- 1Department of Laboratory Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Department of Gastroenterology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Background: Aberrant hypermethylation of the Septin 9 (SEPT9) is an early event in several human cancers, and increasing studies have reported good performance of methylated SEPT9 (mSEPT9) in cancer diagnosis. Recent studies further focused on its value in cancer prognosis, but results are not clearly elucidated.
Methods: A comprehensive search to identify relevant studies about the association between mSEPT9 and cancer prognosis was conducted through the EMBASE, PubMed, and Web of Science databases (up to January 2019). The main outcomes were overall survival (OS) and disease-free survival (DFS). The hazard ratio (HR) and 95% confidence interval (CI) for OS and DFS were extracted from each included study and pooled using a random-effects model.
Results: Ten eligible studies comprising 1,266 cancer patients were included. Results demonstrated that mSEPT9 was associated with poor OS (HR = 2.07, 95% CI = 1.40–3.06). Specially, mSEPT9 detected in preoperative plasma predicted worse OS in cancer patients (HR = 3.25, 95% CI = 1.93–5.48). In addition, we also identified a significant association of mSEPT9 with decreased DFS of cancer (HR = 3.24, 95% CI = 1.81–5.79).
Conclusion: Our meta-analysis supports that mSEPT9 is associated with reduced OS and DFS in cancer patients. Moreover, detection of mSEPT9 using plasma appears to be a convenient and promising way to predict long-term survival of cancer patients.
Introduction
Septins are a conserved group of GTP-binding proteins that play a crucial role in cytokinesis, cytoskeleton, and cell cycle control (Hall and Russell, 2004; Russell and Hall, 2011). As a star member of the Septin gene family, Septin 9 (SEPT9) is located at chromosome 17q25.3 and demonstrates both oncogenic and tumor-suppressive impacts on human cancers (Connolly et al., 2011; Verdier-Pinard et al., 2017). Previous studies have uncovered that methylated SEPT9 (mSEPT9) is associated with tumorigenesis based on transcriptionally silencing due to aberrant hypermethylation of the CpG island within the SEPT9 promoter (Connolly et al., 2011; Wasserkort et al., 2013; Wang et al., 2018). Detection of mSEPT9 has been reported in several cancers, including colorectal cancer (CRC), head and neck squamous cell carcinoma (HNSCC), and gastric cancer (GC) (Lee et al., 2013; Schrock et al., 2017; Song et al., 2018).
Nowadays, the diagnostic significance of mSEPT9 has been elucidated in several cancers, and specially, the mSEPT9 assay (Epi proColon) becomes the first blood-based test approved by U.S. FDA for CRC screening. Some researches further pay attention to the mSEPT9’s prognostic performance on cancer. In 2013, Dietrich et al. detected malignant pleural effusions from 58 cases with various cancers and found that mSEPT9 indicated a poor survival (Dietrich et al., 2013). Subsequently, the association of mSEPT9 with cancer prognosis was investigated in CRC (Lee et al., 2013; Tham et al., 2014; Freitas et al., 2018; Song et al., 2018), GC (Lee et al., 2013), HNSCC (Schrock et al., 2017), and so on (Kuo et al., 2014; Angulo et al., 2016; Branchi et al., 2016; Jung et al., 2016).
To date, however, the prognostic value of mSEPT9 in cancer patients has not yet been methodically elucidated. Herein, we performed a systematic review and meta-analysis to summarize the published data and evaluate the prognostic impact of mSEPT9 on human cancers.
Materials and Methods
Our meta-analysis was conducted based on the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (Moher et al., 2009). The PRISMA 2009 checklist is shown in Supplementary Table S1.
Search Strategy
A comprehensive electronic search was performed via the EMBASE, PubMed, and ISI Web of Science databases through January 2019 without any restriction. The search items were combinations of “SEPT9,” “mSEPT9,” “septin 9,” “prognosis” and “survival.” There was no language restriction.
Criteria of Inclusion and Exclusion
Two independent authors conducted the literature search and study selection. Discrepancies were resolved by discussion. Studies were considered eligible if they met the following criteria: (1) cohort studies for evaluating the prognostic role of mSEPT9 in cancer patients; and (2) studies reporting hazard ratios (HRs) and 95% confidence intervals (CIs) or providing information to estimate HRs. The exclusion criteria were as follows: (1) reviews, meta-analyses, opinion, abstracts, and cellular or animal experiments; and (2) studies with overlapping data. If studies had overlapping data, we kept the one with the larger sample size.
Data Extraction
Two independent authors extracted the following items from each included study: first author, publication year, country, patient number, sampling time, follow-up, cancer type and stage, detection method, and prognostic outcomes. Outcome measures included overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS), and progression-free survival (PFS).
Quality Evaluation
Two authors independently conducted quality evaluation, and discrepancies were resolved by discussion. We used the Newcastle-Ottawa scale (NOS) to assess the quality of each included study, with quality score from 0 to 9 (Supplemental Table S2) (Stang, 2010). Quality evaluation was not an exclusion criterion for eligible studies.
