TaGASR34 with seed dormancy and germination, and found the favorable allele GS34-7Bb associated with higher seed dormancy was infrequently observed in both Chinese and non-Chinese wheat cultivars and thus had good potential to utilize in wheat PHS resistance breeding. These findings provide a theoretical basis for the subsequent study of GASR gene functions in wheat and other crops.
Data Availability Statement
The genome sequences of wheat, rice and Arabidopsis were downloaded from the Ensembl database (http://plants.ensembl.org/index.html), Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/analyses_search_locus.shtml) and PlantTFDB (http://planttfdb.cbi.pku.edu.cn).
Author Contributions
XC and SW Conceived the Study, Put Into Effect the Main Bioinformatics Analyses, and Drafted the Manuscript. DX and Xliu Took Part in the Experiments and Drafting of the Manuscript. Xli, WX, JC, HJ, XM, and JW Processed the Experimental Data and Helped to Draft the Manuscript. HZ, CC, JL, and CM Conceived and Guided the Experiments, and Helped in Coordinating the Project and Drafting the Manuscript. All Authors Read and Accepted the Final Manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31871608), The National key research and development plan “Breeding new wheat varieties with high-yielding, high-quality and water-saving in the south of Huang-Huai River winter wheat area” — the breeding of new wheat germplasm and varieties with resistance to adversity (2017YFD0100703), The China Agriculture Research System (CARS-03), The National Key Research and Development Program of China (2016YFD0101802, 2017YFD0100804), Wheat Genetics and Breeding Research Platform Innovation Team of Anhui’s University (2015-), Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP), and The Agriculture Research System of Anhui province (AHCYTX-02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank for Profs. Jizeng Jia and Xianchun Xia for kindly providing Chinese micro-core wheat collections and foreign germplasms, respectively.
Abbreviations
PHS, Pre-harvest sprouting; GASR, Gibberellin Acid-Stimulated Regulator; SNP, Single nucleotide polymorphism; GI, Germination index; FS, Fielding sprouting; ABA, Abscisic acid; GA, Gibberellin acid; Eth, Ethylene; BR, Brassinolide; IAA, Auxin; ROS, Reactive oxygen species; NO, Nitric oxide; NOS, Nitric oxide synthase; qRT-PCR, quantitative real-time PCR; Ks, number of synonymous substitutions per synonymous site; Ka, number of non-synonymous substitutions per non-synonymous site; CDS, coding sequence; bp, base pair; aa, amino acids; MW, molecular weight; pI, isoelectric point; Da, Dalton.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00980/full#supplementary-material
References
Alonso-Ramírez, A., Rodríguez, D., Reyes, J., Jiménez, J. A., Nicolás, G., López-Climent, M., et al. (2009). Evidence for a role of Gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol. 150, 1335–1344. doi: 10.1104/pp.109.139352
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Appleford, N. E. J., Wilkinson, M. D., Ma, Q., Evans, D. J., Stone, M. C., Pearce, S. P., et al. (2007). Decreased shoot stature and grain α-amylase activity following ectopic expression of a gibberellin 2-oxidase gene in transgenic wheat. J. Exp. Bot. 58, 3213–3226. doi: 10.1093/jxb/erm166
Aubert, D., Chevillard, M., Dorne, A. M., Arlaud, G., Herzog, M. (1998). Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol. Biol. 36, 871–883. doi: 10.1023/A:1005938624418
Barrett, T., Edgar, R. (2006). Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 411, 352–369. doi: 10.1016/S0076-6879(06)11019-8
