- 1Laboratory of Plant Pathology, Department of Agriculture, Crop Production and Rural Environment, University of Thessaly, Volos, Greece
- 2Hellenic Agricultural Organization ‘Demeter’, Institute of Plant Breeding and Genetic Resources, Thessaloniki, Greece
- 3Department of Ichthyology and Aquatic Environment, University of Thessaly, Volos, Greece
During the recent years, High Resolution Melting (HRM) analysis has been developed as a rapid and accurate tool in plant disease diagnostics, species identification and SNP genotyping. This approach has been applied to analyze the genetic diversity in several plant species with molecular markers, including single sequence repeats (SSR). However, no studies have been carried out to investigate the variation of SSR in plant pathogenic fungi by using the HRM technology. In this report, the genetic structure of Venturia inaequalis populations in Greece was investigated for the first time by using six microsatellite markers. The developed HRM protocol was able to generate genotype-specific melting curves, consistent with the haploid nature of the fungus. Unknown samples were genotyped using standard samples as reference controls among multiple runs. Compared to the more complex genome of diploid plants, several limitations were avoided. The shape of the melting curves revealed differences between the genotypes in each SSR marker and showed that all the genotypes could be easily distinguished. The genetic analysis of apple scab populations revealed high genetic variation within the populations (96%), while only 4% of the total gene diversity was attributed to among-population variation. The isolates were grouped into three higher-level populations according to the principal coordinate analysis (PCoA).
Introduction
Apple scab caused by Venturia inaequalis is the most important disease of apples in Greece. The pathogen is able to reproduce both sexually in winter and asexually in spring-summer periods. In order to efficiently protect the final product from apple scab, the growers rely on frequent fungicide applications (up to 16 annually) on 5–9 day-intervals. The polycyclic nature of the disease (MacHardy, 1996), the evolution of races and their ability to overcome the resistance genes in the host (Bus et al., 2011) and also the development of resistance to fungicides (Köller and Wilcox, 2001) are the most important factors that lead to high variation in fungal genome and changes in population genetic structure. Because of the pathogen's ability to adapt to environmental changes in order to survive (McDonald, 1997), many studies have analyzed its genetic structure among populations all over the world (Gladieux et al., 2010; Ebrahimi et al., 2016; Koopman et al., 2017).
Different molecular markers such as random amplified polymorphic DNA (RAPDs), restriction fragment length polymorphisms (RFLPs), amplified fragment length polymorphisms (AFLPs), and microsatellites (or single sequence repeats-SSRs) have been developed to assess the genetic diversity of V. inaequalis in various apple producing countries (Tenzer and Gessler, 1997; Guérin et al., 2004; Xu et al., 2013). Among all molecular markers, SSRs are preferable because of their high reproducibility, amenability to automation and high throughput genotyping (Agarwal et al., 2008). The traditional protocols used for SSR genotyping involve laborious and hazardous post-PCR analysis with polyacrylamide gels followed by staining with ethidium bromide or automated but costly capillary electrophoresis (Ganopoulos et al., 2011).
High-resolution melting (HRM) analysis is a closed-tube method, that measures the fluorescence reduction of intercalating dye in the process of dissociation of double stranded DNA. Over the last years, this method has become increasingly popular due to its simplicity, flexibility, non-destructive nature, superb sensitivity and specificity (Vossen et al., 2009). HRM analysis generates sequence-related melting profiles and can reveal differences in the genotype at the level of a single nucleotide (Chatzidimopoulos et al., 2014a). The application of HRM as a novel quantitative or semi-quantitative technique for the detection of sequence variations in plant fungal diagnostics has recently been reviewed (Zambounis et al., 2015). Furthermore, this approach has been applied widely to analyze genetic diversity with molecular markers, including SSRs, in several plant species (Mackay et al., 2008; Ganopoulos et al., 2011; Distefano et al., 2012). Studies investigating the variation of SSRs in plant pathogenic fungi using the HRM technology, have not been done so far. Since there is no information on the genetic structure of V. inaequalis in Greece, this work was undertaken to genotype scab isolates from major apple growing regions, by developing a rapid and simple HRM analysis protocol.
