- 1School of Biology, Scottish Oceans Institute, University of St Andrews, St Andrews, United Kingdom
- 2Xelect Ltd, St Andrews, United Kingdom
The production of most farmed molluscs, including mussels, oysters, scallops, abalone, and clams, is heavily dependent on natural seed from the plankton. Closing the lifecycle of species in hatcheries can secure independence from wild stocks and enables long-term genetic improvement of broodstock through selective breeding. Genomic techniques have the potential to revolutionize hatchery-based selective breeding by improving our understanding of the characteristics of mollusc genetics that can pose a challenge for intensive aquaculture and by providing a new suite of tools for genetic improvement. Here we review characteristics of the life history and genetics of molluscs including high fecundity, self-fertilization, high genetic diversity, genetic load, high incidence of deleterious mutations and segregation distortion, and critically assess their impact on the design and effectiveness of selective breeding strategies. A survey of the results of current breeding programs in the literature show that selective breeding with inbreeding control is likely the best strategy for genetic improvement of most molluscs, and on average growth rate can be improved by 10% per generation and disease resistance by 15% per generation across the major farmed species by implementing individual or family-based selection. Rapid advances in sequencing technology have resulted in a wealth of genomic resources for key species with the potential to greatly improve hatchery-based selective breeding of molluscs. In this review, we catalog the range of genomic resources currently available for molluscs of aquaculture interest and discuss the bottlenecks, including lack of high-quality reference genomes and the relatively high cost of genotyping, as well as opportunities for applying genomics-based selection.
Introduction
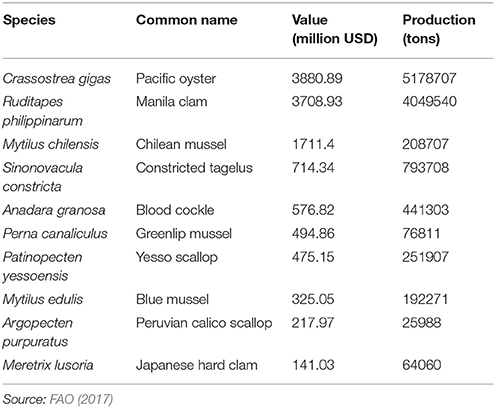
Molluscs comprise a diverse Phylum comprising ten classes and some 85,000-extant species. The majority of the 80-individual species farmed come from the Classes Bivalvia, Cephlapoda, and Gastropoda. Bivalves, namely clam, oyster, scallop and mussel species, dominate in terms of tonnage and economic value (Figure 1A). In total, more than 16 million metric tons of molluscs were farmed worldwide in 2015 with Asia dominating production (Figure 1B; FAO, 2017). The life cycle has been closed for relatively few species, but this does include some of the most valuable species (Table 1). The majority of production, even for the species listed in Table 1, occurs in extensive aquaculture and is dependent on spat harvested from the wild (Boudry, 2009; Astorga, 2014). However, despite its cost-effectiveness, farming based on the collection of wild seed is subject to environmental risk, including depletion of local stocks, disease outbreaks (Boudry et al., 1997, 2004) and the adverse effects of climate change (Waldbusser et al., 2015). The future for industrialized aquaculture of molluscs undoubtedly lies with closing the life cycle of more species and in the development of hatcheries that can provide sufficient production of seed for on-growing in the sea, while allowing selective breeding to improve commercially important traits such as growth, meat yield, disease resistance, and stress tolerance (Boudry et al., 1997; Camara and Symonds, 2014).
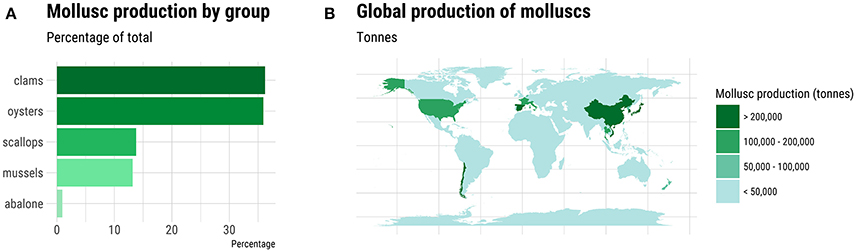
Figure 1. Global production of molluscs in 2015. (A) The percentage of total production for each major group of farmed molluscs. (B) Geographic distribution of major mollusc producing countries. Source: FAO (2017).
Advances in sequencing technology have provided a powerful set of genetic tools which can facilitate the establishment of hatchery-based breeding programs (Yáñez et al., 2015). In recent years, six molluscs of aquaculture significance have had their genomes sequenced and annotated (see Discussion below). Although this compares unfavorably with the more than 20 genomes of aquaculture finfish sequenced, the amount of publically available DNA sequence data for molluscs, as measured by the total number of bases in NCBI's Short Read Archive (SRA) database, has increased dramatically in the last 2 years (Figure 2). The generation of genomic resources for non-model species has been particularly accelerated by the advent of cost effective reduced-representation genome sequencing techniques such as genotyping-by-sequencing (GBS; Elshire et al., 2011) and RAD sequencing (RAD-seq; Baird et al., 2008). The aim of this review is to provide an overview of the genetics and life history of the major farmed molluscs and to discuss the implications for hatchery-based selective breeding. We present a survey of the current state of selective breeding with an emphasis on relevant genetic parameters. Finally, we discuss the application of genomic resources and the use of genetic markers in breeding programs, highlighting some of the bottlenecks and opportunities for increasing production using these new tools.
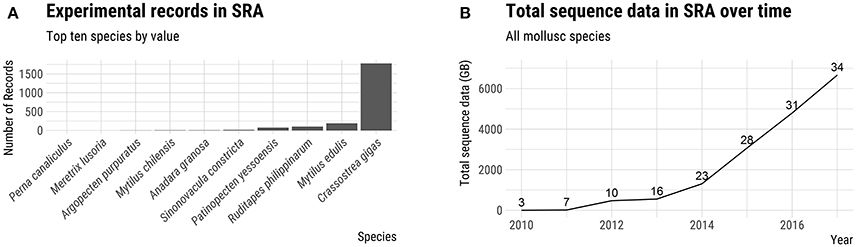
Figure 2. Genomic resources of farmed molluscs. (A) Number of experimental records in NCBI's Short Read Archive (SRA) for top 10 farmed mollusc species by value. (B) Total number of bases (GB) in the SRA for all species of farmed molluscs. The number above the line for each year indicates the number of farmed mollusc species represented in the SRA database. Source: FAO (2017) and NCBI SRA.
Implications of Life History Characteristics for Selective Breeding
Several life history characteristics of molluscs have major implications for selective breeding, including high fecundity and the incidence of hermaphroditism with the potential for self-fertilization. Pacific oysters, for example, can produce tens to hundreds of millions of eggs in a single spawning, resulting in potentially very large family sizes (Quayle, 1988). Diverse reproductive strategies have been documented (Collin, 2013). While most species of clams, mussels, and abalone have separate sexes, clams of the genus Venerupis, Mercenaria, Mya and Spisula, oysters of the genus Crassostrea and Ostrea and mussels including Mytilus can be to various degrees sequential hermaphrodites, whereas most farmed species of scallops are simultaneous (functional) hermaphrodites. Many species display a combination of strategies, with Pacific oysters typically maturing first as males then changing to females (protandric hermaphroditism), but they can also be functionally hermaphroditic at low frequency, and sex-reversal (female to male) can occur depending on environment and food supply (Guo et al., 1998). Individual European flat oysters can go through two or three sex reversal cycles in each spawning season (Joyce et al., 2013). Functional hermaphroditism produces a proportion of self-fertilized gametes, which can quickly elevate levels of inbreeding. For some species of scallops, rates of self-fertilization can average ~20 percent within particular families, and in addition to the effect on levels of inbreeding, failing to account for self-fertilization can result in biased estimates of genetic parameters such as heritability (Martinez, 2007). Finally, extremely high fecundity appears to be a critical factor in explaining several genetic characteristics of mollusc species that directly influence selective breeding (see below).
Implications of Genetic Characteristics for Selective Breeding
Decades of genetic studies involving molluscs have revealed a number of characteristics that differentiate molluscs from other aquaculture species. In particular, molluscs have been shown to exhibit extremely high levels of genetic diversity (Peñaloza et al., 2006; Zhang et al., 2012; Romiguier et al., 2014; Wang et al., 2017), deviations from Hardy-Weinberg proportions in wild populations (i.e., Zouros and Foltz, 1984 and references therein), non-Mendelian segregation of marker loci in pair crosses (Launey and Hedgecock, 2001), high incidence of null alleles at genetic markers (Hedgecock et al., 2004), and correlations between fitness traits and levels of heterozygosity (David, 1998).
High Genetic Diversity
Molluscs exhibit extremely high levels of nuclear genetic diversity relative to many other taxa based on a variety of metrics (English et al., 2000; Bazin et al., 2006; Peñaloza et al., 2006; Sauvage et al., 2007; Zhang et al., 2012; Harrang et al., 2013; Romiguier et al., 2014; Wang et al., 2017), although the ultimate cause of such diversity is not well-understood (Plough, 2016). The neutral theory of evolution predicts that the steady-state level of heterozygosity should scale positively with effective population size (Ne) and mutation rate (Nei and Li, 1979), and it is likely that both factors contribute to the extreme levels of variation observed. The extreme fecundity of molluscs and a tendency toward broadcast spawning (which promotes homogeneity of populations) would favor large effective population sizes, although the high variance in reproductive success commonly observed in molluscs may considerably reduce Ne relative to census size (Hedgecock, 1994; Hedgecock and Pudovkin, 2011). In addition, mutation rates for molluscs may be elevated due to the number of meiosis events necessary to produce millions of eggs during reproduction. For example, Plough et al. (2016) estimated that the mutation rate in the Pacific oyster was 90-fold higher than in Drosophila based on the number and effects of lethal alleles segregating in wild pair crosses. Observations of numerous and highly active transposable elements in molluscan genomes, as well as a high incidence of structural variation, also support the hypothesis of an elevated mutation rate (Zhang et al., 2012; Plough, 2016).
