
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 16 October 2018
Sec. Applied Genetic Epidemiology
Volume 9 - 2018 | https://doi.org/10.3389/fgene.2018.00462
This article is part of the Research TopicThe Genes and Mechanisms Underlying Normal-Range Craniofacial VariationView all 7 articles
Historically, craniofacial genetic research has understandably focused on identifying the causes of craniofacial anomalies and it has only been within the last 10 years, that there has been a drive to detail the biological basis of normal-range facial variation. This initiative has been facilitated by the availability of low-cost hi-resolution three-dimensional systems which have the ability to capture the facial details of thousands of individuals quickly and accurately. Simultaneous advances in genotyping technology have enabled the exploration of genetic influences on facial phenotypes, both in the present day and across human history.
There are several important reasons for exploring the genetics of normal-range variation in facial morphology.
- Disentangling the environmental factors and relative parental biological contributions to heritable traits can help to answer the age-old question “why we look the way that we do?”
- Understanding the etiology of craniofacial anomalies; e.g., unaffected family members of individuals with non-syndromic cleft lip/palate (nsCL/P) have been shown to differ in terms of normal-range facial variation to the general population suggesting an etiological link between facial morphology and nsCL/P.
- Many factors such as ancestry, sex, eye/hair color as well as distinctive facial features (such as, shape of the chin, cheeks, eyes, forehead, lips, and nose) can be identified or estimated using an individual’s genetic data, with potential applications in healthcare and forensics.
- Improved understanding of historical selection and adaptation relating to facial phenotypes, for example, skin pigmentation and geographical latitude.
- Highlighting what is known about shared facial traits, medical conditions and genes.
The facial surface is readily visible and identifiable with a close relationship to the underlying cartilaginous and skeletal structures (Stephan et al., 2005; Wilkinson et al., 2006; De Greef et al., 2006; Al Ali et al., 2014b; Shrimpton et al., 2014). Differences in relative size, shape and spatial arrangement (vertical, horizontal and depth) between the various facial features (e.g., eyes, nose, lips etc.) make each individual human face unique, although closely related individuals such as monozygotic twins have very similar facial structures. Information on an individual’s facial morphology can have several important clinical and forensic applications; informing patient specific models, improving and reducing the need for extensive surgical interventions for craniofacial anomalies/trauma, prediction/reconstruction of the facial form from skeletal remains, and identification of suspects from DNA (Stephan et al., 2005; De Greef et al., 2006; Wilkinson et al., 2006; Beldie et al., 2010; Popat et al., 2010, 2012; Richmond et al., 2012; Al Ali et al., 2014a; Shrimpton et al., 2014; Farnell et al., 2017; Richmond S. et al., 2018).
The development of the face involves a coordinated complex series of embryonic events. Recognizable features of the human face develop around the 4th week of gestation and are closely related to cranial neural crest cells (Marcucio et al., 2015). The facial developmental component processes are listed (Table 1) and the human embryonic sequence of events can be visualized which aids understanding of the movement of the facial processes followed by their fusion (Sharman, 2011).
The facial processes fuse at different times; maxillary – 6 weeks, upper lip – 8 weeks and palate – 12 weeks (O’Rahilly, 1972; Danescu et al., 2015). Molecular studies have shown that the growth, structure and patternation of the facial primordia is controlled by a series of complex interactions that involves many factors such as fibroblast growth factors, sonic hedgehog proteins, bone morphogenetic proteins, homeobox genes Barx1 and Msx1, the distal-less homeobox (Dlx) genes, and local retinoic acid gradients (Barlow et al., 1999; Hu and Helms, 1999; Lee et al., 2001; Ashique et al., 2002; Mossey et al., 2009; Marcucio et al., 2015; Parsons et al., 2015). The fusion between the facial processes depends on a series of events involving cell migration, growth, adhesion, differentiation and apoptosis. Disruptions in the fusion of the facial processes may result in complete or partial clefts of the face, lip and/or palate. The epithelial precursor periderm is involved in cellular adhesions with associated genes IRF6, IKKA, SFN, RIPK4, CRHL3 all of which are under the transcription control of the transcription factor p63 that influences the fusion process and differentiation of the epidermis (Hammond et al., 2017). For detailed embryological development the reader should read the original articles or illustrated reviews (Som and Naidich, 2013, 2014). Post-natally, facial growth tends to follow general somatic growth with periods of steady increments in size interspersed with periods of rapid growth with the peak growth occurring at puberty (Tanner et al., 1966a,b; Bhatia et al., 1979; Kau and Richmond, 2008; Richmond et al., 2009; Richmond S. et al., 2018). The timing, vectors and duration of surges in facial growth tend to be different for males and females and between populations contributing to overall facial variation (Kau et al., 2010; Hopman et al., 2014; Richmond R.C. et al., 2018).
There are many imaging systems available to capture the external facial surface topography such as photography, lasers, photogrammetry, magnetic resonance Imaging (MRI), computerized tomography (CT), and cone beam computerized tomography (CBCT). Many of these techniques have been evaluated in terms of facial coverage, speed of capture, processing time, accuracy, validity and cost (Kovacs et al., 2007; Heike et al., 2010; Kuijpers et al., 2014; Tzou et al., 2014). For an individual who can sit still with a neutral facial posture in natural head position, the speed of capture is not critical. Even with relatively long acquisition times for some photogrammetric, MRI, CT, and CBCT systems, facial landmark reliability of less than 0.5 mm can be achieved (Kau et al., 2005, 2007; Liu et al., 2012). For infants and individuals with unpredictable facial or bodily movements a faster acquisition time will be required although reliability of achieving the same facial posture will be significantly reduced. Facial surfaces that are captured in supine and prone position often show slight differences (Munn and Stephan, 2018).
Assessment and judgment of the face and body can be traced back to the ancient Greeks and Egyptians when mathematical methods such as Fibonacci series and the golden proportion (1:1.618) were applied to art and architecture as a method of defining attractiveness and beauty (Ricketts, 1982). Facial features can be broadly characterized in terms of the size and shape of the whole face and/or its component parts (e.g., big/small head; short/long and wide/thin face, prominent or retrusive chin). Recognition of distinctive facial and lip features such as grooves, nodules, lip demarcation lines has also been reported (Merks et al., 2003; Wilson et al., 2013). To quantify facial features, landmarks have been traditionally used, taken either directly from the face or derived from photographs or radiographs. These landmarks are defined by identifiable/describable facial features, e.g., nasion, inner/outer canthi, commissures that can generate Euclidean distances, angles, and ratios (Farkas et al., 2002, 2004, 2005). One or more facial landmarks can be used to generate principal components, geodesic distances, geodesic arrays, facial shells and signatures which can categorize patterns in facial features (Hammond and Suttie, 2012; Hallgrimsson et al., 2015; Tsagkrasoulis et al., 2017; Abbas et al., 2018). In addition, anthropometric masks have been proposed whereby five landmarks are used to crudely orientate the 3D facial shells which are then non-rigidly mapped on to a template which generates about 10,000 quasi landmarks (Claes et al., 2012). Asymmetry is preserved in some of these techniques. However, if the facial shell is reflected on to the opposite side any facial asymmetry will be lost.
The various acquisition techniques (photographs, MRI, laser and photogrammetry) have been used in different studies and all have identified the PAX3 gene associated with the shape of the nasal root area (Liu et al., 2012; Paternoster et al., 2012; Adhikari et al., 2016; Shaffer et al., 2016; Claes et al., 2018). The use of ordinal and quantitative measures has been explored reporting good correlation with inter-alae and lower lip distances (r = 0.7) and poor association for naso-labial angle (r = 0.16) (Adhikari et al., 2016).
Standardized clinical facial charts/tables/measures are routinely used for newborns (e.g., head circumference, body length) and other specialties such as, ophthalmology and orthodontics. There are many published norms for different racial/population groups used to identify individuals who fall within the normal range and identify any facial dysmorphologies.
The soft tissue facial variation has been explored in a large Caucasian population of 15-year-old children (2514 females and 2233 males) recruited from the Avon Longitudinal Study of Parents and Children (ALSPAC). Face height (28.8%), width of the eyes (10.4%) and prominence of the nose (6.7%) explained 46% of total facial variance (Toma et al., 2012). There were subtle differences between males and females in relation to the relative prominence of the lips, eyes, and nasal bridges including minor facial asymmetries (Toma et al., 2008, 2012; Wilson et al., 2013; Abbas et al., 2018). The dimorphic differences appear to follow similar patterns in different ethnic groups (Farnell et al., 2017).
Facial morphology refers to a series of many different complex traits, each influenced by genetic and environmental factors. In particular, the strong effects that genetic variation can have on facial appearance are highlighted by historical portraits of the European royal family, the Habsburgs (1438–1740). Presumably because of frequent consanguineous marriages, later Habsburg rulers often had extreme facial phenotypes such as the characteristic “Habsburg” jaw (mandibular prognathism). Indeed, the last Habsburg King of Spain, Charles II, was reported to have had difficulties eating and speaking because of facial deformities. The influence of genetic variation is also evident in non-consanguineous families, where dental and facial characteristics are common among siblings and passed on from parents to their offspring (Hughes et al., 2014). Twin studies have historically been employed to explore the relative genetic and environment influence on facial shape exploiting the genetic differences between monozygotic and dizygotic twins (Visscher et al., 2008). Twin studies suggest that 72–81% of the variation of height in boys and 65–86% in girls is due to genetic differences with the environment explaining 5–23% of the variation (Jelenkovic et al., 2011). Similar levels of genetic-environmental contributions have been reported for some facial features. Predominantly genetic influences have been reported for anterior face height, relative prominence of the maxilla and mandible, width of the face/nose, nasal root shape, naso-labial angle, allometry and centroid size (Carels et al., 2001; Carson, 2006; Jelenkovic et al., 2010; Djordjevic et al., 2013a,b, 2016; Cole et al., 2017; Tsagkrasoulis et al., 2017). Substantial heritability estimates for facial attractiveness and sexual dimorphism (0.50–0.70 and 0.40–0.50), respectively (Mitchem et al., 2014), further demonstrate the strong genetic influences on facial phenotypes.
Contrastingly, previous estimates suggest that antero-posterior face height, mandibular body length, ramus height, upper vermillion height, nasal width and maxillary protrusion are more strongly influenced by environmental factors (Jelenkovic et al., 2010; Djordjevic et al., 2016; Sidlauskas et al., 2016; Cole et al., 2017; Tsagkrasoulis et al., 2017). However, it is important to note that heritability estimates for specific traits can be inconsistent for a number of reasons including heterogeneity across study populations, small sample sizes, research designs, acquisition methods and the differing types of analyses employed.
