- Department of Entomology and Agrilife Research, Texas A&M University, College Station, TX, United States
Although much progress has been made in the fight against malaria, the number of people that contract this disease due to the bite of an Anopheline mosquito remains unacceptably high. A better understanding of the relationship between the Plasmodium parasite and its vector is of extreme importance and may allow for the development of new tools to fight the disease. In particular, new genome editing techniques may allow the modification or removal of factors critical for parasite traversal of the vector mosquito. In this review, we aim to highlight what is known about important molecules encoded by the mosquito or parasite involved in this close interaction, focused on a specific window of the parasite life cycle inside the mosquito: from egress and release of the sporozoites from the oocyst into the hemolymph, until salivary gland penetration. While sophisticated molecular tools have already helped to understand aspects of parasite-mosquito interactions, our understanding of this brief period is still limited. Here, we discuss the evidence surrounding essential parasite and mosquito factors relating to sporozoite invasion of the salivary glands, and emphasize the areas where the lack of information is limiting the advance of strategies to manipulate the mosquito in order to block the transmission of malaria parasites.
Introduction
The malaria mortality rate is estimated to have decreased by 60% globally between 2000 and 2015. Such an achievement was possible due to key malaria control interventions such as the distribution of Long Lasting Insecticidal Nets (LLINs), indoor residual spraying, and the use of anti-malarial drugs known as artemisinin-based combination therapies (ACTs). Although progress has been substantial, 219 million cases of malaria were reported in 2017, an increase of about 2 million cases over 2016, along with a still unacceptable death toll of 435,000. As half of the world's population is living in risk areas (World Health Organization, 2018), this highlights the need for sustaining current gains and developing additional control measures.
The mosquito portion of the parasite life cycle starts when an Anopheles female mosquito ingests the sexual stages of the Plasmodium parasite—the microgametocyte (male) and the macrogametocyte (female) during the act of blood feeding from an infected person. In the stomach of the mosquito, the presence of xanthurenic acid and a decrease in the temperature concomitantly with a pH increase, triggers exflagellation of the gametocytes followed by fertilization and formation of a zygote (Aly et al., 2009). The zygote undergoes structural changes, resulting in a motile ookinete that penetrates the midgut epithelium and settles between the epithelial cells and the basal lamina. The ookinete becomes an immobile oocyst responsible for sporogony, a process that generates thousands of sporozoites that are released into the hemolymph of the mosquito. These infective forms invade the mosquito salivary gland and are injected into the vertebrate host initiating another human infection when the mosquito acquires a subsequent bloodmeal (Frischknecht and Matuschewski, 2017).
The successful completion of the mosquito portion of the Plasmodium cycle is dependent on both parasite and mosquito factors critical for the migration of the parasite to the next life stage. Here we review the evidence supporting the involvement of these factors as well as identify gaps in our current knowledge and inconsistencies within the available literature. In particular, we focus on the sporozoite journey starting at oocyst egress until the colonization of the salivary gland by sporozoites. While the evidence for parasite-encoded factors essential for salivary gland invasion is relatively robust due to the long-standing ability to generate loss-of-function parasites, the evidence base for critical mosquito proteins is more variable. Several candidate receptors have been identified, but for each more evidence is required concerning its role and importance in the parasite life cycle. We note that mosquito immune responses against malaria parasites will not be discussed here, and the reader is referred to several excellent recent reviews for discussion of this topic (Marois, 2011; Clayton et al., 2014; Yordanova et al., 2018).
Oocyst Egress
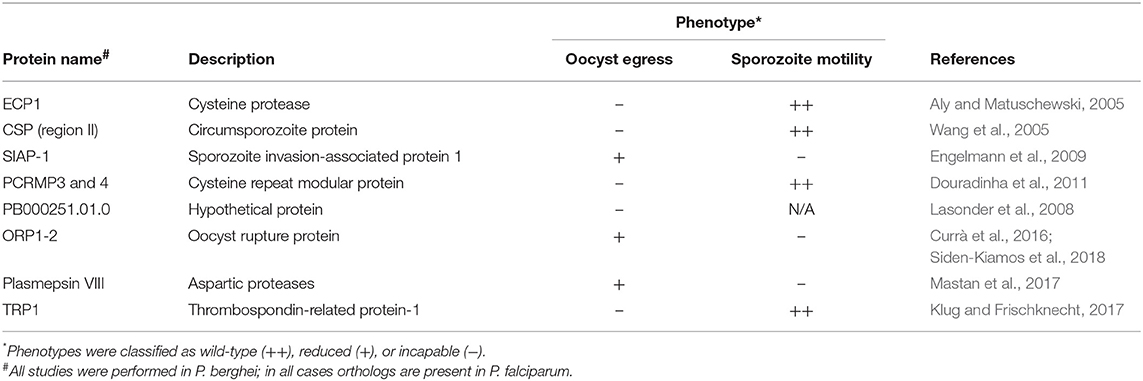
The process of oocyst egress by sporozoites was once thought to occur passively, resulting simply from parasite growth followed by rupture of the oocyst capsule. Gene disruption studies have changed that view. At the present time, at least eight parasite factors from the rodent malaria parasite Plasmodium berghei have been described to have some role in parasite egress from the oocyst, but they display differences in the phenotype of the generated mutants (Table 1). These genes and their corresponding loss of function phenotypes are described below.
The loss of function of ECP1, a cysteine protease, showed that sporozoites can fully develop inside of the oocyst but are unable to exit from it. In addition, the mutant parasites showed an unusual arrangement within the oocyst, oriented in circles under constant gliding locomotion while wild type parasites are arranged radially (Aly and Matuschewski, 2005). Circumsporozoite protein (CSP) mutants having four positively charged amino acids substituted by alanines in conserved region II, present in the C-terminus of the protein, were unable to escape the oocyst, although the mutants displayed normal morphology and motility (Wang et al., 2005). Disruption of the sporozoite invasion-associated protein 1 (SIAP-1) abolished the capacity of parasites to perform gliding motility, a specific type of substrate-dependent locomotion based on Myosin A motor complex; these parasites also did not efficiently egress from the oocyst (Engelmann et al., 2009). Two members of the conserved family Plasmodium cysteine repeat modular protein (PCRMP) are also implicated in parasite egress. Mutants for PCRMP3 or 4 produced oocysts in comparable numbers to WT and displayed normal gliding motility, but salivary glands of mosquitoes infected with both mutant parasites contained no sporozoites. Detailed analyses using electron microscopy revealed that sporozoites failed to effectively egress, as they were visible inside of oocysts at time points when wild type parasites were already detected inside salivary glands (Douradinha et al., 2011). Furthermore, mutants lacking a hypothetical protein (PB000251.01.0) identified in proteomic analyses exhibited similar formation of mature oocysts, producing the same sporozoite numbers as wild type, but displayed a distinct egress pattern as only a few sporozoites were detected in the haemocoel and salivary glands. Although deficient in the egress process, PB000251.01.0 mutant sporozoites were still capable of infecting mice after i.v. inoculation after being mechanically freed from the oocyst (Lasonder et al., 2008).
Two proteins containing a conserved histone-fold domain (HFD) were shown to be essential for oocyst rupture and therefore were named oocyst rupture protein 1 and 2 (ORP1-2). The HFD is found in histones and in proteins involved in transcriptional regulation, and this domain is also known to be responsible for heterodimerization and interaction to a third subunit (Arents and Moudrianakis, 1995). Deletion of these proteins did not impair the progression of the parasite cycle in any stage but the release of sporozoites into the haemolymph. The blocking of egress from the oocyst was not related to an impairment in gliding motility, since mechanical rupture of the oocysts released motile sporozoites. Immunolocalization detected ORP1 in the oocyst capsule during early development, with ORP 2 transported from the cytoplasm to the capsule later on when mature sporozoites had already formed. The authors suggest that the co-localization of both proteins led to a dimerization triggering a change to the capsule allowing the release of sporozoites (Currà et al., 2016). Recently the same group dissected the role of ORP2 and showed that it contains two functional domains that are essential for oocyst egress, the first at the N-terminal (HFD) with the second present in the middle of the protein (Siden-Kiamos et al., 2018).
Aspartic proteases, also referred as plasmepsins, have critical roles in biological processes of the parasite. Most plasmepsins encoded by Plasmodium are expressed in blood stages, being important for hemoglobin catabolism (Banerjee et al., 2002). Plasmepsin VIII is not expressed in blood stages, but has discrete expression in early midgut and salivary gland sporozoites, peaking at day 4 post infection during oocyst development. Loss of function experiments showed that parasites lacking plasmepsin VIII are able to produce oocysts and midgut sporozoites in numbers comparable to wild type parasites, but cannot perform gliding motility and thus have an impaired ability to escape from the oocyst (Mastan et al., 2017).
Lastly, belonging to the thrombospondin-related anonymous protein family, TRP1 has an important role in the egress of sporozoites from the oocyst. Trp1(–) parasites produce oocysts in numbers similar to the WT and show no defect in gliding motility in vitro, however no sporozoites could be detected in the hemolymph, demonstrating that they could not egress from oocysts (Klug and Frischknecht, 2017). In the same work, in vivo imaging of the WT sporozoites egressing from oocysts was performed for the first time. Interestingly, sporozoites were observed escaping the oocyst through different types of egress events, either bursting or budding in a membrane delimited vesicle-like structure. It is not clear what the origin nor function of the associated membrane, but authors suggest that it may be derived from the oocyst wall, but future studies are needed to clarify this event.
Thus far, all of the phenotypes described above and elucidated by reverse genetics were characterized in the rodent malaria parasite P. berghei, in Anopheles mosquitoes. This raises the question as to whether the processes of oocyst egress are conserved in human malaria parasites. Orfano et al. (2016) showed that the mechanism for sporozoite egress from the oocyst differs among Plasmodium species. P. berghei and the avian malaria parasite Plasmodium gallinaceum appear to egress similarly, breaking the oocyst wall. However, human malaria parasites Plasmodium falciparum and Plasmodium vivax share a different mode, using polarized propulsion. This study provides evidence that a complex species-specific mechanism is responsible for this important step and assumptions should be made cautiously when inferring the role of each molecule in this process. A better understanding of different parasite-host combinations could clarify this impasse.
While all of the proteins described above have a substantial impact on sporozoite egress, information on their specific function in the egress process is still missing, as is how they cooperate. Klug and Frischknecht (2017) propose two models for this event, the intracellular pathway and the extracellular pathway. In the former, sporozoites express proteins due to quorum sensing that trigger gliding motility and degradation of the oocyst wall for parasite release. In the latter, proteins expressed by the sporozoite lead to oocyst wall permeability, enabling an inflow of extracellular factors and triggering gliding motility and oocyst wall degradation that stimulates parasite release. One may add that a combination of both might happen and one or the other mechanism may occur in different parasite-host combinations.
