
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
GENERAL COMMENTARY article
Front. Genet., 07 March 2019
Sec. Genomic Assay Technology
Volume 10 - 2019 | https://doi.org/10.3389/fgene.2019.00162
A Commentary on
Leucine-rich Repeat Receptor-Like Gene Screen Reveals that Nicotiana RXEG1 Regulates Glycoside Hydrolase 12 MAMP Detection
by Wang, Y., Xu, Y., Sun, Y., Wang, H., Qi, J., Wan, B., et al. (2018). Nat. Commun. 9:594. doi: 10.1038/s41467-018-03010-8
Innate immunity against pathogen infection by membrane-localized receptors is evolutionarily conserved among eukaryotes (Palma et al., 2007). In plants, innate immunity is essential for defending against harmful microbes that cause dramatic losses in agriculture. Activation of plant immunity is generally determined by pattern-recognition receptors (PRRs). In contrast to mammals, all known plant PRRs are localized at the cell surface (Macho and Zipfel, 2014). There, they detect potentially harmful microbes by recognizing microbe-associated molecular patterns (MAMPs) (Boller and Felix, 2009; Böhm et al., 2014). The diversity and number of MAMPs recognized by individual plant species is likely larger than previously thought (Brunner and Nürnberger, 2012; Zhang et al., 2013; Böhm et al., 2014; Macho et al., 2014). Thus, characterizing PRRs in plants is key to plant-microbe interaction studies.
PRRs such as receptor-like kinases (RLKs) and receptor-like proteins (RLPs) (Shiu et al., 2004; Fischer et al., 2016) play crucial roles in plant immunity, growth, and development. Plant genomes contain hundreds of such receptor-like genes, most of which encode proteins with extracellular leucine-rich repeat (LRR) domains (Shiu and Bleecker, 2003; Shiu et al., 2004; Diévart et al., 2011; Sakamoto et al., 2012), but only a few PRRs with LRR domains have been identified (Sun et al., 2013; Hind et al., 2016; Tang et al., 2017; Wang et al., 2018). Therefore, an efficient approach for quickly identifying PRRs is important for understanding plant innate immunity and developing disease-resistant plants.
Recently, Wang et al. (2018) developed a high-throughput virus-induced gene silencing (VIGS)-based toolkit for characterizing LRR receptor-like genes on a genomic scale (Figure 1A). The approach was demonstrated on the plant model Nicotiana benthamiana, a solanaceous plant and a close relative of tobacco. As a result, 257 tobacco rattle virus–based constructs were generated to silence all 403 identified genes that encode predicted membrane-localized LRR-RLPs and LRR-RLKs in N. benthamiana. Moreover, Wang et al. (2018) successfully identified Response to XEG1 (RXEG1), an LRR-RLP that specifically recognizes the glycoside hydrolase 12 protein XEG1. Their study demonstrated that this genome-wide silencing assay can quickly identify new immune receptors to help mine and utilize crop resistance resources. VIGS has also been successfully developed for monocots (Yuan et al., 2011; Liou et al., 2014; Liu et al., 2016). In view of the importance of monocot cereals such as rice, wheat, and maize—and great yield losses due to microbial attacks—it would be worthwhile to evaluate this method for cereal crops.
