- 1Department of Pediatric Surgery, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou, China
- 2The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
- 3Department of Pediatric Surgery, First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 4Clinical Laboratory Medicine Center of PLA, Xijing Hospital, Air Force Medical University, Xi’an, China
- 5Department of Pathology, Children Hospital and Women Health Center of Shanxi, Taiyuan, China
Background: Hepatoblastoma (HB) is the most common hepatic malignancy in children, accounting for approximately 80% of all childhood liver tumors. Previous genome-wide association studies (GWASs) have found that the LINC00673 rs11655237 C>T polymorphism is associated with the risk of several different adult cancers. However, the association between this polymorphism and HB susceptibility remains unclear.
Methods: We analyzed the association between the LINC00673 rs11655237 C>T polymorphism and HB susceptibility in a hospital-based study of Chinese children. We enrolled 213 HB patients and 958 healthy controls with genotypes determined by TaqMan, and the strength of the association of interest was determined by calculating odds ratios (ORs) and 95% confidence intervals (CIs).
Findings: We found a significant association between the LINC00673 rs11655237 C>T polymorphism and HB risk (CT/TT compared with CC: adjusted OR = 1.40, 95% CI = 1.04–1.88, p = 0.029). Furthermore, stratified analysis indicated that rs11655237 T allele carriers in the following subgroups were more likely to develop HB: children older than 17 months, males, and those with tumors of clinical stages III + IV.
Interpretation: In conclusion, we confirmed that the LINC00673 rs11655237 C>T polymorphism may be associated with HB susceptibility. Prospective studies with larger sample sizes and patients of different ethnicities are needed to validate our findings.
Introduction
Hepatoblastoma (HB) is the most common childhood hepatic malignancy, accounting for approximately 80% of all childhood liver malignancies and 1% of all childhood malignancies (von Schweinitz, 2012). HB is an embryonic tumor that may have originated from pluripotent stem cells in the liver during the embryonic period (Devi et al., 2014). Epidemiological data show that among those under 19 years of age, the incidence rate is 1.2–1.5/106 individuals, with 90% of cases occurring within 3 years of birth and males being more susceptible (Herzog et al., 2000). The clinical symptoms of HB primarily consist of asymptomatic abdominal masses, which may be accompanied by fever, weight loss, anorexia, and obstructive jaundice or acute abdomen caused by tumor rupture (El Asmar and El Rassi, 2016). Alpha-fetoprotein (AFP) can be used as an important biomarker of HB: more than 90% of children with HB have elevated AFP levels, with AFP <100 ng/ml often indicating poor prognosis (Rojas et al., 2014).
The cause of HB remains unclear. It is known that unlike adult liver cancer, its occurrence is not related to hepatitis B virus (HBV) or liver cirrhosis (D’Antiga et al., 2007). Many reports suggest that HB incidence is closely related to Beckwith–Wiedemann Syndrome (BWS), familial adenomatous polyposis (FAP), glycogen storage disease, and fetal alcohol syndrome (Goldman et al., 2010; von Schweinitz, 2012). Premature birth, low body weight, neonatal radiological examination, medication, total parenteral nutrition, and other factors may also contribute to the pathogenesis of HB (Colleen et al., 2006; Hughes and Michels, 2010; El Asmar and El Rassi, 2016).
Non-coding RNAs are generally classified according to length, with those of 200–100,000 nucleotides being referred to as long non-coding RNAs (lncRNAs) (Xuefei et al., 2013). In recent years, genome-wide association studies (GWASs) have found that only 7% of single nucleotide polymorphisms (SNPs) associated with complex diseases or phenotypes are located in protein-coding regions, while the remaining 93% are located in non-coding regions (Jing et al., 2015). GWASs on cancer susceptibility have shown that some relevant SNPs are located in non-coding transcribed regions (Yang et al., 2015; Pan et al., 2016). For instance, the region upstream of the 9p21 locus (encoding the cyclin-dependent kinase inhibitors CDKN2B and CDKN2A and the P53 activator ARF) is associated with tumor susceptibility and is transcribed into lncRNA (Rivand et al., 2017). These findings suggest that genetic variation in lncRNAs may play important roles in tumorigenesis and development (Botti et al., 2018).