From the moment of conception, the parental environment can influence the development of the fetus. Facial development occurs very early at a time when the mother is not always aware that she is pregnant. The developing fetus may be subject to adverse environments at home, in the workplace or through lifestyle activities (smoking, alcohol and drug intake, allergens, paint, pest/weed control, heavy metals, cleaning, body products such as perfumes and creams). Many of these substances can cross the placenta (Naphthalene a volatile polycyclic aromatic hydrocarbon related to solvent emissions is present in household products and pesticides – Mirghani et al., 2015; Nicotine – Wickström, 2007; Drugs and alcohol – Lange et al., 2014). There is evidence to suggest that the effects of some of these substances can also continue post-natally through breast milk fed to the new-born (heavy metals – Caserta et al., 2013; Dioxin – Rivezzi et al., 2013). Some of these early factors such as nictotine and alcohol may potentially influence on early neurological development (Wickström, 2007). Indeed, there is evidence to suggest that high levels of prenatal alcohol exposure can influence facial morphology; individuals with fetal alcohol syndrome disorders can present with facial abnormalities (Hoyme et al., 2016) as well as other developmental anomalies such as caudate nucleus asymmetry and reduced mass of the brain (Suttie et al., 2018). However, the effects of lower levels of prenatal alcohol exposure on facial morphology are less clear (Mamluk et al., 2017; Muggli et al., 2017; Howe et al., 2018c). Similarly, it has been hypothesized that maternal smoking may influence facial morphology and be a risk factor for cleft lip and palate (Xuan et al., 2016) with DNA methylation a possible mediator (Armstrong et al., 2016). However, to date one study has indicated that maternal smoking may interact with the GRID2 and ELAVL2 genes resulting in cleft lip and palate (Beaty et al., 2013). However, previous studies investigating gene-smoking interactions in the etiology of birth defects have produced mixed results (Shi et al., 2008). Another mechanism via which environmental influences can affect facial traits is natural selection, where certain facial traits may have beneficial effects on reproductive fitness. For example, there is evidence that nose shape has been under historical selection in certain climates (Weiner, 1954; Zaidi et al., 2017).
Generally, most modifiable environmental factors have only subtle effects on the face. However, it is important to note that stochastic chance events such as facial trauma, infections, burns, tumors, irradiation and surgical procedures can all have a significant impact on facial development and consequently facial morphology.
The first wave of genetic studies of craniofacial Mendelian traits were based on linkage or candidate gene studies of genetic loci known to be involved in craniofacial development or genetic syndromes affecting the face. Down syndrome, cleft lip and/or palate, Prader-Willi syndrome, and Treacher Collins syndrome can all present with facial abnormalities and genetic loci associated with them have been studied in relation to normal facial development (Boehringer et al., 2011; Brinkley et al., 2016).
Genome-wide association studies (GWAS) have investigated the association between normal facial variation and millions of single nucleotide polymorphisms (SNPs). GWAS studies coupled with high-resolution three-dimensional imaging of the face have enabled the study of the spatial relationship of facial landmarks in great detail. Over the last 6 years there has been significant progress with 9 published GWAS which have identified over 50 loci associated with facial traits (Liu et al., 2012; Paternoster et al., 2012; Adhikari et al., 2016; Cole et al., 2016; Shaffer et al., 2016; Lee et al., 2017; Cha et al., 2018; Claes et al., 2018; Crouch et al., 2018). The genes and broad regional associations are shown in Table 2 (ordered by facial feature and chromosome) and Figure 1 (showing facial region). For detailed information on the biological basis of individual genes, the reader should refer to the original articles. Different facial measures have been applied to facial images obtained from a variety of acquisition systems (2D and 3D). Genes are likely to influence more than one facial trait. For instance, the PAX3 gene is associated with eye to nasion distance, prominence of the nasion and eye width, side walls of the nose, and prominence of nose tip. Similarly, the naso-labial angle will be associated with nose prominence and DCHS2 is linked to both traits.
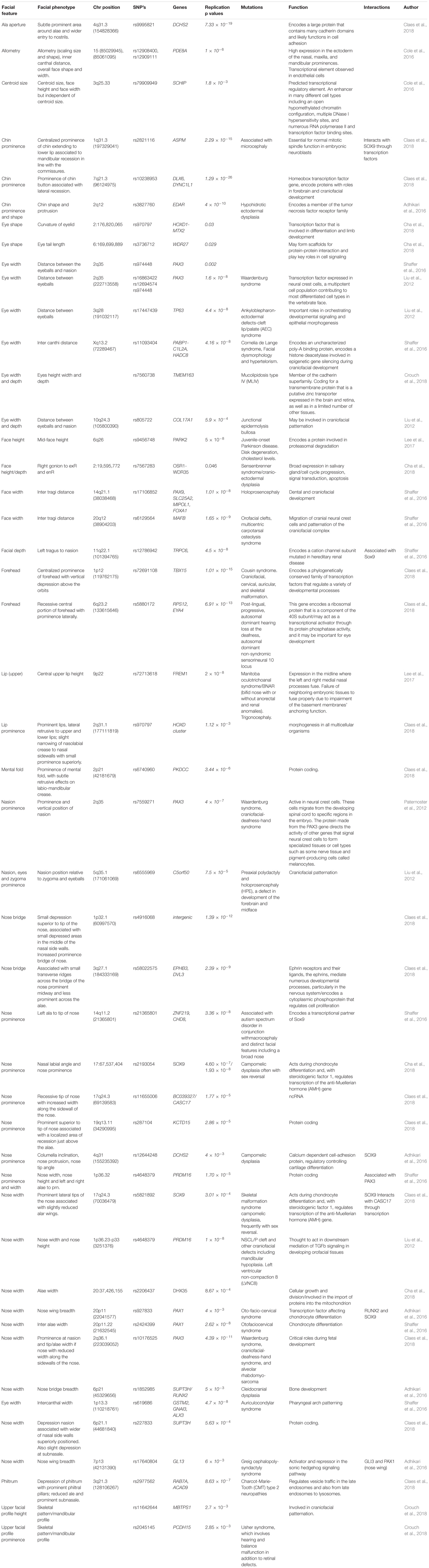
TABLE 2. List of genes and SNP’s associated with normal variation ranked by chromosome position (GWAS).
Some reported genes appear to influence different parts of the face. PRDM16 is linked to the length and the prominence of the nose as well as the width of the alae, SOX9 is thought to be related to the shape of the ala and nose tip, variation in SUPT3H is thought to influence naso-labial angle and shape of the bridge of the nose, while centroid size (squared root of the squared distances of all landmarks of the face from the centroid) and allometry (relationship of size to shape) have been linked to PDE8A and SCHIP17 genes, respectively, (Cole et al., 2016). Eye width and ear – nasion distance and nasion -zygoma – eyes distances are linked to C5orf50. There is some evidence to suggest that there are additive genetic effects on nose shape involving SOX9, DCHS2, CASC17, PAX1, RUNX2, and GL13 and chin shape, SOX9 and ASPM. In addition, it is likely that one or more genes influence the whole shape of the face as well as more localized facial regions (Claes et al., 2018). A significant number of genes are integrally involved in cranial neural crest cells and patternation of the craniofacial complex (e.g., C5orf50, MAFB, and PAX3).
Identifying genetic variants influencing facial phenotypes can lead to improved etiological understanding of craniofacial anomalies, advances in forensic prediction using DNA and testing of evolutionary hypotheses.
Non-syndromic cleft lip/palate (nsCL/P) is a birth defect with a complex etiology, primarily affecting the upper lip and palate (Mossey et al., 2009; Dixon et al., 2011). Previous studies have identified genes associated with both nsCL/P and facial phenotypes; such as variation in MAFB which is associated with face width in normal variation (Beaty et al., 2010, 2013; Boehringer et al., 2011; Liu et al., 2012; Peng et al., 2013; Shaffer et al., 2016). Furthermore, craniofacial sub-phenotypes have been observed in nsCL/P cases and their unaffected family members such as orbicularis oris muscle defects and facial shape differences supporting the existence of nsCL/P related sub-phenotypes (Stanier and Moore, 2004; Marazita, 2007; Neiswanger et al., 2007; Menezes and Vieira, 2008; Weinberg et al., 2009; Aspinall et al., 2014).
The important link between facial variation and nsCL/P is highlighted by a study comparing facial morphologies (linked to genes) of children with nsCL/P and unaffected relatives. There was reduced facial convexity (SNAI1), obtuse nasolabial angles, more protrusive chins (SNAI1, IRF6, MSX1, MAFB), increased lower facial heights (SNAI1), thinner and more retrusive lips and more protrusive foreheads (ABCA4-ARHGAP29, MAFB) in the nsCL/P relatives compared to controls. There was also greater asymmetry in the nsCL/P group (LEFTY1, LEFTY2, and SNAI1) (Miller et al., 2014).
There is evidence that nsCL/P genetic risk variants have an additive effect on philtrum width across the general population. This association suggests that developmental processes relating to normal-variation in philtrum development are also etiologically relevant to nsCL/P, highlighting the shared genetic influences on normal-range facial variation and a cranio-facial anomaly (Howe et al., 2018a).
Similarly, genetic variations associated with normal-range facial differences have been linked to genes involved in Mendelian syndromes such as TBX15 (Cousin syndrome) (Shaffer et al., 2017; Claes et al., 2018), PAX1 (Otofaciocervical syndrome) (Shaffer et al., 2016) and PAX3 (Waardenburg syndrome) (Paternoster et al., 2012). It has been hypothesized that deleterious coding variants may directly cause congenital anomalies while non-coding variants in the same genes influence normal-range facial variation via gene expression pathways (Shaffer et al., 2017; Freund et al., 2018).
Shared genetic pathways may influence both normal-range variation in facial morphology and craniofacial anomalies. Disentangling these shared pathways can improve understanding of the biological processes that are important during embryonic development.
Over time, facial morphology across populations has been influenced by various factors, such as migration, mate-choice, survival and climate, which have contributed to variation in facial phenotypes. Genetic and facial phenotype data can be used to improve understanding of human history.
Ancestry and physical appearance are highly related; it is often possible to infer an individual’s recent ancestry based on physically observable features such as facial structure and skin color. Indeed, previous studies have demonstrated that self-perceived and genetically inferred ancestry are associated with facial morphology, particularly with regards to the shape of the nose (Dawei et al., 1997; Le et al., 2002; Farkas et al., 2005; Claes et al., 2014). Facial morphological differences relating to ancestry are well-characterized when comparing individuals from distinct populations, but distinct differences remain even within more ancestrally homogeneous populations.
Historical migrations, such as the European colonization of Latin America, led to genetic admixture (breeding between individuals from previously isolated populations) (Hellenthal et al., 2014), which greatly influenced the facial morphology of the Latin American population. Indeed, modern day Latin Americans have mixed African, European and Native American ancestry, with genetic admixture highly predictive of physical appearance. For this reason, ancestral markers are often included in facial prediction models (Claes et al., 2014; Ruiz-Linares et al., 2014; Lippert et al., 2017).
Facial phenotypes can influence mate choice and be under selection pressures. These factors can then affect reproductive behavior and lead to population-level changes in facial variation as certain facial phenotypes are favored. Previous studies have suggested that facial features such as attractiveness (Little et al., 2001; Fink and Penton-Voak, 2002), hair color (Wilde et al., 2014; Adhikari et al., 2016; Field et al., 2016; Hysi et al., 2018), eye color (Little et al., 2003; Wilde et al., 2014; Field et al., 2016) and skin pigmentation (Jablonski and Chaplin, 2000, 2010; Wilde et al., 2014; Field et al., 2016) may influence mate choice and/or have been under historical selection. Features related to appearance are also often sexually dimorphic, possibly as a result of sexual and natural selection. For example, significantly more women self-report having blonde and red hair while more men as self-report as having black hair (Hysi et al., 2018).
