- 1National Center for Cool and Cold Water Aquaculture, Agricultural Research Service, United States Department of Agriculture, Kearneysville, WV, United States
- 2Troutlodge, Inc., Sumner, WA, United States
- 3Hendrix Genetics Aquaculture BV/Netherlands, Boxmeer, Netherlands
Bacterial cold water disease (BCWD), caused by Flavobacterium psychrophilum, is an endemic and problematic disease in rainbow trout (Oncorhynchus mykiss) aquaculture. Previously, we have identified SNPs (single nucleotide polymorphisms) associated with BCWD resistance in rainbow trout. The objectives of this study were (1) to validate the SNPs associated with BCWD resistance in a commercial breeding population; and (2) to evaluate retrospectively the accuracy of MAS (marker-assisted selection) for BCWD resistance in this commercial breeding program. Three consecutive generations of the Troutlodge May breeding population were evaluated for BCWD resistance. Based on our previous studies, a panel of 96 SNPs was selected and used to genotype the parents and ten offspring from each of the 138 full-sib families of the 2015 generation, and 37 SNPs associated with BCWD resistance were validated. Thirty-six of the validated SNPs were clustered on chromosomes Omy3, Omy8 and Omy25. Thus, at least three QTL (quantitative trait loci) for BCWD resistance were validated in the 2015 generation. Three SNPs from each QTL region were used for haplotype association analysis. Three haplotypes, Omy3TGG, Omy8GCG and Omy25CGG, were found to be associated with BCWD resistance in the 2015 generation. Retrospective analyses were then performed to evaluate the accuracy of MAS for BCWD resistance using these three favorable haplotypes. The accuracy of MAS was estimated with the Pearson correlation coefficient between the total number of favorable haplotypes in the two parents and the family BCWD survival rates. The Omy8 and Omy25 haplotypes were positively correlated with the family BCWD survival rates across all three generations. The accuracies of MAS using these two haplotypes together were consistently around 0.5, which was equal or greater than the accuracy of the conventional family-based selection in the same generation. In conclusion, we have demonstrated that MAS for BCWD resistance is feasible in this commercial rainbow trout breeding population.
Introduction
Bacterial cold water disease (BCWD), caused by Flavobacterium psychrophilum, is one of the most devastating diseases in rainbow trout (Oncorhynchus mykiss) aquaculture (Nematollahi et al., 2003; Starliper, 2011; Loch and Faisal, 2015). Licensed vaccines for BCWD are not available at present. Use of licensed antibiotics for BCWD treatment increases production costs, and antibiotic-resistant pathogens may emerge. Fortunately, genetic variation for BCWD resistance (Hadidi et al., 2008; Silverstein et al., 2009) has been documented in rainbow trout. Using family-based selection, a rainbow trout line with improved BCWD resistance has been developed (Leeds et al., 2010; Wiens et al., 2013a). However, family-based selection is based on the family breeding values estimated from pedigree relationships and BCWD phenotypes. Evaluation for BCWD resistance in rainbow trout is labor intensive and time consuming. Also, family-based selection cannot exploit within-family genetic variation because disease challenged individuals are excluded as selection candidates, and selection is based on family means obtained from the BCWD performance of siblings of the selection candidates. Thus, selection methods that can utilize within-family variation at an affordable cost are desirable.
Genomic selection (GS), which uses genetic markers covering the whole genome to calculate the genomic estimated breeding values of selection candidates, can improve the accuracy of selection in animal breeding (Meuwissen et al., 2013). Recently, we have demonstrated that GS can substantially increase the accuracy of selection for BCWD resistance in the Troutlodge May spawning population (Vallejo et al., 2017a). While the accuracy of family-based selection for BCWD resistance was 0.36, the accuracy of GS was as high as 0.71 in the same generation. To reduce the cost of genotyping, low-density marker panels were tested for GS. Surprisingly, we have recently found that the accuracy of GS using 70 SNPs (single nucleotide polymorphisms) associated with BCWD resistance is still better than the accuracy of conventional family-based selection in that commercial breeding population (Vallejo et al., 2018). This result indicates that marker-assisted selection (MAS) using only a few SNPs associated with the major QTL (quantitative trait loci) for BCWD resistance might further reduce the cost of genotyping.
It has been documented that MAS can improve the efficiency of selection of quantitative traits (Lande and Thompson, 1990). However, there are only few well-documented examples of MAS in aquaculture breeding programs, although many QTL for diverse traits in aquaculture species were reported in over a decade (Ashton et al., 2017). One of the best known examples is MAS for resistance to infectious pancreatic necrosis virus (IPNV) in Atlantic salmon (Salmo salar) (Houston et al., 2008; Moen et al., 2009, 2015; Gonen et al., 2015). QTL mapping revealed microsatellites associated with a major QTL for IPNV resistance in Atlantic salmon, and the microsatellites were used initially for MAS. Recently, the gene controlling for IPNV resistance has been identified (Moen et al., 2015), and two SNPs closer to the causative gene are used for commercial implementation of MAS for IPNV resistance in Atlantic salmon. Another successful example of MAS in aquaculture species is the development of Japanese flounder (Paralichthys olivaceus) resistant to lymphocystis disease (LD) (Fuji et al., 2007), an iridoviral disease found in many fish species. A major QTL for LD resistance in Japanese flounder was identified by microsatellite QTL mapping (Fuji et al., 2006). The microsatellite linked to the major QTL was used to develop a commercial population of Japanese flounder with no incidence of LD on farm trials while LD incidences were recorded in the control population. Recently, commercial rainbow trout eggs marketed as QTL-innOva® FLAVO (https://aquagen.no/en/), developed by MAS of two QTL for BCWD resistance, have been advertised. However, detailed information about these two QTL has not been published in the scientific literature.
