- 1Department of Ecological Modelling, Helmholtz Centre for Environmental Research – UFZ, Leipzig, Germany
- 2Department of Environmental Microbiology, Helmholtz Centre for Environmental Research – UFZ, Leipzig, Germany
- 3Institute of Biology, Leipzig University, Leipzig, Germany
- 4German Centre for Integrative Biodiversity Research (iDiv), Halle-Jena-Leipzig, Leipzig, Germany
Biodiversity is a key driver of ecosystem functioning, while disturbances are a key driver of biodiversity. Consequently, disturbances crucially influence ecosystem functioning–both directly via affecting ecosystem processes but also indirectly via altering biodiversity. We thus need to disclose the joint relationships between disturbances, biodiversity and functioning (DBF) to understand and predict ecosystem dynamics under realistic conditions. However, biodiversity responses to disturbances have so far insufficiently been studied together with biodiversity effects on functions. For many ecosystems, such integrative exploration of DBF relationships would require too extensive manipulations and observations over unfeasible spatial and temporal scales. We argue that microbial systems offer a bright perspective to overcome these limitations, and present a roadmap for doing so. Microbial systems allow us exposing different, well-characterized communities to multiple, reproducible disturbance regimes, and precisely measuring both biodiversity and associated functions over time. Comprehensive data can be obtained by systematically varying and replicating representative environmental scenarios. These data can further be explored and explained with computational models. Microbial systems thus reveal mechanisms that underlie DBF relationships and allow scrutinizing ecological hypotheses. This advancement of theory will be essential for ecology as a whole. It is particularly relevant in the context of global change, which is expected to promote disturbances as well as loss of biodiversity and functions in many ecosystems.
Disturbances, Biodiversity and Ecosystem Functioning are Inherently Related
Relationships between biodiversity and ecosystem functioning (BEF) are prominently studied in ecology. As biodiversity declines across various ecosystems (Pereira et al., 2010; Bellard et al., 2012), subsequent effects on functioning strongly matter for the future provision of ecosystem services, and human well-being (Hooper et al., 2005; Cardinale et al., 2012). Positive BEF relationships (e.g., Figure 1, right) arise from selection (diverse communities more likely contain and are dominated by functionally important species) and complementarity effects (multiple species occupy different niches or facilitate each other; Loreau et al., 2001).
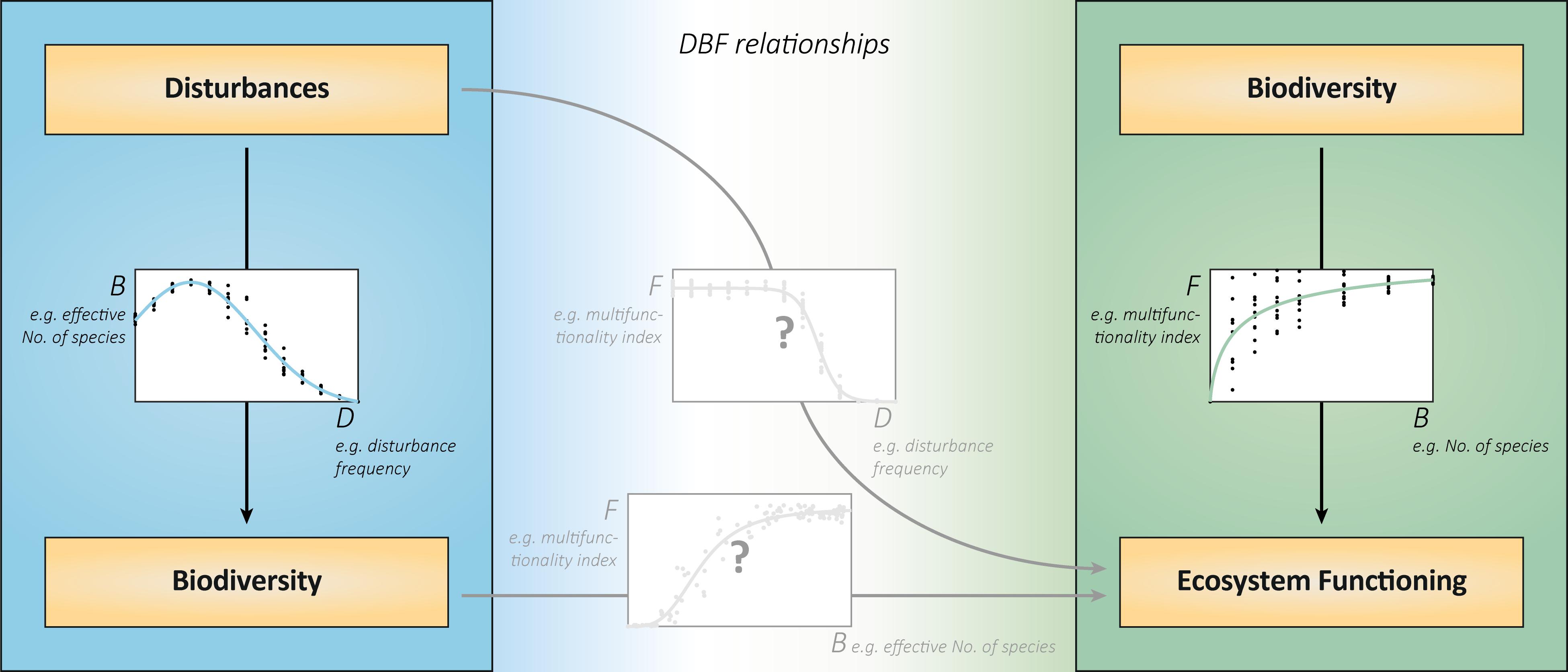
Figure 1. DBF relationships integrate disturbances, biodiversity, and ecosystem functioning. The relationships between disturbances and biodiversity (left) and between biodiversity and ecosystem functions (right) have so far mostly been studied in isolation. The joint analysis will reveal the consequences of disturbances for ecosystem functioning (middle graph). In explaining these consequences, the relationship between realized biodiversity under disturbances and associated functioning (bottom graph) is decisive. This relationship does not show the effects of biodiversity under otherwise equal conditions, but the specific associations of realized biodiversity and ecosystem functioning (BEF) in various disturbance contexts. It is likely to deviate from the relationship between biodiversity of different, randomly assembled communities, and their functioning (right graph). This is because the disturbance regimes will create different, yet realistic, subsets of theoretically possible community compositions than the random artificial assembly. In addition, the associated functioning includes direct effects of disturbances that are not mediated by biodiversity changes. All graphs show virtual data that are consistent with each other. The chosen exemplary metrics of disturbances, biodiversity and ecosystem functioning can be replaced by alternatives. The biodiversity metric “effective number of species” comprises the components richness and evenness and is calculated from species abundances (Tuomisto, 2012).