The possible evolutionary advantages of facial phenotypes have been discussed extensively but anthropological hypotheses can be tested using genetic and facial phenotype data. For example, a masculine face has been hypothesized to be a predictor of immunocompetence (Scott et al., 2013). A previous study tested this hypothesis using 3D facial images and genetic variation in the major histocompatibility complex (MHC) region and found weak evidence to support this (Zaidi et al., 2018). Other possible benefits that have been explored include: the fitness advantages of hair color (Adhikari et al., 2016; Hysi et al., 2018), nasal shape and climate adaptation (Zaidi et al., 2017) and the benefits of darker skin pigmentation (Wilde et al., 2014; Aelion et al., 2016). Strong levels of phenotypic and genotypic spousal assortment have been previously demonstrated for height (Robinson et al., 2017) and similar methods could be applied using facial phenotypes to explore the influences of facial morphology on mate choice.
The premise of reverse genetics is that there is known function of a gene or a group of genes which will create a particular phenotype with a degree of certainty. This has been proposed as a method to build a profile of facial features from a sample of DNA (Claes et al., 2014) but could also be used to determine previous health history or future health risks (Idemyor, 2014). This approach may be appropriate for unique facial characteristics but is more challenging when one or more genes are associated with the variation of facial phenotype (e.g., prominence of the nasal bridge or length of the nose, hair and eye color/tones). The PAX3 gene is associated with the distance between the mid-endocanthion point and surface nasion with a mean distance of 17.5 mm with differing axis values up to 6.7 mm (x), 17.7 mm (y), and 18.9 mm (z). Although, it is known that the PAX3 influences the prominence of the bridge of the nose it is more challenging to know to what extent this influences adjacent facial regions in each individual. In addition, genetic and environmental factors will have subtle influences on the face. Although the molecular understanding of genetic variation influencing facial morphology is improving, the use of DNA as a prediction tool is still a long way off. However, recent studies suggest that DNA has the potential to identify an individual from a small group of possible candidates (Claes et al., 2014; Biedermann et al., 2015; Kayser, 2015). There is clearly a place in forensic science to develop a robust diagnostic tool to determine age, ancestry, appearance, relatedness and sex from DNA samples. One study effectively predicted eye color (85% for brown and 70% for blue), hair color (72% for brown, 63% for blonde, 58% for black, and 48% for red) and ancestry (56%); which are relatively low levels and individually could not be relied on for certain identifications but has greater potential when used collectively (Keating et al., 2013). The prediction of skin color from DNA has also been reported (Chaitanya et al., 2018) and DNA methylation has been demonstrated as a useful predictor of age. Age prediction using methylation techniques have indicated a mean absolute deviation of 5–8 years (Xu et al., 2015; Bocklandt et al., 2011; Hamano et al., 2017)
The determination of facial appearance, health history and future health risk from DNA is has great potential (Claes et al., 2014; Kayser, 2015; Toom et al., 2016) but caution should be expressed with respect to assumptions, interpretation and individual confidentiality as there is a significant threat to an individual in obtaining healthcare insurance (Hallgrimsson et al., 2014; Idemyor, 2014; Toom et al., 2016).
Previously published studies that have identified gene-phenotype associations which provides evidence of associations for complex facial traits which can be integrated into prediction models. The collective use of these techniques to identify the various facial features will increase the robustness of linking the DNA to a likely suspect/candidate.
Table 2 highlights that genetic variants influencing facial morphology can have pleiotropic effects on parts of the body independent to the brain and surrounding craniofacial structures (e.g., cardiovascular, endocrine, gastro-intestinal, central nervous, musculo-skeletal and uro-genital systems). The growing number of GWAS datasets has allowed exploration of the shared genetic influences on different phenotypes (Bulik-Sullivan B. et al., 2015; Pickrell et al., 2016). There is the potential for relationships between medical and facial conditions to be explored using genetic summary data. The limited evidence for genetic correlation between facial and other traits has been reported in Table 3. It is important to note that the strong association between facial morphology and ancestry means that any correlations may be attributable to fine-scale population substructure.
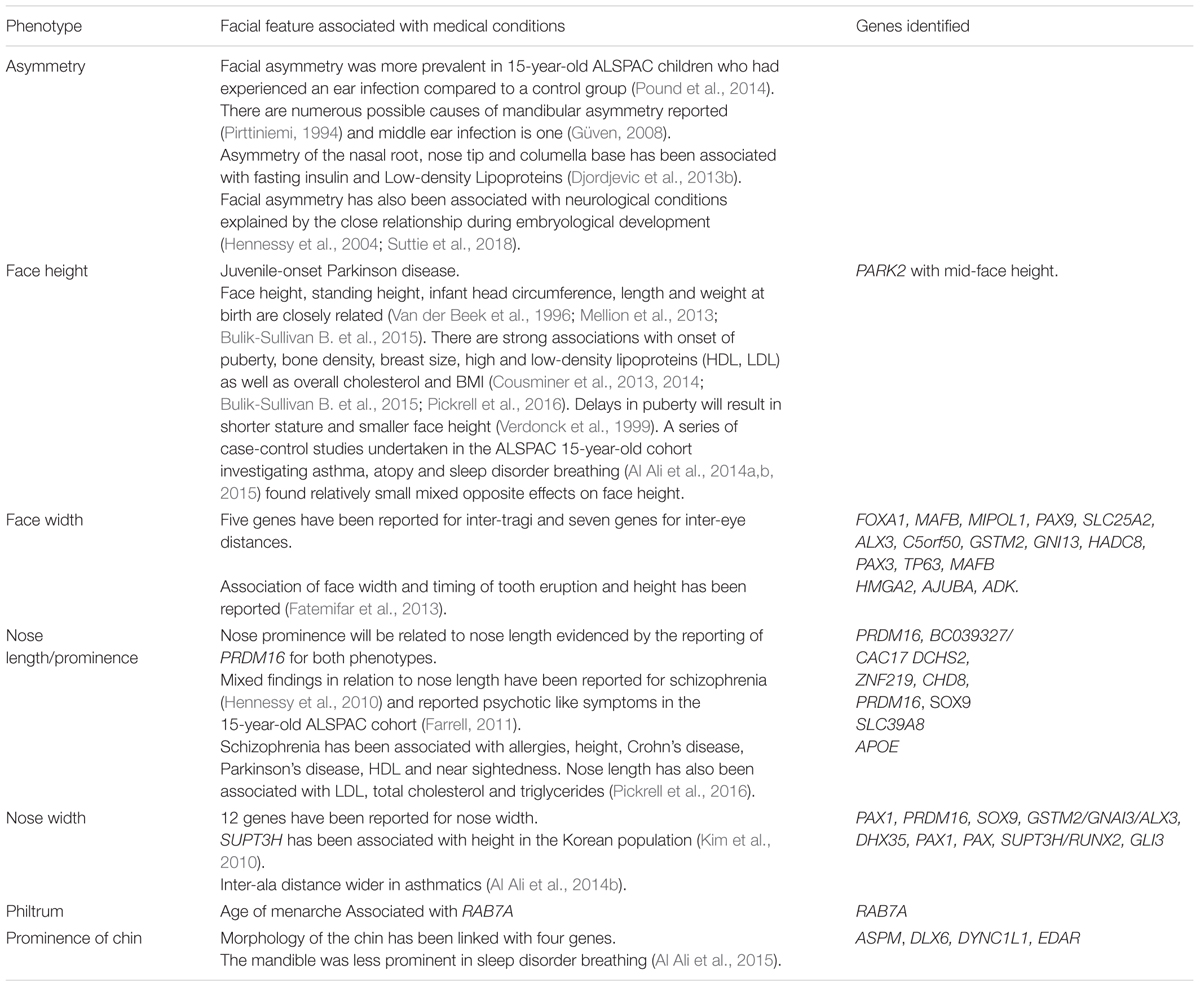
TABLE 3. Reported shared influences of medical conditions, normal facial variation with associated genes.
Normal facial development is dependent on Cranial Neural Crest Cells and correctly spatially positioned and differentiated tissues and structures that influence the shape and morphological features of the face. The disruption of neuro-facial developmental and maturational processes can lead to widespread and long-lasting abnormalities in central nervous system structure and functions and some of these disturbances will also be accompanied with subtle differences in facial features (Hennessy et al., 2010).
Epigenetics refers to mitotically (and perhaps, controversially meiotically) heritable changes in gene expression which are not explained by changes to the DNA base-pair sequence. Epigenetic processes include DNA methylation, histone modification and chromatin remodeling, which can affect gene expression by regulating transcription (Jaenisch and Bird, 2003; Bird, 2007; Gibney and Nolan, 2010; Allis and Jenuwein, 2016). Epigenetic processes are particularly relevant to craniofacial phenotypes because of the general importance of epigenetic gene regulation during embryonic development (Reik, 2007) and their specific role in neural crest development (Hu et al., 2014).
Craniofacial epigenetic studies to date have largely focused on orofacial clefts. Previous epigenome-wide association studies (EWAS) have found evidence of differential DNA methylation between cleft cases and controls (Alvizi et al., 2017), as well as between the different orofacial cleft subtypes (Sharp et al., 2017) implicating the relevance of DNA methylation in craniofacial development. The modifiable nature of epigenetic processes has led to much excitement that these processes may mediate the effect of environmental exposures. The maternal environment is thought to play an important role with regards to orofacial clefts. Previous studies have found strong evidence supporting associations between prenatal smoke exposure (Joubert et al., 2016) and folate supplementation (Richmond R.C. et al., 2018) with differential DNA methylation, but contrastingly there is no clear evidence for an association between prenatal alcohol exposure and DNA methylation (Sharp et al., 2018). In cleft lip tissue, limited evidence was found for an association between LINE-1 methylation and maternal exposures but conclusions were limited by modest sample sizes (Khan et al., 2018). Future work could utilize meditation techniques (Tobi et al., 2018) or Mendelian randomization (Relton and Davey Smith, 2012) to formally investigate the possibility that prenatal exposures influence orofacial cleft risk via epigenetic processes.
Similarly, epigenetic processes may mediate the effects of germline genetic variation. Many of the previously discussed genetic variants associated with facial traits in GWAS reside in non-protein coding regions of the genome with unclear functional relevance. One possibility is that these variants may influence facial phenotypes through gene regulation pathways involving epigenetic processes. Indeed, a previous study demonstrated that a major risk locus for non-syndromic cleft lip/palate (nsCL/P), in a non-coding interval, is involved in the regulation of gene expression in the developing murine face (Uslu et al., 2014) while another study found some evidence that nsCL/P genetic variants may influence nsCL/P risk via changes in DNA methylation and gene expression (Howe et al., 2018b).
Despite the promise of early craniofacial epigenetic studies, there are important caveats worth noting. First, a major issue is that epigenetic modifications can vary across different tissues. Previous studies have used DNA methylation in blood as a proxy for methylation in lip and palate tissues. Despite some evidence for positive correlation between blood and lip tissue DNA methylation (Alvizi et al., 2017; Howe et al., 2018b), the extent to which blood is a suitable proxy is unknown. Furthermore, it is unclear whether the epigenetic profile of lip and palate tissues postnatally are comparable to the same tissues during embryonic development. A previous orofacial cleft GWAS found no clear evidence for enrichment of tissue-specific signals, concluding that this may be attributable to a lack of suitable tissue types (Leslie et al., 2017). Second, when testing causality, epigenetic modifications can vary across the life-course, so it can be difficult to discern the direction of effect between an epigenetic modification and the phenotype. It is therefore important to use causal inference techniques such as epigenetic Mendelian randomization Relton and Davey Smith, 2012) or the Steiger test (Hemani et al., 2017) to orientate the likely directions of effect between phenotypes, epigenetic modifications and gene expression.
