- 1School of Life Science, Shanxi University, Taiyuan, China
- 2Natural History Research Center, Shanghai Natural History Museum, Branch of Shanghai Science and Technology Museum, Shanghai, China
- 3Department of Botany, National Museum of Natural History, Smithsonian Institution, Washington, DC, United States
- 4Key Laboratory of Medicinal Animal and Plant Resources of the Qinghai-Tibetan Plateau in Qinghai Province, School of Life Science, Qinghai Normal University, Xining, China
To explore the origin and evolution of local flora and vegetation, we examined the evolutionary history of Rhus chinensis, which is widely distributed in China’s temperate and subtropical zones, by sequencing three maternally inherited chloroplast DNAs (cpDNA: trnL-trnF, psbA-trnH, and rbcL) and the biparentally inherited nuclear DNA (nuDNA: LEAFY) from 19 natural populations of R. chinensis as well as the ecological niche modeling. In all, 23 chloroplast haplotypes (M1–M23) and 15 nuclear alleles (N1–N15) were detected. The estimation of divergence time showed that the most recent common ancestor dated at 4.2 ± 2.5 million years ago (Mya) from cpDNA, and the initial divergence of genotypes occurred at 4.8 ± 3.6 Mya for the nuDNA. Meanwhile, the multimodality mismatch distribution curves and positive Tajima’s D values indicated that R. chinensis did not experience population expansion after the last glacial maximum. Besides, our study was also consistent with the hypothesis that most refugia in the temperate and subtropical zones of China were in situ during the glaciation.
Introduction
The Quaternary climate oscillations occurred in the past ca. 2.58 million years ago (Mya) have resulted in several glacial and interglacial cycles in the Northern Hemisphere (Shackleton and Opdyke, 1973). These climatic alterations have left imprints in geographical distributions, population structures, and demographic histories of plant and animal species (Abbott et al., 2000; Avise, 2000; Hewitt, 2004, 2011; Qiu et al., 2011, 2013; Wen et al., 2014, 2016), which can be traced by analyses of genetic variations within and between extant populations (Abbott et al., 2000; Johansen and Latta, 2003; Hewitt, 2004). In Europe and North America, the fossil records of plant species and phylogeographic analyses had indicated common patterns of geographical range shifts that plants retreated southward and to lower elevations during glacial periods and while recolonized rapidly the northern areas and higher elevations during the interglacial and postglacial periods (Nason et al., 2002; Petit et al., 2003; Stewart et al., 2010; Sakaguchi et al., 2011; Segovia et al., 2012; Voss et al., 2012; Tzedakis et al., 2013; de Lafontaine et al., 2014). While in China, especially the Qinghai-Tibet Plateau (QTP) and adjacent regions, considerable research achievements have also been attained on inferring the Quaternary phylogeographic histories of plant species based on the approach of population genetics (e.g., Zhang et al., 2005, 2015; Meng et al., 2007; Chen K.M. et al., 2008; Yang et al., 2008; Wang et al., 2009; Opgenoorth et al., 2010; Xu et al., 2010; Qiu et al., 2011; Zou et al., 2012; Wang G.N. et al., 2014; Wen et al., 2014; Liu Y.P. et al., 2015; Wan et al., 2016).
The temperate and subtropical region of China is a model area for studying plant species in response to past climate changes (Chen S.C. et al., 2012; Li X.H. et al., 2012; Qi et al., 2012; Zhao et al., 2013; Fu et al., 2014). Up to date, many phylogeographic studies have been used to elucidate the impacts of the uplifts of the QTP on the climate within the modern-day temperate and subtropical zones, or warm temperate zones in China (e.g., Yellow River Basin, Chen K.M. et al., 2008 and Chen Y.Y. et al., 2008; Yunnan-Guizhou Plateau, Fan et al., 2013; Wang W. et al., 2014; Yangtze River, Sun et al., 2013; Wang H. et al., 2015; Qinling Mountains, Liu J.Q. et al., 2014; Lu et al., 2016; QTP, Liu D. et al., 2015 and Liu Y.P. et al., 2015); i.e., 23.5°–42.0° N and 98.0°–124.0° E (Gao et al., 2003; Shangguan et al., 2009). The results showed that the QTP acted as a barrier against glaciation within the warm temperate zones of China and resulted in the arid climate for thousands of years within the Quaternary period, which has been widely accepted nowadays (Wang et al., 2013; Yu et al., 2013; Meng et al., 2014). Thus, the present warm temperate region probably served as a glacial refugia for plant species in the past time, and this hypothesis has been tested and advanced through phylogeographic studies (e.g., Li Y. et al., 2012; Liu et al., 2012; Qi et al., 2012; Wan et al., 2016). However, it is less well known whether population genetic diversification of plants within the warm temperate zone or within the glacial refugia is due to isolation on a heterogeneous landscape or adaptation and selection along ecological gradients (Su et al., 2015; Zhao et al., 2016). Therefore, more phylogeographic studies of additional plant species within the warm temperate refugial regions are necessary in order to detect their spatial geographic patterns and to assess the underlying causes.
Rhus chinensis belongs to the plant family Anacardiaceae and is a common deciduous tree that is endemic to the warm temperate zone of Asia. It widely occurs at the elevation of 170–2700 m above sea level in Shaanxi, Shanxi, Hebei, Sichuan, Hunnan, and Yunnan of China (Zheng and Min, 1980). Due to its commonality and widespread distribution within the warm temperate zone, R. chinensis is thus an ideal study case for phytogeography within this region. In this study, we used three cpDNA regions (trnL-trnF, psbA-trnH, and rbcL) and one nuDNA region (LEAFY) to examine (1) the genetic diversity and structure of R. chinensis populations in China and (2) how is the demographic history of R. chinensis during the Quaternary climate oscillations, and further to explore the origin and evolution of local flora and vegetation.
Materials and Methods
Population Sampling
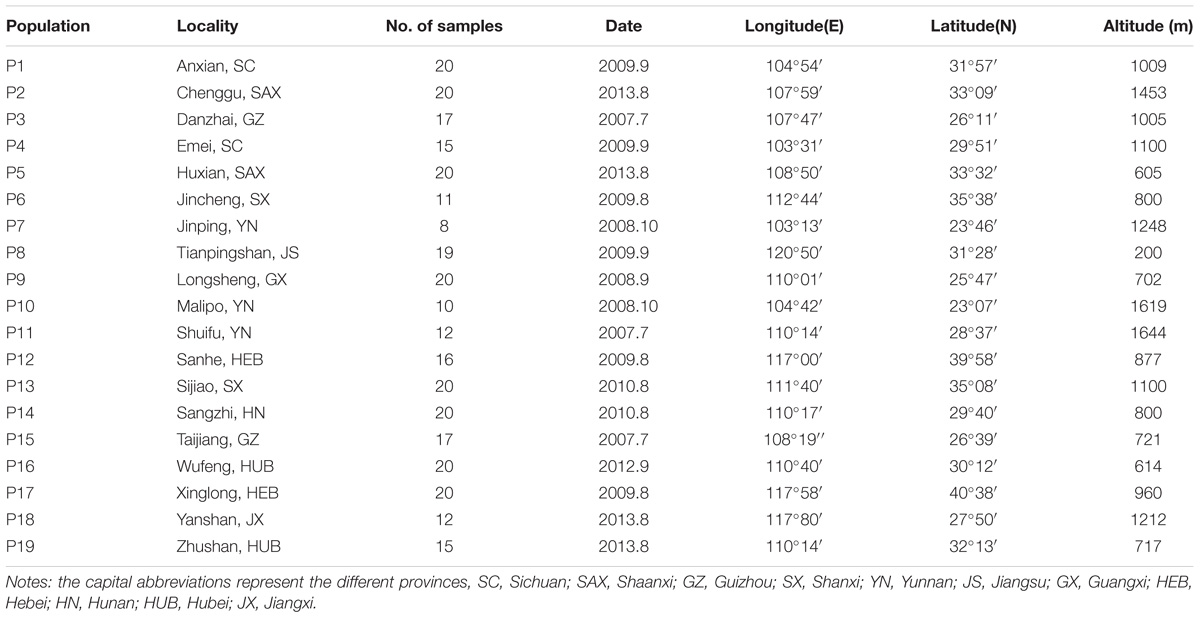
In total, leaf samples of 312 individuals were collected from 19 natural populations of R. chinensis, representing its whole geographic distribution within the warm temperate zone of China (see Figure 1 and Table 1). Eight to 20 individuals were collected for each population, and all individuals were at least 15 m apart. We obtained several voucher specimens for each population, which were deposited at the School of Life Sciences, Shanxi University, Taiyuan, Shanxi, China. The information of latitude, longitude, and altitude of each population were recorded using an Etrex GIS (Garmin, Taiwan, China).