LINC00673 is an lncRNA-encoding gene located on chromosome 17q24.3. It has recently been implicated as an oncogenic molecule with an important role in several physiological processes, such as cell proliferation, differentiation, and apoptosis (Shi et al., 2016; Zhang et al., 2017). Studies have found that LINC00673 plays a key role in the occurrence and development of various malignant tumors, such as pancreatic, liver, gastric, and lung cancer (Shi et al., 2016; Zheng et al., 2016; Ba et al., 2017). Moreover, studies have shown that the LINC00673 rs7214041 polymorphism is significantly associated with the development of pancreatic cancer (Zheng et al., 2016) and neuroblastoma (Zhang et al., 2018). Zhang et al. (2017) reported that the expression of LINC00673 was significantly elevated in liver cancer tissue and correlated with disease progression. LINC00673 may compete with miR-205 as a competing endogenous RNA, inhibiting the expression of miR-205 and promoting the development and progression of liver cancer (Zhang et al., 2017). Ba et al. (2017) reported that LINC00673 is highly expressed in gastric cancer tissue, with expression changes being closely related to lymph node metastasis, distant metastasis, TNM staging, and gastric cancer prognosis. Knockdown of LINC00673 expression can significantly inhibit the proliferation, migration, and invasion of gastric cancer cells (Huang et al., 2017). Ma et al. (2017) found that LINC00673 can bind to EZH2 to inhibit HOAX5 expression, thereby promoting the occurrence and development of non-small cell lung cancer.
Mounting evidence suggests that LINC00673 may participate in the occurrence and development of malignant tumors in various ways. We found that LINC00673 rs11655237 polymorphism was associated with neuroblastoma susceptibility in Chinese population (Zhang et al., 2018). However, to our knowledge, the role of LINC00673 polymorphisms within the context of HB has not yet been reported. Therefore, in this study, we analyzed the correlation between the LINC00673 rs11655237 C>T polymorphism and HB susceptibility.
Materials and Methods
Study Subjects
Hepatoblastoma patients (n = 213) and cancer-free controls (n = 958) were recruited from Guangdong, Henan, Shaanxi, and Shaanxi provinces in China for analysis (Supplementary Table S1). All 213 cases were histopathologically diagnosed with HB. These cases were genetically unrelated to the controls and matched to controls based on age, sex, and ethnicity. Each child’s parent or guardian provided written informed consent for the child’s participation in the study. This subject was approved by the Ethics Committee of Guangzhou Women and Children’s Medical Center.
Genotyping
The hepatoblastoma DNA was extracted from paraffin section. Each subject was genotyped for the LINC00673 rs11655237 C>T polymorphism using the TaqMan platform (Applied Biosystems, Foster City, CA, United States) (Stocker and Ishak, 1994; Hirschman et al., 2005; Rumbajan et al., 2013; He et al., 2016, 2017, 2018). Quality control was performed with positive controls and eight negative controls in each of the 384-well plates. For quality control and to verify the accuracy of the genotyping results, approximately 10% of samples were randomly selected and re-genotyped, and the concordance rate was 100%.
Statistical Analysis
The χ2 test was used to evaluate differences in demographic variables, risk factors, and LINC00673 genotype distributions between the case and control groups. The χ2 test was also used to determine whether the distribution of LINC00673 genotypes was consistent with Hardy–Weinberg equilibrium (HWE). To estimate the strength of the association between the rs11655237 C>T polymorphism and HB risk, unconditional univariate and multivariate logistic regression analyses were performed, adjusting for age and sex, using odds ratios (ORs) and 95% confidence intervals (CIs). Further stratification analysis was performed based on the age, sex, and clinical stage of HB patients with different genotypes. Differences with p-values <0.05 were considered statistically significant. All statistical analyses were two-sided and performed using SAS software (version 9.1; SAS Institute, Cary, NC, United States).
