- 1Naturalis Biodiversity Center, Leiden, Netherlands
- 2Department of Environmental Biology, Institute of Environmental Sciences, Leiden University, Leiden, Netherlands
Mosquito borne diseases pose a threat to human health worldwide. Disease risk is primarily determined by presence and abundance of vector species. A better understanding of mosquito diversity and abundance can direct improved vector control, but this requires a combination of monitoring techniques that yield both rapid and reliable information. Particularly improved larval detection is pivotal to move toward more targeted management with less environmental impact. Current detection methods rely strongly on manual labor and taxonomic expertise, which greatly limits the extent to which these methodologies can be employed. As such, insight in the efficiency of novel, high-throughput vs. traditional sampling techniques is required. We compared the effectiveness of a recently developed environmental DNA (eDNA) approach on water and sediment samples with other commonly used sampling techniques (“dipping” for larvae and adult trapping) in a field study on three Caribbean islands. All sampling methods were employed across a range of ecologically contrasting sites. Species identification was performed both morphologically and molecularly using an in-house developed reference database supplemented with sequences from BOLD and GenBank. Our analysis of water samples from 39 sites shows that eDNA sampling can be more reliable than dipping, yields a higher within-sample richness and produces a subset of the adult community in all sampled water types. Furthermore, for both adults and larvae, our identifications showed complete overlap between morphological and molecular approaches in 133 out of 134 samples. Overall, results from this study provide evidence that both our eDNA-based detection of larvae and our DNA-based identification of larvae and adults present methods that are, although more expensive, as reliable, and for some species even more reliable than the currently used methods. Additionally, our results highlight that a DNA approach can be used to identify larvae of early developmental stages, which generally lack important morphological characteristics. As such it allows for development of efficient disease control strategies, verification of management effectiveness and monitoring of population dynamics.
Introduction
Mosquitoes (Culicidae) present a major risk for human health worldwide (Leslie et al., 2017). They cause hundreds of thousands of deaths annually due to their role as vector for vector-borne diseases (VBD) such as chikungunya, dengue, malaria, yellow fever, and zika (WHO, 2017b). Even though our understanding of these diseases is growing, case incidences are increasing (Risks et al., 2012; WHO, 2017a). Occurrence of VBD is limited by the presence, abundance, and dispersal of their respective vectors (Schaffner and Mathis, 2014). Due to land use change, climate change and increased global trade and travel (Lambin et al., 2001; Patz et al., 2004; Risks et al., 2012), distributions of vector species, especially those of exotic species such as Aedes aegyti and Aedes albopictus are shifting across all continents (Petric et al., 2014). This highlights the importance of monitoring tools that can provide reliable, high-throughput and up-to-date information on species distributions, both for larvae and adults, especially for surveillance near hotspots of travel and trade, such as harbors and airports.
Traditionally, methodologies used for studying the distribution of mosquito larvae and adults rely strongly on morphological identification. This, combined with methodological challenges, greatly limits the extent to which these techniques can be employed. For example, to identify localities where vector control is needed, presence and, ideally, larval habitats of (disease-vectoring) mosquitoes have to be confirmed. Larval identification methods play a crucial role in the detection of these larval habitats because mosquito populations are generally limited by the availability of suitable habitats (Frank et al., 1988; Rejmánková et al., 2013). The detection is usually performed using a “dipping” method (hereafter referred to as dipping), in which larvae and pupa are physically caught and identified (van der Berg and Schaffner, 2016). In doing so, larval habitats can be specifically targeted with mosquito control measures, thus minimizing the impact on the environment. However, dipping can be cumbersome, since the larvae and pupa dive upon visual and auditory disturbance (Becker et al., 2013). Depending on the species, the dive can last up to several minutes, increasing the required sampling effort and possibly decreasing detection probability. In addition to these methodological challenges, samples collected using dipping are typically identified morphologically, which is prone to unresolved or misidentification due to phenotypic plasticity and cryptic species (Jerde et al., 2011; Deiner et al., 2013; Fišer Pečnikar and Buzan, 2014; Mächler et al., 2014). Some characteristics, for example, are only apparent at a certain life stage or gender (Murugan et al., 2016; van der Berg and Schaffner, 2016). This is especially true for culicid larvae, since most characteristics are only visible on the fourth instar (Becker et al., 2013; ECDC, 2014). Likewise, adult sampling is widely used to detect the mosquito species community at a given locality, and is generally carried out using a variety of trapping methods (Becker et al., 2013). Adult sampling may yield higher diversity, since it is independent of larval habitat preference. After trapping, all individuals need to be collected at dawn, thus limiting the number of trapping sites. Afterwards, individuals are sorted to species. In general, identification is performed using morphological keys. This method is therefore almost entirely dependent on taxonomic expertise, which is becoming increasingly difficult to get by Mächler et al. (2014), particularly in the highly biodiverse ecosystems of the tropics. Furthermore (recent), identification keys based on morphology are not available for many regions, with a particular lack of keys in tropical areas where mosquito diversity and mosquito borne disease risk are highest (Rawlins et al., 2008; WHO, 2017b).
Molecular approaches based on environmental DNA (DNA that organisms shed into their environment, hereafter eDNA) or DNA could comprise valuable additions or even alternatives for both larval and adult morphological methods. For larval stages, an eDNA approach can have three main benefits. First, it can greatly reduce the time needed for species collection (Herder et al., 2014) and is non-invasive in the sense that it does not harm the species under investigation (Thomsen and Willerslev, 2015). Second, larvae can—in theory—be detected after adult emergence (Barnes et al., 2014; Schneider et al., 2016) thus allowing for repeated sampling with larger time intervals. Third, detection rates for eDNA may be higher than those of traditional techniques, which has already been observed for taxa such as fishes, amphibians and gastropods (Thomsen et al., 2012; Goldberg et al., 2013; Pilliod et al., 2013; Mächler et al., 2014). A growing number of eDNA studies target macro-invertebrates (Roussel et al., 2015), from which the necessity of eDNA collection based on target species ecology can be inferred (Deiner et al., 2015). Sediment dwelling organisms such as amphipods, for instance, tend to be hard to detect in aquatic samples (Mächler et al., 2014). Despite difficulties with other invertebrate taxa, we expect that eDNA of mosquito larvae can be reliably detected, since these larvae live and molt near the water surface (Rueda, 2008), which can be expected to result in a local accumulation of eDNA, thus allowing for successful eDNA collection (Schneider et al., 2016). For adult mosquitoes, molecular identification would also represent a valuable methodological addition, especially if employed in combination with high-throughput sequencing (also known as next-gen sequencing, hereafter NGS). All individuals within a sample (hereafter bulk sample) can then be analyzed simultaneously, allowing for rapid identification of large numbers of specimens, rendering it less labor intensive and time consuming (Batovska et al., 2016). Moreover, it holds the promise to be less prone to identification mistakes and might even result in a more resolved identification (Fišer Pečnikar and Buzan, 2014), which is especially important for vector control.
However, there are a number of caveats when it comes to using molecular identification. First of all, molecular methods currently remain more costly than traditional methods and may therefore prove to be less readily available for routine surveys. Also, environmental samples in particular are known to contain components that hinder DNA amplification, such as humic substances and polysaccharides (Herder et al., 2014). The amount and variety and therefore the influence of these PCR inhibitors varies across the different types of larval habitats (Wilson, 1997; Schrader et al., 2012). Also, the larval habitats vary in their abiotic properties (Becker et al., 2013), which influences DNA degradation (Strickler et al., 2015). Species in certain water body types may therefore be harder to detect. A better understanding of the factors that determine the reliability and usability of eDNA collection and molecular identification is therefore required to implement these techniques for adequate mosquito vector monitoring.