Although BEF research focused heavily on species richness, awareness increased that additional biodiversity measures, such as evenness or trait diversity, are needed to explain what in a community composition is or is not relevant for functioning (Díaz et al., 2007; Tilman et al., 2014; Gagic et al., 2015). Furthermore, species roles, interactions and, thus, also BEF relationships are often not fixed, but rather depend on the (current) environmental context of an ecosystem (Wellnitz and Poff, 2001; Fetzer et al., 2015). They also vary with the particular function or set of functions examined (Naeem et al., 2012; Schuldt et al., 2018). Therefore, studying multiple functions to assess ecosystem multifunctionality (Byrnes et al., 2014) is needed for a comprehensive analysis of BEF relationships.
In most experimental BEF studies, initial communities are randomly assembled and their biodiversity related to observed functions (e.g., Figure 1, right). However, changes of initial species compositions and abundances in the course of biodiversity experiments are extremely likely. They are typically not random but reflect the community dynamics in the given environmental context (Srivastava and Vellend, 2005). This means that realized community compositions over the course of an experiment, including their temporal changes (Hillebrand et al., 2018), should be analyzed for insights on the actual role of biodiversity for ecosystem functioning, particularly on complementarity effects (Huston, 1997; Rychtecká et al., 2014).
A largely separate field of ecology is the study of disturbances–discrete events that affect populations, resources or the physical environment (Pickett and White, 1985). As most ecosystems are subject to disturbances, their impacts on populations and biodiversity were investigated for a long time (Connell, 1978; Wootton, 1998; White and Jentsch, 2001). The particular form of the relationship between disturbances and biodiversity (e.g., Figure 1, left) varies and is highly debated (Mackey and Currie, 2001; Kershaw and Mallik, 2013). Nonetheless, the considerable influence of disturbances on biodiversity is generally established.
Global change is anticipated to alter, and often increase, the frequency and intensity of disturbances (Miller et al., 2011; Seidl et al., 2011; Newman, 2019). Additionally, combined occurrence of more than one type of disturbances will become more common (Elmqvist et al., 2003; Martínez-Ramos et al., 2016). Hence, ecosystems are subject to combinations and interactions of multiple disturbances with potentially very different characteristics. Such combined disturbances are much less studied than single disturbances and their effects, including “ecological surprises” such as drastic shifts in community compositions and biodiversity, are difficult to predict (Paine et al., 1998; Newman, 2019).
It follows that (multiple) disturbances may affect (multiple) ecosystem functions directly and indirectly. Direct effects are, for instance, the removal of biomass, the reduction of available resources or the suppression of growth and abiotic compound turnover. Indirect effects arise from disturbances driving community compositions and diversity with subsequent effects on functioning. The relationships of disturbances and of realized biodiversity to ecosystem functioning (e.g., Figure 1, center) result from these direct and indirect influences (Loreau, 2010). However, studies addressing the effects of disturbances on both biodiversity and functioning are underrepresented in the current ecological research (Brose and Hillebrand, 2016). This deficiency is even more evident when multiple disturbances and multiple ecosystem functions are taken into account (Naeem et al., 2012; Villnäs et al., 2013). For these reasons, a deeper and more general understanding of DBF relationships requires examining multiple disturbance regimes, resulting community compositions and associated ecosystem functions together.
Additionally, trait-based analyses (McGill et al., 2006; Violle et al., 2007) may elucidate DBF relationships. The composition of communities is strongly influenced by the species traits related to competition, facilitation, and other interactions in a given environment (Tilman, 1982; Suding et al., 2008; Kunstler et al., 2016). Moreover, the effects of disturbances on community composition depend on species response traits, whereas the resulting effects on functioning depend on effect traits (Lavorel and Garnier, 2002; Norberg, 2004). Whether these traits belong to the same or to different species can then explain particular DBF relationships (Larsen et al., 2005; Radchuk et al., 2016). Ultimately, characterizing the patterns and determining the causes of different trait associations within species (e.g., trade-offs due to physiological or energetic constraints; Stearns, 1989) and trait distributions within communities (Garnier et al., 2015) is necessary to generalize the trait-based mechanisms underlying DBF relationships.
Why has ecology not yet comprehensively studied and disentangled DBF relationships more often? For many ecosystems, it is very difficult to manipulate disturbances in a controlled manner without affecting other environmental conditions, let alone combined multiple disturbances. Ample studies of different disturbance regimes, including undisturbed controls, and their long-term impact on biodiversity and (multiple) associated ecosystem functions are, therefore, hardly possible. An additional challenge is to apply the same suite of disturbance regimes to different initial communities and under different environmental contexts. This would be required to investigate how common global change scenarios will impact DBF relationships. Finally, even when such comprehensive manipulations and observations are in principle possible, for example in controlled grassland experiments, their practical realization is extremely demanding due to very high costs and requirements regarding spatial and temporal resolutions and extents. These limitations currently hinder our mechanistic understanding of DBF relationships, adequate theory development (Rillig et al., 2015) and the ability to predict biodiversity and functioning of disturbed ecosystems under global change. Certain recent developments have the potential to alleviate some of these limitations, such as concerted standardized biodiversity experiments across different environmental contexts worldwide (Fraser et al., 2013; Borer et al., 2014) or global inventories of species traits (Jones et al., 2009; Kattge et al., 2011). Another promising perspective for overcoming the limitations is the extensive study of microbial systems.
Microbial Systems Enable Comprehensive and Mechanistic Investigations of DBF Relationships
The general potential of microbial systems for testing and improving ecological theory has been recognized (Jessup et al., 2004; O’Malley et al., 2015; Ketola and Kristensen, 2017; Shade et al., 2018). We argue that they also offer a great opportunity for elucidating DBF relationships and their underlying ecological mechanisms. Exploiting this opportunity is, however, not trivial. It requires reducing the complexity of natural microbial systems in which huge numbers of organisms interact with each other and their (often highly heterogeneous) environment over many spatial and temporal scales. Experimental approaches need to focus on a manageable range of these scales and allow for high reproducibility as well as control, targeted manipulation and measurement of as many relevant factors as possible. Adjustable factors include, for example, the selection and initial composition of microbes studied, the environmental context in which these microbes are kept, and in particular the disturbance regimes applied (Figure 2). Factors to be measured are the community structure and functions, but also traits of the microbes used, including trait distributions, trade-offs, responses to disturbance, and effects on functions. We present a roadmap to comprehensively investigate DBF relationships using microbial systems, which shall facilitate clear interpretation of the results and detection of ecological mechanisms.
