
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol., 09 October 2019
Sec. Biogeography and Macroecology
Volume 7 - 2019 | https://doi.org/10.3389/fevo.2019.00367
This article is part of the Research TopicEcological Development and Functioning of Biological Soil Crusts After Natural and Human DisturbancesView all 16 articles
 Michaeline B. N. Albright1*
Michaeline B. N. Albright1* Rebecca C. Mueller1,2
Rebecca C. Mueller1,2 La Verne Gallegos-Graves1
La Verne Gallegos-Graves1 Jayne Belnap3
Jayne Belnap3 Sasha C. Reed3
Sasha C. Reed3 Cheryl R. Kuske1*
Cheryl R. Kuske1*Dryland grasslands are vast and globally important and, as in all terrestrial ecosystems, soil microbial communities play fundamental roles in regulating dryland ecosystem function. A typical characteristic of drylands is the spatial mosaic of vascular plant cover surrounded by interspace soils, where biological soil crusts (biocrusts)—a complex community of organisms including bacteria, fungi, algae, mosses, and lichens—are common. The implications of this heterogeneity, where plants and biocrust cover co-occur, are often explored in the context of soil fertility and hydrology, but rarely has the impact of these multiple microhabitat types been simultaneously explored to determine the influence on bacterial and fungal communities, key biological players in these ecosystems. Further, our understanding of the temporal dynamics of bacterial and fungal communities in grasslands, and of how these dynamics depend on the microhabitat within the ecosystem, is notably poor. Here we used a temporally and spatially explicit approach to assess bacterial and fungal communities in a grassland on the Colorado Plateau, and to link variation in these communities to edaphic characteristics. We found that microhabitat (e.g., vascular plant rhizosphere, biocrust, and below biocrust) was the strongest driver of differences in bacterial and fungal community richness, diversity, and composition. Microhabitat type also significantly mediated the impact of temporal change in shaping community composition. Taken together, 29% of the variation in bacterial community composition could be explained by microhabitat, date, and microhabitat-by-date interactions, while only 11% of the variation in fungal community composition could be explained by the same factors, suggesting important differences in community assembly processes. Soil microbial communities dictate myriad critical ecosystem functions, thus understanding the factors that control their compostition is crucial to considering and forecasting how terrestrial ecosystems work. Overall, this case study provides insights for future studies on the spatial and temporal dynamics of bacterial and fungal communities in dryland grasslands.
Semiarid, arid, and hyperarid (hereafter, dryland) ecosystems cover over 40% of the terrestrial surface across the globe (Safriel et al., 2005) and ~35% of the western US (Pointing and Belnap, 2012). Dryland grasslands play particularly important roles in drylands: grasslands are home to many endemic plants and animals, are critical for livestock and wildlife grazing, are hotspots of recreation, and are at risk under global change (Maestre et al., 2016). In dryland ecosystems in general, and in the high deserts of the southwestern US in particular, high climate variability is the norm, with temperature and precipitation fluctuating on timescales ranging from hours to seasons to centuries (Sheppard et al., 2002). The high variability in precipitation and temperature makes it a challenging environment for species survival; despite this, a diversity of plants and microorganisms have adapted to these conditions and these organisms have helped shape the unique landscape. This landscape is heterogenous, typically comprised of sparsely distributed plants and numerous types of biological soil crusts (biocrusts) covering the interspace soils among plants (Weber et al., 2016). Biocrusts are diverse and can be formed by cyanobacteria, algae, fungi, lichens, and/or mosses (Weber et al., 2016). The heterogeneity in dryland biota (e.g., vascular plants surrounded by interspace soils; Figure S1) creates a variety of diverse microhabitats, each with distinct soil bacterial and fungal communities (Grondin and Johansen, 1993; Wheeler et al., 1993; Bates et al., 2010, 2012; Steven et al., 2012, 2013, 2014; Yeager et al., 2012; Patzelt et al., 2014; Mueller et al., 2015; McHugh et al., 2017). These bacterial and fungal communities are key players in determining ecosystem functioning by contributing to soil stabilization, hydrology, and nutrient cycling (Belnap, 2006; Maestre et al., 2011, 2016; Bowker et al., 2013).
Our understanding of how microhabitats structure bacterial and fungal communities in drylands is far from complete. However, compared to our knowledge of microhabitat variation, even less is known about temporal variation in bacterial and fungal composition in arid grassland ecosystems. Early studies on seasonal changes in biocrusts used microscopy to look at changes in communities in response to climatic factors (Johansen, 1984; Johansen and Rushforth, 1985). A few more recent studies have tracked temporal variation in bacterial and fungal biomass and composition in drylands using chemical techniques (Bowker et al., 2002; Bell et al., 2009); for example, seasonal differences were observed in the community composition of desert cyanobacterial soil crusts (Bowker et al., 2002; Yeager et al., 2012). However, systematic assessments of dryland grassland bacterial and fungal communities through time are exceedingly rare. Nevertheless, studies from other ecosystems, including agricultural, mesic grasslands, alpine, arctic tundra, and rainforest systems, have shown the potential for dramatic temporal changes in bacterial and/or fungal community composition (Smit et al., 2001; Lipson et al., 2002; Griffiths et al., 2003; Wallenstein et al., 2007; Bjork et al., 2008; Matulich et al., 2015; Smith et al., 2015; Kivlin and Hawkes, 2016). Studies from other ecosystems have also examined the relative impacts of microhabitat variation (most often due to differences in plant composition) compared to the effects of season on bacterial and/or fungal composition (i.e., these studies have simultaneously assessed the control of spatial and temporal variation), with varied results. In both an alpine tundra (Bjork et al., 2008) and a rainforest (Kivlin and Hawkes, 2016), soil microbial community changes over time were of greater magnitude than differences attributed to association with a specific plant community. In contrast, plant composition (tussock or shrub) was a stronger regulator of microbial communities than season in Arctic tundra soils (Wallenstein et al., 2007). The variability in the magnitude of temporal versus microhabitat effects across ecosystems is perhaps not unexpected, as temporal changes at a location may be affected by numerous environmental (e.g., temperature, soil moisture) and ecological (e.g., dispersal) factors, and these factors may be highly variable and location dependent. Nevertheless, tracking seasonal dynamics concurrently with microhabitat controls has the potential to broaden our understanding of controls over the diversity of bacteria and fungi, especially those living in extreme environments (Schadt et al., 2003).
Patchy landscapes with distinct microhabitats caused by the presence or absence of vascular plants or different soil surface covers (Figure S1) are ideal for examining temporal changes in dissimilar environments while keeping other factors, such as climate and land use history, constant. However, to our knowledge no study has simultaneously evaluated how dryland grassland bacterial and fungal communities vary by microhabitat type and through time, leaving interactions between these likely controls unexplored. Here, we assessed how microhabitats and time separately and together impact bacterial and fungal community composition in a semiarid grassland ecosystem. Specifically, we asked: (1) How do changes in community composition across microhabitats compare to changes across season? and (2) Do the temporal community dynamics differ with microhabitat type? We hypothesized that microhabitats would exert greater control over bacterial and fungal community composition than temporal variation. Microhabitats generally vary in pH, soil texture, sunlight/UV exposure, temperature, and other environmental factors that control the composition of the microflora (Steven et al., 2013). Furthermore, we hypothesized that within the interspace microhabitats, the below-biocrust communities would change less than the biocrust communities. This is because surface communities are likely subject both to more extreme climatic changes (e.g., Tucker et al., 2017) and higher rates of microorganism dispersal, which are buffered in subsurface soils. Understanding the relative roles of spatial and temporal variation can provide insights into the drivers of community assembly in extreme environments, and provide a baseline from which to understand the effects of anthropogenic changes on grassland bacterial and fungal communities.
