
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 20 September 2018
Sec. Epigenomics and Epigenetics
Volume 9 - 2018 | https://doi.org/10.3389/fgene.2018.00375
PIWI-LIKE 2, a member of the ARGONAUTE protein family, is exclusively expressed in pre-pachytene and pachytene stages of spermatogenesis. PIWI-LIKE 2 acts in the germ cell development and the silencing of retrotransponsons to maintain the genomic integrity and stem cell character. In the present study we investigated DNA methylation as potential mechanism for the regulation of human PIWI-LIKE 2 expression in cell lines related to spermatozoa precursor cells. We detected a high methylation of the PIWI-LIKE 2 promoter in TCam-2 cells, while in NT2/D1 cells the promoter was hypomethylated. Concordantly, PIWI-LIKE 2 expression is higher in NT2/D1 cells than in TCam-2 cells. By demethylation of the promoter with 5′-Aza-2′-deoxycytidine, PIWI-LIKE 2 expression in TCam-2 was increased, while in NT2/D1 no alterations in PIWI-LIKE 2 expression could be detected. In conclusion, we analyzed the DNA methylation driving PIWI-LIKE 2 expression in undifferentiated germ cell tumors and demonstrated an epigenetic basis for PIWI-LIKE 2 expression in this cell type.
Spermatogenesis is a highly coordinated process that involves mitotic and meiotic divisions, as well as cellular differentiation to produce mature spermatozoa from undifferentiated germline stem cells. Sperm development is associated with the establishment of extensive chromatin and epigenetic changes. This process allows genomic chemical modifications that affect gene expression without altering the underlying nucleotide sequence (Cui et al., 2016). Epigenetic modifications are characterized by the regulation of non-coding RNA, chromatin remodeling, histone modifications and DNA methylation (Stuppia et al., 2015). DNA methylation occurs at the 5′-position of cytosine residues, typically in the context of CpG dinucleotides, which are associated with promoter regions of genes in about 60–80% (Seisenberger et al., 2013; Liyanage et al., 2014). Methylation of CpG sites leads to transcriptional gene silencing in consequence of an altered condensation status of the chromatin. Genomic methylation profiling analysis reveals cell type specific methylation patterns, which result in cell-type specific differential gene expressions and thus differentially regulated tissue-specific processes. Methylation marks for proper male gametogenesis are established during genomic reprogramming in early embryonic development and indicate an exclusive genetic profile of male germ cells compared to somatic tissues (Santos et al., 2002; Bourc’his and Bestor, 2004).
Another factor for successful male germ cell development is the coordinated and timely expression of the members of the PIWI-LIKE gene family, the germ line specific subclade of the Argonaute proteins. These proteins are characterized by the presence of their evolutionarily conserved PAZ (PIWI-ARGONAUTE-ZWILLE) and PIWI (P-element induced wimpy testis) domains (Cox et al., 1998; Cerutti et al., 2000). These structural features function in transcriptional and post-transcriptional control by binding to small RNAs. Thereby PAZ domain is responsible for 3’-end recognition of the bound small RNA, whereas the PIWI domain is involved in mRNA target binding and cleavage (Song et al., 2004; Jinek and Doudna, 2009). PIWI-LIKE proteins are known to bind a distinct class of small RNAs. These small RNAs, called piRNAs (piwi-interacting RNAs), are frequently 24–31 nt in length, map to distinct genomic regions and share a high preference for 5′ Uridine (Aravin et al., 2006, 2007; Grivna et al., 2006). In germline development, PIWI/piRNA complexes mediate the self-renewal of germline stem cells and maintain genomic integrity through suppression of mobile genetic elements and retrotransposons, such as long interspersed nuclear elements-1 (LINE-1) (Unhavaithaya et al., 2009; Marchetto et al., 2013).
The human PIWI subfamily comprises HIWI (PIWI-LIKE 1), HILI (PIWI-LIKE 2), HIWI3 (PIWI-LIKE 3) and HIWI2 (PIWI-LIKE 4). PIWI-LIKE 2 is exclusively expressed in spermatogonia and pre-meiotic spermatocytes (Sasaki et al., 2003). However, it has been demonstrated to be temporarily activated in somatic cells in response to DNA damages (Lim et al., 2013) Furthermore, PIWI-LIKE 2 reveals ectopic expression in several tumor entities, and its intragenically activated products, such as PL2L60A, are expressed in various types of tumor cell lines (Ye et al., 2010; Gainetdinov et al., 2014). Potential regulation mechanisms of PIWI-LIKE 2 expression are scarcely investigated. Normally, MILI (the murine homolog of HILI/PIWI-LIKE 2) is exclusively expressed in the spermatogonia and spermatocytes (Sasaki et al., 2003) and in the female oocytes and supporting cells (Lim et al., 2013).
