
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 20 September 2018
Sec. Nutrigenomics
Volume 9 - 2018 | https://doi.org/10.3389/fgene.2018.00399
This article is part of the Research TopicRegulation and Manipulation of Nutrient-Controlling Genes in CropsView all 6 articles
Rapeseed (Brassica napus L.) is a vital oil crop worldwide. High oleic acid content is a desirable quality trait for rapeseed oil, which makes it more beneficial to human health. However, many germplasm resources with high oleic acid content in rapeseed have not been evaluated with regard to their genotypes, making it difficult to select the best strains with this trait for the breeding of high oleic acid rapeseed variety. This work was to explore the gene-regulation mechanism of this trait using a new super-high oleic acid content (∼85%) line N1379T as genetic material. In this study, the sequences of four homologous fatty acid desaturase (BnFAD2) genes were compared between super-high (∼85%, N1379T) and normal (∼63%) oleic acid content lines. Results showed that there were two single-nucleotide polymorphisms (SNPs) in BnFAD2-1 and BnFAD2-2, respectively, which led to the amino acid changes (E106K and G303E) in the corresponding proteins. Functional analysis of both genes in yeast confirmed that these SNPs were loss-of-function mutations, thus limiting the conversion of oleic acid to linoleic acid and resulting in the considerable accumulation of oleic acid. Moreover, two specific cleaved amplified polymorphic sequences (CAPS) markers for the two SNPs were developed to identify genotypes of each line in the F2 and BC1 populations. Furthermore, these two mutant loci of BnFAD2-1 and BnFAD2-2 genes were positively associated with elevated oleic acid levels and had a similar effect with regard to the increase of oleic acid content. Taken together, these two novel SNPs in two different BnFAD2 genes jointly regulated the high oleic acid trait in this special germplasm. The study provided insight into the genetic regulation involved in oleic acid accumulation and highlighted the use of new alleles of BnFAD2-1 and BnFAD2-2 in breeding high oleic acid rapeseed varieties.
Rapeseed (Brassica napus L.) is one of the most important oil crops across the world. The annual production of rapeseed oil has reached 60–70 million tons, ranks just behind that of soybeans globally (Khan et al., 2018). Similar to other vegetable oils, the fatty acid composition of rapeseed oil is the key trait involved in its utilization mode and range (Napier et al., 2014). After the double-low(low erucic acid and glucosinolates) improvement, rapeseed oil now normally contains ∼7% saturated acids (including palmitic acid C16:0) and stearic acid C18:0), ∼61% oleic acid (C18:1), ∼11% linoleic acid (C18:2), and ∼21% linolenic acid (C18:3) (Sharafi et al., 2015). However, rapeseed oil with higher oleic acid (>75%) has several advantages compared with non-high oleic acid varieties: (1) its anti-oxidative ability provides a longer shelf life, eliminating the need for a hydrogenation process (Browse et al., 1998; Lauridsen et al., 1999; Przybylski et al., 2013); (2) it can decrease low-density lipoprotein levels in humans and then decrease the risk of cardiovascular disease (Chang and Huang, 1998; WHO, 2003); and (3) it is a more cost-effective alternative to olive oil in daily cooking processes, while having a similar function, fatty acid composition, and taste. Therefore, increasing oleic acid content in rapeseed oil has been one of the major objectives for researchers focus on improving rapeseed oil quality.
The fatty acid biosynthesis pathway is a primary metabolic pathway in oil-bearing plants (Ohlrogge and Browse, 1995). Acetyl-CoA is the basic building block of the fatty acid chain and enters the cycle of reduction, dehydration, and reduction to synthesize 16- or 18-carbon products, which are the major (up to 90%) fatty acids of plants. Various desaturases locate in the plastids and the endoplasmic reticulum are responsible for catalyzing these fatty acids to become monounsaturated (palmitoleic acid, C16:1, and C18:1) or polyunsaturated ones (C18:2 and C18:3). Delta-12 fatty acid desaturase 2 (Δ12-FAD2) converts oleic acid precursors to linoleic acid precursors in the lipid biosynthetic pathway (Somerville and Browse, 1991; Ohlrogge and Browse, 1995). The first plant FAD2 gene was identified in Arabidopsis (Okuley et al., 1994) and was soon followed by its discovery in other crops, including soybean (Heppard et al., 1996), sunflower (Hongtrakul et al., 1998), peanut (López et al., 2000), cotton (Pirtle et al., 2001), and maize (Mikkilineni and Rocheford, 2003). The FAD2 gene family is quite large and diverse in plants (Dehghan and Yarizade, 2014). In rapeseed, there are four homologous FAD2 genes (BnFAD2-1, BnFAD2-2, BnFAD2-3, and BnFAD2-4) that are separately distributed on chromosomes A5, C5, A1, and C1 (also named as Bna.A5.FAD2, Bna.C5.FAD2, Bna.A1.FAD2, and Bna.C1.FAD2, respectively). Notably, BnFAD2-1 and BnFAD2-2 are constitutively expressed in all tissues, while BnFAD2-3 and BnFAD2-4 are specifically expressed in the seeds and roots. There are no introns in the BnFAD2-expressing code sequences and they share high identity (>99%) in both their DNA and protein sequences, except for BnFAD2-3. This exception is not altogether surprising as BnFAD2-3 is a pseudogene whose coding sequence is terminated by a stop codon resulting in a frame-shift starting at the 164-bp position (Lee et al., 2013).
The four to six distinct BnFAD2 gene loci were first found in 1997 (Scheffler et al., 1997). Since the close markers linked to these loci in a segregation population were identified, the possible role of BnFAD2 manipulation in oleic acid regulation was considered in rapeseed (Schierholt et al., 2000). Although various high oleic acid (>75%) rapeseed germplasms have been created via mutagenesis (Auld et al., 1992; Schierholt et al., 2001; Hu et al., 2006; Spasibionek, 2006; Yang et al., 2012), only a few have been evaluated at the molecular level. A quantitative trait locus (QTL) was mapped on chromosome N1 (referring to BnFAD2-4 on chromosome C1) in a double haploid (DH) population derived from the DSM100 strain (77% C18:1) (Hu et al., 2006). A single-nucleotide mutation from C to T was identified after sequence analysis of BnFAD2-4, and the allelic markers developed from this mutation were highly correlated to oleic acid content. A similar study was conducted with the high oleic acid variety SW Hickory (78% C18:1) and a 4-bp insertion in the BnaA.FAD2.a sequence (referring to BnFAD2-1 on chromosome A5) was identified that resulted in misreading and premature termination of translation at the 667–669 bp (TGA) position (Yang et al., 2012). Moreover, an ethyl methanesulfonate (EMS) mutagenized population with varying oleic acid content was also established using the high oleic acid (∼75%) cultivar Cabriolet. Some lines in this population were shown to have the nucleotide deletion in BnaA.FAD2.a (again referring to BnFAD2-1 on chromosome A5) as well as other mutations in BnaC.FAD2.a (referring to BnFAD2-2 on chromosome A1) (Wells et al., 2014). Some other loci (except BnFAD2) on genome were also found to have effect on the oleic acid content in rapeseed by the QTL analysis and genome-wide association study (GWAS) methods (Wang et al., 2015; Gacek et al., 2016; Huang et al., 2016;Javed et al., 2016; Qu et al., 2017), but with no further results. In addition, several high oleic acid rapeseed lines have also been created by gene transferring (Peng et al., 2010; Jung et al., 2011; Lee et al., 2016).
We had developed a mutant rapeseed population and found lines with elevated oleic acid content. A stable super-high oleic acid (∼85%) germplasm named N1379T had been screened out. In present study, the BnFAD2 genes of rapeseed lines with high- and normal-oleic acid content were cloned and two single-nucleotide polymorphisms (SNPs) in BnFAD2 genes in the high oleic acid rapeseed line N1379T were identified. The functions of these mutational alleles were also confirmed in yeast. Additionally, the association between genotypes and oleic acid contents was explored. This study would enrich the knowledge of genetic mechanisms of high oleic acid content and facilitate breeding high oleic acid varieties in rapeseed.
The three double-low rapeseed (B. napus L.) lines used in this study were provided by the Jiangsu Academy of Agriculture Sciences (JAAS). N1379T, a stable super-high oleic acid content (∼85%) line, was developed through pedigree selection from an EMS-treated mutagenesis population. The selection procedure was briefly described as follows: plants of the first three generations (M0–M2) from this population underwent open pollination in a greenhouse and lines with the top 50 oleic acid contents were chosen as materials of the next generation. Lines with >80% oleic acid content in generation M3 were performed the microspore culture at the flowering stage and a DH line named N1379T with 85% oleic acid content was selected based on the agronomic traits. Two self-pollinated lines with different genetic backgrounds, 16wh53 and 16wh62, were used as the normal oleic acid content (∼63%) lines.
Two crosses were made to produce the F1 generation (N1379T × 16wh53 and 16wh62 × N1379T) at the experimental field of JAAS (Nanjing, China; 32°02′03″N, 118°52′19″E) in the spring of 2014. The three parents and two F1s were planted independently at the same site in Nanjing as well as in Wuhan at the research field of the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences (Wuhan, China; 32°34′57″N, 114°20′11″E) in the autumn of 2014. The two F2 populations were constructed by self-pollination of the F1 lines from the N1379T × 16wh53 (designated F2-1) and 16wh62 × N1379T (designated F2-2) crosses at the two separate sites in the spring of 2015. At the same time, two backcrossed (BC1) populations were produced by crossing two F1s (as the female parent) separately with 16wh53 and 16wh62 (designated BC1-1 and BC1-2, respectively) at the two locations.
More than 200 seeds were collected from each population and were used for half-seed analysis following the method outlined by Downey and Harvey (1963). Briefly, for a single seed, one cotyledon inside the seed was cut to detect fatty acid composition, while the other cotyledon containing the embryo was left to germinate in a control room. They were then transferred to the field to continue growing as a single F2 or BC1 plant. All the materials planted in the field were managed carefully.