Results
Characteristics of the Study Population
In this study, we enrolled 213 HB cases and 958 healthy controls. The distributions of age, sex, and clinical stages of the study subjects are shown in Supplementary Table S1. No significant differences were observed between HB patients and controls in terms of age (p = 0.105) or sex (p = 0.973). The majority of subjects in both the case and control groups were males, accounting for 60.56% (129/213) and 60.44% (579/958) of subjects, respectively. Forty-two (19.72%), 55 (25.82%), 40 (18.78%), and 15 (7.04%) patients had tumors of clinical stages I, II, III, and IV, respectively, while clinical stage was unknown in 61 cases (28.64%). The 213 HB patients and 958 healthy controls were recruited from the Guangdong, Henan, Shaanxi, and Shaanxi provinces (Supplementary Table S2). Within each province, there were no significant differences in age or sex between HB patients and healthy controls (p > 0.05).
LINC00673 rs11655237 C>T Polymorphism and HB Susceptibility
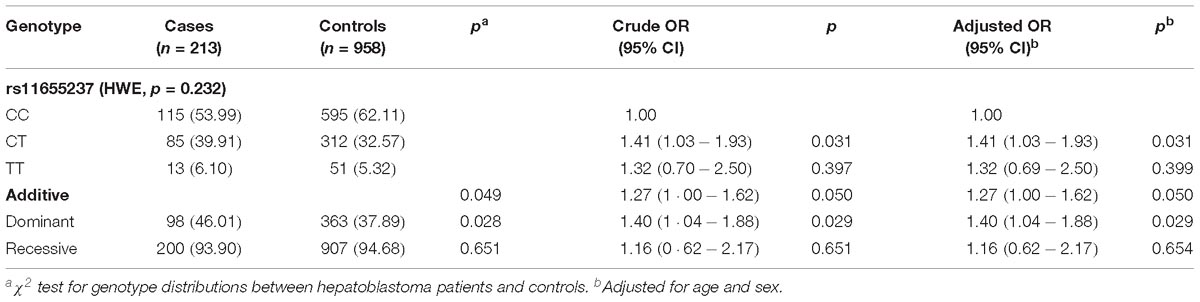
Genotype and allele frequencies of the LINC00673 rs11655237 C>T polymorphism and associations with HB risk are summarized in Table 1. We found that carriage of the rs11655237 T allele was significantly associated with an increased risk of HB (CT compared with CC: adjusted OR = 1.41, 95% CI = 1.03–1.93, p = 0.031; CT/TT compared with CC adjusted OR = 1.40, 95% CI = 1.04–1.88, p = 0.029).
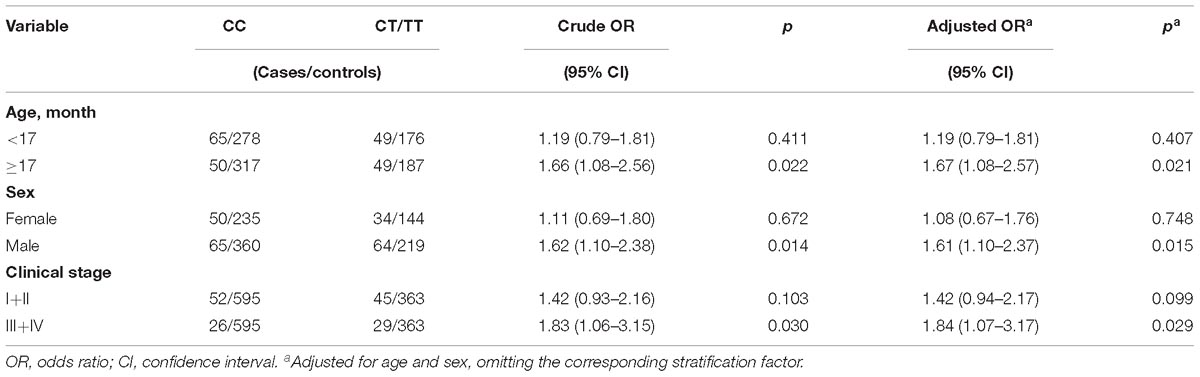
Table 2. Stratification analysis for the association between LINC00673 rs11655237 C>T polymorphism and hepatoblastoma risk.
