- 1Department of Stomatology, The First Affiliated Hospital of Shantou University Medical College, Shantou, China
- 2Cancer Research Center, Shantou University Medical College, Shantou, China
- 3Institute of Precision Cancer and Pathology, Jinan University Medical College, Guangzhou, China
- 4Department of Emergency Medicine, University of Texas MD Anderson Cancer Center, Houston, TX, United States
- 5Department of Endocrine Neoplasia and Hormonal Disorders, University of Texas MD Anderson Cancer Center, Houston, TX, United States
- 6Research Center of Translational Medicine, The Second Affiliated Hospital of Shantou University Medical College, Shantou, China
Periodontitis is the most prevalent inflammatory disease of the periodontium, and is related to oral and systemic health. Exosomes are emerging as non-invasive biomarker for liquid biopsy. We here evaluated the levels of programmed death-ligand 1 (PD-L1) mRNA in salivary exosomes from patients with periodontitis and non-periodontitis controls. The purposes of this study were to establish a procedure for isolation and detection of mRNA in exosomes from saliva of periodontitis patients, to characterize the level of salivary exosomal PD-L1, and to illustrate its clinical relevance. Bioinformatics analysis suggested that periodontitis was associated with an inflammation gene expression signature, that PD-L1 expression positively correlated with inflammation in periodontitis based on gene set enrichment analysis (GSEA) and that PD-L1 expression was remarkably elevated in periodontitis patients versus control subjects. Exosomal RNAs were successfully isolated from saliva of 61 patients and 30 controls and were subjected to qRT-PCR. Levels of PD-L1 mRNA in salivary exosomes were higher in periodontitis patients than controls (P < 0.01). Salivary exosomal PD-L1 mRNA showed significant difference between the stages of periodontitis. In summary, the protocols for isolating and detecting exosomal RNA from saliva of periodontitis patients were, for the first time, characterized. The current study suggests that assay of exosomes-based PD-L1 mRNA in saliva has potential to distinguish periodontitis from the healthy, and the levels correlate with the severity/stage of periodontitis.
Introduction
Periodontitis is one of the most prevalent disease in dentistry, impairing the integrity of the periodontium and leading ultimately to tooth loss. Periodontitis is a chronic inflammatory disease caused by microorganisms colonizing the dentogingival interface (Slots, 2013). Currently, the diagnosis of periodontitis was mostly based on clinical and radiographic evaluations without specific evaluation of the underlying inflammatory response (Buduneli and Kinane, 2011; Baeza et al., 2016). Measuring electrolyte concentration in gingival crevicular fluid (GCF), especially the concentration of sodium, potassium and calcium ions, reflects the clinical status of the periodontal tissues such that the pattern of concentrations of these ions may be used as a potential diagnostic marker for active periodontitis (Koregol et al., 2011).
Given the close relevance of inflammation to periodontitis, immune-regulatory factors have been explored as biomarker of periodontitis (Han et al., 2012; Kimak et al., 2015). Programmed death-ligand 1 (PD-L1), also known as the B7-H1 receptor, plays an important role in cell-mediated immune responses (Gianchecchi et al., 2013). PD-L1 regulates T cell activation and tolerance, and is able to inhibit activated T cell functions and survival (Kim et al., 2005). High expression of PD-L1 in host cells may contribute to the chronicity of inflammatory disorders (Zamani et al., 2016).
PD-L1 is involved in periodontitis. Porphyromonas gingivalis (P. gingivalis) is a keystone pathogen in chronic periodontitis (Groeger et al., 2017), and it induces expression of PD-L1 in malignant and non-malignant oral epithelial cells (Groeger et al., 2017). In periodontitis, P. gingivalis inhibits the synthesis of cytokines and increases humoral responses. This reduces the inflammatory responses related to T- and B-cell activation, and subsequent IFN-γ secretion by a subset of T cells. The T cells that secrete IFN-γ further suppress upregulation of programmed cell death-1 (PD-1)-receptor and its ligand PD-L1 on CD11b+-subset of T cells (Gaddis et al., 2013; Groeger et al., 2017). Interestingly, studies recently have demonstrated elevated PD-L1 levels in saliva from patients with oral cancers or salivary gland carcinoma (Aziz et al., 2015; Goncalves et al., 2015, 2017). Saliva-derived samples have been studied as a source of biomarkers for periodontitis (Baltacioglu et al., 2014; Recker et al., 2015).
Exosomes are small (30—100 nm) membrane-encapsulated vesicles containing nuclei acids and protein cargo, and secreted by eukaryotic cells into the circulation (Tkach and Thery, 2016). The contents of disease cell-derived exosomes may potentially serve as a source of disease biomarkers. Salivary exosomal contents have recently been investigated for diagnosis and prognosis of a wide range of diseases (Michael et al., 2010; Lau et al., 2013; Byun et al., 2015; Machida et al., 2015; Sivadasan et al., 2015; Kim et al., 2017; Zheng et al., 2017; Han et al., 2018). Although detection of PD-L1 mRNA has been reported in periodontitis (Yuan et al., 2015; Zhang et al., 2016), it is unknown whether PD-L1 mRNA can be detected in exosomes of saliva of periodontitis patients and whether the level of salivary exosomal PD-L1 mRNA reflects disease status. We therefore focused on exosomes and saliva, and set to establish a protocol for isolation and detection of exosomes, and exosomal RNA in the saliva of periodontitis patients, and to characterize salivary exosomal PD-L1 as a potential marker for periodontitis.
Materials and Methods
Study Population
This study is a prospective observational investigation at the First Affiliated Hospital of Shantou University. From June, 2017 to June, 2018, 61 periodontitis patients and 30 control subjects were enrolled in the study with informed consent. Each recruited subject was inquired in detail about the medical history and accepted a thorough periodontal examination. Diagnosis of periodontitis was based on assessment of probing pocket depth (PD) and clinical attachment loss (CAL). Patients who fulfilled the requirement of generalized chronic periodontitis according to the classification by the American Academy of Periodontology in 2007 (Slots, 2013) were recruited to the periodontitis group after periodontal examination. The non-periodontitis control group was composed of subjects with no evidence of periodontitis after examination according to the above classification. Smoking and alcohol using status was recorded for all individuals. Exclusion criteria were age <18 years, inability to give informed consent, congenital malformation, chronic diseases (e.g., lip cancer, gingival cancer, carcinoma of the tongue, soft palate carcinoma, jaw bone cancers, cancers of the mouth floor carcinoma, oropharyngeal cancer, salivary gland carcinoma, maxillary sinus carcinoma, cancer occurs in facial ministry skin mucous membrane, epilepsy, cardiac disease, lung disease, renal disease, positive test for human immunodeficiency virus etc.), and history of systemic antibiotic treatment or dental prophylaxis in the previous 6 months. Written informed consents were obtained from all participants in accordance with the principles established by the Helsinki Declaration. This study was approved by the Institutional Review Board of Shantou University Medical College (SUMC) (Shantou, China) under IRB protocol number: #04-070.
Clinical Evaluation
For each individual, PD, CAL and bleeding on probing (BP) values were measured with a periodontal probe. A single calibrated examiner assessed these clinical parameters. Control individuals (control group) did not present any sign or symptom compatible with periodontal disease (PD < 3 mm, CAL < 3 mm and no radiographic evidence of alveolar bone breakdown). On the other hand, the periodontitis group consisted of patients with mild, moderate or severe periodontitis (based on the Classification of Periodontal Diseases and Conditions of Armitage, 2007) (Slots, 2013) with at least one single-rooted tooth with a CAL ≥ 6 mm and probing depth ≥ 5 mm.
Gingival Biopsies
Human gingivae were obtained from all periodontitis patients. Gingiva was harvested during tooth extractions for periodontal reasons in the Department of Stomatology of First Affiliated Hospital of Shantou University. Written informed consent and approval of the Ethics Committee of the SUMC were obtained.
