
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 01 May 2019
Sec. Evolutionary and Population Genetics
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00363
 Hong Nan1,2
Hong Nan1,2 Li-zhi Gao1,3*
Li-zhi Gao1,3*The WRKY gene family plays a vital role in plant development and environment response. Although previous studies suggested that the WRKY genes in carrot (Kuroda type) involved in biotic and abiotic stress responses, the information of WRKY genes in the latest version of the carrot genome (Daucus carota v2.0, Nantes type carrot) and their response to hormone and injury stresses have not been reported. In this study, we performed a genome-wide analysis of WRKYs using a chromosome-scale genome assembly of carrot (Daucus carota subsp. sativus L.). We identified a total of 67 WRKY genes, which were further classified into the three groups. These WRKY genes are unevenly distributed on carrot chromosomes. We found that more than half of them were derived from whole-genome duplication (WGD) events, suggesting that WGDs have played a major role during the evolution of the WRKY gene family. We experimentally ascertained the expression divergence existed between WGD-derived WRKY duplicated gene pairs, which is indicative of functional differentiation between duplicated genes. Our analysis of cis-acting elements indicated that WRKY genes were transcriptionally regulated upon hormone and mechanic injury stresses. Gene expression analyses by qRT-PCR further presented that WRKY genes were involved in hormone and mechanic injury stresses.
Plants often encounter numerous environmental fluctuations containing abiotic and biotic stresses. However, evolutionary alterations have helped them to adapt to these adverse conditions by controlling a network of certain genes through modulating specific transcription factors (TFs). TFs are proteins that can regulate gene expression through binding to specific DNA region adjacent to genes and involve in controlling many important biological processes in the gene transcription regulatory network. In plants, a large number of genes were identified as TFs (Broun, 2004; Yao et al., 2015).
The WRKY family is one of the largest 10 gene families of TFs in higher plants, which has been extensively analyzed in numerous plants since the first WRKY gene was identified in Ipomoea batatas (Ishiguro and Nakamura, 1994; Rushton et al., 2010). The domain of WRKY transcription factor is about 60 residues in length, containing the highly conserved WRKYGQK signature at the N-terminus, and atypical zinc-finger structure at the C-terminus (Eulgem et al., 2000). In a few WRKY proteins, the conserved WRKYGQK amino acid sequences can also be replaced by other various forms, such as WRKYDHK, WRKYDQK, and WRKYGKK (Li M. Y. et al., 2016). And, the zinc-finger structure is either Cx4-5Cx22-23HxH (C2H2) or Cx7Cx23HxC (C2HC). The WRKY TFs can recognize and bind to the W-box promoter cis-element with a consensus C/TTGACC/T sequence (Ciolkowski et al., 2008). Previous studies suggested that the WRKY gene family can be divided into three groups based on the number of WRKY domains and the type of zinc-finger (Eulgem et al., 2000; Yousfi et al., 2016). Group I members have two WRKY domains and a C2H2 zinc-finger type. Group II contains one WRKY domain and a C2H2 (Cx4-5Cx22-23HxH) zinc-finger motif. On the basis of primary amino acid sequence, group II can be further divided into the five subgroups, namely II-a, II-b, II-c, II-d, and II-e. Proteins from group III also have a single WRKY domain but with a C2HC (Cx7Cx23HxC) motif (Eulgem et al., 2000; Guo et al., 2014). Recent studies further reported that the WRKY gene family is more accurately divided into groups I, II (II-a + II-b, II-c, II-d + II-e) and III based on phylogenetic relationships in higher plants (Huang et al., 2015).
WRKY transcription factors play an important role in developmental and physiological processes of plants. For example, the WRKY transcription factor ScWRKY1 isolated from Solanum chacoense was found to be strongly and transiently expressed in fertilized ovules at late torpedo-staged embryos, suggesting a specific role during embryogenesis (Lagac and Matton, 2004). OsWRKY11 in rice was reported to control flowering time and plant height, and the high expression of OsWRKY11 leads to dwarfism and late flowering for both long-day and short-day conditions (Cai et al., 2014). In addition, WRKY genes were also reported to involve in the regulation of diverse biotic and abiotic stresses, such as bacterial (Tao et al., 2009), pathogens (Kim et al., 2008), salinity (Qin et al., 2015), cold (Ramamoorthy et al., 2008), wounding (Wang et al., 2014), heat, and drought (Wu et al., 2009). The transgenic Arabidopsis thaliana lines expressing VqWRKY52 from wild grape displayed strong resistance to powdery mildew and Pseudomonas syringae pv. tomato DC3000 while compared with wild type plants (Wang et al., 2017). Over-expression of VlWRKY48 in Arabidopsis could increase the tolerance of fungal infection and drought stresses (Zhao et al., 2017). In Arabidopsis, AtWRKY30 was greatly induced by salt, drought, H2O2, and mannitol, and overexpression of AtWRKY30 was found to enhance plant tolerance to salinity stress (Scarpeci et al., 2013). GmWRKY21-transgenic Arabidopsis plants were involved in cold stress (Zhou et al., 2008). GhWRKY40, a cotton WRKY gene, was found to play an important role in the wounding- and pathogen-induced responses in transgenic Nicotiana benthamiana (Wang et al., 2014). The grape VlWRKY3 gene was identified to improve the tolerance to salt and drought stresses and resistance to Golovinomyces cichoracearum in transgenic A. thaliana (Guo et al., 2018). Besides, overexpression of OsWRKY11 could increase the tolerance to heat and drought stresses in transgenic rice seedlings (Wu et al., 2009).
WRKY members also play essential roles in signal transduction processes with the involvement of hormones, such as abscisic acid (ABA), salicylic acid (SA), gibberellins (GA), methyl jasmonate (MeJA), and brassinosteroid (BR), which participated in plant immune responses and abiotic stresses. OsWRKY45 was up-regulated by ABA, and overexpression of this gene in Arabidopsis resulted in the enhanced resistance to disease, salt and drought stresses (Qiu and Yu, 2009). In Arabidopsis, WRKY46 was specifically induced by SA, and it coordinated with WRKY70 and WRKY53 in basal resistance against pathogen P. syringae (Hu et al., 2012). AtWRKY12 and AtWRKY13 were involved in the GA signaling regulation of plant flowering time (Li W. et al., 2016). In American ginseng, methyl jasmonate-inducible PqWRKY1 gene was involved in osmotic stress and triterpene ginsenoside biosynthesis (Sun et al., 2013). Additionally, three Arabidopsis WRKY members, AtWRKY46, AtWRKY54, and AtWRKY70, were involved in both BR-regulated plant growth and drought response (Chen et al., 2017).
Carrot (Daucus carota subsp. sativus L.), belonging to the Apiaceae family, is a globally important root crop with great economic values. Its roots contain high quantities of alpha- and beta-carotene, serving as a good source of vitamin K and vitamin B6 (Pinheiro-Santana et al., 1998). Although the WRKY family was preliminarily investigated in a draft genome assembly of carrot (Li M. Y. et al., 2016), tissue-specific expression profiling and the abundance of WRKY genes under ABA, GA, and mechanic injury treatments have not yet been studied. The generation of high-quality genome of carrot at chromosome level (Iorizzo et al., 2016) provides an unprecedented opportunity to perform a genome-wide identification of WRKY transcription factor (TF) family. In this study, we accurately characterized the number, structure, chromosomal locations, and phylogenetic relationships of WRKY TF family throughout the carrot genome. We also performed a genome-wide identification of stress-related cis-elements in promoters of WRKY genes. We comprehensively investigated origins and evolution of the duplicated WRKY genes and the expression atlas of WRKY genes under abiotic stresses across tissues. This study provides an in-depth insight into the evolution and expression of WRKY gene family in carrot.
To comprehensively identify the carrot WRKY genes, genome sequences of this plant were downloaded from the Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#) (Iorizzo et al., 2016), and the WRKY domain (PF03106) was downloaded from Pfam (http://pfam.xfam.org/). All candidate carrot WRKY genes were firstly obtained via searching against the genome with PF03106 file using HMMER3.0 software (http://hmmer.janelia.org/) with parameters as “-E 1e-10 -domE 1e-10.” Then, the conserved domain peptides of the initially identified WRKY members were aligned with MUSCLE (Edgar, 2004) to build carrot-specific HMM file for the WRKY family, and the file was used for the next HMM searches. Finally, the sequences were confirmed using SMART database (http://smart.embl-heidelberg.de/) (Letunic et al., 2011). After manually removing incorrect and redundant predicted proteins, the WRKY gene members were finally identified in carrot.
As a control, the grape protein sequences were downloaded from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#), which were analyzed using the same method as described above. The deduced grape WRKY genes were named as VvWRKY1 to VvWRKY 59 according to Guo et al. (2014). The sequences of Arabidopsis WRKY genes were downloaded from TAIR (https://www.arabidopsis.org/).
The protein sequences of DcsWRKY members were aligned using CLUSTAL method implemented in MEGA and divided into different subgroups according to previous studies (Xie et al., 2005; Xu et al., 2016). In brief, Subgroup I contain two WRKY domains with C2H2 or C2HC. Subgroup II contain one WRKY motif with C2H2 motif and can be divided into five subgroups based on sequences variances in zinc-finger motif, including II-a (CX5CPVKKK(L/V)Q), II-b (CX5CPVRKQVQ), II-c (CX4C), and II-d (CX5CPARKHVE), II-e (CX5CPARK(Q/M)V(E/D). Subgroup III also contain one WRKY motif but with C2HC motif.
To investigate protein properties of the DcsWRKYs, molecular weight (MW) and isoelectric point (PI) were computed using the online ExPASy-ProtParam tool (http://web.expasy.org/protparam/). In order to identify orthologs in A. thaliana for each DcsWRKY gene, we performed BLASTP to search against the well-categorized A. thaliana WRKY sequences with parameters as “E < 1e-15,” and then, the top hit was collected.
To locate positions of DcsWRKY genes on the carrot chromosomes, MapInspect Software (http://www.softsea.com/download/MapInspect.html) was used to investigate the distribution of the putative DcsWRKY members based on the genome annotation (GFF3) file of carrot. The file was obtained from Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html#).