Statistical Analysis
Multivariate-adjusted HRs and 95% CIs were preferentially extracted from each included study, if available. If a study did not report the HR and 95% CI, these measures were extrapolated by the method of Parmar and Tierney (Parmar et al., 1998; Tierney et al., 2007). We used the random-effects model (DerSimonian and Laird) to pool these HRs and 95% CIs and examined the heterogeneity by Cochran’s Q test and I2 statistic (Higgins et al., 2003; Harris et al., 2008). P < 0.10 or I2 > 50% indicates considerable heterogeneity (Higgins and Thompson, 2002). We also performed subgroup analyses to further evaluate the mSEPT9’s prognostic effects based on sample type, sampling time, and cancer type. To assess the stability of pooled results, we applied one-way sensitivity analysis by excluding one study at a time. In addition, the publication bias was examined by Begg’s and Egger’s tests (Begg and Mazumdar, 1994; Egger et al., 1997). All P values were two-sided, and P ≤ 0.05 was considered significant, unless otherwise specified. All statistical analyses were carried out by Stata 12.1 software (College Station, TX, USA).
Results
Study Characteristics
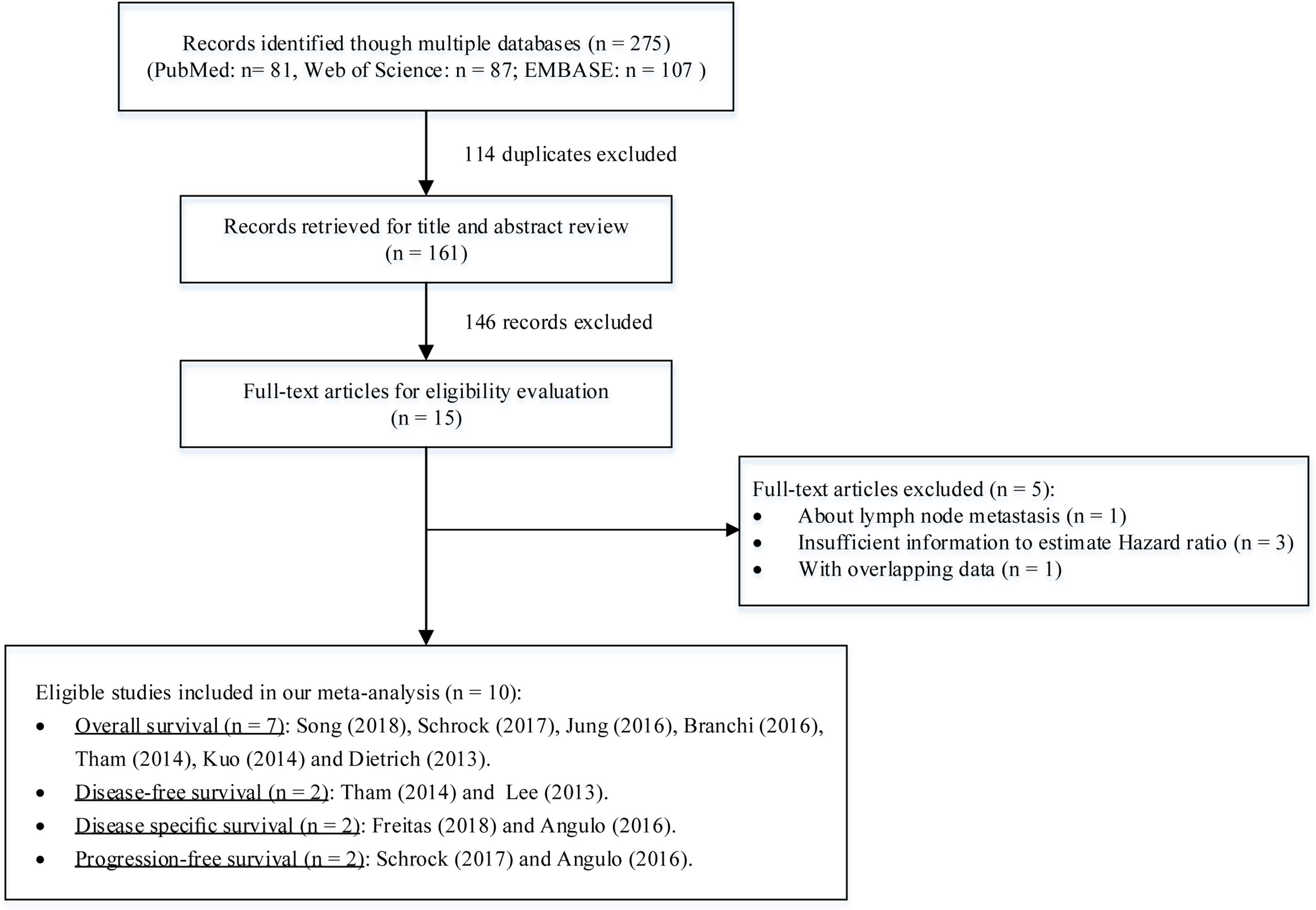
Our search strategy initially obtained 275 records from the PubMed, EMBASE. and Web of Science databases. By title and abstract review, we removed 114 duplicates and 146 records. This large proportion of excluded records consisted of reviews, opinions, conference abstracts, diagnostic studies, in vitro studies, and nonhuman studies. Of the remaining 15 full-text publications, five studies were further excluded because of focusing on lymph node metastasis (Nagata et al., 2017), having overlapping data (de Vos et al., 2017), or insufficient information to estimate HRs and 95% CIs (Perez-Carbonell et al., 2014; Villanueva et al., 2015; Chang et al., 2017). Finally, a total of 10 eligible studies were included for this meta-analysis (Dietrich et al., 2013; Lee et al., 2013; Kuo et al., 2014; Tham et al., 2014; Angulo et al., 2016; Branchi et al., 2016; Jung et al., 2016; Schrock et al., 2017; Freitas et al., 2018; Song et al., 2018) (Figure 1).