Genomic DNA was extracted from fresh leaves at the seedling stage of the three parent lines as well as each F1, F2, and BC1 line using the protocol described by Sambrook and Russell (2001). DNA concentration was determined for each sample using a Nanodrop 2000 (Thermo Scientific Company, United States) and then diluted to 50 ng/μl in ddH2O.
Young seeds of N1379T and 16wh53 were taken from 30-day-old siliques after pollination and put into liquid N2. The frozen seeds were grinded into powders and RNA was isolated using a RNA extraction Kit Promega Z3100 (Promega, United States) according to the manufacturer’s instructions. The quality and purity of the RNA was evaluated with a Bioanalyzer 2100 and RNA 6000 Nano Lab Chip Kit (Agilent, Santa Clara, CA, United States). For each sample, about 50 ng of purified RNA was used for cDNA synthesis following the detailed protocol of PrimeScriptTM 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). The cDNA products were stored until use.
Four BnFAD2 homolog gene sequences (JN992606, JN992607, JN992608, and JN992609, corresponding to BnFAD2-1, BnFAD2-2, BnFAD2-3, and BnFAD2-4), were downloaded from1 GenBank. By comparing these sequences in DNAMAN 6.0 (Lynnon Biosoft, United States), the specific primers P1 and P2 (Table 1) were designed by Primer 5.0 software (PREMIER Biosoft, United States) and used to amplify all of the homologous BnFAD2 genes. For the PCR assay, 1 μl of cDNA was used as the template in 20-μl reaction mixtures containing 1× buffer mix with 0.5 U of pfu enzyme as well as 0.25 μM of each primer (Transgene, China). The PCR conditions were as follows: initial cycle of 4 min at 95°C; followed by 35 cycles of denaturation at 94°C for 30 s, primer annealing at 59°C for 30 s, and extension at 72°C for 1 min, and finally, a single extension cycle of 10 min at 72°C. The amplification products were size-separated on a 1.5% (M/V) agarose gel. The objective bands (∼1.1 kb) were excised and purified using a DNA Gel Extraction Kit (Transgene, China). The products were then ligated separately into pGEM-T vectors for cloning. About 200 single positive colonies for each template were chosen for sequencing (Tsingke Company, Nanjing, China). All of the BnFAD2 sequences of N1379T and 16wh53 were arranged into four groups according to previous results (Lee et al., 2013). Four BnFAD2 homolog genes were downloaded from the reference genome (ZS112). Sequences within the same groups were compared using DNAman 6.0. Each clone was sequenced twice using the forward and reverse primers. Only clones with the same sequences from both primers were analyzed. Nucleotide mutations were recorded only if they appeared more than five times in one group. The DNA sequences were then translated into their amino acids sequences, and a similar comparison was performed with these amino acid sequences.
Allelic primers (HO10, HO11, and HO12) were designed to clone the BnFAD2-1 SNP-containing gene fragments. The HO10–HO12 pair was used for the wild-type (WT) G316, while the HO11–HO12 pair was used for the mutant G316A. Similarly, allelic primers (HO13, HO14, and HO15) were designed for the BnFAD2-2 SNP, with the HO13–HO15 pair being used for the WT G908, and the HO14–HO15 pair being used for the mutant G908A. The primer sequences are shown in Table 1. The genome DNAs from 22 varieties of rapeseed (Supplementary Table S1) in China were used as the PCR template. The PCR assay was conducted in the same manner as that described above, except the annealing temperature, which was 59°C in this analysis. The amplification products were size separated on a 1.5% (M/V) agarose gel and then imaged (Universal Hood II, Bio-rad).
The WT and corresponding mutant genes were enzyme-digested from the pGEM-T vectors and ligated into the expression vector pYES2 (Invitrogen, United States), which were then transformed into the S. cerevisiae strain INVSc1 (Invitrogen) using the electro-stimulation method (Downey and Harvey, 1963). Positive transformants (at least three for each gene) were selected on medium plates lacking uracil (SC-Ura) and were then inoculated in 50-ml SC-Ura liquid media to grow. At the logarithmic phase, the cells were harvested by centrifugation and washed three times with sterile distilled water. The cells were freeze-dried until fatty acid determination.
To amplify the BnFAD2-1 or BnFAD2-2 gene fragments containing the SNP sites, specific primer pairs were designed in Primer 5.0 by utilizing the differences among the four homologous genes. The primer pairs P3–P4 and P5–P6 (Table 1) were used to amplify the pure BnFAD2-1 or BnFAD2-2 gene fragments, respectively. The PCR conditions were the same as those described above. The restriction endonuclease sites on the prospective PCR products were identified using the online tool NEBcutter V2.03. All of the PCR products were analyzed for cleaved amplified polymorphic sequences (CAPS). Enzymatic digestions using BssSαI and HinfI (NEB company) were performed at 37°C in a water-bath for one hour in 20 μl reaction mixtures containing 5 μl of the PCR product, 2 μl of the provided enzyme buffer (10 × ), 0.5 μl of enzyme (10 U), and 12.5 μl ddH2O. After digestion, the products were visualized on 2% agarose gels in 1× TAE buffer. When the band type was the same as the WT, its genotype was marked as uppercase AA or BB (for BnFAD2-1 and BnFAD2-2, respectively). When a mutant type was detected, it was marked as lowercase aa or bb. Furthermore, when both genotypes were detected, they were marked as Aa or Bb.
Determination of the fatty acid profile in the various yeast strains was performed as follows: the yeast powder (0.05 g) of each strain was mixed with 3 ml of KOH–methanol in the 10-ml tubes and incubated at 70°C for 3–5 h. Then, 2 ml of boron trifluoride–methanol solution was added to the mixture, followed by incubation at 70°C for 1.5 h (after adjusting the pH values to 2.0) to achieve methylation. The fatty acid methyl esters (FAMEs) were recovered by mixing with 3-ml n-hexane. The upper liquid layer was isolated for gas chromatography (GC) analysis (Agilent 6400, Redwood City, CA, United States). For determination of the fatty acid profile in the half-seed, the cotyledon sample was crushed using a glass rod and added to a 500-μl mixture of ether: petroleum ether (3:2) at room temperature for 4 h. Then, 250 μl of KOH-methanol was added and incubated at room temperature for 2 h. Tubes were shaken for 1 min after 500 μl of H2O was added. Subsequently, 100 μl of the supernatant was pipetted out for GC analysis. Fatty acids were identified by comparing their retention times against the FAME standards (Sigma Chemicals Co., St. Louis, MO, United States) separated on the same GC machine. The seeds collected from parental lines and each line of the populations were used for oil content analysis by near-infrared spectroscopy.
The data were tabulated in Microsoft Excel and are presented as the means and standard deviations for each diverse rapeseed line (parental, F1, F2, and BC1) at both locations. By following the method of Eshed and Zamir (1996), the additive effect (A) and dominance effect (D) of BnFAD2-1 and BnFAD2-2 in F2 populations were calculated as: A = (phenotype value of mutant homozygotes - phenotype value of WT homozygotes)/2; D = (phenotype value of heterozygotes - (phenotype value of mutant homozygotes + phenotype value of WT homozygotes)/2; and the epistasis effect (I) of two genes was: I = [(phenotype value of aabb genotypes + phenotype value of AABB genotypes) - (phenotype value of aa genotypes + phenotype value of bb genotypes)]/2. Comparisons of the oleic acid content of different genotypes were conducted using one-way ANOVA in Graphpad Prism 7 software (Graphpad Prism Software Inc., San Diego, CA, USA). A p-value less than 0.05 was considered significant. A box-and-whiskers plot was made to show the distribution (min, first quartile, median, third quartile, and max) of the oleic acid content under different genotypes in Graphpad Prism 7.
The sequences of cloned BnFAD2 genes were compared with those from GenBank and from the reference genome. Results showed that the cloned sequences and those from GenBank and reference genome were highly similar and all four BnFAD2 genes shared identical start and termination coding fragments, which allowed us to design the P1-P2 primer pair to clone all of the homologous BnFAD2 genes (Figure 1A; Supplementary Table S2). The positive transformants, 206 and 211, in the T/A clones derived from N1379T and 16wh53, respectively, were sequenced. The BnFAD2 gene sequences of the lines (N1379T/16wh53) were divided into the four groups corresponding to BnFAD2-1 (48/55 sequences), BnFAD2-2 (45/50 sequences), BnFAD2-3 (40/44 sequences), and BnFAD2-4 (73/62 sequences). No sequential differences were found for the BnFAD2-3 and BnFAD2-4 groups between N1379T and 16wh53. However, two SNPs, G316A in the BnFAD2-1 group and G908A in the BnFAD2-2 group, were detected by comparing the two rapeseed lines. These DNA mutations resulted in the amino acid changes E106K and G303E, respectively (Figure 1B).
