- 1Institute of Apicultural Research, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Key Laboratory of Pollinating Insect Biology, Ministry of Agriculture and Rural Affairs of the People’s Republic of China, Beijing, China
- 3Graduate School of Chinese Academy of Agricultural Sciences, Beijing, China
- 4Guangdong Key Laboratory of Animal Conservation and Resource Utilization, Guangdong Public Laboratory of Wild Animal Conservation and Utilization, Guangdong Institute of Applied Biological Resources, Guangzhou, China
Infection and transmission of honey bee viruses pose a serious threat to the pollination services of crops and wild plants, which plays a vital role in agricultural economy and ecology. RNA interference (RNAi) is an effective defense mechanism against commonly occurring viral infections of animals and plants. However, recent studies indicate that the effects of RNAi on the honey bee can induce additional impacts and might not always be effective in suppressing the virus. Moreover, the RNAi responses differed in relation to the developmental stage of the insect and the target tissue used, even though the same method of delivery was used. These results indicate that further analysis and field experiments should be performed to characterize the varying effectiveness of RNAi-based methods for treating honey bee viral infections. In this review, we provide an overview of the current knowledge and the recent progress in RNAi-based anti-viral treatments for honey bees, focusing in particular highlight the role of the dsRNA-delivery method used and its effect on RNAi efficiency and demonstrate the potential practical value of this tool for controlling the virus. We conclude studying the gene function and disease control of honey bee by RNAi technology requires a complex consideration from physiology, genetics to environment.
Introduction
Honey bees are important pollinators of agricultural crops and ecological systems. The honey bee population in European and United States has rapidly deceased in the past few decades and the decrease was associated with microbial infections, parasitic infections, and other biotic or abiotic stress (Cox-Foster et al., 2007; Hou et al., 2014). Honey bee-infecting pathogens as a major impacts have caused severe economic losses by affecting pollination and bee colony population in agricultural and apicultural industry (Aizen et al., 2009). Among the honey bee pathogens, viruses are the majority factors impacted honey bee health but have been poorly characterized (Brutscher et al., 2016). Over 20 honey bee viruses have been identified, some of which cause chronic infection until the bees encounter other stress factors, such as infection with Varroa destructor (Shen et al., 2005; Di Prisco et al., 2011) or Nosema ceranae (Toplak et al., 2013).
Generally, covert infections of honey bee viruses were built in colony that shown no clinical symptoms under the no other stressors. However, there are still a few of viruses that can cause typical signs. Deformed wing virus (DWV), chronic bee paralysis virus (CBPV), black queen cell virus (BQCV), Israeli acute paralysis virus (IAPV), and sacbrood virus (SBV) can make honey bee display the visible symptoms such as deformed wing, paralyzed, black cell and pupae sacbrood. In addition, the viruses establish acute infections such as the infection caused by acute bee paralysis virus (ABPV), which produces apparent symptoms (Azzami et al., 2012). Thence, most of other viruses can be frequently detected in seemingly-health bees and cannot make an accurate conclusion through the phenotypic characteristics. Thus, molecular detection based on the polymerase chain reaction (PCR) technology becomes the conventional means for identifying the bee viruses.
However, although most of honey bee viruses can be detected by PCR, beekeepers can rarely take effectively measures to limit viral infections. Most of the honey bee viruses are positive-sense, single-stranded RNA viruses, which are primarily distributed into Discitrovirus family. The viruses from Discitrovirus family have been shown to readily establish persistent infections and cause large economic losses in the apicultural industry because these viruses are able to replicate efficiently by using internal ribosome entry sites (IRES)-mediated translation mechanism, which is different from the cap-dependent replication mechanism used by most other viruses (Fernández-Miragall et al., 2009). Thus, these viruses are not only difficultly found in host but also there are no effective strategies to control them. However, with the advent of RNA interference (RNAi)-based methodologies, there has been an increasing interest in assessing potential applications of RNAi in controlling virus-mediated diseases and agricultural pests in both laboratory and field (Miller et al., 2008; Hunter et al., 2010; Garbutt et al., 2013; Di Lelio et al., 2014).
In fact, most of insect immune responses are involved in antiviral mechanism of honey bee. Toll, Immune deficiency (Imd), c-Jun N-terminal kinase (JNK) and Janus kinase/Signal Transducer and Activator of Transcription (Jak-STAT) pathways have been confirmed that play a vital role in resistance against virus infection (Brutscher et al., 2015). In addition, several physiological defenses related with antiviral responses of honey bee including melanization, encapsulation, and antimicrobial peptides have been identified (Brutscher et al., 2015). Although all these immune responses contribute to antiviral action, RNAi is still the most broadly defense mechanism in honey bee (Niu et al., 2014).
RNAi was first discovered in transgenic plants (Mathieu and Watts, 1989), followed by the discovery of its prevalence in a wide range species (Mao et al., 2007; Miller et al., 2008; Tian et al., 2009; Garbutt et al., 2013; Ren et al., 2014). RNAi is the major mechanism of antiviral defense, which is a sequence specific and post-transcriptional gene silencing that is triggered by double stranded RNA (dsRNA) (Figure 1A) (Brutscher et al., 2015). RNAi can be applied to interfere with expression of intercellular genes, rendering it a potentially powerful tool for the development of novel insect virus control strategies (Liu et al., 2012). Direct evidence of antiviral function of RNAi has been reported in Drosophila melanogaster (van Rij et al., 2006). Genome analysis shown that honey bees encode RNAi machinery genes, such as dicer-like, Argonaute (Ago) 2 (Elsik et al., 2014). Experiment evidence confirmed that RNAi is systemic in honey bee and found that sid-1 gene was essential for systemically administered dsRNA and gene silencing (Aronstein et al., 2006). RNAi has been used to study developmental gene expression of honey bee larvae (Jarosch et al., 2011; Kamakura, 2011; Wilson and Dearden, 2012), immunity of adults (Ament et al., 2012; Wang et al., 2012), and gene function of honey bee brain (Mustard et al., 2010; Hassani et al., 2012; Louis et al., 2012) as well as the functions of viral components such as the internal ribosome entry site within the intergenic region (IGR-IRES) (Au et al., 2017). In addition, dsRNA treatment has been also used to control honey bee parasites such as N. ceranae (Paldi et al., 2010), ectoparasitic mite V. destructor (Garbian et al., 2012; Campbell et al., 2016) and small hive beetle (Powell et al., 2016). More important, previous studies have demonstrated that RNAi can be used for controlling honey bee viruses and the success of using this treatment method indicates that RNAi could be potentially used for reducing economic losses caused by bee colony-infecting viruses around the world (Evans et al., 2009; Maori et al., 2009; Hunter et al., 2010; Liu et al., 2010; Desai et al., 2012). With the development of RNAi, the applications of dsRNA delivery into honey bees and other insects have been increasingly improved (Jarosch and Moritz, 2011; Jarosch et al., 2011; Hassani et al., 2012). Recently experimental evidence confirmed that RNAi immune response was triggered by Dicer-2 when honey bees were infected by SBV (Fung et al., 2018).