Segregation Distortion and Genetic Load
Since the earliest studies utilizing electrophoretic genetic markers on wild populations of molluscs, two of the most commonly observed phenomena have been deficiencies of heterozygous genotypes relative to expectations of Hardy-Weinberg equilibrium (HWE) and deviation from expected Mendelian ratios (segregation distortion) in pair crosses. These phenomena have been observed across the major groups of cultured molluscs and with a variety of marker types, including allozymes (Zouros and Foltz, 1984; Mallet et al., 1985; Gaffney et al., 1990; Beaumont, 1991; Toro and Vergara, 1995), microsatellites (Bierne et al., 1998; Launey and Hedgecock, 2001), and SNPs (Peñaloza et al., 2006; Vervalle et al., 2013; Hedgecock et al., 2015; Niu et al., 2017). The earliest explanations for these phenomena implicated null alleles, aneuploidy, inbreeding, cryptic population structure (Wahlund effect), or some form of selection (Beaumont, 1991). Null alleles, caused by mutations that produce truncated or inactive allozymes (discussed in Gaffney et al., 1990) or reduce primer binding affinity for amplification of PCR-based markers (McGoldrick et al., 2000), are a likely explanation for at least part of the observed heterozygote deficiencies, and it has been shown in some cases that redesign of PCR primers has eliminated the significant heterozygote deficiencies initially observed (e.g., Hare et al., 1996). Conversely, the incidence of null alleles is often insufficient to account for the observed level of heterozygote deficiency (Beaumont, 1991), and studies that have accounted for null alleles by tracking them through pedigrees have still observed considerable segregation distortion (McGoldrick et al., 2000; Launey and Hedgecock, 2001). A similar argument applies to aneuploidy, which has not typically been observed at high enough levels in molluscs to account for the degree of deviation from HWE or Mendelian ratios (Thiriot-Quiévreux et al., 1992). Further, the fact that segregation distortion is commonly observed in controlled pair crosses (Beaumont, 1991) has eliminated population-level inbreeding and Wahlund effects as a single explanation for the phenomena. Based on these observations, some form of selection is the most likely cause (Beaumont, 1991; Toro and Vergara, 1995). Inbred crosses of European oyster (Bierne et al., 1998) and Pacific oyster (Launey and Hedgecock, 2001) have shown large homozygote deficiencies at microsatellite loci consistent with selection against a large genetic load of deleterious recessive mutations: 15–38 per genome for O. edulis and 8–14 per genome for C. gigas. Both studies also found that genotypic proportions were typically Mendelian at early larval stages and became increasingly distorted with age, which strongly supported viability selection as a cause of the distortions over pre-zygotic possibilities such as meiotic drive. QTL mapping of segregating lethal alleles in inbred C. gigas has demonstrated large homozygote deficiencies and evidence of at least 14–15 recessive or partially recessive viability QTL per genome, and estimated that mortality attributed to viability selection up to the spat stage was ~96%, which was consistent with observations of mortality in hatchery populations (Plough and Hedgecock, 2011). Most recently, Plough et al. (2016) found mortality attributable to viability selection (~99%) in wild crosses of C. gigas to be as high as that observed for inbred crosses, except in this case the genetic load was expressed primarily as selection against additive or partially dominant mutations, resulting in a deficit of heterozygotes. These experimental results from oysters, along with evidence of extremely high levels of genetic diversity in other molluscs, provide a compelling case that a high genetic load is largely responsible for heterozygote deficits in wild populations and segregation distortion in pair crosses (Plough, 2016). One difficult to resolve aspect of this discussion is the fact that there is considerable heterogeneity in the literature with respect to these phenomena, whereby at times segregation distortions in pair crosses are extreme (Launey and Hedgecock, 2001; Wang et al., 2004; Plough and Hedgecock, 2011; Plough et al., 2016) and at other times small or non-significant (Wang et al., 2005; Gutierrez et al., 2017) for the same species. Similarly, reports of segregation distortion sometimes primarily involve homozygote deficits (Launey and Hedgecock, 2001; Plough and Hedgecock, 2011) and at other times primarily heterozygote deficits (Peñaloza et al., 2006; Plough et al., 2016). An attractive feature of the genetic load hypothesis is that it can account for this heterogeneity by way of the fact that expression of genetic load is highly context dependent (Barrett and Charlesworth, 1991; Pekkala et al., 2012). Differences in the overall levels of diversity (and the amount of genetic load) between populations (Lohr and Haag, 2015), the level of inbreeding in a population or individuals in a cross (Barrett and Charlesworth, 1991), levels of dominance and epistasis of the genes involved (Whitlock and Bourguet, 2000), developmental timing of expressed lethals (Plough and Hedgecock, 2011), and the extent to which lethal mutations have been purged from a population (Crnokrak and Barrett, 2002) will all influence the way that genetic load is expressed. For instance, if genetic load in Pacific oyster consists of both partially dominant and recessive mutations, this explains why pair crosses from wild individuals (Plough et al., 2016) would exhibit heterozygote deficiencies (due to initial expression of partially dominant lethals) and inbred crosses (Launey and Hedgecock, 2001; Plough and Hedgecock, 2011) would largely exhibit homozygote deficiencies due to recessive lethals, as partially dominant lethals would likely be purged in previous generations of inbreeding.
Molluscan Genetics and Selective Breeding
A high genetic load, combined with high fecundity, provides the opportunity for large variance in reproductive success among breeders (Hedgecock and Pudovkin, 2011), a phenomenon that has often been observed in molluscs in hatchery settings (Boudry et al., 2002; Lallias et al., 2010). The uneven contribution of parents has the potential to rapidly decrease the Ne of the breeding population, which can lead to both loss of genetic diversity through genetic drift and rapidly increase the level of inbreeding. Both can reduce the long-term response to selection (Lacy, 1987; Meuwissen and Woolliams, 1994). Inbreeding is of particular concern for bivalve aquaculture because inbreeding load—the rate at which fitness declines with increases in inbreeding coefficient (Charlesworth and Willis, 2009)—is likely higher for bivalves than for other animals (Plough, 2016). In support of this, inbreeding depression, typically characterized by reduced growth and increased mortality at early life stages, has been well-documented in cultured molluscs, including in European flat oyster (Bierne et al., 1998; Naciri-Graven et al., 2000), Pacific oyster (McGoldrick and Hedgecock, 1997; Evans et al., 2004), Pacific abalone (Haliotis discus; Deng et al., 2005), and catarina scallop (Argopecten ventricosus; Ibarra et al., 1995). Careful control of inbreeding is thus essential for molluscan selective breeding, and thus methods for optimizing the rate of genetic gain while constraining the rate of inbreeding such as optimal contribution selection (Meuwissen, 1997) and mate selection (Kinghorn, 2011) are important to consider. A further discussion of inbreeding management in aquaculture can be found in Nguyen (2016).
The second important consideration for genetic improvement is whether evidence for non-additive genetic variance for survival and other traits warrants a strong consideration of crossbreeding inbred lines to exploit heterosis as a more optimal breeding strategy than selection without inbreeding, which has become the standard means of genetic improvement for most finfishes (Gjedrem and Rye, 2016). Here, there are both genetic and practical considerations. From a genetic perspective, the more non-additive genetic variation that exists for a trait, the more benefit can be gained by exploiting heterosis. However, it is also important to consider the nature of the non-additive variance. In particular, quantitative genetic theory demonstrates that it is only when there is some level of overdominance with respect to the trait that crossing inbred lines can achieve what selection without inbreeding cannot (Falconer and Mackay, 1996). In general, while direct evidence for heterosis has been observed in molluscs (Cruz and Ibarra, 1997; Hedgecock and Davis, 2007; Deng et al., 2010), other studies have found little or no evidence for heterosis with multiple strains and production systems (e.g., In et al., 2017), and overall there is little evidence for overdominance of relevant traits. Both in studies of heterozygote fitness correlations and direct investigations of heterosis, results have been more consistent with a dominance or epistatic model of heterosis (Hedgecock et al., 1995; David, 1998; Hedgecock and Davis, 2007), suggesting that inbreeding and crossing could be a sub-optimal strategy. One important practical consideration is the difficulty in producing and maintaining inbred lines of molluscs for crossbreeding. Molluscs are not as easily grown, maintained, and propagated as crop species such as maize that have benefited from crossbreeding (Hallauer, 2009), nor is it often possible to crossbreed already established inbred lines or breeds, as is often beneficial in cattle (Gregory and Cundiff, 1980). Thus, while some success has been made with inter-specific hybridization in species such as abalone (Lafarga de la Cruz and Gallardo-Escárate, 2011), the practical difficulties in maintaining and testing enough inbred lines for effective genetic improvement is a barrier for crossbreeding in most molluscs. In summary, selective breeding with effective inbreeding control remains the primary strategy for genetic improvement in molluscs. As is discussed in the following sections, current research indicates large amounts of additive genetic variance for production traits across all important groups of molluscs, and selective breeding programs to date have experienced promising levels of genetic gain per generation. Hereafter, we will discuss selective breeding as selection with control of inbreeding.
Current Status of Selective Breeding Programs
Traits and Genetic Parameters
The aim of breeding programs is to improve one or more traits of commercial importance, such as growth rate, yield, shell pigmentation, temperature tolerance, and disease resistance. Because profitability is one of the key measures of success for breeding programs, existing programs for molluscs have often established breeding goals based on input from industry (Kube et al., 2011; Camara and Symonds, 2014), and the breeding goal often consists of multiple economically important traits to improve simultaneously. Once the traits for improvement have been identified, developing an understanding of the genetic basis of the traits is a crucial step toward implementing selective breeding. Three particularly important genetic parameters of interest are heritability, genetic correlations between traits, and interactions between genotypes and environment (Falconer and Mackay, 1996; Boudry, 2009).
Heritability
Heritability is a measure of the extent to which a phenotype is genetically determined. Narrow-sense heritability, in particular, determines the expected response of a trait to selection. In general, it is difficult to make generalizations about heritability of particular traits because estimates of heritability for the same trait can differ greatly depending on genetic diversity, life stage, and environmental conditions (Gjedrem and Thodesen, 2005). For example, Zheng et al. (2004) found realized heritabilities for larval growth rate in two hatchery stocks of bay scallops (Argopecten irradians) spawned and reared in common conditions to be 0.015 and 0.511 (more than a 30-fold difference), presumably due to very different levels of genetic diversity in the two stocks. Drawing general conclusions from heritability estimates is also confounded by the fact that heritability for the same trait can change dramatically depending on the life stage at which the trait is measured. For example, heritability for shell length was shown to increase with age in red abalone (Haliotis rufescens; Jonasson et al., 1999), but decreased in bay scallops (Zheng et al., 2004), and there is little to no genetic correlation between larval and juvenile traits in Pacific oyster (Ernande et al., 2003). Overall, there are two noteworthy trends. First, most traits of commercial interest in molluscs have been found to have moderate to high narrow-sense heritability (Supplemental File 1), suggesting that additive genetic variance for production traits is generally sufficient for selective breeding. Second, genetic markers are increasingly being used to improve estimates of heritability using family variance components, because molecular parentage assignment allows for families to be reared in common environment, eliminating a potentially confounding source of environmental variation (Vandeputte et al., 2004; Nguyen et al., 2014; Kong et al., 2015).
Genetic Correlations
Knowledge of genetic correlations can often be used to advantage in breeding programs, when a trait that is difficult to measure can be improved by selecting for a correlated trait that is easier to measure (Falconer and Mackay, 1996). Examples in molluscs include genetic correlations between growth and difficult-to-measure traits such as meat yield (de Melo et al., 2016), shell strength (Camara and Symonds, 2014), and disease resistance (Dégremont et al., 2015). A more extensive survey of genetic correlations in molluscs can be found in Supplemental File 1.
Genotype-by-Environment Interactions
Often individuals from centralized breeding programs will be distributed and grown out in many different environmental conditions, and individuals with a particular genotype may exhibit superior phenotypic performance in one environment but not another (Falconer and Mackay, 1996). If significant GxE interactions are present, it may be beneficial to develop multiple selected lines with superior performance in particular environments (Sheridan, 1997). Unfortunately, studies measuring GxE interactions are still mostly lacking for most cultured molluscs (see Supplemental File 1). Experiments in Pacific oyster have revealed weak or no GxE interactions for growth (Evans and Langdon, 2006) and survival (Dégremont et al., 2007), albeit in relatively restricted geographic regions. In contrast, a study involving Eastern oyster found large, significant GxE interactions for both yield and cumulative mortality (Proestou et al., 2016). Breeding studies of silver-lipped pearl oyster have shown minimal GxE interactions for growth (Kvingedal et al., 2010), but larger effects for pearl quality traits (Jerry et al., 2012).
