- 1INGM, National Institute of Molecular Genetics, “Romeo ed Enrica Invernizzi”, Milan, Italy
- 2Dipartimento di Bioscienze, Università Degli Studi Di Milano, Milan, Italy
Ribosomes have been long considered as executors of the translational program. The fact that ribosomes can control the translation of specific mRNAs or entire cellular programs is often neglected. Ribosomopathies, inherited diseases with mutations in ribosomal factors, show tissue specific defects and cancer predisposition. Studies of ribosomopathies have paved the way to the concept that ribosomes may control translation of specific mRNAs. Studies in Drosophila and mice support the existence of heterogeneous ribosomes that differentially translate mRNAs to coordinate cellular programs. Recent studies have now shown that ribosomal activity is not only a critical regulator of growth but also of metabolism. For instance, glycolysis and mitochondrial function have been found to be affected by ribosomal availability. Also, ATP levels drop in models of ribosomopathies. We discuss findings highlighting the relevance of ribosome heterogeneity in physiological and pathological conditions, as well as the possibility that in rate-limiting situations, ribosomes may favor some translational programs. We discuss the effects of ribosome heterogeneity on cellular metabolism, tumorigenesis and aging. We speculate a scenario in which ribosomes are not only executors of a metabolic program but act as modulators.
Introduction
Translation the process by which mRNAs are translated into proteins by ribosomes. Eukaryotic ribosomes are evolutionarily conserved ribozymes constituted by ribosomal proteins (RPs) and rRNAs, whose structure has been spectacularly resolved (Ben-Shem et al., 2011; Klinge et al., 2011; Khatter et al., 2015). Ribosome biogenesis is a massive process occurring in the nucleolus of all cells. Recent progress, combining biochemical techniques, with structural and genetic evidence, has shown that ribosome synthesis is catalyzed and coordinated by more than 200 biogenesis factors. Ribosome biogenesis, therefore, proceeds through precise assembly steps that include several quality checkpoints, both in the nucleus and in the cytoplasm (Kressler et al., 2017; Pena et al., 2017). Furthermore, impairment of these checkpoints leads to defects in maturation that are associated with disease (Narla and Ebert, 2010; Ruggero and Shimamura, 2014).
In the cytoplasm, ribosomes are thought to constitute the hardware of the protein synthesis machinery, which fulfills its activity through four main phases: initiation, elongation, termination, and recycling. The initiation step is one of the most important steps of translation regulation, involving initiation factors, mRNAs, tRNAs, and ribosomes. Briefly, 40S subunits directly bind mRNAs in a way that is dependent on initiation factors and on mRNA structure and, after mRNA binding and scanning to an appropriate start codon, 60S subunits are recruited. Several studies elucidated how translation initiation is affected by alteration in mRNAs-binding factors (Loreni et al., 2014; Chu et al., 2016; Truitt and Ruggero, 2016) and by different features in mRNAs structures, i.e., Untranslated regions (UTRs). Recently, also tRNA has been linked to selective translation, and reprogramming of metabolism since codon reprogramming leads to HIF1α synthesis and an increase of glycolytic factors (Rapino et al., 2018).
Evidences that ribosomes exist in different forms in different cell types or during different stages of development (Milne et al., 1975; Bortoluzzi et al., 2001; Volarevic and Thomas, 2001) have suggested the presence of ribosome heterogeneity. It has been recently demonstrated that mutations in some RPs result in selective translation (Shi and Barna, 2015) and mutations in proteins causing an impairing in ribosome maturation and function, as in the case of ribosomopathies, show a specific mRNA translation signature (Brina et al., 2015; In et al., 2016) In conclusion, in recent years there has been growing evidence that translation is driven by ribosome heterogeneity, manifested as ribosome populations differing in ribosomal components. In this review we discuss ribosome heterogeneity in physiological, and pathological conditions, highlighting the role of translation machinery in driving the last step of the molecular biology central dogma, which elects ribosomes as players in specific mRNAs translation.
Ribosome Heterogeneity in Physiological Conditions May Account for Differential Translation
This topic has been recently discussed (Genuth and Barna, 2018a,b) and we will give a simple summary of some perspectives. Ribosomes are constituted by approximately 80 RPs. For many years now, it is known that the relative abundance of different RPs, in different tissues, or in different growth conditions, may vary (Milne et al., 1975; Bortoluzzi et al., 2001; Volarevic and Thomas, 2001). This is a sine qua non-condition for ribosomal heterogeneity. An obvious alternative explanation for an imbalance of the stoichiometry of RPs within a cell is that RPs perform ribosome-independent functions. An experimental complexity is, therefore, to define whether a differential translation is due to the direct action of heterogeneous ribosomes or to regulatory pathways affected by free RPs. This is the case for RACK1 that was originally isolated as a PKC receptor (Ron et al., 1994; Gallo and Manfrini, 2015). RACK1 is a structural protein of 40S subunits (Gerbasi et al., 2004), involved in several extraribosomal functions (Mamidipudi et al., 2004; Robles et al., 2010; Wehner et al., 2011; Gandin et al., 2013; Fei et al., 2017). RACK1 may affect the efficiency of ribosomes directly (Ceci et al., 2003; Shor et al., 2003; Guo J. et al., 2011; Dobrikov et al., 2018a,b) or indirectly through signaling pathways (Gandin et al., 2013; Volta et al., 2013). In conclusion, data demonstrate that in physiological conditions, ribosomal networks may be more complex than expected and perform choices in translational regulation.
Ribosomal heterogeneity exists in physiological conditions. Accurate proteomics studies have identified sub-stoichiometric relationships within translating polysomes (Shi et al., 2017), showing that ribosomes may preferentially translate specific mRNAs. An experimental validation shows that ribosomes devoid of either RPS25 (eS25) or RpL10A (uL1), in vivo, translate specific mRNAs. Mechanistically, this study shows that the 60S subunits may affect mRNA recruitment through the binding of RPL10A (uL1) to IRES (Internal Ribosome Entry Site) sequences in the 5′UTR (Shi et al., 2017). In monocytes, interferon gamma driven phosphorylation results in RPL13A (uL13) detachment, but here it is still unknown whether ribosomes devoid of RPL13A (uL13) are able to translate selectively (Jia et al., 2012). Furthermore, RPL10 (uL16) R98S mutant leukemia cells are able to survive high oxidative stress levels by increasing IRES-dependent BCL-2 translation (Kampen et al., 2018).
Thus, the concept of a monolithic ribosome (Moore et al., 1968; Yusupova and Yusupov, 2017) may be accompanied by the existence of a more flexible ribosomal platform that performs further tuning on gene expression (Shi and Barna, 2015).
Ribosome Heterogeneity in Pathological Conditions Affects Translation and Gene Expression
Ribosomopathies are inherited diseases caused by the loss of ribosomal component functionality. Some examples of ribosomopathies include Diamond-Blackfan Anemia syndrome (DBA), Shwachman-Diamond syndrome (SDS), Treacher Collins syndrome, 5q-myelodysplastic syndrome, and Dyskeratosis Congenita (DKC). Notably, all of these syndromes are characterized by variably penetrant phenotypes in which specific tissue deficits are found (Narla and Ebert, 2010). Early on, it was shown that DKC1 mutations reduce pseudouridylation and impair IRES mediated translation (Yoon et al., 2006).