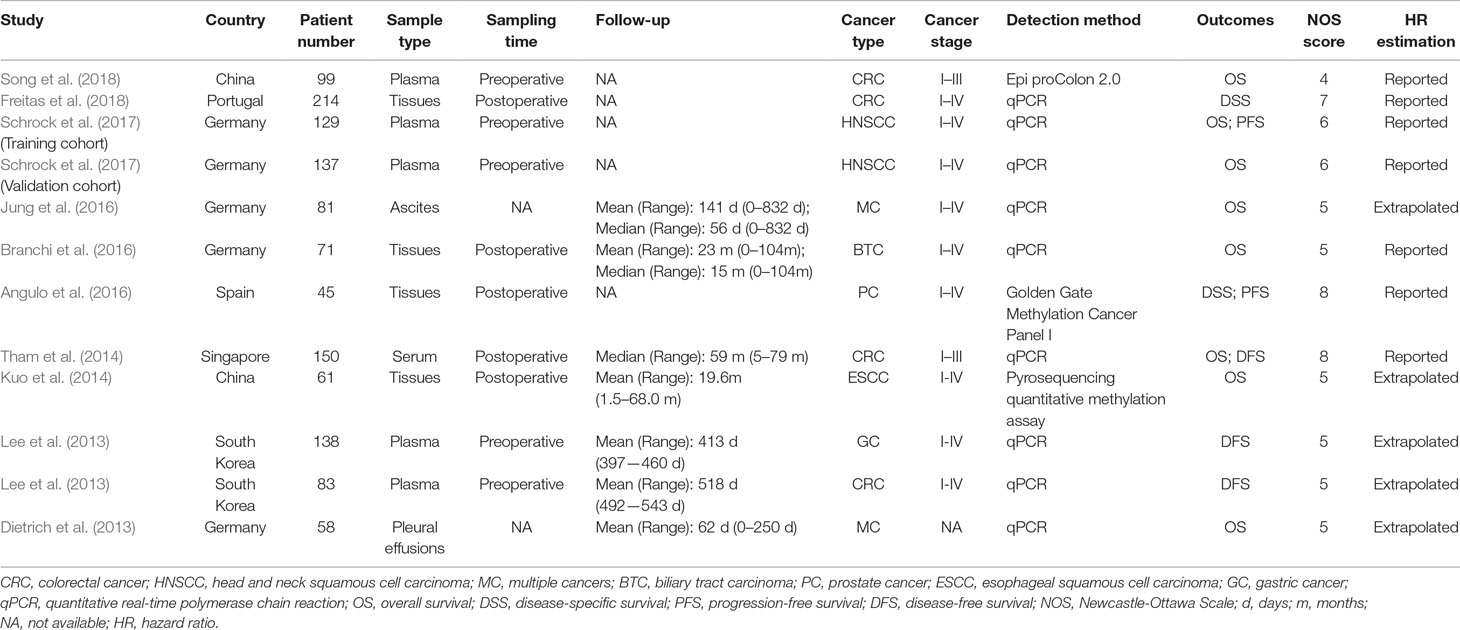
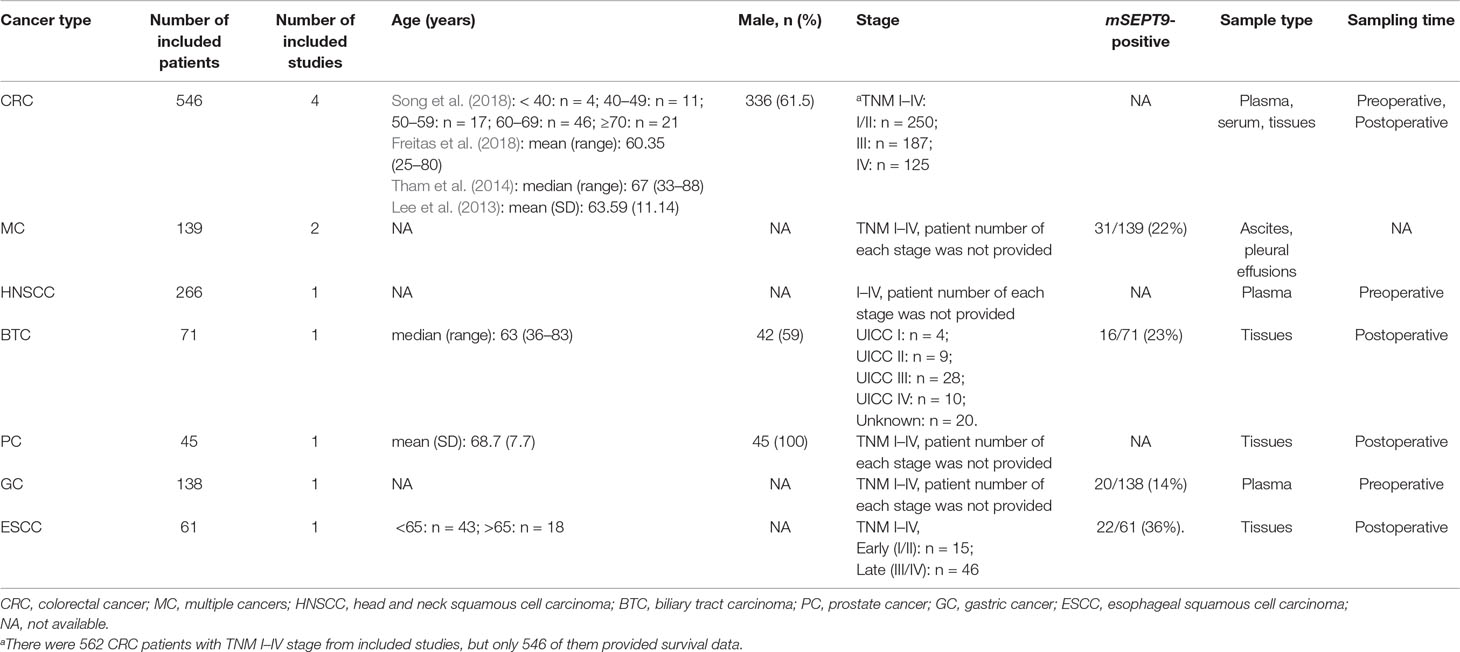
Among these studies including 1,266 cancer patients, seven evaluated the mSEPT9’s prognostic significance on OS (Dietrich et al., 2013; Kuo et al., 2014; Tham et al., 2014; Branchi et al., 2016; Jung et al., 2016; Schrock et al., 2017; Song et al., 2018), two evaluated DFS (Lee et al., 2013; Tham et al., 2014), two evaluated DSS (Angulo et al., 2016; Freitas et al., 2018), and two evaluated on PFS (Angulo et al., 2016; Schrock et al., 2017). There were four studies using plasma or serum, of which three collected preoperative samples (Lee et al., 2013; Schrock et al., 2017; Song et al., 2018) and one collected postoperative samples (Tham et al., 2014). Other studies used tissues (Angulo et al., 2016; Branchi et al., 2016; Freitas et al., 2018), ascites (Jung et al., 2016), or pleural effusions (Dietrich et al., 2013). The cancer type comprised CRC (Lee et al., 2013; Tham et al., 2014; Freitas et al., 2018; Song et al., 2018), GC (Lee et al., 2013), HNSCC (Schrock et al., 2017), biliary tract carcinoma (BTC) (Branchi et al., 2016), prostate cancer (PC) (Angulo et al., 2016), esophageal squamous cell carcinoma (ESCC) (Kuo et al., 2014), and multiple cancers (MC) (Dietrich et al., 2013; Jung et al., 2016). The quality of these studies were assessed by NOS score. More details about characteristics of included studies and cancer patients are summarized in Tables 1 and 2.