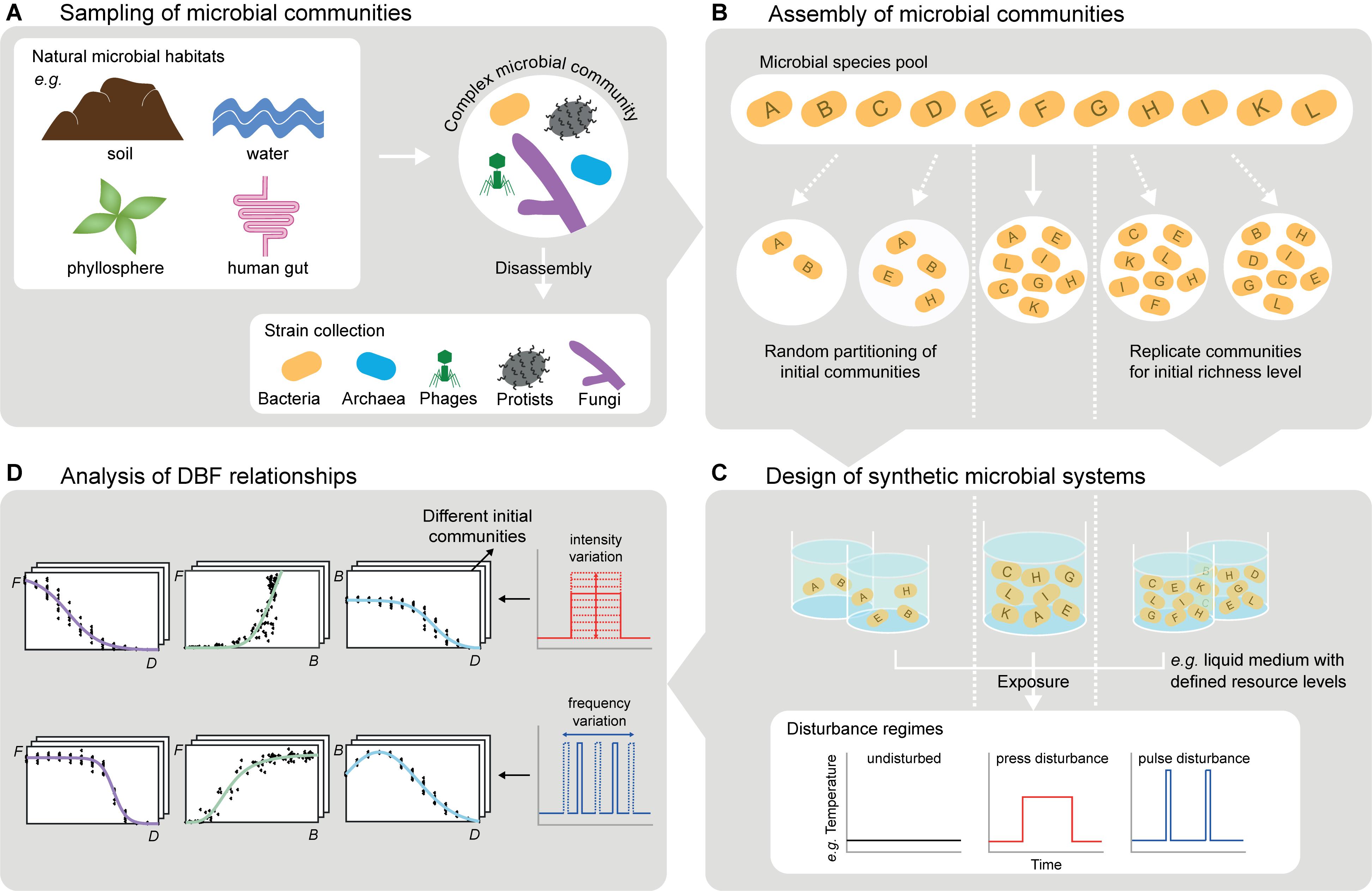
Figure 2. Experimental considerations for studying DBF relationships with microbial systems. (A) Complex microbial communities obtained from environmental samples can be used directly or further be disassembled leading to comprehensive, well-characterized culture collections (cf. section “Choice and Assembly of Microbial Communities”). (B) Such collections enable targeted assembly of initial communities differing e.g., in species richness, evenness, or trait composition. Random partitioning of the species pool allows for varying biodiversity and for replicates with equal biodiversity level but different species (cf. section “Choice and Assembly of Microbial Communities”). (C) Incubated in synthetic microbial ecosystems (e.g., liquid cultures, but also more complex spatially structured systems), the communities can be exposed to disturbance regimes varying in type, frequency and intensity of disturbances, including disturbance combinations (cf. section “Application of Disturbance Regimes”). (D) Monitoring community compositions and (multiple) functions at several points in time reveals DBF relationships. This includes systematic variation of disturbance characteristics (e.g., frequency or intensity) and initial communities (cf. section “Observation of biodiversity and ecosystem functioning”). The experimental observations are a formidable starting point for computational modeling (cf. section “Computational Analysis and Modeling of DBF Relationships”).
Choice and Assembly of Microbial Communities
One approach to generate microbial communities for controlled experiments is direct sampling from environmental microbiomes, which reduces the risk of missing key species. We use the term “species” throughout this section, but the same applies to phylotypes or operational taxonomic units, which are common alternatives when delineating microbial species is impossible. However, environmental sampling does not allow for precise composition and straightforward variation of species richness, evenness, or any compositional demand on traits. Therefore, an alternative approach is targeted assembly of synthetic communities from comprehensive and well-characterized culture collections (Bell et al., 2009a; Wittebolle et al., 2009). To obtain these “species pools” (Figure 2B), complex sampled communities need to be disassembled (Figure 2A), for example by species isolation on selective media, dilution series or flow cytometry-assisted cell sorting (Bai et al., 2015). The species’ traits can then be characterized by cultivation-based or molecular techniques, and classified into interaction, response and effect traits (Haddad et al., 2008; Suding et al., 2008). To select relevant traits from the wealth of microbial morphological, physiological or genetic attributes, the suggestions by Krause et al. (2014) on subsets important for competition (e.g., growth rate, yield, and production of antibiotics) and disturbance responses (e.g., cell and genome size, motility, and reserve materials) provide a basis. Moreover, effect traits depend on the function(s) of interest (e.g., contaminant degradation rate, metabolic and enzymatic potential). The knowledge on species’ traits can guide systematic composition of synthetic communities, differing in richness, evenness or variation of trait expressions, species, or higher taxonomic groups. This is impossible for communities sampled from the environment. However, natural community profiles can serve as templates to be mimicked in assembled synthetic communities.
Both sampled and assembled communities allow studying DBF relationships. They can be placed in controlled versions of their natural environment (Vorholt et al., 2017) or in synthetic microbial systems (Figure 2C) of different environmental complexity (de Roy et al., 2014; Kurkjian, 2019; Tecon et al., 2019). Microbial systems also offer the opportunity to combine experiments in which different realized diversities emerge from communities of the same initial diversity exposed to disturbance regimes with classical BEF experiments, in which communities of different initial diversities are randomly assembled (Figure 2B; Bell et al., 2009b). To realize vast numbers of parallel experiments with different communities, automated pipetting systems can be used (Fetzer et al., 2015). Experiments with only one or two species are recommended to assess each species’ responses to disturbances, effects on functions and pairwise interactions separately, and relate these observations to traits (cf. above).