To investigate phylogenetic relationships of DcsWRKYs and assist their classification, carrot WRKY domain regions together with those from Arabidopsis and grape were aligned with CLUSTAL software (Larkin et al., 2007). The phylogenetic tree was created using Neighbor-joining (NJ) method implemented with MEGA (Tamura et al., 2011). Bootstrap values were calculated for 1,000 iterations. In order to further examine the evolution of DcsWRKY genes, the full-length proteins from all the predicted DcsWRKYs were aligned with CLUSTAL. Hence, a ML (Maximum-Likelihood) phylogenetic tree was constructed. Bootstrap values were also calculated for 1,000 iterations. MEME suite (http://meme-suite.org/tools/meme) (Bailey et al., 2009) was employed to analyze the motifs in each deduced DcsWRKY proteins. And the parameters were set as follows: maximum number of motifs, 10; minimum width, 6; and maximum width, 50.
The upstream 1.5 kb sequences of WRKY genes were extracted with an in-house Perl script, and the cis-elements were identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
For syntenic analysis of DcsWRKY genes, MCScanX (Wang et al., 2012) was employed to detect syntenic gene pairs in the carrot genome. All the proteins were compared against themselves using BLASTP (e-value, 1e-10 and outfmt 6). The BLASTP tabular file and a simplified DcsWRKY gene location file, which contains chromosome name, gene symbol, start location, and end location, were used as an input for MCScanX with default settings to identify syntenic gene pairs, and the gene type were also determined using MCScanX software.
To estimate divergence times of duplicated genes of DcsWRKY, the alignment was performed using MUSCLE and synonymous rate (Ks), non-synonymous rate (Ka), and evolutionary constraint (Ka/Ks) were calculated using the PAML software (Yang, 2007). Divergence times of all duplicated DcsWRKY genes were calculated using the formula T = Ks/2r according to the substitution rate of 6.5 × 10−9 mutations per site per year (Gaut et al., 1996).
Carrot tissues (leaves, storage roots, and stems) were obtained from a carrot individual of Nantes type grown in the greenhouse. To obtain the expression profiles of DcsWRKY genes under hormone treatments, leaves were treated with 300 μM ABA and 100 μM GA. And then, leaves were sampled at 1, 12, 24, and 48 h after the treatments. Carrot leaves sprayed with sterile water were used as a control. The treatment of mechanical damage stress was performed using forceps, and wounded leaf samples were collected at 1, 4, 8, and 12 h after treatments. Leaves without damage treatments collected on different plants were used as a control. All samples were then harvested, frozen immediately in liquid nitrogen, and stored at −80°C for RNA extraction. Three biological replicate samples were contained for each treatment.
Total RNA was extracted from samples using a modified CTAB method. And then, the SuperScript III Reverse Transcriptase kit (Invitrogen) was used to synthesize the first-strand cDNA with 1 μg of total RNA from each sample. Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed on real-time PCR detection system (Bio-Rad). The primer sequences used were designed based on WRKY gene sequences using Primer premier 5.0 software. These sequences were subsequently verified using the BLAST tool at NCBI and a dissociation curve was also analyzed after the PCR reaction to check their specificity. And the gene special primers were listed in Supplementary material (Table S1). Each reaction was carried out in a 10 μL volume, consisting of 5 μL SYBR, 3.6 μL ddH2O, 1 μL template cDNA, and 0.2 μL of each primer. The RT-PCR reaction was performed as follows: 95°C for 30 s, followed by 39 cycles at 95°C for 5 s, and 60°C for 30 s. Then 95°C for 10 s and a melting curve (65–95°C, with 0.5°C increments) was produced to confirm the specificity of amplification. The TUB gene was selected as an internal standard for normalization and three technical replicates were completed for each sample (Li M. Y. et al., 2016). The expression levels of leaves at 0 h were separately used as controls (expression = 1.0) for hormone and wounding treatments. The expression levels at other time points and in other organs were normalized accordingly. Error bars show the standard errors for three independent replicates. The 2−ΔΔCT methods were used to analyze relative transcript abundances (Livak and Schmittgen, 2001). All data were expressed as the mean ± SE after normalization of three independent experiments. One-way ANOVA test was employed using SPSS software (version 18.0) to calculate levels of significance. Statistically significant differences were assessed using LSD (Fisher's least significant difference) test, and P < 0.05 was adopted as the criterion for statistical significance.
We predicted 67 non-redundant WRKY genes from the carrot genome V2.0 (DcsWRKY) using HMM program with open reading frames (ORFs) ranging from 115 to 858 bp. The DcsWRKYs were named as DcsWRKY1 to DcsWRKY67 according to the order of gene IDs (Table 1). As shown in Table 1, the highly conserved domain WRKYGQK was present in 61 DcsWRKY members, whereas the remaining six members contained WRKYGKK, WRKYDQK, and WRKYDHK domains. Of these, WRKYGKK was the most common domain presented in three of the six variants. Slight variations of WRKYGQK domain were also identified in many other plant species, such as broomcorn millet (Yue et al., 2016), cabbage (Brassica oleracea var. captitata) (Yao et al., 2015), grape (Guo et al., 2014), and tomato (Huang et al., 2012). It has been demonstrated that WRKYGQK domain can bind w-box cis-elements to activate the downstream genes. One possible explanation for these WRKY domain variants is an altered binding specificity in the target genes (Guo et al., 2014). For example, NtWRKY12 in tobacco cannot interact with the w-box motif, but it can bind to wk-box element (van Verk et al., 2008). Therefore, it would be interesting to analyze the function and binding specificities of these six proteins with motif variation. In addition, two genes, DcsWRKY17 and DcsWRKY46, were found to lose the Zinc-finger-like motif of C2HH, but the function of zinc-finger-like motif loss still remains undetermined. Protein properties of each DcsWRKY, including molecular weights and theoretical isoelectric points, were analyzed, which were given in Table 1. The molecular weights ranged from 13.54 (DcsWRKY51) to 94.29 kda (DcsWRKY36), with an average of 40.93 kda. The theoretical isoelectric points varied from 4.82 (DcsWRKY25) to 10.04 (DcsWRKY47). These results indicate a high complexity among the WRKY genes in the carrot genome.
Based on the number of WRKY domains and the zinc finger type, the putative WRKY proteins could be divided into three groups, namely, groups I, II, and III (Eulgem et al., 2000). As shown in Table 1 and Figure 1, there were six members in Group I, which contained two WRKYGQK domains, and two zinc-finger motifs of C2H2 type except for DcsWRKY16 and DcsWRKY41 without the zinc-finger structure at C-terminus. Group II, in particular, could be further divided into the five subgroups (II-a, II-b, II-c, II-d, II-e) and contained 3, 10, 16, 10, 14 WRKY members, respectively. Eight members of group III were found in the carrot genome with only one WRKYGQK domain and a zinc-finger motif of C2HC type. Finally, two DcsWRKY genes (DcsWRKY17 and DcsWRKY46) with incomplete structures were not assigned to any of the subgroups. However, based on the phylogenetic tree of WRKY full-protein (Figure 2), they could be classified into subgroup II-d and II-c, respectively. Detailed information about the type of DcsWRKY genes and domains is given in Figure 1. Orthologs are genes in different genomes that originate from a common ancestral gene by speciation and often retain similar functions (Remm et al., 2001), and thus, comparisons between a model species and a less-studied species allow us to understand genomic information of less-studied taxa (Lyons et al., 2008). Orthologs, between A. thaliana and carrot, were detected using BLASTP with e value 1e-15. Our results showed that most WRKY members were found to expand nearly two times more than A. thaliana.
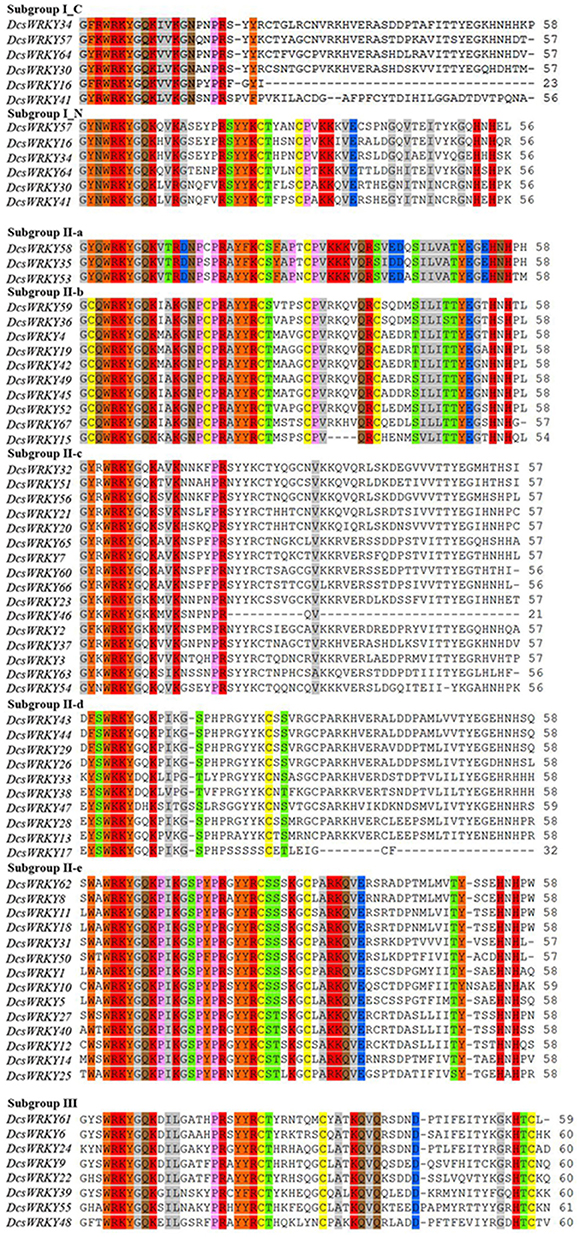
Figure 1. Multiple sequence alignment of the WRKY domain among carrot WRKY genes. The alignment was performed by Clustal W. Dashes indicate gaps. “N” and “C” indicate the N-terminal and C-terminal WRKY domain of a specific WRKY gene.