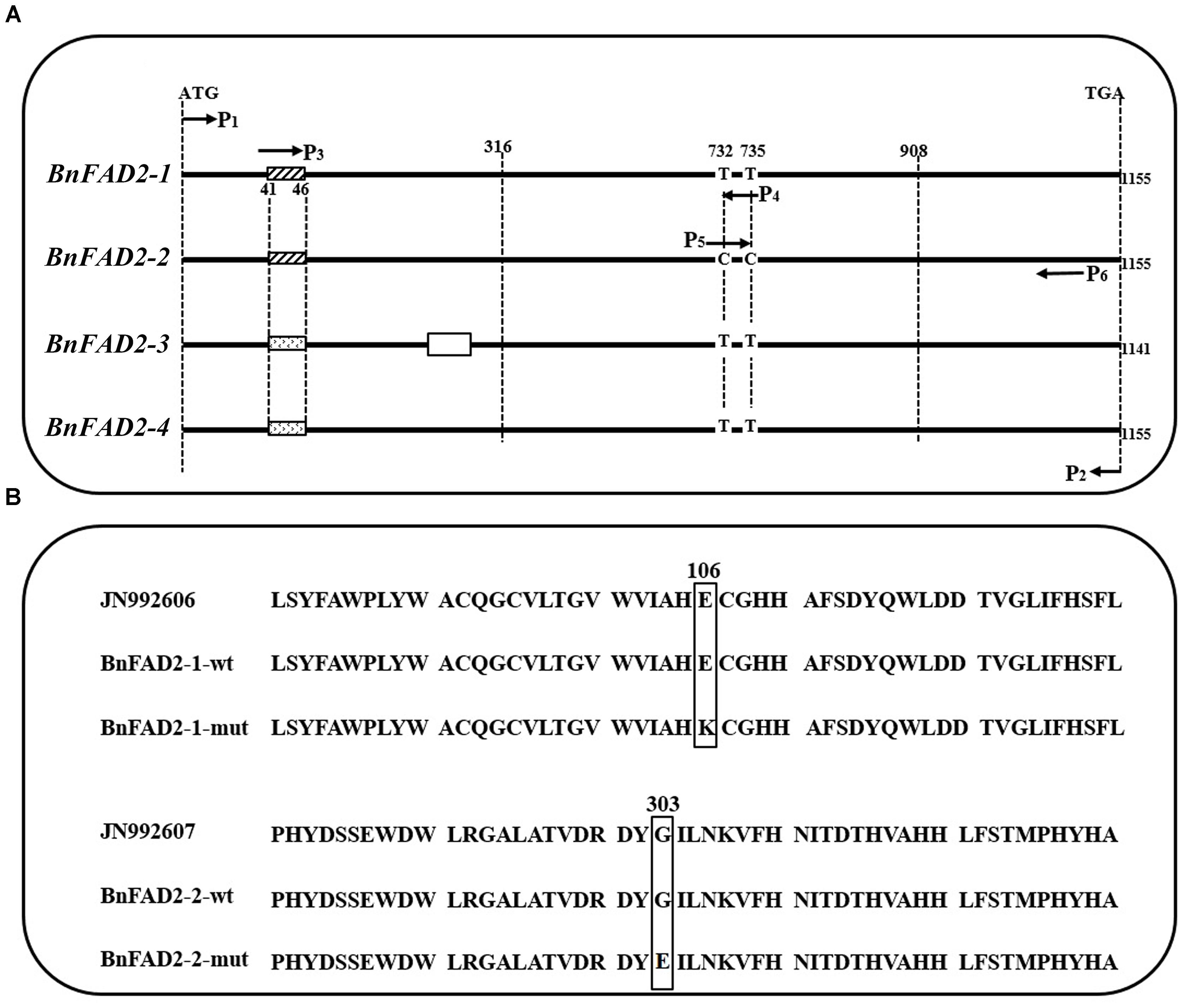
FIGURE 1. Comparison of the four BnFAD2 groups and partial alignments of BnFAD2-1 and BnFAD2-2. (A) Sequence comparison highlighting the differences between BnFAD2-1, BnFAD2-2, BnFAD2-3, and BnFAD2-4. The white box in BnFAD2-3 indicates the missing 15-bp nucleotide sequence; the differently shaded boxes at 41–46 denote nucleotide difference. The BnFAD2 homolog genes have characteristic nucleotides at sites 732 and 735. ATG is the start codon; TGA, the stop codon. (B) Amino acid sequence alignment of the BnFAD2-1 and BnFAD2-2 peptides. The mutant amino acids are bracketed in square frames. The numbers represent the number of residues from the initiation amino acid Met. JN992606 and JN992607 were used as representative sequences (downloaded from GenBank). wt, wild-type; mut, mutant.
Allelic primers were designed to detect these two novel SNPs (G316A and G908A) in 22 registered rapeseed varieties. The HO10–HO12 primer pair (specific to WT G316) amplifies a band for all of the samples, while the HO11–HO12 primer pair (specific to mutant G316A) only amplifies a band from N1379T (Figure 2). Similarly, the HO13–HO15 (WT G908) and HO14–HO15 (mutant G908A) primer pairs identified the mutant only in the high oleic acid line N1379T.
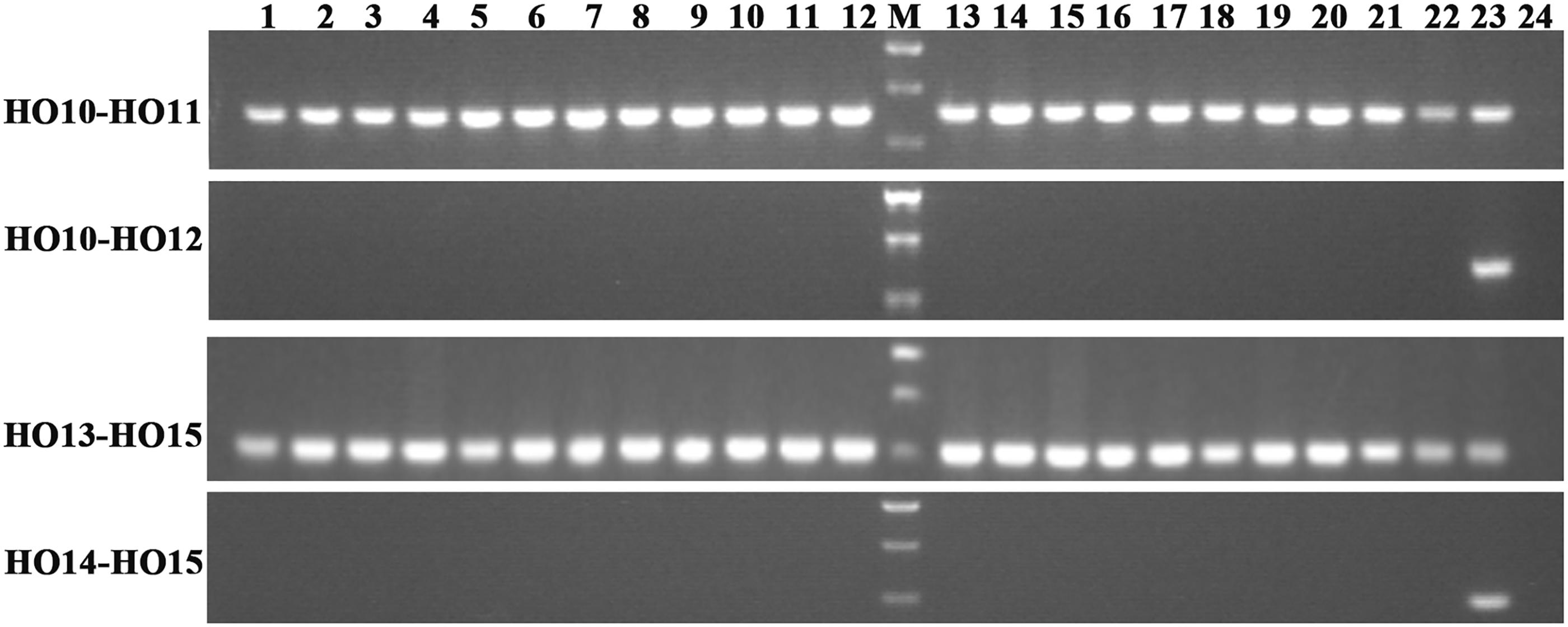
FIGURE 2. Amplification in registered rapeseed varieties using different primer combinations. Lanes 1–22 refer to the numbered varieties listed in Supplementary Table S2. Lane 23, N1379T; Lane 24, H2O (negative control). M, DNA marker. There were three standard bands in the M lane representing the 750, 500, and 250 bp band sizes.
To understand the function of the mutated BnFAD2-1 and BnFAD2-2 (designated BnFAD2-1-mut and BnFAD2-2-mut) genes found in N1379T, these genes and their corresponding WT genes were transformed into the S. cerevisiae strain INVSc1. Their function was evaluated by analyzing the fatty acid profile of the yeast strains. The GC analysis indicated that there was a linoleic acid (C18:2) peak detected in the S. cerevisiae strains transformed with the BnFAD2-1-wt or BnFAD2-2-wt gene (9.8 and 10.07% of the total fatty acid content, respectively) (Figure 3, details of the fatty acid compositions are shown in Supplementary Table S3). However, this peak was not observed in strains transformed with control vector (pYES2), BnFAD2-1-mut, or BnFAD2-2-mut. Additionally, in the BnFAD2-1-wt or BnFAD2-2-wt strains, the palmitoleic acid (C16:1) content decreased from 37 to 7% and the oleic acid (C18:1) content increased from 32 to 48%, while the BnFAD2-1-mut and BnFAD2-2-mut strains had the same fatty acid compositions as the control strains.