Response to Selection
Response to selection for production-related traits may be higher for molluscs than finfish due to their higher fecundity, which allows for a greater intensity of selection (Gjedrem and Thodesen, 2005; Gjedrem and Robinson, 2014). For molluscs, there are numerous studies measuring realized response to selection, which involves phenotypic comparison of selected lines against unselected controls. The majority of studies report response after a single-generation of mass selection, although a few studies encompass multi-generational mass selection (Dégremont et al., 2015) or family-based selection (Liu et al., 2015; de Melo et al., 2016). An extensive but non-exhaustive survey of the literature reporting response to selection for production traits in molluscs revealed that the average response to selection per generation for growth ranged from 6.8% for mussels to 12.1% in oysters (Figure 3). Averaged across studies for all mollusc taxa, the average response to selection for growth was 10.6% per generation. Average response to selection for disease resistance traits was even higher at 15.7% (Figure 3).
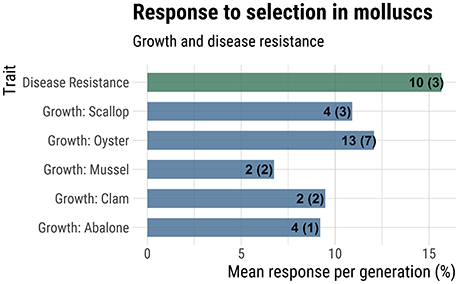
Figure 3. Response to selection for growth and disease resistance. Mean response to selection (% increase in additive genetic value over base population mean) is reported on the x-axis. The numbers at the end of each bar indicate the total number of generations of selection used in the calculation and the number of studies considered (in parentheses). A more complete summary of the data collected can be found in Supplemental File 1, under the tab “Response to Selection”.
Selective Breeding Programs
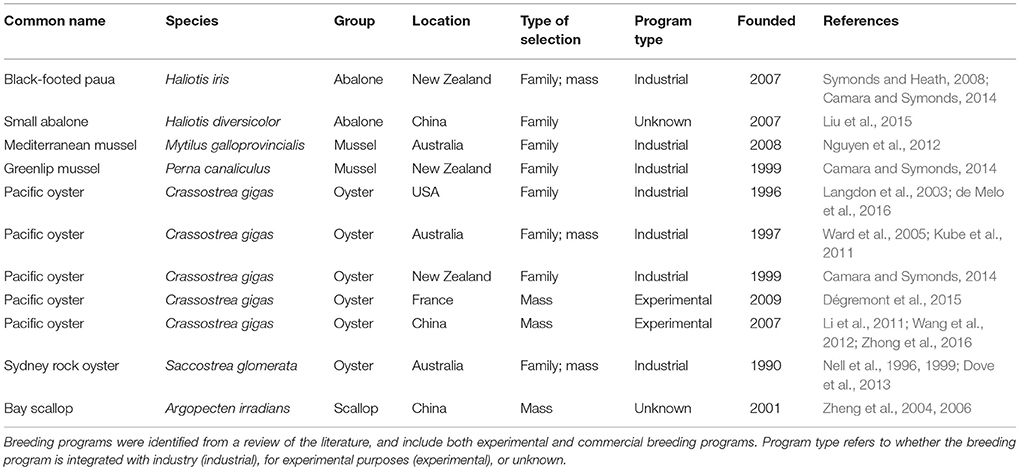
There are several strategies for selective breeding, and the optimal choice depends on a variety of factors, notably the genetic properties of the traits to be selected and the associated costs of implementing a particular strategy. Typically, molluscan breeding programs described in the literature have either utilized mass (individual) selection, in which selection is performed solely based on the phenotypes of individuals (Fjalestad, 2005), or family-based selection, in which selection is performed on the basis of the phenotypic means of families (between-family selection), on deviations of individuals from their respective family means (within-family selection), or a combination of both (combined selection) (Fjalestad, 2005). A number of commercial and experimental molluscan breeding programs have been described in the literature (Table 2).
There are two large-scale, commercially-integrated selective breeding programs for mussel species: a family-based breeding program for blue mussel (Mytilus galloprovincialis) in Australia and a family-based program for greenlip (GreenShell™) mussel (Perna canaliculus) in New Zealand. The breeding program for blue mussel was initiated in Victoria, Australia in 2008 with a founder population of 74 full-sib families (Nguyen et al., 2011). In a trial where individuals were selected based on breeding values for total weight, shell shape, and meat yield, the realized selection responses in these three traits were positive and measured 10% for total weight, 3.0% for shell shape, and 2.3% for meat yield (Nguyen et al., 2014).
Centralized, family-based breeding programs for Pacific oyster have been established in Australia (Ward et al., 2005; Kube et al., 2011), New Zealand (Camara and Symonds, 2014), and the United States (Langdon et al., 2003; de Melo et al., 2016), and experimental mass selection has been reported in France (Dégremont et al., 2015) and China (Li et al., 2011). In a comprehensive analysis, a family-based selection program on the west coast of the United States reported accumulative gains for survival and yield of 15.7 and 19.0%, respectively, following five generations of selection (de Melo et al., 2016).
Recent outbreaks of pathogens such as OsHV-1 in New Zealand (Camara and Symonds, 2014) and France (Dégremont, 2011) have led to increased attention on selective breeding for disease resistance in Pacific oyster. Promisingly, an experimental mass selection program in France demonstrated a 61.8% gain in survival over unselected controls in response to OsHV-1 exposure after four generations (average 15.5% per generation; Dégremont et al., 2015). However, since selection was performed without inbreeding control, genetic gain is likely to be less in a long-term program that minimized inbreeding.
A selection of the main commercial and experimental molluscan breeding programs that have been described in the literature is summarized in Table 2.
Genomic Resources
The application of genomic tools to selective breeding in aquaculture involves two processes: the development of genomic resources for key species and the application of those tools toward selective breeding. In general, the current state of most important species are somewhere in between these two steps: genomic resources have been developed but for the most part are not yet applied to selective breeding. The following section contains a survey of genome scale resources underpinning selective breeding in the main farmed mollusc species. Here, we discuss whole genome sequences, genetic linkage maps, genetic marker panels, and identification of quantitative trait loci (QTL) for key production traits.
Whole Genome Sequences
Declining costs of next-generation DNA sequencing has made it possible to sequence the entire genome of nearly any species in a cost-effective manner. Genome sequences allow genetic markers to be mapped to a specific location in the genome, and when genetic markers associated with particular traits are identified, a genome sequence provides the opportunity to investigate nearby genes or genetic elements to potentially identify causative mutations. As of 2017, there were 22 published, scaffold-level genome sequences for finfish (Yue and Wang, 2017) and at the time of writing only six for molluscs: Pacific oyster (Zhang et al., 2012), pearl oyster (Takeuchi et al., 2012, 2016), Yesso scallop (Wang et al., 2017), Zhikong scallop (Li et al., 2017), Manila clam (Mun et al., 2017), and Eastern oyster (NCBI accession GCA_002022765.4) with a seventh species, blue mussel, (Nguyen et al., 2014; Murgarella et al., 2016) currently at contig level (Supplemental File 2). While at the time of writing it is currently unpublished, the most complete molluscan genome assembled to date is that of the Eastern oyster (NCBI accession GCA_002022765.4). The assembly was performed using 87x long-read PacBio sequencing technology, and the genome was able to be assembled to chromosome level (https://www.ncbi.nlm.nih.gov/assembly/GCF_002022765.2/).
To avoid problems with genome assembly caused by high levels of polymorphism, mollusc genomes have typically been assembled using highly inbred individuals with reduced heterozygosity (Zhang et al., 2012; Wang et al., 2017) or have used modified assembly techniques to account for excess heterozygosity (Takeuchi et al., 2016; Li et al., 2017). As expected, resequencing whole genomes of wild individuals has revealed extremely high diversity at the nucleotide level. The overall rate of SNP and indel polymorphism in a single wild individual was 1.3% for Pacific oyster, 0.81% for Zhikong scallop, and 1.04% for Yesso scallop, which is six to ten times higher than that observed in humans (Zhang et al., 2012; Li et al., 2017; Wang et al., 2017).
One important insight gained from sequencing molluscan genomes is the observation of lineage-specific expansions in gene families related to immune and stress response (Takeuchi et al., 2012, 2016; Zhang et al., 2012; Guo et al., 2015; Li et al., 2017; Mun et al., 2017; Wang et al., 2017), which may be a key evolutionary innovation related to surviving as sessile organisms in often heterogeneous conditions. An illustrative example is the case of heat shock proteins (HSP), which among other functions help to prevent misfolding of proteins under various types of stress (Parsell and Lindquist, 1993). The Pacific oyster genome contains a remarkable 88 heat shock protein 70 (Hsp70) genes, of which 71 clustered together in a phylogenetic analysis, consistent with a species/taxa specific gene expansion (Zhang et al., 2012). HSPs have been found to be upregulated in multiple mollusc species in response to a variety of stressors, including heat-shock (Li et al., 2012; Zhang et al., 2012), viral challenge (He et al., 2015), and exposure to pollutants (Gao et al., 2007), and the expansion of HSP gene families in molluscan lineages suggests that copy number variation in stress-related genes may be an important factor to exploit for selective breeding. Other gene families showing expansion include the C1q gene which has a role in pathogen detection and the activation of the complement system in the innate immune response and has a 12-fold higher copy number in the Manila clam than in humans (Mun et al., 2017). In Zhikong scallop, 270 gene families were found to be significantly expanded relative to other bivalves, including genes involved in neurotransmission, immune responses, signal transduction and xenobiotic metabolism (Li et al., 2017). As copy number variation is increasingly associated with variation in agriculturally important traits (e.g., Wang et al., 2015), further consideration of gene expansions in molluscs may prove to be a fruitful area of research for selective breeding.
Genetic Linkage Maps
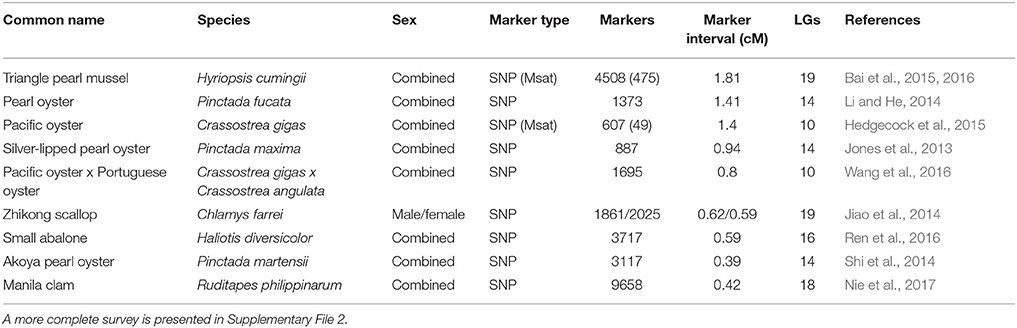
Linkage maps provide a framework of the order and spacing of genetic markers and serve as a starting point for the mapping of quantitative trait loci (QTL) for traits of commercial interest and for the development of marker assisted selection (MAS) programs (Liu and Cordes, 2004). First generation maps typically have been based on amplified fragment length polymorphism (AFLP) markers and/or microsatellites, and whereas dense linkage maps for major species such as Pacific oyster typically have been developed through multiple versions or iterations (Hedgecock et al., 2015 and references therein), equally dense, first generation linkage maps for less developed species have been generated de novo using SNP markers (Jones et al., 2013). Table 3 provides information on high density linkage maps developed recently for major farmed species.