This study was conducted in a semiarid grassland located near Castle Valley, Utah, USA and close to a long-term climate manipulation experiment on the Colroado Plateau (38°40′26.52″ N, 109°24′59.27″ W; Wertin et al., 2015). The site has a history of limited- to no direct anthropogenic impacts prior to establishment of the study and the soils are classified as sandy loam, calcareous, Rizno series (Grand County Soil Survey; Winkler et al., 2019). Air temperature and precipitation values were collected from a weather station located ~200 m from the collection site (Figure S1). The average annual precipitation was 257 mm and the mean annual temperature 14.0°C during our study period (2013–2014). This was similar to longer-term averages (2006–2018) collected at the same weather station, where average annual precipitation was 236 mm and average annual temperature 14.3°C. Soil temperature was not measured at sampling locations, but in drylands, soil, and air temperatures are often highly correlated (Bell et al., 2008).
The site's vegetation was dominated by the native perennial grasses Pleuraphis jamesii (syn. Hilaria jamesii; James' galleta) and Achnatherum hymenoides (syns. Stipa hymenoides and Oryzopsis hymenoides; Indian ricegrass) and the exotic invasive grass Bromus tectorum (cheatgrass). Biocrust communities were dominated by the cyanobacterium Microcoleus vaginatus, the cyanolichens Collema tenax and C. coccophorum, and the moss Syntrichia caninervis. Soils were collected from three soil locations representing distinct general microhabitats: (1) rhizosphere samples were taken from beneath the base of the native perennial grass P. jamesii and the exotic invasive annual grass B. tectorum; (2) biocrust samples were collected from plant interspaces and included three types of biocrusts as characterized by the dominant organisms (lichen, moss, and cyanobacteria); and (3) soils underlying lichen-dominated and cyanobacteria-dominated biocrusts. At each sampling point, four replicates per microhabitat were collected. Microhabitat sampling locations within the site were randomly selected at each time point. Soil samples were taken monthly from April 2013 to April 2014, but due to extreme snow conditions, the study sites were not accessible from December 2013 to February 2014, resulting in a total of 10 sampling timepoints over the 13-month period. In addition to collecting soils for microbial community assessments, samples were concurrently collected from each microhabitat type for soil chemistry. Biocrust samples were collected at 0–0.5 cm, while rhizosphere and below-surface soil samples were collected from 0.5 to 5 cm.
To assess whether dust blowing in the area contained significant amounts of bacteria and fungi that might contribute to soil populations, we also analyzed samples collected from two nearby sites. Sample were collected using “Big Springs Number Eight” (BSNE; Fryrear, 1986) sediment collectors placed 15, 50, and 100 cm above the soil surface (Flagg et al., 2014). Samples from the three heights were pooled for microbial DNA extractions. Dust samples were collected at four timepoints during 2012 (March, July, September) and 2013 (February).
In order to characterize soil chemical differences among microsites, bulk soil chemistry measurements were obtained for composite samples from fall (October and November 2013) and spring (April and May 2014) (e.g., organic matter, total carbon, and nitrogen; Table S1) as soil chemical sampling at all timepoints was beyond the capacity of the project. Spring and late summer/fall are times of year when vascular plants are most active in this region (Wertin et al., 2015), thus April–May and October–November samplings were selected in order to best elucidate differences between plant-associated vs. interspace soil characteristics. Soil chemical analyses were performed by the Colorado State University Soil, Water, and Plant Testing Laboratory (Fort Collins, CO; www.soiltestinglab.colostate.edu) using their standard “routine soil and overburden” approach. Additionally, pH was measured for all individual samples at each timepoint. This was done by suspending 1 g of sample in 10 ml of distilled water. Samples were homogenized using a vortexter, allowed to settle for ~5 min, and pH was measured using a Mettler Toledo pH meter (Mettler-Toledo, LLC, Ohio, USA).
Total nucleic acids were extracted from 0.5 g of soil using the Fast DNA for Soil kit (MP Biomedical) according to the manufacturer's instructions (http://www.mgp.cz/files/kity/FastDNASPINkit.PDF). To quantify changes in soil microbial communities over time and across microhabitats, we examined bacterial and archaeal (hereafter referred to as bacterial) and fungal communities using Illumina high throughput sequencing. Samples were prepared for sequencing on the Illumina MiSeq platform using a two-stage protocol described previously (Mueller et al., 2015). To target the bacterial community, we amplified the V3–V4 region of the 16S ribosomal RNA gene using the F515-R806 primer pair and for the fungal communities, we amplified the LSU D2 hypervariable region using the LR22R-LR3 primer pair (Mueller et al., 2015). Paired-end 250 bp sequences were generated using the Illumina MiSeq platform. A single 12 bp barcode was generated from the combinatorial barcodes and overlapping reads were joined into a single sequence using PEAR (Zhang et al., 2014) with a minimum overlap of 20 bp. Samples were de-multiplexed using QIIME (Caporaso et al., 2010) and any sequence with a mismatch to the barcode or forward primer was removed. Additional quality filtering and OTU clustering was conducted using UPARSE (Edgar, 2013) to remove any sequence with an expected error rate >0.5 and singleton sequence putative chimeras were identified against all de novo sequences and eliminated using UCHIME. OTUs were delineated at 97% sequence similarity for both 16S and LSU sequences. Representative sequences for each OTU bin were classified using the Ribosomal Database Project (RDP) online classifier tool (http://rdp.cme.msu.edu/) against the RDP LSU database and the 16S rRNA gene databases (Wang et al., 2007). Sequences classified to Phylum or Domain with a bootstrap <80% were removed from downstream analysis. OTU tables with taxonomic classifications are provided for fungi and bacteria (Tables S2, S3). Cyanobacteria are included in bacterial OTU analysis in RDP, however, additional classification for Cyanobacteria sequences were also performed using DECIPHER (Murali et al., 2018) with the Genome Taxonomy Database (Parks et al., 2018) (Table S4). Bacterial and Fungal OTU tables were exported for analysis in R (www.R-project.org). Using the OTU tables, we generated a rarefied composition table, randomly drawing the lowest common number of sequences (n = 1,023, fungi and n = 1,663, bacteria). We then calculated a Bray-Curtis distance matrix that was used in the remaining analyses (Oksanen et al., 2018). Richness and shannon diversity of rarified OTU matrix libraries was also calculated in R. Fastq files used in this analysis were deposited to the MG-RAST database (mgp87567, https://www.mg-rast.org/linkin.cgi?project=mgp87567). Data can be downloaded from MG-RAST ID links for bacteria (mgm4824928.3) and fungi (mgm4824929.3). Additionally, we have added a supplemental metadata file for deposited sequence data (Table S5).
To test for differences in bacterial and fungal richness and diversity across microhabitat and sampling date and to estimate the variance explained by each factor, we used a two-way ANOVA design with microhabitat and date as main fixed factors, and a microhabitat-by-date interaction. Differences in pH across microhabitat and date were also assessed using a two-way ANOVA. For significant factors, Tukey's HSD post-hoc tests were used to identify which groups within these factors were significantly different from each other. Using the same factors of microhabitat, date, and microhabitat-by-date, for the multivariate metric of community composition we performed a permutational multivariate analysis of variance (PERMANOVA), using type III partial sums of squares under a reduced model with 999 permutations. We estimated the percent of variation that could be attributed to each significant term for both the ANOVA and PERMANOVA analyses (Quinn and Keough, 2002). Analyses were run separately for bacteria and fungi.
