
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 08 October 2018
Sec. Evolutionary and Population Genetics
Volume 9 - 2018 | https://doi.org/10.3389/fgene.2018.00445
This article is part of the Research Topic Recent Advancements in Honeybee Biology View all 5 articles
DnaJ, also known as Hsp40, plays important roles in maintaining the normal physiological state of an organism under stress conditions by mediating essential processes, such as protein synthesis, degradation, folding and metabolism. However, the exact functions of most DnaJ members are not fully understood in insects. Here, we identified three genes, AccDnaJA1, AccDnaJB12, and AccDnaJC8, in Apis cerana cerana and explored their connection with the environmental stress response. Quantitative real-time PCR results showed that the mRNA levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 were all induced under cold, UV, H2O2 and different pesticides treatment. The expression patterns of AccDnaJB12 and AccDnaJC8 were upregulated by CdCl2 and HgCl2 stress, while the transcriptional levels of AccDnaJA1 were downregulated by CdCl2 and HgCl2 stress. Western blot findings further indicated that AccDnaJB12 protein levels were increased by some stress conditions. Knockdown of each of these three genes downregulated the transcriptional patterns of several stress response-related genes at different levels. Functional analysis further demonstrated that the resistance of A. cerana cerana to lambda-cyhalothrin stress was reduced with knockdown of AccDnaJA1, AccDnaJB12, or AccDnaJC8, indicating that these three genes may be involved in the tolerance to this pesticide. Taken together, these findings indicate that AccDnaJA1, AccDnaJB12, and AccDnaJC8 may play pivotal roles in the stress response by facilitating honeybee survival under some adverse circumstances. To our knowledge, this is the first report that reveals the roles of DnaJ family proteins under different adverse circumstances in A. cerana cerana.
Heat shock proteins (HSPs) are molecular chaperones well known for their function in mediating inappropriate aggregation, folding, misfolding and unfolding of proteins (Kim et al., 2013). These tasks are essential for organisms not only under normal conditions but also under diverse environmental stresses, and the need for HSPs is increased under non-optimal environmental conditions (Rinehart et al., 2007). Based on their molecular weight in kDa, HSPs are divided into six families including small HSP (sHsp), Hsp40, Hsp60, Hsp70, Hsp90, and Hsp100 (Sørensen et al., 2010; Requena et al., 2015). In recent years, the characteristics of each family of HSPs, including Hsp40, had been gradually revealed.
Hsp40 was first identified to stimulate DnaK (an Hsp70 homolog in bacteria) ATPase activity to assist with replication of host cell λ phage DNA (Yochem et al., 1978; Liberek et al., 1988). It is often named DnaJ for the presence of a J domain, which consists of approximately 70 amino acids, in its protein structure (Jiang et al., 2007). In addition, according to the difference in the number and type of domain structure, DnaJ proteins are divided into three classes, DnaJA, DnaJB, and DnaJC. DnaJA contains a J domain, variable C-terminal domain, cysteine-rich region and glycine/phenylalanine-rich region. DnaJB has all the domains presented above except for the cysteine-rich region, while DnaJC only contains a J domain (Vos et al., 2008). Previous studies had indicated that DnaJ usually cooperated with DnaK to participate in diverse cellular biological processes, such as protein export, protein folding, DNA replication and stress responses by relying on ATP hydrolysis (Craig et al., 2006). Fully investigating the role of DnaJ will contribute to understanding the functional mechanism of DnaJ/DnaK in different biological processes under stress conditions.
Recently, some studies revealed the biological function of DnaJ under stressful environmental conditions. For example, overexpression of DnaJA1 in human pancreatic cancer cells represses the stress response capacity of c-Jun, possibly through activation of the DnaK protein, triggering a reduction in cell survival (Stark et al., 2014). Overexpression of DnaJ/DnaK increases the freeze tolerance capacity of Escherichia coli, probably by restraining irreversible protein denaturation (Chow and Tung, 1998). Under acid stress for 0.25 and 1 h, the mRNA levels of DnaJ are increased 2.1- and 35.7-fold, respectively, in Alicyclobacillus acidoterrestris (Jiao et al., 2012). ERdj4, an endoplasmic reticulum DnaJ homolog, can stimulate BiP ATPase activity and is significantly upregulated at both the mRNA and protein levels following endoplasmic reticulum stress in mice (Shen et al., 2002). ERdj5 is also a DnaJ protein in the endoplasmic reticulum, contains a thioredoxin and DnaJ domain and is expressed in response to endoplasmic reticulum stress in humans (Cunnea et al., 2003). Despite the progress achieved in DnaJ research, the functions of many members of DnaJ have not been uncovered in many species, including honeybees.
As one type of Asian honeybee, Apis cerana cerana brings substantial ecosystem services to various terrestrial plants and produces tremendous economic benefits for people. Recently, however, due to the influence of diverse environmental stresses, the honeybee population size had obviously decreased in some regions (Potts et al., 2010; Kulhanek et al., 2017; Li et al., 2018b). Exploring the stress response mechanism at gene expression regulation level will be beneficial to the stress-resistance biology research of honeybees. Furthermore, a recent study reported that insect HSPs played important roles during stress (King and MacRae, 2015), and our previous studies also demonstrated that four sHsp genes (AccHsp22.6, AccHsp23.0, AccHsp24.2, and AccHsp27.6) were involved in the stress response in A. cerana cerana (Liu et al., 2012; Liu Z. et al., 2014; Zhang et al., 2014). These results indicate that HSPs may play key roles in defense against environmental stress in honeybees.
To gain insight into the function of the DnaJ family of HSPs in A. cerana cerana, we identified three DnaJ genes, named AccDnaJA1, AccDnaJB12, and AccDnaJC8, in this study and investigated their response to environmental stress. The results of quantitative real-time PCR (qRT-PCR) analysis revealed that the expression levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 were induced under many stress conditions. Western blot analysis further proved that AccDnaJB12 was upregulated by some stress factors at the protein level. An RNA interference (RNAi) experiment indicated that knockdown of AccDnaJA1, AccDnaJB12, and AccDnaJC8 repressed the transcriptional profiles of several stress response-related genes and decreased the survival ability of A. cerana cerana under lambda-cyhalothrin stress. Taken together, our findings shed light on the stress response mechanisms of AccDnaJA1, AccDnaJB12, and AccDnaJC8 in honeybees.
The online software TFBIND1 and MatInspector database2 were selected to predict the probable cis-acting elements in the 5′-flanking regions of AccDnaJA1 (Gene ID: 107996076), AccDnaJB12 (Gene ID: 108000381), and AccDnaJC8 (Gene ID: 107998514) (Tsunoda and Takagi, 1999).
The honeybee colonies used for the experiment were raised at the College of Animal Science and Technology, Shandong Agricultural University (Tai’an, China). The 15-day-old A. cerana cerana foragers were distinguished by marking the newly emerged honeybees with pigment 15 days in advance. The honeybee colonies were normally managed every day by our beekeeper to maintain a strong and healthy colony.
Recent studies had shown that honeybees were often subjected to multiple environmental stresses, and the foragers (approximately 2–3 weeks old) usually had more opportunity for exposure to adverse circumstances (Potts et al., 2010; Even et al., 2012; Farooqui, 2014; Robertson et al., 2014). In this study, we selected 15-day-old foragers to use for the expression analysis and functional analysis of AccDnaJA1 (GenBank accession no. XP 016909435.1), AccDnaJB12 (GenBank accession no. XP 016916168.1) and AccDnaJC8 (GenBank accession no. XP 016913324.1). The 15-day-old foragers were collected from the strong and healthy A. cerana cerana colonies and divided into 13 groups. Among these groups, 11 groups were randomly exposed to one of the following treatments including 4, 14, and 24°C, ultraviolet radiation (UV), CdCl2, HgCl2, lambda-cyhalothrin, emamectin benzoate, spirodiclofen, avermectin, and paraquat.
For the 4, 14, and 24°C treatments, 3 groups of honeybees were reared in incubators set at 4, 14, and 24°C, respectively, with 70% relative humidity in the dark. For UV treatment, the honeybees were directly irradiated with 30 mJ/cm2 UV using a desk type UV lamp in an incubator (33°C, 70% relative humidity and 24 h dark). With regard to CdCl2 and HgCl2 treatment, honeybees in two groups were injected with 1 μL CdCl2 (2 g/mL) or HgCl2 (2 g/mL), respectively, between the second abdominal segment (A2) and third abdominal segment (A3), which was defined according to the online database3, using a microsyringe. During injected, our operators grasped the two pairs of wings of honeybees by one hand to immobilize them. For pesticide treatment, five groups of honeybees were maintained in incubators that were sprayed with lambda-cyhalothrin (0.05 mg/mL), emamectin benzoate (2 mg/mL), spirodiclofen (2 mg/mL), avermectin (0.05 mg/mL), or paraquat (2 mg/mL) in advance, separately, and kept at 33°C with 70% relative humidity in the dark. The remaining two groups were both used as control groups. Among them, one group was kept untreated and used as the control group for the 4, 14, and 24°C, ultraviolet radiation (UV), lambda-cyhalothrin, emamectin benzoate, spirodiclofen, avermectin, and paraquat treatments. The other group that injected with 1 μL sterilized water between the second abdominal segment (A2) and third abdominal segment (A3) was the control group for the CdCl2 and HgCl2 treatments. All the groups were fed fresh pollen dough and a 30% sucrose solution. Then, the healthy honeybees in each group were sampled at specific times (Supplementary Table S1) and kept in an ultralow temperature freezer (Panasonic, Japan) until use.
Total RNA from whole honeybees was extracted using RNAiso Plus (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The first strand of cDNA was acquired using a HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme, Nanjing, China) following the supplier’s instructions. The qRT-PCR analyses were carried out using SYBR® Premix Ex TaqTM (Tli RNaseH Plus) (TaKaRa, Dalian, China) and a CFX96TM Real-Time System. The efficiency values of all pairs of primers used for qRT-PCR were all within 90–110%. Each pair of qRT-PCR primers had a single peak, and their correlation coefficients (R2) were approaching 1. The specific sequences of qRT-PCR primers are presented in Supplementary Table S2. The β-actin gene (GenBank accession no. HM640276.1) was used as a standard to normalize other gene expression levels as it is stably expressed in honeybees (Lourenço et al., 2008; Yao et al., 2014). For each treatment, at least three biological replicates were performed. The 2-ΔΔCT method and CFX Manager software (version 3.1) were used to analyze the relative levels of gene expression.