Ben-Nissan, G., Lee, J. Y., Borohov, A., Weiss, D. (2004). GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J. 37, 229–238. doi: 10.1046/j.1365-313X.2003.01950.x
Ben-Nissan, G., Weiss, D. (1996). The petunia homologue of tomato gast1: transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol. Biol. 32, 1067–1074. doi: 10.1007/BF00041390
Benech-Arnold, R., Gualano, N., Leymarie, J. D., Corbineau, F. (2006). Hypoxia interferes with ABA metabolism and increases ABA sensitivity in embryos of dormant barley grains. J. Exp. Bot. 57, 1423–1430. doi: 10.1093/jxb/erj122
Berrocal-Lobo, M., Segura, A., Moreno, M., López, G., García-Olmedo, F., Molina, A. (2002). Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 128, 951–961. doi: 10.1104/pp.010685
Bewley, J. D. (1997). Seed germination and dormancy. Plant Cell 9, 1055–1066. doi: 10.1105/tpc.9.7.1055
Blanc, G., Wolfe, K. H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. doi: 10.1016/j.livsci.2009.01.009
Brown, L. K., Wiersma, A. T., Olson, E. L. (2018). Pre-harvest sprouting and α-amylase activity in soft winter wheat. J. Cereal Sci. 79, 311–318. doi: 10.1016/j.jcs.2017.11.016
Bryfczynski, S. (2009). GraphPad: a CS2/CS7 tool for graph creation. DBLP. doi: 10.1145/1566445.1566517
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4, 10. doi: 10.1186/1471-2229-4-10
Chang, C., Feng, J. M., Si, H. Q., Yin, B., Zhang, H. P., Ma, C. X. (2010). Validating a novel allele of viviparous-1 (Vp-1Bf) associated with high seed dormancy of Chinese wheat landrace, Wanxianbaimaizi. Mol. Breeding 25, 517–525. doi: 10.1007/s11032-009-9350-3
Chen, Z., Chen, X., Yan, H., Li, W., Li, Y. Y., Cai, R., et al. (2015). The lipoxygenase Gene Family in Poplar: identification, classification, and expression in response to MeJA treatment. PloS One 10, e0125526. doi: 10.1371/journal.pone.0125526
Cheng, X. R., Xiong, R., Liu, H. L., Wu, M., Chen, F., Yan, H. W., et al. (2018). Basic helix-loop-helix gene family: Genome wide identification, phylogeny, and expression in Moso bamboo. Plant Physiol. Biochem. 132, 104–119. doi: 10.1016/j.plaphy.2018.08.036
Chiang, G. K., Bartsch, M., Barua, D., Nakabayashi, K., Debieu, M., Kronholm, I., et al. (2011). DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol. Ecol. 20, 3336–3349. doi: 10.1111/j.1365-294X.2011.05181.x
Chu, W., Liu, B., Wang, Y., Pan, F., Chen, Z., Yan, H., et al. (2016). Genome-wide analysis of poplar VQ gene family and expression profiling under PEG, NaCl, and SA treatments. Tree Genet. Genomes 12, 124. doi: 10.1007/s11295-016-1082-z
Clerkx, E. J. M., Vries, H. B. D., Ruys, G. J., Groot, S. P. C., Koornneef, M. (2003). Characterization of green seed, an enhancer of abi3-1 in arabidopsis that affects seed longevity. Plant Physiol. 132, 1077–1084. doi: 10.1104/pp.103.022715
Corbineau, F., Poljakoff-Mayber, A., Côme, D. (1991). Responsiveness to abscisic acid of embryos of dormant oat (avena sativa) seeds. Involvement of ABA-inducible proteins. Physiol. Plant. 83, 1–6. doi: 10.1111/j.1399-3054.1991.tb01273.x
Cui, Y. P., Ma, J. J., Liu, G. Y., Wang, N. H., Pei, W. F., Wu, M., et al. (2019). Genome-wide identification, sequence variation, and expression of the Glycerol-3-Phosphate Acyltransferase (GPAT) gene family in Gossypium. Front. Genet. 10, 116. doi: 10.3389/fgene.2019.00116
De la Fuente, J. I., Amaya, I., Castillejo, C., Sánchez-Sevilla, J. F., Quesada, M. A., Botella, M. A., et al. (2006). The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J. Exp. Bot. 10, 2401–2411. doi: 10.1093/jxb/erj213