Human Saliva Collection
Saliva was collected in the morning (8 am to 10 am) from all subjects. During the collection period, subjects were instructed to refrain from eating, drinking or using oral hygiene products for at least 1 h prior to collection, and received no stimulation of salivation. After rinsing the mouth with water, each subject spat 3–5 mL saliva into a 35-mm dish. Subjects were reminded not to cough up mucus, and saliva was collected within 30 min from spitting. These saliva samples were pipetted into 1.5-mL tubes and were kept on ice during processing which did not exceed 60 min. The samples were then centrifuged at 3,000 ×g for 15 min at 4°C to remove cells and cellular fragments. As much supernatant as possible was collected into new 1.5-mL tubes and stored at -80°C.
Gene Set Enrichment Analysis (GSEA)
Gene set enrichment analysis (GSEA; v2.091) was performed to examine the association between gene sets and gene expression (Dong et al., 2017). Periodontitis gene expression profiles from an independent datasets (GSE16134) were collected from NCBI Gene Expression Omnibus (GEO) at http://www.ncbi.nlm.nih.gov/geo/. The expression levels of affected gingival tissue (n = 241) vs. unaffected gingival tissue (n = 69) from GSE16134 was ordered from high to low. We performed GSEA analysis to examine the correlation between periodontitis and the PD-L1 signature pathway, followed by the protocol available at the GSEA website1.
Isolation of Salivary Exosomes
ExoQuickTM exosomes precipitation solution was used for exosomes isolation according to the manufacturer’s instructions (System Biosciences, Mountain View, CA, United States). Briefly, ExoQuick-TCTM solution was added to saliva at a 63/250 ratio and mixed by inverting the tubes several times. Exosomes were precipitated by refrigeration at 4°C overnight, and then collected by centrifugation twice at 1,500 ×g for 30 and 5 min, respectively, in order to remove the supernatant. Supernatant was discarded, and the pellet resuspended in 300 μL TRIZOL for RNA isolation or 20 μL PBS for protein isolation.
RT-qPCR
Total RNA was extracted from exosomes and cell lysates using TRIzol reagent as per the manufacturer’s protocol (ZYMO RESEARCH). RNA was eluted with 25 μL of RNAse-free water. RT-qPCR was performed using an Absolute Blue QPCR SYBR Green Low ROX mix (Thermo Scientific) on an Applied Biosystems’ 7500 real-time PCR system. The Rn value (normalized reporter value) was the fluorescent signal from SYBR Green normalized to the signal of the passive reference dye for a given reaction. No-template and no-RT reactions were performed as negative controls. All assays were performed in 3 separate RTs followed by triplicate qPCR, and the results are shown as the average fold change relative to GAPDH which served as an internal control. Primers for RT-qPCR were:
GAPDH F: 5′-TGCACCACCAACTGCTTAGC-3′
GAPDH R: 5′-GGCATGGACTGTGGTCATGAG-3′
PD-L1 F: 5′-TGCCGACTACAAGCGAATTACTG -3′
PD-L1 R: 5′-CTGCTTGTCCAGATGACTTCGG-3′.
Protein Isolation and Immunoblotting
Exosomal pellets were resuspended in PBS and re-pelleted by centrifugation and then extracted with RIPA buffer (Santa Cruz Biotechnology). Total protein lysates were prepared and analyzed by immunoblotting using anti-ALIX (Cat. No. 2171; Cell Signaling Technology, Beverly, MA, United States), anti-TSG101 (Cat. ab133586; Abcam, Cambridge, United Kingdom), anti-CD63 (Cat. sc-15363; Santa Cruz Biotechnology, CA, United States), anti-CD9 (Cat. sc-9148; Santa Cruz Biotechnology, CA, United States), anti-CD81 (Cat. ab109201; Abcam, Cambridge, United Kingdom), anti-Calnexin (Cat. No. 2433; Cell Signaling Technology, Beverly, MA, United States), anti-LC3 (Cat. No. 2775; Cell Signaling Technology, Beverly, MA, United States), anti-NLRP3 (Cat.19771-1-AP; Proteintech, United States) and anti-NLRP4 (Cat. NB100-56156; Novus Biologicals, United States) as described previously (Gan et al., 2016; Dong et al., 2017).
Transmission Electron Microscopy
Following exosomes isolation, the pellet was washed in PBS, and then subjected to ultracentrifugation at 120,000 ×g for 70 min to re-pellet the exosomes. The exosomes pellets were resuspended in 30 μL PBS, and a 10 μL aliquot of the suspension loaded onto formvar carbon-coated grids and allow to stand for 5 min at room temperature. Next, the exosomes were fixed in 2% paraformaldehyde for 5 min at room temperature and washed thrice with PBS. Excess liquid was drained by gently touching the edge of the grid with clean filter paper. The grid was slightly touched onto a drop of 2% uranyl acetate for 1 min and embedded in a mixture of uranyl acetate (0.8%) and methyl cellulose (0.13%). Excess liquid was drained off, and then the grid was allowed to air dry for several minutes prior to examination under a transmission electron microscope (JEM-1400, Hitachi, Shiga, Japan) (Raposo and Stoorvogel, 2013).
Nanoparticle Tracking Analysis
Total particles in human salivary samples were analyzed by nanoparticle tracking by the A & P Instrument Co., Ltd. (Guangzhou, China), using a NanoSight LM10 system (NanoSight Ltd., Amesbury, United Kingdom). Each sample was diluted in nanoparticle-free PBS and analyzed three times. Data was collected and analyzed using the nanoparticle tracking analysis (NTA) software (RRID: SCR_014239, version 2.3). All measurements were recorded at room temperature.
Cell Culture
All cells were cultured in a sterile incubator maintained at 37°C with 5% CO2. ESCC cells and THP-1 cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Paisley, United Kingdom) or RPMI-1640 medium (Gibco, Paisley, United Kingdom) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Paisley, United Kingdom), 10 mmol/L glutamine, 100 units/ml penicillin (Sigma, St. Louis, MO, United States), and 100 μg/ml streptomycin (Sigma). TE1 cells were provided by Dr. X.C. Xu (The University of Texas MD Anderson Cancer Center, Houston, TX, United States). For LC3 detection, TE1 cells were collected after starvation (i.e., without fetal bovine serum) for 6 h.
Statistical Analysis
All statistical analyses were performed using the SPSS 19.0 statistical software package (SPSS Inc., Chicago, IL, United States) and Prism V6.01 (GraphPad). Summary statistics reporting means and standard errors were stated as appropriate. Statistical methods used included t-test, Pearson correlation and chi-square test.
Results
Bioinformatic Analysis of the Correlation Between Periodontitis and PD-L1
Given the association of periodontitis with inflammation, and the importance of PD-L1 in inflammatory immunity, we analyzed database to test whether periodontitis correlates with PD-L1. GSEA analysis revealed that periodontitis is positively associated with an inflammation signature, and PD-L1 is positively correlated with an inflammation signature in periodontitis (Figure 1A). Analysis of the GEO database (GSE16134) indicated a statistically significant elevation of PD-L1 level in periodontitis patients versus control subjects (Figure 1B). These data support the hypothesis that PD-L1 is positively relevant to periodontitis.
