- 1Graduate School of Regional Innovation Studies, Mie University, Tsu, Japan
- 2Mie University Zebrafish Drug Screening Center, Tsu, Japan
- 3Department of Integrative Pharmacology, Mie University Graduate School of Medicine, Tsu, Japan
- 4Department of Bioinformatics, University Advanced Science Research Promotion Centre, Tsu, Mie, Japan
- 5Department of Molecular Physiology & Biophysics, Vanderbilt University School of Medicine, Nashville, TN, United States
- 6Department of Medical Biochemistry, School of Pharmaceutical Sciences, University of Shizuoka, Shizuoka, Japan
- 7Department of Frontier Materials, Graduate School of Engineering, Nagoya Institute of Technology, Nagoya, Japan
- 8Department of Research Support Platform, Graduate School of Medicine, Osaka City University, Osaka, Japan
Type 2 diabetes mellitus (T2DM) is characterized by persistent hyperglycemia and is influenced by genetic and environmental factors. Optimum T2DM management involves early diagnosis and effective glucose-lowering therapies. Further research is warranted to improve our understanding of T2DM pathophysiology and reveal potential roles of genetic predisposition. We have previously developed an obesity-induced diabetic zebrafish model that shares common pathological pathways with humans and may be used to identify putative pharmacological targets of diabetes. Additionally, we have previously identified several candidate genes with altered expression in T2DM zebrafish. Here, we performed a small-scale zebrafish screening for these genes and discovered a new therapeutic target, centromere protein X (CENPX), which was further validated in a T2DM mouse model. In zebrafish, cenpx knockdown by morpholino or knockout by CRISPR/Cas9 system ameliorated overfeeding-induced hyperglycemia and upregulated insulin level. In T2DM mice, small-interfering RNA-mediated Cenpx knockdown decreased hyperglycemia and upregulated insulin synthesis in the pancreas. Gene expression analysis revealed insulin, mechanistic target of rapamycin, leptin, and insulin-like growth factor 1 pathway activation following Cenpx silencing in pancreas tissues. Thus, CENPX inhibition exerted antidiabetic effects via increased insulin expression and related pathways. Therefore, T2DM zebrafish may serve as a powerful tool in the discovery of new therapeutic gene targets.
Introduction
Along with the epidemic of obesity, there is a parallel increase in the prevalence of diabetes worldwide (Haffner, 2006). Type 2 diabetes mellitus (T2DM) is characterized by persistent hyperglycemia and is primarily caused by pancreatic β-cell dysfunction and insulin resistance in target organs (Chatterjee et al., 2017). It is a complex disorder, and both genetic components, as well as environmental factors, are known to contribute to its pathogenesis (Ding et al., 2015). T2DM is regarded as an inevitably progressive condition with irreversible failure of beta cell function. Initiation of intensive glycemic control measures early in the diabetes course may be associated with a reduction of cardiovascular complications (Group et al., 2008). However, even tighter glucose control does not reduce the incidence of macrovascular diseases, such as nephropathy in patients with T2DM (Ukpds, 1998). Prevention of T2DM is a lifetime task and requires an integrated approach implemented right from the origin of the disease. Therefore, early detection and intervention of impaired glucose tolerance, the preliminary stage of T2DM, have attracted attention as a prophylactic treatment strategy.
T2DM is caused by the alterations of various functional genes owing to their own partial and additive effects, leading to a complex inheritance pattern (Tusie Luna, 2005). The identification of susceptible genes and genetic variants necessitates different methodological approaches. Over the past decade, hundreds of T2DM susceptible loci were captured using genome-wide association studies (GWAS) (Morris et al., 2012; Flannick and Florez, 2016). However, some researchers questioned the biological relevance of these variants in disease or their clinical utility for prognosis or treatment (Goldstein, 2009; Mcclellan and King, 2010). Transcriptomics, or the study of the complete set of RNA transcripts produced by the genome (e.g., via RNA sequencing), is a powerful tool to explore the association between genes and T2DM and through the correlation between the genotype and phenotype (Jenkinson et al., 2016). The strategies exploiting transcriptomics and animal models are expected to discover the novel genetic factors of diabetes that may be crucial for the identification of risk factors, diagnostic indicators (biomarkers), and new therapeutic targets.
Zebrafish is a well-established and widely used model system (Dooley and Zon, 2000) that offers several advantages, including characteristics of vertebrates, high fecundity, rapid development, transparent body in the embryo and larval stage, and a high degree of genetic, anatomical, and physiological similarities to humans. Zebrafish has been traditionally used in developmental biology (Kishi, 2011). In the last decade, it has become a valuable tool to model human diseases, including metabolic diseases (Santoriello and Zon, 2012; Zang et al., 2018). Three approaches are used for the development of a zebrafish obesity model, including an induced model, transgenic line, and mutant line. The induced model is the most common approach, which is carried out by treating zebrafish with a high-fat diet or over-nutrition (Zang et al., 2018). Researchers have designed high-fat zebrafish diets containing chicken egg yolk, vegetable oil, or lard and successfully generated obese zebrafishes (Meguro et al., 2015; Landgraf et al., 2017). Overfeeding with Artemia may also serve as a convenient method to induce obesity in zebrafish (Oka et al., 2010; Tainaka et al., 2011; Hiramitsu et al., 2014; Zang et al., 2014; Zang et al., 2015b). Additionally, zebrafish models of T2DM are also developed with glucose immersion, over-nutrition, and genetic modification (Zang et al., 2018). We have recently created a zebrafish model for both obesity and T2DM by over-nutrition and demonstrated its application in studying human diabetes (Zang et al., 2017). We further performed RNA deep sequencing (RNA-Seq) analysis and identified several candidate genes with altered expression patterns in T2DM zebrafish; these observations have not been reported in studies with humans and rodents (Zang et al., 2017). In the present study, we first carried out a small-scale knockdown screening of these candidate genes using morpholino (MO) antisense oligos to identify their relationships with T2DM. We next performed knockout and knockdown studies in zebrafish and mouse models of T2DM and elucidated that centromeric protein X (CENPX) may be a potential therapeutic target in diabetes.
Materials and Methods
Zebrafish Strains and Maintenance
AB strain and Tg(−1.0ins:EGFP)sc1 strain (referred to as ins-EGFP) (Diiorio et al., 2002) (the Zebrafish International Research Centre, Eugene, OR, USA), and a skeletal muscle insulin-resistant zebrafish transgenic line (zMIR; Tg(acta1:dnIGF1R-EGFP)) (Maddison et al., 2015) were maintained at 28°C with a light/dark cycle of 14/10 h (Westerfield, 2007).
Over-Nutrition-Induced T2DM Zebrafish Model
The T2DM zebrafish model was induced as previously described (Zang et al., 2017). In brief, male healthy adult zebrafishes (4–6 months old) were assigned to either an overfeeding (diet-induced obesity; DIO) or a control group (non-DIO). DIO zebrafishes were fed 120 mg/fish of Otohime B2 (Marubeni Nisshin Feed, Tokyo, Japan) each day (divided into six feedings), whereas non-DIO zebrafishes were fed 20 mg/fish once a day every day. The body weight was measured weekly during the overfeeding treatment. At the experimental endpoint, the blood sample was collected and the fasting blood glucose (FBG) level was measured as previously described (Zang et al., 2013; Zang et al., 2015a).