Association Between mSEPT9 and OS in Cancer Patients
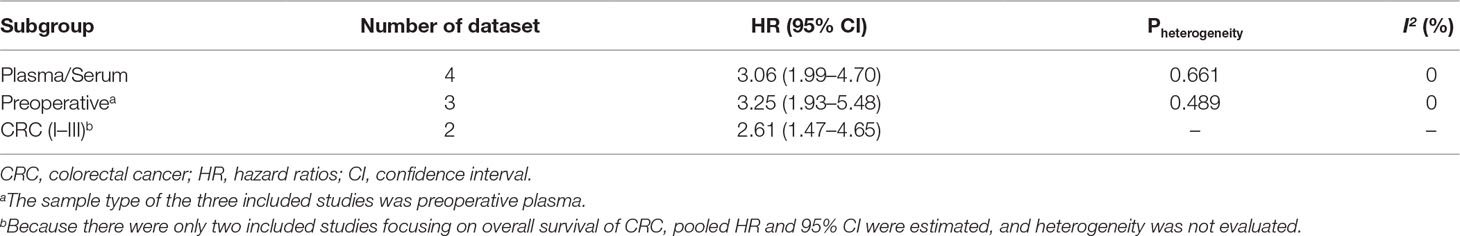
A total of seven studies including 786 cancer patients evaluated the association between mSEPT9 and OS (Dietrich et al., 2013; Kuo et al., 2014; Tham et al., 2014; Branchi et al., 2016; Jung et al., 2016; Schrock et al., 2017; Song et al., 2018). The heterogeneity test showed high heterogeneity among these studies (Pheterogeneity = 0.035, I2 = 53.6%). The pooled HR estimated by a random-effects model was 2.07 (95% CI = 1.40–3.06), suggesting that mSEPT9 was significantly associated with poor OS of cancer (Figure 2A). We further explored the prognostic role of mSEPT9 in specific subgroups (Table 3). Results revealed that patients with mSEPT9 detected in plasma or serum suffered reduced OS than those without (HR = 3.06, 95% CI = 1.99–4.70, Pheterogeneity = 0.661, I2 = 0%). Particularly, mSEPT9 detected in preoperative plasma indicated a 3.25-fold increased risk of worse survival (95% CI = 1.93–5.48, Pheterogeneity = 0.489, I2 = 0%). We also performed a pooled analysis to summarize data from two studies of nonmetastatic CRC (I–III) and found decreased OS in mSEPT9-positive patients (HR = 2.61, 95% CI = 1.47–4.65).
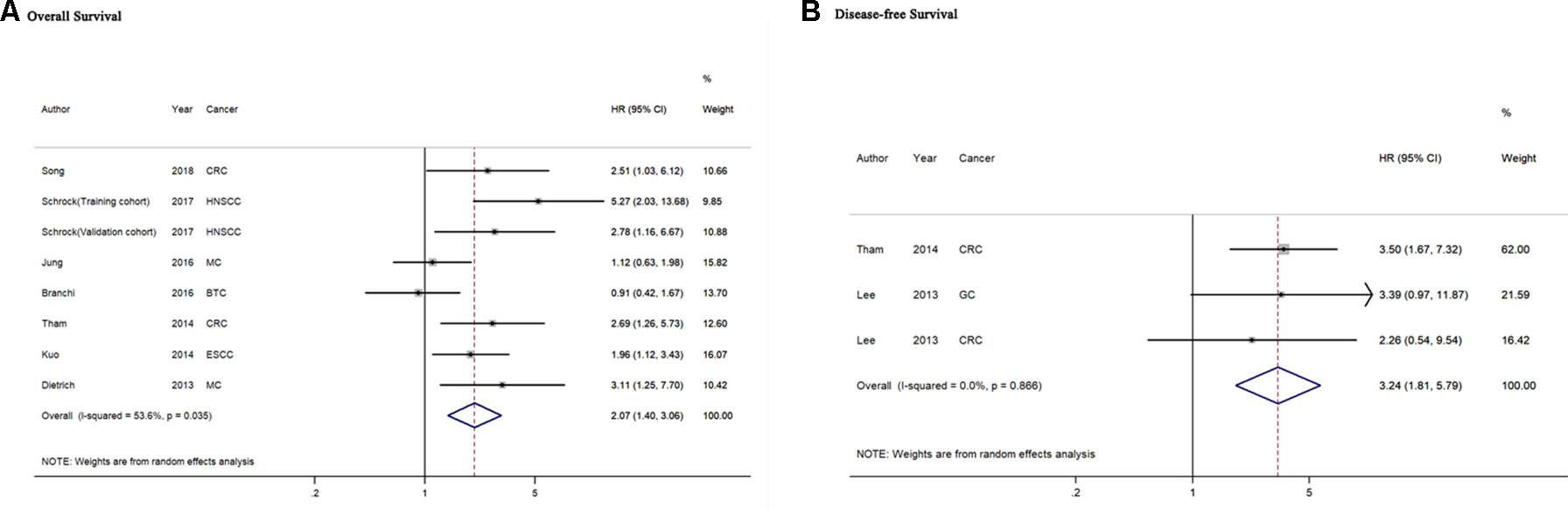
Figure 2 Forest plots for evaluation of the association between mSEPT9 and overall survival (A) or disease-free survival (B) in cancer patients. BTC, biliary tract carcinoma; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; HNSCC, head and neck squamous cell carcinoma; MC, multiple cancers.