In summary, at this portion of the parasite journey, little is known in terms of mosquito molecules influencing the egression of the sporozoite from the oocyst. In the context of the second model, no mosquito factor has yet been described to influence parasite gene expression sufficient to trigger gliding motility or oocyst wall destruction, an area in need of investigation. Pore-forming proteins, expressed in the micronemes of sporozoites, were found to be specifically expressed in oocyst and salivary gland sporozoites (Kaiser et al., 2004a). However, how the parasite may use these proteins to sense the environment and trigger the egress process is a subject that also needs evaluation. On top of that, the fact that some parasites share the same mode of egression but differ from others may be a clue that the host could have molecules influencing this process, as some parasite species can only complete development in specific mosquito species (Alavi et al., 2003). An exchange of the mosquito species and detection of changes in the egression pattern in a specific parasite could provide clues for better understanding of how mosquito factors contribute to this process. Finally, this could be an interesting parasite phase to target with the development of molecules to stop the progression of parasite life cycle, as the oocyst is the last stage in low numbers before bursting into thousands of sporozoites making it harder to develop strategies to contain the parasite.
Specificity of the Salivary Gland Recognition by Sporozoites
The mosquito salivary glands are a paired organ, with each half of the pair composed of one medial and two lateral lobes. The lateral lobes are further divided into proximal and distal regions connected to a main salivary duct (James and Rossignol, 1991). The molecules and mechanisms used by sporozoites to recognize and invade mosquito salivary glands remain poorly understood. The first studies to attempt to address the mechanism of invasion date to the mid 1980's, with the experiments of salivary gland transplants from Rosenberg (1985). Plasmodium knowlesi were shown to be capable of producing oocysts upon feeding to either Anopheles freeborni or Anopheles dirus. However, this parasite was only able to colonize An. dirus salivary glands. Rosenberg performed heterologous salivary gland transplants from these mosquitoes and showed that parasites recognized the salivary gland only from the susceptible species, even when the salivary gland is transplanted to a refractory one (Rosenberg, 1985). This suggested that there are unique parasite-mosquito receptor-ligand interactions that must occur to trigger salivary gland invasion.
Once sporozoites are released from the oocyst, they migrate throughout the entire haemocoel taken by the hemolymph flow (Hillyer et al., 2007). Although in potential contact with all mosquito organs, sporozoites preferentially bind to the distal portions of the salivary gland lobes (Grassi, 1900; Sterling et al., 1973; Golenda et al., 1990; Pimenta et al., 1994). In vitro studies using low-light video microscopy also indicated that a chemotactic attraction may play a role in salivary gland recognition, since sporozoites perform gliding motility toward unheated salivary gland extracts, but glide randomly when exposed to other organs or when the salivary gland extracts were heated at 56°C for 30 min, conditions known to denature high molecular mass proteins and cease the sugar binding activity of carbohydrate-binding proteins (Akaki, 2005).
Furthermore, antibodies directed against salivary gland proteins prevented parasites from invading the salivary glands (Barreau et al., 1995; Brennan et al., 2000) reinforcing the idea of receptor mediated recognition. Touray et al. (1992) demonstrated that salivary gland sporozoites lose their ability to reinvade salivary glands, suggesting the presence of a specific ligand on the parasite surface specific to hemolymph sporozoites. Taken together, these data strongly suggested that sporozoites use receptors on the salivary gland surface to invade this organ. The complete panel of factors or proteins involved in this process require further elucidation, but some of the important sporozoite/mosquito proteins have been characterized and their interactions will be discussed in the next section.
Sporozoite Proteins Involved in Salivary Gland Penetration
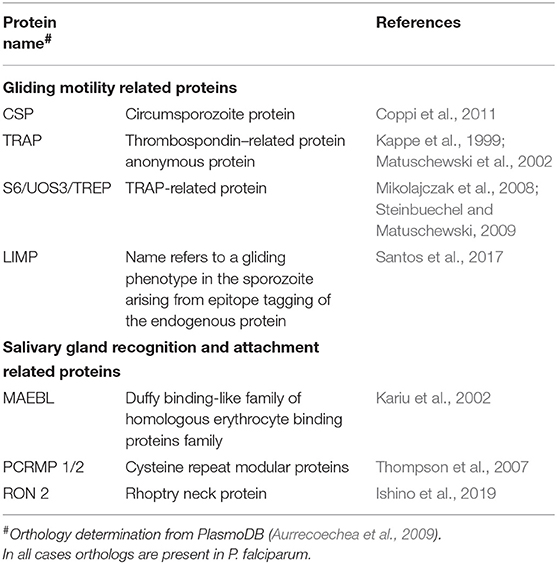
During the Plasmodium life cycle in the invertebrate host, there are major losses along with expansion phases in terms of parasite number. During the sporogonic phase, thousands of sporozoites are formed inside each oocyst, and this phase is the most expansive inside the mosquito. After egress from the oocyst, the mosquito immune system is responsible for clearing sporozoites during their journey through the hemolymph (see Simões et al., 2018 for an extensive review on the role of the mosquito immune response against malaria parasites). Unfortunately, through a combination of immune evasion, immune suppression, or sheer numbers, sporozoites successfully invade in the mosquito salivary gland, ensuring parasite transmission continues. As sporozoites must express proteins responsible for gliding motility, recognition and invasion of the salivary glands, proteomic analyses could elucidate factors of importance during this phase. To date, such analyses have targeted salivary gland sporozoites, since they are of greatest importance for vaccine design (Florens et al., 2002; Hall et al., 2005; Lasonder et al., 2008; Lindner et al., 2013; Swearingen et al., 2016, 2017). However, a number of parasite proteins have been implicated as being necessary for successful invasion of the salivary glands and are discussed next, and summarized on Table 2.
Gliding Motility Related Proteins
CSP
The circumsporozoite protein (CSP) is one of the most studied proteins expressed by the sporozoite. It is also the most abundant surface protein, uniformly covering its membrane (Sterling et al., 1973; Nagasawa et al., 1987; Posthuma et al., 1988; Thathy et al., 2002). CSP expression is stage-specific, it accumulates on oocysts and continues to be translated after sporozoite egress, during salivary gland invasion (Matuschewski et al., 2002), and is still detected on the plasma membrane of early exo-erythrocytic forms in the liver (Hamilton et al., 1988). CSP is a multifunctional protein and is essential for the parasite: knockout of the complete gene in P. berghei showed that parasites are able to produce equal numbers of oocysts as the wild type; however, the number of sporozoites produced was severely reduced (Ménard et al., 1997). The role of CSP is not limited to egress from the oocyst, as it is essential in both salivary gland and hepatocyte invasion. Peptides corresponding to a specific region on CSP N-terminus (formerly called Region I) were shown to bind preferentially to the medial and the distal lateral lobes of the salivary glands (Sidjanski et al., 1997). Additionally, injection of a CSP peptide from the same region into the hemocoel of P. yoelli-infected An. stephensi reduced sporozoite invasion of salivary glands (Myung et al., 2004) and an antibody against CSP was also able reduce invasion (Warburg et al., 1992). Regarding hepatocyte invasion, the C-terminal region of CSP, more specifically a region called RII, is responsible for sporozoite attachment to liver cells, as peptides encompassing this region bound to hepatocytes in vitro and in vivo (Cerami et al., 1992, 1994). In addition, CSP binds to heparan sulfates proteoglycans (HSPG) (Frevert, 2004), which may facilitate sporozoite attachment to hepatocytes (Pinzon-Ortiz et al., 2001). Finally, the conformational state of CSP was shown to be responsible for the migration of the sporozoite through different tissues between the invertebrate and vertebrate hosts. In the mosquito, the N-terminus of CSP is present and masks the adhesive domain present in the C-terminus, allowing the penetration of the salivary glands and transmission. Once in the vertebrate host, proteolytic cleavage of the N-terminal region exposes the adhesive domain on the C-terminus allowing attachment to the liver cells (Coppi et al., 2011). Though CSP is also involved in gliding motility, since disruption of a region on the C-terminus of the protein abolishes motility in P. falciparum parasites (Tewari et al., 2002), collectively these data demonstrate that CSP has a direct role in salivary gland recognition beyond its role in motility.
TRAP
The thrombospondin–related anonymous protein (TRAP) contributes to the invasion of the salivary gland, as it is also essential for gliding motility. TRAP null parasites almost completely lose their ability to invade the salivary glands, as most hemolymph sporozoites failed to associate with the glands. Of those that did, most (>85%) remained on the surface in comparison to 77% of wild-type parasites that had successfully invaded (Sultan et al., 1997). Thus, TRAP appears to be essential for both binding to, and traversing the basal lamina of the salivary glands. Mutations on the extracellular adhesive A domain and the thrombospondin repeat domain reveled that those are the regions important for salivary gland invasion (Wengelnik et al., 1999; Matuschewski et al., 2002), while amino acid substitutions on the cytoplasmic portion of the protein led to a modification on the gliding pattern of mutated parasites (Kappe et al., 1999). TRAP is expressed in the sporozoite micronemes, the secretory organelles of Apicomplexan parasites. Its expression begins in the oocyst, and increases during the salivary gland stages. Expression can also be detected during the erythrocytic stage of the life cycle (Robson et al., 1988; Rogers et al., 1992; Matuschewski et al., 2002). Accordingly, small amounts are exposed to the surface but in the presence of a target cell, large amounts can be detectable at one pole of the parasite, supporting its role in invasion of host cells (Gantt et al., 2000; Bhanot et al., 2003).
S6/UOS3/TREP
S6/UOS3/TREP was first identified in a P. yoelli subtractive hybridization library (Kaiser et al., 2004b), and later its expression and function were characterized in both P. berghei and P. yoelii. S6/UOS3/TREP is specifically expressed in sporozoites and is down regulated during sporozoite maturation: transcripts are highly expressed in oocyst sporozoites but substantially reduced in salivary gland sporozoites (Mikolajczak et al., 2008; Steinbuechel and Matuschewski, 2009). Disruption of S6/UOS3/TREP in P. yoelii or P. berghei resulted in a 90% reduction in parasite numbers found in the salivary glands at 14 days post infection (Mikolajczak et al., 2008; Steinbuechel and Matuschewski, 2009). However, these knockouts also had a negative impact in gliding motility as lower numbers of parasites were associated with trails and the trails were shorter than the wild type controls in gliding motility assays (Combe et al., 2009; Steinbuechel and Matuschewski, 2009). Immunolocalization of S6/UOS3/TREP revealed a punctate appearance and a partial overlapping with TRAP, suggesting that this protein is part of the apical invasive organelles, supporting a potential role in the invasion process (Mikolajczak et al., 2008).