The aim of this study was to identify epigenetic mechanisms that may underlie the differential expression of PIWI-LIKE 2 in germ line and somatic tissues by analyzing the basal promoter methylation of PIWI-LIKE 2 and the effects of a modification of this methylation by 5′-Aza-2′-deoxycytidine treatment in two different in vitro models, TCam-2 and NT2/D1 cells.
Cell lines used for the experiments were the following: TCam-2, a human seminoma cell line with characteristics similar to spermatogonia; and NT2/D1, a human teratocarcinoma cell line with characteristics and gene expression profiles similar to cultured human embryonic stem cells. TCam-2 cells were grown in RPMI 1640 GlutaMAXTM supplemented with 10% fetal calf serum (FCS) and 1% penicillin/streptomycin. NT2/D1 cells were cultivated in DMEM GlutaMAXTM supplemented with 10% FCS and 1% penicillin/streptomycin. The cells were grown as monolayer at 37°C in a 5% CO2 humidified incubator.
For 5′-Aza-2′-deoxycytidine treatment TCam-2 and NT2/D1 cells were plated in a concentration of 4×105 cell in corresponding cultivation medium supplemented with 5′ μM or 10 μM 5′-Aza-2′-deoxycytine (Sigma Aldrich, Taufkirchen, Germany) for 72 h, afterward DNA, RNA and protein was isolated as described. DMSO served as vehicle control.
Genomic DNA was isolated from each cell line with and without treatment of 5′-Aza-2′-deoxycytidine. DNA was isolated using MasterPureTM Complete DNA Purification Kit (Epicentre, United States) according to the manufacturer’s instructions. Extracted DNA was quantified using the BioPhotometer (Eppendorf, Hamburg, Germany) and the purity was determined by OD260/OD280 ratio.
Five hundred nanograms genomic DNA was treated with sodium bisulfite using the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany). Bisulfite treatment converts unmethylated cytosines into uracils while leaving methylated cytosines unmodified. During PCR amplification the generated uracils are converted to thymidine. The methylation specific primers were designed using MethPrimer software1. DNA Primers are listed in Table 1. 50 ng sodium bisulfite treated DNA was used for each PCR reaction. PCR was performed under following conditions: 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. Purified PCR products were cloned into a pCR2.1 vector and ten clones from each sample were submitted to SEQLAB Sequencing Laboratories (Göttingen, Germany) for sequencing analysis. DNA methylation data were analyzed using BiQ Analyzer Software (Max Planck Institute, Munich, Germany).
Different PIWI-LIKE 2 upstream regulating regions were generated using PCR (Primers listed in Table 1). Products were ligated into a pCR2.1 vector according to the manufacturer’s protocol (Life Technologies, Germany). Plasmids were checked for the correct inserts using BigDye® Terminator v1.1 Cycle Sequencing Kit by ZMG. 5 μg Plasmid DNA, including Basic pGL4.10[luc2] vector, were digested with Hind III and XhoI (1 U/μl) for 2h at 37°C. Digest was analyzed using 1% agarose gel and purified using the DNA Gel Extraction Kit (Thermo Fisher, Germany). Fragments were subcloned into pGL4.10[luc2] vector according to the manufacturer’s instructions and validated using sequencing analysis.
One microgram Xho I/HindIII digested DNA was treated for 4 h with M.SssI methylase (4 U/μl, NEB) according to the manufacturer’s protocol. In vitro methylation was controlled by BSTU I digest at 60°C for 1 h. Fragments were ligated into a pGL4.10[luc2] vector (Promega, Mannheim, Germany) and used for luciferase reporter assays.
Total RNA was extracted from cell lines using TRIzol according to the manufacturer’s instructions. 1 μg of the total RNA was reverse transcribed using RevertAid H Minus First Strand cDNA Synthesis Kit (Life Technologies, Germany). Quantitative PCR was performed using the MyiQ Real Time PCR Detection System (Biorad, Germany). PIWI-LIKE 2 (Hs01032719_m1) TaqMan Primers targeting exon 4–5 were used for the detection of PIWI-LIKE 2 expression. GAPDH was used as reference gene (fw: 5′-CAAGGTCATCCATGACAACTTTG-3′ and rv: 5′-GTCCACCACCCTGTTGCTGTAG-3′). The data were normalized to GAPDH levels, and levels of PIWI-LIKE 2 mRNA were determined using the 2-ΔCt method.
Proteins were isolated from TCam-2 cells and NT2/D1 cells by RIPA buffer 50 mM TRIS–HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS + 1 unit of protease inhibitor cocktail (Roche, Mannheim, Germany). The lysates were incubated for 10 min at 4°C, followed by a 10 min centrifugation step at 13,000 rpm, 4°C. The supernatant was used for western blot analysis. The protein concentration was determined by BCA Protein Assay Kit (Biorad, Germany).
