- State Key Laboratory of Developmental Biology of Freshwater Fish, College of Life Sciences, Hunan Normal University, Changsha, China
The autotetraploid fish (4n = 200, RRRR) (abbreviated as 4nRR) resulted from the whole genome duplication of red crucian carp (Carassius auratus red var., 2n = 100, RR) (abbreviated as RCC). During investigation of the influence of polyploidization on organization and evolution of the multigene family of 5S rDNA, molecular organization and chromosomal localization of the 5S rDNA were characterized in autotetraploid fish. By sequence analysis of the coding region (5S) and adjacent non-transcribed spacer (NTS), three distinct 5S rDNA units (type I: 203 bp; type II: 340 bp; and type III: 477bp) were identified and characterized in 4nRR. These 5S rDNA units were inherited from their female parent (RCC), in which obvious base variations in NTS and array recombination of repeat units were found. Using fluorescence in situ hybridization employing different 5S rDNA units as probes, these 5S rDNA clusters were localized in chromosomes of 4nRR, respectively, and showed obvious loss of chromosomal loci (type I and type II). Our data revealed genetic variation of the 5S rDNA multigene family in the genome of autopolyploid fish. Furthermore, results provided new insights into the evolutionary patterns of this vertebrate multigene family.
Introduction
Polyploidy is a significant mode of speciation in eukaryotes (Mallet, 2007; Otto, 2007), especially in vertebrates. Ohno (1970) proposed the genome duplication hypothesis, in which two rounds of whole genome duplication occurred during early vertebrate evolution. Polyploids are generally divided in to categories depending on their chromosomal composition and their manner of formation. The autopolyploids have chromosome sets coming from the genome of one species (e.g., AAAA) and exhibit multivalent pairing during meiosis, while the allopolyploids result from the combination of sets of chromosomes from two or more different taxa (e.g., AABB) and predominantly form bivalent pairings (Comai, 2005). Notably, multivalent pairing may cause meiotic irregularities and result in reduced fertility compared with diploid progenitors (Jackson, 1982; Parisod et al., 2010). Thus, vertebrate autopolyploids are relatively rare compared with allopolyploids, and the influence of autopolyploidization on intragenomic variation is poorly understood.
5S ribosomal RNA (rRNA) is a component of the large ribosomal subunit in all ribosomes. In vertebrates, the 5S ribosomal DNA (5S rDNA) is organized in tandem arrays with repeat units composed of a 120-bp coding sequence (5S) that encodes the 5S rRNA and a highly variable non-transcribed spacer (NTS) (Korn and Brown, 1978; Nielsen et al., 1993; Hallenberg and Frederiksen, 2001; Pasolini et al., 2006). Molecular organization and chromosomal localization of the 5S rDNA have been extensively characterized in bony fish (Iue et al., 1989; Rocco et al., 2005; Qin et al., 2010; Danillo et al., 2011). Polyploidization plays an important role in the evolution of fish. However, the features of the 5S rDNA have been rarely reported in polyploid fish. Previously, we successfully obtained fertile allotetraploid hybrids (4n = 148, RRBB) (abbreviated as 4nRB) from the first generation of Carassius auratus red var. (2n = 100, RR) (♀) × Megalobrama amblycephala (2n = 48, BB) (♂) hybrids (Qin et al., 2014a). The abnormal chromosomal behavior of allotetraploid hybrids during meiosis leads to the formation of autodiploid sperm and autodiploid ova, and the fertilization of these ova by these sperm in turn produces autotetraploid fish (4nRR) (Qin et al., 2014b, 2015a). Autotetraploids produce diploid ova and diploid sperm and maintain the formation of the autotetraploid line (F1–F10), which could be used as a new model system for investigating the influence of polyploidy on the organization and evolution of the multigene family of 5S rDNA. In this paper, molecular organization and chromosomal localization of the 5S rDNA have been characterized in the autotetraploid and their parents (RCC). Obvious loss of chromosomal loci, base variations in NTS, and array recombination of repeat units have been found in the newly established autotetraploidy genomes. Our results extend the knowledge of the influence of polyploidy on the organization and evolution of 5S rDNA of fish, and are also useful in clarifying aspects of vertebrate genome evolution.
Materials and Methods
Source of Samples
All fish were cultured in ponds and fed with artificial feed at the Protection Station of Polyploidy Fish, Hunan Normal University. Fish treatments were carried out according to the regulations for protected wildlife and the Administration of Affairs Concerning Animal Experimentation, and approved by the Science and Technology Bureau of China. Approval from the Department of Wildlife Administration was not required for the experiments conducted in this paper. The fish were deeply anesthetized with 100 mg/L MS-222 (Sigma-Aldrich, St. Louis, MO, United States) before dissection.
Animals and Crosses
During the reproductive season (April to June) in 2012, the first generation (4nRB) of C. auratus red var. (♀) × M. amblycephala (♂) was produced. During the reproductive season (April to June) of 2014, the second generation (4nRR) was produced by self-crossing of 4nRB.
Preparation of Chromosome Spreads
Chromosome counts were performed using kidney tissue from 10 RCC and 10 4nRR. After culture for 1–3 days at a water temperature of 18–22°C, the samples were injected with concanavalin one to three times at a dose of 2–8 mg/g body weight. The interval between injections was 12–24 h. Six hours prior to dissection each sample was injected with colchicine at a dose of 2–4 mg/g body weight. The excised kidney tissue was ground in 0.9% NaCl, followed by hypotonic treatment with 0.075 M KCl at 37°C for 40–60 min and then fixed in 3:1 methanol–acetic acid with three changes. The cells were dropped onto cold, wet slides and stained for 30 min in 4% Giemsa. The shape and number of chromosomes were analyzed under a microscope. For each type of fish, 200 metaphase spreads (20 metaphase spreads from each sample) of chromosomes were analyzed. The preparations were examined under an oil lens at a magnification of 3330×.