The aim of this research is threefold: (i) to explore whether eDNA-based assessments of larval communities in water bodies match with the morphological analysis, thus testing the reliability of eDNA-based assessments of larval mosquito communities; (ii) to explore whether DNA-based identification of adult and larvae matches the morphological identifications, thus determining the usability of molecular identification for high-throughput assessments; (iii) to determine whether DNA-based methods can be used across ecologically contrasting habitats by comparing relationships between larval mosquito community composition, and abiotic properties of the various habitats, using both traditional and DNA-based methods.
To this end, a comparative analysis was carried out to determine the effectiveness of eDNA sampling and molecular identification using a recently developed culicid-specific primer (Krol et al., 2019) vs. traditional sampling and morphological identification for detection of larval, pupal, and adult culicids. We used the mosquito communities of the Caribbean islands Saba, St. Eustatius and St. Maarten (Lesser Antilles) as a study system. These islands are ecologically diverse and have a relatively limited and relatively well-known species pool (Van der Kuyp, 1954), and therefore provide an ideal study system.
Materials and Methods
All field work was conducted during April 2018 on the islands of St. Eustatius, St. Maarten and Saba. Samples of adults, larvae and eDNA were collected in the period of a single week for each of the three islands. For adults, a list of optimal trapping sites was gathered by consulting the local vector control units for knowledge on known larval habitats on each of the three islands. From this list 10 trapping sites per island were selected which cover all available habitats including the urban environment. Only for larvae, aquatic eDNA samples and sedimentary eDNA samples were collected; every water body that was encountered during intensive surveys on the island was included as a sample site. For each water body, we recorded the coordinates (Figure 1) and type (see below). Aquatic eDNA was collected at 36, 17, and 19 sites on St. Eustatius, Saba, and St. Maarten, respectively. Sedimentary eDNA was collected at 6 sites on St. Maarten only.
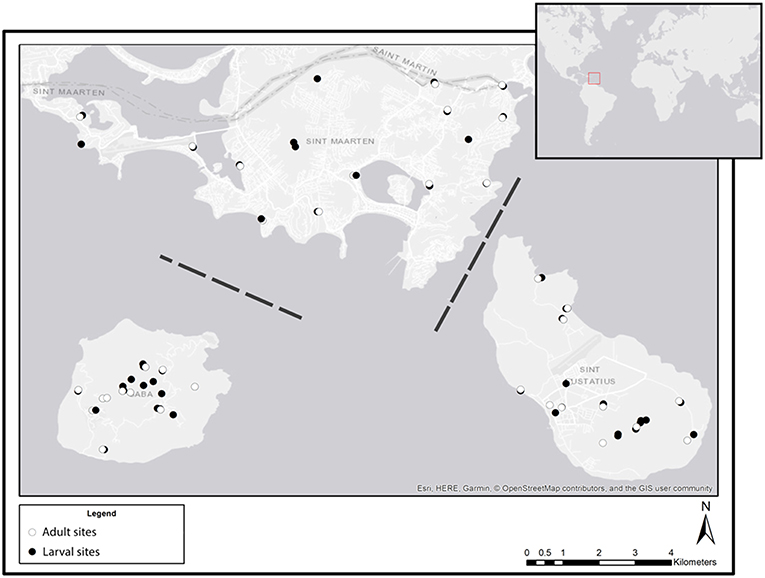
Figure 1. Overview of all sampling sites at each of the three sampled Caribbean islands: St. Maarten (top), Saba (bottom left), and St. Eustatius (bottom right). White symbols indicate sites where adults were sampled, black symbols indicate sites where larval (dipping) and eDNA (water and sediment) samples were collected. Dashed lines indicate that the distance between islands is not to scale (Esri, 2011).
Traditional Sampling of Adults, Larvae, and Pupae
Adult Mosquitoes
Samples of adult mosquitoes, consisting of all individuals caught per trapping method, were collected using Mosquito magnets (Executive), BG-sentinels v2, resting traps, sticky traps, and human-landing catches at each of the sites (Figure 1). The Mosquito magnets were deployed at ground level because the high spatial coverage of the Mosquito magnet is designed to capture mosquitoes from the entire air column, thereby overcoming the stratifying effects of possible host preference (Andreadis and Armstrong, 2007; Harbach, 2007). Placement was ~10 m leeward of larval habitats with a minimum distance of 100 meters between the trapping sites to allow for optimal spatial coverage (Harrington et al., 2005; Epopa et al., 2017; Medeiros et al., 2017). A similar approach was used for the BG-sentinel and resting trap (Burkett-Cadena, 2011). Sample collection encompassed 3 days to yield a representable composition of the mosquito community (Gorsich et al., pers. comm.). To minimize sampling bias which may arise from species-specific variation in lifestyle (as a result of e.g., varying flight times or feeding activity; Harbach, 2007; Panella et al., 2016), adult mosquitoes were collected during 24 h of continuous trapping. All traps were emptied between 5.30 and 7.30 a.m. to prevent the mosquitoes from drying out, which hinders morphological identification. BG-sentinels were baited with BG-lure and a sugar-yeast mixture of which the latter acted as CO2 source as alternative for dry-ice which proved unobtainable on all of the islands. Mosquito magnets were baited with octenol and CO2 using lure and propane combustion, respectively. The latter also served as an electricity source. The use of CO2 and bait is expected to increase yield (Bhalala and Arias, 2009; Hoel et al., 2009; Kweka et al., 2013). All stationary traps were placed out of direct sunlight to prevent captured mosquitoes from drying out. Traps were shielded from rain and wind to prevent damage and optimize the efficiency of the octenol and CO2. Human landing catches (ECDC, 2014) were performed each day at dusk.
Larvae and Pupae
Larvae and pupae were collected by dipping (Becker et al., 2013) in stagnant water bodies such as cisterns, ponds, rock pools, wells, tree holes, pots, and plant containers such as bromeliads. To test how well dipping and eDNA collection perform across a variety of conditions, whilst tackling habitat preference (Becker et al., 2013; Petric et al., 2014), we made an attempt to sample as many different types of water bodies as possible (Harbach, 2007; ECDC, 2014; Richardson and Richardson, 2014; Lebl et al., 2015). The risk of cross contamination was negligible because all sampling locations were spatially separated. Dipping was performed with either a 60 mm diameter sieve, 70 × 50 mm aquarium net or 25 mL pipet, depending on the accessibility of the water body. The larvae and pupae were stored in 96% ethanol. Sticky traps were used after dipping, just above surface level of water bodies that still carried water after sample collection.
eDNA-Based Sampling
Water Samples
Independent of whether larvae and/or pupae were found, aquatic eDNA was collected at all larval sites where >10 mL water could be sampled for a total of 32, 18, and 18 samples for St. Eustatius, Saba, and St. Maarten, respectively. Samples were taken by collecting surface water in steps of 25 mL with a PIPETBOY (Integra Bioscience) without agitating the water. Based on a pilot study, a maximum volume of 200 mL was used to prevent the filters from clogging. Because volume and subsample count varied between sites and are known to influence detection probabilities (Turner et al., 2014), we recorded both parameters at each of the sites. The samples were stored at 4°C until further processing. Within 24 h after collection samples were filtered with a vacuum pump using a Sartorius polycarbonate filter holder and 47 mm 0.2 μm filter membrane (Sartorius-stedim). Filters were stored in 2 mL micro centrifuge tubes containing 900 μL Longmire solution (0.1 M TRIS, 0.1 M EDTA, 0.5% SDS, 10 mM NaCl; Williams et al., 2016) to prevent DNA degradation during storage and transport at ambient temperatures (Renshaw et al., 2015; Williams et al., 2016). After each sample, filter holders and pipets were cleaned by rinsing with bleach (2x) and water (3x) to prevent cross-contamination of samples by destroying the residual DNA. On every island, a negative control (tap water) was filtered and processed as if it was a sample to test for possible contamination between samples.