LIMP
Highly conserved in Plasmodium parasites and related apicomplexans, deletion of LIMP in P. berghei severely impaired gliding motility with large numbers of completely immotile parasites and a corresponding 10-fold reduction in salivary gland invasion (Santos et al., 2017).
Salivary Gland Recognition and Attachment Related Proteins
MAEBL
MAEBL belongs to the Duffy binding-like family of erythrocyte binding proteins (DBL-EBPs) (Kappe et al., 1998). A single copy gene that is well conserved among Plasmodia (Blair et al., 2002), MAEBL is abundantly expressed in immature sporozoites from oocysts, with protein accumulation starting from an apical localization to a uniform distribution on the entire surface of mature oocyst sporozoites. MAEBL is present on hemolymph sporozoites and remains present after salivary gland invasion (Kappe et al., 2001; Blair et al., 2002; Srinivasan et al., 2004). MAEBL is not essential for sporozoite formation, as the number of oocyst sporozoites in P. berghei mutants where the gene was disrupted was comparable to wild type populations. On the other hand, MAEBL-deficient P. berghei yielded very few sporozoites in the salivary glands. This impairment of sporozoite penetration into the salivary gland was not due to a defect in gliding motility, as MAEBL-deficient parasites were able to perform gliding motility as well as the wild type control (Kariu et al., 2002).
PCRMP1/2
The Cysteine Repeat Modular Proteins of Plasmodium are a family of large, multidomain membrane proteins (PCRMP1-4) conserved in Plasmodium and amongst other Aplicomplexan parasites. PCRMP1 is expressed in developing oocysts and peaks in sporulating oocysts, while PCRMP2 transcripts are present in sporulating oocysts and salivary gland sporozoites of P. berghei (Thompson et al., 2007). Both proteins showed a punctate pattern over the surface of salivary gland sporozoites, but not in the impermeable surface of the oocyst wall (Thompson et al., 2007). Examination of the phenotype of single PCRMP1− or 2− deleted parasites revealed that both proteins are essential for sporozoite invasion of the salivary gland. Either PCRMP1− or 2− parasites were able to produce equal numbers of oocysts as the wild type but none of them were found inside of the salivary gland or attached to it, instead they were concentrated in the hemolymph, demonstrating an inability to recognize and invade the salivary glands (Thompson et al., 2007).
RON2
The rhoptry neck protein 2 (RON2) was found to be highly expressed in oocyst sporozoites with transcripts present in hemolymph and salivary gland sporozoites (Ishino et al., 2019). Using a promoter swapping approach to restrict RON2 transcription to the intraerythrocytic stage, RON2-deficient parasites failed to invade the salivary glands as sporozoite numbers recovered from this organ were 20-fold lower in comparison to controls, while the numbers of parasites collected from midgut and hemolymph were the same as the controls (Ishino et al., 2019). When tested for gliding motility, the velocity of circular movement of RON2-deficient sporozoites was comparable to controls, suggesting the machinery for gliding motility is not affected by the lack of RON2 (Ishino et al., 2019).
Summary of Parasite Factors Required for Salivary Gland Invasion
In summary, the process of salivary gland invasion requires gliding motility, attachment and host organ/cell penetration. However, a few questions remain unanswered. Is gliding motility important for recognition/attachment to the salivary gland surface? Or is gliding motility only important for penetration of the salivary glands following recognition? How do each of the proteins discussed above contribute to the recognition/invasion process? Do these proteins have dual functions like those found in TRAP, that are part of the gliding machinery and work as a ligand to the salivary gland, or they have a singular function? Do MAEBL, PCRMP 1/2 and RON2 cooperate to promote parasite attachment and/or penetration of the salivary gland? Do these proteins have the same role in human malaria parasites, since most of the work was done in P. berghei? Although existing data implicate these proteins in salivary gland invasion, a clear mechanistic understanding of how this complex process occurs is still lacking.
Mosquito Factors Required for Salivary Gland Invasion
The presence of parasite proteins required specifically for salivary gland invasion implies the existence of specific molecules on the salivary gland surface encoded by the mosquito. In this section, we discuss the current evidence supporting a role for various putative salivary gland receptors or molecules involved in this interaction. While substantial evidence has accumulated implicating both specific carbohydrate moieties and salivary gland-specific proteins, definitive evidence is lacking in most, if not all, cases.
Carbohydrate-Based Salivary Gland Recognition
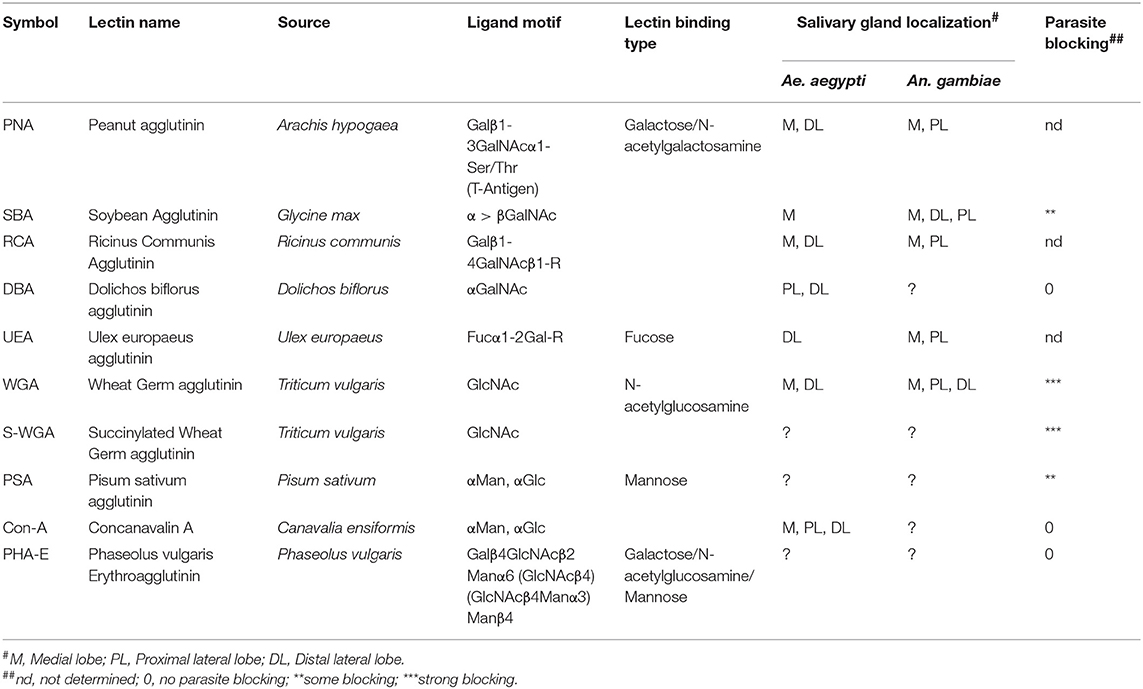
The mosquito basal lamina is rich in glycoconjugates, and different types of lectins (carbohydrate-binding proteins) have been found to bind to the salivary glands with more or less affinity than other organs in Aedes aegypti (Perrone et al., 1986; Barreau et al., 1995). Moreover, different patterns of lectin-binding were found between different strains of An. stephensi and An. albimanus, and between female and male salivary glands of Anopheles gambiae (Mohamed et al., 1991; Andrews et al., 1997). This suggests that the presence of different carbohydrates on the outer surface of the salivary glands may provide a relatively unique marking of tissue identity. What's more, some lectins bind specifically to different portions of the salivary gland lobes. Lectins Peanut Agglutinin (PNA), Ricinus communis Agglutinin (RCA), Soybean Agglutinin (SBA), Ulex europaeus Agglutinin (UEA), and Wheat Germ Agglutinin (WGA) (Table 3) bind the medial and distal-lateral, but not proximal-lateral portions of the salivary gland of the mosquito Ae. aegypti, the known regions for sporozoite penetration (Perrone et al., 1986). Strong differences in the lectin-binding pattern were also found in the mosquito An. gambiae, where PNA, RCA, and UEA were found to bind at the proximal part of lateral and medial lobes and SBA bound to the medial and entire lateral lobes (Ghosh et al., 2009). This pattern is thus distinct from Ae. aegypti, highlighting that salivary gland carbohydrate moieties are distinct in different species. These data collectively indicate an important functional diversity in the salivary gland, in its different regions, between male and female, and mosquito species.
Evidence that carbohydrates may serve as receptor molecules for sporozoites was first provided by Barreau et al. (1995) following the demonstration that specific lectins were able to block parasite invasion of Ae. aegypti salivary gland when co-injected with P. gallinaceum oocyst sporozoites. These investigators tested seven lectins, with succinylated wheat germ agglutinin (S-WGA) and WGA blocking invasion almost completely and both Pisum sativum Agglutinin (PSA) and SBA, partially blocking invasion. No parasite blocking was observed when using Concanavalin A (Con A), Dolichos biflorus Agglutinin (DBA) and Phaseolus vulgaris Erythroagglutinin (PHA-E). This work reinforced the idea that the specific localization of certain carbohydrate moieties, specifically N-acetylglucosamine and mannosyl moieties, are required for parasite penetration of the salivary glands. Table 3 summarizes the lectins used on the experiments mentioned above, as well as their sugar binding preferences and salivary gland binding localization.
If parasite binding to sugars is required for entry into the salivary glands, then treating parasite preparations with an excess of the relevant carbohydrate should inhibit sporozoite binding. In order to evaluate the inhibitory effects of carbohydrates on sporogonic development, An. stephensi mysorensis were allowed to ingest blood meal containing P. vivax gametocytes and mannose, lactose or GalNAc, carbohydrates previously detected on the surface of mosquito salivary glands and midguts (Mohamed et al., 1991; Wilkins and Billingsley, 2001). Sporozoite invasion of salivary glands was completely abolished while only minor perturbations were observed on the development of ookinetes and oocysts, suggesting that those three carbohydrates were able to block sporozoite invasion by binding to their respective ligand on the sporozoite surface used to penetrate the gland (Basseri et al., 2008).