PCR Amplification and Sequencing of 5S rDNA Sequences
Total genomic DNA was isolated from peripheral blood cells according to the standard phenol: chloroform extraction procedure described by Sambrook et al. (1989). To acquire preliminary information on the organization of the 5S rDNA repeat variants, and to test for the possible coexistence of different repeat units in the same array, DNA samples of 3 RCC and 3 4nRR were amplified with primers 5SP1-5SP2R (5′-GCTATGCCCGATCTCGTCTGA-3′ and 5′-CAGGTTGGTAT GGCCGTAAGC-3′) and with primers 5SNT1-5SNT2R (5′-GGCGAGTAGATTGGCTGAACA-3′ and 5′-CAATCTAATCGCCAGTACATTATAT-3′). The PCR reaction was performed in a volume of 25 μL with approximately 20 ng of genomic DNA, 1.5 mM of MgCl2, 200 μM of each dNTP, 0.4 μM of each primer, and 1.25 U of Taq polymerase (Takara). The temperature profile was as follows: an initial denaturation step at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min, with a final extension step at 72°C for 10 min. Amplification products were separated on a 3.0% agarose gel using TBE buffer. The DNA fragments were purified using a gel extraction kit (Sangon) and ligated into pMD18-T (Takara). Plasmids were transformed into Escherichia coli DH5a, propagated, and then purified. The cloned DNA fragments were sequenced using an automated DNA sequencer (ABI PRISM 3730). Sequence homology and variation among the fragments amplified from 3 RCC and 3 4nRR were analyzed using ClustalW software1.
Fluorescence in situ Hybridization
The probes for fluorescence in situ hybridization (FISH) for the 5S gene were constructed for RCC and amplified by PCR using the primers 5′-GCTATGCCCGATCTCGTCTGA-3′ and 5′-CAGGTTGGTATGGCCGTAAGC-3′. The FISH probes were produced by Dig-11-dUTP labeling (using a Nick Translation Kit, Roche, Germany) of purified PCR products. Purified PCR products of 5S rDNA labeled with Dig-11-dUTP (Roche, Germany) were used as probes, and hybridization was performed according to the method described by Yi et al. (2003) with minor modifications. Purified PCR products of 5S rDNA labeled with Dig-11-dUTP (Roche, Germany) were used as probes, and hybridization was performed according to the method described by Yi et al. (2003) with minor modifications. After treatment with 30 μg/ml RNase A in 2 × SSC for 30 min at 37°C, the slides with chromosome metaphase spreads were denatured in 70% deionized formamide/2 × SSC for 2 min at 70°C, dehydrated in a 70, 90, and 100% ethanol series for 5 min each (1 × SSC is 0.15 M NaCl/0.015 M sodium citrate, pH 7.6), and then air-dried. 4 μl of the hybridization mixture (approximately 100 ng of labeled probes, 50% formamide, 10 mg dextran sulfate/ml and 2 × SSC) was denatured for 10 min in boiling water, applied to the air-dried slides carrying denatured metaphase chromosomes under a 22 × 22 mm coverslip, and sealed with rubber cement. The slides were then put in a moist chamber and allowed to incubate overnight at 37°C.
Following overnight incubation, the coverslips were removed and the slides were rinsed at 43°C in: 2 × SSC with 50% formamide, twice, 15 min each; 2 × SSC, 5 min; 1 × SSC, 5 min, then air-dried. The spectrum signals were achieved by application of 8 μl of 5 μg/ml FITC-conjugated antidigoxigenin antibody from sheep (Roche, Germany) and a final incubation in the humidity chamber at 37°C. After a series of washes with TNT (containing 0.1 M Tris–HCl, 0.15 M NaCl, 0.05% Tween 20) at 43°C, the slides were mounted in antifade solution containing 2 μg/ml 4′, 6-diamidino-2-phenylindole (DAPI) for 5 min. Slides were viewed under a Leica inverted CW4000 microscope and a Leica LCS SP2 confocal image system (Leica, Germany). Metaphase spreads of chromosomes were analyzed in 10 RCC and 10 4nRR (20 metaphase spreads in each sample).
Results
Molecular Organization of the 5S rDNA Classes
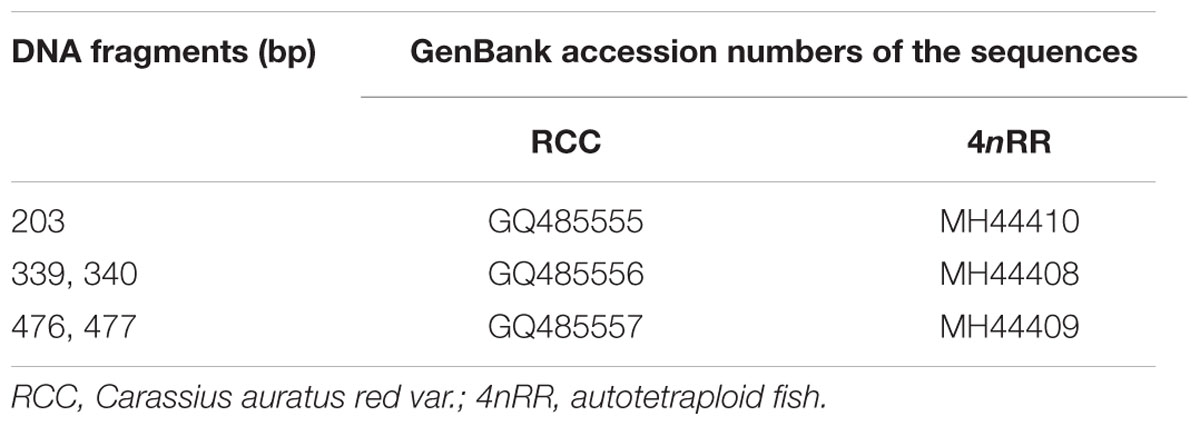
Using the primers 5SP1 and 5SP2R, fragments of approximately 200, 340, and 500 bp were generated from RCC and 4nRR (Figure 1A). All fragments proved to be 5S rDNA sequences, each included the 3′ end of the coding region (pos. 1-21), the whole NTS region, and a large 5′ portion of the coding region of the adjacent unit (pos. 22-120; see Figure 1B). In RCC, the three types of 5S rDNA classes (designated type I: 203 bp; type II: 340 bp; and type III: 477 bp) were characterized by distinct NTS types (designated NTS-I, NTS-II and NTS-III for the 83-, 220-, and 357-bp sequences, respectively; Figure 2). 4nRR had three types of 5S rDNA classes, which were completely inherited from RCC (type I, type II and type III; Figure 2). All 5S rDNA sequences have been submitted to GenBank, and their accession numbers are listed in Table 1.
