
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 05 April 2019
Sec. RNA
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00290
This article is part of the Research Topic Non-Coding RNAs and Human Diseases View all 30 articles
Ribosomes are ribonucleoprotein machines that decode the genetic information embedded in mRNAs into polypeptides. Ribosome biogenesis is tightly coordinated and controlled from the transcription of pre-rRNAs to the assembly of ribosomes. Defects or disorders in rRNA production result in a number of human ribosomopathy diseases. During the processes of rRNA synthesis, non-coding RNAs, especially snoRNAs, play important roles in pre-rRNA transcription, processing, and maturation. Recent research has started to reveal that other long and short non-coding RNAs, including risiRNA, LoNA, and SLERT (among others), are also involved in pre-rRNA transcription and rRNA production. Here, we summarize the current understanding of the mechanisms of non-coding RNA-mediated rRNA generation and regulation and their biological roles.
In humans, there are approximately 300–400 copies of rDNA genes per haploid genome that are distributed over five chromosomes (Henderson et al., 1973; Boisvert et al., 2007). Each rDNA unit is ∼43 kb long. rDNAs are transcribed by RNA polymerase I (Pol I) to generate pre-rRNAs that subsequently undergo multiple modifications and processing steps to remove the external transcribed spacers (ETSs) and internal transcribed spacers (ITSs) to produce mature 18S, 5.8S, and 28S rRNAs (McStay, 2016). Pol I activity is a key determinant for ribosome abundance and is essential for cell growth and proliferation (McStay, 2006; Srivastava et al., 2016). Interestingly, only some rDNA units are transcriptionally active. Uncontrolled rRNA synthesis by dysregulated Pol I is associated with aberrant cell proliferation and oncogenesis (Peltonen et al., 2014; Nguyen le et al., 2015).
Non-coding RNAs play essential roles in rRNA regulation. The small nucleolar RNA (snoRNA) is widely known to guide the nucleotide modifications and processing (Cech and Steitz, 2014; Sloan et al., 2017). Recently, increasingly more studies have started to reveal the roles of other classes of non-coding RNAs in regulating rRNA transcription and nucleolar function (Srivastava et al., 2016; Bazin et al., 2017). In this review, we will focus on recent work investigating how several long non-coding RNAs (lncRNAs) and antisense ribosomal siRNA (risiRNA) regulate rRNA expression and their potential biological roles in anti-stress reactions (Table 1).
Long non-coding RNAs comprise a fast-growing classes of RNA molecules with sizes greater than 200 nt. Most lncRNAs are first transcribed by polymerase II (Pol II), then capped, polyadenylated, and spliced after transcription (Cech and Steitz, 2014). lncRNAs localize in distinct subcellular compartments, including the nucleus, nucleolus and cytoplasm. Nuclear-localized lncRNAs, such as MANTIS and Xist, may function as transcriptional or posttranscriptional regulators or structural scaffolds for nuclear domains (Sun et al., 2017). The nucleolar localization of lncRNAs suggests that they may modulate rRNA transcription and maturation (Cech and Steitz, 2014; Chen, 2016). Several studies revealed that lncRNAs regulate rDNA transcription by altering rDNA epigenetic status or by acting as “decoys” to inhibit transcription factor activity. Interestingly, some lncRNAs might contain short open reading frames that can be translated (Ji et al., 2015).
There are several clusters of tandemly arrayed rDNA genes exist in each mammalian genome, yet not all of these repeats are transcribed. rDNA exists in two types of chromatin – a euchromatic conformation that is actively transcribed and a heterochromatic conformation that is transcriptionally inactive. Silent rDNA repeats are marked by heterochromatic histone modifications and CpG methylation at the rDNA promoter (Schmitz et al., 2010). Silencing of rDNA depends on NoRC, a chromatin-remodeling complex that directs heterochromatin formation. NoRC function requires RNA that is complementary to the rDNA promoter, which is termed as promoter-associated RNAs (pRNA). pRNAs are 150∼300 nt long and are produced from rDNA promoters. TIP5 (TIF interacting protein 5), the large subunit of NoRC, binds to pRNAs. pRNA interacts with regulatory elements in the rDNA promoter, forms a DNA:RNA triplex, and is recognized by the DNA methyltransferase DNMT3b (Figure 1; Mayer et al., 2008; Bierhoff et al., 2010). Thus, the binding of NoRC to the rDNA promoter represses rDNA transcription through recruitment of histone modifying and DNA methylating enzymes (Santoro et al., 2002; Mayer et al., 2006; Guetg et al., 2010).