Morpholino Treatment
The sequences of antisense vivo-morpholino (MO; Gene Tools, Philomath, OR, USA) for cenpx and hmox1 are provided in Supplementary Table S1. MO was intraperitoneally injected at a dose of 15 mg/kg fish weight twice a week during the 8-week experiment period. Additional methods details are given in the Supplementary Material. For the quantitative analysis of insulin level, ins-EGFP zebrafishes were treated with cenpx vivo-MO for 4 weeks. EGFP signaling was quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA) as previously described (Zang et al., 2017).
cenpx sgRNA Microinjection
Zebrafish cenpx mutants were generated using a vector-free method of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 mutagenesis, with some modifications in a previously described report (Varshney et al., 2015). Additional methods details are given in the Supplementary Material. For sgRNA microinjection, zMIR strain embryo was injected with approximately 1 nl solution containing 40–50 pg of cenpx sgRNA, 250 pg of Cas9 protein (PNA Bio, CA, USA), and phenol red at single-cell stage. Injected embryos were raised to adulthood and the adult zebrafishes were assigned to either an overfeeding or a control group. The overfeeding experiment was performed as described above.
Mouse Experiments
Ten-week-old male NSY/Hos mice, an inbred animal model with spontaneous development of T2DM, were purchased from SLC (Shizuoka, Japan). The mice were housed in groups of five in one cage and fed with basal diet (CE-7; CLEA Japan, Tokyo, Japan) or high-fat diet (HFD; 58Y1, Test Diet, Richmond, IN, USA) to induce obesity and diabetes. During the feeding experiment, body weight was measured once a week. Mice were kept fasting for 14 h before withdrawing blood samples for the measurement of fasting levels of blood glucose.
Preparation of Small-Interfering RNA (siRNA)-Lipoplexes and In Vivo Gene Silencing
The siRNA against mouse Cenpx and a scrambled siRNA (negative control) were purchased from Hokkaido System Science Co. (Hokkaido, Japan). The sequences of siRNAs are shown in Supplementary Table S1. The siRNAs were entrapped between the lipid layers of multilayered liposomes by freeze-thawing of lipoplexes comprising polycation liposomes (PCLs) and siRNAs, as previously described (Koide et al., 2016). Freeze-dried siRNA-lipoplexes were diluted with distilled water, and 200 µl of siRNA-lipoplexes (containing 50 µg of siRNAs) or vehicle (0.3 M sucrose) was injected via the tail vein to each NSY/Hos mouse. The siRNA was administered twice per week for 4 weeks. At the end of the experiment, the mice were euthanized with CO2 gas, and liver and pancreas tissues were harvested for immunohistochemistry (IHC) and quantitative polymerase chain reaction (qPCR) analyses.
Fluorescent Immunohistochemistry Staining
Mouse pancreas tissues were fixed with 4% paraformaldehyde and embedded in paraffin. Sections (4 µm) were rehydrated and subjected to antigen retrieval by heating in 0.5% Immunosaver (Nisshin EM, Tokyo, Japan) at 98°C for 45 min. The sections were incubated with a primary antibody against insulin (1:200; Abcam, ab7842, Cambridge, UK) overnight at 4°C. After rinsing with Tris-buffered saline (TBS), the sections were incubated with a fluorescence-conjugated secondary antibody (Goat Anti-Guinea pig IgG H&L, Alexa Fluor® 488; Abcam, ab15015) at room temperature (20°C) for 1 h and mounted with Prolong Glass Antifade Mountant (Abcam, P36984). Images were captured using a BZ-X710 fluorescence microscope (GFP filter; Keyence, Tokyo, Japan). The fluorescence signal of insulin was quantified and normalized with the related islets of Langerhans area using ImageJ.
Insulin Signaling Pathway Analysis by a qPCR Array
Total RNA purification and cDNA synthesis from mouse pancreas tissues were performed as previously described (Zang et al., 2017). A PCR screening array specific for mouse insulin signaling was used (RT2 Profiler PCR Array; QIAGEN PAMM-030ZC) to examine the expression of 89 genes (including five housekeeping genes; Supplementary Table S2). qPCR analysis was performed with ABI StepOnePlus Real-Time PCR System using SYBR Green. Relative gene expression was determined using the ΔCT calculation method. After statistical tests, we performed sub-network enrichment analysis using Pathway Studio 9.0 (Elsevier, Amsterdam, Netherlands) according to our previous study (Zang et al., 2017).
Statistical Analysis
All results are presented as means with their standard errors (SEs). Data were analyzed with GraphPad Prism. P-value ≤ 0.05 was considered statistically significant. Statistical analyses were carried out by Student’s t-test or one-way analysis of variance (ANOVA) with the Bonferroni–Dunn multiple comparison procedure, depending on the number of comparisons.
Results
MO-Mediated Knockdown Screening Identifies cenpx as a Novel Therapeutic Candidate in Diabetic Zebrafish
Based on the altered genes expression in hepatopancreas tissues of T2DM zebrafish (Zang et al., 2017), we performed qPCR analysis on the selective target genes for the subsequent experiments (Supplementary Table S3). We next performed knockdown screening to AB strain zebrafish using vivo-MO to investigate candidates that could reverse blood glucose elevation in T2DM fish. As a result, the knockdown of the genes encoding centromeric protein X (cenpx) and heme oxygenase 1 (hmox1) significantly (P < 0.05) reduced the overfeeding-induced elevation in blood glucose level (Supplementary Figure S1). We focused on cenpx and validated its potential as a novel therapeutic candidate gene for diabetes and homx1 was used as a positive control (Jais et al., 2014).
The T2DM model zebrafish we developed previously was used as wild-type AB strain (Zang et al., 2017). The highest FBG level was approximately 95 ± 9 mg/dl. However, zMIR, a transgenic line of insulin resistance (Maddison et al., 2015), shows higher FBG levels (171 ± 39 mg/dl) after overfeeding to induce T2DM compared with that of the AB strain (P < 0.05). For this reason, we selected zMIR as a strong T2DM zebrafish model in the following experiments. We next confirmed the expression of cenpx in wild-type (AB) and zMIR T2DM fishes after overfeeding. As shown in Figure 1A, the relative mRNA expression level of cenpx in AB and zMIR overfed zebrafish (DIO) was 2.5- and 3-fold higher as compared with their corresponding normally fed zebrafishes (non-DIO), respectively. The basal expression levels of cenpx in the non-DIO zMIR group had no statistically significant difference when compared with non-DIO AB group.