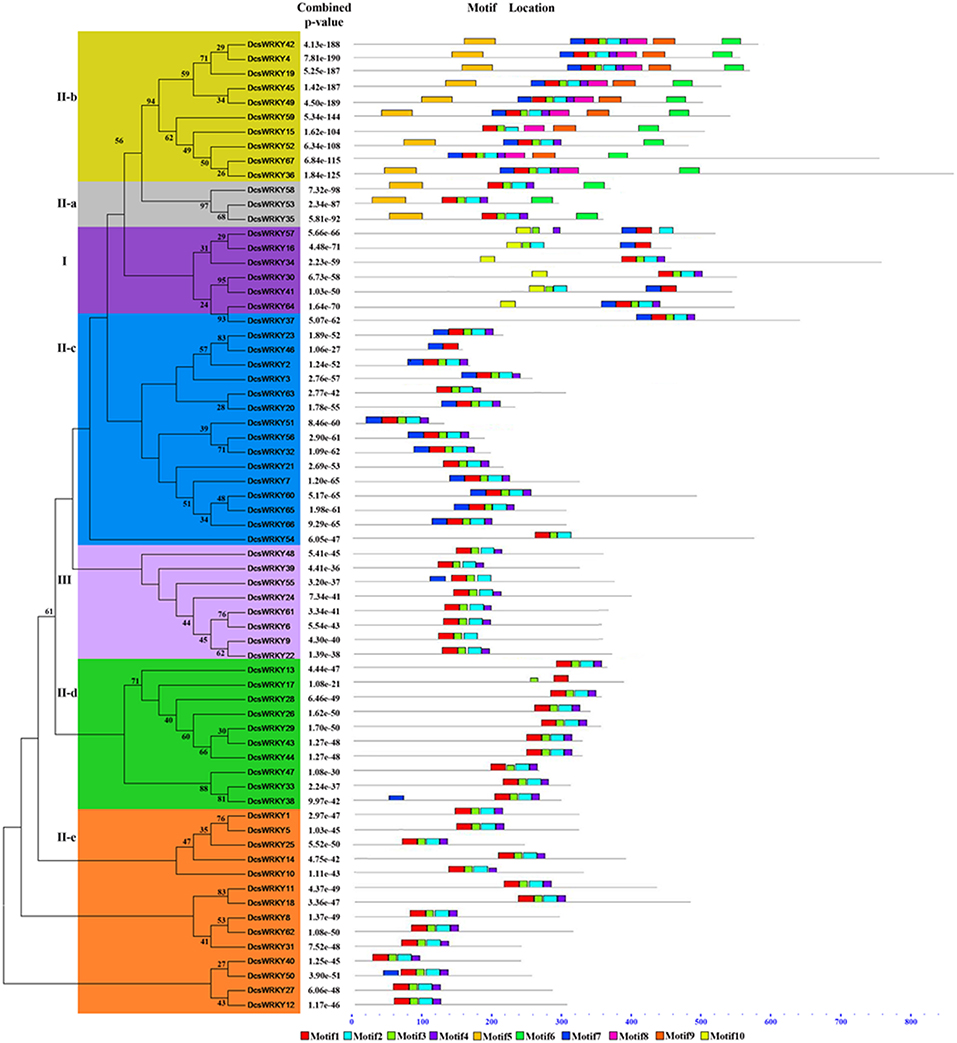
Figure 2. Phylogenetic analysis and conserved motif compositions of the WRKY genes in carrot. Phylogenetic tree was constructed using the maximum-likelihood method based on alignments of complete predicted proteins. The conserved motifs were detected using MEME software and represented by colored boxes. Reliability of the predicted tree was tested using bootstrapping with 1,000 replicates. The length of WRKY proteins can be estimated using the scale at the bottom and conserved motifs were shown in Table 2.
The genomic distribution of DcsWRKY genes was investigated by positioning their approximate positions on each chromosome. As shown in Figure 3, among the nine chromosomes, DcsWRKY genes were unevenly distributed, and the numbers on each chromosome were not related to their sequence lengths (Figure 3, Table S2). Chromosome two harbored the majority of WRKY genes (25.37%), including one members in group I, 13 in group II, and three in group III of the DcsWRKY gene family, followed by 11 genes on chromosome five (one in group I, nine in group II, and one in group III), while chromosome nine only contained three DcsWRKYs (two in group II-c and one in group II-b). Chromosome one, the longest chromosome, only had five in group II of DcsWRKY genes.
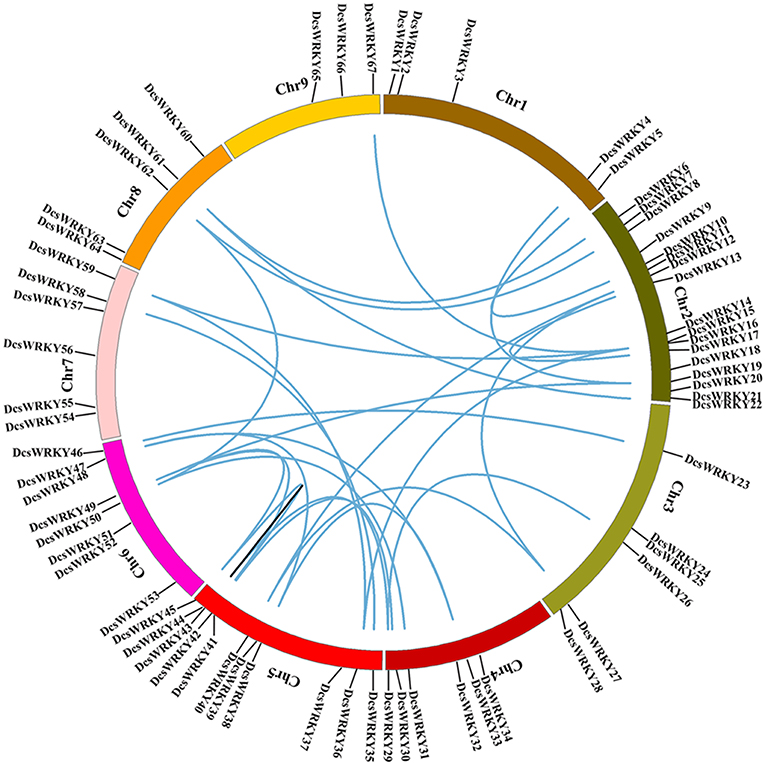
Figure 3. Chromosome distribution and synteny analysis of carrot WRKY genes. Chromosomes 1–9 are shown with different colors. The approximate distribution of each gene is marked with a short black line. Blue curves indicate duplicated DcsWRKY genes, and black lines in the circle indicate tandem duplicated genes.
To better characterize the DcsWRKY gene family, we predicted the conserved motifs in DcsWRKY proteins by MEME online software (Table 2). An unrooted phylogenetic tree was constructed from the 67 DcsWRKY proteins using ML method (Figure 2). In total, 10 distinct motifs were identified, and all DcsWRKY members could be divided into seven large subgroups (i.e., I, II a-e, and III) previously found in higher plants, which shared similar motifs (Xiong et al., 2013; Wen et al., 2014; Chen et al., 2018). As illustrated in Table 2 and Figure 2, after assessing the distribution of the motifs from these subgroups, half of them were identified lying around the WRKY, such as motifs 5–9, while other motifs 1, 2, 3, 4, and 10 were located in the WRKY domain. Overall, the same subgroup shared similar motif compositions, indicating a highly functional conservation. Subgroups II-a and II-b were closely related, and motifs 5 and 6 were uniquely dispersed across them. In most cases, motifs 5 and 6 appeared as a pair, indicating that they are functionally related to the subgroups. Subgroups II-d and II-e were clustered together and contained motifs 1, 2, 3, and 4 except DcsWRKY17. Of these 10 motifs, all groups shared motifs 1, 2, 3, and 4, which partially represented the distribution of the C-terminal domain. Motif 10 was only found in group I, which partially exemplified the distribution of the N-terminal domains.
To understand the diversification and evolution of the DcsWRKY gene family we compared the WRKY members from the two other sequenced plant genomes (grape and Arabidopsis). An unrooted phylogenetic tree was constructed using the Neighbor-Joining (NJ) method implemented with MEGA from the 197 conserved WRKY domains among the three plant species. As shown in Figure 4, the complete WRKY domains were divided into eight subgroups (i.e., I_N, I_C, II a-e, and III), and the WRKY members belonging to the same group from these three species similarly had conserved domain compositions. The ancestral groups I_N and I_C were separately grouped on the phylogenetic tree, while I_C was the basal clade of the phylogenetic tree, which is consistent with the hypothesis that the group I was the oldest group (Wu et al., 2005). In addition, I_C and subgroup II-c were closely clustered, and subgroup II-c appeared polyphyletic.