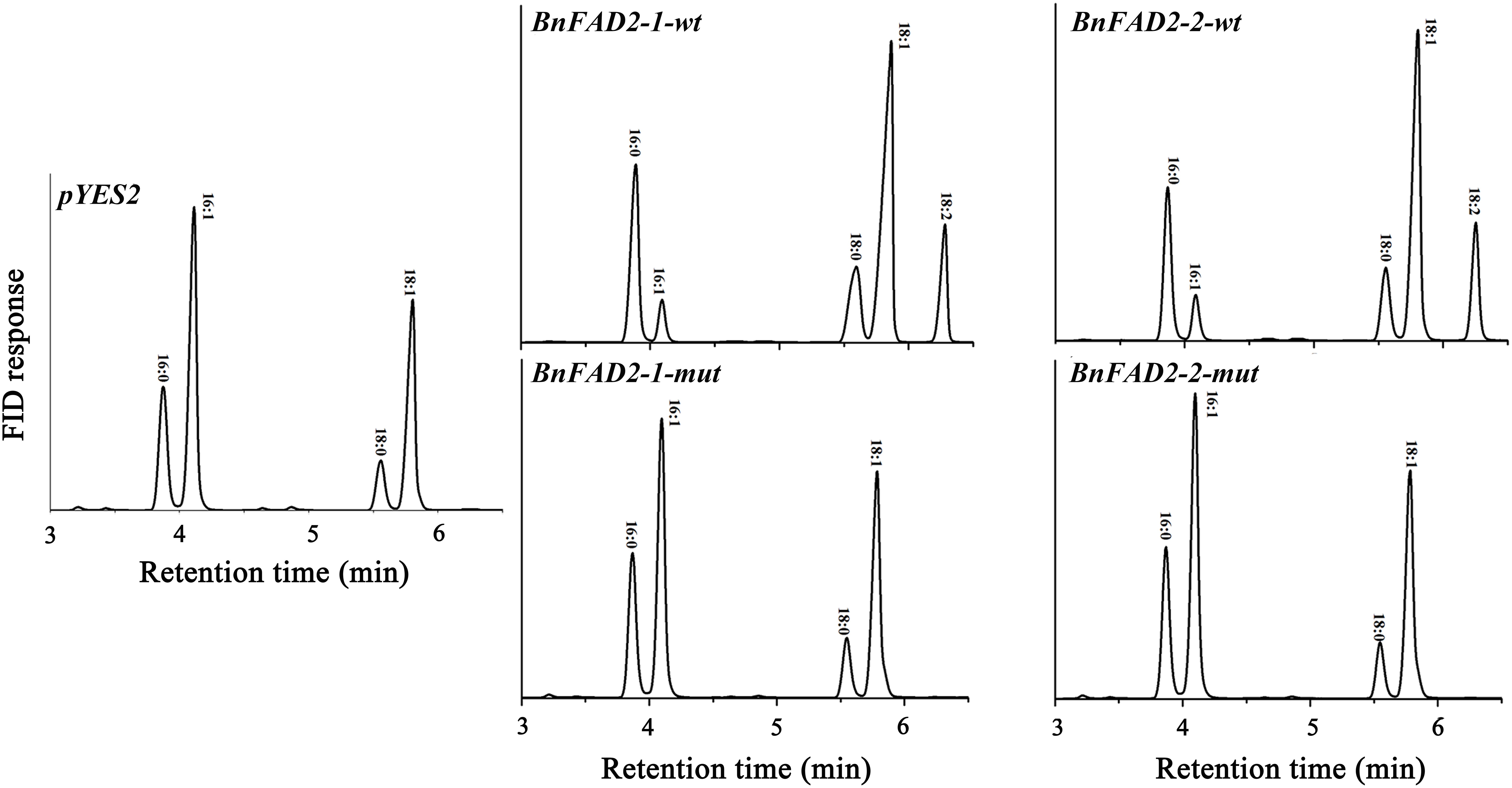
FIGURE 3. Gas chromatograms showing the fatty acid profiles of BnFAD2-transformed yeast. Representative chromatograms highlighting the fatty acid compositions of yeast strains transformed with pYES2, BnFAD2-1-wt, BnFAD2-1-mut, BnFAD2-2-wt, and BnFAD2-2-mut (top to bottom). The x axes represent retention time in minutes, while the y axes represent the detector (FID response). 16:0, palmitic acid; 16:1, palmitoleic acid; 18:0, stearic acid; 18:1, oleic acid.
In order to analyze each SNP, BnFAD2 gene fragments containing each site were amplified separately. A sequence comparison detected two fixed variances inside the open reading frames of the four groups, a different 6-bp nucleotide in the 41–46 bp zone for each of the four groups and two distinct nucleotide mutations (C and C) at the 732 and 735 loci in BnFAD2-2 (Figure 1A). Therefore, two primer pairs (P3–P4, P5–P6) specific for BnFAD2-1 and BnFAD2-2 were designed according to these differences. The nucleotide trace peaks (Figure 4) from the PCR products showed the clear unambiguous line types, indicating that the primer pairs successfully amplified pure fragments of BnFAD2-1 and BnFAD2-2. The sequence differences were easily observed using these sequence peaks. Double peaks were also observed in the hybrid lines.
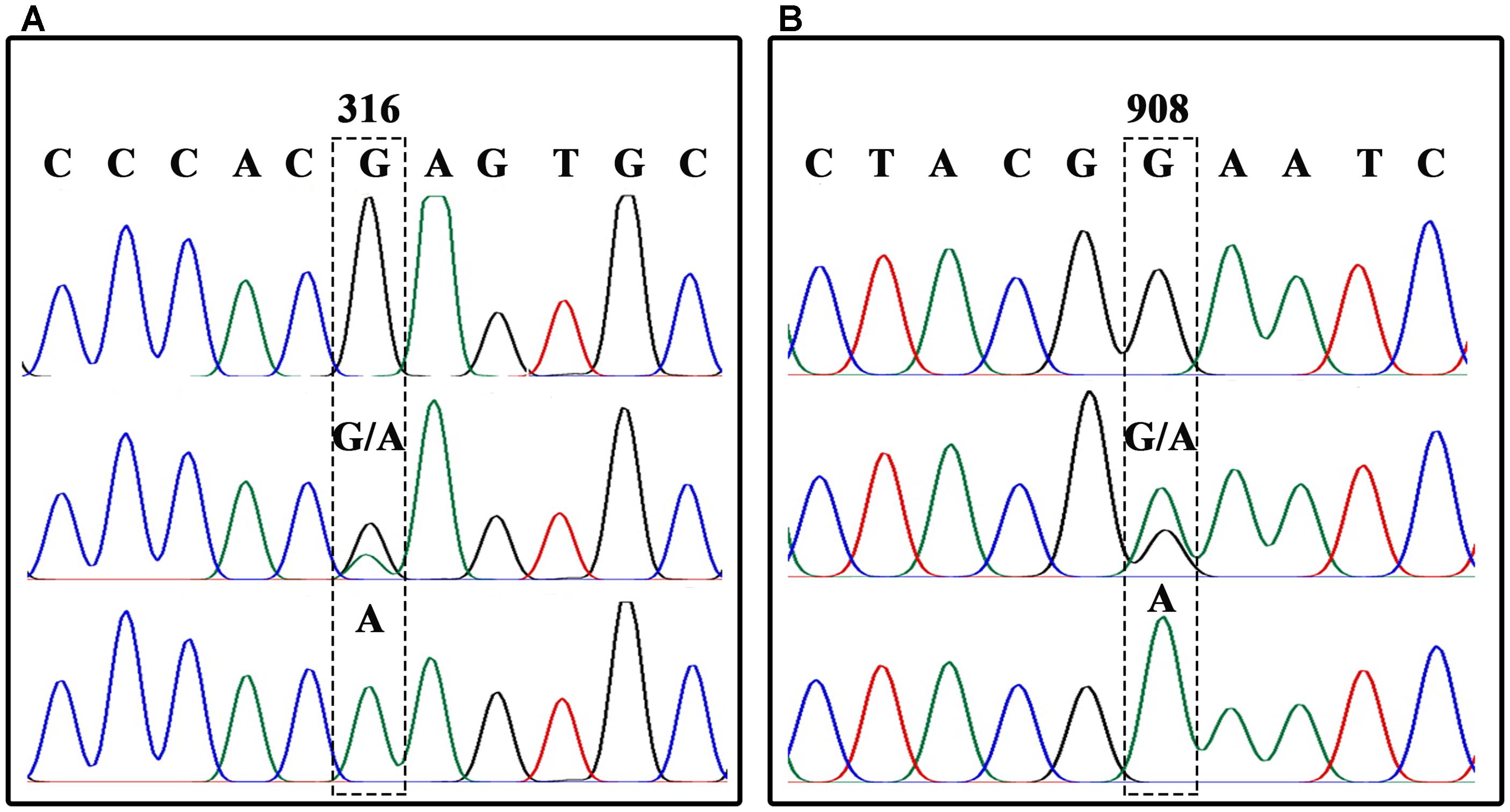
FIGURE 4. Nucleotide trace map of the mutation site fragments in BnFAD2-1 (A) and BnFAD2-2 (B). The upper part of the map shows the wild-type sequences, while the middle and lower parts show the hybrid type and mutation type sequences, respectively. The mutant site of each gene is indicated in a dashed frame and the numbers above the frames represent the nucleotide number when counted from the start codon ATG.
Further analysis showed that the G316A mutation in BnFAD2-1 mediated the loss of the NssSI recognition site, while the G908A mutation in BnFAD2-2 mediated loss of the HinfI recognition site. Therefore, these two restriction enzymes could be used to assess genotypes for these mutants. When using the P3-P4 primer pair for BssSαI, the WT displayed the 295 bp + 436 bp band-type, while the mutant displayed the 731-bp band-type (Figure 5, upper panel). The F1 generation, not surprisingly, showed the 295 bp + 436 bp + 731 bp band type. Alternatively, when using the P5–P6 primer pair for HinfI, the WT displayed the 61 bp + 80 bp + 58 bp + 206 bp band type, while the mutant displayed the 61 bp + 80 bp + 264 bp band type (Figure 5, lower panel). The F1 was again a combination of these, with the 61 bp + 80 bp + 58 bp + 206 bp + 264 bp band type.
