- Institute of Biosciences, University of São Paulo, São Paulo, Brazil
Spiders’ spinning skills have fascinated mankind since ancient times. These animals owe this ability to unique appendages named spinnerets. These structures are connected to silk-producing glands and are responsible for silk extrusion and manipulation, granting spiders the ability to use the threads for functions far beyond web-building. In spite of the relevance of spinnerets, there is scarce knowledge about their development and evolution. Most research on these structures focused on their morphological aspects, or possible applications of the silk for human purposes. In this mini review, we included this literature, but our main purpose is to introduce and discuss the preeminent hypotheses on the origins of spinnerets in light of an evo-devo perspective. We present the available information on genetic pathways involved in spinneret genesis during spider’s embryonic development, evidencing the eminent need for further research in the evolution and development of spinnerets.
Introduction
Fascination with spiders predates classical antiquity. In Greek mythology, there was no weaver comparable to Arachne. She had even dared to say her skills were greater than the goddess Athena’s herself. Offended, Athena challenged the mortal to a weaving competition, after which she destroyed Arachne’s work out of anger and jealousy. Athena also turned Arachne into a tiny animal, condemning her and all her descendants, for she had insulted the gods. Arachne was now a spider, and from that moment on, she would weave for eternity, but not with her once-dexterous hands.
Spiders (Arachnida: Araneae) may not be Arachne’s descendants, but that does not make them any less remarkable. These arthropods make up a highly diversified clade of more than 48,000 described species (World Spider Catalog, 2019). In spite of their notable diversity, spiders share some unique features, such as chelicerae connected to venom glands and modified tarsi in the pedipalps, which allow sperm transfer by males (Coddington and Levi, 1991; Wheeler et al., 2017; Wang et al., 2018). But of all their shared features, spiders are mainly known for their ability to produce and manipulate silk. The major synapomorphy of this clade is its spinning apparatus (Wheeler et al., 2017; Wang et al., 2018), formed by silk-producing glands that lead to appendages called spinnerets, present in the fourth and fifth opisthosomal (abdominal) segments (Pechmann et al., 2010).
Silk is a fibroin, produced by and stored in the silk glands that are like small sacs located inside the opisthosoma. An orb-weaving spider, such as Triconephila clavipes, may possess up to seven types of morphologically and physiologically differentiated silk glands—minor and major ampullate, aciniform, tubuliform, aggregate, piriform, and flagelliform (or coronate) glands. In each of them, distinct patterns of gene expression lead to the production of a different set of proteins (most of them belonging to the spidroin family) that together make up that gland’s silk (Rising and Johansson, 2015; Babb et al., 2017). Each gland leads to a specific spinneret, which opens to the environment in the form of microscopic spigots: terminal projections through which single threads of silk are extruded (Selden et al., 2008; Foelix, 2010). Males of some species possess the so-called epiandrous silk glands, located on the anterior margin of the genital furrow with spigots similar to aciniform glands (Shultz, 1987). They are involved in the production of sperm webs, and hypotheses of homology between them and the other silk glands have long been proposed (Marples, 1967; Shultz, 1987).
The spinning system is central to many ecological aspects of a spider’s life cycle (Shear, 1986; Foelix, 2010; Rising and Johansson, 2015). The threads they produce are strong, highly extensible, and yet lightweight (Rising and Johansson, 2015; Babb et al., 2017). The spigots and the spinnerets can work independently or together in a highly coordinated manner that allows spiders to combine multiple filaments with various arrangements in different ways to produce many kinds of silk for special purposes: build shelters; acquire food; communicate (through pheromones, and vibration); reproduce; disperse (silk balloons and lines); inter alia. Therefore, silk and silk spinning were probably decisive for the group’s success: They have allowed spiders to conquer the terrestrial environment and thrive in it (Rising and Johansson, 2015; Mortimer et al., 2019).