Additional evidence highlighting the use of sugars by sporozoites in the recognition and invasion of salivary glands involves heparan sulfate proteoglycans (HSPG) and chondroitin sulfate proteoglycans (CSPG). It has been suggested that sporozoites initiate attachment on the hepatocyte surface via HSPG through the binding of CSP (Pinzon-Ortiz et al., 2001). CSPGs have also been implicated in the attachment of ookinetes in the apical midgut microvilli, and silencing O-xylosyltransferease (the enzyme responsible for adding β-D-xylosyl to the side chain oxygen of a serine residue of a protein) substantially inhibited parasite development (Dinglasan et al., 2007), indicating a role for these molecules in tissue- binding and parasite life cycle progression. HSPGs and CSPGs were found to be present on the salivary gland surface in adult female An. stephensi mosquitoes, and that the major sporozoite surface protein, CSP, is able to bind to both chondroitin and heparan sulfate (Gantt et al., 1997; Sinnis et al., 2007). Finally, characterization of O-xylosyltransferase 1 (AgOXT1) of Anopheles gambiae confirmed that AgOXT1 could modify serine residues on known HSPG acceptor peptides (Armistead et al., 2011). Knockdown of AgOXT1 also reduced, but did not eliminate invasion of An. gambiae salivary glands by P. falciparum sporozoites (Armistead et al., 2011). This suggests that HSPGs may not be the only molecules that are important in the invasion process. A conditional, tissue specific knockout of AgOXT1 (as null mutants may not be viable), could clarify if a complete blockage of parasite invasion could be achieved, as RNAi-knockdown experiments may not have fully depleted the salivary glands of AgOXT1.
Direct information concerning mosquito protein glycosylation and the genes involved remains scarce. Silencing genes responsible for HS or CS biosynthesis or the core proteins of the proteoglycans, in addition to the enzymes responsible for protein glycosylation specifically in the salivary glands could help elucidate the molecules involved in the initial recognition of salivary gland and those involved in the invasion process. With that in mind, HS is found linked to core proteins like syndecan and glypican (integral membrane and GPI-anchored proteins, respectively) and perlecan proteoglycans (extracellular matrix proteins; Park et al., 2000). Interestingly, syndecan expression is high in salivary glands as shown in a transcriptome analysis of this organ (Valenzuela et al., 2003). To this end, knockout experiments of this specific core protein could be a first step on understanding the actual importance of the proteoglycans in the sporozoite recognition/invasion process.
Protein Receptor-Based Salivary Gland Recognition
SGS1
The first putative receptor described was Ae. aegypti salivary gland surface protein 1 (aaSGS1), a putative receptor for P. gallinaceum sporozoites. Antibodies developed against aaSGS1 were inoculated into infected Ae. aegypti mosquitoes prior to rupture of midgut oocysts, and sporozoites that invaded salivary glands were counted. Anti-aaSGS1 was able to block about 65% of sporozoite invasion in the salivary glands (Korochkina et al., 2006). In addition, SGS1 was found to be expressed on the salivary gland surface, closely associated with the plasma membrane and restricted to the median and distal lateral lobes of the salivary glands, the portions preferentially invaded by sporozoites. Reverse genetic analysis is still needed to evaluate the role of aaSGS1 as a receptor to confirm these findings. Additional experiments such as pull down assays could be performed to discover the corresponding parasite protein that might interact with aaSGS1. While homologs of SGS1 are present in Anopheline mosquitoes and are specifically expressed in salivary glands (Korochkina et al., 2006), it is unclear if these molecules also serve as parasite receptors.
Saglin
In An. gambiae, a protein termed Saglin was described as the receptor for P. falciparum TRAP (Ghosh et al., 2009). Initially, antibodies raised against An. gambiae salivary glands led to the identification of a 100kDa protein, Saglin; biochemical characterization revealed it as a disulphide-bonded homodimer of a 50 kDa protein (Okulate et al., 2007). Monoclonal antibodies against Saglin protein fed to infected females inhibited by 73% the invasion of salivary glands by P. yoelli (Brennan et al., 2000) and 95% by P. falciparum when injected intrathoracically (Ghosh et al., 2009). Furthermore, immunofluorescence microscopy revealed Saglin in the distal lateral lobes of the salivary glands (Brennan et al., 2000). SM1, a synthetic peptide that binds to salivary gland surface and also inhibited parasite invasion of this organ (Ghosh et al., 2001) was found to be a mimotope of the TRAP A domain (Ghosh et al., 2009). This domain harbors an important module called MIDAS; the interaction of TRAP and Saglin was found to be mediated via this module.
While these experiments provide evidence that Saglin is a receptor for sporozoites on the salivary gland surface, recent work has questioned this role (O'Brochta et al., 2019). These authors found Saglin to be expressed in the medial and proximal lateral lobes, not in the distal lateral lobe (Brennan et al., 2000; Ghosh et al., 2009). In addition, ectopic expression of Saglin driven by the strong salivary gland promoter AAPP in the mosquito An. stephensi did not result in an increase of salivary gland colonization by sporozoites (O'Brochta et al., 2019). One explanation could be that Saglin requires a species-specific co-factor to allow parasite invasion; such co-factors would be missing from ecotopic experiments in An. stephensi. Alternatively, a failure to increase invasion could be due to the salivary glands already being completely permissive. Ultimately, genetic evidence such as complete loss-of-function of Saglin, as well as a characterization of any co-factors or interactors will help resolve these questions.
CSPBP
Using a yeast surface display-screening assay, a protein termed CSP Binding Protein (CSPBP) was described as a potential receptor for P. berghei. Anti-CSPBP antibodies were found to label the salivary gland surface, and when fed to infected An. stephensi mosquitoes resulted in a 25 and 90% reduction in P. berghei load in infected salivary glands 14 and 18 days, respectively, after a blood meal (Wang et al., 2013). This data are somewhat curious, as they suggest sporozoites may be cleared from the salivary glands after invasion. Thus, CSPBP may be more important for survival of salivary gland sporozoites than for the invasion process itself. Silencing of CSPBP through RNAi also led to a 75% reduction in sporozoite invasion of salivary glands at day 18 (Wang et al., 2013).
While given the moniker “CSPBP” due to these observations, this protein is a 1:1 ortholog of Drosophila UPF3, and contains a highly conserved SMg4/UPF3 family domain (Figure 1). Interestingly, in members of the An. gambiae complex (Series Pyretophorus) the Upf3 protein contains both polyglycine and polyglutamine stretches interspersed between highly charged regions at the N-terminus (Figure 1). These low complexity regions are not found in other Anopheline species, or Culicine mosquitoes (Figure 1), and were included in the N-terminal antigen used to make the anti-CSPBP antibody (amino acids 1–260) that bound to salivary glands and blocked Plasmodium invasion as described by Wang et al. (2013). Could this be the interaction domain with CSP? Finer scale mapping is needed to determine the specific residues critical for mediating the interaction with CSP, and the role, if any, of these low-complexity regions. We note antibodies recognizing low-complexity regions may cross-react with other components of the extracellular matrix and might also account for observations of anti-CSPBP/UPF3 binding to the salivary gland surface, as well as interactions between CSPBP/Ufp3 and CSP during yeast surface display-screening assays. Performing similar experiments in other major vectors such as An. stephensi or An. funestus could help resolve this.
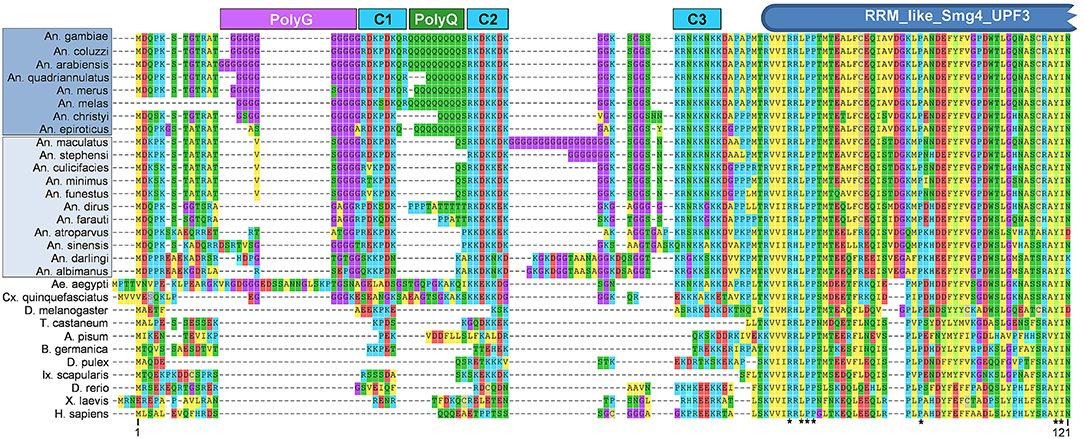
Figure 1. Alignment of N-terminal region of CSPBP/UPF3 across Metazoan taxa. Predicted amino acid sequences of CSPBP/UPF3 from eight species of Anopheles (An.) mosquitoes within Series Pyretophorus (Dark blue highlight), along with 11 other Anopheline species (light blue highlight), Culicine mosquitoes Aedes (Ae.) aegypti, and Culex (Cx.) quinquefasciatus, vinegar fly [Drosophila (D.) melanogaster], red flour beetle [Tribolium (T.) castaneum], pea aphid [Acyrthosiphon (A.) pisum], German cockroach [Blatella (B.) germanica], water flea [Daphia (D.) pulex], lyme disease tick [Ixodes (Ix.) scapularis], zebrafish [Danio (D.) rerio], African clawed frog [Xenopus (X.) laevis], and Homo (H.) sapiens. Protein sequences were aligned using MUSCLE (default parameters) as implemented by MEGA7.0 (Kumar et al., 2016); only the first 121 amino acids (An. gambiae) CSPBP/UPF3 are shown. Polyglycine (PolyG), polyglutamine (PolyQ), and charged (C1, C2, C3) regions are indicated, as is the RRM_like_Smg4_UPF3 domain predicted by NCBI Conserved Domain Search (Marchler-Bauer et al., 2015). *Indicates residues identical in all taxa. Protein sequences were obtained from Vectorbase [AGAP006649-PA, ACOM039391-PA, AARA008524-PA, AQUA004756-PA, AMEM012101-PA, AMEC019700-PA, ACHR004711-PA, AEPI005003-PA, AMAM019987-PA, ASTE011169-PA, ACUA004840-PA, AMIN007750-PA, AFUN004273-PA, ADIR007334-PA, AFAF009144-PA, AATE016492-PA, ASIS017649-PA, ADAC007751-PA, AALB004502-PA, AAEL003735-PA] or NCBI [XP_001847099.1, NP_726375.1, XP_015840521.1, XP_001942591.1, PSN55110.1, EFX71899.1, XP_002404304.1, NP_001189377.1, XP_018102580.1, AAG60690.1].