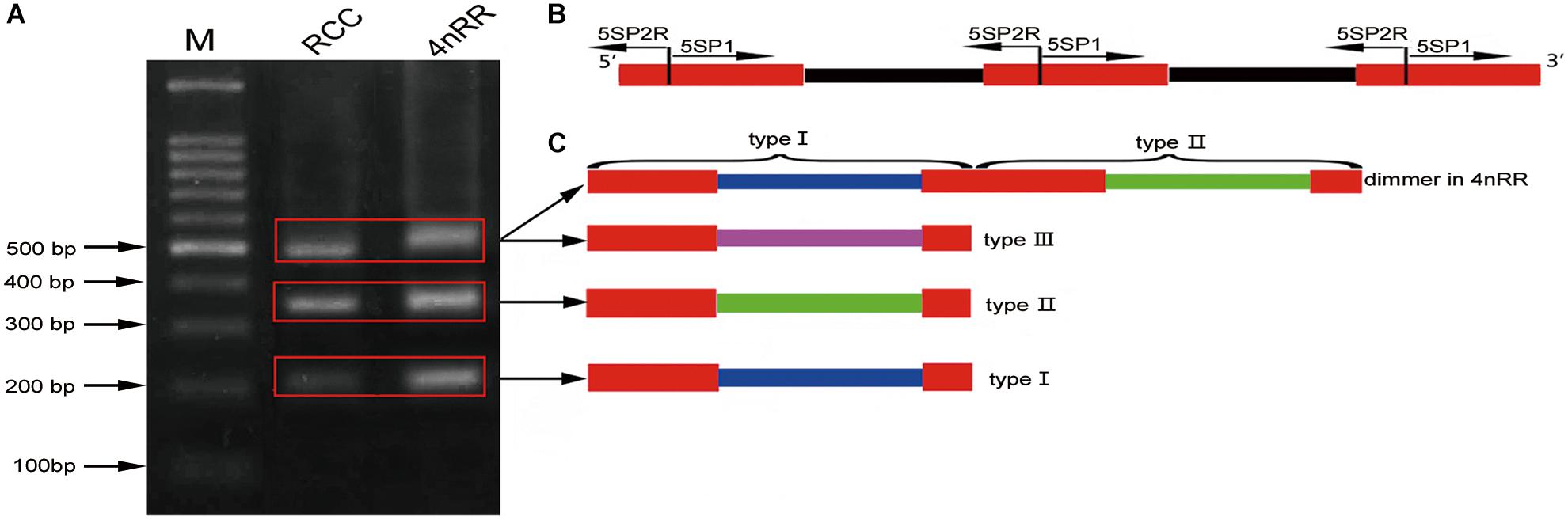
Figure 1. DNA bands amplified from RCC and 4nRR. (A) Using the primer pair 5SP1-5SP2R, DNA fragments were amplified from RCC and 4nRR. M, DNA ladder markers (100 bp increments); lane 1, three DNA fragments (approximately 200, 340, and 500 bp) from RCC; lane 2, three DNA fragments (approximately 200, 340, and 500 bp) from the 4nRR; (B) Arrangement of higher eukaryotic 5S rRNA genes (red) intercalated with non-transcribed DNA segments (NTS; black); (C) Representative sequences of 5S rDNA type I, II, and III from RCC and 4nRR; red indicate 5S rRNA genes; blue, green and purple indicate distinct NTS sequences.
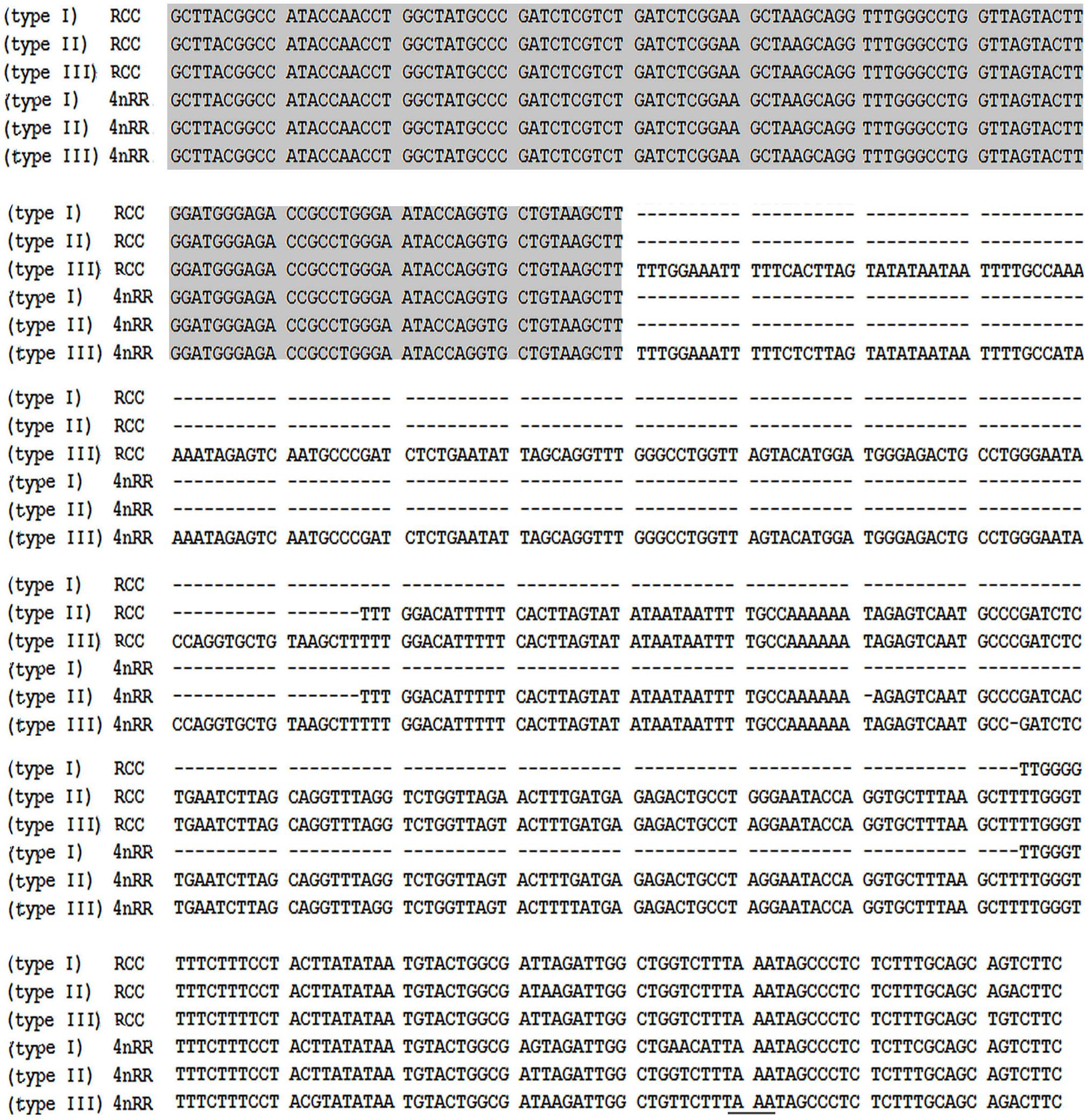
Figure 2. Representative sequences of 5S rDNA from RCC and 4nRR. Complete 5S coding regions are shaded; the NTS upstream TATA elements are underlined.