Application of Disturbance Regimes
A highly controllable approach to expose microbial communities to disturbances (Figure 2C) is to remove part of the community biomass by dilution during serial transfers (Gibbons et al., 2016). This resembles non-specific disturbances, meaning that the removal does not depend on response traits in the communities, while intense disturbances may cause random extinctions (Karakoç et al., 2018). By contrast, specific disturbance responses of different community members may lead to non-random extinctions and are at least as realistic in nature (Srivastava and Vellend, 2005). One option to achieve this is to expose microbes to toxic compounds or resource scarcity for a certain period of time, for instance by cultivating them in miniature membrane bioreactors from which compounds can be washed out or in dialysis bags alternately placed in tanks with or without the compounds (Langenheder et al., 2012; Johnke et al., 2017; Karakoç et al., 2018). Additionally, temperature variation (Jurburg et al., 2017), UV light application (Gibbons et al., 2016) or sonication (Violle et al., 2010) can induce discrete impacts on microbial communities. These disturbances hold the potential to impact certain community members more than others depending on the microbes’ response traits, respectively. They can be applied in different intensities, in recurrent regimes varying in frequency (Figure 2D), and they can be combined in various orders to create multiple disturbance regimes (Jurburg et al., 2017; Calderón et al., 2018).
Observation of Biodiversity and Ecosystem Functioning
We advocate for observations of community compositions and functions at several points in time. From species abundance data various biodiversity metrics can be calculated (Tuomisto, 2012). This includes functional type and trait diversity if the respective traits are characterized (cf. above). The simplest way to measure abundances in microbial communities is to count colony-forming units. Individual strains can be distinguished if they differ in shape or color (Saleem et al., 2012; Karakoç et al., 2018). However, growth-based methods miss bacteria that are in viable but non-culturable states, which is a common strategy to survive unfavorable conditions (Colwell et al., 1996; Robben et al., 2018). Alternatively, microfluidic-based quantitative real-time polymerase chain reaction (PCR) can be used to directly quantify abundances in synthetic microbial communities in high-throughput. Kleyer et al. (2017) applied this method to communities containing up to 11 bacterial strains from various phyla. For more complex communities, composition can be inferred from multiplexed amplicon or metagenome sequencing or from metaproteomics-based methods, including estimates of the community members’ cell numbers or biomass contributions (Delgado-Baquerizo et al., 2016; Kleiner et al., 2017). High-resolution flow cytometry and cytometric pattern analysis are particularly suitable to monitor and quantify the composition of complex microbial communities over time (Koch et al., 2013), for example under short-term disturbances induced by pH and temperature alterations (Liu et al., 2018). Another huge advantage of flow cytometry is that multiple microbial traits are measurable in a short time (Müller and Nebe-von-Caron, 2010).
To measure ecosystem functioning, biomass production is a common proxy used in many BEF studies (Balvanera et al., 2006). For microbial systems, biomass production is a function integrating direct and indirect metabolism to the conversion of substrates into microbial biomass. It can be measured via the optical density of microbial communities (Awasthi et al., 2014; Fetzer et al., 2015). Alternatively, biomass turnover can be assessed by measuring respiration rates (Bell et al., 2005). For a more specific ecosystem function, one can assess the degradation of certain compounds (e.g., contaminants) supplemented to the growth medium (Peter et al., 2011). Feeding stable isotope labeled compounds and analyzing their enrichment in microbial cells can provide information on the community members’ metabolic activity, even at single cell level (Musat et al., 2008). Additional functions to be measured comprise overall substrate utilization, nutrient cycling, abiotic compound production, or pathogen suppression (Bell et al., 2009a; Langenheder et al., 2010; van Elsas et al., 2012). Microbial systems also facilitate the strongly needed measurement of several functions at a time (Miki et al., 2014; Roger et al., 2016). Thus, they let us calculate ecosystem multifunctionality metrics (Byrnes et al., 2014) and investigate how those are influenced by disturbances.
Computational Analysis and Modeling of DBF Relationships
The data obtained from disturbance experiments in microbial systems will yield sets of relationships between different disturbance regimes, biodiversity metrics, and measures of ecosystem functioning (e.g., Figures 1, 2D). Analyzing these DBF relationships’ similarities and disparities for several points in time and different environmental contexts will generate insights on temporal stability and context-dependence of biodiversity responses to (multiple) disturbances and effects on (multiple) ecosystem functions. The data will enable broad tests of ecological hypotheses stating that, for instance, intermediate disturbances may favor biodiversity (Connell, 1978; Sheil and Burslem, 2013), recurrent and multiple disturbances may reinforce each other (Paine et al., 1998; Jurburg et al., 2017) or biodiversity may promote functioning and multifunctionality through species complementarity (Barry et al., 2019) and insurance effects (Yachi and Loreau, 1999; Petchey, 2007). If experiments include monocultures, initial compositions of mixtures are known and each species’ contribution to functioning can be separated (which is common for biomass production, but less common for other functions), partitioning selection and complementarity effects (Loreau and Hector, 2001; Fox, 2005) is applicable to microbial systems (Saleem et al., 2012). More generally, coexistence of species under a certain disturbance regime will indicate their complementarity, and associated measures of functions disclose the role of this complementarity for ecosystem functioning (Rychtecká et al., 2014). Further statistical methods, such as variation partitioning (Legendre, 2007), structural equation modeling (Grace, 2006; Shipley, 2016) or co-occurrence analysis (Faust and Raes, 2012) allow proposing potential causal mechanisms underlying the DBF relationships or potential interactions between microbial community members. The new hypotheses generated in this way should–and with microbial systems they can–be thoroughly tested in further targeted experiments (Berry and Widder, 2014; Eisenhauer et al., 2015; Dormann et al., 2018).
The rich data will reveal further insights when combined with simulation models (Kreft et al., 2013; Song et al., 2014; Vallina et al., 2019). These models should explicitly describe processes such as growth, decay, resource consumption and interactions of microbes, their responses to disturbances and their effects on functions at the level of single populations or even single individuals (Hellweger et al., 2016; Momeni et al., 2017). Thereupon, community dynamics and DBF relationships emerge in the simulations (Smith et al., 2016) not just from the properties of modeled organisms, but also from the complex interactions of all modeled processes. Process definitions and parameter values can be based on measurements of microbial population dynamics in mono- and co-culture experiments and of their traits (cf. above). They can further be obtained by simulating scenarios equal to experiments, and fitting the model definitions such that multiple characteristic “patterns” of simulated communities match those observed in experiments (“pattern-oriented modeling”; Grimm and Railsback, 2012). For microbial systems, multiple experimental patterns at different organizational levels are available and, even better, additional experiments and observations are feasible to test further model predictions (Widder et al., 2016). This allows an iterative interplay between modeling and experiments, whereby the models not only describe the observed patterns, but also reveal the processes and interactions generating them, thus disentangling the complex system dynamics. Once a model is defined, virtual DBF experiments are possible that by far exceed the possibilities with real microbial systems. A huge variety of (single and combined) disturbance regimes, initial community compositions or (heterogeneous) environmental conditions can systematically be simulated, and multiple metrics of biodiversity and functioning investigated including variations in space and time. Ecological hypotheses can be tested, predictions can be made and, most important, DBF relationships can be linked to causal mechanisms and to preconditions for their validity.