The gene regulatory systems are complex and numerous and detailing these regulatory mechanisms has been the goal of the NIH Roadmap Epigenomics Project whereby next generation sequencing technologies (e.g., ChiP seq) are employed to map DNA methylation, histone modifications, chromatin accessibility in a variety of research media such as, animal models (mouse, chicken, zebrafish, frog, and primates) and stem cells and regulated human fetal tissues (Hochheiser et al., 2011; Roosenboom et al., 2016; Van Otterloo et al., 2016). Enhancers have a specific role in the expression of a target gene in different cells, anatomical regions and during different developmental time-points (Visel et al., 2009; Attanasio et al., 2013; Wilderman et al., 2018). The role of enhancers modifying histones, chromatin states are key for normal range craniofacial development and relative position of the various craniofacial tissues. Key transcriptional factors (activators or repressors) have been identified indicating extensive activation during early craniofacial development. These transcriptional factors may be limited to detail the precise facial shape or can be quickly activated in rapid periods of growth and development. Craniofacial enhancers have also been identified acting between the non-coding regions and proposed as a possible instrumental factor in some cleft cases (Wilderman et al., 2018).
The craniofacial region is made up of a series of complex structures which contribute to overall facial shape. Twin studies have indicated that facial shape is mainly due to genetic influences (≈75%) although the percentage variance explained in GWAS studies is extremely low generally explaining less than 2% of the total variance. GWAS may be underestimating and twin and family studies overestimating the levels of heritability. Facial shape and features are the result of mutations, genetic drift, recombination and natural selection. Rare Mendelian mutations, low frequency segregating variants, copy number variants and common variants contribute to complex phenotypes. Genetic interactions or epistasis may also explain the low levels of variance recorded. In addition, there is evidence of pleiotropy, quantitative phenotypes and Mendelian traits all influencing multiple phenotypes suggesting a large number of loci contribute additively to facial variation. Epigenetics focuses on the functional components of the genes and gene activities. The genome is comprised of 3.2 billion nucleotides wrapped in octomeric units of histones (chromatin). Modifications to chromatin through methylation, acetylation, phosphorylation or other processes are known to influence gene expression. Most epigenetic changes are transient and not generally heritable. However, a small proportion of epigenetic changes are transgenerational (Rachdaoui and Sarkar, 2014). However, there is a limited amount of evidence that epigenetic inheritance may carry over multiple generations (Schmidt and Kornfeld, 2016; Gluckman et al., 2007).
Increasing the sample sizes of genetic studies of facial morphology through international collaborations, such as the type II diabetes consortia DIAGRAM (Morris et al., 2012), will help to improve the understanding of genetic associations and shared influences on facial traits (Evans, 2018). The availability of summary statistics on large GWAS studies will also enable the application of quantitative genetics methods to further investigate the genetic architecture of facial morphology. Polygenic risk scores, LD score regression (to reduce confounding biases Bulik-Sullivan B.K. et al., 2015) and Mendelian randomization can provide information on the genetic overlap of facial phenotypes with other genetic traits and the possibility to causally assess the association of risk factors with face development (Smith and Ebrahim, 2003).
Focusing on specific phenotypes and genetic variants in families will identify additional rare variants should be followed-up with a combination of genotyping and deep re-sequencing of the variants or genes of interest in large numbers of cases and controls. The analytical techniques (particularly the bootstrapped response-based imputation modeling (BRIM) and hierarchical spectral clustering analysis) employed by Claes et al. (2014, 2018) provide efficient and valid analyses and arguably more importantly, visual linkages between genetic variants and global shape. In addition, the individual facial traits have yielded impressive levels of significance using a relatively small number of subjects (Evans, 2018). Permutation testing is a valid alternative for more conservative tests such as Bonferroni (Sham and Purcell, 2014). The use of machine-learning and artificial intelligence approaches will be crucial in future GWAS studies to determine patterns and linkages in the numerous large data sets generated and archived related to craniofacial development functional genomics.
There have been nine GWAS studies and it is appropriate to try and integrate their findings through a meta-analysis. Different genetic models, genotyping and imputation techniques have been employed and the between-study heterogeneity should be considered.
Defining facial shape can be undertaken in different ways but it is important to appreciate that there will be associations with not only with other facial features but also body phenotypes and medical conditions. Visualizing and automatic detection of facial phenotypes and determining their prevalence in population groups will facilitate case-control evaluations to determine genetic variants. So far, all GWAS studies have studied the static face but capturing the face during simple facial actions in a population (dynamic movement with or without speech) will enable the exploration of combined neurological and morphological features by assessing both speed and range of movement.
Population cohort studies enables researchers to study the environmental, disease and metabolic risk factors and genetic interactions from pre-birth throughout the lifecourse. Ideally facial images should be captured at birth, 5, 9, 12, 15, and 18 years of age and repeated every 10 years of age to capture facial features. During the pubertal growth period (9–18 years) facial images should be captured more frequently and if studying pubertal influences facial images should be captured at least every 6 months. Acquiring as much information as possible in relation known genetic additive effects, environmental factors and previous medical histories of family members (Grandparents, parents and offspring) will provide further insights into facial relatedness.
With improving knowledge of the controlling mechanisms for normal and abnormal facial development, it is logical to pursue healthcare strategies in the first instance to prevent craniofacial anomalies arising, with discussion of risks with genetic counseling, possibly future gene therapies and the follow up with minimally invasive or non-surgical, scarless procedures to correct craniofacial anomalies such as cleft lip and palate and control vertical and horizontal growth particularly of the upper and lower jaws and nose. Surgical procedures are not always simple as often in CL/P patients there is often insufficient tissues available (epidermis/dermis, cartilage and bone with disrupted orientation of muscle fibers). Prevention may be challenging (other than continually improving environmental conditions and reducing exposure to potential epigenetic factors) as facial development occurs very early in gestation during a period whereby the mother is often unaware she is pregnant. Controlling the mechanisms of normal growth in vivo or alternatively in vitro creating similar morphological tissues with intact innervation, blood and lymphatic systems that could be transplanted later may become reality in the future. Future, environmental epigenetic studies will show whether particular chemicals map to corresponding sensitive genomic regions. It is important to identify early life exposures (particularly conception to birth) that may influence later life health outcomes.
The face develops very early in gestation and facial development is closely related to the cranial neural crest cells. Disruption in early embryological development can lead to wide-ranging effects from subtle neurologic and facial features, which includes asymmetry, to significant impact on facial shape as characterized by a CL/P or in anomalies observed in craniofacial syndromes.
Heritability studies have provided insight into the possible genetic and environmental contributions to face shape. However, the sample sizes and inconsistencies in research design and particularly statistical management have yielded mixed results. Further detail is required on the heritability of facial features with particular attention to inherited pathways of specific facial features in homogenous populations and populations with significant admixture.
From birth to adulthood there are significant body and facial changes. Further work is required to explore the importance of the various biomedical markers and medical conditions (e.g., fasting glucose, cholesterol, asthma, and neurological disorders etc.) on the growth of the face, for example, remodeling of the facial skeleton, spatial changes of the constituent parts of the facial skeleton through sutures, condylar and nasal cartilages as well as the soft tissues, neural and vascular networks. The GWAS studies have provided insights into the genetic influences on facial shape. However, large-scale population studies are needed to identify more genetic variants not only in the context of facial shape but general body development with particularly attention to puberty. With any change in face shape the complex processes and communications at the biological and genome levels need to be identified and explained. The sheer volume of data collected in imaging genetics from images (hundreds of thousands of points), omics datasets (genomics, transcriptomics and cell-specific expression signals etc. – hundreds of millions of sequences) as well as biomarkers for medical conditions generates massive and complex data sets.
The size and heterogeneity of these data sets pose new challenges to efficiently and effectively, store, simplify and analyze the relative interactions and influences for a large number of face shape variables. The aim will be to continually develop and advance existing computerized tools and algorithms to solve these complex problems and this will require a multidisciplinary and internationally based team.
Impressions of an individual’s health are integral to social interactions and judgments are made on the visual appearance of skin, degree of roundness of the face and facial expression (Henderson et al., 2016). There has been significant progress in the first 6 years of GWAS and facial genetics. With increased sample sizes, improved understanding of shared genetic influences on human traits and advancement in techniques there is likely to be significant further progress in the next 6 years. Understanding the face will explain “why we look the way we do” a range of normality and abnormality that will be useful in healthcare applications and forensic science.
SR and LH outlined the overall manuscript. SR and SL wrote the section “Heritability”. LH and AZ wrote the section “Environmental Influences”. SR, ES, SL, and LH wrote the section “Craniofacial Shape Gene Discovery”. LH and SR wrote the section “Estimating Identity”. SR, ES, LH, and SL highlighted the shared facial traits. All authors actively participated in editing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor is currently collaborating with author SR and confirms the absence of any other collaboration.
The authors would like to thank Dr. Ryan Arathimos (University of Bristol) for advice on a draft of the manuscript. The authors would also like to thank the participants of the various studies undertaken.
Abbas, H., Hicks, Y., Marshall, D., Zhurov, A. I., and Richmond, S. (2018). A 3D morphometric perspective for facial gender analysis and classification using geodesic path curvature features. Comput. Vis. Media 4, 17–32. doi: 10.1007/s41095-017-0097-1
Adhikari, K., Fontanil, T., Cal, S., Mendoza-Revilla, J., Fuentes-Guajardo, M., Chacón-Duque, J.-C., et al. (2016). A genome-wide association scan in admixed Latin Americans identifies loci influencing facial and scalp hair features. Nat. Commun. 7:10815. doi: 10.1038/ncomms10815
Aelion, C. M., Airhihenbuwa, C. O., Alemagno, S., Amler, R. W., Arnett, D. K., Balas, A., et al. (2016). The US cancer moonshot initiative. Lancet Oncol. 17, e178–e180. doi: 10.1016/S1470-2045(16)30054-7
Al Ali, A., Richmond, S., Popat, H., Playle, R., Pickles, T., Zhurov, A. I., et al. (2015). The influence of snoring, mouth breathing and apnoea on facial morphology in late childhood: a three-dimensional study. BMJ Open 5:e009027. doi: 10.1136/bmjopen-2015-009027
Al Ali, A., Richmond, S., Popat, H., Toma, A. M., Playle, R., Pickles, T., et al. (2014a). A three-dimensional analysis of the effect of atopy on face shape. Eur. J. Orthod. 36, 506–511. doi: 10.1093/ejo/cjs107
Al Ali, A., Richmond, S., Popat, H., Toma, A. M., Playle, R., Zhurov, A. I., et al. (2014b). The influence of asthma on face shape: a three-dimensional study. Eur. J. Orthod. 36, 373–380. doi: 10.1093/ejo/cjs067
Allis, C. D., and Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17:487. doi: 10.1038/nrg.2016.59
Alvizi, L., Ke, X., Brito, L. A., Seselgyte, R., Moore, G. E., Stanier, P., et al. (2017). Differential methylation is associated with non-syndromic cleft lip and palate and contributes to penetrance effects. Sci. Rep. 7:2441. doi: 10.1038/s41598-017-02721-0
Armstrong, D. A., Green, B. B., Blair, B. A., Guerin, D. J., Litzky, J. F., Chavan, N. R., et al. (2016). Maternal smoking during pregnancy is associated with mitochondrial DNA methylation. Environ. Epigenet 2:dvw020.