The coding region of AccDnaJA11 and part of coding region of AccDnaJB12 were, respectively, fused into the prokaryotic expression vector pET-30a (+) (Novagen, Darmstadt, Germany). Then these two recombinant vectors were, respectively, transformed into Transetta (DE3) chemically competent cells (TransGen Biotech, Beijing, China), and the cell were induced with 0.3 mM isopropyl-1-thio-bgalactopyranoside at 37°C for 10 h during logarithmic growth phase. The 12% SDS-PAGE was used to separate the recombinant proteins (AccDnaJA11 and AccDnaJB12), which was used to inject into the white mouse (Taibang, Tai’an, China) to further acquire anti-AccDnaJA11 and anti-AccDnaJB12, and the remaining operation methods were performed as described as Meng et al. (2010) and Li et al. (2016a).
Total protein from whole honeybee was extracted using a tissue protein extraction kit (ComWin Biotech, Beijing, China). After denaturation, total protein was separated by 12% Tris-glycine gels and electrotransferred onto PVDF membranes (ComWin Biotech, Beijing, China) through a semidry method. Anti-AccDnaJA11 and anti-AccDnaJB12 antibodies [1:500 (v/v) dilution] were used as the primary antibodies, and a horseradish peroxidase-conjugated goat anti-mouse antibody (1:2000 (v/v) dilution, Dingguo Changsheng Biotechnology, Beijing, China) was served as the secondary antibody. The binding reaction was detected using a SageCaptureTM Biological imager (Beijing Sage Creation Science Co., Ltd., Beijing, China) and the ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore Corporation, Billerica, MA, United States).
Primers specific to partial AccDnaJA1, AccDnaJB12, and AccDnaJC8 coding regions were constructed with a T7 promoter sequence at each primer 5′ end. The primer sequences can be found in Supplementary Table S2. EasyTaq® DNA Polymerase (TransGen Biotech, Beijing, China) and a DNA Extraction Kit (Solarbio, Beijing, China) were used to perform PCR amplifications and purify the PCR products following standard procedures, respectively. The PCR amplification procedures were presented in Supplementary Table S3. A T7 RiboMAXTM Express RNAi System (Promega, Madison, United States) and the purified PCR products were used to produce large quantities of dsRNA for AccDnaJA1, AccDnaJB12, and AccDnaJC8 according to the manufacturer’s instructions. As a control, we also produced dsRNA for green fluorescent protein (GFP) (GenBank accession no. U87974) (Wang et al., 2012).
The 15-day-old foragers were divided into four groups (n = 30). Groups 1–3 were fed 7 μg dsRNA AccDnaJA1, dsRNA AccDnaJB12 and dsRNA AccDnaJC8, respectively, and as a control, group 4 was fed 7 μg dsRNA GFP. Then, the four groups were raised in an incubator (34°C, 70% relative humidity and 24 h darkness) and fed fresh pollen dough and a 30% sucrose solution. The healthy honeybees were sampled for verification of RNAi efficiency 2 days after ingestion of dsRNA.
After RNAi of AccDnaJA1, AccDnaJB12, and AccDnaJC8, qRT-PCR was used to detect the mRNA levels of AccANT, AccCDK5r, AccCPR24, AccERR, AccTP1, AccCDK5, AccCYP336A1, AccSp10, AccRBM11, AccAK, AccMTNR1A, and AccAIF3. These genes were proved to be involved in the stress response in our previous studies (Chu et al., 2013; Liu X. et al., 2014; Wang et al., 2014; Chen et al., 2015; Li et al., 2016b, 2018a; Zhang et al., 2016; Zhou et al., 2016; Zhu et al., 2016; Gao et al., 2017; Zhao et al., 2018).
The 15-day-old foragers were divided into four groups (n = 50) fed dsRNA AccDnaJA1, dsRNA AccDnaJB12, dsRNA AccDnaJC8, or dsRNA GFP (7 μg/individual), separately. Two days later, the four groups were all exposed to lambda-cyhalothrin stress as described in the “Diverse Environmental Stress Exposure” section, and then the survival rate of each group was recorded every 0.5 h. At least three biological replicates were performed.
Duncan’s multiple range tests of Statistical Analysis System software (version 9.1) were used to determine the differences between multiple groups. The mean ± SE from three independent experiments was denoted by the standard error bars. Different letters above the error bars were used to present the significant differences. Significant difference and no significant difference between various groups were indicated by the different letter and the same letter, respectively. If there was difference between two groups and the difference was not significant, the overlapped letters were used to indicate statistical significance. ∗P < 0.05 and ∗∗P < 0.01 were determined by the Student’s t-test.
As a histidine-tag fusion protein, AccDnaJA1 was overexpressed in E. coli Transetta (DE3). An SDS-PAGE analysis indicated that this recombinant protein was expressed successfully in Transetta and had obvious stripes between 44.3 and 66.4 kDa, which contained 44.828 kDa AccDnaJA1 and approximately 7 kDa cleavable N- and C-terminal His-tags (Supplementary Figure S1A). The induced overexpression of pET-30a(+) vector Transetta and uninduced overexpression of pET-30a(+)-AccDnaJA1 were used as control.
In regard to the recombinant AccDnaJB12 protein, when its whole coding region was fused into prokaryotic expression vector pET-30a (+), the Transetta (DE3) could not express this recombinant protein. The signal peptide and transmembrane domain usually influence the express ability of E. coli to target heterologous protein, and we found that the protein of AccDnaJB12 had a transmembrane domain near the sit of 250 amino acid. Therefore, we selected the amino acid sequence from 1 to 210 to fuse to prokaryotic expression vector pET-30a (+). The truncated AccDnaJB12 protein is 24.068 kDa, when added to approximately 7 kDa cleavable N- and C-terminal His-tags of the vector pET-30a (+), the recombinant AccDnaJB12 protein is approximately 31 kDa. Consistent with this predicted result, the truncated recombinant AccDnaJB12 protein was successfully expressed (Supplementary Figure S1B).
To preliminarily explore the functions of AccDnaJA1, AccDnaJB12, and AccDnaJC8 with regard to the stress response, partial 5′-flanking regions of AccDnaJA1, AccDnaJB12, and AccDnaJC8 were chosen to predict possible cis-acting elements using TFBIND. We found many putative binding sites bound by stress response-related transcription factors, such as p53, NF-κB, CREB, and AP-1 (Wan et al., 2012; Bagul et al., 2015; Charni-Natan et al., 2018; Chen et al., 2018; Fujimura and Usuki, 2018; Russo et al., 2018), in the 5′-flanking regions of AccDnaJA1, AccDnaJB12, and AccDnaJC8, and a partial list of them is shown in Table 1.
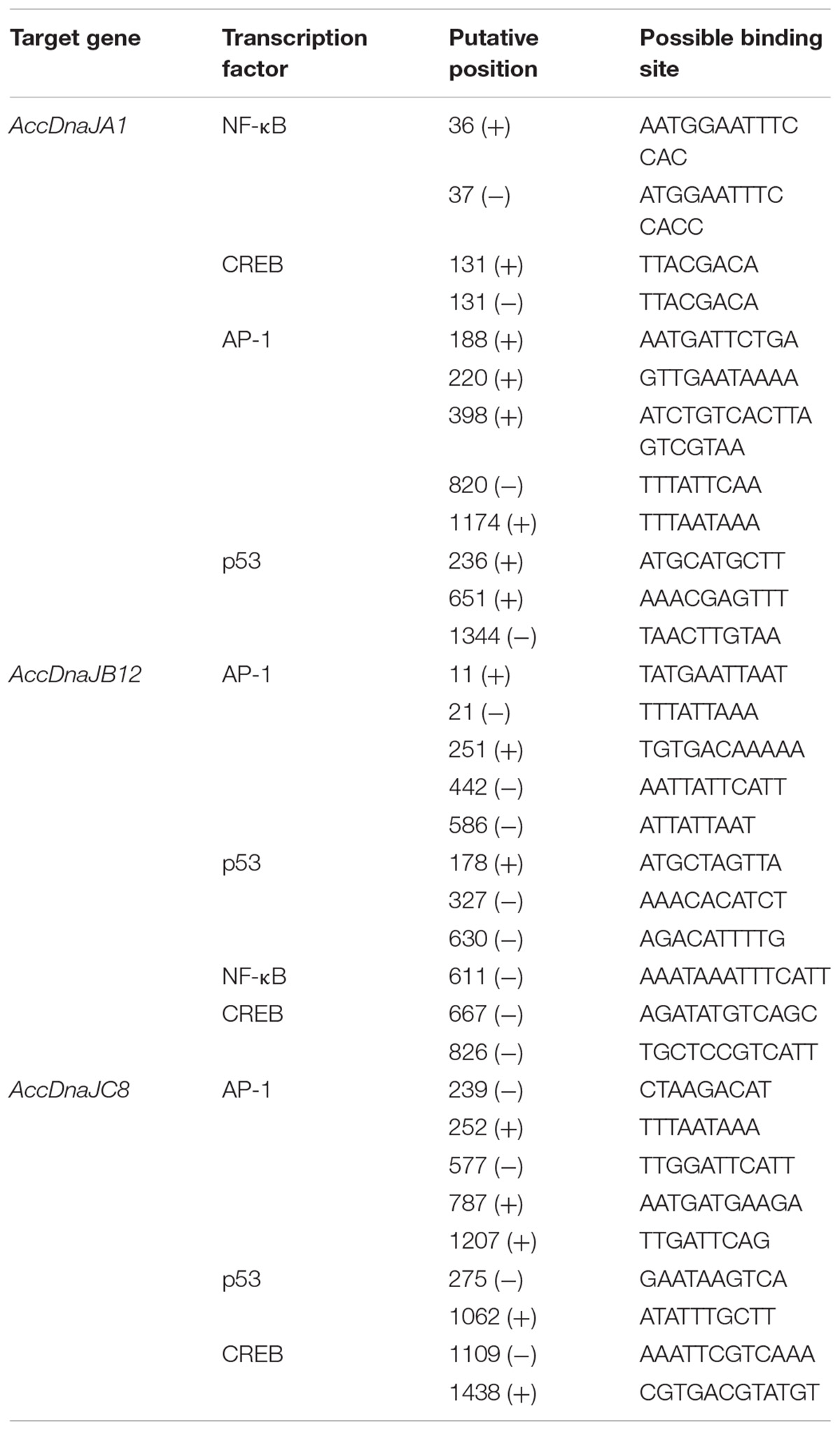
TABLE 1. Predictable stress response-related transcription factors and their binding sites in the 5′ flanking regions of AccDnaJA1, AccDnaJB12, and AccDnaJC8.
