
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 04 December 2018
Sec. Behavioral and Psychiatric Genetics
Volume 9 - 2018 | https://doi.org/10.3389/fgene.2018.00513
This article is part of the Research Topic Genomics and Epigenomics of Alcoholism View all 12 articles
 John Paul Spence1
John Paul Spence1 Jill L. Reiter1
Jill L. Reiter1 Bin Qiu2
Bin Qiu2 Hao Gu2
Hao Gu2 Dawn K. Garcia3
Dawn K. Garcia3 Lingling Zhang2
Lingling Zhang2 Tamara Graves1
Tamara Graves1 Kent E. Williams1
Kent E. Williams1 Paula J. Bice4
Paula J. Bice4 Yi Zou3
Yi Zou3 Zhao Lai4
Zhao Lai4 Weidong Yong2*
Weidong Yong2* Tiebing Liang1*
Tiebing Liang1*Humans show sex differences related to alcohol use disorders (AUD). Animal model research has the potential to provide important insight into how sex differences affect alcohol consumption, particularly because female animals frequently drink more than males. In previous work, inbred strains of the selectively bred alcohol-preferring (P) and non-preferring (NP) rat lines revealed a highly significant quantitative trait locus (QTL) on rat chromosome 4, with a logarithm of the odds score of 9.2 for alcohol consumption. Recently, interval-specific congenic strains (ISCS) were developed by backcrossing the congenic P.NP line to inbred P (iP) rats to further refine the chromosome 4 QTL region. Two ISCS sub-strains, ISCS-A and ISCS-B, were obtained with a narrowed QTL, where the smallest region of overlap consisted of 8.9 Mb in ISCS-B. Interestingly, we found that females from both ISCS lines consumed significantly less alcohol than female iP controls (p < 0.05), while no differences in alcohol consumption were observed between male ISCS and iP controls. RNA-sequencing was performed on the nucleus accumbens of alcohol-naïve female ISCS-B and iP rats, which revealed differentially expressed genes (DEG) with greater than 2-fold change and that were functionally relevant to behavior. These DEGs included down-regulation of Oxt, Asb4, Gabre, Gabrq, Chat, Slc5a7, Slc18a8, Slc10a4, and Ngfr, and up-regulation of Ttr, Msln, Mpzl2, Wnt6, Slc17a7, Aldh1a2, and Gstm2. Pathway analysis identified significant alterations in gene networks controlling nervous system development and function, as well as cell signaling, GABA and serotonin receptor signaling and G-protein coupled receptor signaling. In addition, β-estradiol was identified as the most significant upstream regulator. The expression levels of estrogen-responsive genes that mapped to the QTL interval and have been previously associated with alcohol consumption were measured using RT-qPCR. We found that expression of the Adcyap1r1 gene, encoding the pituitary adenylate cyclase-activating polypeptide type 1 (PAC1) receptor, was upregulated in female ISCS-B compared to female iP controls, while no differences were exhibited in males. In addition, sequence variants in the Adcyap1r1 promoter region showed a differential response to estrogen stimulation in vitro. These findings demonstrate that rat chromosome 4 QTL contains genetic variants that respond to estrogen and are associated with female alcohol consumption.
The development of alcohol use disorders has a strong genetic component. Genetic factors account for more than 50% of the variance in developing alcoholism (Heath et al., 1997; Ducci and Goldman, 2008), and several specific genetic variants are associated with an increased risk for alcoholism (Koss and Goldman, 2000; Dick et al., 2006; Gatti et al., 2010). In addition to genetics, gender-based differences in drug and alcohol use, abuse, and dependence are supported by epidemiological and clinical research (Prescott, 2002; Nolen-Hoeksema, 2004; Ceylan-Isik et al., 2010). Likewise, animal models also display sex-differences in voluntary consumption of drugs and alcohol (Vetter-O’Hagen et al., 2009; Desrivières et al., 2011). However, the mechanisms responsible for sex-specific drinking differences remain largely unknown (Becker and Koob, 2016).
Most sexually dimorphic traits arise through differential gene expression that has different effects on males and females (Dimas et al., 2012; Gershoni and Pietrokovski, 2017). In addition to phenotypic variation that results from the action of sex hormones, gene-by-sex (GxS) interactions involve phenotypic differences that depend on the functional genetic variants (Rawlik et al., 2016). Evidence for sex-specific quantitative traits exist that impact a wide range of complex traits (Karp et al., 2017). Sex-specific quantitative trait loci (QTLs) have also been reported for alcohol preference, ethanol sensitivity, and ethanol locomotor activation in mice and ethanol drinking in rats; however, specific genes mediating these effects have yet to be identified (Melo et al., 1996; Gill et al., 1998; Peirce et al., 1998; Radcliffe et al., 2000; Bice et al., 2006; Vendruscolo et al., 2006; Chesler et al., 2012; DuBose et al., 2013; Vanderlinden et al., 2015)
We and others have used the selectively bred alcohol-preferring (P) and non-preferring (NP) rat model (Li et al., 1991) to investigate the genetic factors involved in alcohol drinking behaviors (Bice et al., 1998; Carr et al., 1998; Liang et al., 2003, 2004; Liang and Carr, 2006). In this model, P rats exhibit several features that are consistent with alcoholism in humans (Cicero, 1979). For example, P rats (1) orally self-administer ethanol in pharmacologically relevant amounts; (2) consume EtOH for its pharmacological effects rather than caloric value or taste; (3) show positive reinforcement; (4) develop tolerance; and (5) exhibit withdrawal symptoms (McBride and Li, 1998; Murphy et al., 2002). QTL analysis of inbred alcohol-preferring/non-preferring (iP/iNP) rats revealed several loci associated with alcohol preference, including a region of chromosome 4 (Chr4) with a LOD score >9.2 (Carr et al., 1998; Bice et al., 2006).
Congenic rat strains were subsequently derived that had transferred the ∼ 130 Mb Chr4 QTL region of the donor strain to the host strain, whereby P.NP designates an iP host rat with the donor iNP Chr4 QTL, and NP.P designates an iNP host rat with the donor iP Chr4 QTL. Alcohol consumption in these reciprocal congenic strains were consistent with the donor strain, such that P.NP rats drank less than P rats and NP.P rats drank more than NP rats (Carr et al., 2006). These and other results confirmed the association of the Chr4 QTL with alcohol consumption, and indicated that multiple loci within this strong QTL may be contributing to the alcohol drinking phenotype (Carr et al., 2006; Spence et al., 2009; Liang et al., 2010). To further refine this Chr4 QTL region, overlapping interval-specific congenic strains (ISCS) were generated by backcrossing the P.NP congenic line with iP rats. This approach resulted in two ISCS lines (Spence et al., 2013). The ISCS-A sub-strain contained ∼ 79 Mb of the NP genomic region between microsatellite markers D4Mgh16 and D4Rat173, while the ISCS-B sub-strain contained ∼ 9 Mb of the NP genomic region between a single nucleotide polymorphism (SNP) in Snca and the marker D4Rat35 (Liang and Carr, 2006; Spence et al., 2013).
Similar to other animal models, female P and iP rats consume more alcohol than the corresponding males (Li et al., 1991; Carr et al., 1998). We hypothesized that a sex-specific QTL existed on rat Chr4 that contributed to the disparate alcohol drinking behavior between males and females. To test this hypothesis, we measured alcohol preference and consumption in male and female P.NP-ISCS rats since they contain a narrowed Chr4 QTL region. To investigate the genetic factors that might be involved in sex-specific drinking phenotypes, we also analyzed transcriptome differences between P.NP-ISCS-B and iP rats in the nucleus accumbens (NAc), a brain region known to play an important role in the reinforcing and rewarding effects of ethanol.
In this study, we report that a sex-specific QTL exists on rat Chr4 that contributes to alcohol consumption. Specifically, we found that replacement of an approximately 9 Mb region of the NP Chr4 locus into the iP genetic background decreased alcohol consumption in female, but not in male ISCS rats when compared to their respective controls. We report that RNA-seq analysis of NAc from female alcohol-naïve P.NP-ISCS and iP rats identified differentially expressed genes (DEGs) related to neuron function, cell signaling, and behavior. In addition, we found that β-estradiol was predicted to be the top upstream regulator of DEGs. Moreover, promoter SNPs in the Chr4 QTL gene Acdyap1r1, which encodes the PAC1 receptor for the neuropeptide pituitary adenylate cyclase-activating polypeptide (PACAP), responded to estrogen stimulation. These findings indicate that Acdyap1r1, a gene previously associated with alcohol abuse in women (Dragan et al., 2017), is a likely candidate gene contributing to female-specific drinking behavior.
Rats were bred and maintained at Indiana University School of Medicine. All animals were housed under 12-h light–dark conditions (7:00 am / 7:00 pm) with free access to laboratory rodent chow and water. The animals used for this study included inbred alcohol-preferring (iP) rats (Lumeng et al., 1977; Bice et al., 1998), congenic P.NP strains (Carr et al., 2006), and ISCS-A and -B (Spence et al., 2013). Heterozygous ISCSB-H F1 animals were generated by crossing ISCS-B with iP rats. The experimental protocol used in this study was reviewed and approved by the Indiana University Institutional Animal Care and Use Committee and was carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals. The animals were maintained in facilities fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Adult alcohol-naïve male and female rats from the two P.NP-ISCS (A and B strains), ISCSB-H, and iP controls were tested for voluntary alcohol drinking using a 2-bottle free-choice protocol, consisting of 10% ethanol or water for 3 weeks, as described previously (Lumeng et al., 1977; Li et al., 1993). Littermates were included from both ISCS-A and ISCS-B strains when possible. Two experiments were conducted by comparing: 1) ISCS-A (23 males, 22 females) vs. iP controls (10 males, 15 females); and 2) ISCS-B (22 males, 23 females), ISCSB-H (20 males, 18 females), and iP control (23 males, 35 females). Both alcohol consumption and alcohol preference were calculated as was described previously (Li et al., 1991). Body weight, but not food intake, was recorded once a week. To exclude the possibility of a sex difference in taste reactivity, ISCS-B rats (8 females, 7 males) were first tested for saccharin (1.03%) and then for quinine (0.5 μM) intake. Consumption data was analyzed by two-way ANOVA followed by Newman-Keuls multiple comparisons test. Results are provided as means and standard error (SE).
Whole brains were extracted from alcohol-naïve adult ISCS-B and iP control rats, and snap frozen in dry ice-bathed isopentane, before brain regions were dissected as previously published (Liang et al., 2004, 2010; Liang and Carr, 2006). The RNA-seq experiment used NAc from female ISCS-B and iP control rats (N = 3 for each group). Tissues were stored at -80°C until RNA isolation. RNA was isolated from the NAc using TRIzol, followed by RNeasy mini-column purification (Qiagen, Valencia, CA, United States). RNA purity was measured using a spectrophotometer (Nanodrop 1000) and the 260/280 absorbance ratios were between 1.8 and 2.0. RNA integrity was measured using an Agilent 2100 Bioanalyzer and all samples had RIN >7.