Comparison of the 120-bp coding region of 5S rDNA with those of RCC and 4nRR revealed great similarity (Figure 3). Nucleotide variation was not detected among the internal control regions (ICRs, i.e., the promoters for transcription) in 4nRR (Figure 3). A comparison of NTS-I revealed six base substitutions among the sequences (Figure 4A). A comparison of NTS-II showed five base substitutions and a deletion-insertion at position -177 (Figure 4B). A comparison of NTS-III elements showed nine base substitutions and a deletion-insertion at position -164 (Figure 4C). The above results indicate that obvious nucleotide variations were found in NTS sequences of 4nRR. In addition, characterization of the NTS-up stream region showed that the TATA control element, the regulatory region for 5S gene transcription, was identifiable in the NTS of RCC and 4nRR (at -29 in all NTS sequences, where it was modified to TAAA; Figure 4), suggesting that all sequences analyzed here were likely to correspond to functional genes.
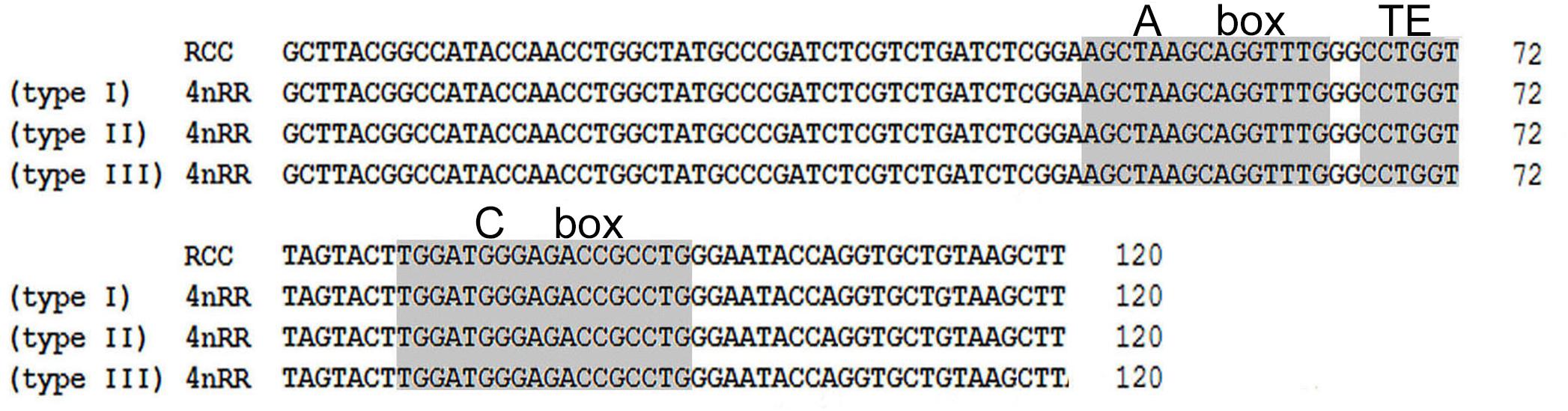
Figure 3. Comparison of 5S coding regions from RCC and 4nRR. Internal control regions of the coding region are shaded.
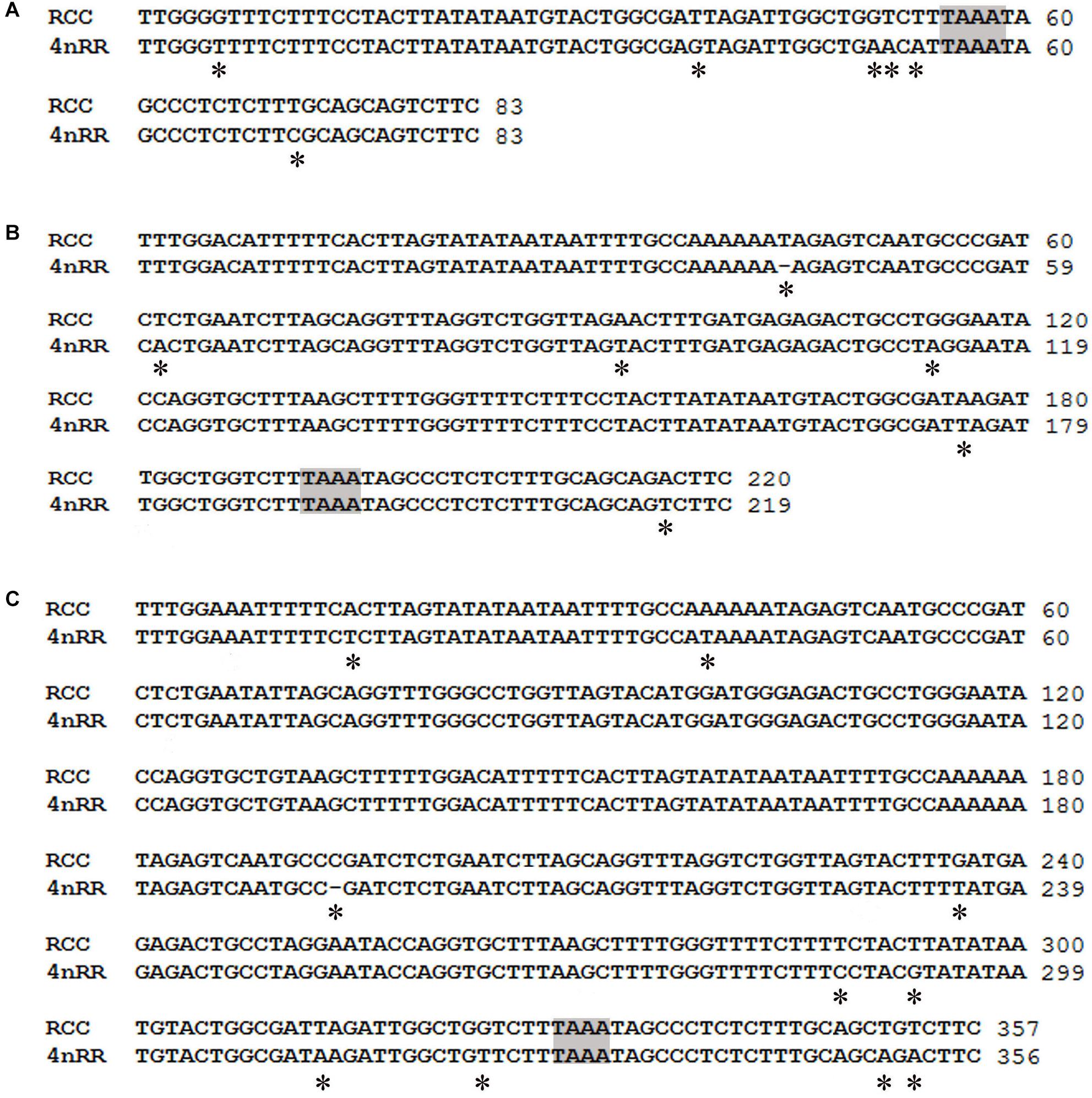
Figure 4. Comparison of the NTS sequences from RCC and 4nRR. (A) NTS-I sequences from RCC and 4nRR; (B) NTS-II from RCC and 4nRR; (C) NTS-III from RCC and 4nRR. The NTS upstream TATA elements are shaded; asterisks mark variable sites in the NTS.