In the past, we have reported several studies of identification of molecular markers associated with BCWD resistance in rainbow trout. Initially, microsatellites were used for QTL mapping, and nine QTL (Vallejo et al., 2013) were identified in the odd-year mapping families of the National Center for Cool and Cold Water Aquaculture (NCCCWA). Similarly, two major QTL (Wiens et al., 2013b) were identified in an even-year NCCCWA mapping family, and the QTL on chromosome Omy19 was validated in a later generation (Vallejo et al., 2014; Liu et al., 2015). Due to the limited number of microsatellites available for genetic mapping, restriction site associated DNA sequencing was later used to identify SNPs associated with the QTL for BCWD resistance (Liu et al., 2015; Palti et al., 2015b). Recently, a 57K SNP genotyping array has been developed in rainbow trout (Palti et al., 2015a), and a reference rainbow trout genome became available (GenBank Assembly Accession GCA_002163495). The development of those resources enabled a genome-wide association study to identify SNPs associated with BCWD resistance (Vallejo et al., 2017b) in two rainbow trout populations, the NCCCWA odd-year population and the 2013 Troutlodge May spawning population. Since the SNPs associated with BCWD resistance were discovered in different mapping populations, and several methods were used for QTL mapping, it is critical to validate the SNPs associated with BCWD resistance before implementation of MAS in a breeding program. The objectives of this study were (1) to validate the SNPs associated with BCWD resistance in the 2015 Troutlodge May spawning population; and (2) to evaluate retrospectively the accuracy of MAS for BCWD resistance in three consecutive generations of the Troutlodge May spawning population.
Materials and Methods
Ethic Statement
The experiments were conducted with the protocols approved by the Institutional Animal Care and Use Committee, National Center for Cool and Cold Water Aquaculture, Agriculture Research Service, United States Department of Agriculture. All efforts were made to minimize suffering and to ensure fish welfare.
Three Consecutive Generations of the Troutlodge May Spawning Strain
Troutlodge, Inc., has four rainbow trout strains (Liu et al., 2017) named by their peak spawning months. All four Troutlodge strains have been under selection for growth but not for BCWD resistance. Three consecutive generations of the odd-year Troutlodge May spawning strain were used in this study (Table 1). In a previous study (Vallejo et al., 2017b) we identified SNPs associated with BCWD resistance in the 2013 generation. In this study, 138 full-sib families in the 2015 generation were used to validate the SNPs associated with BCWD resistance. All three generations were used for retrospective evaluation of MAS for BCWD resistance. Fin clips were collected from all parents and offspring, and were stored in 95% ethanol. DNA was extracted from fin clips using an AutoGenprep 965 automated DNA extractor (Autogen, Holliston, MA, USA) following the manufacturer's instructions.

Table 1. Three consecutive generations of the odd-year Troutlodge May spawning rainbow trout strain used in this study.
BCWD Challenge Experiments
In addition to the two BCWD challenges of the 2013 and 2015 generations described in our previous study (Vallejo et al., 2017a), 120 full-sib families of the 2017 generation were evaluated for BCWD resistance in this study. Eyed eggs of each family were provided by Troutlodge, Inc., and were hatched at the NCCCWA. At 80 days post-hatch, each family was challenged with BCWD following the established protocol described previously in detail (Hadidi et al., 2008). Briefly, 80 fish per family were stocked in two tanks with 40 fish per tank. The fish were challenged by intraperitoneal injection of Flavobacterium psychrophilum strain CSF259-93. Mortalities were collected daily for 21 days after intraperitoneal injection. Survival days (DAYS), the number of days to death post-challenge, were recorded. Survivors at the end of the experiment were assigned a value of 21 days. Each individual fish also had a record of survival status (STATUS). The binary trait STATUS had two classes: fish died during the 21 days period were assigned a value of 2, and fish alive on day 21 post-challenge were assigned a value of 1. The family BCWD survival rate was calculated as the percentage of survivors on day 21 post-challenge.
Genotyping of the 2015 Year-Class Population
SNP Type™ assays (Table S1) were developed for 96 SNPs associated with BCWD resistance identified in our previous studies (Liu et al., 2015; Palti et al., 2015b; Vallejo et al., 2017b). These 96 SNPs were used to genotype the 2015 population consisting of 138 full-sib families. Two parents and ten offspring of each 2015 family including the first five to die fish and five random survivors were genotyped following the protocol described in our previous study (Liu et al., 2016). Briefly, DNA samples were pre-amplified, and the pre-amplified products were diluted and used for genotyping with 96.96 Dynamic Array IFCs (Integrated Fluidic Circuits). The arrays were read using EP1 system, and genotypes were called automatically using Fluidigm SNP genotyping analysis software 4.1 with a confidence threshold of 85. The genotype clusters were examined by eye for each assay, and any wrong calls or no calls were corrected manually. The computer program PedCheck (O'connell and Weeks, 1998) was used to identify genotypes with Mendelian inheritance errors between parents and their offspring.