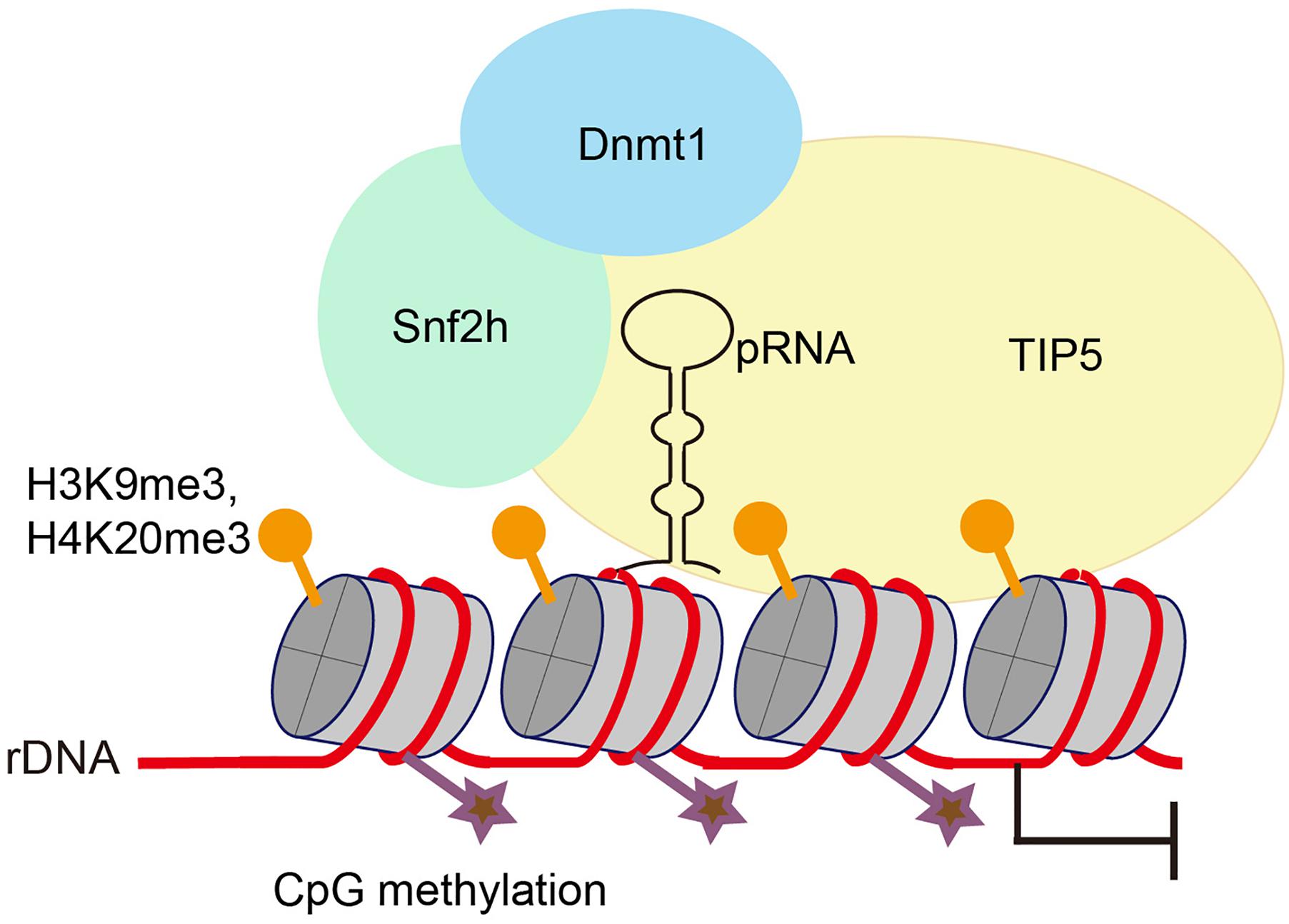
Figure 1. Promoter-associated RNA (pRNA) targets NoRC to nucleolus to promote heterochromatin formation and rDNA silencing. pRNA is complimentary to rRNA genes and folds into a stem-loop structure. TIP5, the core factor of NoRC, recognizes pRNA, facilitates formation of heterochromatin at rRNA genes and promotes transcriptional gene silencing.
Interestingly, by using mature pRNAs to tether heterochromatin at nucleoli in embryonic stem cells, Savić et al. (2014) found that localized heterochromatin condensation of rDNA genes initiates establishment of highly condensed chromatin structures outside of the nucleolus. Meanwhile, the formation of such highly condensed, transcriptionally inactive heterochromatin promotes transcriptional activation of differentiation genes and loss of pluripotency of embryonic stem cells. NoRC safeguards genome stability by triggering heterochromatin formation at telomeres and centromeres (Postepska-Igielska et al., 2013). Whether and how pRNA and NoRC function together to maintain rDNA stability requires further investigation.
When mice are trained with a Morris water maze, both rRNA and pre-rRNA levels are significantly elevated (Li et al., 2018). The trained mice exhibit decreased expression of the lncRNA LoNA in the hippocampus. LoNA is synthesized by Pol II and specifically enriched in nucleolus, and it can suppress the transcription of pre-rRNAs (Figure 2). The 5′ portion of LoNA interacts with nucleolin (NCL), while its 3′ portion contains a snoRNA that binds to fibrillarin (FBL). NCL can remodel rDNA loci, and can, therefore, modulate pre-rRNA transcription (Abdelmohsen and Gorospe, 2012; Durut and Saez-Vasquez, 2015). LoNA binds to NCL and inactivates the chromatin status of rDNA region. FBL is a component of C/D box small nucleolar ribonucleoproteins (snoRNPs), which direct 2′-O-methylation of rRNAs and participate in rRNA processing (Newton et al., 2003). LoNA competes with snoRNAs to bind to FBL, thereby altering the methylation status of rRNAs. By binding to both NCL and FBL proteins, LoNA suppresses rRNA production and alters ribosome heterogeneity (Li et al., 2018).