Array Recombination of the 5S rDNA Repeat Units
Thirty clones of the 500 bp fragment from 4nRR were analyzed, and the sequence analysis revealed that five clones were dimeric 5S rDNA formed by 5S rDNA type I (the 99 bp gene sequence, 83 bp of NTS, and 21 bp gene sequence) and 5S rDNA type II (the 99 bp gene sequence, 220 bp of NTS, and 21 bp gene sequence) (Figure 1C and Supplementary Figure 1). To verify whether the different 5S rDNA classes (type I and type II) were associated within the same tandem array, we designed the primers 5SNT1-5SNT2R. Using these primers, the PCR yielded a single band of 352 bp in 4nRR, but no band in RCC and 4nRB (Figure 5). Sequence analysis revealed that this fragment was formed by 72 bp of the type I (a 51 bp of the NTS and 21 bp gene sequence) and 280 bp of type II (the 99 bp of gene sequence and 181 bp of NTS) (Figure 5 and Supplementary Figure 2). The PCR amplification products of the two primers provided direct evidence to prove that the type I and type II repeats were associated within the same tandem array in 4nRR, suggesting that recombination of chromosomes occurred in the autotetraploid genome.
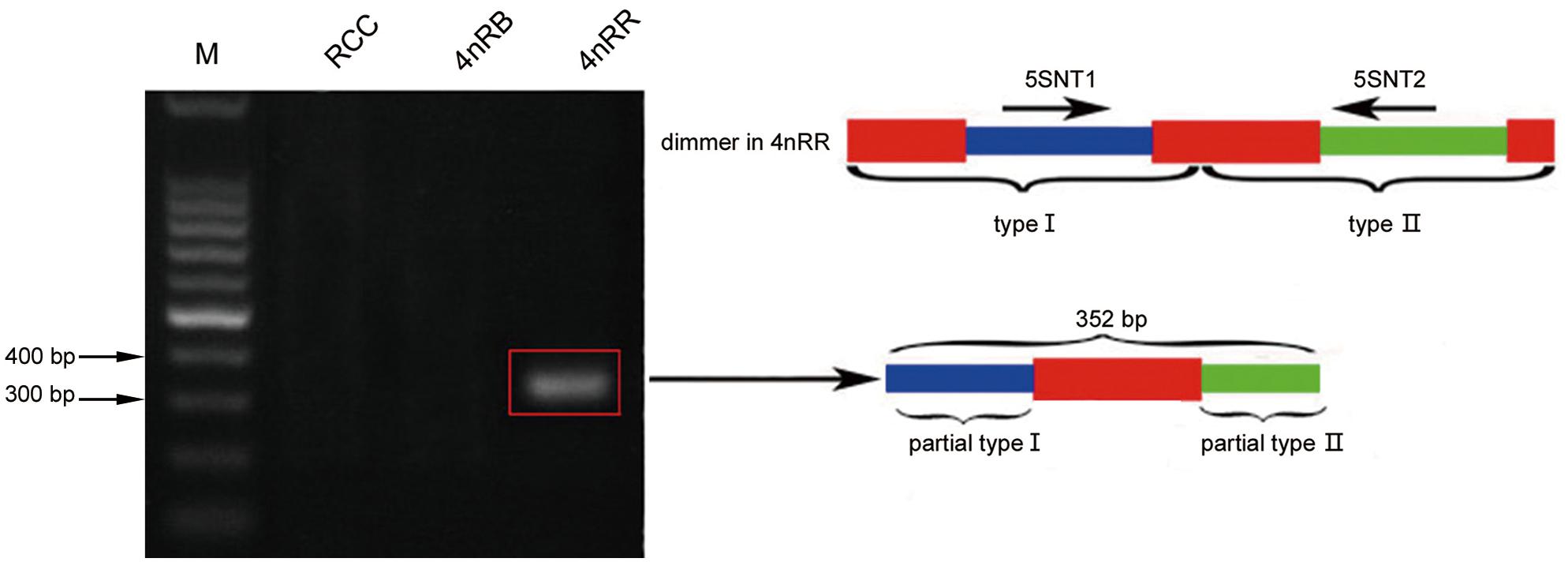
Figure 5. DNA bands amplified from 4nRR. Using the primer pair 5SNT1-5SNT2R, DNA fragments were only amplified from 4nRR (approximately 352 bp); M, DNA ladder markers (100 bp increments); red indicates the 5S rRNA gene, blue indicates NTS of type I, and green indicates NTS of type II.
Chromosomal Loci of 5S rDNA
The hybridization of type I 5S rDNA probes showed eight 5S gene loci in RCC chromosomal metaphases (Figure 6A and Table 2). Sixteen 5S gene loci were expected in 4nRR chromosomal metaphases, but only twelve 5S gene loci were found (Figure 6B and Table 2). Using type II 5S rDNA as a probe, a pair of large 5S gene loci was identified on homologous submetacentric chromosomes in RCC chromosomal metaphases, and a pair of small 5S gene loci was localized on homologous subtelocentric chromosomes (Figure 6C and Table 2). In 4nRR chromosomal metaphases, a pair of large 5S gene loci on a homologous submetacentric chromosome were found and other a pair of large 5S gene loci on a homologous submetacentric chromosome were lost; two pairs of small 5S gene loci was localized on homologous subtelocentric chromosomes (Figure 6D and Table 2). FISH hybridization of the type III 5S rDNA probe to the RCC metaphase chromosomes yielded eight 5S gene loci (Figure 6E and Table 2). As expected, sixteen 5S gene loci were found in 4nRR chromosomal metaphases (Figure 6F and Table 2). The above results indicate that obvious loss of chromosomal loci occurred in 4nRR.