We assessed correlations between environmental parameters, including air temperature, precipitation, and pH, and community metrics (richness, diversity, and composition). For precipitation, we used the cumulative precipitation over the 2 weeks prior to the sampling date (Figure S2). Pearson's correlations were used for univariate community metrics (richness, diversity), while Mantel tests were used for multivariate community metrics. For Mantel tests, each environmental parameter (pH, precipitation, air temperature) was used to generate a Euclidean distance matrix, while Bray-curtis distance matrices were used for bacterial and fungal community composition.
Correlations between temporal distance and community similarity for each individual microhabitat were calculated using Mantel tests. Pairwise temporal distances were measured based on the number of days that separated each sampling time point. All statistical analyses were conducted using the statistical platform R with the packages vegan, phyloseq (McMurdie and Holmes, 2013), picante (Kembel et al., 2010), and ecodist (Goslee and Urban, 2007). P ≤ 0.05 were defined as significant.
Soil chemistry measurements showed significant differences among habitat types (Table S1). Organic matter (OM) ranged from 1.5 to 6.1% and was highest in moss biocrust [5.0% (fall), 6.1% (spring)] and lowest in soils below lichen biocrusts [1.6% (fall), 1.5% (spring)] (Table S1). Total nitrogen (N) concentrations ranged from 0.03 to 0.17% and were higher in the lichen biocrust (0.08–0.17%) and moss biocrust (0.08–0.10%) compared to the other microhabitats. Carbon (C) concentrations were highest in the lichen biocrust [2.5% (fall), 3.5% (spring)]. C:N ratios varied across microhabitat type, with higher C:N ratios in the soil beneath cyanobacteria and lichen biocrusts, and lower C:N ratios in lichen, moss and cyanobacteria biocrusts (Figure S3). The pH was sampled more intensively with measurements taken from each microhabitat at each sampling date. The pH varied across both microhabitats and time, but there was no interaction [two-way ANOVA; microhabitat: F (6, 140) = 86.5, p < 0.001, date: F (9, 140) = 6.7, p < 0.001, microhabitat-by-date: F (54, 140) = 1.3, p = 1.24] (Figures S4, S5).
Bacterial richness differed across microhabitat type, while fungal richness did not [two-way ANOVA; microhabitat: F (7, 143) = 95.1, P < 0.001 (bacteria), F (7, 143) = 1.87, P = 0.08 (fungi), Figures 1A,B]. Bacterial richness was highest in the rhizosphere soils (B. tectorum and P. jamesii) and below lichen biocrusts (Figures 1A,B). Despite the lack of differences in richness for fungi, both bacterial and fungal diversity (Shannon index) differed across microhabitats [two-way ANOVA; microhabitat: F (7, 143) = 46.3, P < 0.001 (bacteria), F (7, 143) = 4.0, P < 0.001 (fungi)]. Bacterial diversity was also highest for the rhizosphere soils and below lichen and was lowest for cyanobacteria biocrust and dust samples (Figure 1C). Bacterial diversity has previously been found as lower in biocrusts compared to surrounding soil communities (Gundlapally and Garcia-Pichel, 2006). Furthermore, cyanobacteria-dominated biocrusts generally are early in the successional sequence of biocrust organisms for these ecosystems (Belnap, 2003) and thus bacterial communities may not be as developed as in other microhabitats. Fungal diversity was lowest in the lichen crust, but similar across all other habitats (Figure 1D).
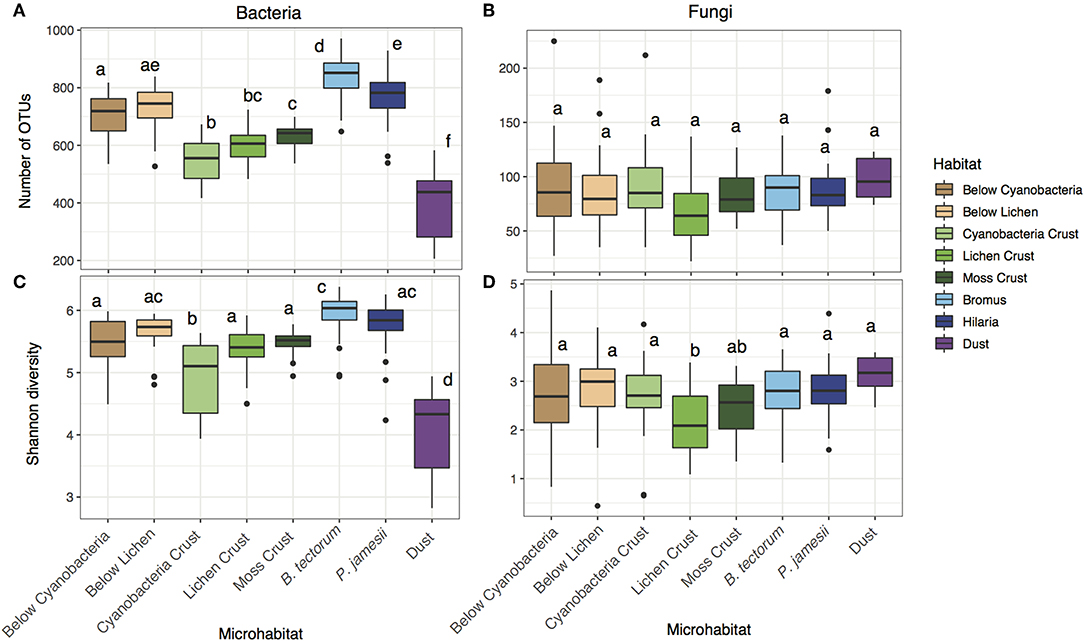
Figure 1. Microhabitat variation in (A) bacterial richness, (B) fungal richness, (C) bacterial diversity, and (D) fungal diversity.
Supporting our first hypothesis, microhabitat type was the strongest driver of differences in community composition for both bacteria and fungi [PERMANOVA; microhabitat: F (7, 143) = 13.3, P = 0.001 (bacteria), F (7, 143) = 5.4, P = 0.001 (fungi), Figure 2]. Overall, microhabitat explained an average 13.3% of estimated variation in bacterial community metrics, and 7.5% of estimated variation for fungal community metrics (Figure 3). Such differences in microbial communities in soils separated by only a few centimeters speaks to the role of microhabitat in structuring community patterns across a range of scales (Maier et al., 2018). Further, the microbial composition, and in particular the bacterial composition of soil associated with the invasive plant species B. tectorum, was distinct from composition associated with the native P. jamesii, as well as the biocrust communities (Figure 1, Figures S5–S8). Plant invasions, such as the spread of B. tectorum, an exotic annual grass that now dominates a large portion of the western US, are changing western U.S. drylands in many ways (Chapin et al., 2000; Belnap and Phillips, 2001; Compagnoni and Adler, 2014; Bradley et al., 2018). Among other influences, B. tectorum has altered plant biodiversity and nutrient cycling (Sperry et al., 2006; Norton et al., 2008), as well as microbial and microfaunal composition (Belnap and Phillips, 2001; Kuske et al., 2002; Belnap et al., 2005). For example, B. tectorum can reduce the ability of fungi to consume N (DeCrappeo et al., 2017). In addition, biocrusts have been shown to enhance B. tectorum growth likely through impacts on soil fertility (Ferrenberg et al., 2018). Our data support the conclusion that invasion by B. tectorum creates a new microhabitat type that helps dictate the extant soil microbial community.