Sediment Samples
The water bodies were, whenever possible, sampled for sedimentary eDNA to allow for the collection of settled eDNA (Turner et al., 2014). Sediment collection consisted of filling a 15 mL falcon tube up to the 10 mL mark (equal to roughly 14 gr.) by scraping the entire depth profile perpendicular to the waterline in a transect ranging from 10 cm under water up to the waterline for 4–6 sub samples at 0.5 m distance from each other. Hereafter all subsamples per sampling location were merged. To each of the samples 5 mL of CTAB buffer was added and subsequently mixed by carefully inverting 2–3 times to prevent DNA degradation during storage and transport (Renshaw et al., 2015). The sediment samples were stored at 4°C until further processing.
Morphological Identification of Larvae and Adults
All larvae and adults from each traditional sample were morphologically identified. Identification was primarily conducted using keys and species descriptions by Belkin et al. (1970), Darsie et al. (2010), and Van der Kuyp (1954).
Construction of DNA Reference Database
An in-house developed reference database for the Cytochrome oxidase I gene (COI) of morphologically identified species was constructed (Supplement 2 in Supplementary Material) to reduce the probability of misidentifications (Virgilio et al., 2010). This database contains species that were likely to occur on Saba, St. Eustatius and St. Maarten, based on the data from “mosquitocatalog.org,” but had insufficient public material on BOLD and GBIF. The dataset was constructed by Sanger sequencing with the primer set jgLCO1490 and jgHCO2198 (Geller et al., 2013). Sequences were obtained by barcoding specimens from the personal collection of Francis Schaffner. The sequences included the species Aedes aegypti, Aedes busckii, Aedes serratus, Aedes taeniorhynchus, Aedes tentius, Aedes tortilis, Anopheles aquasalis, Culex atratus, Culex bisulcatus, Culex idottus, Culex nigripalpus, Culex quinquefasciatus, Deinocerites magnus, Haemagogus chrysochlorus, Haemagogus dyrisolchloratus, Isotomyia perturbans, Limatus durhami, and Psorophora ferox.
DNA Extraction, Amplification, and Sequencing
For each traditional sample containing specimens, either adults or larvae, the right mid leg for adults or the proximal three segments of the larval abdomen were used for DNA extraction, so that the quality of the voucher specimen for morphological analysis was retained. The legs or abdominal segments of all specimens at a given location were merged per sample (hereafter called bulk sample), ground and DNA was extracted conform the Kingfisher's “Machery_Nagel_Tissue_96 KingFisher Flex” protocol for the NucleoMag 96 Tissue kit.
eDNA of water samples was extracted and purified using a PCI protocol developed by Renshaw et al. (2015) followed by a DNeasy blood and tissue kit extraction as clean-up (Supplement 3 in Supplementary Material). This method combines the higher yield of the PCI (Deiner et al., 2015; Goldberg et al., 2016) and the blood and tissue kit inhibitor removal (Zhou et al., 1991) whilst being able to store it at room temperature (Renshaw et al., 2015). eDNA of sediment samples was extracted and purified using the FastDNA soil kit developed by MP Biomedicals. Verification of the quality and quantity of the DNA extracts was performed with a Trinean dropsense 96.
For both bulk samples and eDNA samples, DNA amplification of mini barcodes (154 bp) within the COI region was performed with IonCode labeled culicid primers developed in an earlier study: F: 5′-GGRKCHGGDACWGGDTGAAC-3′; R: 5′-RGATCAWACAAATAAAGGTAWTCGATC-3′ (Krol et al., 2019). Each PCR mixture (20 μL) contained 1.5 μL of DNA solution with 1 μL 10 pM forward and reverse primer in 10 μL 2x environmental master mix (Taqman environmental mix 2.0, Applied Biosystems, Foster City, CA, USA). PCR reactions were performed under thermocycler conditions of 10 min at 95°C, and 40 cycles of 15 s at 95oC, 30 s at 52oC, 40 s at 72oC and 5 m at 72oC in a Bio-rad C1000 touchTM system.
After the PCR, product presence was visually confirmed by gelelectroforesis on E-gel (Invirtogen, Foster City, CA, USA). Samples with product were cleaned by mixing with 18 μL Nucleomag B-beads (Macherey-nagel GmbH & Co, Düren, Germany), incubated for 5 m and placing it on a magnetic rack. Supernatant was removed and the samples were washed two times with 100 μL 80% ethanol and left to air-dry. Thereafter, the samples were taken off the magnetic rack and resuspended in 25 μL Milli-Q. Subsequently DNA concentrations were quantified with the Qiagen Qiaxcel and pooled equimolarly at 26 nM/L with the Qiagility. The pool was diluted to 30 pM/L and subsequent analysis was done conform the IonTorrentTM IonPGMTM Hi-QTM handbook using the BioAnalyzer, Ion OneTouchTM 2 and Iontorrent on a 318TM chip. All 97 bulk samples contained PCR product and were used for the Iontorrent run. Forty seven of 68 water samples and 6 of 10 sediment samples contained PCR product and were used for the Iontorrent run. Samples with undetectable amounts of DNA were not used to prevent dilution of the other samples.
Bioinformatics and Statistical Analysis
Initial assessment of the NGS data was performed with the software packages FastQC v0.11.7 (Andrews, 2010), cutadapt v1.16 (Martin, 2011), prinseq 0.20.4 (Magoč and Salzberg, 2011), FLASH v1.2.11 (Putra et al., 2015), Unoise v10.0.240 (Edgar, 2016), and Vsearch v2.4.3 (Rognes et al., 2016) integrated in the Naturalis Galaxy pipeline.
As in previous studies NGS data were filtered by clipping the primers and, in case of low data quality, by trimming the 3'side of the sequences based on a lower phred-score cut-off of 20 (Deiner et al., 2017). Operational taxonomic units (hereafter OTUs) were after removal of singletons, clustered with both Unoise (alpha 1.5) and Vsearch (threshold: 97% similarity). OTUs with without a read depth of 0.001% of the total amount of reads within at least one sample were discarded to remove artifactual sequences (Alberdi et al., 2018).
Both Unoise and Vsearch clustering algorithms were used since they were expected to yield dissimilar results due to the differences in clustering approach. Also, the authors of Unoise state that Unoise clustering with Iontorrent data may result in inflated abundance of incorrect reads due to sensitivity to barcoding errors. However, no difference in community per sample was detected between the two clustering methods for Bray-curtis similarity on presence-absence data using Past v3 (Figure S1 in Supplementary Material; one-way ANOSIM, R 0.008531, p > 0.1). To reduce required computational power induced by the amount of OTUs, Unoise clustering, which resulted in far fewer OTUs, was used for further analysis.
The optimal Unoise alpha value was determined from the values 0.5, 1.0, 1.5, 2.0, and 3.0 by manually confirming the most parsimonious phylogeny whilst maintaining all Culicomorpha using the neighbor-joining phylogeny (Datasheet S3 in Supplementary Material; ClustalW 2.1) and lowest common ancestor analysis (LCA) (Supplement 5 in Supplementary Material; Megan 6.12.3). LCA was performed using the parameters: min score: 170; max expected: 0.01; min percent identity: 70; top percent: 5; min support percent: 0.0 (off); min support: 1.
Alignment was performed against the internally developed reference database, or if no reference ID > 95% is available against BOLD or thereafter Genbank. If no hit with ID > 95% was found, the optimal hit from the three databases was used. Genbank is used as last resort, since it is known to include misidentifications (Meier et al., 2006).
Hits >98% were accepted as species level identifications and hits >95% as genus level identifications since a culicid specific primer is used (Alberdi et al., 2018). Five Misidentifications were corrected for by manually comparing all species level identifications of species found outside their expected distribution.
The OTU table was transformed into a presence-absence matrix prior to the analysis, since the amount of reads cannot reliably be used as a proxy for biomass within species (Herder et al., 2014) and even more so across species (Goldberg et al., 2016).
Because no mosquitoes were caught using the sticky traps and resting traps, these methods were excluded from further analysis. A total species list was composed using data from the Mosquito Magnet, BG-sentinel, human landing catches, dipping samples, eDNA water samples and eDNA sediment samples.