As a case for study, we will focus our discussion on SDS. Signs of SDS include a peculiar exocrine pancreatic insufficiency, along with neutropenia and variable abnormalities in the skeleton and other organs. In addition, SDS is characterized by a reduction in growth, accompanied by an increased incidence of Acute Myeloid Leukemia, (AML; Dror, 2008). At the ribosomal level, SDS is characterized by the partial loss of free 60S ribosomal subunits due to, in most cases, mutations in the SBDS gene that is necessary for 60S maturation (Boocock et al., 2003; Wong et al., 2011). In a minority of cases, mutations of EFL1p, which acts in synergy with SBDS, have been found (Stepensky et al., 2017; Tan et al., 2018). Overall, the reduced functionality of 60S ribosomes is a common theme for SDS (Warren, 2018). All together these findings generate three questions: (a) how the loss of functionality of ubiquitous 60S ribosomes can generate tissue-specific defects, (b) how specific translational programs can be affected by the lack of 60S subunits, (c) how can we reconcile increased tumor with reduced growth.
Addressing this last question helps to put in the right context the other two. We have recently demonstrated in our lab that cells with mutant Sbds have reduced colony formation ability and are transformed less efficiently by oncogenes (Calamita et al., 2017). In this context, we demonstrated that Sbds deficiency directly acts by reducing the maximal oncogenic and translational capability of cells (Calamita et al., 2017). The paradox of reduced growth associated with tumor predisposition may not necessarily be associated with specific translation in tumor cells, but with a general impairment of tissue homeostasis that favors the appearance of mutant clones. For instance, increased tumor formation is observed in immunocompromised individuals (Verhoeven et al., 2018). To support this interpretation, the relationship between neutropenia and AML was described by different groups (Freedman et al., 2000; Link et al., 2007; Touw and Beekman, 2013). In conclusion, different cell types can be differentially affected by the reduction of RPs, i.e., thresholds can be different depending on the specific cellular demand of ribosomes for translation.
The question of the mechanism by which defects in 60S ribosomes lead to differential translation is more challenging since to our knowledge mRNA selection is driven by 40S subunits, prior to 60S engagement. However, the effects of 60S levels on specific translation are pervasive, and, as described before, IRES mRNA binding can be affected by RPL10 (uL16). In the case of Sbds depletion, characterized by reduced free 60S, two studies have addressed the question of preferential translation performing either microarray (Nihrane et al., 2009), or RNA-Seq on polysomes (Calamita et al., 2017). In addition, a reporter-based study has addressed the effect of SBDS depletion on reinitiation (In et al., 2016). Together, these studies support a model in which the SBDS deficiency reduces free 60S levels diminishing the maximal translational capability, and simultaneously changing translational selectivity. In this context, mRNAs that are intrinsically poorly translated because of uORFs (upstream Open Reading Frames) that require reinitiation are particularly disfavored. Similarly, mouse models have underscored that the reduction of 60S RPs affects the translational program of IRES containing mRNAs (Barna et al., 2008; Kondrashov et al., 2011; Xue et al., 2015).
Finally, mathematical modeling of translation suggests that a quantitative reduction in the translational output may result in strong alterations of specific mRNA translation due to stochastic events (Heinrich and Rapoport, 1980; Mills and Green, 2017). We conclude that some mRNAs can be particularly sensitive to ribosomal availability, and we speculate that this property has been evolutionarily exploited to connect ribosomes with other cellular events. What we still lack is understanding the precise mechanisms.
A Common Theme for the Regulatory Function of Ribosomes?
Metabolic pathways are necessary for converting essential nutrients into energy and macromolecules that sustain cell growth and proliferation. Nutrients and metabolic pathways control all facets of cellular functions. Nutrient and growth factors converge on the translational machinery through signaling pathways that, in turn, regulate the synthesis of ribosomes and the activity of translation factors (Roux and Topisirovic, 2018). Then, translation factors crosstalk to metabolic choices (Biffo et al., 2018). Some well-established observations are the following. mTORC1 controls mitochondrial activity and biogenesis by selectively promoting translation of nucleus-encoded mitochondria-related mRNAs, via inhibition of the eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (4E-BPs; Morita et al., 2013). ROS generation is also controlled partly at the translational level through eIF4E (Truitt et al., 2015). Glutamine metabolism is controlled by eIF4B-mediated translation downstream of mTORC1 pathway (Csibi et al., 2014). eIF3 complex mediates energy metabolism (Shah et al., 2016). Rate-limiting initiation factors that link 60S ribosome biogenesis to translation as eIF6 hierarchically control lipid synthesis and metabolism, through uORF and G/C rich 5′UTR sequences (Brina et al., 2015). eIF5A2 accelerates lipogenesis in hepatocellular carcinoma (Cao et al., 2017). In general, translation and metabolism are dysregulated in a coordinated fashion (Leibovitch and Topisirovic, 2018), and initiation factors may act upstream of metabolic reprogramming (Biffo et al., 2018). The next question is whether ribosomes also control metabolic pathways.
In Zebrafish, rpl11 mutation decreased the glycolytic rate and the lower activity of glycolytic enzymes is rescued by p53 inhibition (Danilova et al., 2011). Moreover, defects, mutations or imbalance of RPs stabilized p53 and changed metabolic flux, specifically by decreasing glycolysis and enhancing aerobic respiration (Deisenroth and Zhang, 2011). Albeit these data do not support a direct crosstalk between ribosome activity and metabolism, they suggest overall that when the translation machinery is perturbed, coordinated pathways involved in cell homeostasis and metabolism are also altered.
Recently, it has been shown that SDS cells display an impairment in Complex IV activity, which causes an oxidative phosphorylation metabolic defect, with a consequent decrease in ATP production (Ravera et al., 2016). The authors suggest an indirect effect of SBDS mutation on energy production levels, indicating a possible role of calcium homeostasis in altering complex IV activity. In our lab we performed a characterization of a cellular model for SDS by immortalizing Mouse Embryonic Fibroblasts (MEFs; Calamita et al., 2017) derived from an SDS mouse model carrying the R126T mutation in homozygosity (SbdsR126T/R126T MEFs) (Tourlakis et al., 2012). Briefly, we established a model for studying SBDS function by retrasducing SbdsR126T/R126T MEFs with either wild-type Sbds (SbdsRESCUE), or mock control (SbdsMOCK) vectors. In this way, we can separate direct events due to a lack of SBDS from indirect effects. We confirmed a decrease in ATP levels associated with Sbds mutation. In addition, our RNA-Seq analysis revealed that genes belonging to complex IV were less expressed when Sbds was mutated (Figure 1). This downregulation could explain an impairment in cytochrome C oxidase activity and a consequent defect in ATP production. Moreover, there is a defect in oxygen consumption rate in SDS cells (Ravera et al., 2016; Calamita et al., 2017), as well as a reduction in the lactate/pyruvate ratio (Calamita et al., 2017). The mechanistic connection between ribosome function and the metabolic effects of its impairment is still to be clarified. Overall, a reduction in ribosomal efficiency seems to associate with a reduction in energy levels and lipid biosynthesis. We suggest that ribosomal capability has coevolved with other cellular functions and, specifically, ribosomes are intimately linked to nutrient levels and cellular growth.
