
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Genet., 12 June 2019
Sec. Epigenomics and Epigenetics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00553
This article is part of the Research TopicEpigenetic Reprogramming and Cancer DevelopmentView all 8 articles
 Maria Prates Rivas1
Maria Prates Rivas1 Talita Ferreira Marques Aguiar1,2
Talita Ferreira Marques Aguiar1,2 Gustavo Ribeiro Fernandes3
Gustavo Ribeiro Fernandes3 Luiz Carlos Caires-Júnior1
Luiz Carlos Caires-Júnior1 Ernesto Goulart1
Ernesto Goulart1 Kayque Alves Telles-Silva1
Kayque Alves Telles-Silva1 Monica Cypriano4
Monica Cypriano4 Silvia Regina Caminada de Toledo4
Silvia Regina Caminada de Toledo4 Carla Rosenberg1
Carla Rosenberg1 Dirce Maria Carraro2
Dirce Maria Carraro2 Cecilia Maria Lima da Costa5
Cecilia Maria Lima da Costa5 Isabela Werneck da Cunha6
Isabela Werneck da Cunha6 Ana Cristina Victorino Krepischi1*
Ana Cristina Victorino Krepischi1*Hepatoblastoma is an embryonal liver tumor carrying few genetic alterations. We previously disclosed in hepatoblastomas a genome-wide methylation dysfunction, characterized by hypermethylation at specific CpG islands, in addition to a low-level hypomethylation pattern in non-repetitive intergenic sequences, in comparison to non-tumoral liver tissues, shedding light into a crucial role for epigenetic dysregulation in this type of cancer. To explore the underlying mechanisms possibly related to aberrant epigenetic modifications, we evaluated the expression profile of a set of genes engaged in the epigenetic machinery related to DNA methylation (DNMT1, DNMT3A, DNMT3B, DNMT3L, UHRF1, TET1, TET2, and TET3), as well as the 5-hydroxymethylcytosine (5hmC) global level. We observed in hepatoblastomas a general disrupted expression of these genes from the epigenetic machinery, mainly UHRF1, TET1, and TET2 upregulation, in association with an enrichment of 5hmC content. Our findings support a model of active demethylation by TETs in hepatoblastoma, probably during early stages of liver development, which in combination with UHRF1 overexpression would lead to DNA hypomethylation and an increase in overall 5hmC content. Furthermore, our data suggest that decreased 5hmC content might be associated with poor survival rate, highlighting a pivotal role of epigenetics in hepatoblastoma development and progression.
Pediatric tumors are inherently different from tumors that develop in adults (Maris and Denny, 2002) since they are supposedly derived from defects of cell differentiation and organogenesis (Anderson, 2006). Embryonal tumors are a special class of childhood cancers characterized by a very early-onset exhibiting histological features that resemble developmental stages of the origin organ (Maris and Denny, 2002).
Cancer development involves the accumulation of genetic mutations over an extended period of time (Hanahan and Weinberg, 2011); however, this model is not suitable for pediatric cancer, which develops in a short temporal window, usually carrying a low number of somatic mutations (Vogelstein et al., 2013; Gröbner et al., 2018). A more comprehensive background takes into account the dysregulation of epigenetic mechanisms (Sandoval and Esteller, 2012), considering the interplay between genetic and epigenetic factors as a key element for tumor development (Soo You and Jones, 2012). Among the epigenetic regulators, DNA methylation, which occurs through the addition of a methyl group to a cytosine (5mC) by DNA methyltransferase enzymes (DNMTs) (Jones, 1999), has been extensively investigated. In tumor cells, well-known DNA methylation disturbances include repression of tumor suppressor genes by promoter hypermethylation, and hypomethylation at repetitive sequences, leading to genomic instability (Łuczak and Jagodzinski, 2006; Rodrigues-Paredez and Esteller, 2011).
Hepatoblastoma is a rare liver embryonal tumor, and the most common form of liver tumor in children (Wiederkehr et al., 2013; López-Terrada et al., 2014), with an incidence of 1-2.4 per million children each year (Paran and La Quaglia, 2009). These tumors carry few genetic alterations, mainly gains of chromosomes 2, 8, 20, and chromosome 18 deletion (Tomlinson and Kappler, 2012; Ozen et al., 2013; Rodrigues et al., 2014), and recurrent CTNNB1 activating mutations; low frequency mutations of other Wnt pathway genes, such as AXIN1, AXIN2, AXIN3 and APC, have also been reported (Tomlinson and Kappler, 2012; Eichenmüller et al., 2014; Jia et al., 2014). In a recent work from our group (Maschietto et al., 2017), the evaluation of hepatoblastoma methylomes disclosed a genome-wide methylation dysfunction, characterized by hypermethylation at specific CpG islands, in addition to a low-level hypomethylation pattern in non-repetitive intergenic sequences, the later finding corroborated by others (Cui et al., 2016).