UPF3 belongs to a family of small, cytoplasmic, RNA binding proteins known to be involved in nonsense mediated mRNA decay (NMD), a mechanism initially described to perform a quality control of mRNAs with truncated open reading frames that contains a premature termination codon. While many proteins involved in NMD are not conserved across taxa, UPF1, UPF2, and UPF3 constitute the evolutionary conserved core set of NMD factors, present in all late-branching eukaryotes (Karousis et al., 2016). In Drosophila, Upf3 loss of function mutants are viable, but display a delay in development whereas mutants for Upf1 and 2 die during the larval stage. Upf3 is known to play more of a peripheral role in the degradation of mRNAs during embryogenesis (Avery et al., 2011). Little is known concerning the NMD pathway in mosquitoes, thus, in addition to a direct role in facilitating parasite invasion, the reduced ability of sporozoites to invade/survive the salivary glands following silencing of CSPBP/Upf3 could be due indirect effects associated with potential global disregulation of NMD. Such disregulation could disrupt the normal homeostasis of salivary gland proteins, immune effectors or energy production. Ultimately, more data is needed on the role of CSPBP/UPF3 in An. gambiae as well as other Anopheline mosquitoes, while potential disruptions to NMD upon CSPBP/UPF3 silencing require investigation. Also, the non-traditional route of secretion to the salivary gland surface as proposed for CSPBP/UPF3 (Wang et al., 2013) remains hypothetical.
PRS1
Plasmodium Responsive Salivary 1 (PRS1) was found to be upregulated in the distal lateral lobes of P. berghei-infected mosquitoes and its expression levels increased proportionally to the number of infecting sporozoites (Rosinski-Chupin et al., 2007). Interestingly, PRS1 was relocated into vesicle-like structures during the invasion process, although the protein did not co-localize with sporozoites, suggesting a role in intracellular response (maybe related to intense protein and/or membrane synthesis for repairing cell damage) rather than a direct interaction with sporozoites. Silencing of PRS1 in An. gambiae salivary glands resulted in a 2-fold decrease in P. falciparum sporozoite numbers (Chertemps et al., 2010). PRS1 belongs to an insect superfamily of genes encoding proteins with DM9 repeat motifs. Recently, the DM9 domain was implicated as a pattern recognition receptor (PRR), with broad microbial binding and agglutination activities in the mollusk Crassostrea gigas (Jiang et al., 2017). Further investigations, possibly using reverse genetics, of this protein as a PRR and sporozoite recognition factor are needed.
ESP
An Epithelial Serine Protease (AgESP) was found to be expressed in the distal region of the lateral and medial lobes on the basal side of the mosquito salivary glands and midgut. Silencing of this protease in An. gambiae mosquitoes led to a decrease in the number of both P. berghei and P. falciparum sporozoites, suggesting a role in sporozoite salivary gland invasion. In addition, knockdown of AgESP resulted in a corresponding decrease in the expression of SRPN6 in the midgut (Rodrigues et al., 2012). SRPN6 is a known marker for Plasmodium midgut and salivary gland invasion, as this protein is induced by rodent and human malaria parasites and its expression correlates with the course of infection (Abraham et al., 2005; Pinto et al., 2008). Knockdown of SRPN6 did not affect AgESP expression, showing that this protein acts upstream of SRPN6 (Rodrigues et al., 2012). The substrate for AgESP remains unknown; a mosquito molecule could be downstream of this reaction like SRPN6 on the midgut or AgESP could be directly acting on the sporozoite; an investigation of protein-protein interactions could be useful to describe the pathway relating AgESP to parasite development.
Glucose Transporter
RNA-seq data used to perform differential expression in the salivary glands of uninfected and Plasmodium-infected Anopheles mosquitoes identified a transmembrane glucose transporter gene as the most abundant upregulated transcript among transport genes (AGAP007752). Although its expression was not limited to the salivary gland, as male Malpighian tubules also expresses this transcript, RNA-seq and qPCR data have confirmed upregulation of this genes in infected female salivary glands (Baker et al., 2011; Pinheiro-Silva et al., 2015). Silencing this gene in An. coluzzii reduced the number P. berghei sporozoites present in the mosquito salivary gland by 44%. This led the authors to suggest that AGAP007752 might be another receptor for sporozoites or have a role in establishing optimal conditions for the parasite, as glucose levels may be critical to supplying the energy needed for subsequent vertebrate infection (Pinheiro-Silva et al., 2015). It is not clear yet if this protein is in fact a receptor and, if yes, what is the sporozoite protein that binds to it and the relevance in the process of sporozoite penetration, as this needs further evaluation.
Summary of Mosquito Factors Required for Sporozoite Invasion of Salivary Glands
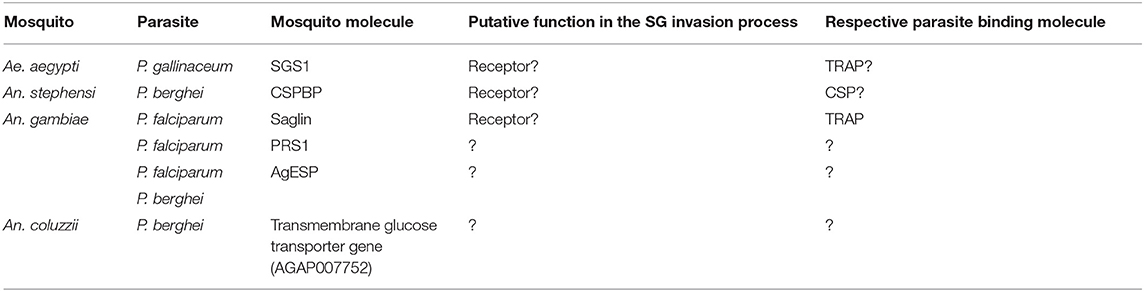
Table 4 summarizes mosquito proteins implicated in salivary gland invasion with their putative function and the respective parasite-binding molecule, if known. We emphasize that different pathways involving protein receptors, glycoproteins and proteoglycans, might mediate sporozoite attachment, and invasion of the salivary gland across multiple species. In one model, sporozoites may first localize to the salivary glands via sugars exposed on the haemocoel side of salivary gland. Subsequently, the parasites would use these or their associated proteoglycans as a bridge to one or more receptors like Saglin, aaSGS1, or AGAP007752 to start the invasion process. The opposite is also possible, with the sporozoite recognizing the salivary glands via a protein receptor and then invading utilizing the proteoglycans or the glycoproteins. We cannot exclude the possibility of sporozoites first attaching on both molecules on a collaborative way (Figure 2), and together this illustrates how little we understand of this critical process in the parasite life cycle. Advances in mosquito genome editing with CRISPR/Cas 9 (Basu et al., 2015; Kistler et al., 2015; Dong et al., 2018) open up the possibility to evaluate in more unambiguous terms each of these putative receptors encoded by the mosquito, and better dissect their role on the process of invasion of the salivary glands by sporozoites.
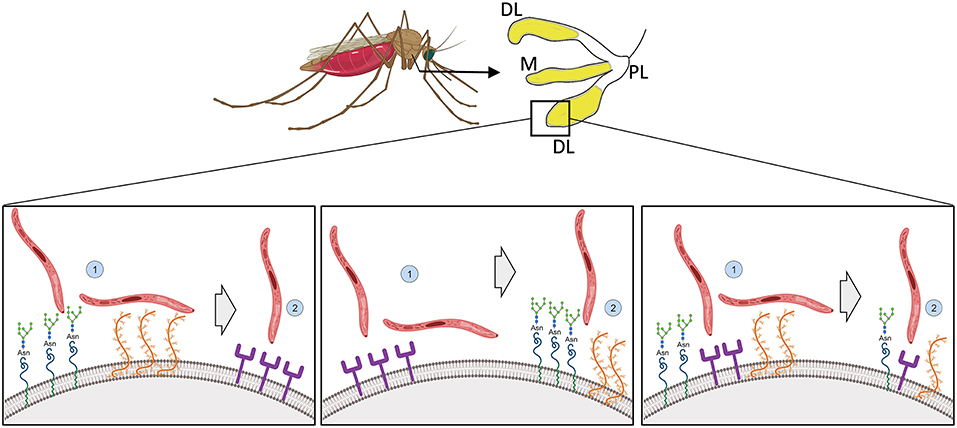
Figure 2. Illustration of possible modes of sporozoite recognition, attachment and invasion of mosquito salivary glands. Sporozoites preferentially invade the distal lateral and medial lobes of mosquito salivary glands (in yellow). How molecules on the salivary gland surface interact with the sporozoite to allow penetration is not known. (Left) Sporozoites recognize and attach to the salivary gland via glycoproteins and proteoglycans and invasion would be completed via protein based receptors. (Middle) Alternatively, the opposite would occur, with the recognition and attachment being done by protein based receptors and the invasion by the glycoproteins and proteoglycans. (Right) Both sugar and peptide-based motifs would be responsible together for recognition, attachment and invasion. Created with Biorender.com.
Conclusions
Sporozoite invasion of the mosquito salivary gland is a complex and multistep process involving recognition, attachment and invasion. Several molecules from the parasites have been shown to play a role in this phase of parasite development, however just a few molecules have been characterized so far from the mosquito and their exact role in this process remain elusive (Figure 3). Although progress is visible, the development of new tools to control malaria based on manipulation of the mosquito genome are hampered by gaps in our knowledge, such as those described here in regards to salivary gland invasion. In the light of modern techniques of molecular manipulation, in vivo imaging and omics, closing these gaps will increase our understanding of mosquito-parasite evolutionary dynamics and could lead to new technologies in order to interrupt malaria transmission.