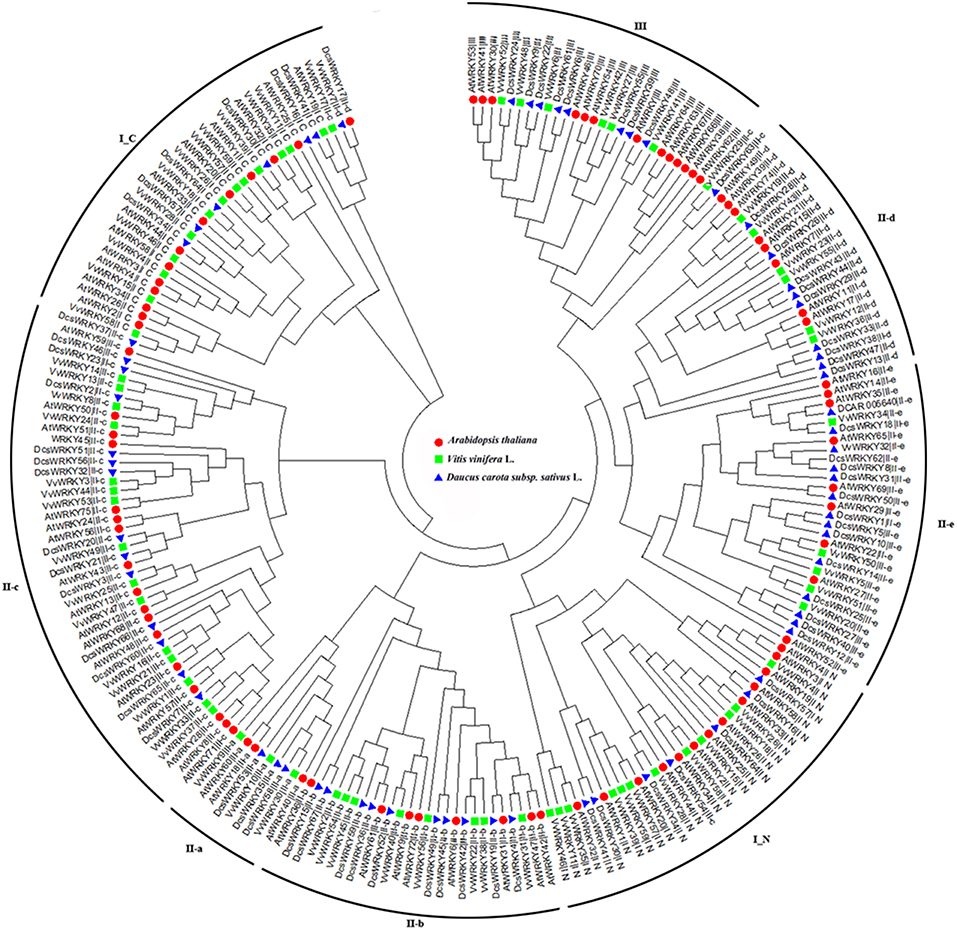
Figure 4. Phylogenetic tree based on WRKY domains from carrot, Arabidopsis, and grape domains of the WRKY gene family. The phylogenetic tree was constructed using Neighbor-Joining method based on 197 WRKY domain sequences. Reliability of the predicted tree was tested using bootstrapping with 1,000 replicates. The tree was divided into eight phylogenetic subgroups, namely I_N, I_C, II-a–II-e, and III. The WRKY genes of Arabidopsis, grape, and carrot were denoted by circles, diamonds, and triangles, respectively.
To investigate the evolution and functional diversification of the WRKY members, we retrieved, and analyzed the upstream 1.5 Kb promoter regions of the DcsWRKY genes. A number of cis-acting regulatory elements, including 9 elements related to plant development and 16 motifs related to stress responses, were analyzed, and the 25 elements were represented in Table S3. Cis-elements related to plant growth and development include light responsive elements (box4, G-box, sp1, and ACE), endosperm expression (skn-1_motif and GCN4_motif), circadian control (circadian), meristem expression (CAT-box), and meristem-specific activation (CCGTCC-box). Cis-elements related to stress responses comprise eight hormone responsive elements (CGTCA-motif, TGACG-motif, ABRE, GARE-motif, TATC-box, p-box, TGA-element, and ERE), wound-responsive element (WUN-motif), anaerobic induction element (ARE), low-temperature responsive element (LTR), and so on. As shown in Table S3, each DcsWRKY gene contained more than one cis-acting regulatory element in their promoter regions, and most DcsWRKY genes contained box4, G-box, skn-1, and MBS motif.
Regarded as the main evolutionary force in both plants and animals, whole-genome duplication (WGD) has been extensively found in most eudicots, following by extensive gene loss (Sémon and Wolfe, 2007). D. carota has experienced two recent species-specific WGD events, Dc-α, and Dc-β, following the earlier γ paleohexaploidy event (~117 MYA) shared by core eudicots (Jiao et al., 2012). Note that these two carrot-specific WGDs occurred approximately 43 and 70 million years ago, respectively. To understand the role of WGD events in the carrot WRKY genes we performed syntenic analysis using MCScanX. To investigate the collinearity of the WRKY gene family, all protein sequences of D. carota subsp. sativus were first identified using BLASTP; the resulting BLASTP hits were then compiled as the input for MCScanX to classify duplicated gene pairs under the default settings. And then, custom Perl scripts were used to collect syntenic gene pairs. According to previous studies (Liu and Ekramoddoullah, 2009; Bi et al., 2016), two or more adjacent homologous genes located on a single chromosome were regarded as tandem duplicated genes, while homologous genes located on different genomics regions or chromosomes were regarded as WGD derived genes or segmentally duplicated genes. The collinear relationships of the duplicated pairs in the DcsWRKY gene family were shown in Table 3. Of the 67 DcsWRKY genes, 27 pairs of WGD duplicated genes (36 DcsWRKY members), and only one pair of tandem duplicated genes (DcsWRKY43 and DcsWRK44) were identified, indicating that the WRKY genes were mainly derived from whole genome duplication events, which acted as a major force to drive the evolution of the DcsWRKY gene family. In many other plants, most WRKY genes were also found to derive from whole genome duplication events, such as cabbage, peanut (Song et al., 2016), and soybean (Yin et al., 2013). As shown in Figure 3, chromosomes two and five had most of WGD-derived DcsWRKY duplicated genes, while chromosome nine only had one (DcsWRKY67). And, the tandem duplicated gene pairs were located on chromosome five. In addition, the two duplicated genes in one pair belonged to the same WRKY group (Table 3), suggesting that duplicated genes may have a similarly conserved function.
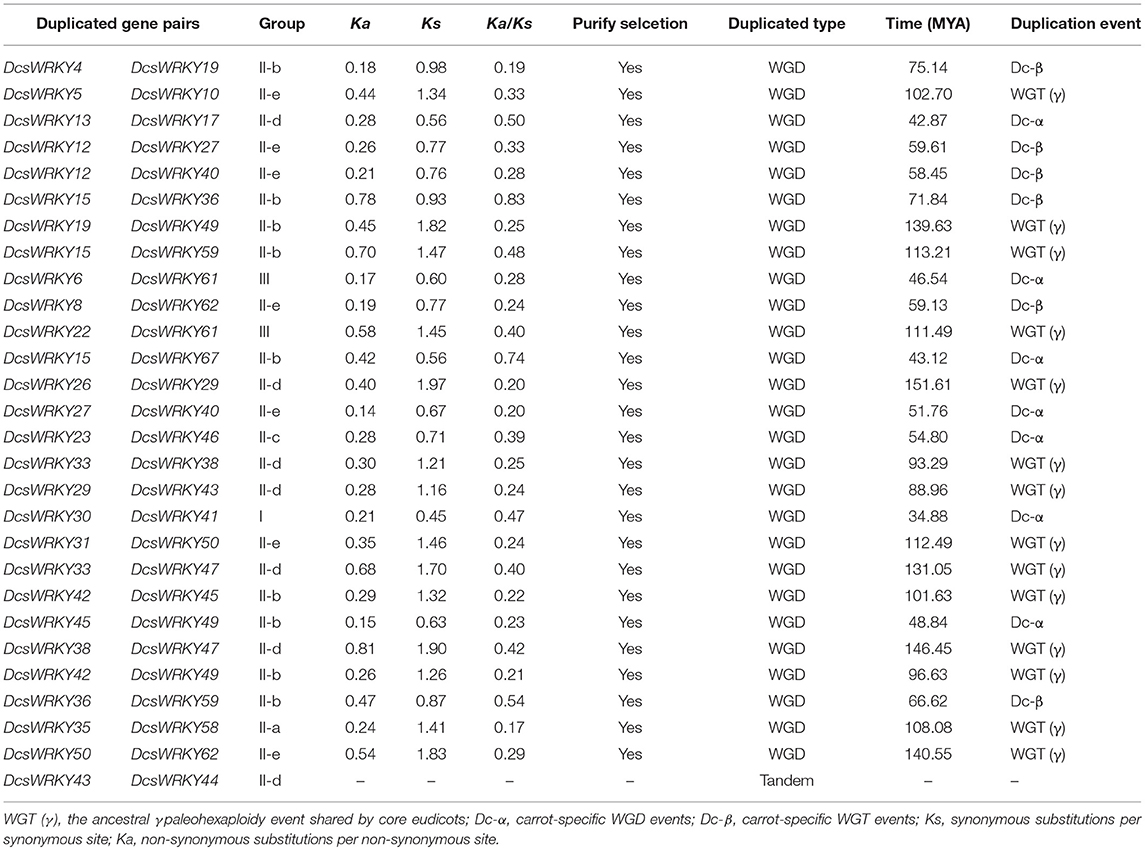
Table 3. Ka/Ks calculation and divergence times of the duplicated DcsWRKY gene pairs in syntenic blocks.
We calculated the synonymous (Ks), non-synonymous (Ka) substitution rate and Ka/Ks ratios of the duplicated WRKY genes in carrot. The Ka values ranged from 0.14 to 0.81, and Ks varied from 0.45 to 1.97. Previous studies suggested that Ka/Ks < 1, Ka/Ks = 1, and Ka/Ks > 1 indicate purifying selection, neutral evolution, and positive selection, respectively (Tang et al., 2013; Song et al., 2016). Of the 28 duplicated gene pairs, all WGD duplicated DcsWRKYs had Ka/Ks < 1, ranging from 0.17 to 0.83. The results suggested purifying selection act on these duplicated gene pairs, which agreed with what observed in peanuts (Song et al., 2016), and B. rapa (Tang et al., 2013).
To time these duplicated DcsWRKY genes, the Ks values were served as proxies for the divergent events, and divergence dates were calculated using the function: Ks/2r. As described in Table 3, divergence dates spanned ~34.88–151.61 million years, suggesting that the duplicated genes in carrot occurred from the gamma polyploidy event. Carrot underwent one γ paleohexaploidy event and two species-specific WGDs, namely Dc-α and Dc-β, which occurred approximately 117, 43, and 70 million years ago, respectively. Thus, there were 14 (~51.9%) duplication events with 22 genes occurred during gamma polyploidy event, while 7 (~25.9%) and 6 (~22.2%) duplication events were identified during Dc-α and Dc-β events, respectively. Moreover, seven DcsWRKY members (DcsWRKY15, DcsWRKY27, DcsWRKY40, DcsWRKY45, DcsWRKY49, DcsWRKY59, DcsWRKY61, and DcsWRKY62) experienced at least two rounds of WGD events, and only the DcsWRKY15 gene involved in all these three WGD events.