Differences in larval detections between dipping and eDNA samples and between different water body types were tested using a one-way ANOSIM: (9,999 permutations) and visualized with Non-metric multidimensional scaling (nMDS) plot using Bray-Curtis similarity in Past v3. The differences were further explored with binomial GLM with log-odds link function, to test for interaction effects between identification method and water body type and (interaction) effects of sample volume and subsample count (Datasheet 2 in Supplementary Material).
Data from the Mosquito Magnet and BG-sentinel was highly unbalanced due to trap failure. Therefore, presence-absence counts of only the Mosquito Magnet, BG-sentinel samples, and dipping samples were used to calculate detection probabilities both overall and per species comparing morphological and molecular determinations via χ2-test using R version 1.1.383 (Datasheet 2 in Supplementary Material).
Results
From all molecular data, 35 of the 255 OTUs were identified to species level. These mainly included species within the family Culicidae, but also other taxa in the Diptera order, and some taxa within the order Crustacea (Supplement 5 in Supplementary Material). The OTUs identified as Culicidae clustered in accordance with the presumed phylogeny (Datasheet 3 in Supplementary Material; Harbach, 2007), indicating that the species level identification at 98% identity was correct. The OTUs within the infraorder Culicomorpha were used for further analysis (Datasheet 1 in Supplementary Material). None of the negative controls contained culicid DNA, thus indicating that no cross contamination occurred during sample processing.
Species Detected by Morphological Analysis
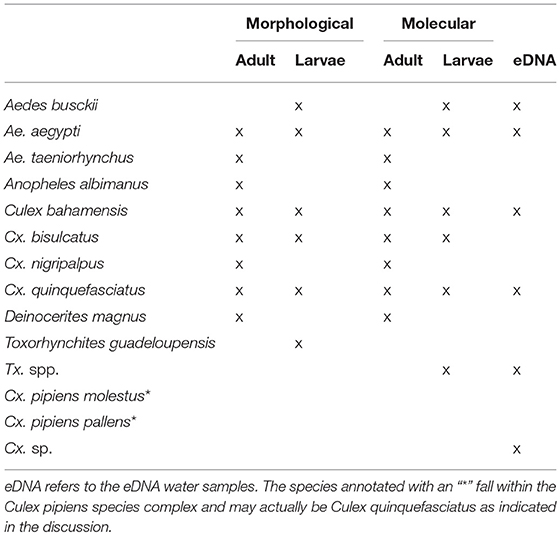
Morphological analysis of the larval samples yielded the following species: Aedes aegypti, Ae. busckii, Culex bahamensis, Cx. bisulcatus, Cx. Quinquefasciatus, and Toxorhynchites guadeloupensis. The adult samples also included the species Aedes taeniorhynchus, Cx. nigripalpus, Deinocerites magnus, and Anopheles albimanus. Molecular analysis of the eDNA and bulk samples resulted in a higher diversity (Table 1).
Species Detected With Molecular Analysis
Molecular analysis of the aquatic eDNA samples yielded the following species: Aedes aegypti, Ae. busckii, Culex bahamensis, Cx. bisulcatus, Cx. pipiens molestus, Cx. quinquefasciatus, Cx. Renatoi, and Toxorhynchites sp. The larval and adult samples also included the species Anopheles albimanus, Aedes taeniorhynchus, Culex bidens, Cx. nigripalpus, Cx. pipiens pallens, and Deinocerites magnus. The species Cx. bidens and Cx. renatoi are likely misidentifications: Cx. bidens = Cx. nigripalpus, Cx. renatoi = Culex sp., which we further elaborate on in the discussion. The corrected species have been used in further analysis.
Molecular analysis of the 6 samples of eDNA from sediment yielded no culicid DNA, which coincided with a lack of larvae and aquatic eDNA at the same locations. The samples did however contain DNA identified as chironomid Chironomus calligraphus.
eDNA Water vs. Dipping
In general, eDNA analysis of water samples resulted in a higher detection rate of larvae than dipping. Of the 68 aquatic samples, 39 contained eDNA of mosquitoes and of the latter 11 samples also contained larvae (Figure 2). Most of the species were detected equally well using both methods (χ2-test: p > 0.28). However, within these samples a significant difference in detection probability was detected for Cx. bisulcatus (χ2 = 7.1842, p <0.05) and Cx. quinquefasciatus (χ2 = 20.651, p < 0.001). Cx. bisulcatus was better detectable by morphology and Cx. quinquefasciatus by eDNA (Table 1). None of the sediment samples contained culicid eDNA. However, none of the larval samples and water samples taken at the same locations contained culicid eDNA, implying absence of mosquitoes in these water bodies.
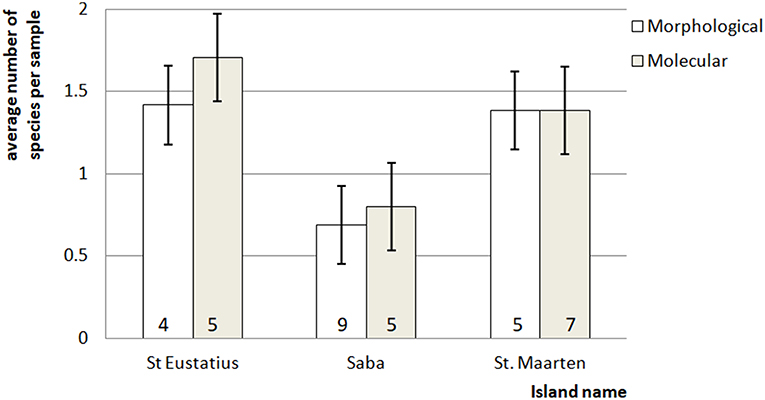
Figure 2. Average number of mosquito species in the bulk samples ± standard error. The number at the bottom of each bar indicates for each island the total number of species detected for that particular method.
The difference in detection chance for Cx. quinquefasciatus and (subsequently) Culex as a whole (NMDS 60% contribution) (Figure S2 in Supplementary Material) resulted in a detected difference between dipping and eDNA-based detections (one-way ANOSIM: R = 0.4093, p < 0.001). This is mainly caused by the fact that Cx. quinquefasciatus was detected over ten times more often in the eDNA samples than in the dipping samples.
Morphological Analysis vs. Molecular Analysis
The molecular and morphological analysis of larval and adult bulk samples performed very similar (Figure 3; Table 1). Differences between morphological and molecular analysis of the larval samples were found for Cx. bahamensis and Cx. bisulcatus. Cx. bahamensis was detected better with molecular analysis (χ2 = 1.1781, p < 0.05) whereas Cx. bisulcatus was detected better with morphological analysis (χ2 = 7.1842, p < 0.001). Differences between morphological and molecular analysis of the adult samples were found for Cx. quinquefasciatus and Culex overall. Cx. quinquefasciatus and Culex spp. were both detected better with molecular identification (χ2-test p < 0.05) (χ2-test p < 0.05) respectively.
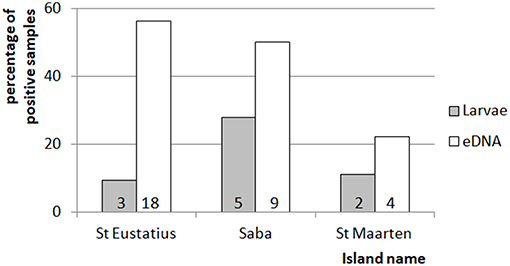
Figure 3. Comparison of between the percentage of positive mosquito samples for the eDNA and traditional dipping method. Indicated is the percentage of times larvae were detected by dipping or eDNA water sampling for each of the islands. The numbers at the bottom of each of the bars indicate the number of detections for the corresponding method and island. The total amount of dipping and eDNA water samples was 32, 18, and 18 for St. Eustatius, Saba, and St. Maarten respectively.
