- 1CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 2University of Chinese Academy of Sciences, Chinese Academy of Sciences, Beijing, China
- 3Department of Botany and Plant Sciences, University of California, Riverside, Riverside, CA, United States
- 4Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
The phylogeographical analysis and ecological niche modeling (ENM) of the widely distributed Northern Hemisphere Sibbaldia procumbens s.l. can help evaluate how tectonic motion and climate change helped shape the current distribution patterns of this species. Three chloroplast regions (the atpI-atpH and trnL-trnF intergenic spacers and the trnL intron) were obtained from 332 (156 from present study and 176 from the previous study) individuals of S. procumbens s.l. An unrooted haplotype network was constructed using the software NETWORK, while BEAST was used to estimate the divergence times among haplotypes. ENM was performed by MAXENT to explore the historical dynamic distribution of S. procumbens s.l. The haplotype distribution demonstrates significant phylogeographical structure (NST > GST; P < 0.01). The best partitioning of genetic diversity by SAMOVA produced three groups, while the time to the most recent common ancestor of all haplotypes was estimated to originate during the Miocene, with most of the haplotype diversity having occurred during the Quaternary. The MAXENT analysis showed S. procumbens s.l. had a wider distribution range during the last glacial maximum and a narrower distribution range during the last interglacial, with predictions into the future showing the distribution range of S. procumbens s.l. shrinking.
Introduction
Intercontinental disjunct distributions of flowering plants are believed to result from the opening and closing of land bridges (Beringia and the North Atlantic) between Eurasia and North America, with both vicariance and long-distance dispersal proposed to play important roles in shaping intercontinental disjunctions (Boufford and Spongberg, 1983; Wen and Shi, 1999; Manos and Donoghue, 2001; Ivanov et al., 2002; Riggins and Seigler, 2012). During the period of about 15–5.5 Ma, due to persistent cooling and aridification, the Beringia was hypothesized to act as the land connection between North America and Eurasia allowing floristic exchange (Riggins and Seigler, 2012; Krasnov et al., 2015). The Bering Strait disappeared during the late Miocene, and reemerged multiple times during Pleistocene glacial periods when sea levels dropped (Marincovich and Gladenkov, 1999; Gladenkov et al., 2002). In comparison, the North Atlantic land bridge is thought to have been the connection between eastern North America and northwestern Europe across Greenland during the late Eocene and Plio-Pleistocene (Qian and Ricklefs, 2000; Ian Milne, 2006).
Large-scale shifts of distribution range are believed to have taken place in response to the development of Quaternary continental ice-sheets, especially the cycles of glacial and interglacial during the Pleistocene (Ujiié and Ujiié, 1999; Zhang et al., 2007; Yang et al., 2008). Many mountain ranges have been strongly glaciated during the Quaternary ice age, the ice-free mountain top and the edge of ice sheet offering the only refugia for alpine plants (Briner et al., 2003; Provan and Bennett, 2008). The refugia were characterized with special, buffering environments, in which species can persist one or more glacial–interglacial cycles (Rowe et al., 2004; Provan and Bennett, 2008). Re-colonization in post-glacial regions may have occurred either through long-distance dispersal from unglaciated areas or from local refugia. Both ways have played important roles in shaping the distribution patterns of species. If dispersal was the predominant mechanism of re-colonization, the species would tolerate a wide range of environmental conditions (Martin et al., 2009). When species survived in local refugia, a certain relict component of this species should exist, mostly endemic and likely with a narrower tolerance range (Martin et al., 2009). High mountain ranges were especially significant for the long-term survival of plants because their height offered sufficient scope for altitudinal shifts when climate changed (Meng et al., 2007; Fu et al., 2016).
The Qinghai-Tibet Plateau (QTP) is the highest and largest plateau in the world, which is the result of collisions between the India and Europe–Asia continents (Dupont-Nivet et al., 2010; Herzschuh et al., 2010). The QTP and adjacent regions harbor hotspots of plant diversity (Myers et al., 2000; Zheng et al., 2002). Abundant precipitation and mountains in this area provide favorable conditions for the formation and differentiation of species (Meng et al., 2007; Ye et al., 2012; Favre et al., 2015). Many plants with intercontinental disjunct distributions have been suggested to originate in the QTP and adjacent regions [e.g., the genus Kelloggia (Nie et al., 2005), Pistacia (Xie et al., 2014), Rhodiola (Zhang et al., 2014), Achlys japonica–Achlys triphylla and Epimedium–Vancouveria of Berberidaceae (Nie et al., 2005; Wang et al., 2007; Zhang et al., 2007), and sect. Quinquefoliae (Hao et al., 2015)].
The genus Sibbaldia (Rosaceae) is widely distributed in Eurasia and North America, and the circumscription of this genus has long been controversial (Muravjova, 1936; Dixit and Panigrahi, 1981; Rajput et al., 1997; Eriksson et al., 2015; Feng et al., 2017). Initially, more than 40 species were included in the genus, but only 10–15 species were kept according to revisions based on morphological characters (Dixit and Panigrahi, 1981; Rajput et al., 1997; Li et al., 2003). More recently, molecular analysis revealed Sibbaldia in the traditional sense is polyphyletic, with the following circumscriptions occurring: S. adpressa and S. sericea were classified in Sibbaldianthe (Rosaceae); S. perpusilloides was included in Chamaecallis (Rosaceae); S. micropetala and S. phanerophlebia were included in Argentina (Rosaceae). S. tenuis, S. pentaphylla, S. tetrandra, S. purpurea, S. omeiensis, and S. sikkimensis were transferred to Potentilla (Rosaceae) (Eriksson et al., 2015; Feng et al., 2017). Finally, Sibbaldia sensu stricto is a small group centered on Sibbaldia procumbens s.l. including Sibbaldia cuneata, Sibbaldia parviflora, and Sibbaldia semiglabra (Rajput et al., 1997; Eriksson et al., 2015; Feng et al., 2017), which presents the intercontinental disjunct distribution between North America and Eurasia.
In the present study, we newly sequenced three chloroplast non-coding regions (atpI-atpH spacer, trnL intron, and trnL-trnF spacer) from 156 S. procumbens s.l. individuals primarily in Eastern Asia, in combination with sequences from populations in North America and Europe (Allen et al., 2015), with the goal (1) to examine the genetic structure of S. procumbens s.l. in the Northern Hemisphere and (2) to investigate the possible historical events that helped shape the current geographical and genetic distribution patterns of S. procumbens s.l.
Materials and Methods
Population Sampling, DNA Isolation, Amplification, and Sequencing
A total of 156 individuals representing 29 populations of S. procumbens s.l. were newly collected from China, Armenia, Austria, Russia, and Japan (Table 1), which covers nearly the entire geographical range of S. procumbens s.l. in Eurasia. Fresh leaves were collected from each individual and dried with silica gel. Total genomic DNA was extracted using the Plant Genomic DNA extraction kit (TIANGEN, Beijing, China).
According to previous studies (Dobes and Paule, 2010; Eriksson et al., 2015), Potentilla tetrandra (Bunge) Hook. f., P. purpurea (Royle) Hook. f., and Sibbaldianthe adpressa (Bunge) Juz were selected as outgroups and sequences were downloaded from NCBI (Table 1). The chloroplast atpI-atpH spacer, the trnL intron and trnL-trnF spacer were amplified from the 156 S. procumbens s.l. individuals using previously published primers. The primers from Shaw et al. (2007) were employed to amplify atpI-atpH; the primers (c and f) from Taberlet et al. (1991) were used to amplify trnL-trnF + trnL intron. The sequences (atpI-atpH spacer, the trnL intron and trnL-trnF spacer regions) of the 176 S. procumbens s.l. individuals from Allen et al. (2015) were also included in our analyses (Table 2).