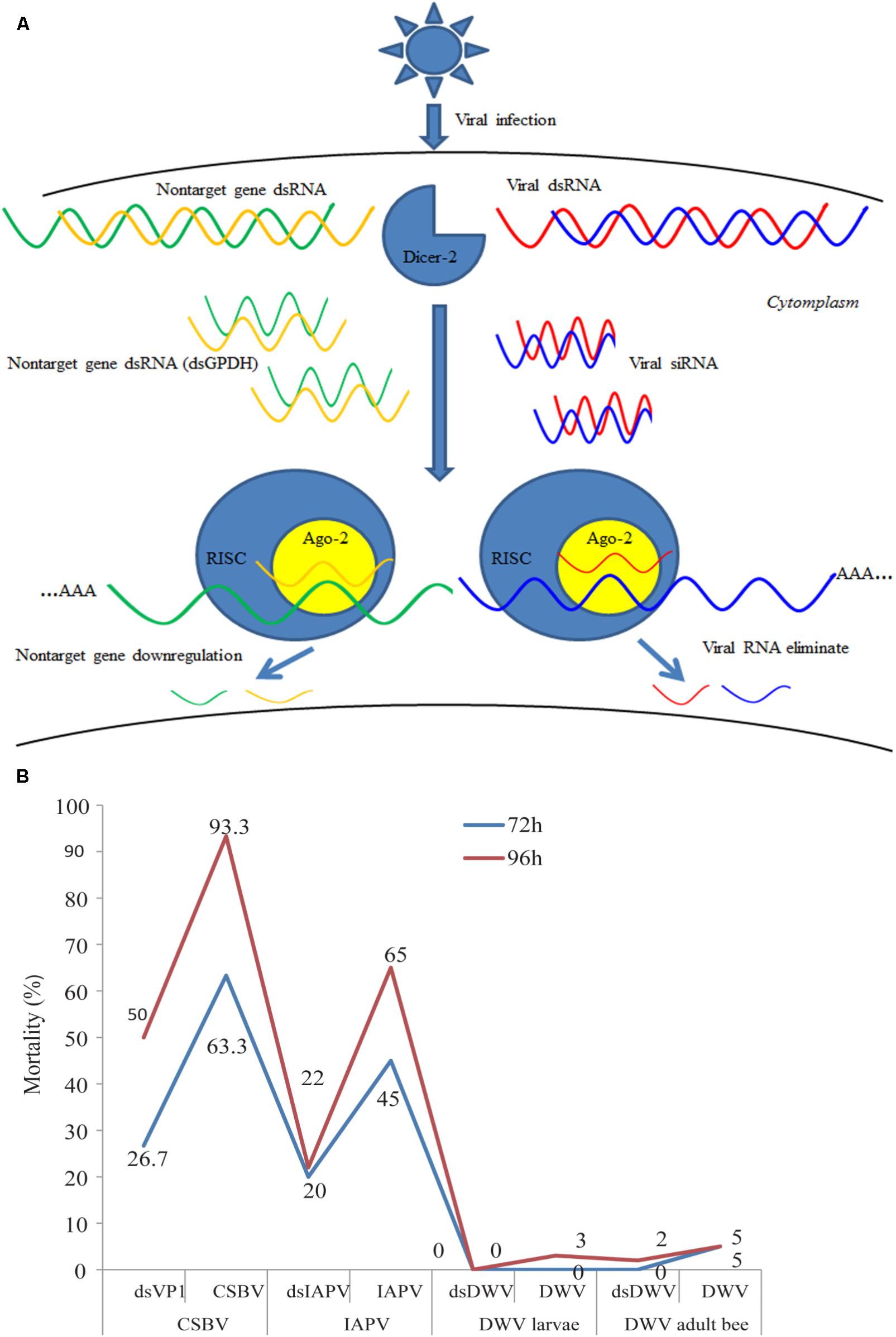
FIGURE 1. Overview of the process of RNAi-mediated gene silencing, possible off-target effects and the mortality of honey bee or larvae treated with RNAi against different honeybee viruses. (A) The short-interfering RNA (siRNA) pathway is one of the major ways for honey bee antiviral defense. Usually, the honey bee RNAi-pathway is induced by Dicer-like cleavage of viral dsRNA into siRNAs. In honey bees, non-specific dsRNA-mediated reduction in virus abundance (Flenniken and Andino, 2013) and degaradation on non-target genes (Jarosch and Moritz, 2012), but the mechanisms of this response have not been fully characterized. AGO2, Argonaute-2; RISC, RNA-induced silencing complex. (B) Bees or larvae were treated with viruses (CSBV, IAPV, and DWV) or target virus-double-stranded RNA (dsVP1, dsIAPV, and dsDWV). The number indicates the percentage of mortality treated with dsRNA and without for different viruses. CSBV, IAPV, DWV, and VP1 mean the Chinese sacbrood virus, Israeli acute paralysis virus, deformed wing virus, and virus protein 1 (Maori et al., 2009; Hunter et al., 2010; Liu et al., 2010; Desai et al., 2012).