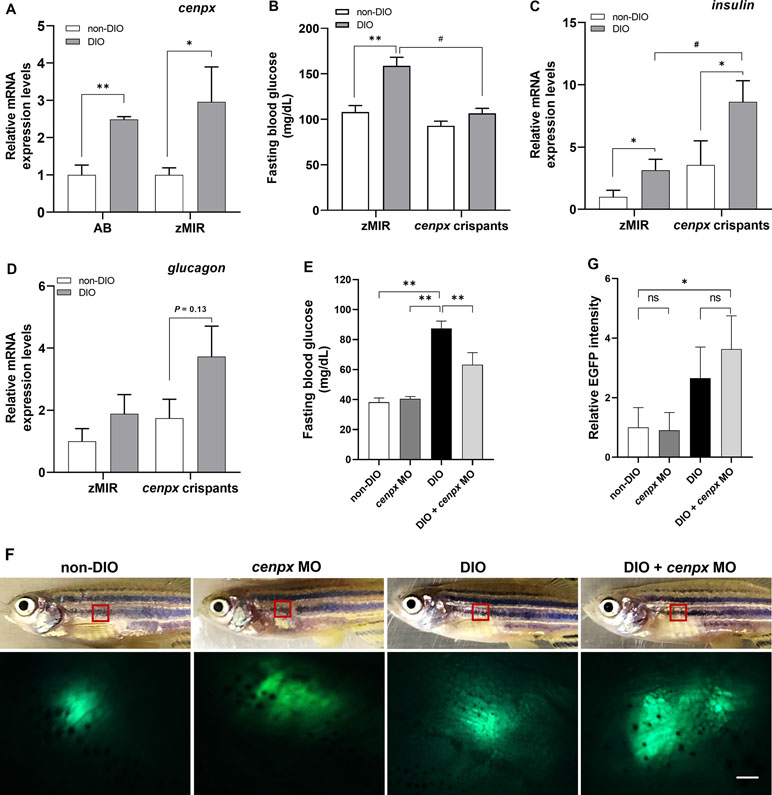
Figure 1 Deletion of cenpx gene suppressed hyperglycemia in diabetic zebrafish. (A) The relative mRNA level of cenpx in the hepatopancreas tissue of AB and zMIR zebrafish. Data are shown as relative levels of cenpx mRNA of AB and zMIR strains to that in their corresponding non-DIO controls. n = 5. *P < 0.05, **P < 0.01 versus the non-DIO group. (B) The fasting blood glucose levels in zMIR zebrafish and cenpx crispants after overfeeding treatment. n = 5. **P < 0.01 versus the non-DIO group; #P < 0.05 versus the DIO group. (C, D) The mRNA levels of insulin and glucagon in cenpx crispants. RNA was isolated from the hepatopancreas of zebrafishes from overfeeding and normal feeding groups. The mRNA levels of insulin and glucagon were compared with that of amylase as a marker for the endocrine pancreas. Data are shown as the relative expression level of insulin and glucagon mRNA to that in non-DIO zMIR. n = 5. *P < 0.05 versus the non-DIO group; #P < 0.05 versus the DIO zMIR group. (E) Fasting blood glucose levels in MO-treated ins-EGFP zebrafish after 4 weeks of normal feeding or overfeeding. Fishes were treated with cenpx MO twice a week. n = 5. **P < 0.01 versus the DIO group; (F) Upper panel are the bright field of ins-EGFP zebrafishes with or without i.p. injection of cenpx MO in DIO. Lower panel are EGFP levels in the exocrine pancreas areas that were monitored by fluorescence stereoscopic microscopy. The red box marks the region displayed in the fluorescence image. Scale bar = 0.5 mm. (G) Graph of relative EGFP intensities from lower panel of F. n = 5. *P < 0.05 versus the non-DIO group.
Knockout of cenpx Ameliorates Diabetes in Zebrafish and Upregulates Insulin Level
To investigate the relationship between cenpx and hyperglycemia, we designed and synthesized three single guide RNAs (sgRNAs) targeted to cenpx to generate gene disruption in F0 zebrafish crispants. After microinjection of the sgRNAs combined with Cas9 protein into zMIR strain, the genome DNA from 24 hpf (hour post fertilization) embryos was extracted and examined for genome editing activity using a heteroduplex mobility assay (Ota et al., 2013). The most effective sgRNA that triggered mutations was sgRNA1 (Supplementary Figure S2). We then selected cenpx sgRNA1 to produce F0 founders. These fishes were raised till the adult stage and then overfed for 4 weeks along with un-injected zMIR zebrafish. The un-injected zMIR zebrafish showed a significant increase in the FBG level (Figure 1B). On the contrary, cenpx crispants showed no elevation in FBG level as compared with the fish on a normal diet. Real-time qPCR analysis using the RNA isolated from the hepatopancreas of zMIR revealed a significant increase in the mRNA level of the insulin gene in the overfed zMIR zebrafish as compared with the fish fed with a normal diet (P < 0.05, Figure 1C). Analysis of the insulin mRNA levels in cenpx crispants showed a statistically significant increase of 2.4-fold in DIO cenpx crispants when compared with that in the non-DIO cenpx crispants (P < 0.05). We also found a significantly higher insulin expression level in DIO cenpx crispants than in the DIO zMIR group (P < 0.05). Additionally, the glucagon expression showed no statistically significant difference in DIO cenpx crispants compared with non-DIO cenpx crispants (P = 0.13; Figure 1D).
To confirm the overproduction of insulin following silencing of the cenpx, we performed intraperitoneal injection of cenpx vivo-MO in the ins-EGFP zebrafish twice a week for 4 weeks and monitored EGFP expression in the pancreas using fluorescence stereoscopic microscopy. The increase in blood glucose levels induced by overfeeding was suppressed upon cenpx MO treatment (P < 0.01), consistent with the observation in AB strain (Figure 1E). The EGFP-positive areas in the pancreas increased in cenpx MO-injected DIO group as compared with the non-DIO group (P < 0.05; Figures 1F, G). There was no significant difference between non-DIO and cenpx MO groups on a non-DIO background, or between DIO and DIO + cenpx MO group.
Knockdown of Cenpx Ameliorates Hyperglycemia in Mouse T2DM Model
We conducted knockdown of Cenpx using siRNA technique in NSY/Hos mice, a spontaneous model of T2DM (Ueda et al., 1995). We designed and synthesized three siRNAs directed against mouse Cenpx gene and investigated the knockdown efficiency in vitro using Hepa 1-6 cells (Supplementary Figure S3). Real-time qPCR analysis showed the siRNA2 effectively inhibited the expression of Cenpx, and therefore siRNA2 was selected for further in vivo experiments. For positive control, Hmox1 siRNA was synthesized according to a previous report (Zhang et al., 2004). siRNA with a scrambled sequence based on Cenpx siRNA was designed and used as the negative control. For siRNA knockdown experiment, mice were fed a normal diet (ND) and a high fat-diet (HFD) and administered with scrambled-, Cenpx-, and Hmox1-siRNAs for 4 weeks. No significant differences in FBG levels and Cenpx mRNA levels in pancreas tissue were found between ND and ND with scrambled siRNA treatment groups, and between HFD and HFD with scrambled siRNA treatment groups (Supplementary Figure S4). The three siRNA-administrated groups fed with HFD exhibited a significant (P < 0.05) increase in body weight as compared with the ND group. No significant difference in body weight was observed between scrambled-, Cenpx-, and Hmox1-siRNA treatment groups (Figure 2A). The FBG levels significantly increased in scrambled siRNA-injected mice fed with HFD (230 ± 10 mg/dl) as compared with the mice in the ND group (146 ± 9 mg/dl) (Figure 2B). In contrast, the FBG levels of Cenpx siRNA- and Hmox1 siRNA-injected mice fed with HFD were 178 ± 11 and 170 ± 8 mg/dl, respectively, which showed a significant decrease compared to that of scrambled siRNA-injected mice fed with HFD (P < 0.05). Quantitative PCR analysis revealed that Cenpx siRNA significantly inhibited the Cenpx mRNA expression level in the pancreas (Figure 2C); however, no silencing effect was observed in the liver tissue (Supplementary Figure S5).