Ashique, A. M., Fu, K., and Richman, J. M. (2002). Endogenous bone morphogenetic proteins regulate outgrowth and epithelial survival during avian lip fusion. Development 129, 4647–4660.
Aspinall, A., Raj, S., Jugessur, A., Marazita, M., Savarirayan, R., and Kilpatrick, N. (2014). Expanding the cleft phenotype: the dental characteristics of unaffected parents of Australian children with non-syndromic cleft lip and palate. Int. J. Paediatr. Dent. 24, 286–292. doi: 10.1111/ipd.12072
Attanasio, C., Nord, A. S., Zhu, Y., Blow, M. J., Li, Z., Liberton, D. K., et al. (2013). Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342:1241006. doi: 10.1126/science.1241006
Barlow, A. J., Bogardi, J. P., Ladher, R., and Francis-West, P. H. (1999). Expression of chick Barx-1 and its differential regulation by FGF-8 and BMP signaling in the maxillary primordia. Dev. Dyn. 214, 291–302. doi: 10.1002/(SICI)1097-0177(199904)214:4<291::AID-AJA2>3.0.CO;2-E
Beaty, T. H., Murray, J. C., Marazita, M. L., Munger, R. G., Ruczinski, I., Hetmanski, J. B., et al. (2010). A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 42, 525–529. doi: 10.1038/ng.580
Beaty, T. H., Taub, M. A., Scott, A. F., Murray, J. C., Marazita, M. L., Schwender, H., et al. (2013). Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum. Genet. 132, 771–781. doi: 10.1007/s00439-013-1283-6
Beldie, L., Walker, B., Lu, Y., Richmond, S., and Middleton, J. (2010). Finite element modelling of maxillofacial surgery and facial expressions–a preliminary study. Int. J. Med. Robot 6, 422–430. doi: 10.1002/rcs.352
Bhatia, S. N., Wright, G. W., and Leighton, B. C. (1979). A proposed multivariate model for prediction of facial growth. Am. J. Orthod. 75, 264–281. doi: 10.1016/0002-9416(79)90274-4
Biedermann, A., Bozza, S., and Taroni, F. (2015). Prediction in forensic science: a critical examination of common understandings. Front. Psychol. 6:737. doi: 10.3389/fpsyg.2015.00737
Bocklandt, S., Lin, W., Sehl, M. E., Sanchez, F. J., Sinsheimer, J. S., Horvath, S., et al. (2011). Epigenetic predictor of age. PLoS One 6:e14821. doi: 10.1371/journal.pone.0014821
Boehringer, S., Van Der Lijn, F., Liu, F., Günther, M., Sinigerova, S., Nowak, S., et al. (2011). Genetic determination of human facial morphology: links between cleft-lips and normal variation. Eur. J. Hum. Genet. 19:1192. doi: 10.1038/ejhg.2011.110
Brinkley, J. F., Fisher, S., Harris, M. P., Holmes, G., Hooper, J. E., Jabs, E. W., et al. (2016). The FaceBase consortium: a comprehensive resource for craniofacial researchers. Development 143, 2677–2688. doi: 10.1242/dev.135434
Bulik-Sullivan, B., Finucane, H. K., Anttila, V., Gusev, A., Day, F. R., Loh, P. R., et al. (2015). An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241. doi: 10.1038/ng.3406
Bulik-Sullivan, B. K., Loh, P. R., Finucane, H. K., Ripke, S., and Yang, J. (2015). Schizophrenia working group of the psychiatric genomics consortium, Patterson N, Daly MJ, Price AL, Neale BMLD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295. doi: 10.1038/ng.3211
Carels, C., Van Cauwenberghe, N., Savoye, I., Willems, G., Loos, R., Derom, C., et al. (2001). A quantitative genetic study of cephalometric variables in twins. Clin. Orthod. Res. 4, 130–140. doi: 10.1034/j.1600-0544.2001.040303.x
Carson, E. A. (2006). Maximum likelihood estimation of human craniometric heritabilities. Am. J. Phys. Anthropol. 131, 169–180. doi: 10.1002/ajpa.20424
Caserta, D., Graziano, A., Lo Monte, G., Bordi, G., and Moscarini, M. (2013). Heavy metals and placental fetal-maternal barrier: a mini-review on the major concerns. Eur. Rev. Med. Pharmacol. Sci. 17, 2198–2206.
Cha, S., Lim, J. E., Park, A. Y., Do, J. H., Lee, S. W., Shin, C., et al. (2018). Identification of five novel genetic loci related to facial morphology by genome-wide association studies. BMC Genomics 19:481. doi: 10.1186/s12864-018-4865-9
Chaitanya, L., Breslin, K., Zuniga, S., Wirken, L., Pospiech, E., Kukla-Bartoszek, M., et al. (2018). The HIrisPlex-S system for eye, hair and skin colour prediction from DNA: introduction and forensic developmental validation. Forensic Sci. Int. Genet. 35, 123–135. doi: 10.1016/j.fsigen.2018.04.004
Claes, P., Liberton, D. K., Daniels, K., Rosana, K. M., Quillen, E. E., Pearson, L. N., et al. (2014). Modeling 3D facial shape from DNA. PLoS Genet. 10:e1004224. doi: 10.1371/journal.pgen.1004224
Claes, P., Roosenboom, J., White, J. D., Swigut, T., Sero, D., Li, J., et al. (2018). Genome-wide mapping of global-to-local genetic effects on human facial shape. Nat. Genet. 50:414. doi: 10.1038/s41588-018-0057-4
Claes, P., Walters, M., and Clement, J. (2012). Improved facial outcome assessment using a 3D anthropometric mask. Int. J. Oral Maxillofac. Surg. 41, 324–330. doi: 10.1016/j.ijom.2011.10.019
Cole, J. B., Manyama, M., Kimwaga, E., Mathayo, J., Larson, J. R., Liberton, D. K., et al. (2016). Genomewide association study of african children identifies association of SCHIP1 and PDE8A with facial size and shape. PLoS Genet. 12:e1006174. doi: 10.1371/journal.pgen.1006174
Cole, J. B., Manyama, M., Larson, J. R., Liberton, D. K., Ferrara, T. M., Riccardi, S. L., et al. (2017). Human facial shape and size heritability and genetic correlations. Genetics 205, 967–978. doi: 10.1534/genetics.116.193185
Cousminer, D. L., Berry, D. J, Timpson, N. J., Ang, W., Thiering, E., Byrne, E. M., et al. (2013). Early growth genetics (EGG) consortium. genome-wide association and longitudinal analyses reveal genetic loci linking pubertal height growth, pubertal timing and childhood adiposity. Hum. Mol. Genet. 22, 2735–2747. doi: 10.1093/hmg/ddt104
Cousminer, D. L., Stergiakouli, E., Berry, D. J., Ang, W., Groen-Blokhuis, M. M., Körner, A., et al. (2014). Early growth genetics consortium. Genome-wide association study of sexual maturation in males and females highlights a role for body mass and menarche loci in male puberty. Hum. Mol. Genet. 23, 4452–4464. doi: 10.1093/hmg/ddu150
Crouch, D. J. M., Winney, B., Koppen, W. P., Christmas, W. J., Hutnik, K., Day, T., et al. (2018). Genetics of the human face: Identification of large-effect single gene variants. Proc. Natl. Acad. Sci. U.S.A. 115, E676–E685. doi: 10.1073/pnas.1708207114
Danescu, A., Mattson, M., Dool, C., Diewert, V. M., and Richman, J. M. (2015). Analysis of human soft palate morphogenesis supports regional regulation of palatal fusion. J. Anat. 227, 474–486. doi: 10.1111/joa.12365
Dawei, W., Guozheng, Q., Mingli, Z., and Farkas, L. G. (1997). Differences in horizontal, neoclassical facial canons in Chinese (Han) and North American Caucasian populations. Aesthetic. Plast. Surg. 21, 265–269. doi: 10.1007/s002669900123
De Greef, S., Claes, P., Vandermeulen, D., Mollemans, W., Suetens, P., and Willems, G. (2006). Large-scale in-vivo Caucasian facial soft tissue thickness database for craniofacial reconstruction. Forensic Sci. Int. 159(Suppl. 1),S126–S146.
Dixon, M. J., Marazita, M. L., Beaty, T. H., and Murray, J. C. (2011). Cleft lip and palate: understanding genetic and environmental influences. Nat. Rev. Genet. 12:167. doi: 10.1038/nrg2933
Djordjevic, J., Jadallah, M., Zhurov, A. I., Toma, A. M., and Richmond, S. (2013a). Three-dimensional analysis of facial shape and symmetry in twins using laser surface scanning. Orthod. Craniofac. Res. 16, 146–160. doi: 10.1111/ocr.12012
Djordjevic, J., Lawlor, D. A., Zhurov, A. I., Toma, A. M., Playle, R., and Richmond, S. (2013b). A population-based cross-sectional study of the association between facial morphology and cardiometabolic risk factors in adolescence. BMJ Open. 3:e002910. doi: 10.1136/bmjopen-2013-002910
Djordjevic, J., Zhurov, A. I., and Richmond, S. (2016). Genetic and environmental contributions to facial morphological variation: a 3D population-based twin study. PLoS One 11:e0162250. doi: 10.1371/journal.pone.0162250
Evans, D. M. (2018). Elucidating the genetics of craniofacial shape. Nat. Genet. 50, 319–321. doi: 10.1038/s41588-018-0065-4
Farkas, L. G., Eiben, O. G., Sivkov, S., Tompson, B., Katic, M. J., and Forrest, C. R. (2004). Anthropometric measurements of the facial framework in adulthood: age-related changes in eight age categories in 600 healthy white North Americans of European ancestry from 16 to 90 years of age. J. Craniofac. Surg. 15, 288–298. doi: 10.1097/00001665-200403000-00027
Farkas, L. G., Katic, M. J., and Forrest, C. R. (2005). International anthropometric study of facial morphology in various ethnic groups/races. J. Craniofacial Surg. 16, 615–646. doi: 10.1097/01.scs.0000171847.58031.9e
Farkas, L. G., Tompson, B. D., Katic, M. J., and Forrest, C. R. (2002). Differences between direct (anthropometric) and indirect (cephalometric) measurements of the skull. J. Craniofac Surg. 13, 105–108; discussion 109–110. doi: 10.1097/00001665-200201000-00024
Farnell, D. J. J., Galloway, J., Zhurov, A., Richmond, S., Perttiniemi, P., and Katic, V. (2017). Initial Results of Multilevel Principal Components Analysis of Facial Shape. Medical Image Understanding and Analysis. Cham: Springer International Publishing, 674–685. doi: 10.1007/978-3-319-60964-5_59
Farrell, K. (2011). A Critical Evaluation of Facial Characteristics and Their Association with Antisocial Behaviour and Psychosis. MSc thesis, Cardiff University, Cardiff.
