
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 24 May 2019
Sec. Behavioral and Evolutionary Ecology
Volume 7 - 2019 | https://doi.org/10.3389/fevo.2019.00178
This article is part of the Research Topic Host Manipulation by Parasites: Honoring David Georges Biron View all 8 articles
Cranberry false blossom disease (CFBD) is caused by a leafhopper-vectored phytoplasma infection. CFBD results in distinctive branching of the upright shoots (witches' broom) and the formation of deformed flowers that fail to produce fruit. This disease is reemerging and poses a serious threat to the cranberry industry. To determine the impact of the disease on host gene expression, we compared transcriptome profiles between plants with CFBD and uninfected cranberry plants. We found that phytoplasma infection induced expression of 132 genes, and suppressed 225 genes, compared to uninfected cranberry plants. Differentially expressed genes between uninfected and infected plants were largely associated with primary and secondary metabolic, defensive, and developmental pathways. Phytoplasma infection increased the expression of genes associated with nutrient metabolism, while suppressing genes associated with defensive pathways. This expression profile change supports the “host manipulation hypothesis,” whereby CFBD enhances host quality for insect vectors, thus promoting phytoplasma transmission.
Phytoplasmas, class Mollicutes, phylum Tenericutes (Hogenhout et al., 2008), are obligate vector-borne bacteria that colonize the phloem tissue of infected plants (Chen, 1971). Collectively, phytoplasmas cause over 700 plant diseases worldwide (Maejima et al., 2014), frequently resulting in devastating losses to agricultural productivity (Lee et al., 2000; Bertaccini, 2007). Due in part to a lack of a rigid cell wall and a minimal genome devoid of coding genes for ATP synthases and sugar uptake, phytoplasmas are highly dependent on their plant hosts (Christensen et al., 2005; Kube et al., 2012). Phytoplasmas are almost exclusively vectored by insects in the order Hemiptera, such as leafhoppers, planthoppers, and psyllids (Weintraub and Beanland, 2006; Hogenhout et al., 2008). When inhabiting the host plant, phytoplasmas rely on the nutrient-rich phloem tissue (Chen, 1971); while in the insect vector, the phytoplasmas rely on the nutrient-rich food sources in the hemolymph (Hogenhout et al., 2008). Phytoplasma-infected plants develop diverse symptoms, such as witches' broom, leaf yellowing or reddening, growth aberrations (proliferations, internode shortening, and stunting), and flower malformations (size reduction, virescence, and phyllody) (Chang, 1998; Namba, 2002). Phytoplasma infection can also cause phloem tissue aberrations, such as extensive phloem necrosis and excess formation of phloem, resulting in swollen veins (Lee et al., 2000).
The physiological, biochemical, and molecular mechanisms of phytoplasma infection are still unclear. Furthermore, the interactions between phytoplasmas, their plant host, and their insect vector are poorly understood. This is partly because phytoplasmas currently cannot be cultured in the laboratory (Mou et al., 2013). Despite the challenges this presents, recent research has demonstrated that phytoplasmas produce effector proteins that interact with plant transcription factors. The phytoplasma can thereby manipulate infected plants developmental processes, phytohormone biosynthesis, and defense responses (Sugio and Hogenhout, 2012). Moreover, some phytoplasma-produced effector proteins were shown to induce phytoplasma-associated disease symptoms in Arabidopsis thaliana L. (Heynh.), such as phyllody, witches' broom, and dwarfism (Hoshi et al., 2009; Sugio and Hogenhout, 2012; Maejima et al., 2014), while an effector protein of aster yellows phytoplasma strain witches' broom was shown to suppress salicylic acid (SA)-mediated defense responses (Lu et al., 2014). Some phytoplasmas harbor plasmids (Lee et al., 2004; Bai et al., 2006; Ishii et al., 2009). The causal agent of Paulownia witches' broom was shown to harbor two plasmids that encode several expressed proteins, some of which were predicted to encode secreted or membrane-localized proteins that may act as effectors (Lin et al., 2009). Candidatus Phytoplasma asteris, onion yellows strain (OY-M), was shown to harbor a plasmid in which ORF3 encodes a membrane protein that is important for adaptation to its insect host (Ishii et al., 2009).
Phytoplasma infection can lead to the production of defense proteins, increased phenolic compounds, and overproduction of hydrogen peroxide in host plants, such as apple and corn (Junqueira et al., 2004; Musetti et al., 2004). The phytoplasma that causes Bois noir disease of grapevine was found to affect host carbohydrate metabolism (Hren et al., 2009). Although the previously mentioned effector protein of aster yellows phytoplasma strain witches' broom suppresses SA-mediated defense responses in Arabidopsis (Lu et al., 2014), phytoplasma infection in apple induced SA-mediated defense responses and suppression of the jasmonic acid (JA) biosynthetic pathway (Musetti et al., 2013). Normally, plants induce SA against biotrophic and hemibiotrophic pathogens (Thomma et al., 1998) and against piercing-sucking insects (War et al., 2012), whereas JA is primarily induced as defense against leaf-chewing insects and necrotrophic pathogens (Liu et al., 2016). The suppression of SA- and/or JA-mediated responses in the phytoplasma-infected plants may facilitate feeding by herbivorous insects. In addition, phytoplasma-encoded effector proteins can increase volatiles that attract the insect vectors (Orlovskis and Hogenhout, 2016). Thus, modification of host plant phytochemical processes (such as nutrient translocation, chemical defense production, and volatile production) not only affects the host plant growth and development, but also influences the host plant-insect vector interaction.
A phytoplasma in the subgroup 16SrIII-Y causes cranberry false blossom disease (CFBD) in the American cranberry (Vaccinium macrocarpon Aiton) (Lee et al., 2014; Polashock et al., 2017). CFBD relies specifically on the blunt-nosed leafhopper (Limotettix vaccinii Van Duzee) to vector the phytoplasma that causes disease (Beckwith and Hutton, 1929). CFBD-affected plants produce bunched upright shoots forming a witches' broom, while leaves turn reddish earlier than uninfected plants in the fall (De Lange and Rodriguez-Saona, 2015), and flowers are malformed, exhibiting an enlarged calyx and shortened, discolored, and streaked petals (Dobroscky, 1931); affected flowers fail to set fruit. Between 1920 and 1940, this disease was a major threat to cranberry production and nearly wiped out the industry in New Jersey, USA. However, CFBD was subsequently controlled with the use of insecticides, removal of infected plants from the fields, and the release of improved cultivars that are less attractive to the insect vector (Chandler et al., 1947). After several years, disease incidence declined, and the disease was only sporadically reported. Recently, the disease has reappeared on many cranberry farms with increasing incidence being reported (Lee et al., 2014).
As a first step to broaden our understanding of the physiological, biochemical, and molecular mechanisms underlying CFBD, we performed transcriptome analysis to determine the phytoplasma-induced alterations in gene expression in infected cranberry. This information will help to further elucidate how changes in gene expression, induced in the host, contribute to vector attraction, potentially promoting transmission of this important reemerging disease.
Healthy (uninfected) runners (V. macrocarpon cv. Crimson Queen) were kindly provided by Integrity Propagation (Chatsworth, NJ, USA), while phytoplasma-infected plants of the same cultivar were collected from a commercial cranberry farm in Chatsworth, NJ. The runners were obtained in November 2016 and 2017 and stored at 10°C until used for propagation. The plants collected in November 2016 were clonally propagated in February 2017 and used for the transcriptional profiling study, whereas the plants collected in 2017 were propagated in February 2018 and were used for the real-time quantitative PCR (RT-qPCR) analysis of key selected genes.
For propagation, stem cuttings (~7 cm) were rooted in 50:50 v/v peat:sand mix in 4 × 4 cm cells. All cuttings were placed in a greenhouse (20 ± 2°C; 70 ± 10% relative humidity; 15:9 light:dark) and fertilized every 3 weeks, with PRO-SOL 20-20-20 N-P-K All Purpose Plant Food (Pro Sol Inc., Ozark, AL, USA) at a rate of 165 ppm N, and watered daily. After rooting, plants of each type (infected and uninfected) were transplanted into 7 × 7 cm pots. To ensure a sufficient number of uniform plants for subsequent experiments, five rooted cuttings were transplanted per pot. Plants were grown in the greenhouse under ambient conditions until they were used in experiments in June.
Prior to experiments, 10 plants (5 plants from infected and uninfected plants) from 10 randomly selected pots were DNA fingerprinted using sequence characterized amplified region (SCAR) markers (Polashock and Vorsa, 2002) to verify that all plants were genetically identical (cv. Crimson Queen). Another 10 plants (5 plants from infected and uninfected plants) from 10 pots were randomly selected and tested for phytoplasma infection by using a nested PCR assay (Lee et al., 2014) to verify that only infected plants were positive for the presence of phytoplasma.