Community Differences Per Water Body Type
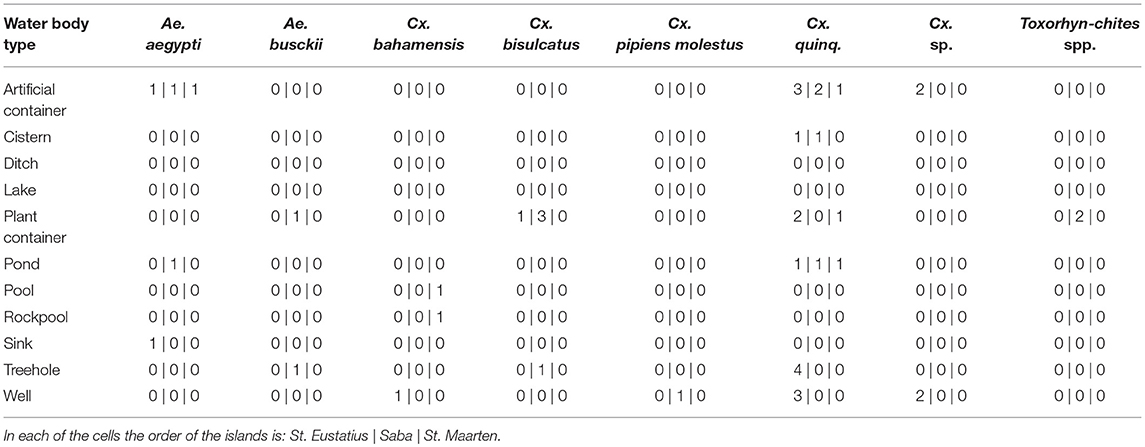
Differences in species community between the different water body types were detected when comparing Bray-curtis similarity over the dipping and eDNA water samples (One-way ANOSIM: R = 0.1991, p < 0.01; Table 2), caused by the habitat types rock pool, plant container and artificial container. The separation is the largest between rock pool and artificial container (R = 1) and moderate between rock pool and plant container (R = 0.2391) and plant container and artificial container (R = 0.2984) (Figure S2B). These effects, when corrected for the influence of volume and subsample count, can be isolated as the result of the lower detection probability of Cx. bisulcatus, which is negatively correlated with the volume (logit ANOVA: Z-val = −2.362, p < 0.05) and Cx. sp. which shows a positive trend toward artificial containers (logit ANOVA: Z-val = 1.799, p < 0.1).
Discussion
In our study, we used a range of sampling and processing methods for detection of Culicidae in a wide variety of habitats on three Caribbean islands: Saba, St. Maarten, and St. Eustatius. Our results suggest that our aquatic eDNA-based approach is as reliable and for certain species even more reliable than dipping. In contrast, eDNA originating from sediments did not result in detection of Culicidae. Although this suggests that this method may not be suitable, the lack of larval detection in the water and dipping samples taken at the same sites implies that no conclusions can be drawn about this method. Species identifications of larval and adult mosquitoes yielded very similar results when comparing morphological identification and DNA from bulk samples, but with some notable exceptions which are discussed below. Finally, we confirm the notion of Krol et al. (2019) that larval detection methods based on eDNA reveal a subset of the adult community, whilst confirming that this originates in part from inherent differences between larval and adult sampling. In our study species that were trapped as adult mosquitoes which were absent from the eDNA samples were also missing in the larval samples. In addition, some species detected in eDNA and dipping samples were absent in adult samples, suggesting that adult and larval sampling yield different yet complimentary parts of the mosquito puzzle.
Necessary Correction Steps in the DNA-Based Identification
Even though molecular identification provides promising results, the molecular analysis initially resulted in some misidentifications, for which manual corrections had to be made. These species included: (i) Culex bidens, (ii) Culex pipiens (var. molestus and pallens), and (iii) Culex renatoi. The former was identified using bulk sample DNA, the latter two were from aquatic eDNA samples. (i) Culex bidens is a species known only from South America, and is therefore an unexpected find. After re-evaluating the OTUs and corresponding BLAST results, it is likely that the OTUs identified as Cx. bidens are misidentified Culex nigripalpus DNA. This species has an identical identity and is congruent with the morphological identification from the sample that the OTUs originate from. (ii) Cx. pipiens var. molestus and pallens were found in samples where morphologically only Cx. quinquefasciatus was identified. However, Cx. quinquefasciatus is part of the Culex pipiens species complex (Harbach, 2012). Since these species are morphologically and molecularly almost identical (Laurito et al., 2013), it is possible that Cx. pipiens is actually present on the islands. (iii) The OTUs identified as Cx. renatoi were derived from aquatic eDNA samples from wells and artificial containers collected in St. Eustatius. They are highly dissimilar compared to the other known Culex species from the island (Cx. bahamensis, Cx. bisulcatus and Cx. quinquefasciatus; identity <93%). And, apart from the Cx. renatoi sequence, there are no sequences available that are similar enough for species identification. Since Cx. renatoi is a species that typically breeds in plant containers and is only known from South America, we consider this a misidentification. This might therefore be a new Culex species for the island, which has not yet been included in the BOLD and Genbank databases. Our results suggest that the reliability of molecular identification, and specifically that of aquatic eDNA sampling, is highly dependent on the quality of the reference library, thus re-emphasizing the previously identified need for more complete global databases (Deiner et al., 2017). All aforementioned misidentifications were corrected for prior to the analysis.
Unidentified OTUs
For all molecular data only 8% of the OTUs could be assigned with certainty to a mosquito species. Of the OTUs that could not be attributed to mosquito species, a large portion was found in environmental samples only (82.9%). The same is true for the OTUs that could not be identified to genus level (95.9%). Therefore, it is likely that these clusters were unidentifiable due to degradation of the DNA and due to the presence of DNA from organisms other than culicids, such as beetles, worms and amphibians. This is supported by the LCA analysis, which shows that a large portion of the unidentified OTUs could not be assigned at all or likely originate from crustaceans and other unknown taxa within the Diptera. Consequently, it is presumed that only a negligible amount of culicid DNA remained unidentified, which is also supported by the detection probabilities of the larval and aquatic samples. There were only two species that were difficult to identify using eDNA: Culex bisulcatus and Toxorhynchites spp. This is most likely caused by a lack of publicly available sequences. For Cx. bisulcatus only one sequence was available originating from our own reference library. DNA originating from bulk samples that, according to morphological identification, contained Cx. bisulcatus, clustered at 97% identity with this sequence. This indicates that genetic variability may indeed play a role in the inability to identify this species. Also for Toxorhynchites spp., a lack of reference sequences is the most likely explanation for the fact that none of the larvae of Toxorhynchites could be identified to species, given that BOLD and GenBank list only sequences for 18 of the 90 known species for this genus (mosquitocatalog.org). Overall, we are convinced that our molecular analysis yields an adequate representation of the observed species communities, both for bulk and aquatic eDNA samples, due to the high similarity when compared with the morphological identifications.
Detection of Larvae Using Dipping vs. eDNA
There were two major differences between sampling of mosquito larvae using dipping and our eDNA approach. First, the analysis of the aquatic samples resulted in a higher diversity (Table S1 in Supplementary Material), which is congruent with previous research comparing traditional and eDNA-based detection (Deiner et al., 2017). Presumably this effect is caused by the inherent biases of the traditional methods (Deiner et al., 2017). Second, the eDNA sampling had a higher probability of detection, which is related to the first result. This difference was mainly caused by Culex quinquefasciatus DNA that was present in water bodies where no larvae were found. Cx. quinquefasciatus was 10 times more often present in aquatic eDNA samples than in the dipping samples. The water bodies where this species was detected included almost every water type, even plant containers. This is uncommon, but has been described before (Frank et al., 1988), thus confirming the generalist nature of the species. The reason for this discrepancy between dipping and eDNA-based detection may be 2-fold: (1) due to our inability to catch larvae using dipping: larvae can dive for several minutes (Becker et al., 2013) rendering them harder to catch, especially in water bodies with lower accessibility such as cisterns and wells, and (2) due to the persistence of eDNA in the aquatic environment. Larval development can be as short as 6–7 days (Becker et al., 2013), but eDNA can persist for weeks (Schneider et al., 2016) and is likely present at the water surface as the larvae spend most time there and also pupate at the water surface. In contrast to the water samples, none of the sediment samples contained culicid eDNA. Some of the samples did, however, contain DNA from chironomids, a closely related family within the infraorder Culicomorpha, indicating that detection was not hindered by PCR inhibition. Although this suggests that sediment samples can potentially be used for monitoring of Culicidae and (phylogenetically related) Diptera, our inability to collect samples at most sites illustrates that it may not be as straightforward as water samples. Overall, we conclude that reliability of aquatic eDNA sampling was higher than dipping, which is mainly due to the underestimation of presence of larvae of Cx. quinquefasciatus, one of the possible disease vectors on the islands.