To examine patterns and expression levels of DcsWRKY genes, 12 carrot WRKY genes in three tissues (storage roots, leaves, and stems) were researched through qRT-PCR experiments. Our results showed that DcsWRKY genes exhibited distinct expression patterns, and tissue-specific expression of WRKY genes was also observed in carrot (Figure 5). For instance, DcsWRKY29 and DcsWRKY57 were particularly highly expressed in leaves but levels of expression were low in roots and stems. DcsWRKY43 exhibited extremely low levels in leaves and roots, whereas it was highly expressed in stems. Previous studies reported that the highly expressed WRKY genes in certain tissues were often found to regulate target genes involved in important processes of growth and development (Yu et al., 2012). The three DcsWRKY genes (DcsWRKY4, DcsWRKY56, and DcsWRKY57) showed no significant expression difference between at least two tissues, suggesting that they likely play an ubiquitous role in carrot. Furthermore, DcsWRKY44 and DcsWRKY61 were constitutively expressed in each tested tissue and shared a similar expression trend, indicating their putative redundant functions in the development and physiological processes of these tissues.
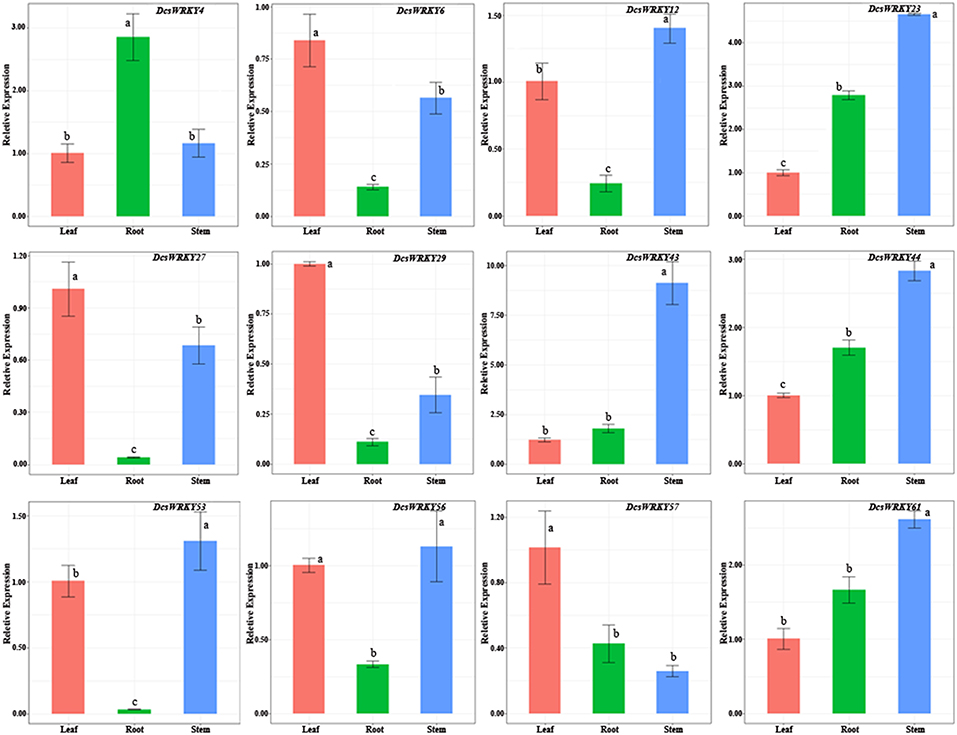
Figure 5. Real-time quantitative PCR expression levels of selected genes in various organs. The y-axis represents the relative expression levels of genes. The x-axis shows different tissues. Error bars represent standard errors of the mean in three biological replicates. One-way ANOVA test (P < 0.05, LSD) was used to statistically evaluate the significance among those samples. The histogram bars labeled with different letters (a, b, c, and d) above histogram bars are significantly different (LSD test, P < 0.05). LSD designates Fisher's least significant difference test.
We also investigated the expression divergence between the duplicated DcsWRKY genes. DcsWRKY6 and DcsWRKY61 were duplicated gene pairs generated during Dc-α event, which are orthologous to AtWRKY41 in Arabidopsis (Tables 1, 3). As shown in Figure 5, DcsWRKY6 was quite lowly expressed in stems, while the expression level of DcsWRK61 was extremely high in stems. Another exemplar pairs of duplicated genes, DcsWRKY29, and DcsWRKY43, were produced during the γ paleohexaploidy event. DcsWRKY29 was up-regulated in leaves and down-regulated in stems, while its paralog DcsWRKY43 was down-regulated in leaves and up-regulated in stems. Previously numerous studies suggested that tissue-specific expression divergence is one of the most important indicators of functional differentiation between duplicated genes, and thus gene duplication plays a key role in the growth of gene networks (Makova and Li, 2003; Li et al., 2005). Thus, the expanded WRKY genes might result in novel biological function after gene duplication events, which are beneficial to regulate various physiological processes by removing their redundancy.
WRKY transcription factors were found to respond to various stresses that may result from the upstream specific cis-elements to regulate gene expressions (Shinozaki et al., 2003; Kim and Zhang, 2004). To understand the roles of these DcsWRKYs in response to abiotic stresses, we exposed ABA and GA to leaves, and performed mechanic injury treatments in carrot. The expression profiles of DcsWRKY genes that contained corresponding cis-elements were examined using qRT-PCR experiments. In hormone treatments, a total of nine DcsWRKY genes with ARRE motif were selected to examine patterns of gene expression under ABA treatment (Figure 6). Our results showed that DcsWRKY5, DcsWRKY7, DcsWRKY18, and DcsWRKY32 were evidently down-regulated by 2-, 10-, 136-, and 12-fold after 1 h, respectively, whereas DcsWRKY4 increased by almost 10 times at 1 h and had a maximum expression level at 48 h. Moreover, the expression level of DcsWRKY58 slightly decreased after 1 h and reached the lowest level after 48 h. We also observed that genes belonging to the same DcWRKY subgroup could show a distinct expression trend, such as DcWRKY5 (II-e), DcsWRKY18 (II-e), and DcsWRKY27 (II-e) (Figure 6).
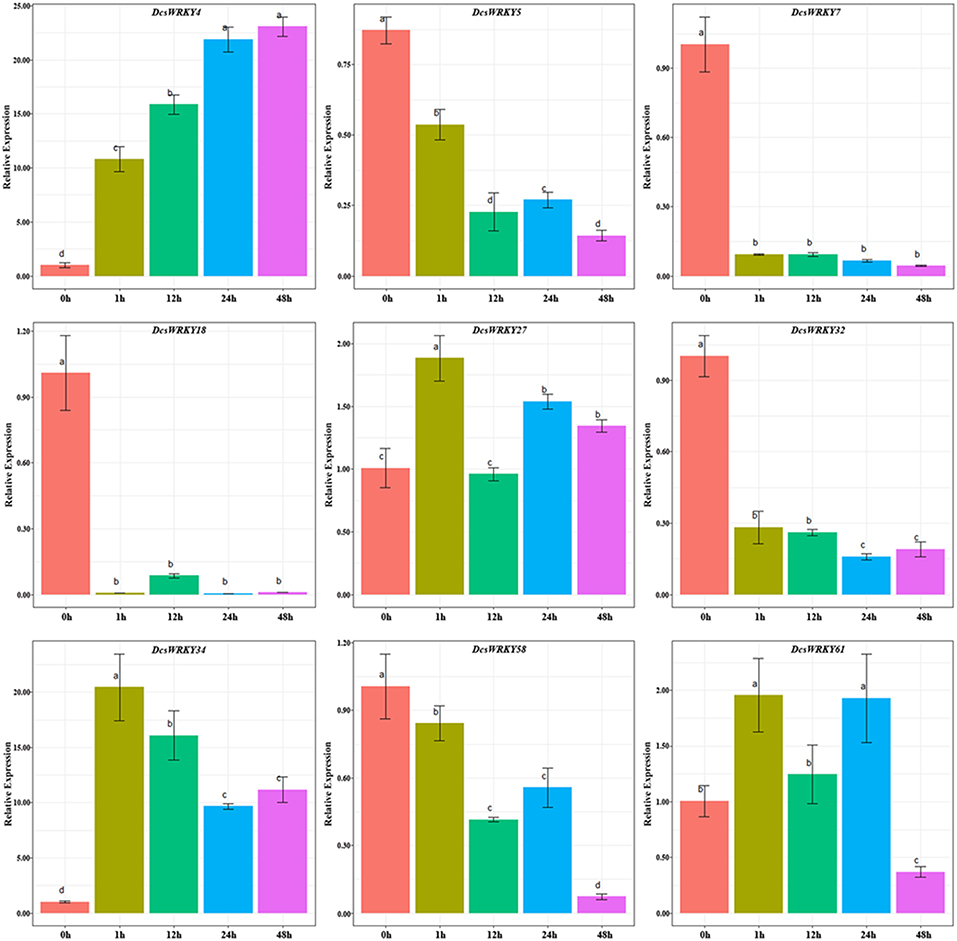
Figure 6. Expression patterns of DcsWRKY genes under ABA treatments. Samples collected at 0, 1, 12, 24, and 48 h after treatment. The expression levels were normalized to control samples. Error bars represent standard errors of the mean in three biological replicates. One-way ANOVA test (P < 0.05, LSD) was used to statistically evaluate the significance among those samples. The histogram bars labeled with different letters (a, b, c, and d) above histogram bars are significantly different (LSD test, P < 0.05). LSD designates Fisher's least significant difference test.