Spinneret Morphology and Diversity
Given their uniqueness and diversity, silk gland, spigot, and spinneret morphologies are largely used as characters for phylogenetic studies (e.g., see Platnick et al., 1991; Griswold et al., 2005; Ramírez, 2014; Magalhães et al., 2017). For instance, spinneret position is one of the most conspicuous features used to split the order Araneae in two suborders: Mesothelae, whose spinning apparatus lies in the middle of the opisthosoma; and Opisthothelae, whose spinnerets are at the posterior region of that tagma (Foelix, 2010; Wheeler et al., 2017). Posterior positioning of the spinnerets in Opisthothelae is due to an expansion of segment 3 of the opisthosoma (O3) and a reduction of the segments posterior to the segment O5. Therefore, although they are visually positioned in different regions, the spinnerets of both suborders are considered homologous structures (Pechmann et al., 2010).
Opisthothelae is further divided into two infraorders: Mygalomorphae and Araneomorphae. Mygalomorphae (tarantulas) constitute a little less than 7% of all Araneae species described (World Spider Catalog, 2019). They have a reduced number of spinnerets compared to Mesothelae, whose representatives bear four pairs (or at least vestiges of them)—what is thought to be the ancestral state for the order Araneae (Marples, 1967). Some Mygalomorph spiders have only one pair of anterior lateral spinnerets positioned in the O4 segment, and the vast majority of the group has posterior lateral and posterior middle spinnerets in segment O5 (Wheeler et al., 2017).
The infraorder Araneomorphae makes up approximately 90% of all described spiders (World Spider Catalog, 2019). They have two pairs of spinnerets in the segment O5, but there is a wide variation of number and morphology on segment O4. Some species bear a pair of spinnerets, but it may be fused as the cribellum or reduced, as the colulus, or completely lost in other species (Hilbrant, 2008; Pechmann et al., 2010; Figure 1).
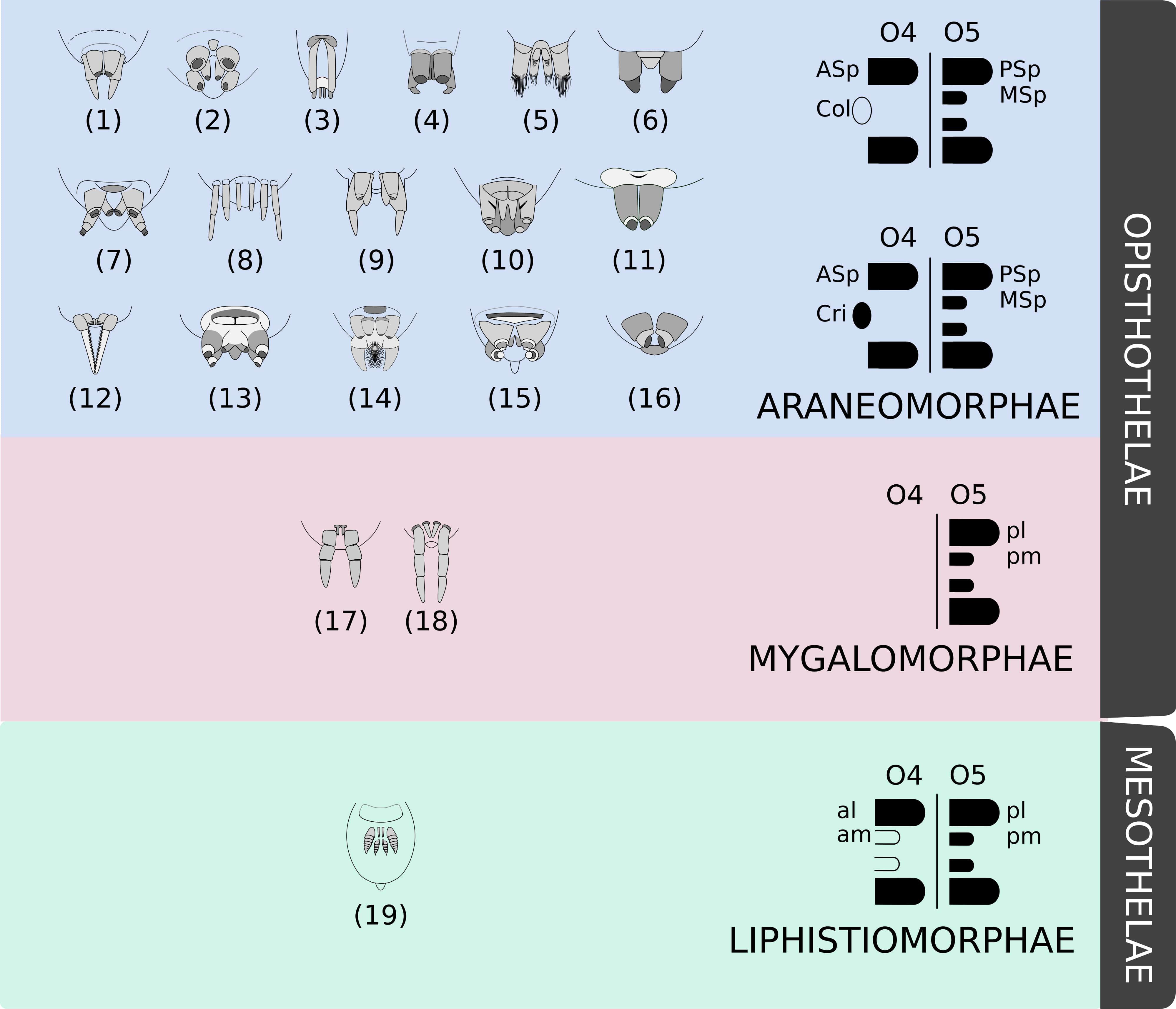