Comparison Between Identification Using DNA Bulk Samples vs. Morphology
In general both identification methods performed comparably, but some differences between molecular and morphological identifications were found. Differences between morphological and molecular analysis of the larval samples were found for Cx. bahamensis and Cx. bisulcatus. Cx. bahamensis was detected more often with molecular analysis. One cause was that most captured larvae were early instars, which are unidentifiable morphologically. A portion of these was kept until they emerged to be identified as adult both morphologically and molecularly, which could be done successfully by both methods. Cx. bisulcatus was identified better by morphological analysis, which is likely due to a lack of reference sequences and will be elaborated on below. Differences between morphological and molecular analysis of the adult samples were found for Cx. quinquefasciatus and Culex overall. Cx. quinquefasciatus was detected more often by molecular analysis, which is reflected in the observed difference in detection probability for the Culex spp.
Differences in Subsample Count
There was no detected difference between the samples in relation to their subsample count.
This is counterintuitive, since eDNA is known to be heterogeneously distributed over the water bodies (Nathan et al., 2014). The cause for absence of this effect may be 2-fold. First, the result of the used subsample volume (25 mL), resulting in a bias toward samples with high subsample count, thereby countering the effects of eDNA heterogeneity. Secondly, we expect there is a correlated effect with habitat preference. The latter would also explain why this parameter still was included in the optimal GLMM model.
Differences Between Water Body Types
A difference in species communities was detected between the water body types originating from the differences between the types plant container and artificial container, rock pool and plant container and rock pool and artificial containers. This is in line with previous studies, showing the existence of species specific habitat preference (Andreadis and Armstrong, 2007; Abella-Medrano et al., 2015). When corrected for sample volume and subsample count, it becomes apparent that the detected difference is caused by Cx. bisulcatus which is negatively affected by higher volumes. This is congruent with the niche of the species, since it mainly inhabits tree holes and plant containers, which have small volumes. Aedes aegypti, shows a positive trend toward bigger water volumes and the Culex previously identified as Cx. renatoi shows a positive trend toward artificial containers. These effects are however not significant, which is likely the result of low sample numbers.
Further Research and Recommendations
Even though the highest mosquito diversity is thought to occur in the dry season (Abella-Medrano et al., 2015), sampling in the wet season may provide a more definitive answer due to the higher mosquito densities. This could also result in a more clear separation of the communities per water body type.
This study confirms the notion that eDNA collection should be tailored toward the ecology of the relevant species to account for the heterogeneous nature of the eDNA. Future research searching to extend the proposed methodology should therefore make a number of adaptations concerning the collection of aquatic eDNA, most notably sampling from specific parts of the watercolumn and collection of sufficient subsamples to cover the heterogeneity of eDNA over the larval habitats. Additionally there is a need for an adequate reference sequence library, as has been previously mentioned. We encourage sequencing of specimens from private/museum collections to supplement current references, which highlights the need for cooperation between institutions to locate and gain access to such material. Furthermore, to gain resolution within species complexes we recommend adding an additional locus to the analysis (e.g., CAD or 16S) (Schneider et al., 2016). Alternatively, other non-molecular approaches such as MALDI-TOF could be considered to augment the molecular analysis, as has been explored by Lawrence et al. (2019).
Conclusion
Results of our study provide evidence that the identification of mosquitoes based on aquatic eDNA using a novel culicid specific primer resulted in reliable detection of culicid larvae and overcomes some of the caveats surrounding dipping. Like dipping, aquatic eDNA collection result in the detection of a subset of the total community and should therefore be combined with adult trapping (e.g., human landing catches) for total culicid diversity assessments. Moreover, our results suggest that molecular identification could be a useful addition, particularly for rapid assessments of total diversity in a sample, overcoming some of the limitations in sample quality, developmental stage, and sampling effort of morphological identification in combination with dipping. In doing so, cryptic communities can be assessed without extensive prior taxonomic knowledge of the present species. However, molecular identification depends strongly on the quality of the reference databases. Therefore, a considerable amount of essential taxonomic work needs to be done before this method can become widely applicable in other regions. Completeness in respect to the expected species should therefore be assessed before implementing the method. Overall, results from this study provide evidence that both our eDNA-based detection of larvae and our DNA-based identification of larvae and adults present methods that are as reliable, and for some species even more reliable than the currently used methods. As such, it allows for development of efficient disease control strategies, verification of management effectiveness and monitoring of population dynamics.
Author Contributions
SB, MS, KD, and BV conceived the idea for this study. SB carried out the measurements together with JV. SB carried out the molecular and bioinformatics work with the help of BV. SB wrote the first draft together with MS. KT and BV commented on the setup and assisted during writing of the paper and revisions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to all people involved in the expedition. Special thanks go to Loes Busscher, Maud Kok, Delia-Maria Goilo, and Emily Khazan for their help during sampling and morphological identification. We would like to thank the laboratory technicians of Naturalis Biodiversity Center for their help during the lab work, especially Frank Stokvis and Kevin Beentjes. We are also grateful to the local governments and vector control units of the three islands for their advice on suitable sampling locations, especially Maria Henry, Gerald Davelaar, Teresa Leslie, Randall Johnson, and Koen Hulshof. We would like to thank Francis Schaffner for supplying mosquitoes for the reference database. Our thanks go to the ErasmusMC for enabling us to use the Mosquito Magnets and to the Gieskes Strijbis fund for funding this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00240/full#supplementary-material
References
Abella-Medrano, C. A., Ibáñez-Bernal, S., MacGregor-Fors, I., and Santiago-Alarcon, D. (2015). Spatiotemporal variation of mosquito diversity (Diptera: Culicidae) at places with different land-use types within a neotropical montane cloud forest matrix. Parasites Vectors 8:487. doi: 10.1186/s13071-015-1086-9
Alberdi, A., Aizpurua, O., Gilbert, M. T. P., and Bohmann, K. (2018). Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol. Evol. 9, 134–147. doi: 10.1111/2041-210X.12849
Andreadis, T. G., and Armstrong, P. M. (2007). A two-year evaluation of elevated canopy trapping for Culex mosquitoes and West Nile virus in an operational surveillance program in the northeastern United States. J. Am. Mosq. Control Assoc. 23, 137–148. doi: 10.2987/8756-971X(2007)23[137:ATEOEC]2.0.CO;2
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed September 24, 2018).
Barnes, M. A., Turner, C. R., Jerde, C. L., Renshaw, M. A., Chadderton, W. L., and Lodge, D. M. (2014). Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48, 1819–1827. doi: 10.1021/es404734p
Batovska, J., Blacket, M. J., Brown, K., and Lynch, S. E. (2016). Molecular identification of mosquitoes (Diptera : Culicidae) in southeastern Australia. Ecol. Evol. 6, 3001–3011. doi: 10.1002/ece3.2095
Becker, N., Dahl, C., Bryant, B., Blair, C. D., Olson, K. E., Clem, R. J., et al. (2013). Mosquitoes and Their Control. Insect Biochemistry and Molecular Biology, Vol. 33. Berlin; Heidelberg: Springer Berlin Heidelberg.