Polymerase chain reactions (PCR) were performed in 50 μl volumes with 10 ng of DNA template, 1 μl of each primer and 44 μl 1.1× PCR Master mix of QINGKE kit multiplex PCR (Qingke, Beijing, China). The PCR protocol included (1) 2 min at 98°C, (2) 30 cycles of 94°C for 30 s, (3) 52°C/50°C for 30 s, (4) 72°C for 30 s, and (5) 72°C for 7 min. PCR products were checked for length and concentrations on 2% agarose gels, and successfully amplified products were sent to Beijing Genomics Institute (Wuhan, Hubei, China) for Sanger sequencing. All newly produced haplotype sequences have been deposited in GenBank (Table 1).
Phylogeographical Analyses
The sequences of each DNA region were aligned using Bioedit (Hall, 1999). The three DNA regions (atpI-atpH spacer, the trnL intron, and trnL-trnF spacer) were concatenated into a single combined sequence for each individual prior to analysis.
The number of haplotypes, haplotype diversity (Grismer et al., 2013), and nucleotide diversity (π) were evaluated with the software DNASP 5.10 (Librado and Rozas, 2009). The distribution of each haplotype was plotted on a map made by ArcGis 10.1 (ESRI, 2012). PERMUT 1.0 (Pons and Petit, 1996) was used to detect the average gene diversity within populations (hS), total gene diversity (Janssens et al., 2016), and the coefficients of differentiation (GST, a population differentiation estimate based solely on haplotype frequencies, and NST, a parameter that considers both haplotype frequencies and their genetic divergence) with 1,000 random permutations. When NST is significantly larger than GST, strong phylogeographic structure is indicated. SAMOVA1.0 (Dupanloup et al., 2002) was used to identify geographically and genetically distinguishable groups. The analysis of molecular variance (AMOVA) was conducted in Arlequin 3.5 (Excoffier and Lischer, 2010).
An unrooted haplotype network was constructed using NETWORK 4.6.11 (Bandelt et al., 1999). Partitioned Bayesian analysis was carried out with MrBayes 3.1.2 (Huelsenbeck and Ronquist, 2001), under the general time reversible substitution model. The best partition scheme and substitution model was determined by PartitionFinder 1.1.1 (Lanfear et al., 2012). We executed one cold and three incrementally heated Monte Carlo Markov chains on two simultaneous runs, sampling every 1,000th generation, with a burn-in of 25%. Indels were included as (0, 1) characters in a separate data partition with software SeqState (Müller, 2005). The divergence time of haplotypes was estimated using BEAST 1.7.5 (Drummond and Rambaut, 2007), following the descriptions by Allen et al. (2015).
Biogeographic Reconstruction
For biogeographic reconstruction, individuals of S. procumbens s.l. were assigned to five areas (labeled A–E) based on their present distribution: Eastern Asia, Central Asia, Western Eurasia, eastern North America, or western North America. Ancestral distributions of S. procumbens s.l. were inferred by BioGeoBEARS (Matzke, 2013, 2014) as implemented in RASP 4.0 (Yu et al., 2015) using 10,000 randomly sampled trees from the posterior distribution generated in the BEAST analysis, all other parameters were set to their defaults.
Ecological Niche Modeling
Ecological niche modeling was carried out in Maxent 3.3.3k (Phillips and Dudík, 2008) to reconstruct potential geographic distribution of S. procumbens s.l. in different historical periods. A total of 225 records (Supplementary Table S1) spread across the distribution range were used for the analysis. Nineteen bioclimatic variables with the 2.5 arc-min spatial resolution were downloaded from WorldClim1, and the variables with | Pearson’s R|≥ 0.7 were excluded; 10 variables (BIO1, annual mean temperature; BIO2, mean diurnal range; BIO5, max temperature of warmest month; BIO7, temperature annual range; BIO8, mean temperature of wettest quarter; BIO9, mean temperature of driest quarter; BIO12, annual precipitation; BIO15, precipitation seasonality; BIO17, precipitation of driest quarter; BIO18, precipitation of warmest quarter) were selected to simulate the past, current, and future suitable areas of S. procumbens s.l. Model validation was performed using Maxent default settings with 10 independent replicates of subsamples. The present distribution of S. procumbens s.l. was established using the bioclimatic variables for the current climate (1960–1990). Future climate scenario data for 2070 (2061–2080) were obtained from global climate models (GCMs), with future climate projections based on IPCC 5th assessment data, with calibration and statistically down-scaling using the data for the “current” climate. The community climate system model 4 (CCSM4) and NCAR-CCSM were chosen for the last glacial maximum (LGM, 0.021 Ma) and the last interglacial (LIG, 0.12–0.14 Ma), respectively.
Results
Sequence Variation and Haplotype Distribution
The total aligned sequence length including all outgroups was 1,712 bp (base pairs). While the aligned sequence of S. procumbens s.l. was 1,563 bp (662 bp for the atpI-atpH spacer, 901 bp for the combined trnL intron and trnL-trnF spacer), with 66 variable characters (including 17 single nucleotide polymorphisms and 14 indels) (Supplementary Table S2) and 45 characters parsimony-informative characters.
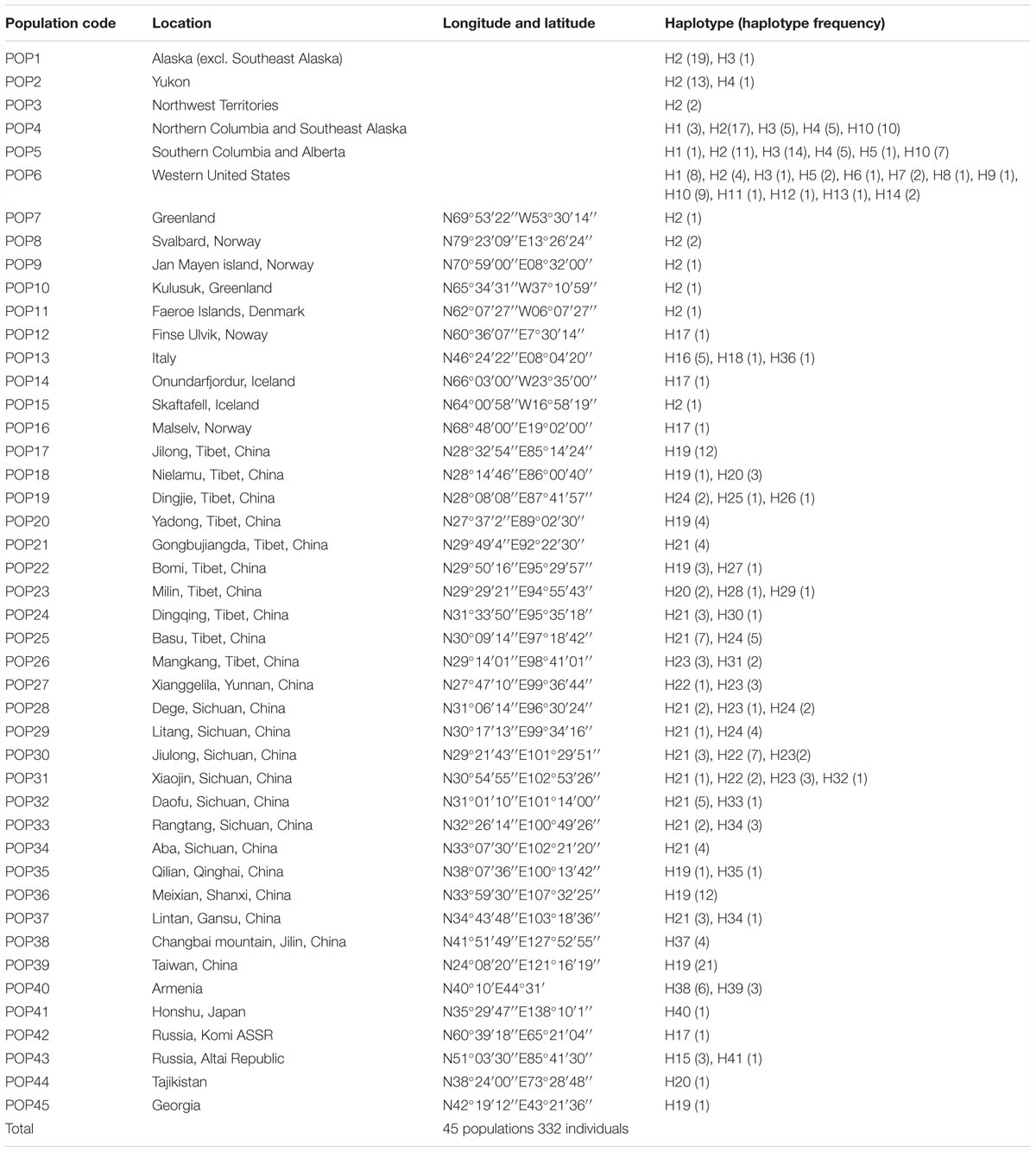
A total of 41 haplotypes were detected, including 19 (Haplotypes 23–41) newly identified haplotypes in this study and 22 previously identified haplotypes by Allen et al. (2015). Haplotypes 1–18 correspond to haplotype A–R in Allen et al. (2015); haplotypes 19–22 correspond to haplotype T, U, V and W in Allen et al. (2015). Among the 28 haplotypes in Eurasia, 16 were found only in a single population, two haplotypes were found in two populations, and five were found in more than two populations (Table 2 and Figure 1). The most frequent and widespread haplotype (H19) was found in 30% of individuals and 17.6% of populations, which was widely distributed in the QTP and adjacent regions of Taiwan and Georgia. Fourteen populations contained only one haplotype (Table 2 and Figure 1). Twelve populations contained two haplotypes, six populations contained three haplotypes, and one population contained four haplotypes (Table 2 and Figure 1).