Genetic Marker Panels for Parentage Assignment
The control of inbreeding in hatchery-based selective breeding programs is critical and requires collection of reliable pedigree information for brood stock. Physical tagging is one method that allows pedigree information to be retained, but it requires families to be reared separately until they are sufficiently large to tag. This requires substantial and expensive infrastructure which may constrain mating designs to a sub-optimal number of families (Vandeputte et al., 2004). Genetic markers can overcome these limitations and allow pedigrees to be constructed in groups of mixed families regardless of family structure.
Microsatellites have been used for parentage assignment since the 1990s. Eight to Fifteen microsatellite loci typically provide >99 percent assignment power for finfish (Vandeputte and Haffray, 2014). The high prevalence of null alleles in molluscs (Hedgecock et al., 2004) adds considerably to the cost of developing and validating parentage assignment panels. For example, after screening 135 microsatellite loci in Australian Blue mussel (Mytilus galloprovincialis) just 10 were found to provide consistent PCR amplification across 74 full-sibling families. Only 62.5% of 2,536 offspring tested could be assigned to single parent pairs using the remaining 10 microsatellites (Nguyen et al., 2011), which is well below the accuracy needed to operate a family based breeding program. Multiplex sets of microsatellites are more efficient in terms of genotyping costs and reduce manual error (Guichoux et al., 2011). By excluding null alleles and using 12 microsatellites each with four to 20 alleles in three multiplex assays, assignment success in the King scallop (Pecten maximus) reached 97% accuracy (Morvezen et al., 2013).
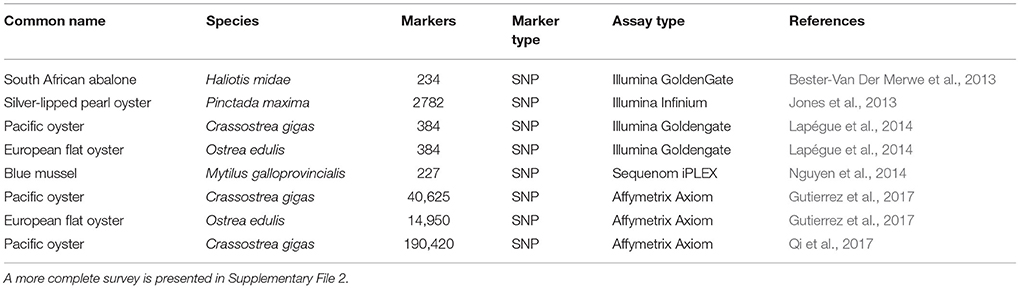
The adoption of NGS methods for SNP discovery and technological advances in SNP genotyping have resulted in SNPs supplanting microsatellites as the genetic markers of choice for parentage assignment (Anderson and Garza, 2006; Strucken et al., 2016). The high degree of polymorphism in mollusc genomes provides a massive number of SNPs for marker design, although the frequency of null alleles incurs additional costs for panel development as with microsatellites. Protein-coding regions of the Pacific oyster genome contain one SNP for every 60 bp sequenced compared to one every 40 bp in non-coding regions (Sauvage et al., 2007). SNP density in the European flat oyster was shown to be similarly high with one SNP in every 76 and 47 bp for coding and non-coding regions, respectively (Harrang et al., 2013). Nguyen et al. (2014) reported that in Mytilus galloprovincialis 179 SNPs could correctly assign 92.5% of offspring to 77 full-sibling families. Several other SNP-based genotyping panels have recently been published for key species (Table 4).
Quantitative Trait Loci
Most economically important traits in aquaculture production are influenced by a large number of small-effect loci, known as quantitative trait loci (QTL). QTL mapping is the process of identifying genetic marker alleles that segregate non-randomly with QTL and positioning those loci in the genome (Lynch and Walsh, 1998). Once QTL for a trait are identified, individuals can be selected for breeding on the basis of marker alleles that segregate with favorable phenotypes (Lande and Thompson, 1990). This strategy, known as marker-assisted selection (MAS), is particularly useful for traits that cannot be measured on selection candidates directly, notably disease resistance or meat-quality traits (Sonesson, 2007a).
QTL mapping in molluscs (as well as terrestrial livestock and finfish) has traditionally relied on linkage-based methods of identifying QTL, in which one or more families are genotyped and scored for a trait of interest, and the co-segregation of markers and trait values are observed for each family (Lynch and Walsh, 1998). While this approach has been used successfully to map QTL in cultured molluscs, it has the disadvantages that marker/QTL associations are only necessarily valid in the families that are genotyped and that QTL are typically not able to be mapped with much precision (Hirschhorn and Daly, 2005). The ability to genotype many thousands of SNP loci has now made possible a different statistical approach to identifying loci associated with traits of interest. Genome-wide association studies (GWAS) identify marker-trait associations at the population rather than the family level by identifying genetic markers that are in linkage disequilibrium with QTL (Hirschhorn and Daly, 2005). While GWAS typically requires many more loci than linkage-based QTL mapping, it is able to provide much more precise locations of QTL, and due to massive improvements in genotyping has begun to be applied in aquaculture (Correa et al., 2015; Gutierrez et al., 2015; Tsai et al., 2015; Vallejo et al., 2017).
The majority of QTL mapping studies in mollusc species have focused on growth, although QTL related to disease resistance (Yu and Guo, 2006; Lallias et al., 2009; Sauvage et al., 2010; Harrang et al., 2015), shell pigmentation (Qin et al., 2007; Petersen et al., 2012; Ge et al., 2014; Zhong et al., 2014), sex (Li et al., 2005; Guo et al., 2012; Jiao et al., 2014), and pearl quality (Jones et al., 2014a,b; Bai et al., 2015, 2016) have also been discovered.
Opportunities for Genomics-Based Selection
Incorporating genomic information into selective breeding programs (hereafter referred to as genomics-based selection) has become an important aspect of genetic improvement in livestock and plant breeding (Meuwissen et al., 2013; Muranty et al., 2014). The principal advantage of incorporating marker information into breeding programs is an increase in the accuracy of predicted breeding values, which in turn causes a proportional increase in the response to selection (Martinez, 2007). Because selection is based on marker genotypes, MAS is useful for traits that are difficult to measure on breeding candidates (Sonesson, 2007b), particularly when markers are linked to large-effect QTL (Robledo et al., 2017). A classic example of MAS in aquaculture is the case of IPN virus resistance in Atlantic salmon, in which a large-effect QTL was discovered in Scottish and Norwegian populations (Houston et al., 2008; Moen et al., 2009), and MAS using markers linked to the locus was subsequently incorporated into commercial breeding programs, substantially reducing the incidence of IPN in commercial facilities (Moen et al., 2015; Robledo et al., 2016).
A more recent development, genomic selection (GS), is the extension of MAS to a genome-wide scale. Rather than targeting specific QTL, GS assumes that genetic markers (typically SNPs) are genotyped at sufficient density such that all QTL are in linkage disequilibrium with at least one marker (Meuwissen et al., 2001), and so in theory can utilize all of the genetic variance in a complex trait (Meuwissen et al., 2013). In practice, a reference population of individuals with both genotypes and phenotypes is used to estimate the effects of each marker on the phenotype of interest, and these effects are used to estimate breeding values for relatives of the reference population (selection candidates) that have been genotyped but have no phenotypes (Meuwissen et al., 2001). Whereas MAS is useful when large-effect loci are present, GS is well-suited for improvement of traits that are under the control of many loci with small effect (Robledo et al., 2017).
MAS and GS have yet to be applied in breeding programs for molluscs, although several studies in other aquaculture species have demonstrated increases in accuracy of breeding value predictions for traits such as growth (Tsai et al., 2015; Nguyen et al., 2018) and disease resistance (Bangera et al., 2017; Robledo et al., 2018). In molluscs there has been significant investment in identifying QTL, and large-effect loci may be applicable to MAS. For example, a QTL explaining as much as 80% of the phenotypic variance for shell pigmentation has been identified in Pacific oyster (Ge et al., 2014). Reduced-representation sequencing techniques such as GBS or RAD-seq will likely be important for future genomics-based selection in molluscs, as such techniques allow for pedigree reconstruction, estimation of trait heritability, genetic map construction, identification of QTL, and estimation of genomic breeding values in the same experiment (Palaiokostas et al., 2016; Robledo et al., 2017). QTL mapping studies involving GBS or RAD-seq have been published for scallops (Jiao et al., 2014), oysters (Li and He, 2014; Shi et al., 2014; Wang et al., 2016), clams (Nie et al., 2017), and abalone (Ren et al., 2016). Recently, the feasibility of GS using 2b-RAD (Wang et al., 2012) was demonstrated in an experiment involving Yesso scallops (Patinopectin yessoensis; Dou et al., 2016). However, while such techniques have considerably reduced the time and expense involved in generating the genomic resources necessary for MAS or GS, the cost of genotyping is still a significant barrier toward further use of these strategies.
Challenges for incorporating genomic approaches in breeding of molluscs include high rates of larval mortality, high incidence of marker segregation distortion, and self-fertilization. A better theoretical understanding of these processes in molluscs will lead to the development of strategies for more optimally balancing control of inbreeding and genetic gain. In addition, the transition from low to high-throughput genotyping technologies will alleviate marker-specific issues such as local segregation distortion, because these markers can be removed from a large dataset with minimal impact on the results. The lack of high quality annotated reference genomes and studies to understand the genetic architecture of important production traits for key species is also a bottleneck for the adoption of genomics-based breeding. Only three of the top ten cultivated molluscan species (Table 1) have had their genomes sequenced to draft level, and the high degree of variability is still a barrier to developing accurate assemblies. Although a reference genome sequence is available for Pacific oyster (Zhang et al., 2012), a significant number of putative assembly errors have been identified (Hedgecock et al., 2015). The low economic value of individual animals is also a challenge for implementation of genomic selection, and novel methods that can reduce the cost of genotyping or reduce the number of markers necessary for genomic selection will be necessary to make this a cost-effective strategy. The development of medium- and high-density SNP arrays for important species, such as those recently developed for Pacific oyster and European flat oyster (Gutierrez et al., 2017) and Pacific oyster (Qi et al., 2017), will make genomic selection more feasible by enabling high-throughput genotyping at low cost per marker. In addition, several ideas have been proposed to reduce genotyping costs for MAS and GS in aquaculture applications, including pooling DNA of test individuals (Sonesson et al., 2010), using low-density genotyping for within-family genomic selection (Lillehammer et al., 2013), and imputation of genotypes using a combination of low- and high-density genotyping (Tsai et al., 2017).
Providing these bottlenecks can be overcome, the gains in accuracy of breeding value predictions that can be obtained using genomic data, along with optimized control of inbreeding, should enable more rapid rates of genetic improvement in cultured molluscs compared to traditional selection schemes.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
IJ is Chief Executive Officer of Xelect Ltd. and CH is a consultant for Xelect Ltd.