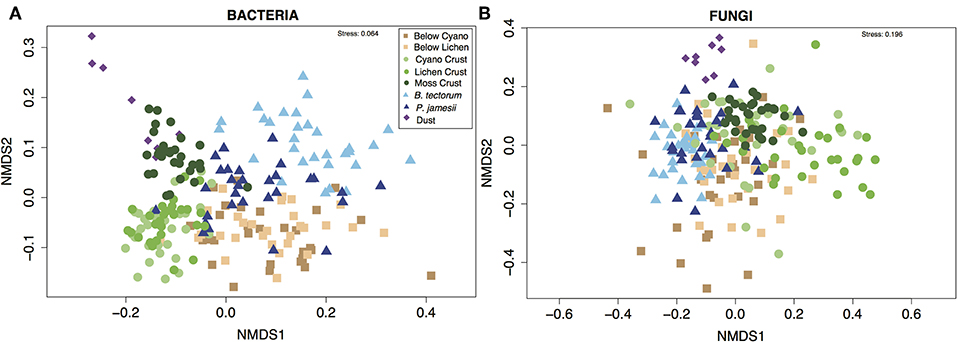
Figure 2. Non-metric multidimensional scaling (NMDS) ordinations showing (A) bacterial and (B) fungal community composition across different microhabitat types.
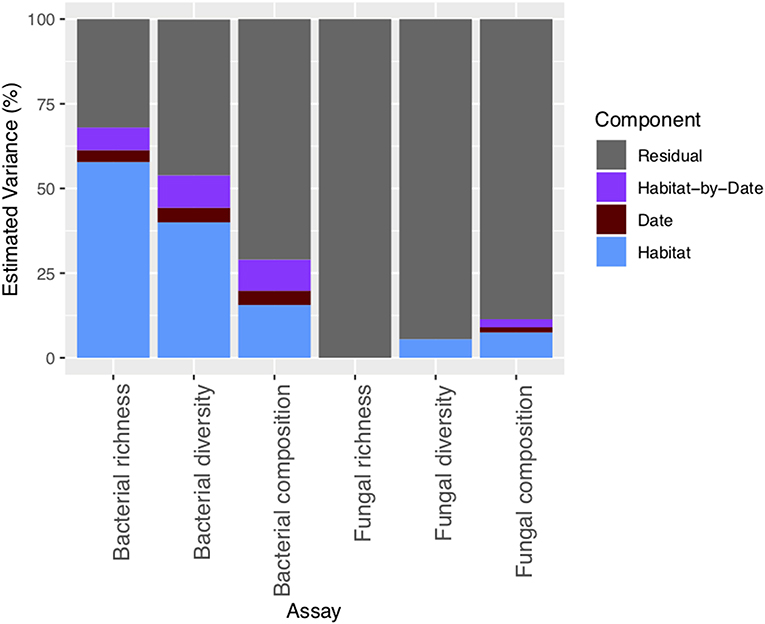
Figure 3. Percentage of estimated variation across bacterial and fungal richness, diversity, and composition metrics explained by the microhabitat, date, and microhabitat-by-date.
Many fungal taxa were common across rhizosphere soils of P. jamesii and B. tectorum (Figures S6, S8, Table S6). In addition, a number of fungal genera were most highly associated with B. tectorum, including Sclerotinia (plant pathogenic fungi), Saccobolus, Zygopleurage, Karstenula (saprotroph), Neofabraea (plant pathogen/saprotroph), Hydropisphaera, and Microsphaeropsis (saprotroph) (Figure S8, Table S6), as well as the bacterial genus Massilia, a copiotrophic root colonizing bacteria (Ofek et al., 2012) (Figure S9, Table S6). In the biocrust communities, the fungal genus Endocarpon was characteristic of cyanobacteria and lichen bicrusts, but was found in much lower abundance in rhizosphere soil communities. Fungal genera Psora, Hymenelia, and Aspicilia were also abundant in lichen biocrusts, while Basidiomycetes were more common in cyanobacteria biocrusts. Moss biocrusts were characterized by Iodophanus, also found in the plant-associated soils, as well as Lamprospora and Umbilicaria. Generally, microbial communities in moss biocrusts were more similar to the rhizosphere soils, which is interesting given that mosses are plants and, like plant roots, have structures called rhizines embedded in the soil (Jones and Dolan, 2012). Mosses in this area fix more CO2 compared with earlier-successional (e.g., cyanobacterial) biocrusts (Tucker et al., 2018), and thus carbon supply to the soil microbial community could be part of what supports a similar community between vascular plant roots and the soils beneath moss rhizines. Differences in the bacterial and fungal composition of biocrusts compared to non-biocrust samples may also be related to the soil depth. This study was limited to coarsely surveying complex communities through molecular techniques, future studies using additional techniques such as microscopy, culturing, and functional assays are needed to confirm the identity of and further characterize the microorganisms forming these complex communities.
Biocrusts (cyanobacteria, lichen, and moss) were dominated by Cyanobacteria and Thaumarchaeota (Nitrososphaera). The most abundant Cyanobacteria was Microcoleus (Phormidiaceae) (Table S4), which has previously been described as the most dominant cyanobacteria in Colorado plateau biocrusts (Garcia-Pichel et al., 2001). Other abundant Cyanobacterial families included Coleofasciculaceae and Nostocaceae (Table S4). The highest relative abundances of Thaumarchaeota were found in the soils underlying the cyanobacteria and lichen crusts (Figure S7), which were the most alkaline microhabitats with the highest C:N ratio (Figures S3, S4). Thaumarchaeota are ammonia-oxidizing archaea, that have previously been reported as significant contributors to biocrusts that may be important in soil crust N cycling (Soule et al., 2009; Marunsenko et al., 2013; Maier et al., 2018). Furthermore, previous work has found that soils ammonia-oxidizing archaea abundances are generally higher in more alkaline and limited soils (Gubry-Rangin et al., 2011). A number of other abundant bacteria genera in this study including Streptomyces, Rubrobacter, Paracoccus, Massilia, Bacillus, and Spirosoma have also been previously characterized in biocrusts from the Southwestern U.S. Colorado plateau region (Gundlapally and Garcia-Pichel, 2006) (Figure S9).
Mineral soil inputs of dust are most often discussed as nutrient inputs but less often considered as inputs of biological (microbial) material. Even so, an abundance of taxa known to play important roles in biocrusts have been documented in indoor and outdoor airborne environments (Despres et al., 2012; Barberan et al., 2015; Warren et al., 2019). We found that both the bacterial and fungal composition of dust were distinct from the soil samples (Figure 2). Furthermore, bacterial richness and diversity were lower in dust samples, while fungal dust samples did not differ from soils (Figure 1). This may be because fungal spores are generally tougher and thus less likely than bacterial cells to be degraded during the time span and harsh conditions of air travel and deposition (Kuske, 2006). Common fungal genera found in dust samples in higher abundances than in the other microhabitats we sampled included Ochrocaldosporium, Thelebolus, Chaetosphaeronema, and Camarosporium (Figure S8, Table S6). Thelebolus is known for ability as a thermophile to survive in extreme temperature conditions (de Hoog et al., 2005), while Camarosporium is known as a fungal plant pathogen (Wanasinghe et al., 2017). Dust samples were dominated by the bacterial genera Zhihengliuella, Oxalicibacterium, and Hymenobacter (Figure S9, Table S7). Hymenobacter has been characterized as a common atmospheric bacterium (Yooseph et al., 2013; Barberan et al., 2015). The most abundant phyla in dust samples was Deinococcus-Thermus (Figure S7), recognized as the most extremophilic phylum of bacteria (Theodorakopoulos et al., 2013).