Sequencing libraries were constructed using the Illumina TruSeq RNA sample preparation protocol. The resulting libraries were sequenced on an Illumina HiSeq 2000 instrument using a standard single-end 50 bp sequencing protocol. The reads were aligned to the reference Rattus norvegicus genome (UCSC Rn 6.0) with TopHat 2 (Trapnell et al., 2010; Kim et al., 2013). No more than 2 mismatches were allowed in the alignment. HTseq was used to count gene expression reads, and DEseq was used to find DEGs after performing median normalization (Anders and Huber, 2010; Anders et al., 2015). DEGs were identified with adjusted p < 0.05 for multiple tests by the Benjamini-Hochberg method for controlling false discovery rate (FDR; Benjamini and Hochberg, 1995) and using a cutoff of sequence read rpmk >1. The RNA-seq data is available at GEO1 with access ID: GSE112399.
Two pathway analyses were used in data exploration. Ingenuity Pathway Analysis (IPA), which builds on manually curated content of the Ingenuity Knowledge database, was applied for predicting significant biological mechanisms and pathways (Qiagen2). The complete data set containing gene identifiers and corresponding expression values was uploaded into the application. Each identifier was mapped to its corresponding object in Ingenuity’s Knowledge Base. An FDR-adjusted p-value cutoff of 0.05 was set to identify molecules whose expression was significantly differentially regulated. These molecules, called Network Eligible molecules, were overlaid onto a global molecular network developed from information contained in Ingenuity’s Knowledge Base. Networks of Network Eligible Molecules were then algorithmically generated based on their connectivity. The Functional Analysis identified the biological functions and/or diseases that were most significant to the data set. Molecules from the dataset that met the FDR-adjusted p-values cutoff of <0.05 and were associated with biological functions and/or diseases in Ingenuity’s Knowledge Base were considered for the analysis. Right-tailed Fisher’s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone. Canonical pathways analysis identified the pathways from the Ingenuity Pathways Analysis library of canonical pathways that were most significant to the data set. Molecules from the data set that met the FDR-adjusted p-values cutoff of p <0.05 and were associated with a canonical pathway in Ingenuity’s Knowledge Base were considered for the analysis. The significance of the association between the data set and the canonical pathway was measured in two ways: (1) A ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway is displayed. (2) Fisher’s exact test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone.
In addition, the Reactome Knowledgebase version 623 was applied to analyze the molecular interaction details of signal transduction, transport, and other cellular processes (Fabregat et al., 2016). Settings for Reactome analysis were DEGs with p-values <0.05 and FC > 2; a total of 212 genes were used for analysis. Pathway hierarchical organization provides Reactome pathways overview and highlights parent-child relationships of overrepresented connections in the pathway.
Reverse transcription quantitative PCR (RT-qPCR) was performed using RNA from a separate set of experimental male and female ISCS-B and iP control (N = 6–8) animals than were utilized for RNA-seq. Each PCR assay was conducted using 6–8 biological replicates, and each cDNA sample was amplified in triplicate in qPCR for the same RT reaction. In brief, 1 μg RNA was reverse transcribed using Superscript III reverse-transcription reagent for first-strand cDNA synthesis (Invitrogen) using random primers. Each 50 μl PCR reaction contained cDNA corresponding to 35 ng of total RNA, SYBR Green Real-Time PCR Master mix (Life Sciences), and primers (5 μM). PCR was performed using the ABI PRISM 7300 Sequence Detection System (ThermoFisher), relative mRNA expression levels were normalized to Gapdh, and the standard curve method was used for data analysis. Vector NTI was used for primer design, and annealing temperatures ranged from 60 to 63°C. The primers used are listed in Supplementary Table S5. T-test was used to analyze the data and statistical significance was set at p < 0.05.
Genomic DNA was isolated from iP and iNP rats using Gentra Puregene Tissue kit (QIAGEN). PCR primers were designed to amplify the Adcyap1r1 promoter region up to 2 kb upstream of the transcription start site; primer sequences are listed in Supplementary Table S5. PCR products were purified using the GenElute PCR Cleanup Kit (Sigma, St. Louis, MO) and ligated upstream of the luciferase gene in the pGL3-basic vector (Invitrogen). Plasmid DNA was isolated using the QIAprep Spin Miniprep kit, and the cloned Adcyap1r1 promoter was sequenced using Indiana University sequencing core service. Genomic DNA sequences were aligned to the Rn.6 reference sequence.
The P and NP reporter plasmids utilized in this study were constructed as previously described (Liang et al., 2003). Human neuroblastoma SK-N-SH cells (ATCC HTB-11) were cultured in Minimal Essential Medium (Invitrogen) containing 7.5% NaHCO3, 2 mM Glut-max, 0.1 mM non-essential amino acids, 1 mM pyruvate, and 10% FBS (Invitrogen, Carlsbad, CA, United States) at 37°C in a humidified 5% CO2 incubator. Twenty-four hours before transfection, 5.0 × 104 cells were plated into individual wells of a 24-well plate. Using Tfx-50 reagent (Promega, Madison, WI, United States), each well was co-transfected with 0.5 μg of the pGL-3 luciferase plasmid plus 4 ng of the CMV Renilla luciferase vector (pRL-CMV, Invitrogen), which was utilized as an internal control for transfection efficiency. The human ER-α expression vector was a kind gift from Dr. Edwin R. Sánchez. The cells were subsequently incubated for 24 h at 37°C and washed with PBS before cell extracts were prepared in passive lysis buffer and assayed for firefly and Renilla luciferase activities using the Dual-Luciferase Reporter Assay System (Promega) in a TD-20/20 Luminometer (Turner BioSystems). The reporter assays were repeated five times in triplicate using plasmids that were independently purified at least twice.
We examined whether the narrowed Chr4 QTL in the ISCS-A and ISCS-B sub-strains affected the drinking phenotype by testing alcohol consumption. In most alcohol drinking animal models, females have higher alcohol consumption than male rats, and this same phenotype was observed in control iP rats (Figure 1). However, we found that alcohol consumption was similar between males and females of both ISCS rat lines (∼3.5 to 4.5 g EtOH/kg BW/day). When ISCS alcohol consumption was compared to the iP background strain, both female ISCS-A and ISCS-B rats consumed 20–30% less alcohol (p < 0.05), while no differences were observed in the male rats (Figure 1). Our findings suggest a sex-specific QTL that maps to the overlapping Chr4 region in these two sub-strains between approximately 83.8 and 92.7 Mb (position reference to RGSC-v3.4).
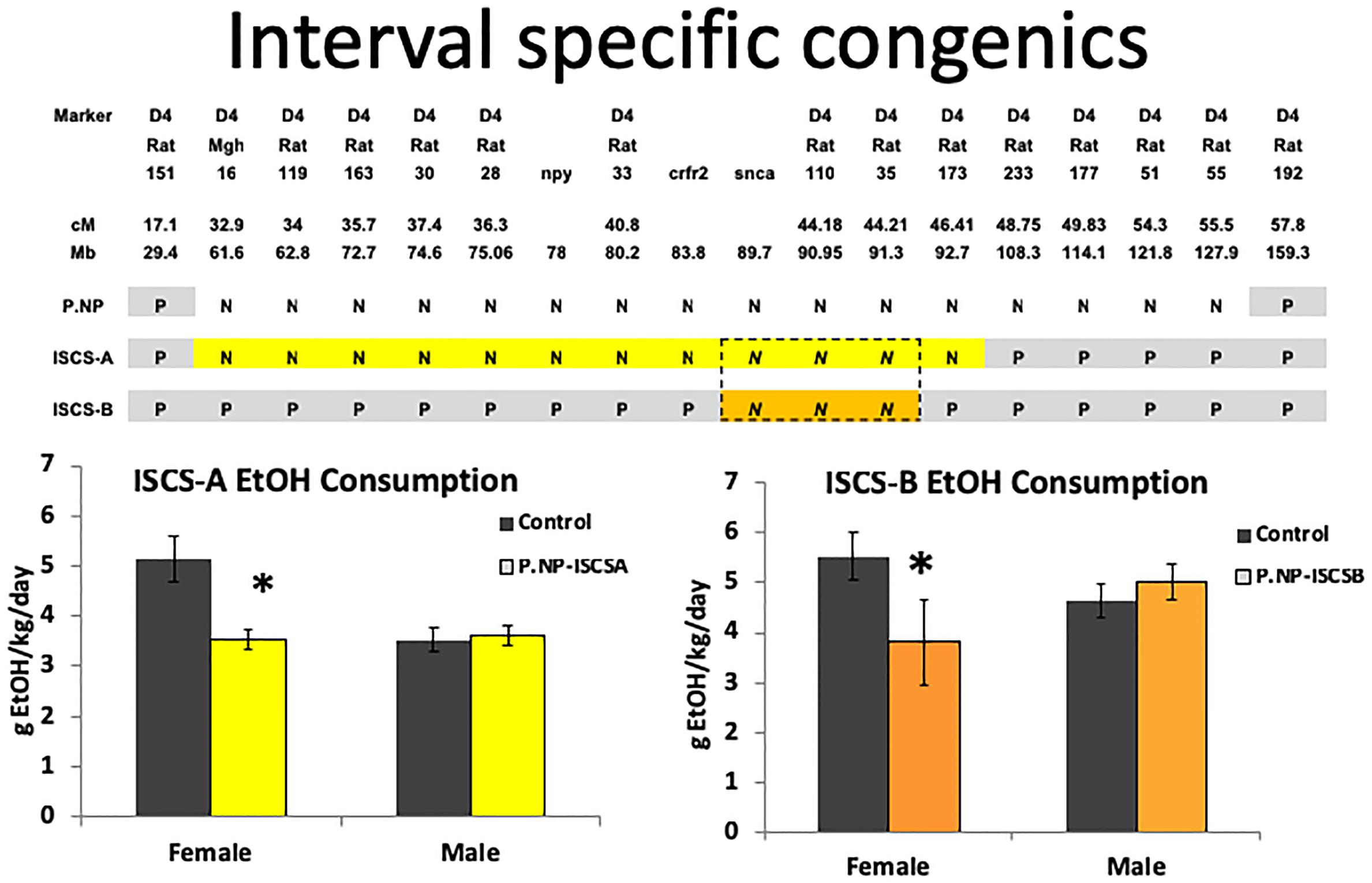
FIGURE 1. P.NP is the congenic strain with the Chr4 QTL region in P rats replaced by the NP Chr4. Backcrossing P.NP with iP rat resulted in interval-specific congenic strains (ISCS). ISCS-A and ISCS-B shared a narrowed Chr4 QTL between 83.8 and 92.7 Mb (region in dash-lined box). Relative to iP control rats, only females in either ISCS consumed less alcohol; alcohol consumption (gEtOH/Kg/day) was calculated and is shown as mean ± SEM. ∗indicates p < 0.05.