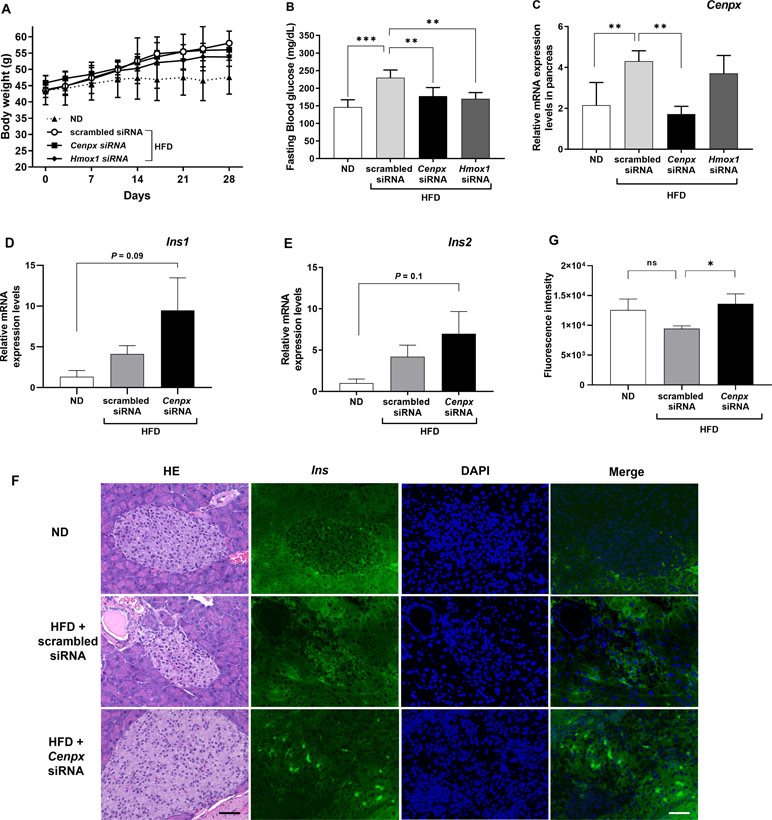
Figure 2 Silencing of Cenpx ameliorates elevated blood glucose levels in a diabetes mouse model. (A) Changes in body weight. (B) Changes in fasting blood glucose levels. (C) Relative mRNA levels in the pancreas from the HFD mice injected with scrambled-, Cenpx-, and Hmox1-siRNAs. The mRNA levels of (D) Insulin 1 (Ins1) and (E) Insulin 2 (Ins2) in HFD mouse treated with Cenpx-siRNA (n = 5). (F) H&E and fluorescent IHC staining for Insulin in ND and HFD mice injected with scrambled siRNA and Cenpx siRNA in the pancreas tissue. Scale bar = 50 µm. (G) Quantification of the fluorescence intensity of insulin in the islets of Langerhans, which was normalized by the related area. n = 10; ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001 versus the scrambled siRNA group.
Consistent with the results observed in the zebrafish model (Figure 1C), the expression of the gene encoding insulin 1 (Ins1) had a trend towards upregulation and that of insulin 2 (Ins2) had no change after Cenpx siRNA administration in the mouse pancreas compared with that of the ND group (P = 0.09 and P = 0.1, respectively; Figure 2D, E). There was no statistically significant difference between scrambled siRNA treated HFD mice and Cenpx siRNA treated HFD mice. The results of fluorescent IHC staining using insulin antibody are shown in Figure 2F. We quantified the fluorescent signal of insulin in each islet of Langerhans and normalized with the related area (Figure 2G). No significant difference was found in insulin level between ND and scrambled siRNA group with HFD treatment. However, we observed a significant increase in the expression of insulin protein in the pancreas tissue of mice from Cenpx siRNA administration HFD group as compared with the mice from the scrambled siRNA HFD group (P < 0.05).
Gene Expression Analysis in the Pancreas of Cenpx Knockdown T2DM Mice
Gene expression analysis using a qPCR array for the insulin signaling pathway was performed to investigate the mechanism in the pancreas tissues of Cenpx siRNA-injected NSY/Hos mice fed with a high-fat diet. Of 84 genes, 17 and 20 genes were upregulated (>1.5-fold) and downregulated (<0.75-fold), respectively, following knockdown of Cenpx by siRNA in T2DM mice as compared with the HFD treated control mice (T2DM mice) (Table 1). We performed a sub-network enrichment analysis for differentially expressed target genes (Kotelnikova et al., 2010) and found several pathways that were altered after Cenpx knockdown (Supplementary Table S4). The top four pathways, including insulin, mammalian target of rapamycin complex 1 (mTOR), leptin, and insulin-like growth factor 1 (IGF1) are shown in Figure 3. Of these, INSULIN signaling pathway was upregulated by knockdown of Cenpx in T2DM mice as compared with the T2DM mice (Supplementary Figure S6A).