Materials and Methods
Isolates
During the 2017 and 2018 spring periods, leaves with distinct spots bearing sporulating lesions of V. inaequalis were collected from five commercial apple orchards in major apple growing areas in Greece. The orchards were located in the areas of Zagora (Central Greece; cv. Scarlett), Kala Nera and Vizitsa (Central Greece; cv. Red Delicious), Veria (Northern Greece; cv. Pink Lady) and Kastoria (Northern Greece; cv. Fuji). Each diseased leaf was transferred to the laboratory in individual paper bags and from each sample a single isolate was made by streaking spores on sterilized potato dextrose agar (PDA) media amended with 100 mg l−1 chloramphenicol, after slight touching a flamed wire loop onto a freshly sporulating lesion. After 24 h of incubation at 20°C, individual germinating conidia were isolated in glass tubes with PDA. A total of 110 single spore isolates (22 from each area) were obtained and stored at 4°C. For nucleic acids extract, fresh subcultures were made from each one of the 110 stored isolates on PDA media and used as inoculum.
Nucleic Acid Preparation
Isolates were grown in 100 mL Erlenmeyer flasks containing 50 mL potato dextrose broth medium (PDB). Flasks were inoculated with 3-mm mycelial plug and incubated in an orbital shaker at 21°C and 150 rpm for 21 days. The globular-like colonies of the liquid culture were obtained from each flask and deposited on sterilized Whatman filter papers to drain the excess moisture. Total nucleic acids from fungal isolates were extracted according to the protocol described by Chatzidimopoulos et al. (2014a). The nucleic acid purity and concentration was estimated on a NanoDrop spectrophotometer (Thermo Scientific) and the final concentration was adjusted to 10 ng μL−1.
Genotyping With High-Resolution Melting Analysis
V. inaequalis isolates were genotyped with microsatellite primer pairs 1tc1g, 1tc1a, vitc1/2, vitc2/D, vica10/154, and vicacg8/42 (Tenzer et al., 1999; Guérin et al., 2004). Nucleic acid extracts (1 μL of each) were used directly in 20-μL reaction volumes containing 1X Kapa HRM Fast qPCR kit (Kapa Biosystems, Cape Town, South Africa), 2.5 mM MgCl2 and 200 nM of each primer. PCR amplification, DNA melting and end point fluorescence level acquiring PCR amplifications were performed on a Rotor-Gene Q thermocycler (Qiagen, Hilden, Germany). To determine the optimal conditions for microsatellite PCR amplification, several runs were conducted in the 36-well carousel with all the primer pairs included. Finally, a rapid PCR protocol was established for all SSRs, under the following conditions: 35 cycles of denaturation at 95°C for 5 s, annealing at 58°C for 20 s, extension at 72°C for 5 s.
High-resolution melting analysis was performed after PCR amplification at the temperature ramping and fluorescence acquisition setting recommended by the manufacturer; that is temperature ramping from 65 to 95°C, rising by 0.1°C/2 s. The melting curves were normalized in between two normalization regions before and after the major fluorescence decrease, representing the melting of the PCR product using the accompanying software (Rotor-gene Q series software v.2.3.1). All samples were plotted according to their melting profiles. Under the difference graph, melting profiles of the samples were compared to that of the core genotype (genotype of the most frequent allele found), which was converted to a horizontal line. To compare the genotype of the samples between PCR-HRM runs (36 were used in each run), reference samples were included. Those samples were genotype-specific and part of the 110 isolates used in the present study. Melting curves plotted within the 80% confidence threshold of the reference curves were considered of the same genotype. To examine the reproducibility of the method, that is consistency of each melting profile, the HRM tests were repeated within 1 week, maintaining the same test conditions, reagent quantities and DNA concentrations.
To verify the validity of the method, representative samples from each genotype were selected for fragment size analysis using polyacrylamide gel electrophoresis (PAGE). The samples were subjected to PCR with the selected SSR primers under the following conditions: 35 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 1 min and a final extension at 72°C for 10 min. Each 25-μL reaction consisted of 10 ng fungal nucleic acid, 200 nM each primer, 10.5 μL DEPC-treated H2O and 12.5 μL TopTaq Master mix 2x (Qiagen). The resulting DNA fragments were resolved in 5% polyacrylamide gel (30% w/w acrylamide/bisacrylamide solution; 29:1) and stained with ethidium bromide. The visualization and determination of the fragment size was performed under a UV-transilluminator at 312 nm following electrophoresis at 40 V.
GenAlEx 6.502b (Peakall and Smouse, 2012) was used to determine genetic structure at hierarchical levels (analysis of molecular variance; AMOVA) and to plot grouping of individuals in the principal coordinates analysis (PCoA). The PCoA procedure in GenAlEx is based on an algorithm published by Orlóci (2013). In the present assay, the standardized option which is based on the conversion of the distance matrix to a covariance matrix was followed. This method divides the respective covariance input by the square root of n-1. At population level, the Nei's standard genetic distance between pairs of populations (Nei, 1972) was calculated for binary data set. This genetic distance matrix is used in GenAlEx for subsequent PCoA analyses involving binary data. PCoA allowed to find and plot the major patterns within the multivariate data set of multiple loci and multiple samples.