GARE motif, TATC box, and p-box are gibberellin-response cis-acting elements (Rogers et al., 1994; Chen T. et al., 2012). In order to understand expression patterns of DcsWRKY genes with them, 12 genes were examined after leaves were sprayed with GA. In this study, GA treatment resulted in a wide variety of DcsWRKY gene expression profiles. As described in Figure 7, gene expression levels of DcsWRKY12, DcsWRKY48, DcsWRKY56, and DcsWRKY60 decreased rapidly by 11, 8, 2.7, and 231 times after 1 h. However, the trend in expression of DcsWRKY58 initially increased and peaked at 1 h, followed by a decrease. In addition, tandem duplicated genes (DcsWRKY43 and DcsWRKY44) had a similar trend under GA treatment: there was an initial increase and reach a maximum level at 12 h, followed by a decrease.
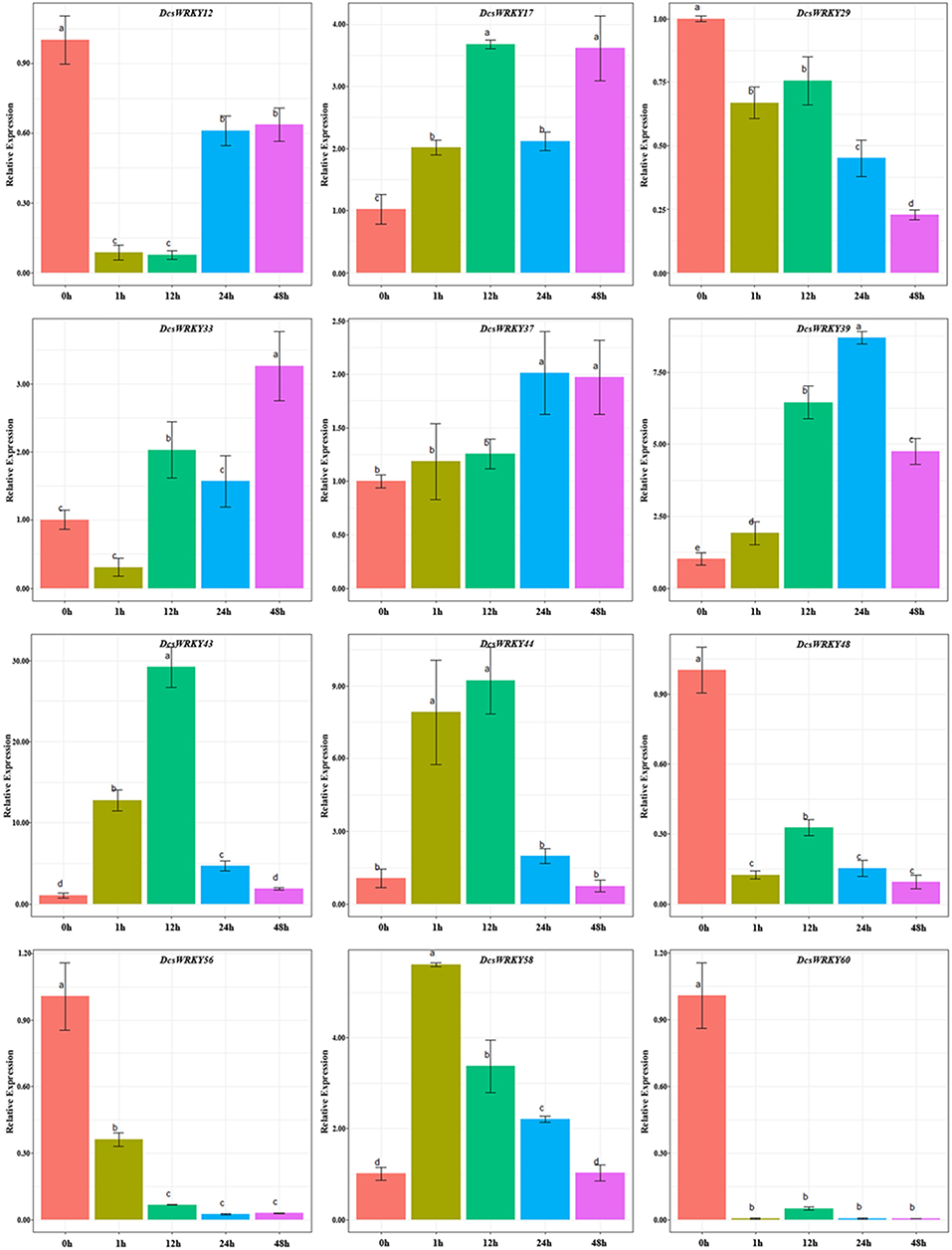
Figure 7. Expression patterns of DcsWRKY genes under GA treatments. Samples collected at 0, 1, 12, 24, and 48 h after treatment. The expression levels were normalized to control samples. Error bars represent standard errors of the mean in three biological replicates. One-way ANOVA test (P < 0.05, LSD) was used to statistically evaluate the significance among those samples. The histogram bars labeled with different letters (a, b, c, and d) above histogram bars are significantly different (LSD test, P < 0.05). LSD designates Fisher's least significant difference test.
The expression patterns of the six carrot WRKY genes with wound-responsive element (WUN-motif) were also investigated through qRT-PCR experiments. Our results showed that these WRKY genes were expressed in distinct behaviors. As shown in Figure 8, after 1 and 4 h treatment of mechanic damage, we observed sharp increase and decrease for some genes, such as DcsWRKY24, DcsWRKY43, DcsWRKY44, and DcsWRKY64. The expression levels of DcsWRKY3 decreased rapidly in leaves after 1 h treatment and then decreased slightly, after 4, 8 and 12 h. The trend in the expression of DcsWRKY5 initially increased and reached a maximum after 4 h and then decreased in 8 and 12 h. Moreover, DcsWRKY43 and DcsWRKY44 also showed a similar expression trend under mechanic injuries.
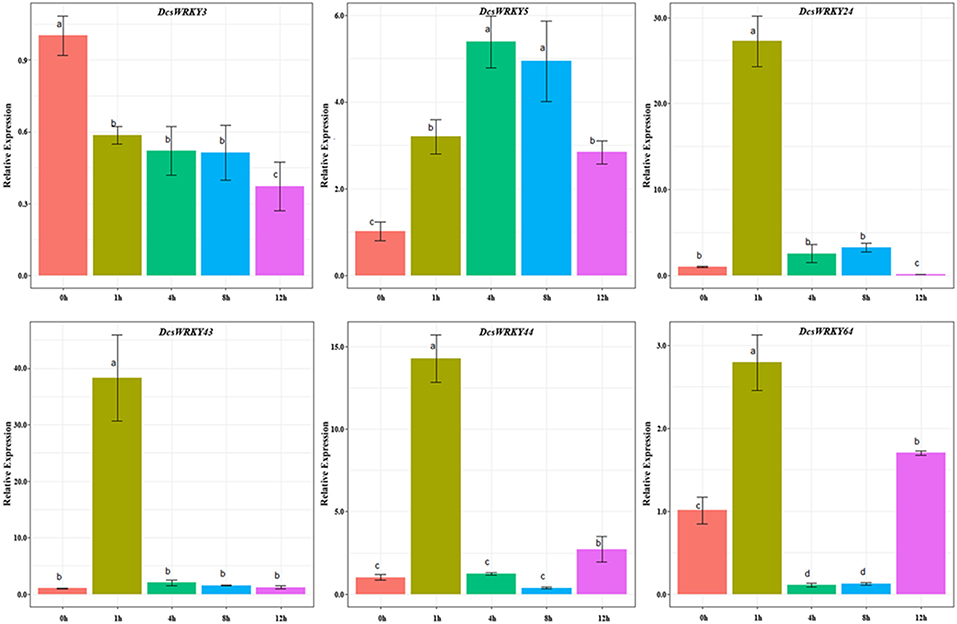
Figure 8. Expression patterns of DcsWRKY genes under mechanic injury treatments. Samples collected at 0, 1, 4, 8, and 12 h after treatment. The expression levels were normalized to control samples. Error bars represent standard errors of the mean in three biological replicates. One-way ANOVA test (P < 0.05, LSD) was used to statistically evaluate the significance among those samples. The histogram bars labeled with different letters (a, b, c, and d) above histogram bars are significantly different (LSD test, P < 0.05). LSD designates Fisher's least significant difference test.
The WRKY gene family, one of the largest transcription factor families, is found to involve in a variety of important functions and have been extensively investigated in many plants (Zhou et al., 2016; Xiao et al., 2017; Yang et al., 2017). In this study, we accurately identified a total of 67 WRKY genes in the high-quality Nantes type carrot genome sequences, which are fewer than that (95 genes, Kuroda type carrot) previously reported in a draft genome assembly of D. carota L. cv. Kuroda (Xu et al., 2014). The chromosome-scale genome assembly has the advantage not only to characterize almost all members of the WRKY gene family but also localize their positions on chromosomes. We found that DcsWRKY genes are unevenly distributed on chromosomes. Chromosome two contained the majority of DcsWRKY genes (up to 17 of 67 DcsWRKY genes), while chromosome nine only had three. Our carrot genome collinearity analysis suggested that WGD events may have played a major role in driving the DcsWRKY gene family evolution, and all DcsWRKY gene pairs are under strong purifying selection. Previous study observed expression patterns of DcWRKY genes at different developmental stages of roots and under heat, cold, salt, and drought stresses (Li M. Y. et al., 2016). In this study, we examined tissue-specific expression profiling of DcsWRKY genes in different tissues and the abundance of WRKY genes under ABA, GA, and mechanic injury treatments. These efforts together shed new light on the evolution and functional divergence of the WRKY gene family in carrot.
Through a genome-wide analysis, a total of 67 DcsWRKY genes were identified in Nantes type carrot. And, our phylogenetic analysis demonstrated that DcsWRKYs could be divided into three major groups, among which group II was further classified into five subgroups, in accordance with the classification of WRKYs in Arabidopsis, rice, grape, tomato, and cucumber (Wu et al., 2005; Huang et al., 2009, 2012; Guo et al., 2014). Comparisons of conserved motifs in DcsWRKY members also supported the classification of DcsWRKY genes, indicating that the WRKY genes, to a certain extent, were functionally conserved among plants.