To determine whether female alcohol consumption was a dominant or recessive trait, we tested alcohol consumption in heterozygous offspring of the ISCS-B line since this sub-strain contained the smallest overlapping genomic region. We found female homozygous ISCS-B animals consumed less alcohol than female heterozygous ISCSB-H (p < 0.05); however, we found no difference between female homozygous iP control and ISCSB-H. The similarity in alcohol consumption between iP and ISCSB-H indicates that the reduction in female drinking is a recessive trait. Additionally, no difference was detected in alcohol consumption between males of any genotypes (Supplementary Figure S1). No sex differences were observed with either saccharin (adjusted p-value = 0.6) or quinine intake (adj p = 0.56), which indicates that differences in alcohol consumption were not associated with taste preferences (Supplementary Figure S2). Furthermore, alcohol consumption was not associated with body weight since we previously showed that there were no differences in body weight between ISCS-B and iP animals (Spence et al., 2013).
Since the only genetic differences between ISCS-B and iP rats existed within a minimum 1.79 Mb and maximum 8.9 Mb region of Chr4, we reasoned that genes in this narrowed Chr4 QTL might impact global gene expression in female rats related to alcohol consumption. To identify transcriptome changes, we performed RNA-seq on the NAc of alcohol-naïve female ISCS-B and iP (control) rats. High quality RNA-seq data were generated as documented by the correlation of biological replicates higher than 0.96. The average read count of each sample was more than 42 million with an average of more than 99.9% mapped reads (Supplementary Table S1).
We found 759 DEGs with the FDR set at p < 0.05; 212 of these DEGs showed FC > 2. The top 30 up- and down-regulated genes with adj p < 0.05 in ISCS-B females are listed in Table 1. Among the down-regulated genes, some are important for neuron functions: Oxt, and Gabre (Donhoffner et al., 2016). Four genes were involved in the cholinergic system: Chat, Slc5a7, Slc18a3, and Lhx8. Other significant down-regulated genes are known to influence neurotransmission, neuron growth, and addiction, including Ngfr, Ntrk1, and Ntsr1 (Gehle and Erwin, 1998; Pandey et al., 2017); and the estrogen receptor gene Esr1 is an important regulator of sex-differences. Among the up-regulated genes, Wnt6 and Cdh1 are associated with neuron development; other up-regulated DEGs are related to detoxification of drug and alcohol metabolism, such as Ptgds and Gstm2. Notably, many of the up-regulated DEGs are related to the development and function of the nervous system. The complete list of DEGs with FC > 2 and FDR adj p < 0.05 can be found in Supplementary Table S3.
To determine the genomic distribution of the DEG identified in the present study, we plotted the negative log-adjusted p-values for each DEG by chromosome (Figure 2). Of the 212 DEGs with FC > 2 (Supplementary Table S3), we found that 13 genes mapped to Chr4, including Tacr1, Chrm2, Neurod6, Slc13a4, and Aqp1; notably, these genes were differentially expressed and associated with drinking differences. Among the Chr4 DEG, Tacr1, which encodes the receptor for tachykinin substance P (also known as neurokinin 1), has been found to be associated with alcohol consumption in both humans and animals (Thorsell et al., 2005; George et al., 2008; Schank et al., 2013).
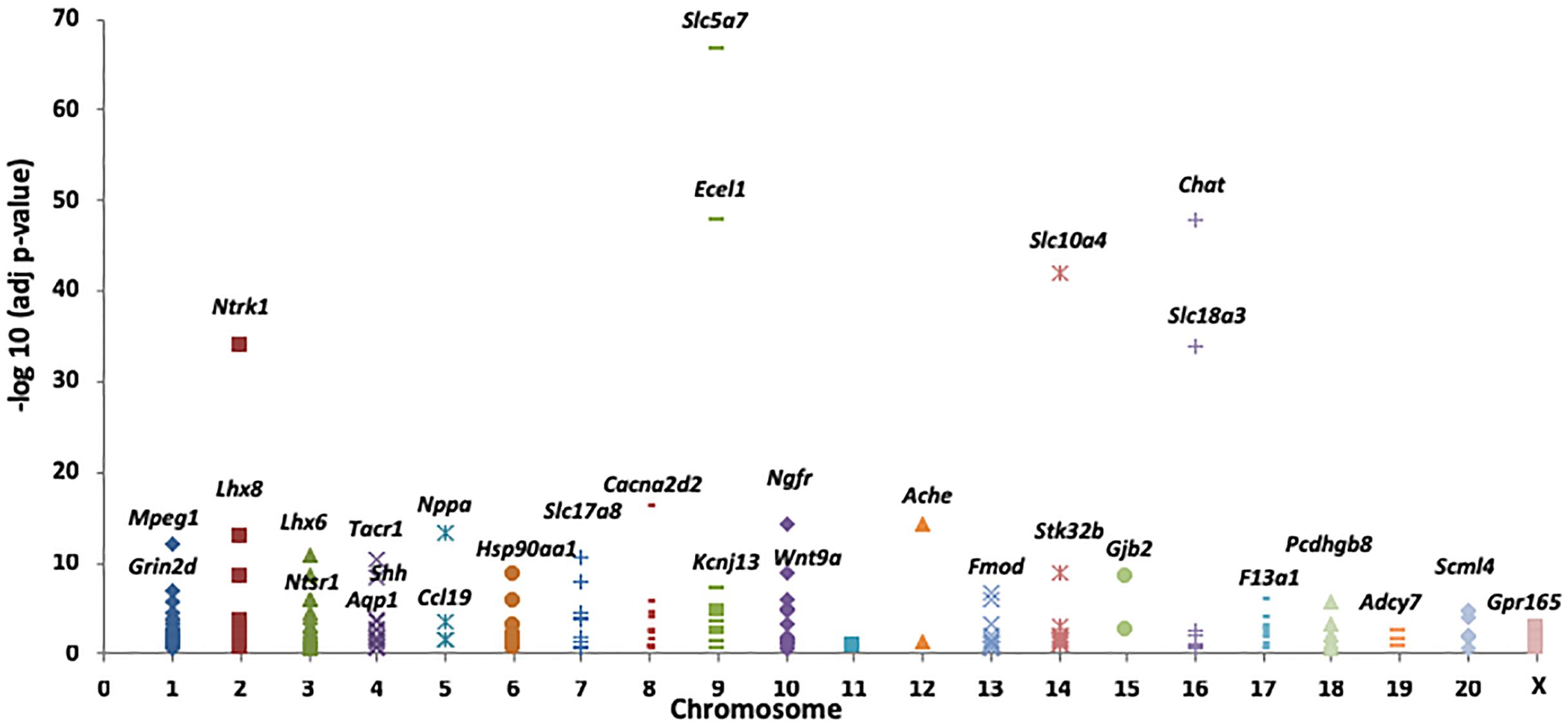
FIGURE 2. A total of 73 differentially expressed genes (DEG) between female iP and ISCS-B rats possess a fold change >2 with p < 0.05. The significance of these genes was plotted by corresponding chromosomes. Genes with the lowest p-values for each chromosome are labeled.
We utilized IPA to predict relevant molecular networks, biological functions, and canonical pathways altered between ISCS-B and iP female rats. The input gene list included 790 DEGs with an FDR-adjusted p-value cutoff of 0.05 (Figure 3 and Supplementary Figure S3). The most significant networks identified were relevant to axonal guidance signaling, cAMP-mediated signaling, and GABA receptor signaling, which are also important for drinking behavior (Figure 3). Genes within these pathways that have been associated with alcohol consumption in previous research studies include Adcy7, Aldh1a2, Chrm2, Grin2d, Nfkb, Gsat4, Tacr1, and Oprm1. Interestingly, the majority of genes in GABA receptor signaling and glutamate degradation III pathways showed reduced expression, while more genes with increased expression were found in pathways involved in axonal guidance, G-protein coupled receptor signaling, and glutathione-redox reactions (Figure 3).
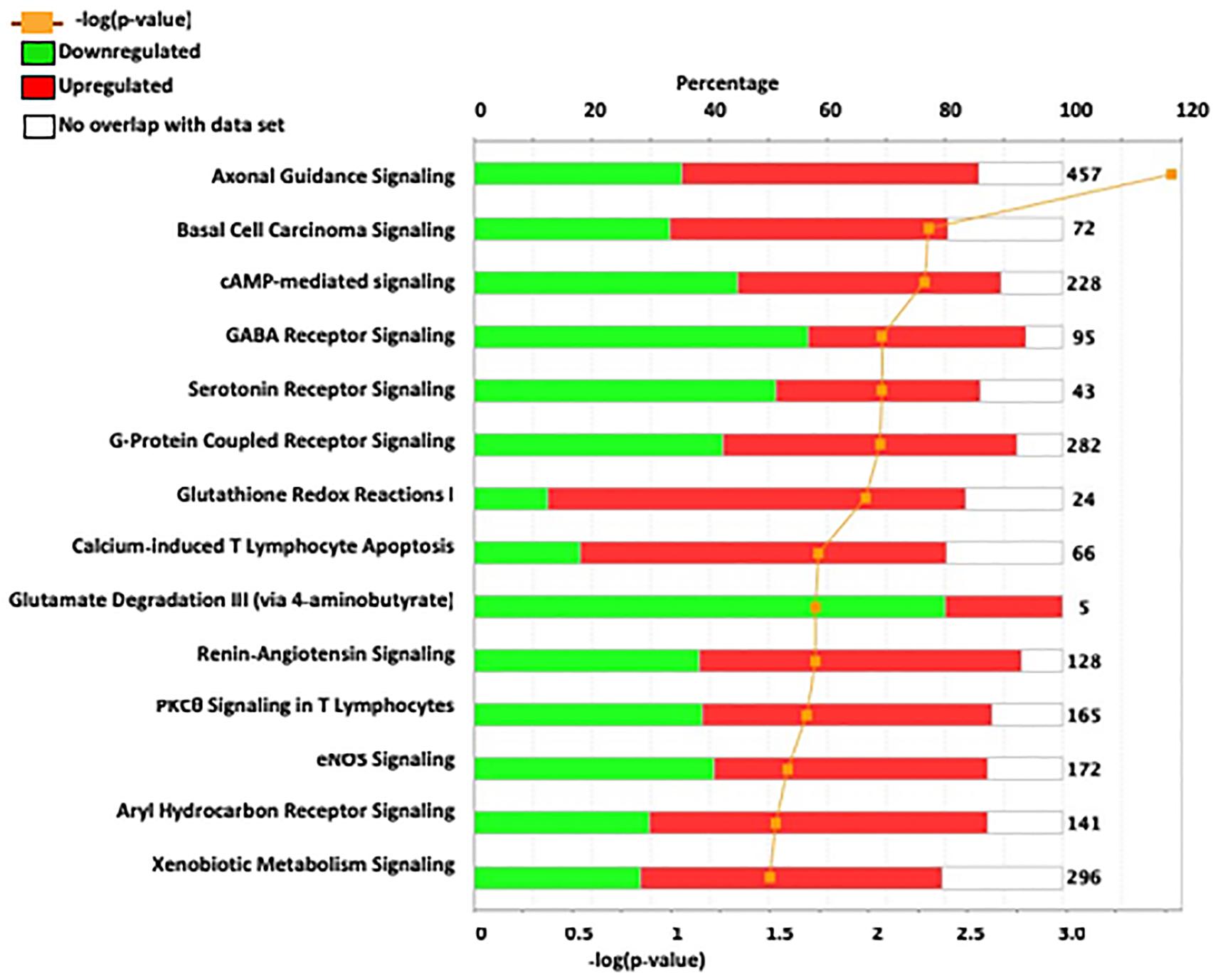
FIGURE 3. In depth analysis of DEG between ISCS-B and iP using IPA. The most significant IPA canonical pathways are listed on the left. The stacked bar chart reveals the percentage of up-regulated (red) and down-regulated (green) genes within each canonical pathway. The numerical value at the right of each bar represents the total number of genes in the canonical pathways. The secondary x-axis (bottom) represents the -log of p-value.