Results
The amplicons with fluorescent dye were melted to generate the melting curves. Each microsatellite marker produced unique melting curves at different temperature range. Figure 1A demonstrates the melting profile of the core genotypes generated by each SSR marker on a single HRM run. A total of 46 genotype-specific HRM profiles were revealed by the six SSR markers used. The most variable marker was vicacg8/42 with ten alleles (HRM curves) per locus. Markers 1tc1g, 1tc1a, and vitc2/D showed eight alleles while vitc1/2 and vica10/154 only six. Polymorphisms within the 110 genotypes from five populations were detected based on the profile of temperature-shifted curves. The difference plots showed relative melting curves with reference to a chosen genotype (baseline; core genotype). Figure 1B shows the normalized HRM melting curves of eight representative V. inaequalis genotypes (maximum number of genotypes found), using the microsatellite marker 1tc1g (only genotype-specific HRM genotypes are shown). The shape of the melting curves revealed the differences between the genotypes under investigation and showed that all the genotypes could be easily distinguished visually by their melting curves. Figure 1C depicts the difference graph of those eight different genotypes produced by marker 1tc1g using the genotype 17KGR1 as the reference. Each one of those genotypes had different fragment size (114–140 bp) as confirmed by separation in polyacrylamide gel after electrophoresis (Figure 3). Melting profiles plotted within the 80% confidence interval of each of these eight genotypes were considered as identical. The results of the other markers used were similar, showing a clear discrimination of most of the genotypes.
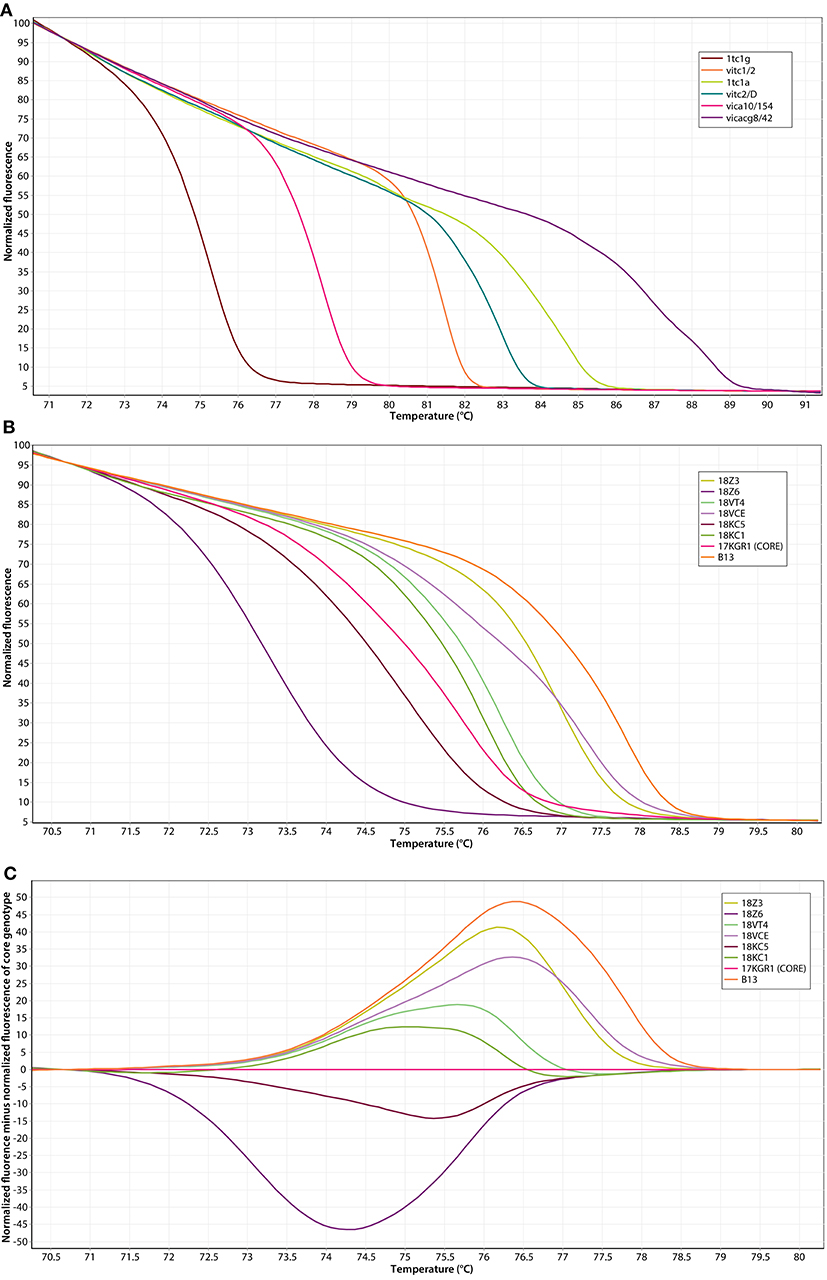
Figure 1. Discrimination of V. inaequalis genotypes using six SSR markers and high resoultion melting (HRM) analysis. (A) HRM profiles of the core genotypes analyzed with the markers 1tc1g, 1tc1a, vitc1/2, vitc2/D, vica10/154, and vicacg8/42. (B) Normalized HRM melting curves of eight unique genotypes using the microsatellite marker 1tc1g. (C) Difference graph of those eight unique genotypes using the 17KGR1 as the reference genotype.