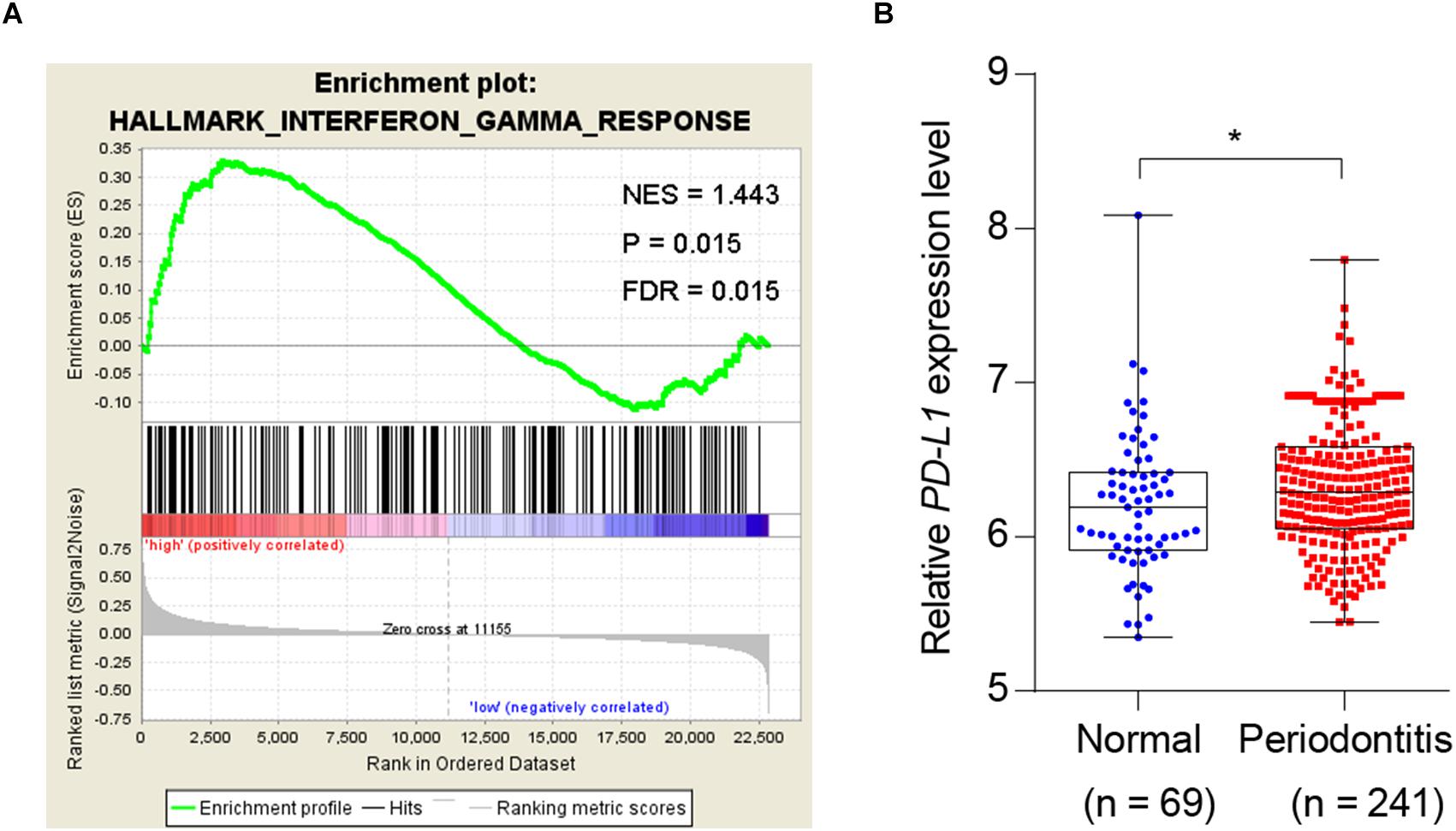
Figure 1. Periodontitis correlated with inflammation and PD-L1 signatures. (A) PD-L1 gene signatures were analyzed by GSEA using the gene sets (GSE 16134) derived from periodontitis patients and normal subjects. FDR = 0.015. (B) Quantified results show the expression level of PD-L1 in the gene sets (GSE 16134). Periodontitis patients (n = 241) compared with normal subjects (n = 69). Error bars indicate SEM. ∗P< 0.05 by Student’s t-test.
Patient Demographics and Clinical Parameters
This pilot study was conducted at Shantou University Medical College. Sixty-one periodontitis patients and 30 control subjects were enrolled in the study (Figure 2). With an average age of 51, 29 male and 32 female patients from the First Affiliated Hospital of Shantou University were recruited. There are also 30 control subjects (14 male and 16 female) were enrolled in this study with an average age of 52. Among demographic and clinicopathological characteristics, age, gender, tobacco use, alcohol use, hypertension and diabetes did not show any significant differences between the two groups (Table 1).
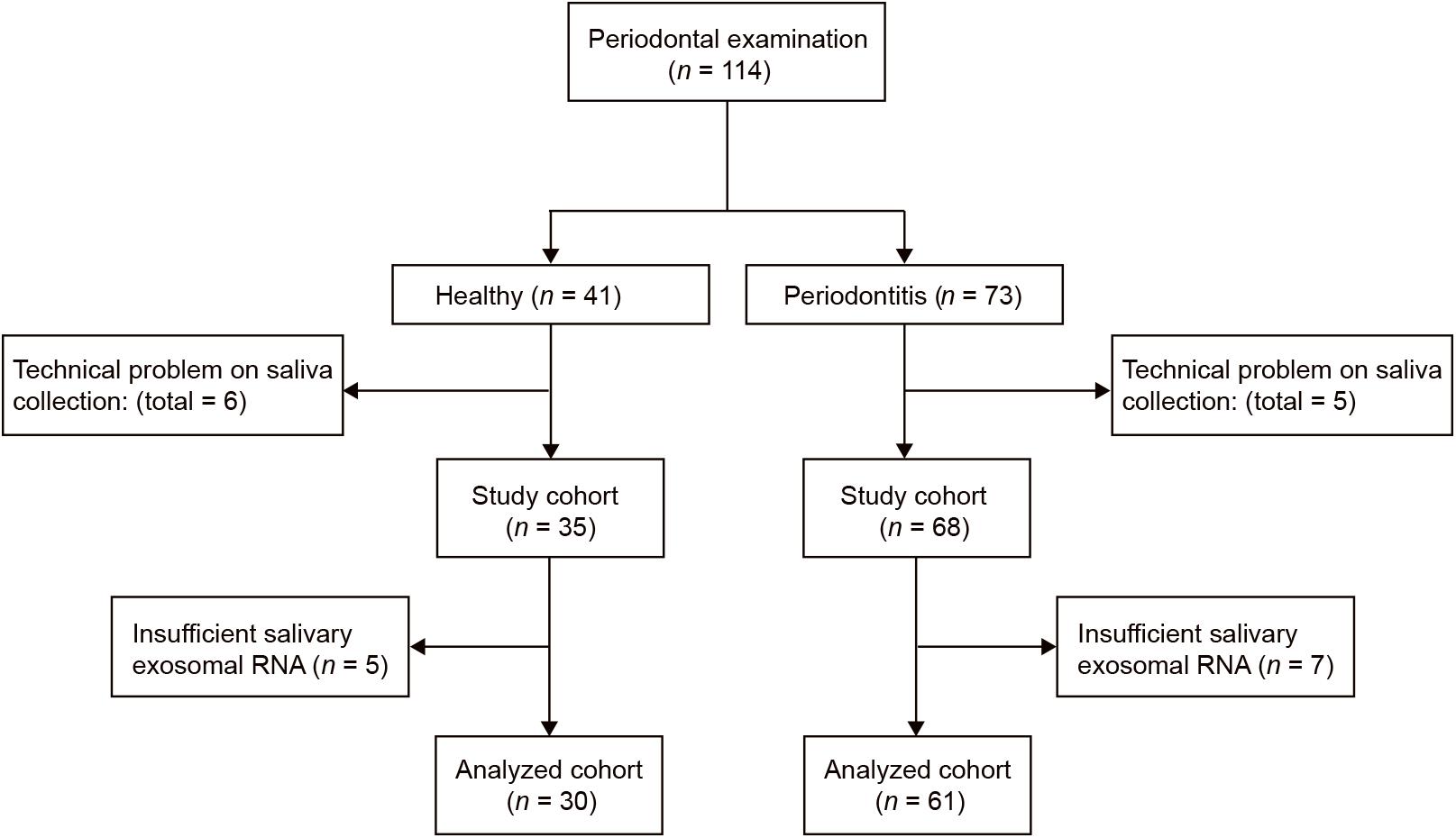
Figure 2. Flow diagram of periodontitis patients and control subjects. Participant numbers are accounted for by the flow diagram for the study.