Stratification Analysis of Association Between LINC00673 rs11655237 C>T Polymorphism and HB Risk
To further evaluate the effect of the LINC00673 rs11655237 C>T polymorphism on HB risk, stratification analysis was further used to evaluate the association between the LINC00673 rs11655237 C>T polymorphism and HB risk in different strata (based on age, sex, and clinical stage) (Table 2). Significant associations was detected in the following subgroups: children older than 17 months (adjusted OR = 1.67; 95% CI = 1.08–2.57, p = 0.021), males (adjusted OR = 1.61; 95% CI = 1.10–2.37, p = 0.015), and patients with tumors of clinical stages III + IV (adjusted OR = 1.84, 95% CI = 1.07–3.17, p = 0.029).
Discussion
The relationship between the LINC00673 rs11655237 C>T polymorphism and the risk of HB was investigated in this hospital-based case-control study. The results of our study showed that the LINC00673 rs11655237 C>T polymorphism was significantly associated with susceptibility to HB. Furthermore, stratified analysis indicated that rs11655237 T allele carriers in the following subgroups were more likely to develop HB: children older than 17 months, males, and patients with tumors of clinical stages III + IV. To our knowledge, we are the first to confirm that the LINC00673 rs11655237 C>T polymorphism is associated with HB susceptibility.
HB is an embryonic tumor that ranks first among childhood liver tumors and seriously endangers children’s health (Herzog et al., 2000). HB and hepatocellular carcinoma are characterized by differences in incidence, age distribution, sex distribution, birth history, genetic basis, and related risk factors (Jeng et al., 2000). Rumbajan et al. (2013) and (Udatsu et al., 2001) showed that in HB patients, some differentially methylated regions (DMRs) exhibited abnormal methylation prior to the development of HB, suggesting that changes in the methylation of DMRs are related to the occurrence of HB. Moreover, studies have shown that the inactivation of the tumor suppressor gene adenomatous polyposis coli (APC), the main function of which is the downregulation of β-catenin, is involved in the occurrence of HB (Li et al., 2014). Indeed, some researchers have used β-catenin as an indicator for evaluating the prognosis of HB (Froberg et al., 2013; Pickard and Williams, 2015).
Over the past decade, it became clear that lncRNAs play an important regulatory role in various processes, including metastasis, imprinting, tumor suppressor dysregulation, and X inactivation (Childs et al., 2015; Gao and Wei, 2017; Yu et al., 2017). In addition, aberrant expression and polymorphisms of lncRNAs are associated with susceptibility to a range of human diseases, including cancer, and these can represent new targets for the diagnosis and treatment of cancer (Amundadottir, 2016). LINC00673 may also act as a tumor suppressor by promoting interaction between protein tyrosine phosphatase, non-receptor type 11 (Ptpn11) and ubiquitin ligase, resulting in degradation of Ptpn11 and lowered oncogenic signaling (Huang et al., 2017). LINC00673 has been shown to be involved in susceptibility to and progression and outcome of many malignancies, acting as either a tumor suppressor or promoter (Wang and Luo, 2018). The relationship between LINC00673 rs11655237 and pancreatic cancer susceptibility in individuals of European descent was identified through GWAS of 9,925 cases of pancreatic cancer and 11,569 controls (Childs et al., 2015). Meanwhile, Zheng et al. (2016) replicated these findings in the Chinese population and found that rs11655237 produced a miR-1231-binding site and interfered with the degradation of PTPN11. Wang and Luo (2018) revealed that the LINC00673 rs11655237 C>T polymorphism is associated with an increased risk of cervical cancer, possibly by downregulating LINC00673 expression in cervical tissues. Zhang et al. (2018) verified that the LINC00673 rs11655237 C>T polymorphism may be associated with neuroblastoma susceptibility.