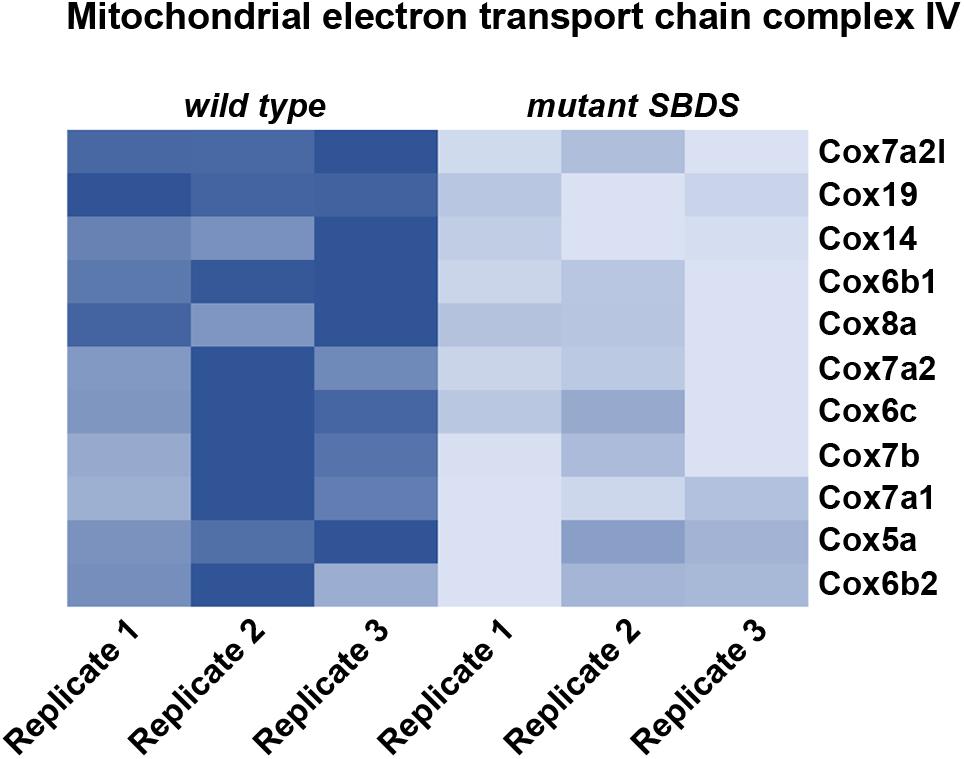
FIGURE 1. Heat map representing relative gene expression levels in a cellular model for Shwachman Diamond Syndrome. We re-infected cells bearing the mutation R126T/R126T (corresponding to one of the most common mutations associated with Shwachman Diamond Syndrome) in the Sbds gene (SbdsR126T/R126T MEFs) with either wild type Sbds (SbdsRESCUE), or mock control (SbdsMOCK). Heat map represents relative gene expression levels of genes associated with mitochondrial electron transport chain complex IV, showing an overall reduction in mutant SbdsMOCK cells, indicating an impairment in ATP production. Heatmap is based on RNASeq raw data available at www.ebi.ac.uk/arrayexpress with accession number ID E-MTAB-5089, and analyzed in our previous work (Calamita et al., 2017).
The connection between ribosomes and growth is indeed strong and well-known. In Drosophila melanogaster the haploinsufficiency of RPs results in the minute phenotype, which includes short and thin bristles and smaller flies (Lambertsson, 1998; Marygold et al., 2007). Moreover, as shown by a myriad of papers, depletion of RPs causes a delay/arrest in cell cycle progression. In several cases, the regulation of growth is associated with ribosome independent function of RPs (Dai and Lu, 2004; Mamidipudi et al., 2004, Dutt et al., 2011; Yao et al., 2016). In other cases, the inhibition of growth has been directly linked to translational control driven by ribosomes (Barna et al., 2008). Depletion of different RPs may result in different types of inhibition of cell cycle progression, in line with the concept of heterogeneity in ribosomes (Badhai et al., 2009). Conversely, nucleolar enlargement grossly equals an increased production of ribosomes and is observed in many cancers (Montanaro et al., 2008). In many models, some heterozygous deletions of RPs reduce tumor growth (Barna et al., 2008; Chen et al., 2014; Wilson-Edell et al., 2014), while some others are associated with cancer development as demonstrated for the first time in zebrafish mutants for RPs in 2004 (Amsterdam et al., 2004). In the last years, several somatic mutations have been linked to tumor progression and belong to both 60S subunits such as RPL5 (uL18) and RPL10 (uL16) (De Keersmaecker et al., 2013), RPL 11 (uL5) (Tzoneva et al., 2013; Fancello et al., 2017), RPL22 (eL22) (Rao et al., 2012) and RPL 23 (uL23) (Fancello et al., 2017) and to 40S subunits such as RPS15 (uS19) (Landau et al., 2015; Ljungstrom et al., 2016), RPS27 (eS27) (Dutton-Regester et al., 2014) and RPSA (uS2) (Fancello et al., 2017). On the contrary, RPs overexpression has been also identified in cancer progression (Artero-Castro et al., 2011; Guo X. et al., 2011; Yang et al., 2016). Several recent reviews provide a comprehensive discussion on how, in some cases, loss of RPs contributes to cancer (Sulima et al., 2017; Genuth and Barna, 2018a; Pelletier et al., 2018).
The ribosomal apparatus also appears to affect longevity. Alterations in ribosomal protein expression result in an extension of eukaryotic lifespan (Hansen et al., 2007; Steffen et al., 2008).
In short, the persistent link between ribosomal function in growth and metabolism makes us speculate that there may be a yet-to-be-unveiled mechanistic connection. We favor a model in which mRNAs important for cell cycle progression or for key metabolic pathways contain UTRs that have coevolved with the translational machinery in order to be preferentially translated in conditions of optimal ribosomal capability. In this context, ribosomal heterogeneity may further tune the cell′s translational capabilities.
Mitochondrial Ribosomes
Several mitochondrial ribosome proteins are also involved in different cellular processes, such as cell cycle, apoptosis and mitochondrial homeostasis regulation. Mutations in mt-RPs genes are associated with mitochondrial dysfunctions and disorders (Saada et al., 2007; Smits et al., 2011; Serre et al., 2013; Menezes et al., 2015; Richman et al., 2015). For instance, mutant MRPS16 (bS16m) causes mitochondrial respiratory chain disorders (Miller et al., 2004) and loss of MRPL10 (uL10m) diminished mitochondrial respiration and intracellular ATP levels (Li et al., 2016). In addition, a recent study claims the regulation of cytoplasmic protein homeostasis by mitochondrial translation (Suhm et al., 2018). These studies elucidate the fact that a crosstalk between the cytoplasmic and the mitochondrial ribosomal machinery may be present.
Conclusion
Ribosomes have been long considered as monolithic structures ensuring mRNAs translation in a passive way. Nowadays, it has been well established that ribosomes can affect not only mRNA selection but also other fundamental processes such as cell growth and lately, cell homeostasis and metabolism (Figure 2). There is an increasing number of studies evidencing that the inter-correlation between ribosomes and metabolic pathways leads to a common cellular phenotype. Since ribosomes are a rate-limiting component of the translational program, further studies are needed to elucidate specific molecular mechanisms by which ribosome heterogeneity, supported by the translational apparatus, sustain cell growth and metabolic homeostasis.