Dong, L., Wang, F., Tao, L., Dong, Z., Li, A., Jing, R., et al. (2014). Natural variation of TaGASR7-A1 affects grain length in common wheat under multiple cultivation conditions. Mol. Breeding 34, 937–947. doi: 10.1007/s11032-014-0087-2
Fan, S., Zhang, D., Zhang, L., Gao, C., Xin, M., Tahir, M. M., et al. (2017). Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Genomics 18, 827. doi: 10.1186/s12864-017-4213-5
Finch-Savage, W. E., Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x
Finkelstein, R., Reeves, W., Ariizumi, T. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi: 10.1146/annurev.arplant.59.032607.092740
Finkelstein, R. R., Gampala, S. S. L., Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14, S15. doi: 10.1105/tpc.010441
Furukawa, T., Sakaguchi, N., Shimada, H. (2006). Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet. Syst. 81, 171–180. doi: 10.1266/ggs.81.171
Gao, Y. M., Liu, H., Wang, Y., Li, F., Xiang, Y. (2017). Genome-wide identification of PHD-finger genes and expression pattern analysis under various treatments in moso bamboo (Phyllostachys edulis). Plant Physiol. Biochem. 123, S0981942817304357. doi: 10.1016/j.plaphy.2017.12.034
Graeber, K., Nakabayashi, K., Miatton, E., Leubner-Metzger, G., Soppe, W. J. J. (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35, 1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x
Herzog, M., Dorne, A. M., Grellet, F. (1995). GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol. Biol. 27, 743–752. doi: 10.1007/BF00020227
He, H., Vidigal, D. D. S., Snoek, L. B., Schnabel, S., Nijveen, H., Hilhorst, H., et al. (2014). Interaction between parental environment and genotype affects plant and seed performance in arabidopsis. J. Exp. Bot. 65, 6603–6615. doi: 10.1093/jxb/eru378
Hu, M. J., Zhang, H. P., Cao, J. J., Zhu, X. F., Wang, S. X., Jiang, H., et al. (2016). Characterization of an IAA-glucose hydrolase gene TaTGW6 associated with grain weight in common wheat (Triticum aestivum L.). Mol. Breeding 36, 25. doi: 10.1007/s11032-016-0449-z
Huang, X., Zhang, X., Gong, Z., Yang, S., Shi, Y. (2016). ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J. 89354, –365. doi: 10.1111/tpj.13389
Ibrahim, E. A. (2016). Seed priming to alleviate salinity stress in germinating seeds. J. Plant Physiol. 192, 38–46. doi: 10.1016/j.jplph.2015.12.011
Jacobsen, S. E., Olszewsk, N. E. (1993). Mutations at the spindly locus of arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896. doi: 10.1105/tpc.5.8.887
Jiang, H., Zhao, L. X., Chen, X. J., Cao, J. J., Wu, Z. Y., Liu, K., et al. (2018). A novel 33-bp insertion in the promoter of TaMFT-3A is associated with pre-harvest sprouting resistance in common wheat. Mol. Breeding 38, 69. doi: 10.1007/s11032-018-0830-1
Jorge, V., Fregene, M. A., Duque, M. C., Bonierbale, M. W., Tohme, J., Verdier, V. (2000). Genetic mapping of resistance to bacterial blight disease in cassava ( Manihot esculenta Crantz). TAG Theor Appl Genet 101, 865–872. doi: 10.1007/s001220051554
Kendall, S. L., Hellwege, A., Marriot, P., Whalley, C., Graham, I. A., Penfield, S. (2011). Induction of dormancy in arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23, 2568–2580. doi: 10.1105/tpc.111.087643
Kiana, T., Brady, S. M., Ryan, A., Eugene, L., Provart, N. J. (2005). The botany array resource: e-northerns, expression angling, and promoter analyses. Plant J. 43, 153–163. doi: 10.1111/j.1365-313x.2005.02437.x
Kim, Y. J., Kim, J. Y., Seok Yoon, J. S., Yeon Kim, D. Y., Hong, M. J., Seo, Y. W. (2016). Characterization of 4 TaGAST genes during spike development and seed germination and their response to exogenous phytohormones in common wheat. Mol. Biol. Rep. 43, 1435–1449. doi: 10.1007/s11033-016-4077-9