According to the inter-population AMOVA analysis of the SSR-HRM markers, there were significant (p < 0.001) genetic differences among the five Venturia inaequalis populations (Table 1). AMOVA also revealed a high percentage of variation due to population subdivision (96%) while only 4% of the total gene diversity was attributed to among-population differentiation. The genetic relationship was further validated by the principal coordinate analysis (PCoA), in which the individual distance matrix was used as input. Coordinates 1 and 2 explained 9.33 and 8.68% of the variance for panel A (Figure 2). Concerning panel B (PCoA for five populations), the population distance matrix was used as input, in which coordinates 1 and 2 explained 51.81 and 26.33% of the variance (Figure 2).
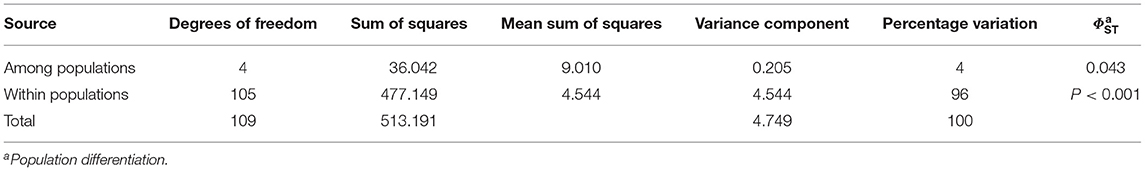
Table 1. Analysis of molecular variance results for five V. inaequalis populations based on SSR-HRM markers.
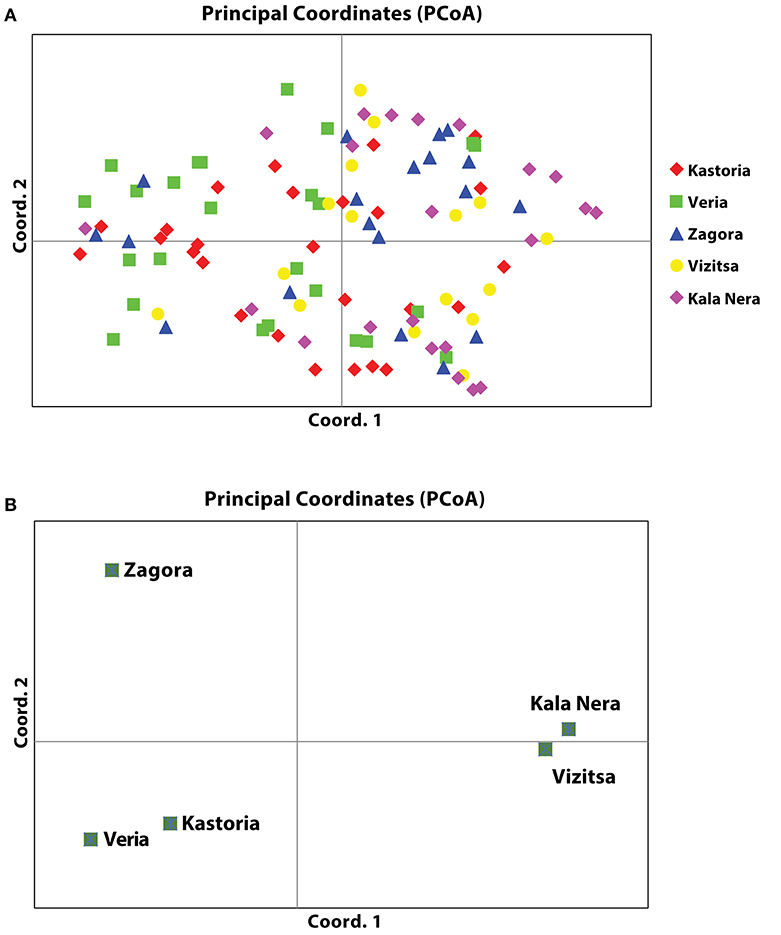
Figure 2. Genetic relationships of five populations of V. inaequalis based on SSR-HRM markers. (A) Principal coordinate analysis (PCoA) of 110 individuals in five populations of V. inaequalis based on 46 polymorphic SSR-HRM loci. 18.01% of total variance accumulated on the first two components (axis 1 = 9.33%; axis 2 = 8.68%). (B) Collapsed PCoA depicting relationships among populations (axis 1 = 51.81%; axis 2 = 26.33%). Samples included are indicated after population name.