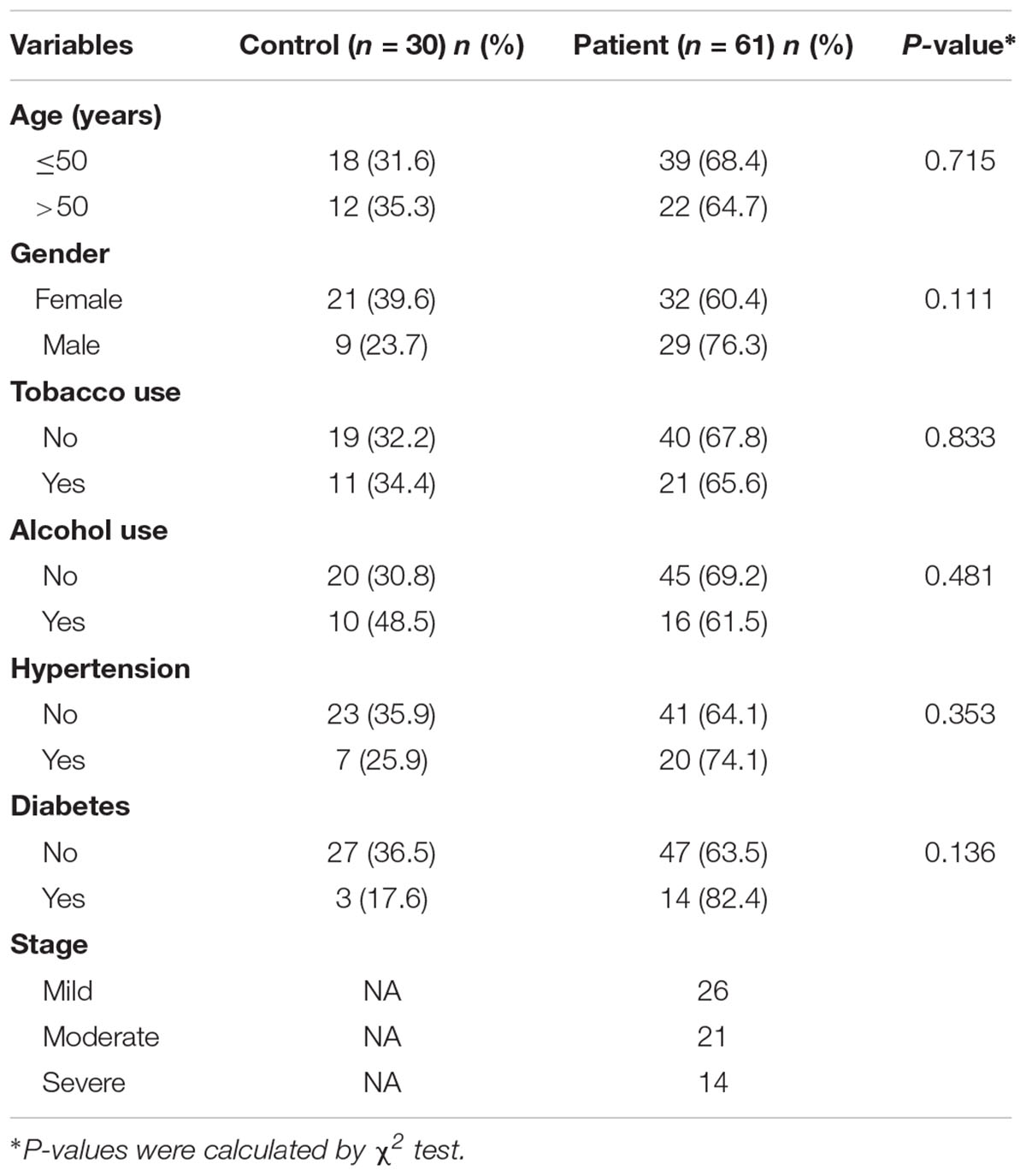
Table 1. The demographics and clinicopathological characteristics of the periodontitis patients and control subjects.
Extraction and Characterization of Exosomes From Human Saliva
Isolation of exosomes from human saliva was confirmed by transmission electron microscopy (TEM) (i.e., spherical membrane-bound particles with diameters between 30 and 100 nm) (Thery et al., 2009; Raposo and Stoorvogel, 2013; Figure 3A) and nanoparticle tracking analysis showing that human exosomes had an average diameter of 95 nm (Filipe et al., 2010; Webber and Clayton, 2013; Figure 3B). Immunoblotting of exosomal markers (ALIX, TSG101, CD63, CD9, CD81) and the intracellular protein that is not present in exosomes (Calnexin) (Mathivanan and Simpson, 2009; Taylor and Gercel-Taylor, 2011; Lotvall et al., 2014; Figure 3C) were performed for further confirmation. For eliminating the possibility of contamination by autophagosomes where significant amount of PD-L1 would be found, LC3 II, the marker of autophagosome was evaluated by immunoblotting; salivary exosomes samples did not contain LC3 while lysate of TE1 cells cultured in DMEM without serum for 6 h (positive control) showed the presence of LC3 II (Figure 3C). In addition, the marker of endoplasmic reticulum, calnexin, and the markers of inflammasomes were also analyzed by immunoblotting to exclude contamination of exosomes. The immunoblotting showed that calnexin, NLRP3 and NLRP4 could be detected in positive control but not in salivary exosomes samples (Figure 3C). Thus, exosomes were successfully isolated from saliva of periodontitis patients without significant contamination by autophagosomes, endoplasmic reticulum or inflammasomes.
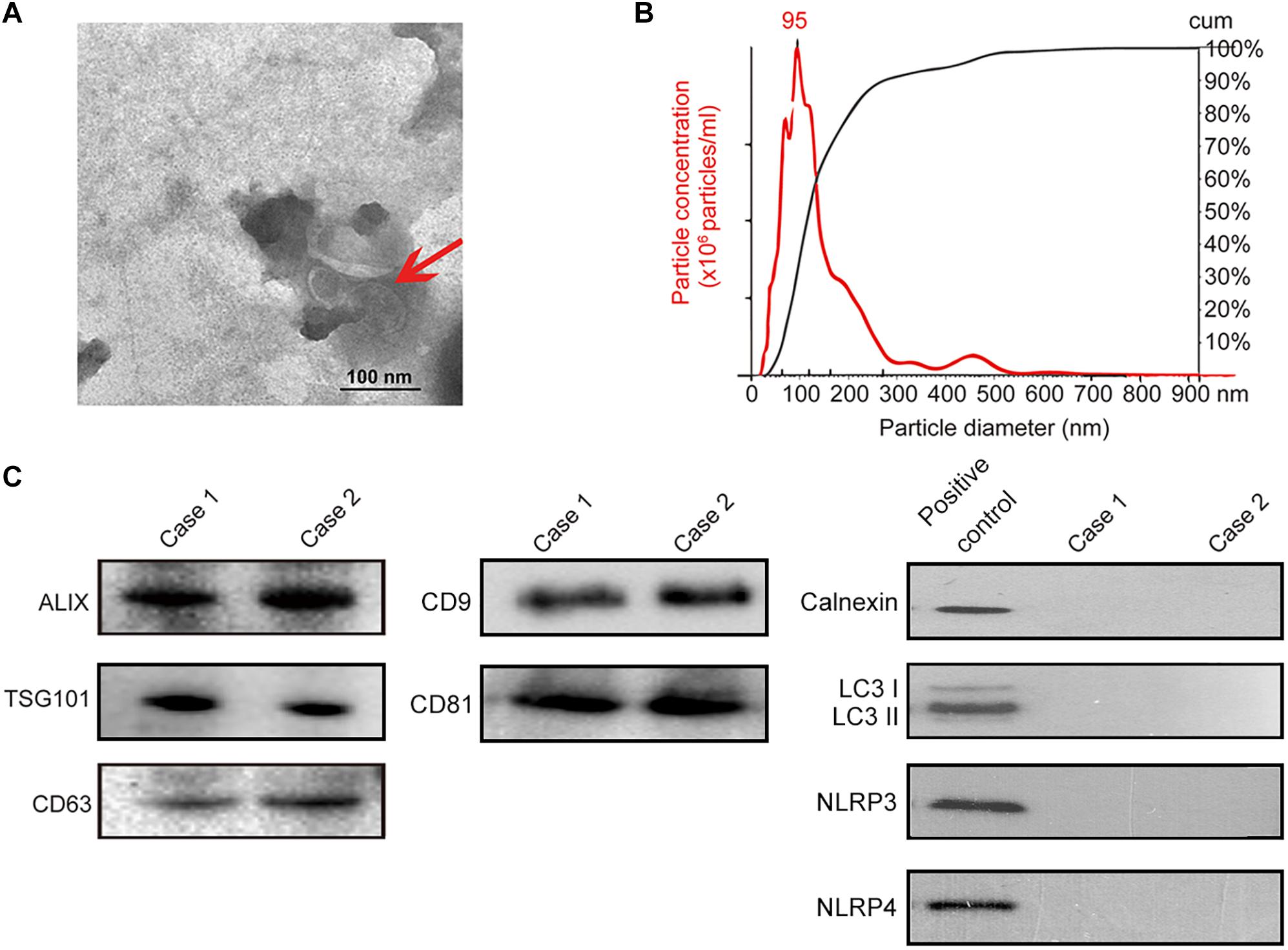
Figure 3. Identification of salivary exosomes in periodontitis patients and normal subjects. (A) Transmission electron microscopy of exosomes isolated from human saliva. Scale bar: 100 nm. (B) Exosomes concentration and size distribution by NanoSight analysis of human saliva. (C) Immunoblotting showed the exosomal membrane markers (ALIX, TSG101 CD63, CD9 and CD81), the intracellular protein Calnexin, the marker of autophagosome LC3 and markers of inflammasome (NLRP3 and NLRP4) in exosomes isolated from the saliva of one normal subject (case 01) and one periodontitis patient (case 02). Positive control for Calnexin was TE1 cells, and positive control for LC3 was TE1 cells after starvation for 6 h. Positive control for NLRP3 and NLRP4 was THP-1 cells.