Genotyping of Parents of 2013 and 2017 Populations
Parents of the 2013 generation were genotyped with the rainbow trout 57K SNP array as described in our previous publications (Palti et al., 2015a; Vallejo et al., 2017a). Briefly, genotyping was performed by a commercial genotyping service provider (GeneSeek/Neogen, Inc., Lincoln, NE), and the genotype data were filtered with our in-house bioinformatics pipeline to remove low quality genotypes. A customized Illumina SNP bead-array including 10K rainbow trout SNPs was used to genotype parents of the 2017 generation. The genotyping service for the Illumina array was also provided by GeneSeek/Neogen, Inc., Lincoln, NE.
Family-Based Association Analysis for BCWD Resistance
The program PLINK 1.9 (Chang et al., 2015) was used for family-based association analysis to identify SNPs associated with BCWD resistance (P < 0.01) in the 2015 generation. The procedure QFAM was used to analyze the phenotypic data DAYS, and the PERM option was used to account for the dependence between related individuals. The procedure TDT (transmission disequilibrium test) was used to analyze the binary phenotype STATUS. SNPs with MAF (minor allele frequency) smaller than 0.05 were excluded from the association analyses.
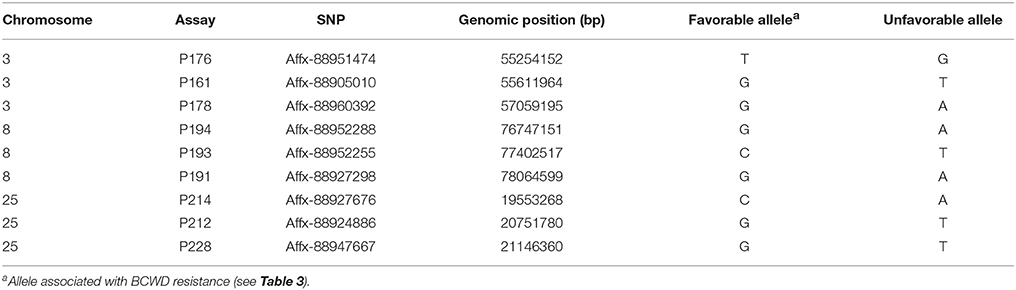
Haplotype association analysis was performed using the program PLINK 1.07 (Purcell et al., 2007). Three SNPs (Table 2) from each QTL region were selected for haplotype analysis following these criteria: (1) they were associated with the BCWD resistance (see results); (2) their MAFs are larger than 0.2, which ensures that these SNPs are informative; and (3) they span a genomic region smaller than 2 Mb. The option—hap-impute was used to impute the haplotypes of parents and offspring of the 2015 generation, and the option—mhf 0.1 was used to filter out haplotypes with a frequency less than 0.1. The output files were then used to identify haplotypes associated with DAYS or STATUS (P < 0.01) using the same procedures described above to identify SNPs associated with BCWD resistance. The haplotypes associated with BCWD resistance are referred as favorable haplotypes hereafter.
Evaluation of MAS for BCWD Resistance
Haplotypes of the three QTL regions were reconstructed for all parents of the three generations using the default setting of program Beagle version 4.1 (Browning and Browning, 2007). The number of favorable haplotypes in each parent was counted for each QTL region. All the statistical analyses described below were performed using R version 3.4.3 (R Core Team, 2017). The accuracy of MAS was defined as the Pearson correlation coefficient between the total number of favorable haplotypes in the two parents and the family BCWD survival rates, similar to the approach we previously used for estimating accuracies of GS and traditional family-based selection (Vallejo et al., 2017a). The R function cor.test was used to test if the correlation coefficient is significantly different from 0 (P < 0.05). The families in each generation were grouped by the total number of favorable haplotypes in the two parents. After removing the haplotype groups with less than 10 families, analysis of variance was performed to test if the number of favorable haplotypes was associated with family BCWD survival rates. Then, R function TukeyHSD was used to compare the mean family BCWD survival rates between the haplotype groups.
Accuracy of Family-Based Selection for BCWD Resistance
The estimated breeding values (EBV) of parents of the 2017 generation were estimated using BCWD survival phenotypes from the 2013 and 2015 disease challenges and pedigree records spanning eight generations. The records of BCWD survival STATUS were fit into pedigree-based BLUP (best linear unbiased prediction) threshold model using the computer program BLUPF90 (Misztal et al., 2015) as we have previously described (Vallejo et al., 2017a). The accuracy of family-based selection was estimated with the Pearson correlation coefficient between mid-parent EBV and the family BCWD survival rates.