DNA hydroxymethylation is a kind of DNA modification (Münzel et al., 2011) originated by 5mC conversion to 5-hydroxymethylcytosine (5hmC), in a process catalyzed by proteins encoded by TET genes. The 5hmC is an intermediate residue of the active demethylation process and seems to be related to pluripotency maintenance in stem cells and transcriptional activation (Wu and Zhang, 2011; Ko et al., 2011).
To elucidate the underlying mechanisms resulting in DNA methylation loss in hepatoblastomas, we evaluated the expression profiles of the DNMT and TET genes as well as the global level of 5hmC in a hepatoblastomas cohort.
Nineteen hepatoblastoma samples (HBs) and eight paired non-tumoral liver tissue samples were obtained from hepatoblastoma patients submitted to surgical resection in two pediatric cancer institutions (A. C. Camargo Cancer Center and GRAACC, São Paulo, Brazil). This study was carried out in accordance with the recommendations of Research Ethics Committee - A.C. Camargo Cancer Center, Brazil (registration number 1987/14), with written informed consent from all parents/guardian of participants. Clinical features of this HB cohort are described in Supplementary Table 1. All patients received pre-surgery chemotherapy according to the SIOPEL protocol1 and COG protocol2.
Microfluidics-based electrophoresis (Bioanalyzer, Agilent Technologies; CA, United States) was performed to verify quality, and only RNA samples with RNA Integrity Number (RIN) > 7.0 were used for gene expression analysis. The cDNA was synthesized with the High Capacity RNA-to-cDNA kit (Applied Biosystems, EUA) according to standard procedures. The experiment was designed and interpreted according to recommendations contained in International Guideline MIQE (The Minimum Information for Publication of Quantitative Real-Time PCR Experiments). The suitable housekeeping gene was chosen using the geNorm algorithm (Vandesompele et al., 2002) after expression analysis of ACTB, GAPDH, B2M genes and 18S ribosomal RNA (18S rRNA). Data were normalized using the expression values of the housekeeping gene 18S rRNA, and all reactions were performed with three technical replicates. The delta-delta Ct (ΔΔCt) method was used for data analysis (Livak and Schmittgen, 2001). The Kruskal Wallis post hoc Dunn test with Bonferroni correction was used for statistical analysis in the GraphPad Prism 7 software.
Nanodrop (Thermo Fisher Scientific, United States) was used to access quantity and integrity of DNA samples. The 5hmC global level quantification of 18 HBs and eigth paired non-tumoral liver tissue samples was carried out using the MethylFlash Hydroxymethylated DNA 5-hmC Quantification Kit (Epigentek, United States). The values of each sample are given as percentages relative to internal controls of the kit, according to the manufacturer’s recommendation. The t-test was used for the statistical analysis.
Gene expression analysis of five DNA methylation genes (DNMT1, DNMT3A, DNMT3B, DNMT3L, and UHRF1) revealed up-regulation of DNMT3A (fold-change mean: 7.0; p-value: 0.024) and DNMT1 (fold-change mean: 3.6; p-value: 0.012) expression in the group of 19 HBs as compared to eight paired non-tumoral liver tissues, while DNMT3B and DNMT3L expression values were not significantly altered (Figure 1). Data also showed a very significant increase in UHRF1 expression in tumor samples (fold-change mean: 20.4; p-value: 0.015).
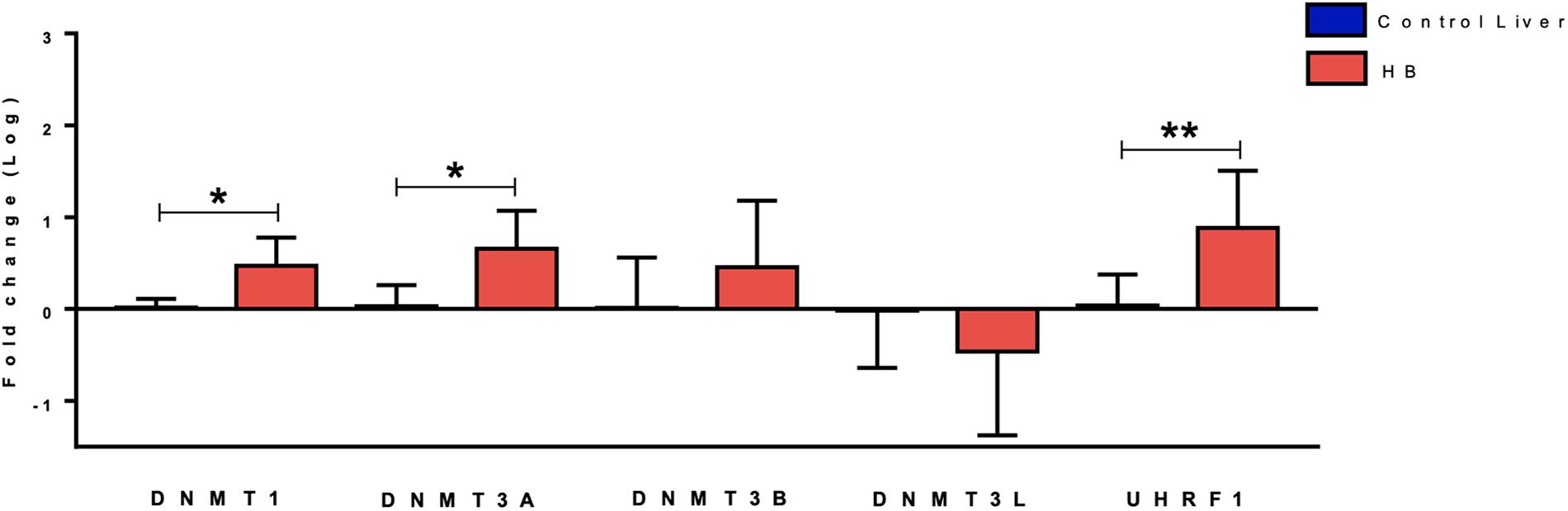
Figure 1. Expression analysis of DNA methylation genes in hepatoblastomas. DNMT1, DNMT3A and UHRF1 upregulation in 19 tumor samples compared with eight non-tumoral liver samples as evidenced by RT-qPCR. Control Liver: non-tumoral liver tissue samples; HB: hepatoblastoma samples. Endogenous gene: 18S rRNA. The Kruskal Wallis post hoc Dunn test with Bonferroni correction was used for statistical analysis. ∗p-value < 0.05, ∗∗p-value < 0.01.
