- 1Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 2Women's Reproductive Health Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 3Department of Medical Genetics, Faculty of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 4Research Laboratories, Bambino Gesù Children's Hospital-IRCCS, Rome, Italy
- 5Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
- 6Department of Medical Genetics, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
Background: Accumulating evidence suggests that functional dysregulations of miRNAs, especially miR-196a-2 and miR-149, in cancers could be attributed to polymorphisms in miRNA sequences. This study was aimed at clarifying the association of mir-196a-2 rs11614913 and mir-149 rs2292832 with cancer risk by performing an updated meta-analysis of genetic association studies.
Methods: PubMed, Embase, Scopus, and ScienceDirect databases were searched until 9 April 2018 to identify eligible studies. Studies should meet the following criteria to be included in the meta-analysis: evaluation of genetic association between rs11614913 and/or rs2292832 and susceptibility to cancer; A case-control design; Written in English; Availability of sufficient data for estimating odds ratio (OR) and its 95% confidence interval (95%CI). Studies that met the following criteria were excluded: review articles, meta-analysis, abstracts or conference papers; duplicate publications; studies on animals or cell-lines; studies without a case-control design; studies that did not report genotype frequencies. Pooled ORs and 95% CIs were estimated using a total of 111 studies (41,673 cases and 49,570 controls) for mir-196a rs11614913 and 44 studies (15,954 cases and 19,594 controls) for mir-149 rs2292832. Stratified analysis according to quality scores, genotyping method, ethnicity, broad cancer category and cancer type was also performed.
Results: Mir-196a-2 rs11614913 T allele was associated with decreased cancer risk in overall population. The association was only significant in Asians but not Caucasians. In subgroup analysis, significant associations were found in high quality studies, gynecological cancers, ovarian, breast, and hepatocellular cancer. Mir-149 rs2292832 was not associated with cancer risk in overall population and there were no differences between Asians and Caucasians. However, the T allele was associated with a decrease risk of gastrointestinal tract cancers under the heterozygote model and an increased risk of colorectal cancer under the recessive model.
Conclusions: The present meta-analysis suggests that mir-196a-2 rs11614913 may contribute to the risk of cancer especially in Asians. Mir-149 rs2292832 may modulate the risk of gastrointestinal tract cancers especially colorectal cancer. This study had some limitations such as significant heterogeneity in most contrasts, limited number of studies enrolling Africans or Caucasians ancestry and lack of adjustment for covariates and environmental interactions.
Introduction
Despite remarkable recent progress in clinical management, diagnosis and treatment, cancer has remained one of the major causes of death worldwide. According to the recent World Health Organization (WHO) report, about one in six deaths were caused by cancer in 2015. It is predicted that cancer-related death will increase up to 13.2 million by 2030 worldwide (Ferlay et al., 2010; Bray et al., 2012). Complex genetic and environmental risk factors and also interactions between these components contribute to the etiopathology of different cancers. Until recent years, much effort has been devoted to link the alteration of protein coding genes to tumorigenesis. However, latest evidence has demonstrated the emerging role of noncoding RNAs in cancer development and, especially, introduced microRNAs (miRNAs) as new players in pathobiology of cancers (Peng and Croce, 2016). MiRNAs are short noncoding functional RNAs that are involved in the regulation of transcriptome (Ha and Kim, 2014). They modulate important cellular processes both in normal physiology and disease state and are involved in almost all cellular processes altered during tumorigenesis (Osada and Takahashi, 2007; Li et al., 2009). Human mir-196a (MIR196A2, HGNC:31568) and mir-149 (MIR149, HGNC: 31536) are well-studied miRNAs that may function either as oncomiRs, by targeting tumor suppressor genes, or as tumor suppressors, by targeting oncogenes, in different conditions (Lu et al., 2016; He J. et al., 2018; Ow et al., 2018). It has been shown that single nucleotide polymorphism (SNP) in miRNA genes, such as hsa-mir-196a-2 rs11614913 and hsa-mir-149 rs2292832, may influence their functions through altering miRNA expression, maturation and/or efficiency of targeting and, thereby, contribute to the risk of cancer (Hu et al., 2008; Hoffman et al., 2009; Tu et al., 2012; Nariman-Saleh-Fam et al., 2016, 2017). Several association studies in a range of populations evaluated the contribution of mir-196a-2 rs11614913 and mir-149 rs2292832 to cancer risk; but results are inconclusive. Therefore, this study was aimed at clarifying the association of mir-196a-2 rs11614913 and mir-149 rs2292832 with cancer risk by performing an updated meta-analysis of genetic association studies.
Materials and Methods
Publication Search
To identify all potentially eligible publications, PubMed, Embase, Scopus and ScienceDirect databases were searched, with respect to specific search tips of each database, using following keywords. (“microRNA 196a2” OR “miRNA-196a2” OR “mir-196a2” OR “mir196a” OR “mir-196a-2” OR “pre-mir-196a” OR “pre-mir196a” OR “196a” OR “rs11614913”) OR (“microRNA 149” OR “miRNA-149” OR “mir-149” OR “mir149” OR “pre-mir-149" OR “pre-mir149” OR “rs2292832”) AND (“single nucleotide polymorphism” OR “SNP” OR “variant” OR “variation” OR “polymorphism” OR “mutation” OR “locus”) AND (“neoplasm” OR “cancer” OR “tumor” OR “carcinoma” OR “sarcoma” OR “lymphoma” OR “adenoma” OR “leukemia” OR “leucemia” OR “malignancy” OR “malignance” OR “malignant” OR “glioma”). Last search was performed on 9 April 2018. References of the relevant literature and review articles were also evaluated to identify all potentially eligible articles. This meta-analysis carried out in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Liberati et al., 2009).
Inclusion and Exclusion Criteria
Studies should meet the following criteria to be included in the meta-analysis: (1) evaluation of genetic association between rs11614913 and/or rs2292832 and susceptibility to cancer; (2) a case-control design; (3) Written in English; (4) Availability of sufficient data for estimating odds ratio (OR) and its 95% confidence interval (95%CI). Studies that met the following criteria were excluded: (1) review articles, meta-analysis, abstracts or conference papers; (2) duplicate publications; (3) studies on animals or cell-lines; (4) studies without a case-control design (5) studies that did not report genotype frequencies.
Data Extraction
Data was extracted from each eligible study and manually checked. Then, items were recorded for each eligible study: the first author, publication year, category of cancer, type of cancer, country, ethnicity, source of controls, genotyping method, number of subjects in the case and the control groups, genotype counts for each SNP in the case and the control groups. A broad cancer category was assigned for each study according to the following scheme: gastrointestinal tract cancers (GI, including gastric, esophageal, colorectal, bladder, pancreatic, or hepatocellular cancers), head and neck squamous cell carcinoma (HNC, including oral, non-oral, and nasopharyngeal cancers), gynecologic cancer (GyC, including endometrial, ovarian, and cervical cancers), hematological malignancies (HM, including leukemia and lymphomas), urogenital cancers (UG, including prostate, renal cell, and bladder cancers), or other cancers.
Quality Assessment
The quality of each study was assessed using quality assessment criteria for genetic association studies used elsewhere (Thakkinstian et al., 2011; Xue et al., 2015). This score is based on seven items including representativeness of cases, representativeness of controls, ascertainment of cancer case, control selection, genotyping examination, Hardy-Weinberg Equilibrium (HWE) status in controls, and total sample size. Quality scores ranged from 0 (lowest) to 15 (highest). Studies which were scored equal to or less than eight were regarded as low quality, while those with scores of greater than eight were regarded as high quality.
Statistical Analysis
The Meta package for R was used to perform meta-analysis (Schwarzer, 2007). Association of rs11614913 and rs2292832 with cancer was estimated by calculating pooled ORs and their 95% CIs assuming homozygote, heterozygote, dominant, recessive, and allelic models. Heterogeneity was assessed using the Chi-squared based Q test (Lau et al., 1997). In the presence of a significant heterogeneity (i.e., P-value of Q-test < 0.05 or I2 > 50%), the random effect (RE) model (DerSimonian and Laird, 1986) was used to calculate pooled ORs and 95% CIs. Otherwise, the fixed effect (FE) model was used (Mantel and Haenszel, 1959). Significance of the pooled OR was determined by the Z-test (P < 0.05 was considered significant). In cases of remarkable heterogeneity (i.e., I2 > 50%), the potential sources of heterogeneity across studies was explored using univariate meta-regression and stratified analysis. Moreover, subgroup analyses based on genotyping method, study quality, ethnicities, broad cancer categories, and cancer types were carried out. To assess consistency of results and influence of each study on the pooled OR, sensitivity analysis was done by omitting one study at a time and recalculating summary OR and 95% CI. Publication bias was evaluated by the Begg's rank correlation test of funnel plot asymmetry (Begg and Mazumdar, 1994) the “Trim and Fill” approach was used to correct for asymmetry in cases of significant rank correlation test (Duval and Tweedie, 2000a,b). All P-values were two-sided and P-value < 0.05 was considered statistically significant. All statistical analyses were performed in R (version 3.3.1).
Dealing With HWD (Departure From Hardy-Weinberg Equilibrium)
Departure from Hardy-Weinberg equilibrium (HWE) may be caused by a range of factors, among which genotyping error is more importantly relevant to the association study context. Currently there is no consensus on the way of handling association studies with the controls not in HWE, but it has been recommended that such studies should not be excluded from meta-analysis (Minelli et al., 2008). However, sensitivity analysis should be performed to evaluate the possible effects of such studies on the pooled estimates (Attia et al., 2003; Thakkinstian et al., 2005; Zintzaras and Lau, 2008; Wang X. B. et al., 2014). In the present meta-analysis, the following approach with regards to HWE-deviated studies was followed. Departure of genotype distributions from HWE (i.e., HWD) in the control group of each study was evaluated using the Chi-squared or the exact goodness of fit test. Meta-analyses, including the overall and subgroup analyses, were performed considering all eligible studies including HWD studies. However, to evaluate possible impacts of HWE-deviated studies, HWD sensitivity analysis was performed by evaluating the influence of excluding these studies on point estimates and identifying the influenced genotype contrasts. In cases that excluding HWD studies altered the result of meta-analysis, ORs of such studies were adjusted for HWE deviation by means of incorporating the HWE-expected genotype counts in the control group as recommended (Trikalinos et al., 2006; Zintzaras et al., 2006; Zintzaras, 2008; Zintzaras and Lau, 2008; Srivastava and Srivastava, 2012) and the HWD-adjusted pooled ORs were calculated in genotype contrasts.
Results
Study Characteristics
The process of selecting eligible studies is depicted in Figure 1. A total of 1,645 articles were found from different sources outlined in materials and methods and screened by reading titles and abstracts. A total of 1,509 articles were excluded in which 577 articles were duplicates, 114 articles were abstracts or conference meetings, 86 articles were meta-analysis, 404 were review articles, 7 articles were not written in English, 26 articles were related to other diseases, 36 articles were related to other genes, or polymorphisms and 259 more articles had either obvious irrelevant study design or irrelevant disease/gene. The full text of the remaining 136 articles were evaluated and 9 more articles were also excluded as they did not have either sufficient data to calculate ORs and 95%CIs (n: 4) or an association study design (n: 5). Finally, a total of 127 eligible articles remained (Horikawa et al., 2008; Yang et al., 2008, 2016; Hoffman et al., 2009; Hu et al., 2009, 2013; Tian et al., 2009; Catucci et al., 2010; Christensen et al., 2010; Dou et al., 2010; Kim et al., 2010, 2012; Kontorovich et al., 2010; Li et al., 2010, 2014; Liu et al., 2010, 2013, 2014; Okubo et al., 2010; Peng et al., 2010; Qi et al., 2010, 2014, 2015; Srivastava et al., 2010, 2017; Wang et al., 2010, 2013, 2016; Akkiz et al., 2011; George et al., 2011; Hong et al., 2011; Jedlinski et al., 2011; Mittal et al., 2011; Vinci et al., 2011, 2013; Zhan et al., 2011; Zhou et al., 2011, 2014; Alshatwi et al., 2012; Chen et al., 2012; Chu et al., 2012, 2014; Hezova et al., 2012; Linhares et al., 2012; Min et al., 2012; Tu et al., 2012; Zhang M. et al., 2012; Zhang M. W. et al., 2012; Zhu et al., 2012; Ahn et al., 2013; Han et al., 2013; Huang et al., 2013, 2017; Lv et al., 2013; Ma et al., 2013; Pavlakis et al., 2013; Umar et al., 2013; Wei et al., 2013, 2014; Zhang et al., 2013; Bansal et al., 2014; Dikeakos et al., 2014; Du et al., 2014; Hao et al., 2014; Kou et al., 2014; Kupcinskas et al., 2014a,b; Omrani et al., 2014; Parlayan et al., 2014; Pu et al., 2014; Qu et al., 2014; Roy et al., 2014; Tong et al., 2014; Wang N. et al., 2014; Wang R. et al., 2014; Wang X. H. et al., 2014; Deng et al., 2015; Dikaiakos et al., 2015; Dong et al., 2015; He et al., 2015; Li T. et al., 2015; Liu, 2015; Li X. et al., 2015; Martin-Guerrero et al., 2015; Nikolić et al., 2015; Pratedrat et al., 2015; Sodhi et al., 2015; Sushma et al., 2015; Yan et al., 2015; Yin et al., 2015, 2016, 2017; Dai et al., 2016; Gu and Tu, 2016; Hashemi et al., 2016; Jiang et al., 2016; Li H. et al., 2016; Li J. et al., 2016; Li M. et al., 2016; Miao et al., 2016; Morales et al., 2016; Ni and Huang, 2016; Peckham-Gregory et al., 2016; Shen et al., 2016; Song et al., 2016; Su et al., 2016; Sun et al., 2016; Toraih et al., 2016a,b; Zhang L. H. et al., 2016; Zhao et al., 2016; Afsharzadeh et al., 2017; Bodal et al., 2017; Cîmpeanu et al., 2017; Poltronieri-Oliveira et al., 2017; Rakmanee et al., 2017; Rogoveanu et al., 2017; Tandon et al., 2017; Zhang E. et al., 2017; Abdel-Hamid et al., 2018; Damodaran et al., 2018; Doulah et al., 2018; He J. et al., 2018; He Y. et al., 2018; Mashayekhi et al., 2018; Minh et al., 2018; Ranjbar et al., 2018). Characteristics of the included studies are tabulated in Tables 1, 2.
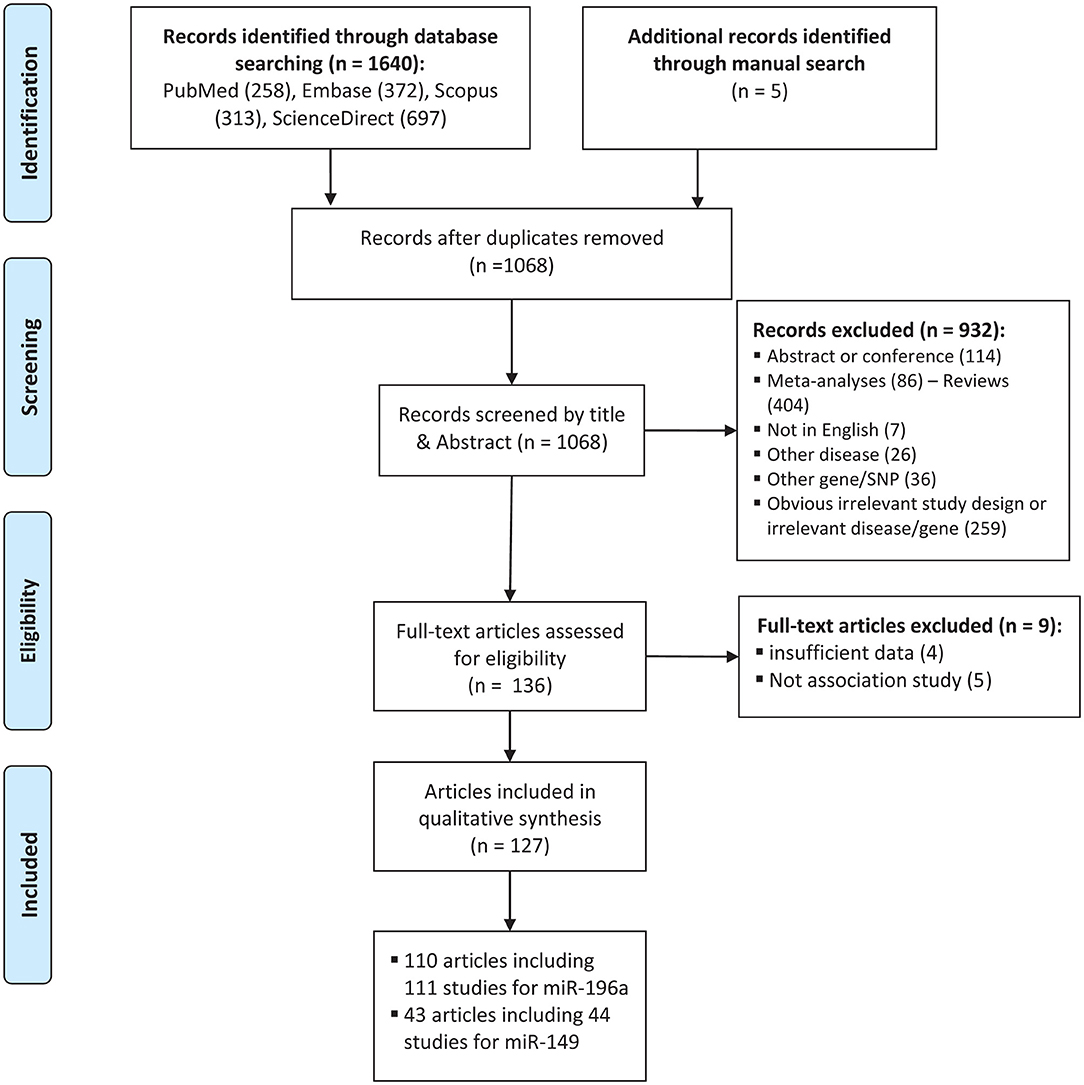
Figure 1. Flowchart of the identification of eligible studies for meta-analysis of cancer risk associated with mir-196a-2 rs11614913 and mir-149 rs2292832.