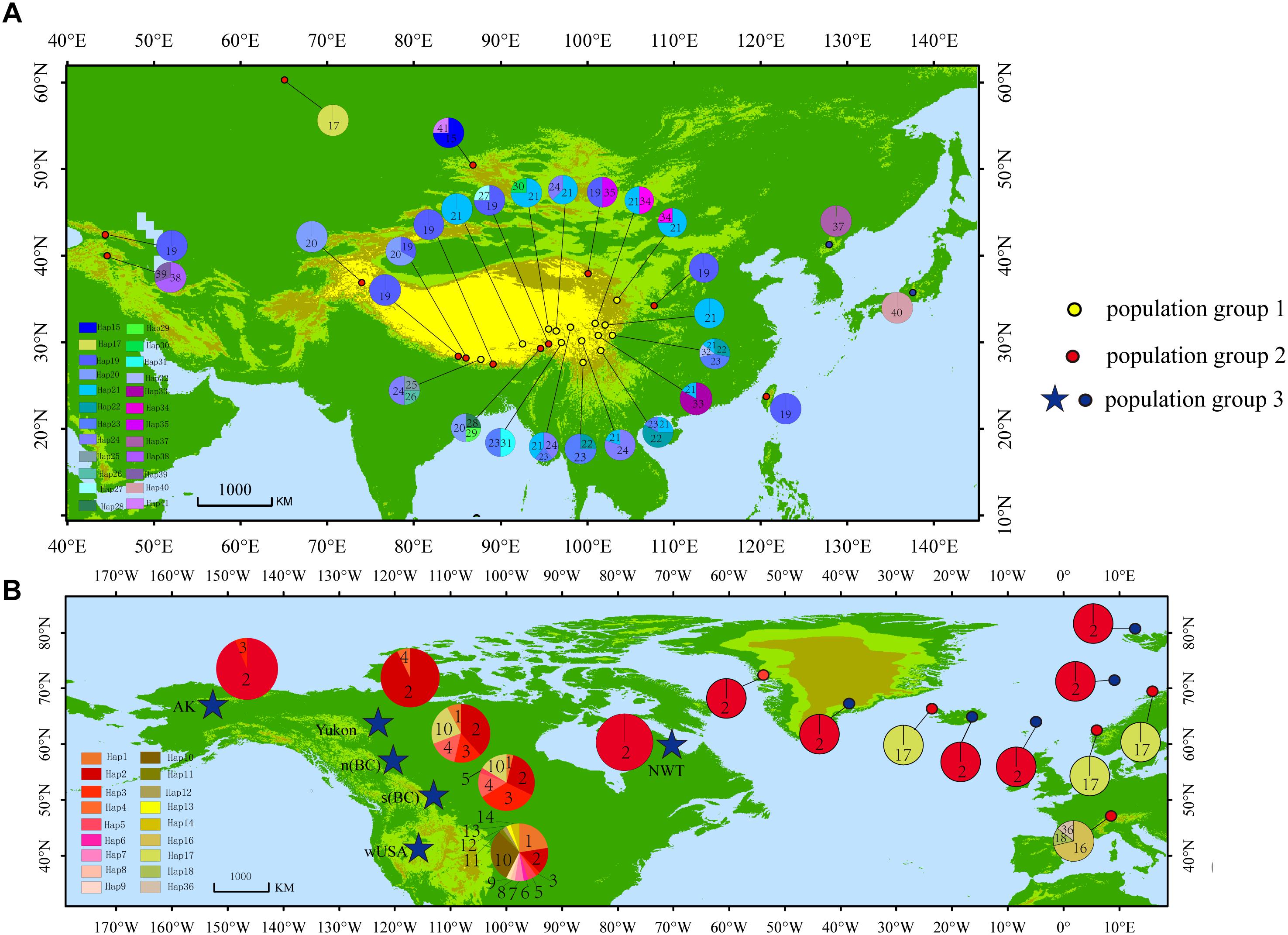
Figure 1. Distribution of haplotypes. (A) Haplotype distribution in Asia. (B) Haplotype distribution in North America and Europe [Modified according to Allen et al. (2015)].
Genetic Diversity and Structure
The haplotype diversity (Janssens et al., 2016) and the nucleotide diversity (π) was 0.8977 and 0.6 × 10-2, respectively, for all populations in the Northern Hemisphere. The haplotype diversity and nucleotide diversity for populations in Eurasia was 0.8394 and 0.46 × 10-2, respectively. For the populations in Eurasia, hS and hT were 0.426 and 0.871. The coefficients of differentiation measured over the 39 populations in Eurasia were GST = 0.515, NST = 0.826; and the Permutation test revealed that NST was significantly higher than GST (P < 0.01), which indicates phylogeographic structure for Eurasian populations. AMOVA results showed that a large proportion (92.97%) of the chloroplast variation was among populations (Table 3). The SAMOVA analysis identified three population groups (Figures 1, 2).
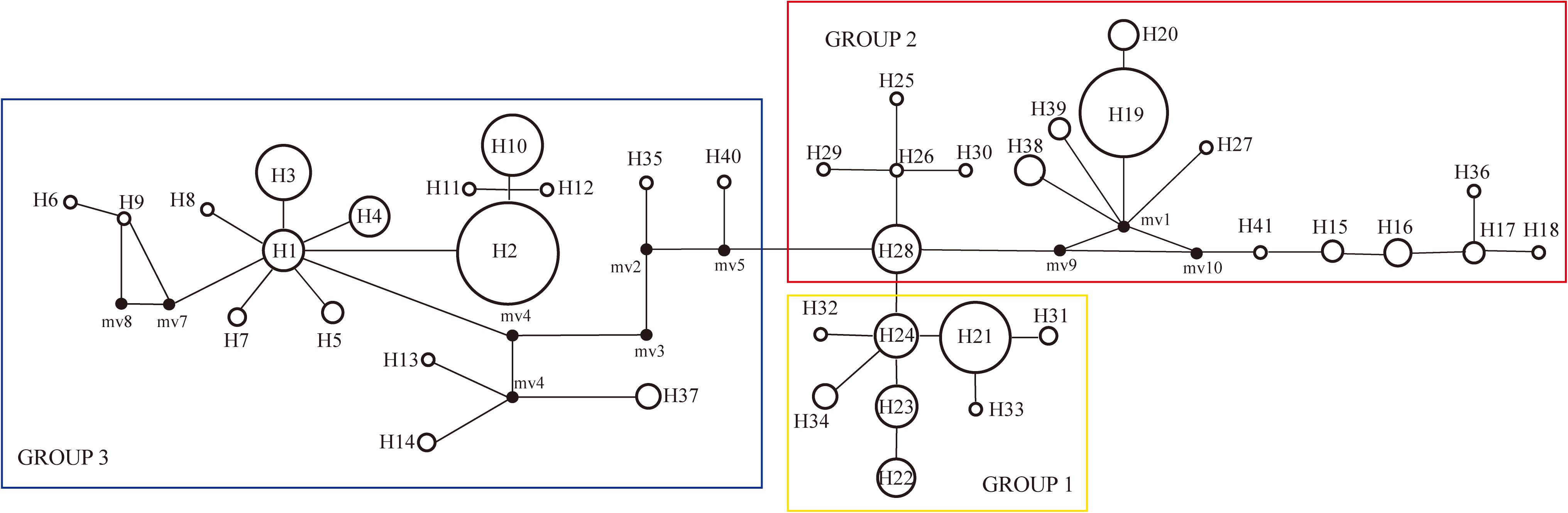
Figure 2. Unrooted haplotype network with SAMOVA defined groups, the size of the circles represents the frequency of haplotypes, with larger circles representing haplotypes found more frequently than smaller circles.