Precipitation and pH were the two environmental parameters that significantly correlated with changes in microbial diversity in the grassland microhabitats, although these relationships were relatively weak. Both bacterial and fungal richness and diversity were positively correlated with pH [R2 = 0.34, p < 0.001 (bacteria richness) and R2 = 0.13, p = 0.05 (bacteria diversity), R2 = 0.15, p = 0.03 (fungal richness), and R2 = 0.21, p = 0.003 (fungal diversity)]. pH ranged from 6.9 to 8.4 where more alkaline soils had greater richness and diversity (Figure 1, Figure S4). Bacterial and fungal composition were significantly correlated with pH (Mantel test; R = 0.21, p = 0.001 and R = 0.06, p = 0.03, respectively). Considering climate controls, bacterial diversity was positively correlated with precipitation (R2 = 0.14, p = 0.04), while both fungal richness and diversity were negatively correlated with precipitation (R2 = −0.15, p = 0.04 and R2 = −0.14, p = 0.05, respectively). These results are consistent with previous studies showing that, across global soils, changes in pH are correlated with changes in bacterial community composition (Fierer and Jackson, 2006; Rousk et al., 2010), but manipulative studies would be needed to test for causation.
Bacterial richness and diversity [two-way ANOVA; date: F(13, 143) = 4.5, P < 0.001 (richness), F(13, 140) = 4.0, P < 0.001 (diversity)] showed significant temporal variability, as well as a significant microhabitat-by-date interaction [two-way ANOVA; microhabitat-by-date: F(54, 143) = 1.8, P < 0.001 (bacterial richness), F(54, 143) = 1.8, P < 0.001 (bacterial diversity)]. In contrast, fungal richness and diversity showed no significant temporal variability. Both bacterial and fungal composition changed significantly across the sampling dates [PERMANOVA; date: F(13, 143) = 2.9, P < 0.001 (bacteria), F(13, 143) = 1.5, P = 0.001 (fungi)] (Figures S6B, S7B); however overall sampling date explained much less of the variation in community composition than microhabitat (Figure 3). Some fungal communities may be more stable over time than bacteria because of their extensive hyphal structures that allow them to regulate moisture availability which can change rapidly over time in dryland ecosystems (Bapiri et al., 2010; Yuste et al., 2011; Barnard et al., 2013). However, some soil fungi have also been shown to be sensitive to moisture conditions (Meisner et al., 2018).
For both bacteria and fungi, a significant microhabitat-by-date interaction was observed [PERMANOVA; microhabitat-by-date: F(54, 143) = 1.5, P < 0.001 (bacteria), F(54, 143) = 1.1, P = 0.006 (fungi)]. Bacterial and fungal composition were both significantly correlated with temporal distance (time between sampling points) for four out of the seven microhabitats, including cyanobacteria biocrust, below lichen, and plant-associated soil (B. tectorum and P. jamesii) (Table 1). In addition, bacterial composition was significantly correlated with temporal distance for the microhabitat moss biocrust (Table 1). These results partially support our second hypothesis that biocrust communities would change the most, and below-biocrust communities would change the least, over time. Surface soils typically experience greater climatic variability (Tucker et al., 2017), which may drive stronger temporal changes in the cyanobacteria biocrust communities. However, lichen and moss biocrust composition did not change significantly over time. This higher stability, despite the location on the surface may be because these biocrusts occur in later successional stages than cyanobacteria biocrusts (Belnap, 2003) and thus may be better established and more resistant to fluctuations in moisture and temperature. Other morphological characteristics of moss and lichen biocrusts may also increase resilience. The rhizosphere communities also showed a temporal correlation with bacterial community composition. The strongest correlation was with B. tectorum, which is an annual invasive C3 grass that can germinate in September, expand roots over the winter, and then grow between March and April (Belnap and Phillips, 2001). The perennial C4 grass P. jamesii's rhizophere microbial composition was also significantly correlated with time, but more weakly than B. tectorum. Temporal changes in these rhisozphere bacterial communities are likely associated with both plant life-cycles and development stage, which have been shown to influence rhizosphere communities (Houlden et al., 2008; Philippot et al., 2013; Li et al., 2014).
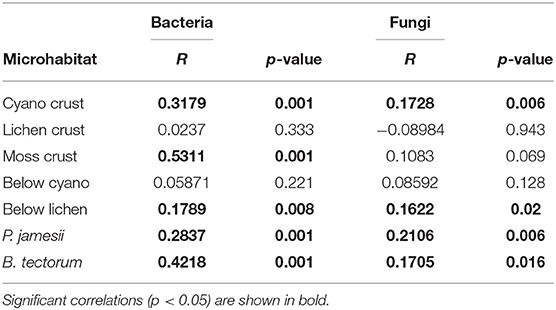
Table 1. Correlations between community composition in each microhabitat and temporal distance (Mantel tests).
Taken together, ~29% of the variation in bacterial community composition could be explained by microhabitat, date, and microhabitat-by-date interaction, while only around 11% of variation in fungal community composition could be explained by the same factors (Figure 3). The low amount of variation explained by the environmental factors tested suggests that either fungal composition is driven by environmental factors not measured in the current study or that fungal assembly is more stochastic. Studies in other ecosystems have found that fungal communities exhibit highly stochastic assembly processes compared to bacteria, where environmental selection processes are the stronger drivers (Schmidt et al., 2014; Powell et al., 2015; Jiang et al., 2018). Our data suggest that these patterns may hold in dryland ecosystems as well.
Dryland grasslands are currently highly impacted by a variety of global changes, including altered precipitation, increased temperature and nutrient deposition, and physical disruption (Maestre et al., 2016; Plaza et al., 2018). Dryland grasslands are typically resource limited and can show low resistance and resilience to both abiotic and biotic perturbations (Barger et al., 2006; Belnap and Sherrod, 2009; Kuske et al., 2012; D'Odorico et al., 2013; Steven et al., 2018), thus the spatial heterogeneity in these landscapes is unlikely to remain static. The differences in composition across microhabitats in this study highlight the importance of spatial heterogeneity in shaping the overall diversity of microbial communities in grassland ecosystems. The microhabitats can be very close to one another, distanced by only a few centimeters or less, yet the communities observed here showed strong variation among locations and across time. While this study only examined microhabitats within a single grassland site for a single year, we expect that across larger spatial scales (multiple sites), we will see similar strong effects of microhabitat variation. The role of microhabitat heterogeneity and climatic variability in shaping diverse dryland landscapes remains an important question, and future studies that explore this variation and its consistency (or lack thereof) across multiple sites would be extremely valuable. This case study of the temporal and spatial structure of bacterial and fungal communities is one step to understanding the variability and assembly controls over microbial composition in drylands.
MA analyzed the data and wrote the first draft. RM analyzed the sequence data. LG-G conducted DNA extraction, PCR amplification, and prepared information for sequencing. JB, CK, and SR conceived the study and the monthly sampling.