Conclusion
We see a great perspective for microbial experiments, especially when combined with computational models, for analyzing, understanding and generalizing the shapes, causes and consequences of DBF relationships. Thus, they can substantially contribute to developing ecological theory that integrates disturbances, biodiversity, and ecosystem functions. This integration is needed and will ultimately improve our ability to predict and manage the fate and functioning of ecosystems under global change.
Author Contributions
TB, AW, and AC conceived the perspective. TB and AW wrote the manuscript. All authors critically revised the manuscript and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Volker Grimm, Robert Ptacnik, and the reviewers for highly valuable comments on the manuscript.
References
Awasthi, A., Singh, M., Soni, S. K., Singh, R., and Kalra, A. (2014). Biodiversity acts as insurance of productivity of bacterial communities under abiotic perturbations. ISME J. 8, 2445–2452. doi: 10.1038/ismej.2014.91
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528:16192. doi: 10.1038/nature16192
Balvanera, P., Pfisterer, A. B., Buchmann, N., He, J. S., Nakashizuka, T., Raffaelli, D., et al. (2006). Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol. Lett. 9, 1146–1156. doi: 10.1111/j.1461-0248.2006.00963.x
Barry, K. E., Mommer, L., van Ruijven, J., Wirth, C., Wright, A. J., Bai, Y., et al. (2019). The future of complementarity: disentangling causes from consequences. Trends Ecol. Evol. 34, 167–180. doi: 10.1016/j.tree.2018.10.013
Bell, T., Gessner, M. O., Griffiths, R. I., McLaren, J. R., Morin, P. J., van der Heijden, M., et al. (2009a). “Microbial biodiversity and ecosystem functioning under controlled conditions and in the wild,” in Biodiversity, Ecosystem Functioning, and Human Wellbeing: an Ecological and Economic Perspective, eds S. Naeem, D. E. Bunker, A. Hector, M. Loreau, and C. Perrings, (Oxford: Oxford University Press), 121–133. doi: 10.1093/acprof:oso/9780199547951.003.0009
Bell, T., Lilley, A. K., Hector, A., Schmid, B., King, L., and Newman, J. A. (2009b). A linear model method for biodiversity-ecosystem functioning experiments. Am. Nat. 174, 836–849. doi: 10.1086/647931
Bell, T., Newman, J. A., Silverman, B. W., Turner, S. L., and Lilley, A. K. (2005). The contribution of species richness and composition to bacterial services. Nature 436, 1157–1160. doi: 10.1038/nature03891
Bellard, C., Bertelsmeier, C., Leadley, P., Thuiller, W., and Courchamp, F. (2012). Impacts of climate change on the future of biodiversity. Ecol. Lett. 15, 365–377. doi: 10.1111/j.1461-0248.2011.01736.x
Berry, D., and Widder, S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 5:219. doi: 10.3389/fmicb.2014.00219
Borer, E. T., Harpole, W. S., Adler, P. B., Lind, E. M., Orrock, J. L., Seabloom, E. W., et al. (2014). Finding generality in ecology: a model for globally distributed experiments. Methods Ecol. Evol. 5, 65–73. doi: 10.1111/2041-210x.12125
Brose, U., and Hillebrand, H. (2016). Biodiversity and ecosystem functioning in dynamic landscapes. Phil. Trans. R. Soc. B Biol. Sci. 371:20150267. doi: 10.1098/rstb.2015.0267
Byrnes, J. E. K., Gamfeldt, L., Isbell, F., Lefcheck, J. S., Griffin, J. N., Hector, A., et al. (2014). Investigating the relationship between biodiversity and ecosystem multifunctionality: challenges and solutions. Methods Ecol. Evol. 5, 111–124. doi: 10.1111/2041-210x.12143
Calderón, K., Philippot, L., Bizouard, F., Breuil, M.-C., Bru, D., and Spor, A. (2018). Compounded disturbance chronology modulates the resilience of soil microbial communities and N-cycle related functions. Front. Microbiol. 9:2721. doi: 10.3389/fmicb.2018.02721
Cardinale, B. J., Duffy, J. E., Gonzalez, A., Hooper, D. U., Perrings, C., Venail, P., et al. (2012). Biodiversity loss and its impact on humanity. Nature 486, 59–67. doi: 10.1038/nature11148
Colwell, R. R., Brayton, P., Herrington, D., Tall, B., Huq, A., and Levine, M. M. (1996). Viable but non-culturable Vibrio cholerae O1 revert to a cultivable state in the human intestine. World J. Microbiol. Biotechnol. 12, 28–31. doi: 10.1007/bf00327795
Connell, J. H. (1978). Diversity in tropical rain forests and coral reefs. Science 199, 1302–1310. doi: 10.1126/science.199.4335.1302
de Roy, K., Marzorati, M., Van den Abbeele, P., Van de Wiele, T., and Boon, N. (2014). Synthetic microbial ecosystems: an exciting tool to understand and apply microbial communities. Environ. Microbiol. 16, 1472–1481. doi: 10.1111/1462-2920.12343
Delgado-Baquerizo, M., Giaramida, L., Reich, P. B., Khachane, A. N., Hamonts, K., Edwards, C., et al. (2016). Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J. Ecol. 104, 936–946. doi: 10.1111/1365-2745.12585
Díaz, S., Lavorel, S., de Bello, F., Quetier, F., Grigulis, K., and Robson, M. (2007). Incorporating plant functional diversity effects in ecosystem service assessments. Proc. Natl. Acad. Sci. U.S.A. 104, 20684–20689. doi: 10.1073/pnas.0704716104
Dormann, C. F., Bobrowski, M., Dehling, D. M., Harris, D. J., Hartig, F., Lischke, H., et al. (2018). Biotic interactions in species distribution modelling: 10 questions to guide interpretation and avoid false conclusions. Glob. Ecol. Biogeogr. 27, 1004–1016. doi: 10.1111/geb.12759
Eisenhauer, N., Bowker, M. A., Grace, J. B., and Powell, J. R. (2015). From patterns to causal understanding: structural equation modeling (SEM) in soil ecology. Pedobiologia 58, 65–72. doi: 10.1016/j.pedobi.2015.03.002
Elmqvist, T., Folke, C., Nyström, M., Peterson, G., Bengtsson, J., Walker, B., et al. (2003). Response diversity, ecosystem change, and resilience. Front. Ecol. Environ. 1:488–494. doi: 10.2307/3868116
Faust, K., and Raes, J. (2012). Microbial interactions: from networks to models. Nat. Rev. Microbiol. 10, 538–550. doi: 10.1038/nrmicro2832
Fetzer, I., Johst, K., Schäwe, R., Banitz, T., Harms, H., and Chatzinotas, A. (2015). The extent of functional redundancy changes as species’ roles shift in different environments. Proc. Natl. Acad. Sci. U.S.A. 112, 14888–14893. doi: 10.1073/pnas.1505587112
Fox, J. W. (2005). Interpreting the, selection effect’ of biodiversity on ecosystem function. Ecol. Lett. 8, 846–856. doi: 10.1111/j.1461-0248.2005.00795.x
Fraser, L. H., Henry, H. A., Carlyle, C. N., White, S. R., Beierkuhnlein, C., Cahill, J. F. Jr., et al. (2013). Coordinated distributed experiments: an emerging tool for testing global hypotheses in ecology and environmental science. Front. Ecol. Environ. 11:147–155. doi: 10.1890/110279
Gagic, V., Bartomeus, I., Jonsson, T., Taylor, A., Winqvist, C., Fischer, C., et al. (2015). Functional identity and diversity of animals predict ecosystem functioning better than species-based indices. Proc. R. Soc. B-Biol. Sci. 282:20142620. doi: 10.1098/rspb.2014.2620
Garnier, E., Navas, M.-L., and Grigulis, K. (2015). Plant Functional Diversity. Organism Traits, Community Structure, and Ecosystem Properties. Oxford: Oxford University Press.