The hydroxymethylation process was evaluated by assessing the expression level of the three TET genes, and all of them were found to be significantly upregulated in hepatoblastomas as compared to non-tumoral liver tissue (Figure 2A): TET1 (fold-change mean: 21.5; p-value: 0.009), TET2 (fold-change mean: 7.15; p-value: 0.009), and TET3 (fold-change mean: 4.3; p-value: 0.024). Given the observed TET genes upregulation in HBs, the global content of 5hmC was evaluated in tumors and control hepatic tissues. Regardless of the variability in total 5hmC content seen across HBs, an overall enrichment of approximately 2.5× (p-value 4.2e-05; t-test) was observed in tumors (mean: 0.33%; SD: 0.15) relative to non-tumoral livers (mean: 0.13%; SD: 0.07) (Figure 2B). Statistical analysis based on the clinical data showed that patients with overall survival ≤ 5 years exhibited a 5hmC content lower than the mean of the tumor group (p-value 0.02; Fisher’s test) (Figure 3), closer to the level that was detected in the group of non-tumoral liver tissues.
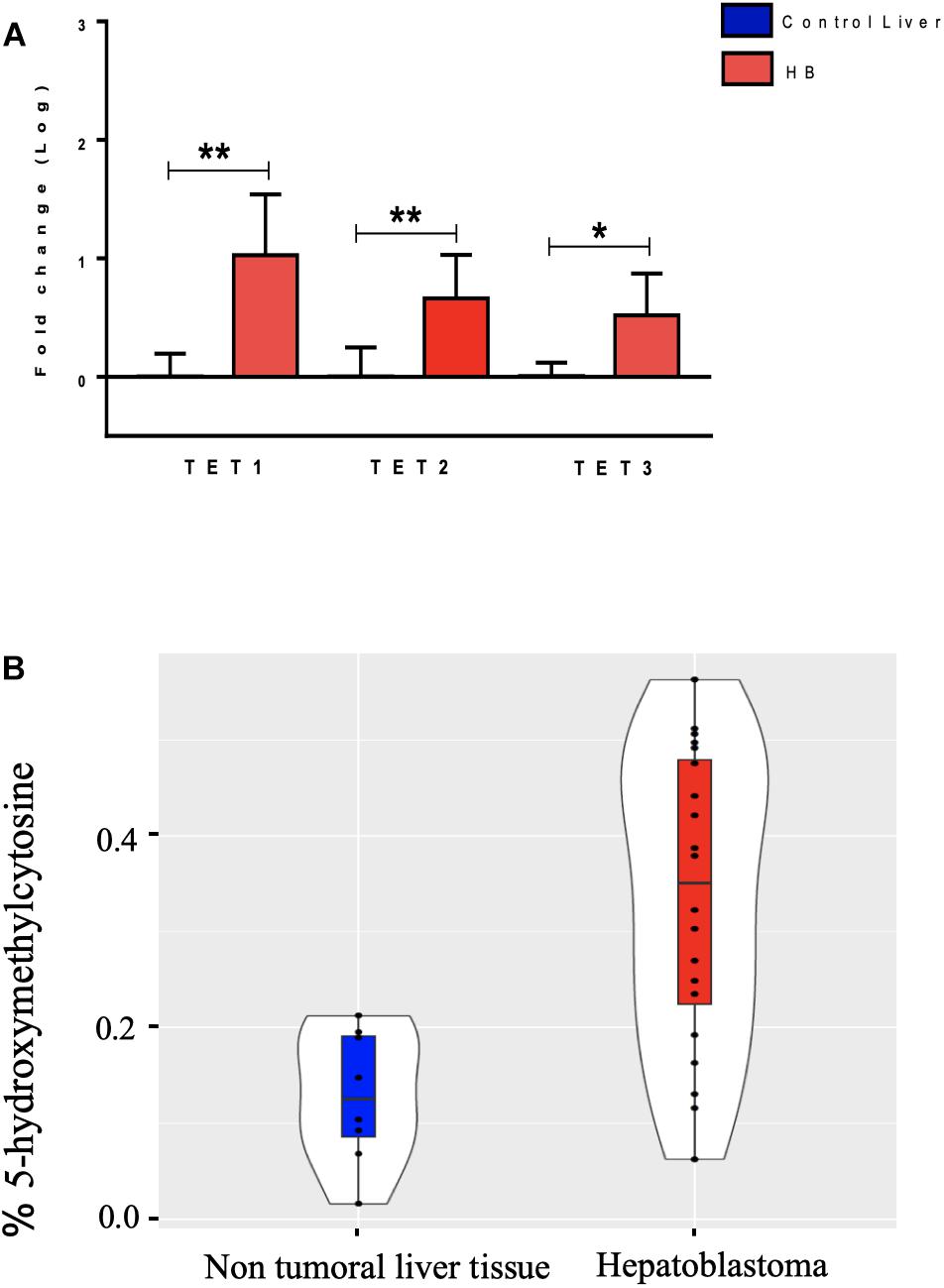
Figure 2. Analysis of DNA hydroxymethylation in hepatoblastomas. (A) Expression analysis of DNA hydroxymethylation genes in hepatoblastomas. Upregulation of three genes of the TET family in 19 tumor samples compared with eight non-tumoral liver samples as evidenced by RT-qPCR. Control Liver: non-tumoral liver tissue samples; HB: hepatoblastoma samples. Endogenous gene: 18S rRNA. The Kruskal Wallis post hoc Dunn test with Bonferroni correction was used for statistical analysis. ∗p-value < 0.05; ∗∗p-value < 0.01. (B) 5hmC global level enrichment in hepatoblastoma samples. Violin plots showing the overall level of 5hmC in percentage in hepatoblastomas samples (red) and non-tumoral liver tissues (blue). Hepatoblastoma samples mean: 0.33%, SD: 0.15; non-tumoral liver tissue samples mean: 0.13%; SD: 0.07. The t-test was used for the statistical analysis. p-value: 4.2e-05.