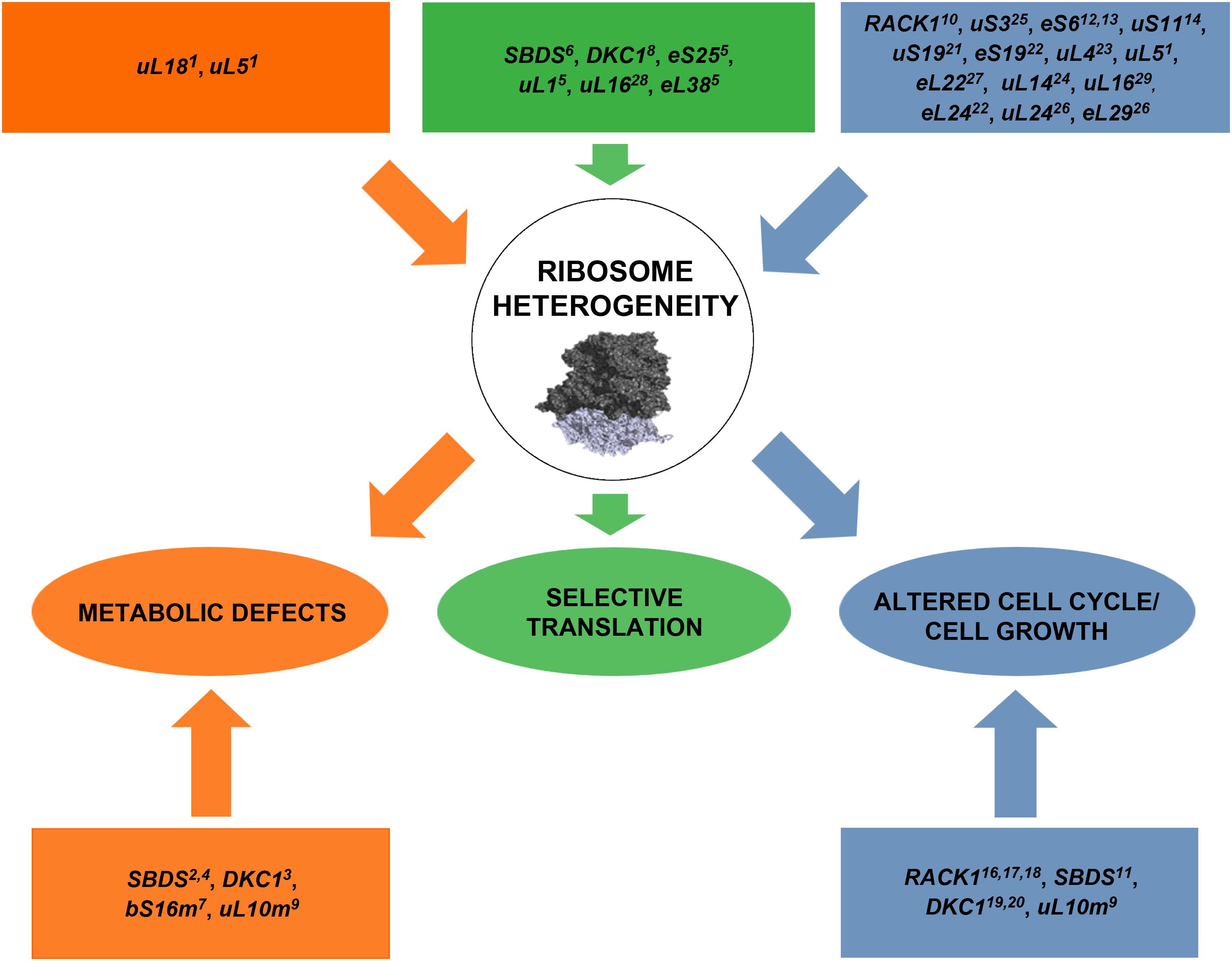
FIGURE 2. A schematic model representing the list of genes whose mutations perturb ribosome machinery. The color indicates the associated phenotype, specified in ovals in the third row. In some cases, the effect is associated or supposed to be associated with extra-ribosomal functions of mutated genes (listed in squares in the fourth row). Briefly, alterations in ribosome biogenesis and/or mutations in ribosomal proteins are responsible for metabolic changes, abnormal cell cycle progression/cell growth, and selective translation. Ribosomal subunits adapted from 40S (Lomakin and Steitz, 2013) PDB code 5ANB to 60S (Weis et al., 2015) PDB code 4KZX. Orange color indicates the flux of alterations converging to metabolic defects, green color the flux converging to selective translation and blue color the one converging to altered cell cycle/cell growth. The exploration of effects of RP lesions on cell cycle, translation, and cell metabolism is a highly active area of research and novel effects of RP lesions still need to be discovered. 1Danilova et al., 2011; 2Calamita et al., 2017; 3Angrisani et al., 2018; 4Ravera et al., 2016; 5Shi et al., 2017; 6In et al., 2016; 7Miller et al., 2004; 8Yoon et al., 2011; 9Li et al., 2016; 10Ceci et al., 2012; 11Menne et al., 2007; 12Volarevic et al., 2000; 13Sulic et al., 2005; 14Dutt et al., 2011; 15Teng et al., 2013; 16Hermanto et al., 2002; 17Mamidipudi et al., 2004; 18Fei et al., 2017; 19Alawi and Lin, 2013; 20Ge et al., 2010; 21Yao et al., 2016; 22Badhai et al., 2009; 23He et al., 2016; 24Dai et al., 2004; 25Yoon et al., 2006; 26Li et al., 2012; 27Stadanlick et al., 2011; 28Kampen et al., 2018; 29De Keersmaecker et al., 2013.
Author Contributions
SB and PC reviewed and edited the manuscript. SB, PC, GG, and AS reviewed the literature. SB, PC, and AM wrote the manuscript. GG conceived and prepared figures, and edited the manuscript. All authors contributed, read, and approved the manuscript.
Funding
This work was supported by Grant ERC TRANSLATE 338999 and IG 2014 AIRC to SB. PC was supported by Fondazione Umberto Veronesi.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize for the excellent works that could not be cited due to space constraints.