For protein analysis, we used the Novex Mini Cell Electrophoresis (Life Technologies, Germany) and Mini Trans-Blot system (BioRad, Germany). Protein was prepared by standard protocols and electrophoresed at 125 mA for 60 min. Gels were blotted onto a polyvinylidene fluoride membrane in a BioRad blotting chamber for 2 h at 200 V at 4°C according to published protocols. After the membrane had been blocked in PBSTM (phosphate-buffered saline, 0.1% v/v Tween 20, 5% low fat milk powder), it was incubated in a solution containing primary antibodies raised against PIWI-LIKE 2 (ab181340, 1:5000, Abcam, Germany) or ß-ACTIN (AC-15 mouse antibody, 1:10,000, Sigma-Aldrich, Germany) at 4°C overnight. Secondary antibody (anti-rabbit-horseradish peroxidase [HRP], DAKO, Germany) incubation in a 1:10,000 dilution was applied for 1 h at RT. Finally, the membrane was incubated in 1 ml Amersham ECL prime Western Blot detection system and the signal was detected by using Kodak X-Ray film (Kodak, Germany).
The PIWI-LIKE 2 fragments used in the luciferase reporter assays were amplified by PCR (primers including XhoI and HindIII recognition sites listed in Table 1). PCR was carried out at 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 2 min. pGL4.10[luc2] vector (Promega, Mannheim, Germany) containing the luciferase gene under control of the PIWI-LIKE 2 fragments was transfected in 1 × 104 TCam2 or NT2/D1 cells at a ratio of 3:2 (μl transfection reagent: μg DNA). FuGeneHD (Roche, Germany) was used as transfection reagent. Additionally pGL4.10[luc2] PIWI-LIKE 2 constructs methylated by M.SssI were transfected. Transfection of empty pGL4.10[luc2] vector and untreated cells were used as additional controls. Transfections were conducted as co-transfections with pGL4.74 empty vector containing a constitutive expressed Renilla fly as normalization control in a ratio of 1:50 (Renilla fly: Luciferase fly). 48 h later, cells were lysed for 20 min in 50 μl 1× lysis buffer. Luciferase activity was measured after addition of the luciferase assay buffer by a luminometer in a 96-well plate.
Firstly, we analyzed the region upstream of PIWI-LIKE 2 transcription start site (TSS) to identify a putative promoter as well as target sites for CpG methylations that might contribute to gene expression regulation. Using PromoterScan1.7 Software2 we identified the region +4 bp to +254 bp adjacent to the PIWI-LIKE 2 TSS containing a putative promoter. Further analysis of the selected region using MethPrimer Software3 showed the occurrence of 41 CpG dinucleotides set in a CpG island. All of these CpG sites were located within the -300 to +300 bp region relative to the TSS of PIWI-LIKE 2 (Figure 1A). We investigated the methylation status of the PIWI-LIKE 2 promoter using bisulfite sequencing. The basal levels of PIWI-LIKE 2 promoter methylation differ in the analyzed cell lines. In the seminoma cell line TCam-2, the PIWI-LIKE 2 promoter is heavily methylated (85%), whereas in the pluripotent embryonal carcinoma cell line NT2/D1, the PIWI-LIKE 2 promoter exhibits a low promoter methylation (22%) (Figure 1B).
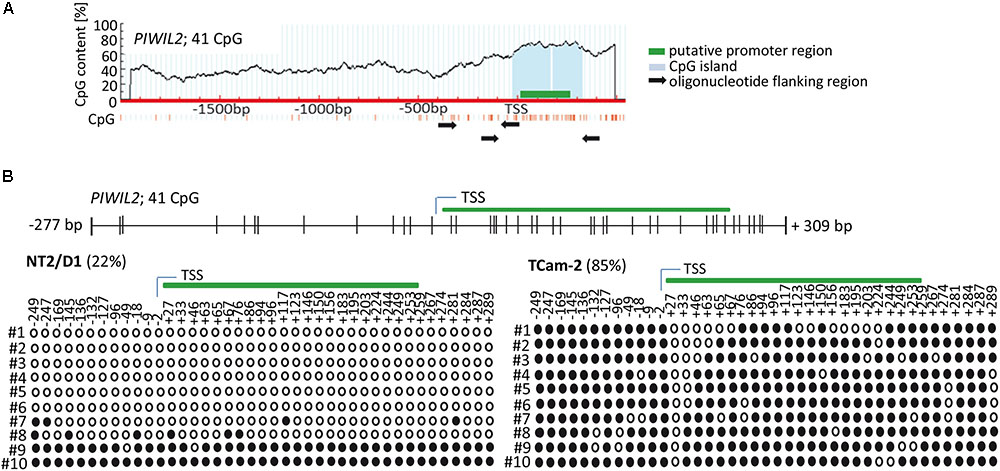
FIGURE 1. PIWI-LIKE 2 promoter analysis (A) schematic diagram of CpG dinucleotide density across the genomic PIWI-LIKE 2 locus. Green box indicates putative promoter region. Light blue box shows the predicted CpG island. Black arrows designate the location of the oligonucleotides used for bisulfite sequencing PCR. (B) Upper panel shows the location of the investigated CpG dinucleotides according to the transcription start site (TSS) of PIWI-LIKE 2. Lower panel shows basic PIWI-LIKE 2 methylation level of the investigated CpG dinucleotides in NT2/D1 and TCam-2 cells. Filled circles indicate methylated CpGs, empty circles indicate demethylated CpGs. #1-10 refers to the different clones analyzed per PIWI-LIKE 2 promoter fragment.