To determine whether AccDnaJA1, AccDnaJB12, and AccDnaJC8 responded to cold (4, 14, and 24°C), heavy metal (CdCl2 and HgCl2) and UV stress at the transcriptional level, qRT-PCR was carried out, and the β-actin gene was used as an internal control. As shown in Figure 1A, the mRNA levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 were little changed under normal condition within 5 h, while they were all upregulated and reached their highest levels at 1, 1, and 3 h, respectively, under 4°C stress (Figure 1B). At 14°C, AccDnaJA1, AccDnaJB12, and AccDnaJC8 were all increased and reached their maximum levels at 1 h (Figure 1C). Under stress of 24°C, all three genes reached a peak at 4 h (Figure 1D). Under UV stress, AccDnaJA1, AccDnaJB12, and AccDnaJC8 were all upregulated with a peak at 2 h, and then all were gradually downregulated (Figure 1E). The expression levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 were nearly not changed when honeybees were injected sterilized water (Figure 1F). However, when honeybees were injected with HgCl2, AccDnaJB12 was induced and reached the highest transcript level at 2 h, and AccDnaJA1 and AccDnaJC8 was repressed in response to HgCl2 stress (Figure 1G). With CdCl2 treatment, AccDnaJA1 was downregulated, while AccDnaJB12 and AccDnaJC8 were both upregulated at 3 h (Figure 1H). These results suggest that AccDnaJA1, AccDnaJB12, and AccDnaJC8 can respond to the above six stress conditions to a certain degree at transcriptional level.
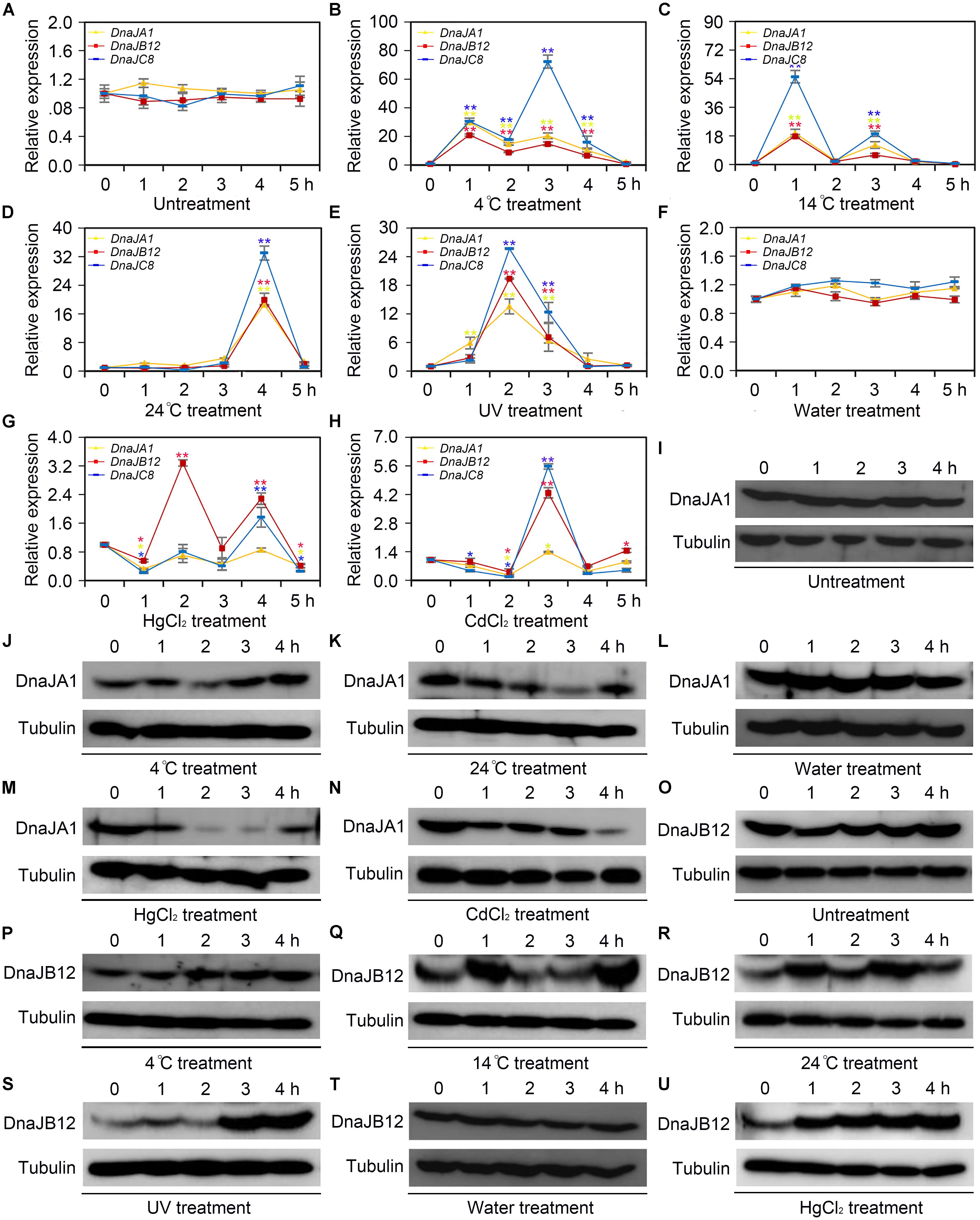
FIGURE 1. Transcription and translation levels analysis of AccDnaJA1, AccDnaJB12, and AccDnaJC8 under cold, UV and heavy metal stress. (A–H) qRT-PCR was performed to reveal the mRNA levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8. (A) qRT-PCR results of the control group of 4°C (B), 14°C (C), 24°C (D), and UV (E). (F) qRT-PCR results of the control group of HgCl2 (G) and CdCl2 (H). Mean ± SE of three replicates of six individuals each. ∗P < 0.05, ∗∗P < 0.01 by the Student’s t-test. Yellow, red, and blue asterisk above the error bars indicate significant differences of AccDnaJA1, AccDnaJB12, and AccDnaJC8, respectively. The β-actin gene was used as an internal control. (I–U) Western blot analysis of AccDnaJA1 and AccDnaJB12. Equivalent quality of total protein from Apis cerana cerana was loaded for each sample. The target proteins were immunoblotted with anti-AccDnaJA1 or anti-AccDnaJB12. Tubulin was used as a control. (I) The control group of AccDnaJA1 under 4°C (J) and 24°C (K) treatment. (L) The control group of AccDnaJA1 under HgCl2 (M) and CdCl2 (N) treatment. (O) Western blot results of the control group of AccDnaJB12 under 4°C (P), 4°C (Q), and 24°C (R) treatment. (T) Western blot results of the control group of AccDnaJB12 under HgCl2 (U) treatment.
To investigate the protein levels of AccDnaJA1 under some stress conditions, western blot was carried out. The protein profiles of AccDnaJA1 were not altered under normal condition (Figure 1I), while were increased under 4°C stress (Figure 1J). The protein levels of AccDnaJA1 were downregulated under 24°C stress (Figure 1K), and were not obviously regulated under 14°C stress (Supplementary Figure S2). The expression levels of AccDnaJA1 were not changed when honeybees were injected sterilized water, while were inhibited under both HgCl2 and CdCl2 stress (Figures 3L–N), which was consistent with the results from qRT-PCR. These findings suggest that AccDnaJA1 may play a role in various stress conditions at different degrees.
Then, we used western blot to examine the protein levels of AccDnaJB12 under some stress conditions. The protein levels of AccDnaJB12 were nearly not altered under normal condition (Figure 1O). The expression levels of AccDnaJB12 were increased under treatment at 4, 14, and 24°C (Figures 1P–R). UV stress caused an increase in AccDnaJB12 levels at 3.0 and 4.0 h (Figure 1S). When injected sterilized water, the protein levels of AccDnaJB12 were not changed (Figure 1T), while injected with HgCl2 triggered a continuous upregulation of the protein levels of AccDnaJB12 from 1.0 to 4.0 h (Figure 1U). These findings indicate that AccDnaJB12 may play a pivotal role in some environmental stress response.
When collecting water, pollen and nectar, honeybees are often suffered from pesticides used in agriculture (Goulson et al., 2015). To explore the mRNA levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 compared to the β-actin gene under various pesticide stresses, qRT-PCR was performed. The mRNA levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 were not obviously changed under normal condition (Figure 2A). In response to lambda-cyhalothrin treatment, AccDnaJA1, AccDnaJB12, and AccDnaJC8 were all upregulated with the highest mRNA levels at 1.0, 1.5 and 1.5 h, respectively (Figure 2B). Both AccDnaJA1, and AccDnaJC8 were increased from 1 to 5 h under paraquat stress, and AccDnaJB12 was also upregulated from 1 to 5 h except at 2 h when honeybees were subjected to paraquat stress (Figure 2C). Compared to the effects of the above two pesticide stresses, emamectin benzoate had only a slightly influence on the transcriptional levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 (Figure 2D). As presented in Figure 2E, all three genes were induced with a peak at 2 h by spirodiclofen stress. Under avermectin stress, AccDnaJA1, AccDnaJB12, and AccDnaJC8 were all upregulated continuously from 1.0 to 4.5 h (Figure 2F).