Figure 1. Examples of the diversity of spinneret morphology. Drawings adapted from Brescovit et al. (2002) and Joqué and Dippeaar-Schoeman (2007) showing examples of morphologies of some spider species belonging to different families: (1) Miturgidae, (2) Corinnidae, (3) Gnaphosidae, (4) Clubionidae, (5) Prodidomidae, (6) Senoculidae, (7) Ctenidae, (8) Hahniidae, (9) Agielenidae, (10) Amaurobiidae, (11) Zodariidae, (12) Hersiliidae, (13) Titanoedidae, (14) Oecobidae, (15) Uloboridae, (16) Palpimanidae, (17) Paratropididae, (18) Nemesiidae, and (19) Liphistiidae. These examples are not representative of the familial diversity of Araneae. Mesothelae spiders (green) possess four pairs of spinnerets (at least vestiges) in the middle of the abdomen, whilst Opisthotelae (pink and blue) possess them in the end of the opisthosoma. Mygalomorphae (pink) have a reduced number of spinnerets and Araneomorphae (blue) present a remarkable diversity of organizations (as the colulus or the cribellum). Schematics were based on Hilbrant (2008). Abbreviations: al, anterior lateral spinneret; am, anterior median spinneret; ASp, anterior spinneret; Col, colulus; Cri, cribellum; MSp, median spinneret; pl, posterior lateral spinneret; pm, posterior median spinneret; PSp, posterior spinneret.
Spinneret Origins
The origin of silk production precedes the evolution of spinnerets (Shultz, 1987). This is supported by fossils described by Selden et al. (2008) that bear spigot-like structures and were found with silk threads, but had no signs of spinneret-like appendages. They were then classified in a newly proposed order named Uraraneida, and the authors uphold the vision of spinnerets as spider-exclusive features. Huang et al. (2018) disagree with this exclusivity hypothesis, arguing that another spinneret-bearing fossil (described as Chimerarachne yingi by Wang et al., 2018) may be classified as some sort of protospider not belonging to Araneae.
Whether specimens like this are to be included in the order Araneae or not is open to discussion; but, no matter the conclusion, spiders remain the only extant lineage that possesses spinnerets. These appendages must then go back at least to the last common ancestor of Araneae (Selden et al., 2008; Garwood et al., 2016; Wang et al., 2018), which lived prior to or during the Carboniferous period, given the oldest spider fossil is approximately 305 million years old (Selden et al., 2014).