Compared with carrot (67 DcsWRKYs; genome size 473 Mb), a comparable number of WRKYs were identified in Arabidopsis (72; genome size 125 Mb), although the number of WRKYs was fewer in cucumber (55; genome size 367 Mb) and grape (59; genome size 487 Mb) and more in tomato (81; genome size 900 Mb) and rice (103; genome size 389 Mb) (International Rice Genome Sequencing, 2005; Jaillon et al., 2007; Huang et al., 2009, 2012; Ling et al., 2011; Tomato Genome, 2012; Guo et al., 2014; Iorizzo et al., 2016; Zhou et al., 2016). These results indicate that the number of WRKY genes may not be associated with the genome size. Moreover, the subgroup distribution of WRKY genes among plant species were also different. As showed in Figure 4, numbers of subgroups II-b, II-d, and II-e in carrot are larger than those of A. thaliana and grape, suggesting that carrot WRKYs of the three subgroups might have experienced linage-specific amplification. In this study, the collinearity analysis of DcsWRKY genes also support the conjecture and showed that more than half of the subgroup II-b, II-d, and II-e were undergo duplication events. Major WRKY members of subgroup II-b and II-e expand during Dc-α or Dc-β events, and most of subgroup II-d expand during γ paleohexaploidy event.
Gene duplication is an important driving force during plant evolution, which plays a central role in gene family expansion (Makova and Li, 2003; Li et al., 2005). Previous studies have demonstrated that gene duplication largely accounts for new gene functions (Ohno, 1970). In this study, we found 36 DcsWRKY genes were segmentally duplicated but only two genes were tandemly duplicated, indicating that segmental duplications make a great contribution to the expansion of DcsWRKY genes. Since tissue-specific expression patterns can provide clues for gene functional divergence during the evolution (Yao et al., 2015), we validated the expression of DcsWRKY genes in leaves, roots and stems through qRT-PCR. All tested 12 genes showed different expression levels, of which DcsWRKY43 and DcsWRKY44, for example, were tandemly duplicated and showed a similar expression trend but DcsWRKY43 was extremely highly expressed in stems. Moreover, DcsWRKY29 and DcsWRKY43 were WGD-derived duplicated gene pairs, which were expressed highly in leaves and lowly in stems, and lowly in leaves and highly in stems, respectively. Our results indicate that the expanded WRKY genes might result in novel biological function to remove their genetic redundancy and functional divergence might have occurred after gene duplication.
Previous studies founded that WRKY genes have complex regulatory networks in biotic and abiotic stresses and hormone responses (Chen L. et al., 2012; Phukan et al., 2016). In order to provide a foundation for further study of WRKY genes in carrot, the expression patterns of DcsWRKYs containing the corresponding stress signal cis-regulatory elements were evaluated at different time-points in response to ABA, GA as well as mechanic injury treatments. Consistent with previous studies (Guo et al., 2014; Wang et al., 2015), the expression levels of WRKY genes were rapidly induced by ABA and GA treatments within a few hours, indicating that these genes may play an important role in stress responses in carrot. ABA is a stress hormone and plays essential roles in plant responses to abiotic stresses (Agarwal et al., 2011; Chen L. et al., 2012). In our study, DcsWRKY58 (II-a) was involved in the ABA signal transduction pathway which showing down-regulated after ABA treatment and reaching the lowest level after 48 h (Figure 6). Previous studies showed that AtWRKY40 (homologous to DcsWRKY58) acts as a protein interacting with ABA receptors and is involved in biotic and abiotic stresses (Zou et al., 2007; Chen L. et al., 2012). This result could provide support for our findings that DcsWRKY58 not only participates in ABA signal transduction pathway, but also responses to biotic and abiotic stresses. Li M. Y. et al. (2016) showed that DcWRKY31 (II-a) in Kuroda type carrot whose homologous gene in Arabidopsis is also AtWRKY40, were found to be evidently up-regulated under drought, salt and pathogenic stresses. Thus, we can further indicate that DcsWRKY58 in Nantes type carrot might was involved in drought, salt, and pathogenic stresses and regulated abiotic stress responses depending on ABA signaling pathway. Another example was DcsWRKY5 (II-e) in this study and DcWRKY18 (II-e) (Li M. Y. et al., 2016) which were both homologous to AtWRKY22 (AT4G01250.1). Gibberellin (GA) also plays an important role in biotic stresses and plant disease resistance responses. ABF1 (AfWRKY1) and ABF2 (AfWRKY2) had been implicated in GA and ABA signaling and were involved in seed germination (Rushton et al., 2012). Rice OsWRKY71 and OsWRKY51 were found to act as regulators of ABA-inducible pathway and GA-repressible pathway in aleurone cell (Xie et al., 2006). Our study showed that DcsWRKY genes with GA gibberellin-response cis-acting elements had a variety of response patterns after GA treatments, indicating that they might be induced by hormones. Wounding is a common damage for plants and presents a constant threat to plants survival. It not only physically damages plant, but also provides pathways for pathogen invasion. However, there were only a few reports about transcriptional abundance of WRKY genes under wounding treatment. In Arabidopsis, AtWRKY15, AtWRKY22, and AtWRKK33 were induced by wounding stress (Cheong et al., 2002). In this study, carrot homologs of these three WRKYs (DcsWRKY43/DcsWRKY44, DcsWRKY5, and DcsWRKY64) were also induced by wounding treatments, suggesting the potential functions of these DcsWRKY genes in mechanic injury stresses. Additionally, it was found that genes belonging to the same subgroup showed a distinct expression trend, such as DcWRKY5 (II-e), DcsWRKY18 (II-e), and DcsWRKY27 (II-e) in ABA treatments, DcWRKY17 (II-d), DcsWRKY29 (II-d), and DcsWRKY33 (II-d) in GA treatments. The different expression trends of DcsWRKY genes under abiotic stresses suggested that they might respond to abiotic stresses through different genetic networks. It has been reported that the co-expressed/co-responsive genes are likely to have common regulatory motifs in their promoters and are possibly regulated by a common set of TFs (Liu et al., 2005). DcsWRKY43 and DcsWRKY44 showed similar expression trend under GA and mechanic damage treatments and had the same cis-acting regulatory elements in upstream promoter regions, suggesting that these two genes may be regulated by a common set of TFs under stresses. Our expression analysis indicated that DcsWRKY genes were expressed in a tissue-specific behavior, some of which were in response to hormone signals, and mechanic injury stresses. This comprehensive analysis will enhance our understanding of the evolution and functional diversification of the WRKY gene.
HN performed data analysis, experiments, and drafted the manuscript. LG served as the principal investigator, facilitated the project, and revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We appreciate the reviewers for their comments on this manuscript. This study was supported by the Key Project of the Natural Science Foundation of Yunnan Province (2015FA030) and Yunnan Innovation Team Project (to LG).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00363/full#supplementary-material
Agarwal, P., Reddy, M. P., and Chikara, J. (2011). WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 38, 3883–3896. doi: 10.1007/s11033-010-0504-5
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bi, C., Xu, Y., Ye, Q., Yin, T., and Ye, N. (2016). Genome-wide identification and characterization of WRKY. gene family in Salix suchowensis. Peerj 4:e2437. doi: 10.7717/peerj.2437
Broun, P. (2004). Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 7, 202–209. doi: 10.1016/j.pbi.2004.01.013
Cai, Y., Chen, X., Xie, K., Xing, Q., Wu, Y., Li, J., et al. (2014). Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS ONE 9:e102529. doi: 10.1371/journal.pone.0102529
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2018). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. doi: 10.1080/07352689.2018.1441103
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., and Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chen, T., Yang, Q., Gruber, M., Kang, J., Sun, Y., Ding, W., et al. (2012). Expression of an alfalfa (Medicago sativa L.) ethylene response factor gene MsERF8 in tobacco plants enhances resistance to salinity. Mol. Biol. Rep. 39, 6067–6075. doi: 10.1007/s11033-011-1421-y
Cheong, Y. H., Chang, H. S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. doi: 10.1104/pp.002857
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., and Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Gaut, B. S., Morton, B. R., McCaig, B. C., and Clegg, M. T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. U.S.A. 93, 10274–10279. doi: 10.1073/pnas.93.19.10274
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., et al. (2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 65, 1513–1528. doi: 10.1093/jxb/eru007
Guo, R., Qiao, H., Zhao, J., Wang, X., Tu, M., Guo, C., et al. (2018). The grape VlWRKY3 gene promotes abiotic and biotic stress tolerance in transgenic Arabidopsis thaliana. Front. Plant Sci. 9:545. doi: 10.3389/fpls.2018.00545
Hu, Y., Dong, Q., and Yu, D. (2012). Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185−186, 288–297. doi: 10.1016/j.plantsci.2011.12.003
Huang, S., Gao, Y., Liu, J., Peng, X., Niu, X., Fei, Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics 287, 495–513. doi: 10.1007/s00438-012-0696-6
Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275–1281. doi: 10.1038/ng.475
Huang, X., Li, K., Xu, X., Yao, Z., Jin, C., and Zhang, S. (2015). Genome-wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress. BMC Genomics 16:1104. doi: 10.1186/s12864-015-2233-6
International Rice Genome Sequencing, Project.P. (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Iorizzo, M., Ellison, S., Senalik, D., Zeng, P., Satapoomin, P., Huang, J., et al. (2016). A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 48, 657–666. doi: 10.1038/ng.3565
Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the the 5′upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571.