Discussion
To genotype V. inaequalis populations originating from the main Greek apple producing areas, six different microsatellite markers were analyzed using HRM analysis. The method was found capable of discriminating all the genotypes in the flanking region and a reference pattern for genotyping samples was developed. HRM has proven to be a time-efficient and labor-saving methodology that facilitates high throughput analysis (Chatzidimopoulos et al., 2014a). In the present assay a large number of unknown samples was genotyped by using reference melting curves in each PCR-HRM run. Among different runs the reference plots were constant, maintaining the same melting peaks. Furthermore, it has been shown that samples can be screened for sequence variations with different primers in just a single run, provided that primers melting temperatures and reagents are similar.
The selection of SSRs in coupling with HRM analysis is more preferable than RAPDs or RFLPs because of the shorter amplicon size. It is well-documented that amplicons with size smaller than 300 bp are separated very accurately by HRM analysis (Reed and Wittwer, 2004). In plants, the HRM-SSR approach has been successfully used for genotyping and authenticity testing (Mackay et al., 2008; Ganopoulos et al., 2011; Distefano et al., 2012; Xanthopoulou et al., 2014). The development and validation of an HRM protocol to discriminate SSR genotypes in plant pathogenic fungi has not yet been reported. The HRM is mainly used in species identification like Alternaria and Aspergillus (Garganese et al., 2018; Xanthopoulou et al., 2019) or SNP detection for resistance like in Botrytis cinerea and Colletotrichum spp. (Chatzidimopoulos et al., 2014b; Forcelini et al., 2018). The data of the present assay suggest that HRM is most suitable for SSR genetic analysis in the haploid fungus V. inaequalis.
Compared to the limitations found in diploid organisms, SSR genotyping with HRM is much simpler in plant pathogenic fungi. Distefano et al. (2012) found many variants in flanking sequences of Citrus species. The occurrence of homoplasy, the large number of alleles among species and the presence of SNPs in the flanking region are all factors that contribute to the generation of inconsistent melting profiles. Those limitations were also observed by Yang et al. (2014) in SSR-HRM genotyping in sheep. In the present assay, the melting curves were genotype-specific and one melting peak per isolate was generated in each SSR locus, consistent with the haploid nature of V. inaequalis (Koopman et al., 2017). To validate the HRM profiles, the size of some of the amplicons was determined on agarose or polyacrylamide gels (Figure 3). However, in most agarose gels the bands were very similar in size despite the fact that the amplicons showed different melting profiles in HRM analysis. It has been demonstrated that the potential resolving power of the HRM approach is much greater than conventional gel or melting curve analysis (Ganopoulos et al., 2011). Furthermore, Distefano et al. (2012) showed that HRM is even more sensitive than capillary electrophoresis fragment size analysis. However, it has been shown that alleles of the same size are not always characterized by identical sequence (Distefano et al., 2012). In the present assay several PCR products were sequenced to correlate the HRM curves with a specific genotype but the chromatogram data contained many ambiguous base calls. The possible presence of SNPs would generate a different melting profile, which would correspond to a different genotype. Ganopoulos et al. (2011) have shown that melting curves from different amplicons can be differentiated on the basis of shape even when they define the same Tm values and concluded that the shape of the HRM curves is more informative than melting curve analysis. Those are the main reasons for which HRM was selected over other techniques to genotype the apple scab populations.