For the transcriptional profiling study, total RNA was extracted from leaves of three uninfected plants and three infected plants from six different pots. Total RNA was also extracted from five plants from each group (uninfected and infected) for RT-qPCR analysis of selected genes. Briefly, fresh cranberry leaves, approximately 50–75 mg, and two 5-mm stainless steel beads were added to 2-mL Safe-Lock tubes (Eppendorf North America, Hauppauge, NY) filled with 800 μL cetyltrimethylammonium bromide buffer. Samples were ground for 1 min at 30 Hz with a TissueLyser II instrument (Qiagen, Germantown, MD). The sample suspensions were extracted with 700 μL chloroform and then centrifuged at 11,000 g for 5 min. Supernatants were transferred to new tubes. For nucleic acid precipitation, 0.7 volumes of isopropanol were added to each tube, incubated on ice for 10 min, and centrifuged at 13,000 g for 5 min. Pellets were washed in 500 μL of 70% ethanol and centrifuged at maximum speed for 1 min and then all remaining ethanol was removed from the tubes by using a micropipette. Remaining pellets were resuspended in 400 μL of RNase-free water, and the RNA was precipitated by the addition of 100 μL of 10 M lithium chloride and an overnight incubation on ice. After the overnight incubation, the samples were centrifuged at 13,000 g for 5 min. The pellets were resuspended in 400 μL RNase-free water. Forty microliters of 3 M sodium acetate and 880 μL of 100% ethanol were added for RNA reprecipitation. After a 10-min incubation on ice, the samples were centrifuged at 13,000 g for 10 min. Pellets were washed with 70% ethanol and resuspended in 50 μL RNase-free water. The RNA concentrations were determined using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) with absorbance ratios of A260/280 and A260/230 nm.
RNA was sent to the Novogene Corporation (Sacramento, CA) for library construction and sequencing. Three independent cDNA libraries were constructed for each treatment (uninfected and infected). Prior to library construction, all samples were tested for RNA integrity and determined to be of sufficient quality, as determined by Novogene, using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). After passing the quality-control procedures, mRNA was enriched using oligo(dT) beads. The mRNA was randomly fragmented, and the first-strand cDNA was synthesized using random hexamers and reverse transcriptase. Then, the second strand was synthesized with a custom Illumina second-strand synthesis buffer (Illumina Inc, San Diego, CA) with DNA polymerase I, dNTPs, and RNase H. The double-stranded cDNA fragments were purified, end repaired, poly-A tailed, adapter ligated, size selected, and PCR enriched. Library concentration was quantified using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA), then diluted to 1 ng/μL before checking insert size on an Agilent 2100, and quantified to greater accuracy by quantitative PCR. Libraries were sequenced using the Illumina HiSeq platform. Clean reads were obtained for sequence assembly after removing adaptors and reads of poor quality. Transcriptome de novo assembly was carried out with Trinity (Grabherr et al., 2011). The assembled transcriptome was annotated using BLAST, applied by Novogene (Sacramento, CA, USA), using seven databases, namely, NCBI non-redundant protein sequences, NCBI nucleotide sequences, Protein family, Eukaryotic Orthologous Groups/Cluster of Orthologous Groups of protein, SwissProt, Kyoto Encyclopedia of Genes and Genome (KEGG), and Gene Ontology (GO). The evaluation threshold was 1e-5 for NCBI non-redundant protein sequences, NCBI nucleotide sequences, and SwissProt databases; 1e-3 for Eukaryotic Orthologous Groups; 0.01 for Protein family; 1e-6 for GO; and 1e-10 for KEGG. Corset (Davidson and Oshlack, 2014) was used for categorical clustering of de novo-assembled contigs, while individual reads were aligned with RSEM (Li and Dewey, 2011). Differentially expressed genes (DEGs) between uninfected and infected cranberry plants were identified using DESeq (Anders and Huber, 2010).
In 2018, five plants from each treatment (uninfected and infected) were selected for RT-qPCR. Total RNA was extracted from each sample by using the method described above. The total RNA was treated with Optizyme DNaseI (Fisher Scientific, Hampton, New Hampshire, USA) to remove any residual DNA. The cDNA was synthesized using 1 μg of RNA and the Superscript Vilo cDNA synthesis kit as per the manufacturer's (Invitrogen) protocol.
Based on the transcriptome data and de novo annotation, six candidate target genes were selected for RT-qPCR. The genes and the primers for the selected genes are listed in Table 1. The expression of the actin and RNA helicase 8 genes were used as the endogenous controls (Rodriguez-Saona et al., 2013), while the dehydrodolichyl diphosphate synthase 6 gene was used as the interplate calibrator. The primers of all candidate genes and endogenous controls were designed using PrimerQuest (Integrated DNA Technologies Inc., Skokie, IL, USA).
RT-qPCR reactions were performed using the Power SYBR Green PCR Master Mix (Applied Biosystem, Foster City, CA, USA) according to manufacturer's directions. The reactions were run on a QuantStudio 5 RT-qPCR System (Applied Biosystems) with the following conditions: 50°C for 2 min; 95°C for 10 min; and 40 cycles at 95°C for 15 s and 58°C for 1 min, with melting curve set at 95°C for 15 s, 60°C for 1 min, and 95°C for 1 s. There were three technical replicates for each sample. Relative expression levels were calculated using the ΔΔCT method using the QuantStudio Design & Analysis Software version 1.4.3 (Applied Biosystems). The relative expression data were tested for normality and for homogeneity of variance to check the parametric assumption requirement. If needed, data were log10 transformed before using analysis of variance (ANOVA); otherwise, non-parametric tests such as the Mann-Whitney U test were used. All parametric and non-parametric tests were performed using IBM SPSS Statistics package version 24 (IBM, Armonk, NY, USA).
All cranberry plants were of the same cultivar (Crimson Queen), as determined using SCAR markers (data not shown). The false blossom-infected plants used for these experiments tested positive for phytoplasma by using the nested PCR assay described above. Uninfected plants were all negative (data not shown).
An average of 58,605,833 reads were generated for uninfected plants, while infected plants averaged 59,361,545 reads. The raw reads were filtered to remove reads of low quality, leaving an average of 55,074,020 (Q20: 97.7%, GC content: 46.08%) and 55,502,498 clean reads (Q20: 97.82%, GC content: 45.88%) for the uninfected and infected transcriptomes, respectively. The clean reads were de novo assembled using Trinity, resulting in 131,404 total unigenes with a size distribution consisting of 32,050 unigenes of 200–500 bp, 36,313 unigenes of 500–1,000 bp, 34,553 unigenes of 1000–2,000 bp, and 28,488 unigenes of ≥2,000 bp. The average length of unigenes was 1,346 bp with an N50 of 2,024 bp. Contigs were BLASTed against seven databases, applied by Novogene, to achieve comprehensive gene functional annotation. Of the 131,404 unigenes, 63.59% (83,564 unigenes) were able to be annotated using at least one database.
Of the total unigenes, 43.83% (57,602 unigenes) were annotated with Blast2GO version 2.5 (Götz et al., 2008), based on the protein annotation results of NCBI non-redundant protein sequences and Protein family databases. After annotation, unigenes were grouped into three main GO domains namely, Biological Process, Cellular Component, and Molecular Function (Figure 1). The predominant GO terms were cellular process, metabolic process, binding, catalytic activity, and single-organism process.
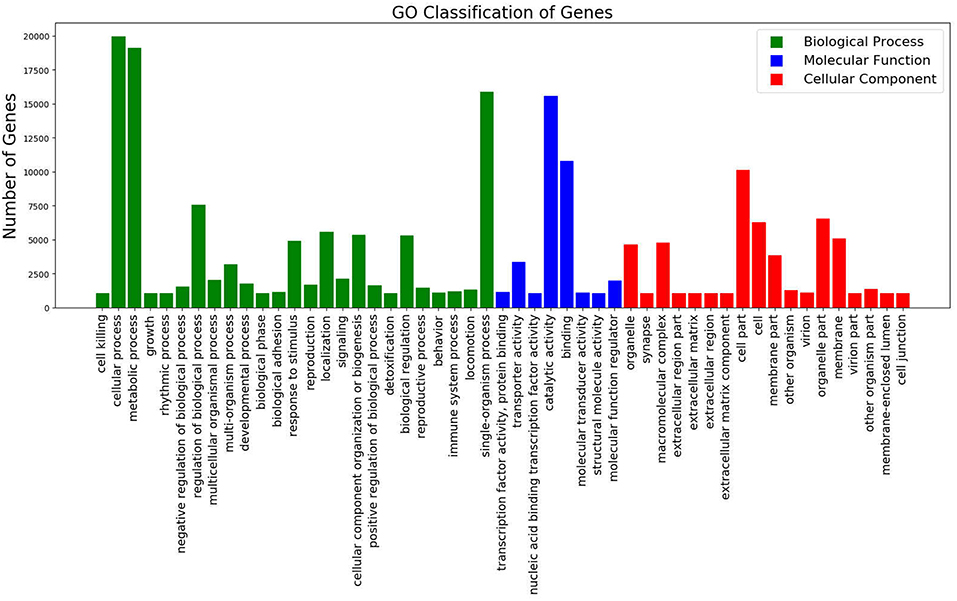
Figure 1. GO Classification of Genes. Total counts of GO terms associated with all annotated unigenes classified in 3 categories; Biological Process (Green), Molecular Function (Blue), and Cellular Component (Red).
A large proportion of genes in the GO category biological process were associated with “cellular process,” “metabolic process,” and “single-organism metabolic process.” In the molecular function category, “catalytic activity,” “binding” and “transporter activity” were of the highest proportion. Of the genes categorized as cellular components, “cell part,” “membrane part” and “organelle part” were the most enriched terms (Figure 1).
For Eukaryotic Orthologous Groups (KOG) classification, 26,863 unigenes were annotated using the NCBI gene orthologous relationships. Of the KOG categories “general function prediction only” is the largest category in which most genes were categorized, followed by “post-translational modification, protein turnover, and chaperones” and the group “translation, ribosomal structure, and biogenesis.”
The 30,744 unigenes were annotated through the KEGG Orthology database and were placed into 129 KEGG pathways. The most represented pathways were “carbohydrate metabolism,” “translation,” and “folding, sorting and degradation.”