Results
Validation of SNPs Associated With BCWD Resistance
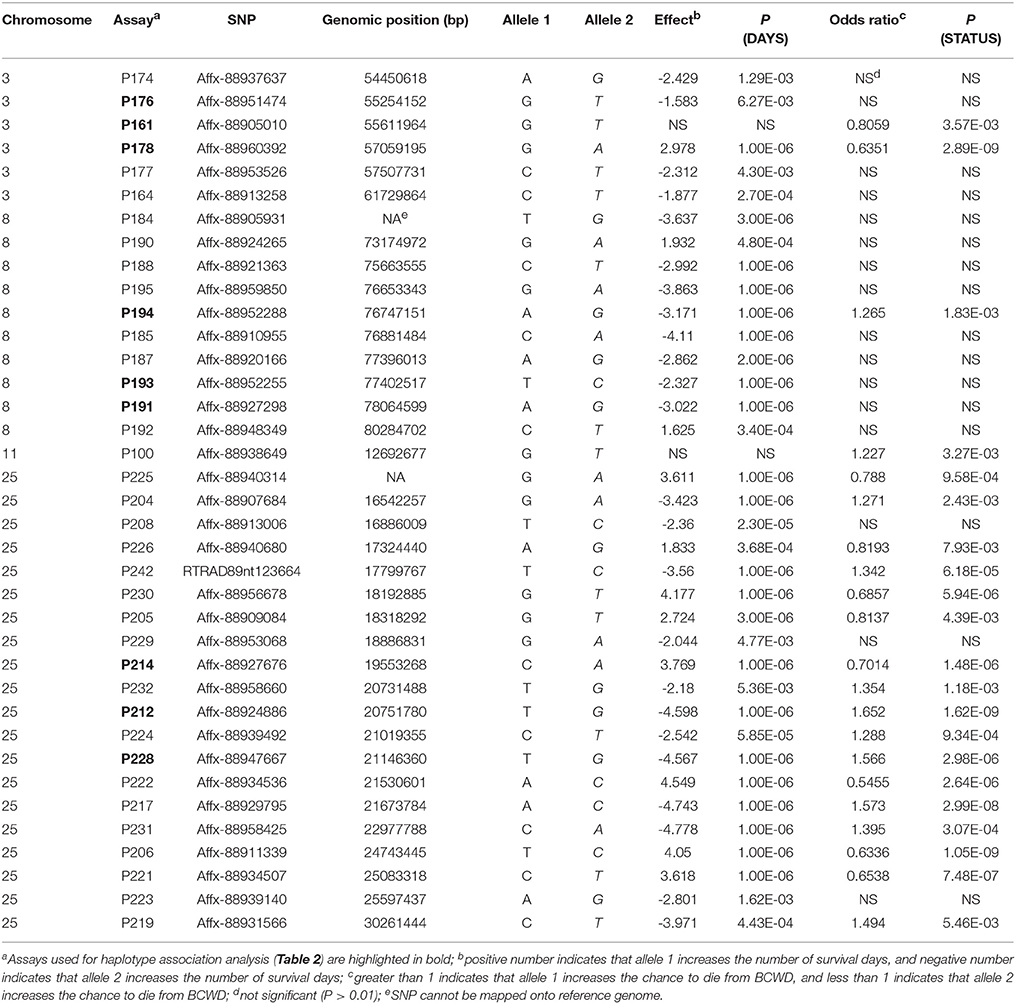
Among the 96 SNP assays used to genotype the 2015 parents and offspring, six assays (Table S1) were removed from further analyses due to difficulties to score the genotypes or more than five Mendelian inheritance errors were identified between the genotypes of parents and their offspring. Eight SNPs (Table S1) were also removed from the association analyses because their MAFs were smaller than 0.05. Of the remaining 82 SNPs, 37 SNPs were associated with BCWD resistance in the 2015 generation (Table 3). These 37 SNPs were located on chromosomes Omy3, Omy8, Omy11 and Omy25. Since only one of the 37 SNPs was located on chromosome Omy11, and this SNP was only significant for STATUS, we focused on the other three chromosomes in the remaining analyses. For simplicity, we will refer to each of the three chromosomal regions as a QTL region hereafter.
Haplotypes Associated With BCWD Resistance
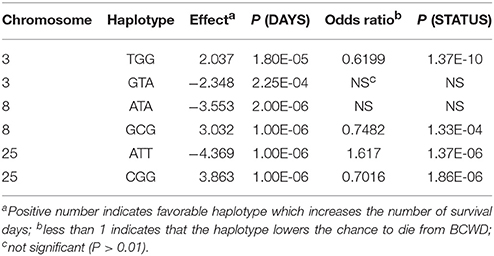
Three SNPs (Table 2) from each QTL region were used for haplotype association analysis in the 2015 generation, and the results are presented in Table 4. For each QTL region, we identified one haplotype associated with BCWD resistance (increases BCWD survival days or lowers the chance to die from BCWD) and one haplotype associated with BCWD susceptibility (decreases BCWD survival days or increases the chance to die from BCWD). Since one of the goals of rainbow trout breeding is to improve BCWD resistance, we focused on the haplotypes associated with BCWD resistance and not the haplotypes associated with BCWD susceptibility. Consistent with the single SNP association analysis, the BCWD resistant haplotypes had the combination of the favorable SNP alleles associated with BCWD resistance (Table 3). To explicitly reference each haplotype, we added its chromosomal name ahead of its allele combination. Thus, the three haplotypes associated with BCWD resistance were referred to as Omy3TGG, Omy8GCG and Omy25CGG, respectively.
MAS for BCWD Resistance
The haplotypes of each parent were reconstructed for each QTL region, and then we evaluated retrospectively the accuracies of MAS for BCWD resistance using the favorable haplotype for each QTL region (Table 5, Figure S1). Both haplotypes Omy8GCG and Omy25CGG were significantly correlated with the family BCWD survival rates across the three generations. However, haplotype Omy3TGG was only significantly correlated with the family BCWD survival rates in the 2013 generation. Thus, we decided to use the favorable haplotypes Omy8GCG and Omy25CGG together for MAS for BCWD resistance. The accuracies of MAS were consistently around 0.5 across the three generations (Table 5), which was higher than the accuracy of family-based selection (0.36) in the 2015 generation we reported previously (Vallejo et al., 2017a), and was similar to the accuracy of family-based selection (0.48) in the 2017 generation (Table 5).