Fatemifar, G., Hoggart, C. J., Paternoster, L., Kemp, J. P., Prokopenko, I., Horikoshi, M., et al. (2013). Genome-wide association study of primary tooth eruption identifies pleiotropic loci associated with height and craniofacial distances. Hum. Mol. Genet. 22, 3807–3817. doi: 10.1093/hmg/ddt231
Field, Y., Boyle, E. A., Telis, N., Gao, Z., Gaulton, K. J., Golan, D., et al. (2016). Detection of human adaptation during the past 2000 years. Science 354, 760–764. doi: 10.1126/science.aag0776
Fink, B., and Penton-Voak, I. (2002). Evolutionary psychology of facial attractiveness. Curr. Direc. Psychol. Sci. 11, 154–158. doi: 10.1111/1467-8721.00190
Freund, M. K., Burch, K., Shi, H., Mancuso, N., Kichaev, G., Garske, K. M., et al. (2018). Phenotype-specific enrichment of Mendelian disorder genes near GWAS regions across 62 complex traits. bioRxiv
Gibney, E., and Nolan, C. (2010). Epigenetics and gene expression. Heredity 105:4. doi: 10.1038/hdy.2010.54
Gluckman, P. D., Hanson, M. A., and Beedle, A. S. (2007). Non-genomic transgenerational inheritanceof disease risk. Bioessays 29, 145–154. doi: 10.1002/bies.20522
Güven, O. A. (2008). clinical study on temporomandibular joint ankylosis in children. J. Craniofac. Surg. 19, 1263–1269. doi: 10.1097/SCS.0b013e3181577b1b
Hallgrimsson, B., Mio, W., Marcucio, R. S., and Spritz, R. (2014). Let’s face it–complex traits are just not that simple. PLoS Genet. 10:e1004724. doi: 10.1371/journal.pgen.1004724
Hallgrimsson, B., Percival, C. J., Green, R., Young, N. M., Mio, W., and Marcucio, R. (2015). Morphometrics, 3D imaging, and craniofacial development. Curr. Top. Dev. Biol. 115, 561–597. doi: 10.1016/bs.ctdb.2015.09.003
Hamano, Y., Manabe, S., Morimoto, C., Fujimoto, S., and Tamaki, K. (2017). Forensic age prediction for saliva samples using methylation-sensitive high resolution melting: exploratory application for cigarette butts. Sci. Rep. 7:10444. doi: 10.1038/s41598-017-10752-w
Hammond, N. L., Dixon, J., and Dixon, M. J. (2017). Periderm: Life-cycle and function during orofacial and epidermal development. Semin. Cell Dev. Biol. [Epub ahead of print].
Hammond, P., and Suttie, M. (2012). Large-scale objective phenotyping of 3D facial morphology. Hum. Mutat. 33, 817–825. doi: 10.1002/humu.22054
Heike, C. L., Upson, K., Stuhaug, E., and Weinberg, S. M. (2010). 3D digital stereophotogrammetry: a practical guide to facial image acquisition. Head Face Med. 6:18. doi: 10.1186/1746-160X-6-18
Hellenthal, G., Busby, G. B., Band, G., Wilson, J. F., Capelli, C., Falush, D., et al. (2014). A genetic atlas of human admixture history. Science 343, 747–751. doi: 10.1126/science.1243518
Hemani, G., Tilling, K., and Smith, G. D. (2017). Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 13:e1007081. doi: 10.1371/journal.pgen.1007081
Henderson, A. J., Holzleitner, I. J., Talamas, S. N., and Perrett, D. I. (2016). Perception of health from facial cues. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150380. doi: 10.1098/rstb.2015.0380
Hennessy, R. J., Baldwin, P. A., Browne, D. J., Kinsella, A., and Waddington, J. L. (2010). Frontonasal dysmorphology in bipolar disorder by 3D laser surface imaging and geometric morphometrics: comparisons with schizophrenia. Schizophr. Res. 122, 63–71. doi: 10.1016/j.schres.2010.05.001
Hennessy, R. J., Lane, A., Kinsella, A., Larkin, C., O’Callaghan, E., and Waddington, J. L. (2004). 3D morphometrics of craniofacial dysmorphology reveals sex-specific asymmetries in schizophrenia. Schizophr. Res. 67, 261–268. doi: 10.1016/j.schres.2003.08.003
Hochheiser, H., Aronow, B. J., Artinger, K., Beaty, T. H., Brinkley, J. F., Chai, Y., et al. (2011). The FaceBase consortium: a comprehensive program to facilitate craniofacial research. Dev. Biol. 355, 175–182. doi: 10.1016/j.ydbio.2011.02.033
Hopman, S. M., Merks, J. H., Suttie, M., Hennekam, R. C., and Hammond, P. (2014). Face shape differs in phylogenetically related populations. Eur. J. Hum. Genet. 22, 1268–1271. doi: 10.1038/ejhg.2013.289
Howe, L. J., Lee, M. K., Sharp, G. C., Smith, G. D., St Pourcain, B., Shaffer, J. R., et al. (2018a). Investigating the shared genetics of non-syndromic cleft lip/palate and facial morphology. PLoS Genet. 14:e1007501. doi: 10.1371/journal.pgen.1007501
Howe, L. J., Richardson, T. G., Arathimos, R., Alvizi, L., Passos-Bueno, M.-R., Stanier, P., et al. (2018b). DNA methylation mediates genetic liability to non-syndromic cleft lip/palate. bioRxiv
Howe, L. J., Sharp, G. C., Hemani, G., Zuccolo, L., Richmond, S., and Lewis, S. J. (2018c). Prenatal alcohol exposure and facial morphology in a UK cohort. bioRxiv.
Hoyme, H. E., Kalberg, W. O., Elliott, A. J., Blankenship, J., Buckley, D., Marais, A. S., et al. (2016). Updated Clinical Guidelines for Diagnosing Fetal Alcohol Spectrum Disorders. Pediatrics 138:e20154256. doi: 10.1542/peds.2015-4256
Hu, D., and Helms, J. A. (1999). The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development 126, 4873–4884.
Hu, N., Strobl-Mazzulla, P. H., and Bronner, M. E. (2014). Epigenetic regulation in neural crest development. Dev. Biol. 396, 159–168. doi: 10.1016/j.ydbio.2014.09.034
Hughes, T. E., Townsend, G. C., Pinkerton, S. K., Bockmann, M. R., Seow, W. K., Brook, A. H., et al. (2014). The teeth and faces of twins: providing insights into dentofacial development and oral health for practising oral health professionals. Aust. Dent. J. 59(Suppl. 1), 101–116. doi: 10.1111/adj.12101
Hysi, P. G., Valdes, A. M., Liu, F., Furlotte, N. A., Evans, D. M., Bataille, V., et al. (2018). Genome-wide association meta-analysis of individuals of European ancestry identifies new loci explaining a substantial fraction of hair color variation and heritability. Nat. Genet. 50, 652–656. doi: 10.1038/s41588-018-0100-5
Idemyor, V. (2014). Genomic medicine: health care issues and the unresolved ethical and social dilemmas. Am. J. Ther. 21, 548–553. doi: 10.1097/MJT.0b013e3182583bd1
Jablonski, N. G., and Chaplin, G. (2000). The evolution of human skin coloration. J. Hum. Evol. 39, 57–106. doi: 10.1006/jhev.2000.0403
Jablonski, N. G., and Chaplin, G. (2010). Human skin pigmentation as an adaptation to UV radiation. Proc. Natl. Acad. Sci. U.S.A. 107(Suppl. 2),8962–8968. doi: 10.1073/pnas.0914628107
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33:245.
Jelenkovic, A., Ortega-Alonso, A., Rose, R. J., Kaprio, J., Rebato, E., and Silventoinen, K. (2011). Genetic and environmental influences on growth from late childhood to adulthood: a longitudinal study of two Finnish twin cohorts. Am. J. Hum. Biol. 23, 764–773. doi: 10.1002/ajhb.21208
Jelenkovic, A., Poveda, A., Susanne, C., and Rebato, E. (2010). Common genetic and environmental factors among craniofacial traits in Belgian nuclear families: comparing skeletal and soft-tissue related phenotypes. Homo 61, 191–203. doi: 10.1016/j.jchb.2009.10.003
Joubert, B. R., Felix, J. F., Yousefi, P., Bakulski, K. M., Just, A. C., Breton, C., etal. (2016). DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am. J. Hum. Genet. 98, 680–696. doi: 10.1016/j.ajhg.2016.02.019
Kau, C. H., and Richmond, S. (2008). Three-dimensional analysis of facial morphology surface changes in untreated children from 12 to 14 years of age. Am. J. Orthod. Dentofacial Orthop. 134, 751–760. doi: 10.1016/j.ajodo.2007.01.037
Kau, C. H., Richmond, S., Incrapera, A., English, J., and Xia, J. J. (2007). Three-dimensional surface acquisition systems for the study of facial morphology and their application to maxillofacial surgery. Int. J. Med. Robot 3, 97–110. doi: 10.1002/rcs.141
Kau, C. H., Richmond, S., Zhurov, A., Ovsenik, M., Tawfik, W., Borbely, P., et al. (2010). Use of 3-dimensional surface acquisition to study facial morphology in 5 populations. Am. J. Orthod. Dentofacial Orthop. 137(4 Suppl):S56.e1-9; discussion S56-7.