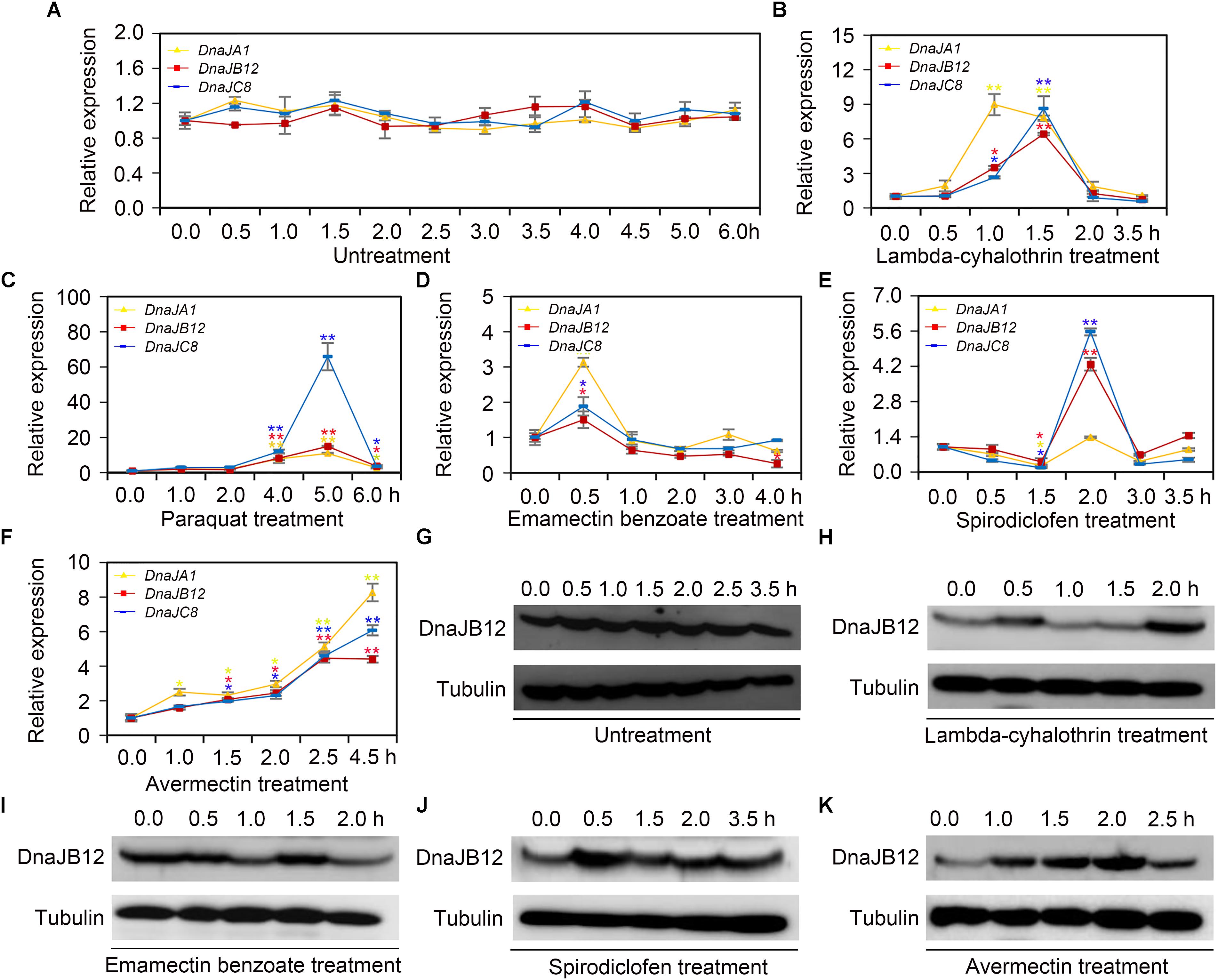
FIGURE 2. The relative expression of AccDnaJA1, AccDnaJB12, and AccDnaJC8 under several pesticide stresses. (A–F) The mRNA levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 under normal condition (A), and lambda-cyhalothrin (B), paraquat (C), emamectin benzoate (D), spirodiclofen (E), and avermectin (F) treatment. The expression patterns of AccDnaJA1, AccDnaJB12, and AccDnaJC8 were normalized to that of the β-actin gene. The data are shown as the mean ± SE from at least three replicates of six individuals each. Yellow, red, and blue asterisk above the error bars indicate significant differences of AccDnaJA1, AccDnaJB12, and AccDnaJC8, respectively (∗P < 0.05, ∗∗P < 0.01 by the Student’s t-test). (G–K) The protein levels of AccDnaJB12 under normal (G), lambda-cyhalothrin (H), emamectin benzoate (I), spirodiclofen (J), and avermectin (K) conditions. The control was tubulin.
Then we used western blot to detect the protein levels of AccDnaJB12 under some pesticide stresses. As shown in Figure 2G, the translation levels of AccDnaJB12 were not significantly regulated under normal condition from 0 h to 3.5 h, while were increased at 0.5 and 2.0 h under lambda-cyhalothrin stress (Figure 2H). When A. cerana cerana was exposed to emamectin benzoate stress, the AccDnaJB12 protein levels were repressed at 1.0 and 2.0 h (Figure 2I). Though the AccDnaJB12 protein levels were induced in response to both the spirodiclofen and avermectin treatments, it seemed that the upregulation varied by stress condition as spirodiclofen triggered maximum protein expression at 0.5 h and avermectin caused maximum protein expression at 2.0 h (Figures 2J,K).
qRT-PCR was used to verify the efficiency of RNAi-mediated gene silencing. After 2 days of feeding dsRNA AccDnaJA1, AccDnaJB12, or AccDnaJC8 to the honeybees, the transcriptional levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 compared to that of the β-actin gene were tested. As presented in Figure 3A, AccDnaJA was successfully silenced compared to the expression level in the control group fed dsRNA GFP. AccDnaJB12 and AccDnaJC8 were also knocked down compared with the expression levels in the dsRNA GFP-fed group (Figures 3B,C).
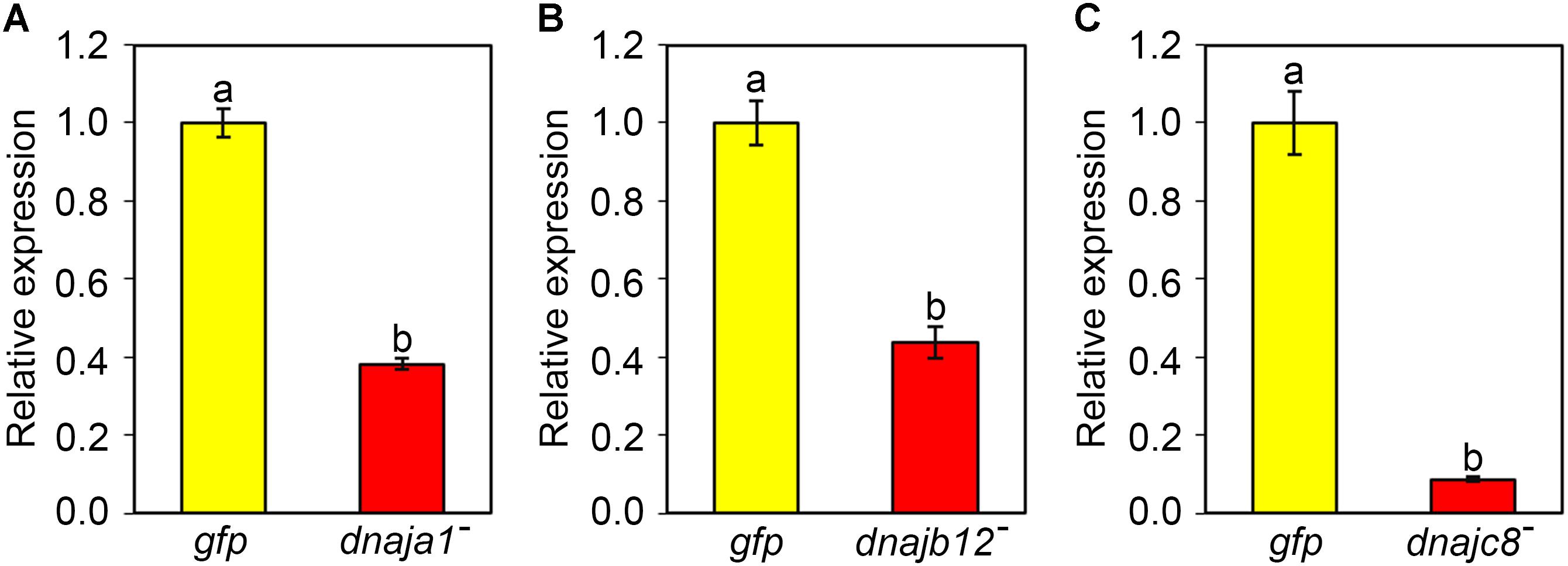
FIGURE 3. Efficiency of RNAi on the transcriptional levels of AccDnaJA1 (A), AccDnaJB12 (B), and AccDnaJC8 (C). For validation, total RNA was acquired from 15-day-old foragers after feeding with 7 μg dsRNA AccDnaJA1, dsRNA AccDnaJB12, dsRNA AccDnaJC8, or dsRNA GFP for 2 days. qRT-PCR was performed to examine the mRNA profiles of AccDnaJA1, AccDnaJB12, and AccDnaJC8 normalized against β-actin. The abbreviations dnaja1-, dnajb12-, dnajc8-, and gfp indicate AccDnaJA1 knockdown, AccDnaJB12 knockdown, AccDnaJC8 knockdown and GFP control, respectively. Mean ± SE of three replicates of six individuals each. Various letters above bars suggest significant differences between two groups (p < 0.01) based on Duncan’s multiple range tests.