Next, we tested whether the basal PIWI-LIKE 2 mRNA expression correlates to the CpG promoter methylation in both analyzed human cell lines. mRNA expression was analyzed using qRT-PCR, and cT (cycle threshold) values from three independent experiments were taken to assess mean PIWI-LIKE 2 mRNA expressions calculated by the 2-ΔCt method with normalization to GAPDH mRNA expression. We found that the expression of PIWI-LIKE 2 mRNA was 52 times higher in NT2/D1 cells (2-ΔCtmean = 5.14∗10-4) compared to TCam-2 cells (2-ΔCtmean = 9.77∗10-6; p = 0.01) (Figure 2A). This difference in mRNA expression could be explained by a different methylation status of both cell lines. Furthermore, the induction of PIWI-LIKE 2 expression by the demethylating agent 5′-Aza-2′-deoxycytidine (5AzadC) was analyzed in the used cell lines. Treatment of TCam-2 cells with 10 μM 5AzadC led to a 50 times enhanced expression of PIWI-LIKE 2 mRNA (2-ΔCtmean = 4.89∗10-4; p = 0.027) compared to the basic mRNA expression. Protein expression of PIWI-LIKE 2 was measured in TCam2 after 72 h incubation with and without 5AzadC (Figure 2B). PIWI-LIKE 2 protein expression was increased with rising 5AzadC concentration (5 μM–10 μM; lane 3,4) compared to the vehicle control (lane 2). An increase of protein products with 130 kDa, 110 kDa, 80 kDa, and 60 kDa could be detected. Different molecular weights could point out on PIWI-LIKE 2 splice variants of different length and modifications. Neither PIWI-LIKE 2 mRNA expression (2-ΔCtmean = 5.51E-04) nor protein expression (not shown) was changed after treatment with 5AzadC in NT2/D1 cell line. Next, we examined the impact of 5AzadC on PIWI-LIKE 2 promoter methylation status in NT2/D1 and TCam-2 cell lines. Treatment with 5AzadC led to a decrease in methylation of the CpG dinucleotides located within the -130 to +65 bp region in TCam-2 and reduced the overall methylation of the investigated promoter segment from 85 to 73% (Figure 3). NT2/D1 cell line exhibited no decrease in methylation status of the analyzed promoter site. These results suggest that basal methylation of PIWI-LIKE 2 depends on the origin of the cell line. Treatment of cells with 5-AzadC allows a partial demethylation of the PIWI-LIKE 2 promoter. Of interest, knockdown of PIWI-LIKE 2 in either NT2/D1 or TCam-2 cells resulted in a significant reduction of proliferation (-48.2 and –19.6%, respectively, compared to untreated control) and cell vitality (-62.9 and -30.6%, respectively) and in a significant induction of apoptosis (+1351% and + 716%, respectively; see Supplementary Figure 1, 2).
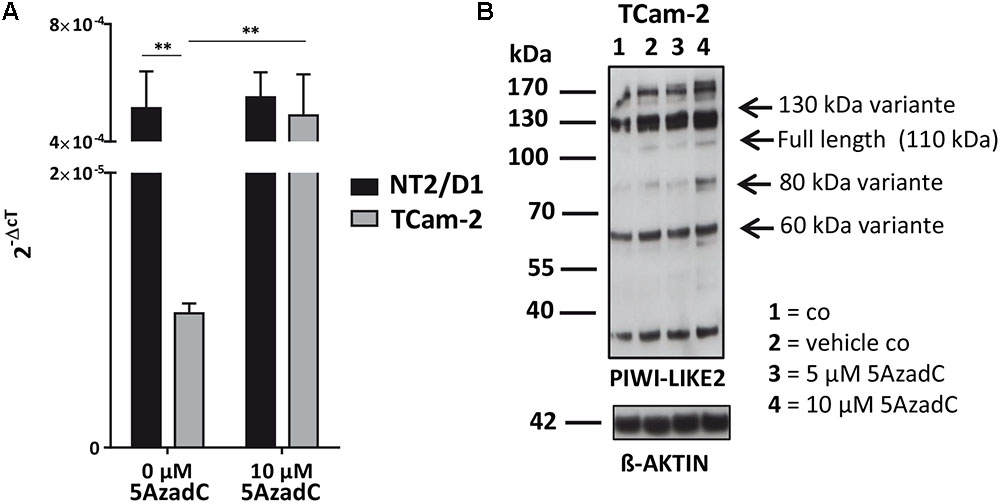
FIGURE 2. PIWI-LIKE 2 mRNA and protein expression analysis (A). qRT-PCR was performed without and with 5AzadC treatment after 72 h in NT2/D1 and TCam-2 cells. The data shown are means ± SD of three independent experiments. ∗P < 0.05, Student’s t-test. (B) Western Blot analysis of PIWI-LIKE 2 was conducted after treatment of TCam-2 cells without and with 5AzadC for 72 h. Untreated cells and DMSO treated cells served as control. ß-ACTIN was the loading control. ∗∗P < 0.01.