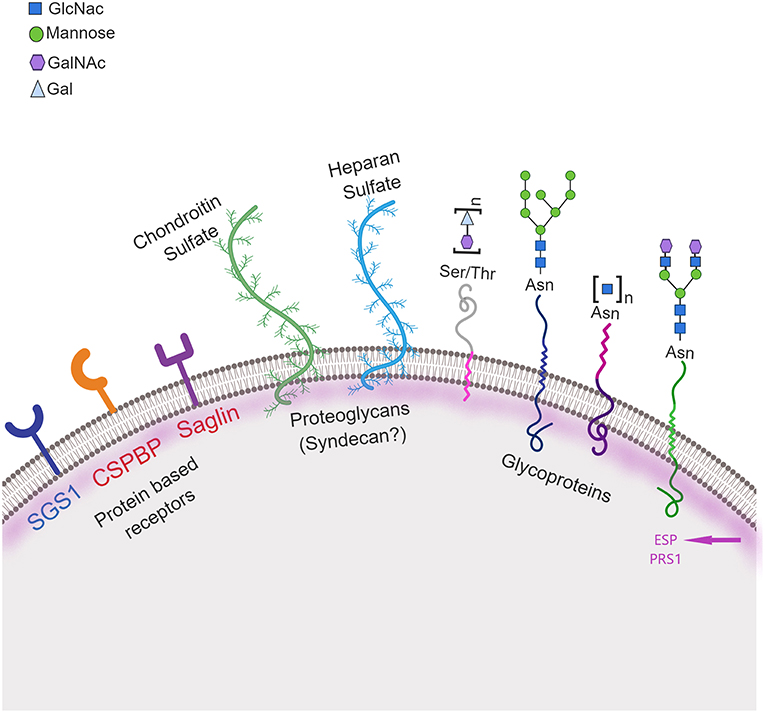
Figure 3. Schematic representation of mosquito molecules involved in salivary gland penetration by sporozoites. Three classes of molecules on the mosquito salivary gland surface may be involved in recognition, attachment and invasion by the parasite: protein receptor-based, including: CSPBP—circumsporozoite binding protein and Saglin, both identified in An. gambiae (in red) and SGS1 identified in Ae. aegypti (in blue); glycoproteins including proteins containing carbohydrate moieties, specifically N-acetylglucosamine (GlcNac), mannosyl, N-acetylgalactosamine (GalNac), and galactose (Gal) moieties; and proteoglycans including both chondroitin and heparan sulfate. Two other proteins, ESP—Epithelial Serine Protease and PRS1—Plasmodium Responsive Salivary 1 are also involved in this process. Ser, Serine; Thr, Threonine; Asn, Asparagine. Created with Biorender.com.
Author Contributions
BK and ZA conceived of the topic and edited the manuscript to produce the final version. BK drafted the initial manuscript.
Funding
This work was supported by Texas A&M Agrilife Research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abraham, E. G., Pinto, S. B., Ghosh, A., Vanlandingham, D. L., Budd, A., Higgs, S., et al. (2005). An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 102, 16327–16332. doi: 10.1073/pnas.0508335102
Akaki, M. (2005). A chemotactic response facilitates mosquito salivary gland infection by malaria sporozoites. J. Exp. Biol. 208, 3211–3218. doi: 10.1242/jeb.01756
Alavi, Y., Arai, M., Mendoza, J., Tufet-Bayona, M., Sinha, R., Fowler, K., et al. (2003). The dynamics of interactions between Plasmodium and the mosquito: a study of the infectivity of Plasmodium berghei and Plasmodium gallinaceum, and their transmission by Anopheles stephensi, Anopheles gambiae, and Aedes aegypti. Int. J. Parasitol. 33, 933–943. doi: 10.1016/S0020-7519(03)00112-7
Aly, A. S. I., and Matuschewski, K. (2005). A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J. Exp. Med. 202, 225–230. doi: 10.1084/jem.20050545
Aly, A. S. I., Vaughan, A. M., and Kappe, S. H. I. (2009). Malaria parasite development in the mosquito and infection of the mammalian host. Annu. Rev. Microbiol. 63, 195–221. doi: 10.1146/annurev.micro.091208.073403
Andrews, L., Laughinghouse, A., and Sina, B. J. (1997). Lectin binding characteristics of male and female salivary gland proteins of Anopheles gambiae: identification and characterization of female specific glycoproteins. Insect Biochem. Mol. Biol. 27, 159–166. doi: 10.1016/S0965-1748(96)00081-1
Arents, G., and Moudrianakis, E. N. (1995). The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. U.S.A. 92, 11170–11174. doi: 10.1073/pnas.92.24.11170
Armistead, J. S., Wilson, I. B. H., van Kuppevelt, T. H., and Dinglasan, R. R. (2011). A role for heparan sulfate proteoglycans in Plasmodium falciparum sporozoite invasion of anopheline mosquito salivary glands. Biochem. J. 438, 475–483. doi: 10.1042/BJ20110694
Aurrecoechea, C., Brestelli, J., Brunk, B. P., Dommer, J., Fischer, S., Gajria, B., et al. (2009). PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543. doi: 10.1093/nar/gkn814
Avery, P., Vicente-Crespo, M., Francis, D., Nashchekina, O., Alonso, C. R., and Palacios, I. M. (2011). Drosophila Upf1 and Upf2 loss of function inhibits cell growth and causes animal death in a Upf3-independent manner. RNA 17, 624–638. doi: 10.1261/rna.2404211
Baker, D. A., Nolan, T., Fischer, B., Pinder, A., Crisanti, A., and Russell, S. (2011). A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics. 12:296. doi: 10.1186/1471-2164-12-296
Banerjee, R., Liu, J., Beatty, W., Pelosof, L., Klemba, M., and Goldberg, D. E. (2002). Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc. Natl. Acad. Sci. U.S.A. 99, 990–995. doi: 10.1073/pnas.022630099
Barreau, C., Touray, M., Pimenta, P. F., Miller, L. H., and Vernick, K. D. (1995). Plasmodium gallinaceum: sporozoite invasion of Aedes aegypti salivary glands is inhibited by anti-gland antibodies and by lectins. Exp. Parasitol. 81, 332–343. doi: 10.1006/expr.1995.1124
Basseri, H. R., Doosti, S., Akbarzadeh, K., Nateghpour, M., Whitten, M. M., and Ladoni, H. (2008). Competency of Anopheles stephensi mysorensis strain for Plasmodium vivax and the role of inhibitory carbohydrates to block its sporogonic cycle. Malar. J. 7:131. doi: 10.1186/1475-2875-7-131
Basu, S., Aryan, A., Overcash, J. M., Samuel, G. H., Anderson, M. A. E., Dahlem, T. J., et al. (2015). Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 112, 4038–4043. doi: 10.1073/pnas.1502370112
Bhanot, P., Frevert, U., Nussenzweig, V., and Persson, C. (2003). Defective sorting of the thrombospondin-related anonymous protein (TRAP) inhibits Plasmodium infectivity. Mol. Biochem. Parasitol. 126, 263–273. doi: 10.1016/S0166-6851(02)00295-5
Blair, P. L., Kappe, S. H. I., Maciel, J. E., Balu, B., and Adams, J. H. (2002). Plasmodium falciparum MAEBL is a unique member of the ebl family. Mol. Biochem. Parasitol. 122, 35–44. doi: 10.1016/S0166-6851(02)00067-1
Brennan, J. D., Kent, M., Dhar, R., Fujioka, H., and Kumar, N. (2000). Anopheles gambiae salivary gland proteins as putative targets for blocking transmission of malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 97, 13859–13864. doi: 10.1073/pnas.250472597
Cerami, C., Frevert, U., Sinnis, P., Takacs, B., Clavijo, P., Santos, M. J., et al. (1992). The basolateral domain of the hepatocyte plasma membrane bears receptors for the circumsporozoite protein of Plasmodium falciparum sporozoites. Cell 70, 1021–1033. doi: 10.1016/0092-8674(92)90251-7
Cerami, C., Frevert, U., Sinnis, P., Takacs, B., and Nussenzweig, V. (1994). Rapid clearance of malaria circumsporozoite protein (CS) by hepatocytes. J. Exp. Med. 179, 695–701. doi: 10.1084/jem.179.2.695
Chertemps, T., Mitri, C., Perrot, S., Sautereau, J., Jacques, J. C., Thiery, I., et al. (2010). Anopheles Gambiae PRS1 modulates Plasmodium development at both midgut and salivary gland steps. PLoS ONE 5:e11538. doi: 10.1371/journal.pone.0011538
Clayton, A. M., Dong, Y., and Dimopoulos, G. (2014). The anopheles innate immune system in the defense against malaria infection. J. Innate Immun. 6, 169–181. doi: 10.1159/000353602
Combe, A., Moreira, C., Ackerman, S., Thiberge, S., Templeton, T. J., and Ménard, R. (2009). TREP, a novel protein necessary for gliding motility of the malaria sporozoite. Int. J. Parasitol. 39, 489–496. doi: 10.1016/j.ijpara.2008.10.004
Coppi, A., Natarajan, R., Pradel, G., Bennett, B. L., James, E. R., Roggero, M. A., et al. (2011). The malaria circumsporozoite protein has two functional domains, each with distinct roles as sporozoites journey from mosquito to mammalian host. J. Exp. Med. 208, 341–356. doi: 10.1084/jem.20101488
Currà, C., Gessmann, R., Pace, T., Picci, L., Peruzzi, G., Varamogianni-Mamatsi, V., et al. (2016). Release of Plasmodium sporozoites requires proteins with histone-fold dimerization domains. Nat. Commun. 7:13846. doi: 10.1038/ncomms13846
Dinglasan, R. R., Alaganan, A., Ghosh, A. K., Saito, A., van Kuppevelt, T. H., and Jacobs-Lorena, M. (2007). Plasmodium falciparum ookinetes require mosquito midgut chondroitin sulfate proteoglycans for cell invasion. Proc. Natl. Acad. Sci. U.S.A. 104, 15882–15887. doi: 10.1073/pnas.0706340104
Dong, Y., Simões, M. L., Marois, E., and Dimopoulos, G. (2018). CRISPR/Cas9 -mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection. PLoS Pathog. 14:e1006898. doi: 10.1371/journal.ppat.1006898
Douradinha, B., Augustijn, K. D., Moore, S. G., Ramesar, J., Mota, M. M., Waters, A. P., et al. (2011). Plasmodium cysteine repeat modular proteins 3 and 4 are essential for malaria parasite transmission from the mosquito to the host. Malar. J. 10:71. doi: 10.1186/1475-2875-10-71
Engelmann, S., Silvie, O., and Matuschewski, K. (2009). Disruption of plasmodium sporozoite transmission by depletion of sporozoite invasion-associated protein 1. Eukaryot. Cell 8, 640–648. doi: 10.1128/EC.00347-08
Florens, L., Washburn, M. P., Raine, J. D., Anthony, R. M., Grainger, M., Haynes, J. D., et al. (2002). A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526. doi: 10.1038/nature01107
Frevert, U. (2004). Malaria circumsporozoite protein binds to heparan sulfate proteoglycans associated with the surface membrane of hepatocytes. J. Exp. Med. 177, 1287–1298. doi: 10.1084/jem.177.5.1287
Frischknecht, F., and Matuschewski, K. (2017). Plasmodium sporozoite biology. Cold Spring Harb. Perspect. Med. 7:a025478. doi: 10.1101/cshperspect.a025478
Gantt, S., Persson, C., Rose, K., Birkett, A. J., Abagyan, R., and Nussenzweig, V. (2000). Antibodies against thrombospondin-related anonymous protein do not inhibit Plasmodium sporozoite infectivity in vivo. Infect. Immun. 68, 3667–3673. doi: 10.1128/IAI.68.6.3667-3673.2000
Gantt, S. M., Clavijo, P., Bai, X., Esko, J. D., and Sinnis, P. (1997). Cell adhesion to a motif shared by the malaria circumsporozoite protein and thrombospondin is mediated by its glycosaminoglycan-binding region and not by CSVTCG. J. Biol. Chem. 272, 19205–19213. doi: 10.1074/jbc.272.31.19205