Acknowledgments
This review of genetic resources and selective breeding in molluscs was carried out as part of the European Marine Biological Research Infrastructure Cluster (EMBRIC) project funded by the European Union's Horizon 2020 research and innovation program under grant agreement No 654008.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00253/full#supplementary-material
References
Anderson, E. C., and Garza, J. C. (2006). The power of single-nucleotide polymorphisms for large-scale parentage inference. Genetics 172, 2567–2582. doi: 10.1534/genetics.105.048074
Astorga, M. P. (2014). Genetic considerations for mollusk production in aquaculture: current state of knowledge. Front. Genet. 5:435. doi: 10.3389/fgene.2014.00435
Bai, Z., Han, X., Luo, M., Lin, J., Wang, G., and Li, J. (2015). Constructing a microsatellite-based linkage map and identifying QTL for pearl quality traits in triangle pearl mussel (Hyriopsis cumingii). Aquaculture 437, 102–110. doi: 10.1016/j.aquaculture.2014.11.008
Bai, Z.-Y., Han, X.-K., Liu, X.-J., Li, Q.-Q., and Li, J.-L. (2016). Construction of a high-density genetic map and QTL mapping for pearl quality-related traits in Hyriopsis cumingii. Sci. Rep. 6:32608. doi: 10.1038/srep32608
Baird, N. A., Etter, P. D., Atwood, T. S., Currey, M. C., Shiver, A. L., Lewis, Z. A., et al. (2008). Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376. doi: 10.1371/journal.pone.0003376
Bangera, R., Correa, K., Lhorente, J. P., Figueroa, R., and Yáñez, J. M. (2017). Genomic predictions can accelerate selection for resistance against Piscirickettsia salmonis in Atlantic salmon (Salmo salar). BMC Genomics 18:121. doi: 10.1186/s12864-017-3487-y
Barrett, S. C., and Charlesworth, D. (1991). Effects of a change in the level of inbreeding on the genetic load. Nature 352, 522–524. doi: 10.1038/352522a0
Bazin, E., Gle, S., and Galtier, N. (2006). Mitochondrial genetic diversity in animals. Science 312, 570–572. doi: 10.1126/science.1122033
Beaumont, A. R. (1991). Genetic studies of laboratory reared mussels, Mytilus edulis: heterozygote deficiencies, heterozygosity and growth. Biol. J. Linn. Soc. 44, 273–285. doi: 10.1111/j.1095-8312.1991.tb00620.x
Bester-Van Der Merwe, A., Blaauw, S., Du Plessis, J., and Roodt-Wilding, R. (2013). Transcriptome-wide single nucleotide polymorphisms (SNPs) for abalone (Haliotis midae): validation and application using goldengate medium-throughput genotyping assays. Int. J. Mol. Sci. 14, 19341–19360. doi: 10.3390/ijms140919341
Bierne, N., Launey, S., Naciri-Graven, Y., and Bonhomme, F. (1998). Early effect of inbreeding as revealed by microsatellite analyses on Ostrea edulis larvae. Genetics 148, 1893–1906.
Boudry, P. (2009). “Genetic variation and selective breeding in hatchery-propagated molluscan shellfish,” in New Technologies in Aquaculture: Improving Production Efficiency, Quality and Environmental Management, eds G. Burnell and G. Allan (Cambridge: Woodhead Publishing Ltd), 87–108.
Boudry, P., Barré, M., and Gérard, A. (1997). “Genetic Improvement and Selection in Shellfish: a Review Based on Oyster Research and Production,” in Proceedings of the TECAM Seminar on Genetics and Breeding in the Mediterranean Aquaculture Species CIHEAM-FAO, (Zaragoza), 28–29.
Boudry, P., Collet, B., Cornette, F., Hervouet, V., and Bonhomme, F. (2002). High variance in reproductive success of the Pacific oyster (Crassostrea gigas, Thunberg) revealed by microsatellite-based parentage analysis of multifactorial crosses. Aquaculture 204, 283–296. doi: 10.1016/S0044-8486(01)00841-9
Boudry, P., Dégremont, L., Taris, N., Mccombie, H., Haffray, P., and Ernande, B. (2004). Genetic variability and selective breeding for traits of aquacultural interest in the Pacific oyster (Crassostrea gigas). Bull. Aquacult. Assoc. Can. 104, 12–18.
Camara, M. D., and Symonds, J. (2014). Genetic improvement of New Zealand aquaculture species: programmes, progress and prospects. N. Z. J. Mar. Freshwater Res. 48, 466–491. doi: 10.1080/00288330.2014.932291
Charlesworth, D., and Willis, J. H. (2009). The genetics of inbreeding depression. Nat. Rev. Genetics 10, 783–796. doi: 10.1038/nrg2664
Collin, R. (2013). Phylogenetic patterns and phenotypic plasticity of molluscan sexual systems. Integr. Comp. Biol. 53, 723–735. doi: 10.1093/icb/ict076
Correa, K., Lhorente, J. P., López, M. E., Bassini, L., Naswa, S., Deeb, N., et al. (2015). Genome-wide association analysis reveals loci associated with resistance against Piscirickettsia salmonis in two Atlantic salmon (Salmo salar L.) chromosomes. BMC Genomics 16:854. doi: 10.1186/s12864-015-2038-7
Crnokrak, P., and Barrett, S. C. (2002). Perspective: purging the genetic load: a review of the experimental evidence. Evolution 56, 2347–2358. doi: 10.1111/j.0014-3820.2002.tb00160.x
Cruz, P., and Ibarra, A. M. (1997). Larval growth and survival of two catarina scallop (Argopecten circularis, Sowerby, 1835) populations and their reciprocal crosses. J. Exp. Mar. Biol. Ecol. 212, 95–110. doi: 10.1016/S0022-0981(96)02742-6
David, P. (1998). Heterozygosity–fitness correlations: new perspectives on old problems. Heredity 80, 531–537. doi: 10.1046/j.1365-2540.1998.00393.x
de Melo, C. M. R., Durland, E., and Langdon, C. (2016). Improvements in desirable traits of the Pacific oyster, Crassostrea gigas, as a result of five generations of selection on the West Coast, USA. Aquaculture 460, 105–115. doi: 10.1016/j.aquaculture.2016.04.017
Dégremont, L. (2011). Evidence of herpesvirus (OsHV-1) resistance in juvenile Crassostrea gigas selected for high resistance to the summer mortality phenomenon. Aquaculture 317, 94–98. doi: 10.1016/j.aquaculture.2011.04.029
Dégremont, L., Ernande, B., Bédier, E., and Boudry, P. (2007). Summer mortality of hatchery-produced Pacific oyster spat (Crassostrea gigas). I. Estimation of genetic parameters for survival and growth. Aquaculture 262, 41–53. doi: 10.1016/j.aquaculture.2006.10.025
Dégremont, L., Nourry, M., and Maurouard, E. (2015). Mass selection for survival and resistance to OsHV-1 infection in Crassostrea gigas spat in field conditions: response to selection after four generations. Aquaculture 446, 111–121. doi: 10.1016/j.aquaculture.2015.04.029
Deng, Y., Liu, X., Zhang, G., and Guo, X. (2005). Inbreeding depression and maternal effects on early performance of Pacific abalone. N. Am. J. Aquac. 67, 231–236. doi: 10.1577/A04-021.1
Deng, Y., Liu, X., Zhang, G., and Wu, F. (2010). Heterosis and combining ability: a diallel cross of three geographically isolated populations of Pacific abalone Haliotis discus hannai Ino. Chin. J. Oceanol. Limnol. 28, 1195–1199. doi: 10.1007/s00343-010-9903-7
Dou, J., Li, X., Fu, Q., Jiao, W., Li, Y., Li, T., et al. (2016). Evaluation of the 2b-RAD method for genomic selection in scallop breeding. Sci. Rep. 6:19244. doi: 10.1038/srep19244
Dove, M. C., Nell, J. A., and O'Connor, W. A. (2013). Evaluation of the progeny of the fourth-generation Sydney rock oyster Saccostrea glomerata (Gould, 1850) breeding lines for resistance to QX disease (Marteilia sydneyi) and winter mortality (Bonamia roughleyi). Aquac. Res. 44, 1791–1800. doi: 10.1111/are.12012
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379. doi: 10.1371/journal.pone.0019379
English, L. J., Maguire, G. B., and Ward, R. D. (2000). Genetic variation of wild and hatchery populations of the Pacific oyster, Crassostrea gigas (Thunberg), in Australia. Aquaculture 187, 283–298. doi: 10.1016/S0044-8486(00)00321-5
Ernande, B., Clobert, J., McCombie, H., and Boudry, P. (2003). Genetic polymorphism and trade-offs in the early life-history strategy of the Pacific oyster, Crassostrea gigas (Thunberg, 1795): a quantitative genetic study. Evolution 16, 399–414. doi: 10.1046/j.1420-9101.2003.00543.x
Evans, F., Matson, S., Brake, J., and Langdon, C. (2004). The effects of inbreeding on performance traits of adult Pacific oysters (Crassostrea gigas). Aquaculture 230, 89–98. doi: 10.1016/j.aquaculture.2003.09.023
Evans, S., and Langdon, C. (2006). Effects of genotype × environment interactions on the selection of broadly adapted Pacific oysters (Crassostrea gigas). Aquaculture 261, 522–534. doi: 10.1016/j.aquaculture.2006.07.022
Falconer, D. S., and Mackay, T. F. C. (1996). Introduction to Quantitative Genetics, 4th Edn. Harlow: Pearson Education Limited.
FAO (2017). Fisheries and Aquaculture Software. FishStatJ - Software for Fishery Statistical Time Series [online]. Available online at: http://www.fao.org/fishery/statistics/software/fishstatj/en
Fjalestad, K. T. (2005). “Selection methods,” in Selection and Breeding Programs in Aquaculture, ed T. Gjedrem (Dordrecht: Springer), 159–170.
Gaffney, P. M., Scott, T. M., Koehn, R. K., and Diehl, W. J. (1990). Interrelationships of heterozygosity, growth rate and heterozygote deficiencies in the coot clam, Mulinia lateralis. Genetics 124, 687–699.
Gao, Q., Song, L., Ni, D., Wu, L., Zhang, H., and Chang, Y. (2007). cDNA cloning and mRNA expression of heat shock protein 90 gene in the haemocytes of Zhikong scallop Chlamys farreri. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 147, 704–715. doi: 10.1016/j.cbpb.2007.04.010
Ge, J., Li, Q., Yu, H., and Kong, L. (2014). Identification and mapping of a SCAR marker linked to a locus involved in shell pigmentation of the Pacific oyster (Crassostrea gigas). Aquaculture 434, 249–253. doi: 10.1016/j.aquaculture.2014.08.027
Gjedrem, T., and Robinson, N. (2014). Advances by selective breeding for aquatic species: a review. Agric. Sci. 5, 1152–1158. doi: 10.4236/as.2014.512125
Gjedrem, T., and Rye, M. (2016). Selection response in fish and shellfish: a review. Rev. Aquacult. 10, 1–12. doi: 10.1111/raq.12154
Gjedrem, T., and Thodesen, J. (2005). “Selection,” in Selection and Breeding Programs in Aquaculture, ed T. Gjedrem (Dordrecht: Springer), 89–110.