References
Alawi, F., and Lin, P. (2013). Dyskerin localizes to the mitotic apparatus and is required for orderly mitosis in human cells. PLoS One 8:e80805. doi: 10.1371/journal.pone.0080805
Amsterdam, A., Sadler, K. C., Lai, K., Farrington, S., Bronson, R. T., Lees, J. A., et al. (2004). Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2:E139. doi: 10.1371/journal.pbio.0020139
Angrisani, A., Matrone, N., Belli, V., Vicidomini, R., Di Maio, N., Turano, M., et al. (2018). A functional connection between dyskerin and energy metabolism. Redox Biol. 14, 557–565. doi: 10.1016/j.redox.2017.11.003
Artero-Castro, A., Castellvi, J., García, A., Hernández, J., Ramón y Cajal, S., and Lleonart, M. E. (2011). Expression of the ribosomal proteins Rplp0, Rplp1, and Rplp2 in gynecologic tumors. Hum. Pathol. 42, 194–203. doi: 10.1016/j.humpath.2010.04.020
Badhai, J., Fröjmark, A. S., Davey, E. J., Schuster, J., and Dahl, N. (2009). Ribosomal protein S19 and S24 insufficiency cause distinct cell cycle defects in Diamond-Blackfan anemia. Biochim. Biophys. Acta 1792, 1036–1042. doi: 10.1016/j.bbadis.2009.08.002
Barna, M., Pusic, A., Zollo, O., Costa, M., Kondrashov, N., Rego, E., et al. (2008). Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975. doi: 10.1038/nature07449
Ben-Shem, A., Garreau de Loubresse, N., Melnikov, S., Jenner, L., Yusupova, G., and Yusupov, M. (2011). The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334, 1524–1529. doi: 10.1126/science.1212642
Biffo, S., Manfrini, N., and Ricciardi, S. (2018). Crosstalks between translation and metabolism in cancer. Curr. Opin. Genet. Dev. 48, 75–81. doi: 10.1016/j.gde.2017.10.011
Boocock, G. R., Morrison, J. A., Popovic, M., Richards, N., Ellis, L., Durie, P. R., et al. (2003). Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 33, 97–101. doi: 10.1038/ng1062
Bortoluzzi, S., d’Alessi, F., Romualdi, C., and Danieli, G. A. (2001). Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics 17, 1152–1157. doi: 10.1093/bioinformatics/17.12.1152
Brina, D., Miluzio, A., Ricciardi, S., Clarke, K., Davidsen, P. K., Viero, G., et al. (2015). eIF6 coordinates insulin sensitivity and lipid metabolism by coupling translation to transcription. Nat. Commun. 6:8261. doi: 10.1038/ncomms9261
Calamita, P., Miluzio, A., Russo, A., Pesce, E., Ricciardi, S., Khanim, F., et al. (2017). SBDS-deficient cells have an altered homeostatic equilibrium due to translational inefficiency which explains their reduced fitness and provides a logical framework for intervention. PLoS Genet. 13:e1006552. doi: 10.1371/journal.pgen.1006552
Cao, T. T., Lin, S. H., Fu, L., Tang, Z., Che, C. M., Zhang, L. Y., et al. (2017). Eukaryotic translation initiation factor 5A2 promotes metabolic reprogramming in hepatocellular carcinoma cells. Carcinogenesis 38, 94–104. doi: 10.1093/carcin/bgw119
Ceci, M., Gaviraghi, C., Gorrini, C., Sala, L. A., Offenhauser, N., Marchisio, P. C., et al. (2003). Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426, 579–584. doi: 10.1038/nature02160
Ceci, M., Welshhans, K., Ciotti, M. T., Brandi, R., Parisi, C., Paoletti, F., et al. (2012). RACK1 is a ribosome scaffold protein for beta-actin mRNA/ZBP1 complex. PLoS One 7:e35034. doi: 10.1371/journal.pone.0035034
Chen, B., Zhang, W., Gao, J., Chen, H., Jiang, L., Liu, D., et al. (2014). Downregulation of ribosomal protein S6 inhibits the growth of non-small cell lung cancer by inducing cell cycle arrest, rather than apoptosis. Cancer Lett. 354, 378–389. doi: 10.1016/j.canlet.2014.08.045
Chu, J., Cargnello, M., Topisirovic, I., and Pelletier, J. (2016). Translation initiation factors: reprogramming protein synthesis in cancer. Trends Cell Biol. 26, 918–933. doi: 10.1016/j.tcb.2016.06.005
Csibi, A., Lee, G., Yoon, S. O., Tong, H., Ilter, D., Elia, I., et al. (2014). The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Curr. Biol. 24, 2274–2280. doi: 10.1016/j.cub.2014.08.007
Dai, M. S., and Lu, H. (2004). Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482. doi: 10.1074/jbc.M403722200
Dai, M. S., Zeng, S. X., Jin, Y., Sun, X. X., David, L., and Lu, H. (2004). Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 24, 7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004
Danilova, N., Sakamoto, K. M., and Lin, S. (2011). Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br. J. Haematol. 152, 217–228. doi: 10.1111/j.1365-2141.2010.08396.x
De Keersmaecker, K., Atak, Z. K., Li, N., Vicente, C., Patchett, S., Girardi, T., et al. (2013). Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 45, 186–190. doi: 10.1038/ng.2508
Deisenroth, C., and Zhang, Y. (2011). The ribosomal protein-Mdm2-p53 pathway and energy metabolism: bridging the gap between feast and famine. Genes Cancer 2, 392–403. doi:10.1177/1947601911409737
Dobrikov, M. I., Dobrikova, E. Y., and Gromeier, M. (2018a). Ribosomal RACK1:protein kinase C βII modulates intramolecular interactions between unstructured regions of eukaryotic initiation factor 4G (eIF4G) that control eIF4E and eIF3 binding. Mol. Cell. Biol. 38, 1–11. doi: 10.1128/MCB.00306-18
Dobrikov, M. I., Dobrikova, E. Y., and Gromeier, M. (2018b). Ribosomal RACK1:protein kinase C βII phosphorylates eukaryotic initiation factor 4G1 at S1093 to modulate cap-dependent and -independent translation initiation. Mol. Cell. Biol. doi: 10.1128/MCB.00304-18
Dror, Y. (2008). Shwachman-Diamond syndrome: implications for understanding the molecular basis of leukaemia. Expert Rev. Mol. Med. 10:e38. doi: 10.1017/S1462399408000938
Dutt, S., Narla, A., Lin, K., Mullally, A., Abayasekara, N., Megerdichian, C., et al. (2011). Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117, 2567–2576. doi: 10.1182/blood-2010-07-295238
Dutton-Regester, K., Gartner, J. J., Emmanuel, R., Qutob, N., Davies, M. A., Gershenwald, J. E., et al. (2014). A highly recurrent RPS27 5’UTR mutation in melanoma. Oncotarget 5, 2912–2917. doi: 10.18632/oncotarget.2048
Fancello, L., Kampen, K. R., Hofman, I. J., Verbeeck, J., and De Keersmaecker, K. (2017). The ribosomal protein gene RPL5 is a haploinsufficient tumor suppressor in multiple cancer types. Oncotarget 8, 14462–14478. doi: 10.18632/oncotarget.14895
Fei, L., Ma, Y., Zhang, M., Liu, X., Luo, Y., Wang, C., et al. (2017). RACK1 promotes lung cancer cell growth via an MCM7/RACK1/ Akt signaling complex. Oncotarget 8, 40501–40513. doi: 10.18632/oncotarget.17120
Freedman, M. H., Bonilla, M. A., Fier, C., Bolyard, A. A., Scarlata, D., Boxer, L. A., et al. (2000). Myelodysplasia syndrome and acute myeloid leukemia in patients with congenital neutropenia receiving G-CSF therapy. Blood 96, 429–436.