The effects of RNAi of AccDnaJA1, AccDnaJB12, and AccDnaJC8 on the transcription levels of several stress response genes were investigated using qRT-PCR, and the β-actin gene was used as an internal control. As shown in Figure 4A, the mRNA levels of AccANT, AccCDK5r, AccCPR24, AccERR, AccTP1, AccCDK5, AccCYP336A1, AccRBM11, AccAK, and AccMTNR1A were inhibited when AccDnaJA1 was knocked down, while the transcriptional levels of AccSp10 were slightly upregulated. When AccDnaJB12 was silenced, the expression levels of AccANT, AccCPR24, AccERR, AccTP1, AccCDK5, AccRBM11, AccAK, and AccMTNR1A were downregulated, and the transcriptional levels of AccCYP336A1 were upregulated approximately 1.7-fold (Figure 4B). Knockdown of AccDnaJC8 decreased the expression profiles of AccANT, AccTP1, AccCDK5, AccSp10, AccRBM11, and AccAK (Figure 4C). These findings indicate that the mRNA profiles of several stress response genes are influenced by knockdown of AccDnaJA1, AccDnaJB12, and AccDnaJC8, and AccDnaJA1, AccDnaJB12, and AccDnaJC8 may play important roles in stress response.
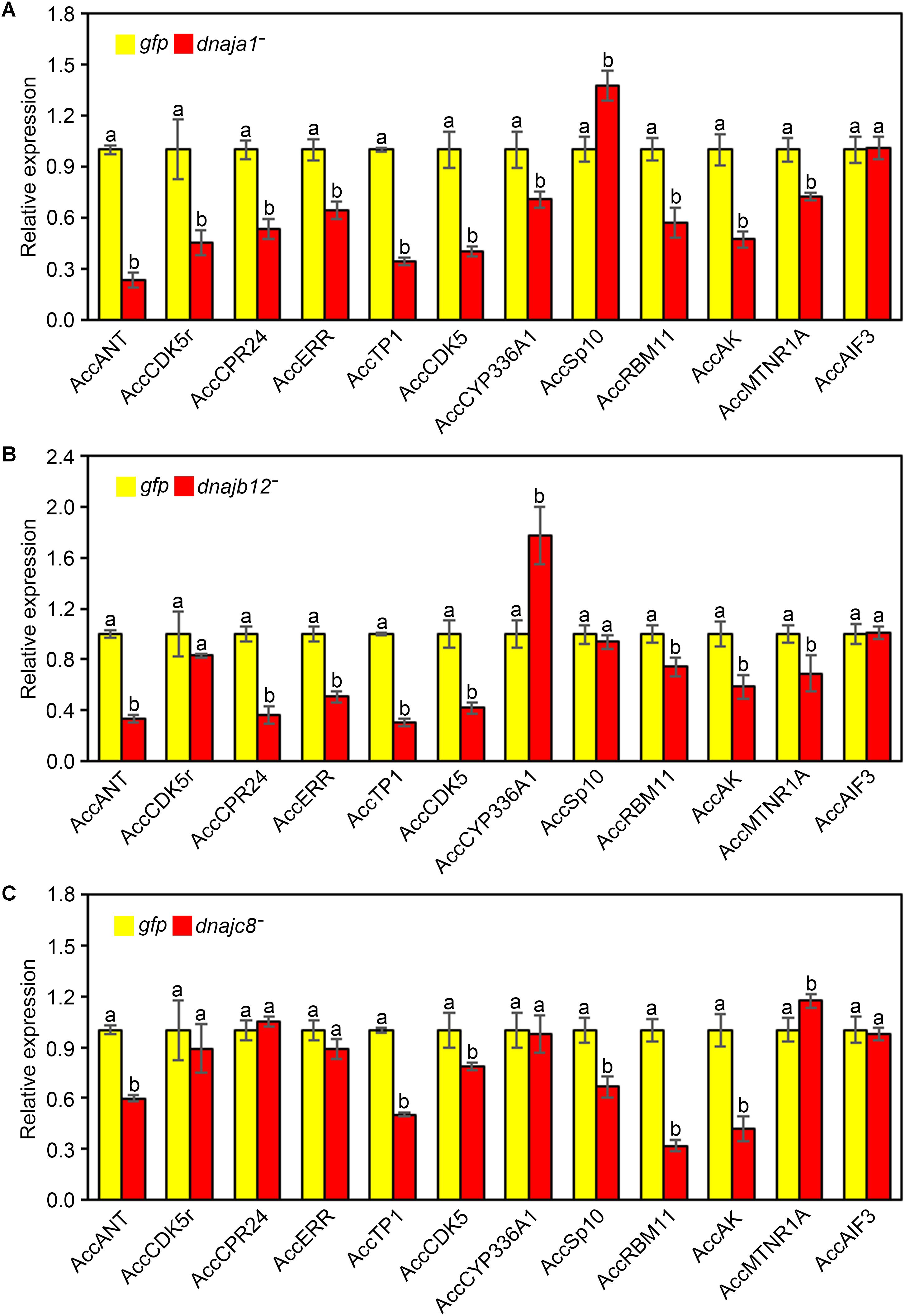
FIGURE 4. Effects of AccDnaJA1, AccDnaJB12, and AccDnaJC8 knockdown on the other stress response genes. Relative mRNA levels of several stress response genes were investigated by qRT-PCR compared to that of the β-actin gene when silencing AccDnaJA1 (A), AccDnaJB12 (B), and AccDnaJC8 (C). AccDnaJA1 knockdown, AccDnaJB12 knockdown, AccDnaJC8 knockdown, and GFP control were abbreviated as dnaja1-, dnajb12-, dnajc8-, and gfp, respectively. Mean ± SD are given (n = 3 with six individuals each). Significant differences between two groups were presented by various letters above the bars according to Duncan’s multiple range tests.
To further demonstrate the role of AccDnaJA1, AccDnaJB12, and AccDnaJC8 in the stress conditions, a functional analysis was performed using the RNAi experiment. After 2 days of feeding with dsRNA AccDnaJA1, AccDnaJB12, or AccDnaJC8, the honeybees were exposed to lambda-cyhalothrin stress, and we found that the removal of AccDnaJA1, AccDnaJB12, or AccDnaJC8 decreased the survival ability of A. cerana cerana to a certain degree compared with the dsRNA GFP treatment group (Figure 5). These results suggest that AccDnaJA1, AccDnaJB12, and AccDnaJC8 are involved in lambda-cyhalothrin tolerance in honeybee.
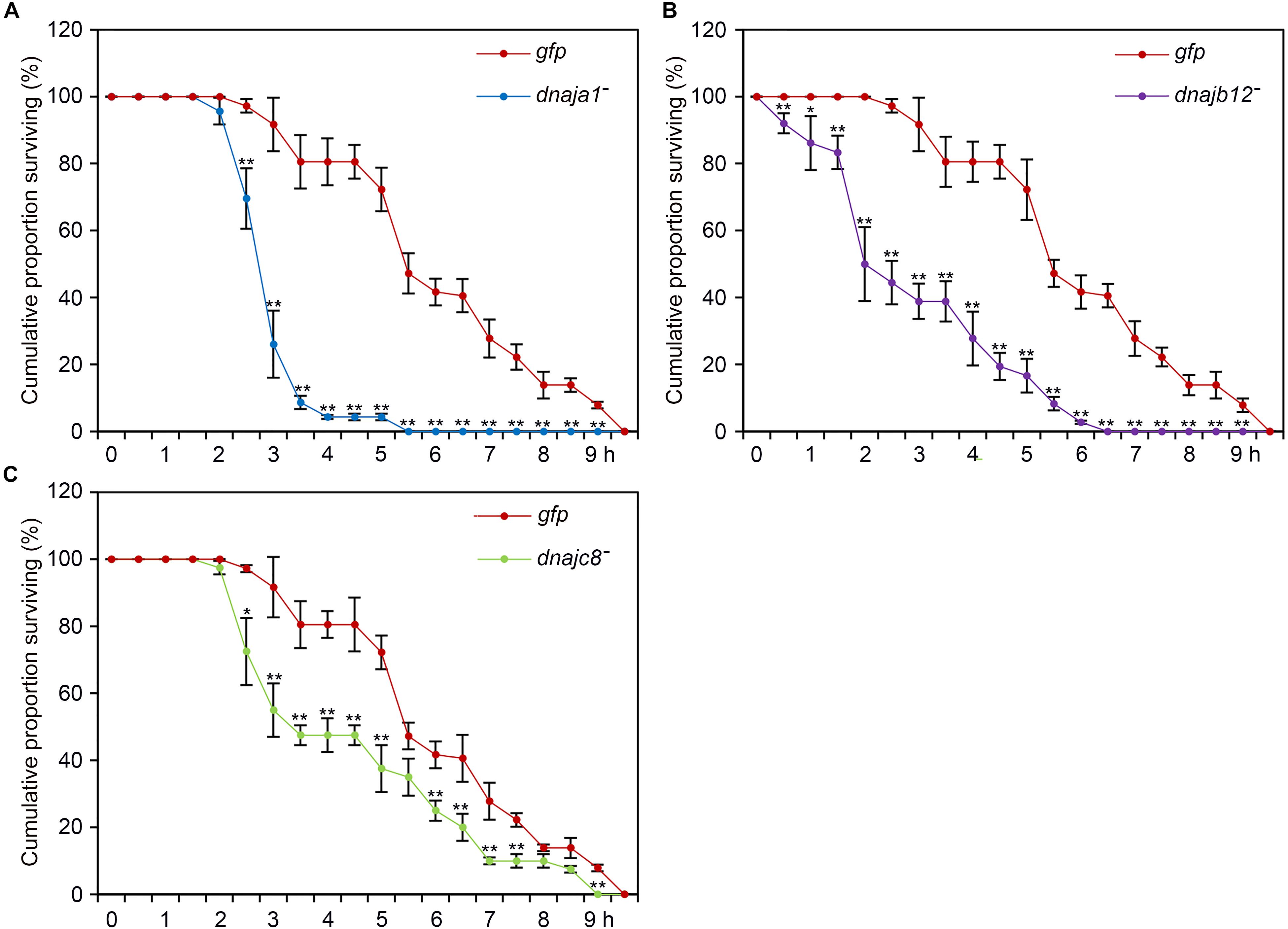
FIGURE 5. Lambda-cyhalothrin stress tolerance of Apis cerana cerana after RNAi AccDnaJA1 (A), AccDnaJB12 (B), and AccDnaJC8 (C). Survival rate of honeybees after a 10 h exposure to lambda-cyhalothrin when knockdown of AccDnaJA1, AccDnaJB12, and AccDnaJC8 was recorded. The abbreviations dnaja1-, dnajb12-, dnajc8-, and gfp indicate AccDnaJA1 RNAi, AccDnaJB12 RNAi, AccDnaJC8 RNAi, and GFP control, respectively. Mean ± SE of three replicates of 50 individuals each. The significant differences of AccDnaJA1, AccDnaJB12, and AccDnaJC8 were indicated by asterisk, respectively (∗P < 0.05 and ∗∗P < 0.01 by the Student’s t-test).