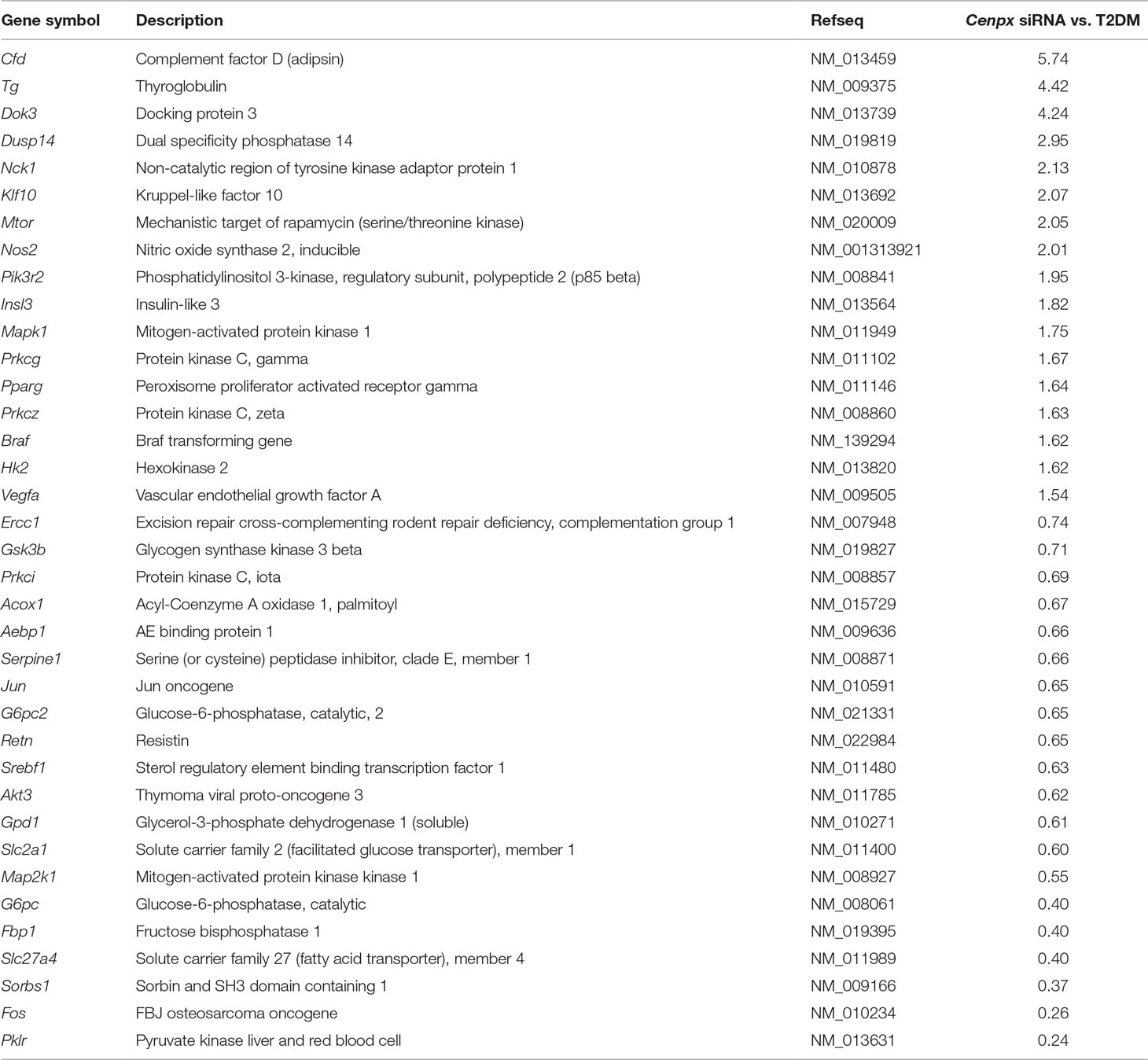
Table 1 Fold change in the levels of gene expression in pancreas of Cenpx siRNA-treated T2DM mouse compared to those in pancreas of a T2DM control mouse.
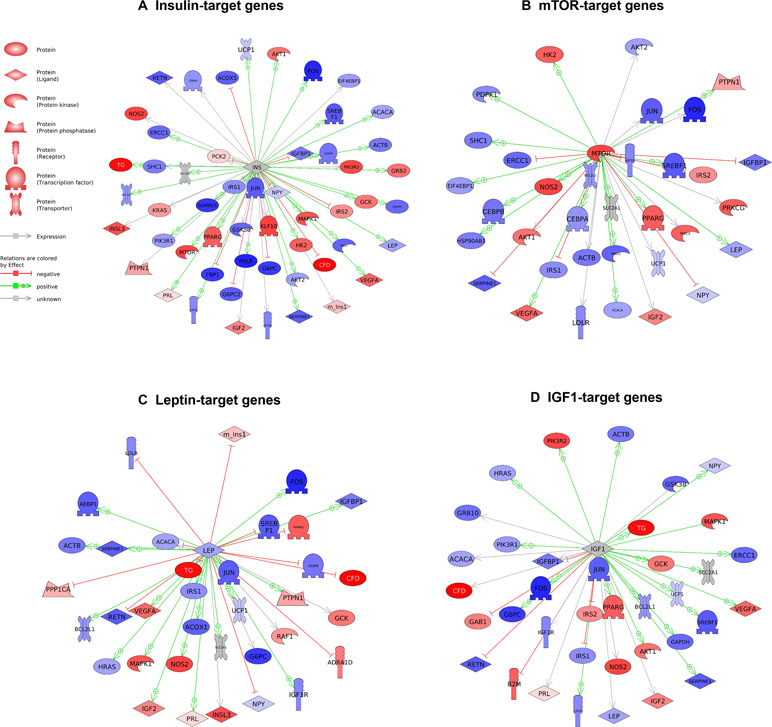
Figure 3 Pathways activated by Cenpx knockdown in the mouse pancreas tissue. (A) Insulin-target gene pathway. (B) mTOR-target gene pathway. (C) Leptin-target gene pathway. (D) IGF1-target gene pathway. Red and blue denote genes with increased and decreased expression, respectively, in Cenpx knockdown in HFD mice as compared with control HFD mice. Grey indicates the genes that were undetected in the gene expression analysis.
Discussion
Knockdown of Cenpx by siRNA in mice significantly decreased the levels of the Cenpx mRNA in the pancreas, but there was no change in the liver tissue (Figure 2C and Supplementary Figure S5). These data suggest the specificity of siRNA for pancreatic tissue, most likely due to pharmacokinetic properties of siRNA. Huang and colleagues have reported that intravenously administered siRNA exhibits remarkable accumulation in the glandular tissues, including the pancreas (Huang et al., 2016). The half-life of siRNA in the pancreas was 2.3-fold longer than that in liver tissue (30.3 h vs. 13 h, respectively). Therefore, to sustain the effect of siRNA-mediated knockdown, we administered the siRNA twice per week, and the effects of Cenpx siRNA in the liver were hardly observed due to a shorter half-life of siRNA in liver than pancreas tissues.
Current treatment modalities for T2DM mainly focus on lowering blood glucose level via lifestyle modification or anti-diabetic drugs that stimulate insulin secretion from the pancreas or enhance the peripheral tissue sensitivity (Chatterjee et al., 2017). However, patients with diabetes may require multiple medications to avoid chronic hyperglycemia-caused complications for considerable heterogeneity in the therapeutic response (Maccallum, 2014). Therefore, clarification of the onset mechanism of T2DM and identification of novel therapeutic genes are desirable for more effective and safer treatment of this disease. GWAS is a primary approach for the identification of genetic variants associated with disease risk (Mccarthy et al., 2008). In the past decade, 144 genetic variants at 129 susceptibility loci have been reported to be associated with T2DM (Morris et al., 2012; Flannick and Florez, 2016). However, only a small proportion (∼10%) from this large number of T2DM-associated variants identified by GWAS could explain the heritability of T2DM, suggesting that much of the variants are still undiscovered. Recently, with the high cost and low clinical utility of GWAS (Goldstein, 2009; Mcclellan and King, 2010), new strategies that could identify the causal variants of T2DM were proposed, including next-generation sequencing (NGS) studies and/or post-GWAS experiments, including gene function study, animal models, and clinical studies (Mcclellan and King, 2010). Here, we combined transcriptomic study (NGS), gene function study (gene expression analysis, CRISPR/Cas9, and RNA interference technique) and animal models to discover new predictors of T2DM development. As a result, CENPX was identified as a novel therapeutic target for T2DM. Our results demonstrate the suitability of this combinational genetic study for new drug target discovery in T2DM and resolution of the questions disputed with GWAS.