Ko, C. B., Woo, Y. M., Lee, D. J., Lee, M. C., Kim, C. S. (2007). Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol. Biochem. (Paris) 45, 722–728. doi: 10.1016/j.plaphy.2007.07.010
Koornneef, M., Van der veen, J. H. (1980). Induction and analysis of gibberellin sensitive mutants in arabidopsis thaliana (Ll.) heynh. Theor. Appl. Genet. 58, 257–263. doi: 10.1007/BF00265176
Koornneef, M., Reuling, G., Karssen, C. M. (1984). The isolation and characterization of abscisic acid-insensitive mutants of arabidopsis thaliana.. Physiol. Plant. 61, 377–383. doi: 10.1111/j.1399-3054.1984.tb06343.x
Kotilainen, M. (1999). GEG participates in the regulation of cell and organ shape during corolla and carpel development in . Plant Cell 11, 1093–1104. doi: 10.1105/tpc.11.6.1093
Kucera, B., Cohn, M. A., Leubner-Metzger, G. (2005). Plant hormone interactions during seed dormancy release and germination. seed Sci. Res.. doi: 10.1079/SSR2005218
Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., et al. (2004). The arabidopsis cytochrome P450 CYP707a encodes aba 8’-hydroxylases: key enzymes in ABA catabolism. Embo J. 23, 1647–1656. doi: 10.1038/sj.emboj.7600121
Lee, S., Cheng, H., King, K. E., Wang, W., Peng, J. (2002). Gibberellin regulates arabidopsis seed germination via rgl2, a gai/rga-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658. doi: 10.1101/gad.969002
Leymarie, J., Robayo-Romero, M. E., Gendreau, E., Benech-Arnold, R. L., Corbineau, F. (2008). Involvement of ABA in induction of secondary dormancy in barley (Hordeum vulgare L.) seeds. Plant Cell Physiol. 49, 1830–1838. doi: 10.1093/pcp/pcn164
Li, Q., Li, L., Liu, Y., Lv, Q., Zhang, H., Zhu, J., et al. (2017). Influence of TaGW2-6a on seed development in wheat by negatively regulating gibberellin synthesis. Plant Sci. 263, 226. doi: 10.1016/j.plantsci.2017.07.019
Ling, H. Q., Zhao, S., Liu, D., Wang, J., Sun, H., Zhang, C., et al. (2013). Draft genome of the wheat A-genome progenitor Triticum urartu. Sci. Found. China 496, 87–90. doi: 10.1038/nature11997
Liu, H., Wu, M., Zhu, D., Pan, F., Wang, Y., Wang, Y., et al. (2017). Genome-wide analysis of the AAAP gene family in moso bamboo (Phyllostachys edulis). BMC Plant Biol. 17, 29. doi: 10.1186/s12870-017-0980-z
Liu, Z. H., Zhu, L., Shi, H. Y., Chen, Y., Zhang, J. M., Zheng, Y., et al. (2013). Cotton GASL genes encoding putative gibberellin-regulated proteins are involved in response to GA signaling in fiber development. Mol. Biol. Rep. 40, 4561–4570. doi: 10.1007/s11033-013-2543-1
Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Mares, D. J. (1983). Preservation of dormancy in freshly harvested wheat grain. Aust. J. Agric. Res. 34, 33–38. doi: 10.1071/AR9830033
Mares, D. J., Mrva, K. (2001). Mapping quantitative trait loci associated with variation in grain dormancy in Australian wheat. Aust. J. Agric. Res. 52, 1257–1265. doi: 10.1071/ar01049
Marta, B. L., Ana, S., Manuel, M. (2002). Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 128, 951–961. doi: 10.1104/pp.010685
Moyano-Cañete, E., Bellido, M. L., García-Caparrós, N., Medina-Puche, L., Amil-Ruiz, F., González-Reyes, J. A., et al. (2013). FaGAST2, a strawberry ripening-related gene, acts together with faGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 54, 218–236. doi: 10.1093/pcp/pcs167
Nakamura, S., Abe, F., Kawahigashi, H., Nakazono, K., Tagiri, A., Matsumoto, T. (2011). A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23, 3215–3229. doi: 10.1105/tpc.111.088492
Nambara, E., Marion-Poll, A. (2003). ABA action and interactions in seeds. Trends Plant Sci. 8, 213–217. doi: 10.1016/s1360-1385(03)00060-8