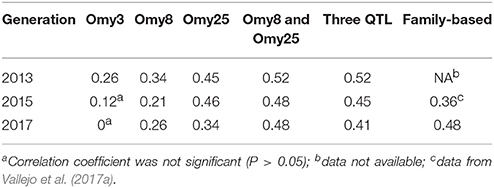
Table 5. Accuracies of MAS and family-based selection in three generations of the odd-year Troutlodge May spawning strain.
To further demonstrate that favorable haplotypes Omy8GCG and Omy25CGG can be used for MAS for BCWD resistance, we grouped the families by the total number of favorable haplotypes in the two parents. As shown in Figure 1, the mean family BCWD survival rates increased in all three generations with an increase in total number of favorable haplotypes in the two parents. The families with the highest number of favorable parental haplotypes had a significantly higher mean family BCWD survival rate than the families with the least number of favorable parental haplotypes in each generation. The magnitude of difference in percentage points was 19.8, 24.2 and 20.1 for 2013, 2015 and 2017 generations, respectively.
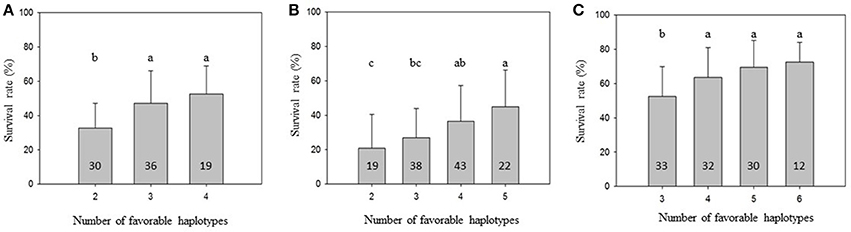
Figure 1. Mean family BCWD survival rate with an error bar. The families in each population were grouped by the total number of favorable haplotypes Omy8GCG and Omy25CGG in the two parents. Groups not sharing the same letter above the error bar have significantly different mean BCWD survival rates (Tukey's HSD, P < 0.05). The number in each column indicates the number of families in each haplotype group. (A) 2013 generation, (B) 2015 generation; (C) 2017 generation.
Discussion
BCWD is one of the most devastating diseases in rainbow trout aquaculture. Previously, we have identified SNPs associated with BCWD resistance in rainbow trout (Liu et al., 2015; Palti et al., 2015b; Vallejo et al., 2017b), although we have found that QTL and SNPs associated with BCWD resistance can vary between populations. In this study, 37 SNPs associated with BCWD resistance were validated in the 2015 year-class population, and we have demonstrated that MAS for BCWD resistance in rainbow trout is feasible in three consecutive generations of the odd-year Troutlodge May spawning strain.
Validation of QTL for BCWD Resistance in Rainbow Trout
It is essential to validate the genetic markers associated with the traits of interest before implementation of MAS in breeding programs. Among the 96 SNPs associated with BCWD resistance identified in our previous studies (Liu et al., 2015; Palti et al., 2015b; Vallejo et al., 2017b), 37 SNPs were validated in the 2015 generation in this study. All validated SNPs, except one, were clustered on chromosomes Omy3, Omy8 and Omy25. Thus, at least three QTL for BCWD resistance were validated in the 2015 year-class population. This result is not unexpected because these three QTL had the largest effects in the 2013 year-class population (Vallejo et al., 2017b). Although 10 SNPs on chromosome Omy19 were included in the SNP panel, none of them was associated with BCWD resistance in the 2015 year-class population. This result is not surprising either because the QTL on Omy19 was identified and validated in even-year NCCCWA families (Wiens et al., 2013b; Vallejo et al., 2014; Liu et al., 2015). This result is also consistent with previous reports of population specific QTL for BCWD resistance (Vallejo et al., 2013, 2017b; Campbell et al., 2014; Palti et al., 2015b). Certainly, some QTL identified in our previous studies could be false positive, which might be another reason that some SNPs could not be validated in this study.
Molecular Breeding for BCWD Resistance in Rainbow Trout
In this report, we have demonstrated through retrospective analysis that MAS for BCWD resistance is feasible in three consecutive generations of the odd-year Troutlodge May spawning strain. Three SNPs from each of the two major QTL regions on chromosomes Omy8 and Omy25 were used to reconstruct the haplotypes of all parents for the three consecutive generations. The mean family BCWD survival rates increased in all three generations with the increase of the total number of favorable haplotypes in the two parents. The estimated accuracies of MAS using these two favorable haplotypes together were consistently around 0.5 across all three generations of the odd-year Troutlodge May spawning strain, which was equal or greater than the accuracy of traditional family-based selection in the same generation. We are currently evaluating whether these SNPs and haplotypes are also associated with BCWD resistance in other rainbow trout aquaculture strains. Campbell et al. (2014) identified 12 SNPs associated with BCWD resistance in another commercial rainbow trout population. But, none of the 12 SNPs was located on chromosome Omy25, and the only significant SNP on chromosome Omy8 was located in a region different from the Omy8 QTL targeted in this study. Thus, MAS with the favorable haplotypes reported in this study may not work in the population used by Campbell et al. (2014).