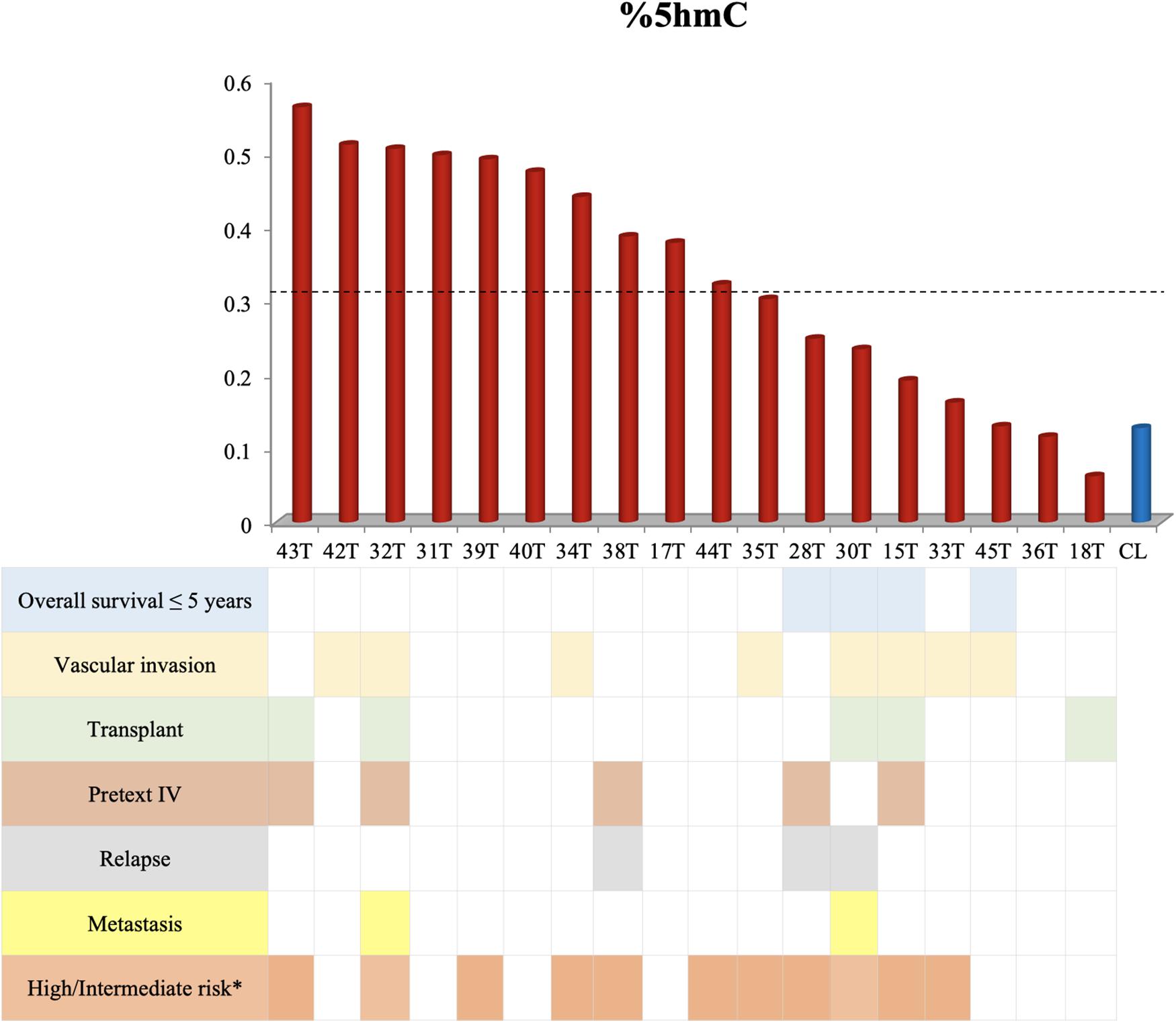
Figure 3. Decreased overall survival is associated with lower levels of 5hmC. This figure shows the 5hmC global content of 18 tumor samples in the columns and their respective clinical data below. Tumor samples with overall survival ≤ 5 years present reduced 5hmC content, below the mean of the tumor group (0.33% - represented by the dashed line) and close to the level of non-tumoral hepatic tissues (CL). The Fisher’s test was used for the statistical analysis. p-value 0.02. CL: non-tumoral liver tissue samples; T: hepatoblastoma tumor samples. ∗According to the CHIC criteria (Meyers et al., 2017).
We previously reported changes in the methylation profile of hepatoblastomas, with global hypomethylation in intergenic non-repetitive sequences, as well as hypermethylation at CpG islands, affecting genes associated with tumor suppression, lipid metabolism, and liver differentiation (Maschietto et al., 2017). Except for the UHRF1 gene (Beck et al., 2018), here we disclosed for the first time dysregulated expression of genes from the DNA methylation machinery that could be related to the detected epigenetic disturbance, such as upregulation of DNMT3A, DNMT1, UHRF1, and the TET family of genes.
The interaction between UHRF1/DNMT1 plays a pivotal role in the inheritance of DNA methylation patterns, even though the details of this complex network remain under analysis. Thus, UHRF1 is considered an essential epigenetic regulator for DNA methylation maintenance (Bostick et al., 2007). It is known that UHRF1/DNMT1 are involved in the maintenance of the hypermethylation of tumor suppressor gene promoters in cancer (Alhosin et al., 2016; Ashraf et al., 2017); in particular, overexpression of UHRF1 was recently reported to silence specific tumor suppressor genes in hepatoblastomas (Beck