Salivary Exosomal PD-L1 mRNA Was Elevated in Periodontitis Patients
We next determined the salivary exosomal and gingival PD-L1 mRNA expression, in 61 periodontitis patients and 30 control subjects, by qPCR analysis. As shown in Figure 4A, 45 of 61 cases (73.8%) had increased salivary exosomal PD-L1 expression compared with the mean salivary exosomal PD-L1 expression in control subjects. Mean salivary exosomal PD-L1 expression in the periodontitis patients was found to be about 10-fold higher when compared with the paired control subjects (P< 0.001). Similarly, mean gingival PD-L1 mRNA expression in the periodontitis patients was more than 10-fold higher than that in control subjects (Figure 4B, P< 0.001). More importantly, salivary exosomal PD-L1 mRNA levels highly correlated with gingival PD-L1 mRNA levels in periodontitis patients (r = 0.800 and P< 0.001, Pearson’s correlation test; Figure 4B). Collectively, our results strongly suggested that salivary exosomal PD-L1 mRNA could be a feasible biomarker of periodontitis.
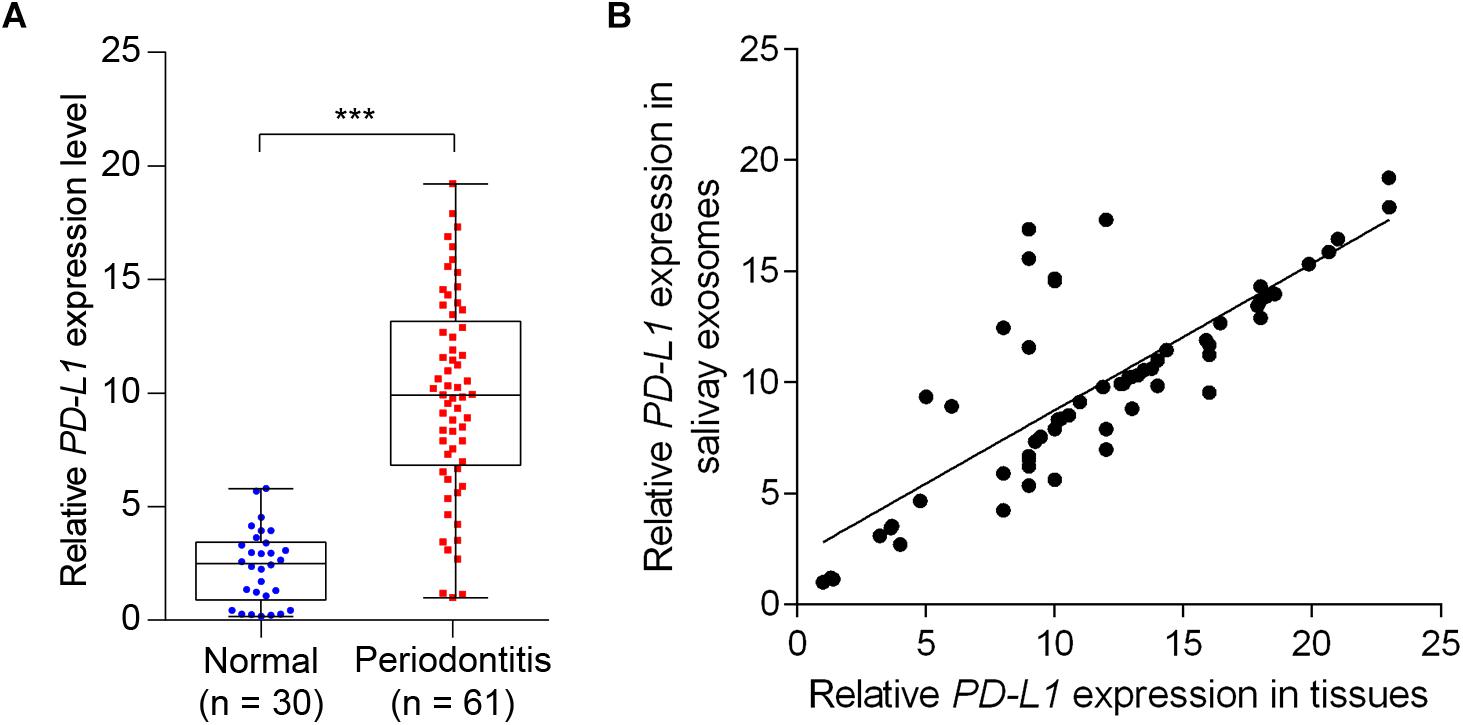
Figure 4. Detection of salivary exosomal PD-L1 in periodontitis patients and normal subjects. (A) Salivary exosomal PD-L1 was measured by RT-qPCR in periodontitis patients (n = 61) and normal controls (n = 30). Error bars indicate SEM. ∗∗∗P <0.001 by Student’s t-test. (B) Salivary exosomal PD-L1 correlated with periodontal tissue PD-L1 expression in periodontitis patients (Pearson’s correlation test).
Clinical Relevance of Salivary Exosomal PD-L1 mRNA in Periodontitis Patients
We further assessed the association between salivary exosomal PD-L1 mRNA and clinical parameters in 61 periodontitis patients. The median value (i.e., relative value of salivary exosomal PD-L1 mRNA = 9.90) was chosen to classify patients into high-(n = 31) and low-PD-L1 (n = 30) groups (Table 2). High PD-L1 expression was only associated with advanced stage (P = 0.005; χ2 test; Table 2), and PD-L1 expression was not found to be statistically significantly associated with other clinicopathological parameters (Table 2). Our results demonstrated that salivary exosomal PD-L1 mRNA could reflect the stage of periodontitis, suggesting that PD-L1 may be relevant to the disease progression.