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Jiao, Y., Leebens-Mack, J., Ayyampalayam, S., Bowers, J. E., McKain, M. R., McNeal, J., et al. (2012). A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13:R3. doi: 10.1186/gb-2012-13-1-r3
Kim, C. Y., and Zhang, S. (2004). Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J. 38, 142–151. doi: 10.1111/j.1365-313X.2004.02033.x
Kim, K.-C., Lai, Z., Fan, B., and Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371. doi: 10.1105/tpc.107.055566
Lagac,é, M., and Matton, D. P. (2004). Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta 219, 185–189. doi: 10.1007/s00425-004-1253-2
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Letunic, I., Doerks, T., and Bork, P. (2011). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Li, M. Y., Xu, Z. S., Tian, C., Huang, Y., Wang, F., and Xiong, A. S. (2016). Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci. Rep. 6:23101. doi: 10.1038/srep23101
Li, W., Wang, H., and Yu, D. (2016). Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant 9, 1492–1503. doi: 10.1016/j.molp.2016.08.003
Li, W. H., Yang, J., and Gu, X. (2005). Expression divergence between duplicate genes. Trends Genet. 21, 602–607. doi: 10.1016/j.tig.2005.08.006
Ling, J., Jiang, W., Zhang, Y., Yu, H., Mao, Z., Gu, X., et al. (2011). Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 12:471. doi: 10.1186/1471-2164-12-471
Liu, F., Vantoai, T., Moy, L. P., Bock, G., Linford, L. D., and Quackenbush, J. (2005). Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 137, 1115–1129. doi: 10.1104/pp.104.055475
Liu, J. J., and Ekramoddoullah, A. K. (2009). Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome 52, 77–88. doi: 10.1139/G08-106
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lyons, E., Pedersen, B., Kane, J., Alam, M., Ming, R., Tang, H., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with Rosids. Plant Physiol. 148, 1772–1781. doi: 10.1104/pp.108.124867
Makova, K. D., and Li, W. H. (2003). Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 13, 1638–1645. doi: 10.1101/gr.1133803
Ohno, S. (ed.). (1970). “The creation of a new gene from a redundant duplicate of an old gene,” in Evolution by Gene Duplication (Berlin; Heidelberg: Springer), 71–82.
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Pinheiro-Santana, H. M., Stringheta, P. C., Brandao, S. C. C., Paez, H. H., and Queiroz, V. M. V. D. (1998). Evaluation of total carotenoids, a- and b-carotene in carrots (Daucus carota L.) during home processing. Food Sci. Tech- Brazil 18, 39–44. doi: 10.1590/S0101-20611998000100009
Qin, Y., Tian, Y., and Liu, X. (2015). A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophysical Biophys. Res. Co. 464, 428–433. doi: 10.1016/j.bbrc
Qiu, Y., and Yu, D. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65, 35–47. doi: 10.1016/j.envexpbot.2008.07.002
Ramamoorthy, R., Jiang, S.-Y., Kumar, N., Venkatesh, P. N., and Ramachandran, S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 49, 865–879. doi: 10.1093/pcp/pcn061
Remm, M., Storm, C. E. V., and Sonnhammer, E. L. L. (2001). Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052. doi: 10.1006/jmbi.2000.5197
Rogers, J. C., Lanahan, M. B., and Rogers, S. W. (1994). The cis-acting gibberellin response complex in high-pl [alpha]-amylase gene promoters (requirement of a coupling element for high-level transcription). Plant Physiol. 105, 151–158. doi: 10.1104/pp.105.1.151
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signaling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Scarpeci, T. E., Zanor, M. I., Mueller-Roeber, B., and Valle, E. M. (2013). Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 83, 265–277. doi: 10.1007/s11103-013-0090-8
Sémon, M., and Wolfe, K. H. (2007). Consequences of genome duplication. Curr. Opin. Genet. Dev. 17, 505–512. doi: 10.1016/j.gde.2007.09.007
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/s1369-5266(03)00092-x
Song, H., Wang, P., Lin, J. Y., Zhao, C., Bi, Y., and Wang, X. (2016). Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 7, 534. doi: 10.3389/fpls.2016.00534
Sun, Y., Niu, Y., Xu, J., Li, Y., Luo, H., Zhu, Y., et al. (2013). Discovery of WRKY transcription factors through transcriptome analysis and characterization of a novel methyl jasmonate-inducible PqWRKY1 gene from Panax quinquefolius. Plant Cell Tiss. Org. 114, 269–277. doi: 10.1007/s11240-013-0323-1
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, J., Wang, F., Hou, X. L., Wang, Z., and Huang, Z. N. (2013). Genome-wide fractionation and identification of WRKY transcription factors in Chinese cabbage (Brassica rapa ssp. pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol. Biol. Rep. 32, 781–795. doi: 10.1007/s11105-013-0672-2
Tao, Z., Liu, H., Qiu, D., Zhou, Y., Li, X., Xu, C., et al. (2009). A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 151, 936–948. doi: 10.1104/pp.109.145623
Tomato Genome, C. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
van Verk, M. C, Pappaioannou, D, Neeleman, L, Bol, J. F, and Linthorst, H. J. M. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995. doi: 10.1104/pp.107.112789
Wang, X., Guo, R., Tu, M., Wang, D., Guo, C., Wan, R., et al. (2017). Ectopic expression of the wild grape WRKY transcription factor VqWRKY52 in Arabidopsis thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen Botrytis cinerea. Front. Plant Sci. 8:97. doi: 10.3389/fpls.2017.00097
Wang, X., Yan, Y., Li, Y., Chu, X., Wu, C., and Guo, X. (2014). GhWRKY40, a multiple stress-responsive cotton WRKY gene, plays an important role in the wounding response and enhances susceptibility to ralstonia solanacearum infection in transgenic Nicotiana benthamiana. PLoS ONE 9:e93577. doi: 10.1371/journal.pone.0093577
Wang, X., Zeng, J., Li, Y., Rong, X., Sun, J., Sun, T., et al. (2015). Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 6:615. doi: 10.3389/fpls.2015.00615
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wen, F., Zhu, H., Li, P., Jiang, M., Mao, W., Ong, C., et al. (2014). Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res. 21, 327–339. doi: 10.1093/dnares/dst060
Wu, K. L., Guo, Z. J., Wang, H. H., and Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xiao, Y., Zhou, L., Lei, X., Cao, H., Wang, Y., Dou, Y., et al. (2017). Genome-wide identification of WRKY genes and their expression profiles under different abiotic stresses in Elaeis guineensis. PLoS ONE 12:e0189224. doi: 10.1371/journal.pone.0189224
Xie, Z., Zhang, Z. L., Zou, X., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Xie, Z., Zhang, Z. L., Zou, X., Yang, G., Komatsu, S., and Shen, Q. J. (2006). Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 46, 231–242. doi: 10.1111/j.1365-313X.2006.02694.x
Xiong, W., Xu, X., Zhang, L., Wu, P., Chen, Y., Li, M., et al. (2013). Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas L.). Gene 524, 124–132. doi: 10.1016/j.gene.2013.04.047
Xu, H., Watanabe, K. A., Zhang, L., and Shen, Q. J. (2016). WRKY transcription factor genes in wild rice Oryza nivara. DNA Res. 23, 311–323. doi: 10.1093/dnares/dsw025
Xu, Z. S., Tan, H. W., Wang, F., Hou, X. L., and Xiong, A. S. (2014). CarrotDB: a genomic and transcriptomic database for carrot. Database (Oxford) 2014:bau096. doi: 10.1093/database/bau096
Yang, Y., Zhou, Y., Chi, Y., Fan, B., and Chen, Z. (2017). Characterization of soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Sci. Rep. 7:17804. doi: 10.1038/s41598-017-18235-8
Yang, Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yao, Q. Y., Xia, E. H., Liu, F. H., and Gao, L. Z. (2015). Genome-wide identification and comparative expression analysis reveal a rapid expansion and functional divergence of duplicated genes in the WRKY gene family of cabbage, Brassica oleracea var. capitata. Gene 557, 35–42. doi: 10.1016/j.gene.2014.12.005
Yin, G., Xu, H., Xiao, S., Qin, Y., Li, Y., Yan, Y., et al. (2013). The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events andsubsequent divergent selection among subgroups. BMC Plant Biol. 13:148. doi: 10.1186/1471-2229-13-148
Yousfi, F. E., Makhloufi, E., Marande, W., Ghorbel, A. W., Bouzayen, M., and Bergès, H. (2016). Comparative analysis of WRKY genes potentially involved in salt stress responses in Triticum turgidum L. ssp. durum. Front. Plant Sci. 7:2034. doi: 10.3389/fpls.2016.02034
Yu, F., Huaxia, Y., Lu, W., Wu, C., Cao, X., and Guo, X. (2012). GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 12:144. doi: 10.1186/1471-2229-12-144
Yue, H., Wang, M., Liu, S., Du, X., Song, W., and Nie, X. (2016). Transcriptomewide identification and expression profiles of the WRKY transcription factor family in Broomcorn millet (Panicum miliaceum L.). BMC Genomics 17:343. doi: 10.1186/s12864-016-2677-3
Zhao, J., Zhang, X., Guo, R., Wang, Y., Guo, C., Li, Z., et al. (2017). Over-expression of a grape WRKY transcription factor gene, VlWRKY48, in Arabidopsis thaliana increases disease resistance and drought stress tolerance. Plant Cell Tiss. Org. 132, 359–370. doi: 10.1007/s11240-017-1335-z
Zhou, H., Li, Y., Zhang, Q., Ren, S., Shen, Y., Qin, L., et al. (2016). Genome-wide analysis of the expression of WRKY family genes in different developmental stages of wild strawberry (Fragaria vesca) fruit. PLoS ONE 11:e0154312. doi: 10.1371/journal.pone.0154312
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Keywords: WRKY transcription factor, carrot, whole genome duplication, expression divergence, hormone and mechanic stresses
Citation: Nan H and Gao L (2019) Genome-Wide Analysis of WRKY Genes and Their Response to Hormone and Mechanic Stresses in Carrot. Front. Genet. 10:363. doi: 10.3389/fgene.2019.00363
Received: 15 December 2018; Accepted: 05 April 2019;
Published: 01 May 2019.
Edited by:
Tian Tang, Sun Yat-sen University, ChinaReviewed by:
Xiping Wang, Northwest A&F University, ChinaCopyright © 2019 Nan and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-zhi Gao, TGdhb2dlbm9taWNzQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.