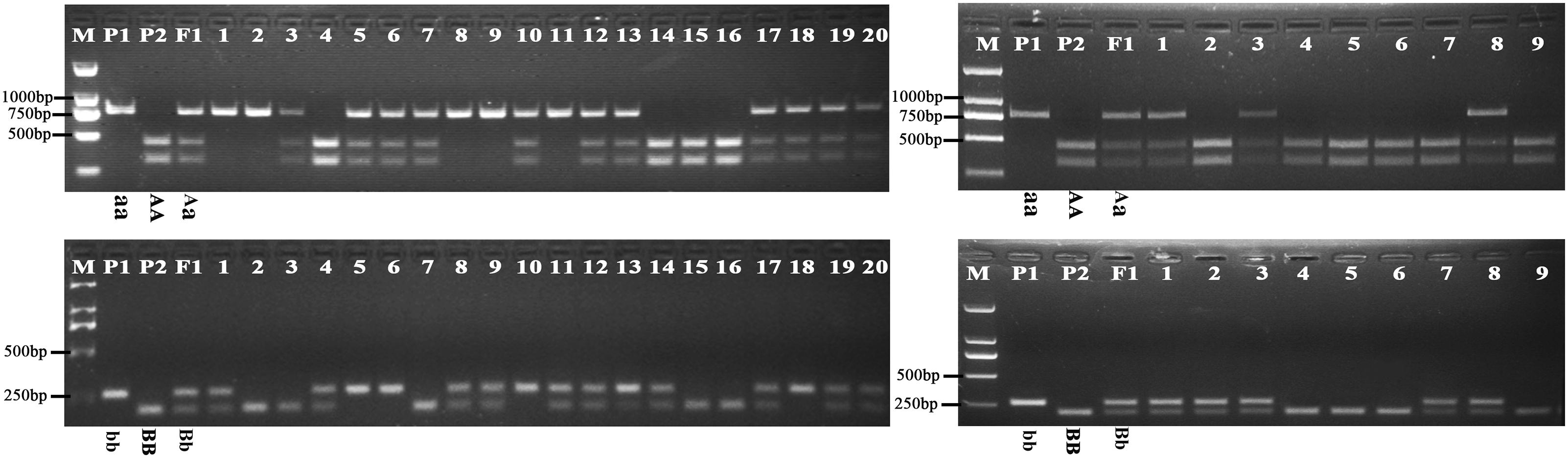
FIGURE 5. Genotyping of the segregation lines using CAPS markers. The genotype of each line is shown under its enzyme-digested band. The left panel shows the band types for the P1, P2, F1, and F2-1 populations digested by BssSαI (upper) and HinfI (lower) restriction enzymes, and no. 1–20 refer to the single lines from the F2-1 population. The right panel shows the band types for the P1, P2, F1, and BC1-1 populations digested by BssSαI (upper) and HinfI (lower) restriction enzymes, and no.1-9 refer to the single lines from the BC1-1 population. M, DNA marker; P1, mutant line N1379T; P2, normal line 16wh53. The 60, 80, and 58 bp bands produced by HinfI appear as a single band on the gel because of their very similar lengths; however, these could be differentiated during our analysis. There were three standard bands in the M lane representing the 750, 500, and 250 bp band sizes.
The oleic acid content and genotypes were determined for all of the lines in the segregated populations (two F2s and two BC1s) (Figure 5). The parents 16wh53 and 16wh62 producing normal (∼63%) oleic acid content contained the BnFAD2-1-wt and BnFAD2-2-wt (AABB) genotype. By contrast, the N1379T line had the double BnFAD2-1-mut and BnFAD2-2-mut (aabb) genotype. Notably, lines with the same genotype as the parental strains had similar oleic acid contents. However, oleic acid content segregation was observed in the F2 and BC1 populations from the two sites (Table 2). There were significant differences for oleic (C18:1), linoleic (C18:2), and linolenic (C18:3) acids between different genotypes in segregation populations, while no differences in oil content existed between genotypes. Generally, F2 and BC1 lines with more mutant alleles had higher oleic acid content. Indeed, the average oleic acid content of lines with the aabb or AABB genotype in the F2 population were ∼85% and ∼63%, respectively, similar to the parental lines. The increase in oleic acid content was companied by a decrease in linoleic and linolenic acids, while the oil contents of genotype lines were without significant change. A similar situation was observed in population F2-2 and BC1-2 (Supplementary Table S4).
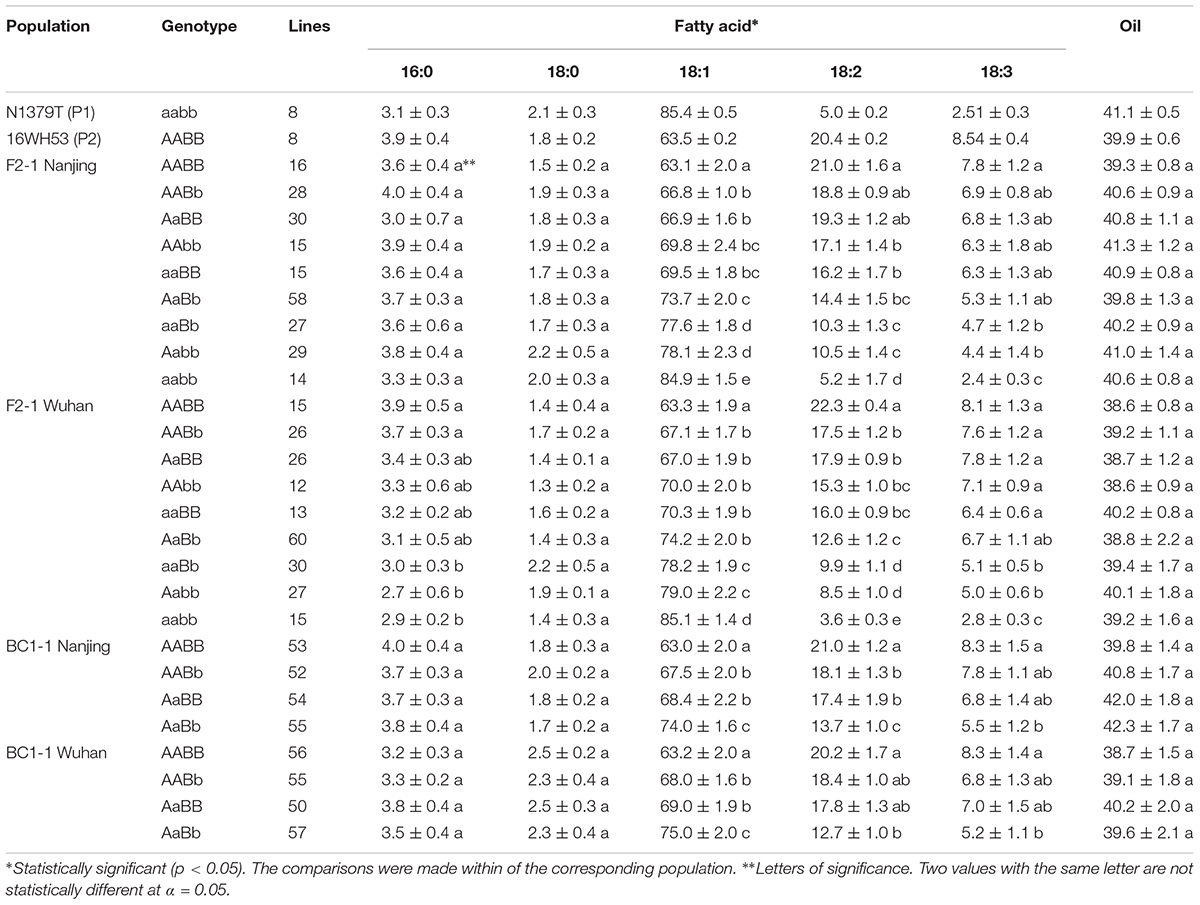
TABLE 2. Average fatty acid percent and oil content of each genotype in the parental lines and the segregated populations at two locations.
The genetic effects of additive, dominance, and epistasis in F2 populations of these two mutant BnFAD2 alleles were calculated by using the classic genetic method (Table 3). All the effects from each population were positively functioned. The additive effect was the major effect of the alleles, much higher than other two effects (dominance and epistasis effect). The BnFAD2-1-mut and BnFAD2-2-mut alleles had a very similar genetic effect on the oleic acid content in the four F2 populations. The inheritance of the oleic acid content in N1379T was mainly controlled by the additive effect.
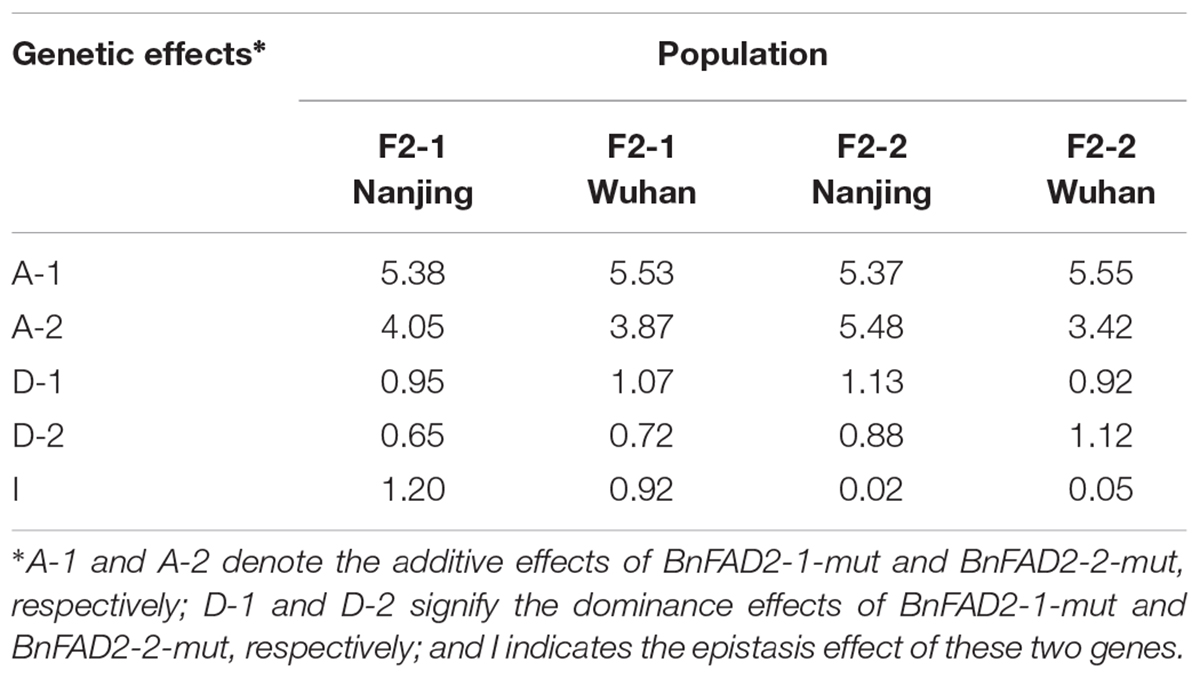
TABLE 3. Genetic effects of the addition, dominance, and epistasis of these two mutant alleles in F2 populations.