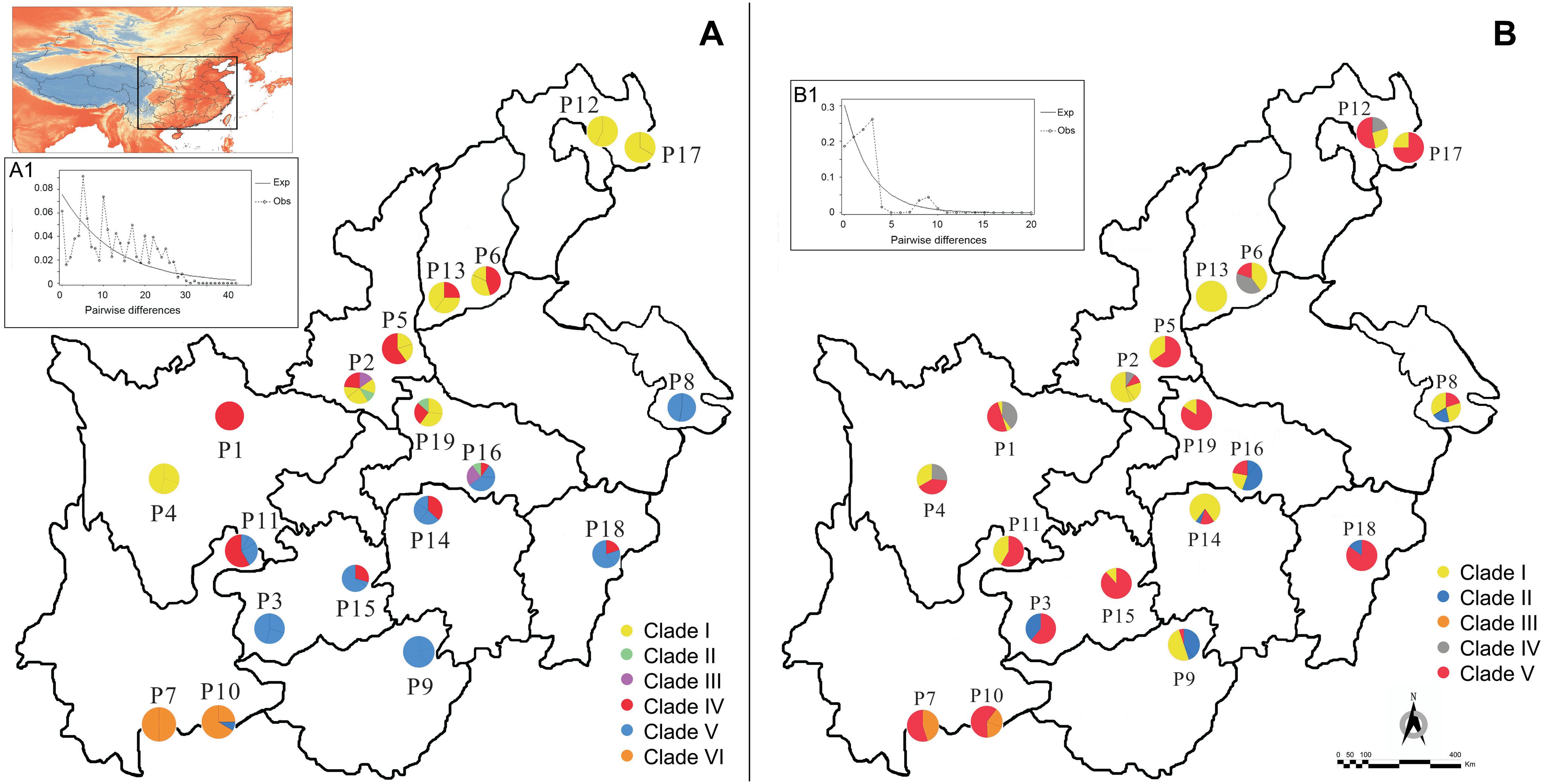
Figure 1. Geographic distribution of haplotypes and genotypes for Rhus chinensis based on cpDNA (A) and nuDNA (B). Multimodality mismatch distribution curves of cpDNA and nuDNA in the overall populations are shown in A1 and B1.
The species from Anacardiaceae were used as outgroups. The cpDNA sequences of four species were downloaded from GenBank, Rhus virens (EF682861, KF664327, KF664558), Rhus typhina (AY640446, HQ427036, HQ590236), Rhus glabra (AY640440, KF664325, KX397919), and Pistacia vera (EF193139, KF664307, AJ235786). There are no nuclear sequences for the above four species in GenBank, so we used another two species Mangifera indica (GU338039) and P. chinensis (KC174710) as the outgroups in the nuDNA analysis.
DNA Sequencing
Total genomic DNAs from approximately 20 mg of silica gel-dried leaf materials were extracted using a Plant Genomic DNA kit (Tiangen Biotech, Beijing, China), and three cpDNA fragments (trnL-trnF, psbA-trnH, and rbcL) and one nuclear gene (LEAFY) were amplified and sequenced by the following primers: trnL-trnF (5′-CGAAATCGGTAGAGGCTACG-3′; 5′-ATTTGAACTGGTGACACGAG-3′; Taberlet et al., 1991), psbA-trnH (5′-GTTATGCATFAACGTAATGCTC-3′; 5′-CGCGCATGGTGGATTCACAAATC-3′; Sang et al., 1997), rbcL (5′-ATGTCACCACAAACAGAGAC-3′; 5′-TCAAATTCAAACTTGATTTCTTTC-3′; Little and Barrington, 2003), LEAFY (5′-TACACGGCAGCGAAGATAGC-3′; 5′-CTAGAAGCAGCGGCACTATTG-3′; Oh and Potter, 2003). Polymerase chain reaction (PCR) was performed in a volume of 50 μL and each reaction contained 30–50 ng genomic DNA, 25 μL amplification reaction mixture (PCR mix kit, Tiangen Biotech, Beijing, China), and 20 μmol/L primers, and under the following conditions: initial denaturation at 94°C for 3 min, 35 cycles of 30 s at 94°C, 30 s at 54–60°C, 90 s at 72°C, and a final extension step of 7 min at 72°C. All the qualified PCR products were sent to Majorbio Bio-pharm Technology Co., Ltd. (Shanghai) for sequencing.
Data Analysis
We aligned sequences with Clustal_X (Thompson et al., 1997) and coded indels following the method of Simmons and Ochoterena (2000). Indels within mononucleotide repeat regions were deleted for phylogenetic analyses, because the homology of these indels could not be verified (Chen S.C. et al., 2012).