Gibbons, S. M., Scholz, M., Hutchison, A. L., Dinner, A. R., Gilbert, J. A., and Coleman, M. L. (2016). Disturbance regimes predictably alter diversity in an ecologically complex bacterial system. mBio 7:e001372-16. doi: 10.1128/mBio.01372-16
Grace, J. B. (2006). Structural Equation Modeling and Natural Systems. Cambridge: Cambridge University Press.
Grimm, V., and Railsback, S. F. (2012). Pattern-oriented modelling: a ‘multiscope’ for predictive systems ecology. Philos. Trans. R. Soc. B Biol. Sci. 367, 298–310. doi: 10.1098/rstb.2011.0180
Haddad, N. M., Holyoak, M., Mata, T. M., Davies, K. F., Melbourne, B. A., and Preston, K. (2008). Species’ traits predict the effects of disturbance and productivity on diversity. Ecol. Lett. 11, 348–356. doi: 10.1111/j.1461-0248.2007.01149.x
Hellweger, F. L., Clegg, R. J., Clark, J. R., Plugge, C. M., and Kreft, J. U. (2016). Advancing microbial sciences by individual-based modelling. Nat. Rev. Microbiol. 14, 461–471. doi: 10.1038/nrmicro.2016.62
Hillebrand, H., Blasius, B., Borer, E. T., Chase, J. M., Downing, J. A., Eriksson, B. K., et al. (2018). Biodiversity change is uncoupled from species richness trends: consequences for conservation and monitoring. J. Appl. Ecol. 55, 169–184. doi: 10.1111/1365-2664.12959
Hooper, D. U., Chapin, F. S., Ewel, J. J., Hector, A., Inchausti, P., Lavorel, S., et al. (2005). Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75, 3–35. doi: 10.1890/04-0922
Huston, M. A. (1997). Hidden treatments in ecological experiments: re-evaluating the ecosystem function of biodiversity. Oecologia 110, 449–460. doi: 10.1007/s004420050180
Jessup, C. M., Kassen, R., Forde, S. E., Kerr, B., Buckling, A., Rainey, P. B., et al. (2004). Big questions, small worlds: microbial model systems in ecology. Trends Ecol. Evol. 19, 189–197. doi: 10.1016/j.tree.2004.01.008
Johnke, J., Boenigk, J., Harms, H., and Chatzinotas, A. (2017). Killing the killer: predation between protists and predatory bacteria. FEMS Microbiol. Lett. 364:fnx089. doi: 10.1093/femsle/fnx089
Jones, K. E., Bielby, J., Cardillo, M., Fritz, S. A., O’Dell, J., Orme, C. D. L., et al. (2009). PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648–2648. doi: 10.1890/08-1494.1
Jurburg, S. D., Nunes, I., Brejnrod, A., Jacquiod, S., Priemé, A., Sørensen, S. J., et al. (2017). Legacy effects on the recovery of soil bacterial communities from extreme temperature perturbation. Front. Microbiol. 8:1832. doi: 10.3389/fmicb.2017.01832
Karakoç, C., Radchuk, V., Harms, H., and Chatzinotas, A. (2018). Interactions between predation and disturbances shape prey communities. Sci. Rep. 8:2968. doi: 10.1038/s41598-018-21219-x
Kattge, J., Díaz, S., Lavorel, S., Prentice, I. C., Leadley, P., Bönisch, G., et al. (2011). TRY − a global database of plant traits. Glob. Change Biol. 17, 2905–2935. doi: 10.1111/j.1365-2486.2011.02451.x
Kershaw, H. M., and Mallik, A. U. (2013). Predicting plant diversity response to disturbance: applicability of the intermediate disturbance hypothesis and mass ratio hypothesis. Crit. Rev. Plant Sci. 32, 383–395. doi: 10.1080/07352689.2013.791501
Ketola, T., and Kristensen, T. N. (2017). Experimental approaches for testing if tolerance curves are useful for predicting fitness in fluctuating environments. Front. Ecol. Evol. 5:129. doi: 10.3389/fevo.2017.00129
Kleiner, M., Thorson, E., Sharp, C. E., Dong, X., Liu, D., Li, C., et al. (2017). Assessing species biomass contributions in microbial communities via metaproteomics. Nat. Comm. 8:1558. doi: 10.1038/s41467-017-01544-x
Kleyer, H., Tecon, R., and Or, D. (2017). Resolving species level changes in a representative soil bacterial community using microfluidic quantitative PCR. Front. Microbiol. 8:2017. doi: 10.3389/fmicb.2017.02017
Koch, C., Fetzer, I., Harms, H., and Müller, S. (2013). CHIC—an automated approach for the detection of dynamic variations in complex microbial communities. Cytom. Part A 83A, 561–567. doi: 10.1002/cyto.a.22286
Krause, S., Le Roux, X., Niklaus, P. A., van Bodegom, P. M., Lennon, J. T., Bertilsson, S., et al. (2014). Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front. Microbiol. 5:251. doi: 10.3389/fmicb.2014.00251
Kreft, J.-U., Plugge, C. M., Grimm, V., Prats, C., Leveau, J. H. J., Banitz, T., et al. (2013). Mighty small: observing and modeling individual microbes becomes big science. Proc. Natl. Acad. Sci. U.S.A. 110, 18027–18028. doi: 10.1073/pnas.1317472110
Kunstler, G., Falster, D., Coomes, D. A., Hui, F., Kooyman, R. M., Laughlin, D. C., et al. (2016). Plant functional traits have globally consistent effects on competition. Nature 529, 204–207. doi: 10.1038/nature16476
Kurkjian, H. M. (2019). The metapopulation microcosm plate: a modified 96-well plate for use in microbial metapopulation experiments. Methods Ecol. Evol. 10, 162–168. doi: 10.1111/2041-210x.13116
Langenheder, S., Bulling, M. T., Prosser, J. I., and Solan, M. (2012). Role of functionally dominant species in varying environmental regimes: evidence for the performance-enhancing effect of biodiversity. BMC Ecol. 12:14. doi: 10.1186/1472-6785-12-14