The Factors Affecting Efficiency of RNAi
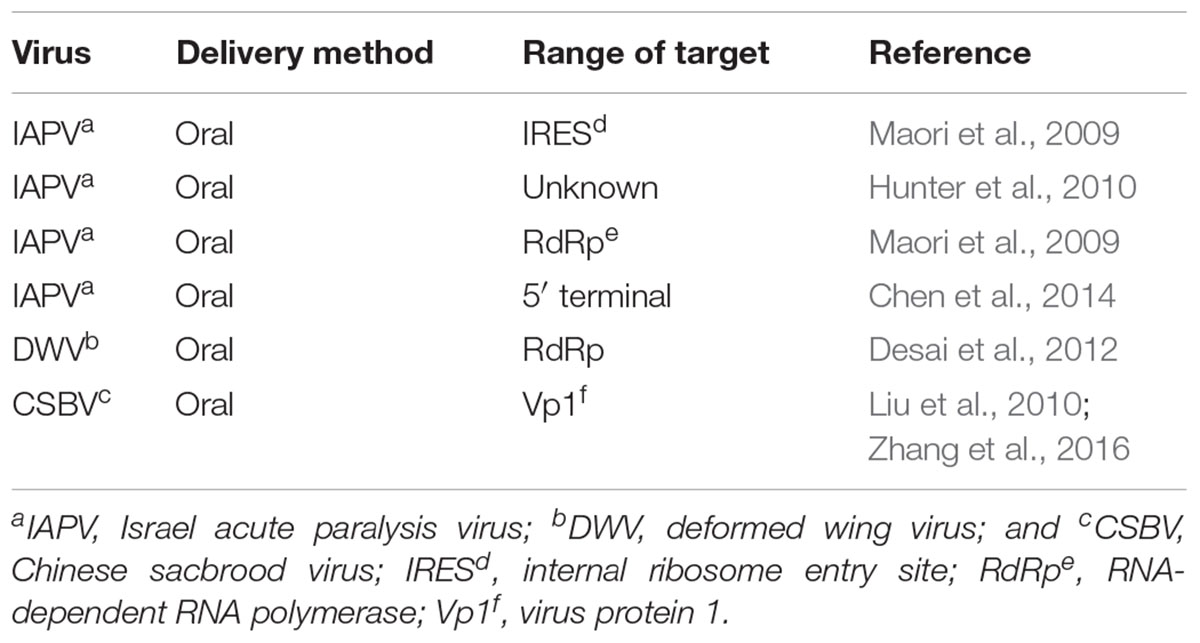
Although the RNAi has been commonly used in honey bee viruses (Table 1), there are more challenges associated with dsRNA delivery in honey bees than in other insects due to the lack of bee cell cultivation system (Maori et al., 2009; Hunter et al., 2010; Liu et al., 2010; Desai et al., 2012; Chen et al., 2014; Zhang et al., 2016). The efficiency of RNAi delivery can be influenced by several factors, which can act alone or in combination. Some of the influencing factors include the life stage of the target insect, stability of the target gene, target tissue site, and dsRNA quantity (Flenniken and Andino, 2013). The oral delivery of dsRNA of a non-target gene, dsRNA-GFP, to honey bee larvae caused changes in expression level of approximately 1400 genes, which account for 10% honey bee genes (Nunes et al., 2013). Moreover, molecular mechanisms underlying the RNAi-based antiviral effect in honey bees have not been fully characterized, and little is known about the optimal RNAi delivery method for treating honey bees in different development stages, castle, and aims (Niu et al., 2014).
The Way for Delivery of RNAi
The methods of dsRNA delivery can influence the success of RNAi treatment. The soaking way is suitable only for certain insect cells and tissues as well as for specific insects of developmental stages that readily absorb dsRNA from the solution, and therefore, it is rarely used (Scott et al., 2013). Typically, two primarily dsRNA delivery methods are used: orally or via injection. Both methods have been used to control honey bee disease, although new delivery methods are under development (Jarosch and Moritz, 2011; Jarosch et al., 2011; Hassani et al., 2012). RNAi uptake by cells can occur via passive or active pathways (Whangbo and Hunter, 2008). The responses of cell receptors to these two delivery methods are considerably different and lead to significant differences in effectiveness of the RNAi treatments. For example, injection of dsRNA into the body cavity of locust had a higher sensitivity than that induced by oral dsRNA administration, and four dsRNase in gut juice of the locust can affect the sensitivity of RNAi (Wynant et al., 2014).
For injection delivery, cuticular damage caused stimulates immune function which can further complicate the interpretation of the results (Katoch et al., 2013). In order to avoid or reduce the effects induced by sample manipulation or RNAi injection, Nunes and Simões (2009) used a non-invasive method by using a vitellogenin RNAi system that involved administration of dsRNA to second instar larvae of honey bee. The data indicated that about 60% of treated larvae could develop into adult stage and that approximately 90% of vitellogenin transcripts in worker bees were silenced as compared to those of the untreated control group. Even though the same method of dsRNA delivery was used, the RNAi responses differed.
Delivery methods of RNAi can yield false positive results. Although adult worker bees are highly sensitive to the used delivery method, invasive delivery methods (such as injection) can induce the anticipated responses, which could then activate cellular or humoral actions related to physiology and survival (Nunes and Simões, 2009; Flenniken and Andino, 2013). In addition, recent studies showed that the mortality rate of RNAi-treated honey bees was correlated to the type of dsRNA delivery methods used (rather than the presence of RNAi) and found that the bee mortality was caused by detrimental effects of tissue damage in embryos and larvae of honey bee (Amdam et al., 2003; Aronstein and Saldivar, 2005).
A study has shown that silencing of vitellogenin gene will cause the honey bee workers into extremely earlier forages and leading to behavior maturation (Antonio et al., 2008). To understand better the interaction between different genes, Wang et al. (2013) developed an injection protocol for knockdown the two genes simultaneously, vitellogenin (vg) and ultraspiracle (usp), and found that vg plays a key role among the vg, usp and juvenile hormone (JH) during the process of behavioral maturation. However, Dearden et al. (2009) tried to inject the dsRNA into embryos but not applied any genes in practice.
The Difference of Target Tissues or Genes and Time Injected
RNAi application and efficacy remains variable between genes, organisms and life stages, even insect species. Moreover, gene knockdown efficacy varies in different insect species depending on transcript level of target gene, protein turnover rates and dsRNA uptake efficiency by cells or organs. For instance, the effects obtained by injection of dsRNA on D. melanogaster and Manduca sexta have only been achieved in hemocytes compared to other tissues (Scott et al., 2013). In mosquitoes, most tissues can be reached through injection of dsRNA but depending on genes and dose-dependent in central nervous system (Biessmann et al., 2010). The sensitivity and effectiveness of RNAi vary and depend on the intrinsic characteristics of the target species, as well as the site of target tissue (Xavier, 2010). A few of insects, including the desert locusts and red flour beetle, are amenable to systemic RNAi gene silencing (Miller et al., 2008; Wynant et al., 2014). In contrast, insects such as tobacco hornworm and silk moth are not amenable to systemic RNAi gene silencing (Miller et al., 2008; Xavier, 2010). In order to understand the factors influencing the varied responses amongst different tissues, a study on migratory locust (an agricultural insect pest) was conducted by injecting dsRNA and analyzing the responses in various locust tissues (Ren et al., 2014). The results showed that the locust ovaries were completely insensitive to dsRNA. While further study showed that the injected dsRNA was absent in the follicle cells and oocytes and, the lack of uptake may be the primary factor for the ineffective RNAi response in locust ovaries. These findings reveal the tissue-dependent variability in responses to RNAi.