To understand what factors might be driving these DEGs, we used IPA upstream analysis to predict the top transcriptional regulators. The top three upstream regulators were β-estradiol, α-synuclein, and β-catenin. Other additional relevant upstream regulators are included in Table 2. Among these predicted upstream regulators, our findings suggest that the hormones β-estradiol, progesterone, as well as dihydrotestosterone, likely contribute to differences in sex-specific gene expression.
In addition to IPA analysis, DEGs with p-values <0.05, FC > 2 were used as input for Reactome data analysis, which is focused on molecular interactions within cells. The results of this analysis showed that the signal transduction pathways involving peptide ligand-binding receptors of class A/1 had the lowest p-values, indicating the significance of these interactions (Figure 4). We also found interesting regulator effect networks in our dataset. For example, Ngf and Raf1 play roles in dopamine and coordination, and ASCL1, estrogen receptor, and NGF are regulators of cell movement in neurons. More networks can be found in the (Supplementary Table S4).
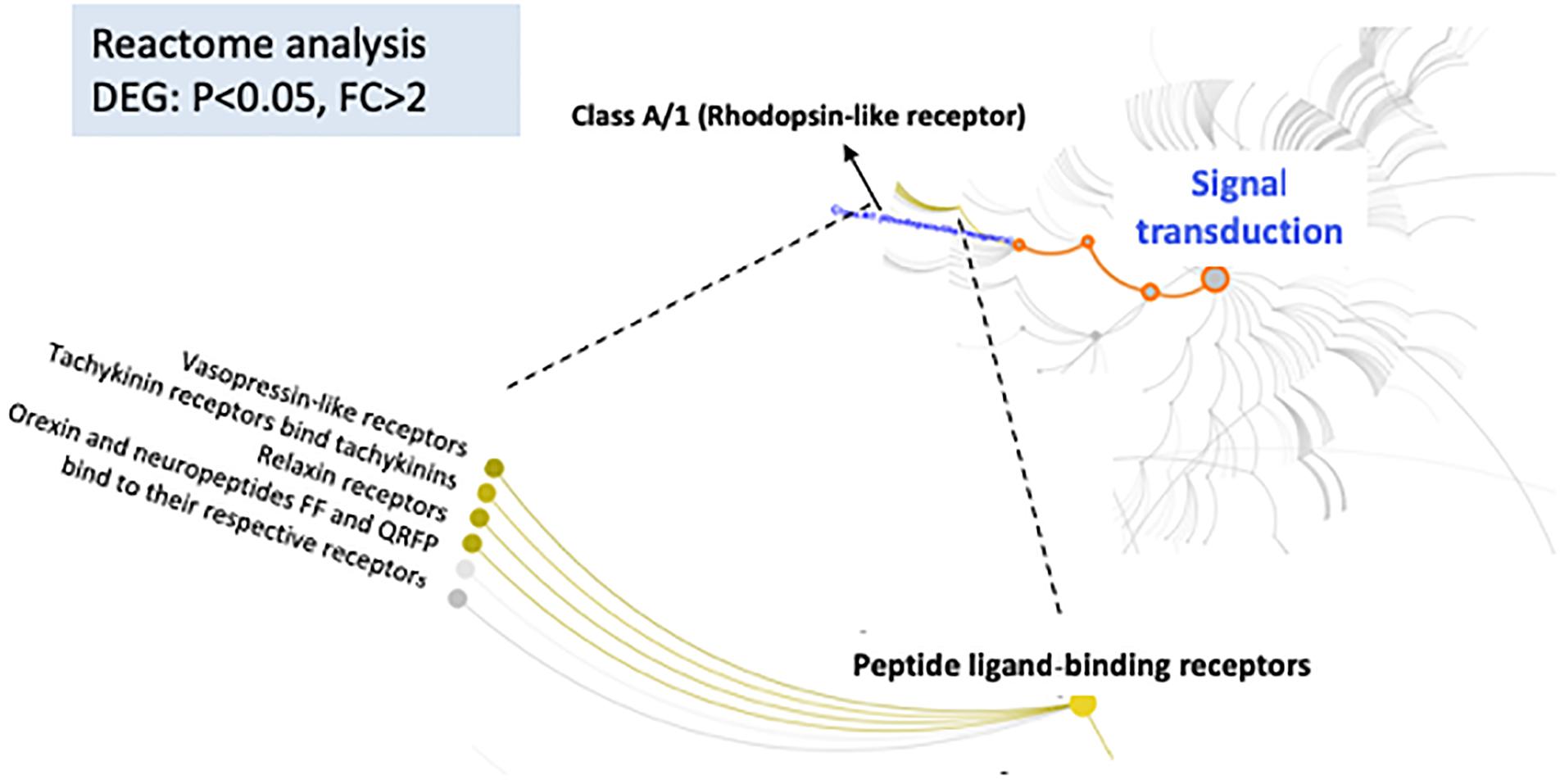
FIGURE 4. Reactome database was used to analyze DEG relationships organized into biological pathways and processes (https://reactome.org/dev/). It reveals significant alterations in signal transduction pathways between female iP and ISCS-B rats. Color-highlighted edges indicate over-represented interactions.
To determine whether the DEG identified in female NAc were sex-specific or also were differentially expressed in males, mRNA expression levels of selected DEGs were measured using RT-qPCR in both male and female ISCS-B and iP NAc tissue. The genes selected for measurement either showed a high fold change between the ISCS-B and iP RNA-seq data (e.g., Lhx8, Ngfr, Slc5a7), or were located in the Chr4 QTL and had been shown to be associated with alcohol consumption (e.g., Adcyap1r1, Adcy7, Snca, and Tacr1) or were reported previously to be differentially expressed between P and NP rats (e.g., Ppm1K and Aqp1). RT-qPCR was performed using RNA isolated from a different set of ISCS-B and iP rats. Figure 5 displays the fold-change (ISCS-B/iP ratio) and Supplementary Table S2 includes relative gene expression levels and t-test p-values between ISCS-B and iP animals for both males and females. Among these genes, Chat, Lhx8, Ngfr, Slc18a3, and Tacr1 were found to be differentially expressed in both male and female ISCS-B compared to iP rats; however, differential expression of Adcy7 and Slc5a7 was observed between ISCS-B and iP males (p = 0.009 and 0.002, respectively), but no differences were found in females. Interestingly, Nap1l5, which is a maternally imprinted gene (Smith et al., 2003), was up-regulated in ISCS-B compared to iP females, but not males, which suggests that altered expression might be related to sex-specific differences in epigenetic regulation. Of particular relevance, Adcyap1r1, which has been associated with drinking in women (Dragan et al., 2017), was also up-regulated in ISCS-B compared to iP females, but was not significantly different in males (Figures 6A,B).
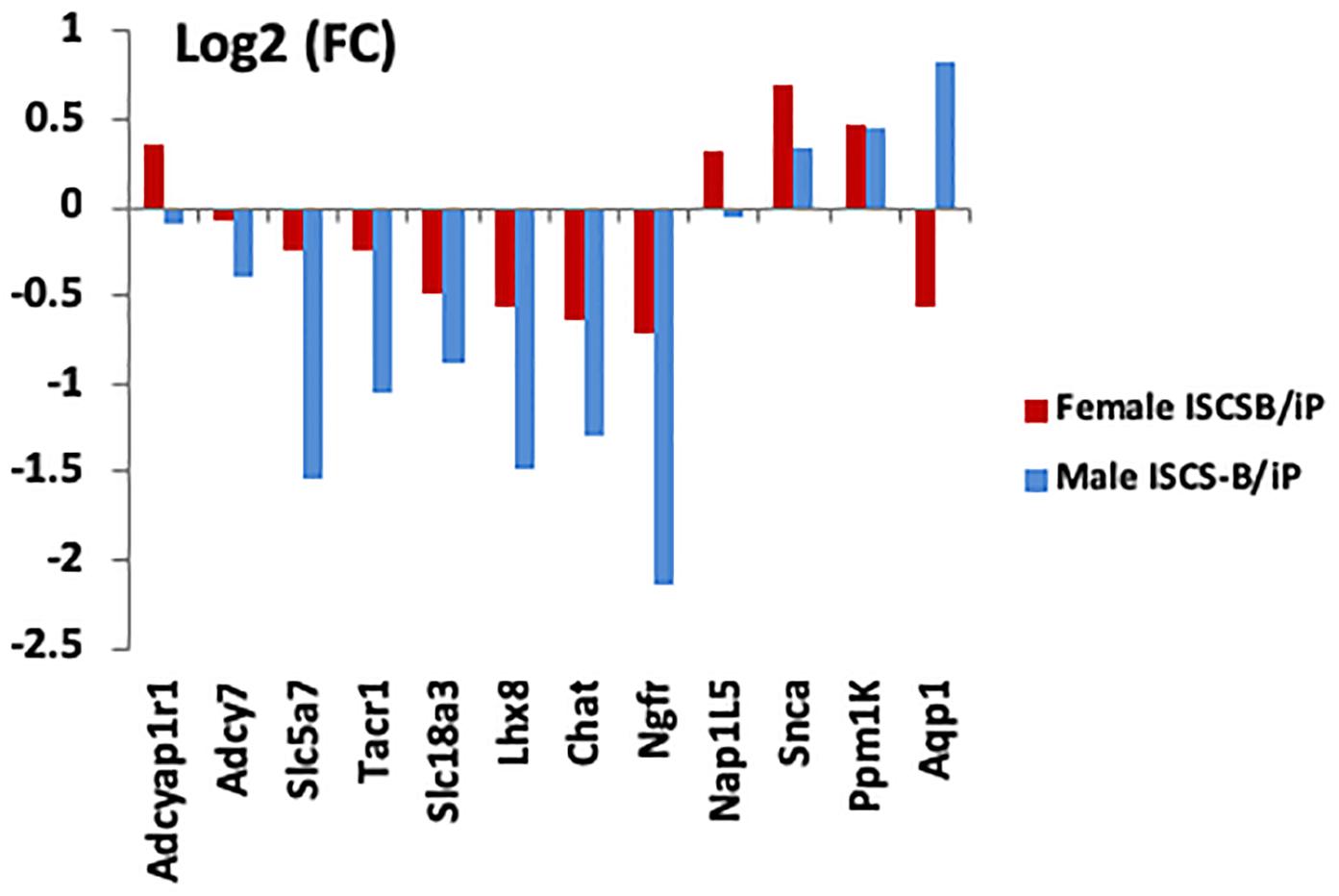
FIGURE 5. RT-qPCR comparisons of gene expression between male and female iP and ISCS-B rats. Chat, Slc18a3, Lhx8, and Ngfr were confirmed to be differentially expressed in both male and female ISCS-B compared to iP rats. Adcyap1r1 and Nap1L5 were upregulated in female ISCS-B compared to iP animals, but no difference was found in males.