PartitionFinder determined one partition (trnL-trnF + trnL intron + atpI-atpH) as the best scheme. The phylogenetic analysis of all haplotypes revealed three clades (Figure 3). The early diverging clade (clade 1) includes eight haplotypes, H21–H24 and H31–H34, which are all endemic to the QTP and adjacent regions. Clade 2 consists of 11 haplotypes (H15–H20, H25–H30, H36–H41), which are endemic to Asia with a wider current distribution range than clade 1. The haplotypes in clade 3 form two small subclades; the first subclade consists of 15 haplotypes (H1–H14, H37), with H1 and H3–H14 distributed in North America, with H2 having a wide distribution around North America and extending east to Northwest Europe. H37 is distributed in the Changbai Mountains in the northeast of China; the second subclade includes seven haplotypes, which are mostly distributed in Europe (H15–H18, H36, H41), with only H35 distributed in the Qilian Mountains (north of the QTP) in China.
Figure 3. Phylogenetic relationships among haplotypes. Numbers below branches denote posterior probability. Number above nodes indicates the estimated time to the most recent common ancestor. The haplotypes with blue color branches represent the haplotypes from Allen et al. (2015).
The divergence of S. procumbens s.l. was estimated to be 5.75 Ma (95% HPD 4.09–7.49 Ma); the split between clade 2 and clade 3 was dated to be around 4.92 Ma (95% HPD 3.56–6.43 Ma); subsequent divergence within clades 2 and 3 was around 1.55 Ma (95% HPD 0.83–2.43 Ma) and 3.09 Ma (95% HPD 2.05–4.29 Ma), respectively.
RASP analysis resolved Eastern Asia as the most likely ancestral region of S. procumbens s.l. (Figure 4). Fifteen dispersal and four vicariance events were identified.
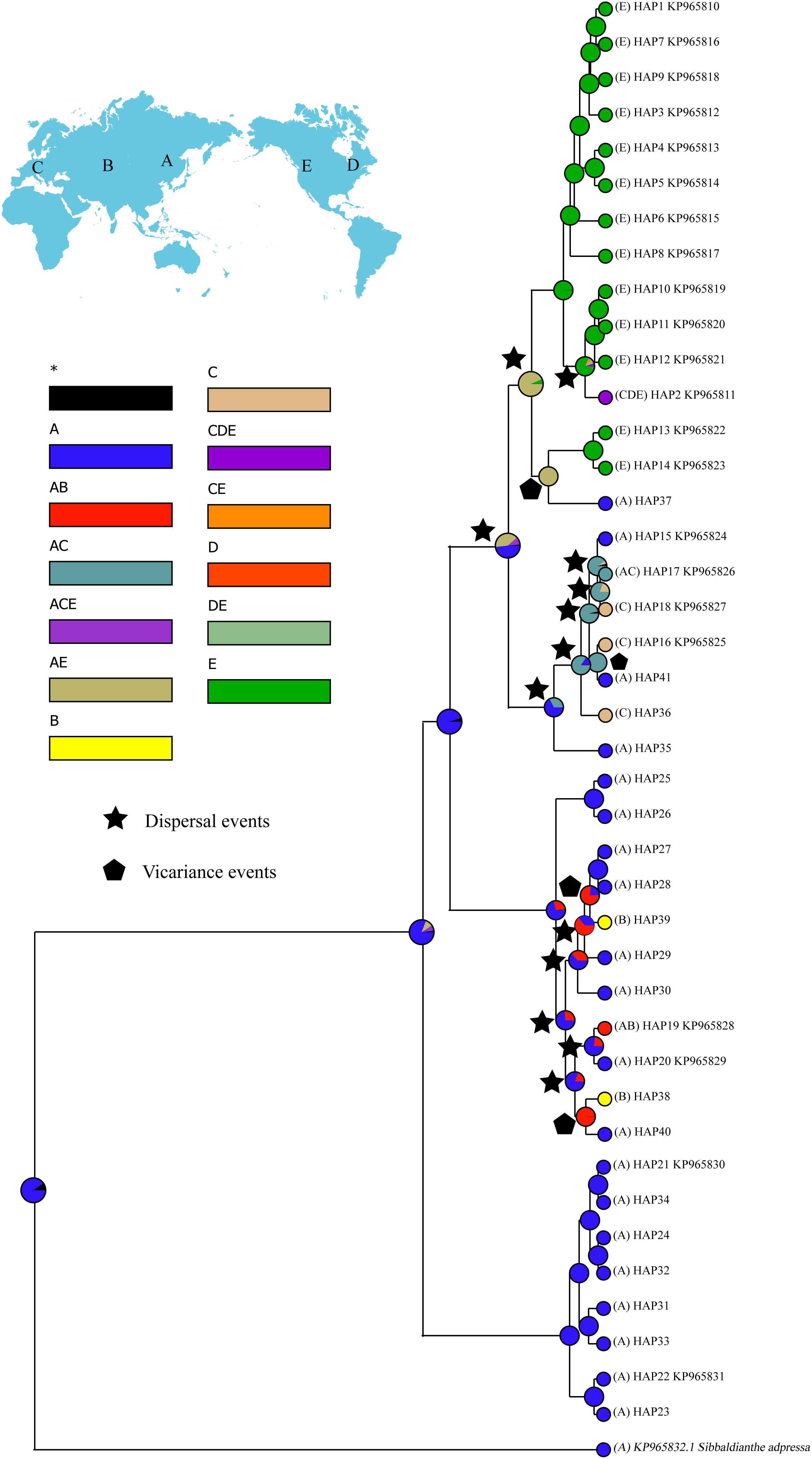
Figure 4. Ancestral distributions reconstructed by RASP. A, Eastern Asia; B, Central Asia; C, Western Eurasia; D, eastern North America; E, western North America.
Past, Present, and Future Ecological Niche Models
The AUC (area under curve) value for the potential distribution of S. procumbens s.l. was high (0.978 for future, 0.979 for LGM, and 0.95 for LIG), indicating good predictive model performance. The predicted distribution under current conditions was generally similar to its observed distribution across the Northern Hemisphere, with western North America and the QTP as the main distribution areas, and sparse distribution in Greenland, the Mediterranean, and Taiwan. At the LGM, the range of S. procumbens s.l. showed an expansion in distribution to the Caucasus Mountains and north of the Mediterranean (the Alps and Scandinavian Peninsula); the expansion was also detected in the Western Rocky Mountains and the Eastern Appalachian Mountains (Figure 5). At the LIG, the range of S. procumbens s.l. shrunk to the tallest mountain peaks in the QTP, western North America, and Scandinavia (Figure 5). The predicted distribution of S. procumbens s.l. in the future (2061–2080) is contracted, mainly concentrated in the QTP and adjacent regions (Figure 5).