Heat shock proteins play roles as molecular chaperones in stress conditions by binding abnormally folded proteins, mediating the detrimental influence of misfolding and facilitating the return of these proteins to their native conformations at the time a favorable environment once again prevails (Feder and Hofmann, 1999; Rinehart et al., 2007). Though extensive studies have suggested that some members of the HSPs are generally expressed during periods of environmental stress, the expression regulation models of many DnaJ family genes are still unclear (Cheong et al., 2013; King and MacRae, 2015). In this study, we presented that the transcriptional levels of three DnaJ genes (AccDnaJA1, AccDnaJB12, and AccDnaJC8) were regulated under different environmental stresses, and western blot assays further supported our results. Furthermore, knockdown of AccDnaJA1, AccDnaJB12, or AccDnaJC8 inhibited the expression of several stress response-related genes and increased the mortality rate of honeybees under lambda-cyhalothrin treatment. Our results thus reveal that AccDnaJA1, AccDnaJB12, and AccDnaJC8 may play important roles in defense against adversity.
Accurate analysis of the role of a gene needs prediction of transcription factor binding ability to its regulatory regions. The 5′-flanking regions of AccDnaJA1, AccDnaJB12, and AccDnaJC8 included many binding sites for NF-κB, CREB, AP-1, and p53 (Table 1), all of which are well-known transcription factors related to stress response. For example, the stress response pathway TIF-IA-NF-κB plays important roles in human colorectal tumors exposed to aspirin and chemopreventive agents (Chen et al., 2018). The activation of the p38 MAPK-CREB signaling pathway under MeHg-induced oxidative stress is conducive to neuronal cell death and cerebrocortical neuronal hyperactivity (Fujimura and Usuki, 2018). Fra1, a member of the transcription factor family of AP-1, is associated with metal treatment response in Paracentrotus lividus embryos (Russo et al., 2018). In HepG2 cells, p53 can change the different secretome of the liver and can influence whole organism homeostasis and liver function under various stress stimuli (Charni-Natan et al., 2018). Therefore, AccDnaJA1, AccDnaJB12, and AccDnaJC8 may be involved in the stress response in A. cerana cerana.
Previous data had suggested that low temperature might cause chilling injury in insects induced by protein conformational changes and membrane phase transitions, and minor temperature changes triggering cold damage are possibly far less predictable (Ramlov, 2000; Sinclair et al., 2003). To cope with the injury from cold, insects use a series of strategies, including upregulated expression of HSPs (Rinehart et al., 2007). For example, Drosophila melanogaster Hsp90, Hsp83, Hsp68, Hsp40, Hsp27, Hsp26, Hsp23, and Hsp22 are cold inducible, and knockdown of Hsp23 and Hsp22 by RNAi perturbs D. melanogaster recovery from low-temperature injury (Colinet et al., 2010; Colinet and Hoffmann, 2012). In our study, under cold stress, AccDnaJA1, AccDnaJB12, and AccDnaJC8 were all increased at the transcriptional level or the protein level (Figures 1B–D). Honeybees usually congregate to acquire body heat to address cold stress at the behavioral level. The inducible expression of AccDnaJA1, AccDnaJB12, and AccDnaJC8 may be a kind of decision to defend against cold stress at the gene expression regulation level.
Recent studies demonstrated that heavy metal and UV stress significantly influenced HSP expression and thereby affected the fitness and resistance of a natural population of organisms (Sørensen et al., 2010; Chen and Zhang, 2015). The majority of sHSPs present expression variation in response to multiple heavy metals in Plutella xylostella (Chen and Zhang, 2015). Hsp70 displays particular sensitivity to zinc and iron exposure, while Hsp60 shows the most sensitive response to manganese and cadmium treatments in Litopenaeus vannamei (Qian et al., 2012). The transcription levels of Hsp27 are induced by UV stress in mouse skin tumors (Kiriyama et al., 2001). In addition, UV is likely to induce HSP expression, which may provide an adaptive response accompanied by increased exposure to UV radiation (Trautinger et al., 1996). In our study, AccDnaJA1 was suppressed by both HgCl2 and CdCl2 treatment, while AccDnaJB12 and AccDnaJC8 were induced by HgCl2 and CdCl2 treatment (Figures 1G,H). The western blot experiment further proved our results (Figures 1M,N). The mRNA levels of AccDnaJA1 and AccDnaJC8 were upregulated by UV stress (Figure 1E), and both the transcription and translation of AccDnaJB12 were increased under UV conditions (Figures 1E,S). These findings suggest that HSPs, at least AccDnaJA1, AccDnaJB12, and AccDnaJC8, are involved in heavy metal and UV stress.
Herbicides and insecticides are usually used in agriculture by humans to defend against ruderal or injurious insects. However, they also cause obvious damage to honeybees. Herbicides decrease honeybees’ food resource diversity, whereas insecticides directly kill honeybees by influencing their immunocompetence, behavior and antioxidant ability when interacting with different pathogens (Sanchez-Bayo et al., 2016; Li et al., 2018b). A previous study also proved that exposure to field-realistic doses of neonicotinoid pesticides decreased honeybee health in Canada’s corn-growing regions by using realistic experiments (Tsvetkov et al., 2017). Furthermore, recent studies reported that HSPs were involved in pesticide stress. The mRNA levels of Hsp27 are significantly increased when Chironomus riparius is exposed to triclosan and bisphenol stress (Martinez-Paz et al., 2014). The expression profiles of 13 sHSPs are increased under chlorfenapyr and beta-cypermethrin stress in P. xylostella (Chen and Zhang, 2015). Our results indicated that the mRNA levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 were all increased by pesticide stress (Figures 2B–F), and western blot results showed that the protein levels of AccDnaJB12 were upregulated under lambda-cyhalothrin, spirodiclofen, and avermectin stresses (Figures 2H,J,K). These results suggest that AccDnaJA1, AccDnaJB12, and AccDnaJC8 may play an essential role in resisting pesticide stress in honeybees.
Notably, the transcriptional levels and translational levels of and AccDnaJA1 AccDnaJB12 were not exactly the same under some stress conditions (Figures 1, 2). Differences in the transcriptional levels and translational levels of invE were also found by a previous study in Shigella sonnei (Mitobe et al., 2009). The following explanations may be considered for this difference. First, the transcription and translation of genes are regulated by different signaling pathways. The transcription of genes is mainly affected by core promoter and enhancer (Dikstein, 2011), and the translation of genes are governed by various factors, such as mRNA structure, RNA biding protein, 5′ or 3′ untranslated regions (Wang et al., 2015). For example, the interaction between HBV pregenomic RNA 5′ terminal redundancy and RNA-binding motif protein 24 inhibits the translation of core protein, while the interaction between RNA-binding motif protein 24 and the 3′ terminal redundancy increased HBV RNA stability (Yao et al., 2018). Second, multiple adverse circumstances can upregulate the mRNA levels of a gene but not enough to affect translation. Third, the protein can be degraded and accumulated in the body. For example, when the transcription of a gene is induced, the decreased protein expression levels may exist due to protein degradation.
RNAi is an effective tool for loss-of-function studies across eukaryotes, especially for organisms in which transgenosis is too hard to realize (Hannon, 2002; Fellmann and Lowe, 2014). Knockdown by RNAi also presents the importance of HSPs for the survival of insects in response to adverse circumstances (Rinehart et al., 2007; Fellmann and Lowe, 2014). To further demonstrate the connection of AccDnaJA1, AccDnaJB12, and AccDnaJC8 with pesticide stress, RNAi was used. Functional analysis proved that separate knockdown of AccDnaJA1, AccDnaJB12, and AccDnaJC8 decreased the survival rate of A. cerana cerana under lambda-cyhalothrin stress (Figure 5), suggesting that these three genes may protect honeybees from lambda-cyhalothrin stress to some extent. Furthermore, knockdown of AccDnaJA1, AccDnaJB12, or AccDnaJC8 decreased the transcriptional levels of several stress response-related genes (Figure 4). These results further indicate that these three DnaJ genes play an essential role during the stress response for A. cerana cerana.
It has been indicated that the roles of individual proteins from the same HSP subfamily often differ in a variety of environmental conditions (King and MacRae, 2015). In this paper, the differential expression of AccDnaJA1, AccDnaJB12, or AccDnaJC8 in different stressors may suggest their functional differences to some extent. The differences of specific molecular mechanisms of AccDnaJA1, AccDnaJB12, or AccDnaJC8 in response to adverse circumstances are likely to contribute to their differential expression under stress conditions. In addition, these three genes were induced or repressed at different levels under multiple environmental stresses (Figures 1, 2), indicating that their regulation roles in response various stressors might have distinction to a certain degree. Transgenic technology, RNA-seq and proteomics can be used for further investigating the reason of the differential expression of AccDnaJA1, AccDnaJB12, and AccDnaJC8 in environmental stress in the future.