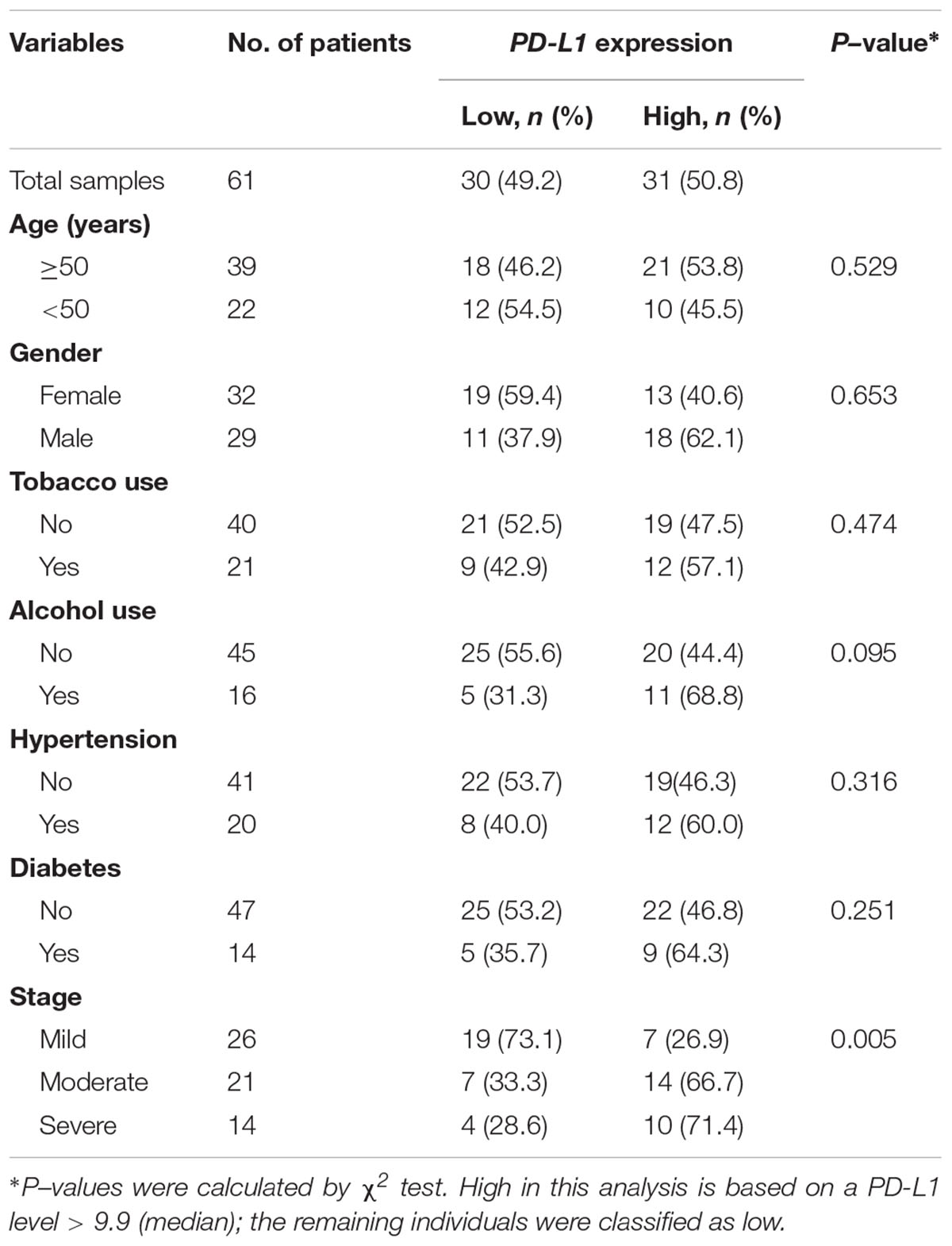
Table 2. The clinicopathological characteristics related to PD-L1 expression in periodontitis patients.
Discussion
We characterized PD-L1 mRNA expression in exosomes derived from saliva of periodontitis patients, and have evaluated the clinical relevance of the levels of salivary exosomes PD-L1 mRNA in the disease. One of the main findings was that the level of salivary exosomes PD-L1 mRNA in periodontitis patients is highly distinct relative to non-periodontitis controls. Moreover, high level of salivary exosomes PD-L1 was associated with an advanced stage of periodontitis, suggesting it can reflect disease progression. This is the first to establish a procedure of detection of saliva-based exosomal PD-L1 in disease, and the first salivary exosomal biomarker for periodontitis. In the majority of previous investigations of periodontitis biomarker, the specimens are either gingival tissues or gingival crevicular fluids (GCF) (Buduneli and Kinane, 2011; Ebersole et al., 2014; Jagannathan et al., 2014; Kebschull et al., 2014). However, sample collection and procedures for both gingival tissues and GCF are challenging: gingival tissue biopsy involves acquisition from invasive and limited tissue, and GCF sample collection involves sampling of a minute amount of fluid on filter paper strips, which requires a longer sampling time. Saliva-based assay overcomes these forgoing problems, as shown by our current study which involves an easy, non-invasive, and rapid collection of salivary specimens. We also demonstrate that mRNA can be extracted well from salivary exosomes, supporting the notion that exosomes-derived samples prevent mRNA from degradation (Kim et al., 2005; Soung et al., 2017). To our knowledge, this is the first to detect PD-L1 in exosomes and in saliva in periodontitis. The procedure described in the current study may be used for detecting mRNAs in oral diseases as well as in other systemic diseases.
PD-L1 has been reported to play an important role in a wide range of cancers and inflammation-originated diseases including periodontitis (Ota et al., 2015; Clark et al., 2016; Concha-Benavente et al., 2016; Zhang et al., 2016; Groeger et al., 2017). The role of PD-L1 to inhibit destruction of inflammatory tissues has been reported (Scandiuzzi et al., 2014). In two previous reports, it has been shown that PD-L1 expression in periodontitis tissues is increased in mild and moderate periodontitis (Yuan et al., 2015; Zhang et al., 2016). However, it is unclear whether PD-L1 expression correlates with disease status of chronic periodontitis. It has been also reported that expression of PD-1 and PD-L1 in peripheral CD4+ T lymphocytes and CD8+ T lymphocyte of chronic periodontitis patients was upregulated (Zhu et al., 2014). High expression of PD-L1 in host cells may contribute to the chronicity of inflammatory disorders (Zamani et al., 2016). The previous work showed that the PD-L1 expression in different compartments may be different and have different roles in inflammatory disease. Therefore, further correlation between PD-L1 and periodontitis are worthy to investigated. In our current study, it suggests that assay of exosome-based PD-L1 mRNA in saliva has potential to distinguish periodontitis from the healthy, and the levels correlate with the severity/stage of periodontitis. The increased expression of PD-L1 in severe periodontitis might due to the requirement of the body for more PD-L1 to inhibit destruction of inflammatory tissues. It has been shown that tumor cells upregulate PD-L1 expression to evade host immune response and thereby to maintain disease (Flies et al., 2016; Wang et al., 2016; Lakin et al., 2017). Moreover, the potential for PD-L1 to serve as a disease biomarker has also been revealed (Colwell, 2015; Fusi et al., 2015). PD-L1 is upregulated in cancers, such as glioblastoma (Wang et al., 2016), ovarian cancer (Abiko et al., 2013), lung cancer and oral carcinoma (Kim et al., 2005; Ota et al., 2015), and upregulated PD-L1 predicts worse survival. In periodontitis, PD-L1 has been detected in gingival tissues, gingival crevicular fluid (GCF) and blood (Olsen et al., 2016; Zhang et al., 2016; Groeger et al., 2017). Our analysis of published database validated that periodontitis specimens harbor high levels of PD-L1. Although detection of PD-L1 has been widely reported in tissues and blood (Fusi et al., 2015; Wang et al., 2016; Groeger et al., 2017), there are no publications for PD-L1 detection in salivary exosomes. We show that PD-L1 is present in exosomes of saliva from periodontitis patients, suggesting the possibility that PD-L1 is enriched in exosomes. Our results are consistent with a recent study that exosomes released from melanoma cells were found to carry a remarkable amount of PD-L1 on their surfaces (Chen et al., 2018). We here described a more convenient procedure for isolation and detection of PD-L1 in salivary exosomes. Since PD-L1 involves many diseases including cancers (i.e., oral cancers) and immune diseases (Gianchecchi et al., 2013; Dai et al., 2014; Roemer et al., 2016), the protocol established in current study may provide a platform for easy and rapid assay of PD-L1 in variety of applications. The limitation of the study was the relatively small size of the patient cohorts. Further investigations using larger and multiple cohorts are worthy to be performed to validate the findings of the current studies.
In summary, we described the procedure for isolating mRNA from exosomes derived from saliva of periodontitis patients. Our further assay showed that exosomal PD-L1 in saliva was enriched in periodontitis and was associated with advanced stage of disease. These findings are worthy to validate in further investigations with expanding samples.
Author Contributions
HZ conceived and designed the experiments. JY and DY provided patients and the study materials. YL, KL, XX, ZY, HD, and ZJ performed the in vitro experiments about patient specimen analysis, bioinformatics assay, and analyzed data. JY, YL, XX, S-CY, and HZ interpreted data and wrote the manuscript. JY, S-CY, DY, and HZ contributed to discussion and reviewed the manuscript.