This study was supported through a Science Focus Area grant to CK from the U.S. Department of Energy, Biological and Environmental Research Division, United States (2018LANLF260). JB and SR were supported by the U.S. Department of Energy Terrestrial Ecosystem Sciences Program (DE-SC-0008168). Any trade, product, or firm name is used for descriptive purposes only and does not imply endorsement by the US Government. The Los Alamos LA-UR number for this publication is LA-UR-18-30421.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Dr. Theresa McHugh for valuable suggestions on a previous version of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00367/full#supplementary-material
Bapiri, A., Baath, E., and Rousk, J. (2010). Drying-rewetting cycles affect fungal and bacterial growth differently in an arable soil. Microb. Ecol. 60, 419–428. doi: 10.1007/s00248-010-9723-5
Barberan, A., Ladau, J., Leff, J. W., Pollard, K. S., Menninger, H. L., Dunn, R. R., et al. (2015). Continental-scale distributions of dust-associated bacteria and fungi. Proc. Natl. Acad. Sci. U.S.A. 112, 5756–5761. doi: 10.1073/pnas.1420815112
Barger, N. N., Herrick, J. E., Van Zee, J., and Belnap, J. (2006). Impacts of biological soil crust disturbance and composition on C and N loss from water erosion. Biogeochemistry 77, 247–263. doi: 10.1007/s10533-005-1424-7
Barnard, R. L., Osborne, C. A., and Firestone, M. K. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7, 2229–2241. doi: 10.1038/ismej.2013.104
Bates, S. T., Nash, T. H., and Garcia-Pichel, F. (2012). Patterns of diversity for fungal assemblages of biological soil crusts from the southwestern United States. Mycologia 104, 353–361. doi: 10.3852/11-232
Bates, S. T., Nash, T. H., Sweat, K. G., and Garcia-Pichel, F. (2010). Fungal communities of lichen-dominated biological soil crusts: diversity, relative microbial biomass, and their relationship to disturbance and crust cover. J. Arid. Environ. 74, 1192–1199. doi: 10.1016/j.jaridenv.2010.05.033
Bell, C., McIntyre, N., Cox, S., Tissue, D., and Zak, J. (2008). Soil microbial responses to temporal variations of moisture and temperature in a Chihuahuan Desert Grassland. Microb. Ecol. 56, 153–167. doi: 10.1007/s00248-007-9333-z
Bell, C. W., Acosta-Martinez, V., McIntyre, N. E., Cox, S., Tissue, D. T., and Zak, J. C. (2009). Linking microbial community structure and function to seasonal differences in soil moisture and temperature in a Chihuahuan desert grassland. Microb. Ecol. 58, 827–842. doi: 10.1007/s00248-009-9529-5
Belnap, J. (2003). The world at your feet: desert biological soil crusts. Front. Ecol. Environ. 1, 181–189. doi: 10.1890/1540-9295(2003)001[0181:TWAYFD]2.0.CO;2
Belnap, J. (2006). The potential roles of biological soil crusts in dryland hydrologic cycles. Hydrol. Process 20, 3159–3178. doi: 10.1002/hyp.6325
Belnap, J., and Phillips, S. L. (2001). Soil biota in an ungrazed grassland: response to annual grass (Bromus tectorum) invasion. Ecol. Appl. 11, 1261–1275. doi: 10.1890/1051-0761(2001)011[1261:SBIAUG]2.0.CO;2
Belnap, J., Phillips, S. L., Sherrod, S. K., and Moldenke, A. (2005). Soil biota can change after exotic plant invasion: does this affect ecosystem processes? Ecology 86, 3007–3017. doi: 10.1890/05-0333
Belnap, J., and Sherrod, S. K. (2009). Soil amendment effects on the exotic annual grass Bromus tectorum L. and facilitation of its growth by the native perennial grass Hilaria jamesii (Torr.) Benth. Plant. Ecol. 201, 709–721. doi: 10.1007/s11258-008-9463-5
Bjork, R. G., Bjorkman, M. P., Andersson, M. X., and Klemedtsson, L. (2008). Temporal variation in soil microbial communities in Alpine tundra. Soil Biol. Biochem. 40, 266–268. doi: 10.1016/j.soilbio.2007.07.017
Bowker, M. A., Maestre, F. T., and Mau, R. L. (2013). Diversity and patch-size distributions of biological soil crusts regulate dryland ecosystem multifunctionality. Ecosystems 16, 923–933. doi: 10.1007/s10021-013-9644-5
Bowker, M. A., Reed, S. C., Belnap, J., and Phillips, S. L. (2002). Temporal variation in community composition, pigmentation, and F-v/F-m of desert cyanobacterial soil crusts. Microb. Ecol. 43, 13–25. doi: 10.1007/s00248-001-1013-9
Bradley, B. A., Curtis, C. A., Fusco, E. J., Abatzoglou, J. T., Balch, J. K., Dadashi, S., et al. (2018). Cheatgrass (Bromus tectorum) distribution in the intermountain Western United States and its relationship to fire frequency, seasonality, and ignitions. Biol. Invasions 20, 1493–1506. doi: 10.1007/s10530-017-1641-8
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chapin, F. S., Zavaleta, E. S., Eviner, V. T., Naylor, R. L., Vitousek, P. M., Reynolds, H. L., et al. (2000). Consequences of changing biodiversity. Nature 405, 234–242. doi: 10.1038/35012241
Compagnoni, A., and Adler, P. B. (2014). Warming, competition, and Bromus tectorum population growth across an elevation gradient. Ecosphere 5, 1–34. doi: 10.1890/ES14-00047.1
de Hoog, G. S., Gottlich, E., Platas, G., Genilloud, O., Leotta, G., and van Brummelen, J. (2005). Evolution, taxonomy and ecology of the genus Thelebolus in Antarctica. Stud. Mycol. 51, 33–76. Available online at: https://www.scopus.com/record/display.uri?eid=2-s2.0-23644444615&origin=inward&txGid=2ca79eb9bbbc660cb4b5d3fbe6dfc4fb
DeCrappeo, N. M., DeLorenze, E. J., Giguere, A. T., Pyke, D. A., and Bottomley, P. J. (2017). Fungal and bacterial contributions to nitrogen cycling in cheatgrass-invaded and uninvaded native sagebrush soils of the western USA. Plant Soil 416, 271–281. doi: 10.1007/s11104-017-3209-x
Despres, V. R., Huffman, J. A., Burrows, S. M., Hoose, C., Safatov, A. S., Buryak, G., et al. (2012). Primary biological aerosol particles in the atmosphere: a review. Tellus B 64:15598. doi: 10.3402/tellusb.v64i0.15598
D'Odorico, P., Bhattachan, A., Davis, K. F., Ravi, S., and Runyan, C. W. (2013). Global desertification: drivers and feedbacks. Adv. Water Resour. 51, 326–344. doi: 10.1016/j.advwatres.2012.01.013
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Ferrenberg, S., Faist, A. M., Howell, A., and Reed, S. C. (2018). Biocrusts enhance soil fertility and Bromus tectorum growth, and interact with warming to influence germination. Plant Soil 429, 77–90. doi: 10.1007/s11104-017-3525-1
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631. doi: 10.1073/pnas.0507535103
Flagg, C. B., Neff, J. C., Reynolds, R. L., and Belnap, J. (2014). Spatial and temporal patterns of dust emissions (2004-2012) in semi-arid landscapes, southeastern Utah, USA. Aeolian Res. 15, 31–43. doi: 10.1016/j.aeolia.2013.10.002
Garcia-Pichel, F., Lopez-Cortes, A., and Nubel, U. (2001). Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl. Environ. Microb. 67, 1902–1910. doi: 10.1128/AEM.67.4.1902-1910.2001
Goslee, S. C., and Urban, D. L. (2007). The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22, 1–19. doi: 10.18637/jss.v022.i07
Griffiths, R. I., Whiteley, A. S., O'Donnell, A. G., and Bailey, M. J. (2003). Influence of depth and sampling time on bacterial community structure in an upland grassland soil. FEMS Microbiol. Ecol. 43, 35–43. doi: 10.1111/j.1574-6941.2003.tb01043.x
Grondin, A. E., and Johansen, J. R. (1993). Microbial spatial heterogeneity in microbiotic crusts in Colorado National Monument 1. Algae. Great Basin Nat. 53, 24–30.