Association Between mSEPT9 and DFS in Cancer Patients
Two included studies comprising three datasets of 371 cancer patients reported the association of mSEPT9with DFS of cancer(Lee et al., 2013; Tham et al., 2014). The heterogeneity test showed no heterogeneity among these studies (Pheterogeneity = 0.866, I2 = 0%). The pooled HR of the aforementioned studies was 3.24 (95% CI = 1.81–5.79), indicating that mSEPT9 predicted for worse DFS in cancer patients (Figure 2B). Subgroup analysis failed to be performed because of the limited number of relevant studies.
Association Between mSEPT9 and DSS/PFS in Cancer Patients
Only two studies reported the association of mSEPT9 with DSS in cancer patients (Angulo et al., 2016; Freitas et al., 2018). Angulo et al. identified that SEPT9 was hypermethylated in PC patients with a decreased DSS (HR = 7.64, 95% CI = 2.35–24.82) (Angulo et al., 2016). However, Freitas et al. reported that mSEPT9 independently indicated an increased DSS in CRC patients (HR = 0.67, 95% CI = 0.47–0.97), and specially, mSEPT9 was associated with a better DSS in colon cancer (HR = 0.47, 95% CI = 0.28–0.81) (Freitas et al., 2018).
For the mSEPT9’s prognostic role in PFS, Angulo et al. focusing on PC (HR = 2.52, 95% CI = 1.17–5.39) and Schrock et al. focusing on HNSCC (HR = 1.19, 95% CI = 1.10–1.56) both showed a significant association between mSEPT9 and poor PFS of patients (Angulo et al., 2016; Schrock et al., 2017).
Sensitivity Analyses and Publication Bias
Sensitivity analyses suggested that our pooled results were quite stable for both OS (Supplementary Figure S1A) and DFS (Supplementary Figure S1B). We observed a borderline significant publication bias in meta-analysis for OS (PEgger’s test = 0.048, PBegg’s test = 0.063). Therefore, we conducted a trim-and-fill analysis and found that despite publication bias, the adjusted pooled HR consistently demonstrated a significant association between mSEPT9 and OS (HR = 1.61, 95% CI = 1.09–2.38, Supplementary Figure S2). There was no obvious publication bias for meta-analysis for DFS (PEgger’s test = 0.443, PBegg’s test = 0.296).
Discussion
Several studies have investigated the association between mSEPT9 and prognosis in human cancers, but results are uncertain due to the limited sample size and various cancer types. Herein, we conducted a systematic review and meta-analysis and supported that mSEPT9 significantly predicted for worse cancer prognosis.
By systematic literature search, rigorous screening, and analysis, we identified that mSEPT9-positive cancer patients would suffer two-fold risk of decreased OS. Further subgroup analysis supported this result. Sensitivity analysis and trim-and-fill analysis guaranteed the robustness of our results. Specially, mSEPT9 detected in preoperative plasma significantly indicated a worse OS, implying a convenient and promising way to predict long-term survival of cancer patients. In addition, our meta-analysis also supported that mSEPT9 was significantly associated with poor DFS of cancer. Sensitivity analysis suggested that the result was stable, and Cochran’s Q test and I2 statistic did not indicate considerable heterogeneity. The aforementioned results all suggested that mSEPT9 could be a good prognostic biomarker for cancer patients. Traditionally, serum tumor markers (i.e., CEA, CA19-9) are used for screening and prognosis prediction, but their performance is still unsatisfactory. Previous studies have confirmed the excellent property of mSEPT9 in early diagnosis of several cancers and have clearly elucidated the potential mechanisms (Church et al., 2014; Koch et al., 2018; Pan et al., 2018). Now we provide evidence to support that mSEPT9 also could be a promising biomarker for cancer prognosis, which can be combined with traditional tumor biomarkers to greatly improve prognosis prediction in the future.
There were several limitations in our work. First, our results strongly supported that mSEPT9 could be a prognostic indicator of OS and DFS for human cancer, but there were not enough studies for subgroup analysis to fully clarify its impact on different cancer types, sampling times, and pathological stages. Second, there were only two included studies about DSS and PFS. The limited number of studies impeded us to conduct a meta-analysis to evaluate the impact of mSEPT9 on DSS and PFS. Last, some included studies did not provide multivariate-adjusted HRs, so we used unadjusted HRs instead. These unadjusted HRs were possibly influenced by potential confounders in the original studies. When we pooled them into a meta-analysis, the influence might be magnified and lead to a risk of bias on the pooled results. More studies with elaborate design should be conducted to verity our results and further explore more detailed impacts of mSEPT9 on cancer prognosis.
Conclusion
Our meta-analysis suggests that mSEPT9 could predict for worse OS and DFS in cancer patients. Specially, patients with detection of mSEPT9 in preoperative plasma would suffer significantly decreased OS of cancer. To the best of our knowledge, this is the first meta-analysis providing robust evidence that mSEPT9 could be a promising biomarker for cancer prognosis.