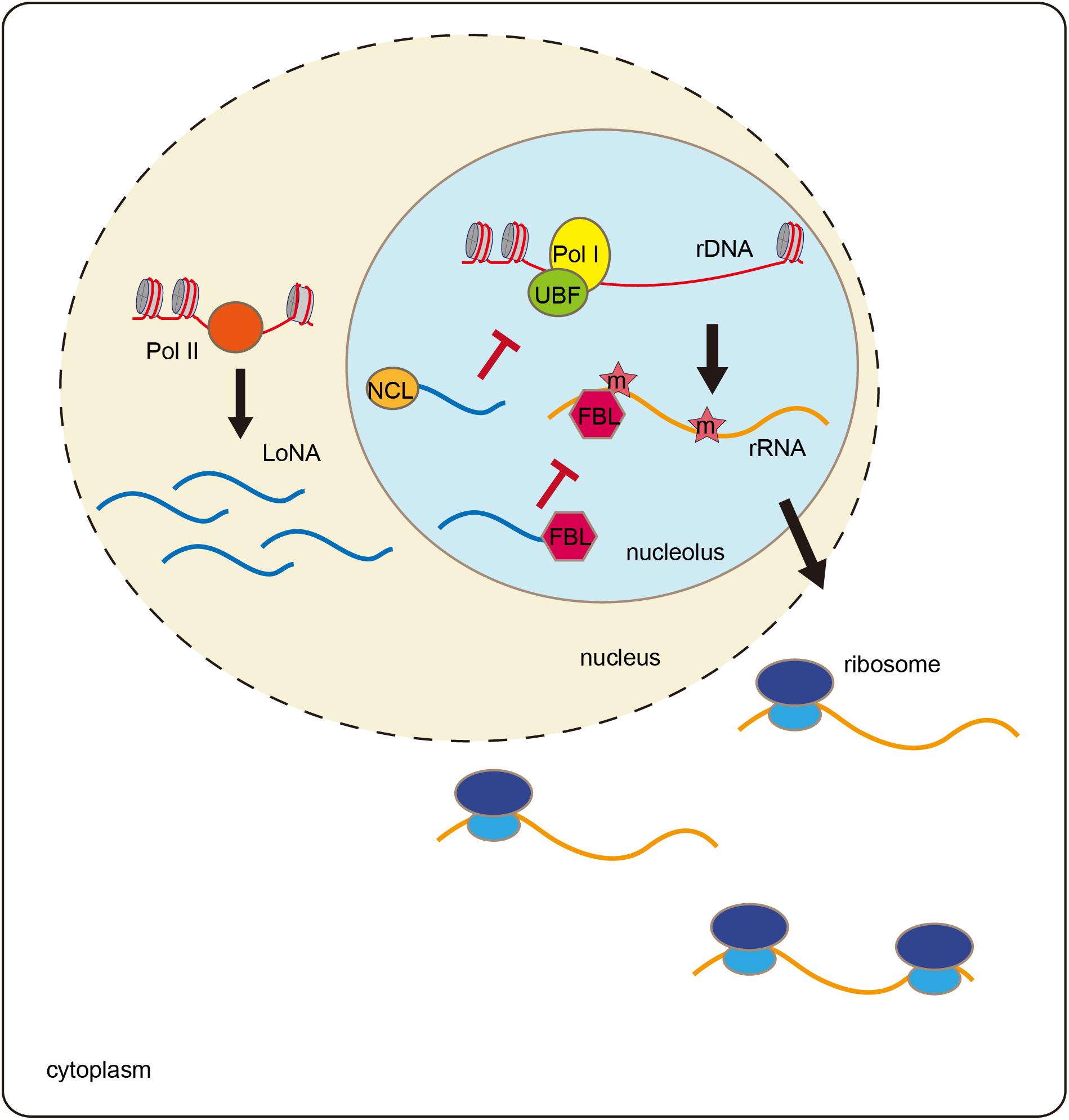
Figure 2. LoNA modulates rRNA and promotes learning and memory. LoNA is synthesized by Pol II. LoNA interacts with nucleolin (NCL) and inactivates the chromatin status of rDNA region via reducing the loading of UBF and Pol I to rDNA loci. Meanwhile, LoNA competes with snoRNAs to bind to FBL, thereby altering the methylation status of rRNAs. Therefore, by binding to both NCL and FBL proteins, LoNA suppresses rRNA production and alters ribosome heterogeneity.
Nucleolar stress is accompanied by decreased rRNA synthesis and failures in ribosome biogenesis and functions, which are considered to be cellular stress events associated with aging and neurodegenerative diseases (Boulon et al., 2010; Wu et al., 2018). In patients with Alzheimer disease (AD), rRNA production decreases (Dönmez-Altuntaş et al., 2005). In an AD animal model, LoNA expression is significantly increased in the mouse brain, which is accompanied by decreased rRNA levels (Li et al., 2018). Mice lacking LoNA are more efficient in locating the hidden platform in Morris water maze tests than are the control animals. LoNA-deficient AD mice show rescue of the learning and memory defects compared to the control animals in Morris water mazes and in object context discrimination behavioral tests. These results suggest that hippocampal LoNA is involved in learning and memory and may represent a potential therapeutic target for AD treatment.
SLERT is a box H/ACA snoRNA-ended lncRNA (Xing et al., 2017). SLERT contains 694 nt and is highly expressed in many human cell lines, especially in human embryonic stem cells and ovarian carcinoma cells. SLERT mainly accumulates in the nucleolus, and its localization depends on its box H/ACA snoRNA ends. SLERT depletion results in decreased levels of the 18S and 28S rRNAs, indicating that SLERT promotes rRNA production.
Mass spectrometry (MS) data of SLERT-associated proteins identified DDX21, a DEAD-box RNA helicase that is involved in multiple steps of ribosome biogenesis (Holmström et al., 2008; Calo et al., 2015; Sloan et al., 2015). SLERT depletion enhances the interaction between DDX21 and Pol I by tightening the DDX21 rings surrounding Pol I complexes, thereby suppressing rDNA transcription (Xing et al., 2017). Dysregulated rRNA synthesis by Pol I is associated with uncontrolled cancer cell proliferation (Nguyen le et al., 2015). The interaction between SLERT and DDX21, therefore, represents a potential therapeutic target for future anti-cancer drug discovery (Peltonen et al., 2014).
Upon stress, cells utilize various strategies to suppress rDNA transcription to promote survival, for example, by inactivating certain transcription factors and inducing chromatin remodeling (Bierhoff et al., 2014; Holland et al., 2016). Furthermore, a class of lncRNAs, PAPAS, is expressed to inhibit pre-rRNA transcription (Figure 3). PAPAS is transcribed by RNA polymerase II from a fraction of the rDNA units in an antisense orientation, and, therefore, it is called “promoter and pre-rRNA antisense” (PAPAS) (Bierhoff et al., 2010, 2014). PAPAS comprises a heterogeneous population of 12–16 kb lncRNAs that are complementary to both the pre-rRNA coding region and the rDNA promoter. PAPAS responds to distinct stresses and modulates pre-rRNA synthesis accordingly.