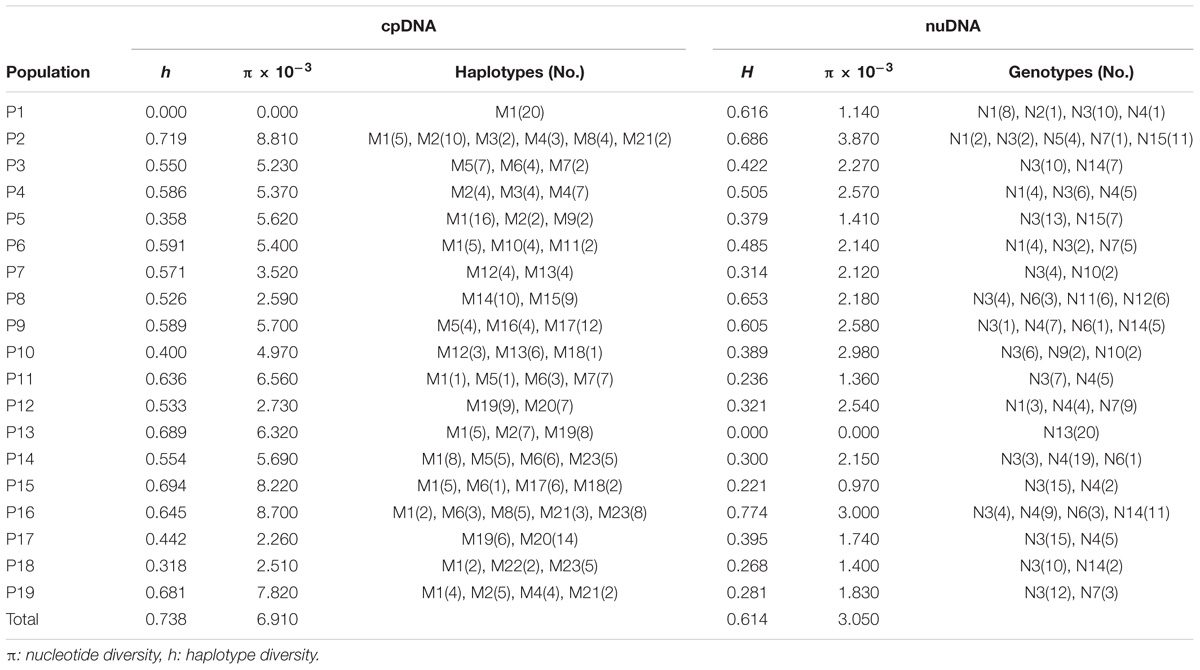
The levels of inter- and intra-population genetic diversity (h: haplotype diversity and π: nucleotide diversity) were calculated for the cpDNA and nuDNA using DnaSP version 5.0 (Rozas et al., 2003). We compared GST and NST using the U-statistic, which is approximated by a Gaussian variable by taking into account the covariance between GST and NST, and a one-sided test (Pons and Petit, 1996). The former considers only haplotype frequencies while NST also takes into account differences between haplotypes. When NST is larger than GST, phylogeographic structure is obvious, which indicates that closely related haplotypes were found more often in the same area than less closely related haplotypes (Pons and Petit, 1996). We also estimated genetic differentiation among all populations with AMOVA and inferred population growth and expansion according to Tajima’s D using Arlequin version 3.0 (Excoffier et al., 2005), with 1000 random permutations to test for significance of partitions. Genealogical relationships among cpDNA and nuDNA haplotypes were constructed using TCS version 1.21 (Clement et al., 2000).
The phylogenetic relationships among haplotypes and genotypes of cpDNA and nuDNA were reconstructed with Bayesian inference (BI) methods in MrBayes version 3.1.2 (Ronquist and Huelsenbeck, 2003). We applied the best fit model, GTR + I + G, which was inferred by Modeltest 3.7 under the Akaike information criterion (Posada and Crandall, 1998; Ronquist and Huelsenbeck, 2003). The BI consisted of two parallel runs with four incrementally heated chains and three million generations sampled every 1000 generation. The output was diagnosed for convergence using TRACER v.1.3 (Rambaut and Drummond, 2007), and summary statistics and trees were generated using the last two million generation in MrBayes version 3.1.2 (Ronquist and Huelsenbeck, 2003). In order to distinguish the haplotypes and genotypes clearly, the branches with high bootstrap value (>0.95) were classified as new clades based on the phylogenetic trees (Porter et al., 2005; Pyron, 2011; Ye et al., 2017).
The divergence times within R. chinensis were estimated using a molecular clock and fossil data. Three fossils of Rhus were used to calibrate the node ages of R. typhina and R. glabra (6.0 Mya), R. typhina and R. virens (38.1 Mya), and R. typhina and P. vera (49.1 Mya) for cpDNA data, respectively (Yi et al., 2004), while one fossil (49.1 Mya) was used as the divergence time between Rhus and Pistacia for nuDNA data. Both the strict and relaxed molecular clock rates were tested in MEGA6 (Tamura et al., 2013) using the BI summary tree, and they could not be rejected for either the cpDNA or nuDNA data. Therefore, the strict and relaxed clocks were both applied to the two datasets in the BASEML and MCMCTREE programs of PAML (Yang, 2007), and used our BI summary tree as the guide tree.
Ecological Niche Modeling
We compared the current distributions of R. chinensis with its inferred distributions during the last glacial maximum (LGM; ∼21,000 years BP) with ecological niche modeling in Maxent version 3.3.3 (Phillips et al., 2006). To perform this modeling, we first obtained the geocoordinates of 73 occurrence data of R. chinensis from the Chinese Virtual Herbarium1 and Global Biodiversity Information Facility2. Subsequently, we constructed the models using 19 bioclimatic variables from the WorldClim database3 (Hijmans et al., 2005) representing the present (averaged from 1950) and the LGM according to the Community Climate System Model (CCSM; Collins et al., 2006). We employed 20 replicates based on 80% of the distribution coordinates for training and 20% for testing, and adopted the model with the best AUC values (Phillips et al., 2006). We performed a jackknife test to estimate the percent contributions of bioclimatic variables to the prediction for the distributional models. Meanwhile, we also employed the “10 percentile presence” threshold logistic approach as determined by Maxent in order to distinguish the threshold between suitable and unsuitable habitats for further analyses. We drew Graphics for each predicted SDM using DIVA-GIS 7.5 (Hijmans et al., 2005).
Results
Genetic Diversity and Structure
Aligned cpDNA dataset consisted of 2051 bp with 70 nucleotide substitutions and two indels. We detected 23 different haplotypes (M1–M23) based on combined cpDNA dataset from 19 populations. The LEAFY gene region varied from 412 to 645 bp and had an aligned length of 682 bp, which contained 14 nucleotide substitutions. Our sequences of LEAFY comprised 15 genotypes (N1–N15). Based on cpDNA and nuDNA sequences, the total haplotype diversity of R. chinensis was estimated to be 0.738 and 0.614, and the total nucleotide diversity was inferred to be 6.910 × 10−3 and 3.050 × 10−3, respectively (Table 2). We found the highest levels of haplotype and nucleotide diversity in four populations: P2, P11, P14, and P16 (Table 2). The most widespread haplotypes and genotypes were M1 (in 11 of 19 populations, cpDNA) and N3 (in 18 of 19 populations, nuDNA; Table 2), respectively. Based on cpDNA and nuDNA sequences, M1 and N3 were the primary haplotype and genotype, respectively (Figure 1).
AMOVA analysis indicated that genetic variation in R. chinensis was greater within populations than among them (P < 0.01; Table 3). The mismatch distribution (Figures 1A1,B1) and positive values of Tajima’s D value (1.19, 0.05 < P < 0.10 for cpDNA; 2.37, P < 0.01 for nuDNA) of all populations rejected a sudden expansion model, and positive Tajima’s D may indicate population admixture. Phylogeographic structure is not obvious at the species level for both sets of genetic markers. For the cpDNA data, NST (0.382) was slightly higher than GST (0.375), while for the LEAFY, the difference between the two indices was not significant (NST = 0.321, GST = 0.319, P > 0.05).
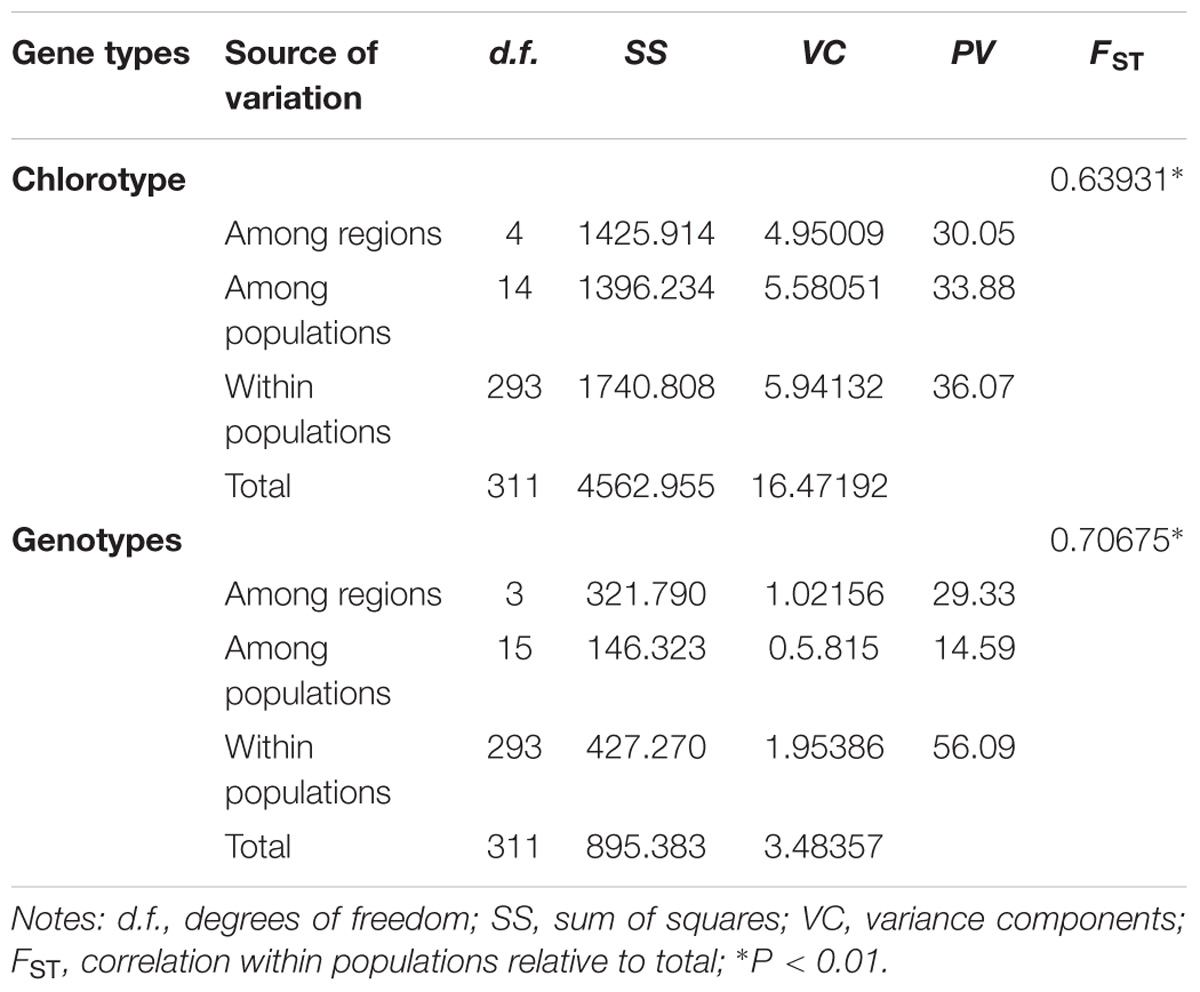
Table 3. Analysis of molecular variance (AMOVA) of Rhus chinensis populations based on nucleotide sequences.