et al., 2018). Indeed, enhanced UHRF1 expression was identified in all cancers so far examined (Ashraf et al., 2017). Therefore, it is possible that DNMT1 and DNMT3A upregulation observed in the present study contributes to the hypermethylation at specific CpG islands we previously reported in hepatoblastomas. Nevertheless, it has also been suggested that impaired interaction between UHRF1 and DNMT1 could be the origin of the global hypomethylation in cancer cells (Hervouet et al., 2010; Pacaud et al., 2014). Moreover, a different line of evidence revealed that UHRF1 overexpression also acts as a negative regulator by degradation of the de novo DNA methylation protein, DNMT3A, as an additional mechanism leading to widespread DNA hypomethylation in cancer cells (Jia et al., 2016). Complementary to these findings, Mudbhary et al. (2014) demonstrated in both zebrafish hepatocytes and hepatocellular carcinoma that UHRF1 overexpression promotes DNMT1 destabilization and de-localization, driving DNA hypomethylation; further, UHRF1 increased expression was already associated with poor prognosis in hepatocellular carcinoma, contributing to cell proliferation and metastasis (Liu et al., 2017). These data further support the hypothesis that disruption of normal cooperation of DNA methylases and UHRF1 can play a dual role in the patterns of DNA methylation in cancer cells, which mechanisms remain partially elusive. In combination with our results, these evidences suggest that, at least in liver cancer, including hepatoblastomas, the disclosed UHRF1 overexpression may result in inefficient or impaired DNA methylation process, irrespective of the DNMTs expression level, contributing to the global low level hypomethylation pattern in non-repetitive intergenic sequences reported in hepatoblastomas (Cui et al., 2016; Maschietto et al., 2017) and hepatocellular carcinoma (Shen et al., 2013).