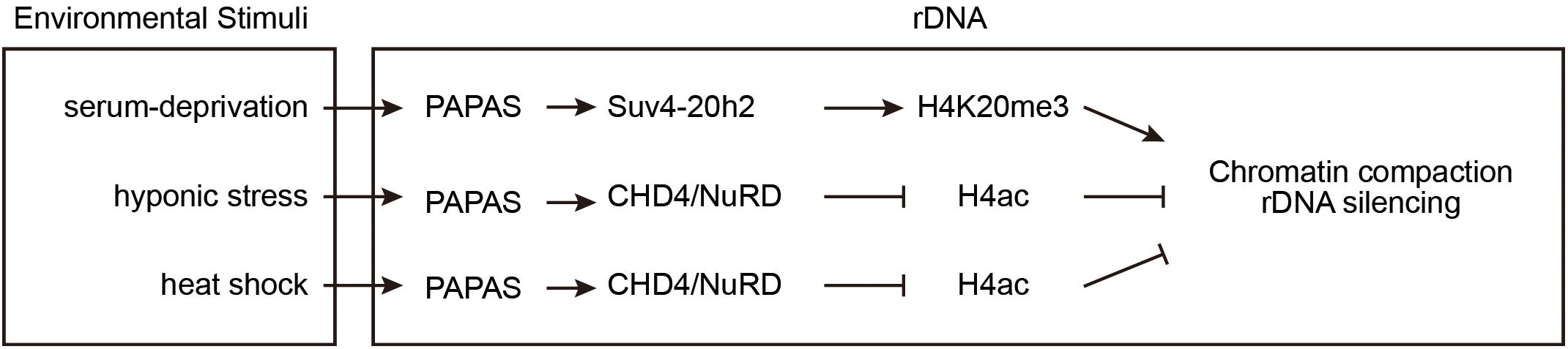
Figure 3. PAPAS responds to environmental stimuli to suppress rDNA transcription. At rDNA clusters, the lncRNA PAPAS directs Suv4-20h2 for repressive mark H4K20me3 deposition after growth factor deprivation. Upon hypotonic and heat-shock stresses, PAPAS recruits NuRD to rDNA and induces a nucleosomal “off” position to repress rDNA transcription.
In density-arrested or serum-deprived cells, pre-rRNA synthesis is suppressed and rDNA is enriched with H4K20me3 marks, while PAPAS is upregulated. After starved cells are refed with serum, the levels of H4K20me3 modifications and PAPAS decrease. RNA immunoprecipitation (RIP) experiments revealed that quiescence-induced PAPAS recruits Suv4-20h2 to transcription-competent rRNA genes to trigger H4K20me3 modification and chromatin compaction. Furthermore, siRNA-mediated knockdown of endogenous PAPAS decreased the level of H4K20me3 modifications, but not the level of H3K9me3 modifications (Bierhoff et al., 2014).
Hypo-osmotic stress also upregulates PAPAS and inhibits rDNA transcription. However, unlike serum deprivation, hypo-osmotic shock does not increase the Suv4-20h2 occupancy and H4K20me3 abundance at rDNA loci, but rather induces the degradation of Suv4-20h2 (Zhao et al., 2016a). The Mi-2/nucleosome remodeling and deacetylase factor (NuRD) is a multisubunit protein complex containing the HDAC1 histone deacetylase and the ATP-dependent remodeling enzyme CHD4 (Xue et al., 1998; Torchy et al., 2015). Upon hypo-osmotic stress, the elevated PAPAS associates with CHD4/NuRD and recruits them to rDNA regions where they deacetylate histone H4, remodel the promoter-bound nucleosomes, and reinforce transcriptional repression (Zhao et al., 2016a).
Similar to hypotonicity, heat shock also increases PAPAS expression, induces the degradation of Suv4-20h2, recruits NuRD to rDNA, and turns off transcription of pre-rRNA (Zhao et al., 2016b). Recent studies revealed the molecular mechanism of how PAPAS recruits CHD4/NuRD to rDNA. CHD4 is an RNA-binding protein that associates with both DNA and RNA via its N-terminal PHD and chromo-domains. Heat-shock elicits CHD4 dephosphorylation to facilitate its association with PAPAS (Zhao et al., 2018). PAPAS binds to the adjacent rDNA sequence via the formation of a DNA-RNA triplex, thereby directing CHD4/NuRD to rDNA, where it remodels the chromatin into a transcription refractory state (Zhao et al., 2016b, 2018).
Unlike other Pol I-transcribed rRNAs, 5S rRNAs are transcribed by Pol III. 5S rRNA genes are clustered as tandem repeats with intergenic sequences, and they are located on distinct chromosomes (Ciganda and Williams, 2011). A number of studies have revealed Pol II binding sites adjacent to Pol III-transcribed genes, including the 5S rRNA genes (Oler et al., 2010; Hu et al., 2012). These cryptic Pol II transcripts may therefore modulate the transcription of neighboring 5S rRNAs.
Hu et al. (2016) identified a lncRNA, 5S rRNA overlapped transcripts (5S-OT), that is transcribed by RNA polymerase II and is complementary to the 5S rRNA. 5′ and 3′ rapid amplification of cDNA ends (RACE) experiments demonstrated that this transcript contains 847 and 354 nt in mice and humans, respectively. Chromatin immunoprecipitation (ChIP) experiments indicated that Pol II binds to the promoter at the 5S-OT transcription start site in both mouse and human cells. Inhibition of Pol II with α-amanitin results in a decreased level of 5S-OT transcripts, which further leads to a reduction of nascent 5S rRNAs. Consistently, knocking down 5S-OT by siRNAs also inhibits the production of nascent 5S rRNAs. It was suggested that in mammalian cells, the lncRNA 5S-OT associates with 5S rDNA clusters where it promotes the transcription of 5S rRNAs, thus providing a mechanism to couple Pol II and Pol III transcription.