Ghosh, A. K., Devenport, M., Jethwaney, D., Kalume, D. E., Pandey, A., Anderson, V. E., et al. (2009). Malaria parasite invasion of the mosquito salivary gland requires interaction between the Plasmodium TRAP and the Anopheles saglin proteins. PLoS Pathog. 5:e1000265. doi: 10.1371/journal.ppat.1000265
Ghosh, A. K., Ribolla, P. E., and Jacobs-Lorena, M. (2001). Targeting Plasmodium ligands on mosquito salivary glands and midgut with a phage display peptide library. Proc. Natl. Acad. Sci. U.S.A. U. S. A. 98, 13278–13281. doi: 10.1073/pnas.241491198
Golenda, C. F., Starkweather, W. H., and Wirtz, R. A. (1990). The distribution of circumsporozoite protein (CS) in Anopheles stephensi mosquitoes infected with Plasmodium falciparum malaria. J. Histochem. Cytochem. 38, 475–481. doi: 10.1177/38.4.2181019
Grassi, G. B. (1900). Studi di uno Zoologo sulla Malaria. Roma: Mem Rend Accad Lincei. doi: 10.5962/bhl.title.37999
Hall, N., Karras, M., Raine, J. D., Carlton, J. M., Kooij, T. W. A., Berriman, M., et al. (2005). A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307, 82–86. doi: 10.1126/science.1103717
Hamilton, A. J., Suhrbier, A., Nicholas, J., and Sinden, R. E. (1988). Immunoelectron microscopic localization of circumsporozoite antigen in the differentiating exoerythrocytic trophozoite of Plasmodium berghei. Cell Biol. Int. Rep. 12, 123–129. doi: 10.1016/0309-1651(88)90126-9
Hillyer, J. F., Barreau, C., and Vernick, K. D. (2007). Efficiency of salivary gland invasion by malaria sporozoites is controlled by rapid sporozoite destruction in the mosquito haemocoel. Int. J. Parasitol. 37, 673–681. doi: 10.1016/j.ijpara.2006.12.007
Ishino, T., Murata, E., Tokunaga, N., Baba, M., Tachibana, M., Thongkukiatkul, A., et al. (2019). Rhoptry neck protein 2 expressed in Plasmodium sporozoites plays a crucial role during invasion of mosquito salivary glands. Cell. Microbiol. 21:e12964. doi: 10.1111/cmi.12964
James, A. A., and Rossignol, P. A. (1991). Mosquito salivary glands: parasitological and molecular aspects. Parasitol. Today 7, 267–271. doi: 10.1016/0169-4758(91)90092-3
Jiang, S., Wang, L., Huang, M., Jia, Z., Weinert, T., Warkentin, E., et al. (2017). DM9 domain containing protein functions as a pattern recognition receptor with broad microbial recognition spectrum. Front. Immunol. 8:1607. doi: 10.3389/fimmu.2017.01607
Kaiser, K., Camargo, N., Coppens, I., Morrisey, J. M., Vaidya, A. B., and Kappe, S. H. I. (2004a). A member of a conserved Plasmodium protein family with membrane-attack complex/perforin (MACPF)-like domains localizes to the micronemes of sporozoites. Mol. Biochem. Parasitol. 133, 15–26. doi: 10.1016/j.molbiopara.2003.08.009
Kaiser, K., Matuschewski, K., Camargo, N., Ross, J., and Kappe, S. H. I. (2004b). Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol. Microbiol. 51, 1221–1232. doi: 10.1046/j.1365-2958.2003.03909.x
Kappe, S., Bruderer, T., Gantt, S., Fujioka, H., Nussenzweig, V., and Ménard, R. (1999). Conservation of a gliding motility and cell invasion machinery in Apicomplexan parasites. J. Cell Biol. 147, 937–943. doi: 10.1083/jcb.147.5.937
Kappe, S. H., Gardner, M. J., Brown, S. M., Ross, J., Matuschewski, K., Ribeiro, J. M., et al. (2001). Exploring the transcriptome of the malaria sporozoite stage. Proc. Natl. Acad. Sci. U.S.A. 98, 9895–9900. doi: 10.1073/pnas.171185198
Kappe, S. H. I., Noe, A. R., Fraser, T. S., Blair, P. L., and Adams, J. H. (1998). A family of chimeric erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 95, 1230–1235. doi: 10.1073/pnas.95.3.1230
Kariu, T., Yuda, M., Yano, K., and Chinzei, Y. (2002). MAEBL is essential for malarial sporozoite infection of the mosquito salivary gland. J. Exp. Med. 195, 1317–1323. doi: 10.1084/jem.20011876
Karousis, E. D., Nasif, S., and Mühlemann, O. (2016). Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip. Rev. RNA 7, 661–682. doi: 10.1002/wrna.1357
Kistler, K. E., Vosshall, L. B., and Matthews, B. J. (2015). Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 11, 51–60. doi: 10.1016/j.celrep.2015.03.009
Klug, D., and Frischknecht, F. (2017). Motility precedes egress of malaria parasites from oocysts. Elife 6:e19157. doi: 10.7554/eLife.19157
Korochkina, S., Barreau, C., Pradel, G., Jeffery, E., Li, J., Natarajan, R., et al. (2006). A mosquito-specific protein family includes candidate receptors for malaria sporozoite invasion of salivary glands. Cell. Microbiol. 8, 163–175. doi: 10.1111/j.1462-5822.2005.00611.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lasonder, E., Janse, C. J., Van Gemert, G. J., Mair, G. R., Vermunt, A. M. W., Douradinha, B. G., et al. (2008). Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog. 4:e1000195. doi: 10.1371/journal.ppat.1000195
Lindner, S. E., Swearingen, K. E., Harupa, A., Vaughan, A. M., Sinnis, P., Moritz, R. L., et al. (2013). Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol. Cell. Proteomics 12, 1127–1143. doi: 10.1074/mcp.M112.024505
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 45:D200–D203. doi: 10.1093/nar/gku1221
Marois, E. (2011). The multifaceted mosquito anti-Plasmodium response. Curr. Opin. Microbiol. 14, 429–435. doi: 10.1016/j.mib.2011.07.016
Mastan, B. S., Narwal, S. K., Dey, S., Kumar, K. A., and Mishra, S. (2017). Plasmodium berghei plasmepsin VIII is essential for sporozoite gliding motility. Int. J. Parasitol. 47, 239–245. doi: 10.1016/j.ijpara.2016.11.009
Matuschewski, K., Nunes, A. C., Nussenzweig, V., and Ménard, R. (2002). Plasmodium sporozoite invasion into insect and mammalian cells is directed by the same dual binding system. EMBO J. 21, 1597–1606. doi: 10.1093/emboj/21.7.1597
Ménard, R., Sultan, A. A., Cortes, C., Altszuler, R., Van Dijk, M. R., Janse, C. J., et al. (1997). Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature 385, 336–339. doi: 10.1038/385336a0
Mikolajczak, S. A., Silva-Rivera, H., Peng, X., Tarun, A. S., Camargo, N., Jacobs-Lorena, V., et al. (2008). Distinct malaria parasite sporozoites reveal transcriptional changes that cause differential tissue infection competence in the mosquito vector and mammalian host. Mol. Cell. Biol. 28, 6196–6207. doi: 10.1128/MCB.00553-08
Mohamed, H. A., Ingram, G. A., Molyneux, D. H., and Sawyer, B. V. (1991). Use of fluorescein-labelled lectin binding of salivary glands to distinguish between Anopheles stephensi and An. Albimanus species and strains. Insect Biochem. 21, 767–773. doi: 10.1016/0020-1790(91)90118-X
Myung, J. M., Marshall, P., and Sinnis, P. (2004). The Plasmodium circumsporozoite protein is involved in mosquito salivary gland invasion by sporozoites. Mol. Biochem. Parasitol. 133, 53–59. doi: 10.1016/j.molbiopara.2003.09.002
Nagasawa, H., Procell, P. M., Atkinson, C. T., Campbell, G. H., Collins, W. E., and Aikawa, M. (1987). Localization of circumsporozoite protein of Plasmodium ovale in midgut oocysts. Infect. Immun. 55, 2928–2932.
O'Brochta, D. A., Alford, R., Harrell, R., Aluvihare, C., Eappen, A. G., Li, T., et al. (2019). Is Saglin a mosquito salivary gland receptor for Plasmodium falciparum? Malar. J. 18:2. doi: 10.1186/s12936-018-2634-5
Okulate, M. A., Kalume, D. E., Reddy, R., Kristiansen, T., Bhattacharyya, M., Chaerkady, R., et al. (2007). Identification and molecular characterization of a novel protein Saglin as a target of monoclonal antibodies affecting salivary gland infectivity of Plasmodium sporozoites. Insect Mol. Biol. 16, 711–722. doi: 10.1111/j.1365-2583.2007.00765.x
Orfano, A. S., Nacif-Pimenta, R., Duarte, A. P. M., Villegas, L. M., Rodrigues, N. B., Pinto, L. C., et al. (2016). Species-specific escape of Plasmodium sporozoites from oocysts of avian, rodent, and human malarial parasites. Malar. J. 15:394. doi: 10.1186/s12936-016-1451-y
Park, P. W., Reizes, O., and Bernfield, M. (2000). Cell surface heparan sulfate proteoglycans: selective regulators of ligand-receptor encounters. J. Biol. Chem. 275, 29923–29926. doi: 10.1074/jbc.R000008200
Perrone, J. B., DeMaio, J., and Spielman, A. (1986). Regions of mosquito salivary glands distinguished by surface lectin-binding characteristics. Insect Biochem. 16, 313–318. doi: 10.1016/0020-1790(86)90041-7
Pimenta, P. F., Touray, M., and Miller, L. (1994). The journey of malaria sporozoites in the mosquito salivary gland. J. Eukaryot. Microbiol. 41, 608–624. doi: 10.1111/j.1550-7408.1994.tb01523.x
Pinheiro-Silva, R., Borges, L., Coelho, L. P., Cabezas-Cruz, A., Valdés, J. J., Do Rosário, V., et al. (2015). Gene expression changes in the salivary glands of Anopheles coluzzii elicited by Plasmodium berghei infection. Parasites Vectors 8:485. doi: 10.1186/s13071-015-1079-8
Pinto, S. B., Kafatos, F. C., and Michel, K. (2008). The parasite invasion marker SRPN6 reduces sporozoite numbers in salivary glands of Anopheles gambiae. Cell. Microbiol. 10, 891–898. doi: 10.1111/j.1462-5822.2007.01091.x
Pinzon-Ortiz, C., Friedman, J., Esko, J., and Sinnis, P. (2001). The binding of the circumsporozoite protein to cell surface heparan sulfate proteoglycans is required for plasmodium sporozoite attachment to target cells. J. Biol. Chem. 276, 26784–26791. doi: 10.1074/jbc.M104038200
Posthuma, G., Meis, J. F., Verhave, J. P., Hollingdale, M. R., Ponnudurai, T., Meuwissen, J. H., et al. (1988). Immunogold localization of circumsporozoite protein of the malaria parasite Plasmodium falciparum during sporogony in Anopheles stephensi midguts. Eur. J. Cell Biol. 46, 18–24.