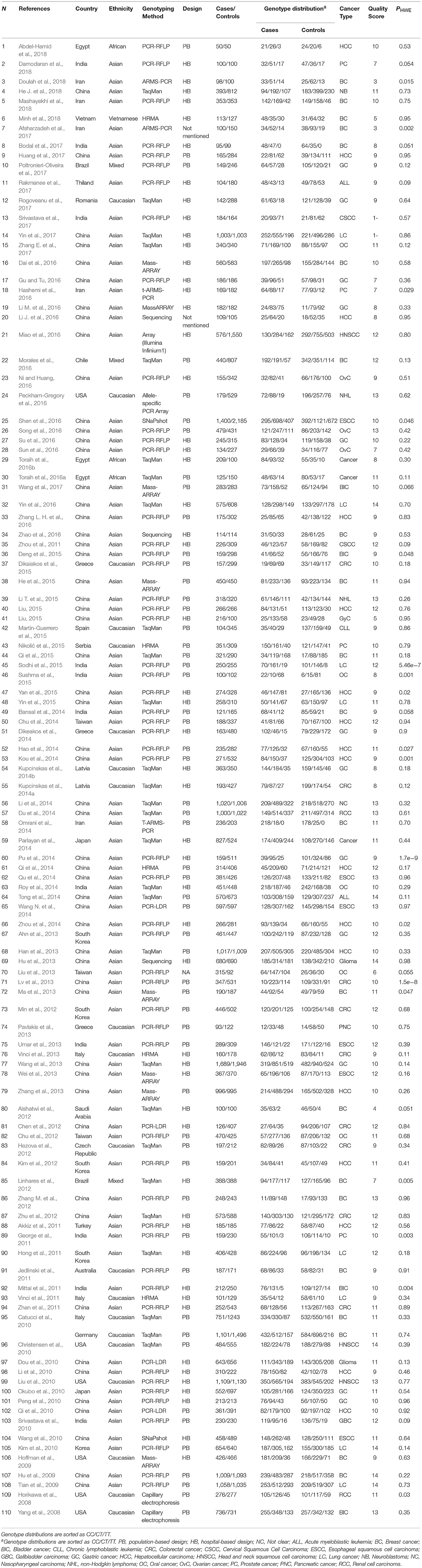
Table 1. Main characteristics of studies evaluating cancer risk associated with miR-196a-2 rs11614913 C/T which included in the current meta-analysis.
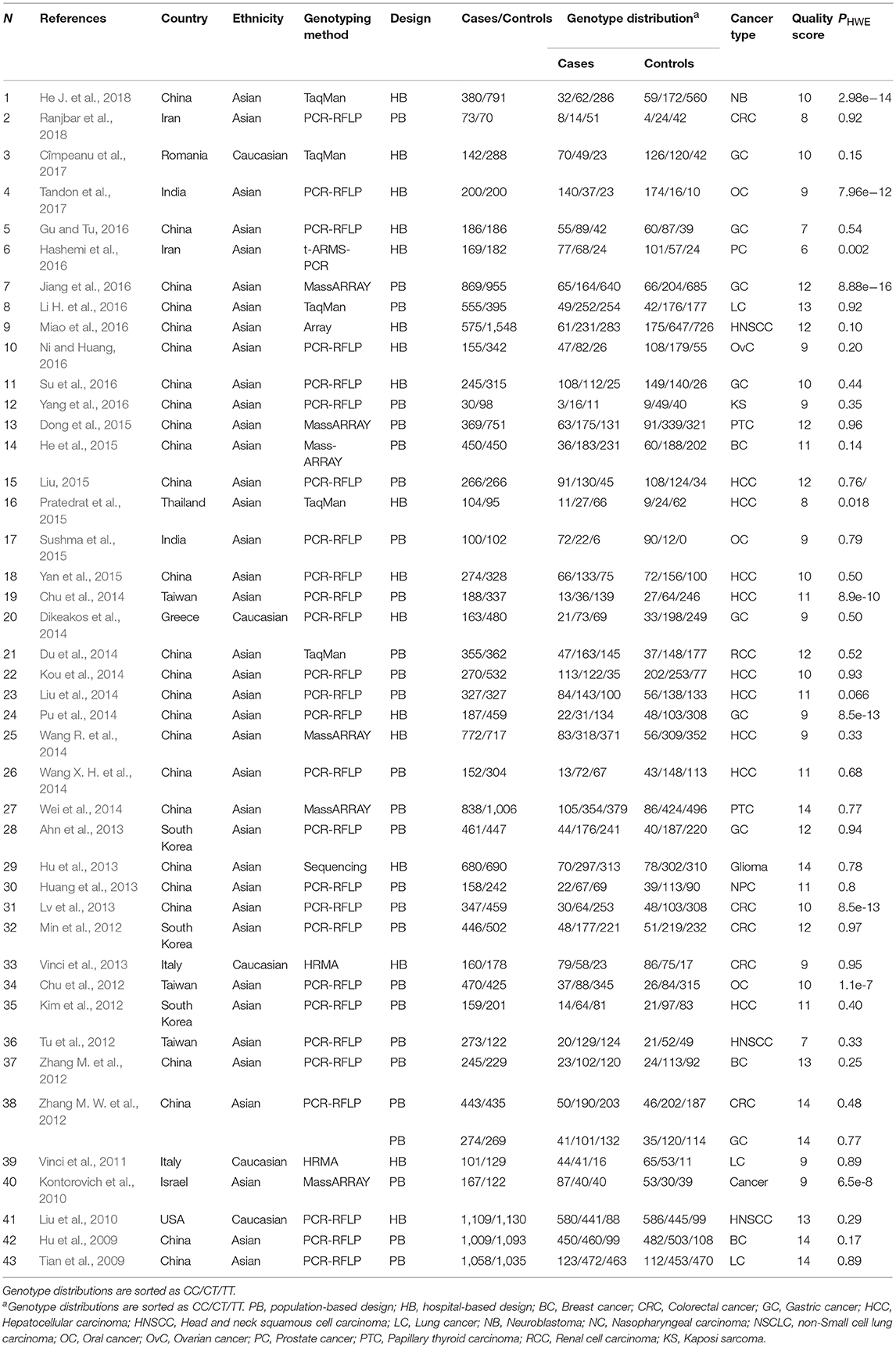
Table 2. Main characteristics of studies evaluating cancer risk associated with miR-149 rs2292832 C/T which included in the current meta-analysis.
A total of 110 articles which included 111 studies (41,673 cases and 49,570 controls) evaluated the association of miR-196a-2 rs11614913 and cancer risk (Table 1). The article by Catucci et al. included two studies on separate populations (Catucci et al., 2010). In a study on head and neck carcinoma the genotype frequencies were not reported in the paper and data were retrieved by contacting the authors (Christensen et al., 2010). The final meta-analysis of mir-196a-2 rs11614913 and cancer risk included 111 studies (including 41,673 patients and 49,570 controls), among which 93 were scored greater than eight in the quality assessment and regarded as high quality studies. In 93 out of 111 studies, the genotype distribution of rs11614913 in the control group was concordant with HWE. Mir-196a-2 rs11614913 were genotype using a range of techniques with the most common being PCR-RFLP (n = 53). With regard to the ethnicity, 85 studies were performed in Asians, 20 were performed in Caucasians and the remaining six studies were included either Africans or individuals from different ancestries (mixed ancestry). In four studies the patients were subgrouped into multiple cancer types. Namely, Liu evaluated both ovarian and endometrial cancers (Liu, 2015), Parlayan evaluated gastric, lung, colorectal, prostate, and acute leukemia (Parlayan et al., 2014), Toraih studied both GI and non-GI cancers in a study (Toraih et al., 2016b) and hepatic and renal cancers in another study (Toraih et al., 2016a). For these studies genotype distribution of all patients were used to calculate point estimates in the overall analysis. However, in subgroup analysis, as these studies were assigned to more than one subgroup, the genotype distribution of patients with the relevant cancer category/type was used for pooling data. When studies were subgrouped according to the broad cancer category, there were 49 gastrointestinal tract cancers (GI), nine head and neck squamous cell carcinoma (HNSCC), six gynecologic cancers (GyC), six hematological malignancies (HM), 12 urogenital cancers (UG), and 34 other cancers. When studies subgrouped according to cancer type, there were 21 breast cancer (BC), 18 hepatocellular carcinoma (HCC), 13 gastric cancer (GC), ten colorectal cancer (CRC), nine lung cancer (LC), four bladder cancer (BlC), five prostate cancer (PC), six oral carcinoma (OC), four ovarian cancer (OvC), six esophageal cancer (ESCC), and 22 other cancer types.
Moreover, 43 articles comprising 44 studies (15,954 cases and 19,594 controls) evaluated the association of mir-149 rs2292832 and cancer risk (Table 2), among which 39 studies were evaluated as being high quality (quality score > 8). The genotype distribution of rs2292832 in the control groups of 34 studies were in agreement with HWE. The main genotyping technique was PCR-RFLP (29 studies). Most studies (n: 39) were performed in Asian populations and only few studies (n: 5) had focused on Caucasians. According to the broad cancer category, there were 22 GI studies, seven HNC studies and 15 studies on other cancer types. When studies were subgrouped by cancer type, there were four studies on BC, nine studies on HCC, eight studies on GC, five studies on CRC, three studies on LC and 15 studies on other types of cancer.
Quantitative Synthesis
Association of mir-196a2 rs11614913 and Cancer Risk
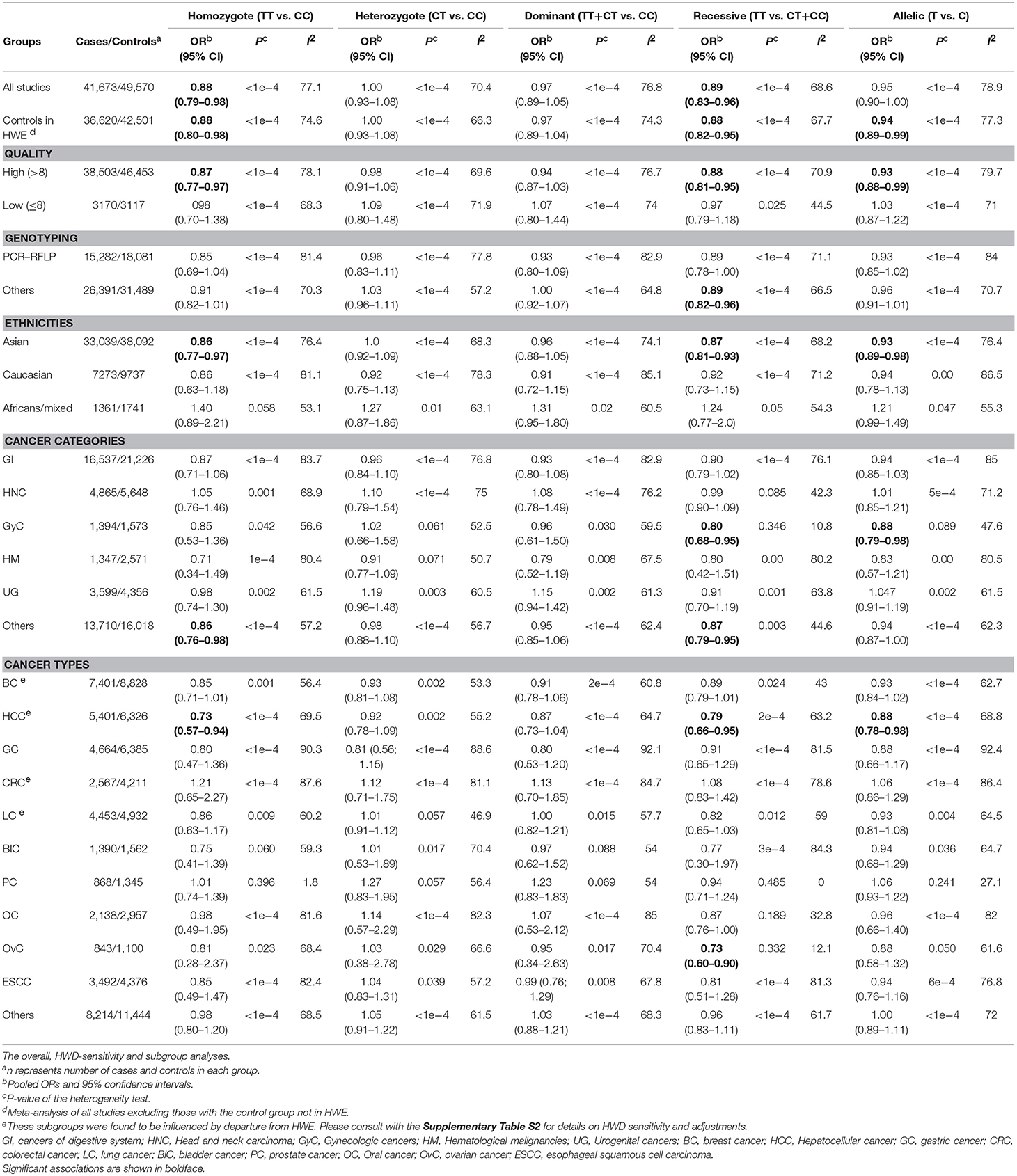
Statistically significant associations between mir-196a2 rs11614913 and cancer risk were observed assuming the homozygote (TT vs. CC, ORRE [95% CI]: 0.88 [0.79–0.98], P: 0.027) and the recessive (TT vs. CC+CT, ORRE [95% CI]: 0.89 [0.83–0.95], P: 0.001) models (Table 3). Mir-196a-2 rs11614913 was not associated with cancer risk in the heterozygote and dominant models and there was a non-significant borderline association in the allele contrast (Table 3). Supplementary Figure S1 shows the forest plots of association of mir-196a-2 rs11614913 and cancer risk in five models. The results of subgroup analysis for mir-196a-2 rs11614913 are shown in Table 4. Decreased risk of cancer was found in high quality studies under the homozygote (TT vs. CC, ORRE [95% CI]: 0.87 [0.77–0.97], P: 0.017), the recessive (TT vs. CC+CT, ORRE [95% CI]: 0.88 [0.81–0.95], P: 0.001) and the allele contrasts (T vs. C, ORRE [95% CI]: 0.93 [0.88–0.99], P: 0.020). In subgroup analysis by genotyping method, the only significant association was observed under the recessive model for studies which used a method other than PCR-RFLP (TT vs. CC+CT, ORRE [95% CI]: 0.89 [0.82–0.96], P: 0.007). When sub-grouped by ethnicity, decreased risks of cancer under the homozygote (TT vs. CC, ORRE [95% CI]: 0.86 [0.77–0.97], P: 0.016), the recessive (TT vs. CC+CT, ORRE [95% CI]: 0.87 [0.81–0.93] P: 0.0004) and the allelic (T vs. C, ORRE [95% CI]: 0.93 [0.89–0.98], P: 0.015) models were found only in Asians but not in Caucasians or the African/mixed ancestry subgroups (Table 4, Figure 2). Subgrouping by broad cancer categories indicated that mir-196a-2 rs11614913 was associated by a decreased risk of gynecologic cancer (GyC) assuming the recessive model (TT vs. CC+CT, ORFE [95% CI]: 0.80 [0.68–0.95], P: 0.010) and the allelic contrast (T vs. C, ORFE [95% CI]: 0.88 [0.79–0.98], P: 0.021) (Figure 3). No significant findings were observed for gastrointestinal, head and neck, hematological or urogenital cancers (Table 4). Supplementary Figure S2 presents the forest plots for subgroups according to the broad cancer categories. Further subgrouping by cancer type revealed significant association of mir-196a-2 rs11614913 with hepatocellular carcinoma (Figure 4) under the homozygote model (TT vs. CC, ORRE [95% CI]: 0.73[0.57–0.94], P: 0.017), the recessive model (TT vs. CC+CT, ORRE [95% CI]: 0.79 [0.66–0.95], P: 0.017) and the allele contrast (T vs. C, ORFE [95% CI]: 0.88 [0.78–0.98], P: 0.030), and with ovarian cancer (Figure 5) under the recessive model (TT vs. CC+CT, ORFE [95% CI]: 0.73[0.60–0.90], P: 0.003). Supplementary Figure S3 presents the forest plots for subgroups according to cancer type.