The conversion of methylated cytosine to 5hmC, mediated by TET enzymes, is the first step in the DNA demethylation active pathway (Wu and Zhang, 2011; Ko et al., 2011; Rasmussen and Helin, 2016). In adult cancer, a decrease in TET expression was reported to reduce the 5hmC level in solid and hematological tumors (Lian et al., 2012; Liu et al., 2013; Rawłuszko-Wieczorek et al., 2015; Rasmussen and Helin, 2016). However, the activity of these enzymes in pediatric tumors is poorly explored. Despite the limited size of our cohort, here we showed an increased expression of TET genes in hepatoblastomas, suggesting the occurrence of an active demethylation mechanism. Studies with animal models described TET overexpression in embryonic development stages repressing differentiation (Ito et al., 2010; Rawłuszko-Wieczorek et al., 2015; Rasmussen and Helin, 2016), as well as in cellular reprogramming of IPS cells (Bagci and Fisher, 2013). Thus, we can speculate that TET upregulation in hepatoblastomas indicates the occurrence of a blockage of liver differentiation, which corroborates the long-standing hypothesis of repression of the differentiation pathway of the origin organ leading to embryonal tumors genesis (Maris and Denny, 2002). A functional role for the observed TET overexpression was substantiated by the enrichment of the 5hmC content in these tumors, then partially explaining the DNA hypomethylation described in hepatoblastoma, in addition to the possible impaired de novo methylation caused by the observed UHRF1 overexpression. An additional analysis regarding the underlying nature of the DNA sequences with 5hmC enrichment in hepatoblastomas (if intergenic non-repetitive or CpG islands) would add valuable information, and should be made in the future. Remarkably, in hepatocellular carcinoma, a reduced 5hmC content was directly associated with lower overall survival and poor prognosis (Liu et al., 2013). Our findings also suggested that lower levels of 5hmC in hepatoblastomas could be associated with decreased overall survival, highlighting the 5hmC content as a potential biomarker of risk for these tumors. However, these preliminary results require further validation in a larger cohort of hepatoblastomas.
This study addressed possible mechanisms leading to the hypomethylation pattern displayed by hepatoblastomas. Our data revealed disturbance in the expression of genes related to the DNA methylation machinery and enrichment of the 5hmC content in hepatoblastomas. As hepatoblastomas exhibit the lowest mutation rate reported so far for pediatric solid tumors (Gröbner et al., 2018), this analysis of genes of the epigenetic machinery provides evidence that epigenetic disruption is probably key for the onset of this liver embryonal tumor, and highlights the 5hmC content as a potential biomarker for poor overall survival.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
This study was carried out in accordance with the recommendations of Research Ethics Committee – A.C. Camargo Cancer Center, Brazil, registration number 1987/14. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved and all the samples were collected and extracted in the A.C.Camargo Cancer Center Bank of Macromolecules, following the technical and ethical procedures of the institution (Campos et al., 2012; Olivieri et al., 2014).
MR and AK conceived the study and participated in its design. MR, LC-J, EG, and KT-S performed the experiments and analyzed the data. GF performed statistical analysis. TA, CdC, IdC, MC, SdT, and DC evaluated patients, collected biological samples, and revised clinical data. MR and AK wrote the manuscript. CR and AK critically revised the manuscript. All authors have read and approved the final version of the manuscript.
This project was funded by the Fundação de Amparo à Pesquisa do Estado de São Paulo – FAPESP (Project CEPID – Human Genome and Stem Cell Research Center – 2013/08028-1). MR and TA were supported by scholarships FAPESP 2016/23462-8 and 2016/04785-0, respectively. EG and LC-J were also supported by FAPESP (2015/14821-1 and 2017/16283-2, respectively).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the patients and their families for their precious collaboration.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00553/full#supplementary-material
Alhosin, M., Omran, Z., Zamzami, M., Al-Malki, A. L., Choudhry, H., Mousli, M., et al. (2016). Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J. Exp. Clin. Cancer Res. 35:174. doi: 10.1186/s13046-016-0453-455
Anderson, L. M. (2006). Environmental genotoxicants/carcinogens and childhood cancer: bridgeable gaps in scientific knowledge. Mut. Res. 608, 136–156. doi: 10.1016/j.mrgentox.2006.02.016
Ashraf, W., Ibrahim, A., Alhosin, M., Zaayter, L., Ouararhni, K., Papin, C., et al. (2017). The epigenetic integrator UHRF1: on the road to become a universal biomarker for cancer. Oncotarget 8, 51946–51962. doi: 10.18632/oncotarget.17393
Bagci, H., and Fisher, A. G. (2013). DNA demethylation in pluripotency and reprogramming: the role of tet proteins and cell division. Cell Stem Cell 13, 265–269. doi: 10.1016/j.stem.2013.08.005
Beck, A., Trippel, F., Wagner, A., Joppien, S., Felle, M., Vokuhl, C., et al. (2018). Overexpression of UHRF1 promotes silencing of tumor suppressor genes and predicts outcome in hepatoblastoma. Clin. Epigenet. 10:27. doi: 10.1186/s13148-018-0462-7
Bostick, M., Kim, J. K., Estève, P. O., Clark, A., Pradhan, S., and Jacobsen, S. E. (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764. doi: 10.1126/science.1147939
Campos, A. H. J. F. M., Silva, A. A., De Carvalho Mota, L. D., Olivieri, E. R., Prescinoti, V. C., Patrão, D., et al. (2012). The value of a tumor bank in the development of cancer research in Brazil: 13 years of experience at the A C Camargo Hospital. Biopreserv. Biobank. 10, 168–173. doi: 10.1089/bio.2011.0032.