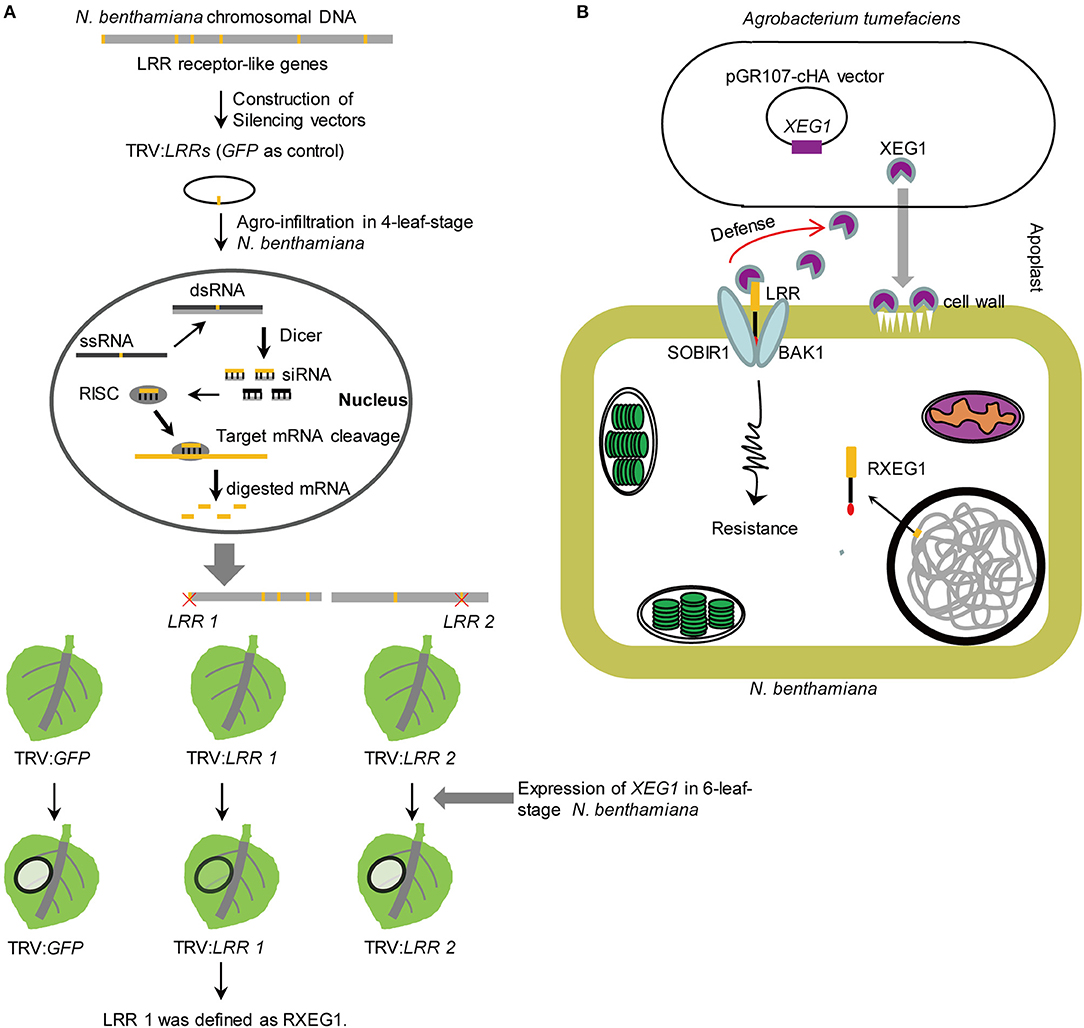
Figure 1. Identification of RXEG1 in N. benthamiana. (A) VIGS-based approach for identification of PRRs. The 403 LRR receptor-like genes, which encode proteins with both LRR and TM domains, were identified in N. benthamiana genome. LRR receptor-like genes were cloned into the modified gene silencing vector pTRV. Agrobacterium tumefaciens strain carrying 257 silencing vectors inflitrated into 4-leaf-stage N. benthamiana. Co-expressing GH12 protein XEG1 that was identified from Phytophthora sojae in 6-leaf-stage TRV scilencing N. benthamiana. XEG1-included cell death was compromised in leaves treated with RXEG1. (B) A possible model of RXEG1's role in defense to XEG1. Phytophthora infects plants early by secreting a glycolytic enzyme XEG1 to attack the cell wall. The receptor protein RXEG1, identified by Wang et al. (2018), can recognize XEG1 of Phytophthora infection. Subsequently, RXEG1 was found to be a key factor for cell necrosis and defense response after XEG1 recognition. Activation of RXEG1 could significantly improve plant resistance to Phytophthora infection. This figure was made based on Wang et al. (2018) and a news report (http://news.njau.edu.cn/2018/0210/c70a92967/page.htm).