CENPX, also referred to as FANCM-interacting histone-fold protein 2 (MHF2), is localized at the centromeres throughout the cell cycle (Amano et al., 2009). It was recently classified as an integral component of the Fanconi anemia (FA) core complex, which is required for DNA damage response and repair (Yan et al., 2010). CENPX is a component of three different complexes, namely: 1) CENPS-CENPX complex, essential for the stable assembly of the outer kinetochore (Amano et al., 2009; Perpelescu and Fukagawa, 2011); 2) CENP-T-W-S-X complex, that binds to supercoiled DNA and plays an important role in kinetochore assembly (Nishino et al., 2012); 3) CENPX and CENPS, Fanconi Anemia M (FANCM)-associated proteins (Singh et al., 2010; Yan et al., 2010; Nishino et al., 2012). MHF1 (CENPS) and MHF2 form MHF1–MHF2 heterodimers and interact with each other to form an (MHF1–MHF2)2 tetramer that preferentially binds to branched DNA and interacts with FANCM to facilitate the repair of branched DNA (Fox et al., 2014; Zafar et al., 2017). Taken together, CENPX is essential to protect genome stability and plays a role in DNA damage repair. Although DNA damage is thought to be a major contributing factor for T2DM, little is known about the potential contribution of DNA repair disturbance in diabetes (Blasiak et al., 2004; Shimizu et al., 2014). In T2DM, evidence of increased oxidative DNA damage and decreased DNA repair activity has been reported (Tabak et al., 2011; Manoel-Caetano et al., 2012). However, our results revealed an increase in cenpx expression level in diabetic zebrafish, and the knockdown or knockout of Cenpx expression exerts a protective effect in diabetic zebrafish and mouse models. We hypothesize that CENPX may participate in T2DM by altering the expression of the genes responsible for glucose homeostasis rather than performing any role in DNA repair. For instance, insulin action may be negatively regulated by CENPX under a hyperglycemic state. Therefore, CENPX deletion (or reduced expression) may result in the upregulation of insulin expression and consequently activation of downstream pathways.
To clarify our hypothesis, we predicted the therapeutic pathways induced by Cenpx knockdown in the pancreas tissue of T2DM mice using qPCR array. We found insulin signaling to be a major pathway (Figure 1) as per the results observed in zebrafish, though other obesity- or diabetes-related pathways may also be affected following Cenpx knockdown. The mTOR pathway, the second most affected pathway (Supplementary Table S4), is known to form two distinct multiprotein complexes known as mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) (Sengupta et al., 2010; Oh and Jacinto, 2011). It is shown to be highly involved in insulin signaling (Yoon, 2017). The mTOR signaling pathway is dysregulated in human T2DM (Um et al., 2004; Newgard et al., 2009), and the loss of mTORC1 signaling results in the impairment of β-cell homeostasis and insulin processing (Blandino-Rosano et al., 2017). Intriguingly, we observed that the EIF4EBP2 pathway acting downstream of mTORC1, which is also a known regulator of beta cell proliferation in the pancreas, was also upregulated after Cenpx knockdown in T2DM mice (Supplementary Figure S6B) (Blandino-Rosano et al., 2017). Consistent with these results in mice, cenpx knockdown (morphant) increased the number of beta cells in zebrafish (Figure 1). Leptin and IGF1 pathways are strongly connected with insulin signaling (Hakuno and Takahashi, 2018; Marques-Oliveira et al., 2018) and were upregulated after Cenpx knockdown in T2DM mice. These transcriptome results are reasonably associated with the phenotypes of CENPX knockdown mice, indicating that the pathophysiological contribution of CENPX would be common in all vertebrate animals. However, the direct relationship between CENPX expression and these pathways is still unclear. Future studies are warranted to confirm these findings to provide a better understanding of the research in this field.
In the present study, we discovered CENPX as a new therapeutic target against T2DM using zebrafish and mouse models. We performed cenpx knockout experiments using CRISPR/Cas9 genome editing technology in T2DM zebrafish and Cenpx knockdown experiments using siRNA technique in the T2DM mouse model. Inhibition of CENPX expression in zebrafish and mouse diabetic models led to the amelioration of hyperglycemia through the induction of insulin secretion. To the best of our knowledge, no report has been published on the assessment of expression or activity of CENPX in obesity, glucose homeostasis, and diabetes mellitus. Our findings highlight CENPX inhibition as a potential therapeutic strategy for diabetes control.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
All animal procedures were approved by the Ethics Committee of Mie University and were performed according to the Japanese Animal Welfare Regulation “Act on Welfare and Management of Animals” (Ministry of Environment of Japan) and complied with international guidelines.
Author Contributions
LZ and YS conceived and designed the research; performed the experiments; analyzed the data; interpreted the results of the experiments; prepared the figures; and drafted, edited, revised, and approved the final version of the manuscript. HN, AO, HK, NO, DT, and MS performed the experiments and analyzed the data. WC revised and approved the final version of the manuscript. NN takes sole responsibility for the integrity, accuracy, and analysis of the data, and has full access to the data.