Phylogeography and Divergence Time
In the cpDNA phylogeny, eight haplotypes (M2–M4, M9–M11, and M19–M20) were clustered into clade I (Figure 2A). M21 and M8 were separately defined into clade II and clade III (Figure 2A). M1 independently belonged to clade IV, which was probably the ancestral haplotype (Figure 2A). Clade V included ten haplotypes (M5–M7, M14–M18, and M22–M23), while clade VI only contained two haplotypes (M12–M13) (Figure 2A). Based on nuDNA data, five clades were redefined (Figure 2B). Nine genotypes (N2, N4–N5, N7–N8, N11–N13, and N15) were clustered into clade I (Figure 2B). Clade II and III, each included two genotypes, namely, N6 and N14 existed in clade II, while N9 and N10 occurred in clade III (Figure 2B). N1 and N3 were individually defined into clade IV and clade V (Figure 2B). In addition, the divergence time of R. chinensis estimated with a strict molecular clock was highly consistent with that based on a relaxed molecular clock. According to the cpDNA phylogenetic tree, the crown age of R. chinensis was dated to be 4.2 ± 2.5 Mya, when the clade VI (P7 and P10–P11, Yunnan populations) split from all other clades (Figure 2A). Similarly, for the LEAFY gene, the crown age of R. chinensis was dated to be 4.8 ± 3.6 Mya (Figure 2B). Additionally, the clade III (P7 and P10–P11, Yunnan populations) diverged from clade I and clade II at 3.8 ± 3.0 Mya (Figure 2B).
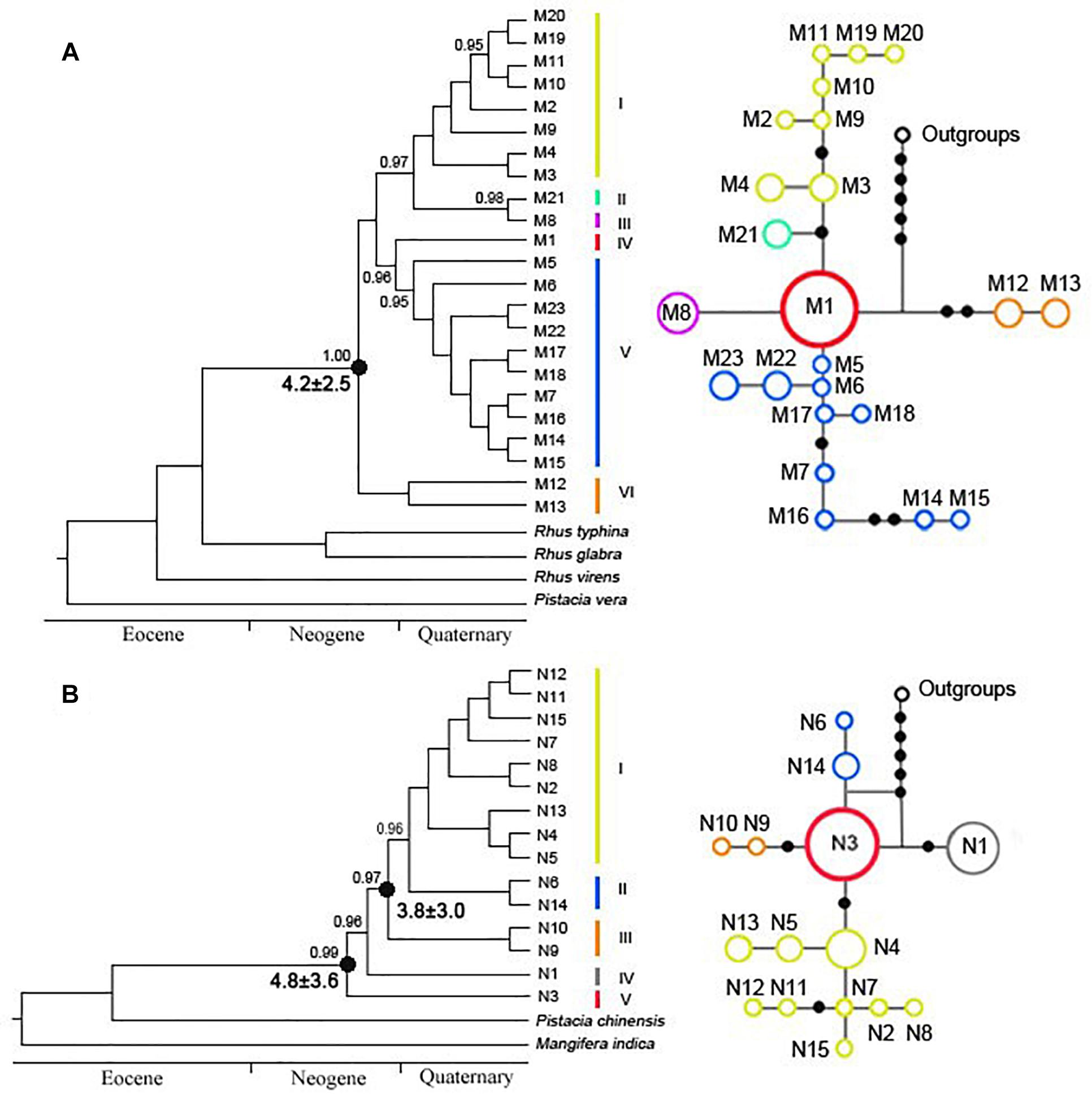
Figure 2. Phylogenetic trees of Rhus chinensis calculated by Bayesian inference (BI) of (A) cpDNA and (B) nuDNA. Numbers above the branches indicate the bootstrap values (>0.95). Clades of cpDNA and nuDNA representing haplotypes and genotypes are discussed in the text.
Ecological Niche Modeling
The inferred past (LGM) and current distributions of R. chinensis are illustrated in (Figure 3). The AUC values based on both training and test presence data for the present and at the LGM were all higher than expected (not shown), which demonstrated good model performance. It was notable that the current distribution model indicated that R. chinensis mainly occurred in the warm temperate zone of China, which also suggested that it should occur in the same region during the LGM period (Figure 3A). In the comparison with the two simulated distributions, the LGM distribution model predicted that the species was mainly concentrated in Yunnan and central China including Shaanxi, Sichuan, Hubei, and Jiangxi provinces, and it had slightly shrunk in these regions during the LGM period (Figure 3B).