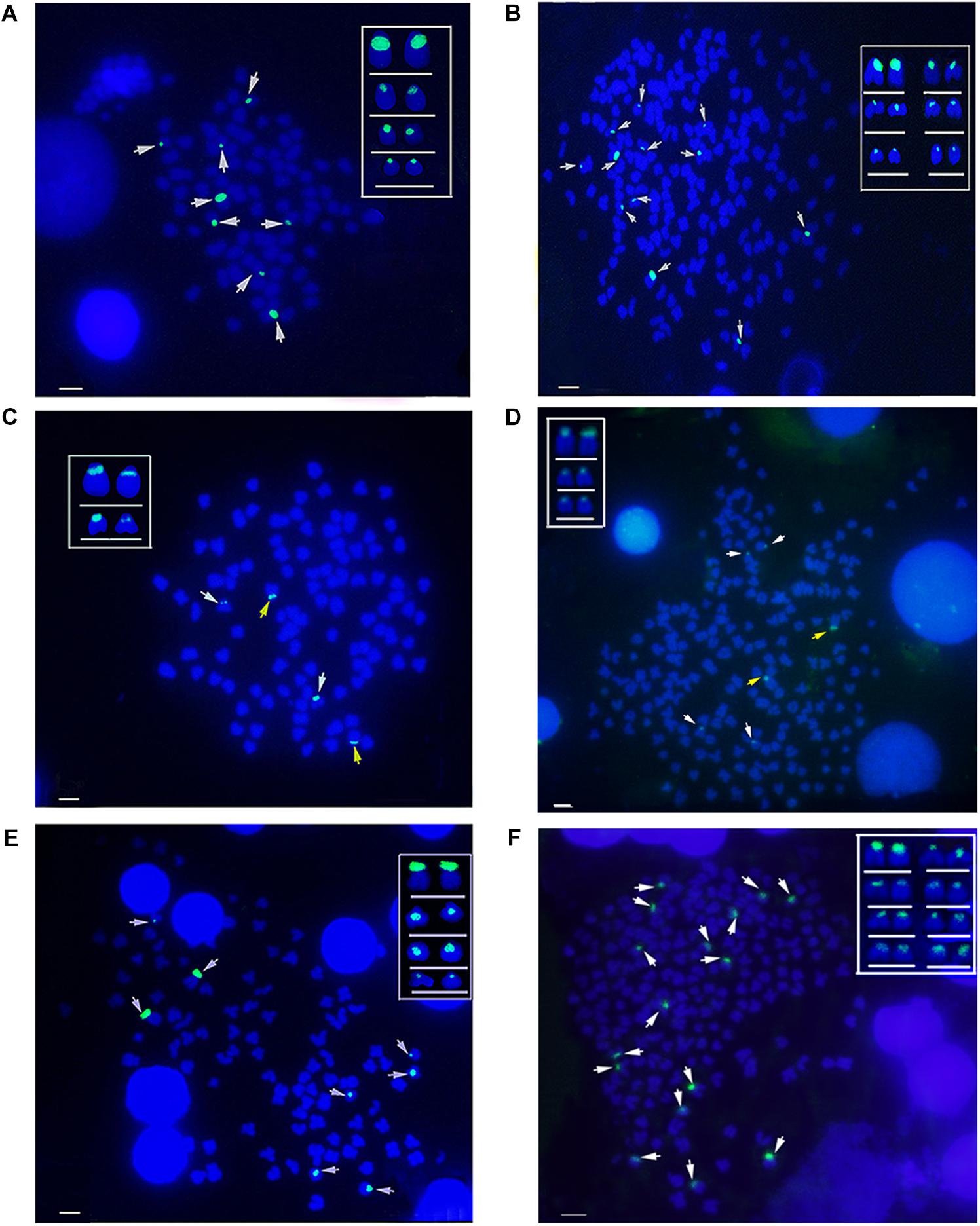
Figure 6. Examination of hybridizing signals by FISH in RCC and 4nRR. (A) Type I as probe showed eight 5S gene loci (white arrows) in RCC; (B) Type I as probe showed twelve 5S gene loci of type I (white arrows) in 4nRR; (C) Type II as probe showed two big (yellow arrows) and two small 5S gene loci (white arrows) in RCC; (D) Type II as probe showed two big (yellow arrows) and four small 5S gene loci (white arrows) in 4nRR; (E) Type III as probe showed eight 5S gene loci (white arrows) in RCC; (F) Type III as probe sixteen 5S gene loci (white arrows) in 4nRR. Bar = 3 μm.
Discussion
The evolution of 5S rDNA is driven by birth-and-death processes with strongly purifying selection (Nei and Rooney, 2005; Pinhal et al., 2011; Vizoso et al., 2011), which can lead to the existence of different types of NTS (Pinhal et al., 2011). In teleosts, two distinct 5S rDNA classes are characterized by distinct NTS types and base substitutions in the 5S rRNA gene (Pendas et al., 1994; Moran et al., 1996; Martins et al., 2000; Wasko et al., 2001; Pinhal et al., 2009). Thus, possession of two 5S rDNA classes seems to be a general trend for the organization of these sequences in the genomes of fish (Martins and Galetti, 2001). As ancient polyploidy fish, RCC have undergone an additional round of whole-genome duplication (Qin et al., 2016b). The origin of genic variants has been attributed to events of genome duplication followed by processes that result in the divergence of the duplicated sequences. Thus, RCC possess three distinct 5S rDNA classes that are characterized by distinct types of NTS (Qin et al., 2010). In the current study, 4nRR derived from the distant hybridization of C. auratus red var. (2n = 100, RR) (♀) × M. amblycephala (2n = 48, BB) (♂), possess four sets of RCC-derived chromosomes and exhibit stability in chromosome number (or ploidy) over consecutive generations (F1–F10) (Qin et al., 2014b). 4nRR have three distinct 5S rDNA classes that are completely inherited from RCC, but no new types of 5S rDNA class were found, suggesting that divergence of the duplicated 5S rDNA sequences were not fully formed in the early generations of the autotetraploid fish.
Because of incompatibility between parental chromosomes, allopolyploidization can increase genomic changes (Pontes et al., 2004). Our previous study revealed the influence of allopolyploidy on 5S rDNA in fish, including parental genome specific loss, substitutions, and insertions-deletions in the NTS sequence (Qin et al., 2010, 2016a). Theoretically, homologous chromosomes should have high compatibility in autotetraploids. In this paper, however, obvious base variation and insertions-deletions of NTS were also observed in 4nRR, suggesting that autotetraploidization could lead to genetic variation in newly established autotetraploid genomes. Although there was genetic variation in NTS of 5S rDNA, all sequences analyzed here were likely to correspond to functional genes, because they exhibited all the necessary features for correct gene expression: three ICRs (box A, internal element, and box C), a TATA control element, and a T-rich tail.