In this study, 213 HB patients and 958 cancer-free controls from four different provinces across China were genotyped to evaluate the association between the LINC00673 rs11655237 C>T polymorphism and the risk of HB. Our results showed that the LINC00673 rs11655237 C>T polymorphism may indeed affect HB susceptibility. However, HB is a multifactorial disease that may also be affected by environmental factors and genetic background. Our research is limited by the lack of valuable information on these other aspects, including dietary intake and the living environment of the parents. Selection bias is another obvious possible confounding factor, and the study population should not be considered to represent the entire Chinese population. Moreover, this study investigated only one polymorphism, and other polymorphisms of LINC00673, alone or in combination, should be investigated. Finally, environmental factors that may interact with the LINC00673 polymorphism were not investigated. The relationship between this polymorphism and patients outcome was not investigated, due to insufficient outcome data. To better elucidate the relationship of the LINC00673 polymorphism with susceptibility to HB, future studies should be designed to avoid these shortcomings as much as possible. The expression of LINC00673 may need to be tested.
Conclusion
In conclusion, our study is the first to analyze the correlation between the LINC00673 rs11655237 C>T polymorphism and HB risk among the Chinese population. Our results confirmed that the LINC00673 rs11655237 C>T polymorphism may have significant effects on HB risk in the Chinese population.
Ethics Statement
Each child’s parent or guardian provided written informed consent for the child’s participation in the study. This subject was approved by the Ethics Committee of Guangzhou Women and Children’s Medical Center.
Author Contributions
TY, JL, JZ, YX, SL, and HX conceptualized and designed the study, drafted the initial manuscript, and reviewed and revised the manuscript. YW, TT, JY, JP, CH, and YY designed the data collection instruments, collected data, carried out the initial analyses, and reviewed and revised the manuscript. YZ and JH conceptualized and designed the study, coordinated and supervised data collection, and critically reviewed the manuscript for important intellectual content. All authors approved the final manuscript as submitted and agreed to be accountable for all aspects of the work.
Funding
This study was funded by the National Natural Science Foundation of China (Nos. 81602199 and 81802333), the Guangzhou Science Technology and Innovation Commission (No. 201607010395), and the Natural Science Foundation of Guangdong Province, China (Nos. 2016A030313496 and 2018A030310053). The funding body has no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00506/full#supplementary-material
References
Amundadottir, L. T. (2016). Pancreatic cancer genetics. Int. J. Biol. Sci. 12, 314–325. doi: 10.7150/ijbs.15001
Ba, M. C., Long, H., Cui, S. Z., Gong, Y. F., Yan, Z. F., Wu, Y. B., et al. (2017). Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget 8, 95542–95553. doi: 10.18632/oncotarget.20980
Botti, G., Collina, F., Scognamiglio, G., Aquino, G., Cerrone, M., Liguori, G., et al. (2018). LncRNA HOTAIR polymorphisms association with cancer susceptibility in different tumor types. Curr. Drug Targets 19, 1220–1226. doi: 10.2174/1389450118666170622091940
Childs, E. J., Mocci, E., Campa, D., Bracci, P. M., Gallinger, S., Goggins, M., et al. (2015). Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat. Genet. 47, 911–916. doi: 10.1038/ng.3341
Colleen, C. M., Mark, S. B., Maria, J. S., Philip, C. N., and Michael, S. Z. (2006). Maternal and infant birth characteristics and hepatoblastoma. Am. J. Epidemiol. 163:818. doi: 10.1093/aje/kwj104