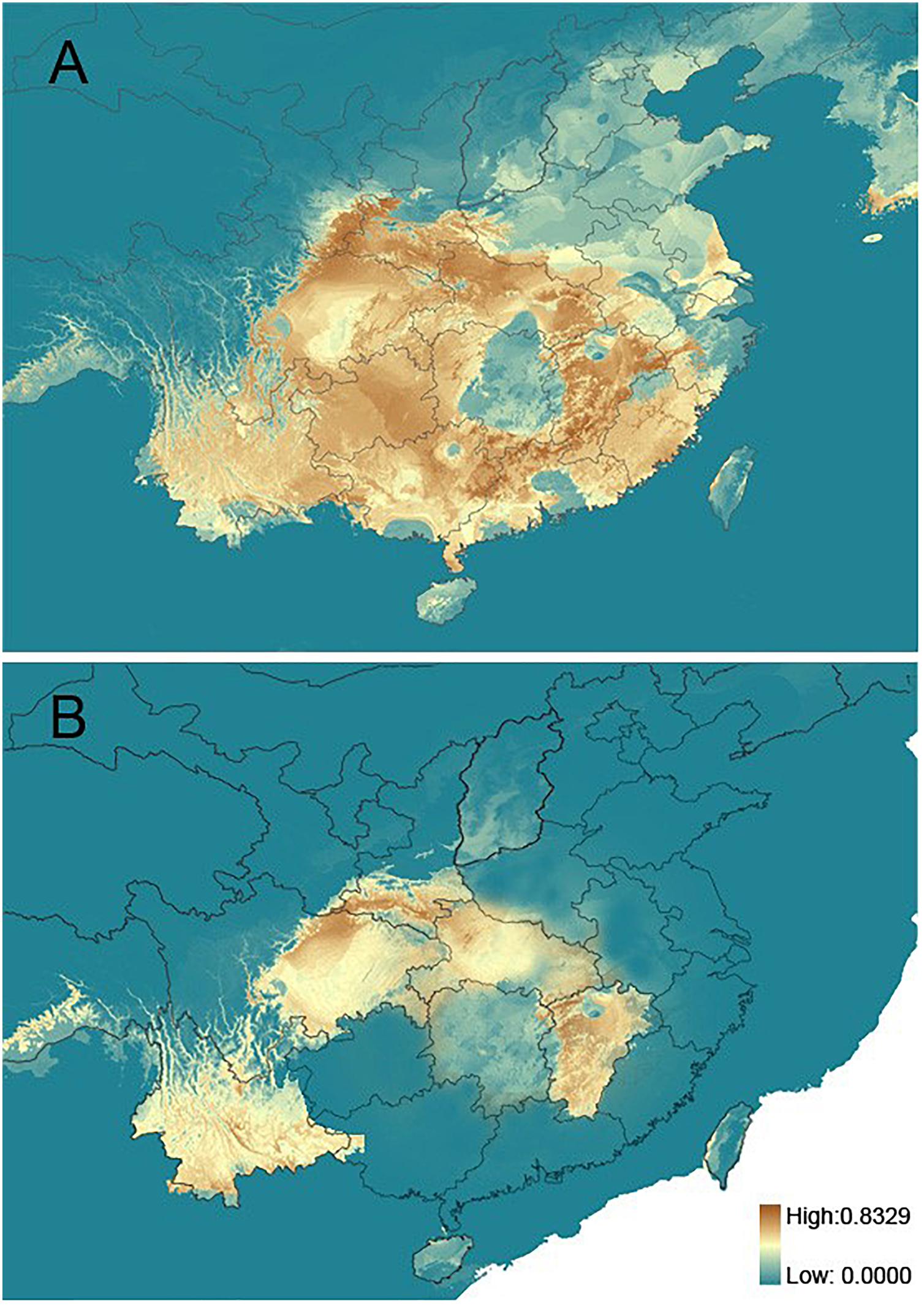
Figure 3. Maps showing the potential distribution by ENM. (A) The present. (B) Last glacial maximum.
Discussion
We did not detect a clear phylogeographic structure among the 19 populations of R. chinensis sampled in the present study. We found a somewhat lower differentiation among R. chinensis populations (NST = 0.382 for cpDNA, NST = 0.321 for nuDNA) compared to sympatric species such as Platycarya strobilacea (Chen S.C. et al., 2012) and Cotinus coggygria (Wang W. et al., 2014). Limited phylogeographic structure within a metapopulation may be due to high levels of geneflow and/or of geophysical connectedness (Avise et al., 1987). High levels of gene flow among R. chinensis populations may be due to the seed dispersal mechanism, which has been implicated in high levels of gene flow in many other plant species (e.g., Lopez et al., 2007; Song et al., 2013; Johnson et al., 2017). R. chinensis can produce 1000 seeds per plant on average, and the seeds are dispersed by animals, including mammals and birds, and by water (Huang and Qiu, 1994; Wang W. et al., 2014). Therefore, it is possible that relatively limited population differentiation may be due to the movement of seeds, including maternal and bi-parental genetic material, throughout the warm temperate zone. Geophysical connectedness within the range of R. chinensis may also be responsible for high levels of gene flow among populations. Stated another way, there may be limited barriers to dispersal. In the distributional area of R. chinensis, no obvious geographic barriers have been observed. Therefore, R. chinensis does not appear to be geographically isolated, allowing ecological niche modeling to be used in the assessment of species status (Li X.H. et al., 2012; Liu L. et al., 2014; Wang W. et al., 2014). Ecological niche models suggested the suitable habitats of R. chinensis were continuous in the present time while compressed during the LGM period, demonstrating multiple possible isolated glacial refugia (Figure 3). The response to impact of cold and warm times on the distribution of R. chinensis was validated in the simulation of ecological niche modeling, although we only used the simulated environment of current and LGM period (Figure 3). This pattern of range shifts indicated a likely scenario of repeated glacial compressions followed by interglacial expansions for R. chinensis during the Quaternary climatic oscillations. It is interesting that the geographic distribution of the cpDNA haplotypes differs from the nuDNA genotypes (Figure 2). Mismatch distributions between organellar DNA haplotypes and nuclear DNA genotypes have been reported in other groups such as Sophora davidii (Fan et al., 2013), Cycas diannanensis (Liu J. et al., 2015), and Osteomeles schwerinae (Wang Z.W. et al., 2015). Therefore, we thought that the forest birds and mammals were known as seed dispersers for many species in Anacardiaceae (Wang W. et al., 2014), which might have directly impacted the genetic structure with biparental inheritance.
The populations originated from Yunnan occurred at the China–Vietnam border and split from other clades at 4.2 ± 2.5 and 3.8 ± 3.0 Mya according to the cpDNA (clade VI) and nuDNA (clade III), respectively (Figures 2A,B). Early diverging populations in Yunnan have been detected in other genera or species such as Ceratotropis (3.62 Mya, Javadi et al., 2011), Incarvillea sinensis (4.4 Mya, Chen S. et al., 2012), and Stuckenia filiformis (3.93 Mya, Du and Wang, 2016). Within these species, the uplift of the QTP has been implicated as the main mechanisms of driving diversifications, but the estimated divergences were more recent than the last phase of the uplift (7–8 and 13–15 Mya; Harrison et al., 1992; Shi et al., 1998; Spicer et al., 2003). So, we thought that the geographical isolation of Yunnan populations was caused by the isolation of the QTP uplift in late Pliocene. Furthermore, the suitable climate in the temperate and subtropical zone could have subsequently facilitated the Pliocene-Pleistocene diversification of R. chinensis into different eco-geographic populations (Javadi et al., 2011).
Previous phylogeographic studies have widely supported hypotheses that climatic changes during the LGM forced plants into refugia within Central China, where they were protected by the QTP from the brunt of the ice age (Tian et al., 2009; Liu et al., 2012). After the glaciers retreated, the plants expanded their ranges rapidly (Hewitt, 2000; Li Z.H. et al., 2012; Qi et al., 2012). Our results showed that the range of R. chinensis had increased since the LGM (Figure 3) but did not support a rapid expansion based on the mismatch distribution (Figure 1) and Tajima’s D (1.19, 0.05 < P < 0.10 for cpDNA; 2.37, P < 0.01 for nuDNA). Refugia in the warm temperate China may have been dominated by evergreen forest or temperate deciduous forest during the LGM (Liu, 1988). Thus, southern Shaanxi, northern Sichuan, Yunnan, and Jiangxi could have supported R. chinensis during the LGM and been its main center of diversity. Just as P. strobilacea (Chen S.C. et al., 2012), Cercidiphyllum (Qi et al., 2012), and C. coggygria (Wang W. et al., 2014), the plants were slightly affected and were able to survive in situ at the period of the glaciation. So, the characterized phylogeographic structure of R. chinensis was consistent with the second hypothesis, which was that they survived in situ and occupied multiple localized glacial refugia during the glaciation.