Furthermore, human 5S-OT contains an antisense Alu element at its 3′ end (Hu et al., 2016). Alu is a primate-specific transposable element. The Alu element in the human 5S-OT gene belongs to the AluY subfamily (Batzer and Deininger, 2002). In human cells, 5S-OT regulates alternative splicing of multiple genes in trans via Alu/anti-Alu pairing with targeted genes and by interacting with the splicing factor U2AF65.
Therefore, the lncRNA 5S-OT modifies 5S rRNA and mRNAs via cis and trans mechanisms, respectively. Since 5S-OT is relatively conserved in eukaryotes from fission yeast to humans, it will be interesting to examine whether similar mechanisms are applicable in other organisms.
The gene silencing capacity of small interfering RNAs (siRNAs) was first described in Caenorhabditis elegans two decades ago (Fire et al., 1991). siRNAs silence complementary nucleic acids in both the cytoplasm and nucleus. Previous research has focused on the mechanism of siRNA-dependent regulation of mRNAs. In the cytoplasm, siRNAs can direct the degradation of targeted RNAs and inhibit protein translation (Ipsaro and Joshua-Tor, 2015). In the nucleus, siRNAs can guide heterochromatin formation and inhibit transcription elongation (Feng and Guang, 2013; Rechavi and Lev, 2017). Here, we will summarize our recent work that begins to illustrate the function of siRNAs in the regulation of ribosomal RNAs (Figure 4; Zhou et al., 2017a; Zhu et al., 2018).
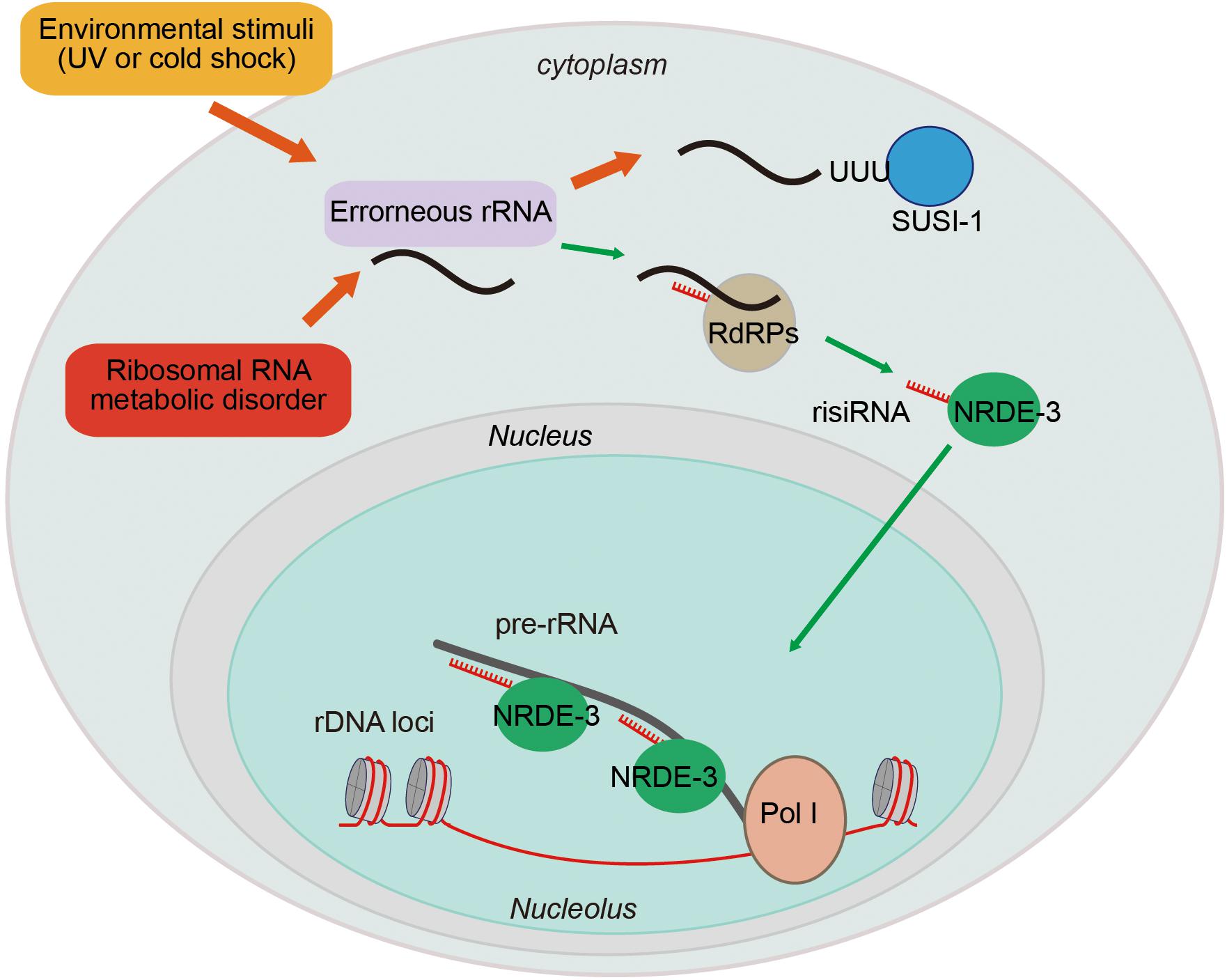
Figure 4. Ribosomal siRNAs silence pre-rRNAs via the nuclear RNAi pathway. rRNA biogenesis is a multistep process, including a series of pre-rRNA processing, folding, and modification steps. Endogenous metabolic stresses and environmental stimuli can cause RNA processing disorders, leading to the accumulation of erroneous RNAs. This accumulation leads to the recruitment of RdRPs to synthesize risiRNA, subsequently turning on the nuclear RNAi-mediated gene silencing pathway to inhibit pre-rRNA expression.