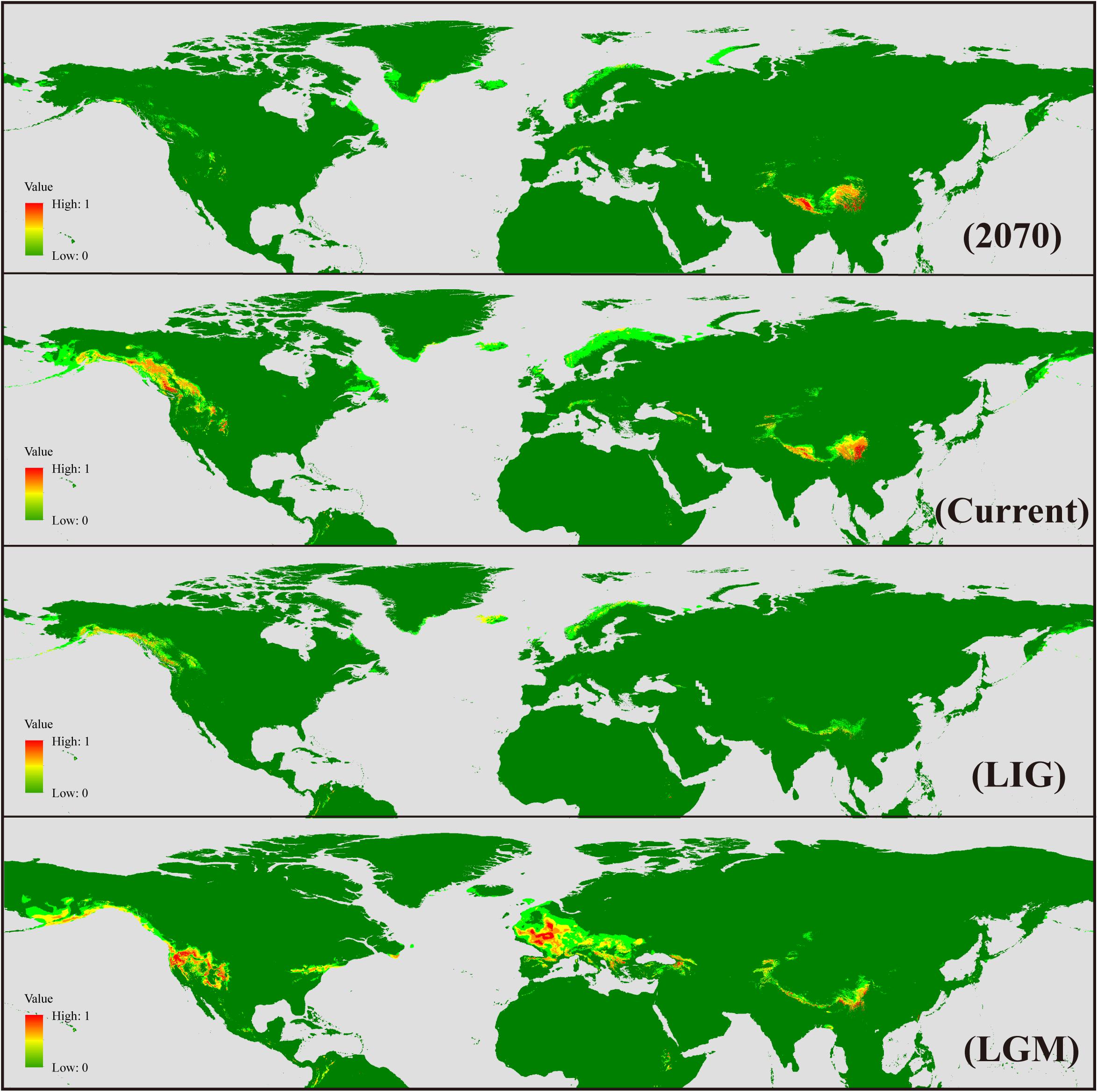
Figure 5. Geographical distribution models showing climatic suitability for Sibbaldia procumbens s.l. for 2070, present time, last interglacial (LIG), and last glacial maximum (LGM).
Discussion
Haplotype Divergence During Neogene
and Quaternary
Climate change and tectonic motion are thought as important factors driving species formation and differentiation (Bruch et al., 2006; Fortelius et al., 2006; Johnson et al., 2006). According to our estimation, the diversification of S. procumbens s.l. is likely to have begun around 5.75 Ma (95% HPD 4.09–7.49 Ma) in the late Miocene (Figure 3). A global second cooling happened 11.6–5.3 Ma (Molnar et al., 2010), which could have driven the initial divergence of S. procumbens s.l. In addition, the early diverging haplotypes (clade 1 in Figure 3) of S. procumbens s.l. are distributed in the QTP (Figure 1), and the fifth stage uplift of the Himalayas occurred about 7–5 Ma (Harrison et al., 1992; Yin, 2006), which may have been another factor driving the initial diversification within S. procumbens s.l.
The divergence between the Asia clade (clade 2) and the North America + Europe clade (clade 3) was estimated to be around the early Pliocene (Figure 3). This divergence likely resulted from the spread of S. procumbens s.l. from the QTP to other regions. A series of mountain ranges extending from the Himalayas are thought to have facilitated the spread and diversification of this species (Allen et al., 2015).
The migration of S. procumbens s.l. from Asia into North America via the Bering Strait was estimated to be around 2.36 Ma (95% HDP 1.55–3.29 Ma), which generally coincides with the estimation by Allen et al. (2015). The reduction of the sea level between Asia and Alaska during the late Pliocene may have facilitated the spread of many species (Vermeij, 1991; Berggren, 1995). Long-distance dispersal could also play an important role in the distribution expansion of S. procumbens s.l. (Givnish et al., 2004; Nathan, 2006; Mummenhoff and Franzke, 2007); since the seeds are small achenes (Coker, 1966; Li et al., 2003) which can be transmitted with wind or germinate after passage through the digestive tract of birds and other herbivores (Myers et al., 2004; Nathan, 2006; Mummenhoff and Franzke, 2007).
Quaternary glacial–interglacial cycles played important roles in the formation and differentiation of species (Nie et al., 2005; Meng et al., 2007; Yang et al., 2008; Wang et al., 2009). Periodic climatic fluctuations in the Quaternary were considered one of the main factors that formed geographical patterns of modern alpine plants (Yang et al., 2008; Wang et al., 2009; Zhang et al., 2014). In the current study, we found the greatest differentiation of S. procumbens s.l. occurred during the Quaternary (Figure 3), which indicate the glacial–interglacial cycles are likely factors contributing to the recent diversification within this species.
In general, our data suggest that it is likely the rapid landmass uplifts of the QTP and the global cooling during the Neogene that promoted the speciation and spread of S. procumbens s.l., while the Quaternary glacial–interglacial cycles have driven the recent differentiation of this species.
Spread of Sibbaldia procumbens s.l.
Even though the fossil evidence of S. procumbens s.l. is rare, our phylogenetic analysis (Figure 3) resolved an early diverging clade composed of eight haplotypes which were all distributed in the QTP and adjoining regions suggesting the origin of the species occurred in the QTP or adjoining regions. This corresponds with the results of RASP analysis (Figure 4), which indicate Eastern Asia as the most likely ancestral region of S. procumbens s.l. The QTP and adjoining regions is one of the world’s most important centers of biodiversity due to its high species richness and abundance of endemic species (Myers et al., 2000; Luo et al., 2016). This region has been suggested as an important origin and differentiation area for taxa with intercontinental disjunct distribution (Nie et al., 2005; Wang et al., 2007; Zhang et al., 2014).