The VIGS-based approach for identification of PRRs has advantages over methods that rely on map-based cloning and Arabidopsis T-DNA insertion lines (Zipfel et al., 2006; Jehle et al., 2013; Zhang et al., 2013; Albert et al., 2015): (1) VIGS can be easily performed on N. benthamiana, which are amenable to highly efficient VIGS and protein expression (within one month); and (2) VIGS in N. benthamiana avoids gene function redundancy and allows for simultaneous silencing of multiple homologous genes (Wang et al., 2018). Thus, silencing efficiency can be evaluated by analyzing gene expression and alterations in plant growth. The silencing efficiency of 230 LRR receptor-like genes (i.e., 43 LRR-RLP and 187 LRR-RLK genes) is > 60% (Wang et al., 2018).
In addition to identifying and characterizing PRRs, elucidating the mechanisms by which PRRs perceive microbial attack will significantly advance our understanding of plant innate immunity. The comprehensive and intensive work of Wang et al. (2018) revealed how N. benthamiana recognizes XEG1, a widely distributed MAMP in microbial taxa (Figure 1B). When microbes attack plants, XEG1 is secreted early into the infected plants to degrade xyloglucan and β-glucan in plant cell walls (Ma et al., 2015; Gui et al., 2017). Wang et al. (2018) demonstrated that RXEG1 specifically recognizes XEG1, associates XEG1 via the LRR domain in the apoplast, and forms a complex with the LRR receptor-like kinases BAK1 and SOBIR1 to transduce the XEG1-induced defense signal. The mode of action for RXEG1 will help elucidate the mechanisms of disease resistance and regulation of various crops in the future. RXEG1 in N. benthamiana can identify XEG1 family proteins secreted by various microbes. Therefore, RXEG1 could potentially be used to protect a broad range of plants, especially other solanaceous plant species such tomatoes, whereby high disease resistance might be achieved through genetic engineering or by spraying.
Wang et al. (2018) identified a new PRR (RXEG1) and its associated PKs. Further experimentation is needed to reveal ligand binding specificity, structural requirements, and modifications of RXEG1. More specifically, studies are needed to reveal how dynamics and protein composition are controlled during immune receptor complex assembly, and which intracellular hubs are involved in transforming various microbial signal inputs into a generic plant immune response.
Currently, the recognition of certain MAMPs remains restricted to solanaceous plants (Wang et al., 2016, 2018; Franco-Orozco et al., 2017). For example, RXEG1-like genes or proteins have been found in various dicots, but not in monocots. Silencing LRR receptor candidates for identification of MAMP recognition receptors in cereal crops is a straightforward approach that would advance MAMP recognition and improve crop resistance.
WW drafted the manuscript. SW and WW revised the manuscript. SW draw Figure 1. All authors contributed to the writing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Work in our lab was supported by the National Natural Science Foundation of China (31771700 and 31701343). We thank Dr. Jinghua Zhang and Dr. Hui Liu for valuable discussion in drawing Figure 1.
Albert, I., Böhm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1:15140. doi: 10.1038/nplants.2015.140
Böhm, H., Albert, I., Fan, L., Reinhard, A., and Nürnberger, T. (2014). Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 20, 47–54. doi: 10.1016/j.pbi.2014.04.007
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Brunner, F., and Nürnberger, T. (2012). Identification of immunogenic microbial patterns takes the fast lane. Proc. Natl. Acad. Sci. U.S.A. 109, 4029–4030. doi: 10.1073/pnas.1201444109
Diévart, A., Gilbert, N., Droc, G., Attard, A., Gourgues, M., Guiderdoni, E., et al. (2011). Leucine-rich repeat receptor kinases are sporadically distributed in eukaryotic genomes. BMC Evol. Biol. 11:367. doi: 10.1186/1471-2148-11-367
Fischer, I., Diévart, A., Droc, G., Dufayard, J. F., and Chantret, N. (2016). Evolutionary dynamics of the leucine-rich repeat receptor-like kinase (LRR-RLK) subfamily in Angiosperms. Plant Physiol. 170, 1595–1610. doi: 10.1104/pp.15.01470