Funding
This work was supported by JSPS KAKENHI (grant numbers 15K19074, 15KK0305, and 18K08240).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Ms. Masako Inoue for handling and care of zebrafish, and Ms. Takako Taguchi and Ms. Azusa Kato for their secretarial assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00693/full#supplementary-material
References
Amano, M., Suzuki, A., Hori, T., Backer, C., Okawa, K., Cheeseman, I. M., et al. (2009). The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 186, 173–182. doi: 10.1083/jcb.200903100
Blandino-Rosano, M., Barbaresso, R., Jimenez-Palomares, M., Bozadjieva, N., Werneck-De-Castro, J. P., Hatanaka, et al. (2017). Loss of mTORC1 signalling impairs beta-cell homeostasis and insulin processing. Nat. Commun. 8, 16014. doi: 10.1038/ncomms16014
Blasiak, J., Arabski, M., Krupa, R., Wozniak, K., Zadrozny, M., Kasznicki, J., et al. (2004). DNA damage and repair in type 2 diabetes mellitus. Mutat. Res. 554, 297–304. doi: 10.1016/j.mrfmmm.2004.05.011
Chatterjee, S., Khunti, K., Davies, M. J. (2017). Type 2 diabetes. Lancet 389, 2239–2251. doi: 10.1016/S0140-6736(17)30058-2
Diiorio, P. J., Moss, J. B., Sbrogna, J. L., Karlstrom, R. O., Moss, L. G. (2002). Sonic hedgehog is required early in pancreatic islet development. Dev. Biol. 244, 75–84. doi: 10.1006/dbio.2002.0573
Ding, D., Chong, S., Jalaludin, B., Comino, E., Bauman, A. E. (2015). Risk factors of incident type 2-diabetes mellitus over a 3-year follow-up: results from a large Australian sample. Diabetes Res. Clin. Pract. 108, 306–315. doi: 10.1016/j.diabres.2015.02.002
Dooley, K., Zon, L. I. (2000). Zebrafish: a model system for the study of human disease. Curr. Opin. Genet. Dev. 10, 252–256. doi: 10.1016/S0959-437X(00)00074-5
Flannick, J., Florez, J. C. (2016). Type 2 diabetes: genetic data sharing to advance complex disease research. Nat. Rev. Genet. 17, 535–549. doi: 10.1038/nrg.2016.56
Fox, D., 3rd, Yan, Z., Ling, C., Zhao, Y., Lee, D. Y., Fukagawa, T., et al. (2014). The histone-fold complex MHF is remodeled by FANCM to recognize branched DNA and protect genome stability. Cell Res. 24, 560–575. doi: 10.1038/cr.2014.42
Goldstein, D. B. (2009). Common genetic variation and human traits. N. Engl. J. Med. 360, 1696–1698. doi: 10.1056/NEJMp0806284
Group, A. C., Patel, A., Macmahon, S., Chalmers, J., Neal, B., Billot, L., et al. (2008). Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N. Engl. J. Med. 358, 2560–2572. doi: 10.1056/NEJMoa0802987
Haffner, S. M. (2006). Relationship of metabolic risk factors and development of cardiovascular disease and diabetes. Obesity (Silver Spring) 14 Suppl 3, 121S–127S. doi: 10.1038/oby.2006.291
Hakuno, F., Takahashi, S. I. (2018). IGF1 receptor signaling pathways. J. Mol. Endocrinol. 61, T69–T86. doi: 10.1530/JME-17-0311
Hiramitsu, M., Shimada, Y., Kuroyanagi, J., Inoue, T., Katagiri, T., Zang, L., et al. (2014). Eriocitrin ameliorates diet-induced hepatic steatosis with activation of mitochondrial biogenesis. Sci. Rep. 4, 3708. doi: 10.1038/srep03708
Huang, Y., Cheng, Q., Ji, J. L., Zheng, S., Du, L., Meng, L., et al. (2016). Pharmacokinetic behaviors of intravenously administered siRNA in glandular tissues. Theranostics 6, 1528–1541. doi: 10.7150/thno.15246
Jais, A., Einwallner, E., Sharif, O., Gossens, K., Lu, T. T., Soyal, S. M., et al. (2014). Heme oxygenase-1 drives metaflammation and insulin resistance in mouse and man. Cell 158, 25–40. doi: 10.1016/j.cell.2014.04.043
Jenkinson, C. P., Goring, H. H., Arya, R., Blangero, J., Duggirala, R., Defronzo, R. A. (2016). Transcriptomics in type 2 diabetes: bridging the gap between genotype and phenotype. Genom. Data 8, 25–36. doi: 10.1016/j.gdata.2015.12.001
Kishi, S. (2011). The search for evolutionary developmental origins of aging in zebrafish: a novel intersection of developmental and senescence biology in the zebrafish model system. Birth Defects Res. C Embryo Today 93, 229–248. doi: 10.1002/bdrc.20217
Koide, H., Okamoto, A., Tsuchida, H., Ando, H., Ariizumi, S., Kiyokawa, C., et al. (2016). One-step encapsulation of siRNA between lipid-layers of multi-layer polycation liposomes by lipoplex freeze-thawing. J. Control. Release 228, 1–8. doi: 10.1016/j.jconrel.2016.01.032
Kotelnikova, E., Yuryev, A., Mazo, I., Daraselia, N. (2010). Computational approaches for drug repositioning and combination therapy design. J. Bioinform. Comput. Biol. 8, 593–606. doi: 10.1142/S0219720010004732
Landgraf, K., Schuster, S., Meusel, A., Garten, A., Riemer, T., Schleinitz, D., et al. (2017). Short-term overfeeding of zebrafish with normal or high-fat diet as a model for the development of metabolically healthy versus unhealthy obesity. BMC Physiol. 17, 4. doi: 10.1186/s12899-017-0031-x
Maccallum, L. (2014). Optimal medication dosing in patients with diabetes mellitus and chronic kidney disease. Can. J. Diabetes 38, 334–343. doi: 10.1016/j.jcjd.2014.04.006
Maddison, L. A., Joest, K. E., Kammeyer, R. M., Chen, W. (2015). Skeletal muscle insulin resistance in zebrafish induces alterations in beta-cell number and glucose tolerance in an age- and diet-dependent manner. Am. J. Physiol. Endocrinol. Metab. 308, E662–E669. doi: 10.1152/ajpendo.00441.2014
Manoel-Caetano, F. S., Xavier, D. J., Evangelista, A. F., Takahashi, P., Collares, C. V., Puthier, D., et al. (2012). Gene expression profiles displayed by peripheral blood mononuclear cells from patients with type 2 diabetes mellitus focusing on biological processes implicated on the pathogenesis of the disease. Gene 511, 151–160. doi: 10.1016/j.gene.2012.09.090
Marques-Oliveira, G. H., Silva, T. M., Lima, W. G., Valadares, H. M. S., Chaves, V. E. (2018). Insulin as a hormone regulator of the synthesis and release of leptin by white adipose tissue. Peptides 106, 49–58. doi: 10.1016/j.peptides.2018.06.007
Mccarthy, M. I., Abecasis, G. R., Cardon, L. R., Goldstein, D. B., Little, J., Ioannidis, J. P., et al. (2008). Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 9, 356–369. doi: 10.1038/nrg2344
Mcclellan, J., King, M. C. (2010). Genetic heterogeneity in human disease. Cell 141, 210–217. doi: 10.1016/j.cell.2010.03.032
Meguro, S., Hasumura, T., Hase, T. (2015). Body Fat Accumulation in zebrafish is induced by a diet rich in fat and reduced by supplementation with green tea extract. PLoS One 10. doi: 10.1371/journal.pone.0120142