Funding
This work was supported by the following funding agencies: National Natural Science Foundation of China (81572876, 81773087, 81071736, and 30973508) to HZ; Li Ka Shing Foundation Grant for Joint Research Program between Shantou University and Technion-Israel Institute of Technology (43209504) to HZ, Shantou University, Guangdong Province.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the dentists and patients who participated in these studies.
Footnotes
References
Abiko, K., Mandai, M., Hamanishi, J., Yoshioka, Y., Matsumura, N., Baba, T., et al. (2013). PD-L1 on tumor cells is induced in ascites and promotes peritoneal dissemination of ovarian cancer through CTL dysfunction. Clin. Cancer Res. 19, 1363–1374. doi: 10.1158/1078-0432.CCR-12-2199
Aziz, S., Ahmed, S. S., Ali, A., Khan, F. A., Zulfiqar, G., Iqbal, J., et al. (2015). Salivary immunosuppressive cytokines IL-10 and IL-13 are significantly elevated in oral squamous cell carcinoma patients. Cancer Invest. 33, 318–328. doi: 10.3109/07357907.2015.1041642
Baeza, M., Garrido, M., Hernandez-Rios, P., Dezerega, A., Garcia-Sesnich, J., Strauss, F., et al. (2016). Diagnostic accuracy for apical and chronic periodontitis biomarkers in gingival crevicular fluid: an exploratory study. J. Clin. Periodontol. 43, 34–45. doi: 10.1111/jcpe.12479
Baltacioglu, E., Yuva, P., Aydin, G., Alver, A., Kahraman, C., Karabulut, E., et al. (2014). Lipid peroxidation levels and total oxidant/antioxidant status in serum and saliva from patients with chronic and aggressive periodontitis. oxidative stress index: a new biomarker for periodontal disease? J. Periodontol. 85, 1432–1441. doi: 10.1902/jop.2014.130654
Buduneli, N., and Kinane, D. F. (2011). Host-derived diagnostic markers related to soft tissue destruction and bone degradation in periodontitis. J. Clin. Periodontol. 38(Suppl. 11), 85–105. doi: 10.1111/j.1600-051X.2010.01670.x
Byun, J. S., Hong, S. H., Choi, J. K., Jung, J. K., and Lee, H. J. (2015). Diagnostic profiling of salivary exosomal microRNAs in oral lichen planus patients. Oral Dis. 21, 987–993. doi: 10.1111/odi.12374
Chen, G., Huang, A. C., Zhang, W., Zhang, G., Wu, M., Xu, W., et al. (2018). Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560, 382–386. doi: 10.1038/s41586-018-0392-8
Clark, C. A., Gupta, H. B., Sareddy, G., Pandeswara, S., Lao, S., Yuan, B., et al. (2016). Tumor-Intrinsic PD-L1 signals regulate cell growth, pathogenesis, and autophagy in ovarian cancer and melanoma. Cancer Res. 76, 6964–6974. doi: 10.1158/0008-5472.CAN-16-0258
Colwell, J. (2015). Is PD-L1 expression a biomarker of response? Cancer Discov. 5:1232. doi: 10.1158/2159-8290.CD-ND2015-004
Concha-Benavente, F., Srivastava, R. M., Trivedi, S., Lei, Y., Chandran, U., Seethala, R. R., et al. (2016). Identification of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNgamma that induce PD-L1 expression in head and neck cancer. Cancer Res. 76, 1031–1043. doi: 10.1158/0008-5472.CAN-15-2001
Dai, S., Jia, R., Zhang, X., Fang, Q., and Huang, L. (2014). The PD-1/PD-Ls pathway and autoimmune diseases. Cell Immunol. 290, 72–79. doi: 10.1016/j.cellimm.2014.05.006
Dong, H., Ma, L., Gan, J., Lin, W., Chen, C., Yao, Z., et al. (2017). PTPRO represses ERBB2-driven breast oncogenesis by dephosphorylation and endosomal internalization of ERBB2. Oncogene 36, 410–422. doi: 10.1038/onc.2016.213
Ebersole, J. L., Kirakodu, S., Novak, M. J., Stromberg, A. J., Shen, S., Orraca, L., et al. (2014). Cytokine gene expression profiles during initiation, progression and resolution of periodontitis. J. Clin. Periodontol. 41, 853–861. doi: 10.1111/jcpe.12286
Filipe, V., Hawe, A., and Jiskoot, W. (2010). Critical evaluation of nanoparticle tracking analysis (NTA) by nanosight for the measurement of nanoparticles and protein aggregates. Pharm. Res. 27, 796–810. doi: 10.1007/s11095-010-0073-2
Flies, A. S., Lyons, A. B., Corcoran, L. M., Papenfuss, A. T., Murphy, J. M., Knowles, G. W., et al. (2016). PD-L1 Is not constitutively expressed on tasmanian devil facial tumor cells but is strongly upregulated in response to IFN-gamma and can be expressed in the tumor microenvironment. Front. Immunol. 7:581. doi: 10.3389/fimmu.2016.00581
Fusi, A., Festino, L., Botti, G., Masucci, G., Melero, I., Lorigan, P., et al. (2015). PD-L1 expression as a potential predictive biomarker. Lancet Oncol. 16, 1285–1287. doi: 10.1016/S1470-2045(15)00307-1
Gaddis, D. E., Maynard, C. L., Weaver, C. T., Michalek, S. M., and Katz, J. (2013). Role of TLR2-dependent IL-10 production in the inhibition of the initial IFN-gamma T cell response to Porphyromonas gingivalis. J. Leukoc. Biol. 93, 21–31. doi: 10.1189/jlb.0512220
Gan, J., Ke, X., Jiang, J., Dong, H., Yao, Z., Lin, Y., et al. (2016). Growth hormone-releasing hormone receptor antagonists inhibit human gastric cancer through downregulation of PAK1-STAT3/NF-kappaB signaling. Proc. Natl. Acad. Sci. U.S.A. 113, 14745–14750. doi: 10.1073/pnas.1618582114
Gianchecchi, E., Delfino, D. V., and Fierabracci, A. (2013). Recent insights into the role of the PD-1/PD-L1 pathway in immunological tolerance and autoimmunity. Autoimmun. Rev. 12, 1091–1100. doi: 10.1016/j.autrev.2013.05.003
Goncalves, A. S., Arantes, D. A., Bernardes, V. F., Jaeger, F., Silva, J. M., Silva, T. A., et al. (2015). Immunosuppressive mediators of oral squamous cell carcinoma in tumour samples and saliva. Hum. Immunol. 76, 52–58. doi: 10.1016/j.humimm.2014.11.002
Goncalves, A. S., Mosconi, C., Jaeger, F., Wastowski, I. J., Aguiar, M. C. F., Silva, T. A., et al. (2017). Overexpression of immunomodulatory mediators in oral precancerous lesions. Hum. Immunol. 78, 752–757. doi: 10.1016/j.humimm.2017.09.003
Groeger, S., Jarzina, F., Mamat, U., and Meyle, J. (2017). Induction of B7-H1 receptor by bacterial cells fractions of Porphyromonas gingivalis on human oral epithelial cells: B7-H1 induction by Porphyromonas gingivalis fractions. Immunobiology 222, 137–147. doi: 10.1016/j.imbio.2016.10.011
Han, D. H., Shin, H. S., Kim, M. S., Paek, D., and Kim, H. D. (2012). Group of serum inflammatory markers and periodontitis-metabolic syndrome coexistence in Koreans. J. Periodontol. 83, 612–620. doi: 10.1902/jop.2011.110304
Han, Y., Jia, L., Zheng, Y., and Li, W. (2018). Salivary exosomes: emerging roles in systemic disease. Int. J. Biol. Sci. 14, 633–643. doi: 10.7150/ijbs.25018
Jagannathan, R., Lavu, V., and Rao, S. R. (2014). Comparison of the proportion of non-classic (CD14+CD16+) monocytes/macrophages in peripheral blood and gingiva of healthy individuals and patients with chronic periodontitis. J. Periodontol. 85, 852–858. doi: 10.1902/jop.2013.120658
Kebschull, M., Demmer, R. T., Grun, B., Guarnieri, P., Pavlidis, P., and Papapanou, P. N. (2014). Gingival tissue transcriptomes identify distinct periodontitis phenotypes. J. Dent. Res. 93, 459–468. doi: 10.1177/0022034514527288
Kim, J., Shin, H., and Park, J. (2017). RNA in salivary extracellular vesicles as a possible tool for systemic disease diagnosis. J. Dent. Res. 96, 938–944. doi: 10.1177/0022034517702100
Kim, J. W., Wieckowski, E., Taylor, D. D., Reichert, T. E., Watkins, S., and Whiteside, T. L. (2005). Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin. Cancer Res. 11, 1010–1020.