Birkeland (2012) and Allen et al. (2015) proposed S. procumbens s.l. migrated both eastward and westward from Asia. The distribution patterns and phylogenetic analysis of haplotypes (Figures 1–3), and a series of dispersal events identified by the RASP analysis (Figure 4) support this proposal. More specifically, S. procumbens s.l. likely originated from the QTP and adjacent regions in the uplift phase during the late Miocene. The global cooling during this period provided a wider habitat, likely promoting the spread of this species, and the high mountain ranges extending from the QTP provided corridors for the spread outward: (1) to the west, S. procumbens s.l. dispersed along the Himalayas to the West Pamir Mountains (Tajikistan), across the northern Iranian Plateau to the Caucasus, continued to spread westward to the Balkan and Carpathian Mountains, then along the Alpine Mountains to the Scandinavian Mountains, finally arriving to lands and islands in the North Atlantic Ocean, and even to eastern North America and (2) to the east, the species likely first spread to the mountain ranges north of QTP (e.g., the Qilian Mountains and Kunlun Mountains), and moved further along the mountain range to the Tian Shan and Altai Mountains, then extended south and east to Siberia, finally reaching western North America via the Bering Strait. The North American clade was further divided into two subclades (Figure 3), the first is widespread and presents high haplotype diversity and the second consists of individuals in California and the Changbai Mountains. This supports the hypothesis of Allen et al. (2015) that S. procumbens s.l. shifted far southward after arriving to North America.
Additionally, S. procumbens s.l. could have also spread along the Qilian Mountains, Helan Mountains, Yin Mountains, and Yan Mountains to the Changbai Mountains in the northeast of China, and later arriving to Japan. A series of mountains (e.g., Qinling Mountains, Daba Mountains, Dabie Mountains, and Wuyi Mountains) could have been the route for the spread of S. procumbens s.l. from the QTP to Taiwan. Currently we cannot rule out the possibility that S. procumbens s.l. found in Taiwan are from Japan, or vice versa.
Refugia in Eastern Asia
Refugia are geographical areas of sheltered topography that provided suitable stable microclimates allowing species to persist throughout climatic oscillations (Rowe et al., 2004; Provan and Bennett, 2008). The refugia can be identified from fossils, paleo environmental data (Comes and Kadereit, 1998; Petit et al., 2002), and phylogeographic information (Stehlik, 2003; Tribsch and Schönswetter, 2003; Provan and Bennett, 2008; Eidesen et al., 2013). Multiple Pleistocene refugia for S. procumbens s.l. in North America and Europe have been reported (California and the Southern Rocky Mountains; Eidesen et al., 2013; Allen et al., 2015). Similarly, several refugia in Asia for S. procumbens s.l. during the LGM have been suggested in the current study.
As indicated by Allen et al. (2015) and Eidesen et al. (2007), genetic differences between populations in different regions suggest long separation through glacial intervals. In Asia, scattered distribution of S. procumbens s.l. are found in the Changbai Mountains in the northeast of China and in Japan. Both regions possess unique private haplotypes, implying both regions have been geographically separated for one or more glacial cycles, and could have served as refugia for S. procumbens s.l. during the LGM.
High haplotype diversity was discovered in multiple populations (Bomi, Dingqing, Mangkang, Xiaojin, and Daofu) in the eastern QTP (Table 1 and Figures 1, 4), implying micro-refugia may have existed along the mountains. We hypothesize S. procumbens s.l. originated in the eastern QTP and then colonized other mountain ranges surrounding the QTP. The mountains with high elevation provided the conditions necessary for differentiation of the species. There is only one haplotype (Haplotype 19) in the island of Taiwan; while haplotype 19 is also widespread across Asia. This implies Taiwan may have been recolonized by S. procumbens s.l. during interglacial periods, rather than serving as one refugium for S. procumbens s.l. during the LGM.
Conclusion
Our study implies current geographic and genetic distribution of S. procumbens s.l. is likely to have been shaped by changing climates in both the Neogene and Quaternary periods. Multiple regions in Eastern Asia may have served as important refugia for S. procumbens s.l. during extreme climatic events. Long-distance dispersal and vicariance may have played an important role in shaping extant distribution patterns of S. procumbens s.l. The distribution range of S. procumbens s.l. appeared to have experienced expansion and contraction during the LGM and LIG, respectively; in the future, when the global climate becomes warmer with rising carbon dioxide, the distribution of S. procumbens s.l. will shrink and be limited to the QTP and adjacent regions.
Data Availability
The datasets generated for this study can be found in National Center for Biotechnology Information, Please see Table 1 in the manuscript.
Author Contributions
Y-XS, H-CW, and HS conceived and designed this study. H-JZ, TF, and XZ performed the experiments and analyzed the data. H-JZ, JL, and Y-XS wrote the manuscript. TD, A-PM, and TF aided in filed collections. All authors read and approved the final manuscript.
Funding
This work was supported by the Key Projects of the Joint Fund of the National Natural Science Foundation of China (U1802232), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA20050203), the National Key R&D Program of China (2017YFC0505200), and grants-in-aid from the Major Program of the National Natural Science Foundation of China (31590823).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00201/full#supplementary-material
Footnotes
References
Allen, G. A., Marr, K. L., McCormick, L. J., and Hebda, R. J. (2015). Geographical origins, migration patterns and refugia of Sibbaldia procumbens, an arctic–alpine plant with a fragmented range. J. Biogeogr. 42, 1665–1676. doi: 10.1111/jbi.12543
Bandelt, H. J., Forster, P., and Rohl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Berggren, W. A. (1995). Geochronology, time scales and global stratigraphic correlation. SEPM Spec. Publ. 54:386. doi: 10.2110/pec.95.54
Birkeland, S. (2012). Rare to be Warm in Svalbard: an Ecological and Genetic Snapshot of Four Red Listed Plant Species. Master thesis, Centre for Ecological and Evolutionary Synthesis, University of Oslo, Oslo.
Boufford, D. E., and Spongberg, S. A. (1983). Eastern Asian-eastern North American phytogeographical relationships-a history from the time of Linnaeus to the twentieth century. Ann. Mo. Bot. Gard. 70, 423–439. doi: 10.2307/2992081
Briner, J., Miller, G., Davis, P. T., Bierman, P., and Caffee, M. (2003). Last Glacial Maximum ice sheet dynamics in Arctic Canada inferred from young erratics perched on ancient tors. Quat. Sci. Rev. 22, 437–444. doi: 10.1016/S0277-3791(03)00003-9
Bruch, A., Utescher, T., Mosbrugger, V., Gabrielyan, I., and Ivanov, D. (2006). Late Miocene climate in the circum-Alpine realm—a quantitative analysis of terrestrial palaeofloras. Palaeogeogr. Palaeoclimatol. Palaeoecol. 238, 270–280. doi: 10.1016/j.palaeo.2006.03.028
Comes, H. P., and Kadereit, J. W. (1998). The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci. 3, 432–438. doi: 10.1016/S1360-1385(98)01327-2
Dixit, B., and Panigrahi, G. (1981). Revision of the genus Sibbaldia L.(Rosaceae) in India. Proc. Plant Sci. 90, 253–272.