Gubry-Rangin, C., Hai, B., Quince, C., Engel, M., Thomson, B. C., James, P., et al. (2011). Niche specialization of terrestrial archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. U.S.A. 108, 21206–21211. doi: 10.1073/pnas.1109000108
Gundlapally, S. R., and Garcia-Pichel, F. (2006). The community and phylogenetic diversity of biological soil crusts in the Colorado Plateau studied by molecular fingerprinting and intensive cultivation. Microb. Ecol. 52, 345–357. doi: 10.1007/s00248-006-9011-6
Houlden, A., Timms-Wilson, T. M., Day, M. J., and Bailey, M. J. (2008). Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol. Ecol. 65, 193–201. doi: 10.1111/j.1574-6941.2008.00535.x
Jiang, Y. L., Lei, Y. B., Yang, Y., Korpelainen, H., Niinemets, U., and Li, C. Y. (2018). Divergent assemblage patterns and driving forces for bacterial and fungal communities along a glacier forefield chronosequence. Soil Biol. Biochem. 118, 207–216. doi: 10.1016/j.soilbio.2017.12.019
Johansen, J. R. (1984). Response of soil algae to a hundred-year storm in the Great Basin Desert, U.S.A. Phykos 23, 51–54.
Johansen, J. R., and Rushforth, S. R. (1985). Cryptogamic soil crusts - seasonal-variation in algal populations in the Tintic Mountains, Juab County, Utah. Great Basin Nat. 45, 14–21.
Jones, V. A. S., and Dolan, L. (2012). The evolution of root hairs and rhizoids. Ann. Bot. London 110, 205–212. doi: 10.1093/aob/mcs136
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Kivlin, S. N., and Hawkes, C. V. (2016). Temporal and spatial variation of soil bacteria richness, composition, and function in a neotropical rainforest. PLoS ONE 11:e0159131. doi: 10.1371/journal.pone.0159131
Kuske, C. R. (2006). Current and emerging technologies for the study of bacteria in the outdoor air. Curr. Opin. Biotech. 17, 291–296. doi: 10.1016/j.copbio.2006.04.001
Kuske, C. R., Ticknor, L. O., Miller, M. E., Dunbar, J. M., Davis, J. A., Barns, S. M., et al. (2002). Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microb. 68, 1854–1863. doi: 10.1128/AEM.68.4.1854-1863.2002
Kuske, C. R., Yeager, C. M., Johnson, S., Ticknor, L. O., and Belnap, J. (2012). Response and resilience of soil biocrust bacterial communities to chronic physical disturbance in arid shrublands. ISME J. 6, 886–897. doi: 10.1038/ismej.2011.153
Li, X. Z., Rui, J. P., Mao, Y. J., Yannarell, A., and Mackie, R. (2014). Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 68, 392–401. doi: 10.1016/j.soilbio.2013.10.017
Lipson, D. A., Schadt, C. W., and Schmidt, S. K. (2002). Changes in soil microbial community structure and function in an alpine dry meadow following spring snow melt. Microb. Ecol. 43, 307–314. doi: 10.1007/s00248-001-1057-x
Maestre, F. T., Bowker, M. A., Canton, Y., Castillo-Monroy, A. P., Cortina, J., Escolar, C., et al. (2011). Ecology and functional roles of biological soil crusts in semi-arid ecosystems of Spain. J. Arid Environ. 75, 1282–1291. doi: 10.1016/j.jaridenv.2010.12.008
Maestre, F. T., Eldridge, D. J., Soliveres, S., Kefi, S., Delgado-Baquerizo, M., Bowker, M. A., et al. (2016). Structure and functioning of dryland ecosystems in a changing world. Annu. Rev. Ecol. Evol. Syst. 47, 215–237. doi: 10.1146/annurev-ecolsys-121415-032311
Maier, S., Tamm, A., Wu, D. M., Caesar, J., Grube, M., and Weber, B. (2018). Photoautotrophic organisms control microbial abundance, diversity, and physiology in different types of biological soil crusts. ISME J. 12, 1032–1046. doi: 10.1038/s41396-018-0062-8
Marunsenko, Y., Bates, S. T., Anderson, I., Johnson, S., Soule, T., and Garcia-Pichel, F. (2013). Ammonia-oxidizing archaea and bacteria are structured by geography in biological soil crusts across North American arid lands. Ecol. Process. 2:9. doi: 10.1186/2192-1709-2-9
Matulich, K. L., Weihe, C., Allison, S. D., Amend, A. S., Berlemont, R., Goulden, M. L., et al. (2015). Temporal variation overshadows the response of leaf litter microbial communities to simulated global change. ISME J. 9, 2477–2489. doi: 10.1038/ismej.2015.58
McHugh, T. A., Morrissey, E. M., Mueller, R. C., Gallegos-Graves, L. V., Kuske, C. R., and Reed, S. C. (2017). Bacterial, fungal, and plant communities exhibit no biomass or compositional response to two years of simulated nitrogen deposition in a semiarid grassland. Environ. Microbiol. 19, 1600–1611. doi: 10.1111/1462-2920.13678
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
Meisner, A., Jacquiod, S., Snoek, B. L., ten Hooven, F. C., and van der Putten, W. H. (2018). Drought legacy effects on the composition of soil fungal and prokaryote communities. Front. Microbiol. 9:294. doi: 10.3389/fmicb.2018.00294
Mueller, R. C., Belnap, J., and Kuske, C. R. (2015). Soil bacterial and fungal community responses to nitrogen addition across soil depth and microhabitat in an arid shrubland. Front. Microbiol. 6:891. doi: 10.3389/fmicb.2015.00891
Murali, A., Bhargava, A., and Wright, E. S. (2018). IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6:140. doi: 10.1186/s40168-018-0521-5
Norton, U., Mosier, A. R., Morgan, J. A., Derner, J. D., Ingram, L. J., and Stahl, P. D. (2008). Moisture pulses, trace gas emissions and soil C and N in cheatgrass and native grass-dominated sagebrush-steppe in Wyoming, USA. Soil Biol. Biochem. 40, 1421–1431. doi: 10.1016/j.soilbio.2007.12.021
Ofek, M., Hadar, Y., and Minz, D. (2012). Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS ONE 7:e40117. doi: 10.1371/journal.pone.0040117
Oksanen, J., Blanchet, F., Friendly, F., Kindt, R., Legendre, P., McGlinn, D., et al. (2018). vegan: Community Ecology Package. Available online at: https://cran.r-project.org/web/packages/vegan/vegan.pdf
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Patzelt, D. J., Hodac, L., Friedl, T., Pietrasiak, N., and Johansen, J. R. (2014). Biodiversity of soil cyanobacteria in the hyper-arid Atacama Desert, Chile. J. Phycol. 50, 698–710. doi: 10.1111/jpy.12196
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Plaza, C., Zaccone, C., Sawicka, K., Mendez, A. M., Tarquis, A., Gasco, G., et al. (2018). Soil resources and element stocks in drylands to face global issues. Sci. Rep. 8:13788. doi: 10.1038/s41598-018-32229-0
Pointing, S. B., and Belnap, J. (2012). Microbial colonization and controls in dryland systems. Nat. Rev. Microbiol. 10:551. doi: 10.1038/nrmicro2831
Powell, J. R., Karunaratne, S., Campbell, C. D., Yao, H., Robinson, L., and Singh, B. K. (2015). Deterministic processes vary during community assembly for ecologically dissimilar taxa. Nat. Commun. 6:8444. doi: 10.1038/ncomms9444
Quinn, G. P., and Keough, M. J. (2002). Experimental Design and Data Analysis for Biologists. Cambridge; New York, NY: Cambridge University Press. doi: 10.1017/CBO9780511806384
Rousk, J., Baath, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Safriel, U., Adeel, Z., Niemeijer, D., Puigdefabregas, J., White, R., Lal, R., et al. (2005). “Dryland systems,” in Ecosystems and Human Well-Being: Current State and Trends, eds R. Hassan, R. Scholes, and N. Ash (Washington, DC: Island Press), 623–652.