Morris, A. P., Voight, B. F., Teslovich, T. M., Ferreira, T., Segre, A. V., Steinthorsdottir, V., et al. (2012). Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990. doi: 10.1038/ng.2383
Newgard, C. B., An, J., Bain, J. R., Muehlbauer, M. J., Stevens, R. D., Lien, L. F., et al. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9, 311–326. doi: 10.1016/j.cmet.2009.02.002
Nishino, T., Takeuchi, K., Gascoigne, K. E., Suzuki, A., Hori, T., Oyama, T., et al. (2012). CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148, 487–501. doi: 10.1016/j.cell.2011.11.061
Oh, W. J., Jacinto, E. (2011). mTOR complex 2 signaling and functions. Cell Cycle 10, 2305–2316. doi: 10.4161/cc.10.14.16586
Oka, T., Nishimura, Y., Zang, L., Hirano, M., Shimada, Y., Wang, Z., et al. (2010). Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 10, 21. doi: 10.1186/1472-6793-10-21
Ota, S., Hisano, Y., Muraki, M., Hoshijima, K., Dahlem, T. J., Grunwald, D. J., et al. (2013). Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells 18, 450–458. doi: 10.1111/gtc.12050
Perpelescu, M., Fukagawa, T. (2011). The ABCs of CENPs. Chromosoma 120, 425–446. doi: 10.1007/s00412-011-0330-0
Santoriello, C., Zon, L. I. (2012). Hooked! Modeling human disease in zebrafish. J. Clin. Invest. 122, 2337–2343. doi: 10.1172/JCI60434
Sengupta, S., Peterson, T. R., Sabatini, D. M. (2010). Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322. doi: 10.1016/j.molcel.2010.09.026
Shimizu, I., Yoshida, Y., Suda, M., Minamino, T. (2014). DNA damage response and metabolic disease. Cell Metab. 20, 967–977. doi: 10.1016/j.cmet.2014.10.008
Singh, T. R., Saro, D., Ali, A. M., Zheng, X. F., Du, C. H., Killen, M. W., et al. (2010). MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell 37, 879–886. doi: 10.1016/j.molcel.2010.01.036
Tabak, O., Gelisgen, R., Erman, H., Erdenen, F., Muderrisoglu, C., Aral, H., et al. (2011). Oxidative lipid, protein, and DNA damage as oxidative stress markers in vascular complications of diabetes mellitus. Clin. Invest. Med. 34, E163–E171. doi: 10.25011/cim.v34i3.15189
Tainaka, T., Shimada, Y., Kuroyanagi, J., Zang, L., Oka, T., Nishimura, Y., et al. (2011). Transcriptome analysis of anti-fatty liver action by Campari tomato using a zebrafish diet-induced obesity model. Nutr. Metab. (Lond.) 8, 88. doi: 10.1186/1743-7075-8-88
Tusie Luna, M. T. (2005). Genes and type 2 diabetes mellitus. Arch. Med. Res. 36, 210–222. doi: 10.1016/j.arcmed.2005.03.004
Ueda, H., Ikegami, H., Yamato, E., Fu, J., Fukuda, M., Shen, G., et al. (1995). The NSY mouse: a new animal model of spontaneous NIDDM with moderate obesity. Diabetologia 38, 503–508. doi: 10.1007/BF00400717
Ukpds (1998). Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). UK Prospective Diabetes Study (UKPDS) Group. Lancet 352, 837–853. doi: 10.1016/S0140-6736(98)07019-6
Um, S. H., Frigerio, F., Watanabe, M., Picard, F., Joaquin, M., Sticker, M., et al. (2004). Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205. doi: 10.1038/nature02866
Varshney, G. K., Pei, W., Lafave, M. C., Idol, J., Xu, L., Gallardo, V., et al. (2015). High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 25, 1030–1042. doi: 10.1101/gr.186379.114
Westerfield, M. (2007). “THE ZEBRAFISH BOOK,” in A guide for the laboratory use of zebrafish (Danio rerio), 5th Edition (Eugene: University of Oregon Press).
Yan, Z., Delannoy, M., Ling, C., Daee, D., Osman, F., Muniandy, P. A., et al. (2010). A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell 37, 865–878. doi: 10.1016/j.molcel.2010.01.039
Yoon, M. S. (2017). The role of mammalian target of rapamycin (mTOR) in insulin signaling. Nutrients 9, 1176. doi: 10.3390/nu9111176
Zafar, F., Okita, A. K., Onaka, A. T., Su, J., Katahira, Y., Nakayama, J. I., et al. (2017). Regulation of mitotic recombination between DNA repeats in centromeres. Nucleic Acids Res. 45, 11222–11235. doi: 10.1093/nar/gkx763
Zang, L., Maddison, L. A., Chen, W. (2018). Zebrafish as a model for obesity and diabetes. Front. Cell Dev. Biol. 6, 91. doi: 10.3389/fcell.2018.00091
Zang, L., Shimada, Y., Nishimura, N. (2017). Development of a novel zebrafish model for type 2 diabetes mellitus. Sci. Rep. 7, 1461. doi: 10.1038/s41598-017-01432-w
Zang, L., Shimada, Y., Kawajiri, J., Tanaka, T., Nishimura, N. (2014). Effects of Yuzu (Citrus junos Siebold ex Tanaka) peel on the diet-induced obesity in a zebrafish model. J. Funct. Foods 10, 499–510. doi: 10.1016/j.jff.2014.08.002
Zang, L. Q., Shimada, Y., Nishimura, Y., Tanaka, T., Nishimura, N. (2013). A novel, reliable method for repeated blood collection from aquarium fish. Zebrafish 10, 425–432. doi: 10.1089/zeb.2012.0862
Zang, L. Q., Shimada, Y., Nishimura, Y., Tanaka, T., Nishimura, N. (2015a). Repeated blood collection for blood tests in adult zebrafish. J. Vis. Exp. (102), e53272. doi: 10.3791/53272
Zang, L. Q., Shimada, Y., Tanaka, T., Nishimura, N. (2015b). Rhamnan sulphate from Monostroma nitidum attenuates hepatic steatosis by suppressing lipogenesis in a diet-induced obesity zebrafish model. J. Funct. Foods 17, 364–370. doi: 10.1016/j.jff.2015.05.041
Keywords: centromere protein X, type 2 diabetes mellitus, insulin, zebrafish model, CRISPR/Cas9, gene silencing, therapeutic gene target
Citation: Zang L, Shimada Y, Nakayama H, Chen W, Okamoto A, Koide H, Oku N, Dewa T, Shiota M and Nishimura N (2019) Therapeutic Silencing of Centromere Protein X Ameliorates Hyperglycemia in Zebrafish and Mouse Models of Type 2 Diabetes Mellitus. Front. Genet. 10:693. doi: 10.3389/fgene.2019.00693
Received: 19 March 2019; Accepted: 02 July 2019;
Published: 29 July 2019.
Edited by:
Lawrence Todd Reiter, University of Tennessee Health Science Center (UTHSC), United StatesReviewed by:
Jin Liang, Cornell University, United StatesGautam Kao, University of Gothenburg, Sweden
Copyright © 2019 Zang, Shimada, Nakayama, Chen, Okamoto, Koide, Oku, Dewa, Shiota and Nishimura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqing Zang, bGlxaW5nMDMxMEBnbWFpbC5jb20=; bGlxaW5nQGRvYy5tZWRpYy5taWUtdS5hYy5qcA==
 Liqing Zang
Liqing Zang Yasuhito Shimada2,3,4
Yasuhito Shimada2,3,4 Wenbiao Chen
Wenbiao Chen Takehisa Dewa
Takehisa Dewa