Kimak, A., Strycharz-Dudziak, M., Bachanek, T., and Kimak, E. (2015). Lipids and lipoproteins and inflammatory markers in patients with chronic apical periodontitis. Lipids Health Dis. 14:162. doi: 10.1186/s12944-015-0156-5
Koregol, A. C., More, S. P., Nainegali, S., Kalburgi, N., and Verma, S. (2011). Analysis of inorganic ions in gingival crevicular fluid as indicators of periodontal disease activity: a clinico-biochemical study. Contemp. Clin. Dent. 2, 278–282. doi: 10.4103/0976-237X.91788
Lakin, N., Rulach, R., Nowicki, S., and Kurian, K. M. (2017). Current advances in checkpoint inhibitors: lessons from non-central nervous system cancers and potential for glioblastoma. Front. Oncol. 7:141. doi: 10.3389/fonc.2017.00141
Lau, C., Kim, Y., Chia, D., Spielmann, N., Eibl, G., Elashoff, D., et al. (2013). Role of pancreatic cancer-derived exosomes in salivary biomarker development. J. Biol. Chem. 288, 26888–26897. doi: 10.1074/jbc.M113.452458
Lotvall, J., Hill, A. F., Hochberg, F., Buzas, E. I., Di Vizio, D., Gardiner, C., et al. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the international society for extracellular vesicles. J. Extracell. Vesicles 3:26913. doi: 10.3402/jev.v3.26913
Machida, T., Tomofuji, T., Ekuni, D., Maruyama, T., Yoneda, T., Kawabata, Y., et al. (2015). MicroRNAs in salivary exosome as potential biomarkers of aging. Int. J. Mol. Sci. 16, 21294–21309. doi: 10.3390/ijms160921294
Mathivanan, S., and Simpson, R. J. (2009). ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000. doi: 10.1002/pmic.200900351
Michael, A., Bajracharya, S. D., Yuen, P. S., Zhou, H., Star, R. A., Illei, G. G., et al. (2010). Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 16, 34–38. doi: 10.1111/j.1601-0825.2009.01604.x
Olsen, I., Taubman, M. A., and Singhrao, S. K. (2016). Porphyromonas gingivalis suppresses adaptive immunity in periodontitis, atherosclerosis, and Alzheimer’s disease. J. Oral Microbiol. 8:33029. doi: 10.3402/jom.v8.33029
Ota, K., Azuma, K., Kawahara, A., Hattori, S., Iwama, E., Tanizaki, J., et al. (2015). Induction of PD-L1 expression by the EML4-ALK oncoprotein and downstream signaling pathways in non-small cell lung cancer. Clin. Cancer Res. 21, 4014–4021. doi: 10.1158/1078-0432.CCR-15-0016
Raposo, G., and Stoorvogel, W. (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383. doi: 10.1083/jcb.201211138
Recker, E. N., Brogden, K. A., Avila-Ortiz, G., Fischer, C. L., Pagan-Rivera, K., Dawson, D. V., et al. (2015). Novel biomarkers of periodontitis and/or obesity in saliva-An exploratory analysis. Arch. Oral. Biol. 60, 1503–1509. doi: 10.1016/j.archoralbio.2015.07.006
Roemer, M. G., Advani, R. H., Ligon, A. H., Natkunam, Y., Redd, R. A., Homer, H., et al. (2016). PD-L1 and PD-L2 genetic alterations define classical hodgkin lymphoma and predict outcome. J. Clin. Oncol. 34, 2690–2697. doi: 10.1200/JCO.2016.66.4482
Scandiuzzi, L., Ghosh, K., Hofmeyer, K. A., Abadi, Y. M., Lazar-Molnar, E., Lin, E. Y., et al. (2014). Tissue-expressed B7-H1 critically controls intestinal inflammation. Cell Rep. 6, 625–632. doi: 10.1016/j.celrep.2014.01.020
Sivadasan, P., Gupta, M. K., Sathe, G. J., Balakrishnan, L., Palit, P., Gowda, H., et al. (2015). Human salivary proteome–a resource of potential biomarkers for oral cancer. J. Proteomics 127(Pt A), 89–95. doi: 10.1016/j.jprot.2015.05.039
Slots, J. (2013). Periodontology: past, present, perspectives. Periodontol 2000 62, 7–19. doi: 10.1111/prd.12011
Soung, Y. H., Ford, S., Zhang, V., and Chung, J. (2017). Exosomes in cancer diagnostics. Cancers 9:8. doi: 10.3390/cancers9010008
Taylor, D. D., and Gercel-Taylor, C. (2011). Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin. Immunopathol. 33, 441–454. doi: 10.1007/s00281-010-0234-8
Thery, C., Ostrowski, M., and Segura, E. (2009). Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593. doi: 10.1038/nri2567
Tkach, M., and Thery, C. (2016). Communication by extracellular vesicles: where we are and where we need to go. Cell 164, 1226–1232. doi: 10.1016/j.cell.2016.01.043
Wang, Z., Zhang, C., Liu, X., Sun, L., Li, G., Liang, J., et al. (2016). Molecular and clinical characterization of PD-L1 expression at transcriptional level via 976 samples of brain glioma. Oncoimmunology 5:e1196310. doi: 10.1080/2162402X.2016.1196310
Webber, J., and Clayton, A. (2013). How pure are your vesicles? J. Extracell. Vesicles 2:19861. doi: 10.3402/jev.v2i0.19861
Yuan, W., Wang, X., Zhang, J., Zhou, W., Feng, Y., Chen, J., et al. (2015). [Programmed death ligand 1 negatively regulates inflammatory response of chronic periodontitis]. Hua Xi Kou Qiang Yi Xue Za Zhi 33,366–369.
Zamani, M. R., Aslani, S., Salmaninejad, A., Javan, M. R., and Rezaei, N. (2016). PD-1/PD-L and autoimmunity: a growing relationship. Cell Immunol. 310, 27–41. doi: 10.1016/j.cellimm.2016.09.009
Zhang, J., Wang, C. M., Zhang, P., Wang, X., Chen, J., Yang, J., et al. (2016). Expression of programmed death 1 ligand 1 on periodontal tissue cells as a possible protective feedback mechanism against periodontal tissue destruction. Mol. Med. Rep. 13, 2423–2430. doi: 10.3892/mmr.2016.4824
Zheng, X., Chen, F., Zhang, Q., Liu, Y., You, P., Sun, S., et al. (2017). Salivary exosomal PSMA7: a promising biomarker of inflammatory bowel disease. Protein Cell 8, 686–695. doi: 10.1007/s13238-017-0413-7
Keywords: immune checkpoint, exosomes, saliva, chronic periodontal disease, biomarker, disease stage
Citation: Yu J, Lin Y, Xiong X, Li K, Yao Z, Dong H, Jiang Z, Yu D, Yeung S-CJ and Zhang H (2019) Detection of Exosomal PD-L1 RNA in Saliva of Patients With Periodontitis. Front. Genet. 10:202. doi: 10.3389/fgene.2019.00202
Received: 17 September 2018; Accepted: 26 February 2019;
Published: 14 March 2019.
Edited by:
Cesar Wong, Hong Kong Polytechnic University, Hong KongReviewed by:
Fatah Kashanchi, George Mason University, United StatesHF Tsang, Hong Kong Adventist Hospital, Hong Kong
Copyright © 2019 Yu, Lin, Xiong, Li, Yao, Dong, Jiang, Yu, Yeung and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Zhang, aGFvbGFiY2FuY2VyY2VudGVyQDE2My5jb20=
†Shared co-first authorship
 Jialiang Yu1†
Jialiang Yu1† Sai-Ching Jim Yeung
Sai-Ching Jim Yeung Hao Zhang
Hao Zhang