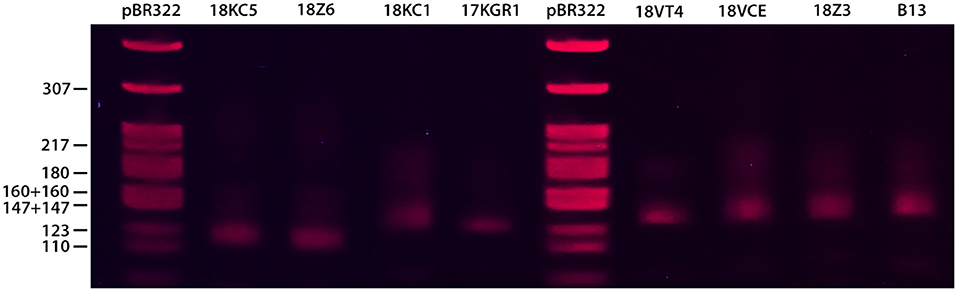
Figure 3. Fragment analysis of the eight reference genotypes used in HRM analysis with SSR marker 1tc1g in polyacrylamide gel following electrophoresis and staining with ethidium bromide.
In the present assay, HRM ensured reliable interpretations of the melting curve profiles of the genotypes because the SSRs showed low complexity in the flanking region. However, one important limitation of the present assay was the formation of stutter products due to slipped strand mispairing. Such issues are quite common in microsatellite genotyping (Butler, 2015). The consequence of this is a shortened PCR product, smaller than the primary amplicon (SSR allele) by a single repeat unit. A similar limitation was also noted by Tenzer et al. (1999) in a V. inaequalis population genetics study. The formation of two amplicons was quite visible in melting curve analysis performed by Rotor-gene Q software. To overcome this issue, genotypes were aligned to the appropriate reference genotypes by comparing a) the melting curve shape of the mispaired sample with the standard reference curve (more obvious in HRM difference plotting) and b) the melting peaks in melting curve analysis. Usually, those two parameters were correlated (similar melting curve shapes had similar melting peaks).
The genetic analysis of apple scab populations originated from different regions in Greece revealed high genetic variation within the populations (96%) while only 4% of the total gene diversity was attributed to among-population differentiation. Those results are in accordance with previous studies in V. inaequalis populations conducted in Europe, Asia and Africa (Gladieux et al., 2010; Ebrahimi et al., 2016; Koopman et al., 2017). PCoA grouped the isolates into three populations. Isolates from the areas of Kastoria and Veria that are in the same latitude (Northern Greece) grouped together. The other three populations originated from different mountainsides of the same area. However, PCoA showed that one population was distant from the other two which were grouped together. Zagora is an isolated area and separated from Kala Nera and Vizitsa by the Pelion mountain. On previous studies it has been shown that mountains are physical barriers that prevent the pathogen to migrate between different areas (Gladieux et al., 2010). Despite the fact that Kastoria and Veria are also separated by mountains, the populations showed little differentiation. Most of the apple cultivars grown on those areas are originated from the same area and the transportation of trees from nurseries may move inoculum from the one area to another (Koopman et al., 2017).
Another factor that can explain the genetic diversity of the populations in different regions is the selection of resistant subpopulations due to fungicide pressure. In Mediterranean areas the pathogen reproduces sexually and control strategies are targeting the primary infections with ascospores in spring. A recent survey in the same areas of sampling revealed the occurrence of resistant strains of V. inaequalis to several fungicides including dodine, difenoconazole, cyprodinil, and trifloxystrobin (Chatzidimopoulos et al., 2018). Interestingly, the sensitivity levels of the populations to the fungicides were highly correlated to the groups formed by PCoA. The populations of the pathogen in Kastoria and Veria showed similar resistance levels to the fungicides tested while populations from the other areas had different sensitivities. The different timing and programmes of fungicide applications combined with the recombination opportunities may lead to high variation in fungal genome among different areas (Boehm et al., 2003).
Conclusion
It was shown that SSR genotyping in V. inaequalis with HRM analysis can be a fast and cost-effective tool (HRM requires a fluorescent stain and typical PCR reagents), suitable to replace gel and capillary electrophoresis. The developed HRM protocol generated genotype-specific melting curves, consistent with the haploid nature of the fungus. For the first time unknown samples were genotyped using standard samples as reference controls among multiple runs. The genetic background of V. inaequalis isolates, originated from major apple growing areas, was recorded for the first time in Greece and showed high genetic variation within these populations.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
MC, FL, and EV: conceived and designed the experiments. MC, FL, ON, and EM-D: performed the experiments. MC and IG: analyzed the data. MC, FL, PP, and EV: contributed reagents, materials, analysis tools. MC and IG (partly): wrote the paper.