Ogbonnaya, F. C., Imtiaz, M., Ye, G., Hearnden, P. R., Hernandez, E., Eastwood, R. F., et al. (2008). Genetic and QTL analysis of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor. Appl. Genet. 116, 891–902. doi: 10.1007/s00122-008-0712-8
Rodríguez, M. V., Margineda, M., González-Martín, J. F., Insausti, P., Benech-Arnold, R. L. (2001). Predicting preharvest sprouting susceptibility in barley: a model based on temperature during grain filling. Agron. J. 93, 1071–1079. doi: 10.2134/agronj2001.9351071x
Roxrud, I., Lid, S. E., Fletcher, J. C., Schmidt, E. D., Opsahl-Sorteberg, H. G. (2007). GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 48, 471–483. doi: 10.1093/pcp/pcm016
Rubinovich, L., Weiss, D. (2010). The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 64, 1018–1027. doi: 10.1111/j.1365-313X.2010.04390.x
Segura, A., Moreno, M., Madueño, F., Molina, A., García-Olmedo, F. (1999). Snakin-1, a peptide from potato that is active against plant pathogens. Mol. Plant Microbe Interact. 12, 16–23. doi: 10.1094/MPMI.1999.12.1.16
Shi, L., Gast, R. T., Gopalraj, M., Olszewski, N. E. (1992). Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J. 2, 153–159. doi: 10.1111/j.1365-313x.1992.00153.x
Shi, L., Olszewski, N. E. (1998). Gibberellin and abscisic acid regulate GAST1 expression at the level of transcription. Plant Mol. Biol. 38, 1053–1060. doi: 10.1023/A:1006007315718
Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y., Tang, S. Y., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Gen. 9, e1003577. doi: 10.1371/journal.pgen.1003577
Shu, K., Zhou, W., Yang, W. (2017). APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol. 217, 977–983. doi: 10.1111/nph.14880
Sturn, A., Quackenbush, J., Trajanoski, Z. (2002). Genesis: cluster analysis of microarray data. Bioinformatics 18, 207–208. doi: 10.1093/bioinformatics/18.1.207
Sun, T., Wang, Y., Wang, M., Li, T., Zhou, Y., Wang, X., et al. (2015). Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 15, 269. doi: 10.1186/s12870-015-0657-4
Sun, S., Wang, H., Yu, H., Zhong, C., Zhang, X., Peng, J., et al. (2013). GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 64, 1637–1647. doi: 10.1093/jxb/ert021
Taylor, B. H., Scheuring, C. F. (1994). A molecular marker for lateral root initiation: The RSI-1 gene of tomato (Lycopersicon esculentum Mill) is activated in early lateral root primordia. Mol. Gen. Gen. 243, 148–157. doi: 10.1007/bf00280311
Thompson, A. J., Jackson, A. C., Symonds, R. C., Mulholland, B. J., Dadswell, A. R., Blake, P. S., et al. (2000). Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 23, 363–374. doi: 10.1046/j.1365-313x.2000.00789.x
Tomoyuki, F., Norihiro, S., Hiroaki, S. (2006). Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet. Syst. 81, 171–180. doi: 10.1266/ggs.81.171
Trethowan, R. M. (1995). Evaluation and selection of bread wheat (Triticum aestivum L.) for preharvest sprouting tolerance. Aust. J. Agric. Res. 46, 463–474. doi: 10.1071/ar9950463
Voorrips, R. E. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Walker-Simmons, M. K. (1987). ABA levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol. 84, 61–66. doi: 10.1104/pp.84.1.61
Wang, L., Wang, Z., Xu, Y. Y., Joo, S. H., Kim, S. K., Xue, Z., et al. (2009). OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 57, 498–510. doi: 10.1111/j.1365-313X.2008.03707.x
Wang, Y., Feng, L., Zhu, Y., Li, Y., Yan, H., Xiang, Y. (2015). Comparative genomic analysis of the WRKY III gene family in populus, grape, arabidopsis and rice. Biol. Direct 10, 48. doi: 10.1186/s13062-015-0076-3