Kau, C. H., Richmond, S., Zhurov, A. I., Knox, J., Chestnutt, I., Hartles, F., et al. (2005). Reliability of measuring facial morphology with a 3-dimensional laser scanning system. Am. J. Orthod. Dentofacial Orthop. 128, 424–430. doi: 10.1016/j.ajodo.2004.06.037
Kayser, M. (2015). Forensic DNA Phenotyping: predicting human appearance from crime scene material for investigative purposes. Forensic Sci. Int. Genet. 18, 33–48. doi: 10.1016/j.fsigen.2015.02.003
Keating, B., Bansal, A. T., Walsh, S., Millman, J., Newman, J., Kidd, K., et al. (2013). First all-in-one diagnostic tool for DNA intelligence: genome-wide inference of biogeographic ancestry, appearance, relatedness, and sex with the Identitas v1 Forensic Chip. Int. J. Legal Med. 127, 559–572. doi: 10.1007/s00414-012-0788-1
Khan, M. F. J., Little, J., Mossey, P. A., Steegers-Theunissen, R. P., Autelitano, L., Lombardo, I., et al. (2018). Evaluating LINE-1 methylation in cleft lip tissues and its association with early pregnancy exposures. Epigenomics 10, 105–113. doi: 10.2217/epi-2017-0081
Kim, J. J., Lee, H. I., Park, T., Kim, K., Lee, J. E., Cho, N. H., et al. (2010). Identification of 15 loci influencing height in a Korean population. J. Hum. Genet. 55, 27–31. doi: 10.1038/jhg.2009.116
Kovacs, L., Eder, M., Hollweck, R., Zimmermann, A., Settles, M., Schneider, A., et al. (2007). Comparison between breast volume measurement using 3D surface imaging and classical techniques. Breast 16, 137–145. doi: 10.1016/j.breast.2006.08.001
Kuijpers, M. A., Chiu, Y. T., Nada, R. M., Carels, C. E., and Fudalej, P. S. (2014). Three-dimensional imaging methods for quantitative analysis of facial soft tissues and skeletal morphology in patients with orofacial clefts: a systematic review. PLoS One 9:e93442. doi: 10.1371/journal.pone.0093442
Lange, S., Shield, K., Koren, G., Rehm, J., and Popova, S. (2014). A comparison of the prevalence of prenatal alcohol exposure obtained via maternal self-reports versus meconium testing: a systematic literature review and meta-analysis. BMC Pregn. Childbirth 14:127. doi: 10.1186/1471-2393-14-127
Le, T. T., Farkas, L. G., Ngim, R. C., Levin, L. S., and Forrest, C. R. (2002). Proportionality in Asian and North American Caucasian faces using neoclassical facial canons as criteria. Aesthetic. Plast. Surg. 26, 64–69. doi: 10.1007/s00266-001-0033-7
Lee, M. K., Shaffer, J. R., Leslie, E. J., Orlova, E., Carlson, J. C., Feingold, E., et al. (2017). Genome-wide association study of facial morphology reveals novel associations with FREM1 and PARK2. PLoS One 12:e0176566. doi: 10.1371/journal.pone.0176566
Lee, S. H., Fu, K. K., Hui, J. N., and Richman, J. M. (2001). Noggin and retinoic acid transform the identity of avian facial prominences. Nature 414, 909–912. doi: 10.1038/414909a
Leslie, E. J., Carlson, J. C., Shaffer, J. R., Butali, A., Buxó, C. J., Castilla, E. E., et al. (2017). Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum. Genet. 136, 275–286. doi: 10.1007/s00439-016-1754-7
Lippert, C., Sabatini, R., Maher, M. C., Kang, E. Y., Lee, S., Arikan, O., et al. (2017). Identification of individuals by trait prediction using whole-genome sequencing data. Proc. Natl. Acad. Sci. U.S.A. 114, 10166–10171. doi: 10.1073/pnas.1711125114
Little, A. C., Burt, D. M., Penton-Voak, I. S., and Perrett, D. I. (2001). Self-perceived attractiveness influences human female preferences for sexual dimorphism and symmetry in male faces. Proc. R. Soc. Lond. B Biol. Sci. 268, 39–44. doi: 10.1098/rspb.2000.1327
Little, A. C., Penton-Voak, I. S., Burt, D. M., and Perrett, D. I. (2003). Investigating an imprinting-like phenomenon in humans: partners and opposite-sex parents have similar hair and eye colour. Evol. Hum. Behav. 24, 43–51. doi: 10.1016/S1090-5138(02)00119-8
Liu, F., Van Der Lijn, F., Schurmann, C., Zhu, G., Chakravarty, M. M., Hysi, P. G., et al. (2012). A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet. 8:e1002932. doi: 10.1371/journal.pgen.1002932
Mamluk, L., Edwards, H. B., Savovic, J., Leach, V., Jones, T., Moore, T. H. M., et al. (2017). Low alcohol consumption and pregnancy and childhood outcomes: time to change guidelines indicating apparently ‘safe’ levels of alcohol during pregnancy? A systematic review and meta-analyses. BMJ Open 7:e015410. doi: 10.1136/bmjopen-2016-015410
Marazita, M. (2007). Subclinical features in non-syndromic cleft lip with or without cleft palate (CL/P): review of the evidence that subepithelial orbicularis oris muscle defects are part of an expanded phenotype for CL/P∗. Orthodont. Craniofac. Res. 10, 82–87. doi: 10.1111/j.1601-6343.2007.00386.x
Marcucio, R., Hallgrimsson, B., and Young, N. M. (2015). Facial morphogenesis: physical and molecular interactions between the brain and the face. Curr. Top. Dev. Biol. 115, 299–320. doi: 10.1016/bs.ctdb.2015.09.001
Mellion, Z. J., Behrents, R. G., and Johnston, L. E. Jr. (2013). The pattern of facial skeletal growth and its relationship to various common indexes of maturation. Am. J. Orthod. Dentofacial Orthop. 143, 845–854. doi: 10.1016/j.ajodo.2013.01.019
Menezes, R., and Vieira, A. R. (2008). Dental anomalies as part of the cleft spectrum. Cleft. Palate. Craniofac. J. 45, 414–419. doi: 10.1597/07-064.1
Merks, J. H., van Karnebeek, C. D., Caron, H. N., and Hennekam, R. C. (2003). Phenotypic abnormalities: terminology and classification. Am. J. Med. Genet. A 123a, 211–230. doi: 10.1002/ajmg.a.20249
Miller, S. F., Weinberg, S. M., Nidey, N. L., Defay, D. K., Marazita, M. L., Wehby, G. L., et al. (2014). Exploratory genotype-phenotype correlations of facial form and asymmetry in unaffected relatives of children with non-syndromic cleft lip and/or palate. J. Anat. 224, 688–709. doi: 10.1111/joa.12182
Mirghani, H., Osman, N., Dhanasekaran, S., Elbiss, H. M., and Bekdache, G. (2015). Transplacental transfer of 2-naphthol in human placenta. Toxicol. Rep. 2, 957–960. doi: 10.1016/j.toxrep.2015.05.011
Mitchem, D. G., Purkey, A. M., Grebe, N. M., Carey, G., Garver-Apgar, C. E., Bates, T. C., et al. (2014). Estimating the sex-specific effects of genes on facial attractiveness and sexual dimorphism. Behav. Genet. 44, 270–281. doi: 10.1007/s10519-013-9627-5
Morris, A. P., Voight, B. F., Teslovich, T. M., Ferreira, T., Segre, A. V., Steinthorsdottir, V., et al. (2012). Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990. doi: 10.1038/ng.2383
Mossey, P. A., Little, J., Munger, R. G., Dixon, M. J., and Shaw, W. C. (2009). Cleft lip and palate. Lancet 374, 1773–1785. doi: 10.1016/S0140-6736(09)60695-4
Muggli, E., Matthews, H., Penington, A., Claes, P., O’Leary, C., Forster, D., et al. (2017). Association between prenatal alcohol exposure and craniofacial shape of children at 12 Months of Age. JAMA Pediatr. 171, 771–780. doi: 10.1001/jamapediatrics.2017.0778
Munn, L., and Stephan, C. N. (2018). Changes in face topography from supine-to-upright position-And soft tissue correction values for craniofacial identification. Forensic Sci. Int. 289, 40–50. doi: 10.1016/j.forsciint.2018.05.016
Neiswanger, K., Weinberg, S. M., Rogers, C. R., Brandon, C. A., Cooper, M. E., Bardi, K. M., et al. (2007). Orbicularis oris muscle defects as an expanded phenotypic feature in nonsyndromic cleft lip with or without cleft palate. Am. J. Med. Genet. Part A 143, 1143–1149. doi: 10.1002/ajmg.a.31760
Parsons, T. E., Downey, C. M., Jirik, F. R., Hallgrimsson, B., and Jamniczky, H. A. (2015). Mind the gap: genetic manipulation of basicranial growth within synchondroses modulates calvarial and facial shape in mice through epigenetic interactions. PLoS One 10:e0118355. doi: 10.1371/journal.pone.0118355
Paternoster, L., Zhurov, A. I., Toma, A. M., Kemp, J. P., Pourcain, B. S., Timpson, N. J., et al. (2012). Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am. J. Hum. Genet. 90, 478–485. doi: 10.1016/j.ajhg.2011.12.021
Peng, S., Tan, J., Hu, S., Zhou, H., Guo, J., Jin, L., et al. (2013). Detecting genetic association of common human facial morphological variation using high density 3D image registration. PLoS Comput. Biol. 9:e1003375. doi: 10.1371/journal.pcbi.1003375
Pickrell, J. K., Berisa, T., Liu, J. Z., Segurel, L., Tung, J. Y., and Hinds, D. A. (2016). Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717. doi: 10.1038/ng.3570
Pirttiniemi, P. M. (1994). Associations of mandibular and facial asymmetries–a review. Am. J. Orthod. Dentofacial Orthop. 106, 191–200. doi: 10.1016/S0889-5406(94)70038-9
Popat, H., Richmond, S., and Drage, N. A. (2010). New developments in: three-dimensional planning for orthognathic surgery. J. Orthod. 37, 62–71. doi: 10.1179/14653121042885
Popat, H., Richmond, S., Marshall, D., and Rosin, P. L. (2012). Three-dimensional assessment of functional change following Class 3 orthognathic correction–a preliminary report. J. Craniomaxillofac. Surg. 40, 36–42. doi: 10.1016/j.jcms.2010.12.005
Pound, N., Lawson, D. W., Toma, A. M., Richmond, S., Zhurov, A. I., and Penton-Voak, I. S. (2014). Facial fluctuating asymmetry is not associated with childhood ill-health in a large British cohort study. Proc Biol Sci. 281:20141639. doi: 10.1098/rspb.2014.1639
Rachdaoui, N., and Sarkar, D. K. (2014). Transgenerational epigenetics and brain disorders. Int. Rev. Neurobiol. 115, 51–73. doi: 10.1016/B978-0-12-801311-3.00002-0
Reik, W. (2007). Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447:425. doi: 10.1038/nature05918
Relton, C. L., and Davey Smith, G. (2012). Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int. J. Epidemiol. 41, 161–176. doi: 10.1093/ije/dyr233
Richmond, S., Toma, A. M., and Zhurov, A. I. (2009). New perspectives on craniofacial growth. Orthod. Fr. 80, 359–369. doi: 10.1051/orthodfr/2009033
Richmond, S. A., Ali, A. M., Beldi, L., Chong, Y. T., Cronin, A., Djordjevic, J., et al. (2012). Detailing Patient Specific Modeling to Aid Clinical Decision Making. E LBP. Dordrecht: Springer.
Richmond, R. C., Sharp, G. C., Herbert, G., Atkinson, C., Taylor, C., Bhattacharya, S., et al. (2018). The long-term impact of folic acid in pregnancy on offspring DNA methylation: follow-up of the Aberdeen Folic Acid Supplementation Trial (AFAST). Int. J. Epidemiol. 47, 928–937. doi: 10.1093/ije/dyy032
Richmond, S., Wilson-Nagrani, C., Zhurov, A. I., Farnell, D., Galloway, J., Mohd Ali, A. S., Pirttiniemi, P., Katic, V. (2018). “Factors influencing facial shape,” in Evidenced-Based Orthodontics. eds G. J. Huang, R. S., K.W.L. Vig (Hoboken, NJ: Wiley-Blackwell).
Ricketts, R. M. (1982). The biologic significance of the divine proportion and Fibonacci series. Am. J. Orthod. 81, 351–370. doi: 10.1016/0002-9416(82)90073-2
Rivezzi, G., Piscitelli, P., Scortichini, G., Giovannini, A., Diletti, G., Migliorati, G., et al. (2013). A general model of dioxin contamination in breast milk: results from a study on 94 women from the Caserta and Naples areas in Italy. Int. J. Environ. Res. Public Health 10, 5953–5970. doi: 10.3390/ijerph10115953
Robinson, M. R., Kleinman, A., Graff, M., Vinkhuyzen, A. A., Couper, D., Miller, M. B., et al. (2017). Genetic evidence of assortative mating in humans. Nat. Hum. Behav. 1:0016.
Roosenboom, J., Hens, G., Mattern, B. C., Shriver, M. D., and Claes, P. (2016). Exploring the underlying genetics of craniofacial morphology through various sources of knowledge. Biomed. Res. Int. 2016:3054578. doi: 10.1155/2016/3054578
Ruiz-Linares, A., Adhikari, K., Acuña-Alonzo, V., Quinto-Sanchez, M., Jaramillo, C., Arias, W., et al. (2014). Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 10:e1004572. doi: 10.1371/journal.pgen.1004572
Schmidt, E., and Kornfeld, J. W. (2016). Decoding lamarck-transgenerational control of metabolism by noncoding RNAs. Pflugers. Arch. 468, 959–969. doi: 10.1007/s00424-016-1807-8
Scott, I. M., Clark, A. P., Boothroyd, L. G., and Penton-Voak, I. S. (2013). Do men’s faces really signal heritable immunocompetence? Behav. Ecol. 24, 579–589.