MAS for a minor QTL may not be productive although successful examples of MAS for major QTL in aquaculture species were reported (Houston et al., 2008, 2012; Moen et al., 2009). We previously reported that the QTL on chromosome Omy3 was not a major QTL explaining only up to 2% of the genetic variance for BCWD resistance in the 2013 year-class population (Vallejo et al., 2017b). In this study, six SNPs on chromosome Omy3 were associated with BCWD resistance in the 2015 year-class population, and a haplotype on Omy3, associated with BCWD resistance, was also identified in the 2015 year-class population. However, the effect of this QTL was not as large as the other two QTL on chromosomes Omy8 and Omy25. The total number of Omy3 favorable haplotypes in the two parents was not significantly correlated with the family BCWD survival rates in both the 2015 and 2017 generations. In the 2013 generation, the correlation coefficient for this minor QTL was significant, but the accuracy of MAS remained the same with or without this Omy3 QTL (Table 5).
The successful MAS reported in this study provides a less expensive alternative for molecular breeding for BCWD resistance in rainbow trout. Previously, we have demonstrated that GS can substantially improve the accuracy of selection for BCWD resistance in rainbow trout (Vallejo et al., 2017a). The accuracy of GS for BCWD resistance in rainbow trout can be as high as 0.71 in the 2015 generation (Vallejo et al., 2017a), which is higher than the accuracy of MAS reported in this study. However, those results were based on one generation of GS, and the accuracy of GS depends on many factors such as experimental design, size of reference population, number of SNPs, and statistical models used to estimate the SNP effects (Vallejo et al., 2017a). Comparing to GS, MAS for BCWD resistance has several advantages. (1) The genotyping cost for MAS is much lower. Only six SNPs were used for retrospective MAS in this study. Thus, the genotyping cost of MAS is considerably lower than that of GS, which typically requires tens of thousands SNPs covering the whole genome. Recently, we found that the accuracy of GS for BCWD resistance in the 2015 generation with only 70 of the 96 SNPs used in this study can remain as high as that of the 57K SNP array (Vallejo et al., 2018). Nonetheless, the genotyping cost with six SNPs is still cheaper than that with 70 SNPs. (2) MAS, once established, does not require a reference population, and is solely based on the genotypes of candidate breeders. GS requires both genotypes and phenotypes from a reference population (or training sample) in each generation of selection. In addition to the added cost of genotyping the reference population, collecting BCWD phenotypes is labor intensive and requires a special fish challenge facility due to biosecurity constraints. (3) MAS can be used to directly select egg production fish to produce customized eggs with improved BCWD resistance. Genotyping of a large number of egg production fish for genomic selection is cost prohibitive and very complicated for logistical reasons. Therefore, the choice to implement MAS or GS for BCWD resistance in breeding programs depends on many factors including the breeding program goals, the resources available, the anticipated economic gains from the genetic improvement and the options available for maximizing the economic gains.
Comparison of MAS and Family-Based Selection for BCWD Resistance
The accuracy of MAS was equal or greater than the accuracy of family-based selection in the same generation. The accuracy of MAS in the 2015 generation was 0.48, which was higher than 0.36, the accuracy of family-based selection in the same generation reported in our previous study (Vallejo et al., 2017a). The accuracies of selection in the 2017 generation were 0.48 for both MAS and the conventional family-based selection. The improved accuracy of family-based selection in the 2017 generation was mainly because BCWD phenotypes in two generations, 2013 and 2015, were available and used to estimate EBV of parents for the 2017 generation. Although there was not a large difference between these two methods in terms of the estimated accuracy of selection, MAS might still be more attractive in practice because MAS does not require obtaining BCWD phenotypes in each generation. However, the extent of improvement on BCWD resistance with MAS alone could be limited because only the two major QTL for BCWD resistance were targeted for MAS in this study. We reported previously that both major and minor QTL control BCWD resistance in the odd-year Troutlodge May spawning strain (Vallejo et al., 2017b). To maximize and sustain the genetic improvement for BCWD resistance through selective breeding over multiple generations, it would be necessary to integrate MAS and family-based selection to select both major and minor QTL for BCWD resistance in this commercial population.
Conclusion
The accuracies of MAS for BCWD resistance using SNP haplotypes from two major QTL regions in the odd-year Troutlodge May spawning strain were consistently around 0.5 across three generations, which was equal or greater than the accuracy of conventional family-based genetic merit predictions in the same generation. Therefore, we have demonstrated in this report that MAS for BCWD resistance in rainbow trout is feasible in this commercial breeding population.