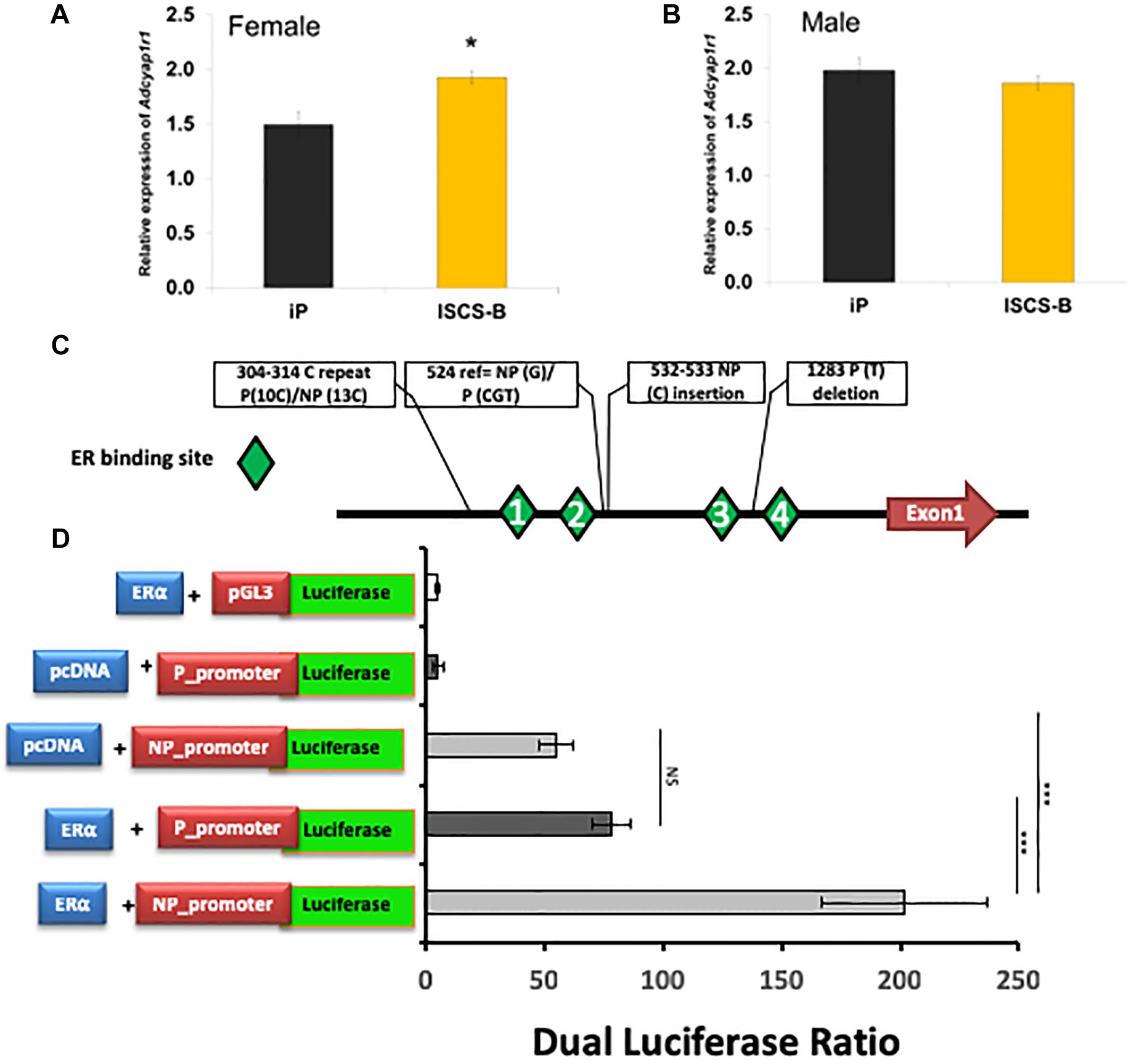
FIGURE 6. (A) Adcyap1r1 expression is increased in female ISCS-B relative to iP rats. (B) Male ISCS-B rats demonstrate no significant difference from iP in Adcyap1r1 expression. (C) A schematic of the Adcyap1r1 promoter sequence demonstrating ER binding sites and polymorphisms between iP and iNP. (D) ERα transactivation of both variants of the Adcyap1r1 promoter enhanced luciferase activity, but the iNP promoter exhibited significantly more luciferase activity than iP.
In addition to RNA-seq, we also performed GenBank and literature searches for genes that mapped to the Chr4 interval region and that had evidence of an association with alcohol consumption or response to estrogen. This approach identified several estrogen-responsive genes, including Adcyap1r1. Importantly, the Adcyap1r1 genotype was associated with alcohol abuse in women (Dragan et al., 2017), and it also has been shown to respond to alcohol treatment (Koh et al., 2006). In addition, the human ADCYAP1R1 gene contains a SNP in an estrogen response element (ERE) that was associated with PTSD only in females (Ressler et al., 2011). Therefore, we selected this gene for further mechanistic investigation to determine whether an ERE also existed in the rat Adcyap1r1 promoter, and if any genetic variants existed between iP and iNP strains that might affect estrogen receptor α (ERα) binding. Estrogen receptor binding site prediction was performed using Genomtix4, which revealed four potential EREs within 2 kb upstream of the Adcyap1r1 transcription start site (Figure 6C), which was designated as + 1. We cloned and sequenced a 2 kb region of the Adcyap1r1 promoter from genomic DNA of both iP and ISCS-B, which contained the Adcyap1r1 iNP Chr4 sequence. In addition to confirming the presence of ERE consensus sites, we identified five sequence variants, including two that mapped near predicted EREs at nt 485–494 (#1) and nt 1303–1310 (#4) (Table 3 and Figure 6).
To evaluate the potential of these genomic variants to act as cis-elements in ERα transactivation of Adcyap1r1 gene expression, we performed in vitro dual luciferase promoter assays. The iP and iNP promoter regions of Adcyap1r1 were cloned into the pGL3 vector separately, and tested with and without dual ERα expression from the pcDNA vector. When empty pcDNA vector was transfected, the NP promoter showed higher luciferase activity compared to the P promoter. This effect could result from an endogenous estrogen effect or from the action of other transcription factors. As shown in Figure 6, luciferase expression was significantly upregulated by ERα, when the Adcyap1r1 promoter was transcribed from the iNP compared to the iP genome. This result indicated that the iNP variant was more sensitive to ERα binding than the iP variant and suggested that these variants could contribute to the observed sex-differences in alcohol consumption between the iP and iNP strains.
Our results revealed a sex-specific QTL for alcohol consumption on rat Chr4 and point to a potential role for the gene Acdyap1r1 in female-specific alcohol drinking behavior in the P rat model of AUD. Specifically, we identified DEG’s in the NAc that were associated with the transferred QTL region in ISCS-B rats. These DEG’s were predicted to affect gene networks controlling nervous system development and function, as well as drug metabolism and behavior. Notably, the top predicted upstream regulator of DEG’s in female NAc was β-estradiol. Importantly, we found that sequence variants in the Acdyap1r1 promoter showed differential activation by the estrogen receptor.
Females drink more alcohol than males in most, if not all, selectively bred lines of rodent models for alcohol consumption, including the iP rats used in this study (Li and Lumeng, 1984; Lancaster and Spiegel, 1992; McBride and Li, 1998; Chester et al., 2006). In the present study, we found that when a relatively small region of the iNP genome was substituted in the iP Chr4 QTL region, homozygous female ISCS rats consumed less alcohol (g EtOH/kg BW/day) and they exhibited similar alcohol consumption compared to iP control animals (Figure 1); this effect was not detected in their male counterparts. The reduced alcohol consumption in ISCS females was not due to body weight since we previously demonstrated that ISCS-B animals do not differ in body weight from the iP line (Spence et al., 2013). Additionally, because heterozygous female ISCSB-H animals consumed similar amounts of alcohol as iP controls, the sex-specific QTL we identified is a recessive trait.
The sex-specific drinking phenotype identified in the current research suggests that genetic variation from the NP genome within the ISCS-B congenic region likely contributes to decreased alcohol consumption in females in the P and NP model. We previously created the ISCS lines that decreased the background heterogeneity found in the P.NP congenic strain and simultaneously narrowed the QTL region (Spence et al., 2009; Spence et al., 2013; Baud and Flint, 2017). The sole difference within the entire genomes of ISCS-B and iP is a 1.7–8.9 Mb genomic segment, or less than approximately 0.3% of the 2719 Mb rat genome (Jensen-Seaman et al., 2004). Clearly, this ISCS-B genomic region contains variants from the NP donor, which modify gene expression via cis-elements or interact with trans-acting factors (e.g., estrogen receptor), thereby altering gene networks that significantly affect female alcohol consumption (Figure 6). We hypothesized that many interactions exist between genes within this 1.7–8.9 Mb genomic region and genes outside this region, and replacement of this region would result in trans-regulation via cis-acting elements.
Adcy7 has been associated with female-specific ethanol consumption in mice (Pronko et al., 2010; Cruz et al., 2011; Desrivières et al., 2011). IPA showed that ADCY7, ADCY, and Creb are involved in drug metabolism and behavior. Our finding indicated that the adenylate cyclase (AC) signaling pathway was enriched with genes associated with female alcohol consumption or response to estrogen regulation (Figure 7). We emphasize that upregulation of Adcyap1r1 may affect AC, and downstream genes such as PAK, ERK and BDNF, which could also play important roles in neuron protection.