Robson, K. J. H., Hall, J. R. S., Jennings, M. W., Harris, T. J. R., Marsh, K., Newbold, C. I., et al. (1988). A highly conserved amino-acid sequence in thrombospondin, properdin and in proteins from sporozoites and blood stages of a human malaria parasite. Nature 335, 79–82. doi: 10.1038/335079a0
Rodrigues, J., Oliveira, G. A., Kotsyfakis, M., Dixit, R., Molina-Cruz, A., Jochim, R., et al. (2012). An epithelial serine protease, AgESP, is required for plasmodium invasion in the mosquito Anopheles gambiae. PLoS ONE 7:e35210. doi: 10.1371/journal.pone.0035210
Rogers, W. O., Malik, A., Mellouk, S., Nakamura, K., Rogers, M. D., Szarfman, A., et al. (1992). Characterization of Plasmodium falciparum sporozoite surface protein 2. Proc. Natl. Acad. Sci. U.S.A. 89, 9176–9180. doi: 10.1073/pnas.89.19.9176
Rosenberg, R. (1985). Inability of Plasmodium knowlesi sporozoites to invade Anopheles freeborni salivary glands. Am. J. Trop. Med. Hyg. 34, 687–691. doi: 10.4269/ajtmh.1985.34.687
Rosinski-Chupin, I., Briolay, J., Brouilly, P., Perrot, S., Gomez, S. M., Chertemps, T., et al. (2007). SAGE analysis of mosquito salivary gland transcriptomes during Plasmodium invasion. Cell. Microbiol. 9, 708–724. doi: 10.1111/j.1462-5822.2006.00822.x
Santos, J. M., Egarter, S., Zuzarte-Luís, V., Kumar, H., Moreau, C. A., Kehrer, J., et al. (2017). Malaria parasite LIMP protein regulates sporozoite gliding motility and infectivity in mosquito and mammalian hosts. Elife 6:e24109. doi: 10.7554/eLife.24109
Siden-Kiamos, I., Pace, T., Klonizakis, A., Nardini, M., Garcia, C. R. S., and Currà, C. (2018). Identification of Plasmodium berghei Oocyst Rupture Protein 2 (ORP2) domains involved in sporozoite egress from the oocyst. Int. J. Parasitol. 48, 1127–1136. doi: 10.1016/j.ijpara.2018.09.004
Sidjanski, S. P., Vanderberg, J. P., and Sinnis, P. (1997). Anopheles stephensi salivary glands bear receptors for region I of the circumsporozoite protein of Plasmodium falciparum. Mol. Biochem. Parasitol. 90, 33–41. doi: 10.1016/S0166-6851(97)00124-2
Simões, M. L., Caragata, E. P., and Dimopoulos, G. (2018). Diverse host and restriction factors regulate mosquito–pathogen interactions. Trends Parasitol. 34, 603–616. doi: 10.1016/j.pt.2018.04.011
Sinnis, P., Coppi, A., Toida, T., Toyoda, H., Kinoshita-Toyoda, A., Xie, J., et al. (2007). Mosquito heparan sulfate and its potential role in malaria infection and transmission. J. Biol. Chem. 282, 25376–25384. doi: 10.1074/jbc.M704698200
Srinivasan, P., Abraham, E. G., Ghosh, A. K., Valenzuela, J., Ribeiro, J. M. C., Dimopoulos, G., et al. (2004). Analysis of the Plasmodium and anopheles transcriptomes during oocyst differentiation. J. Biol. Chem. 279, 5581–5587. doi: 10.1074/jbc.M307587200
Steinbuechel, M., and Matuschewski, K. (2009). Role for the Plasmodium sporozoite-specific transmembrane protein S6 in parasite motility and efficient malaria transmission. Cell. Microbiol. 11, 279–288. doi: 10.1111/j.1462-5822.2008.01252.x
Sterling, C. R., Aikawa, M., and Vanderberg, J. P. (1973). The passage of Plasmodium berghei sporozoites through the salivary glands of Anopheles stephensi: an electron microscope study. J. Parasitol. 59, 593–605. doi: 10.2307/3278847
Sultan, A. A., Thathy, V., Frevert, U., Robson, K. J. H., Crisanti, A., Nussenzweig, V., et al. (1997). TRAP is necessary for gliding motility and infectivity of Plasmodium sporozoites. Cell 90, 511–522. doi: 10.1016/S0092-8674(00)80511-5
Swearingen, K. E., Lindner, S. E., Flannery, E. L., Vaughan, A. M., Morrison, R. D., Patrapuvich, R., et al. (2017). Proteogenomic analysis of the total and surface-exposed proteomes of Plasmodium vivax salivary gland sporozoites. PLoS Negl. Trop. Dis. 11:e0005791. doi: 10.1371/journal.pntd.0005791
Swearingen, K. E., Lindner, S. E., Shi, L., Shears, M. J., Harupa, A., Hopp, C. S., et al. (2016). Interrogating the Plasmodium sporozoite surface: identification of surface-exposed proteins and demonstration of glycosylation on CSP and TRAP by mass spectrometry-based proteomics. PLOS Pathog. 12:e1005606. doi: 10.1371/journal.ppat.1005606
Tewari, R., Spaccapelo, R., Bistoni, F., Holder, A. A., and Crisanti, A. (2002). Function of region I and II adhesive motifs of Plasmodium falciparum circumsporozoite protein in sporozoite motility and infectivity. J. Biol. Chem. 277, 47613–47618. doi: 10.1074/jbc.M208453200
Thathy, V., Fujioka, H., Gantt, S., Nussenzweig, R., Nussenzweig, V., and Ménard, R. (2002). Levels of circumsporozoite protein in the Plasmodium oocyst determine sporozoite morphology. EMBO J. 21, 1586–1596. doi: 10.1093/emboj/21.7.1586
Thompson, J., Fernandez-Reyes, D., Sharling, L., Moore, S. G., Eling, W. M., Kyes, S. A., et al. (2007). Plasmodium cysteine repeat modular proteins 1-4: complex proteins with roles throughout the malaria parasite life cycle. Cell. Microbiol. 9, 1466–1480. doi: 10.1111/j.1462-5822.2006.00885.x
Touray, M. G., Warburg, A., Laughinghouse, A., Krettli, A. U., and Miller, L. H. (1992). Developmentally regulated infectivity of malaria sporozoites for mosquito salivary glands and the vertebrate host. J. Exp. Med. 175, 1607–1612. doi: 10.1084/jem.175.6.1607
Valenzuela, J. G., Francischetti, I. M. B., Pham, V. M., Garfield, M. K., and Ribeiro, J. M. C. (2003). Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. Biol. 33, 717–732. doi: 10.1016/S0965-1748(03)00067-5
Wang, J., Zhang, Y., Zhao, Y. O., Li, M. W. M., Zhang, L., Dragovic, S., et al. (2013). Anopheles gambiae circumsporozoite protein-binding protein facilitates plasmodium infection of mosquito salivary glands. J. Infect. Dis. 208, 1161–1169. doi: 10.1093/infdis/jit284
Wang, Q., Fujioka, H., and Nussenzweig, V. (2005). Exit of plasmodium sporozoites from oocysts is an active process that involves the circumsporozoite protein. PLoS Pathog. 1:e9. doi: 10.1371/journal.ppat.0010009
Warburg, A., Touray, M., Krettli, A. U., and Miller, L. H. (1992). Plasmodium gallinaceum: antibodies to circumsporozoite protein prevent sporozoites from invading the salivary glands of Aedes aegypti. Exp. Parasitol. 75, 303–307. doi: 10.1016/0014-4894(92)90215-V
Wengelnik, K., Spaccapelo, R., Naitza, S., Robson, K. J. H., Janse, C. J., Bistoni, F., et al. (1999). The A-domain and the thrombospondin-related motif of Plasmodium falciparum TRAP are implicated in the invasion process of mosquito salivary glands. EMBO J. 18, 5195–5204. doi: 10.1093/emboj/18.19.5195
Wilkins, S., and Billingsley, P. F. (2001). Partial characterization of oligosaccharides expressed on midgut microvillar glycoproteins of the mosquito, Anopheles stephensi Liston. Insect Biochem. Mol. Biol. 31, 937–948. doi: 10.1016/S0965-1748(01)00040-6
Keywords: mosquito, sporozoite, salivary gland, plasmodium, SGS1, saglin, carbohydrate-binding
Citation: Kojin BB and Adelman ZN (2019) The Sporozoite's Journey Through the Mosquito: A Critical Examination of Host and Parasite Factors Required for Salivary Gland Invasion. Front. Ecol. Evol. 7:284. doi: 10.3389/fevo.2019.00284
Received: 20 May 2019; Accepted: 15 July 2019;
Published: 14 August 2019.
Edited by:
Jayme A. Souza-Neto, São Paulo State University, BrazilReviewed by:
Eric Calvo, National Institutes of Health, United StatesEric Marois, Institut National de la Santé et de la Recherche Médicale (INSERM), France
Copyright © 2019 Kojin and Adelman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bianca B. Kojin, YmlidXJpbmlAdGFtdS5lZHU=
 Bianca B. Kojin
Bianca B. Kojin Zach N. Adelman
Zach N. Adelman