Data Availability
All datasets analyzed for this study are included in the manuscript and the Supplementary Files.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions
HX and YL designed the study and revised the manuscript. NS designed the study, summarized the data, and wrote the manuscript. TW, DL, and YZ performed literature search, collected data, and performed some analysis. All authors read and approved the final manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00887/full#supplementary-material
Supplementary Figure S1 | Sensitivity analysis for overall survival (A) and disease-free survival (B).
Supplementary Figure S2 | Begg's funnel plots of overall survival before (A) and after (B) performing trim-and-fill analysis.
Supplementary Table S1 | PRISMA 2009 Checklist.
Supplementary Table S2 | Quality evaluation of included studies by Newcastle-Ottawa Scale.
References
Angulo, J. C., Andres, G., Ashour, N., Sanchez-Chapado, M., Lopez, J. I., Ropero, S. (2016). Development of castration resistant prostate cancer can be predicted by a DNA hypermethylation profile. J. Urol. 195 (3), 619–626. doi: 10.1016/j.juro.2015.10.172
Begg, C. B., Mazumdar, M. (1994). Operating characteristics of a rank correlation test for publication bias. Biometrics 50 (4), 1088–1101. doi: 10.2307/2533446
Branchi, V., Schaefer, P., Semaan, A., Kania, A., Lingohr, P., Kalff, J. C., et al. (2016). Promoter hypermethylation of SHOX2 and SEPT9 is a potential biomarker for minimally invasive diagnosis in adenocarcinomas of the biliary tract. Clin. Epigenet. 8, 133. doi: 10.1186/s13148-016-0299-x
Chang, W. L., Lai, W. W., Kuo, I. Y., Lin, C. Y., Lu, P. J., Sheu, B. S., et al. (2017). A six-CpG panel with DNA methylation biomarkers predicting treatment response of chemoradiation in esophageal squamous cell carcinoma. J. Gastroenterol. 52 (6), 705–714. doi: 10.1007/s00535-016-1265-2
Church, T. R., Wandell, M., Lofton-Day, C., Mongin, S. J., Burger, M., Payne, S. R., et al. (2014). Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 63 (2), 317–325. doi: 10.1136/gutjnl-2012-304149
Connolly, D., Abdesselam, I., Verdier-Pinard, P., Montagna, C. (2011). Septin roles in tumorigenesis. Biol. Chem. 392 (8-9), 725–738. doi: 10.1515/BC.2011.073
de Vos, L., Gevensleben, H., Schrock, A., Franzen, A., Kristiansen, G., Bootz, F., et al. (2017). Comparison of quantification algorithms for circulating cell-free DNA methylation biomarkers in blood plasma from cancer patients. Clin. Epigenet. 9, 125. doi: 10.1186/s13148-017-0425-4
Dietrich, D., Jung, M., Puetzer, S., Leisse, A., Holmes, E. E., Meller, S., et al. (2013). Diagnostic and prognostic value of SHOX2 and SEPT9 DNA methylation and cytology in benign, paramalignant and malignant pleural effusions. PLoS One 8 (12), e84225. doi: 10.1371/journal.pone.0084225
Egger, M., Davey Smith, G., Schneider, M., Minder, C. (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ 315 (7109), 629–634. doi: 10.1136/bmj.315.7109.629
Freitas, M., Ferreira, F., Carvalho, S., Silva, F., Lopes, P., Antunes, L., et al. (2018). A novel DNA methylation panel accurately detects colorectal cancer independently of molecular pathway. J. Transl. Med. 16 (1), 45. doi: 10.1186/s12967-018-1415-9
Hall, P. A., Russell, S. E. (2004). The pathobiology of the septin gene family. J. Pathol. 204 (4), 489–505. doi: 10.1002/path.1654
Harris, R. J., Bradburn, M. J., Deeks, J. J., Harbord, R. M., Altman, D. G., Sterne, J. A. C. (2008). metan: fixed- and random-effects meta-analysis. Stata J. 8 (1), 3–28. doi: 10.1177/1536867X0800800102
Higgins, J. P., Thompson, S. G. (2002). Quantifying heterogeneity in a meta-analysis. Stat. Med. 21 (11), 1539–1558. doi: 10.1002/sim.1186
Higgins, J. P., Thompson, S. G., Deeks, J. J., Altman, D. G. (2003). Measuring inconsistency in meta-analyses. BMJ 327 (7414), 557–560. doi: 10.1136/bmj.327.7414.557
Jung, M., Putzer, S., Gevensleben, H., Meller, S., Kristiansen, G., Dietrich, D. (2016). Diagnostic and prognostic value of SHOX2 and SEPT9 DNA methylation and cytology in benign, paramalignant, and malignant ascites. Clin. Epigenet. 8, 24. doi: 10.1186/s13148-016-0192-7