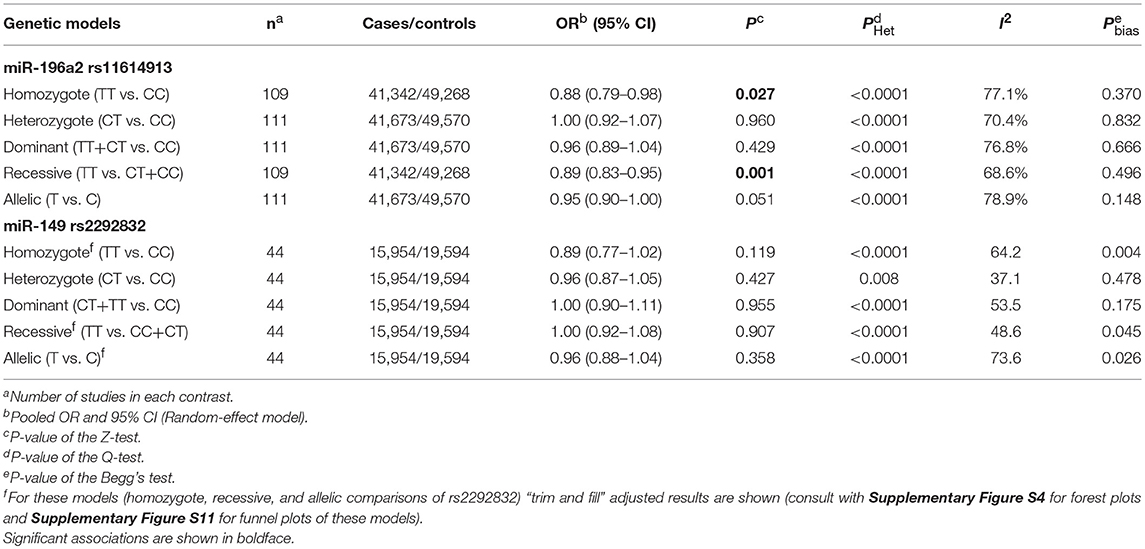
Table 3. Summary of the results of meta-analysis of cancer risk associated with miR-196a-2 rs11614913 and miR-149 rs2292832.
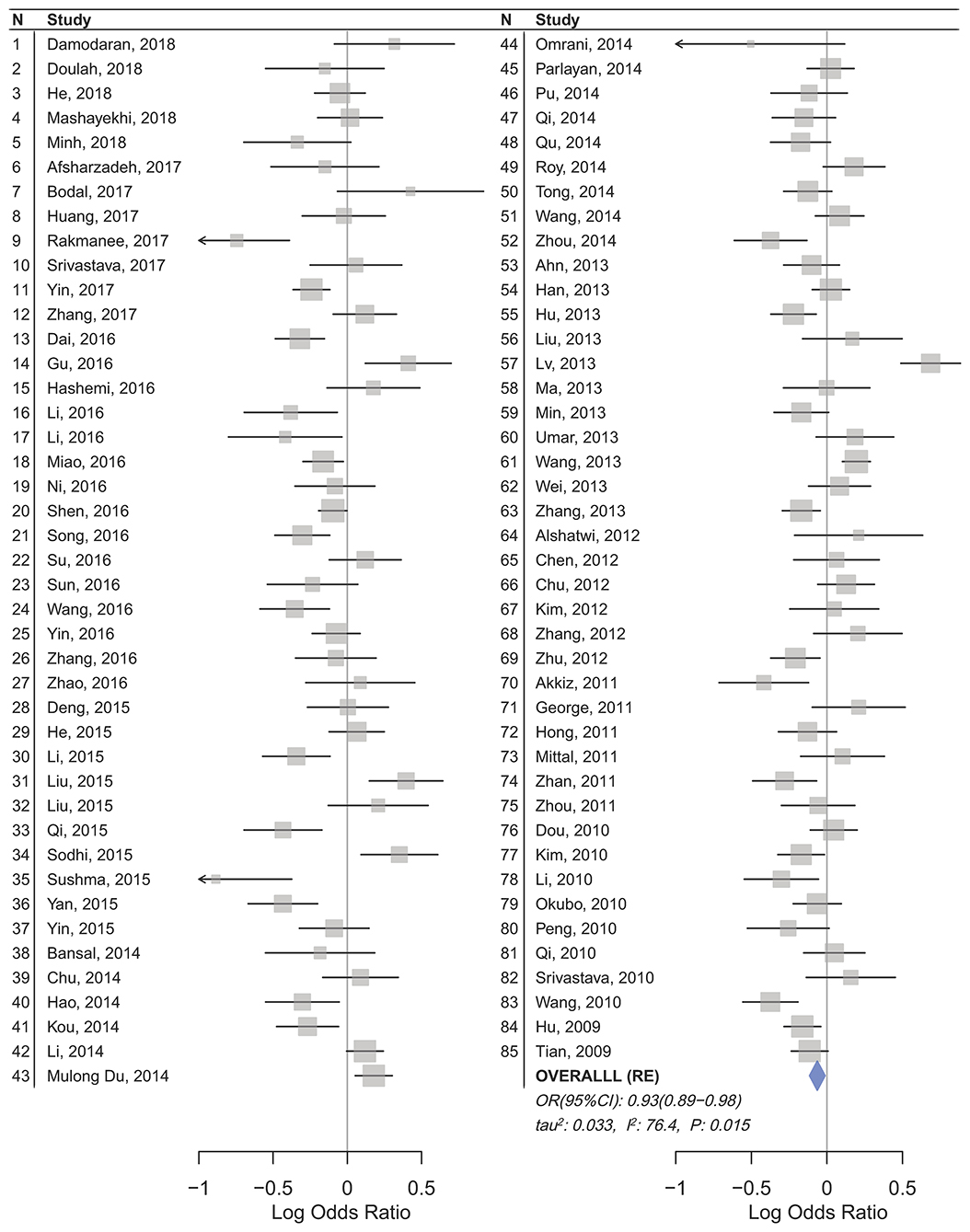
Figure 2. Forest plot of cancer risk associated with mir-196a-2 rs11614913 in the Asian subgroup (Allele contrast T vs. C). The plot is designed in a side-by-side mode to represent 85 studies in the Asian subgroup. The result of meta-analysis is shown beneath the second column.
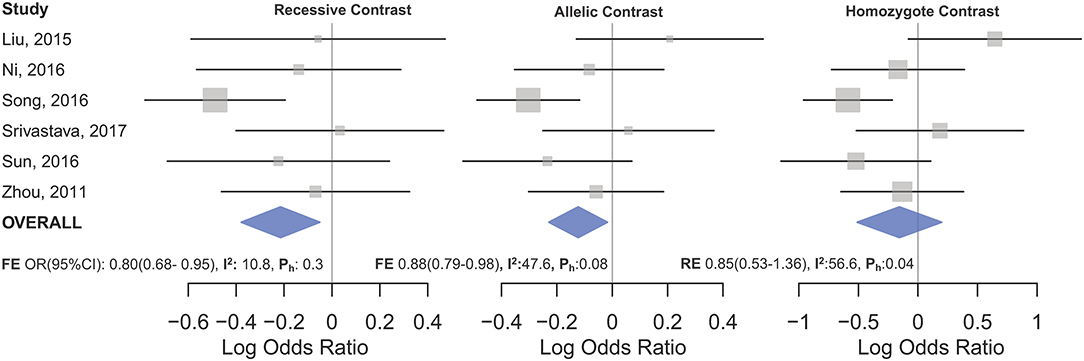
Figure 3. Forest plot of cancer risk associated with mir-196a-2 rs11614913 in the gynecological cancers subgroup. From left to right: recessive (TT vs. CT+CC), allelic (T vs. C), and homozygote (TT vs. CC) contrast.
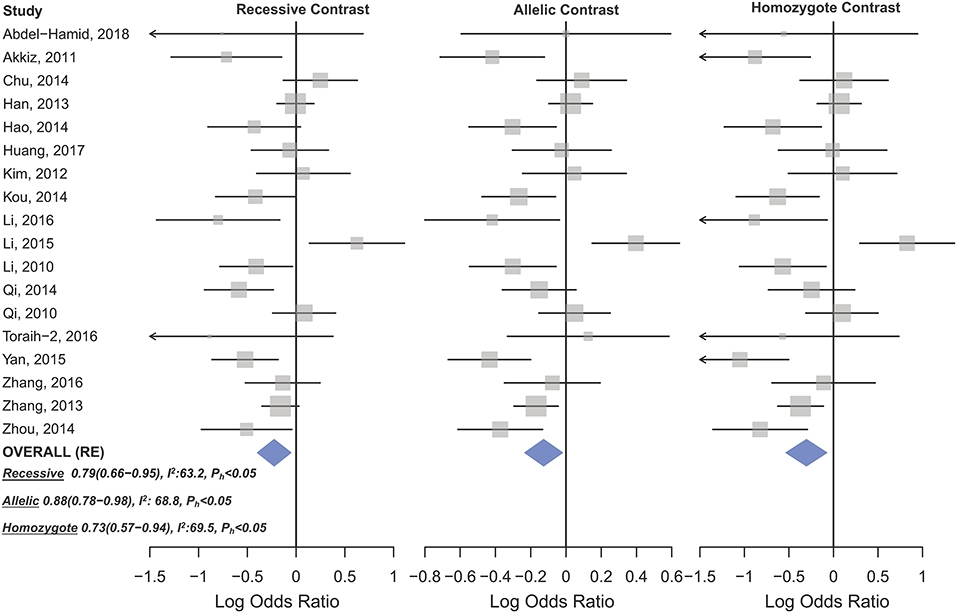
Figure 4. Forest plot of cancer risk associated with mir-196a-2 rs11614913 in the hepatocellular cancer subgroup. From left to right: recessive (TT vs. CT+CC), allelic (T vs. C), and homozygote (TT vs. CC) contrast.
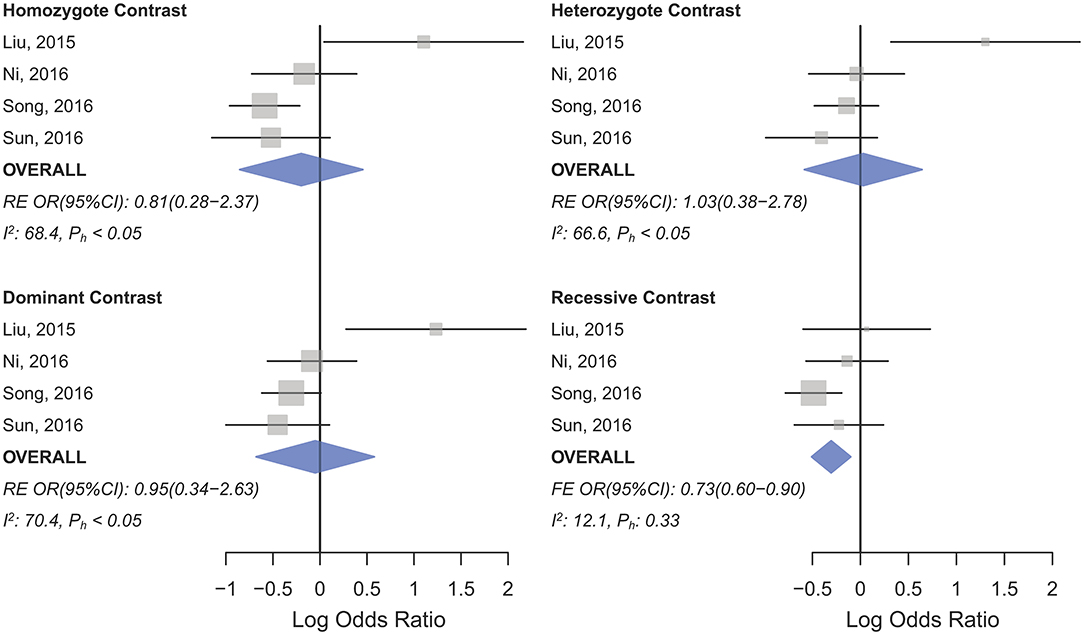
Figure 5. Forest plot of cancer risk associated with mir-196a-2 rs11614913 in the ovarian cancer subgroup. Top left: homozygote (TT vs. CC) contrast; Top right: heterozygote comparison (CT vs. CC); Bottom left: dominant comparison (CT+TT vs. CC); Bottom right: recessive comparison (TT vs. CT+CC).
HWD sensitivity analysis (i.e., excluding studies with controls deviated from HWE) revealed stable results in the overall analysis under the homozygote, heterozygote, dominant, and recessive models (Table 4). However, excluding HWD studies made the borderline allele contrast statistically significant (Table 4). Moreover, excluding HWE violating studies had no dramatic effects on subgroup meta-analyses using quality of studies, genotyping methods, the ethnicity and the broad cancer category (Supplementary Table S1). In meta-analysis subgrouped by cancer type, the results were also stable for gastric, bladder, oral, ovarian, prostate, and esophageal cancer subgroups after excluding HWD studies (Supplementary Table S1). However, excluding such studies altered the results for the breast, hepatocellular, colorectal, and lung cancer subgroups. Therefore, for these subgroups, pooled ORs were estimated to account for departures from HWE (denoted as HWD-adjusted ORs) (Supplementary Table S2). When corrected for HWD, mir-196a-2 rs11614913 was found to be significantly associated with breast cancer under the homozygote (TT vs. CC, HWD-adjusted ORRE [95% CI]: 0.75 [0.61–0.93], P: 0.011) and recessive (TT vs. CC+CT, HWD-adjusted ORRE [95% CI]: 0.84 [0.71–0.98], P: 0.030) models (Figure 6). The association with hepatocellular cancer under the homozygote and the recessive models was remained significant after adjustment for HWD (TT vs. CC, HWD-adjusted ORRE [95% CI]: 0.69 [0.53–0.91], P: 0.011 and TT vs. CC+CT, HWD-adjusted ORRE [95% CI]: 0.72 [0.57–0.90], P: 0.008). Furthermore, adjustment for HWD confirmed that mir-196a-2 rs11614913 is not associated with colorectal or lung cancer assuming any genetic model (Supplementary Table S2).
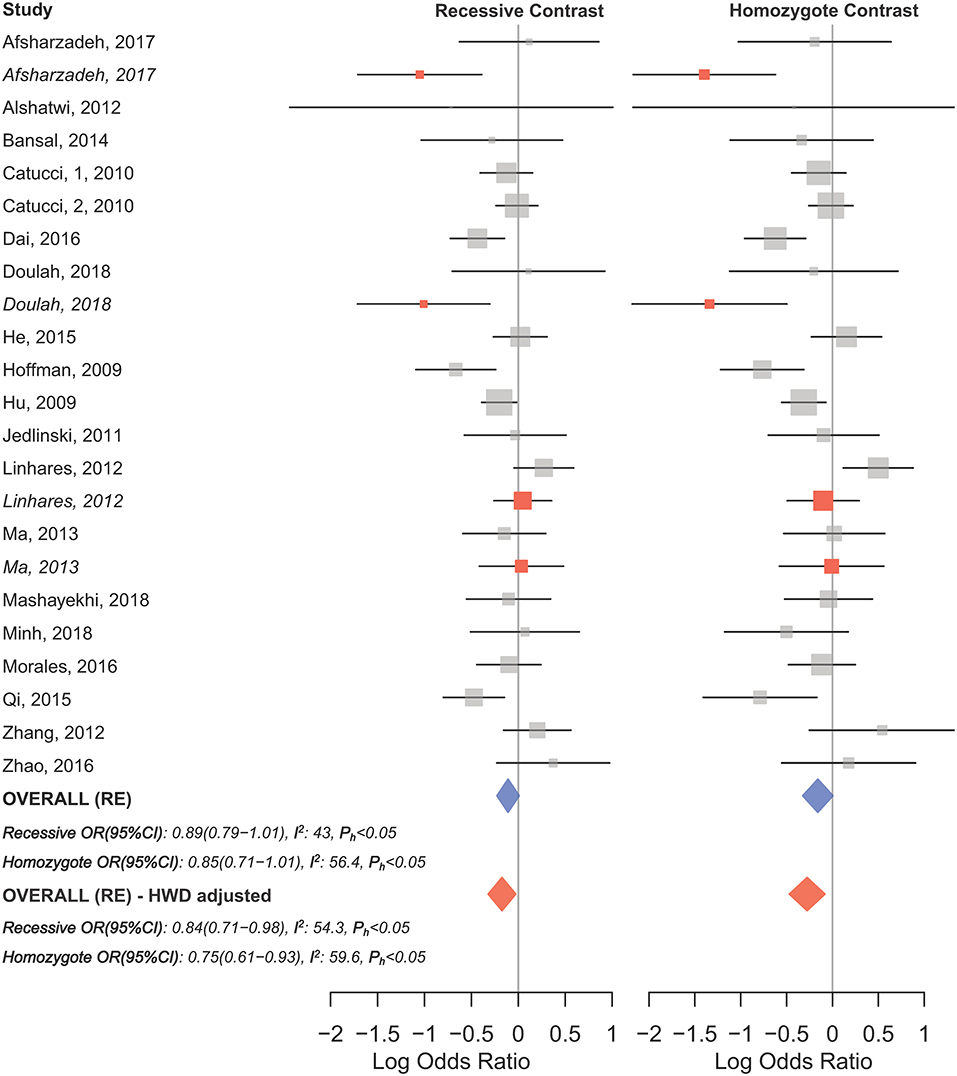
Figure 6. Forest plot of the original and HWD-adjusted meta-analysis of breast cancer risk and mir-196a-2 rs11614913 under the recessive (TT vs. CT+CC) and homozygote (TT vs. CC) models. Red boxes represent HWD-adjusted log ORs and 95%CIs of studies with HWE-deviated controls (i.e., Linhares et al., 2012; Ma et al., 2013; Afsharzadeh et al., 2017; Doulah et al., 2018). The blue and red diamonds represent the estimated pooled effects of the original and HWD-adjusted meta-analysis, respectively.