Langenheder, S., Bulling, M. T., Solan, M., and Prosser, J. I. (2010). Bacterial biodiversity-ecosystem functioning relations are modified by environmental complexity. PLoS One 5:e10834. doi: 10.1371/journal.pone.0010834
Larsen, T. H., Williams, N. M., and Kremen, C. (2005). Extinction order and altered community structure rapidly disrupt ecosystem functioning. Ecol. Lett. 8, 538–547. doi: 10.1111/j.1461-0248.2005.00749.x
Lavorel, S., and Garnier, E. (2002). Predicting changes in community composition and ecosystem functioning from plant traits: revisiting the Holy Grail. Funct. Ecol. 16, 545–556. doi: 10.1046/j.1365-2435.2002.00664.x
Legendre, P. (2007). Studying beta diversity: ecological variation partitioning by multiple regression and canonical analysis. J. Plant Ecol. 1, 3–8. doi: 10.1093/jpe/rtm001
Liu, Z., Cichocki, N., Bonk, F., Günther, S., Schattenberg, F., Harms, H., et al. (2018). Ecological stability properties of microbial communities assessed by flow cytometry. mSphere 3:e00564-17. doi: 10.1128/mSphere.00564-17
Loreau, M. (2010). Linking biodiversity and ecosystems: towards a unifying ecological theory. Philos. Trans. R. Soc. B Biol. Sci. 365, 49–60. doi: 10.1098/rstb.2009.0155
Loreau, M., and Hector, A. (2001). Partitioning selection and complementarity in biodiversity experiments. Nature 412, 72–76. doi: 10.1038/35083573
Loreau, M., Naeem, S., Inchausti, P., Bengtsson, J., Grime, J. P., Hector, A., et al. (2001). Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294, 804–808. doi: 10.1126/science.1064088
Mackey, R. L., and Currie, D. J. (2001). The diversity-disturbance relationship: is it generally strong and peaked? Ecology 82, 3479–3492. doi: 10.2307/2680166
Martínez-Ramos, M., Ortiz-Rodríguez, I. A., Piñero, D., Dirzo, R., and Sarukhan, J. (2016). Anthropogenic disturbances jeopardize biodiversity conservation within tropical rainforest reserves. Proc. Natl. Acad. Sci. U.S.A. 113, 5323–5328. doi: 10.1073/pnas.1602893113
McGill, B. J., Enquist, B. J., Weiher, E., and Westoby, M. (2006). Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21, 178–185. doi: 10.1016/j.tree.2006.02.002
Miki, T., Yokokawa, T., and Matsui, K. (2014). Biodiversity and multifunctionality in a microbial community: a novel theoretical approach to quantify functional redundancy. Proc. R. Soc. B Biol. Sci. 281:20132498. doi: 10.1098/rspb.2013.2498
Miller, A. D., Roxburgh, S. H., and Shea, K. (2011). How frequency and intensity shape diversity-disturbance relationships. Proc. Natl. Acad. Sci. U.S.A. 108, 5643–5648. doi: 10.1073/pnas.1018594108
Momeni, B., Xie, L., and Shou, W. (2017). Lotka-Volterra pairwise modeling fails to capture diverse pairwise microbial interactions. eLife 6:e25051. doi: 10.7554/eLife.25051
Müller, S., and Nebe-von-Caron, G. (2010). Functional single-cell analyses: flow cytometry and cell sorting of microbial populations and communities. FEMS Microbiol. Rev. 34, 554–587. doi: 10.1111/j.1574-6976.2010.00214.x
Musat, N., Halm, H., Winterholler, B., Hoppe, P., Peduzzi, S., Hillion, F., et al. (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 17861–17866. doi: 10.1073/pnas.0809329105
Naeem, S., Duffy, J. E., and Zavaleta, E. (2012). The functions of biological diversity in an age of extinction. Science 336, 1401–1406. doi: 10.1126/science.1215855
Newman, E. A. (2019). Disturbance ecology in the anthropocene. Front. Ecol. Evol. 7:147. doi: 10.3389/fevo.2019.00147
Norberg, J. (2004). Biodiversity and ecosystem functioning: a complex adaptive systems approach. Limnol. Oceanogr. 49, 1269–1277. doi: 10.4319/lo.2004.49.4_part_2.1269
O’Malley, M. A., Travisano, M., Velicer, G. J., and Bolker, J. A. (2015). How do microbial populations and communities function as model systems? Q. Rev. Biol. 90, 269–293. doi: 10.1086/682588
Paine, T. R., Tegner, J. M., and Johnson, A. E. (1998). Compounded perturbations yield ecological surprises. Ecosystems 1, 535–545. doi: 10.1007/s100219900049
Pereira, H. M., Leadley, P. W., Proenca, V., Alkemade, R., Scharlemann, J. P. W., Fernandez-Manjarres, J. F., et al. (2010). Scenarios for global biodiversity in the 21st century. Science 330, 1496–1501. doi: 10.1126/science.1196624
Petchey, O. L. (2007). “Effects of environmental variability on ecological communities: testing the insurance hypothesis of biodiversity in aquatic microcosms,” in The Impact of Environmental Variability on Ecological Systems, eds D. A. Vasseur, and K. S. Mccann, (Dordrecht: Springer Netherlands), 179–196. doi: 10.1007/978-1-4020-5851-6_9
Peter, H., Beier, S., Bertilsson, S., Lindstrom, E. S., Langenheder, S., and Tranvik, L. J. (2011). Function-specific response to depletion of microbial diversity. ISME J. 5, 351–361. doi: 10.1038/ismej.2010.119
Pickett, S. T. A., and White, P. S. (1985). The Ecology of Natural Disturbance and Patch Dynamics. New York, NY: Academic Press.