Differential expression analysis between uninfected and infected plants showed 356 DEGs. Infected plants showed an upregulation of 131 genes and downregulation of 225 genes, as compared with uninfected plants. Two genes expressed only in infected plants appear to be derived from the phytoplasma, as determined by BLAST. As noted in Table 1, one was similar to wheat blue dwarf phytoplasma plasmid (pWBD2), suggesting that the phytoplasma that causes CBFD in this experiment might harbor a plasmid. The other gene was identified as encoding an unknown phytoplasma protein. The DEGs mapped in the KEGG database showed that the pathways most affected (q-value, ≤0.1) by CFBD were “cutin, suberin, wax biosynthesis,” “carotenoid biosynthesis,” and “fatty acid elongation” (Figure 2). Specifically, the genes related to cutin, suberin, wax biosynthesis, fatty acid omega-hydrolase gene (CYP86A1), and alcohol-forming fatty acyl-CoA reductase gene (FAR) were downregulated. The carotenoid biosynthesis pathway showed two different forms (sequences) of the (+) -abscisic acid 8′-hydroxylase, one of which was upregulated while the other was downregulated. For fatty acid elongation, the very-long-chain (3R)-3-hydroxyacyl-CoA dehydrogenase gene (PAS2) was downregulated, as well as the 3-ketoacyl-CoA synthase gene.
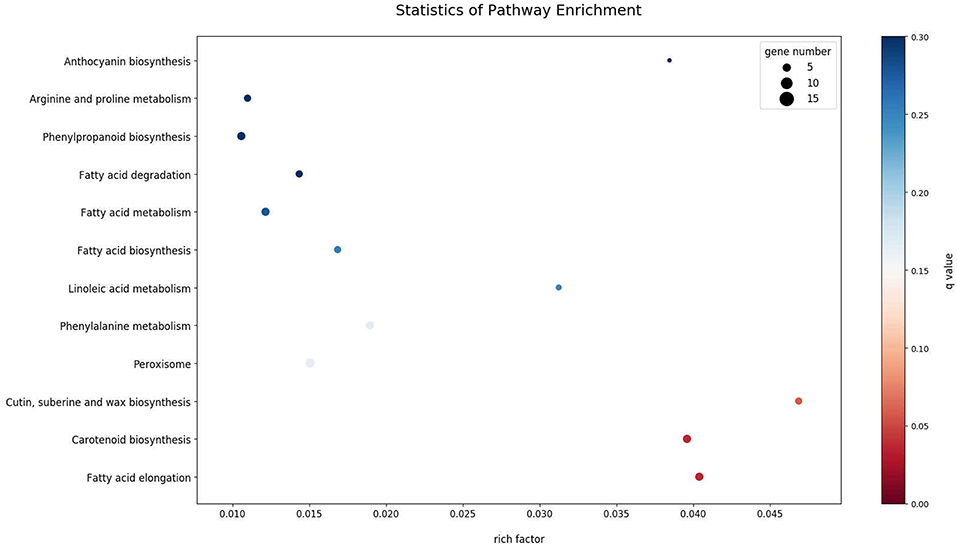
Figure 2. KEGG Enrichment Scatter Plot of the 15 most enriched pathways. The 15 most significantly DEG-enriched pathways (padj < 0.05) are displayed on the y-axis and their associated level of enrichment is displayed along the x-axis. The degree of KEGG enrichment is measured by rich factor; described as the ratio of the number of DEGs in the pathway compared to the total number of genes found in the pathway. The greater the rich factor, the greater the degree of gene enrichment. The q-value is the p-value adjusted to sample distribution (also referred to as padj) and indicates significance of pathway enrichment. Smaller q-values indicate more significant levels of enrichment. Dot size represents the number of different genes and the color indicates the q-value.
Plant defenses, in part, rely on changes in gene expression in a myriad of biological pathways. The list of genes potentially involved in defense can include those that relate to molecular signaling, physical defenses (such as cell wall strengthening), and chemical defense (secondary metabolite biosynthesis). In this study, many of the genes related to plant defense were downregulated in phytoplasma-infected plants relative to uninfected plants (Figure 3). For example, two members of the ATP-binding cassette transporter (ABC) family (ABCC and ABCG) and a predicted endochitinase A were expressed only in uninfected plants (i.e., they were downregulated in infected plants). In contrast, some genes such as those related to mismatch repair (MSH1 and RAD7), a gene in the glycosyl hydrolase family, and a GTP pyrophosphokinase were expressed primarily in infected plants.
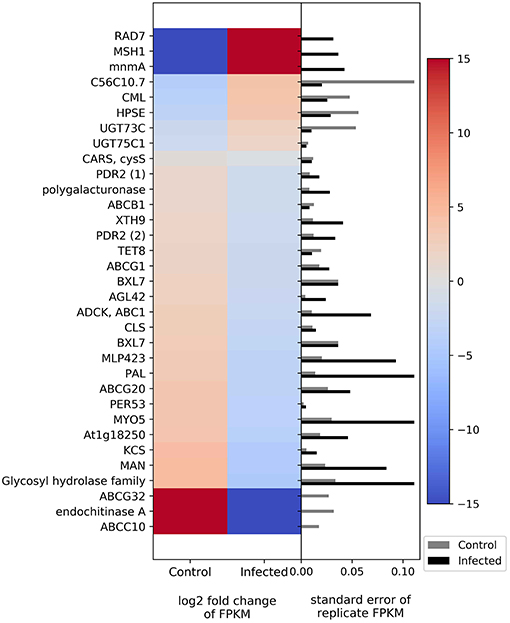
Figure 3. Heatmap of DEGs related to defense. The left side of the plot is the heatmap of log2 fold change of FPKM between all infected and uninfected groups. The right side of the plot is the standard error for all replicates divided by the average FPKM for the uninfected and infected groups. See Supplemental Table 2 for gene names associated with all abbreviations.
Molecular signaling genes related to plant defense include calcium-binding protein calmodulin-like protein 44 (CML), ABC in various families and members, and cysteinyl-tRNA synthase. CML is upregulated in phytoplasma-infected plants relative to uninfected plants, whereas those in the ABC family show downregulation in phytoplasma-infected plants. Cysteinyl-tRNA synthase is slightly downregulated in infected plants. Genes related to cell wall and wax synthesis include xyloglucosyl transferase, beta-D-xylosidase7, and 3-ketoacyl-CoA synthase. These genes were all downregulated in infected plants. The genes associated with plant secondary compound biosynthesis include enzymes such as anthocyanidin 3-O-glucoside 5-O-glucosyltransferase, UDP-glucosyl transferase 73C, phenylalanine ammonia-lyase (PAL), ent-copalyl diphosphate synthase, and peroxidase. Anthocyanidin 3-O-glucoside 5-O-glucosyltransferase and UDP-glucosyltransferase 73C were slightly upregulated in phytoplasma-infected plants relative to uninfected plants, whereas PAL, ent-copalyl diphosphate synthase, and peroxidase were slightly downregulated in infected plants.
The transcriptome results show differential expression of two genes involved in photosynthesis; cytochrome b599 was upregulated in infected plants, whereas photosystem II reaction center I protein was downregulated (Figure 4). Genes related to carbohydrate metabolism were also differentially regulated (Figure 4). The genes that show upregulation in infected plants include fructokinase, raffinose synthase, and malate dehydrogenase, whereas the genes that show downregulation relative to uninfected plants include polygalacturonase, xyloglucosyl transferase, laccase, galacturonosyltransferase, mannan endo-1,4-beta-mannosidase, and glycosyltransferase.
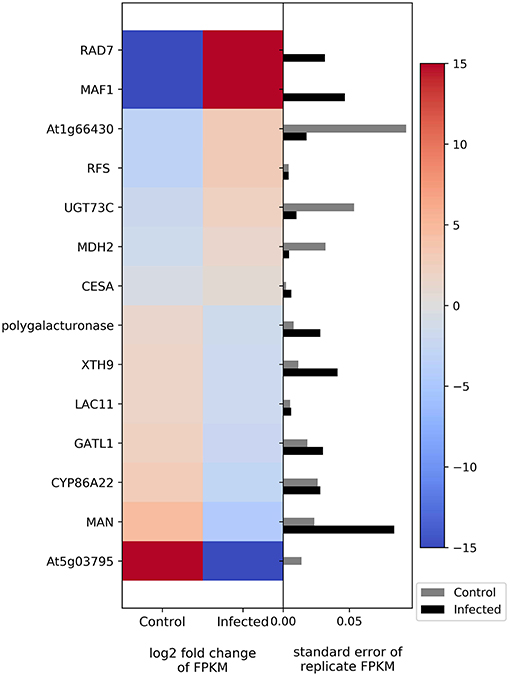
Figure 4. Heatmap of DEGs related to photosynthesis and carbohydrate metabolism. The left side of the plot is the heatmap of log2 fold change of FPKM between all infected and uninfected groups. The right side of the plot is the standard error for all replicates divided by the average FPKM for the uninfected and infected groups. See Supplemental Table 2 for gene names associated with all abbreviations.
The genes AP2-like factor (AP2) and (+)-abscisic acid 8′-hydroxylase (E1.14.13.93) are related to the plant hormones ethylene and abscisic acid, respectively. Both genes show upregulation in infected plants (Figure 5). Other genes related to flowering and development, such as FAR, suppressor of overexpression of CONSTANS 1, circadian clock coupling factor ZGT, phytochrome A, and transcription factor TGA, show downregulation (Figure 5).