Dobes, C., and Paule, J. (2010). A comprehensive chloroplast DNA-based phylogeny of the genus Potentilla (Rosaceae): implications for its geographic origin, phylogeography and generic circumscription. Mol. Phylogenet. Evol. 56, 156–175. doi: 10.1016/j.ympev.2010.03.005
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. doi: 10.1186/1471-2148-7-214
Dupanloup, I., Schneider, S., and Excoffier, L. (2002). A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 11, 2571–2581. doi: 10.1046/j.1365-294X.2002.01650.x
Dupont-Nivet, G., Lippert, P. C., Van Hinsbergen, D. J., Meijers, M. J., and Kapp, P. (2010). Palaeolatitude and age of the Indo–Asia collision: palaeomagnetic constraints. Geophys. J. Int. 182, 1189–1198. doi: 10.1111/j.1365-246X.2010.04697.x
Eidesen, P., Alsos, I., Popp, M., Stensrud,Ø., Suda, J., and Brochmann, C. (2007). Nuclear vs. plastid data: complex Pleistocene history of a circumpolar key species. Mol. Ecol. 16, 3902–3925. doi: 10.1111/j.1365-294X.2007.03425.x
Eidesen, P. B., Ehrich, D., Bakkestuen, V., Alsos, I. G., Gilg, O., Taberlet, P., et al. (2013). Genetic roadmap of the Arctic: plant dispersal highways, traffic barriers and capitals of diversity. New Phytol. 200, 898–910. doi: 10.1111/nph.12412
Eriksson, T., Lundberg, M., Töpel, M., Östensson, P., and Smedmark, J. E. (2015). Sibbaldia: a molecular phylogenetic study of a remarkably polyphyletic genus in Rosaceae. Plant Syst. Evol. 301, 171–184. doi: 10.1007/s00606-014-1063-3
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Favre, A., Packert, M., Pauls, S. U., Jahnig, S. C., Uhl, D., Michalak, I., et al. (2015). The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. Camb. Philos. Soc. 90, 236–253. doi: 10.1111/brv.12107
Feng, T., Moore, M. J., Yan, M. H., Sun, Y. X., Zhang, H. J., Meng, A. P., et al. (2017). Phylogenetic study of the tribe Potentilleae (Rosaceae), with further insight into the disintegration of Sibbaldia. J. Syst. Evol. 55, 177–191. doi: 10.1111/jse.12243
Fortelius, M., Eronen, J., Liu, L., Pushkina, D., Tesakov, A., Vislobokova, I., et al. (2006). Late Miocene and Pliocene large land mammals and climatic changes in Eurasia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 238, 219–227. doi: 10.1016/j.palaeo.2006.03.042
Fu, P.-C., Gao, Q.-B., Zhang, F.-Q., Xing, R., Khan, G., Wang, J.-L., et al. (2016). Responses of plants to changes in Qinghai–Tibetan Plateau and glaciations: evidence from phylogeography of a Sibiraea (Rosaceae) complex. Biochem. Syst. Ecol. 65, 72–82. doi: 10.1016/j.bse.2016.01.006
Givnish, T. J., Millam, K. C., Evans, T. M., Hall, J. C., Chris Pires, J., Berry, P. E., et al. (2004). Ancient vicariance or recent long-distance dispersal? inferences about phylogeny and South American–African disjunctions in Rapateaceae and Bromeliaceae based on ndh F sequence data. Int. J. Plant Sci. 165, S35–S54. doi: 10.1086/421067
Gladenkov, A. Y., Oleinik, A. E., Marincovich, L., and Barinov, K. B. (2002). A refined age for the earliest opening of Bering Strait. Palaeogeogr. Palaeoclimatol. Palaeoecol. 183, 321–328. doi: 10.1016/S0031-0182(02)00249-3
Grismer, L. L., Wood, P. L. Jr., Anuar, S., Muin, M. A., Quah, E. S., McGuire, J. A., et al. (2013). Integrative taxonomy uncovers high levels of cryptic species diversity in Hemiphyllodactylus Bleeker, 1860 (Squamata: Gekkonidae) and the description of a new species from Peninsular Malaysia. Zool. J. Linn. Soc. 169, 849–880. doi: 10.1111/zoj.12064
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser, 41, 95–98.
Hao, Z.-Z., Liu, Y.-Y., Nazaire, M., Wei, X.-X., and Wang, X.-Q. (2015). Molecular phylogenetics and evolutionary history of sect. Quinquefoliae (Pinus): Implications for Northern Hemisphere biogeography. Mol. Phylogen. Evol. 87, 65–79. doi: 10.1016/j.ympev.2015.03.013
Harrison, T. M., Copeland, P., Kidd, W., and Yin, A. (1992). Raising tibet. Science 255, 1663–1670. doi: 10.1126/science.255.5052.1663
Herzschuh, U., Birks, H. J. B., Ni, J., Zhao, Y., Liu, H., Liu, X., et al. (2010). Holocene land-cover changes on the Tibetan Plateau. Holocene 20, 91–104. doi: 10.1177/0959683609348882
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Ian Milne, R. (2006). Northern Hemisphere plant disjunctions: a window on tertiary land bridges and climate change? Ann. Bot. 98, 465–472. doi: 10.1093/aob/mcl148
Ivanov, D., Ashraf, A. R., Mosbrugger, V., and Palamarev, E. (2002). Palynological evidence for Miocene climate change in the Forecarpathian Basin (Central Paratethys. NW Bulgaria). Palaeogeogr. Palaeoclimatol. Palaeoecol. 178, 19–37. doi: 10.1016/S0031-0182(01)00365-0
Janssens, S. B., Vandelook, F., De Langhe, E., Verstraete, B., Smets, E., Vandenhouwe, I., et al. (2016). Evolutionary dynamics and biogeography of Musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of Southeast Asia. New Phytol. 210, 1453–1465. doi: 10.1111/nph.13856
Johnson, W. E., Eizirik, E., Pecon-Slattery, J., Murphy, W. J., Antunes, A., Teeling, E., et al. (2006). The late Miocene radiation of modern Felidae: a genetic assessment. Science 311, 73–77. doi: 10.1126/science.1122277
Krasnov, B. R., Shenbrot, G. I., and Khokhlova, I. S. (2015). Historical biogeography of fleas: the former Bering Land Bridge and phylogenetic dissimilarity between the Nearctic and Palearctic assemblages. Parasitol. Res. 114, 1677–1686. doi: 10.1007/s00436-015-4349-7
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Li, C., Ikeda, H., and Ohba, H. (2003). Potentilla, Comarum, Sibbaldia, Chamaerhodos, Fragaria and Duchesnea. Flora of China. St. Louis: Missouri Botanical Garden, 291–338.
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Luo, D., Yue, J. P., Sun, W. G., Xu, B., Li, Z. M., Comes, H. P., et al. (2016). Evolutionary history of the subnival flora of the Himalaya-Hengduan Mountains: first insights from comparative phylogeography of four perennial herbs. J. Biogeogr. 43, 31–43. doi: 10.1111/jbi.12610
Manos, P. S., and Donoghue, M. J. (2001). Progress in Northern Hemisphere Phytogeography: an Introduction. Chicago, IL: The University of Chicago Press.
Marincovich, L. Jr., and Gladenkov, A. Y. (1999). Evidence for an early opening of the Bering Strait. Nature 397, 149–151. doi: 10.1038/16446
Martin, P., De Broyer, C., Fiers, F., Michel, G., Sablon, R., and Wouters, K. (2009). Biodiversity of Belgian groundwater fauna in relation to environmental conditions. Freshw. Biol. 54, 814–829. doi: 10.1111/j.1365-2427.2008.01993.x
Matzke, N. J. (2013). BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts. R Package, Version 0.2. Available at: http://CRAN.R-project.org/package=BioGeoBEARS.
Matzke, N. J. (2014). Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 63, 951–970. doi: 10.1093/sysbio/syu056
Meng, L., Yang, R., Abbott, R. J., Miehe, G., Hu, T., and Liu, J. (2007). Mitochondrial and chloroplast phylogeography of Picea crassifolia Kom. (Pinaceae) in the Qinghai-Tibetan Plateau and adjacent highlands. Mol. Ecol. 16, 4128–4137. doi: 10.1111/j.1365-294X.2007.03459.x
Molnar, P., Boos, W. R., and Battisti, D. S. (2010). Orographic controls on climate and paleoclimate of Asia: thermal and mechanical roles for the Tibetan plateau. Annu. Rev. Earth Plant Sci. 38, 77–102. doi: 10.1146/annurev-earth-040809-152456
Mummenhoff, K., and Franzke, A. (2007). Gone with the bird: late tertiary and quaternary intercontinental long-distance dispersal and allopolyploidization in plants. Syst. Biodivers. 5, 255–260. doi: 10.1017/S1477200007002393
Muravjova, D. (1936). The genus Sibbaldia L. and its species. Acta Inst. Bot. Acad. Sci. URSS Sér. 1, 217–241.