Belkin, J. N., Heinemann, S. J., and Page, W. A. (1970). The Culicidae of Jamaica. Contrib. Amer. Ent. Inst. 6, 1–458. doi: 10.1007/s13398-014-0173-7.2
Bhalala, H., and Arias, J. R. (2009). The Zumba mosquito trap and BG-Sentinel trap: novel surveillance tools for host-seeking mosquitoes. J. Am. Mosq. Control Assoc. 25, 134–139. doi: 10.2987/08-5821.1
Burkett-Cadena, N. D. (2011). A wire-frame shelter for collecting resting mosquitoes. J. Am. Mosq. Control Assoc. 27, 153–155. doi: 10.2987/10-6076.1
Darsie, R. F., Taylor, D. S., Prusak, Z. A., and Verna, T. N. (2010). Checklist of the mosquitoes of the Bahamas with three additions to its fauna and keys to the adult females and fourth instars. J. Am. Mosq. Control Assoc. 26, 127–134. doi: 10.2987/09-5982.1
Deiner, K., Bik, H. M., Mächler, E., Seymour, M., Lacoursière-Roussel, A., Altermatt, F., et al. (2017). Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895. doi: 10.1111/mec.14350
Deiner, K., Knapp, R. A., Boiano, D. M., and May, B. (2013). Increased accuracy of species lists developed for alpine lakes using morphology and cytochrome oxidase I for identification of specimens. Mol. Ecol. Resour. 13, 820–831. doi: 10.1111/1755-0998.12130
Deiner, K., Walser, J.-C., Mächler, E., and Altermatt, F. (2015). Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA. Biol. Conserv. 183, 53–63. doi: 10.1016/j.biocon.2014.11.018
ECDC (2014). Guidelines for the Surveillance of Native Mosquitoes in Europe. Euro Surveillance : Bulletin Européen sur les Maladies Transmissibles = European Communicable Disease Bulletin, Vol. 17.
Edgar, R. C. (2016). UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv. 081257. doi: 10.1101/081257
Epopa, P. S., Millogo, A. A., Collins, C. M., North, A., Tripet, F., Benedict, M. Q., et al. (2017). The use of sequential mark-release-recapture experiments to estimate population size, survival and dispersal of male mosquitoes of the Anopheles gambiae complex in Bana, a west African humid savannah village. Parasites Vectors 10, 1–15. doi: 10.1186/s13071-017-2310-6
Esri. (2011). World Light Gray Base [basemap]. Available online at: https://www.arcgis.com/home/item.html?id=ed712cb1db3e4bae9e85329040fb9a49 (accessed October 10, 2018).
Fišer Pečnikar, Ž., and Buzan, E. V. (2014). 20 years since the introduction of DNA barcoding: from theory to application. J. Appl. Genet. 55, 43–52. doi: 10.1007/s13353-013-0180-y
Frank, J. H., Stewart, J. P., and Watson, D. A. (1988). Mosquito larvae in axils of the imported Bromeliad Billbergia pyramidalis in Southern Florida. Fla. Entomol. 71, 33–43. doi: 10.2307/3494890
Geller, J., Meyer, C., Parker, M., and Hawk, H. (2013). Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol. Ecol. Resour. 13, 851–861. doi: 10.1111/1755-0998.12138
Goldberg, C. S., Sepulveda, A., Ray, A., Baumgardt, J., and Waits, L. P. (2013). Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshw. Sci. 32, 792–800. doi: 10.1899/13-046.1
Goldberg, C. S., Turner, C. R., Deiner, K., Klymus, K. E., Thomsen, P. F., Murphy, M. A., et al. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 7, 1299–1307. doi: 10.1111/2041-210X.12595
Harbach, R. E. (2007). The Culicidae (Diptera): a review of taxonomy, classification and phylogeny. Zootaxa 1668, 591–638. doi: 10.1017/CBO9781107415324.004
Harbach, R. E. (2012). Culex pipiens: species versus species complex – taxonomic history and perspective. J. Am. Mosq. Control Assoc. 28, 10–23. doi: 10.2987/8756-971X-28.4.10
Harrington, L. C., Scott, T. W., Lerdthusnee, K., Coleman, R. C., Costero, A., Clark, G. G., et al. (2005). Dispersal of the dengue vector Aedes aegypti within and between rural communities. Am. J. Trop. Med. Hyg. 72, 209–220. doi: 10.4269/ajtmh.2005.72.209
Herder, J., Valentini, A., Bellemain, E., Dejean, T., van Delft, J., Thomsen, P. F., et al. (2014). Environmental DNA - A Review of the Possible Applications for the Detection of (invasive) Species. Stichting RAVON Report 2013 104, 21, 1789–1793. doi: 10.13140/RG.2.1.4002.1208
Hoel, D. F., Kline, D. L., and Allan, S. A. (2009). Evaluation of six mosquito traps for collection of Aedes albopictus and associated mosquito species in a suburban setting in North Central Florida1. J. Am. Mosq. Control Assoc. 25, 47–57. doi: 10.2987/08-5800.1
Jerde, C. L., Mahon, A. R., Chadderton, W. L., and Lodge, D. M. (2011). “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 4, 150–157. doi: 10.1111/j.1755-263X.2010.00158.x
Krol, L., Van der Hoorn, B., Gorsich, E. E., Trimbos, K., van Bodegom, P. M., and Schrama, M. (2019). How does eDNA compare to traditional trapping? Detecting mosquito communities in South-African freshwater ponds. Front. Ecol. Evol. doi: 10.3389/fevo.2019.00260
Kweka, E. J., Owino, E. A., Lee, M.-C., Dixit, A., Himeidan, Y. E., and Mahande, A. M. (2013). Efficacy of resting boxes baited with Carbon dioxide versus CDC light trap for sampling mosquito vectors: a comparative study. Global Health Perspect. 1, 11–18. doi: 10.5645/ghp2013.01.01.03
Lambin, E. F., Turner, B. L., Geist, H. J., Agbola, S. B., Angelsen, A., Folke, C., et al. (2001). The causes of land-use and land-cover change : moving beyond the myths. 11, 261–269. doi: 10.1016/S0959-3780(01)00007-3
Laurito, M., de Oliveira, T. M. P., Almirón, W. R., and Sallum, M. A. M. (2013). COI barcode versus morphological identification of Culex (Culex) (Diptera: Culicidae) species: a case study using samples from Argentina and Brazil. Mem. Inst. Oswaldo Cruz 108, 110–122. doi: 10.1590/0074-0276130457
Lawrence, A. L., Batovska, J., Webb, C. E., Lynch, S. E., Blacket, M. J., Jan, Š., et al. (2019). Accurate identification of Australian mosquitoes using protein profiling. Parasitology 146, 462–471. doi: 10.1017/S0031182018001658
Lebl, K., Zittra, C., Silbermayr, K., Obwaller, A., Berer, D., Brugger, K., et al. (2015). Mosquitoes (Diptera: Culicidae) and their relevance as disease vectors in the city of Vienna, Austria. Parasitol. Res. 114, 707–713. doi: 10.1007/s00436-014-4237-6
Leslie, T. E., Carson, M., Coeverden, E., van De Klein, K., Braks, M., and Krumeich, A. (2017). An analysis of community perceptions of mosquito-borne disease control and prevention in Sint Eustatius, Caribbean Netherlands. Glob. Health Action 10:1350394. doi: 10.1080/16549716.2017.1350394
Mächler, E., Deiner, K., Steinmann, P., and Altermatt, F. (2014). Utility of environmental DNA for monitoring rare and indicator macroinvertebrate species. Freshw. Sci. 33, 1174–1183. doi: 10.1086/678128
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10. doi: 10.14806/ej.17.1.200
Medeiros, M. C. I., Boothe, E. C., Roark, E. B., and Hamer, G. L. (2017). Dispersal of male and female Culex quinquefasciatus and Aedes albopictus mosquitoes using stable isotope enrichment. PLoS Negl. Trop. Dis. 11:e0005347. doi: 10.1371/journal.pntd.0005347