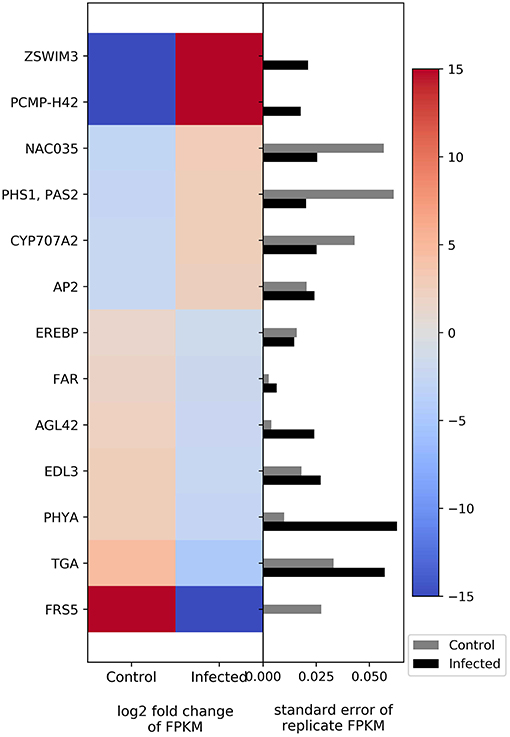
Figure 5. Heatmap of DEGs related to flowering and development. The left side of the plot is the heatmap of log2 fold change of FPKM between all infected and uninfected groups. The right side of the plot is the standard error for all replicates divided by the average FPKM for the uninfected and infected groups. See Supplemental Table 2 for gene names associated with all abbreviations.
The target genes (Table 1) were selected to verify differences in the expression of phytoplasma-infected plants, relative to uninfected plants, by RT-qPCR. The genes were selected to be representative of the three functional groups described above (defense, photosynthesis and carbohydrate metabolism, and flowering and development). The genes selected were fructokinase-6 (FX), CML, PAL, and AP2. The expression of FX, which is related to carbohydrate metabolism, was significantly (p ≤ 0.01) increased in infected plants (Figure 6). The CML-encoding gene, which is involved in defense signaling, was significantly (p ≤ 0.05) upregulated in infected plants. The expression of the PAL-encoding gene, which is involved in the early stages of secondary chemical defense synthesis, was lower in the infected plants in the transcriptome data (Figure 3), but the difference in expression by RT-qPCR was not significant (p = 0.897) (Figure 6). The expression of the AP2-encoding gene, which is involved in floral meristem development, was significantly (p ≤ 0.01) increased in infected plants (Figure 6).
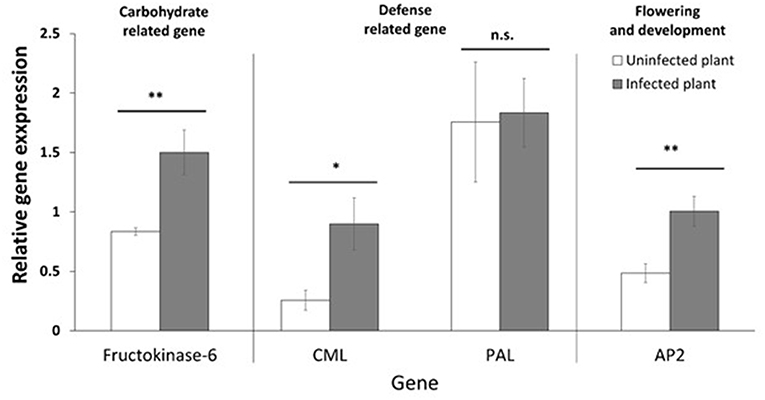
Figure 6. Relative gene expression (mean, ±SE) of selected genes. Asterisk indicates statistically significant differences (*p ≤ 0.05, **p ≤ 0.01). n.s. = not significant (p > 0.05).
We also selected two genes that appear to be encoded by the phytoplasma. One has similarity to wheat blue dwarf phytoplasma plasmid (pWBD2), and the other (Unknown Phyt) encodes a protein with similarity to “hypothetical protein CPX 001471” (from Candidatus Phytoplasma pruni, accession number KOR75512.1) and “hypothetical protein” from Vaccinium witches' broom phytoplasma (accession number WP 017193546.1). These two genes were shown in the transcriptome data to be expressed only in false blossom-infected plants, and RT-qPCR data confirmed their expression only in infected plants (data not shown). Either one or both of these genes may encode an effector and this possibility should be further explored.
Cranberry false blossom disease (CFBD), caused by a phytoplasma, is a serious threat to the cranberry industry. This disease causes physiological and biochemical changes in cranberry that influence the host-vector (blunt-nosed leafhopper) interaction (Pradit et al., 2019). Here, we show the changes in the cranberry transcriptome associated with CFBD. Surprisingly, of the more than 131,000 unigenes in the transcriptome, the expression of only 356 genes were influenced by phytoplasma infection. Phytoplasma infection induced the expression of 132 genes and suppressed the expression of 225 genes, relative to the uninfected, healthy plants. Other studies found phytoplasma infection influences the expression of more than 2,000 genes in grapevine (1,326 upregulated genes and 1,130 downregulated genes), Mexican lime tree (1,943 upregulated genes and 862 downregulated genes), and Chinese jujube (2,070 upregulated and 2,196 downregulated genes) (Hren et al., 2009; Mardi et al., 2015; Fan et al., 2017).
The stage of infection by phytoplasma, as well as the plant tissue collected and timing of collection of those tissues, can influence the differences in quality and quantity of gene expression detected. In Jujube witches' broom, the stage wherein the plant showed weak symptoms had the greatest number of DEGs, whereas the primary and late stage of phytoplasma infection had a fewer number of DEGs (Wang et al., 2018). In this study, the plants were approximately 4 months old when the leaf tissue was collected for the experiments. At the time tissue was collected, the infected plants were smaller in size and bushy (shorter internodes) relative to the uninfected plants. The plants were considered to be young and, therefore, at the primary stage of symptom development. In the field, severe symptom development (witches' broom) occurs in plants that have been infected for many years. Moreover, the titer of the bacteria in the plant can also play a role in gene expression. Early in the growing season, jujube trees with Jujube witches' broom have high bacterial density in the root but very low in the petiole, whereas when they are actively growing during the season, a high density of phytoplasma is found in the petiole (Yi et al., 2001). Here, we considered our plants to be early in the growing season, and this may be one reason why the number of DEGs detected seems relatively low. Furthermore, the most dramatic morphological change in cranberry with CFBD is the formation of deformed flowers. Thus, the transcriptome of developing flower tissues is likely to exhibit a high number of DEGs. It is also possible that the DEGs detected here might differ in number and/or the magnitude of differential expression in other cranberry cultivars.
We considered three general groups when examining the DEGs, namely, defense related, photosynthesis and carbohydrate metabolism related, and flowering and development related, as changes in these groups are likely to affect plant-vector interactions and possibly disease spread.
The defense mechanisms of plants can be separated into physical and chemical defenses (Bennett and Wallsgrove, 1994; Will and van Bel, 2006). The physical defenses induce changes in the physical structure of the plant, whereas the chemical defenses range from changing phytohormone signaling, such as JA or SA, to secondary plant chemical production and accumulation (Freeman and Beattie, 2008).
The first physical barrier for the plant pathogen is the plant cell wall, a rigid cellulose-based support. Cell walls are important for maintaining cell structure, intercellular communication (Clarke et al., 1985), and defense against pathogens (Underwood, 2012; Malinovsky et al., 2014). High-molecular-weight polysaccharides, including cellulose, hemicellulose (typically xyloglucan or xylan), and pectin, are the main components of plant cell walls. Phytoplasma infection results in the structural modification of host plants, such as cell wall enhancement (Rudzinska-Langwald and Kaminska, 2001). The genes that are related to cell wall modification in this study, such as xyloglucan:xyloglucosyl transferase and beta-D-xylosidase 7, tended to be suppressed in the infected plant (Figure 3). Xyloglucan is a structural polysaccharide of the primary cell wall (Levy et al., 1991). The xyloglucan:xyloglucosyl transferase, also known as xyloglucan endotransglycosylase/hydrolase, is an enzyme involved in cell wall elongation and reconstruction. Increased xyloglucan endotransglycosylase/hydrolase activity at the infection site and adjacent tissue were observed in tomato when parasitized by dodder (Albert et al., 2004). The beta-D-xylosidase releases xylose from xylan-containing oligosaccharides (Rahman et al., 2003) and is potentially involved in secondary cell wall hemicellulose metabolism. The suppression of this gene in plants can facilitate the invasion of disease into the cell of the host plant (Minic, 2007; Lippmann et al., 2009).
Phytoplasmas can be detected in most organs of phytoplasma-infected plants. These bacteria colonize the sieve tubes of phloem and seem to be in close contact with the plasma membrane (Marcone, 2009). Membrane proteins of the phytoplasma are considered a factor that plays a role in colonization and infection of the host cell and triggers changes in the chemical metabolism of the host plant (Malembic-Maher et al., 2005). Moreover, phytoplasmas express effector proteins, specific molecules that alter the host, to facilitate successful invasion of the plant (Sugio et al., 2011). Our transcriptome and RT-qPCR results show expression of at least one gene that may be on a plasmid of the pathogen and one “hypothetical protein” that may encode effectors.