Though we may infer when spiders and their spinnerets arose, there is still no consensus on their morphological origins. It is difficult to define which structures found in other arthropods would be homologous to spinnerets or if there is even homology to any other known feature. Some hypotheses have been proposed, five of which will be detailed. Back in the 19th century, Jaworowski (1896 apud Hilbrant, 2008) had proposed that spinnerets would be locomotor appendages, similar to the prosoma legs, which lost their primary function. As a matter of fact, the developmental study by Pechmann and Prpic (2009) has demonstrated that the posterior spinneret in Acanthoscurria geniculata presents a leg-like gene expression profile—what was interpreted as evidence of serial homology between spinnerets and legs. Moreover, this hypothesis is further supported by the fact that spinnerets are derived from embryonic limb buds similar to the ones from which legs are formed (Hilbrant, 2008).
Shultz (1987) commented on the similarities between spinnerets and legs, but presented a series of arguments that link the origins of the spinning apparatus to reproductive needs. Based on analyses within the Araneae order and on comparisons between these animals and other arachnids and non-chelicerate arthropods that also utilize silk, he argued that the silk-spinning system was primarily related to the protection of genital products, such as eggs or spermatophores.
Marples (1967), on the other hand, argued that spinnerets could be modified ventral appendages that became specialized as four pairs of spinnerets in the ancestor of all spiders. This hypothesis was based on the observation that gland-associated appendages are commonly found in arthropods and their relatives. The median spinnerets would be the first to evolve by modification from the original glands. The lateral spinnerets would have then originated from a paedomorphosis process that resulted in the retention of the limb-bud of the associated ancestral appendage.
Damen et al. (2002) put forward another hypothesis. They hypothesized the existence of a structure in chelicerates, which would have a shared gene expression pattern to other arthropod features. This was based on previous work by Averof and Cohen (1997) that had demonstrated that the pdm/nubbin and apterous genes were similarly expressed in both crustacean epipods (gills) and insect wings—a result considered to be an evidence of homology. Damen et al. (2002) found out that those genes were expressed in the book gills of horseshoe crab (Limulus polyphemus) embryos. And in spider Cuppienius salei embryos, they showed that both genes are expressed specifically in the book lungs, tubular tracheae and spinnerets primordia. Such findings led them to propose a possible homology of the spinning appendages with the wings and gills of their insect and crustacean counterparts, respectively.
Whilst Sharma (2017) has argued that there is not enough evidence to infer such deep homology, Selden et al. (2008) has followed these assumptions and considered spinnerets to be homologous to the horseshoe crab gills. The latter claim no other arachnid order bears a structure to which homology hypotheses may be traced and propose a possible pathway by which ventral appendages of the opisthosoma might have reappeared. Based on fossil evidence, they have suggested that spinnerets may be the result of gene expression reactivation in ventral plates of a uraraneid-like spider ancestor. Such plates found in uraraneid Attercopus fimbriungis are thought to be appendage-derived (as are the plates observed in modern scorpions) and would give rise to a segmented appendage if genes such as Distal-less (Dll) had their expression reinitiated. When this process happened, the spigots present on the plates would have been carried and a spinneret would be formed.
Clarke et al. (2015) has taken this hypothesis further. They have found that a spider-specific gene duplication has occurred, and that gene families with genes that are expressed only in silk glands are more likely to retain the paralog sequences that emerged from this event. They then discuss the possibility that this duplication event, followed by paralog retention, may have created a genetic background that allowed, or facilitated the reactivation as proposed by Selden et al. (2008).
Genes and Development
Genetic and developmental research in an evo-devo framework may be the keys to shedding light on the origins and evolution of spinnerets (Selden et al., 2008). Scarce information on genetic pathways involved in spinneret embryonic development is available hitherto, although the participation of a few genes in this process have been studied, but their function is not always known, as most results are limited to descriptions of expression patterns (Pechmann et al., 2010).
The role of Notch signaling genes in appendage segmentation seems to be conserved in all arthropods (Prpic and Damen, 2009). However, these genes have also been shown to play a role in the caudal lobe establishment and opisthosoma patterning in Parasteatoda tepidariorum. Interference RNA experiments resulted in a non-functional caudal lobe when Delta, Notch, and Suppressor of Hairless orthologs were knocked down (Oda et al., 2007).