Gallo, S., and Manfrini, N. (2015). Working hard at the nexus between cell signaling and the ribosomal machinery: an insight into the roles of RACK1 in translational regulation. Translation 3:e1120382. doi: 10.1080/21690731.2015.1120382
Gandin, V., Senft, D., Topisirovic, I., and Ronai, Z. A. (2013). RACK1 function in cell motility and protein synthesis. Genes Cancer 4, 369–377. doi: 10.1177/1947601913486348
Ge, J., Rudnick, D. A., He, J., Crimmins, D. L., Ladenson, J. H., Bessler, M., et al. (2010). Dyskerin ablation in mouse liver inhibits rRNA processing and cell division. Mol. Cell. Biol. 30, 413–422. doi: 10.1128/MCB.01128-09
Genuth, N. R., and Barna, M. (2018a). Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genet. 19, 431–452. doi: 10.1038/s41576-018-0008-z
Genuth, N. R., and Barna, M. (2018b). The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell 71, 364–374. doi: 10.1016/j.molcel.2018.07.018
Gerbasi, V. R., Weaver, C. M., Hill, S., Friedman, D. B., and Link, A. J. (2004). Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 24, 8276–8287. doi: 10.1128/MCB.24.18.8276-8287.2004
Guo, J., Wang, S., Valerius, O., Hall, H., Zeng, Q., Li, J. F., et al. (2011). Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 155, 370–383. doi: 10.1104/pp.110.160663
Guo, X., Shi, Y., Gou, Y., Li, J., Han, S., Zhang, Y., et al. (2011). Human ribosomal protein S13 promotes gastric cancer growth through down-regulating p27(Kip1). J. Cell Mol. Med. 15, 296–306. doi: 10.1111/j.1582-4934.2009.00969.x
Hansen, M., Taubert, S., Crawford, D., Libina, N., Lee, S. J., and Kenyon, C. (2007). Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110. doi: 10.1111/j.1474-9726.2006.00267.x
He, X., Li, Y., Dai, M. S., and Sun, X. X. (2016). Ribosomal protein L4 is a novel regulator of the MDM2-p53 loop. Oncotarget 7, 16217–16226. doi: 10.18632/oncotarget.7479
Heinrich, R., and Rapoport, T. A. (1980). Mathematical modelling of translation of mRNA in eucaryotes; steady state, time-dependent processes and application to reticulocytes. J. Theor. Biol. 86, 279–313. doi: 10.1016/0022-5193(80)90008-9
Hermanto, U., Zong, C. S., Li, W., and Wang, L. H. (2002). RACK1, an insulin-like growth factor I (IGF-I) receptor-interacting protein, modulates IGF-I-dependent integrin signaling and promotes cell spreading and contact with extracellular matrix. Mol. Cell. Biol. 22, 2345–2365. doi: 10.1128/MCB.22.7.2345-2365.2002
In, K., Zaini, M. A., Müller, C., Warren, A. J., von Lindern, M., and Calkhoven, C. F. (2016). Shwachman-Bodian-Diamond syndrome (SBDS) protein deficiency impairs translation re-initiation from C/EBPα and C/EBPβ mRNAs. Nucleic Acids Res. 44, 4134–4146. doi: 10.1093/nar/gkw005
Jia, J., Arif, A., Willard, B., Smith, J. D., Stuehr, D. J., Hazen, S. L., et al. (2012). Protection of extraribosomal RPL13a by GAPDH and dysregulation by S-nitrosylation. Mol. Cell 47, 656–663. doi: 10.1016/j.molcel.2012.06.006
Kampen, K. R., Sulima, S. O., Verbelen, B., Girardi, T., Vereecke, S., Rinaldi, G., et al. (2018). The ribosomal RPL10 R98S mutation drives IRES-dependent BCL-2 translation in T-ALL. Leukemia doi: 10.1038/s41375-018-0176-z [Epub ahead of print].
Khatter, H., Myasnikov, A. G., Natchiar, S. K., and Klaholz, B. P. (2015). Structure of the human 80S ribosome. Nature 520, 640–645. doi: 10.1038/nature14427
Klinge, S., Voigts-Hoffmann, F., Leibundgut, M., Arpagaus, S., and Ban, N. (2011). Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334, 941–948. doi: 10.1126/science.1211204
Kondrashov, N., Pusic, A., Stumpf, C. R., Shimizu, K., Hsieh, A. C., Ishijima, J., et al. (2011). Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397. doi: 10.1016/j.cell.2011.03.028
Kressler, D., Hurt, E., and Bassler, J. (2017). A puzzle of life: crafting ribosomal subunits. Trends Biochem. Sci. 42, 640–654. doi: 10.1016/j.tibs.2017.05.005
Lambertsson, A. (1998). The minute genes in Drosophila and their molecular functions. Adv. Genet. 38, 69–134. doi: 10.1016/S0065-2660(08)60142-X
Landau, D. A., Tausch, E. A., Taylor-Weiner, N., Stewart, C., Reiter, J. G., Bahlo, J., et al. (2015). Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530. doi: 10.1038/nature15395
Leibovitch, M., and Topisirovic, I. (2018). Dysregulation of mRNA translation and energy metabolism in cancer. Adv. Biol. Regul. 67, 30–39. doi: 10.1016/j.jbior.2017.11.001
Li, C., Ge, M., Yin, Y., Luo, M., and Chen, D. (2012). Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol. Cell. Biochem. 370, 127–139. doi: 10.1007/s11010-012-1404-x
Li, H. B., Wang, R. X., Jiang, H. B., Zhang, E. D., Tan, J. Q., Xu, H. Z., et al. (2016). Mitochondrial ribosomal protein L10 associates with cyclin B1/Cdk1 activity and mitochondrial function. DNA Cell Biol. 35, 680–690. doi: 10.1089/dna.2016.3271
Link, D. C., Kunter, G., Kasai, Y., Zhao, Y., Miner, T., McLellan, M. D., et al. (2007). Distinct patterns of mutations occurring in de novo AML versus AML arising in the setting of severe congenital neutropenia. Blood 110, 1648–1655. doi: 10.1182/blood-2007-03-081216
Ljungstrom, V., Cortese, D., Young, E., Pandzic, T., Mansouri, L., Plevova, K., et al. (2016). Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood 127, 1007–1016. doi: 10.1182/blood-2015-10-674572
Lomakin, I. B., and Steitz, T. A. (2013). The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 500, 307–311. doi: 10.1038/nature12355
Loreni, F., Mancino, M., and Biffo, S. (2014). Translation factors and ribosomal proteins control tumor onset and progression: how? Oncogene 33, 2145–2156. doi: 10.1038/onc.2013.153
Mamidipudi, V., Zhang, J., Lee, K. C., and Cartwright, C. A. (2004). RACK1 regulates G1/S progression by suppressing Src kinase activity. Mol. Cell. Biol. 24, 6788–6798. doi: 10.1128/MCB.24.15.6788-6798.2004
Marygold, S. J., Roote, J., Reuter, G., Lambertsson, A., Ashburner, M., Millburn, G. H., et al. (2007). The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8:R216. doi: 10.1186/gb-2007-8-10-r216
Menezes, M. J., Guo, Y., Zhang, J., Riley, L. G., Cooper, S. T., Thorburn, D. R., et al. (2015). Mutation in mitochondrial ribosomal protein S7 (MRPS7) causes congenital sensorineural deafness, progressive hepatic and renal failure and lactic acidemia. Hum. Mol. Genet. 24, 2297–2307. doi: 10.1093/hmg/ddu747
Menne, T. F., Goyenechea, B., Sanchez-Puig, N., Wong, C. C., Tonkin, L. M., Ancliff, P. J., et al. (2007). The Shwachman-Bodian-diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 39, 486–495. doi: 10.1038/ng1994
Miller, C., Saada, A., Shaul, N., Shabtai, N., Ben-Shalom, E., Shaag, A., et al. (2004). Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann. Neurol. 56, 734–738. doi: 10.1002/ana.20282
Mills, E. W., and Green, R. (2017). Ribosomopathies: there’s strength in numbers. Science 358:eaan2755. doi: 10.1126/science.aan2755
Milne, A. N., Mak, W. W., and Wong, J. T. (1975). Variation of ribosomal proteins with bacterial growth rate. J. Bacteriol. 122, 89–92.