Association of mir-149 rs2292832 and Cancer Risk
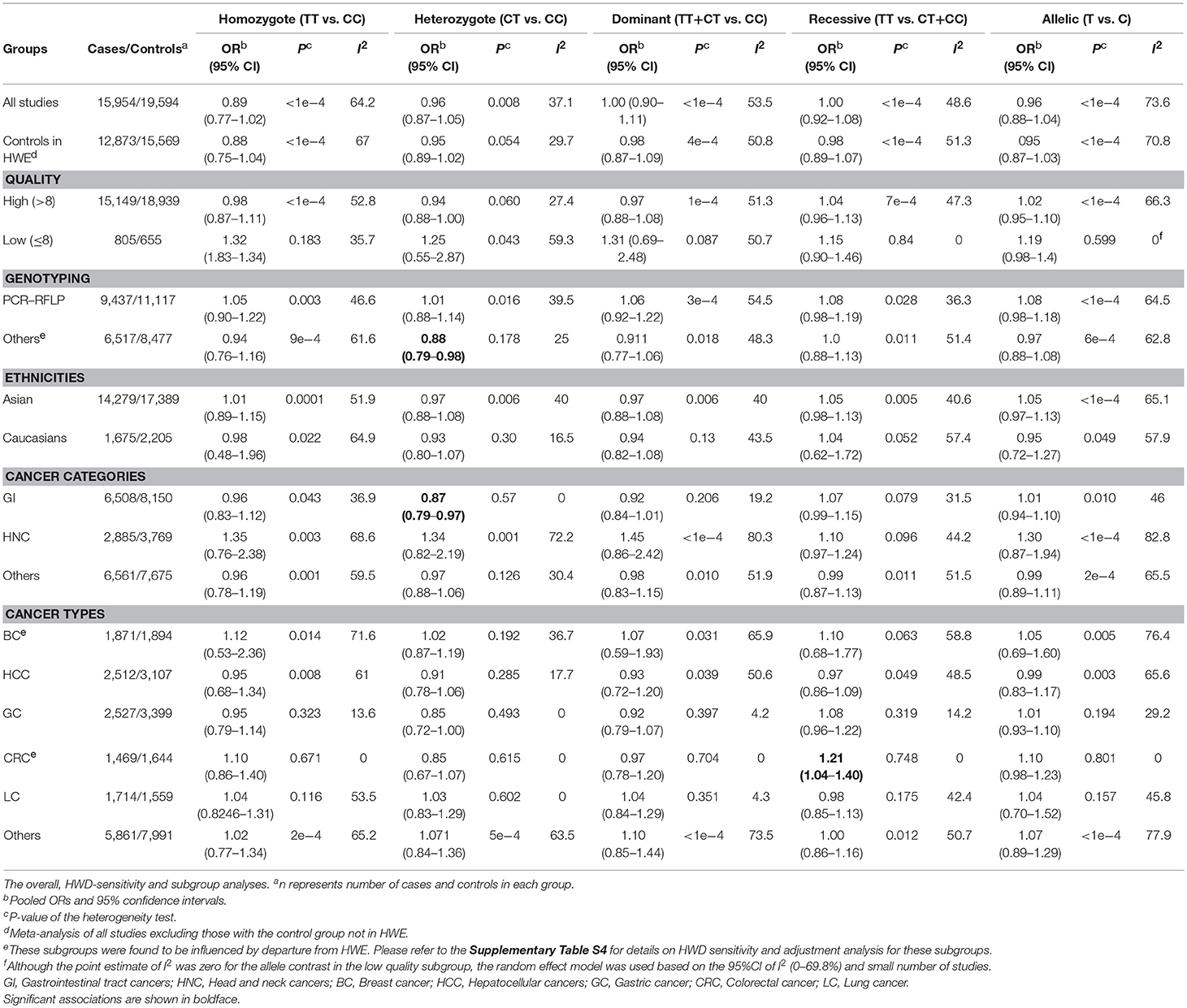
The overall analysis showed no significant association with cancer risk under any genetic model (Table 3). Supplementary Figure S4 shows the forest plots for the association of mir-149 rs2292832 and cancer risk under different genetic models. However, in subgroup analyses (Table 5) significant association of rs2292832 with cancer risk was observed in studies which used a genotyping method other than PCR-RFLP (CT vs. CC, ORFE [95% CI]: 0.88 [0.79–0.98], P: 0.025). When subgrouped by broad cancer category, a decreased risk of gastrointestinal tract cancers was found in the heterozygote model (Figure 7A, CT vs. CC, ORFE [95% CI]: 0.87 [0.79–0.97], P: 0.011). Subgrouping by cancer type, however, revealed an increased risk of colorectal cancer for individuals carrying TT genotype compared to those who carry at least one C allele (Figure 7B, TT vs. CT+CC, ORFE [95% CI]: 1.21 [1.04–1.40], P: 0.011). No significant association was observed for other comparisons (Table 5). Supplementary Figures S5, S6 show the forest plots for subgroup analysis according to the broad cancer category and cancer type, respectively. Sensitivity analysis revealed that HWD studies had no significant effect on point estimates in the overall meta-analysis of mir-149 rs2292832 and cancer risk, and still no significant association was observed in overall analysis (Table 5). Moreover, most subgroup analyses were also stable after removing HWD studies (Supplementary Table S3). However, removing studies with HWD controls influenced comparisons in three subgroups: (i) non-PCR-RFLP subgroup (heterozygote model); (ii) the breast cancer subgroup (recessive model); (iii) the colorectal cancer subgroup (recessive model). Therefore, for these subgroups, HWE-expected genotype distributions in controls were used for pooling ORs (denoted as HWD-adjusted OR) (Supplementary Table S4). Adjusting for HWD in these subgroups confirmed the results of original analyses and showed that rs2292832 is associated with cancer risk in non-RFLP subgroup under the heterozygote model (Figure 8, CT vs. CC, HWD-adjusted ORRE [95% CI]: 0.68 [0.48–0.98], P: 0.040) and with colorectal cancer risk under the recessive model (TT vs. CT+CC, HWD-adjusted ORFE [95% CI]: 1.29 [1.11–1.50], P: 0.0007). No association with breast cancer risk was identified after adjusting for HWD (Supplementary Table S4).
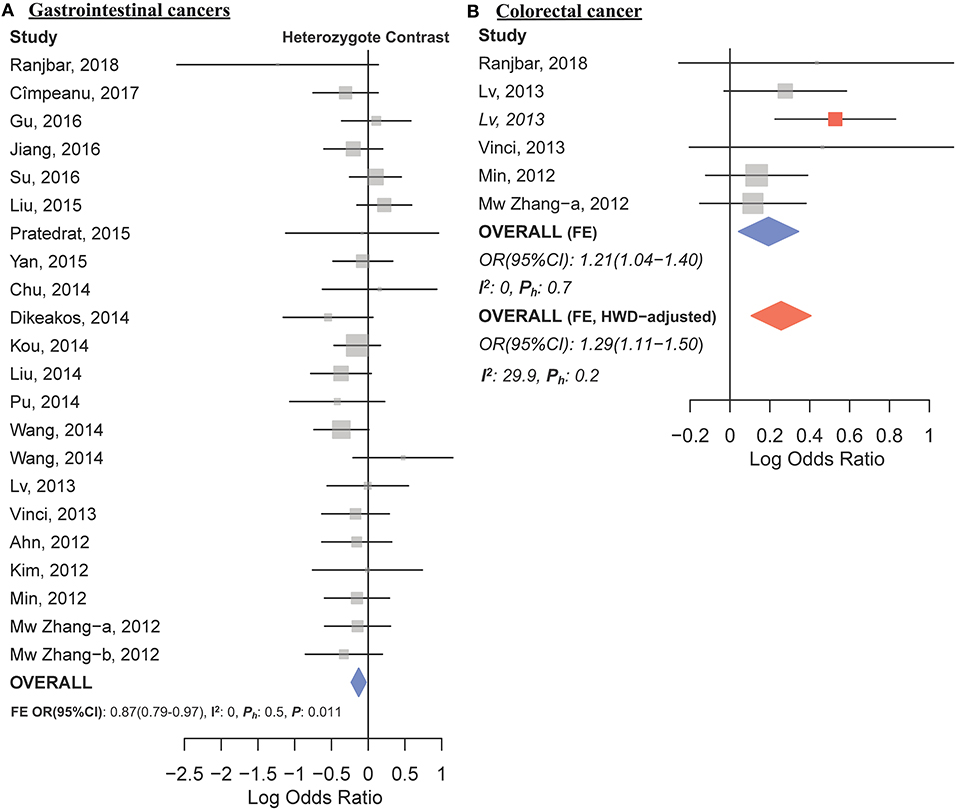
Figure 7. Forest plots of cancer risk associated with mir-149 rs2292832; (A) In the gastrointestinal cancers subgroup assuming the heterozygote model (CT vs. CC). (B) In the colorectal cancer subgroup assuming the recessive model (TT vs. CT+CC). The red box shows the HWD-adjusted log OR (95%CI) for the study by Lv et al. (2013). The blue and red diamonds represent the estimated pooled effect of the original and HWD-adjusted meta-analysis.
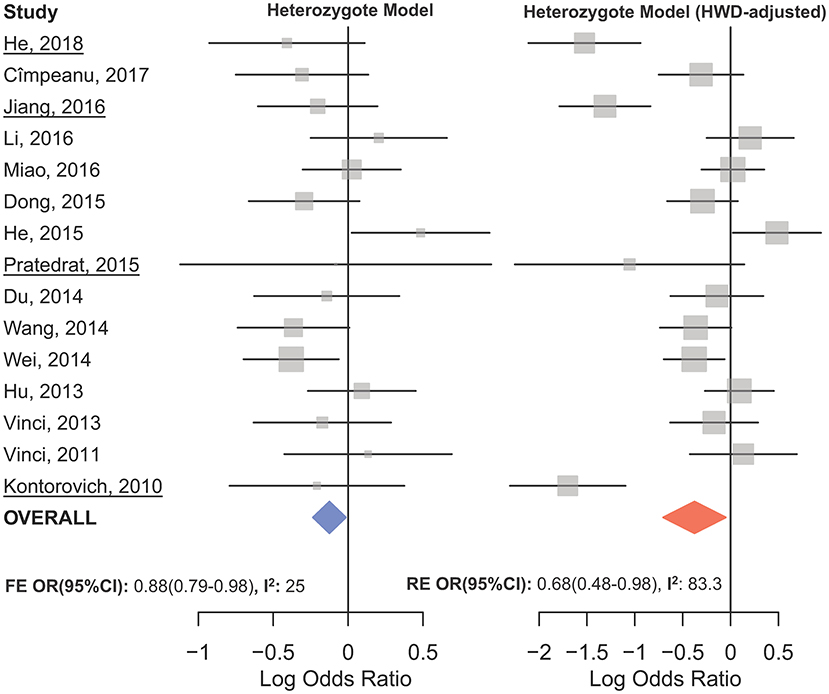
Figure 8. Forest plot of the original (Left) and HWD-adjusted (Right) meta-analysis of cancer risk associated with mir-149 rs2292832 in subgroup of studies which used a non-RFLP genotyping method assuming the heterozygote model. In the right plot, HWD-adjusted log ORs and 95%CI were used for the underlined studies (i.e., He, 2018; Kontorovich et al., 2010; Pratedrat et al., 2015; Jiang et al., 2016).
Heterogeneity, Meta-regression, and Sensitivity Analysis
Heterogeneity was evaluated for both polymorphisms in all genetic models (Tables 3, 4, 5). Significant between study heterogeneity was observed in the overall estimation under all genetic models for mir-196a-2 rs11614913 and consequently random effect model was used. Univariate meta-regression using cancer type, country, ethnicity, the quality of study (either high or low), genotyping method, source of controls (PB or HB) or HWE was performed to identify potential sources of heterogeneity. For mir-196a-2, meta-regression showed that at least a part of the observed between study heterogeneity in the heterozygote (R2: 24.23%, P: 0.007) and dominant (R2: 19.86%, P: 0.028) models could be attributed to the country moderator. However, there was still significant unaccounted heterogeneity even after correcting for the effect of country moderator (Heterozygote I2: 63.78 and dominant I2: 72.05, P for test of residual heterogeneity < 0.0001). Moreover, Galbraith plot analysis demonstrated three studies (Lv et al., 2013; Wang et al., 2013; Dikeakos et al., 2014) as the most extreme outliers in all genetic models that account for a considerable portion of the observed heterogeneities (Supplementary Figure S7). Excluding these studies led to a 11.6% reduction of I2 in the homozygote model (from 77.1 to 65.5%), a 12.8% reduction in the heterozygote model (from 70.6 to 57.8%), a 12.3% reduction in the dominant model (from 76.9 to 64.6%), an 9.1% reduction in the recessive model (from 68.8 to 59.7%) and a 11.3% reduction in the allelic model (from 79 to 67.7%). However, excluding these studies did not alter any genotypic contrast and results were comparable to the overall analyses (data not shown). Sensitivity analysis by omitting one study at a time revealed that no individual study significantly influenced the genotype contrasts (Supplementary Figure S8). In the allele contrast, omitting no single study dramatically influenced pooled OR or its 95%CI. However, given that the original 95%CI was borderline (0.90–1.00), omitting some studies lead the upper limit of 95%CI to fall slightly below one (Supplementary Figure S8-e).
Statistically significant heterogeneity was also observed in the overall analysis of miR-149 rs2292832 and cancer risk and, consequently, RE model was used to estimate pooled OR (Table 3). Subgrouping by study level moderators led to a reduction in heterogeneity in some subgroups (Table 5). However, univariate meta-regression showed no statistically significant source of heterogeneity (All P > 0.05). Sensitivity analysis by omitting one study at a time revealed no single influential study (Supplementary Figure S9).
Publication Bias
Rank correlation test of the mir-196a-2 rs11614913 Begger's funnel plot asymmetry revealed no statistically significant evidence of publication bias in any contrast (Table 3 and Supplementary Figure S10). However, rank correlation test for asymmetry of mir-149 rs2292832 funnel plots showed statistically significant results in the homozygote, recessive and the allelic contrasts (Table 3 and Supplementary Figure S11). Consequently, the “trim and fill” approach (Duval and Tweedie, 2000a,b) employed to correct for funnel plot asymmetry arising from publication bias in these models. The results of overall analysis using original studies or trim and fill method under the three models were comparable (see Supplementary Figure S4 to compare forest plots of the original studies vs. trim-and-fill method and Supplementary Figure S11 for funnel plots). After excluding studies with controls deviating from HWE in sensitivity analysis, rank correlation test was still significant in the mentioned three contrasts.