Antisense ribosomal siRNAs (risiRNAs) are widely present in various organisms. In Schizosaccharomyces pombe lacking Cid14, rRNAs become substrates for the RNAi pathway, giving rise to siRNAs targeting rRNA (Buhler et al., 2008). In Neurospora crassa, DNA damage induces the expression of the Argonaute protein QDE-2 and a class of RNAs that interact with it (qiRNAs) from the ribosomal DNA locus (Lee et al., 2009).
In C. elegans, upon exposure to low temperature treatment or ultraviolet (UV) light, risiRNAs accumulate (Zhou et al., 2017b). risiRNAs are complementary to the 18S and 26S rRNAs, contain 22 nt, and start with a 5′ guanosine. risiRNAs belong to a class of 22G-RNAs that are synthesized by the RNA-dependent RNA polymerases (RdRPs) in C. elegans. risiRNAs associate with the Argonaute protein NRDE-3 and translocate to nucleolus, where they suppress pre-rRNA expression (Zhou et al., 2017b; Zhu et al., 2018).
Ribosomal siRNAs act to surveil erroneous rRNAs and maintain rRNA homeostasis. Misprocessed rRNAs are usually detected and degraded by multiple surveillance machineries, including the exosome and Trf4/Air2/Mtr4p polyadenylation (TRAMP) complexes (Schmidt and Butler, 2013). The exonuclease SUSI-1 (ceDIS3L2) is involved in the 3′–5′ degradation of oligouridylated rRNA fragments (Astuti et al., 2012; Zhou et al., 2017b). When the rRNA modification or processing steps are disrupted, or upon cold shock treatment or UV exposure, erroneous rRNAs are oligouridylated and recognized by RdRPs to generate risiRNAs via a poorly understood mechanism (Zhou et al., 2017b; Zhu et al., 2018). risiRNAs in turn silence rRNAs via the RNAi machinery to prohibit the accumulation of erroneous rRNAs.
Downregulation of rRNA transcription is one of the major strategies to preserve cellular homeostasis upon encountering stress conditions and to limit energy consumption under unfavorable conditions (Parlato and Liss, 2014). The risiRNA/RNAi-directed feedback loop, therefore, may compensate for dysfunctions in the exoribonuclease-engaged degradation of erroneous rRNAs. Consistently, when rRNA modification and processing steps are defective, the animals grow more slowly because of the presence of risiRNAs (Zhu et al., 2018).
Ribosome biogenesis is tightly coordinated and controlled from the transcription of pre-rRNAs to the assembly of ribosomes, a process that is influenced by many developmental programs and environmental stress challenges (McStay, 2016; Sloan et al., 2017). Cells respond to these signals by modulating the transcription, processing, maturation of rRNAs and the assembly and usage of ribosomes. Defects or disorders in any of these steps lead to a number of human diseases. In addition to protein factors, small and long regulatory RNAs also play important roles in the regulation of pre-rRNA transcription and rRNA maturation. The regulatory RNAs may act to sense developmental signals and environmental stresses. For example, PAPAS can sense nutrition deprivation, heat-shock, and hypo-osmotic stresses. risiRNAs respond to cold shock and UV and further down regulate pre-rRNA transcription. risiRNAs also surveil the fidelity and precision of rRNA modifications and processing to avoid the accumulation of erroneous rRNAs expressed during the development of organisms. More interestingly, the lncRNA LoNA is involved in learning and memory by modulating rRNA transcription.
Many questions still remain to be addressed to fully understand the mechanisms and roles of non-coding RNAs in anti-stress pathways and rRNA regulation. For example, how do these lncRNAs surveil distinct environmental stresses? Cold shock could induce untemplated addition of oligouridylation at the 3′ ends of 26S rRNAs to elicit the production of risiRNAs (Zhou et al., 2017b). Heat-shock elicits CHD4 dephosphorylation to facilitate its association with PAPAS (Zhao et al., 2018). Were these surveillance mechanisms conserved among different organisms during evolution? Do poikilotherms and homeotherms use similar mechanisms to sense temperature alterations in the environment? Beside nuclear non-coding RNAs, cytoplasmic lncRNAs are frequently bound to and degraded at ribosomes (Carlevaro-Fita et al., 2016). Whether these cytoplasmic lncRNAs can in turn regulate rRNAs need further investigation. In particular, what are the biological roles of non-coding RNAs in regulating rRNAs during developmental processes? With new emerging technologies, many novel discoveries will help to answer these important questions.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by grants from the National Key R&D Program of China (2018YFC1004500 and 2017YFA0102903), the National Natural Science Foundation of China (Nos. 31671346, 91640110, 31870812, and 31871300), the Major/Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology (2017FXZY005), and CAS Interdisciplinary Innovation Team.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to the members of the SG’s lab for their comments.