As the different alleles segregated into the F2 and BC1 populations, the effects of each BnFAD2 allele on the oleic acid content were determined (Figure 6). Lines with different alleles of BnFAD2-1 and BnFAD2-2 were separated by CAPS markers, and then the oleic acid contents of the same-genotypes lines were calculated in both the F2 and BC1 populations. Our results showed that both of the BnFAD2-1 and BnFAD2-2 had a significant and equal effect on the oleic acid content individually, not just together. For example, compared with the oleic acid content of AA-genotype (homozygous BnFAD2-1-wt) lines, the aa-genotype (homozygous BnFAD2-1-mut) lines had a 10.68% increase in oleic acid content in the F2 population, and the Aa-genotype (heterozygous BnFAD2-1) lines had an increase of 6.5%. This is similar to what was observed for the Aa-genotype lines in the BC1 population. In the F2 populations, the average oleic acid content of the bb-genotype (homozygous BnFAD2-2-mut) lines was ∼78%, a 10% increase compared to the BB-genotype (homozygous BnFAD2-2-wt). Furthermore, the same genotype had similar effects in the different populations, with the average oleic acid content of the Aa and Bb genotypes in the F2 population being ∼6.69% higher than that of the AA and BB genotypes. The average oleic acid content of the Aa and Bb genotypes in the BC1 population was similarly 6.33% higher than that of the AA and BB genotypes.
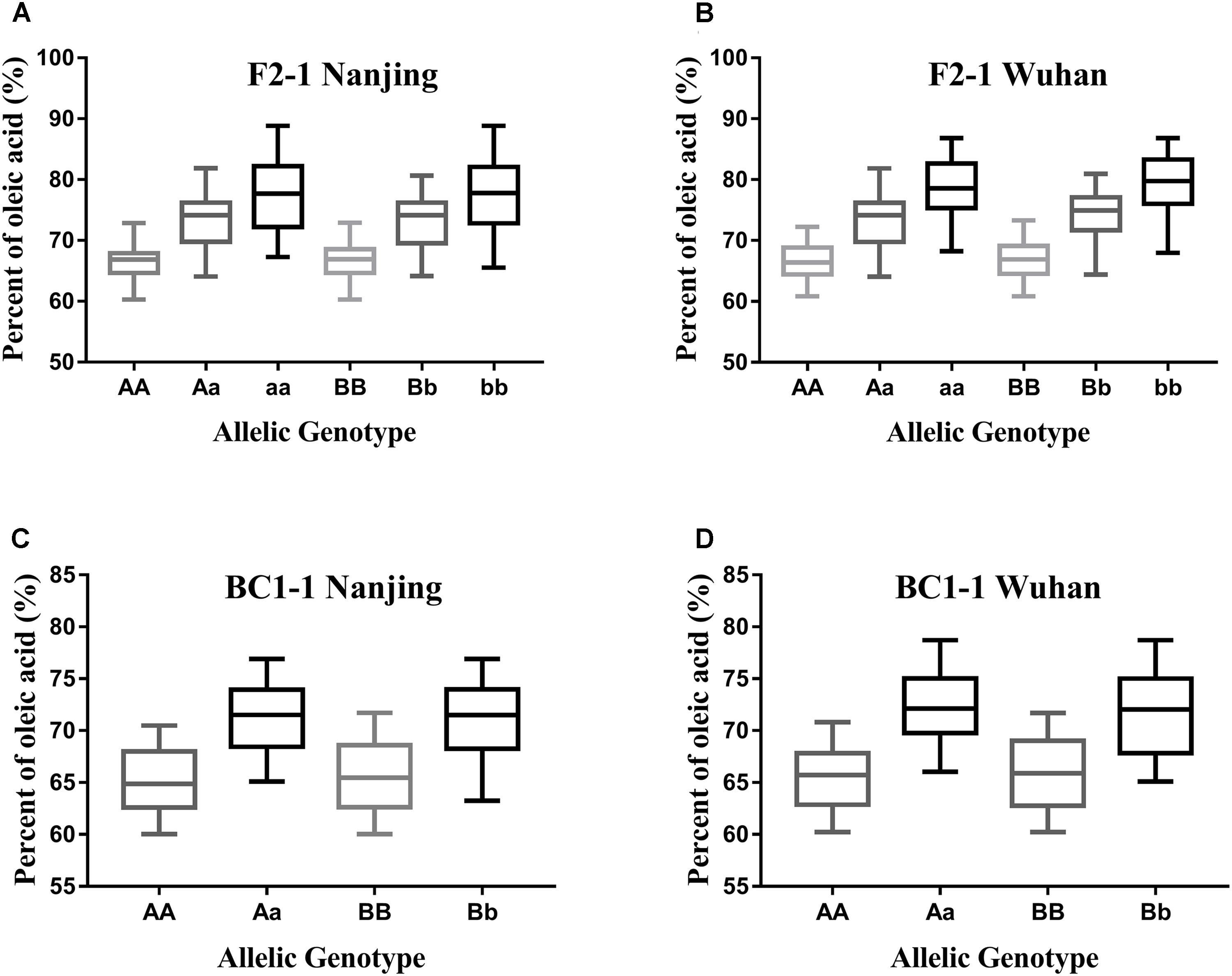
FIGURE 6. Distribution of the oleic acid content for lines with different allelic genotypes in the segregation populations. Genotype effects mediated by the BnFAD2-1 (three genotypes: AA, Aa, and aa) and BnFAD2-2 (three genotypes: BB, Bb, and bb) genes are shown for the F2 population (A,B) and BC1 population (C,D) at two sites (Nanjing and Wuhan).
Beneficial fatty acids facilitate the utilization of rapeseed oil (Napier et al., 2014), and several high oleic acid rapeseed lines have been created by mutagenesis (Auld et al., 1992; Schierholt et al., 2001; Hu et al., 2006; Spasibionek, 2006; Yang et al., 2012). These include N1379T, the stable super-high (∼85%) oleic acid line that was developed by conventional methods. Compared with other vegetable oils, high oleic acid rapeseed oil is a cost-effective alternative for home-cooking. Considering its other features, such as the high oil content (45∼50%) in seeds of rapeseed, it is not surprising that the area of rapeseed cultivation and consumer demand for rapeseed oil have increased (Fu et al., 2016). Unfortunately, our understanding of the genetics and molecular mechanisms underlying high oleic acid traits is limited. In this study, we evaluated mutations in the BnFAD2 genes in the high oleic acid line (N1379T) to determine the effects of functional loss of BnFAD2 on oleic acid content.
Notably, we were not the first to identify a relationship between BnFAD2 mutations and oleic acid content in rapeseed seeds (Hu et al., 2006; Yang et al., 2012); however, aside from BnFAD2 genes, no other reported QTLs seem to have a connection with oleic acid content. This is particularly true for the transgenic lines with high oleic acid levels (>80%), which were produced by transferring the antisense BnFAD2 gene fragments into normal oleic acid rapeseed lines (Peng et al., 2010; Jung et al., 2011; Lee et al., 2016). Our results confirmed that the high oleic acid trait in N1379T was controlled by two mutant BnFAD2 genes (BnFAD2-1 and BnFAD2-2). To our knowledge, this is the first study to provide genetic and molecular evidences of two-gene-controlled high oleic acid trait in rapeseed.
Although it is not altogether surprising that mutations in plant FAD2 reduce its enzymatic activity and enhance oleic acid content, not all mutations would result in these changes. The position of the mutation in the BnFAD2 gene is, therefore, very important. Δ12-FAD2 is a trans-membrane protein with three conserved histidine-rich motifs (also called histidine boxes, H-boxes), which form the active center of the enzyme (Shanklin et al., 1994). Different plant species, especially oilseed crops, have multiple FAD2 genes/proteins with highly similar sequences and structures (Dehghan and Yarizade, 2014). For rapeseed, the three H-boxes in BnFAD2 are 105-HECGHHAF-111, 137-WKYSHRRHH, and 315-HVAHHLFS-323 (Tanhuanpää et al., 1998). Mutations that occur near the H-boxes have a high probability of affecting BnFAD2 enzyme activity. In the present study, one of the detected mutations was E106K in BnFAD2-1, which located within the first H-box. The other mutation site was G303E in BnFAD2-2, near the third H-box motif. Therefore, it is not surprising that these SNP mutations resulted in loss of BnFAD2 function. However, it is not a requirement for the SNP mutation to occur inside or near the H-boxes to be effective. In fact, a series of SNPs were created in a mutagenic oleic acid-enhanced soybean population in which most of the mutations were not in or near the H-boxes (Wells et al., 2014). It was demonstrated that the Ser-185 of GmFAD2-1, while being outside of the H-boxes, played a key role in regulating post-translational modifications that directly affected FAD2-1 enzyme activity (Tang et al., 2005). In the present study, Ser-185 was also not in the H-boxes of the BnFAD2 enzymes. While the investigation of BnFAD2 regulatory elements inside and/or outside of the genes was outside the scope of this study, future work should explore this regulation in order to more fully explain the relationship between gene mutation and the observed elevation in oleic acid content (Xiao et al., 2014).