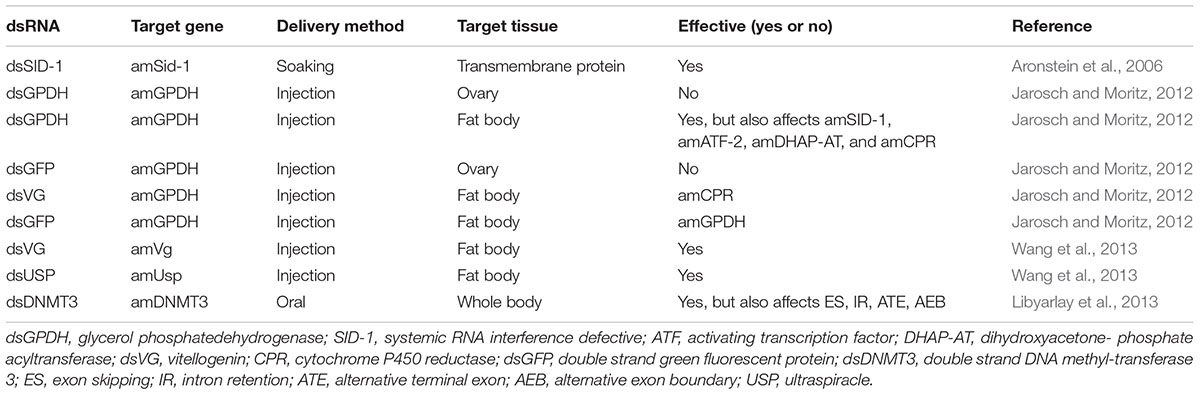
Although RNAi-based methods are commonly used to conduct functional studies of genes, the responses to RNAi treatments drastically vary among different species and tissues. As described by Mutti et al. (2011), they applied RNAi to knockdown the insulin receptor substrate (IRS) and target of rapamycin (TOR) in larvae reared on queen diet to investigated how the nutrition and JH signaling determine the caste of honey bee, and the results showed that knockdown the IRS and TOR will induce the different additional effects in transcriptome, proteome, and total lipid level. Analysis of the systemic effect of RNAi on honey bee demonstrated that abdominal application (injection) of small interfering RNA (siRNA) resulted in gene silencing of primarily the fat body tissue and the other tissue was not amenable to the RNAi treatment with this delivery method (Wang X.B. et al., 2010; Jarosch and Moritz, 2011). Similarly, hemocytes of D. melanogaster have been shown to have lower sensitivity to dsRNA than that shown by fat body (Miller et al., 2008; Xavier, 2010). While, when employed RNAi to knock down the DNA methyl-transferase 3 of honey bee, it caused wide and diverse changes in fat tissue (Libyarlay et al., 2013).
Evaluation of the effect of RNAi treatment at the mRNA and protein expression levels showed that the level of gene suppression by RNAi was directly influenced by the quantity of dsRNA used and the circadian rhythm of the bees (Katoch et al., 2013). The dsRNA injected into the hemolymph relies on the circulation system to carry them to the target sites. However, hemolymph has a heavy impact on the dsRNA and the impact varies amongst different species and target tissues. For example, significant reduction in silencing of Relish in honey bee heads showed that the silencing effect of dsRNA in tissues was discontinuous at the site of injection, abdominal hemocoel (Schlüns and Crozier, 2007). Apart from that, although the effects of RNAi treatment may be the same at the mRNA and protein level, the dsRNA injected will have the effect only in the morning (not evening) at the protein level (Leboulle et al., 2013).
The difference in susceptibility to degradation of dsRNA may be influenced by the size and quantity of dsRNA. A previous study demonstrated that the RNAi efficiency of long dsRNA (>69 bp) was higher than that of short dsRNA (31 bp) (Miller et al., 2012). In addition, the effect of RNAi is dose dependent. Wang et al. (2013) found newly emerged honey bee could well-accept 4 μL dsRNA, while the mortality rapidly increased when more than 4 μL dsRNA was injected. They suggested two or more days injection strategy may be more suitable than the single injection for an experiment which requires higher amount of injection volume. Although there were no reports about the efficiency of the old adult bees feed with dsRNA, emerging bees are used usually to perform RNAi experiment after artificially infected by certain virus, which means that the immunity response is determined on a relatively short period (Smet et al., 2016). As shown in Figure 1B, there was a significant difference in the mortality rate of virus-infected honey bees and virus-infected larvae after treatment with dsRNA against different viruses. Particularly, the mortality of honey bees treated with CSBV was 63.3% after 72 h post-treatment, whereas the mortality rate of DWV was 0% (Maori et al., 2009; Hunter et al., 2010; Liu et al., 2010; Desai et al., 2012). Therefore, even though the effects of different sizes of dsRNA have not been identified in honey bee, further investigations have to be made. Thus, several studies have reported the tissue-dependent variability in effectiveness of RNAi-mediated gene silencing and the findings are summarized in Table 2.
Potential Effects of Other Honey Bee Pathogens
Some pathogens of honey bee will possibly impact expected results. Experiment studies have confirmed that seemingly healthy bees can harbor several diseases, including viral infections (Todd et al., 2007). For example, Chen et al. (2004) revealed that a large number of emergent honey bees were simultaneously infected by multiple viruses such as DWV, SBV, and Kashmir bee virus (KBV). Moreover, when inoculating mix of several viruses of IAPV, SBV, KBV, DWV, and BQCV to cell and adult bees, the results showed that IAPV was rapidly increase to higher level than others even SBV was the main component of the mixture (Carrillo-Tripp et al., 2016). In addition, bees often harbor mixed infections caused by several viruses along with other pathogens such as Nosema apis (Todd et al., 2007). Thus, other pathogens might cause unexpected results. For example, RNAi was used to silence prophenoloxidase, which was considered as a resistance to American foulbrood (AFB), and found that no difference between RNAi treated and untreated groups (Chan, 2012). In addition, the viruses are not easily been detected and leading to unexpected results if they built covert infection at lower level (de Miranda et al., 2010). Therefore, the effectiveness of RNAi treatment against viral infections may be reduced by the prevalence of other pathogens or stresses.
Suppression of Viral RNAi Suppressor
Some plant and animal viruses have developed an effective strategy during the course of evolution with the host. For example, Cucumber mosaic virus has been shown to encode a 2b suppressor that inhibits Arabidopsis Ago1 cleavage activity to counter plant defense (Zhang et al., 2006). Furthermore, suppressors, including 2b, not only bind Ago protein but can also bind dsRNA and siRNA in vitro (Wang et al., 2006; Wang Y. et al., 2010). Subsequently, 1A, an insect virus suppressor of Cricket Paralysis virus (CrPV), was shown to bind to Ago-2 to inhibit slicing of mRNA in vitro (Nayak et al., 2010). In addition, virus suppressors, such as P6 of Cauliflower mosaic virus and B2 of Flock house virus, also bind other proteins or RNA components of RNAi to inhibit the RNAi (Haas et al., 2008; Ruiz-Ferrer and Voinnet, 2009). Based on analysis of viral suppressor of RNAi (VSR) of Drosophila C virus and CrPV, DvExNPGP is representative majorly conserved motif of Dicistroviridae family, which has the ability to express virus suppressor protein (van Rij et al., 2006; Nayak et al., 2010). Likewise, sequence analysis showed that several honey bee viruses including IAPV, KBV and ABPV, also contain a DvExNPGP motif at the 5′ terminus of their genomes, and demonstrated these honey bee viruses might encode a VSR and experiment confirmed the level of IAPV was reduced when silenced IAPV-encoded putative suppressor of RNAi (Chen et al., 2014). Apart from virus suppressors, other mechanisms that enable interference with RNAi and prevent spread of RNA-mediated defense signal have also been identified. For example, p25, a viral movement protein of potato virus X, has been characterized as an effector suppressing anti-viral, the possibility should not be dismissed (Voinnet et al., 2010).