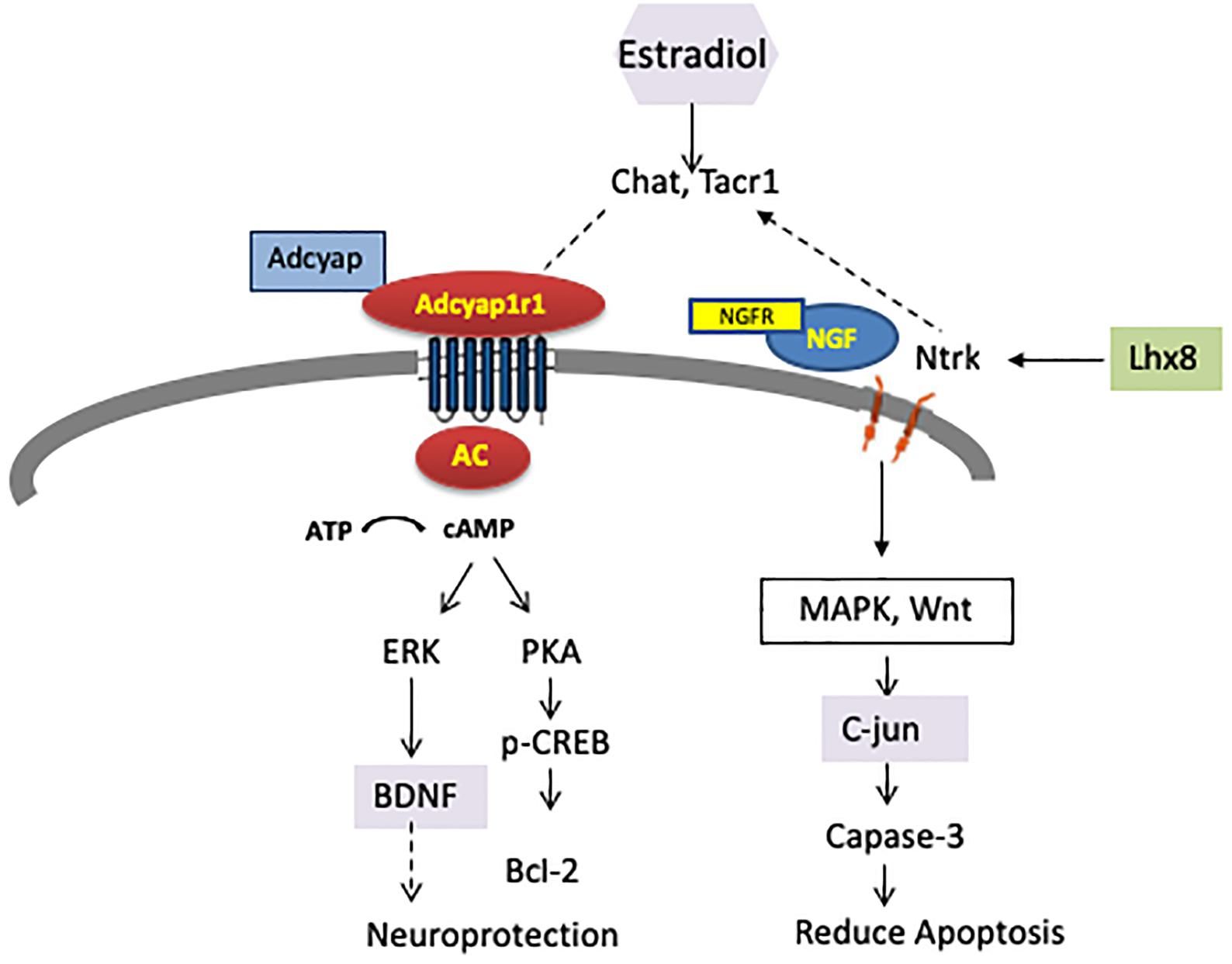
FIGURE 7. DEGs were enriched in two pathways which affect neuron function and are also regulated by estrogen receptor (ER). Adenylate cyclase (AC) and downstream signaling affecting neuroprotection was upregulated, while NtrK, Ngfr, and Lhx8 signaling affecting apoptosis were downregulated.
Hormones affect gene expression at both cis- (e.g., in the narrowed Chr4 region) and trans- (elsewhere) locations. Through data mining and literature searching, we identified multiple genes in the Chr4 QTL (e.g., Aqp1, Abcg2, Adcyap1r1, Npy, and Snca) that are regulated by estrogen directly or indirectly. Bioinformatics analysis also predicted upstream regulators of the DEGs which indicated that sex hormones are likely to play an important role (Table 3). Hormones, such as β-estradiol, target multiple DEGs, including Tacr1 on Chr4, and Chat, Nts, Nupr1, Ogn, Ttr, and Oprm1 on other chromosomes. Importantly, all these genes have relatively high fold change between ISCS-B and iP rats. In addition, β-estradiol is also predicted to target Aldh1a2 and affect alcohol metabolism. Esr1 encoding the estrogen receptor α is significantly decreased in ISCS-B NAc. Interestingly, β-estradiol is a regulator of the Chat and Tacr1 genes.
We hypothesized that the sex-dependent differences in gene expression were related to promoter variants between the iP and ISCS-B lines, altering the binding affinity for trans-acting factors (e.g., estrogen receptor) involved in gene transcription (Figure 8). We show that genetic variants in the Adcyap1r1 promoter respond to estrogen stimulation, thereby providing one possible mechanism for sex-specific differences in alcohol consumption. In humans, ADCYAP1R1 has been associated with female alcohol consumption (Dragan et al., 2017). Pituitary adenylyl cyclase-activating polypeptide (ADCYAP)-ADCYAP1R1 pathway is regulated by estrogen and is involved in abnormal fear responses underlying PTSD (Ramikie and Ressler, 2016). Our finding that SNPs in the promoter region of the NP Adcyap1r1 allele are more sensitive to estrogen stimulation in vitro suggests that alcohol consumption is mediated, in part, by estrogen regulation of ADCYAP1R1 in the P and NP model, providing a potential explanation for why alcohol consumption was decreased in female ISCS lines. Moreover, ADCYAP was found to be co-localized with ChAT in nerve fibers (Drescher et al., 2006) and it is possible that Adcyap1r1 could also interact with ChAT, thereby providing a potential mechanism for how increased ADCYAP1R1 expression might promote neuroprotection (Figure 8). In addition, pathway analysis showed an increase of glutathione-redox reactions I, conveying a similar neuroprotection function.
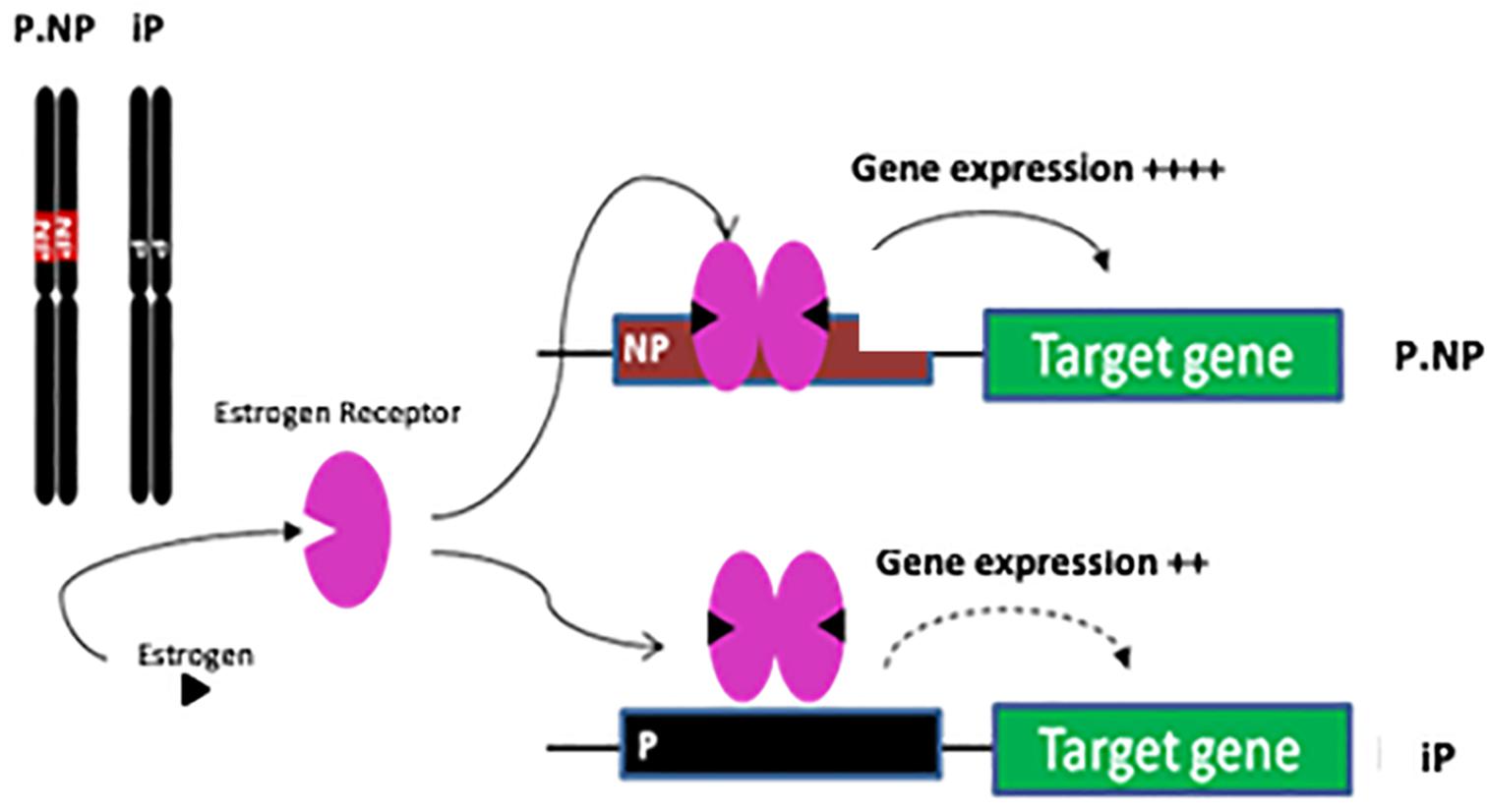
FIGURE 8. Schematic of genetic and estrogen receptor interaction affecting gene expression. The only difference between congenic P.NP and iP is on Chr4 region. Compared with P genomic sequence, the genetic difference in NP is proposed to lead to stronger binding of estrogen receptor in promoters of targeted genes resulting in increased gene expression.
In this research, we observed that genes related to cholinergic function were reduced more than 4-fold in ISCS-B relative to iP, including Chat, Slc18a3, Slc5a7, and LIM homeobox 8. Consistent with our findings, previous studies found higher expression of Chat, Slc18a3, and Slc5a7 in the NAc shell of adult P rats compared to NP rats (McBride et al., 2013). Further, cocaine treatment and withdrawal are also associated with increased ChAT, Slc5a7, and Slc18a3 expression in the NAc (Eipper-Mains et al., 2013). Others also have reported that alcohol consumption reduces ChAT in NAc, resulting in fewer basal forebrain cholinergic neurons (Jamal et al., 2007; Coleman et al., 2011; Pereira et al., 2014). Thus, the cholinergic neuron function in the NAc deserves further investigation with regard to its involvement in female drinking. Another significant finding is the down-regulation of Ngfr and Lhx8, resulting in alterations of Ntrk1, which might regulate downstream MAPK signaling and Wnt (Figure 8).