Montanaro, L., Treré, D., and Derenzini, M. (2008). Nucleolus, ribosomes, and cancer. Am. J. Pathol. 173, 301–310. doi: 10.2353/ajpath.2008.070752
Moore, P. B., Traut, R. R., Noller, H., Pearson, P., and Delius, H. (1968). Ribosomal proteins of Escherichia coli. II. Proteins from the 30 s subunit. J. Mol. Biol. 31, 441–461. doi: 10.1016/0022-2836(68)90420-8
Morita, M., Gravel, S. P., Chénard, V., Sikström, K., Zheng, L., Alain, T., et al. (2013). mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711. doi: 10.1016/j.cmet.2013.10.001
Narla, A., and Ebert, B. L. (2010). Ribosomopathies: human disorders of ribosome dysfunction. Blood 115, 3196–3205. doi: 10.1182/blood-2009-10-178129
Nihrane, A., Sezgin, G., Dsilva, S., Dellorusso, P., Yamamoto, K., Ellis, S. R., et al. (2009). Depletion of the Shwachman-Diamond syndrome gene product, SBDS, leads to growth inhibition and increased expression of OPG and VEGF-A. Blood Cells Mol. Dis. 42, 85–91. doi: 10.1016/j.bcmd.2008.09.004
Pelletier, J., Thomas, G., and Volarevic, S. (2018). Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer 18, 51–63. doi: 10.1038/nrc.2017.104
Pena, C., Hurt, E., and Panse, V. G. (2017). Eukaryotic ribosome assembly, transport and quality control. Nat. Struct. Mol. Biol. 24, 689–699. doi: 10.1038/nsmb.3454
Rao, S., Lee, S. Y., Gutierrez, A., Perrigoue, J., Thapa, R. J., Tu, Z., et al. (2012). Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 120, 3764–3773. doi: 10.1182/blood-2012-03-415349
Rapino, F., Delaunay, S., Rambow, F., Zhou, Z., Tharun, L., De Tullio, P., et al. (2018). Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 558, 605–609. doi: 10.1038/s41586-018-0243-7
Ravera, S., Dufour, C., Cesaro, S., Bottega, R., Faleschini, M., Cuccarolo, P., et al. (2016). Evaluation of energy metabolism and calcium homeostasis in cells affected by Shwachman-Diamond syndrome. Sci. Rep. 6:25441. doi: 10.1038/srep25441
Richman, T. R., Ermer, J. A., Davies, S. M., Perks, K. L., Viola, H. M., Shearwood, A. M., et al. (2015). Mutation in MRPS34 compromises protein synthesis and causes mitochondrial dysfunction. PLoS Genet. 11:e1005089. doi: 10.1371/journal.pgen.1005089
Robles, M. S., Boyault, C., Knutti, D., Padmanabhan, K., and Weitz, C. J. (2010). Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science 327, 463–466. doi: 10.1126/science.1180067
Ron, D., Chen, C. H., Caldwell, J., Jamieson, L., Orr, E., and Mochly-Rosen, D. (1994). Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc. Natl. Acad. Sci. U.S.A. 91, 839–843. doi: 10.1073/pnas.91.3.839
Roux, P. P., and Topisirovic, I. (2018). Signaling pathways involved in the regulation of mRNA translation. Mol. Cell. Biol. 38:e00070-18. doi: 10.1128/MCB.00070-18
Ruggero, D., and Shimamura, A. (2014). Marrow failure: a window into ribosome biology. Blood 124, 2784–2792. doi: 10.1182/blood-2014-04-526301
Saada, A., Shaag, A., Arnon, S., Dolfin, T., Miller, C., Fuchs-Telem, D., et al. (2007). Antenatal mitochondrial disease caused by mitochondrial ribosomal protein (MRPS22) mutation. J. Med. Genet. 44, 784–786. doi: 10.1136/jmg.2007.053116
Serre, V., Rozanska, A., Beinat, M., Chretien, D., Boddaert, N., Munnich, A., et al. (2013). Mutations in mitochondrial ribosomal protein MRPL12 leads to growth retardation, neurological deterioration and mitochondrial translation deficiency. Biochim. Biophys. Acta 1832, 1304–1312. doi: 10.1016/j.bbadis.2013.04.014
Shah, M., Su, D., Scheliga, J. S., Pluskal, T., Boronat, S., Motamedchaboki, K., et al. (2016). A transcript-specific eIF3 complex mediates global translational control of energy metabolism. Cell Rep. 16, 1891–1902. doi: 10.1016/j.celrep.2016.07.006
Shi, Z., and Barna, M. (2015). Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 31, 31–54. doi: 10.1146/annurev-cellbio-100814-125346
Shi, Z., Fujii, K., Kovary, K. M., Genuth, N. R., Rost, H. L., Teruel, M. N., et al. (2017). Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 67, 71–83.e7. doi: 10.1016/j.molcel.2017.05.021
Shor, B., Calaycay, J., Rushbrook, J., and McLeod, M. (2003). Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J. Biol. Chem. 278, 49119–49128. doi: 10.1074/jbc.M303968200
Smits, P., Saada, A., Wortmann, S. B., Heister, A. J., Brink, M., Pfundt, R., et al. (2011). Mutation in mitochondrial ribosomal protein MRPS22 leads to Cornelia de Lange-like phenotype, brain abnormalities and hypertrophic cardiomyopathy. Eur. J. Hum. Genet. 19, 394–399. doi: 10.1038/ejhg.2010.214
Stadanlick, J. E., Zhang, Z., Lee, S. Y., Hemann, M., Biery, M., Carleton, M. O., et al. (2011). Developmental arrest of T cells in Rpl22-deficient mice is dependent upon multiple p53 effectors. J. Immunol. 187, 664–675. doi: 10.4049/jimmunol.1100029
Steffen, K. K., MacKay, V. L., Kerr, E. O., Tsuchiya, M., Hu, D., Fox, L. A., et al. (2008). Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133, 292–302. doi: 10.1016/j.cell.2008.02.037
Stepensky, P., Chacón-Flores, M., Kim, K. H., Abuzaitoun, O., Bautista-Santos, A., Simanovsky, N., et al. (2017). Mutations in. J. Med. Genet. 54, 558–566. doi: 10.1136/jmedgenet-2016-104366
Suhm, T., Kaimal, J. M., Dawitz, H., Peselj, C., Masser, A. E., Hanzén, S., et al. (2018). Mitochondrial translation efficiency controls cytoplasmic protein homeostasis. Cell Metab. 27, 1309–1322.e6. doi: 10.1016/j.cmet.2018.04.011
Sulic, S., Panic, L., Barkic, M., Mercep, M., Uzelac, M., and Volarevic, S. (2005). Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 19, 3070–3082. doi: 10.1101/gad.359305