Gregory, K. E., and Cundiff, L. V. (1980). Crossbreeding in beef cattle: evaluation of systems. J. Anim. Sci. 51, 1224–1242. doi: 10.2527/jas1980.5151224x
Guichoux, E., Lagache, L., Wagner, S., Chaumeil, P., Léger, P., Lepais, O., et al. (2011). Current trends in microsatellite genotyping. Mol. Ecol. Resour. 11, 591–611. doi: 10.1111/j.1755-0998.2011.03014.x
Guo, X., Hedgecock, D., Hershberger, W. K., Cooper, K., and Allen, S. K. (1998). Genetic determinants of protandric sex in the Pacific oyster, Crassostrea gigas Thunberg. Evolution 52, 394–402. doi: 10.1111/j.1558-5646.1998.tb01640.x
Guo, X., He, Y., Zhang, L., Lelong, C., and Jouaux, A. (2015). Immune and stress responses in oysters with insights on adaptation. Fish Shellfish Immunol. 46, 107–119. doi: 10.1016/j.fsi.2015.05.018
Guo, X., Li, Q., Wang, Q. Z., and Kong, L. F. (2012). Genetic Mapping and QTL analysis of growth-related traits in the pacific oyster. Mar. Biotechnol. 14, 218–226. doi: 10.1007/s10126-011-9405-4
Gutierrez, A. P., Turner, F., Gharbi, K., Talbot, R., Lowe, N. R., Peñaloza, C., et al. (2017). Development of a medium density combined-species SNP array for Pacific and European oysters (Crassostrea gigas and Ostrea edulis). G3 7, 2209–2218. doi: 10.1534/g3.117.041780
Gutierrez, A. P., Yáñez, J. M., Fukui, S., Swift, B., and Davidson, W. S. (2015). Genome-wide association study (GWAS) for growth rate and age at sexual maturation in Atlantic Salmon (Salmo salar). PLoS ONE 10:e0119730. doi: 10.1371/journal.pone.0119730
Hallauer, A. R. (2009). Corn Breeding. Iowa State Research Farm Progress Reports, 475. Available online at:http://lib.dr.iastate.edu/farms_reports
Hare, M. P., Karl, S. A., and Avise, J. C. (1996). Anonymous nuclear DNA markers in the American oyster and their implications for the heterozygote deficiency phenomenon in marine bivalves. Mol. Biol. Evol. 13, 334–345. doi: 10.1093/oxfordjournals.molbev.a025593
Harrang, E., Heurtebise, S., Faury, N., Robert, M., Arzul, I., and Lapègue, S. (2015). Can survival of European flat oysters following experimental infection with Bonamia ostreae be predicted using QTLs? Aquaculture 448, 521–530. doi: 10.1016/j.aquaculture.2015.06.019
Harrang, E., Lapègue, S., Morga, B., and Bierne, N. (2013). A high load of non-neutral amino-acid polymorphisms explains high protein diversity despite moderate effective population size in a marine bivalve with sweepstakes reproduction. G3 3, 333–341. doi: 10.1534/g3.112.005181
He, Y., Jouaux, A., Ford, S. E., Lelong, C., Sourdaine, P., Mathieu, M., et al. (2015). Transcriptome analysis reveals strong and complex antiviral response in a mollusc. Fish Shellfish Immunol. 46, 131–144. doi: 10.1016/j.fsi.2015.05.023
Hedgecock, D. (1994). “Does variance in reproductive success limit effective population sizes of marine organisms?” in Genetics and Evolution of Aquatic Organisms, ed A. R. Beaumont (London: Chapman and Hall), 122–134.
Hedgecock, D., and Davis, J. P. (2007). Heterosis for yield and crossbreeding of the Pacific oyster Crassostrea gigas. Aquaculture 272, 17–29. doi: 10.1016/j.aquaculture.2007.07.226
Hedgecock, D., Li, G., Hubert, S., Bucklin, K., and Ribes, V. (2004). Widespread null alleles and poor cross-species amplification of microsatellite DNA loci cloned from the Pacific oyster, Crassostrea gigas. J. Shellfish Res. 23, 379–386.
Hedgecock, D., McGoldrick, D. J., and Bayne, B. L. (1995). Hybrid vigor in Pacific oysters: an experimental approach using crosses among inbred lines. Aquaculture 137, 285–298. doi: 10.1016/0044-8486(95)01105-6
Hedgecock, D., and Pudovkin, A. I. (2011). Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull. Mar. Sci. 87, 971–1002. doi: 10.5343/bms.2010.1051
Hedgecock, D., Shin, G., Gracey, A. Y., Den Berg, D. V., and Samanta, M. P. (2015). Second-generation linkage maps for the Pacific Oyster Crassostrea gigas reveal errors in assembly of genome scaffolds. G3 5, 2007–2019. doi: 10.1534/g3.115.019570
Hirschhorn, J. N., and Daly, M. J. (2005). Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108. doi: 10.1038/nrg1521
Houston, R. D., Haley, C. S., Hamilton, A., Guy, D. R., Tinch, A. E., Taggart, J. B., et al. (2008). Major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar). Genetics 178, 1109–1115. doi: 10.1534/genetics.107.082974
Ibarra, A. M., Cruz, P., and Romero, B. A. (1995). Effects of inbreeding on growth and survival of self-fertilized catarina scallop larvae, Argopecten circularis. Aquaculture 134, 37–47. doi: 10.1016/0044-8486(95)00022-T
In, V., Van, S. V., Van, O'Connor, W., Van, P. T., Dove, M., Knibb, W., et al. (2017). Are strain genetic effect and heterosis expression altered with culture system and rearing environment in the Portuguese oyster (Crassostrea angulata)? Aquac. Res. 48, 4058–4069. doi: 10.1111/are.13227
Jerry, D. R., Kvingedal, R., Lind, C. E., Evans, B. S., Taylor, J. J., and Safari, A. E. (2012). Donor-oyster derived heritability estimates and the effect of genotypexenvironment interaction on the production of pearl quality traits in the silver-lip pearl oyster, Pinctada maxima. Aquaculture 338–341, 66–71. doi: 10.1016/j.aquaculture.2012.02.001
Jiao, W., Fu, X., Dou, J., Li, H., Su, H., Mao, J., et al. (2014). High-resolution linkage and quantitative trait locus mapping aided by genome survey sequencing: building up an integrative genomic framework for a bivalve mollusc. DNA Res. 21, 85–101. doi: 10.1093/dnares/dst043
Jonasson, J., Stefansson, S. E., Gudnason, A., and Steinarsson, A. (1999). Genetic variation for survival and shell length of cultured red abalone (Haliotis rufescens) in Iceland. J. Shellfish Res. 18, 621–625.
Jones, D. B., Jerry, D. R., Khatkar, M. S., Moser, G., Raadsma, H. W., Taylor, J. J., et al. (2014a). Determining genetic contributions to host oyster shell growth: quantitative trait loci and genetic association analysis for the silver-lipped pearl oyster, Pinctada maxima. Aquaculture 434, 367–375. doi: 10.1016/j.aquaculture.2014.08.040
Jones, D. B., Jerry, D. R., Khatkar, M. S., Moser, G., Raadsma, H. W., Taylor, J. J., et al. (2014b). Quantitative trait loci and genetic association analysis reveals insights into complex pearl quality traits in donor silver-lipped pearl oysters. Aquaculture 434, 476–485. doi: 10.1016/j.aquaculture.2014.08.038
Jones, D. B., Jerry, D. R., Khatkar, M. S., Raadsma, H. W., and Zenger, K. R. (2013). A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: a valuable resource for gene localisation and marker-assisted selection. BMC Genomics 14:810. doi: 10.1186/1471-2164-14-810
Joyce, A., Holthuis, T. D., Charrier, G., and Lindegarth, S. (2013). Experimental effects of temperature and photoperiod on synchrony of gametogenesis and sex ratio in the European Oyster Ostrea edulis (Linnaeus). J. Shellfish Res. 32, 447–458. doi: 10.2983/035.032.0225
Kinghorn, B. P. (2011). An algorithm for efficient constrained mate selection. Genet. Sel. Evol. 43. doi: 10.1186/1297-9686-43-4
Kong, N., Li, Q., Yu, H., and Kong, L. F. (2015). Heritability estimates for growth-related traits in the Pacific oyster (Crassostrea gigas) using a molecular pedigree. Aquac. Res. 46, 499–508. doi: 10.1111/are.12205
Kube, P. D., Cunningham, M., Dominik, S., Parkinson, S., Henshall, J., Finn, B., et al. (2011). Enhancement of the Pacific oyster Selective Breeding Program. Hobart, TAS: CSIRO Marine; Atmospheric Research, Australian Seafood Industries P/L, Seafood CRC; Fisheries Research; Development Corporation (FRDC).
Kvingedal, R., Evans, B. S., Lind, C. E., Taylor, J. J., Dupont-Nivet, M., and Jerry, D. R. (2010). Population and family growth response to different rearing location, heritability estimates and genotype × environment interaction in the silver-lip pearl oyster (Pinctada maxima). Aquaculture 304, 1–6. doi: 10.1016/j.aquaculture.2010.02.035
Lacy, R. C. (1987). Society for conservation biology loss of genetic diversity from managed populations: interacting effects of drift, loss of genetic diversity from managed populations: interacting effects of drift, mutation, immigration, selection, and population su. Conserv. Biol. 1, 143–158. doi: 10.1111/j.1523-1739.1987.tb00023.x
Lafarga de la Cruz, F., and Gallardo-Escárate, C. (2011). Intraspecies and interspecies hybrids in Haliotis: natural and experimental evidence and its impact on abalone aquaculture. Rev. Aquacult. 3, 74–99. doi: 10.1111/j.1753-5131.2011.01045.x
Lallias, D., Gomez-Raya, L., Haley, C. S., Arzul, I., Heurtebise, S., Beaumont, A. R., et al. (2009). Combining two-stage testing and interval mapping strategies to detect QTL for resistance to Bonamiosis in the european flat oyster Ostrea edulis. Mar. Biotechnol. 11, 570–584. doi: 10.1007/s10126-008-9173-y
Lallias, D., Taris, N., Boudry, P., Bonhomme, F., and Lapègue, S. (2010). Variance in the reproductive success of flat oyster Ostrea edulis L. assessed by parentage analyses in natural and experimental conditions. Genet. Res. 92, 175–187. doi: 10.1017/S0016672310000248
Lande, R., and Thompson, R. (1990). Efficiency of multistage marker-assisted selection in the improvement of multiple quantitative traits. Genetics 124, 743–756.
Langdon, C., Evans, F., Jacobson, D., and Blouin, M. (2003). Yields of cultured Pacific oysters Crassostrea gigas Thunberg improved after one generation of selection. Aquaculture 220, 227–244. doi: 10.1016/S0044-8486(02)00621-X
Lapégue, S., Harrang, E., Heurtebise, S., Flahauw, E., Donnadieu, C., Gayral, P., et al. (2014). Development of SNP-genotyping arrays in two shellfish species. Mol. Ecol. Resour. 14, 820–830. doi: 10.1111/1755-0998.12230
Launey, S., and Hedgecock, D. (2001). High genetic load in the pacific oyster Crassostrea gigas. Genetics 159, 255–265.