Meier, R., Shiyang, K., Vaidya, G., and Ng, P. K. L. (2006). DNA Barcoding and Taxonomy in Diptera : A Tale of High Intraspecific Variability and Low Identification Success. Syst. Biol. 55, 715–728. doi: 10.1080/10635150600969864
Murugan, K., Vadivalagan, C., Karthika, P., Panneerselvam, C., Paulpandi, M., Subramaniam, J., et al. (2016). DNA barcoding and molecular evolution of mosquito vectors of medical and veterinary importance. Parasitol. Res. 115, 107–121. doi: 10.1007/s00436-015-4726-2
Nathan, L. M., Simmons, M., Wegleitner, B. J., Jerde, C. L., and Mahon, A. R. (2014). Quantifying environmental DNA signals for aquatic invasive species across multiple detection platforms. Environ. Sci. Technol. 48, 12800–12806. doi: 10.1021/es5034052
Panella, N. A., Crockett, R. J. K., Biggerstaff, B. J., and Komar, N. (2016). The Centers for Disease Control and Prevention resting trap: a novel device for collecting resting mosquitoes. J. Am. Mosq. Control Assoc. 27, 323–325. doi: 10.2987/09-5900.1
Patz, J. A., Daszak, P., Tabor, G. M., Aguirre, A. A., Pearl, M., Epstein, J., et al. (2004). Unhealthy landscapes : policy recommendations on land use change and infectious disease emergence. Environ. Health Perspect. 1092, 1092–1098. doi: 10.1289/ehp.6877
Petric, D., Bellini, R., Scholte, E. J., Rakotoarivony, L. M., and Schaffner, F. (2014). Monitoring population and environmental parameters of invasive mosquito species in Europe. Parasites Vectors 7, 1–14. doi: 10.1186/1756-3305-7-187
Pilliod, D. S., Goldberg, C. S., Arkle, R. S., Waits, L. P., and Richardson, J. (2013). Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can. J. Fish. Aquat. Sci. 70, 1123–1130. doi: 10.1139/cjfas-2013-0047
Putra, C. A., Hikmatullah, D., Prawiradilaga, D. M., and Harris, J. B. C. (2015). Surveys at Bagan Percut, Sumatra, reveal its international importance to migratory shorebirds and breeding herons. Kukila 18, 46–59. doi: 10.1093/bioinformatics/btr026
Rawlins, S. C., Hinds, A., and Rawlins, J. M. (2008). Malaria and its vectors in the Caribbean: the continuing challenge of the disease forty-five years after eradication from the islands. West Indian Med. J. 57, 462–469.
Rejmánková, E., Grieco, J., and Achee, N. (2013). World' s Largest Science, Technology and Medicine Open Access Book Publisher. INTECH.
Renshaw, M. A., Olds, B. P., Jerde, C. L., Mcveigh, M. M., and Lodge, D. M. (2015). The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol-chloroform-isoamyl alcohol DNA extraction. Mol. Ecol. Resour. 15, 168–176. doi: 10.1111/1755-0998.12281
Richardson, B. A., and Richardson, M. J. (2014). Bromeliad invertebrate communities on Saba, Netherlands Antilles. Caribbean Naturalist 14, 1–12. Available online at: https://www.eaglehill.us/CANAonline/cana-n14-2014.shtml
Risks, P. H., Options, C., Medlock, J. M., Hansford, K. M., Schaffner, F., and Versteirt, V. (2012). A review of the invasive mosquitoes in europe, public health risks, and control options. 12, 435–447. doi: 10.1089/vbz.2011.0814
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. Peer J. 4:e2584. doi: 10.7717/peerj.2584
Roussel, J.-M. M., Paillisson, J.-M. M., Tréguier, A., and Petit, E. (2015). The downside of eDNA as a survey tool in water bodies. J. Appl. Ecol. 52, 823–826. doi: 10.1111/1365-2664.12428
Rueda, L. M. (2008). Global diversity of mosquitoes (Insecta: Diptera: Culicidae) in freshwater. Freshwater Animal Diversity Assessment. eds E. V. Balian, C. Lévêque, H. Segers, and K. Martens (Dordrecht: Springer Netherlands), 595, 477–487.
Schaffner, F., and Mathis, A. (2014). Dengue and dengue vectors in the WHO European region: past, present, and scenarios for the future. Lancet Infect. Dis. 14, 1271–1280. doi: 10.1016/S1473-3099(14)70834-5
Schneider, J., Valentini, A., Dejean, T., Montarsi, F., Taberlet, P., Glaizot, O., et al. (2016). Detection of invasive mosquito vectors using environmental DNA (eDNA) from water samples. PLoS ONE 11:e0162493. doi: 10.1371/journal.pone.0162493
Schrader, C., Schielke, A., Ellerbroek, L., and Johne, R. (2012). PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 113, 1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x
Strickler, K. M., Fremier, A. K., and Goldberg, C. S. (2015). Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 183, 85–92. doi: 10.1016/j.biocon.2014.11.038
Thomsen, P. F., Kielgast, J., Iversen, L. L., Wiuf, C., Rasmussen, M., Gilbert, M. T. P., et al. (2012). Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 21, 2565–2573. doi: 10.1111/j.1365-294X.2011.05418.x
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA - an emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Turner, C. R., Barnes, M. A., Xu, C. C. Y., Jones, S. E., Jerde, C. L., and Lodge, D. M. (2014). Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods Ecol. Evol. 5, 676–684. doi: 10.1111/2041-210X.12206
van der Berg, H., and Schaffner, F. (2016). Training Curriculum Invasive Mosquitoes and (re-)Emerging Vector-Borne Diseases in the WHO European Region. WHO.
Van der Kuyp, E. (1954). Mosquitoes of the Netherlands Antilles and their hygienic importance. Studies Fauna Curacao Other Caribbean Islands 23, 36–114.
Virgilio, M., Backeljau, T., Nevado, B., and De Meyer, M. (2010). Comparative performances of DNA barcoding across insect orders. BMC Bioinformatics 11:206. doi: 10.1186/1471-2105-11-206
WHO (2017b). Vector-Borne Diseases. Geneva: World Health Organization (WHO). Available online at: https://www.who.int/news-room/fact-sheets/detail/vector-borne-diseases (accessed January 16, 2018).
Williams, K. E., Huyvaert, K. P., and Piaggio, A. J. (2016). No filters, no fridges : a method for preservation of water samples for eDNA analysis. BMC Res. Notes 9:298. doi: 10.1186/s13104-016-2104-5
Wilson, I. G. (1997). Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63, 3741–3751.
Keywords: Aedes aegypti, biomonitoring, Culex quinquefasciatus, Dutch Leeward Isles, eDNA, mosquito communities, mosquitoes, vector-borne diseases
Citation: Boerlijst SP, Trimbos KB, Van der Beek JG, Dijkstra KDB, Van der Hoorn BB and Schrama M (2019) Field Evaluation of DNA Based Biodiversity Monitoring of Caribbean Mosquitoes. Front. Ecol. Evol. 7:240. doi: 10.3389/fevo.2019.00240
Received: 25 January 2019; Accepted: 11 June 2019;
Published: 02 July 2019.
Edited by:
David Andrew Bohan, INRA Centre Dijon Bourgogne Franche-Comté, FranceReviewed by:
Konstantinos Demertzis, Democritus University of Thrace, GreeceCameron Ewart Webb, University of Sydney, Australia
Copyright © 2019 Boerlijst, Trimbos, Van der Beek, Dijkstra, Van der Hoorn and Schrama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sam P. Boerlijst, c2FtYm9lcmxpanN0QHlhaG9vLmNvbQ==
†These authors share last authorship
 Sam P. Boerlijst
Sam P. Boerlijst Krijn B. Trimbos
Krijn B. Trimbos Jordy G. Van der Beek
Jordy G. Van der Beek Klaas Douwe B. Dijkstra1
Klaas Douwe B. Dijkstra1 Berry B. Van der Hoorn
Berry B. Van der Hoorn Maarten Schrama
Maarten Schrama