Koch, A., Joosten, S. C., Feng, Z., De Ruijter, T. C., Draht, M. X., Melotte, V., et al. (2018). Analysis of DNA methylation in cancer: location revisited. Nat. Rev. Clin. Oncol. 15 (7), 459–466. doi: 10.1038/s41571-018-0004-4
Kuo, I. Y., Chang, J. M., Jiang, S. S., Chen, C. H., Chang, I. S., Sheu, B. S., et al. (2014). Prognostic CpG methylation biomarkers identified by methylation array in esophageal squamous cell carcinoma patients. Int. J. Med. Sci. 11 (8), 779–787. doi: 10.7150/ijms.7405
Lee, H. S., Hwang, S. M., Kim, T. S., Kim, D. W., Park, D. J., Kang, S. B., et al. (2013). Circulating methylated septin 9 nucleic acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl. Oncol. 6 (3), 290–296. doi: 10.1593/tlo.13118
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6 (7), e1000097. doi: 10.1371/journal.pmed.1000097
Nagata, H., Kozaki, K. I., Muramatsu, T., Hiramoto, H., Tanimoto, K., Fujiwara, N., et al. (2017). Genome-wide screening of DNA methylation associated with lymph node metastasis in esophageal squamous cell carcinoma. Oncotarget 8 (23), 37740–37750. doi: 10.18632/oncotarget.17147
Pan, Y., Liu, G., Zhou, F., Su, B., Li, Y. (2018). DNA methylation profiles in cancer diagnosis and therapeutics. Clin. Exp. Med. 18 (1), 1–14. doi: 10.1007/s10238-017-0467-0
Parmar, M. K., Torri, V., Stewart, L. (1998). Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat. Med. 17 (24), 2815–2834. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8
Perez-Carbonell, L., Balaguer, F., Toiyama, Y., Egoavil, C., Rojas, E., Guarinos, C., et al. (2014). IGFBP3 methylation is a novel diagnostic and predictive biomarker in colorectal cancer. PLoS One 9 (8), e104285. doi: 10.1371/journal.pone.0104285
Russell, S. E., Hall, P. A. (2011). Septin genomics: a road less travelled. Biol. Chem. 392 (8-9), 763–767. doi: 10.1515/BC.2011.079
Schrock, A., Leisse, A., de Vos, L., Gevensleben, H., Droge, F., Franzen, A., et al. (2017). Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: an observational prospective cohort study. Clin. Chem. 63 (7), 1288–1296. doi: 10.1373/clinchem.2016.270207
Song, L., Guo, S., Wang, J., Peng, X., Jia, J., Gong, Y., et al. (2018). The blood mSEPT9 is capable of assessing the surgical therapeutic effect and the prognosis of colorectal cancer. Biomark Med. 12 (9), 961–973. doi: 10.2217/bmm-2018-0012
Stang, A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25 (9), 603–605. doi: 10.1007/s10654-010-9491-z
Tham, C., Chew, M., Soong, R., Lim, J., Ang, M., Tang, C., et al. (2014). Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer 120 (20), 3131–3141. doi: 10.1002/cncr.28802
Tierney, J. F., Stewart, L. A., Ghersi, D., Burdett, S., Sydes, M. R. (2007). Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8, 16. doi: 10.1186/1745-6215-8-16
Verdier-Pinard, P., Salaun, D., Bouguenina, H., Shimada, S., Pophillat, M., Audebert, S., et al. (2017). Septin 9_i2 is downregulated in tumors, impairs cancer cell migration and alters subnuclear actin filaments. Sci. Rep. 7, 44976. doi: 10.1038/srep44976
Villanueva, A., Portela, A., Sayols, S., Battiston, C., Hoshida, Y., Mendez-Gonzalez, J., et al. (2015). DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 61 (6), 1945–1956. doi: 10.1002/hep.27732
Wang, Y., Chen, P. M., Liu, R. B. (2018). Advance in plasma SEPT9 gene methylation assay for colorectal cancer early detection. World J. Gastrointest. Oncol. 10 (1), 15–22. doi: 10.4251/wjgo.v10.i1.15
Keywords: cancer, Septin 9 (SEPT9), methylation, prognosis, biomarker, meta-analysis
Citation: Shen N, Wang T, Li D, Zhu Y, Xie H and Lu Y (2019) Hypermethylation of the SEPT9 Gene Suggests Significantly Poor Prognosis in Cancer Patients: A Systematic Review and Meta-Analysis. Front. Genet. 10:887. doi: 10.3389/fgene.2019.00887
Received: 18 February 2019; Accepted: 22 August 2019;
Published: 19 September 2019.
Edited by:
John Frederick Pearson, University of Otago, New ZealandReviewed by:
Parvin Mehdipour, Tehran University of Medical Sciences, IranHaiwei Mou, Cold Spring Harbor Laboratory, United States
Copyright © 2019 Shen, Wang, Li, Zhu, Xie and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanjun Lu, anVueWFubHVfMjAwMEAxNjMuY29t; Huaping Xie, cmFpbmVyeGlAMTI2LmNvbQ==
 Na Shen
Na Shen Ting Wang1
Ting Wang1 Yanjun Lu
Yanjun Lu