Specifically regarding spinnerets, Damen et al. (2002) provided evidence of pdm/nubbin and apterous expression in their primordia. Pechmann and Prpic (2009) used in situ hybridization to show that Dll, dachshund, extradenticle, and homothorax are also expressed in those appendages during spider ontogeny. Their main aim was to discover whether those genes, previously shown to participate in appendage development in araneomorph spiders (Prpic et al., 2003), were also expressed in mygalomorphs. An impressive observation is that Dll expression is a marker of which spinneret limb buds are to be retained and become fully formed spinnerets in adult spiders. In A. geniculata embryos, pairs of spinneret primordia are observed in both O4 and O5 segments, but only the latter expresses Dll during embryonic development and grow into spinnerets. On the other hand, in C. salei and P. tepidariorum, both pairs express Dll and later became functional appendages (Pechmann and Prpic, 2009).
Hilbrant (2008) analyzed morphological and genetic aspects of C. salei embryonic development. He found evidence of gene expression of homologs of wingless, omb, H15, pairberry, and activator protein-2 (AP-2). A highlight is made for AP-2, a transcription factor that is initially expressed in the spinneret limb buds in both O4 and O5, but vanishes from O4 in later stages. The limb bud in segment O5 then divides into two portions to give rise to both median and posterior spinnerets, while the limb in segment O4 generates only the anterior spinnerets, which are shorter. AP-2 is thought to be involved in cell proliferation, but Hilbrant (2008) claims that this process was not increased in segment O5 limb bud. Therefore, distinct gene expression patterns may underlie the morphological differences observed in adult spiders, but by means other than cell proliferation.
Finally, Hox genes Abdominal-A and Ultrabithorax are also expressed in the segments from which spinnerets emerge (Abzhanov et al., 1999; Hughes and Kaufman, 2002); however, they have been shown to foreclose appendage development in the abdomen of insects by repressing Dll expression (Vachon et al., 1992; Abzhanov et al., 1999). Pechmann et al. (2010) proposed that there might be a mechanism, maybe exclusive to spiders, by which those genes may be co-expressed.
Schwager et al. (2017) have shown that almost all genes in the Hox cluster are duplicated in the P. tepidariorum genome. This must be true for other spiders as well, given that they present evidences that this duplication event probably preceded the divergence of spiders and scorpions, and involved the entire genome of this hypothetical ancestor. In the same study, the authors also showed that the paralogs of each duplicated Hox gene have expression profiles that differs spatially and temporally, what has been interpreted as evidence of neo-functionalization.
Conclusion and Future Directions
The arachnologist Michael R. Gray had said in 1978 that “the evolution of spider silk has been an event comparable in importance to the evolution of flight in the insects, or warmbloodedness in the vertebrates” (apud Foelix, 2010). Yet, the research on the origins and evolution of spinnerets is quite sparse, and knowledge gaps are largely open.
Hypotheses on the origins that have been tested with genetic data are interesting, and they represent the growing importance of developmental data in understanding the evolution of complex morphological characters in an evo-devo approach, but they must be further explored. Additional genetic and developmental research is vital to find a more comprehensive and explanatory theory for spinneret evolution. In particular, research on whether spinnerets are homologous to either wings and gills, legs, or even other features should be pursued.
To do so, groundwork must be built, and we are already running behind. In the NCBI database, searched on October 29, 2019, there were only 9 spider genomes, which represents less than 0.02% of the described Araneae species. It is crucial that we gather genomic information to assemble a great dataset to compare spiders to other arachnids (that are not well represented in NCBI as well, totalizing 25 genomes, excluding spiders), and arthropods as a whole, to look for Araneae-specific genes that might be related to spinnerets.
There are also other possibilities for the origins of spinnerets other than Araneae-specific genes. It is known that genetic pathways may be co-opted as their participant genes acquire new functions (Brakefield, 2006), and a process alike has been described for spiders (Setton and Sharma, 2018). This could result in a new morphological feature, as has been suggested to have happened to firefly lanterns Stansbury and Moczek (2014) and Schwager et al. (2017) have already discussed this possibility for spider Hox genes. The latter, however, state that functional studies must be done to further test these ideas.