Myers, J. A., Vellend, M., Gardescu, S., and Marks, P. (2004). Seed dispersal by white-tailed deer: implications for long-distance dispersal, invasion, and migration of plants in eastern North America. Oecologia 139, 35–44. doi: 10.1007/s00442-003-1474-2
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Da Fonseca, G. A., and Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853–858. doi: 10.1038/35002501
Nathan, R. (2006). Long-distance dispersal of plants. Science 313, 786–788. doi: 10.1126/science.1124975
Nie, Z. L., Wen, J., Sun, H., and Bartholomew, B. (2005). Monophyly of Kelloggia Torrey ex Benth.(Rubiaceae) and evolution of its intercontinental disjunction between western North America and eastern Asia. Am. J. Bot. 92, 642–652. doi: 10.3732/ajb.92.4.642
Petit, R. J., Brewer, S., Bordács, S., Burg, K., Cheddadi, R., Coart, E., et al. (2002). Identification of refugia and post-glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. For. Ecol. Manage. 156, 49–74. doi: 10.1016/S0378-1127(01)00634-X
Phillips, S. J., and Dudík, M. (2008). Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31, 161–175. doi: 10.1111/j.0906-7590.2008.5203.x
Pons, O., and Petit, R. J. (1996). Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237–1245.
Provan, J., and Bennett, K. (2008). Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 23, 564–571. doi: 10.1016/j.tree.2008.06.010
Qian, H., and Ricklefs, R. E. (2000). Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 407, 180–182. doi: 10.1038/35025052
Rajput, M. T. M., Saleha Tahir, S., Hussain, S., and Spongberg, S. (1997). The genus Sibbaldia (Rosaceae). Pakistan. J. Bot. 29, 1–38.
Riggins, C. W., and Seigler, D. S. (2012). The genus Artemisia (Asteraceae: Anthemideae) at a continental crossroads: molecular insights into migrations, disjunctions, and reticulations among Old and New World species from a Beringian perspective. Mol. Phylogenet. Evol. 64, 471–490. doi: 10.1016/j.ympev.2012.05.003
Rowe, K. C., Heske, E. J., Brown, P. W., and Paige, K. N. (2004). Surviving the ice: Northern refugia and postglacial colonization. Proc. Natl. Acad. Sci. U.S.A. 101, 10355–10359. doi: 10.1073/pnas.0401338101
Shaw, J., Lickey, E. B., Schilling, E. E., and Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288. doi: 10.1073/pnas.0401338101
Stehlik, I. (2003). Resistance or emigration? Response of alpine plants to the ice ages. Taxon 52, 499–510. doi: 10.2307/3647448
Taberlet, P., Gielly, L., Pautou, G., and Bouvet, J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105–1109. doi: 10.1007/bf00037152
Tribsch, A., and Schönswetter, P. (2003). In search for Pleistocene refugia for mountain plants: patterns of endemism and comparative phylogeography confirm palaeo-environmental evidence in the Eastern European Alps. Taxon 52, 477–497. doi: 10.2307/3647447
Ujiié, H., and Ujiié, Y. (1999). Late Quaternary course changes of the Kuroshio Current in the Ryukyu Arc region, northwestern Pacific Ocean. Mar. Micropaleontol. 37, 23–40. doi: 10.1016/S0377-8398(99)00010-9
Vermeij, G. J. (1991). Anatomy of an invasion: the trans-Arctic interchange. Paleobiology 17, 281–307. doi: 10.1017/s0094837300010617
Wang, L., Abbott, R. J., Zheng, W., Chen, P., Wang, Y., and Liu, J. (2009). History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Mol. Ecol. 18, 709–721. doi: 10.1111/j.1365-294X.2008.04055.x
Wang, W., Chen, Z.-D., Liu, Y., Li, R.-Q., and Li, J.-H. (2007). Phylogenetic and biogeographic diversification of Berberidaceae in the northern hemisphere. Syst. Bot. 32, 731–742. doi: 10.1600/036364407783390791
Wen, J., and Shi, S. (1999). A phylogenetic and biogeographic study of Hamamelis (Hamamelidaceae), an eastern Asian and eastern North American disjunct genus. Biochem. Syst. Ecol. 27, 55–66. doi: 10.1016/s0305-1978(98)00067-2
Xie, L., Yang, Z.-Y., Wen, J., Li, D.-Z., and Yi, T.-S. (2014). Biogeographic history of Pistacia (Anacardiaceae), emphasizing the evolution of the Madrean-Tethyan and the eastern Asian-Tethyan disjunctions. Mol. Phylogen. Evol. 77, 136–146. doi: 10.1016/j.ympev.2014.04.006
Yang, F. S., Li, Y. F., Ding, X., and Wang, X. Q. (2008). Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan Plateau and its correlation with the Quaternary climate change. Mol. Ecol. 17, 5135–5145. doi: 10.1111/j.1365-294X.2008.03976.x
Ye, J., Chen, Z., Liu, B., Qin, H., and Yang, Y. (2012). Disjunct distribution of vascular plants between southwestern area and Taiwan area in China. Biodivers. Sci. 20, 482–494. doi: 10.3724/sp.j.1003.2012.13056
Yin, A. (2006). Cenozoic tectonic evolution of the Himalayan orogen as constrained by along-strike variation of structural geometry, exhumation history, and foreland sedimentation. Earth Sci. Rev. 76, 1–131. doi: 10.1016/j.earscirev.2005.05.004
Yu, Y., Harris, A. J., Blair, C., and He, X. (2015). RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Zhang, J.-Q., Meng, S.-Y., Allen, G. A., Wen, J., and Rao, G.-Y. (2014). Rapid radiation and dispersal out of the Qinghai-Tibetan Plateau of an alpine plant lineage Rhodiola (Crassulaceae). Mol. Phylogen. Evol. 77, 147–158. doi: 10.1016/j.ympev.2014.04.013
Zhang, M.-L., Uhink, C. H., and Kadereit, J. W. (2007). Phylogeny and biogeography of Epimedium/Vancouveria (Berberidaceae): Western North American-East Asian disjunctions, the origin of European mountain plant taxa, and East Asian species diversity. Syst. Bot. 32, 81–92. doi: 10.1600/036364407780360265
Keywords: Sibbaldia procumbens s.l., chloroplast regions, phylogeography, ecological niche modeling, Qinghai-Tibet Plateau
Citation: Zhang H-J, Feng T, Landis JB, Deng T, Zhang X, Meng A-P, Sun H, Wang H-C and Sun Y-X (2019) Molecular Phylogeography and Ecological Niche Modeling of Sibbaldia procumbens s.l. (Rosaceae). Front. Genet. 10:201. doi: 10.3389/fgene.2019.00201
Received: 23 October 2018; Accepted: 26 February 2019;
Published: 13 March 2019.
Edited by:
Renchao Zhou, Sun Yat-sen University, ChinaReviewed by:
Joana Isabel Robalo, Instituto Universitàrio de Ciências Psicològicas, Sociais e da Vida, PortugalSilvia Perea, Museo Nacional de Ciencias Naturales (MNCN), Spain
Copyright © 2019 Zhang, Feng, Landis, Deng, Zhang, Meng, Sun, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hang Sun, c3VuaGFuZ0BtYWlsLmtpYi5hYw== Heng-Chang Wang, aGN3YW5nQHdiZ2Nhcy5jbg== Yan-Xia Sun, c3VueWFueGlhQHdiZ2Nhcy5jbg==
†These authors have contributed equally to this work
 Hua-Jie Zhang
Hua-Jie Zhang Tao Feng
Tao Feng Jacob B. Landis
Jacob B. Landis Tao Deng
Tao Deng Xu Zhang
Xu Zhang Ai-Ping Meng
Ai-Ping Meng Hang Sun
Hang Sun Heng-Chang Wang
Heng-Chang Wang Yan-Xia Sun
Yan-Xia Sun