Possible Affects From Genetically Modified Plants
The energy resource of honey bee is major from flowering plants, fruits, or crops and wild plants secreted honeydew. However, genetically modified plants and animals are being increasingly used for pest control or disease prevention. A number of novel approaches for RNAi-based pest control for plants have also been studied (Tian et al., 2009; Li et al., 2011; Zhu et al., 2011). To identify the potential effects of Bt crops, Vélez et al. (2016) employed the dsRNA of Diabrotica virgifera virgifera ATPase and found that RNAi had still impact on larval development and adult life span of honey bee, although there was no significant difference between treatment and control groups. However, despite the development of transgenic plants by using RNAi seems promising, the effect of the transgenic plants on honey bees has not been fully characterized. Moreover, the effect of genetically modified plant components on the dsRNA delivered to the honey bees is also poorly understood.
Future Perspectives
Although the uses of RNAi for controlling viruses hold a significant promise, it is still in its infancy in honey bee and has its limitations and possible risk (Burand and Hunter, 2013). Multiple virus infection is very common in honey bee colonies even in one bee (de Miranda et al., 2010). Different virus strains or highly similar viruses in genome could be present at the same time in such field isolates as DWV and Varroa destructor virus (VDV), or among IAPV, Kashmir bee virus (KBV) and Kakugo virus (KV). Even if purified virus was from experimental infection honey bee samples, it still might host several viruses (Carrillo-Tripp et al., 2016). Therefore, vsiRNAs from siRNA pathway of various viruses can be produced. In addition, it is still unknown about siRNA response of multiple virus infection because there are no infectious clones for single virus to use (Niu et al., 2014). Therefore, it might not get exactly the expected results from siRNA pathway in bees and progress to impact the use of dsRNA in beekeeping practice.
Although considerable progress has been achieved in developing RNAi-based treatments for controlling honey bees viruses, several important questions remain to be answered. First, RNAi-based approaches should include utilization of next generation sequencing technology and the methodology used to identify novel potential target genes (Wang et al., 2011). Previous studies have demonstrated that dsRNA can produce off-target effects that have physiology, developmental, and reproductive consequences in the target organism (Jarosch and Moritz, 2012).
The analysis of honey bee hemolymph components in detail is essential to design an effective RNAi strategy. The stability of dsRNA in the target insects may vary due to the differences in the types of extracellular enzymes secreted into various organs. For example, DNAse/RNAse activity in lepidopteran species can affect the RNAi effectiveness (Liu et al., 2010; Allen and Walker, 2012). In addition, dsRNA was rapidly degraded after it was injected into M. sexta, whereas dsRNA injected in B. germanica persisted for a longer time period (Garbutt et al., 2013). This gap can be alleviated by systematic analysis of molecular physiological basis of RNAi mechanisms in honey bee will facilitate the application of RNAi for resolve of gene function.
Although a number of studies have been performed to assess the application of RNAi in honey bees, the efficiency of gene silencing through the various developmental stages of the honey bee have not been thoroughly characterized. The type of target tissue/organ and the specific development stage in which RNAi responses are obtained indicate not only the characteristics of the examined genes, but can also indicate the functional and developmental role of the target genes. Typically, RNAi is used to target the following three insect developmental stages: egg, larva, and adult. The developmental stage used for RNAi treatment may result in varying responses. For example, injection of dsRNA in pupae and adults of Athalia rosae lead to higher RNAi treatment efficiency than that obtained by using eggs, and the results showed that application of dsRNA via injection into the mid to late larval stages did not yield different results (Yoshiyama et al., 2013). Furthermore, another study showed that RNAi treatment begins to have effects in the larvae infected by CSBV of Apis cerana 12 h after oral application of dsRNA (Liu et al., 2010).
For systematic RNAi application, the size and quantity of dsRNA used should be considered. Based on the findings of the previous studies, we speculate that there may have been two factors that may have influenced the results of RNAi treatment in some studies. The first factor may have been inefficient dsRNA uptake or no response of intracellular RNAi machinery (Nunes and Simões, 2009). The second factor may be related to the optimum quantity of dsRNA that needs to be administered to the bees for obtaining gene silencing. Studies indicate that dsRNA uptake is inefficient in the ovaries of locust (Table 2). Since injection of dsRNA into honey bees is not a convenient and practical method, future research should focus on developing methods that enable efficient uptake of dsRNA by the target tissues and also enable the dsRNA to persist in vivo after oral application. In addition, study is required to address several questions, including the role and interactions of siRNA from other pathogens with the host RNAi machinery. Taken together, we conclude that much works have to be done to make the RNAi-based treatment strategy become reliably effective tool to study gene functions and gene mechanisms of honey bees.
Author Contributions
CH and QD conceived this manuscript. DY, XX, HZ, SY, XW, and DZ participated in the writing, reviewing, and critical analysis of this manuscript. CH and QD coordinated the manuscript.
Funding
The study was supported by the National Natural Science Foundation of China (Grant No. 31572471) and the Agricultural Science and Technology Innovation Program (Grant No. CAAS-ASTIP-2018-IAR).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a shared affiliation, though no other collaboration, with several of the authors (DY, XX, SY, XW, DZ, QD, and CH).