Radchuk, V., De Laender, F., van den Brink, P. J., and Grimm, V. (2016). Biodiversity and ecosystem functioning decoupled: invariant ecosystem functioning despite non-random reductions in consumer diversity. Oikos 125, 424–433. doi: 10.1111/oik.02220
Rillig, M. C., Kiessling, W., Borsch, T., Gessler, A., Greenwood, A. D., Hofer, H., et al. (2015). Biodiversity research: data without theory—theory without data. Front. Ecol. Evol. 3:20. doi: 10.3389/fevo.2015.00020
Robben, C., Fister, S., Witte, A. K., Schoder, D., Rossmanith, P., and Mester, P. (2018). Induction of the viable but non-culturable state in bacterial pathogens by household cleaners and inorganic salts. Sci. Rep. 8:15132. doi: 10.1038/s41598-018-33595-5
Roger, F., Bertilsson, S., Langenheder, S., Osman, O. A., and Gamfeldt, L. (2016). Effects of multiple dimensions of bacterial diversity on functioning, stability and multifunctionality. Ecology 97, 2716–2728. doi: 10.1002/ecy.1518
Rychtecká, T., Lanta, V., Weiterová, I., and Lepš, J. (2014). Sown species richness and realized diversity can influence functioning of plant communities differently. Naturwissenschaften 101, 637–644. doi: 10.1007/s00114-014-1198-7
Saleem, M., Fetzer, I., Dormann, C. F., Harms, H., and Chatzinotas, A. (2012). Predator richness increases the effect of prey diversity on prey yield. Nat. Comm. 3:1305. doi: 10.1038/ncomms2287
Schuldt, A., Assmann, T., Brezzi, M., Buscot, F., Eichenberg, D., Gutknecht, J., et al. (2018). Biodiversity across trophic levels drives multifunctionality in highly diverse forests. Nat. Comm. 9:2989. doi: 10.1038/s41467-018-05421-z
Seidl, R., Schelhaas, M.-J., and Lexer, M. J. (2011). Unraveling the drivers of intensifying forest disturbance regimes in Europe. Glob. Change Biol. 17, 2842–2852. doi: 10.1111/j.1365-2486.2011.02452.x
Shade, A., Dunn, R. R., Blowes, S. A., Keil, P., Bohannan, B. J. M., Herrmann, M., et al. (2018). Macroecology to unite all life, large and small. Trends Ecol. Evol. 33, 731–744. doi: 10.1016/j.tree.2018.08.005
Sheil, D., and Burslem, D. F. R. P. (2013). Defining and defending Connell’s intermediate disturbance hypothesis: a response to Fox. Trends Ecol. Evol. 28, 571–572. doi: 10.1016/j.tree.2013.07.006
Shipley, B. (2016). Cause and Correlation in Biology. A User’s Guide to Path Analysis, Structural Equations and Causal Inference With R. Cambridge: Cambridge University Press.
Smith, S. L., Vallina, S. M., and Merico, A. (2016). Phytoplankton size-diversity mediates an emergent trade-off in ecosystem functioning for rare versus frequent disturbances. Sci. Rep. 6:34170. doi: 10.1038/srep34170
Song, H.-S., Cannon, W., Beliaev, A., and Konopka, A. (2014). Mathematical modeling of microbial community dynamics: a methodological review. Processes 2, 711–752. doi: 10.3390/pr2040711
Srivastava, D. S., and Vellend, M. (2005). Biodiversity-ecosystem function research: is it relevant to conservation? Annu. Rev. Ecol. Evol. Syst. 36, 267–294. doi: 10.1146/annurev.ecolsys.36.102003.152636
Stearns, S. C. (1989). Trade-offs in life-history evolution. Funct. Ecol. 3, 259–268. doi: 10.2307/2389364
Suding, K. N., Lavorel, S., Chapin, F. S., Cornelissen, J. H. C., Díaz, S., Garnier, E., et al. (2008). Scaling environmental change through the community-level: a trait-based response-and-effect framework for plants. Glob. Change Biol. 14, 1125–1140. doi: 10.1111/j.1365-2486.2008.01557.x
Tecon, R., Mitri, S., Ciccarese, D., Or, D., van der Meer, J. R., and Johnson, D. R. (2019). Bridging the holistic-reductionist divide in microbial ecology. mSystems 4:e00265-18. doi: 10.1128/mSystems.00265-18
Tilman, D. (1982). Resource Competition and Community Structure. Princeton, NJ: Princeton University Press.
Tilman, D., Isbell, F., and Cowles, J. M. (2014). Biodiversity and ecosystem functioning. Annu. Rev. Ecol. Evol. Syst. 45, 471–493. doi: 10.1146/annurev-ecolsys-120213-091917
Tuomisto, H. (2012). An updated consumer’s guide to evenness and related indices. Oikos 121, 1203–1218. doi: 10.1111/j.1600-0706.2011.19897.x
Vallina, S., Martinez-Garcia, R., Smith, S., and Bonachela, J. (2019). “Models in microbial ecology,” in Encyclopedia of Microbiology, ed. T. M. Schmidt, (Cambridge, MA: Academic Press), 211–246.
van Elsas, J. D., Chiurazzi, M., Mallon, C. A., Elhottova, D., Kristufek, V., and Salles, J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 109, 1159–1164. doi: 10.1073/pnas.1109326109
Villnäs, A., Norkko, J., Hietanen, S., Josefson, A. B., Lukkari, K., and Norkko, A. (2013). The role of recurrent disturbances for ecosystem multifunctionality. Ecology 94, 2275–2287. doi: 10.1890/12-1716.1
Violle, C., Navas, M. L., Vile, D., Kazakou, E., Fortunel, C., Hummel, I., et al. (2007). Let the concept of trait be functional! Oikos 116, 882–892. doi: 10.1111/j.0030-1299.2007.15559.x
Violle, C., Pu, Z., and Jiang, L. (2010). Experimental demonstration of the importance of competition under disturbance. Proc. Natl. Acad. Sci. U.S.A. 107, 12925–12929. doi: 10.1073/pnas.1000699107
Vorholt, J. A., Vogel, C., Carlström, C. I., and Müller, D. B. (2017). Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22, 142–155. doi: 10.1016/j.chom.2017.07.004
Wellnitz, T., and Poff, N. L. (2001). Functional redundancy in heterogeneous environments: implications for conservation. Ecol. Lett. 4, 177–179. doi: 10.1046/j.1461-0248.2001.00221.x
White, P. S., and Jentsch, A. (2001). “The search for generality in studies of disturbance and ecosystem dynamics,” in Progress in Botany, eds K. Esser, U. Lüttge, J. W. Kadereit, and W. Beyschlag, (Berlin: Springer), 399–450. doi: 10.1007/978-3-642-56849-7_17
Widder, S., Allen, R. J., Pfeiffer, T., Curtis, T. P., Wiuf, C., Sloan, W. T., et al. (2016). Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 10, 2557–2568. doi: 10.1038/ismej.2016.45
Wittebolle, L., Marzorati, M., Clement, L., Balloi, A., Daffonchio, D., Heylen, K., et al. (2009). Initial community evenness favours functionality under selective stress. Nature 458, 623–626. doi: 10.1038/nature07840
Wootton, J. T. (1998). Effects of disturbance on species diversity: a multitrophic perspective. Am. Nat. 152, 803–825. doi: 10.1086/286210
Keywords: ecological theory, trait-based ecology, microcosm experiments, multifunctionality, environmental change, computational modeling
Citation: Banitz T, Chatzinotas A and Worrich A (2020) Prospects for Integrating Disturbances, Biodiversity and Ecosystem Functioning Using Microbial Systems. Front. Ecol. Evol. 8:21. doi: 10.3389/fevo.2020.00021
Received: 26 June 2019; Accepted: 24 January 2020;
Published: 18 February 2020.
Edited by:
Robert Ptacnik, Wasser Cluster Lunz, AustriaReviewed by:
S. Lan Smith, Japan Agency for Marine-Earth Science and Technology, JapanXin Jing, KU Leuven, Belgium
Copyright © 2020 Banitz, Chatzinotas and Worrich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Banitz, dGhvbWFzLmJhbml0ekB1ZnouZGU=; Anja Worrich, YW5qYS53b3JyaWNoQHVmei5kZQ==
 Thomas Banitz
Thomas Banitz Antonis Chatzinotas
Antonis Chatzinotas Anja Worrich
Anja Worrich