Conclusion
We used cpDNA and nuDNA sequences, and ecological niche modeling to investigate the evolutionary history of R. chinensis distributed in the warm temperate zone of China. The cpDNA and nuDNA data separately revealed six and five clades corresponding to the geographic regions. The divergence among haplotypes and genotypes of R. chinensis occurred at the Pliocene based on cpDNA and nuDNA data. Our ENMs showed enlarged potential distributions in the present compared to LGM, but we did not detect a sudden demographic expansion after the glaciation according to the molecular data. Our results suggest that R. chinensis was not affected by glacial cycles seriously and survived in situ and occupied a few main refugia.
Author Contributions
ZR conceived and designed the research. YL and YZ collected the samples, performed the experiments, and conducted data analyses. XS and ZR drafted the manuscript. JW polished the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 31870366, 31170359, 31800310, and 31600301), the Key Laboratory of Medicinal Animal and Plant Resources of the Qinghai-Tibet Plateau in Qinghai Province (Grant No. 2017-ZJ-Y13), the National High Technology Research and Development “863” Program (Grant No. 2014AA021802), and Shanxi International Science and Technology Cooperation Project (2018).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank two reviewers for their constructive comments.
Footnotes
References
Abbott, R. J., Smith, L. C., Milne, R. I., Crawford, R. M., Wolff, K., and Balfour, J. (2000). Molecular analysis of plant migration and refugia in the Arctic. Science 289, 1343–1346. doi: 10.1126/science.289.5483.1343
Avise, J. C. (2000). Phylogeography: the History and Formation of Species. Cambridge: Harvard University Press.
Avise, J. C., Arnold, J., Ball, R. M., Bermingham, E., Lamb, T., Neeigel, E., et al. (1987). Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 18, 489–522. doi: 10.1146/annurev.es.18.110187.002421
Chen, K. M., Abbott, R. J., Milne, R. I., Tian, X. M., and Liu, J. Q. (2008). Phylogeography of Pinus tabulaeformis Carr. (Pinaceae), a dominant species of coniferous forest in northern China. Mol. Ecol. 17, 4276–4288. doi: 10.1111/j.1365-294X.2008.03911.x
Chen, S., Xing, Y., Tao, S., Zhou, Z., Dilcher, E. D. L., and Soltis, D. E. (2012). Phylogeographic analysis reveals significant spatial genetic structure of Incarvillea sinensis as a product of mountain building. BMC Plant Biol. 12:58. doi: 10.1186/1471-2229-12-58
Chen, S. C., Zhang, L., Zeng, J., Shi, F., Yang, H., Mao, Y. R., et al. (2012). Geographic variation of chloroplast DNA in Platycarya strobilacea (Juglandaceae). J. Syst. Evol. 50, 374–385. doi: 10.1111/j.1759-6831.2012.00210.x
Chen, Y. Y., Li, X. L., Yin, L. Y., and Li, W. (2008). Genetic diversity of the threatened aquatic plant Ottelia alismoides, in the Yangtze river. Aquat. Bot. 88, 10–16. doi: 10.1016/j.aquabot.2007.08.002
Clement, M., Posada, D., and Crandall, K. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x
Collins, W. D., Bitz, C. M., Blackmon, M. L., Bonan, G. B., Bretherton, C. S., Carton, J. A., et al. (2006). The Community Climate System Model version 3 (CCSM3). J. Clim. 19, 2122–2143. doi: 10.1175/JCLI3761.1
de Lafontaine, G., Amasifuen Guerra, C. A., Ducousso, A., Sánchez-Goñi, M. F., and Petit, R. J. (2014). Beyond skepticism: uncovering cryptic refugia using multiple lines of evidence. New Phytol. 204, 450–454. doi: 10.1111/nph.13089
Du, Z. Y., and Wang, Q. F. (2016). Allopatric divergence of Stuckenia filiformis (Potamogetonaceae) on the Qinghai-Tibet Plateau and its comparative phylogeography with S. pectinata in China. Sci. Rep. 6:20883. doi: 10.1038/srep20883
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50. doi: 10.1177/117693430500100003
Fan, D. M., Yue, J. P., Nie, Z. L., Li, Z. M., Comes, H. P., and Sun, H. (2013). Phylogeography of Sophora davidii (Leguminosae) across the ’Tanaka-Kaiyong Line’, an important phytogeographic boundary in Southwest China. Mol. Ecol. 22, 4270–4288. doi: 10.1111/mec.12388
Fu, Z. Z., Li, Y. H., Zhang, K. M., and Li, Y. (2014). Molecular data and ecological niche modeling reveal population dynamics of widespread shrub Forsythia suspensa (Oleaceae) in China’s warm-temperate zone in response to climate during the Pleistocene. BMC Evol. Biol. 14:114. doi: 10.1186/1471-2148-14-114
Gao, Q., Peng, S. L., Zhao, P., Zeng, X. P., Cai, X., Yu, M., et al. (2003). Explanation of vegetation succession in subtropical southern China based on ecophysiological characteristics of plant species. Tree Physiol. 23, 641–648. doi: 10.1093/treephys/23.9.641
Harrison, T. M., Copeland, P., Kidd, W. S., and Yin, A. (1992). Raising Tibet. Science 255, 1663–1670. doi: 10.1126/science.255.5052.1663
Hewitt, G. M. (2000). The genetic legacy of the Quaternary ice ages. Nature 405, 907–913. doi: 10.1038/35016000
Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 183–195. doi: 10.1098/rstb.2003.1388
Hewitt, G. M. (2011). Quaternary phylogeography: the roots of hybrid zones. Genetica 139, 617–638. doi: 10.1007/s10709-9547-3
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G., and Jarvis, A. (2005). Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. doi: 10.1002/joc.1276
Huang, J. D., and Qiu, F. B. (1994). A study on propagation of Rhus chinensis by seeds. Guihaia 14, 85–89.
Javadi, F., Tun, Y. T., Kawase, M., Guan, K., and Yamaguchi, H. (2011). Molecular phylogeny of the subgenus Ceratotropis (genus Vigna, Leguminosae) reveals three eco-geographical groups and Late Pliocene-Pleistocene diversification: evidence from four plastid DNA region sequences. Ann. Bot. 108, 367–380. doi: 10.1093/aob/mcr141
Johansen, A. D., and Latta, R. G. (2003). Mitochondrial haplotype distribution, seed dispersal and patterns of post glacial expansion of ponderosa pine. Mol. Ecol. 12, 293–298. doi: 10.1046/j.1365-294X.2003.01723.x
Johnson, J. S., Gaddis, K. D., Cairns, D. M., and Krutovsky, K. V. (2017). Seed dispersal at alpine treeline: an assessment of seed movement within the alpine treeline ecotone. Ecosphere 8:e01649. doi: 10.1002/ecs2.1649
Li, X. H., Shao, J. W., Lu, C., Zhang, X. P., and Qiu, Y. X. (2012). Chloroplast phylogeography of a temperate tree Pteroceltis tatarinowii (Ulmaceae) in China. J. Syst. Evol. 50, 325–333. doi: 10.1111/j.1759-6831.2012.00203.x
Li, Y., Yan, H. F., and Ge, X. J. (2012). Phylogeographic analysis and environmental niche modeling of widespread shrub Rhododendron simsii in China reveals multiple glacial refugia during the last glacial maximum. J. Syst. Ecol. 50, 362–373. doi: 10.1111/j.1759-6831.2012.00209.x
Li, Z. H., Zou, J. B., Mao, K. S., Lin, K., Li, H. P., Liu, J. Q., et al. (2012). Population genetic evidence for complex evolutionary histories of four high altitude juniper species in the Qinghai-Tibetan Plateau. Evolution 66, 831–845. doi: 10.1111/j.1558-5646.2011.01466.x