It was important to verify the sequences and function of WT and mutant BnFAD2-1 and BnFAD2-2. Therefore, we transformed the genes into yeast, which have been widely used for the functional characterization of FADs. AtFAD2 was the first FAD gene to be functionally expressed in S. cerevisiae and resulted in a successful linoleic acid production (Covello and Reed, 1996). This not only confirmed desaturase activity but also proved that yeast could be a suitable host for functional oilseed gene expression. Since then, many FAD2 genes have been cloned from different oilseeds (e.g., peanut, cotton, sunflower, soybean, olive, etc.) and tested using yeast (Jung et al., 2000; Martínez-Rivas et al., 2001; Pirtle et al., 2001; Hernandez et al., 2005; Tang et al., 2005). In fact, BnFAD2-1 and BnFAD2-2 in WT were also previously confirmed to have catalytic activity in yeast (Xiao et al., 2010; Lee et al., 2013). In our investigation, both the WT and corresponding mutant gene were expressed in yeast to identify mutation-mediated changes in enzyme function. Our results showed that both mutant genes were non-functional and could not catalyze oleic acid to linoleic acid. Furthermore, it showed that both BnFAD2-1 and BnFAD2-2 contributed equally to linoleic acid formation as similar levels were observed in yeast for both genes. Yeast strains transformed with BnFAD2-1-mut and BnFAD2-2-mut genes had similar fatty acid compositions to pYES2 strains, indicating these two mutant genes were not functional. Meanwhile, a decrease in palmitoleic acid (C16:1) and increases in oleic (C18:1) and linoleic (C18:2) acid were observed in the BnFAD2-1 and BnFAD2-2 strains, possibly because these two genes synthesized linoleic acid (C18:2) by catalyzing oleic acid (C18:1), which activated the flow from palmitoleic acid (C16:1) to oleic acid (C18:1). With the palmitoleic acid (C16:1) continued to be processed to form oleic acid (C18:1) in the rapeseed, oleic acid would accumulate, because the mutant BnFAD2-1 and BnFAD2-2 could not convert this to linoleic acid (C18:2). Although this work highlighted a definitive role for these mutated sequences in the observed increase in oleic acid, questions still remained concerning these genes. For example, additional work was required to determine the contribution of each BnFAD2 in the accumulation of oleic acid (C18:1) in rapeseed seeds and whether these genes compensate for each other. Furthermore, future studies should determine if the plant can continue to grow if all of the BnFAD2 genes lose their function at the same time as well as identify other biological pathways oleic acid (C18:1) participates in (Guan et al., 2012). While our work provided a solid foundation for continued investigation, answering these outstanding questions would assist in further exploring the roles of C18:1 in rapeseed development.
Our results proved that the mutations were in the constitutive-expressing BnFAD2, which were in accordance with the significant increase of oleic acid content in both vegetable tissues and seeds of N1379T (Jianqin et al., 2012; Lee et al., 2013). Fatty acids are essential components of plant membranes, and increased oleic acid level will cause a concomitant decrease in polyunsaturated fatty acids. This will limit the general mobilization of membranes inside tissues under low temperatures (Upchurch, 2008). For example, a FAD2 mutant in Arabidopsis failed to undergo normal growth during development at low temperatures (Miquel et al., 1993; Miquel and Browse, 1994). Furthermore, another report suggested that high oleic acid expression in rapeseed would likely be accompanied with reduced yield and lower germination rates (Schierholt and Becker, 2001, 2011). Thus, although it was beyond the scope of the present study, a comprehensive exploration was required to evaluate the effects of increased oleic acid on the yield of N1379T.
Ultimately, one of the goals of this work was to detect genetic markers linked to higher oleic acid levels and utilize these markers to improve breeding decisions. To perform marker-assisted selection (MAS), the homologous and heterozygous genotypes must be clearly distinguishable. It is particularly difficult to design specific primers to identify and amplify the mutant SNP sites in BnFAD2 genes, as they have very similar DNA sequences (Lee et al., 2013). In this study, we developed CAPS markers for both these SNP sites and successfully genotyped lines using PCR and electrophoresis. The CAPS markers were stable and can unambiguously differentiate the parent and hybrid genotypes, allowing for mass selection. Moreover, the two mutated BnFAD2 genes are located on different chromosomes (A5 and C5), suggesting that they have no linkage effects on each other. Our genotyping system would allow candidate lines with the desired allelic genes to be screened via CAPS marker-driven PCR, making it easy to select high oleic acid-expressing rapeseed varieties for breeding. In addition, genetic effect analysis revealed that the additive effect contributed much more to oleic acid content than the other two effects (dominance and epistasis effect). As the additive effect is the inheritable factor, the trait of high oleic acid content in N1379T can be easily transferred to other varieties and utilized in breeding.
In conclusion, we demonstrated that two separate SNPs in the homologous BnFAD2-1 and BnFAD2-2 genes induced loss of enzyme function of both enzymes and were responsible for the super-high oleic acid (85%) content in N1379T. Furthermore, during our investigation we designed CAPS markers to effectively detect the genotypes of these alleles, thus providing a MAS system to use for the selection of this trait. This study provided a foundation on using N1379T as a new valuable trait resource for developing the high oleic acid rapeseed varieties in future.
WL designed the experiments, processed the molecular biology and data analysis, and wrote the manuscript. JG determined the fatty acids profiles. MH did the sequences comparison and designed the primers. SC and LC processed part of PCR assays. JZ revised the manuscript. HP selected the germplasm, developed the populations, took part in the experimental design, and finalized the manuscript.
This work was supported by the National Key Research and Development Program of China (2016YFD0101300), National Natural Science Foundation of China (31870519), the Research Funding of Jiangsu Academy of Agricultural Sciences (6111618), the earmarked fund for China Agriculture Research System (CARS-12), and Jiangsu Collaborative Innovation Center for Modern Crop Production. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors wish to thank Dr. Guangyuan Lu at the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences for helping the part work of the population development and David Brunton at the University of Adelaide for revising the manuscript. The authors would also like to thank Editage (www.editage.cn) for English language editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00399/full#supplementary-material
TABLE S1 | Rapeseed varieties tested.
TABLE S2 | Comparison of the cloned BnFAD2 homolog gene sequences with those downloaded from GenBank and with the reference genome.
TABLE S3 | Percentage of each fatty acid in Saccharomyces cerevisiae transferred with different BnFAD2 genes.
TABLE S4 | Average fatty acid and oil content percentages of each genotype in the parental lines and the segregated populations (F2-2 and BC1-2) at two locations.
Auld, D. L., Heikkinen, M. K., Erickson, D. A., Sernyk, J. L., and Romero, J. E. (1992). Rapeseed mutants with reduced levels of polyunsaturated fatty acids and increased levels of oleic acid. Crop Sci. 32, 657–662. doi: 10.2135/cropsci1992.0011183X003200030016x
Browse, J., Spychalla, J., Okuley, J., and Lightner, J. (1998). Altering the Fatty Acid Composition of Vegetable Oils. New York, NY: Cambridge University Press.
Chang, N. W., and Huang, P. C. (1998). Effects of the ratio of polyunsaturated and monounsaturated fatty acid to saturated fatty acid on rat plasma and liver lipid concentrations. Lipids 33, 481–487. doi: 10.1007/s11745-998-0231-9
Covello, P. S., and Reed, D. W. (1996). Functional expression of the extraplastidial Arabidopsis thaliana oleate desaturase gene (FAD2) in Saccharomyces cerevisiae. Plant Physiol. 111, 223–226. doi: 10.1104/pp.111.1.223
Dehghan, N. F., and Yarizade, K. (2014). Bioinformatics study of delta-12 fatty acid desaturase 2 (FAD2) gene in oilseeds. Mol. Biol. Rep. 41, 5077–5087. doi: 10.1007/s11033-014-3373-5
Downey, R. K., and Harvey, B. L. (1963). Method of breeding for oil quality in rape. Can. J. Plant Sci. 43, 271–275. doi: 10.4141/cjps63-054
Eshed, Y., and Zamir, D. (1996). Less-than-additive epistatic interaction of quantitative trait loci in tomato. Genetics 143, 1807–1817.
Fu, D. H., Jiang, L. Y., Mason, A. S., Xiao, M. L., Zhu, L. R., Li, L. Z., et al. (2016). Research progress and strategies for multifunctional rapeseed: a case study of China. J. Integr. Agric. 15, 1673–1684. doi: 10.1016/S2095-3119(16)61384-9
Gacek, K., Bayer, P. E., Bartkowiak-Broda, I., Szala, L., Bocianowski, J., Edwards, D., et al. (2016). Genome-wide association study of genetic control of seed fatty acid biosynthesis in Brassica napus. Front. Plant Sci. 7:2062. doi: 10.3389/fpls.2016.02062
Guan, M., Li, X., and Guan, C. (2012). Microarray analysis of differentially expressed genes between Brassica napus strains with high- and low-oleic acid contents. Plant Cell Rep. 31, 929–943. doi: 10.1007/s00299-011-1213-9
Heppard, E. P., Kinney, A. J., Stecca, K. L., and Miao, G. H. (1996). Developmental and growth temperature regulation of two different microsomal omega-6 desaturase genes in soybeans. Plant Physiol. 110, 311–319. doi: 10.1104/pp.110.1.311
Hernandez, M. L., Mancha, M., and Martinez-Rivas, J. M. (2005). Molecular cloning and characterization of genes encoding two microsomal oleate desaturases (FAD2) from olive. Phytochemistry 66, 1417–1426. doi: 10.1016/j.phytochem.2005.04.004
Hongtrakul, V., Slabaugh, M. B., and Knapp, S. J. (1998). A seed specific Δ-12 oleate desaturase gene is duplicated, rearranged, and weakly expressed in high oleic acid sunflower lines. Crop Sci. 38, 1245–1249. doi: 10.2135/cropsci1998.0011183X003800050022x
Hu, X., Sullivan Gilbert, M., Gupta, M., and Thompson, S. A. (2006). Mapping of the loci controlling oleic and linolenic acid contents and development of fad2 and fad3 allele-specific markers in canola (Brassica napus L.). Theor. Appl. Genet. 113, 497–507. doi: 10.1007/s00122-006-0315-1
Huang, X. Q., Huang, T., Hou, G. Z., Li, L., Hou, Y., and Lu, Y. H. (2016). Identification of QTLs for seed quality traits in rapeseed (Brassica napus L.) using recombinant inbred lines (RILs). Euphytica 210, 1–16. doi: 10.1007/s10681-016-1675-5
Javed, N., Geng, J., Tahir, M., Mcvetty, P. B. E., Li, G., and Duncan, R. W. (2016). Identification of QTL influencing seed oil content, fatty acid profile and days to flowering in Brassica napus L. Euphytica 207, 191–211. doi: 10.1007/s10681-016-1675-5
Jianqin, G., Huiming, P., Weihua, L., Maolong, H., and Cunkou, Q. (2012). Dynamics of oleic acid contents in organs of high-oleic rapeseed lines. Chin. J. Oil Crop Sci. 34, 359–365.