Abdelmohsen, K., and Gorospe, M. (2012). RNA-binding protein nucleolin in disease. RNA Biol. 9, 799–808. doi: 10.4161/rna.19718
Astuti, D., Morris, M. R., Cooper, W. N., Staals, R. H., Wake, N. C., Fews, G. A., et al. (2012). Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat. Genet. 44, 277–284. doi: 10.1038/ng.1071
Batzer, M. A., and Deininger, P. L. (2002). Alu repeats and human genomic diversity. Nat. Rev. Genet. 3, 370–379. doi: 10.1038/nrg798
Bazin, J., Baerenfaller, K., Gosai, S. J., Gregory, B. D., Crespi, M., and Bailey-Serres, J. (2017). Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc. Natl. Acad. Sci. U.S.A. 114, E10018–E10027. doi: 10.1073/pnas.1708433114
Bierhoff, H., Dammert, M. A., Brocks, D., Dambacher, S., Schotta, G., and Grummt, I. (2014). Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell 54, 675–682. doi: 10.1016/j.molcel.2014.03.032
Bierhoff, H., Schmitz, K., Maass, F., Ye, J., and Grummt, I. (2010). Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb. Symp. Quant. Biol. 75, 357–364. doi: 10.1101/sqb.2010.75.060
Boisvert, F. M., van Koningsbruggen, S., Navascues, J., and Lamond, A. I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585. doi: 10.1038/nrm2184
Boulon, S., Westman, B. J., Hutten, S., Boisvert, F. M., and Lamond, A. I. (2010). The nucleolus under stress. Mol. Cell 40, 216–227. doi: 10.1016/j.molcel.2010.09.024
Buhler, M., Spies, N., Bartel, D. P., and Moazed, D. (2008). TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct. Mol. Biol. 15, 1015–1023. doi: 10.1038/nsmb.1481
Calo, E., Flynn, R. A., Martin, L., Spitale, R. C., Chang, H. Y., and Wysocka, J. (2015). RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518, 249–253. doi: 10.1038/nature13923
Carlevaro-Fita, J., Rahim, A., Guigo, R., Vardy, L. A., and Johnson, R. (2016). Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 22, 867–882. doi: 10.1261/rna.053561.115
Cech, T. R., and Steitz, J. A. (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94. doi: 10.1016/j.cell.2014.03.008
Chen, L. L. (2016). Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 41, 761–772. doi: 10.1016/j.tibs.2016.07.003
Ciganda, M., and Williams, N. (2011). Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip. Rev. RNA 2, 523–533. doi: 10.1002/wrna.74
Dönmez-Altuntaş, H., Akalin, H., Karaman, Y., Demirtaş, H., Imamoğlu, N., and Ozkul, Y. (2005). Evaluation of the nucleolar organizer regions in Alzheimer’s disease. Gerontology 51, 297–301. doi: 10.1159/000086365
Durut, N., and Saez-Vasquez, J. (2015). Nucleolin: dual roles in rDNA chromatin transcription. Gene 556, 7–12. doi: 10.1016/j.gene.2014.09.023
Feng, X., and Guang, S. (2013). Small RNAs, RNAi and the inheritance of gene silencing in Caenorhabditis elegans. J. Genet. Genomics 40, 153–160. doi: 10.1016/j.jgg.2012.12.007
Fire, A., Albertson, D., Harrison, S. W., and Moerman, D. G. (1991). Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle. Development 113, 503–514.
Guetg, C., Lienemann, P., Sirri, V., Grummt, I., Hernandez-Verdun, D., Hottiger, M. O., et al. (2010). The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 29, 2135–2146. doi: 10.1038/emboj.2010.17
Henderson, A. S., Warburton, D., and Atwood, K. C. (1973). Letter: ribosomal DNA connectives between human acrocentric chromosomes. Nature 245, 95–97. doi: 10.1038/245095b0
Holland, M. L., Lowe, R., Caton, P. W., Gemma, C., Carbajosa, G., Danson, A. F., et al. (2016). Early-life nutrition modulates the epigenetic state of specific rDNA genetic variants in mice. Science 353, 495–498. doi: 10.1126/science.aaf7040
Holmström, T. H., Mialon, A., Kallio, M., Nymalm, Y., Mannermaa, L., Holm, T., et al. (2008). c-Jun supports ribosomal RNA processing and nucleolar localization of RNA helicase DDX21. J. Biol. Chem. 283, 7046–7053. doi: 10.1074/jbc.M709613200
Hu, S., Wang, X., and Shan, G. (2016). Insertion of an Alu element in a lncRNA leads to primate-specific modulation of alternative splicing. Nat. Struct. Mol. Biol. 23, 1011–1019. doi: 10.1038/nsmb.3302
Hu, S., Wu, J., Chen, L., and Shan, G. (2012). Signals from noncoding RNAs: unconventional roles for conventional pol III transcripts. Int. J. Biochem. Cell Biol. 44, 1847–1851. doi: 10.1016/j.biocel.2012.07.013
Ipsaro, J. J., and Joshua-Tor, T. (2015). From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 22, 20–28. doi: 10.1038/nsmb.2931
Ji, Z., Song, R., Regev, A., and Struhl, K. (2015). Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. eLife 4:e08890. doi: 10.7554/eLife.08890
Lee, H. C., Chang, S. S., Choudhary, S., Aalto, A. P., Maiti, M., Bamford, D. H., et al. (2009). qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459, 274–277. doi: 10.1038/nature08041
Li, D., Zhang, J., Wang, M., Li, X., Gong, H., Tang, H., et al. (2018). Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat. Commun. 9:1726. doi: 10.1038/s41467-018-04072-4
Mayer, C., Neubert, M., and Grummt, I. (2008). The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 9, 774–780. doi: 10.1038/embor.2008.109
Mayer, C., Schmitz, K. M., Li, J., Grummt, I., and Santoro, R. (2006). Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 22, 351–361. doi: 10.1016/j.molcel.2006.03.028
McStay, B. (2006). Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 20, 1207–1214. doi: 10.1101/gad.1436906