Little, D. P., and Barrington, D. S. (2003). Major evolutionary events in the origin and diversification of the fern genus Polystichum (Dryopteridaceae). Am. J. Bot. 90, 508–514. doi: 10.3732/ajb.90.3.508
Liu, D., Hou, F., Liu, Q., Zhang, X., Yan, T., and Song, Z. (2015). Strong population structure of Schizopygopsis chengi and the origin of S. chengi baoxingensis revealed by mtDNA and microsatellite markers. Genetica 143, 73–84. doi: 10.1007/s10709-015-9815-8
Liu, J., Zhou, W., and Gong, X. (2015). Species delimitation, genetic diversity and population historical dynamics of Cycas diannanensis (Cycadaceae) occurring sympatrically in the Red River region of China. Front. Plant Sci. 6:696. doi: 10.3389/fpls.2015.00696
Liu, J. Q., Duan, Y. W., Hao, G., Ge, X. J., and Sun, H. (2014). Evolutionary history and underlying adaptation of alpine plants on the Qinghai-Tibet Plateau. J. Syst. Evol. 52, 241–249. doi: 10.1111/jse.12094
Liu, J. Q., Sun, Y. S., Ge, X. J., Gao, L. M., and Qiu, Y. X. (2012). Phylogeographic studies of plants in China: advances in the past and directions in the future. J. Syst. Evol. 50, 267–275. doi: 10.1111/j.1759-6831.2012.00214.x
Liu, K. B. (1988). Quaternary history of the temperate forests of China. Quat. Sci. Rev. 7, 1–20. doi: 10.1016/0277-3791(88)90089-3
Liu, L., Hao, Z. Z., Liu, Y. Y., Wei, X. X., Cun, Y. Z., and Wang, X. Q. (2014). Phylogeography of Pinus armandii and its relatives: heterogeneous contributions of geography and climate changes to the genetic differentiation and diversification of Chinese White Pines. PLoS One 9:e85920. doi: 10.1371/journal.pone.0085920
Liu, Y. P., Su, X., He, Y. H., Han, L. M., Huang, Y. Y., and Wang, Z. Z. (2015). Evolutionary history of Orinus thoroldii (Poaceae), endemic to the western Qinghai-Tibetan Plateau in China. Biochem. Syst. Ecol. 59, 159–167. doi: 10.1016/j.bse.2015.01.014
Lopez, S., Rousset, F., Shaw, F. H., Shaw, R. G., and Ronce, O. (2007). Migration load in plants: role of pollen and seed dispersal in heterogeneous landscapes. J. Evol. Biol. 21, 294–309. doi: 10.1111/j.1420-9101.2007.01442.x
Lu, Q. X., Zhu, J. N., Dan, Y., and Xu, X. W. (2016). Genetic and geographical structure of boreal plants in their southern range: phylogeography of Hippuris vulgaris in China. BMC Evol. Biol. 16:34. doi: 10.1186/s12862-016-0603-6
Meng, H. H., Gao, X. Y., Huang, J. F., and Zhang, M. L. (2014). Plant phylogeography in arid Northwest China: retrospectives and perspectives. J. Syst. Evol. 52, 1–16. doi: 10.1111/jse.12088
Meng, L. H., Yang, R., Abbott, R. J., Miehe, G., Hu, T. H., and Liu, J. Q. (2007). Mitochondrial and chloroplast phylogeography of Picea crassifolia Kom. (Pinaceae) in the Qinghai-Tibetan Plateau and adjacent highlands. Mol. Ecol. 16, 4128–4137. doi: 10.1111/j.1365-294X.2007.03459.x
Nason, J. D., Hamrick, J. L., and Fleming, T. H. (2002). Historical vicariance and postglacial colonization effects on the evolution of genetic structure in Lophocereus, a Sonoran Desert columnar cactus. Evolution 56, 2214–2226. doi: 10.1111/j.0014-3820.2002.tb00146.x
Oh, S. H., and Potter, D. (2003). Phylogenetic utility of the second intron of LEAFY in Neillia and Stephanandra (Rosaceae) and implications for the origin of Stephanandra. Mol. Phylogenet. Evol. 29, 203–215. doi: 10.1016/S1055-7903(03)00093-9
Opgenoorth, L., Vendramin, G. G., Mao, K. S., Miehe, G., Miehe, S., Liepelt, S., et al. (2010). Tree endurance on the Tibetan Plateau marks the world’s highest known tree line of the Last Glacial Maximum. New Phytol. 185, 332–342. doi: 10.1111/j.1469-8137.2009.03007.x
Petit, R., Aguinagalde, I., de Beaulieu, J. L., Bittkau, C., Brewer, S., Cheddadi, R., et al. (2003). Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300, 1563–1565. doi: 10.1126/science.1083264
Phillips, S. J., Anderson, R. P., and Schapire, R. E. (2006). Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259. doi: 10.1016/j.ecolmodel.2005.03.026
Pons, O., and Petit, R. J. (1996). Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237–1245.
Porter, M. L., Perez-Losada, M., and Crandall, K. A. (2005). Model-based multi-locus estimation of decapod phylogeny and divergence times. Mol. Phylogenet. Evol. 37, 355–369. doi: 10.1007/BF02524290
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Pyron, R. A. (2011). Divergence time estimation using fossils as terminal taxa and the origins of Lissamphibia. Syst. Biol. 60, 466–481. doi: 10.1093/sysbio/syr047
Qi, X. S., Chen, C., Comes, H. P., Sakaguchi, S., Liu, Y. H., Tanaka, N., et al. (2012). Molecular data and ecological niche modeling reveal a highly dynamic evolutionary history of the East Asian tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytol. 196, 617–630. doi: 10.1111/j.1469-8137.2012.04242.x
Qiu, Y. X., Fu, C. X., and Comes, H. P. (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244. doi: 10.1016/j.ympev.2011.01.012
Qiu, Y. X., Liu, Y. F., Kang, M., Yi, G. M., and Huang, H. W. (2013). Spatial and temporal population genetic variation and structure of Nothotsuga longibracteata (Pinaceae), a relic conifer species endemic to subtropical China. Genet. Mol. Biol. 36, 598–607. doi: 10.1590/S1415-47572013000400019
Rambaut, A., and Drummond, A. J. (2007). Tracer. Available at: http://beast.bio.ed.ac.uk/tracer
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rozas, J., Sánchez-DelBarrio, J. C., Messeguer, X., and Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. doi: 10.1079/9780851994758.0139
Sakaguchi, S., Takeuchi, Y., Yamasaki, M., Sakurai, S., and Isagi, Y. (2011). Lineage admixture during postglacial range expansion is responsible for the increased gene diversity of Kalopanax septemlobus in a recently colonised territory. Heredity 107, 338–348. doi: 10.1038/hdy.2011.20
Sang, T., Crawford, D. J., and Stuessy, T. F. (1997). Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 84, 1120–1120. doi: 10.2307/2446155
Segovia, R. A., Pérez, M. F., and Hinojosa, L. F. (2012). Genetic evidence for glacial refugia of the temperate tree Eucryphia cordifolia (Cunoniaceae) in southern South America. Am. J. Bot. 99, 121–129. doi: 10.3732/ajb.1100013
Shackleton, N. J., and Opdyke, N. D. (1973). Oxygen isotope and palaeomagnetic stratigraphy of Equatorial Pacific core V28-238: oxygen isotope temperatures and ice volumes on a 105 year and 106 year scale. Quat. Res. 3, 39–55. doi: 10.1016/0033-5894(73)90052-5
Shangguan, T. L., Li, J. P., and Guo, D. G. (2009). Advance in mountain vegetation ecology in the warm-temperate zone of China. J. Mount. Sci. 27, 129–139.
Shi, Y. F., Li, J. J., and Li, B. Y. (1998). Uplift and Environmental Changes of Qinghai-Tibetan Plateau in the late Cenozoic. Guangzhou: Guangdong Science and Technology Press.