Discussion
The possible contribution of miRNAs, especially mir-196a-2 and mir-149, to the risk of cancer has stimulated great attention in recent years. Many studies evaluated the functional alterations of these micro-regulators in a wide range of cancers. Accumulating evidence suggests that, at least a part of functional dysregulations of miRNAs in cancers could be attributed to polymorphisms in miRNA sequences (Hu et al., 2008; Hoffman et al., 2009; Tu et al., 2012; Ghaedi et al., 2015; Nariman-Saleh-Fam et al., 2016, 2017). Two mature miRNAs, miR-196a-5p and miR-196a-3p, are generated from the stem-loop structure of hsa-mir-196a-2 (Kozomara and Griffiths-Jones, 2014) with the studied polymorphism, rs11614913, residing in the 3′ arm (Figure 9A). This polymorphism, therefore, may potentially alter miRNA processing and also binding to related target mRNAs (Hoffman et al., 2009) (Figure 9B). Previous studies have shown that the expression level of mature miR-196a-3p was higher in CC carriers with lung cancer compared to CT and TT individuals (Hu et al., 2008). More evidences have been provided by Hoffman et al. (2009) who observed elevated expression of mature mir-196a-2 forms in MCF-7 cells transfected with pre-mir-196a-C vector compared with cells transfected with pre-mir-196a-T vector. The potential of rs11614913 in influencing targeting function of mir-196a-2 has also been documented by whole-genome expression microarrays which found different numbers of dysregulated mRNAs after transfecting cells with pre-mir-196a-C or pre-mir-196a-T vector (Hoffman et al., 2009). Hsa-mir-149 also generates two mature miRNAs (miR-149-5p and miR-149-3p) and the studied polymorphism, rs2292832, does not reside in the mature sequence of neither miR-149-5p or miR-149-3p (Figure 10A). Therefore, it has been hypothesized that rs2292832 is not a structure-shifting polymorphism for pri-mir-149 or pre-mir-149 (Wei et al., 2014). However, Tu et al. reported that the T allele may disrupt the maturation process compared with the C allele and, consequently, decrease miR-149 expression (Tu et al., 2012) (Figure 10B) in head and neck squamous cell carcinoma patients.
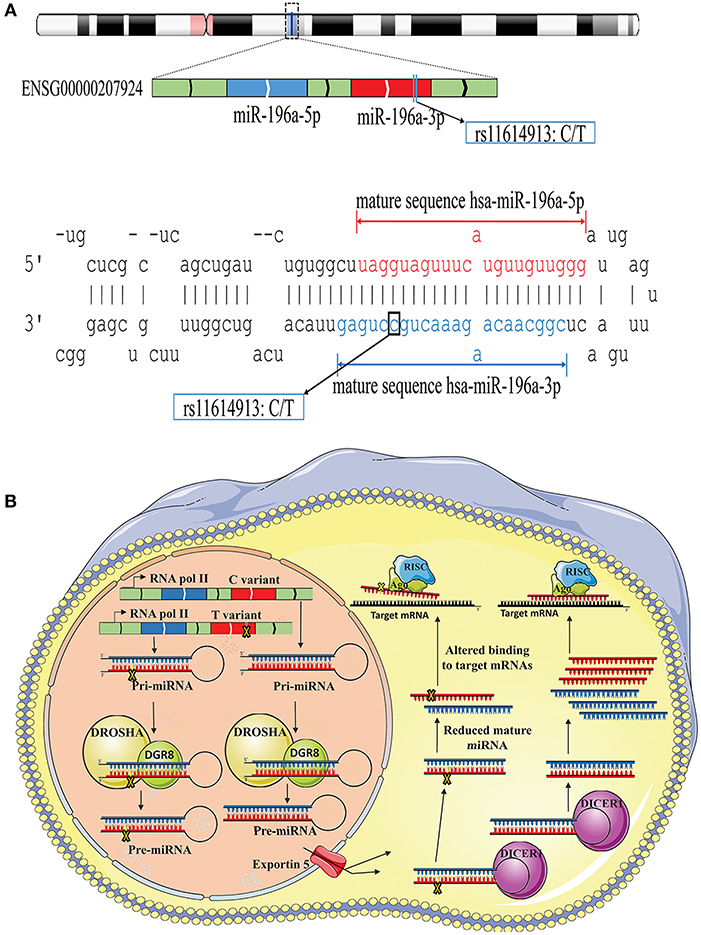
Figure 9. Schematic illustration of hsa-mir-196a-2 locus and effect of rs11614913 on its processing and targeting. (A) Top: Hsa-mir-196-a-2 generates two mature miRNAs, miR-196a-5p and mir-196a-3p, and rs11614913 lies in miR-196a-3p at GRCh37 chr12: 54,385,599. Bottom: Stem-loop structure of hsa-mir-196, including mature miR-196-5p (Red) and miR-196-3p (Blue) sequences. (B) Rs11614913 alters miRNA processing and/or binding to related target mRNAs.
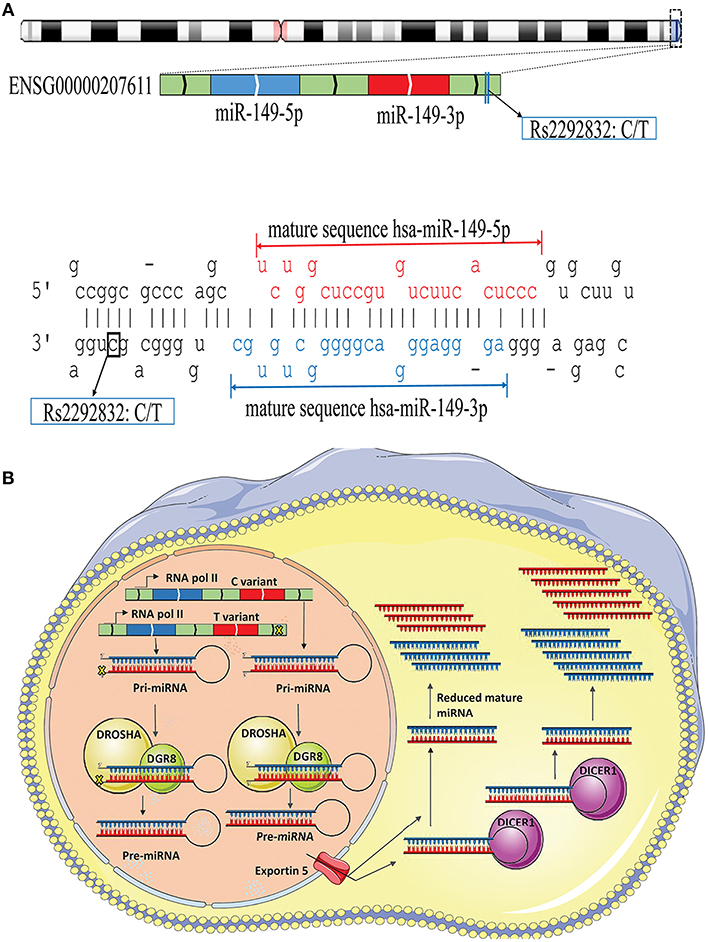
Figure 10. Schematic illustration of hsa-mir-149 locus and effect of rs2292832 on its processing. (A) Top: Hsa-mir-149 generates two mature miRNAs (miR-149-5p and miR-149-3p), and rs2292832 resides in pre-miRNA sequence, but not in either mature miRNAs, at GRCh37 chr2: 241,395,593. Bottom: Stem-loop structure of hsa-mir-149, including mature miR-149-5p (Red), and miR-149-3p (Blue) sequences. (B) Rs12292832 may alter miRNA processing and decrease mature miR-149 (−5p and −3p forms) expression.
Increasing number of association studies evaluating miRNA polymorphisms and cancer risk with contradictory results merits the need for comprehensive systematic reviews and meta-analyses. Several meta-analyses have evaluated the risk of cancer associated with mir-196a-2 rs11614913 or mir-149 rs2292832 (Chu et al., 2011; Zhang H. et al., 2012; Feng et al., 2016; Yan et al., 2017; Liu et al., 2018). However, the conclusion of these studies with regards to the subgroup analysis and the significant genetic model varies due, at least in part, to differences in the number of studies included or in the methodology. Moreover, several recent genetic association studies have not been included in previously published meta-analyses. Therefore, it was necessary to perform an updated meta-analysis with larger number of studies to clarify the association of mir-196a-2 rs11614913 or mir-149 rs2292832 with cancer risk. Therefore, compared to the previous meta-analysis, we included more studies in the analyses. The present meta-analysis also evaluated and corrected for the possible influence of departure of the control group of association studies from HWE. Although checking for departure from HWE has been recommended, currently there is no consensus about how HWD studies should be handled in meta-analysis (Minelli et al., 2008). Result of simulations suggests no advantage for excluding these studies (Minelli et al., 2008). However, sensitivity analysis to detect any possible bias imposed by such studies and/or using HWE-expected counts instead of the observed genotype frequencies have been recommended and implemented in several studies (Attia et al., 2003; Thakkinstian et al., 2005; Trikalinos et al., 2006; Zintzaras et al., 2006; Zintzaras, 2008; Zintzaras and Lau, 2008; Srivastava and Srivastava, 2012; Wang X. B. et al., 2014). The current study noticed that most analyses, especially those with sufficiently large number of studies, were not influenced by excluding HWD studies. However, as it is rationally expected, excluding HWD studies from some subgroup with relatively small number of studies may influence the analysis, and therefore in such situations adjusted analyses were preferred.
For mir-196a-2 rs11614913, the previous largest meta-analysis, conducted by Liu et al. (2018), included 84 studies compared to 111 studies in the present meta-analysis. By including 41,673 patients and 49,570 control subjects, the present meta-analysis showed a decreased risk of cancer in the homozygote and the recessive models (Table 3). Although, the association was not significant in allele contrast, the OR and 95%CI of the allele contrast were borderline and influenced by excluding some individual studies. Excluding HWE-deviated or low quality studies yielded significant associations under allelic model. As there is, currently, no way to adjust allele frequencies for departure from HWE, the possibility that HWD studies may bias the allele contrast cannot be rolled out and a definite conclusion cannot be drawn under allelic model. Apart from the allele contrast, the results of other genetic models were statistically stable and not influenced by removing any single study, HWE deviated or low quality studies. The results also suggest that mir-196a-2 rs11614913 may pose an ethnic dependent effect on cancer risk as associations with cancer were only observed in Asians. However, it should be noted that most studies enrolled Asian patients, mainly Chinese patients, and the number of studies involving other ethnicities were relatively small. Moreover, different minor allele frequencies (MAF) may partly contribute to the observed differences among ethnicities (Average MAF in Asians T: 0.501 ± 0.127, Caucasians T: 0.410 ± 0.1, Others T: 0.338 ± 0.066). In-line with previous studies, the current meta-analysis also confirmed that rs11614913 is associated with decreased risks of hepatocellular cancer under three genetic models (Liu et al., 2018) and that it may not modulate risk of urogenital cancers (Wang et al., 2017). The results of meta-analysis of all studies, subgroup analysis by ethnicity and hepatocellular cancer are in agreement with findings of the previous largest meta-analysis (Liu et al., 2018). However, the increase in the number of analyzed studies led to discrepancies with regards to conclusions in some subgroup analysis. (i) In contrast to the studies by Liu and Pan (Pan et al., 2017; Liu et al., 2018), the present meta-analysis did not find a significant association with head and neck carcinoma in any genetic model. This discrepancy may be attributed to the number of studies included in meta-analyses [nine studies in the present meta-analysis compared to four studies in the meta-analysis by Liu et al. (2018)]. Furthermore, differences in defining head and neck cancer may also explain different conclusions drawn from the present study and the study by Pan and colleagues (Pan et al., 2017). They included esophageal cancer as a type of head and neck cancer, whereas we considered it as a type of gastrointestinal tract cancers (according to the ICD-10-CM C15-C26). (ii) Additionally, the present meta-analysis found significant associations between mir-196a-2 rs11614913 and decreased risks of gynecologic cancers (especially ovarian cancer), which have not been reported in any previous meta-analysis. Interestingly, low heterogeneity was observed in the gynecological cancers subgroup assuming the two significant contrasts (i.e., recessive and allelic). (iii) Although previous meta-analyses (Yan et al., 2017; Zhang H. et al., 2017; Liu et al., 2018) failed to find a significant association between mir-196a-2 rs11614913 and breast cancer, the current study showed, by incorporating more association studies and performing HWD sensitivity analysis, that adjusting for departure from HWE may reveal significant associations under the homozygote and recessive contrasts (Figure 6 and Supplementary Table S2). (iv) Moreover, contradictory to previous meta-analyses, no association with gastric (Yan et al., 2017), colorectal (Xie et al., 2015; Yan et al., 2017) or lung cancer (Ren et al., 2016; Yan et al., 2017; Liu et al., 2018), was found. Apart from larger sample sizes and correcting for HWD, sometimes this discrepancy in results may also be related to methodological differences in the design, specifically the inclusion criteria, of meta-analyses. As a case in point, studies by Hu et al. (2008) and Yoon et al. (2012) did not meet the inclusion criteria of our study, as they deal with the survival or recurrence risk of lung cancer patients with approaches that differed from routine case-control genetic association studies. However, we noticed that these studies were included in a previous meta-analysis (Liu et al., 2018).
The current meta-analysis also showed that mir-149 rs2292832 is not associated with risk of cancer in any genetic model and the results were statistically reliable, as summary effects were not influenced by excluding any single study, HWE-deviated or low quality studies. No differences in cancer risk was observed between ethnicities. Similar to miR-196a-2 polymorphism, the Asian subgroup comprised a large proportion of studies and relatively few studies with limited sample sizes were performed in Caucasians. Therefore, a definite conclusion cannot be drawn in Caucasians and more studies are needed further clarify the association of this SNP with cancer risk in Caucasians. The results of overall analysis were comparable to previous meta-analyses (Li L. et al., 2015; Feng et al., 2016). By pooling the results of 22 studies, this meta-analysis found a decreased risk of gastrointestinal tract (GI) cancers for individuals who carry the CT genotype compared to those with the CC genotype in a heterozygote model. Interestingly, there was no significant heterogeneity in the GI subgroup assuming the heterozygote model indicating the reliability of meta-analysis in this subgroup. Previous meta-analyses yielded different results with regard to GI cancers. A previous meta-analysis of seven studies on GI cancers suggested a marginally elevated risk under the recessive model (TT vs. CT+CC), while another pooling of 10 studies found a borderline decreased risk for the CT vs. TT contrast. Although no significant association was identified in the head and neck cancers subgroup, it should be noted that significant heterogeneity was present in all models except the recessive contrast and number of samples were relatively small. For colorectal cancer the association was in reverse direction and an increased risk was observed in individuals who carry the TT genotype compared with subject who carry at-least one C allele (Table 5). A similar association based on three studies was previously reported (Rong et al., 2017), but not reproduced in other meta-analyses (Li et al., 2013; Feng et al., 2016). Taken together, the current results based on five studies suggest an increased risk for colorectal cancer that was stable after correcting for departure from HWE. Although no significant heterogeneity was detected in the colorectal cancer subgroup under any genetic model, it should be noted that the limited number of studies may influence heterogeneity evaluation and more definite conclusion may be drawn by analyzing larger sample sizes. In the case of breast cancer, a previous meta-analysis of three studies found a significant association (Feng et al., 2016). We found no significant association in the original and HWD-adjusted analysis. However, number of studies in the colorectal and breast cancer subgroups are relatively limited and results should be interpreted with caution. More studies with large sample sizes are needed for a definite conclusion.
However, the present study has some limitations. First, significant heterogeneity was present in most analyses especially for mir-196a-2 polymorphism. We, therefore, used random effect model and performed meta-regression; but no significant source of heterogeneity was observed for most analyses, suggesting that other unknown study level moderators may contribute to the heterogeneity. Second: The molecular mechanisms underlying association of these miRNAs-SNP with risk of cancer are complex and might be strongly affected by different genetic background as well as other masked variables. This, in turn, may limit the efficacy of the overall analysis especially in the case of miR-196a-2 rs11614913. Stratified analyses based on a specific cancer category or a cancer type may help to reduce this heterogeneity and, therefore, are considered to be more reliable. Third, this study was based on unadjusted ORs of the original studies and no adjustment for covariates like age and gender or interaction with environmental factors were done and this fact may also potentially contribute to the between study heterogeneity. Fourth, some limitations such as language restriction or lack of access to the genotype counts of mir-196a-2 rs11614913 in four studies with insufficiently reported data may bring in publication bias. The trim and fill method has been shown to reduce the bias in estimates in the presence of publication bias and heterogeneity (Peters et al., 2007). However, it has been recommended that this method should be considered as sensitivity analysis as we cannot be sure that asymmetry in funnel plot is truly caused by publication bias (Peters et al., 2007). Although rank correlation test of funnel plots of mir-149 rs2292832 was significant in three genetic models raising the possibility of publication bias, adjusting for such a bias using trim-and-fill method did not afford any change in analysis of overall studies in these models (Supplementary Figure S4). Fifth, number of studies in some subgroup analyses was limited and, consequently, results of such analysis should be interpreted with caution. Most studies were performed enrolling Asian patients and the number of studies on Caucasians or Africans was limited. Therefore, more association studies with larger sample sizes on Africans and Caucasians are needed to make precise estimations of cancer risk associated with the studied polymorphisms. Assigning ethnicity to each study population could be another limitation of meta-analysis of association studies as each ethnicity may regroup several sub-populations with somewhat different genetic background. Sixth, the control groups of association studies were not uniformly defined and non-differential misclassification bias may have occurred.