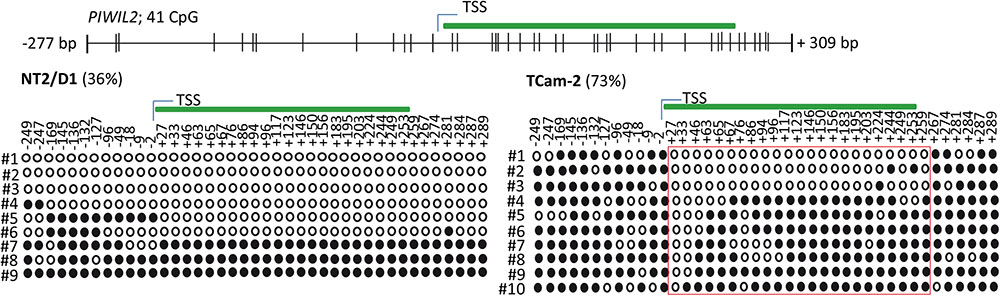
FIGURE 3. PIWI-LIKE 2 promoter CpG methylation analysis. Upper panel shows the location of the investigated CpG dinucleotides according to the TSS of PIWI-LIKE 2. Lower panel shows PIWI-LIKE 2 methylation level of the investigated CpG dinucleotides in NT2/D1 and TCam-2 cells after treatment with 5AzadC for 72h. Red boxes indicate CpG dinucleotides with prominent changes in methylation status. Filled circles indicate methylated CpG’s, empty circles indicate demethylated CpGs. #1-10 refers to the different clones analyzed per PIWI-LIKE 2 promoter fragment.
Next, we addressed the question whether the PIWI-LIKE 2 promoter can be activated in vitro and if the activation could be silenced by methylation of CpG-sites within this sequence. Therefore, six promoter fragments of different length and location were generated (A-F; see Figure 4). We assumed the region 2000 bp downstream of the TSS of the PIWI-LIKE 2 full length variant to potentially regulate PIWI-LIKE 2 transcription (Fragment A). Furthermore, we designed shortened promoter fragments containing up to 600 bp downstream of the TSS. Promoter fragment E represented the region around the predicted CpG island and putative promoter site (-300 bp to +300 bp). Fragments were cloned 5′ of a luciferase gene and transfected into the embryonal carcinoma cell line NT2/D1 and the seminoma cell line TCam-2. A constitutive Renilla luciferase expressing pGL4.74 plasmid was co-transfected for normalization. Luciferase activity was strongly induced by the PIWI-LIKE 2 full length promoter fragment compared to the empty reporter construct. After 48 h of transfection there was a 35fold (p = 0.007) increase in TCam-2 and 10fold activation in NT2/D1 (p = 0.001). The shortened promoter fragment D leaded to an induction as well. There was a three fold induction in TCam-2 (p = 0.049) and a 6.5fold increasing of luciferase activity in NT2/D1 (p = 0.034). The CpG island containing fragment E showed an eightfold activation of luciferase expression in NT2/D1 (p = 0.0002) and a threefold activation in TCam-2 (p = 0.014). Transfection of in vitro methylated promoter fragments resulted in a reduced luciferase activity for all fragments.