Franco-Orozco, B., Berepiki, A., Ruiz, O., Gamble, L., Griffe, L. L., Wang, S., et al. (2017). A new proteinaceous pathogen-associated molecular pattern (PAMP) identified in Ascomycete fungi induces cell death in Solanaceae. N. Phytol. 214, 1657–1672. doi: 10.1111/nph.14542
Gui, Y. J., Chen, J. Y., Zhang, D. D., Li, N. Y., Li, T. G., Zhang, W. Q., et al. (2017). Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 19, 1914–1932. doi: 10.1111/1462-2920.13695
Hind, S. R., Strickler, S. R., Boyle, P. C., Dunham, D. M., Bao, Z., O'Doherty, I. M., et al. (2016). Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2:16128. doi: 10.1038/nplants.2016.128
Jehle, A. K., Lipschis, M., Albert, M., Fallahzadeh-Mamaghani, V., Fürst, U., Mueller, K., et al. (2013). The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 25, 2330–2340. doi: 10.1105/tpc.113.110833
Liou, M. R., Huang, Y. W., Hu, C. C., Lin, N. S., and Hsu, Y. H. (2014). A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol. J. 12, 330–343. doi: 10.1111/pbi.12140
Liu, N., Xie, K., Jia, Q., Zhao, J., Chen, T., Li, H., et al. (2016). Foxtail mosaic virus-induced gene silencing in monocot plants. Plant Physiol. 171, 1801–1807. doi: 10.1104/pp.16.00010
Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., et al. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
Macho, A. P., Schwessinger, B., Ntoukakis, V., Brutus, A., Segonzac, C., Roy, S., et al. (2014). A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343, 1509–1512. doi: 10.1126/science.1248849
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Palma, K., Zhao, Q., Cheng, Y. T., Bi, D., Monaghan, J., Cheng, W., et al. (2007). Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21, 1484–1493. doi: 10.1101/gad.1559607
Sakamoto, T., Deguchi, M., Brustolini, O. J., Santos, A. A., Silva, F. F., and Fontes, E. P. (2012). The tomato RLK superfamily: phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense. BMC Plant Biol. 12:229. doi: 10.1186/1471-2229-12-229
Shiu, S. H., and Bleecker, A. B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530–543. doi: 10.1104/pp.103.021964
Shiu, S. H., Karlowski, W. M., Pan, R., Tzeng, Y. H., Mayer, K. F., and Li, W. H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
Sun, Y., Li, L., Macho, A. P., Han, Z., Hu, Z., Zipfel, C., et al. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628. doi: 10.1126/science.1243825
Tang, D., Wang, G., and Zhou, J. M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Wang, L., Albert, M., Einig, E., Fürst, U., Krust, D., Felix, G., et al. (2016). The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat. Plants 2:16185. doi: 10.1038/nplants.2016.185
Wang, Y., Xu, Y., Sun, Y., Wang, H., Qi, J., Wan, B., et al. (2018). Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun. 9:594. doi: 10.1038/s41467-018-03010-8
Yuan, C., Li, C., Yan, L., Jackson, A. O., Liu, Z., Han, C., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6:e26468. doi: 10.1371/journal.pone.0026468
Zhang, W., Fraiture, M., Kolb, D., Löffelhardt, B., Desaki, Y., Boutrot, F. F., et al. (2013). Arabidopsis receptor-like protein 30 and receptor-like kinase suppressor of BIR-1/EVERSGHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241. doi: 10.1105/tpc.113.117010
Keywords: genome-wide VIGS screen, leucine-rich repeat (LRR) receptor-like gene (protein), microbe-associated molecular patterns (MAMPs), Nicotiana benthamiana, pattern-recognition receptors (PRRs), plant immune response, response to XEG1 (RXEG1) protein, virus-induced gene silencing (VIGS)
Citation: Wu S, Wang W, Xu X and Wu X (2019) Commentary: Leucine-rich Repeat Receptor-Like Gene Screen Reveals that Nicotiana RXEG1 Regulates Glycoside Hydrolase 12 MAMP Detection. Front. Genet. 10:162. doi: 10.3389/fgene.2019.00162
Received: 14 March 2018; Accepted: 14 February 2019;
Published: 07 March 2019.
Edited by:
Emmanouil Dermitzakis, Université de Genève, SwitzerlandReviewed by:
Jiannis Ragoussis, McGill University, CanadaCopyright © 2019 Wu, Wang, Xu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Wang, d2FuZ3dlaUBoZW5hdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.