In conclusion, this meta-analysis showed that mir-196a-2 rs11614913 T allele is associated with decreased cancer risk in overall population, high quality studies and studies on Asian populations. It is also associated with a decreased risk of gynecological cancers, ovarian, breast and hepatocellular cancer. Mir-149 rs2292832 was not associated with cancer risk in overall population, high quality studies, Asians or Caucasians. However, the T allele was associated with a decrease risk of gastrointestinal tract cancers under the heterozygote model and an increased risk of colorectal cancer under the recessive model.
Author Contributions
MB and AM conceived the original idea and supervised the project. JC, ZN-S-F, and ZS contributed to the literature search and data management. MB, JC, and ZN-S-F wrote the manuscript with support from all authors. MB contributed to the data analysis, interpretation of results, and data visualization with inputs from AM. EO and ZS assisted with data visualization. All authors provided critical feedback, discussed the results, and contributed to the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Karl Kelsey and Dr. Brock Christensen for providing the genotype counts of mir-196a-2 rs11614913 for their study (Christensen et al., 2010).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00186/full#supplementary-material
Abbreviations
BC, breast cancer; BlC, bladder cancer; CRC, colorectal cancer; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; GI, cancers of digestive system; GyC, Gynecologic cancers; HCC, Hepatocellular cancer; HM, Hematological malignancies; HNC, Head and neck carcinoma; HWE, Hardy-Weinberg equilibrium; HWD, Hardy-Weinberg deviation; LC, lung cancer; UG, Urogenital cancers; OC, Oral cancer; OR, Odds ratio; OvC, ovarian cancer; PC, prostate cancer.
References
Abdel-Hamid, M., Elshaer, S., and Darwish, A. (2018). Association of MicroRNA related single nucleotide polymorphisms 196A-2 and 499 with the risk of hepatocellular carcinoma in Egyptian patients. Meta Gene. 16, 139–142. doi: 10.1016/j.mgene.2018.02.007
Afsharzadeh, S. M., Mohaddes Ardebili, S. M., Seyedi, S. M., Karimian Fathi, N., and Mojarrad, M. (2017). Association between rs11614913, rs3746444, rs2910164 and occurrence of breast cancer in Iranian population. Meta Gene. 11, 20–25. doi: 10.1016/j.mgene.2016.11.004
Ahn, D. H., Rah, H., Choi, Y. K., Jeon, Y. J., Min, K. T., Kwack, K., et al. (2013). Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the korean population. Mol. Carcinogene. 52, 39–51. doi: 10.1002/mc.21962
Akkiz, H., Bayram, S., Bekar, A., Akgöllü, E., and Ülger, Y. (2011). A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J Viral Hepatitis. 18, e399–e407. doi: 10.1111/j.1365-2893.2010.01414.x
Alshatwi, A. A., Shafi, G., Hasan, T. N., Syed, N. A., Al-Hazzani, A. A., Alsaif, M. A., et al. (2012). Differential expression profile and genetic variants of MicroRNAs sequences in breast cancer patients. PLoS ONE 7:030049. doi: 10.1371/journal.pone.0030049
Attia, J., Thakkinstian, A., and D'Este, C. (2003). Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J. Clin. Epidemiol. 56, 297–303. doi: 10.1016/S0895-4356(03)00011-8
Bansal, C., Sharma, K. L., Misra, S., Srivastava, A. N., Mittal, B., and Singh, U. S. (2014). Common genetic variants in pre-microRNAs and risk of breast cancer in the North Indian population. Ecancermedicalscience 8:473. doi: 10.3332/ecancer.2014.473
Begg, C. B., and Mazumdar, M. (1994). Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101. doi: 10.2307/2533446
Bodal, V. K., Sangwan, S., Bal, M. S., Kaur, M., Sharma, S., and Kaur, B. (2017). Association between microrna 146a and microrna 196a2 genes polymorphism and breast cancer risk in north indian women. Asian Pac. J. Cancer Prev. 18, 2345–2348. doi: 10.22034/APJCP.2017.18.9.2345
Bray, F., Jemal, A., Grey, N., Ferlay, J., and Forman, D. (2012). Global cancer transitions according to the Human Development Index (2008-2030): a population-based study. Lancet Oncol. 13, 790–801. doi: 10.1016/S1470-2045(12)70211-5
Cîmpeanu, R. A., Popescu, D. M., Burada, F., Cucu, M. G., Gheonea, D. I., Ioana, M., et al. (2017). miR-149 rs2292832 C>T polymorphism and risk of gastric cancer. Roman. J. Morphol. Embryol. 58, 125–129.
Catucci, I., Yang, R., Verderio, P., Pizzamiglio, S., Heesen, L., Hemminki, K., et al. (2010). Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum. Mutat. 31, E1052–E1057. doi: 10.1002/humu.21141
Chen, H., Sun, L. Y., Chen, L. L., Zheng, H. Q., and Zhang, Q. F. (2012). A variant in microRNA-196a2 is not associated with susceptibility to and progression of colorectal cancer in Chinese. Inter. Med. J. 42, e115–e119. doi: 10.1111/j.1445-5994.2011.02434.x
Christensen, B. C., Avissar-Whiting, M., Ouellet, L. G., Butler, R. A., Nelson, H. H., McClean, M. D., et al. (2010). Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin. Cancer Res. 16, 3713–3720. doi: 10.1158/1078-0432.CCR-10-0657
Chu, H., Wang, M., Shi, D., Ma, L., Zhang, Z., Tong, N., et al. (2011). Hsa-miR-196a2 Rs11614913 polymorphism contributes to cancer susceptibility: evidence from 15 case-control studies. PLoS ONE 6:e18108. doi: 10.1371/journal.pone.0018108
Chu, Y. H., Hsieh, M. J., Chiou, H. L., Liou, Y. S., Yang, C. C., Yang, S. F., et al. (2014). MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS ONE 9:089930. doi: 10.1371/journal.pone.0089930
Chu, Y. H., Tzeng, S. L., Lin, C. W., Chien, M. H., Chen, M. K., and Yang, S. F. (2012). Impacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancer. PLoS ONE 7:39777. doi: 10.1371/journal.pone.0039777
Dai, Z. M., Kang, H. F., Zhang, W. G., Li, H. B., Zhang, S. Q., Ma, X. B., et al. (2016). The associations of single nucleotide polymorphisms in miR196a2, miR-499, and miR-608 with breast cancer susceptibility: a STROBE-compliant observational study. Medicine 95:e2826. doi: 10.1097/MD.0000000000002826
Damodaran, M., Paul, S. F. D., and Venkatesan, V. (2018). Genetic polymorphisms in miR-146a, miR-196a2 and miR-125a genes and its association in prostate cancer. Pathol. Oncol. Res. 1–8. doi: 10.1007/s12253-018-0412-x [Epub ahead of print].
Deng, S., Wang, W., Li, X., and Zhang, P. (2015). Common genetic polymorphisms in pre-microRNAs and risk of bladder cancer. World J. Surg. Oncol. 13:297. doi: 10.1186/s12957-015-0683-6
DerSimonian, R., and Laird, N. (1986). Meta-analysis in clinical trials. Control Clin. Trials 7, 177–188. doi: 10.1016/0197-2456(86)90046-2
Dikaiakos, P., Gazouli, M., Rizos, S., Zografos, G., and Theodoropoulos, G. E. (2015). Evaluation of genetic variants in miRNAs in patients with colorectal cancer. Cancer Biomarkers 15, 163–168. doi: 10.3233/CBM-140449
Dikeakos, P., Theodoropoulos, G., Rizos, S., Tzanakis, N., Zografos, G., and Gazouli, M. (2014). Association of the miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek population. Mol. Biol. Rep. 41, 1075–1080. doi: 10.1007/s11033-013-2953-0
Dong, G., Zhang, R., Xu, J., and Guo, Y. (2015). Association between microRNA polymorphisms and papillary thyroid cancer susceptibility. Int. J. Clin. Exp. Pathol. 8, 13450–13457.
Dou, T., Wu, Q., Chen, X., Ribas, J., Ni, X., Tang, C., et al. (2010). A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J. Cancer Res. Clin. Oncol. 136, 1853–1859. doi: 10.1007/s00432-010-0844-5
Doulah, A., Salehzadeh, A., and Mojarrad, M. (2018). Association of single nucleotide polymorphisms in miR-499 and miR-196a with susceptibility to breast cancer. Trop. J. Pharmaceut. Res. 17, 319–323. doi: 10.4314/tjpr.v17i2.17
Du, M., Lu, D., Wang, Q., Chu, H., Tong, N., Pan, X., et al. (2014). Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis 35, 1629–1635. doi: 10.1093/carcin/bgu082
Duval, S., and Tweedie, R. (2000a). A Nonparametric “Trim and Fill” Method of Accounting for Publication Bias in Meta-Analysis. J. Am. Stat. Assoc. 95, 89–98. doi: 10.2307/2669529
Duval, S., and Tweedie, R. (2000b). Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 56, 455–463. doi: 10.1111/j.0006-341X.2000.00455.x
Feng, Y., Duan, F., Song, C., Zhao, X., Dai, L., and Cui, S. (2016). Systematic evaluation of cancer risk associated with rs2292832 in miR-149 and rs895819 in miR-27a: a comprehensive and updated meta-analysis. Oncotarget 7, 22368–22384. doi: 10.18632/oncotarget.8082
Ferlay, J., Shin, H. R., Bray, F., Forman, D., Mathers, C., and Parkin, D. M. (2010). Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917. doi: 10.1002/ijc.25516
George, G. P., Gangwar, R., Mandal, R. K., Sankhwar, S. N., and Mittal, R. D. (2011). Genetic variation in microRNA genes and prostate cancer risk in North Indian population. Mol. Biol. Rep. 38, 1609–1615. doi: 10.1007/s11033-010-0270-4
Ghaedi, H., Bastami, M., Zare-Abdollahi, D., Alipoor, B., Movafagh, A., Mirfakhraie, R., et al. (2015). Bioinformatics prioritization of SNPs perturbing microRNA regulation of hematological malignancy-implicated genes. Genomics 106, 360–366. doi: 10.1016/j.ygeno.2015.10.004
Gu, J. Y., and Tu, L. (2016). Investigating the role of polymorphisms in miR-146a,−149, and−196a2 in the development of gastric cancer. Genetics Mol. Res. 15:gmr.15027839. doi: 10.4238/gmr.15027839
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15:509. doi: 10.1038/nrm3838
Han, Y., Pu, R., Han, X., Zhao, J., Zhang, Y., Zhang, Q., et al. (2013). Associations of pri-miR-34b/c and pre-miR-196a2 polymorphisms and their multiplicative interactions with Hepatitis B virus mutations with hepatocellular carcinoma risk. PLoS ONE 8:058564. doi: 10.1371/journal.pone.0058564
Hao, Y. X., Wang, J. P., and Zhao, L. F. (2014). Associations between three common MicroRNA polymorphisms and hepatocellular carcinoma risk in Chinese. Asian Pac. J. Cancer Prevent. 14, 6601–6604. doi: 10.7314/APJCP.2013.14.11.6601
Hashemi, M., Moradi, N., Ziaee, S. A., Narouie, B., Soltani, M. H., Rezaei, M., et al. (2016). Association between single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and miR-149 and prostate cancer risk in a sample of Iranian population. J. Adv. Res. 7, 491–498. doi: 10.1016/j.jare.2016.03.008
He, B., Pan, Y., Xu, Y., Deng, Q., Sun, H., Gao, T., et al. (2015). Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumor Biol. 36, 4575–4582. doi: 10.1007/s13277-015-3102-2
He, J., Zou, Y., Liu, X., Zhu, J., Zhang, J., Zhang, R., et al. (2018). Association of common genetic variants in pre-microRNAs and neuroblastoma susceptibility: a two-center study in chinese children. Mol. Ther. Nucleic Acids 11, 1–8. doi: 10.1016/j.omtn.2018.01.003
He, Y., Yu, D., Zhu, L., Zhong, S., Zhao, J., and Tang, J. (2018). miR-149 in human cancer: a systemic review. J. Cancer 9, 375–388. doi: 10.7150/jca.21044
Hezova, R., Kovarikova, A., Bienertova-Vasku, J., Sachlova, M., Redova, M., Vasku, A., et al. (2012). Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J. Gastroenterol. 18, 2827–2831. doi: 10.3748/wjg.v18.i22.2827
Hoffman, A. E., Zheng, T., Yi, C., Leaderer, D., Weidhaas, J., Slack, F., et al. (2009). microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 69, 5970–5977. doi: 10.1158/0008-5472.CAN-09-0236
Hong, Y. S., Kang, H. J., Kwak, J. Y., Park, B. L., You, C. H., Kim, Y. M., et al. (2011). Association between MicroRNA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean population. J. Prevent. Med. Public Health 44, 125–130. doi: 10.3961/jpmph.2011.44.3.125
Horikawa, Y., Wood, C. G., Yang, H., Zhao, H., Ye, Y., Gu, J., et al. (2008). Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res. 14, 7956–7962. doi: 10.1158/1078-0432.CCR-08-1199
Hu, E., Wang, D., Zhang, X., Li, J., Hu, Y., Gong, H., et al. (2013). Four common polymorphisms in microRNAs and the risk of adult glioma in a Chinese case-control study. J. Mol. Neurosci. 51, 933–940. doi: 10.1007/s12031-013-9980-0
Hu, Z., Chen, J., Tian, T., Zhou, X., Gu, H., Xu, L., et al. (2008). Genetic variants of miRNA sequences and non–small cell lung cancer survival. J. Clin. Invest. 118, 2600–2608. doi: 10.1172/JCI34934
Hu, Z., Liang, J., Wang, Z., Tian, T., Zhou, X., Chen, J., et al. (2009). Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 30, 79–84. doi: 10.1002/humu.20837
Huang, G. L., Lu, Y., Pu, X. X., He, Y. X., Chen, M. L., Li, Y. Z., et al. (2013). Association study between miR-149 gene polymorphism and nasopharyngeal carcinoma. Biomed. Rep. 1, 599–603. doi: 10.3892/br.2013.97
Huang, Y., Sheng, S., Chen, B., Lin, R., Yang, J., and Hao, B. (2017). MiR-146a genetic polymorphism contributes to the susceptibility to hepatocellular carcinoma in a Chinese population. Int. J. Clin. Exp. Pathol. 10, 1833–1839.
Jedlinski, D. J., Gabrovska, P. N., Weinstein, S. R., Smith, R. A., and Griffiths, L. R. (2011). Single nucleotide polymorphism in hsa-mir-196a-2 and breast cancer risk: a case control study. Twin Res. Hum. Genetics 14, 417–421. doi: 10.1375/twin.14.5.417
Jiang, J., Jia, Z. F., Cao, D. H., Wu, Y. H., Sun, Z. W., and Cao, X. Y. (2016). Association of the MIR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. Future Oncol. 12, 2215–2226. doi: 10.2217/fon-2016-0224
Kim, M. J., Yoo, S. S., Choi, Y. Y., and Park, J. Y. (2010). A functional polymorphism in the pre-microRNA-196a2 and the risk of lung cancer in a Korean population. Lung Cancer 69, 127–129. doi: 10.1016/j.lungcan.2010.04.015
Kim, W. H., Min, K. T., Jeon, Y. J., Kwon, C.-I., Ko, K. H., Park, P. W., et al. (2012). Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene 504, 92–97. doi: 10.1016/j.gene.2012.05.014
Kontorovich, T., Levy, A., Korostishevsky, M., Nir, U., and Friedman, E. (2010). Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int. J. Cancer 127, 589–597. doi: 10.1002/ijc.25065
Kou, J. T., Fan, H., Han, D., Li, L., Li, P., Zhu, J., et al. (2014). Association between four common microRNA polymorphisms and the risk of hepatocellular carcinoma and HBV infection. Oncol. Lett. 8, 1255–1260. doi: 10.3892/ol.2014.2257
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.1093/nar/gkt1181
Kupcinskas, J., Bruzaite, I., Juzenas, S., Gyvyte, U., Jonaitis, L., Kiudelis, G., et al. (2014a). Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci. Rep. 4:5993. doi: 10.1038/srep05993
Kupcinskas, J., Wex, T., Link, A., Leja, M., Bruzaite, I., Steponaitiene, R., et al. (2014b). Gene polymorphisms of micrornas in helicobacter pylori-induced high risk atrophic gastritis and gastric cancer. PLoS ONE 9:087467. doi: 10.1371/journal.pone.0087467
Lau, J., Ioannidis, J. P., and Schmid, C. H. (1997). Quantitative synthesis in systematic reviews. Ann. Intern. Med. 127, 820–826. doi: 10.7326/0003-4819-127-9-199711010-00008
Li, H., Ren, Y., Xia, L., Qu, R., Kong, L., Yin, Z., et al. (2016). Association of microrna-149 polymorphism with lung cancer risk in Chinese non-smoking female: a case-control study. PLoS ONE 11:163626. doi: 10.1371/journal.pone.0163626
Li, J., Cheng, G., and Wang, S. (2016). A single-nucleotide polymorphism of miR-196a2T>C rs11614913 is associated with hepatocellular carcinoma in the chinese population. Genetic Test. Mol. Biomarkers 20, 213–215. doi: 10.1089/gtmb.2015.0271
Li, L., Liu, T., Li, Z., Zhang, L., and Zhang, Z. (2015). The miR-149 rs2292832 T/C polymorphism may decrease digestive cancer susceptibility: an updated meta-analysis. Int. J. Clin. Exp. Med. 8, 15351–15361.