Author Contributions
YP and SL conceived and planned the study; YP, JE, TL and GW coordinated, supervised, and performed the disease challenge experiments; RV calculated the estimated breeding values of parents of 2017 generation; GG involved in the selection of SNPs used in this study; KM provided eyed eggs for all three Troutlodge populations and fin clips of all parents; AH provided the genotypes of parents of 2017 generation; SL performed the data analysis and drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
KM was employed by Troutlodge, Inc., and author AH was employed by Hendrix Genetics Aquaculture BV/Netherlands.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by Agricultural Research Service CRIS projects 8082-32000-006 and 8082-31000-012. The authors would like to thank Roseanna Long and Kristy Shewbridge for their technical assistance to collect fin clips, extract DNA and genotype the samples. We also thank Ryan Lipscomb, Clayton Birkett, Jenea McGowan, Josh Kretzer, Travis Moreland, and Keira Osbourn for fish rearing and help with the disease challenge experiments. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
References
Ashton, D. T., Ritchie, P. A., and Wellenreuther, M. (2017). Fifteen years of quantitative trait loci studies in fish: challenges and future directions. Mol. Ecol. 26, 1465–1476. doi: 10.1111/mec.13965
Browning, S. R., and Browning, B. L. (2007). Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097. doi: 10.1086/521987
Campbell, N. R., Lapatra, S. E., Overturf, K., Towner, R., and Narum, S. R. (2014). Association mapping of disease resistance traits in rainbow trout using restriction site associated DNA sequencing. G3-Genes Genom. Genet. 4, 2473–2481. doi: 10.1534/g3.114.014621
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. doi: 10.1186/S13742-015-0047-8
Fuji, K., Hasegawa, O., Honda, K., Kumasaka, K., Sakamoto, T., and Okamoto, N. (2007). Marker-assisted breeding of a lymphocystis disease-resistant Japanese flounder (Paralichthys olivaceus). Aquaculture 272, 291–295. doi: 10.1016/j.aquaculture.2007.07.210
Fuji, K., Kobayashi, K., Hasegawa, O., Coimbra, M. R. M., Sakamoto, T., and Okamoto, N. (2006). Identification of a single major genetic locus controlling the resistance to lymphocystis disease in Japanese flounder (Paralichthys olivaceus). Aquaculture 254, 203–210. doi: 10.1016/j.aquaculture.2005.11.024
Gonen, S., Baranski, M., Thorland, I., Norris, A., Grove, H., Arnesen, P., et al. (2015). Mapping and validation of a major QTL affecting resistance to pancreas disease (salmonid alphavirus) in Atlantic salmon (Salmo salar). Heredity 115, 405–414. doi: 10.1038/hdy.2015.37
Hadidi, S., Glenney, G. W., Welch, T. J., Silverstein, J. T., and Wiens, G. D. (2008). Spleen size predicts resistance of rainbow trout to Flavobacterium psychrophilum challenge. J. Immunol. 180, 4156–4165. doi: 10.4049/jimmunol.180.6.4156
Houston, R. D., Davey, J. W., Bishop, S. C., Lowe, N. R., Mota-Velasco, J. C., Hamilton, A., et al. (2012). Characterisation of QTL-linked and genome-wide restriction site-associated DNA (RAD) markers in farmed Atlantic salmon. BMC Genomics 13:244. doi: 10.1186/1471-2164-13-244
Houston, R. D., Haley, C. S., Hamilton, A., Guyt, D. R., Tinch, A. E., Taggart, J. B., et al. (2008). Major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar). Genetics 178, 1109–1115. doi: 10.1534/genetics.107.082974
Lande, R., and Thompson, R. (1990). Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124, 743–756.
Leeds, T. D., Silverstein, J. T., Weber, G. M., Vallejo, R. L., Palti, Y., Rexroad, C. E., et al. (2010). Response to selection for bacterial cold water disease resistance in rainbow trout. J. Anim. Sci. 88, 1936–1946. doi: 10.2527/jas.2009-2538
Liu, S., Palti, Y., Gao, G., and Rexroad, C. E. (2016). Development and validation of a SNP panel for parentage assignment in rainbow trout. Aquaculture 452, 178–182. doi: 10.1016/j.aquaculture.2015.11.001
Liu, S., Palti, Y., Martin, K. E., Parsons, J. E., and Rexroad, C. E. (2017). Assessment of genetic differentiation and genetic assignment of commercial rainbow trout strains using a SNP panel. Aquaculture 468, 120–125. doi: 10.1016/j.aquaculture.2016.10.004
Liu, S., Vallejo, R. L., Palti, Y., Gao, G., Marancik, D. P., Hernandez, A. G., et al. (2015). Identification of single nucleotide polymorphism markers associated with bacterial cold water disease resistance and spleen size in rainbow trout. Front. Genet. 6:298. doi: 10.3389/Fgene.2015.00298
Loch, T. P., and Faisal, M. (2015). Emerging flavobacterial infections in fish: a review. J. Adv. Res. 6, 283–300. doi: 10.1016/j.jare.2014.10.009
Meuwissen, T., Hayes, B., and Goddard, M. (2013). Accelerating improvement of livestock with genomic selection. Annu. Rev. Anim. Biosci. 1, 221–237. doi: 10.1146/annurev-animal-031412-103705
Misztal, I., Tsuruta, S., Lourenco, D., Aguilar, I., Legarra, A., and Vitezica, Z. (2015). Manual for BLUPF90 Family of Programs. Available online at: http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=blupf90_all2.pdf (June 18, 2015).
Moen, T., Baranski, M., Sonesson, A. K., and Kjøglum, S. (2009). Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics 10:368. doi: 10.1186/1471-2164-10-368
Moen, T., Torgersen, J., Santi, N., Davidson, W. S., Baranski, M., Odegard, J., et al. (2015). Epithelial cadherin determines resistance to infectious pancreatic necrosis virus in atlantic salmon. Genetics 200, 1313–1326. doi: 10.1534/genetics.115.175406
Nematollahi, A., Decostere, A., Pasmans, F., and Haesebrouck, F. (2003). Flavobacterium psychrophilum infections in salmonid fish. J. Fish Dis. 26, 563–574. doi: 10.1046/j.1365-2761.2003.00488.x
O'connell, J., and Weeks, D. (1998). PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63, 259–266.