References
Aizen, M. A., Garibaldi, L. A., Cunningham, S. A., and Klein, A. M. (2009). How much does agriculture depend on pollinators? Lessons from long-term trends in crop production. Ann. Bot. 103, 1579–1588. doi: 10.1093/aob/mcp076
Allen, M. L., and Walker, W. B. (2012). Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect Physiol. 58, 391–396. doi: 10.1016/j.jinsphys.2011.12.014
Amdam, G. V., Guidugli, K. R., Norberg, K., and Omholt, S. W. (2003). Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA. BMC Biotechnol. 3:1. doi: 10.1186/1472-6750-3-1
Ament, S. A., Wang, Y., Chen, C. C., Blatti, C. A., Hong, F., Liang, Z. S., et al. (2012). The transcription factor Ultraspiracle influences honey bee social behavior and behavior-related gene expression. PLoS Genet. 8:e1002596. doi: 10.1371/journal.pgen.1002596
Antonio, D. S. M., Guiduglilazzarini, K. R., do Nascimento, A. M., Simões, Z. L., and Hartfelder, K. (2008). RNAi-mediated silencing of vitellogenin gene function turns honeybee (Apis mellifera) workers into extremely precocious foragers. Naturwissenschaften 10, 953–961. doi: 10.1007/s00114-008-0413-9
Aronstein, K., Pankiw, T., and Saldivar, E. (2006). SID-I is implicated in systemic gene silencing in the honey bee. J. Apicul. Res. 45, 20–24. doi: 10.1080/00218839.2006.11101307
Aronstein, K., and Saldivar, E. (2005). Characterization of a honey bee Toll related receptor gene Am18w and its potential involvement in antimicrobial immune defense. Apidologie 36, 3–14. doi: 10.1051/apido:2004062
Au, H. H. T., Elspass, V. M., and Jan, E. (2017). Functional insights into the adjacent stem-loop in honey bee dicistroviruses that promotes IRES-mediated translation and viral infection. J. Virol. 92:e01725-17. doi: 10.1128/JVI.01725-17
Azzami, K., Ritter, W., Tautz, J., and Beier, H. (2012). Infection of honey bees with acute bee paralysis virus does not trigger humoral or cellular immune responses. Arch. Virol. 157, 689–702. doi: 10.1007/s00705-012-1223-0
Biessmann, H., Andronopoulou, E., Biessmann, M. R., Douris, V., Dimitratos, S. D., Eliopoulos, E., et al. (2010). The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS One 5:e9471. doi: 10.1371/journal.pone.0009471
Brutscher, L. M., Daughenbaugh, K. F., and Flenniken, M. L. (2015). Antiviral defense mechanisms in honey bees. Curr. Opin. Insect Sci. 10, 71–82. doi: 10.1016/j.cois.2015.04.016
Brutscher, L. M., McMenamin, A. J., and Flenniken, M. L. (2016). The buzz about honey bee viruses. PLoS Pathog. 12:e1005757. doi: 10.1371/journal.ppat.1005757
Burand, J. P., and Hunter, W. B. (2013). RNAi: future in insect management. J. Invertebr. Pathol. 112, S68–S74. doi: 10.1016/j.jip.2012.07.012
Campbell, E. M., Budge, G. E., Watkins, M., and Bowman, A. S. (2016). Transcriptome analysis of the synganglion from the honey bee mite, Varroa destructor and RNAi knockdown of neural peptide targets. Insect Biochem. Mol. Biol. 1, 116–126. doi: 10.1016/j.ibmb.2015.12.007
Carrillo-Tripp, J., Dolezal, A. G., Goblirsch, M. J., Miller, W. A., Toth, A. L., and Bonning, B. C. (2016). In vivo and in vitro infection dynamics of honey bee viruses. Sci. Rep. 6:22265. doi: 10.1038/srep22265
Chan, M. Y. (2012). Development and application of honey bee in vitro systems. PLoS One 8:e69831. doi: 10.1371/journal.pone.0069831
Chen, Y., Zhao, Y., Hammond, J., Hsu, H., Evans, J., and Feldlaufer, M. (2004). Multiple virus infections in the honey bee and genome divergence of honey bee viruses. J. Inverteb. Pathol. 87, 84–93. doi: 10.1016/j.jip.2004.07.005
Chen, Y. P., Pettis, J. S., Corona, M., Chen, W. P., Li, C. J., Spivak, M., et al. (2014). Israeli acute paralysis virus: epidemiology, pathogenesis and implications for honey bee health. PLoS Pathog. 10:e1004261. doi: 10.1371/journal.ppat.1004261
Cox-Foster, D. L., Holmes, E. C., Palacios, G., Evans, J. D., Moran, N. A., Quan, P. L., et al. (2007). A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287. doi: 10.1126/science.1146498
de Miranda, J. R., Cordoni, G., and Budge, G. (2010). The acute bee paralysis virus-Kashmir bee virus-Israeli acute paralysis virus complex. J. Invertebr. Pathol. 103, S30–S47. doi: 10.1016/j.jip.2009.06.014
Dearden, P. K., Duncan, E. J., and Wilson, M. J. (2009). RNA interference (RNAi) in honeybee (Apis mellifera) embryos. Cold Spring Harb. Protoc. 6:rot5228. doi: 10.1101/pdb.prot5228
Desai, S. D., Eu, Y. J., Whyard, S., and Currie, R. W. (2012). Reduction in deformed wing virus infection in larval and adult honey bees (Apis mellifera L.) by double-stranded RNA ingestion. Insect Mol. Biol. 21, 446–455. doi: 10.1111/j.1365-2583.2012.01150.x
Di Lelio, I., Varricchio, P., Di Prisco, G., Marinelli, A., Lasco, V., Caccia, S., et al. (2014). Functional analysis of an immune gene of Spodoptera littoralis by RNAi. J. Insect Physiol. 64, 90–97. doi: 10.1016/j.jinsphys.2014.03.008
Di Prisco, G., Pennacchio, F., Caprio, E., Boncristiani, H. F., Evans, J. D., and Chen, Y. (2011). Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 92, 151–155. doi: 10.1099/vir.0.023853-0
Elsik, C. G., Worley, K. C., Bennett, A. K., Beye, M., Camara, F., Childers, C. P., et al. (2014). Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15:86. doi: 10.1186/1471-2164-15-86
Evans, J. D., Chen, Y. P., Pettis, J., and Williams, V. (2009). Bee cups: single-use cages for honey bee experiments. J. Apicul. Res. 4, 300–302. doi: 10.1080/00218839.2009.11101548
Fernández-Miragall, O., Quinto, S. L., and Martínez-Salas, E. (2009). Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 139, 172–182. doi: 10.1016/j.virusres.2008.07.009
Flenniken, M. L., and Andino, R. (2013). Non-specific dsRNA-mediated antiviral response in the honey bee. PLoS One 8:e77263. doi: 10.1371/journal.pone.0077263
Fung, E., Hill, K., Hogendoorn, K., Glatz, R. V., Napier, K. R., Bellgard, M. I., et al. (2018). De novo, assembly of honey bee RNA viral genomes by tapping into the innate insect antiviral response pathway. J. Invertebr. Pathol. 152, 38–47. doi: 10.1016/j.jip.2018.01.002
Garbian, Y., Maori, E., Kalev, H., Shafir, S., and Sela, I. (2012). Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathog. 8:e1003035. doi: 10.1371/journal.ppat.1003035