Li, L., Sheng, Y., Lv, L., and Gao, J. (2013). The association between two MicroRNA variants (miR-499, miR-149) and gastrointestinal cancer risk: a meta-analysis. PLOS ONE 8:e81967. doi: 10.1371/journal.pone.0081967
Li, M., Li, R. J., Bai, H., Xiao, P., Liu, G. J., Guo, Y. W., et al. (2016). Association between the pre-miR-196a2 rs11614913 polymorphism and gastric cancer susceptibility in a Chinese population. Genetics Mol. Res. 15:gmr.15027516. doi: 10.4238/gmr.15027516
Li, M., Marin-Muller, C., Bharadwaj, U., Chow, K.-H., Yao, Q., and Chen, C. (2009). MicroRNAs: control and loss of control in human physiology and disease. World J. Surg. 33, 667–684. doi: 10.1007/s00268-008-9836-x
Li, P., Yan, H., Zhang, H., Yu, L., Wang, Z., Zhai, Y., et al. (2014). A functional polymorphism in MIR196A2 is associated with risk and progression of nasopharyngeal carcinoma in the Chinese population. Genetic Test. Mol. Biomarkers 18, 149–155. doi: 10.1089/gtmb.2013.0400
Li, T., Niu, L., Wu, L., Gao, X., Li, M., Liu, W., et al. (2015). A functional polymorphism in microRNA-196a2 is associated with increased susceptibility to non-Hodgkin lymphoma. Tumor Biol. 36, 3279–3284. doi: 10.1007/s13277-014-2957-y
Li, X., Li, K., and Wu, Z. (2015). Association of four common SNPs in microRNA polymorphisms with the risk of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 8, 9560–9566.
Li, X.-D., Li, Z.-G., Song, X.-X., and Liu, C.-F. (2010). A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosis. Pathology 42, 669–673. doi: 10.3109/00313025.2010.522175
Liberati, A., Altman, D. G., Tetzlaff, J., Mulrow, C., Gøtzsche, P. C., Ioannidis, J. P. A., et al. (2009). The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 339:b2700. doi: 10.1136/bmj.b2700
Linhares, J. J., Azevedo, M. Jr, Siufi, A. A., de Carvalho, C. V., Wolgien, M. D. C. G. M., Noronha, E. C., et al. (2012). Evaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancer. BMC Med. Genetics 13:119. doi: 10.1186/1471-2350-13-119
Liu, C. J., Tsai, M. M., Tu, H. F., Lui, M. T., Cheng, H. W., and Lin, S. C. (2013). MiR-196a overexpression and mir-196a2 gene polymorphism are prognostic predictors of oral carcinomas. Ann. Surg. Oncol. 20, S406–S414. doi: 10.1245/s10434-012-2618-6
Liu, M. F., Chen, W. Q., He, Y. Z., and Gu, Y. L. (2014). Role of miR-149C>T polymorphisms on the risk of hepatocellular carcinoma in aChinese population. Genetics Mol. Res. 13, 7184–7189. doi: 10.4238/2014.September.5.4
Liu, X. Y. (2015). Association of SNPS in miR-146a, miR-196a2 and miR-499 with risk of endometrial/ovarian cancer. Clin. Chem. Lab. Med. 53:S400. doi: 10.1093/abbs/gmv042
Liu, Y., He, A., Liu, B., Zhong, Y., Liao, X., Yang, J., et al. (2018). rs11614913 polymorphism in miRNA-196a2 and cancer risk: an updated meta-analysis. OncoTargets Ther. 11, 1121–1139. doi: 10.2147/OTT.S154211
Liu, Z., Li, G., Wei, S., Niu, J., El-Naggar, A. K., Sturgis, E. M., et al. (2010). Genetic variants in selected pre-microrna genes and the risk of squamous cell carcinoma of the head and neck. Cancer 116, 4753–4760. doi: 10.1002/cncr.25323
Lu, Y.-C., Chang, J. T., Chan, E.-C., Chao, Y.-K., Yeh, T.-S., Chen, J.-S., et al. (2016). miR-196, an emerging cancer biomarker for digestive tract cancers. J. Cancer 7, 650–655. doi: 10.7150/jca.13460
Lv, M., Dong, W., Li, L., Zhang, L., Su, X., Wang, L., et al. (2013). Association between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese population. J. Cancer Res. Clin. Oncol. 139, 1405–1410. doi: 10.1007/s00432-013-1456-7
Ma, F., Zhang, P., Lin, D., Yu, D., Yuan, P., Wang, J., et al. (2013). There is no association between microRNA gene polymorphisms and risk of triple negative breast cancer in a Chinese Han population. PLoS ONE 8:e60195. doi: 10.1371/journal.pone.0060195
Mantel, N., and Haenszel, W. (1959). Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748.
Martin-Guerrero, I., Gutierrez-Camino, A., Lopez-Lopez, E., Bilbao-Aldaiturriaga, N., Pombar-Gomez, M., Ardanaz, M., et al. (2015). Genetic variants in MiRNA processing genes and Pre-MiRNAs are associated with the risk of chronic lymphocytic leukemia. PLoS ONE 10:0118905. doi: 10.1371/journal.pone.0118905
Mashayekhi, S., Saeidi Saedi, H., Salehi, Z., Soltanipour, S., and Mirzajani, E. (2018). Effects of miR-27a, miR-196a2 and miR-146a polymorphisms on the risk of breast cancer. Br. J. Biomed. Sci. 75, 76–81. doi: 10.1080/09674845.2017.1399572
Miao, L., Wang, L., Zhu, L., Du, J., Zhu, X., Niu, Y., et al. (2016). Association of microRNA polymorphisms with the risk of head and neck squamous cell carcinoma in a Chinese population: a case-control study. Chin. J. Cancer 35:77. doi: 10.1186/s40880-016-0136-9
Min, K. T., Kim, J. W., Jeon, Y. J., Jang, M. J., Chong, S. Y., Oh, D., et al. (2012). Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol. Carcinogenesis 51, E65–E73. doi: 10.1002/mc.21849
Minelli, C., Thompson, J. R., Abrams, K. R., Thakkinstian, A., and Attia, J. (2008). How should we use information about HWE in the meta-analyses of genetic association studies? Int. J. Epidemiol. 37, 136–146. doi: 10.1093/ije/dym234
Minh, T. T. H., Thanh, N. T. N., Van Thiep, T., and Hue, N. T. (2018). “Association between single nucleotide polymorphism Rs11614913 (C>T) on Mir-196a2 and breast cancer in vietnamese population,” in 6th International Conference on the Development of Biomedical Engineering in Vietnam (BME6), Vol. 63, eds T. Vo Van, T. Nguyen Le, and T. Nguyen Duc, BME 2017. IFMBE Proceedings (Singapore: Springer).
Mittal, R. D., Gangwar, R., George, G. P., Mittal, T., and Kapoor, R. (2011). Investigative role of Pre-MicroRNAs in bladder cancer patients: a case-control study in North India. DNA Cell Biol. 30, 401–406. doi: 10.1089/dna.2010.1159
Morales, S., Gulppi, F., Gonzalez-Hormazabal, P., Fernandez-Ramires, R., Bravo, T., Reyes, J. M., et al. (2016). Association of single nucleotide polymorphisms in Pre-miR-27a, Pre-miR-196a2, Pre-miR-423, miR-608 and Pre-miR-618 with breast cancer susceptibility in a South American population. BMC Genetics 17:109. doi: 10.1186/s12863-016-0415-0
Nariman-Saleh-Fam, Z., Bastami, M., Somi, M. H., Behjati, F., Mansoori, Y., Daraei, A., et al. (2017). miRNA-related polymorphisms in miR-423 (rs6505162) and PEX6 (rs1129186) and risk of esophageal squamous cell carcinoma in an iranian cohort. Genet. Test. Mol. Biomarkers 21, 382–390. doi: 10.1089/gtmb.2016.0346
Nariman-Saleh-Fam, Z., Bastami, M., Somi, M. H., Samadi, N., Abbaszadegan, M. R., Behjati, F., et al. (2016). In silico dissection of miRNA targetome polymorphisms and their role in regulating miRNA-mediated gene expression in esophageal cancer. Cell Biochem. Biophys. 74, 483–497. doi: 10.1007/s12013-016-0754-5
Ni, J., and Huang, Y. (2016). Role of polymorphisms in miR-146a, miR-149, miR-196a2 and miR-499 in the development of ovarian cancer in a Chinese population. Int. J. Clin. Exp. Pathol. 9, 5706–5711.
Nikolić, Z., Savić Pavićević, D., Vucić, N., Cidilko, S., Filipovic, N., Cerovic, S., et al. (2015). Assessment of association between genetic variants in microRNA genes hsa-miR-499, hsa-miR-196a2 and hsa-miR-27a and prostate cancer risk in Serbian population. Exp. Mol. Pathol. 99, 145–150. doi: 10.1016/j.yexmp.2015.06.009
Okubo, M., Tahara, T., Shibata, T., Yamashita, H., Nakamura, M., Yoshioka, D., et al. (2010). Association between common genetic variants in Pre-microRNAs and gastric cancer risk in japanese population. Helicobacter 15, 524–531. doi: 10.1111/j.1523-5378.2010.00806.x
Omrani, M., Hashemi, M., Eskandari-Nasab, E., Hasani, S. S., Mashhadi, M. A., Arbabi, F., et al. (2014). Hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomarkers Med. 8, 259–267. doi: 10.2217/bmm.13.118
Osada, H., and Takahashi, T. (2007). MicroRNAs in biological processes and carcinogenesis. Carcinogenesis 28, 2–12. doi: 10.1093/carcin/bgl185
Ow, S. H., Chua, P. J., and Bay, B. H. (2018). miR-149 as a Potential Molecular Target for Cancer. Curr. Med. Chem. 25, 1046–1054. doi: 10.2174/0929867324666170718102738
Pan, W., Wu, C., Su, Z., Duan, Z., Li, L., Mi, F., et al. (2017). Genetic polymorphisms of non-coding RNAs associated with increased head and neck cancer susceptibility: a systematic review and meta-analysis. Oncotarget 8, 62508–62523. doi: 10.18632/oncotarget.20096
Parlayan, C., Ikeda, S., Sato, N., Sawabe, M., Muramatsu, M., and Arai, T. (2014). Association analysis of single nucleotide polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer in elderly Japanese: a case-control study. Asian Pac. J. Cancer Prevent. 15, 2101–2107. doi: 10.7314/APJCP.2014.15.5.2101
Pavlakis, E., Papaconstantinou, I., Gazouli, M., Theodosopoulos, T., Karamanolis, G., Genatas, K., et al. (2013). MicroRNA gene polymorphisms in pancreatic cancer. Pancreatology 13, 273–278. doi: 10.1016/j.pan.2013.02.005
Peckham-Gregory, E. C., Thapa, D. R., Martinson, J., Duggal, P., Penugonda, S., Bream, J. H., et al. (2016). MicroRNA-related polymorphisms and non-Hodgkin lymphoma susceptibility in the Multicenter AIDS Cohort Study. Cancer Epidemiol. 45, 47–57. doi: 10.1016/j.canep.2016.09.007
Peng, S., Kuang, Z., Sheng, C., Zhang, Y., Xu, H., and Cheng, Q. (2010). Association of MicroRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Digest. Dis. Sci. 55, 2288–2293. doi: 10.1007/s10620-009-1007-x
Peng, Y., and Croce, C. M. (2016). The role of MicroRNAs in human cancer. Signal Transduct. Targeted Ther. 1:15004. doi: 10.1038/sigtrans.2015.4
Peters, J. L., Sutton, A. J., Jones, D. R., Abrams, K. R., and Rushton, L. (2007). Performance of the trim and fill method in the presence of publication bias and between-study heterogeneity. Stat. Med. 26, 4544–4562. doi: 10.1002/sim.2889
Poltronieri-Oliveira, A. B., Madeira, F. F., Nunes, D. B. S. M., Rodrigues, G. H., Lopes, B. C., Manoel-Caetano, F. S., et al. (2017). Polymorphisms of miR-196a2 (rs11614913) and miR-605 (rs2043556) confer susceptibility to gastric cancer. Gene Rep. 7, 154–163. doi: 10.1016/j.genrep.2017.04.006
Pratedrat, P., Sopipong, W., Makkoch, J., Praianantathavorn, K., Chuaypen, N., Tangkijvanich, P., et al. (2015). Single nucleotide polymorphisms in miR-149 (rs2292832) and miR-101-1 (rs7536540) are not associated with hepatocellular carcinoma in thai patients with Hepatitis B virus infection. Asian Pac. J. Cancer Prevent. 16, 6457–6461. doi: 10.7314/APJCP.2015.16.15.6457
Pu, J. Y., Dong, W., Zhang, L., Liang, W. B., Yang, Y., and Lv, M. L. (2014). No association between single nucleotide polymorphisms in pre-mirnas and the risk of gastric cancer in Chinese population. Iran. J. Basic Med. Sci. 17, 128–133. doi: 10.22038/IJBMS.2014.2246
Qi, J. H., Wang, J., Chen, J., Shen, F., Huang, J. T., Sen, S., et al. (2014). High-resolution melting analysis reveals genetic polymorphisms in MicroRNAs confer hepatocellular carcinoma risk in Chinese patients. BMC Cancer 14:643. doi: 10.1186/1471-2407-14-643
Qi, P., Dou, T.-h., Geng, L., Zhou, F.-g., Gu, X., Wang, H., et al. (2010). Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum. Immunol. 71, 621–626. doi: 10.1016/j.humimm.2010.02.017
Qi, P., Wang, L., Zhou, B., Yao, W. J., Xu, S., Zhou, Y., et al. (2015). Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet. Mol. Res. 14, 6289–6296. doi: 10.4238/2015.June.11.2
Qu, Y., Qu, H., Luo, M., Wang, P., Song, C., Wang, K., et al. (2014). MicroRNAs related polymorphisms and genetic susceptibility to esophageal squamous cell carcinoma. Mol. Genet. Genom. 289, 1123–1130. doi: 10.1007/s00438-014-0873-x
Rakmanee, S., Pakakasama, S., Hongeng, S., Sanguansin, S., Thongmee, A., and Pongstaporn, W. (2017). Increased risk of Thai childhood acute lymphoblastic leukemia with the MiR196a2 T > C polymorphism. Asian Pac. J. Cancer Prevent. 18, 1117–1120. doi: 10.22034/APJCP.2017.18.4.1117
Ranjbar, R., Chaleshi, V., Aghdaei, H. A., and Morovvati, S. (2018). Investigating the association between miR-608 rs4919510 and miR-149 rs2292832 with Colorectal Cancer in Iranian Population. Microrna 7, 100–106 doi: 10.2174/2211536607666180206145540
Ren, Y.-G., Zhou, X.-M., Cui, Z.-G., and Hou, G. (2016). Effects of common polymorphisms in miR-146a and miR-196a2 on lung cancer susceptibility: a meta-analysis. J. Thoracic Dis. 8, 1297–1305. doi: 10.21037/jtd.2016.05.02
Rogoveanu, I., Burada, F., Cucu, M. G., Vere, C. C., Ioana, M., and Cîmpeanu, R. A. (2017). Association of microRNA polymorphisms with the risk of gastric cancer in a Romanian population. J. Gastrointest. Liver Dis. 26, 231–238. doi: 10.15403/jgld.2014.1121.263.rog
Rong, G.-Q., Zhang, X.-M., Chen, B., Yang, X.-D., Wu, H.-R., and Gong, W. (2017). MicroRNA gene polymorphisms and the risk of colorectal cancer. Oncol. Lett. 13, 3617–3623. doi: 10.3892/ol.2017.5885
Roy, R., De Sarkar, N., Ghose, S., Paul, R. R., Pal, M., Bhattacharya, C., et al. (2014). Genetic variations at microRNA and processing genes and risk of oral cancer. Tumour Biol. 35, 3409–3414. doi: 10.1007/s13277-013-1450-3
Shen, F., Chen, J., Guo, S., Zhou, Y., Zheng, Y., Yang, Y., et al. (2016). Genetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han population. Tumor Biol. 37, 4777–4784. doi: 10.1007/s13277-015-4268-3
Sodhi, K. K., Bahl, C., Singh, N., Behera, D., and Sharma, S. (2015). Functional genetic variants in pre-MIR-146a and 196a2 genes are associated with risk of lung cancer in North Indians. Future Oncol. 11, 2159–2173. doi: 10.2217/fon.15.143
Song, Z. S., Wu, Y., Zhao, H. G., Liu, C. X., Cai, H. Y., Guo, B. Z., et al. (2016). Association between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancer. Oncol. Lett. 11, 194–200. doi: 10.3892/ol.2015.3877
Srivastava, K., and Srivastava, A. (2012). Comprehensive review of genetic association studies and meta-analyses on miRNA polymorphisms and cancer risk. PLoS ONE 7:e50966. doi: 10.1371/journal.pone.0050966
Srivastava, K., Srivastava, A., and Mittal, B. (2010). Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J. Hum. Genet. 55, 495–499. doi: 10.1038/jhg.2010.54
Srivastava, S., Singh, S., Fatima, N., Mittal, B., and Srivastava, A. N. (2017). Pre-microrna gene polymorphisms and risk of cervical squamous cell carcinoma. J. Clin. Diagnostic Res. 11, GC01–GC04. doi: 10.7860/JCDR/2017/25361.10543
Su, R., Li, W., and Luo, R. (2016). Association between miR-146a, miR-149, miR-196a2 and miR-499 gene polymorphisms and the susceptibility to gastric cancer in a Chinese population. Int. J. Clin. Exp. Pathol. 9, 2192–2199.