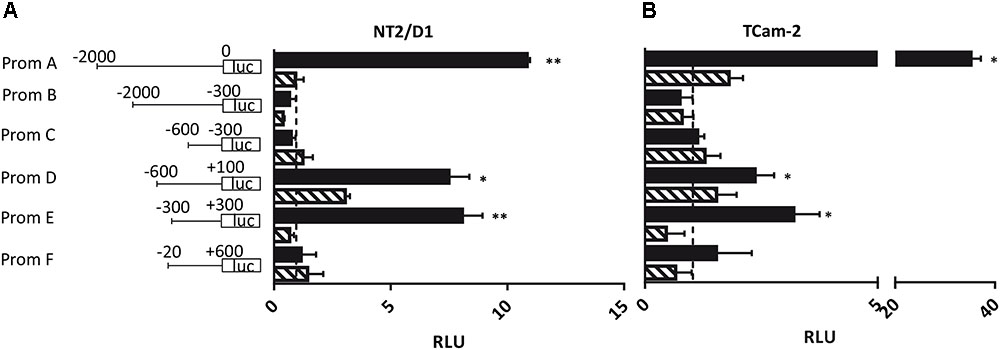
FIGURE 4. Luciferase assay in NT2/D1 (A) and TCam-2 cells (B). The luciferase reporter assays were performed 48 h after transfection with the indicated PIWI-LIKE 2 promoter fragments of different length and location and a renilla transfection control plasmid, which was co-transfected. An empty luciferase vector served as control and was set to an expression of 1 (dashed line). Respective PIWI-LIKE 2 promoter constructs after in vitro methylation were also transfected. The data shown are means ± SD of three independent experiments. ∗P < 0.05, ∗∗P < 0.01, Student’s t-test.
DNA methylation is highly dynamic in mammalian germ cells during development. Maternal and paternal genomes are differentially marked and must be properly reprogrammed before the primordial germ cell enters the germinal ridge (Santos et al., 2002; Sasaki and Matsui, 2008; Messerschmidt et al., 2014). DNA methylation as epigenetic modification of the mammalian genome has widespread influences on gene expression. PIWI-LIKE genes are known to be involved in germ cell specification and maturation. MIWI2 (the murine homolog of human PIWI-LIKE 4) is expressed in the pre-pachytene phase of spermatogenesis during the period of de novo methylation. It has been detected in the nucleus, where it acts directly on transposable elements repression on DNA level via the piRNA metabolic process. Inactivation of MIWI2 leads to male sterility due to an early meiotic arrest, which correlates with retrotransposon derepression (Carmell et al., 2007; Aravin et al., 2008; Fazio et al., 2011). PIWI-LIKE 1 and PIWI-LIKE 2 associate with primary piRNAs in the cytoplasm and are required for the PIWI-LIKE 4 nuclear localization and association with secondary piRNAs antisense (Aravin et al., 2006, 2007; Reuter et al., 2011). The piRNA process acts upstream of known mediators of the DNA methylation. Besides their function in transposable elements repression, piRNAs are probably involved in other processes during meiosis, such as translation regulation (Thomson and Lin, 2009).
PIWI-LIKE 2 is completely repressed in somatic cells and, during development, silenced at post-meiotic stages. In this study, we wanted to investigate promoter DNA methylation as potential mechanism for expression control of PIWI-LIKE 2. A high density of CpG sites spanning the region at about -300 bp to +300 bp relative to the PIWI-LIKE 2 full length TSS makes an epigenetic regulation conceivable. PIWI-LIKE 2 promoter is hypomethylated (22%) in NT2/D1, but highly methylated in TCam-2. Concomitantly, NT2/D1 shows a higher basal mRNA expression. These results suggest CpG methylation status of PIWI-LIKE 2 correlates with its expression. Neither CpG methylation nor mRNA and protein expression was changed in NT2/D1 after demethylation treatment via 5′-Aza-2′-deoxycytidine (5AzadC). This indicates an open chromatin status on the PIWI-LIKE 2 promoter in this cell line, which is not altered and enables a higher PIWI-LIKE 2 expression in comparison to TCam-2. However, it is of note that although most of the clones analyzed in NT2/D1 were hypomethylated, 2 of 10 (basal methylation state) or 3 of 9 (after 5AzadC treatment) were hypermethylated in a way which is comparable to T-Cam2. This observation seems to stand in contradiction to the proposed hypomethylation of the PIWI-LIKE 2 promoter in cell types or lines with a higher differentiation potential. However, one may speculate that during the cultivation and treatment of the NT2/D1, in single cells the differentiation process was induced unintentionally. The observed hypermethylation of the PIWI-LIKE 2 promoter may in this context be another indication for silencing of PIWI-LIKE 2 expression as an very early event during germ cell differentiation.