Li, J., He, Q., Sun, H., and Liu, X. (2012). Acclimation-dependent expression of heat shock protein 70 in Pacific abalone (Haliotis discus hannai Ino) and its acute response to thermal exposure. Chin. J. Oceanol. Limnol. 30, 146–151. doi: 10.1007/s00343-012-1026-x
Li, L., Xiang, J., Liu, X., Zhang, Y., Dong, B., and Zhang, X. (2005). Construction of AFLP-based genetic linkage map for Zhikong scallop, Chlamys farreri Jones et Preston and mapping of sex-linked markers. Aquaculture 245, 63–73. doi: 10.1016/j.aquaculture.2004.12.015
Li, Q., Wang, Q., Liu, S., and Kong, L. (2011). Selection response and realized heritability for growth in three stocks of the Pacific oyster Crassostrea gigas. Fish. Sci. 77, 643–648. doi: 10.1007/s12562-011-0369-0
Li, Y., and He, M. (2014). Genetic mapping and QTL analysis of growth-related traits in Pinctada fucata using restriction-site associated DNA sequencing. PLoS ONE 9:e111707. doi: 10.1371/journal.pone.0111707
Li, Y., Sun, X., Hu, X., Xun, X., Zhang, J., Guo, X., et al. (2017). Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat. Commun. 8:1721. doi: 10.1038/s41467-017-01927-0
Lillehammer, M., Meuwissen, T. H., and Sonesson, A. K. (2013). A low-marker density implementation of genomic selection in aquaculture using within-family genomic breeding values. Genet. Sel. Evol. 45:39. doi: 10.1186/1297-9686-45-39
Liu, J., Lai, Z., Fu, X., Wu, Y., Bao, X., Hu, Z., et al. (2015). Genetic parameters and selection responses for growth and survival of the small abalone Haliotis diversicolor after four generations of successive selection. Aquaculture 436, 58–64. doi: 10.1016/j.aquaculture.2014.10.046
Liu, Z. J., and Cordes, J. F. (2004). DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238, 1–37. doi: 10.1016/j.aquaculture.2004.05.027
Lohr, J. N., and Haag, C. R. (2015). Genetic load, inbreeding depression, and hybrid vigor covary with population size: an empirical evaluation of theoretical predictions. Evolution 69, 3109–3122. doi: 10.1111/evo.12802
Lynch, M., and Walsh, B. (1998). Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer, 980.
Mallet, A. L., Zouros, E., Gartner-Kepkay, K. E., Freeman, K. R., and Dickie, L. M. (1985). Marine biology. Mar. Biol. 87, 165–172. doi: 10.1007/BF00539424
Martinez, V. (2007). Marker-assisted selection in fish and shellfish breeding schemes,” in Current Status and Future Perspectives in Crops, Livestock, Forestry, and Fish, ed E. P. Guimaraes (Rome: FAO), 329–362.
McGoldrick, D. J., and Hedgecock, D. (1997). Fixation, segregation and linkage of allozyme loci in inbred families of the Pacific oyster Crassostrea gigas (Thunberg): implications for the causes of inbreeding depression. Genetics 146, 321–334.
McGoldrick, D. J., Hedgecock, D., English, L. J., Baoprasertkul, P., and Ward, R. D. (2000). The transmission of microsatellite alleles in Australian and North American stocks of the Pacific oyster (Crassostrea gigas): selection and null alleles. J. Shellf. Res. 19, 779–788.
Meuwissen, T. H. (1997). Maximizing the response of selection with a predefined rate of inbreeding. J. Anim. Sci 75, 934–940. doi: 10.2527/1997.754934x
Meuwissen, T., Hayes, B., and Goddard, M. (2013). Accelerating improvement of livestock with genomic selection. Ann. Rev. Animal Biosci. 1, 221–237. doi: 10.1146/annurev-animal-031412-103705
Meuwissen, T., Hayes, B. J., and Goddard, M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829.
Meuwissen, T. H., and Woolliams, J. A. (1994). Effective sizes of livestock populations to prevent a decline in fitness. Theor. Appl. Genet. 89, 1019–1026. doi: 10.1007/BF00224533
Moen, T., Baranski, M., Sonesson, A. K., and Kjøglum, S. (2009). Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics 10:368. doi: 10.1186/1471-2164-10-368
Moen, T., Torgersen, J., Santi, N., Davidson, W. S., Baranski, M., Ødegård, J., et al. (2015). Epithelial cadherin determines resistance to infectious pancreatic necrosis virus in Atlantic salmon. Genetics 200, 1313–1326. doi: 10.1534/genetics.115.175406
Morvezen, R., Cornette, F., Charrier, G., Guinand, B., Egue, S., Boudry, P., et al. (2013). Multiplex PCR sets of novel microsatellite loci for the great scallop Pecten maximus and their application in parentage assignment. Aquat. Living Resour. 26, 207–213. doi: 10.1051/alr/2013052
Mun, S., Kim, Y.-J., Markkandan, K., Shin, W., Oh, S., Woo, J., et al. (2017). The whole-genome and transcriptome of the Manila clam (Ruditapes philippinarum). Genome Biol. Evol. 9, 1487–1498. doi: 10.1093/gbe/evx096
Muranty, H., Jorge, V., Bastien, C., Lepoittevin, C., Bouffier, L., and Sanchez, L. (2014). Potential for marker-assisted selection for forest tree breeding: lessons from 20 years of MAS in crops. Tree Genet. Genom. 10, 1491–1510. doi: 10.1007/s11295-014-0790-5
Murgarella, M., Puiu, D., Novoa, B., Figueras, A., Posada, D., and Canchaya, C. (2016). Correction: a first insight into the genome of the filter-feeder mussel Mytilus galloprovincialis. PLoS ONE 11:e0160081. doi: 10.1371/journal.pone.0160081
Naciri-Graven, Y., Launey, S., Lebayon, N., Gerard, A., and Baud, J.-P. (2000). Influence of parentage upon growth in Ostrea edulis : evidence for inbreeding depression. Genet. Res. 76, 159–168. doi: 10.1017/S0016672300004663
Nei, M., and Li, W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273. doi: 10.1073/pnas.76.10.5269
Nell, J. A., Sheridan, A. K., and Smith, I. R. (1996). Progress in a Sydney rock oyster, Saccostrea commercialis (Iredale and Roughley), breeding program. Aquaculture 144, 295–302. doi: 10.1016/0044-8486(96)01328-2
Nell, J. A., Smith, I. R., and Sheridan, A. K. (1999). Third generation evaluation of Sydney rock oyster Saccostrea commercialis (Iredale and Roughley) breeding lines. Aquaculture 170, 195–203. doi: 10.1016/S0044-8486(98)00408-6
Nguyen, T. T. T., Hayes, B., and Ingram, B. A. (2012). Progress on Selective Breeding Program for Blue Mussel in Victoria. Fisheries Victoria Research Report Series No. 60. Queenscliff, VIC.
Nguyen, N. H. (2016). Genetic improvement for important farmed aquaculture species with a reference to carp, tilapia and prawns in Asia: achievements, lessons and challenges. Fish Fish. 17, 483–506. doi: 10.1111/faf.12122
Nguyen, N. H., Premachandra, H. K. A., Kilian, A., and Knibb, W. (2018). Genomic prediction using DArT-Seq technology for yellowtail kingfish Seriola lalandi. BMC Genomics 19:107. doi: 10.1186/s12864-018-4493-4
Nguyen, T. T., Hayes, B. J., Guthridge, K., Ab Rahim, E. S., and Ingram, B. A. (2011). Use of a microsatellite-based pedigree in estimation of heritabilities for economic traits in Australian blue mussel, Mytilus galloprovincialis. J. Anim. Breeding Genet. 128, 482–490. doi: 10.1111/j.1439-0388.2011.00948.x
Nguyen, T. T. T., Hayes, B. J., and Ingram, B. A. (2014). Genetic parameters and response to selection in blue mussel (Mytilus galloprovincialis) using a SNP-based pedigree. Aquaculture 420–421, 295–301. doi: 10.1016/j.aquaculture.2013.11.021
Nie, H., Yan, X., Huo, Z., Jiang, L., Chen, P., Liu, H., et al. (2017). Construction of a High-density genetic map and quantitative trait locus mapping in the Manila clam Ruditapes philippinarum. Sci. Rep. 7:229. doi: 10.1038/s41598-017-00246-0
Niu, D., Du, Y., Wang, Z., Xie, S., Nguyen, H., Dong, Z., et al. (2017). Construction of the first high-density genetic linkage map and analysis of quantitative trait loci for growth-related traits in Sinonovacula constricta. Mar. Biotechnol. 19, 488–496. doi: 10.1007/s10126-017-9768-2
Palaiokostas, C., Ferraresso, S., Franch, R., Houston, R. D., and Bargelloni, L. (2016). Genomic prediction of resistance to pasteurellosis in Gilthead sea bream (Sparus aurata) Using 2b-RAD Sequencing. G3 6, 3693–3700. doi: 10.1534/g3.116.035220
Parsell, D. A., and Lindquist, S. (1993). The function of heat -shock proteins in stress tolerance: degradation and reactiv ation of damaged proteins. Annu. Rev. Genet. 27, 437–496. doi: 10.1146/annurev.ge.27.120193.002253
Pekkala, N., Knott, K. E., Kotiaho, J. S., and Puurtinen, M. (2012). Inbreeding rate modifies the dynamics of genetic load in small populations. Ecol. Evol. 2, 1791–1804. doi: 10.1002/ece3.293
Peñaloza, C., Bishop, S., Toro, J., and Houston, R. (2006). “RAD sequencing reveals genome-wide heterozygote deficiency in pair crosses of the Chilean mussel Mytilus spp.,” in 10th World Congress of Genetics Applied to Livestock Production, 2–4. Available online at: https://www.asas.org/docs/default-source/wcgalp-proceedings-oral/275_paper_9207_manuscript_964_0.pdf?sfvrsn=2
Petersen, J. L., Baerwald, M. R., Ibarra, A. M., and May, B. (2012). A first-generation linkage map of the Pacific lion-paw scallop (Nodipecten subnodosus): initial evidence of QTL for size traits and markers linked to orange shell color. Aquaculture 350–353, 200–209. doi: 10.1016/j.aquaculture.2012.03.039
Plough, L. V. (2016). Genetic load in marine animals: a review. Curr. Zool. 62, 567–579. doi: 10.1093/cz/zow096
Plough, L. V., and Hedgecock, D. (2011). Quantitative trait locus analysis of stage-specific inbreeding depression in the pacific oyster Crassostrea gigas. Genetics 189, 1473–1486. doi: 10.1534/genetics.111.131854
Plough, L. V., Shin, G., and Hedgecock, D. (2016). Genetic inviability is a major driver of type III survivorship in experimental families of a highly fecund marine bivalve. Mol. Ecol. 25, 895–910. doi: 10.1111/mec.13524
Proestou, D. A., Vinyard, B. T., Corbett, R. J., Piesz, J., Allen, S. K., Small, J. M., et al. (2016). Performance of selectively-bred lines of eastern oyster, Crassostrea virginica, across eastern US estuaries. Aquaculture 464, 17–27. doi: 10.1016/j.aquaculture.2016.06.012
Qi, H., Song, K., Li, C., Wang, W., Li, B., Li, L., et al. (2017). Construction and evaluation of a high-density SNP array for the Pacific oyster (Crassostrea gigas). PLoS ONE 12:e0174007. doi: 10.1371/journal.pone.0174007
Qin, Y., Liu, X., Zhang, H., Zhang, G., and Guo, X. (2007). Identification and mapping of amplified fragment length polymorphism markers linked to shell color in bay scallop, Argopecten irradians irradians (Lamarck, 1819). Mar. Biotechnol. 9, 66–73. doi: 10.1007/s10126-006-6076-7
Quayle, D. B. (1988). Pacific Oyster Culture in British Columbia. Ottawa: Canadian Bulletin of Fisheries; Aquatic Sciences.