Autopolyploids are traditionally used to demonstrate multivalent pairing multivalent pairing during meiosis. However, the coexistence of four homologous chromosome sets does not result in multivalent formation during meiosis in 4nRR, and diploid-like chromosome pairing was restored (Qin et al., 2019). The presence of two distinct 5S rDNA sequence types organized in different chromosomal regions or even on different chromosomes has been described for several fish (Pendas et al., 1994; Moran et al., 1996; Sajdak et al., 1998; Martins et al., 2002; Rodrigues et al., 2012; Qin et al., 2015b). In the current study, the different 5S rDNA classes (type I and type II) were associated within the same tandem array in 4nRR. In addition, type I and type II 5S rDNA clusters were localized in the chromosomes of 4nRR, and showed obvious loss of chromosomal loci. These findings are clear evidence that elimination of repetitive sequences and recombination of chromosomes occurred in newly established autotetraploid genomes. A positive linear relationship was found between increased bivalent pairing and elimination of specific, low-copy DNA sequences (Wendel, 2000). Thus, we speculate that the elimination of DNA sequences or recombination of chromosomes might generate immediate divergence between homologous chromosomes, providing a physical basis for diploid-like chromosome pairing in 4nRR.
Ethics Statement
Fish treatments were carried out according to the regulations for protected wildlife and the Administration of Affairs Concerning Animal Experimentation, and approved by the Science and Technology Bureau of China. Approval from the Department of Wildlife Administration was not required for the experiments conducted in this manuscript. The fish were deeply anesthetized with 100 mg/L MS-222 (Sigma-Aldrich, St. Louis, MO, United States) before dissection.
Author Contributions
QQ and SL designed the experiments. QL, CW, LC, YZ, HQ, and CZ performed the experiments. QQ and QL performed the statistical analysis. QQ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was financially supported by grants from the Natural Science Foundation of Hunan Province for Distinguished Young Scholars (Grant No. 2017JJ1022), the National Natural Science Foundation of China (Grant Nos. 31430088 and 31210103918), the Major Program of the Educational Commission of Hunan Province (Grant No. 17A133), the State Key Laboratory of Developmental Biology of Freshwater Fish, the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (20134486), the Earmarked Fund for China Agriculture Research System (CARS-45), and the Construction Project of Key Disciplines of Hunan Province and China.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to sincerely thank many researchers who help to complete this manuscript, including Drs. Yao Zhanzhou and Zhao Rurong.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00437/full#supplementary-material
Abbreviations
4nRB, allotetraploid hybrids; 4nRR, autotetraploid fish; BSB, blunt snout bream; FISH, fluorescence in situ hybridization; ICRs, internal control regions; NTS, non-transcribed spacer; PCR, polymerase chain reaction; RCC, red crucian carp.
Footnotes
References
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Danillo, P., Yoshimura, T. S., Araki, C. S., and Martins, C. (2011). The 5S rDNA family evolves through concerted and birth-and-death evolution in fish genomes: an example from freshwater stingrays. BMC Evol. Biol. 11:151. doi: 10.1186/1471-2148-11-151
Hallenberg, C., and Frederiksen, S. (2001). Effect of mutations in the upstream promoter on the transcription of human 5S rRNA genes. Biochim. Biophys. Acta 1520, 169–173. doi: 10.1016/S0167-4781(01)00264-0
Iue, S., Shostak, N. G., Kuprianova, N. S., Serenkova, T. I., Fel’Gengauér, P. E., Gimalov, F., et al. (1989). Intragenomic polymorphism of the primary structure of 5s rRNA gene variants of the loach (misgurnus fossilis l.). Determination of the transcriptional activity. Mol. Biol. 23, 1295–1308.
Jackson, R. C. (1982). Polyploidy and diploidy: new perspectives on chromosome pairing and its evolutionary implications. Am. J. Bot. 69, 1512–1523. doi: 10.1002/j.1537-2197.1982.tb13400.x
Korn, L. J., and Brown, D. D. (1978). Nucleotide sequence of Xenopus borealis oocyte 5S DNA: comparison of sequences that flank several related eucaryotic genes. Cell 15, 1145–1156. doi: 10.1016/0092-8674(78)90042-9
Martins, C., and Galetti, P. M. Jr. (2001). Two 5S rDNA arrays in neotropical fish species: is it a general rule for fishes? Genetica 111, 439–446. doi: 10.1023/A:1013799516717
Martins, C., Wasko, A. P., Oliveira, C., Porto-Foresti, F., Parise-Maltempi, P. P., Wright, J. M., et al. (2002). Dynamics of 5S rDNA in the tilapia (Oreochromis niloticus) genome: repeat units, inverted sequences, pseudogenes and chromosome loci. Cytogenet. Genome Res. 98, 78–85. doi: 10.1159/000068542
Martins, C., Wasko, A. P., Oliveira, C., and Wright, J. M. (2000). Nucleotide sequence of 5s rDNA and localization of the ribosomal RNA genes to metaphase chromosomes of the tilapiine cichlid fish, oreochromis niloticus. Hereditas 133, 39–46. doi: 10.1111/j.1601-5223.2000.00039.x
Moran, P., Martinez, J. L., Garcia-Vazquez, E., and Pendas, A. M. (1996). Sex chromosome linkage of 5S rDNA in rainbow trout (Oncorhynchus mykiss). Cytogenet. Genome Res. 75, 145–150. doi: 10.1159/000134466
Nei, M., and Rooney, A. P. (2005). Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39, 121–152. doi: 10.1146/annurev.genet.39.073003.112240
Nielsen, J. N., Hallenberg, C., Frederiksen, S., Sørensen, P. D., and Lomholt, B. (1993). Transcription of human 5S rRNA genes is influenced by an upstream DNA sequence. Nucleic Acids Res. 21, 3631–3636. doi: 10.1093/nar/21.16.3631