Genes involved in signal transduction pathways, such as CML and ABC in various families and members, can be important in sensing and responding to phytoplasma infection. CML is an important Ca2+ sensor, which plays a significant role in plant tolerance to several biotic and abiotic stress (Ali et al., 2003). Specifically, calcium/calmodulin-mediated regulation plays a role in plant phytohormones, including SA and glucosinolate (Cheval et al., 2013). In this study, phytoplasma infection increased CML expression (log2Fold change = 3.768, Figure 3), suggesting an induction of phytohormone-mediated signaling.
Functional ABC transporters are located in cell membranes and act as ATP-driven pumps. ABC transporters move various chemical substrates, such as lipids, phytohormones, and heavy metals, through the plasma membrane (Kretzschmar et al., 2011). ABC transporters are known to respond to abiotic and biotic stresses and play important roles in detoxification (Lu et al., 1998), hormone transport (Geisler et al., 2005; Kuromori et al., 2010), and chemical defense (Badri et al., 2009). In rice, nearly half of the ABCG members have a positive response to JA and SA, the phytohormones associated with herbivore and pathogen defense (Moons, 2008). NpPDR1, an ABCG protein of Tex-Mex tobacco, was implicated in pathogen defense and was shown to be involved in active terpenoid transport in plants (Jasinski et al., 2001). Both JA and SA promote NpPDR1 expression, supporting the association with defense signaling pathways (Jasinski et al., 2001). In this study, the genes encoding ABC transporters tended to be downregulated in phytoplasma-infected plants, whereas in coconut palm, phytoplasma infection caused upregulation of ABC transporter genes (Nejat et al., 2015). It is possible that the dampening of plant defenses by some phytoplasmas, such as the cause of CFBD, suppresses ABC transporter expression.
In this study, we hypothesized that phytoplasma infection would influence the expression of genes related to phytohormone signaling because ethylene, JA, and SA play a central role in the regulation of defense responses. SA is generally stimulated by pathogens (Thomma et al., 1998) and feeding by piercing-sucking insects (War et al., 2012). In this study, genes directly related to phytohormones, such as JA, SA, and cytokinin, were not influenced by phytoplasma infection. These results are consistent with our measurement of the phytohormone levels in CFBD-infected plants vs. uninfected plants that show no significant differences (NP, unpublished data). The effect of phytoplasma infection on the expression of genes related to JA and SA signaling in plants is quite variable. Some studies show upregulation in both hormones (Mou et al., 2013; Mardi et al., 2015; Wang et al., 2018), whereas others show upregulation in SA and downregulation in JA (Sugio et al., 2011; Musetti et al., 2013), or vice versa (i.e., suppression of SA and induction of JA; Lu et al., 2014).
Other hormones, such as auxins, abscisic acid (ABA), and gibberellin (GA), play roles in plant development and growth regulation. ABA not only regulates the growth and developmental processes in plants but also acts as in adaptive responses to environmental stresses (Saito et al., 2004). GAs are plant hormones that regulate various developmental processes, including stem elongation, flowering, flower development, and leaf and fruit senescence. Recently, ABA and GA were reported as a key regulators of plant immunity (Denancé et al., 2013). In this study, we found two forms (sequences) of the gene that encodes an (+)-abscisic acid 8′-hydroxylase that is involved in ABA synthesis. One form is upregulated, while the other was down regulated (Supplementary Table 1). Lower ABA levels were shown in phytoplasma-infected Euphorbia coerulescens and Orbea gigantea plants (Omar et al., 2014). In the Jujube plant, the genes related to ABA synthesis were also downregulated (Wang et al., 2018). It has been suggested that ABA is involved in the growth of axillary buds due to the loss of apical dominance (Dewir et al., 2015). This may be a factor in the excessive branching (witches' broom formation) induced by some phytoplasmas, including the causal agent of CFBD.
The ent-copalyl diphosphate synthase gene, which is involved in the GA biosynthetic pathway (Prisic et al., 2004), was downregulated in infected plants. In coconut palm and tomato plant, the overexpression of GA 2 oxidase, an enzyme in the GA catabolic pathway, resulted in lower GA levels in infected plants (Ding et al., 2013; Nejat et al., 2015). The reduction of GA levels might be associated with the formation of abnormal flowers in cranberry with CFBD and other plants in which deformed flowers are a symptom of phytoplasma infection.
Plant secondary compounds, such as flavonoids, lignin, alkaloids, and terpenoids, play important roles in the defense mechanisms of plants. During the infection, plants commonly use phenylpropanoid compounds to synthesize flavonoids, lignin, and phytoalexins. The 4-coumarate-CoA ligase and PAL are key enzymes in this metabolic pathway (Dixon et al., 2002). In our study, 4-coumarate-CoA ligase and PAL are downregulated (Figure 3 and Supplementary Table 1) in infected plants relative to uninfected plants. The downregulation of these genes can result in lower levels of phenolic compounds in infected cranberry plants. In fact, Pradit et al. (2019) reported lower levels of proanthocyanidins, a class of polyphenols, in CFBD-infected than uninfected plants. In contrast, phytoplasma infection in apple and corn plants causes an increase in phenolic compounds and overproduction of hydrogen peroxide (Junqueira et al., 2004; Musetti et al., 2004).
Only two genes involved in photosynthesis, cytochrome b599, an important component of photosystem II, and photosystem II reaction center I protein, were highly influenced by phytoplasma infection in cranberry. Phytoplasma infection causes an inhibition of the entire electron transport chain in photosystem II in grape leaves (Bertamini and Nedunchezhian, 2001), significantly depresses photosystem I activity in Paulownia leaves (Mou et al., 2013), and downregulates the genes associated with the electron transport chain, such as ferredoxin, photosystem I, and photosystem II, in jujube (Wang et al., 2018). The grana and stroma lamellae, structures of chloroplasts, were destroyed, and original lamellae were formed in the infected jujube leaves (Xue et al., 2018). Our results show the suppression of photosystem II reaction center I protein gene expression, while the gene for cytochrome b599 showed induction, so the effects of CFBD on photosynthesis are unclear. The stage of infection is important in the expression levels of photosynthetic- and chlorophyll-related genes (Wang et al., 2018). Chlorophyll degradation in the later infection stages contributes to the yellowing and/or reddening symptoms of phytoplasma infection.
Photosynthetic products are first used for sucrose synthesis, and a complex enzyme system is used in the carbohydrate metabolism in plants (Hassid and Putman, 1950; Stein and Granot, 2018; Xue et al., 2018). Phytoplasma infection results in an accumulation of sugar in Catharanthus roseus (Lepka et al., 1999) and in coconut palm (Maust et al., 2003), whereas sugar content was reduced in maize (Junqueira et al., 2004). In jujube, sugar metabolism was downregulated at the first infection stage but then upregulated in the actively infecting stage (Wang et al., 2018; Xue et al., 2018). In our study, the genes associated with carbohydrate metabolism were both up- and downregulated (Figure 4). Interestingly, the genes that show upregulation include fructokinase and malate dehydrogenase (MDH). Fructokinase phosphorylates fructose to form fructose 6-phosphate, which is then used for glycolysis (Stein and Granot, 2018). Fructose is the preferential energy source for Spiroplasma citri, a plant-pathogenic bacterium in the same taxonomic class as phytoplasmas (André et al., 2005). MDH is involved in the oxidation of malate to form oxaloacetate. This enzyme acts in the citric acid cycle and is involved in energy release from carbohydrates and other substrates. Thus, phytoplasma infection seems to interfere with photosynthesis while increasing the metabolism of carbohydrates.
Cranberry plants with CBFD display abnormalities in plant growth and flower formation (Dobroscky, 1931; De Lange and Rodriguez-Saona, 2015). In this study, we expected some changes in the expression of flowering and development genes. We found that CFBD induces the expression of zinc finger SWIM domain-containing protein 3 and pentatricopeptide repeat-containing protein (PPR), while suppressing protein FAR1-related sequence (FRS). The zinc finger proteins are a super family of proteins involved in protein-protein interactions and are associated with many activities of plant growth and development, photosynthesis, and resistance mechanisms for biotic and abiotic stress (Feurtado et al., 2011; Lu et al., 2011). The pentatricopeptide repeat-containing protein can alter RNA sequences, turnover, processing. or translation. It is targeted to mitochondria or chloroplasts. This protein affects organelle biogenesis and function and, consequently, has an effect on photosynthesis, respiration, plant development, and environmental responses (Barkan and Small, 2014). FRS is the family of proteins that influence the transcription process. FRS family members play multiple roles in cellular processes, including light signal transduction, circadian rhythm and flowering, shoot meristem and floral development, plant immunity, ABA response, and chlorophyll biosynthesis (Ma and Li, 2018). Together, these proteins (zinc finger SWIM domain-containing protein 3, pentatricopeptide repeat-containing protein, and FRS) have a potentially wide range of functions in plant cells, including growth and development and response to biotic stress. Alterations in their expression patterns due to CFBD may be partly responsible for the visible symptoms (witches' broom and development of malformed flowers) of the disease.
Higher expression was noted in NAC domain-containing protein, PHS1, PAS2, and AP2. NAC is a transcription factor that acts as a floral repressor, controlling flowering time. NAC is normally expressed in response to cold temperatures (Nuruzzaman et al., 2013). PHS1 and PAS2 are synthesized by an endoplasmic reticulum-localized elongase multiprotein complex. PHS1 and PAS2 are involved in multiple biological processes, especially proliferation control of meristematic and, non-meristematic cells and cell dedifferentiation and proliferation. PHS1 and PAS2 expression can be enhanced by cytokinins, leading to callus-like structure development of the apical part of seedlings (Bellec et al., 2002; Bach et al., 2008). AP2 or APETALA2 is a gene and a member of a large family of transcription factors that play various roles throughout the plant lifecycle. The function of AP2 is to control floral organ identity determination and to respond to biotic or abiotic stresses (Riechmann and Meyerowitz, 1998). Increasing NAC gene expression can repress flower formation, whereas PHS1, PAS2, and AP2 manipulate the organ development at the apical part of plant. Thus, this set of genes may also play a role in the bushy appearance and/or flower abnormality in CFBD-affected plants.