Garbutt, J. S., Bellés, X., Richards, E. H., and Reynolds, S. E. (2013). Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: evidence from Manduca sexta and Blattella germanica. J. Insect Physiol. 59, 171–178. doi: 10.1016/j.jinsphys.2012.05.013
Haas, G., Azevedo, J., Moissiard, G., Geldreich, A., Himber, C., Bureau, M., et al. (2008). Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J. 27, 2102–2112. doi: 10.1038/emboj.2008.129
Hassani, A. K., Schuster, S., Dyck, Y., Demares, F., Leboulle, G., and Armengaud, C. (2012). Identification, localization and function of glutamate-gated chloride channel receptors in the honeybee brain. Eur. J. Neurosci. 36, 2409–2420. doi: 10.1111/j.1460-9568.2012.08144.x
Hou, C. S., Hadassah, R., Yossi, S., and Chejanovsky, N. (2014). Dynamics of the presence of Israeli acute paralysis virus in honey bee colonies with colony collapse disorder. Viruses 6, 2012–2027. doi: 10.3390/v6052012
Hunter, W., Ellis, J., Vanengelsdorp, D., Hayes, J., Westervelt, D., Glick, E., et al. (2010). Large-scale Field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLoS Pathog. 6:e1001160. doi: 10.1371/journal.ppat.1001160
Jarosch, A., and Moritz, R. A. (2011). Systemic RNA-interference in the honeybee Apis mellifera: tissue dependent uptake of fluorescent siRNA after intra-abdominal application observed by laser-scanning microscopy. J. Insect Physiol. 57, 851–857. doi: 10.1016/j.jinsphys.2011.03.013
Jarosch, A., and Moritz, R. A. (2012). RNA interference in honeybees: off-target effects caused by dsRNA. Apidologie 43, 128–138. doi: 10.1007/s13592-011-0092-y
Jarosch, A., Stolle, E., Crewe, R. M., and Moritz, R. A. (2011). Alternative splicing of a single transcription factor drives selfish reproductive behavior in honeybee workers (Apis mellifera). Proc. Natl. Acad. Sci. U.S.A. 108, 15282–15287. doi: 10.1073/pnas.1109343108
Kamakura, M. (2011). Royalactin induces queen differentiation in honeybees. Nature 473, 478–483. doi: 10.1038/nature10093
Katoch, R., Sethi, A., Thakur, N., and Murdock, L. L. (2013). RNAi for insect control: current perspective and future challenges. Appl. Biochem. Biotechnol. 4, 847–873. doi: 10.1007/s12010-013-0399-4
Leboulle, G., Niggebrügge, C., Roessler, R., Briscoe, A. D., Menzel, R., and Ibarra, N. H. D. (2013). Characterisation of the RNA interference response against the long-wavelength receptor of the honeybee. Insect Biochem. Mol. Biol. 43, 959–969. doi: 10.1016/j.ibmb.2013.07.006
Li, X., Zhang, M., and Zhang, H. (2011). RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS One 6:e17788. doi: 10.1371/journal.pone.0017788
Libyarlay, H., Li, Y., Stroud, H., Feng, S., Newman, T. C., Kaneda, M., et al. (2013). RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. U.S.A. 110, 12750–12755. doi: 10.1073/pnas.1310735110
Liu, J., Swevers, L., Iatrou, K., Huvenne, H., and Smagghe, G. (2012). Bombyx mori DNA/RNA non-specific nuclease: expression of isoforms in insect culture cells, subcellular localization and functional assays. J. Insect Physiol. 58, 1166–1176. doi: 10.1016/j.jinsphys.2012.05.016
Liu, X., Zhang, Y., Yan, X., and Han, R. (2010). Prevention of Chinese sacbrood virus infection in Apis cerana using RNA interference. Curr. Microbiol. 61, 422–428. doi: 10.1007/s00284-010-9633-2
Louis, T., Musso, P. Y., de Oliveira, S. B., Garreau, L., Giurfa, M., Raymond, V., et al. (2012). Amelα8 subunit knockdown in the mushroom body vertical lobes impairs olfactory retrieval in the honeybee, Apis mellifera. Eur. J. Neurosci. 36, 3438–3450. doi: 10.1111/j.1460-9568.2012.08261.x
Mao, Y. B., Cai, W. J., Wang, J. W., Hong, G. J., Tao, X. Y., Wang, L. J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotech. 25, 1307–1313. doi: 10.1038/nbt1352
Maori, E., Paldi, N., Shafir, S., Kalev, H., Tsur, E., Glick, E., et al. (2009). IAPV, a bee-affecting virus associated with colony collapse disorder can be silenced by dsRNA ingestion. Insect Mol. Biol. 18, 55–60. doi: 10.1111/j.1365-2583.2009.00847.x
Mathieu, P., and Watts, G. (1989). Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 3, 643–649.
Miller, S., Brown, S., and Tomoyasu, Y. (2008). Larval RNAi in Drosophila? Dev. Genes Evol. 218, 505–510. doi: 10.1007/s00427-008-0238-8
Miller, S., Miyata, K., Brown, S., and Tomoyasu, Y. (2012). Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS One 7:e47431. doi: 10.1371/journal.pone.0047431
Mustard, J. A., Pham, P. M., and Smith, B. H. (2010). Modulation of motor behavior by dopamine and the D1-like dopamine receptor AmDOP2 in the honey bee. J. Insect Physiol. 56, 422–430. doi: 10.1016/j.jinsphys.2009.11.018
Mutti, N. S., Dolezal, A. G., Wolschin, F., Mutti, J. S., Gill, K. S., and Amdam, G. V. (2011). IRS and TOR nutrient-signaling pathways act via juvenile hormone to influence honey bee caste fate. J. Exp. Biol. 1, 3977–3984. doi: 10.1242/jeb.061499
Nayak, A., Berry, B., Tassetto, M., Kunitomi, M., Acevedo, A., Deng, C., et al. (2010). Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 17, 547–554. doi: 10.1038/nsmb.1810
Niu, J., Meeus, I., Cappelle, K., Piot, N., and Smagghe, G. (2014). The immune response of the small interfering RNA pathway in the defense against bee viruses. Curr. Opin. Insect Sci. 6, 22–27. doi: 10.1016/j.ibmb.2015.12.006
Nunes, M. F., Aleixo, A. C., Barchuk, A. R., Bomtorin, A. D., Grozinger, C. M., and Simões, Z. P. (2013). Non-target effects of green fluorescent protein (GFP)-derived double-stranded RNA (dsRNA-GFP) used in honey bee RNA interference (RNAi) assays. Insects 4, 90–103. doi: 10.3390/insects4010090
Nunes, M. F., and Simões, Z. P. (2009). A non-invasive method for silencing gene transcription in honeybees maintained under natural conditions. Insect Biochem. Mol. Biol. 39, 157–160. doi: 10.1016/j.ibmb.2008.10.011
Paldi, N., Glick, E., Oliva, M., Zilberberg, Y., Aubin, L., Pettis, J., et al. (2010). Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl. Environ. Microbiol. 76, 5960–5964. doi: 10.1128/AEM.01067-10
Powell, M. E., Bradish, H. M., Gatehouse, J. A., and Fitches, E. C. (2016). Systemic RNAi in the small hive beetle (Aethina tumida Murray, Coleoptera: Nitidulidae), a serious pest of the European honey bee (Apis mellifera). Pest Manag. Sci. 1, 53–63. doi: 10.1002/ps.4365