Regarding this proposal, if genomic information is associated to transcriptomics, it is possible to compare between spiders and other animals not only the expression patterns of candidate genes, as it has been done (e.g., Damen et al., 2002), but also their expression levels during development. This framework must provide more information on the processes that may have underlain the emergence of spinnerets. This data would allow us to go beyond spinneret origins and understand how they evolved. One may then infer that spinnerets are highly plastic structures that changed substantially throughout millions of years of evolution, sometimes convergently, as has been shown by molecular phylogenies (Wheeler et al., 2017; Fernández et al., 2018). That opens questions such as: Is this diversification a result of genetic drift or natural selection? Is there a conserved genetic developmental pathway underlying distinct spinneret ontogeny? Or are there distinct pathways correlating with different morphologies?
Describing the dynamics of the genes during spider development, and the impact of variations that have accumulated during the course of evolution in spinneret morphology and function are key to go forward in understanding their history. Given the ecological importance of these appendages and the many unanswered questions about their development, function, and evolution, any discovery has the potential to transform our understanding of the conquest of the terrestrial environment by arthropods.
Author Contributions
All authors searched and reviewed the literature, provided critical feedback and helped shape the manuscript. PM-M took the lead in writing the manuscript with input from NL-M-H and TT.
Funding
This work was supported by Conselho Nacional de Pesquisa e Desenvolvimento Tecnológico—CNPq (NL#142192/2017-1) and Fundação de Amparo à Pesquisa do Estado de São Paulo—FAPESP (PM#2018/06719-0, NL #2017/19616-2, and #2019/12282-7). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001, and the Institute of Biosciences, USP, through the first Interdepartmental Grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank V. A. S. Cunha, B. Genevcius, R. D. Monfardini, and M. G. Honda, members of our team, and Dr. Prashant Sharma, from the University of Wisconsin-Madison, for their comments and suggestions. We would also like to thank the two reviewers for their helpful comments on earlier versions of the manuscript.
References
Abzhanov, A., Popadic, A., and Kaufman, T. C. (1999). Chelicerate Hox genes and the homology of arthropod segments. Evol. Dev. 1, 77–89. doi: 10.1046/j.1525-142x.1999.99014.x
Averof, M., and Cohen, S. M. (1997). Evolutionary origin of insect wings from ancestral gills. Nature 385:627. doi: 10.1038/385627a0
Babb, P. L., Lahens, N. F., Correa-Garhwal, S. M., Nicholson, D. N., Kim, E. J., Hogenesch, J. B., et al. (2017). The Nephila clavipes genome highlights the diversity of spider silk genes and their complex expression. Nat. Genet. 49:895. doi: 10.1038/ng.3852
Brakefield, P. M. (2006). Evo-devo and constraints on selection. Trends Ecol. Evol. 21, 362–368. doi: 10.1016/j.tree.2006.05.001
Brescovit, A. D., Bonaldo, A. B., Bertani, R., and Rheims, C. A. (2002). Araneae. Amazonian Arachnida and Myriapoda. Sofia: Pensoft, 303–344.
Clarke, T. H., Garb, J. E., Hayashi, C. Y., Arensburger, P., and Ayoub, N. A. (2015). Spider transcriptomes identify ancient large-scale gene duplication event potentially important in silk gland evolution. Genome Biol. Evol. 7, 1856–1870. doi: 10.1093/gbe/evv110
Coddington, J. A., and Levi, H. W. (1991). Systematics and evolution of spiders (Araneae). Annu. Rev. Ecol. Syst. 22, 565–592. doi: 10.1146/annurev.es.22.110191.003025
Damen, W. G., Saridaki, T., and Averof, M. (2002). Diverse adaptations of an ancestral gill: a common evolutionary origin for wings, breathing organs, and spinnerets. Curr. Biol. 12, 1711–1716. doi: 10.1016/S0960-9822(02)01126-0
Fernández, R., Kallal, R. J., Dimitrov, D., Ballesteros, J. A., Arnedo, M. A., Giribet, G., et al. (2018). Phylogenomics, diversification dynamics, and comparative transcriptomics across the spider tree of life. Curr. Biol. 28, 1489–1497. doi: 10.1016/j.cub.2018.03.064
Garwood, R. J., Dunlop, J. A., Selden, P. A., Spencer, A. R. T., Atwood, R. C., Vo, N. T., et al. (2016). Almost a spider: a 305-million-year-old fossil arachnid and spider origins. Proc. R. Soc. B 283:20160125. doi: 10.1098/rspb.2016.0125
Griswold, C. E., Ramirez, M. J., Coddington, J. A., and Platnick, N. I. (2005). Atlas of phylogenetic data for entelegyne spiders (Araneae: Araneomorphae: Entelegynae) with comments on their phylogeny. Proc. Calif. Acad. Sci. 56, 1–324.