Phytoplasma infection caused downregulation on FARs, AGL42, EID1-like F-box protein 3, phytochrome A, and transcription factor TGA. FARs play an important role in long-chain fatty acid alcohol metabolism, which can be found in the root, seed coat, and wound-induced leaf tissue (Doan et al., 2009; Domergue et al., 2010). AGL42 interacts genetically with CONSTANS 1 and FLOWERING LOCUS T. The two interacting components (CONSTANS 1 and Flowering T) have been shown to regulate flowering in Arabidopsis (Yoo et al., 2005). EID1-like F-box protein 3 is an F-box protein that functions as a negative regulator in phytochrome A-specific light signaling ubiquitin ligase complexes. Phytochrome A is important in light sensing for plant flowering (Marrocco et al., 2006). Transcription factor TGAs are implicated as regulators of pathogenesis-related genes. Transcription factor TGA proteins plays a role not only in defense against pathogens but also in processes involved in plant development (Zander et al., 2012). Thus, CFBD suppresses genes related to flowering, development, and defenses.
Phytoplasmas require insects as vectors. For phytophagous insects, nitrogen content is an important factor in food selection (Minkenberg and Ottenheim, 1990). The DEGs show both up- and downregulation of amino acid metabolism (Supplementary Table 1). Arginine and proline metabolism show upregulation, whereas alanine, glycine, serine, and threonine metabolism were suppressed by phytoplasma infection. Pradit et al. (2019) reported higher nitrogen content in CFBD-infected compared to uninfected plants. Thus, there is some manipulation of nitrogen metabolism in infected plants, but it is unclear how these changes in gene expression affect leaf nitrogen content and availability.
In this study, we tested the expression of selected target genes by RT-qPCR. The genes we selected include those related to defense (CML and PAL), carbohydrate metabolism (FK), and flowering (AP2). The relative gene expression data show that phytoplasma infection significantly induced the expression of CML, FK, and AP2 in the host plant, but there was no significant effect on PAL expression. This confirms that in cranberry with CFBD, defense signaling, carbohydrate metabolism, and flowering were impacted. PAL expression was lower in infected plants in the transcriptome data (Figure 3), suggesting a dampening of the phenylpropanoid pathway defense response. However, the RT-qPCR data (Figure 6) show PAL expression was not significantly impacted. Changes in PAL expression could be transient and “missed” in the samples used for RT-qPCR. Alternatively, changes in this pathway and its products (phenylpropanoids) may occur primarily downstream of PAL.
Cranberry false blossom disease was found to influence the expression of 358 unigenes (132 upregulated and 225 downregulated) in the leaves of young cranberry plants. The DEGs show that phytoplasma infection influences plant defense, photosynthesis and carbohydrate metabolism, and flowering and development of cranberry plants. Interestingly, the JA and SA phytohormone pathways are not influenced by this phytoplasma, Thus, the defense responses induced by the JA and SA signaling pathways appear to be uninduced in infected plants. Furthermore, the genes associated with secondary plant metabolism were suppressed in the phytoplasma-infected plant, while expression of carbohydrate metabolism genes was enhanced. Phytoplasma infection also caused the induction of malformed flowers and, thus, no fruit load, which may also contribute to nutrient availability and attraction of the leafhopper vector of this CFBD. Pradit et al. (2019) showed higher nutrient content, reduced defenses, and enhanced performance of phytophagous insects on plants with CFBD as compared with uninfected plants. Together, our data support the “host manipulation hypothesis” (Eigenbrode et al., 2018), wherein substantial manipulation of host plant gene expression facilitates the survival of the bacteria in the host plant and fosters dispersal of this vector-borne pathogen by depressing plant defenses while enhancing nutritional benefits to insect herbivores. It has long been known that some cranberry cultivars are less attractive to this insect vector (Wilcox and Beckwith, 1933). The next step in this research is to see how universal the changes described herein are across other cranberry cultivars, and specifically, what differences might affect vector attraction.
The datasets generated for this study can be found in NCBI Bioproject ID PRJNA542435.
NP and CR-S conceived the project. JP assisted in development of the project and directed the transcriptome and RT-qPCR work. JK provided bioinformatics support and analyses. NP, JP, and JK wrote the paper, with editing and suggestions from CR-S.
This work was supported by a Thai Royal scholarship to NP, hatch project no. NJ08140 and NJ08252 to CR-S, and funds from the New Jersey Blueberry and Cranberry Research Council, Inc., Cape Cod Cranberry Growers Association, and Ocean Spray Cranberries, Inc. to CR-S.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Kristia Adams for all the help and assistance during the molecular analyses.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00178/full#supplementary-material
Albert, M., Werner, M., Proksch, P., Fry, S. C., and Kaldenhoff, R. (2004). The cell wall-modifying xyloglucan endotransglycosylase/hydrolase LeXTH1 is expressed during the defense reaction of tomato against the plant parasite Cuscuta reflexa. Plant Biol. 6, 402–407. doi: 10.1055/s-2004-817959
Ali, G. S., Reddy, V. S., Lindgren, P. B., Jakobek, J. L., and Reddy, A. S. N. (2003). Differential expression of genes encoding calmodulin-binding proteins in response to bacterial pathogens and inducers of defense responses. Plant Mol. Biol. 51, 803–815. doi: 10.1023/A:1023001403794
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
André, A., Maucourt, M., Moing, A., Rolin, D., and Renaudin, J. (2005). Sugar import and phytopathogenicity of Spiroplasma citri: glucose and fructose play distinct roles. Mol. Plant Microbe Interact. 18, 33–42. doi: 10.1094/MPMI-18-0033
Bach, L., Michaelson, L. V., Haslam, R., Bellec, Y., Gissot, L., Marion, J., et al. (2008). The very-long-chain hydroxy fatty acyl-CoA dehydratase Pasticcino2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. U.S.A. 105, 14727–14731. doi: 10.1073/pnas.0805089105
Badri, D. V., Quintana, N., El Kassis, E. G., Kim, H. K., Choi, Y. H., Sugiyama, A., et al. (2009). An ABC Transporter mutation alters root exudation of phytochemicals that povoke an overhaul of natural soil microbiota. Plant Physiol. 151, 2006–2017. doi: 10.1104/pp.109.147462
Bai, X., Zhang, J., Ewing, A., Miller, S. A., Jancso Radek, A., Shevchenko, D. V., et al. (2006). Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188:3682. doi: 10.1128/JB.188.10.3682-3696.2006
Barkan, A., and Small, I. (2014). Pentatricopeptide repeat proteins in plants. Ann. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Beckwith, C. S., and Hutton, S. B. (1929). Cranberry false blossom and the blunt-nosed leafhopper. New Jersey Agric. Exp. St. Bull. 491, 1–16.
Bellec, Y., Harrar, Y., Butaeye, C., Darnet, S., Bellini, C., and Faure, J.-D. (2002). Pasticcino2 is a protein tyrosine phosphatase-like involved in cell proliferation and differentiation in Arabidopsis. Plant J. 32, 713–722. doi: 10.1046/j.1365-313X.2002.01456.x
Bennett, R. N., and Wallsgrove, R. M. (1994). Secondary metabolites in plant defense mechanisms. New Phytol. 127, 617–633. doi: 10.1111/j.1469-8137.1994.tb02968.x
Bertaccini, A. (2007). Phytoplasmas: diversity, taxonomy, and epidemiology. Front Biosci. 12:673–689. doi: 10.2741/2092
Bertamini, M., and Nedunchezhian, N. (2001). Effects of phytoplasma [Stolbur-Subgroup (Bois Noir-BN)] on photosynthetic pigments, saccharides, ribulose 1,5-bisphosphate carboxylase, nitrate and nitrite reductases, and photosynthetic activities in field-grown grapevine (Vitis vinifera L. cv. Chardonnay) leaves. Photosynthetica 39, 119–122. doi: 10.1023/A:1012412406727
Chandler, F. B., Wilcox, R. B., Bain, H. F., Bergman, H. F., and Dermen, H. (1947). Cranberry breeding investigation of the U.S. Dept. of Agriculture. Cranberries 12, 6–9.