In TCam-2, 5AzadC treatment leads to a significant increase of PIWI-LIKE 2 mRNA and protein, but a comparable low overall promoter demethylation (85–73%). Interestingly, almost only CpG dinucleotides spanning the PIWI-LIKE 2 promoter region from -130 nt to +65 nt are partially demethylated. Either this region is relevant for PIWI-LIKE 2 expression regulation or a splice variant could potentially be activated, which is not regulated by the investigated 41 CpG islands and has another promoter site. Gainetdinov et al. (2014) demonstrated the presence of 60 kDa (PL2L60A) and 80 kDa (PL2L80A) isoforms of PIWI-LIKE 2 in testicular cancer cell lines including NT2/D1. Furthermore, they identified alternative TSSs within the PIWI-LIKE 2 sequence. These were mapped to exon 5 and exon 7 and can be activated in vitro. Beyond, there is evidence for the existence of PIWI-LIKE 2 splice variants by isoforms with 50 kDa (PL2L50) and 40 kDa (PL2L40) (Ye et al., 2010). Recently, an intragenic promoter in intron 10 of the PIWI-LIKE 2 genomic sequence regulating a 60 kDa isoform was identified in human cells and verified in luciferase reporter assays (Liu et al., 2017). Here, we show that the full length PIWI-LIKE 2 promoter and promoter fragments surrounding the TSS of full length PIWI-LIKE 2 are able to drive luciferase expression in human cell lines. The activation was markedly reduced after in vitro methylation of these fragments. The data indicate that in humans DNA methylation is able to induce epigenetically silencing of PIWI-LIKE 2 expression. Furthermore, it suggests the region around the TSS and exon 1 is subject of epigenetic regulation.
Our data exhibit a high mRNA expression but low overall CpG demethylation in TCam-2. Furthermore, we observed a decreased proliferation and cell vitality and an increased apoptosis induction upon suppression of PIWI-LIKE 2 expression in both NT2/D1 and TCam-2. Analogously, insufficient PIWI-LIKE 2 expression is associated with male infertility in mouse and human (Kuramochi-Miyagawa et al., 2004; Kuramochi-Miyagawa et al., 2008; Heyn et al., 2012). In human, the dysfunction was associated with the hypermethylation of PIWI-LIKE 2 promoter and its interacting factor TDRD1 and resulted in a disrupted production of piRNAs and a hypomethylation of the LINE-1 repetitive sequences in patients affected with spermatogenic arrest (Heyn et al., 2012). An proposed association of single nucleotide polymorphisms (SNPs) in the PIWI-LIKE 2 gene with spermatogenic failure (Gu et al., 2010) has not been re-analyzed in other male infertility patient cohorts so far.
In conclusion, PIWI-LIKE 2 is essential for the germline integrity and self-renewal of stem cells. Therefore, it may be an interesting target for the prediction of fertilization rates and embryo development in assisted reproductive techniques (ART). Thus, the methylation profile of PIWI-LIKE 2 and its corresponding expression could potentially provide further predictive information for clinical decisions.
MG performed the experiments, analyzed the data, and drafted the manuscript. TG and HB conceived the study, assisted in drafting the manuscript, and reviewed the data and the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Ariane Klemm for the support of this work. We acknowledge the financial support of the Open Access Publication Fund of the Martin-Luther-University Halle-Wittenberg.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00375/full#supplementary-material
Aravin, A. A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207. doi: 10.1038/nature04916
Aravin, A. A., Sachidanandam, R., Bourc’his, D., Schaefer, C., Pezic, D., Fejes-Toth, K., et al. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799. doi: 10.1016/j.molcel.2008.09.003
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K., and Hannon, G. J. (2007). Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747. doi: 10.1126/science.1142612
Bourc’his, D., and Bestor, T. H. (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99. doi: 10.1038/nature02886
Carmell, M. A., Girard, A., van de Kant, H. J. G., Bourc’his, D., Bestor, T. H., de Rooij, D. G., et al. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514. doi: 10.1016/j.devcel.2007.03.001
Cerutti, L., Mian, N., and Bateman, A. (2000). Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25, 481–482. doi: 10.1016/S0968-0004(00)01641-8
Cox, D. N., Chao, A., Baker, J., Chang, L., Qiao, D., and Lin, H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727. doi: 10.1101/gad.12.23.3715
Cui, X., Jing, X., Wu, X., Yan, M., Li, Q., Shen, Y., et al. (2016). DNA methylation in spermatogenesis and male infertility. Exp. Ther. Med. 12, 1973–1979. doi: 10.3892/etm.2016.3569
Fazio, S., Bartonicek, N., Di Giacomo, M., Abreu-Goodger, C., Sankar, A., Funaya, C., et al. (2011). The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259–263. doi: 10.1038/nature10547