Funding
This research project was funded by the State Scholarships Foundation (I.K.Y.) under the Action Support of Postdoctoral Researchers in the framework of Operational Programme Human Resources Development, Education and Lifelong Learning with priority axes 6, 8, 9. It was co-funded by the European Social Fund and by National Resources through the National Strategic Reference Framework 2014–2020 (NSRF 2014–2020).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agarwal, M., Shrivastava, N., and Padh, H. (2008). Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 27, 617–631. doi: 10.1007/s00299-008-0507-z
Boehm, E. W. A., Freeman, S., Shabi, E., and Michailides, T. J. (2003). Microsatellite primers indicate the presence of asexual populations of Venturia inaequalis in coastal Israeli apple orchards. Phytoparasitica 31, 236–251. doi: 10.1007/BF02980833
Bus, V. G., Rikkerink, E. H., Caffier, V., Durel, C.-E., and Plummer, K. M. (2011). Revision of the nomenclature of the differential host-pathogen interactions of Venturia inaequalis and Malus. Annu. ReV. Phytopathol. 49, 391–413. doi: 10.1146/annurev-phyto-072910-095339
Butler, J. M. (2015). “Chapter 3—STR alleles and amplification artifacts,” in Advanced Topics in Forensic DNA Typing: Interpretation, ed J. M. Butler (San Diego, CA: Academic Press), 47–86. doi: 10.1016/B978-0-12-405213-0.00003-8
Chatzidimopoulos, M., Ganopoulos, I., Madesis, P., Vellios, E., Tsaftaris, A., and Pappas, A. C. (2014a). High-resolution melting (HRM) analysis for rapid detection and characterization of Botrytis cinerea phenotypes resistant to fenhexamid and boscalid. Plant Pathol. 63, 1336–1343. doi: 10.1111/ppa.12210
Chatzidimopoulos, M., Ganopoulos, I., Vellios, E., Madesis, P., Tsaftaris, A., and Pappas, A. C. (2014b). Development of a two-step high-resolution melting (HRM) analysis for screening sequence variants associated with resistance to the QoIs, benzimidazoles and dicarboximides in airborne inoculum of Botrytis cinerea. FEMS Microbiol. Lett. 360, 126–131. doi: 10.1111/1574-6968.12594
Chatzidimopoulos, M., Lioliopoulou, F., and Vellios, E. K. (2018). “Characterization and distribution of fungicide resistant Venturia inaequalis phenotypes in Greece,” in 19th Hellenic Phytopathological Congress: Hellenic Phytopathological Society (Athens), 160.
Distefano, G., Caruso, M., La Malfa, S., Gentile, A., and Wu, S.-B. (2012). High resolution melting analysis is a more sensitive and effective alternative to gel-based platforms in analysis of SSR—An example in Citrus. PLoS ONE 7:e44202. doi: 10.1371/journal.pone.0044202
Ebrahimi, L., Fotuhifar, K.-B., Javan Nikkhah, M., Naghavi, M.-R., and Baisakh, N. (2016). Population genetic structure of apple scab (Venturia inaequalis (Cooke) G. Winter) in Iran. PLoS ONE 11:e0160737. doi: 10.1371/journal.pone.0160737
Forcelini, B. B., Lee, S., Oliveira, M. S., and Peres, N. A. (2018). Development of figh-throughput SNP genotyping assays for rapid detection of strawberry Colletotrichum species and the G143A mutation. Phytopathology 108, 1501–1508. doi: 10.1094/PHYTO-04-18-0128-R
Ganopoulos, I., Argiriou, A., and Tsaftaris, A. (2011). Microsatellite high resolution melting (SSR-HRM) analysis for authenticity testing of protected designation of origin (PDO) sweet cherry products. Food Control 22, 532–541. doi: 10.1016/j.foodcont.2010.09.040
Garganese, F., Ippolito, A., di Rienzo, V., Lotti, C., Montemurro, C., and Sanzani, S. M. (2018). A new high-resolution melting assay for genotyping Alternaria species causing citrus brown spot. J. Sci. Food Agric. 98, 4578–4583. doi: 10.1002/jsfa.8986
Gladieux, P., Zhang, X.-G., Róldan-Ruiz, I., Caffier, V., Leroy, T., Devaux, M., et al. (2010). Evolution of the population structure of Venturia inaequalis, the apple scab fungus, associated with the domestication of its host. Mol. Ecol. 19, 658–674. doi: 10.1111/j.1365-294X.2009.04498.x
Guérin, F., Franck, P., Loiseau, A., Devaux, M., and Le Cam, B. (2004). Isolation of 21 new polymorphic microsatellite loci in the phytopathogenic fungus Venturia inaequalis. Mol. Ecol. Notes 4, 268–270. doi: 10.1111/j.1471-8286.2004.00637.x
Köller, W., and Wilcox, W. F. (2001). Evidence for the predisposition of fungicide-resistant Isolates of Venturia inaequalis to a preferential selection for resistance to other fungicides. Phytopathology 91, 776–781. doi: 10.1094/PHYTO.2001.91.8.776
Koopman, T. A., Meitz-Hopkins, J. C., Bester-van der Merwe, A. E., Tobutt, K. R., Bester, C., and Lennox, C. L. (2017). Genetic diversity and gene flow of four South African Venturia inaequalis (apple scab) populations. Phytopathology 107, 455–462. doi: 10.1094/PHYTO-07-16-0279-R
MacHardy, W. E. (1996). Apple Scab: Biology, Epidemiology, and Management. St. Paul, MN: American Phytopathological Society (APS Press).