We speculate that a potential mechanism for sex-dependent differences in gene expression is that promoter variants between the iP and ISCS-B lines alter the binding affinity for trans-acting factors, such as estrogen receptor (Figure 6), resulting in different levels of mRNA expression between female ISCS-B and iP controls. Because two interrelated pathways were found to be enriched for genes associated with female alcohol consumption or response to estrogen regulation (Figure 7); therefore, we further speculate that upregulation of Adcyap1r1 may affect adenylyl cyclase signaling resulting in neuron protection and downregulation of Ntrk and Ngf signaling resulting in reduced apoptosis. These data suggest that AC and cholinergic function may play important roles in female alcohol consumption. Additionally, BDNF, C-jun and estradiol are all significant upstream regulators that affect DEG expression and are functionally important in these pathways. Together, these findings demonstrate a potential role for estrogen in the upregulation of Adcyap1r1 expression that is associated with a sex-specific QTL for alcohol consumption on rat chromosome 4.
TL along with JPS, JLR, and WY designed the research and wrote the manuscript. JPS, TG, KEW, and PJB performed the animal model development, alcohol consumption test, and molecular analysis in the research. WY along with BQ, HG, and LZ performed cloning and transient transfection. DKG and ZL executed the RNA-seq. YZ along with TL, WY, and JPS analyzed the data. All authors have read and given final approval for the manuscript.
This research was supported by grants from CAMS Innovation Fund for Medical Sciences (CIFMS) (2017-I2M-3-015), the National Science Foundation of China (Nos. 81700751 and 2013CB945001), the National Institutes of Health NIH/NIAAA, R01 AA10707, P60AA007611, and R24AA015512. We appreciate the funding from Biomedical Research Grant of the Indiana University School of Medicine that allowed us to initiate this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to express our appreciation to Michelle Johnson and Judy Powers who provided technical support, Dr. Lucinda Carr, Dr. Lawrence Lumeng, and Indiana Alcohol Research Center, for their support in initiating this long-term project and providing useful discussion of this research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00513/full#supplementary-material
FIGURE S1 | Female ISCS-B rats demonstrated less consumption than iP and ISCSB-H. Males demonstrated no differences between strains. Female heterozygotes (ISCSB-H) showed a similar level of alcohol consumption when compared to iP controls.
FIGURE S2 | Female and male ISCS-B rats exhibited similar consumption of either saccharin or quinine indicating no taste difference between sexes.
FIGURE S3 | Differentially expressed genes were plotted by gene expression and fold change. The top five Ingenuity Pathway Analysis networks, diseases and bio functions are listed.
TABLE S1 | RNA-seq reads of each sample.
TABLE S2 | Gene expression in the nucleus accumbens of both male and female rats using RT-qPCR.
TABLE S3 | Differentially expressed gene list.
TABLE S4 | Significant top regulator effect networks.
TABLE S5 | List of primer sequences.
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Baud, A., and Flint, J. (2017). Identifying genes for neurobehavioural traits in rodents: progress and pitfalls. Dis. Model Mech. 10, 373–383. doi: 10.1242/dmm.027789
Becker, J. B., and Koob, G. F. (2016). Sex differences in animal models: focus on addiction. Pharmacol. Rev. 68, 242–263. doi: 10.1124/pr.115.011163
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–290.
Bice, P., Foroud, T., Bo, R., Castelluccio, P., Lumeng, L., Li, T. K., et al. (1998). Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm. Genome 9, 949–955. doi: 10.1007/s003359900905
Bice, P. J., Foroud, T., Carr, L. G., Zhang, L., Liu, L., Grahame, N. J., et al. (2006). Identification of QTLs influencing alcohol preference in the High Alcohol Preferring (HAP) and Low Alcohol Preferring (LAP) mouse lines. Behav. Genet. 36, 248–260. doi: 10.1007/s10519-005-9019-6
Carr, L. G., Foroud, T., Bice, P., Gobbett, T., Ivashina, J., Edenberg, H., et al. (1998). A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol. Clin. Exp. Res. 22, 884–887. doi: 10.1111/j.1530-0277.1998.tb03883.x
Carr, L. G., Habegger, K., Spence, J. P., Liu, L., Lumeng, L., and Foroud, T. (2006). Development of congenic rat strains for alcohol consumption derived from the alcohol-preferring and nonpreferring rats. Behav. Genet. 36, 285–290. doi: 10.1007/s10519-005-9021-z
Ceylan-Isik, A. F., McBride, S. M., and Ren, J. (2010). Sex difference in alcoholism: who is at a greater risk for development of alcoholic complication? Life Sci. 87, 133–138. doi: 10.1016/j.lfs.2010.06.002
Chesler, E. J., Plitt, A., Fisher, D., Hurd, B., Lederle, L., Bubier, J. A., et al. (2012). Quantitative trait loci for sensitivity to ethanol intoxication in a C57BL/6Jx129S1/SvImJ inbred mouse cross. Mamm. Genome 23, 305–321. doi: 10.1007/s00335-012-9394-2
Chester, J. A., de Paula Barrenha, G., DeMaria, A., and Finegan, A. (2006). Different effects of stress on alcohol drinking behaviour in male and female mice selectively bred for high alcohol preference. Alcohol Alcohol. 41, 44–53. doi: 10.1093/alcalc/agh242
Cicero, T. (1979). “A critique of animal analogues of alcoholism,” in Biochemistry and Pharmacology of Ethanol, Vol. 2, eds E. Majchrowicz and E. P. Noble (New York, NY: Plenum Press), 533–560.
Coleman, L. G. Jr., He, J., Lee, J., Styner, M., and Crews, F. T. (2011). Adolescent binge drinking alters adult brain neurotransmitter gene expression, behavior, brain regional volumes, and neurochemistry in mice. Alcohol. Clin. Exp. Res. 35, 671–688. doi: 10.1111/j.1530-0277.2010.01385.x
Cruz, M. T., Bajo, M., Maragnoli, M. E., Tabakoff, B., Siggins, G. R., and Roberto, M. (2011). Type 7 adenylyl cyclase is involved in the ethanol and CRF sensitivity of GABaergic synapses in mouse central amygdala. Front. Neurosci. 4:207. doi: 10.3389/fnins.2010.00207
Desrivières, S., Pronko, S. P., Lourdusamy, A., Ducci, F., Hoffman, P. L., Wodarz, N., et al. (2011). Sex-specific role for adenylyl cyclase type 7 in alcohol dependence. Biol. Psychiatry 69, 1100–1108. doi: 10.1016/j.biopsych.2011.01.037
Dick, D. M., Bierut, L., Hinrichs, A., Fox, L., Bucholz, K. K., Kramer, J., et al. (2006). The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behav. Genet. 36, 577–590. doi: 10.1007/s10519-005-9041-8
Dimas, A. S., Nica, A. C., Montgomery, S. B., Stranger, B. E., Raj, T., Buil, A., et al. (2012). Sex-biased genetic effects on gene regulation in humans. Genome Res. 22, 2368–2375. doi: 10.1101/gr.134981.111
Donhoffner, M. E., Goings, S. P., Atabaki, K., and Wood, R. I. (2016). Intracerebroventricular oxytocin self-administration in female rats. J. Neuroendocrinol. 28. doi: 10.1111/jne.12416
Dragan, W. L., Czerski, P. M., and Dragan, M. (2017). PAC1 receptor (ADCYAP1R1) genotype and problematic alcohol use in a sample of young women. Neuropsychiatr. Dis. Treat. 13, 1483–1489. doi: 10.2147/NDT.S137331
Drescher, M. J., Drescher, D. G., Khan, K. M., Hatfield, J. S., Ramakrishnan, N. A., Abu-Hamdan, M. D., et al. (2006). Pituitary adenylyl cyclase-activating polypeptide (PACAP) and its receptor (PAC1-R) are positioned to modulate afferent signaling in the cochlea. Neuroscience 142, 139–164. doi: 10.1016/j.neuroscience.2006.05.065
DuBose, C. S., Chesler, E. J., Goldowitz, D., and Hamre, K. M. (2013). Use of the expanded panel of BXD mice narrow QTL regions in ethanol-induced locomotor activation and motor incoordination. Alcohol. Clin. Exp. Res. 37, 170–183. doi: 10.1111/j.1530-0277.2012.01865.x
Ducci, F., and Goldman, D. (2008). Genetic approaches to addiction: genes and alcohol. Addiction 103, 1414–1428. doi: 10.1111/j.1360-0443.2008.02203.x
Eipper-Mains, J. E., Kiraly, D. D., Duff, M. O., Horowitz, M. J., McManus, C. J., Eipper, B. A., et al. (2013). Effects of cocaine and withdrawal on the mouse nucleus accumbens transcriptome. Genes Brain Behav. 12, 21–33. doi: 10.1111/j.1601-183X.2012.00873.x
Fabregat, A., Sidiropoulos, K., Garapati, P., Gillespie, M., Hausmann, K., Haw, R., et al. (2016). The reactome pathway knowledgebase. Nucleic Acids Res. 44, D481–D487. doi: 10.1093/nar/gkv1351
Gatti, D. M., Zhao, N., Chesler, E. J., Bradford, B. U., Shabalin, A. A., Yordanova, R., et al. (2010). Sex-specific gene expression in the BXD mouse liver. Physiol. Genomics 42, 456–468. doi: 10.1152/physiolgenomics.00110.2009
Gehle, V. M., and Erwin, V. G. (1998). Common quantitative trait loci for alcohol-related behaviors and CNS neurotensin measures: voluntary ethanol consumption. Alcohol. Clin. Exp. Res. 22, 401–408. doi: 10.1097/00000374-199804000-00016
George, D. T., Gilman, J., Hersh, J., Thorsell, A., Herion, D., Geyer, C., et al. (2008). Neurokinin 1 receptor antagonism as a possible therapy for alcoholism. Science 319, 1536–1539. doi: 10.1126/science.1153813
Gershoni, M., and Pietrokovski, S. (2017). The landscape of sex-differential transcriptome and its consequent selection in human adults. BMC Biol. 15:7. doi: 10.1186/s12915-017-0352-z
Gill, K., Desaulniers, N., Desjardins, P., and Lake, K. (1998). Alcohol preference in AXB/BXA recombinant inbred mice: gender differences and gender-specific quantitative trait loci. Mamm. Genome 9, 929–935. doi: 10.1007/s003359900902
Heath, A. C., Bucholz, K. K., Madden, P. A., Dinwiddie, S. H., Slutske, W. S., Bierut, L. J., et al. (1997). Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol. Med. 27, 1381–1396. doi: 10.1017/S0033291797005643
Jamal, M., Ameno, K., Ikuo, U., Kumihashi, M., Wang, W., and Ijiri, I. (2007). Ethanol and acetaldehyde: in vivo quantitation and effects on cholinergic function in rat brain. Novartis Found. Symp. 285, 137–141, discussion 141–144, 198–199.