Otto, S. P. (2007). The evolutionary consequences of polyploidy. Cell 131, 452–462. doi: 10.1016/j.cell.2007.10.022
Parisod, C., Holderegger, R., and Brochmann, C. (2010). Evolutionary consequences of autopolyploidy. New Phytol. 186, 5–17. doi: 10.1111/j.1469-8137.2009.03142.x
Pasolini, P., Costagliola, D., Rocco, L., and Tinti, F. (2006). Molecular organization of 5S rDNAs in Rajidae (Chondrichthyes): structural features and evolution of piscine 5S rRNA genes and nontranscribed intergenic spacers. J. Mol. Evol. 62, 564–574. doi: 10.1007/s00239-005-0118-z
Pendas, A. M., Moran, P., Freije, J. P., and Garcia-Vazquez, E. (1994). Chromosomal mapping and nucleotide sequence of two tandem repeats of Atlantic salmon 5S rDNA. Cytogenet. Cell Genet. 67, 31–36. doi: 10.1159/000133792
Pinhal, D., Araki, C. S., Gadig, O. B. F., and Martins, C. (2009). Molecular organization of 5s rdna in sharks of the genus Rhizoprionodon: insights into the evolutionary dynamics of 5s rdna in vertebrate genomes. Genet. Res. 91, 61–72. doi: 10.1017/s0016672308009993
Pinhal, D., Yoshimura, T. S., Araki, C. S., and Martins, C. (2011). The 5s rDNA family evolves through concerted and birth-and-death evolution in fish genomes: an example from freshwater stingrays. BMC Evol. Biol. 11:151. doi: 10.1186/1471-2148-11-151
Pontes, O., Neves, N., Silva, M., Lewis, M. S., Madlung, A., Comai, L., et al. (2004). Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid arabidopsis suecica genome. Proc. Natl. Acad. Sci. U.S.A. 101, 18240–18245. doi: 10.1073/pnas.0407258102
Qin, Q., Cao, L., Wang, Y., Ren, L., Liu, Q., Zhou, Y., et al. (2019). Rapid genomic and genetic changes in the first generation of autotetraploid lineages derived from distant hybridization of Carassius auratus, red var. (♀) × Megalobrama amblycephala, (♂). Mar. Biotechnol. 21, 139–149. doi: 10.1007/s10126-018-9859-8
Qin, Q., He, W., Liu, S., Wang, J., Xiao, J., and Liu, Y. (2010). Analysis of 5S rDNA organization and variation in polyploid hybrids from crosses of different fish subfamilies. J. Exp. Zool. B Mol. Dev. Evol. 314, 403–411. doi: 10.1002/jez.b.21346
Qin, Q., Lai, Z., Cao, L., Xiao, Q., Wang, Y., and Liu, S. (2016a). Rapid genomic changes in allopolyploids of Carassius auratus red var. (♀) × Megalobrama amblycephala (♂). Sci. Rep. 6:34417. doi: 10.1038/srep34417
Qin, Q., Wang, J., Hu, M., Huang, S., and Liu, S. (2016b). Autotriploid origin of Carassius auratus as revealed by chromosomal locus analysis. Sci. China Life Sci. 59, 622–626. doi: 10.1007/s11427-016-5040-7
Qin, Q., Wang, J., Dai, J., Wang, Y., Liu, Y., and Liu, S. (2015a). Induced All-female autotriploidy in the allotetraploids of Carassius auratus red var. (♀) × Megalobrama amblycephala (♂). Mar. Biotechnol. 17, 604–612. doi: 10.1007/s10126-015-9647-7
Qin, Q., Wang, J., Wang, Y., Liu, Y., and Liu, S. (2015b). Organization and variation analysis of 5S rDNA in gynogenetic offspring of Carassius auratus red var. (♀) × Megalobrama amblycephala (♂). BMC Genet. 16:26. doi: 10.1186/s12863-015-0186-z
Qin, Q., Wang, Y., Wang, J., Dai, J., Liu, Y., and Liu, S. (2014a). Abnormal chromosome behavior during meiosis in the allotetraploid of Carassius auratus red var. (♀) × Megalobrama amblycephala (♂). BMC Genet. 15:95. doi: 10.1186/s12863-014-0095-6
Qin, Q., Wang, Y., Wang, J., Dai, J., Xiao, J., Hu, F., et al. (2014b). The autotetraploid fish derived from hybridization of Carassius auratus red var. (Female) × Megalobrama amblycephala (Male). Biol. Reprod. 91:93. doi: 10.1095/biolreprod.114.122283
Rocco, L., Costagliola, D., Fiorillo, M., Tinti, F., and Stingo, V. (2005). Molecular and chromosomal analysis of ribosomal cistrons in two cartilaginous fish, Taeniura lymma and Raja montagui (Chondrichthyes, Batoidea). Genetica 123, 245–253. doi: 10.1007/s10709-004-2451-3
Rodrigues, D. S., Rivera, M., and Lourenço, L. B. (2012). Molecular organization and chromosomal localization of 5S rDNA in AmazonianEngystomops (Anura, Leiuperidae). BMC Genet. 13:17. doi: 10.1186/1471-2156-13-17
Sajdak, S. L., Reed, K. M., and Phillips, R. B. (1998). Intraindividual and interspecies variation in the 5S rDNA of coregonid fish. J. Mol. Evol. 46, 680–688. doi: 10.1007/PL00006348
Sambrook, J., Fritsch, E., and Maniatis, T. (1989). Molecular cloning: a laboratory manual+ cold spring harbor. Immunology 49:411.
Vizoso, M., Vierna, J., González-Tizón, A. M., and Martínez-Lage, A. (2011). The 5S rDNA gene family in mollusks: characterization of transcriptional regulatory regions, prediction of secondary structures, and long-term evolution, with special attention to Mytilidae mussels. J. Hered. 102, 433–447. doi: 10.1093/jhered/esr046
Wasko, A. P., Martins, C., Wright, J. M., and Galetti, P. M. Jr. (2001). Molecular organization of 5S rDNA in fishes of the genus Brycon. Genome 44, 893–902. doi: 10.1139/gen-44-5-893
Wendel, J. F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42, 225–249. doi: 10.1007/978-94-011-4221-2_12
Keywords: autotetraploid line, distant hybridization, 5S rDNA, FISH, chromosomal loci
Citation: Qin QB, Liu QW, Wang CQ, Cao L, Zhou YW, Qin H, Zhao C and Liu SJ (2019) Molecular Organization and Chromosomal Localization Analysis of 5S rDNA Clusters in Autotetraploids Derived From Carassius auratus Red Var. (♀) × Megalobrama amblycephala (♂). Front. Genet. 10:437. doi: 10.3389/fgene.2019.00437
Received: 08 September 2018; Accepted: 29 April 2019;
Published: 15 May 2019.
Edited by:
Ancha Baranova, George Mason University, United StatesReviewed by:
Lenin Arias Rodriguez, Universidad Juárez Autónoma de Tabasco, MexicoJingou Tong, Institute of Hydrobiology (CAS), China
Copyright © 2019 Qin, Liu, Wang, Cao, Zhou, Qin, Zhao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: ShaoJun Liu, bHNqQGh1bm51LmVkdS5jbg==
 QinBo Qin
QinBo Qin ShaoJun Liu
ShaoJun Liu