Shaffer, J. R., Li, J., Lee, M. K., Roosenboom, J., Orlova, E., Adhikari, K., et al. (2017). Multiethnic GWAS reveals polygenic architecture of earlobe attachment. Am. J. Hum. Genet. 101, 913–924. doi: 10.1016/j.ajhg.2017.10.001
Shaffer, J. R., Orlova, E., Lee, M. K., Leslie, E. J., Raffensperger, Z. D., Heike, C. L., et al. (2016). Genome-wide association study reveals multiple loci influencing normal human facial morphology. PLoS Genet. 12:e1006149. doi: 10.1371/journal.pgen.1006149
Sham, P. C., and Purcell, S. M. (2014). Statistical power and significance testing in large-scalegenetic studies. Nat. Rev. Genet. 15, 335–346. doi: 10.1038/nrg3706
Sharman, N. (2011). “Inside the human body,” in Creation, ed. N. Sharman (London: BBC One, BBC), 2 minutes.
Sharp, G. C., Arathimos, R., Reese, S. E., Page, C. M., Felix, J., Küpers, L. K., et al. (2018). Maternal alcohol consumption and offspring DNA methylation: findings from six general population-based birth cohorts. Epigenomics 10, 27–42. doi: 10.2217/epi-2017-0095
Sharp, G. C., Ho, K., Davies, A., Stergiakouli, E., Humphries, K., McArdle, W., et al. (2017). Distinct DNA methylation profiles in subtypes of orofacial cleft. Clin. Epigenet. 9:63. doi: 10.1186/s13148-017-0362-2
Shi, M., Wehby, G. L., and Murray, J. C. (2008). Review on genetic variants and maternal smoking in the etiology of oral clefts and other birth defects. Birth Defects Res. C Embryo Today 84, 16–29. doi: 10.1002/bdrc.20117
Shrimpton, S., Daniels, K., de Greef, S., Tilotta, F., Willems, G., Vandermeulen, D., et al. (2014). A spatially-dense regression study of facial form and tissue depth: towards an interactive tool for craniofacial reconstruction. Forensic Sci. Int. 234, 103–110. doi: 10.1016/j.forsciint.2013.10.021
Sidlauskas, M., Salomskiene, L., Andriuskeviciute, I., Sidlauskiene, M., Labanauskas, Z., Vasiliauskas, A., et al. (2016). Heritability of mandibular cephalometric variables in twins with completed craniofacial growth. Eur. J. Orthod. 38, 493–502. doi: 10.1093/ejo/cjv062
Smith, G. D., and Ebrahim, S. (2003). ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? 2003. Int. J. Epidemiol. 32, 1–22. doi: 10.1093/ije/dyg070
Som, P. M., and Naidich, T. P. (2013). Illustrated review of the embryology and development of the facial region, part 1: early face and lateral nasal cavities. AJNR Am. J. Neuroradiol. 34, 2233–2240. doi: 10.3174/ajnr.A3415
Som, P. M., and Naidich, T. P. (2014). Illustrated review of the embryology and development of the facial region, part 2: late development of the fetal face and changes in the face from the newborn to adulthood. AJNR Am. J. Neuroradiol. 35, 10–18. doi: 10.3174/ajnr.A3414
Stanier, P., and Moore, G. E. (2004). Genetics of cleft lip and palate: syndromic genes contribute to the incidence of non-syndromic clefts. Hum. Mol. Genet. 13(Suppl. 1), R73–R81. doi: 10.1093/hmg/ddh052
Stephan, C., Norris, R., and Henneberg, M. (2005). Does sexual dimorphism in facial soft tissue depths justify sex distinction in craniofacial identification? J. Forensic Sci. 50, 513–508. doi: 10.1520/JFS2004251
Suttie, M., Wozniak, J. R., Parnell, S. E., Wetherill, L., Mattson, S. N., Sowell, E. R., et al. (2018). Combined face-brain morphology and associated neurocognitive correlates in fetal alcohol spectrum disorders. Alcohol. Clin. Exp. Res. 42, 1769–1782. doi: 10.1111/acer.13820
Tanner, J. M., Whitehouse, R. H., and Takaishi, M. (1966a). Standards from birth to maturity for height, weight, height velocity, and weight velocity: British children, 1965. I. Arch. Dis. Child 41, 454–471. doi: 10.1136/adc.41.219.454
Tanner, J. M., Whitehouse, R. H., and Takaishi, M. (1966b). Standards from birth to maturity for height, weight, height velocity, and weight velocity: British children, 1965. II. Arch. Dis. Child 41, 613–635. doi: 10.1136/adc.41.220.613
Tobi, E. W., Slieker, R. C., Luijk, R., Dekkers, K. F., Stein, A. D., Xu, K. M., et al. (2018). DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci. Adv. 4:eaao4364.
Toma, A. M., Zhurov, A., Playle, R., and Richmond, S. (2008). A three-dimensional look for facial differences between males and females in a British-Caucasian sample aged 151/2 years old. Orthod. Craniofac. Res. 11, 180–185. doi: 10.1111/j.1601-6343.2008.00428.x
Toma, A. M., Zhurov, A. I., Playle, R., Marshall, D., Rosin, P. L., and Richmond, S. (2012). The assessment of facial variation in 4747 British school children. Eur. J. Orthod. 34, 655–664. doi: 10.1093/ejo/cjr106
Toom, V., Wienroth, M., M’Charek, A., Prainsack, B., Williams, R., Duster, T., et al. (2016). Approaching ethical, legal and social issues of emerging forensic DNA phenotyping (FDP) technologies comprehensively: reply to ’Forensic DNA phenotyping: predicting human appearance from crime scene material for investigative purposes’ by Manfred Kayser. Forensic Sci. Int. Genet. 22, e1–e4. doi: 10.1016/j.fsigen.2016.01.010
Tsagkrasoulis, D., Hysi, P., Spector, T., and Montana, G. (2017). Heritability maps of human face morphology through large-scale automated three-dimensional phenotyping. Sci. Rep. 7:45885. doi: 10.1038/srep45885
Tzou, C. H., Artner, N. M., Pona, I., Hold, A., Placheta, E., Kropatsch, W. G., et al. (2014). Comparison of three-dimensional surface-imaging systems. J. Plast. Reconstr. Aesthet Surg. 67, 489–497. doi: 10.1016/j.bjps.2014.01.003
Uslu, V. V., Petretich, M., Ruf, S., Langenfeld, K., Fonseca, N. A., Marioni, J.C., et al. (2014). Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat. Genet. 46, 753–758. doi: 10.1038/ng.2971
Van der Beek, M. C., Hoeksma, J. B., and Prahl-Andersen, B. (1996). Vertical facial growth and statural growth in girls: a longitudinal comparison. Eur. J. Orthod. 18, 549–555. doi: 10.1093/ejo/18.6.549
Van Otterloo, E., Williams, T., and Artinger, K. B. (2016). The old and new face of craniofacial research: How animal models inform human craniofacial genetic and clinical data. Dev. Biol. 415, 171–187. doi: 10.1016/j.ydbio.2016.01.017
Verdonck, A., Gaethofs, M., Carels, C., and de Zegher, F. (1999). Effect of low-dose testosterone treatment on craniofacial growth in boys with delayed puberty. Eur. J. Orthod. 21, 137–143. doi: 10.1093/ejo/21.2.137
Visel, A., Rubin, E. M., and Pennacchio, L. A. (2009). Genomic views of distant-acting enhancers. Nature 461, 199–205. doi: 10.1038/nature08451
Visscher, P. M., Hill, W. G., and Wray, N. R. (2008). Heritability in the genomics era–concepts and misconceptions. Nat. Rev. Genet. 9, 255–266. doi: 10.1038/nrg2322
Weinberg, S., Naidoo, S., Bardi, K., Brandon, C., Neiswanger, K., Resick, J., et al. (2009). Face shape of unaffected parents with cleft affected offspring: combining three-dimensional surface imaging and geometric morphometrics. Orthodont. Craniofacial Res. 12, 271–281. doi: 10.1111/j.1601-6343.2009.01462.x
Weiner, J. S. (1954). Nose shape and climate. Am. J. Phys. Anthropol. 12, 615–618. doi: 10.1002/ajpa.1330120412
Wickström, R. (2007). Effects of nicotine during pregnancy: human and experimental evidence. Curr. Neuropharmacol. 5, 213–222. doi: 10.2174/157015907781695955
Wilde, S., Timpson, A., Kirsanow, K., Kaiser, E., Kayser, M., Unterländer, M., et al. (2014). Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc. Natl. Acad. Sci. U.S.A. 111, 4832–4837. doi: 10.1073/pnas.1316513111
Wilderman, A., VanOudenhove, J., Kron, J., Noonan, J. P., and Cotney, J. (2018). High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Rep. 23, 1581–1597. doi: 10.1016/j.celrep.2018.03.129
Wilkinson, C., Rynn, C., Peters, H., Taister, M., Kau, C. H., and Richmond, S. (2006). A blind accuracy assessment of computer-modeled forensic facial reconstruction using computed tomography data from live subjects. Forensic Sci. Med. Pathol. 2, 179–187. doi: 10.1007/s12024-006-0007-9
Wilson, C., Playle, R., Toma, A., Zhurov, A., Ness, A., and Richmond, S. (2013). The prevalence of lip vermilion morphological traits in a 15-year-old population. Am. J. Med. Genet. A 161a, 4–12. doi: 10.1002/ajmg.a.35515
Xu, C., Qu, H., Wang, G., Xie, B., Shi, Y., Yang, Y., et al. (2015). A novel strategy for forensic age prediction by DNA methylation and support vector regression model. Sci. Rep. 5:17788. doi: 10.1038/srep17788
Xuan, Z., Zhongpeng, Y., Yanjun, G., Jiaqi, D., Yuchi, Z., Bing, S., et al. (2016). Maternal active smoking and risk of oral clefts: a meta-analysis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 122, 680–690. doi: 10.1016/j.oooo.2016.08.007
Zaidi, A. A., Mattern, B. C., Claes, P., McEcoy, B., Hughes, C., and Shriver, M. D. (2017). Investigating the case of human nose shape and climate adaptation. PLoS Genet. 13:e1006616. doi: 10.1371/journal.pgen.1006616
Keywords: 3D imaging, admixture, ancestry, facial variation, geometric morphometrics, facial genetics, facial phenotyping, genetic-environmental influences
Citation: Richmond S, Howe LJ, Lewis S, Stergiakouli E and Zhurov A (2018) Facial Genetics: A Brief Overview. Front. Genet. 9:462. doi: 10.3389/fgene.2018.00462
Received: 06 July 2018; Accepted: 20 September 2018;
Published: 16 October 2018.
Edited by:
Peter Claes, KU Leuven, BelgiumReviewed by:
Hui-Qi Qu, Children’s Hospital of Philadelphia, United StatesCopyright © 2018 Richmond, Howe, Lewis, Stergiakouli and Zhurov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen Richmond, cmljaG1vbmRzQGNhcmRpZmYuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.