Hilbrant, M. (2008). Development and Evolution of the Spider Silk Production System. Dissertation, University of Cologne, Köln.
Huang, D., Hormiga, G., Cai, C., Su, Y., Yin, Z., Xia, F., et al. (2018). Origin of spiders and their spinning organs illuminated by mid-Cretaceous amber fossils. Nat. Ecol. Evol. 2:623. doi: 10.1038/s41559-018-0475-9
Hughes, C. L., and Kaufman, T. C. (2002). Hox genes and the evolution of the arthropod body plan1. Evol. Dev. 4, 459–499. doi: 10.1046/j.1525-142X.2002.02034.x
Jaworowski, A. (1896). Die Entwicklung des Spinnapparates bei Trochosa singoriensis, etc. Jena. Zeischr. f. Naturg. 30, 39–74.
Joqué, R., and Dippeaar-Schoeman, A. S. (2007). Spider Families of the World. Second Edition. Tervuren: Peeters nv.
Magalhães, I. L., Brescovit, A. D., and Santos, A. J. (2017). Phylogeny of Sicariidae spiders (Araneae: Haplogynae), with a monograph on Neotropical sicarius. Zool. J. Linn. Soc. 179, 767–864.
Marples, B. J. (1967). The spinnerets and epiandrous glands of spiders. Zool. J. Linn. Soc. 46, 209–222. doi: 10.1111/j.1096-3642.1967.tb00504.x
Mortimer, B., Soler, A., Wilkins, L., and Vollrath, F. (2019). Decoding the locational information in the orb web vibrations of Araneus diadematus and Zygiella x-notata. J. R. Soc. Interface 16:20190201. doi: 10.1098/rsif.2019.0201
Oda, H., Nishimura, O., Hirao, Y., Tarui, H., Agata, K., and Akiyama-Oda, Y. (2007). Progressive activation of Delta-Notch signaling from around the blastopore is required to set up a functional caudal lobe in the spider Achaearanea tepidariorum. Development 134, 2195–2205. doi: 10.1242/dev.004598
Pechmann, M., Khadjeh, S., Sprenger, F., and Prpic, N. M. (2010). Patterning mechanisms and morphological diversity of spider appendages and their importance for spider evolution. Arthropod. Struct. Dev. 39, 453–467. doi: 10.1016/j.asd.2010.07.007
Pechmann, M., and Prpic, N. M. (2009). Appendage patterning in the South American bird spider Acanthoscurria geniculata (Araneae: Mygalomorphae). Dev. Genes Evol. 219, 189–198. doi: 10.1007/s00427-009-0279-7
Platnick, N. I., Coddington, J. A., Forster, R. R., and Griswold, C. E. (1991). Spinneret morphology and the phylogeny of haplogyne spiders (Araneae, Araneomorphae). Am. Museum Novitates 3016, 1–76.