Jung, J. H., Kim, H., Go, Y. S., Lee, S. B., Hur, C. G., Kim, H. U., et al. (2011). Identification of functional BrFAD2-1 gene encoding microsomal delta-12 fatty acid desaturase from Brassica rapa and development of Brassica napus containing high oleic acid contents. Plant Cell Rep. 30, 1881–1892. doi: 10.1007/s00299-011-1095-x
Jung, S., Powell, G., Moore, K., and Abbott, A. (2000). The high oleate trait in the cultivated peanut [Arachis hypogaea L]. II. Molecular basis and genetics of the trait. Mol. Gen. Genet. 263, 806–811. doi: 10.1007/s004380000243
Khan, S., Anwar, S., Kuai, J., Noman, A., Shahid, M., Din, M., et al. (2018). Alteration in yield and oil quality traits of winter rapeseed by lodging at different planting density and nitrogen rates. Sci. Rep. 8:634. doi: 10.1038/s41598-017-18734-8
Lauridsen, C., Nielsen, J. H., Henckel, P., and Sorensen, M. T. (1999). Antioxidative and oxidative status in muscles of pigs fed rapeseed oil, vitamin E, and copper. J. Anim. Sci. 77, 105–115. doi: 10.2527/1999.771105x
Lee, K. R., In Sohn, S., Jung, J. H., Kim, S. H., Roh, K. H., Kim, J. B., et al. (2013). Functional analysis and tissue-differential expression of four FAD2 genes in amphidiploid Brassica napus derived from Brassica rapa and Brassica oleracea. Gene 531, 253–262. doi: 10.1016/j.gene.2013.08.095
Lee, K. R., Kim, E. H., Roh, K. H., Kim, J. B., Kang, H. C., Go, Y. S., et al. (2016). High-oleic oilseed rapes developed with seed-specific suppression of FAD2 gene expression. Appl. Biol. Chem. 59, 669–676. doi: 10.1007/s13765-016-0208-1
López, Y., Nadaf, H. L., Smith, O. D., Connell, J. P., Reddy, A. S., and Fritz, A. K. (2000). Isolation and characterization of the Δ12-fatty acid desaturase in peanut (Arachis hypogaea L.) and search for polymorphisms for the high oleate trait in Spanish market-type lines. Theor. Appl. Genet. 101, 1131–1138. doi: 10.1007/s001220051589
Martínez-Rivas, J. M., Sperling, P., Lühs, W., and Heinz, E. (2001). Spatial and temporal regulation of three different microsomal oleate desaturase genes (FAD2) from normal-type and high-oleic varieties of sunflower (Helianthus annuus L.). Mol. Breed. 8, 159–168. doi: 10.1023/A:1013324329322
Mikkilineni, V., and Rocheford, T. R. (2003). Sequence variation and genomic organization of fatty acid desaturase-2 (FAD2) and fatty acid desaturase-6 (fad6) cDNAs in maize. Theor. Appl. Genet. 106, 1326–1332. doi: 10.1007/s00122-003-1190-7
Miquel, M., James, D., Dooner, H., and Browse, J. (1993). Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc. Natl. Acad. Sci. U.S.A. 90, 6208–6212. doi: 10.1073/pnas.90.13.6208
Miquel, M. F., and Browse, J. A. (1994). High-oleate oilseeds fail to develop at low temperature. Plant Physiol. 106, 421–427. doi: 10.1104/pp.106.2.421
Napier, J. A., Haslam, R. P., Beaudoin, F., and Cahoon, E. B. (2014). Understanding and manipulating plant lipid composition: metabolic engineering leads the way. Curr. Opin. Plant Biol. 19, 68–75. doi: 10.1016/j.pbi.2014.04.001
Ohlrogge, J., and Browse, J. (1995). Lipid biosynthesis. Plant Cell 7, 957–970. doi: 10.2307/3870050
Okuley, J., Lightner, J., Feldmann, K., Yadav, N., Lark, E., and Browse, J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6, 147–158. doi: 10.2307/3869682
Peng, Q., Hu, Y., Wei, R., Zhang, Y., Guan, C., Ruan, Y., et al. (2010). Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds. Plant Cell Rep. 29, 317–325. doi: 10.1007/s00299-010-0823-y
Pirtle, I. L., Kongcharoensuntorn, W., Nampaisansuk, M., Knesek, J. E., Chapman, K. D., and Pirtle, R. M. (2001). Molecular cloning and functional expression of the gene for a cotton Delta-12 fatty acid desaturase (FAD2). Biochim. Biophys. Acta 1522, 122–129. doi: 10.1016/S0167-4781(01)00312-8
Przybylski, R., Gruczynska, E., and Aladedunye, F. (2013). Performance of regular and modified canola and soybean oils in rotational frying. J. Am. Oil Chem. Soc. 90, 1271–1280. doi: 10.1007/s11746-013-2278-0
Qu, C., Jia, L., Fu, F., Zhao, H., Lu, K., Wei, L., et al. (2017). Genome-wide association mapping and Identification of candidate genes for fatty acid composition in Brassica napus L. using SNP markers. BMC Genomics 18:232. doi: 10.1186/s12864-017-3607-8
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Scheffler, J. A., Sharpe, A. G., Schmidt, H., Sperling, P., Parkin, I. A. P., Lühs, W., et al. (1997). Desaturase multigene families of Brassica napus arose through genome duplication. Theor. Appl. Genet. 94, 583–591. doi: 10.1007/s001220050454
Schierholt, A., and Becker, H. C. (2001). Environmental variability and heritability of high oleic acid content in winter oilseed rape. Plant Breed. 120, 63–66. doi: 10.1046/j.1439-0523.2001.00549.x
Schierholt, A., and Becker, H. C. (2011). Influence of oleic acid content on yield in winter oilseed rape. Crop Sci. 51, 1973–1979. doi: 10.2135/cropsci2010.07.0411
Schierholt, A., Becker, H. C., and Ecke, W. (2000). Mapping a high oleic acid mutation in winter oilseed rape (Brassica napus L.). Theor. Appl. Genet. 101, 897–901. doi: 10.1007/s001220051559
Schierholt, A., Rücker, B., and Becker, H. C. (2001). Inheritance of high oleic acid mutations in winter oilseed rape (Brassica napus L.). Crop Sci. 41, 1444–1449. doi: 10.2135/cropsci2001.4151444x
Shanklin, J., Whittle, E., and Fox, B. G. (1994). Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33, 12787–12794. doi: 10.1021/bi00209a009
Sharafi, Y., Majidi, M. M., Goli, S. H., and Rashidi, F. (2015). Oil content and fatty acids composition in Brassica species. Int. J. Food Prop. 18, 2145–2154. doi: 10.1080/10942912.2014.968284
Somerville, C., and Browse, J. (1991). Plant lipids: metabolism, mutants, and membranes. Science 252, 80–87. doi: 10.1126/science.252.5002.80
Spasibionek, S. (2006). New mutants of winter rapeseed (Brassica napus L.) with changed fatty acid composition. Plant Breed. 125, 259–267. doi: 10.1111/j.1439-0523.2006.01213.x
Tang, G. Q., Novitzky, W. P., Carol Griffin, H., Huber, S. C., and Dewey, R. E. (2005). Oleate desaturase enzymes of soybean: evidence of regulation through differential stability and phosphorylation. Plant J. 44, 433–446. doi: 10.1111/j.1365-313X.2005.02535.x
Tanhuanpää, P., Vilkki, J., and Vihinen, M. (1998). Mapping and cloning of FAD2 gene to develop allele-specific PCR for oleic acid in spring turnip rape (Brassica rapa ssp. oleifera). Mol. Breed. 4, 543–550. doi: 10.1023/A:1009642317634
Upchurch, R. G. (2008). Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 30, 967–977. doi: 10.1007/s10529-008-9639-z
Wang, X., Long, Y., Yin, Y., Zhang, C., Gan, L., Liu, L., et al. (2015). New insights into the genetic networks affecting seed fatty acid concentrations in Brassica napus. BMC Plant Biol. 15:91. doi: 10.1186/s12870-015-0475-8
Wells, R., Trick, M., Soumpourou, E., Clissold, L., Morgan, C., Werner, P., et al. (2014). The control of seed oil polyunsaturate content in the polyploid crop species Brassica napus. Mol. Breed. 33, 349–362. doi: 10.1007/s11032-013-9954-5
WHO (2003). Diet, Nutrition and the Prevention of Chronic Diseases. Geneva: World Health Organization.
Xiao, G., Zhang, Z. Q., Wu, X.-M., Tan, T. L., and Guan, C. Y. (2010). Cloning and characterization of Six FAD2 pseudogenes of Brassica napus. Acta Agron. Sin. 36, 435–441. doi: 10.1016/S1875-2780(09)60038-9
Xiao, G., Zhang, Z. Q., Yin, C. F., Liu, R. Y., Wu, X. M., Tan, T. L., et al. (2014). Characterization of the promoter and 5′-UTR intron of oleic acid desaturase (FAD2) gene in Brassica napus. Gene 545, 45–55. doi: 10.1016/j.gene.2014.05.008
Keywords: rapeseed (Brassica napus L.), BnFAD2, single-nucleotide polymorphism, oleic acid, homolog genes, genotyping
Citation: Long W, Hu M, Gao J, Chen S, Zhang J, Cheng L and Pu H (2018) Identification and Functional Analysis of Two New Mutant BnFAD2 Alleles That Confer Elevated Oleic Acid Content in Rapeseed. Front. Genet. 9:399. doi: 10.3389/fgene.2018.00399
Received: 11 July 2018; Accepted: 31 August 2018;
Published: 20 September 2018.
Edited by:
Shuijin Hua, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Yang Zhu, University of Pennsylvania, United StatesCopyright © 2018 Long, Hu, Gao, Chen, Zhang, Cheng and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiming Pu, cHVodWltaW5nQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.