Chang, C.-J. (1998). Pathogenicity of aster yellows phytoplasma and Spiroplasma citri on periwinkle. Phytopathology 88, 1347–1350. doi: 10.1094/PHYTO.1998.88.12.1347
Chen, T. A. (1971). Mycoplasmalike organisms in sieve tube elements of plants infected with blueberry stunt and cranberry false blossom. Phytopathology 61, 233–236. doi: 10.1094/Phyto-61-233
Cheval, C., Aldon, D., Galaud, J.-P., and Ranty, B. (2013). Calcium/calmodulin-mediated regulation of plant immunity. Biochim. Biophys. Acta Mol. Cell Res. 1833, 1766–1771. doi: 10.1016/j.bbamcr.2013.01.031
Christensen, N. M., Axelsen, K. B., Nicolaisen, M., and Schulz, A. (2005). Phytoplasmas and their interactions with hosts. Trends Plant Sci. 10, 526–535. doi: 10.1016/j.tplants.2005.09.008
Clarke, A. E., Anderson, M. A., Bacic, T., Harris, P. J., and Mau, S.-L. (1985). Molecular basis of cell recognition during fertilization in higher plants. J. Cell Sci. 1985(Suppl. 2), 261–285. doi: 10.1242/jcs.1985.Supplement_2.14
Davidson, N. M., and Oshlack, A. (2014). Corset: enabling differential gene expression analysis for de novoassembled transcriptomes. Genome Biol. 15:410. doi: 10.1186/s13059-014-0410-6
De Lange, E. S., and Rodriguez-Saona, C. (2015). Blunt-nosed Leafhopper: A Vector of Cranberry False Blossom Disease. Rutgers Coorperative Extension. Factsheet 1248. Available online at: https://njaes.rutgers.edu/fs1248/ (accessed January, 29, 2019).
Denancé, N., Sánchez-Vallet, A., Goffner, D., and Molina, A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4:155. doi: 10.3389/fpls.2013.00155
Dewir, Y. H., El Mahrouk, M. E., Hafez, Y. M., Rihan, H. Z., Sáez, C. A., and Fuller, M. P. (2015). Antioxidative capacity and electrolyte leakage in healthy versus phytoplasma infected tissues of Euphorbia coerulescens and Orbea gigantea. J. Plant Physiol. Pathol. 3. doi: 10.4172/2329-955X.1000139
Ding, Y., Wei, W., Wu, W., Davis, R. E., Jiang, Y., Lee, I. M., et al. (2013). Role of gibberellic acid in tomato defence against potato purple top phytoplasma infection. Ann. Appl. Biol. 162, 191–199. doi: 10.1111/aab.12011
Dixon, R. A., Achnine, L., Kota, P., Liu, C.-J., Reddy, M. S. S., and Wang, L. (2002). The phenylpropanoid pathway and plant defense—a genomics perspective. Mol. Plant Pathol. 3, 371–390. doi: 10.1046/j.1364-3703.2002.00131.x
Doan, T. T. P., Carlsson, A. S., Hamberg, M., Bülow, L., Stymne, S., and Olsson, P. (2009). Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J. Plant Physiol. 166, 787–796. doi: 10.1016/j.jplph.2008.10.003
Dobroscky, I. D. (1931). Studies on cranberry false blossom disease and its insect vector. Contrib. Boyce Thomps. Inst. 3, 59–83.
Domergue, F., Vishwanath, S. J., Joubès, J., Ono, J., Lee, J. A., Bourdon, M., et al. (2010). Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 153, 1539–1554. doi: 10.1104/pp.110.158238
Eigenbrode, S. D., Bosque-Pérez, N. A., and Davis, T. S. (2018). Insect-borne plant pathogens and their vectors: ecology, evolution, and complex interactions. Annu. Rev. Entomol. 63, 169–191. doi: 10.1146/annurev-ento-020117-043119
Fan, X.-P., Liu, W., Qiao, Y.-S., Shang, Y.-J., Wang, G.-P., Tian, X., et al. (2017). Comparative transcriptome analysis of Ziziphus jujuba infected by jujube witches' broom phytoplasmas. Sci. Hortic. 226, 50–58. doi: 10.1016/j.scienta.2017.08.026
Feurtado, J. A., Huang, D., Wicki-Stordeur, L., Hemstock, L. E., Potentier, M. S., Tsang, E. W. T., et al. (2011). The Arabidopsis C2H2 Zinc finger indeterminate domain1/enhydrous promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23, 1772–1794. doi: 10.1105/tpc.111.085134
Freeman, B. C., and Beattie, G. A. (2008). An overview of plant defenses against pathogens and herbivores. Plant Health Instructor 94. doi: 10.1094/PHI-I-2008-0226-01
Geisler, M., Blakeslee, J. J., Bouchard, R., Lee, O. R., Vincenzetti, V., Bandyopadhyay, A., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44(2), 179–194. doi: 10.1111/j.1365-313X.2005.02519.x
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hassid, W. Z., and Putman, E. W. (1950). Transformation of sugars in plants. Ann. Rev. Plant Physiol. 1, 109–124. doi: 10.1146/annurev.pp.01.060150.000545
Hogenhout, S. A., Oshima, K., Ammar, E.-D., Kakizawa, S., Kingdom, H. N., and Namba, S. (2008). Phytoplasmas: bacteria that manipulate plants and insects. Mol. Plant Pathol. 9, 403–423. doi: 10.1111/j.1364-3703.2008.00472.x
Hoshi, A., Oshima, K., Kakizawa, S., Ishii, Y., Ozeki, J., Hashimoto, M., et al. (2009). A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc. Natl. Acad. U.S.A. 106, 6416–6421. doi: 10.1073/pnas.0813038106
Hren, M., Nikolic, P., Rotter, A., Blejec, A., Terrier, N., Ravnikar, M., et al. (2009). 'Bois noir' phytoplasma induces significant reprogramming of the leaf transcriptome in the field grown grapevine. BMC Genomics 10:460. doi: 10.1186/1471-2164-10-460
Ishii, Y., Kakizawa, S., Hoshi, A., Maejima, K., Kagiwada, S., Yamaji, Y., et al. (2009). In the non-insect-transmissible line of onion yellows phytoplasma (OY-NIM), the plasmid-encoded transmembrane protein ORF3 lacks the major promoter region. Microbiology 155, 2058–2067. doi: 10.1099/mic.0.027409-0
Jasinski, M., Stukkens, Y., Degand, H., Purnelle, B., Marchand-Brynaert, J., and Boutry, M. (2001). A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13, 1095–1107. doi: 10.1105/tpc.13.5.1095
Junqueira, A., Bedendo, I., and Pascholati, S. (2004). Biochemical changes in corn plants infected by the maize bushy stunt phytoplasma. Physiol. Mol. Plant Pathol. 65, 181–185. doi: 10.1016/j.pmpp.2005.01.005
Kretzschmar, T., Burla, B., Lee, Y., Martinoia, E., and Nagy, R. (2011). Functions of ABC transporters in plants. Essays Biochem. 50:145. doi: 10.1042/bse0500145
Kube, M., Mitrovic, J., Duduk, B., Rabus, R., and Seemüller, E. (2012). Current view on phytoplasma genomes and encoded metabolism. Sci. World J. 2012:25. doi: 10.1100/2012/185942
Kuromori, T., Miyaji, T., Yabuuchi, H., Shimizu, H., Sugimoto, E., Kamiya, A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. 107:2361. doi: 10.1073/pnas.0912516107
Lee, I.-M., Davis, R. E., and Gundersen-Rindal, D. E. (2000). Phytoplasma: phytopathogenic mollicutes. Ann. Rev. Microbiol. 54, 221–255. doi: 10.1146/annurev.micro.54.1.221
Lee, I.-M., Gundersen-Rindal, D. E., Davis, R. E., Bottner, K. D., Marcone, C., and Seemüller, E. (2004). ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. Int. J. Syst. Evol. Microbiol. 54, 1037–1048. doi: 10.1099/ijs.0.02843-0
Lee, I.-M., Polashock, J., Bottner-Parker, K. D., Bagadia, P. G., Rodriguez-Saona, C., Zhao, Y., et al. (2014). New subgroup 16SrIII-Y phytoplasmas associated with false-blossom diseased cranberry (Vaccinium macrocarpon) plants and with known and potential insect vectors in New Jersey. Eur. J. Plant Pathol. 139, 399–406. doi: 10.1007/s10658-014-0396-7
Lepka, P., Stitt, M., Moll, E., and SeemÜLler, E. (1999). Effect of phytoplasmal infection on concentration and translocation of carbohydrates and amino acids in periwinkle and tobacco. Physiol. Mol. Plant Pathol. 55, 59–68. doi: 10.1006/pmpp.1999.0202
Levy, S., York, W. S., Stuike-Prill, R., Meyer, B., and Staehelin, L. A. (1991). Simulations of the static and dynamic molecular conformations of xyloglucan. The role of the fucosylated sidechain in surface-specific sidechain folding. Plant J. 1, 195–215. doi: 10.1111/j.1365-313X.1991.00195.x
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Lin, C., Zhou, T., Li, H., Fan, Z., Li, Y., Piao, C., et al. (2009). Molecular characterisation of two plasmids from paulownia witches'-broom phytoplasma and detection of a plasmid-encoded protein in infected plants. Eur. J. Plant Pathol. 123, 321–330. doi: 10.1007/s10658-008-9369-z
Lippmann, R., Kaspar, S., Rutten, T., Melzer, M., Kumlehn, J., Matros, A., et al. (2009). Protein and metabolite analysis reveals permanent induction of stress defense and cell regeneration processes in a tobacco cell suspension culture. Int. J. Mo.l Sci. 10:7. doi: 10.3390/ijms10073012
Liu, L., Sonbol, F.-M., Huot, B., Gu, Y., Withers, J., Mwimba, M., et al. (2016). Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7:13099. doi: 10.1038/ncomms13099
Lu, Y., Hall, D. A., and Last, R. L. (2011). A small Zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23, 1861–1875. doi: 10.1105/tpc.111.085456