Wang, Y., Liu, H., Zhu, D., Gao, Y., Yan, H., Xiang, Y. (2017). Genome-wide analysis of VQ motif-containing proteins in Moso bamboo (Phyllostachys edulis). Planta 246, 165. doi: 10.1007/s00425-017-2693-9
Wigoda, N., Ben-Nissan, G., Granot, D., Schwartz, A., Weiss, D. (2006). The gibberellin-induced, cysteine-rich protein gip2 from petunia hybrida exhibits in planta antioxidant activity. Plant J. 48, 796–805. doi: 10.1111/j.1365-313X.2006.02917.x
Wilkins, O., Nahal, H., Foong, J., Provart, N. J., Campbell, M. M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149, 981–993. doi: 10.1104/pp.108.132795
Wu, S., Wu, M., Dong, Q., Jiang, H., Cai, R., Xiang, Y. (2016). Genome-wide identification, classification and expression analysis of the PHD-finger protein family in Populus trichocarpa.. Gene 575, 75. doi: 10.1016/j.gene.2015.08.042
Yamauchi, Y., Ogawa, M., Kuwahara, A., Hanada, A., Kamiya, Y. J., Yamaguchi, S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of arabidopsis thaliana seeds. Plant Cell 16, 367–378. doi: 10.1105/tpc.018143
Yamauchi, Y., Takeda-Kamiya, N., Hanada, A., Ogawa, M., Kuwahara, A., Seo, M., et al. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 48, 555–561. doi: 10.1093/pcp/pcm023
Yoshimoto, K., Jikumaru, Y., Kamiya, Y., Kusano, M., Consonni, C., Panstruga, R., et al. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927. doi: 10.4161/auto.6.1.10843
Zhang, S. C., Wang, X. J. (2008). Expression pattern of GASA,downstream genes of DELLA, in Arabidopsis. China Sci. Bull. 53, 3839–3846. doi: 10.1007/s11434-008-0525-9
Zhao, P., Wang, D., Wang, R., Kong, N., Zhang, C., Yang, C., et al. (2018). Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress. BMC Genomics 19, 61. doi: 10.1186/s12864-018-4443-1
Zhong, C. M., Xu, H., Siting, Y., Wang, S., Li, L., Zhang, S., et al. (2015). Gibberellic acid-stimulated Arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis.. Plant Physiol. 169, 2288. doi: 10.1104/pp.15.00858
Zhang, S., Xu, R., Luo, X., Jiang, Z., Shu, H. (2013). Genome-wide identifcation and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene 531, 377–387. doi: 10.1016/j.gene.2013.07.107
Zhang, S., Wang, X. (2011). Overexpression of gasa5 increases the sensitivity of arabidopsis to heat stress. J. Plant Physiol. 168, 2093–2101. doi: 10.1016/j.jplph.2011.06.010
Zhang, S., Yang, C., Peng, J., Sun, S., Wang, X. (2009). GASA5, a regulator of flowering time and stem growth in arabidopsis thaliana.. Plant Mol. Biol. 69, 745–759. doi: 10.1007/s11103-009-9452-7
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. doi: 10.1038/ncomms12617
Keywords: common wheat, seed dormancy, GASR, GA, ABA
Citation: Cheng X, Wang S, Xu D, Liu X, Li X, Xiao W, Cao J, Jiang H, Min X, Wang J, Zhang H, Chang C, Lu J and Ma C (2019) Identification and Analysis of the GASR Gene Family in Common Wheat (Triticum aestivum L.) and Characterization of TaGASR34, a Gene Associated With Seed Dormancy and Germination. Front. Genet. 10:980. doi: 10.3389/fgene.2019.00980
Received: 12 May 2019; Accepted: 13 September 2019;
Published: 18 October 2019.
Edited by:
Mehar Hasan Asif, National Botanical Research Institute (CSIR), IndiaReviewed by:
Ashutosh Pandey, Bielefeld University, GermanyAjay Kumar Pandey, National Agri-Food Biotechnology Institute, India
Kai Shu, Sichuan Agricultural University, China
Copyright © 2019 Cheng, Wang, Xu, Liu, Li, Xiao, Cao, Jiang, Min, Wang, Zhang, Chang, Lu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiping Zhang, emhocDIwQDE2My5jb20=; Chuanxi Ma, bWFjaHVhbnhpQGFoYXUuZWR1LmNu
†These authors have contributed equally to this work
 Xinran Cheng
Xinran Cheng Shengxing Wang†
Shengxing Wang†