Cui, X., Liu, B., Zheng, S., Dong, K., and Dong, R. (2016). Genome-wide analysis of DNA methylation in hepatoblastoma tissues. Oncol. Lett. 12, 1529–1534. doi: 10.3892/ol.2016.4789
Eichenmüller, M., Trippel, F., Kreuder, M., Beck, A., Schwarzmayr, T., Häberle, B., et al. (2014). The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J. Hepatol. 61, 1312–1320. doi: 10.1016/j.jhep.2014.08.009
Gröbner, S. N., Worst, B. C., Weischenfeldt, J., Buchhalter, I., Kleinheinz, K., Rudneva, V. A., et al. (2018). The landscape of genomic alterations across childhood cancers. Nature 555, 321–327. doi: 10.1038/nature25480
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell Press 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Hervouet, E., Lalier, L., Debien, E., Cheray, M., Geairon, A., Rogniaux, H., et al. (2010). Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS One 5:e11333. doi: 10.1371/journal.pone.0011333
Ito, S., D’Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. doi: 10.1038/nature09303
Jia, D., Dong, R., Jing, Y., Xu, D., Wang, Q., Chen, L., et al. (2014). Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology 60, 1686–1696. doi: 10.1002/hep.27243
Jia, Y., Li, P., Fang, L., Zhu, H., Xu, L., Cheng, H., et al. (2016). Negative regulation of DNMT3A de novo DNA methylation by frequently overexpressed UHRF family proteins as a mechanism for widespread DNA hypomethylation in cancer. Cell Discov. 2:16007. doi: 10.1038/celldisc.2016.7
Jones, P. A. (1999). The DNA methylation paradox. Trends Genet. 15, 34–37. doi: 10.1016/S0168-9525(98)01636-9
Ko, M., Bandukwala, H. S., An, J., Lamperti, E. D., Thompson, E. C., Hastie, R., et al. (2011). Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl. Acad. Sci. U.S.A. 108, 14566–14571. doi: 10.1073/pnas.1112317108
Lian, C. G., Xu, Y., Ceol, C., Wu, F., Larson, A., Dresser, K., et al. (2012). Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150, 1135–1146. doi: 10.1016/j.cell.2012.07.033
Liu, C., Liu, L., Chen, X., Shen, J., Shan, J., Xu, Y., et al. (2013). Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One 8:e62828. doi: 10.1371/journal.pone.0062828
Liu, X., Ou, H., Xiang, L., Li, X., Huang, Y., and Yang, D. (2017). Elevated UHRF1 expression contributes to poor prognosis by promoting cell proliferation and metastasis in hepatocellular carcinoma. Oncotarget 8, 10510–10522. doi: 10.18632/oncotarget.14446
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Terrada, D., Alaggio, R., de Dávila, M. T., Czauderna, P., Hiyama, E., Katzenstein, H., et al. (2014). Towards an international pediatric liver tumor consensus classification: proceedings of the Los Angeles COG liver tumors symposium. Mod. Pathol. 27, 472–491. doi: 10.1038/modpathol.2013.8
Łuczak, M. W., and Jagodzinski, P. P. (2006). The role of DNA methylation in cancer development. Folia Histochem. Cytobiol. 44, 143–154.