D’Antiga, L., Vallortigara, F., Cillo, U., Talenti, E., Rugge, M., Zancan, L., et al. (2007). Features predicting unresectability in hepatoblastoma. Cancer 110, 1050–1058. doi: 10.1002/cncr.22876
Devi, L. P., Kumar, R., Handique, A., and Kumar, M. (2014). Hepatoblastoma–a rare liver tumor with review of literature. J. Gastrointest. Cancer 45(Suppl. 1), 261–264. doi: 10.1007/s12029-014-9659-y
El Asmar, A., and El Rassi, Z. (2016). Hepatoblastoma in childhood, long term survival achieved: 2 case reports and literature review. Int. J. Surg. Case Rep. 21, 55–58. doi: 10.1016/j.ijscr.2016.02.019
Froberg, J. E., Yang, L., and Lee, J. T. (2013). Guided by RNAs: x-inactivation as a model for lncRNA function. J. Mol. Biol. 425, 3698–3706. doi: 10.1016/j.jmb.2013.06.031
Gao, P., and Wei, G. H. (2017). Genomic insight into the role of lncRNA in cancer susceptibility. Int. J. Mol. Sci. 18:E1239. doi: 10.3390/ijms18061239
Goldman, L. J., Nodal, C., and Jimenez, E. (2010). Successful airway control with the laryngeal mask in an infant with beckwith-wiedemann syndrome and hepatoblastoma for central line catheterization. Paediatr. Anaesth. 10, 445–448. doi: 10.1046/j.1460-9592.2000.00548.x
He, J., Wang, F., Zhu, J., Zhang, R., Yang, T., Zou, Y., et al. (2016). Association of potentially functional variants in the XPG gene with neuroblastoma risk in a chinese population. J. Cell. Mol. Med. 20, 1481–1490. doi: 10.1111/jcmm.12836
He, J., Zou, Y., Liu, X., Zhu, J., Zhang, J., Zhang, R., et al. (2018). Association of common genetic variants in pre-microRNAs and neuroblastoma susceptibility: a two-center study in chinese children. Mol. Ther. Nucleic Acids 11, 1–8. doi: 10.1016/j.omtn.2018.01.003
He, J., Zou, Y., Wang, T., Zhang, R., Yang, T., Zhu, J., et al. (2017). Genetic variations of GWAS-identified genes and neuroblastoma susceptibility: a replication study in southern chinese children. Transl. Oncol. 10, 936–941. doi: 10.1016/j.tranon.2017.09.008
Herzog, C. E., Andrassy, R. J., and Eftekhari, F. (2000). Childhood cancers: hepatoblastoma. Oncologist 5, 445–453. doi: 10.1634/theoncologist.5-6-445
Hirschman, B. A., Pollock, B. H., and Tomlinson, G. E. (2005). The spectrum of APC mutations in children with hepatoblastoma from familial adenomatous polyposis kindreds. J. Pediatr. 147, 263–266. doi: 10.1016/j.jpeds.2005.04.019
Huang, M., Hou, J., Wang, Y., Xie, M., Wei, C., Nie, F., et al. (2017). Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol. Ther. 25, 1014–1026. doi: 10.1016/j.ymthe.2017.01.017
Hughes, L. J., and Michels, V. V. (2010). Risk of hepatoblastoma in familial adenomatous polyposis. Am. J. Med. Genet. 43, 1023–1025. doi: 10.1002/ajmg.1320430621
Jeng, Y. M., Wu, M. Z., Mao, T. L., Chang, M. H., and Hsu, H. C. (2000). Somatic mutations of beta-catenin play a crucial role in the tumorigenesis of sporadic hepatoblastoma. Cancer Lett. 152, 45–51. doi: 10.1016/s0304-3835(99)00433-4
Jing, G., Wei, L., Jiayou, Z., Xiaoping, M., and An-Yuan, G. (2015). lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 43, D181–D186. doi: 10.1093/nar/gku1000
Li, H., Yu, B., Li, J., Su, L., Yan, M., Zhu, Z., et al. (2014). Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 5, 2318–2329.
Ma, C., Wu, G., Zhu, Q., Liu, H., Yao, Y., Yuan, D., et al. (2017). Long intergenic noncoding RNA 00673 promotes non-small-cell lung cancer metastasis by binding with EZH2 and causing epigenetic silencing of HOXA5. Oncotarget 8, 32696–32705. doi: 10.18632/oncotarget.16158
Pan, W., Liu, L., Wei, J., Ge, Y., Zhang, J., Chen, H., et al. (2016). A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol. Carcinog. 55, 90–96. doi: 10.1002/mc.22261