Schadt, C. W., Martin, A. P., Lipson, D. A., and Schmidt, S. K. (2003). Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301, 1359–1361. doi: 10.1126/science.1086940
Schmidt, S. K., Nemergut, D. R., Darcy, J. L., and Lynch, R. (2014). Do bacterial and fungal communities assemble differently during primary succession? Mol. Ecol. 23, 254–258. doi: 10.1111/mec.12589
Sheppard, P. R., Comrie, A. C., Packin, G. D., Angersbach, K., and Hughes, M. K. (2002). The climate of the US Southwest. Clim. Res. 21, 219–238. doi: 10.3354/cr021219
Smit, E., Leeflang, P., Gommans, S., van den Broek, J., van Mil, S., and Wernars, K. (2001). Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microb. 67, 2284–2291. doi: 10.1128/AEM.67.5.2284-2291.2001
Smith, A. P., Marin-Spiotta, E., and Balser, T. (2015). Successional and seasonal variations in soil and litter microbial community structure and function during tropical postagricultural forest regeneration: a multiyear study. Global Change Biol. 21, 3532–3547. doi: 10.1111/gcb.12947
Soule, T., Anderson, I. J., Johnson, S. L., Bates, S. T., and Garcia-Pichel, F. (2009). Archaeal populations in biological soil crusts from arid lands in North America. Soil Biol. Biochem. 41, 2069–2074. doi: 10.1016/j.soilbio.2009.07.023
Sperry, L. J., Belnap, J., and Evans, R. D. (2006). Bromus tectorum invasion alters nitrogen dynamics in an undisturbed arid grassland ecosystem. Ecology 87, 603–615. doi: 10.1890/05-0836
Steven, B., Belnap, J., and Kuske, C. R. (2018). Chronic physical disturbance substantially alters the response of biological soil crusts to a wetting pulse, as characterized by metatranscriptomic sequencing. Front. Microbiol. 9:2382. doi: 10.3389/fmicb.2018.02382
Steven, B., Gallegos-Graves, L., Belnap, J., and Kuske, C. R. (2013). Dryland soil microbial communities display spatial biogeographic patterns associated with soil depth and soil parent material. FEMS Microbiol. Ecol. 86, 101–113. doi: 10.1111/1574-6941.12143
Steven, B., Gallegos-Graves, L., Yeager, C. M., Belnap, J., Evans, R. D., and Kuske, C. R. (2012). Dryland biological soil crust cyanobacteria show unexpected decreases in abundance under long-term elevated CO2. Environ. Microbiol. 14, 3247–3258. doi: 10.1111/1462-2920.12011
Steven, B., Gallegos-Graves, L. V., Yeager, C., Belnap, J., and Kuske, C. R. (2014). Common and distinguishing features of the bacterial and fungal communities in biological soil crusts and shrub root zone soils. Soil Biol. Biochem. 69, 302–312. doi: 10.1016/j.soilbio.2013.11.008
Theodorakopoulos, N., Bachar, D., Christen, R., Alain, K., and Chapon, V. (2013). Exploration of Deinococcus-Thermus molecular diversity by novel group-specific PCR primers. Microbiologyopen 2, 862–872. doi: 10.1002/mbo3.119
Tucker, C. L., Ferrenberg, S., and Reed, S. C. (2018). Climatic sensitivity of dryland soil CO2 fluxes differs dramatically with biological soil crust successional state. Ecosystems 22, 15–32. doi: 10.1007/s10021-018-0250-4
Tucker, C. L., McHugh, T. A., Howell, A., Gill, R., Weber, B., Belnap, J., et al. (2017). The concurrent use of novel soil surface microclimate measurements to evaluate CO2 pulses in biocrusted interspaces in a cool desert ecosystem. Biogeochemistry 135, 239–249. doi: 10.1007/s10533-017-0372-3
Wallenstein, M. D., McMahon, S., and Schimel, J. (2007). Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol. Ecol. 59, 428–435. doi: 10.1111/j.1574-6941.2006.00260.x
Wanasinghe, D. N., Hyde, K. D., Jeewon, R., Crous, P. W., Wijayawardene, N. N., Jones, E. B. G., et al. (2017). Phylogenetic revision of Camarosporium (Pleosporineae, Dothideomycetes) and allied genera. Stud. Mycol. 87, 207–256. doi: 10.1016/j.simyco.2017.08.001
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Warren, S. D., St Clair, L. L., and Leavitt, S. D. (2019). Aerobiology and passive restoration of biological soil crusts. Aerobiologia 35, 45–56. doi: 10.1007/s10453-018-9539-1
Weber, B., Burkhard, B., and Belnap, J. (2016). Biological Soil Crusts: An Organizing Principle in Drylands, Vol. 226. Basel: Springer International Publishing. doi: 10.1007/978-3-319-30214-0
Wertin, T. M., Reed, S. C., and Belnap, J. (2015). C-3 and C-4 plant responses to increased temperatures and altered monsoonal precipitation in a cool desert on the Colorado Plateau, USA. Oecologia 177, 997–1013. doi: 10.1007/s00442-015-3235-4
Wheeler, C. C., Flechtner, V. R., and Johansen, J. R. (1993). Microbial spatial heterogeneity in microbiotic crusts in Colorado National Monument 2. Bacteria. Great Basin Nat. 53, 31–39.
Winkler, D. E., Grossiord, C., Belnap, J., Howell, A., Ferrenberg, S., Smith, H., et al. (2019). Earlier plant growth helps compensate for reduced carbon fixation after 13 years of warming. Func. Ecol. doi: 10.1111/1365-2435.13432. [Epub ahead of print].
Yeager, C. M., Kuske, C. R., Carney, T. D., Johnson, S. L., Ticknor, L. O., and Belnap, J. (2012). Response of biological soil crust diazotrophs to season, altered summer precipitation, and year-round increased temperature in an arid grassland of the Colorado Plateau, USA. Front. Microbiol. 3:358. doi: 10.3389/fmicb.2012.00358
Yooseph, S., Andrews-Pfannkoch, C., Tenney, A., McQuaid, J., Williamson, S., Thiagarajan, M., et al. (2013). A metagenomic framework for the study of airborne microbial communities. PLoS ONE 8:e81862. doi: 10.1371/journal.pone.0081862
Yuste, J. C., Penuelas, J., Estiarte, M., Garcia-Mas, J., Mattana, S., Ogaya, R., et al. (2011). Drought-resistant fungi control soil organic matter decomposition and its response to temperature. Global Change Biol. 17, 1475–1486. doi: 10.1111/j.1365-2486.2010.02300.x
Keywords: biological soil crusts, diversity, temporal dynamics, fungi, bacteria
Citation: Albright MBN, Mueller RC, Gallegos-Graves LV, Belnap J, Reed SC and Kuske CR (2019) Interactions of Microhabitat and Time Control Grassland Bacterial and Fungal Composition. Front. Ecol. Evol. 7:367. doi: 10.3389/fevo.2019.00367
Received: 09 January 2019; Accepted: 17 September 2019;
Published: 09 October 2019.
Edited by:
Steven D. Warren, Rocky Mountain Research Station, United States Forest Service, United StatesReviewed by:
Jeffrey R. Johansen, John Carroll University, United StatesCopyright © 2019 Albright, Mueller, Gallegos-Graves, Belnap, Reed and Kuske. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheryl R. Kuske, a3Vza2VAbGFubC5nb3Y=; Michaeline B. N. Albright, bWFsYnJpZ2h0QGxhbmwuZ292
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.