Palti, Y., Gao, G., Liu, S., Kent, M. P., Lien, S., Miller, M. R., et al. (2015a). The development and characterization of a 57K single nucleotide polymorphism array for rainbow trout. Mol. Ecol. Resour. 15, 662–672. doi: 10.1111/1755-0998.12337
Palti, Y., Vallejo, R. L., Gao, G., Liu, S., Hernandez, A. G., Rexroad, C. E. 3rd., et al. (2015b). Detection and validation of qtl affecting bacterial cold water disease resistance in rainbow trout using restriction-site associated DNA sequencing. PLoS ONE 10:e0138435. doi: 10.1371/journal.pone.0138435
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M., Sham, P.C., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
R Core Team (2017). R: A Language and Environment for Statistical computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org
Silverstein, J. T., Vallejo, R. L., Palti, Y., Leeds, T. D., Rexroad, C. E., Welch, T. J., et al. (2009). Rainbow trout resistance to bacterial cold-water disease is moderately heritable and is not adversely correlated with growth. J. Anim. Sci. 87, 860–867. doi: 10.2527/jas.2008-1157
Starliper, C. E. (2011). Bacterial coldwater disease of fishes caused by Flavobacterium psychrophilum. J. Adv. Res. 2, 97–108. doi: 10.1016/j.jare.2010.04.001
Vallejo, R. L., Leeds, T. D., Gao, G., Parsons, J. E., Martin, K. E., Evenhuis, J. P., et al. (2017a). Genomic selection models double the accuracy of predicted breeding values for bacterial cold water disease resistance compared to a traditional pedigree-based model in rainbow trout aquaculture. Genet. Select. Evol. 49:17. doi: 10.1186/s12711-017-0293-6
Vallejo, R. L., Liu, S., Gao, G., Fragomeni, B. O., Hernandez, A. G., Leeds, T. D., et al. (2017b). Similar genetic architecture with shared and unique quantitative trait loci for bacterial cold water disease resistance in two rainbow trout breeding populations. Front. Genet. 8:156. doi: 10.3389/Fgene.2017.00156
Vallejo, R. L., Palti, Y., Liu, S., Evenhuis, J. P., Gao, G., Rexroad, C. E. 3rd., et al. (2013). Detection of qtl in rainbow trout affecting survival when challenged with Flavobacterium psychrophilum. Mar. Biotechnol. 16, 349–360. doi: 10.1007/s10126-013-9553-9
Vallejo, R. L., Palti, Y., Liu, S. X., Marancik, D. P., and Wiens, G. D. (2014). Validation of linked QTL for bacterial cold water disease resistance and spleen size on rainbow trout chromosome Omy19. Aquaculture 432, 139–143. doi: 10.1016/j.aquaculture.2014.05.003
Vallejo, R. L., Silva, R. M. O., Evenhuis, J. P., Gao, G., Liu, S., Parsons, J. E., et al. (2018). Accurate genomic predictions for BCWD resistance in rainbow trout using low-density SNP panels: Evidence that strong long-range LD is a major contributing factor. J. Anim. Breed. Genet. 135, 263–274. doi: 10.1111/jbg.12335
Wiens, G. D., Lapatra, S. E., Welch, T. J., Evenhuis, J. P., Rexroad, C. E., and Leeds, T. D. (2013a). On-farm performance of rainbow trout (Oncorhynchus mykiss) selectively bred for resistance to bacterial cold water disease: effect of rearing environment on survival phenotype. Aquaculture 388, 128–136. doi: 10.1016/j.aquaculture.2013.01.018
Wiens, G. D., Vallejo, R. L., Leeds, T. D., Palti, Y., Hadidi, S., Liu, S., et al. (2013b). Assessment of genetic correlation between bacterial cold water disease resistance and spleen index in a domesticated population of rainbow trout: identification of QTL on chromosome Omy19. PLoS ONE 8:e75749. doi: 10.1371/journal.pone.0075749
Keywords: rainbow trout, bacterial cold water disease, haplotype, SNP, MAS, QTL, Flavobacterium psychrophilum
Citation: Liu S, Vallejo RL, Evenhuis JP, Martin KE, Hamilton A, Gao G, Leeds TD, Wiens GD and Palti Y (2018) Retrospective Evaluation of Marker-Assisted Selection for Resistance to Bacterial Cold Water Disease in Three Generations of a Commercial Rainbow Trout Breeding Population. Front. Genet. 9:286. doi: 10.3389/fgene.2018.00286
Received: 15 March 2018; Accepted: 11 July 2018;
Published: 03 August 2018.
Edited by:
Ross Houston, University of Edinburgh, United KingdomReviewed by:
JUNHONG XIA, Sun Yat-sen University, ChinaDirk-Jan De Koning, Swedish University of Agricultural Sciences, Sweden
Copyright © 2018 Liu, Vallejo, Evenhuis, Martin, Hamilton, Gao, Leeds, Wiens and Palti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sixin Liu, c2l4aW4ubGl1QGFycy51c2RhLmdvdg==
 Sixin Liu
Sixin Liu Roger L. Vallejo
Roger L. Vallejo Jason P. Evenhuis
Jason P. Evenhuis Kyle E. Martin
Kyle E. Martin Alastair Hamilton
Alastair Hamilton Guangtu Gao
Guangtu Gao Timothy D. Leeds
Timothy D. Leeds Gregory D. Wiens
Gregory D. Wiens Yniv Palti
Yniv Palti