Mackay, J. F., Wright, C. D., and Bonfiglioli, R. G. (2008). A new approach to varietal identification in plants by microsatellite high resolution melting analysis: application to the verification of grapevine and olive cultivars. Plant Methods 4:8. doi: 10.1186/1746-4811-4-8
McDonald, B. A. (1997). The population genetics of fungi: tools and techniques. Phytopathology 87, 448–453. doi: 10.1094/PHYTO.1997.87.4.448
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Reed, G. H., and Wittwer, C. T. (2004). Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 50, 1748–1754. doi: 10.1373/clinchem.2003.029751
Tenzer, I., degli Ivanissevich, S., Morgante, M., and Gessler, C. (1999). Identification of microsatellite markers and their application to population genetics of Venturia inaequalis. Phytopathology 89, 748–753. doi: 10.1094/PHYTO.1999.89.9.748
Tenzer, I., and Gessler, C. (1997). Subdivision and genetic structure of four populations of Venturia inaequalis in Switzerland. Eur. J. Plant Pathol. 103, 565–571. doi: 10.1023/A:1008636913211
Vossen, R. H., Aten, E., Roos, A., and den Dunnen, J. T. (2009). High-Resolution Melting Analysis (HRMA)—more than just sequence variant screening. Hum. Mutat. 30, 860–866. doi: 10.1002/humu.21019
Xanthopoulou, A., Ganopoulos, I., Koubouris, G., Tsaftaris, A., Sergendani, C., Kalivas, A., et al. (2014). Microsatellite high-resolution melting (SSR-HRM) analysis for genotyping and molecular characterization of an Olea europaea germplasm collection. Plant Genet. Resour. 12, 273–277. doi: 10.1017/S147926211400001X
Xanthopoulou, A., Ganopoulos, I., Tryfinopoulou, P., Panagou, E. Z., Osanthanunkul, M., Madesis, P., et al. (2019). Rapid and accurate identification of black aspergilli from grapes using high-resolution melting (HRM) analysis. J. Sci. Food Agric. 99, 309–314. doi: 10.1002/jsfa.9189
Xu, X., Harvey, N., Roberts, A., and Barbara, D. (2013). Population variation of apple scab (Venturia inaequalis) within mixed orchards in the UK. Eur. J. Plant Pathol. 135, 97–104. doi: 10.1007/s10658-012-0068-4
Yang, M., Yue, Y. J., Guo, T. T., Han, J. L., Liu, J. B., Guo, J., et al. (2014). Limitation of high-resolution melting curve analysis for genotyping simple sequence repeats in sheep. Genet. Mol. Res. 13, 2645–2653. doi: 10.4238/2014.April.8.7
Keywords: HRM, SSR, Venturia inaequalis, apple scab, population genetics
Citation: Chatzidimopoulos M, Ganopoulos I, Moraitou-Daponta E, Lioliopoulou F, Ntantali O, Panagiotaki P and Vellios EK (2019) High-Resolution Melting (HRM) Analysis Reveals Genotypic Differentiation of Venturia inaequalis Populations in Greece. Front. Ecol. Evol. 7:489. doi: 10.3389/fevo.2019.00489
Received: 20 July 2019; Accepted: 29 November 2019;
Published: 13 December 2019.
Edited by:
Denis Baurain, University of Liège, BelgiumReviewed by:
Frank Dunemann, Julius Kühn-Institut, GermanyClara Valero, University of São Paulo, Brazil
Copyright © 2019 Chatzidimopoulos, Ganopoulos, Moraitou-Daponta, Lioliopoulou, Ntantali, Panagiotaki and Vellios. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Chatzidimopoulos, bXhhdHppZEBhZ3IudXRoLmdy
 Michael Chatzidimopoulos
Michael Chatzidimopoulos Ioannis Ganopoulos
Ioannis Ganopoulos Eleni Moraitou-Daponta1
Eleni Moraitou-Daponta1 Fenia Lioliopoulou
Fenia Lioliopoulou Olga Ntantali
Olga Ntantali Evangelos K. Vellios
Evangelos K. Vellios