Simmons, M. P., and Ochoterena, H. (2000). Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 49, 369–381. doi: 10.1093/sysbio/49.2.369
Song, B., Zhang, Z. Q., Stöcklin, J., Yang, Y., Niu, Y., Chen, J. G., et al. (2013). Multifunctional bracts enhance plant fitness during flowering and seed development in Rheum nobile (Polygonaceae), a giant herb endemic to the high Himalayas. Oecologia 172, 359–370. doi: 10.1007/s00442-012-2518-2
Spicer, R., Harris, N. B. W., Widdowson, M., Herman, A. B., Guo, S. X., Valdes, P. J., et al. (2003). Constant elevation of southern Tibet over the past 15 million years. Nature 421, 622–624. doi: 10.1038/nature01356
Stewart, J. R., Lister, A. M., Barnes, I., and Dalen, L. (2010). Refugia revisited: individualistic responses of species in space and time. Proc. R. Soc. Lond. B Biol. Sci. 277, 661–671. doi: 10.1098/rspb.2009.1272
Su, X., Wu, G. L., Li, L. L., and Liu, J. Q. (2015). Species delimitation in plants using the Qinghai-Tibetan Plateau endemic Orinus (Poaceae: Tridentinae) as an example. Ann. Bot. 116, 35–48. doi: 10.1093/aob/mcv062
Sun, Y. S., Abbott, R. J., Li, L., Li, L. L., Zou, J. B., and Liu, J. Q. (2013). Evolutionary history of purple cone spruce (Picea purpurea) in the Qinghai-Tibet plateau: homoploid hybrid origin and Pleistocene expansion. Mol. Ecol. 23, 343–359. doi: 10.1111/mec.12599
Taberlet, P., Gielly, L., Pautou, G., and Bouvet, J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105–1109. doi: 10.1007/BF00037152
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tian, B., Liu, R. R., Wang, L. Y., Qiu, Q., Chen, K. M., and Liu, J. Q. (2009). Phylogeographic analyses suggest that deciduous species (Ostryopsis davidiana Decne., Betulaceae) survived in northern China during the Last Glacial Maximum. J. Biogeogr. 36, 2148–2155. doi: 10.1111/j.1365-2699.2009.02157.x
Tzedakis, P. C., Emerson, B. C., and Hewitt, G. M. (2013). Cryptic or mystic? Glacial tree refugia in northern Europe. Trends Ecol. Evol. 28, 696–704. doi: 10.1016/j.tree.2013.09.001
Voss, N., Eckstein, R. L., and Durka, W. (2012). Range expansion of a selfing polyploid plant despite widespread genetic uniformity. Ann. Bot. 110, 585–593. doi: 10.1093/aob/mcs117
Wan, D. S., Feng, J. J., Jiang, D. C., Mao, K. S., Duan, Y. W., Miehe, G., et al. (2016). The Quaternary evolutionary history, potential distribution dynamics, and conservation implications for a Qinghai-Tibet Plateau endemic herbaceous perennial, Anisodus tanguticus (Solanaceae). Ecol. Evol. 6, 1977–1995. doi: 10.1002/ece3.2019
Wang, G. N., He, X. Y., Miehe, G., and Mao, K. S. (2014). Phylogeography of the Qinghai-Tibet Plateau endemic alpine herb Pomatosace filicula (Primulaceae). J. Syst. Evol. 52, 1–14. doi: 10.1111/jse.12089
Wang, H., Dai, J., Zheng, J., and Ge, Q. (2015). Temperature sensitivity of plant phenology in temperate and subtropical regions of China from 1850 to 2009. Int. J. Climatol. 35, 913–922. doi: 10.1002/joc.4026
Wang, L. Y., Abbott, R. J., Zheng, W., Chen, P., Wang, Y. J., and Liu, J. Q. (2009). History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Mol. Ecol. 18, 709–721. doi: 10.1111/j.1365-294X.2008.04055.x
Wang, Q., Abbott, R. J., Yu, Q. S., Lin, K., and Liu, J. Q. (2013). Pleistocene climate change and the origin of two desert plant species, Pugionium cornutum and Pugionium dolabratum (Brassicaceae), in Northwest China. New Phytol. 199, 277–287. doi: 10.1111/nph.12241
Wang, W., Tian, C. Y., Li, Y. H., and Li, Y. (2014). Molecular data and ecological niche modeling reveal the phylogeographic pattern of Cotinus coggygria (Anacardiaceae) in China’s warm-temperate zone. Plant Biol. 16, 1114–1120. doi: 10.1111/plb.12157
Wang, Z. W., Chen, S. T., Nie, Z. L., Zhang, J. W., Zhou, Z., Deng, T., et al. (2015). Climatic factors drive population divergence and demography: insights based on the phylogeography of a riparian plant species endemic to the Hengduan mountains and adjacent regions. PLoS One 10:e0145014. doi: 10.1371/journal.pone.0145014
Wen, J., Nie, Z. L., and Ickert-Bond, S. M. (2016). Intercontinental disjunctions between eastern Asia and western North America in vascular plants highlight the biogeographic importance of the Bering land bridge from late Cretaceous to Neogene. J. Syst. Evol. 54, 469–490. doi: 10.1111/jse.12222
Wen, J., Zhang, J. Q., Nie, Z. L., Zhong, Y., and Sun, H. (2014). Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 5:4. doi: 10.3389/fgene.2014.00004
Xu, T., Abbott, R. J., Milne, R. I., Mao, K., Du, F. K., Wu, G., et al. (2010). Phylogeography and allopatric divergence of cypress species (Cupressus L.) in the Qinghai-Tibetan Plateau and adjacent regions. BMC Evol. Biol. 10:194. doi: 10.1186/1471-2148-10-194
Yang, F. S., Li, Y. F., Ding, X., and Wang, X. Q. (2008). Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan Plateau and its correlation with the Quaternary climate change. Mol. Ecol. 17, 5135–5145. doi: 10.1111/j.1365-294X.2008.03976.x
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Ye, J. W., Bai, W. N., Bao, L., Wang, T. M., Wang, H. F., and Ge, J. P. (2017). Sharp genetic discontinuity in the aridity-sensitive Lindera obtusiloba (Lauraceae): solid evidence supporting the Tertiary floral subdivision in East Asia. J. Biogeogr. 44, 2082–2095. doi: 10.1111/jbi.13020
Yi, T., Miller, A. J., and Wen, J. (2004). Phylogenetic and biogeographic diversification of Rhus (Anacardiaceae) in the Northern Hemisphere. Mol. Phylogenet. Evol. 33, 861–879. doi: 10.1016/j.ympev.2004.07.006
Yu, Q. S., Wang, Q., Wu, G. L., Ma, Y. Z., He, X. Y., Wang, X., et al. (2013). Genetic differentiation and delimitation of Pugionium dolabratum and Pugionium cornutum (Brassicaceae). Plant Syst. Evol. 299, 1355–1365. doi: 10.1007/s00606-013-0800-3
Zhang, J. Q., Meng, S. Y., Wen, J., and Rao, G. Y. (2015). DNA barcoding of Rhodiola (Crassulaceae): a case study on a group of recently diversified medicinal plants from the Qinghai-Tibetan Plateau. PLoS One 10:e0119921. doi: 10.1371/journal.pone.0119921
Zhang, Q., Chiang, T. Y., George, M., Liu, J. Q., and Abbott, R. J. (2005). Phylogeography of the Qinghai-Tibetan Plateau endemic Juniperus przewalskii (Cupressaceae) inferred from chloroplast DNA sequence variation. Mol. Ecol. 14, 3513–3524. doi: 10.1111/j.1365-294X.2005.02677.x
Zhao, C., Wang, C. B., Ma, X. G., Liang, Q. L., and He, X. J. (2013). Phylogeographic analysis of a temperate-deciduous forest restricted plant (Bupleurum longiradiatum Turcz.) reveals two refuge areas in China with subsequent refugial isolation promoting speciation. Mol. Phylogenet. Evol. 68, 628–643. doi: 10.1016/j.ympev.2013.04.007
Zhao, J. L., Gugger, P. F., Xia, Y. M., and Li, Q. J. (2016). Ecological divergence of two closely related Roscoea species associated with late Quaternary climate change. J. Biogeogr. 43, 1990–2001. doi: 10.1111/jbi.12809
Zou, J. B., Peng, X. L., Li, L., Liu, J. Q., Miehe, G., and Opgenoorth, L. (2012). Molecular phylogeography and evolutionary history of Picea likiangensis in the Qinghai-Tibetan Plateau inferred from mitochondrial and chloroplast DNA sequence variation. J. Syst. Evol. 50, 341–350. doi: 10.1111/j.1759-6831.2012.00207.x
Keywords: Rhus chinensis, evolutionary history, DNA sequences, China’s temperate and subtropical zone, ecological niche modeling
Citation: Liang Y, Zhang Y, Wen J, Su X and Ren Z (2019) Evolutionary History of Rhus chinensis (Anacardiaceae) From the Temperate and Subtropical Zones of China Based on cpDNA and Nuclear DNA Sequences and Ecological Niche Model. Front. Genet. 10:171. doi: 10.3389/fgene.2019.00171
Received: 04 June 2018; Accepted: 15 February 2019;
Published: 05 March 2019.
Edited by:
Zhonghu Li, Northwest University, ChinaReviewed by:
James C. Crabbe, Wolfson College, United KingdomDong-rui Jia, Yunnan University, China
Copyright © 2019 Liang, Zhang, Wen, Su and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhumei Ren, em1yZW5Ac3h1LmVkdS5jbg== Xu Su, eHVzdTg1Mjc5NzJAMTI2LmNvbQ==
 Yukang Liang1
Yukang Liang1 Xu Su
Xu Su Zhumei Ren
Zhumei Ren