Collectively, our work identified three DnaJ genes (AccDnaJA1, AccDnaJB12, and AccDnaJC8) in A. cerana cerana. The expression levels of AccDnaJA1, AccDnaJB12, and AccDnaJC8 in response to diverse environmental stresses indicate that these three genes may play key roles in stress-related defense mechanisms. Knockdown of these three genes repressed the mRNA levels of several stress response genes and increased the mortality rate of honeybees to a kind of pesticide (lambda-cyhalothrin) stress, which further demonstrated the roles of AccDnaJA1, AccDnaJB12, and AccDnaJC8 in antistress circumstances. These results may be helpful for further unraveling the roles of the insect DnaJ family in stressful environmental conditions in future.
XG and BX planned and supervised the experiments. GL, HaZ, XZ, and YZ performed the experiments. GL, HaZ, HuZ, and XY analyzed the data. GL, BX, and XG wrote the paper. All authors approved the final version of the manuscript.
This work was financially supported by the Funds of Shandong Province “Double Tops” Program, the Shandong Province Modern Agricultural Technology System Innovation Team Special Fund (No. SDAIT-24-04), the National Natural Science Foundation of China (No. 31572470), and the earmarked fund for the China Agriculture Research System (No. CARS-44).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00445/full#supplementary-material
FIGURE S1 | The expression of the recombinant AccDnaJA1 or AccDnaJB12. Recombinant AccDnaJA1 (A) and AccDnaJB12 (B) were separated by 12% SDS-PAGE. Lanes 1: protein molecular weight marker; Lanes 2–3: overexpression of pET-30a(+) and uninduced overexpression of pET-30a(+)-target protein, respectively; Lanes 4–9: induced overexpression target protein. The sites of target genes were marked with a red box.
FIGURE S2 | The protein levels of AccDnaJA1 under 14°C treatment. The target proteins were immunoblotted with anti-AccDnaJA1 and tubulin was used as a control.
TABLE S1 | The collection times after treatment for the experimental groups.
TABLE S2 | Primer sequences used in this study.
TABLE S3 | PCR amplification procedures used in this study.
Bagul, P. K., Deepthi, N., Sultana, R., and Banerjee, S. K. (2015). Resveratrol ameliorates cardiac oxidative stress in diabetes through deacetylation of NFkB-p65 and histone 3. J. Nutr. Biochem. 26, 1298–1307. doi: 10.1016/j.jnutbio.2015.06.006
Charni-Natan, M., Solomon, H., Molchadsky, A., Jacob-Berger, A., Goldfinger, N., and Rotter, V. (2018). Various stress stimuli rewire the profile of liver secretome in a p53-dependent manner. Cell Death Dis. 9:647. doi: 10.1038/s41419-018-0697-4
Chen, J., Lobb, I. T., Morin, P., Novo, S. M., Simpson, J., Kennerknecht, K., et al. (2018). Identification of a novel TIF-IA-NF-kappaB nucleolar stress response pathway. Nucleic Acids Res. 46, 6188–6205. doi: 10.1093/nar/gky455
Chen, X., Yao, P., Chu, X., Hao, L., Guo, X., and Xu, B. (2015). Isolation of arginine kinase from Apis cerana cerana and its possible involvement in response to adverse stress. Cell Stress Chaperones 20, 169–183. doi: 10.1007/s12192-014-0535-2
Chen, X., and Zhang, Y. (2015). Identification of multiple small heat-shock protein genes in Plutella xylostella (L.) and their expression profiles in response to abiotic stresses. Cell Stress Chaperones 20, 23–35. doi: 10.1007/s12192-014-0522-7
Cheong, K., Choi, J., Choi, J., Park, J., Jang, S., and Lee, Y. H. (2013). Eukaryotic DNAJ/K Database: a comprehensive phylogenomic analysis platform for the dnaj/k family. Genomics Inform. 11, 52–54. doi: 10.5808/GI.2013.11.1.52
Chow, K. C., and Tung, W. L. (1998). Overexpression of dnaK/dnaJ and groEL confers freeze tolerance to Escherichia coli. Biochem. Biophys. Res. Commun. 253, 502–505. doi: 10.1006/bbrc.1998.9766
Chu, X., Lu, W., Zhang, Y., Guo, X., Sun, R., and Xu, B. (2013). Cloning, expression patterns, and preliminary characterization of AccCPR24, a novel RR-1 type cuticle protein gene from Apis cerana cerana. Arch. Insect Biochem. Physiol. 84, 130–144. doi: 10.1002/arch.21132
Colinet, H., and Hoffmann, A. A. (2012). Comparing phenotypic effects and molecular correlates of developmental, gradual and rapid cold acclimation responses in Drosophila melanogaster. Funct. Ecol. 26, 84–93. doi: 10.1111/j.1365-2435.2011.01898.x
Colinet, H., Lee, S. F., and Hoffmann, A. (2010). Knocking down expression of Hsp22 and Hsp23 by RNA interference affects recovery from chill coma in Drosophila melanogaster. J. Exp. Biol. 213(Pt 24), 4146–4150. doi: 10.1242/jeb.051003
Craig, E. A., Huang, P., Aron, R., and Andrew, A. (2006). The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 156, 1–21.
Cunnea, P. M., Miranda-Vizuete, A., Bertoli, G., Simmen, T., Damdimopoulos, A. E., Hermann, S., et al. (2003). ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 278, 1059–1066. doi: 10.1074/jbc.M206995200
Dikstein, R. (2011). The unexpected traits associated with core promoter elements. Transcription 2, 201–206. doi: 10.4161/trns.2.5.17271
Even, N., Devaud, J. M., and Barron, A. B. (2012). General stress responses in the honey bee. Insects 3, 1271–1298. doi: 10.3390/insects3041271
Farooqui, T. (2014). Oxidative stress and age-related olfactory memory impairment in the honeybee Apis mellifera. Front. Genet. 5:60. doi: 10.3389/fgene.2014.00060
Feder, M. E., and Hofmann, G. E. (1999). Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282. doi: 10.1146/annurev.physiol.61.1.243
Fellmann, C., and Lowe, S. W. (2014). Stable RNA interference rules for silencing. Nat. Cell Biol. 16, 10–18. doi: 10.1038/ncb2895
Fujimura, M., and Usuki, F. (2018). Methylmercury induces oxidative stress and subsequent neural hyperactivity leading to cell death through the p38 MAPK-CREB pathway in differentiated SH-SY5Y cells. Neurotoxicology 67, 226–233. doi: 10.1016/j.neuro.2018.06.008
Gao, L., Wang, H., Liu, Z., Liu, S., Zhao, G., Xu, B., et al. (2017). The initial analysis of a serine proteinase gene (AccSp10) from Apis cerana cerana: possible involvement in pupal development, innate immunity and abiotic stress responses. Cell Stress Chaperones 22, 867–877. doi: 10.1007/s12192-017-0818-5
Goulson, D., Nicholls, E., Botias, C., and Rotheray, E. L. (2015). Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347:1255957. doi: 10.1126/science.1255957
Jiang, J., Maes, E. G., Taylor, A. B., Wang, L., Hinck, A. P., Lafer, E. M., et al. (2007). Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28, 422–433. doi: 10.1016/j.molcel.2007.08.022
Jiao, L., Fan, M., Hua, C., Wang, S., and Wei, X. (2012). Expression of DnaJ gene in Alicyclobacillus acidoterrestris under stress conditions by quantitative real-time PCR. J. Food Sci. 77, M446–M451. doi: 10.1111/j.1750-3841.2012.02790.x
Kim, Y. E., Hipp, M. S., Bracher, A., Hayer-Hartl, M., and Hartl, F. U. (2013). Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355. doi: 10.1146/annurev-biochem-060208-092442
King, A. M., and MacRae, T. H. (2015). Insect heat shock proteins during stress and diapause. Annu. Rev. Entomol. 60, 59–75. doi: 10.1146/annurev-ento-011613-162107
Kiriyama, M. T., Oka, M., Takehana, M., and Kobayashi, S. (2001). Expression of a small heat shock protein 27 (HSP27) in mouse skin tumors induced by UVB-irradiation. Biol. Pharm. Bull. 24, 197–200. doi: 10.1248/bpb.24.197
Kulhanek, K., Steinhauer, N., Rennich, K., Caron, D. M., Sagili, R. R., Pettis, J. S., et al. (2017). A national survey of managed honey bee 2015–2016 annual colony losses in the USA. J. Apic. Res. 56, 328–340. doi: 10.1080/00218839.2017.1344496
Li, G., Zhang, Y., Ni, Y., Wang, Y., Xu, B., and Guo, X. (2018a). Identification of a melatonin receptor type 1A gene (AccMTNR1A) in Apis cerana cerana and its possible involvement in the response to low temperature stress. Naturwissenschaften 105:24. doi: 10.1007/s00114-018-1546-0
Li, G., Zhao, H., Liu, Z., Wang, H., Xu, B., and Guo, X. (2018b). The wisdom of honeybee defenses against environmental stresses. Front. Microbiol. 9:722. doi: 10.3389/fmicb.2018.00722
Li, G., Jia, H., Wang, H., Yan, Y., Guo, X., Sun, Q., et al. (2016a). A typical RNA-binding protein gene (AccRBMA11) in Apis cerana cerana: characterization of AccRBM1111 and its possible involvement in development and stress responses. Cell Stress Chaperones 21, 1005–1019. doi: 10.1007/s12192-016-0725-1
Li, G., Zhao, H., Wang, H., Guo, X., Guo, X., Sun, Q., et al. (2016b). Characterization of a decapentapletic gene (AccDpp) from Apis cerana cerana and its possible involvement in development and response to oxidative stress. PLoS One 11:e0149117. doi: 10.1371/journal.pone.0149117
Liberek, K., Georgopoulos, C., and Zylicz, M. (1988). Role of the Escherichia coli DnaK and DnaJ heat shock proteins in the initiation of bacteriophage lambda DNA replication. Proc. Natl. Acad. Sci. U.S.A. 85, 6632–6636. doi: 10.1073/pnas.85.18.6632