Ren, P., Peng, W., You, W., Huang, Z., Guo, Q., Chen, N., et al. (2016). Genetic mapping and quantitative trait loci analysis of growth-related traits in the small abalone Haliotis diversicolor using restriction-site-associated DNA sequencing. Aquaculture 454, 163–170. doi: 10.1016/j.aquaculture.2015.12.026
Robledo, D., Matika, O., Hamilton, A., and Houston, R. D. (2018). Genome-wide association and genomic selection for resistance to Amoebic Gill Disease in Atlantic salmon. G3 8, 1195–1203. doi: 10.1534/g3.118.200075
Robledo, D., Palaiokostas, C., Bargelloni, L., Martínez, P., and Houston, R. (2017). Applications of genotyping by sequencing in aquaculture breeding and genetics. Rev. Aquacult. 9, 1–13. doi: 10.1111/raq.12193
Robledo, D., Taggart, J. B., Ireland, J. H., McAndrew, B. J., Starkey, W. G., Haley, C. S., et al. (2016). Gene expression comparison of resistant and susceptible Atlantic salmon fry challenged with Infectious Pancreatic Necrosis virus reveals a marked contrast in immune response. BMC Genomics 17:279. doi: 10.1186/s12864-016-2600-y
Romiguier, J., Gayral, P., Ballenghien, M., Bernard, A., Cahais, V., Chenuil, A., et al. (2014). Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 515, 261–263. doi: 10.1038/nature13685
Sauvage, C., Bierne, N., Lapègue, S., and Boudry, P. (2007). Single nucleotide polymorphisms and their relationship to codon usage bias in the Pacific oyster Crassostrea gigas. Gene 406, 13–22. doi: 10.1016/j.gene.2007.05.011
Sauvage, C., Boudry, P., De Koning, D. J., Haley, C. S., Heurtebise, S., and Lapègue, S. (2010). QTL for resistance to summer mortality and OsHV-1 load in the Pacific oyster (Crassostrea gigas). Anim. Genet. 41, 390–399. doi: 10.1111/j.1365-2052.2009.02018.x
Sheridan, A. K. (1997). Genetic improvement of oyster production - a critique. Aquaculture 153, 165–179. doi: 10.1016/S0044-8486(97)00024-0
Shi, Y., Wang, S., Gu, Z., Lv, J., Zhan, X., Yu, C., et al. (2014). High-density single nucleotide polymorphisms linkage and quantitative trait locus mapping of the pearl oyster, Pinctada fucata martensii Dunker. Aquaculture 434, 376–384. doi: 10.1016/j.aquaculture.2014.08.044
Sonesson, A. K. (2007a). “Possibilities for marker-assisted selection in fish breeding schemes,” in Marker-Assisted Selection: Current Status and Future Application in Crops, Livestock, Forestry and Fish, ed. E. P. Guimaraes (Rome: FAO), 309–328.
Sonesson, A. K. (2007b). Within-family marker-assisted selection for aquaculture species. Genet. Sel. Evol. 39, 301–317. doi: 10.1186/1297-9686-39-3-301
Sonesson, A. K., Meuwissen, T. H. E., and Goddard, M. (2010). The use of communal rearing of families and DNA pooling in aquaculture genomic selection schemes. Genet. Select. Evol. 42:41. doi: 10.1186/1297-9686-42-41
Strucken, E. M., Lee, S. H., Lee, H. K., Song, K. D., Gibson, J. P., and Gondro, C. (2016). How many markers are enough? Factors influencing parentage testing in different livestock populations. J. Anim. Breed. Genet. 133, 13–23. doi: 10.1111/jbg.12179
Symonds, J. E., and Heath, P. (2008). Getting picky with paua: selective breeding to improve productivity. Water Atmos. 16, 20–21.
Takeuchi, T., Kawashima, T., Koyanagi, R., Gyoja, F., Tanaka, M., Ikuta, T., et al. (2012). Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res. 19, 117–130. doi: 10.1093/dnares/dss005
Takeuchi, T., Koyanagi, R., Gyoja, F., Kanda, M., Hisata, K., Fujie, M., et al. (2016). Bivalve-specific gene expansion in the pearl oyster genome: implications of adaptation to a sessile lifestyle. Zool. Lett. 2:3. doi: 10.1186/s40851-016-0039-2
Thiriot-Quiévreux, C., Pogson, G. H., and Zouros, E. (1992). Genetics of growth rate variation in bivalves: aneuploidy and heterozygosity effects in a Crassostrea gigas family. Genome 35, 39–45. doi: 10.1139/g92-007
Toro, J. E., and Vergara, A. M. (1995). Evidence for selection against heterozygotes: post-settlement excess of allozyme heterozygosity in a cohort of the Chinean oyster, Ostrea chinensis Philippi, 1845. Biol. Bull. 188, 117–119. doi: 10.2307/1542076
Tsai, H.-Y., Hamilton, A., Tinch, A. E., Guy, D. R., Gharbi, K., Stear, M. J., et al. (2015). Genome wide association and genomic prediction for growth traits in juvenile farmed Atlantic salmon using a high density SNP array. BMC Genomics 16:969. doi: 10.1186/s12864-015-2117-9
Tsai, H.-Y., Matika, O., Edwards, S. M., Antolín–Sánchez, R., Hamilton, A., Guy, D. R., et al. (2017). Genotype imputation to improve the cost-efficiency of genomic selection in farmed Atlantic Salmon. G3 7, 1377–1383. doi: 10.1534/g3.117.040717
Vallejo, R. L., Liu, S., Gao, G., Fragomeni, B. O., Hernandez, A. G., Leeds, T. D., et al. (2017). Similar genetic architecture with shared and unique quantitative trait loci for bacterial cold water disease resistance in two rainbow trout breeding populations. Front. Genet. 8:156. doi: 10.3389/fgene.2017.00156
Vandeputte, M., and Haffray, P. (2014). Parentage assignment with genomic markers: a major advance for understanding and exploiting genetic variation of quantitative traits in farmed aquatic animals. Front. Genet. 5:432. doi: 10.3389/fgene.2014.00432
Vandeputte, M., Kocour, M., Mauger, S., Dupont-Nivet, M., De Guerry, D., Rodina, M., et al. (2004). Heritability estimates for growth-related traits using microsatellite parentage assignment in juvenile common carp (Cyprinus carpio L.). Aquaculture 235, 223–236. doi: 10.1016/j.aquaculture.2003.12.019
Vervalle, J., Hepple, J.-A., Jansen, S., Plessis, J. D., Wang, P., Rhode, C., et al. (2013). Integrated linkage map of Haliotis midae linnaeus based on microsatellite and SNP markers. J. Shellfish Res. 32, 89–103. doi: 10.2983/035.032.0115
Waldbusser, G. G., Hales, B., Langdon, C. J., Haley, B. A., Schrader, P., Brunner, E. L., et al. (2015). Saturation-state sensitivity of marine bivalve larvae to ocean acidification. Nat. Clim. Chang. 5, 273–280. doi: 10.1038/nclimate2479
Wang, J., Li, L., and Zhang, G. (2016). A high-density SNP genetic linkage map and QTL analysis of growth-related traits in a hybrid family of oysters (Crassostrea gigas × Crassostrea angulata) using genotyping-by-sequencing. G3 6, 1417–1426. doi: 10.1534/g3.116.026971
Wang, L., Song, L., Chang, Y., Xu, W., Ni, D., and Guo, X. (2005). A preliminary genetic map of Zhikong scallop (Chlamys farreri Jones et Preston 1904). Aquac. Res. 36, 643–653. doi: 10.1111/j.1365-2109.2005.01268.x
Wang, S., Bao, Z., Pan, J., Zhang, L., Yao, B., Zhan, A., et al. (2004). AFLP linkage map of an intraspecific cross in Chlamys farreri. J. Shellfish Res. 23, 491–499.
Wang, S., Meyer, E., McKay, J. K., and Matz, M. V. (2012). 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat. Methods 9, 808–810. doi: 10.1038/nmeth.2023
Wang, S., Zhang, J., Jiao, W., Li, J., Xun, X., Sun, Y., et al. (2017). Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1:0120. doi: 10.1038/s41559-017-0120
Wang, Y., Xiong, G., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Ward, R. D., Thompson, P. A., Appleyard, S. A., Swan, A. A., and Kube, P. D. (2005). Sustainable Genetic Improvement of Pacific Oysters in Tasmania and South Australia. Canberra, ACT: Fisheries Research and Development Corporation final report.
Whitlock, M. C., and Bourguet, D. (2000). Factors affecting the genetic load in Drosophila: synergistic epistasis and correlations among fitness components. Evolution 54, 1654–1660. doi: 10.1111/j.0014-3820.2000.tb00709.x
Yáñez, J. M., Newman, S., and Houston, R. D. (2015). Genomics in aquaculture to better understand species biology and accelerate genetic progress. Front. Genet. 6:128. doi: 10.3389/fgene.2015.00128
Yu, Z., and Guo, X. (2006). Identification and mapping of disease-resistance QTLs in the eastern oyster, Crassostrea virginica Gmelin. Aquaculture 254, 160–170. doi: 10.1016/j.aquaculture.2005.10.016
Yue, G. H., and Wang, L. (2017). Current status of genome sequencing and its applications in aquaculture. Aquaculture 468, 337–347. doi: 10.1016/j.aquaculture.2016.10.036
Zhang, G., Fang, X., Guo, X., Li, L., Luo, R., Xu, F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zheng, H., Zhang, G., Liu, X., and Guo, X. (2006). Sustained response to selection in an introduced population of the hermaphroditic bay scallop Argopecten irradians irradians Lamarck (1819). Aquaculture 255, 579–585. doi: 10.1016/j.aquaculture.2005.11.037
Zheng, H., Zhang, G., Liu, X., Zhang, F., and Guo, X. (2004). Different responses to selection in two stocks of the bay scallop, Argopecten irradians irradians Lamarck (1819). J. Exp. Mar. Biol. Ecol. 313, 213–223. doi: 10.1016/j.jembe.2004.04.015
Zhong, X., Feng, D., Yu, H., Kong, L., and Li, Q. (2016). Genetic variation and breeding signature in mass selection lines of the pacific oyster (Crassostrea gigas) assessed by SNP markers. PLoS ONE 11:e0150868. doi: 10.1371/journal.pone.0150868
Zhong, X., Li, Q., Guo, X., Yu, H., and Kong, L. (2014). QTL mapping for glycogen content and shell pigmentation in the Pacific oyster Crassostrea gigas using microsatellites and SNPs. Aquacult. Internat. 22, 1877–1889. doi: 10.1007/s10499-014-9789-z
Keywords: aquaculture, genomics, SNP genotyping, heritability, marker assisted selection, genomic selection, molluscs, selective breeding
Citation: Hollenbeck CM and Johnston IA (2018) Genomic Tools and Selective Breeding in Molluscs. Front. Genet. 9:253. doi: 10.3389/fgene.2018.00253
Received: 27 November 2017; Accepted: 25 June 2018;
Published: 18 July 2018.
Edited by:
Nguyen Hong Nguyen, University of the Sunshine Coast, AustraliaReviewed by:
Yniv Palti, Cool and Cold Water Aquaculture Research (USDA-ARS), United StatesScott Newman, Genus (United Kingdom), United Kingdom
Pierre Boudry, Institut Français de Recherche pour l'Exploitation de la Mer (IFREMER), France
Copyright © 2018 Hollenbeck and Johnston. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher M. Hollenbeck, Y2gyNjNAc3QtYW5kcmV3cy5hYy51aw==
Ian A. Johnston, aWFuLmpvaG5zdG9uQHhlbGVjdC5jby51aw==
 Christopher M. Hollenbeck
Christopher M. Hollenbeck Ian A. Johnston
Ian A. Johnston