Prpic, N. M., and Damen, W. G. (2009). Notch-mediated segmentation of the appendages is a molecular phylotypic trait of the arthropods. Dev. Biol. 326, 262–271. doi: 10.1016/j.ydbio.2008.10.049
Prpic, N. M., Janssen, R., Wigand, B., Klingler, M., and Damen, W. G. (2003). Gene expression in spider appendages reveals reversal of exd/hth spatial specificity, altered leg gap gene dynamics, and suggests divergent distal morphogen signaling. Dev. Biol. 264, 119–140. doi: 10.1016/j.ydbio.2003.08.002
Ramírez, M. J. (2014). The morphology and phylogeny of dionychan spiders (Araneae: Araneomorphae). Bull. Am. Museum Nat. History 2014, 1–375. doi: 10.1206/821.1
Rising, A., and Johansson, J. (2015). Toward spinning artificial spider silk. Nat. Chem. Biol. 11:309. doi: 10.1038/nchembio.1789
Schwager, E. E., Sharma, P. P., Clarke, T., Leite, D. J., Wierschin, T., Pechmann, M., et al. (2017). The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 15:62. doi: 10.1186/s12915-017-0399-x
Selden, P. A., Shcherbakov, D. E., Dunlop, J. A., and Eskov, K. Y. (2014). Arachnids from the Carboniferous of Russia and Ukraine, and the permian of kazakhstan. Paläontol. Z. 88, 297–307. doi: 10.1007/s12542-013-0198-9
Selden, P. A., Shear, W. A., and Sutton, M. D. (2008). Fossil evidence for the origin of spider spinnerets, and a proposed arachnid order. Proc. Natl. Acad Sci. U.S.A. 105, 20781–20785. doi: 10.1073/pnas.0809174106
Setton, E. V., and Sharma, P. P. (2018). Cooption of an appendage-patterning gene cassette in the head segmentation of arachnids. Proc. Natl. Acad Sci. U.S.A. 115, E3491–E3500. doi: 10.1073/pnas.1720193115
Sharma, P. P. (2017). Chelicerates and the conquest of land: a view of arachnid origins through an evo-devo spyglass. Integr. Comp. Biol. 57, 510–522. doi: 10.1093/icb/icx078
Shultz, J. W. (1987). The origin of the spinning apparatus in spiders. Biol. Rev. 62, 89–113. doi: 10.1111/j.1469-185X.1987.tb01263.x
Stansbury, M. S., and Moczek, A. P. (2014). The function of Hox and appendage-patterning genes in the development of an evolutionary novelty, the Photuris firefly lantern. Proc. R. Soc. B Biol. Sci. 281:20133333. doi: 10.1098/rspb.2013.3333
Vachon, G., Cohen, B., Pfeifle, C., McGuffin, M. E., Botas, J., and Cohen, S. M. (1992). Homeotic genes of the Bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell 71, 437–450. doi: 10.1016/0092-8674(92)90513-C
Wang, B., Dunlop, J. A., Selden, P. A., Garwood, R. J., Shear, W. A., Müller, P., et al. (2018). Cretaceous arachnid Chimerarachne yingi gen. et sp. nov. illuminates spider origins. Nat. Ecol. Evol. 2:614. doi: 10.1038/s41559-017-0449-3
Wheeler, W. C., Coddington, J. A., Crowley, L. M., Dimitrov, D., Goloboff, P. A., Griswold, C. E., et al. (2017). The spider tree of life: phylogeny of Araneae based on target-gene analyses from an extensive taxon sampling. Cladistics 33, 574–616. doi: 10.1111/cla.12182
Keywords: Araneae, evo-devo, gene expression, homology, morphological novelty, silk, spigots
Citation: Mariano-Martins P, Lo-Man-Hung N and Torres TT (2020) Evolution of Spiders and Silk Spinning: Mini Review of the Morphology, Evolution, and Development of Spiders’ Spinnerets. Front. Ecol. Evol. 8:109. doi: 10.3389/fevo.2020.00109
Received: 17 December 2019; Accepted: 03 April 2020;
Published: 22 May 2020.
Edited by:
Alessandro Minelli, University of Padua, ItalyReviewed by:
Peter Michalik, University of Greifswald, GermanyGustavo Hormiga, The George Washington University, United States
Copyright © 2020 Mariano-Martins, Lo-Man-Hung and Torres. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pedro Mariano-Martins, cG1hcmlhbm84M0BnbWFpbC5jb20=; Tatiana Teixeira Torres, dHR0b3JyZXNAaWIudXNwLmJy
 Pedro Mariano-Martins
Pedro Mariano-Martins Nancy Lo-Man-Hung
Nancy Lo-Man-Hung Tatiana Teixeira Torres
Tatiana Teixeira Torres