Maris, J. M., and Denny, C. T. (2002). Focus on embryonal malignancies. Cancer Cell 2, 447–450. doi: 10.1016/S1535-6108(02)00206-4
Maschietto, M., Rodrigues, T. C., Kashiwabara, A. Y., de Araujo, ÉS. S., Marques Aguiar, T. F., da Costa, C. M. L., et al. (2017). DNA methylation landscape of hepatoblastomas reveals arrest at early stages of liver differentiation and cancer-related alterations. Oncotarget 8, 97871–97889. doi: 10.18632/oncotarget.14208
Meyers, R. L., Maibach, R., Hiyama, E., Häberle, B., Krailo, M., Rangaswami, A., et al. (2017). Risk-stratified staging in paediatric hepatoblastoma: a unified analysis from the Children’s hepatic tumors international collaboration. Lancet Oncol. 18, 122–131. doi: 10.1016/S1470-2045(16)30598-8
Mudbhary, R., Hoshida, Y., Chernyavskaya, Y., Jacob, V., Villanueva, A., Fiel, M. I., et al. (2014). UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 25, 196–209. doi: 10.1016/j.ccr.2014.01.003
Münzel, M., Globisch, D., and Carell, T. (2011). 5-Hydroxymethylcytosine, the sixth base of the genome. Angew. Chem. Int. Ed. Engl. 50, 6460–6468. doi: 10.1002/anie.201101547
Olivieri, E. H. R., Franco, L. de A., Pereira, R. G., Mota, L. D. C., Campos, A. H. J. F. M., and Carraro, D. M. (2014). Biobanking practice: RNA storage at low concentration affects integrity. Biopreserv. Biobank. 12, 46–52. doi: 10.1089/bio.2013.0056
Ozen, C., Yildiz, G., Dagcan, A. T., Cevik, D., Ors, A., Keles, U., et al. (2013). Genetics and epigenetics of liver cancer. New Biotechnol. 30, 381–384. doi: 10.1016/j.nbt.2013.01.007
Pacaud, R., Brocard, E., Lalier, L., Hervouet, E., Vallette, F. M., and Cartron, P. F. (2014). The Dnmt1/PCNA/UHRF1 disruption induces tumorigenesis characterized by similar genetic and epigenetic signatures. Sci. Rep. 18:4230. doi: 10.1038/srep04230
Paran, T. S., and La Quaglia, M. P. (2009). “Hepatic Tumors in Childhood,” in Pediatric Surgery, eds P. Puri and M. Höllwarth (Heidelberg: Springer), 727–735. doi: 10.1007/978-3-540-69560-8_75
Rasmussen, K. D., and Helin, K. (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750. doi: 10.1101/gad.276568.115
Rawłuszko-Wieczorek, A. A., Siera, A., and Jagodziński, P. P. (2015). TET proteins in cancer: current ‘state of the art’. Crit. Rev. Oncol. Hematol. 96, 425–436. doi: 10.1016/j.critrevonc.2015.07.008
Rodrigues, T. C., Fidalgo, F., da Costa, C. M., Ferreira, E. N., da Cunha, I. W., Carraro, D. M., et al. (2014). Upregulated genes at 2q24 gains as candidate oncogenes in hepatoblastomas. Future Oncol. 10, 2449–2457. doi: 10.2217/fon.14.149
Rodrigues-Paredez, M., and Esteller, M. (2011). Cancer epigenetics reaches mainstream oncology. Nat. Med. 17, 330–339. doi: 10.1038/nm.2305
Sandoval, J., and Esteller, M. (2012). Cancer epigenomics: beyond genomics. Curr. Opin. Genet. Dev. 22, 50–55. doi: 10.1016/j.gde.2012.02.008
Shen, J., Wang, S., Zhang, Y. J., Wu, H. C., Kibriya, M. G., Jasmine, F., et al. (2013). Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using infinium human methylation 450 beadchips. Epigenetics 8, 34–43. doi: 10.4161/epi.23062
Soo You, J., and Jones, P. A. (2012). Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22, 9–20. doi: 10.1016/j.ccr.2012.06.008
Tomlinson, G. E., and Kappler, R. (2012). Genetics and epigenetics of hepatoblastoma. Pediatr. Blood Cancer 59, 785–792. doi: 10.1002/pbc.24213
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1–0034.11.
Vogelstein, B., Papadopoulos, N., Velculescu, V. E., Zhou, S., Diaz, L. A. Jr., and Kinzler, K. W. (2013). Cancer genome landscapes. Science 339, 1546–1558. doi: 10.1126/science.1235122
Wiederkehr, J. C., Coelho, I. M., Avilla, S. G., Wiederkehr, B. A., and Wiederkehr, H. A. (2013). Liver tumors in infancy. Hepatic Surg. 18, 423–460. doi: 10.5772/51764
Keywords: hepatoblastoma, epigenetics, hydroxymethylation, active demethylation, DNA hypomethylation
Citation: Rivas MP, Aguiar TFM, Fernandes GR, Caires-Júnior LC, Goulart E, Telles-Silva KA, Cypriano M, de Toledo SRC, Rosenberg C, Carraro DM, da Costa CML, da Cunha IW and Krepischi ACV (2019) TET Upregulation Leads to 5-Hydroxymethylation Enrichment in Hepatoblastoma. Front. Genet. 10:553. doi: 10.3389/fgene.2019.00553
Received: 14 March 2019; Accepted: 24 May 2019;
Published: 12 June 2019.
Edited by:
Carolina Vicente-Dueñas, Instituto de Investigación Biomédica de Salamanca (IBSAL), SpainReviewed by:
Gokul Gopinath, Texas A&M University Baylor College of Dentistry, United StatesCopyright © 2019 Rivas, Aguiar, Fernandes, Caires-Júnior, Goulart, Telles-Silva, Cypriano, de Toledo, Rosenberg, Carraro, da Costa, da Cunha and Krepischi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Cristina Victorino Krepischi, YW5hLmtyZXBpc2NoaUBpYi51c3AuYnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.