McStay, B. (2016). Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev. 30, 1598–1610. doi: 10.1101/gad.283838.116
Newton, K., Petfalski, E., Tollervey, D., and Caceres, J. F. (2003). Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Mol. Cell. Biol. 23, 8519–8527. doi: 10.1128/MCB.23.23.8519-8527.2003
Nguyen le, X. T., Raval, A., Garcia, J. S., and Mitchell, B. S. (2015). Regulation of ribosomal gene expression in cancer. J. Cell Physiol. 230, 1181–1188. doi: 10.1002/jcp.24854
Oler, A. J., Alla, R. K., Roberts, D. N., Wong, A., Hollenhorst, P. C., Chandler, K. J., et al. (2010). Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 17, 620–628. doi: 10.1038/nsmb.1801
Parlato, R., and Liss, B. (2014). How Parkinson’s disease meets nucleolar stress. Biochim. Biophys. Acta 1842, 791–797. doi: 10.1016/j.bbadis.2013.12.014
Peltonen, K., Colis, L., Liu, H., Trivedi, R., Moubarek, M. S., Moore, H. M., et al. (2014). A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 25, 77–90. doi: 10.1016/j.ccr.2013.12.009
Postepska-Igielska, A., Krunic, D., Schmitt, N., Greulich-Bode, K. M., Boukamp, P., and Grummt, I. (2013). The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 14, 704–710. doi: 10.1038/embor.2013.87
Rechavi, O., and Lev, I. (2017). Principles of transgenerational small RNA inheritance in Caenorhabditis elegans. Curr. Biol. 27, R720–R730. doi: 10.1016/j.cub.2017.05.043
Santoro, R., Li, J., and Grummt, I. (2002). The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32, 393–396. doi: 10.1038/ng1010
Savić, N., Bär, D., Leone, S., Frommel, S. C., Weber, F. A., Vollenweider, E., et al. (2014). lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15, 720–734. doi: 10.1016/j.stem.2014.10.005
Schmidt, K., and Butler, J. S. (2013). Nuclear RNA surveillance: role of TRAMP in controlling exosome specificity. Wiley Interdiscip. Rev. RNA 4, 217–231. doi: 10.1002/wrna.1155
Schmitz, K. M., Mayer, C., Postepska, A., and Grummt, I. (2010). Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269. doi: 10.1101/gad.590910
Sloan, K. E., Leisegang, M. S., Doebele, C., Ramirez, A. S., Simm, S., Safferthal, C., et al. (2015). The association of late-acting snoRNPs with human pre-ribosomal complexes requires the RNA helicase DDX21. Nucleic Acids Res. 43, 553–564. doi: 10.1093/nar/gku1291
Sloan, K. E., Warda, A. S., Sharma, S., Entian, K. D., Lafontaine, D. L. J., and Bohnsack, M. T. (2017). Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152. doi: 10.1080/15476286.2016.1259781
Srivastava, R., Srivastava, R., and Ahn, S. H. (2016). The epigenetic pathways to ribosomal DNA silencing. Microbiol. Mol. Biol. Rev. 80, 545–563. doi: 10.1128/MMBR.00005-16
Sun, Q., Hao, Q., and Prasanth, K. V. (2017). Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 34, 142–157. doi: 10.1016/j.tig.2017.11.005
Torchy, M. P., Hamiche, A., and Klaholz, B. P. (2015). Structure and function insights into the NuRD chromatin remodeling complex. Cell. Mol. Life Sci. 72, 2491–2507. doi: 10.1007/s00018-015-1880-8
Wu, J., Jiang, X., Li, Y., Zhu, T., Zhang, J., Zhang, Z., et al. (2018). PHA-4/FoxA senses nucleolar stress to regulate lipid accumulation in Caenorhabditis elegans. Nat. Commun. 9:1195. doi: 10.1038/s41467-018-03531-2
Xing, Y. H., Yao, R. W., Zhang, Y., Guo, C. J., Jiang, S., Xu, G., et al. (2017). SLERT regulates DDX21 rings associated with Pol I transcription. Cell 169, 664–678.e16. doi: 10.1016/j.cell.2017.04.011
Xue, Y., Wong, J., Moreno, G. T., Young, M. K., Côté, J., and Wang, W. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861. doi: 10.1016/S1097-2765(00)80299-3
Zhao, Z., Dammert, M. A., Grummt, I., and Bierhoff, H. (2016a). lncRNA-Induced nucleosome repositioning reinforces transcriptional repression of rRNA genes upon hypotonic stress. Cell Rep. 14, 1876–1882. doi: 10.1016/j.celrep.2016.01.073
Zhao, Z., Dammert, M. A., Hoppe, S., Bierhoff, H., and Grummt, I. (2016b). Heat shock represses rRNA synthesis by inactivation of TIF-IA and lncRNA-dependent changes in nucleosome positioning. Nucleic Acids Res. 44, 8144–8152. doi: 10.1093/nar/gkw496
Zhao, Z., Senturk, N., Song, C., and Grummt, I. (2018). lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 32, 836–848. doi: 10.1101/gad.311688.118
Zhou, X., Chen, X., Wang, Y., Feng, X., and Guang, S. (2017a). A new layer of rRNA regulation by small interference RNAs and the nuclear RNAi pathway. RNA Biol. 14, 1492–1498. doi: 10.1080/15476286.2017.1341034
Zhou, X., Feng, X., Mao, H., Li, M., Xu, F., Hu, K., et al. (2017b). RdRP-synthesized antisense ribosomal siRNAs silence pre-rRNA via the nuclear RNAi pathway. Nat. Struct. Mol. Biol. 24, 258–269. doi: 10.1038/nsmb.3376
Keywords: rRNA, non-coding RNA, PAPAS, SLERT, 5S-OT, risiRNA, LoNA
Citation: Yan Q, Zhu C, Guang S and Feng X (2019) The Functions of Non-coding RNAs in rRNA Regulation. Front. Genet. 10:290. doi: 10.3389/fgene.2019.00290
Received: 29 September 2018; Accepted: 18 March 2019;
Published: 05 April 2019.
Edited by:
Thomas S. Wingo, Emory University, United StatesReviewed by:
Fabrizio Loreni, University of Rome Tor Vergata, ItalyCopyright © 2019 Yan, Zhu, Guang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouhong Guang, c2d1YW5nQHVzdGMuZWR1LmNu Xuezhu Feng, ZmVuZ3h6QHVzdGMuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.