Ren, D., Cai, Z., Song, J., Wu, Z., and Zhou, S. (2014). dsRNA uptake and persistence account for tissue-dependent susceptibility to RNA interference in the migratory locust, Locusta migratoria. Insect Mol. Biol. 23, 175–184. doi: 10.1111/imb.12074
Ruiz-Ferrer, V., and Voinnet, O. (2009). Roles of plant small RNAs in biotic stress responses. Ann. Rev. Plant Biol. 60, 485–510. doi: 10.1146/annurev.arplant.043008.092111
Schlüns, H., and Crozier, R. H. (2007). Relish regulates expression of antimicrobial peptide genes in the honeybee, Apis mellifera, shown by RNA interference. Insect Mol. Biol. 16, 753–759. doi: 10.1111/j.1365-2583.2007.00768.x
Scott, J. G., Michel, K., Bartholomay, L. C., Siegfried, B. D., Hunter, W. B., Smagghe, G., et al. (2013). Towards the elements of successful insect RNAi. J. Insect Physiol. 59, 1212–1221. doi: 10.1016/j.jinsphys.2013.08.014
Shen, M., Cui, L., Ostiguy, N., and Cox-Foster, D. (2005). Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic Varroa mite. J. Gen. Virol. 86, 2281–2289. doi: 10.1099/vir.0.80824-0
Smet, L. D., Ravoet, J., Wenseleers, T., and de Graaf, D. C. (2016). Expression of key components of theRNAi machinery are suppressed in Apis mellifera that suffer a high virus infection. Entomol. Sci. 20, 1–11. doi: 10.1111/ens.12227
Tian, H., Peng, H., Yao, Q., Chen, H., Xie, Q., Tang, B., et al. (2009). Developmental control of a Lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One 4:e6225. doi: 10.1371/journal.pone.0006225
Todd, J. H., Miranda, J. R. D., and Ball, B. V. (2007). Incidence and molecular characterization of viruses found in dying New Zealand honey bee (Apis mellifera) colonies infested with Varroa destructor. Apidologie 4, 354–367. doi: 10.1051/apido:2007021
Toplak, I., Urška, J. C., Katherine, A., and Aleš, G. (2013). Chronic bee paralysis virus and Nosema ceranae experimental co-infection of winter honey bee workers (Apis mellifera L.). Viruses 5, 2282–2297. doi: 10.3390/v5092282
van Rij, R. P., Saleh, M. C., Berry, B., Foo, C., Houk, A., Antoniewski, C., et al. (2006). The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20, 2985–2995. doi: 10.1101/gad.1482006
Vélez, A. M., Jurzenski, J., Matz, N., Zhou, X., Wang, H., Ellis, M., et al. (2016). Developing an in vivo toxicity assay for RNAi risk assessment in honey bees, Apis mellifera L. Chemosphere 144, 1083–1090. doi: 10.1016/j.chemosphere.2015.09.068
Voinnet, O., Lederer, C., and Baulcombe, D. C. (2010). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103, 157–167. doi: 10.1016/S0092-8674(00)00095-7
Wang, X. B., Wu, Q., Ito, T., Cillo, F., Li, W. X., Chen, X., et al. (2010). RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 484–489. doi: 10.1073/pnas.0904086107
Wang, Y., Mutti, N. S., Ihle, K. E., Siegel, A., Dolezal, A. G., Kaftanoglu, O., et al. (2010). Down-regulation of honey bee IRS gene biases behavior toward food rich in protein. PLoS Genet. 6:e1000896. doi: 10.1371/journal.pgen.1000896
Wang, X. H., Aliyari, R., Li, W. X., Li, H. W., Kim, K., Carthew, R., et al. (2006). RNA interference directs innate immunity against viruses in adult Drosophila. Science 312, 452–454. doi: 10.1126/science.1125694
Wang, Y., Baker, N., and Amdam, G. V. (2013). RNAi-mediated double gene knockdown and gustatory perception measurement in honey bees (Apis mellifera). J. Vis. Exp. 77:e50446. doi: 10.3791/50446
Wang, Y., Brent, C. S., Fennern, E., and Amdam, G. V. (2012). Gustatory perception and fat body energy metabolism are jointly affected by vitellogenin and juvenile hormone in honey Bees. PLoS Genet. 8:e1002779. doi: 10.1371/journal.pgen.1002779
Wang, Y., Zhang, H., Li, H., and Miao, X. (2011). Second-generation sequencing supply an effective way to screen RNAi targets in large scale for potential application in pest insect control. PLoS One 6:e18644. doi: 10.1371/journal.pone.0018644
Whangbo, J. S., and Hunter, C. P. (2008). Environmental RNA interference. Trends Genet. 24, 297–305. doi: 10.1016/j.tig.2008.03.007
Wilson, M. J., and Dearden, P. K. (2012). Pair-rule gene orthologues have unexpected maternal roles in the honeybee (Apis mellifera). PLoS One 7:e46490. doi: 10.1371/journal.pone.0046490
Wynant, N., Duressa, T. F., Santos, D., Van Duppen, J., Proost, P., Huybrechts, R., et al. (2014). Lipophorins can adhere to dsRNA, bacteria and fungi present in the hemolymph of the desert locust: a role as general scavenger for pathogens in the open body cavity. J. Insect Physiol. 64, 7–13. doi: 10.1016/j.jinsphys.2014.02.010
Xavier, B. (2010). Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol. 55, 111–128. doi: 10.1146/annurev-ento-112408-085301
Yoshiyama, N., Tojo, K., and Hatakeyama, M. (2013). A survey of the effectiveness of non-cell autonomous RNAi throughout development in the sawfly, Athalia rosae (Hymenoptera). J. Insect Physiol. 59, 400–407. doi: 10.1016/j.jinsphys.2013.01.009
Zhang, J., Zhang, Y., and Han, R. (2016). The high-throughput production of dsRNA against sacbrood virus for use in the honey bee Apis cerana (Hymenoptera: Apidae). Virus Genes 5, 698–705. doi: 10.1007/s11262-016-1346-6
Zhang, X., Yuan, Y. R., Pei, Y., Lin, S. S., Tuschl, T., Patel, D. J., et al. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. doi: 10.1101/gad.1495506
Keywords: RNAi, honey bee viruses, sensitivity of RNAi, gene function, Apis mellifera
Citation: Yang D, Xu X, Zhao H, Yang S, Wang X, Zhao D, Diao Q and Hou C (2018) Diverse Factors Affecting Efficiency of RNAi in Honey Bee Viruses. Front. Genet. 9:384. doi: 10.3389/fgene.2018.00384
Received: 21 May 2018; Accepted: 27 August 2018;
Published: 11 September 2018.
Edited by:
Jianke Li, Institute of Apiculture Research (CAAS), ChinaCopyright © 2018 Yang, Xu, Zhao, Yang, Wang, Zhao, Diao and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingyun Diao, ZHF5dW4xQDEyNi5jb20= Chunsheng Hou, aG91Y2h1bnNoZW5nQGNhYXMuY24=
†These authors have contributed equally to this work
 Dahe Yang
Dahe Yang Xiang Xu
Xiang Xu Hongxia Zhao
Hongxia Zhao Sa Yang
Sa Yang Xinling Wang1,2
Xinling Wang1,2 Chunsheng Hou
Chunsheng Hou