Pickard, M. R., and Williams, G. T. (2015). Molecular and cellular mechanisms of action of tumour suppressor GAS5 lncRNA. Genes 6, 484–499. doi: 10.3390/genes6030484
Rivand, M., Khorrami, M. S., Fiuji, H., Shahidsales, S., Hasanzadeh, M., Jazayeri, M. H., et al. (2017). The 9p21 locus: a potential therapeutic target and prognostic marker in breast cancer. J. Cell. Physiol. 233, 5170–5179. doi: 10.1002/jcp.26332
Rojas, Y., Guillerman, R. P., Zhang, W., Vasudevan, S. A., Nuchtern, J. G., and Thompson, P. A. (2014). Relapse surveillance in AFP-positive hepatoblastoma: re-evaluating the role of imaging. Pediatr. Radiol. 44, 1275–1280. doi: 10.1007/s00247-014-3000-6
Rumbajan, J. M., Maeda, T., Souzaki, R., Mitsui, K., Higashimoto, K., Nakabayashi, K., et al. (2013). Comprehensive analyses of imprinted differentially methylated regions reveal epigenetic and genetic characteristics in hepatoblastoma. BMC Cancer 13:608. doi: 10.1186/1471-2407-13-608
Shi, X., Ma, C., Zhu, Q., Yuan, D., Ming, S., Gu, X., et al. (2016). Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget 7, 25558–25575. doi: 10.18632/oncotarget.8338
Udatsu, Y., Kusafuka, T., Kuroda, S., Miao, J., and Okada, A. (2001). High frequency of β-catenin mutations in hepatoblastoma. Pediatr. Surg. Int. 17, 508–512. doi: 10.1007/s003830000576
von Schweinitz, D. (2012). Hepatoblastoma: recent developments in research and treatment. Semin. Pediatr. Surg. 21, 21–30. doi: 10.1053/j.sempedsurg.2011.10.011
Wang, Y., and Luo, T. (2018). LINC00673 rs11655237 polymorphism is associated with increased risk of cervical cancer in a chinese population. Cancer Control 25:1073274818803942. doi: 10.1177/1073274818803942
Xuefei, S., Ming, S., Hongbing, L., Yanwen, Y., and Yong, S. (2013). Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 339, 159–166. doi: 10.1016/j.canlet.2013.06.013
Yang, C., Tang, R., Ma, X., Wang, Y., Luo, D., Xu, Z., et al. (2015). Tag SNPs in long non-coding RNA H19 contribute to susceptibility to gastric cancer in the chinese han population. Oncotarget 6, 15311–15320.
Yu, J., Yan, L., Gong, Z., Zhang, S., Guo, C., Li, X., et al. (2017). Overexpression long non-coding RNALINC00673is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget 8, 16621–16632. doi: 10.18632/oncotarget.14200
Zhang, L. G., Zhou, X. K., Zhou, R. J., Lv, H. Z., and Li, W. P. (2017). Long non-coding RNA LINC00673 promotes hepatocellular carcinoma progression and metastasis through negatively regulating miR-205. Am. J. Cancer Res. 7, 2536–2544.
Zhang, Z., Chang, Y., Jia, W., Zhang, J., Zhang, R., Zhu, J., et al. (2018). LINC00673 rs11655237 C > T confers neuroblastoma susceptibility in chinese population. Biosci. Rep. 38:BSR20171667. doi: 10.1042/BSR20171667
Keywords: hepatoblastoma, cancer susceptibility, LINC00673, liver malignancy, genetic association analysis
Citation: Yang T, Li J, Wen Y, Tan T, Yang J, Pan J, Hu C, Yao Y, Zhang J, Xin Y, Li S, Xia H, He J and Zou Y (2019) LINC00673 rs11655237 C>T Polymorphism Impacts Hepatoblastoma Susceptibility in Chinese Children. Front. Genet. 10:506. doi: 10.3389/fgene.2019.00506
Received: 31 January 2019; Accepted: 07 May 2019;
Published: 24 May 2019.
Edited by:
Ihab Younis, Carnegie Mellon University in Qatar, QatarReviewed by:
Maria Gazouli, National and Kapodistrian University of Athens, GreeceTania Lee Slatter, University of Otago, New Zealand
Copyright © 2019 Yang, Li, Wen, Tan, Yang, Pan, Hu, Yao, Zhang, Xin, Li, Xia, He and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing He, aGVqaW5nMTk4Mzc0QGdtYWlsLmNvbQ==; aGVqaW5nQGd3Y21jLm9yZw==; Yan Zou, Mzc4MzE5Njk2QHFxLmNvbQ==; bW9ua251dEAxMjYuY29t
 Tianyou Yang1
Tianyou Yang1 Jing Pan
Jing Pan Huimin Xia
Huimin Xia Jing He
Jing He Yan Zou
Yan Zou