Jensen-Seaman, M. I., Furey, T. S., Payseur, B. A., Lu, Y., Roskin, K. M., Chen, C. F., et al. (2004). Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 14, 528–538. doi: 10.1101/gr.1970304
Karp, N. A., Mason, J., Beaudet, A. L., Benjamini, Y., Bower, L., Braun, R. E., et al. (2017). Prevalence of sexual dimorphism in mammalian phenotypic traits. Nat. Commun. 8:15475. doi: 10.1038/ncomms15475
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Koh, P. O., Won, C. K., and Ho, J. H. (2006). Ethanol decreases the expression of pituitary adenylate cyclase activating polypeptide in rat testes. J. Vet. Med. Sci. 68, 635–637. doi: 10.1292/jvms.68.635
Koss, M. P., and Goldman, D. (2000). Genetic factors and alcoholism. Am. J. Public Health 90:1799. doi: 10.2105/AJPH.90.11.1799
Lancaster, F. E., and Spiegel, K. S. (1992). Sex differences in pattern of drinking. Alcohol 9, 415–420. doi: 10.1016/0741-8329(92)90041-8
Li, T. K., and Lumeng, L. (1984). Alcohol preference and voluntary alcohol intakes of inbred rat strains and the National Institutes of Health heterogeneous stock of rats. Alcohol. Clin. Exp. Res. 8, 485–486. doi: 10.1111/j.1530-0277.1984.tb05708.x
Li, T. K., Lumeng, L., and Doolittle, D. P. (1993). Selective breeding for alcohol preference and associated responses. Behav. Genet. 23, 163–170. doi: 10.1007/BF01067421
Li, T. K., Lumeng, L., Doolittle, D. P., and Carr, L. G. (1991). Molecular associations of alcohol-seeking behavior in rat lines selectively bred for high and low voluntary ethanol drinking. Alcohol Alcohol. Suppl. 1, 121–124.
Liang, T., and Carr, L. G. (2006). Regulation of alpha-synuclein expression in alcohol-preferring and – non preferring rats. J. Neurochem. 99, 470–482. doi: 10.1111/j.1471-4159.2006.04111.x
Liang, T., Habegger, K., Spence, J. P., Foroud, T., Ellison, J. A., Lumeng, L., et al. (2004). Glutathione S-transferase 8-8 expression is lower in alcohol-preferring than in alcohol-nonpreferring rats. Alcohol. Clin. Exp. Res. 28, 1622–1628. doi: 10.1097/01.ALC.0000145686.79141.57
Liang, T., Kimpel, M. W., McClintick, J. N., Skillman, A. R., McCall, K., Edenberg, H. J., et al. (2010). Candidate genes for alcohol preference identified by expression profiling in alcohol-preferring and -nonpreferring reciprocal congenic rats. Genome Biol. 11:R11. doi: 10.1186/gb-2010-11-2-r11
Liang, T., Spence, J., Liu, L., Strother, W. N., Chang, H. W., Ellison, J. A., et al. (2003). alpha-Synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and –nonpreferring rats. Proc. Natl. Acad. Sci. U.S.A. 100, 4690–4695. doi: 10.1073/pnas.0737182100
Lumeng, L., Hawkins, T. D., and Li, T. (1977). “New strains of rats with alcohol preference and nopreference,” in Alcohol and Aldehyde Metabolizing System, Vol. III, eds R. G. Thurman, J. R. Williamson, H. Drott, and B. S. Chance (New York, NY: Academic Press), 537–544. doi: 10.1016/B978-0-12-691403-0.50056-2
McBride, W. J., Kimpel, M. W., McClintick, J. N., Ding, Z. M., Hyytia, P., Colombo, G., et al. (2013). Gene expression within the extended amygdala of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Alcohol 47, 517–529. doi: 10.1016/j.alcohol.2013.08.004
McBride, W. J., and Li, T. K. (1998). Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. Crit. Rev. Neurobiol. 12, 339–369. doi: 10.1615/CritRevNeurobiol.v12.i4.40
Melo, J. A., Shendure, J., Pociask, K., and Silver, L. M. (1996). Identification of sex-specific quantitative trait loci controlling alcohol preference in C57BL/ 6 mice. Nat. Genet. 13, 147–153. doi: 10.1038/ng0696-147
Murphy, J. M., Stewart, R. B., Bell, R. L., Badia-Elder, N. E., Carr, L. G., McBride, W. J., et al. (2002). Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav. Genet. 32, 363–388. doi: 10.1023/A:1020266306135
Nolen-Hoeksema, S. (2004). Gender differences in risk factors and consequences for alcohol use and problems. Clin. Psychol. Rev. 24, 981–1010. doi: 10.1016/j.cpr.2004.08.003
Pandey, S., Badve, P. S., Curtis, G. R., Leibowitz, S. F., and Barson, J. R. (2017). Neurotensin in the posterior thalamic paraventricular nucleus: inhibitor of pharmacologically relevant ethanol drinking. Addict. Biol. doi: 10.1111/adb.12546 [Epub ahead of print].
Peirce, J. L., Derr, R., Shendure, J., Kolata, T., and Silver, L. M. (1998). A major influence of sex-specific loci on alcohol preference in C57Bl/6 and DBA/2 inbred mice. Mamm. Genome 9, 942–948. doi: 10.1007/s003359900904
Pereira, P. A., Neves, J., Vilela, M., Sousa, S., Cruz, C., and Madeira, M. D. (2014). Chronic alcohol consumption leads to neurochemical changes in the nucleus accumbens that are not fully reversed by withdrawal. Neurotoxicol. Teratol. 44, 53–61. doi: 10.1016/j.ntt.2014.05.007
Prescott, C. A. (2002). Sex differences in the genetic risk for alcoholism. Alcohol Res. Health 26, 264–273.
Pronko, S. P., Saba, L. M., Hoffman, P. L., and Tabakoff, B. (2010). Type 7 adenylyl cyclase-mediated hypothalamic-pituitary-adrenal axis responsiveness: influence of ethanol and sex. J. Pharmacol. Exp. Ther. 334, 44–52. doi: 10.1124/jpet.110.166793
Radcliffe, R. A., Bohl, M. L., Lowe, M. V., Cycowski, C. S., and Wehner, J. M. (2000). Mapping of quantitative trait loci for hypnotic sensitivity to ethanol in crosses derived from the C57BL/6 and DBA/2 mouse strains. Alcohol. Clin. Exp. Res. 24, 1335–1342. doi: 10.1111/j.1530-0277.2000.tb02101.x
Ramikie, T. S., and Ressler, K. J. (2016). Stress-related disorders, pituitary adenylate cyclase-activating peptide (PACAP)ergic system, and sex differences. Dialogues Clin. Neurosci. 18, 403–413.
Rawlik, K., Canela-Xandri, O., and Tenesa, A. (2016). Evidence for sex-specific genetic architectures across a spectrum of human complex traits. Genome Biol. 17:166. doi: 10.1186/s13059-016-1025-x
Ressler, K. J., Mercer, K. B., Bradley, B., Jovanovic, T., Mahan, A., Kerley, K., et al. (2011) Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature 470, 492–497. doi: 10.1038/nature09856
Schank, J. R., Tapocik, J. D., Barbier, E., Damadzic, R., Eskay, R. L., Sun, H., et al. (2013). Tacr1 gene variation and neurokinin 1 receptor expression is associated with antagonist efficacy in genetically selected alcohol-preferring rats. Biol. Psychiatry 73, 774–781. doi: 10.1016/j.biopsych.2012.12.027
Smith, R. J., Dean, W., Konfortova, G., and Kelsey, G. (2003). Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res. 13, 558–569. doi: 10.1101/gr.781503
Spence, J. P., Lai, D., Shekhar, A., Carr, L. G., Foroud, T., and Liang, T. (2013). Quantitative trait locus for body weight identified on rat chromosome 4 in inbred alcohol-preferring and -nonpreferring rats: potential implications for neuropeptide Y and corticotrophin releasing hormone 2. Alcohol 47, 63–67. doi: 10.1016/j.alcohol.2012.10.005
Spence, J. P., Liang, T., Liu, L., Johnson, P. L., Foroud, T., Carr, L. G., et al. (2009). From QTL to candidate gene: a genetic approach to alcoholism research. Curr. Drug Abuse Rev. 2, 127–134. doi: 10.2174/1874473710902020127
Thorsell, A., Slawecki, C. J., Khoury, A., Mathe, A. A., and Ehlers, C. L. (2005). Effect of social isolation on ethanol consumption and substance P/neurokinin expression in Wistar rats. Alcohol 36, 91–97. doi: 10.1016/j.alcohol.2005.07.003
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van, Baren MJ, et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Vanderlinden, L. A., Saba, L. M., Bennett, B., Hoffman, P. L., and Tabakoff, B. (2015). Influence of sex on genetic regulation of “drinking in the dark” alcohol consumption. Mamm. Genome 26, 43–56. doi: 10.1007/s00335-014-9553-8
Vendruscolo, L. F., Terenina-Rigaldie, E., Raba, F., Ramos, A., Takahashi, R. N., and Mormede, P. (2006). Evidence for a female-specific effect of a chromosome 4 locus on anxiety-related behaviors and ethanol drinking in rats. Genes Brain Behav. 5, 441–450. doi: 10.1111/j.1601-183X.2005.00177.x
Keywords: alcohol use disorder, sex-difference, congenic rat model, nucleus accumbens, RNA-seq, Adcyap1r1
Citation: Spence JP, Reiter JL, Qiu B, Gu H, Garcia DK, Zhang L, Graves T, Williams KE, Bice PJ, Zou Y, Lai Z, Yong W and Liang T (2018) Estrogen-Dependent Upregulation of Adcyap1r1 Expression in Nucleus Accumbens Is Associated With Genetic Predisposition of Sex-Specific QTL for Alcohol Consumption on Rat Chromosome 4. Front. Genet. 9:513. doi: 10.3389/fgene.2018.00513
Received: 07 March 2018; Accepted: 12 October 2018;
Published: 04 December 2018.
Edited by:
Kristin Hamre, The University of Tennessee Health Science Center, United StatesReviewed by:
Camron D. Bryant, Boston University, United StatesCopyright © 2018 Spence, Reiter, Qiu, Gu, Garcia, Zhang, Graves, Williams, Bice, Zou, Lai, Yong and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tiebing Liang, dGxpYW5nQGl1LmVkdQ== Weidong Yong, d3lvbmdAY25pbGFzLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.