Sun, X. C., Zhang, A. C., Tong, L. L., Wang, K., Wang, X., Sun, Z. Q., et al. (2016). miR-146a and miR-196a2 polymorphisms in ovarian cancer risk. Genet. Mol. Res. 15:gmr.15038468. doi: 10.4238/gmr.15038468
Sushma, P. S., Jamil, K., Uday Kumar, P., Satyanarayana, U., Ramakrishna, M., and Triveni, B. (2015). Genetic variation in microRNAs and risk of oral squamous cell carcinoma in South Indian population. Asian Pac. J. Cancer Prevent. 16, 7589–7594. doi: 10.7314/APJCP.2015.16.17.7589
Tandon, D., Dewangan, J., Srivastava, S., Garg, V. K., and Rath, S. K. (2017). MiRNA genetic variants: as potential diagnostic biomarkers for oral cancer. Pathol. Res. Pract. 214, 281–289. doi: 10.1016/j.prp.2017.10.002
Thakkinstian, A., McElduff, P., D'Este, C., Duffy, D., and Attia, J. (2005). A method for meta-analysis of molecular association studies. Stat. Med. 24, 1291–1306. doi: 10.1002/sim.2010
Thakkinstian, A., McKay, G. J., McEvoy, M., Chakravarthy, U., Chakrabarti, S., Silvestri, G., et al. (2011). Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am. J. Epidemiol. 173, 1365–1379. doi: 10.1093/aje/kwr025
Tian, T., Shu, Y., Chen, J., Hu, Z., Xu, L., Jin, G., et al. (2009). A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol. Biomarkers Prevent. 18, 1183–1187. doi: 10.1158/1055-9965.EPI-08-0814
Tong, N., Xu, B., Shi, D., Du, M., Li, X., Sheng, X., et al. (2014). Hsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese children. Mutat. Res. 759, 16–21. doi: 10.1016/j.mrfmmm.2013.11.004
Toraih, E. A., Fawz, M. S., Elgazzaz, M. G., Hussein, M. H., Shehata, R. H., and Daoud, H. G. (2016a). Combined genotype analyses of precursor miRNA196a2 and 499a variants with hepatic and renal cancer susceptibility a preliminary study. Asian Pac. J. Cancer Prevent. 17, 3369–3375.
Toraih, E. A., Fawzy, M. S., Mohammed, E. A., Hussein, M. H., and EL-Labban, M. M. (2016b). MicroRNA-196a2 biomarker and targetome network analysis in solid tumors. Mol. Diagn. Ther. 20, 559–577. doi: 10.1007/s40291-016-0223-2
Trikalinos, T. A., Salanti, G., Khoury, M. J., and Ioannidis, J. P. (2006). Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am. J. Epidemiol. 163, 300–309. doi: 10.1093/aje/kwj046
Tu, H. F., Liu, C. J., Chang, C. L., Wang, P. W., Kao, S. Y., Yang, C. C., et al. (2012). The association between genetic polymorphism and the processing efficiency of miR-149 affects the prognosis of patients with head and neck squamous cell carcinoma. PLoS ONE 7:051606. doi: 10.1371/journal.pone.0051606
Umar, M., Upadhyay, R., Prakash, G., Kumar, S., Ghoshal, U. C., and Mittal, B. (2013). Evaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancer. Mol. Carcinogenesis 52, 10–18. doi: 10.1002/mc.21931
Vinci, S., Gelmini, S., Mancini, I., Malentacchi, F., Pazzagli, M., Beltrami, C., et al. (2013). Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods 59, 138–146. doi: 10.1016/j.ymeth.2012.09.002
Vinci, S., Gelmini, S., Pratesi, N., Conti, S., Malentacchi, F., Simi, L., et al. (2011). Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin. Chem. Lab. Med. 49, 2073–2080. doi: 10.1515/CCLM.2011.708
Wang, J., Zhang, Y., Zhang, Y., and Chen, L. (2016). Correlation between miRNA-196a2 and miRNA-499 polymorphisms and bladder cancer. Int. J. Clin. Exp. Med. 9, 20484–20488.
Wang, K., Guo, H., Hu, H., Xiong, G., Guan, X., Li, J., et al. (2010). A functional variation in pre-microRNA-196a is associated with susceptibility of esophageal squamous cell carcinoma risk in Chinese Han. Biomarkers 15, 614–618. doi: 10.3109/1354750X.2010.505299
Wang, N., Li, Y., Zhou, R. M., Wang, G. Y., Wang, C. M., Chen, Z. F., et al. (2014). Hsa-miR-196a2 functional SNP is associated with the risk of ESCC in individuals under 60 years old. Biomarkers 19, 43–48. doi: 10.3109/1354750X.2013.866164
Wang, R., Zhang, J., Ma, Y., Chen, L., Guo, S., Zhang, X., et al. (2014). Association study of miR149 rs2292832 and miR608 rs4919510 and the risk of hepatocellular carcinoma in a largescale population. Mol. Med. Rep. 10, 2736–2744. doi: 10.3892/mmr.2014.2536
Wang, S., Tao, G., Wu, D., Zhu, H., Gao, Y., Tan, Y., et al. (2013). A functional polymorphism in MIR196A2 is associated with risk and prognosis of gastric cancer. Mol. Carcinogenesis 52, 87–95. doi: 10.1002/mc.22017
Wang, X. B., Cui, N. H., Yang, J., Qiu, X. P., Gao, J. J., Yang, N., et al. (2014). Angiotensin-converting enzyme insertion/deletion polymorphism is not a major determining factor in the development of sporadic alzheimer disease: evidence from an updated meta-analysis. PLoS ONE 9:e111406. doi: 10.1371/journal.pone.0111406
Wang, X. H., Wang, F. R., Tang, Y. F., Zou, H. Z., and Zhao, Y. Q. (2014). Association of miR-149C>T and miR-499A>G polymorphisms with the risk of hepatocellular carcinoma in the Chinese population. Genetics Mol. Res. 13, 5048–5054. doi: 10.4238/2014.July.4.20
Wang, Y.-H., Hu, H.-N., Weng, H., Chen, H., Luo, C.-L., Ji, J., et al. (2017). Association between polymorphisms in MicroRNAs and risk of urological cancer: a meta-analysis based on 17,019 subjects. Front. Physiol. 8:325. doi: 10.3389/fphys.2017.00975
Wei, J., Zheng, L., Liu, S., Yin, J., Wang, L., Wang, X., et al. (2013). MiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese population. Hum. Immunol. 74, 1199–1205. doi: 10.1016/j.humimm.2013.06.012
Wei, W. J., Lu, Z. W., Li, D. S., Wang, Y., Zhu, Y. X., Wang, Z. Y., et al. (2014). Association of the miR-149 Rs2292832 polymorphismwith papillary thyroid cancer risk and clinicopathologic characteristics in a Chinese population. Int. J. Mol. Sci. 15, 20968–20981. doi: 10.3390/ijms151120968
Xie, M., Li, Y., Wu, J., and Wu, J. (2015). A risk of digestive tract neoplasms susceptibility in miR-146a and miR-196a2. Famil. Cancer 14, 229–239. doi: 10.1007/s10689-014-9776-6
Xue, W., Zhu, M., Wang, Y., He, J., and Zheng, L. (2015). Association between PLCE1 rs2274223 A > G polymorphism and cancer risk: proof from a meta-analysis. Sci. Rep. 5:7986. doi: 10.1038/srep07986
Yan, P., Xia, M., Gao, F., Tang, G., Zeng, H., Yang, S., et al. (2015). Predictive role of miR-146a rs2910164 (C>G), miR-149 rs2292832 (T>C), miR-196a2 rs11614913 (T>C) and miR-499 rs3746444 (T>C) in the development of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 8, 15177–15183.
Yan, W., Gao, X., and Zhang, S. (2017). Association of miR-196a2 rs11614913 and miR-499 rs3746444 polymorphisms with cancer risk: a meta-analysis. Oncotarget 8, 114344–114359. doi: 10.18632/oncotarget.22547
Yang, H., Dinney, C. P., Ye, Y., Zhu, Y., Grossman, H. B., and Wu, X. (2008). Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 68, 2530–2537. doi: 10.1158/0008-5472.CAN-07-5991
Yang, H., Lu, Q. L., Wu, X. J., Ma, H. Y., Qu, Y. Y., Zhang, D. Z., et al. (2016). Association of genetic variations in miR-146a rs2910164 and mir-149 rs11614913 with the development of classic Kaposi sarcoma. Genetics Mol. Res. 15:gmr.15048855. doi: 10.4238/gmr15048855
Yin, Z., Cui, Z., Guan, P., Li, X., Wu, W., Ren, Y., et al. (2015). Interaction between polymorphisms in pre-miRNA genes and cooking oil fume exposure on the risk of lung cancer in Chinese non-smoking female population. PLoS ONE 10:0128572. doi: 10.1371/journal.pone.0128572
Yin, Z., Cui, Z., Ren, Y., Xia, L., Li, H., and Zhou, B. (2017). MiR-196a2 and lung cancer in Chinese non-smoking females: a genetic association study and expression analysis. Oncotarget 8, 70890–70898. doi: 10.18632/oncotarget.20174
Yin, Z., Cui, Z., Ren, Y., Xia, L., Wang, Q., Zhang, Y., et al. (2016). Association between polymorphisms in pre-miRNA genes and risk of lung cancer in a Chinese non-smoking female population. Lung Cancer 94, 15–21. doi: 10.1016/j.lungcan.2016.01.013
Yoon, K.-A., Yoon, H., Park, S., Jang, H.-J., Zo, J. I., Lee, H.-S., et al. (2012). The prognostic impact of microRNA sequence polymorphisms on the recurrence of patients with completely resected non–small cell lung cancer. J. Thoracic Cardiovasc. Surg. 144, 794–807. doi: 10.1016/j.jtcvs.2012.06.030
Zhan, J.-f., Chen, L.-h., Chen, Z.-x., Yuan, Y.-w., Xie, G.-z., Sun, A.-m., et al. (2011). A Functional Variant in MicroRNA-196a2 Is Associated with Susceptibility of Colorectal Cancer in a Chinese Population. Arch. Med. Res. 42, 144–148. doi: 10.1016/j.arcmed.2011.04.001
Zhang, E., Xu, Z., Duan, W., Huang, S., and Lu, L. (2017). Association between polymorphisms in premiRNA genes & risk of oral squamous cell cancer in a Chinese population. PLoS ONE 12:0176044. doi: 10.1371/journal.pone.0176044
Zhang, H., Su, Y.-L., Yu, H., and Qian, B.-Y. (2012). Meta-Analysis of the Association between Mir-196a-2 Polymorphism and Cancer Susceptibility. Cancer Biol. Med. 9, 63–72. doi: 10.3969/j.issn.2095-3941.2012.01.012
Zhang, H., Zhang, Y., Yan, W., Wang, W., Zhao, X., Ma, X., et al. (2017). Association between three functional microRNA polymorphisms (miR-499 rs3746444, miR-196a rs11614913 and miR-146a rs2910164) and breast cancer risk: a meta-analysis. Oncotarget 8, 393–407. doi: 10.18632/oncotarget.13426
Zhang, J., Wang, R., Ma, Y. Y., Chen, L. Q., Jin, B. H., Yu, H., et al. (2013). Association between single nucleotide polymorphisms in miRNA196a-2 and miRNA146a and susceptibility to hepatocellular carcinoma in a Chinese population. Asian Pac. J. Cancer Prevent. 14, 6427–6431. doi: 10.7314/APJCP.2013.14.11.6427
Zhang, L. H., Hao, B. B., Zhang, C. Y., Dai, X. Z., and Zhang, F. (2016). Contributions of polymorphisms in mir146a, mir196a, and mir499 to the development of hepatocellular carcinoma. Genet. Mol. Res. 15:gmr.15038582. doi: 10.4238/gmr.15038582
Zhang, M., Jin, M., Yu, Y., Zhang, S., Wu, Y., Liu, H., et al. (2012). Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur. J. Cancer Care 21, 274–280. doi: 10.1111/j.1365-2354.2011.01308.x
Zhang, M. W., Jin, M. J., Yu, Y. X., Zhang, S. C., Liu, B., Jiang, X., et al. (2012). Associations of lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene polymorphisms with gastrointestinal cancer risk. Mol. Carcinog 51(Suppl. 1), E21–31. doi: 10.1002/mc.20863
Zhao, H., Xu, J., Zhao, D., Geng, M., Ge, H., Fu, L., et al. (2016). Somatic Mutation of the SNP rs11614913 and Its Association with Increased MIR 196A2 Expression in Breast Cancer. DNA Cell Biol. 35, 81–87. doi: 10.1089/dna.2014.2785
Zhou, B., Dong, L. P., Jing, X. Y., Li, J. S., Yang, S. J., Wang, J. P., et al. (2014). Association between miR-146aG>C and miR-196a2C>T polymorphisms and the risk of hepatocellular carcinoma in a Chinese population. Tumor Biol. 35, 7775–7780. doi: 10.1007/s13277-014-2020-z
Zhou, B., Wang, K., Wang, Y., Xi, M., Zhang, Z., Song, Y., et al. (2011). Common genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinoma. Mol. Carcinogenesis 50, 499–505. doi: 10.1002/mc.20740
Zhu, L., Chu, H., Gu, D., Ma, L., Shi, D., Zhong, D., et al. (2012). A functional polymorphism in miRNA-196a2 is associated with colorectal cancer risk in a Chinese population. DNA Cell Biol. 31, 349–353. doi: 10.1089/dna.2011.1348
Zintzaras, E. (2008). Variance estimation of allele-based odds ratio in the absence of Hardy-Weinberg equilibrium. Eur. J. Epidemiol. 23, 323–326. doi: 10.1007/s10654-008-9242-6
Zintzaras, E., Koufakis, T., Ziakas, P. D., Rodopoulou, P., Giannouli, S., and Voulgarelis, M. (2006). A meta-analysis of genotypes and haplotypes of methylenetetrahydrofolate reductase gene polymorphisms in acute lymphoblastic leukemia. Eur. J. Epidemiol. 21, 501–510. doi: 10.1007/s10654-006-9027-8
Keywords: microRNA, polymorphism, meta-analysis, cancer, mir-196a-2, mir-149
Citation: Choupani J, Nariman-Saleh-Fam Z, Saadatian Z, Ouladsahebmadarek E, Masotti A and Bastami M (2019) Association of mir-196a-2 rs11614913 and mir-149 rs2292832 Polymorphisms With Risk of Cancer: An Updated Meta-Analysis. Front. Genet. 10:186. doi: 10.3389/fgene.2019.00186
Received: 29 July 2018; Accepted: 19 February 2019;
Published: 15 March 2019.
Edited by:
Yujing Li, Emory University, United StatesReviewed by:
Peter Igaz, Semmelweis University, HungaryGraziella Curtale, The Scripps Research Institute, United States
Copyright © 2019 Choupani, Nariman-Saleh-Fam, Saadatian, Ouladsahebmadarek, Masotti and Bastami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Masotti, YW5kcmVhLm1hc290dGlAb3BiZy5uZXQ=
Milad Bastami, YmFzdGFtaW1AdGJ6bWVkLmFjLmly
†These authors have contributed equally to this work
 Jalal Choupani
Jalal Choupani Ziba Nariman-Saleh-Fam
Ziba Nariman-Saleh-Fam Zahra Saadatian
Zahra Saadatian Elaheh Ouladsahebmadarek
Elaheh Ouladsahebmadarek Andrea Masotti
Andrea Masotti Milad Bastami
Milad Bastami