Liu, X., Lu, W., Zhang, Y., Xu, B., and Guo, X. (2014). Cloning and characterization of an adenine nucleotide translocator gene in Apis cerana cerana, (Hymenoptera: Apidae). Appl. Entomol. Zool. 49, 77–88. doi: 10.1007/s13355-013-0227-x
Liu, Z., Xi, D., Kang, M., Guo, X., and Xu, B. (2012). Molecular cloning and characterization of Hsp27.6: the first reported small heat shock protein from Apis cerana cerana. Cell Stress Chaperones 17, 539–551. doi: 10.1007/s12192-012-0330-x
Liu, Z., Yao, P., Guo, X., and Xu, B. (2014). Two small heat shock protein genes in Apis cerana cerana: characterization, regulation, and developmental expression. Gene 545, 205–214. doi: 10.1016/j.gene.2014.05.034
Lourenço, A. P., Mackert, A., Cristino, A. D. S., and Simões, Z. L. P. (2008). Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39, 372–385. doi: 10.1371/journal.pone.0155640
Martinez-Paz, P., Morales, M., Martin, R., Martinez-Guitarte, J. L., and Morcillo, G. (2014). Characterization of the small heat shock protein Hsp27 gene in Chironomus riparius (Diptera) and its expression profile in response to temperature changes and xenobiotic exposures. Cell Stress Chaperones 19, 529–540. doi: 10.1007/s12192-013-0479-y
Meng, F., Zhang, L., Kang, M. J., Guo, X., and Xu, B. H. (2010). Molecular characterization, immunohistochemical localization and expression of a ribosomal protein L17 gene from Apis cerana cerana. Arch. Insect Biochem. Physiol. 75, 121–138. doi: 10.1002/arch.20386
Mitobe, J., Morita-Ishihara, T., Ishihama, A., and Watanabe, H. (2009). Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiol. 9:110. doi: 10.1186/1471-2180-9-110
Potts, S. G., Biesmeijer, J. C., Kremen, C., Neumann, P., Schweiger, O., and Kunin, W. E. (2010). Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353. doi: 10.1016/j.tree.2010.01.007
Qian, Z., Liu, X., Wang, L., Wang, X., Li, Y., Xiang, J., et al. (2012). Gene expression profiles of four heat shock proteins in response to different acute stresses in shrimp, Litopenaeus vannamei. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 156, 211–220. doi: 10.1016/j.cbpc.2012.06.001
Ramlov, H. (2000). Aspects of natural cold tolerance in ectothermic animals. Hum. Reprod. 15(Suppl. 5), 26–46. doi: 10.1093/humrep/15.suppl_5.26
Requena, J. M., Montalvo, A. M., and Fraga, J. (2015). Molecular chaperones of Leishmania: central players in many stress-related and -unrelated physiological processes. Biomed. Res. Int. 2015:301326. doi: 10.1155/2015/301326
Rinehart, J. P., Li, A., Yocum, G. D., Robich, R. M., Hayward, S. A., and Denlinger, D. L. (2007). Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc. Natl. Acad. Sci. U.S.A. 104, 11130–11137. doi: 10.1073/pnas.0703538104
Robertson, A. J., Trost, B., Scruten, E., Robertson, T., Mostajeran, M., Connor, W., et al. (2014). Identification of developmentally-specific kinotypes and mechanisms of varroa mite resistance through whole-organism, kinome analysis of honeybee. Front. Genet. 5:139. doi: 10.3389/fgene.2014.00139
Russo, R., Bonaventura, R., Chiaramonte, M., Costa, C., Matranga, V., and Zito, F. (2018). Response to metals treatment of Fra1, a member of the AP-1 transcription factor family, in P. lividus sea urchin embryos. Mar. Environ. Res. 139, 99–112. doi: 10.1016/j.marenvres.2018.05.003
Sanchez-Bayo, F., Goulson, D., Pennacchio, F., Nazzi, F., Goka, K., and Desneux, N. (2016). Are bee diseases linked to pesticides? - A brief review. Environ. Int. 8, 7–11. doi: 10.1016/j.envint.2016.01.009
Shen, Y., Meunier, L., and Hendershot, L. M. (2002). Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 277, 15947–15956. doi: 10.1074/jbc.M112214200
Sinclair, B. J., Vernon, P., Klok, C. J., and Chown, S. L. (2003). Insects at low temperatures: an ecological perspective. Trends Ecol. Evol. 18, 257–262. doi: 10.1016/j.jtherbio.2016.06.026
Sørensen, J. G., Kristensen, T. N., and Loeschcke, V. (2010). The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6, 1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x
Stark, J. L., Mehla, K., Chaika, N., Acton, T. B., Xiao, R., Singh, P. K., et al. (2014). Structure and function of human DnaJ homologue subfamily a member 1 (DNAJA1) and its relationship to pancreatic cancer. Biochemistry 53, 1360–1372. doi: 10.1021/bi401329a
Trautinger, F., Kindas-Mugge, I., Knobler, R. M., and Honigsmann, H. (1996). Stress proteins in the cellular response to ultraviolet radiation. J. Photochem. Photobiol. B 35, 141–148. doi: 10.1016/S1011-1344(96)07344-7
Tsunoda, T., and Takagi, T. (1999). Estimating transcription factor bindability on DNA. Bioinformatics 15, 622–630. doi: 10.1093/bioinformatics/15.7.622
Tsvetkov, N., Samson-Robert, O., Sood, K., Patel, H. S., Malena, D. A., Gajiwala, P. H., et al. (2017). Chronic exposure to neonicotinoids reduces honey bee health near corn crops. Science 356, 1395–1397. doi: 10.1126/science.aam7470
Vos, M. J., Hageman, J., Carra, S., and Kampinga, H. H. (2008). Structural and functional diversities between members of the human HSPB. Biochemistry 47, 7001–7011. doi: 10.1021/bi800639z
Wan, Q., Whang, I., and Lee, J. (2012). Molecular and functional characterization of HdHSP20: a biomarker of environmental stresses in disk abalone Haliotis discus discus. Fish Shellfish Immunol. 33, 48–59. doi: 10.1016/j.fsi.2012.03.034
Wang, Y., Brent, C. S., Fennern, E., and Amdam, G. V. (2012). Gustatory perception and fat body energy metabolism are jointly affected by vitellogenin and juvenile hormone in honey bees. PLoS Genet. 8:e1002779. doi: 10.1371/journal.pgen.1002779
Wang, F., Zhang, Y., Yao, P., Guo, X., Li, H., and Xu, B. (2014). Molecular identification and stress response of the apoptosis-inducing factor gene 3 (AccAIF3) from Apis cerana cerana. Apidologie 45, 685–700. doi: 10.1007/s13592-014-0285-2
Wang, Z., Sun, X., Zhao, Y., Guo, X., Jiang, H., Li, H., et al. (2015). Evolution of gene regulation during transcription and translation. Genome Biol. Evol. 7, 1155–1167. doi: 10.1093/gbe/evv059
Yao, P., Chen, X., Yan, Y., Liu, F., Zhang, Y., Guo, X., et al. (2014). Glutaredoxin 1, glutaredoxin 2, thioredoxin 1, and thioredoxin peroxidase 3 play important roles in antioxidant defense in Apis cerana cerana. Free Radic. Biol. Med. 68, 335–346. doi: 10.1016/j.freeradbiomed.2013.12.020
Yao, Y., Yang, B., Cao, H., Zhao, K., Yuan, Y., Chen, Y., et al. (2018). RBM24 stabilizes hepatitis B virus pregenomic RNA but inhibits core protein translation by targeting the terminal redundancy sequence. Emerg. Microbes Infect. 7:86. doi: 10.1038/s41426-018-0091-4
Yochem, J., Uchida, H., Sunshine, M., Saito, H., Georgopoulos, C. P., and Feiss, M. (1978). Genetic analysis of two genes, dnaJ and dnaK, necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol. Gen. Genet. 164, 9–14. doi: 10.1007/BF00267593
Zhang, Y., Liu, Y., Guo, X., Li, Y., Gao, H., Guo, X., et al. (2014). Shsp22.6, an intronless small heat shock protein gene, is involved in stress defence and development in Apis cerana cerana. Insect Biochem. Mol. Biol. 53, 1–12. doi: 10.1016/j.ibmb.2014.06.007
Zhang, Y. Y., Guo, X. L., Liu, Y. L., Liu, F., Wang, H. F., Guo, X. Q., et al. (2016). Functional and mutational analyses of an omega-class glutathione S-transferase (GSTO2) that is required for reducing oxidative damage in Apis cerana cerana. Insect Mol. Biol. 25, 470–486. doi: 10.1111/imb.12236
Zhao, G., Wang, C., Wang, H., Gao, L., Liu, Z., Xu, B., et al. (2018). Characterization of the CDK5 gene in Apis cerana cerana (AccCDK5) and a preliminary identification of its activator gene. Cell Stress Chaperones 23, 13–28. doi: 10.1007/s12192-017-0820-y
Zhou, Y., Wang, F., Liu, F., Wang, C., Yan, Y., Guo, X., et al. (2016). Cloning and molecular identification of triosephosphate isomerase gene from Apis cerana cerana, and its role in response to various stresses. Apidologie 47, 792–804. doi: 10.1007/s13592-016-0436-8
Keywords: Apis cerana cerana, environmental stresses, DnaJ, expression analysis, RNA interference, functional analysis
Citation: Li G, Zhao H, Zhang X, Zhang Y, Zhao H, Yang X, Guo X and Xu B (2018) Environmental Stress Responses of DnaJA1, DnaJB12 and DnaJC8 in Apis cerana cerana. Front. Genet. 9:445. doi: 10.3389/fgene.2018.00445
Received: 12 July 2018; Accepted: 14 September 2018;
Published: 08 October 2018.
Edited by:
Jianke Li, Institute of Apiculture Research (CAAS), ChinaReviewed by:
Angelique Christine Paulk, Harvard Medical School, United StatesCopyright © 2018 Li, Zhao, Zhang, Zhang, Zhao, Yang, Guo and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingqi Guo, eHFndW9Ac2RhdS5lZHUuY24= Baohua Xu, Ymh4dUBzZGF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.