Gainetdinov, I. V., Skvortsova, Y. V., Stukacheva, E. A., Bychenko, O. S., Kondratieva, S. A., Zinovieva, M. V., et al. (2014). Expression profiles of PIWI-LIKE 2 short isoforms differ in testicular germ cell tumors of various differentiation subtypes. PLoS One 9:e112528. doi: 10.1371/journal.pone.0112528
Grivna, S. T., Beyret, E., Wang, Z., and Lin, H. (2006). A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714. doi: 10.1101/gad.1434406
Gu, A., Ji, G., Shi, X., Long, Y., Xia, Y., Song, L., et al. (2010). Genetic variants in Piwi-interacting RNA pathway genes confer susceptibility to spermatogenic failure in a Chinese population. Hum. Reprod. 25, 2955–2961. doi: 10.1093/humrep/deq274
Heyn, H., Ferreira, H. J., Bassas, L., Bonache, S., Sayols, S., Sandoval, J., et al. (2012). Epigenetic disruption of the PIWI pathway in human spermatogenic disorders. PLoS One 7:e47892. doi: 10.1371/journal.pone.0047892
Jinek, M., and Doudna, J. A. (2009). A three-dimensional view of the molecular machinery of RNA interference. Nature 457, 405–412. doi: 10.1038/nature07755
Kuramochi-Miyagawa, S., Kimura, T., Ijiri, T. W., Isobe, T., Asada, N., Fujita, Y., et al. (2004). Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131, 839–849. doi: 10.1242/dev.00973
Kuramochi-Miyagawa, S., Watanabe, T., Gotoh, K., Totoki, Y., Toyoda, A., Ikawa, M., et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917. doi: 10.1101/gad.1640708
Lim, S. L., Tsend-Ayush, E., Kortschak, R. D., Jacob, R., Ricciardelli, C., Oehler, M. K., et al. (2013). Conservation and expression of PIWI-interacting RNA pathway genes in male and female adult gonad of amniotes. Biol. Reprod. 89:136. doi: 10.1095/biolreprod.113.111211
Liu, S.-S., Liu, N., Liu, M.-Y., Sun, L., Xia, W.-Y., Lu, H.-M., et al. (2017). An unusual intragenic promoter of PIWI-LIKE 2 contributes to aberrant activation of oncogenic PL2L60. Oncotarget 8, 46104–46120. doi: 10.18632/oncotarget.17553
Liyanage, V. R., Jarmasz, J. S., Murugeshan, N., Del Bigio, M. R., Rastegar, M., and Davie, J. R. (2014). DNA modifications: function and applications in normal and disease States. Biology 3, 670–723. doi: 10.3390/biology3040670
Marchetto, M. C. N., Narvaiza, I., Denli, A. M., Benner, C., Lazzarini, T. A., Nathanson, J. L., et al. (2013). Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503, 525–529. doi: 10.1038/nature12686
Messerschmidt, D. M., Knowles, B. B., and Solter, D. (2014). DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 28, 812–828. doi: 10.1101/gad.234294.113
Reuter, M., Berninger, P., Chuma, S., Shah, H., Hosokawa, M., Funaya, C., et al. (2011). Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264–267. doi: 10.1038/nature10672
Santos, F., Hendrich, B., Reik, W., and Dean, W. (2002). Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241, 172–182. doi: 10.1006/dbio.2001.0501
Sasaki, H., and Matsui, Y. (2008). Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 9, 129–140. doi: 10.1038/nrg2295
Sasaki, T., Shiohama, A., Minoshima, S., and Shimizu, N. (2003). Identification of eight members of the Argonaute family in the human genome. Genomics 82, 323–330. doi: 10.1016/S0888-7543(03)00129-0
Seisenberger, S., Peat, J. R., Hore, T. A., Santos, F., Dean, W., and Reik, W. (2013). Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20110330. doi: 10.1098/rstb.2011.0330
Song, J.-J., Smith, S. K., Hannon, G. J., and Joshua-Tor, L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. doi: 10.1126/science.1102514
Stuppia, L., Franzago, M., Ballerini, P., Gatta, V., and Antonucci, I. (2015). Epigenetics and male reproduction: the consequences of paternal lifestyle on fertility, embryo development, and children lifetime health. Clin. Epigenetics 7:120. doi: 10.1186/s13148-015-0155-4
Thomson, T., and Lin, H. (2009). The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 25, 355–376. doi: 10.1146/annurev.cellbio.24.110707.175327
Unhavaithaya, Y., Hao, Y., Beyret, E., Yin, H., Kuramochi-Miyagawa, S., Nakano, T., et al. (2009). MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 284, 6507–6519. doi: 10.1074/jbc.M809104200
Keywords: epigenetics, PIWI-LIKE 2, germ cell tumors, promoter methylation, spermatogenesis
Citation: Giebler M, Greither T and Behre HM (2018) Differential Regulation of PIWI-LIKE 2 Expression in Primordial Germ Cell Tumor Cell Lines by Promoter Methylation. Front. Genet. 9:375. doi: 10.3389/fgene.2018.00375
Received: 23 April 2018; Accepted: 24 August 2018;
Published: 20 September 2018.
Edited by:
Rui Henrique, IPO Porto, PortugalReviewed by:
Alexandra M. Lopes, Universidade do Porto, PortugalCopyright © 2018 Giebler, Greither and Behre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hermann M. Behre, aGVybWFubi5iZWhyZUBtZWRpemluLnVuaS1oYWxsZS5kZQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.