Sulima, S. O., Hofman, I. J. F., De Keersmaecker, K., and Dinman, J. D. (2017). How ribosomes translate cancer. Cancer Discov. 7, 1069–1087. doi: 10.1158/2159-8290.CD-17-0550
Tan, Q. K., Cope, H., Spillmann, R. C., Stong, N., Jiang, Y. H., McDonald, M. T., et al. (2018). Further evidence for the involvement of EFL1 in a shwachman–diamond-like syndrome and expansion of the phenotypic features. Cold Spring Harb. Mol. Case Stud. 4:a003046. doi: 10.1101/mcs.a003046
Teng, T., Mercer, C. A., Hexley, P., Thomas, G., and Fumagalli, S. (2013). Loss of tumor suppressor RPL5/RPL11 does not induce cell cycle arrest but impedes proliferation due to reduced ribosome content and translation capacity. Mol. Cell. Biol. 33, 4660–4671. doi: 10.1128/MCB.01174-13
Tourlakis, M. E., Zhong, J., Gandhi, R., Zhang, S., Chen, L., Durie, P. R., et al. (2012). Deficiency of Sbds in the mouse pancreas leads to features of Shwachman-Diamond syndrome, with loss of zymogen granules. Gastroenterology 143, 481–492. doi: 10.1053/j.gastro.2012.04.012
Touw, I. P., and Beekman, R. (2013). Severe congenital neutropenia and chronic neutrophilic leukemia: an intriguing molecular connection unveiled by oncogenic mutations in CSF3R. Haematologica 98, 1490–1492. doi: 10.3324/haematol.2013.090571
Truitt, M. L., Conn, C. S., Shi, Z., Pang, X., Tokuyasu, T., Coady, A. M., et al. (2015). Differential requirements for eIF4E dose in normal development and cancer. Cell 162, 59–71. doi: 10.1016/j.cell.2015.05.049
Truitt, M. L., and Ruggero, D. (2016). New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 16, 288–304. doi: 10.1038/nrc.2016.27
Tzoneva, G., Perez-Garcia, A., Carpenter, Z., Khiabanian, H., Tosello, V., Allegretta, M., et al. (2013). Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat. Med. 19, 368–371. doi: 10.1038/nm.3078
Verhoeven, D., Stoppelenburg, A. J., Meyer-Wentrup, F., and Boes, M. (2018). Increased risk of hematologic malignancies in primary immunodeficiency disorders: opportunities for immunotherapy. Clin. Immunol. 190, 22–31. doi: 10.1016/j.clim.2018.02.007
Volarevic, S., Stewart, M. J., Ledermann, B., Zilberman, F., Terracciano, L., Montini, E., et al. (2000). Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288, 2045–2047. doi: 10.1126/science.288.5473.2045
Volarevic, S., and Thomas, G. (2001). Role of S6 phosphorylation and S6 kinase in cell growth. Prog. Nucleic Acid Res. Mol. Biol. 65, 101–127. doi: 10.1016/S0079-6603(00)65003-1
Volta, V., Beugnet, A., Gallo, S., Magri, L., Brina, D., Pesce, E., et al. (2013). RACK1 depletion in a mouse model causes lethality, pigmentation deficits and reduction in protein synthesis efficiency. Cell. Mol. Life Sci. 70, 1439–1450. doi: 10.1007/s00018-012-1215-y
Warren, A. J. (2018). Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv. Biol. Regul. 67, 109–127. doi: 10.1016/j.jbior.2017.09.002
Wehner, P., Shnitsar, I., Urlaub, H., and Borchers, A. (2011). RACK1 is a novel interaction partner of PTK7 that is required for neural tube closure. Development 138, 1321–1327. doi: 10.1242/dev.056291
Weis, F., Giudice, E., Churcher, M., Jin, L., Hilcenko, C., Wong, C. C., et al. (2015). Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat. Struct. Mol. Biol. 22, 914–919. doi: 10.1038/nsmb.3112
Wilson-Edell, K. A., Kehasse, A., Scott, G. K., Yau, C., Rothschild, D. E., Schilling, B., et al. (2014). RPL24: a potential therapeutic target whose depletion or acetylation inhibits polysome assembly and cancer cell growth. Oncotarget 5, 5165–5176. doi: 10.18632/oncotarget.2099
Wong, C. C., Traynor, D., Basse, N., Kay, R. R., and Warren, A. J. (2011). Defective ribosome assembly in Shwachman-Diamond syndrome. Blood 118, 4305–4312. doi: 10.1182/blood-2011-06-353938
Xue, S., Tian, S., Fujii, K., Kladwang, W., Das, R., and Barna, M. (2015). RNA regulons in Hox 5’ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38. doi: 10.1038/nature14010
Yang, S., Cui, J., Yang, Y., Liu, Z., Yan, H., Tang, C., et al. (2016). Over-expressed RPL34 promotes malignant proliferation of non-small cell lung cancer cells. Gene 576(1 Pt 3), 421–428. doi: 10.1016/j.gene.2015.10.053
Yao, Y., Liu, Y., Lv, X., Dong, B., Wang, F., Li, J., et al. (2016). Down-regulation of ribosomal protein S15A inhibits proliferation of human glioblastoma cells in vivo and in vitro via AKT pathway. Tumour Biol. 37, 4979–4990. doi: 10.1007/s13277-015-4323-0
Yoon, A., Peng, G., Brandenburger, Y., Zollo, O., Xu, W., Rego, E., et al. (2006). Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 312, 902–906. doi: 10.1126/science.1123835
Yoon, I. S., Chung, J. H., Hahm, S. H., Park, M. J., Lee, Y. R., Ko, S. I., et al. (2011). Ribosomal protein S3 is phosphorylated by Cdk1/cdc2 during G2/M phase. BMB Rep. 44, 529–534. doi: 10.5483/BMBRep.2011.44.8.529
Keywords: ribosomal proteins, ribosomopathies, ribosome heterogeneity, metabolism, Shwachman-diamond syndrome, eIF6, RACK1
Citation: Calamita P, Gatti G, Miluzio A, Scagliola A and Biffo S (2018) Translating the Game: Ribosomes as Active Players. Front. Genet. 9:533. doi: 10.3389/fgene.2018.00533
Received: 09 August 2018; Accepted: 22 October 2018;
Published: 15 November 2018.
Edited by:
Chiara Gamberi, Concordia University, CanadaReviewed by:
Kim De Keersmaecker, KU Leuven, BelgiumGary Loughran, University College Cork, Ireland
Copyright © 2018 Calamita, Gatti, Miluzio, Scagliola and Biffo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Piera Calamita, Y2FsYW1pdGFAaW5nbS5vcmc=; Stefano Biffo, YmlmZm9AaW5nbS5vcmc=
 Piera Calamita
Piera Calamita Guido Gatti1,2
Guido Gatti1,2 Annarita Miluzio
Annarita Miluzio Alessandra Scagliola
Alessandra Scagliola Stefano Biffo
Stefano Biffo