Lu, Y.-P., Li, Z.-S., Drozdowicz, Y. M., Hörtensteiner, S., Martinoia, E., and Rea, P. A. (1998). AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione s-conjugates and chlorophyll catabolites: functional comparisons with AtMRP1. Plant Cell 10:267. doi: 10.1105/tpc.10.2.267
Lu, Y.-T., Li, M.-Y., Cheng, K.-T., Tan, C. M., Su, L.-W., Lin, W.-Y., et al. (2014). Transgenic plants that express the phytoplasma effector SAP11 show altered phosphate starvation and defense responses. Plant Physiol. 164, 1456–1469. doi: 10.1104/pp.113.229740
Ma, L., and Li, G. (2018). FAR1-RELATED SEQUENCE (FRS) and FRS-RELATED FACTOR (FRF) family proteins in Arabidopsis growth and development. Front. Plant Sci. 9:692. doi: 10.3389/fpls.2018.00692
Maejima, K., Oshima, K., and Namba, S. (2014). Exploring the phytoplasmas, plant pathogenic bacteria. J. Gen. Plant Pathol. 80, 210–221. doi: 10.1007/s10327-014-0512-8
Malembic-Maher, S., Gall, F. L., Danet, J.-L., Borne, F. D. Bové, J.-M., and Garnier-Semancik, M. (2005). Transformation of tobacco plants for single-chain antibody expression via apoplastic and symplasmic routes, and analysis of their susceptibility to stolbur phytoplasma infection. Plant Sci. 168, 349–358. doi: 10.1016/j.plantsci.2004.08.008
Malinovsky, F. G., Fangel, J. U., and Willats, W. G. T. (2014). The role of the cell wall in plant immunity. Front. Plant Sci. 5:178. doi: 10.3389/fpls.2014.00178
Marcone, C. (2009). “Movement of phytoplasmas and the development of disease in the plant,” in Phytoplasmas: Genomes, Plant Hosts and Vectors, ed P. G. Weintraub and P. Jones (Wallingford: CABI), 114–131. doi: 10.1079/9781845935306.0114
Mardi, M., Karimi Farsad, L., Gharechahi, J., and Salekdeh, G. H. (2015). In-depth transcriptome sequencing of Mexican Lime trees infected with Candidatus Phytoplasma aurantifolia. PLoS ONE 10:e0130425. doi: 10.1371/journal.pone.0130425
Marrocco, K., Zhou, Y., Bury, E., Dieterle, M., Funk, M., Genschik, P., et al. (2006). Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J. 45, 423–438. doi: 10.1111/j.1365-313X.2005.02635.x
Maust, B. E., Espadas, F., Talavera, C., Aguilar, M., Santamaría, J. M., and Oropeza, C. (2003). Changes in carbohydrate metabolism in coconut palms infected with the lethal yellowing phytoplasma. Phytopathology 93, 976–981. doi: 10.1094/PHYTO.2003.93.8.976
Minic, Z. (2007). Physiological roles of plant glycoside hydrolases. Planta 227:723. doi: 10.1007/s00425-007-0668-y
Minkenberg, O. P. J. M., and Ottenheim, J. J. G. W. (1990). Effect of leaf nitrogen content of tomato plants on preference and performance of a leafmining fly. Oecologia 83, 291–298. doi: 10.1007/BF00317551
Moons, A. (2008). Transcriptional profiling of the PDR gene family in rice roots in response to plant growth regulators, redox perturbations and weak organic acid stresses. Planta 229, 53–71. doi: 10.1007/s00425-008-0810-5
Mou, H.-Q., Lu, J., Zhu, S.-F., Lin, C.-L., Tian, G.-Z., Xu, X., et al. (2013). Transcriptomic analysis of Paulownia infected by Paulownia witches'-broom phytoplasma. PLoS ONE 8:e77217. doi: 10.1371/journal.pone.0077217
Musetti, R., di Toppi, L. S., Ermacora, P., and Favali, M. A. (2004). Recovery in apple trees infected with the apple proliferation phytoplasma: an ultrastructural and biochemical study. Phytopathology 94, 203–208. doi: 10.1094/PHYTO.2004.94.2.203
Musetti, R., Farhan, K., De Marco, F., Polizzotto, R., Paolacci, A., Ciaffi, M., et al. (2013). Differentially-regulated defense genes in Malus domestica during phytoplasma infection and recovery. Eur. J. Plant Pathol. 136, 13–19. doi: 10.1007/s10658-012-0147-6
Namba, S. (2002). Molecular biological studies on phytoplasmas. J. Gen. Plant Pathol. 68, 257–259. doi: 10.1007/PL00013086
Nejat, N., Cahill, D. M., Vadamalai, G., Ziemann, M., Rookes, J., and Naderali, N. (2015). Transcriptomics-based analysis using RNA-Seq of the coconut (Cocos nucifera) leaf in response to yellow decline phytoplasma infection. Mol. Genet. Genomics 290, 1899–1910. doi: 10.1007/s00438-015-1046-2
Nuruzzaman, M., Sharoni, A. M., and Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248. doi: 10.3389/fmicb.2013.00248
Omar, A. F., Dewir, Y. H., and El-Mahrouk, M. E. (2014). Molecular identification of phytoplasmas in fasciated cacti and succulent species and associated hormonal perturbation. J. Plant Interac. 9, 632–639. doi: 10.1080/17429145.2014.882421
Orlovskis, Z., and Hogenhout, S. A. (2016). A bacterial parasite effector mediates insect vector attraction in host plants independently of developmental changes. Front. Plant Sci. 7:885. doi: 10.3389/fpls.2016.00885
Polashock, J. J., Caruso, F. L., Averill, A. L., and Schilder, A. C. (2017). “False Blossom,” in Compendium of Blueberry, Cranberry, and Lingonberry Diseases and Pests, eds J. J. Polashock, F. L. Caruso, A. L. Averill, and A. C. Schilder (St. Paul, MN: APS Press), 123–124. doi: 10.1094/9780890545386
Polashock, J. J., and Vorsa, N. (2002). Development of SCAR markers for DNA fingerprinting and germplasm analysis of American cranberry. J. Am. Soc. Horti. Sci. 127, 677–684. doi: 10.21273/JASHS.127.4.677
Pradit, N., Mescher, M., Wang, Y., Vorsa, N., and Rodriguez-Saona, C. (2019). Phytoplasma infection of cranberries benefits non-vector phytophagous insects. Front. Ecol. Evol. 7:181. doi: 10.3389/fevo.2019.00181
Prisic, S., Xu, M., Wilderman, P. R., and Peters, R. J. (2004). Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 136, 4228–4236. doi: 10.1104/pp.104.050567
Rahman, A. K. M. S., Sugitani, N., Hatsu, M., and Takamizawa, K. (2003). A role of xylanase, α-L-arabinofuranosidase, and xylosidase in xylan degradation. Can. J. Microbiol. 49, 58–64. doi: 10.1139/w02-114
Riechmann, J. L., and Meyerowitz, E. M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 33–46.
Rodriguez-Saona, C., Polashock, J., and Malo, E. A. (2013). Jasmonate-mediated induced volatiles in the American cranberry, Vaccinium macrocarpon: from gene expression to organismal interactions. Front. Plant Sci. 4:115. doi: 10.3389/fpls.2013.00115
Rudzinska-Langwald, A., and Kaminska, M. (2001). Ultrastructural changes in aster yellows phytoplasma affected Limonium sinuatum Mill. plants II. Pathology of cortex parenchyma cells. Acta Soc. Bot. Pol. 70, 273–279. doi: 10.5586/asbp.2001.035
Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., et al. (2004). Arabidopsis CYP707A encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134, 1439–1449. doi: 10.1104/pp.103.037614
Stein, O., and Granot, D. (2018). Plant fructokinases: evolutionary, developmental, and metabolic aspects in sink tissues. Front. Plant Sci. 9:339. doi: 10.3389/fpls.2018.00339
Sugio, A., and Hogenhout, S. A. (2012). The genome biology of phytoplasma: modulators of plants and insects. Curr. Opin. Microbiol. 15, 247–254. doi: 10.1016/j.mib.2012.04.002
Sugio, A., MacLean, A. M., Kingdom, H. N., Grieve, V. M., Manimekalai, R., and Hogenhout, S. A. (2011). Diverse targets of phytoplasma effectors: from plant development to defense against insects. Ann. Rev. Phytopathol. 49, 175–195. doi: 10.1146/annurev-phyto-072910-095323
Thomma, B. P. H. J., Eggermont, K., Penninckx, I. A. M. A., Mauch-Mani, B., Vogelsang, R., Cammue, B. P. A., et al. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 95, 15107–15111. doi: 10.1073/pnas.95.25.15107
Underwood, W. (2012). The plant cell wall: a dynamic barrier against pathogen invasion. Front. Plant Sci. 3:85. doi: 10.3389/fpls.2012.00085
Wang, H., Ye, X., Li, J., Tan, B., Chen, P., Cheng, J., et al. (2018). Transcriptome profiling analysis revealed co-regulation of multiple pathways in jujube during infection by ‘Candidatus Phytoplasma ziziphi’. Gene 665, 82–95. doi: 10.1016/j.gene.2018.04.070
War, A. R., Paulraj, M. G., Ahmad, T., Buhroo, A. A., Hussain, B., Ignacimuthu, S., et al. (2012). Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 7, 1306–1320. doi: 10.4161/psb.21663
Weintraub, P. G., and Beanland, L. (2006). Insect vectors of phytoplasmas. Ann. Rev. Entomol. 51, 91–111. doi: 10.1146/annurev.ento.51.110104.151039
Wilcox, R. B., and Beckwith, C. S. (1933). A factor in the varietal resistance of cranberries to the false blossom disease. J. Agric. Res. 47, 583–590.
Will, T., and van Bel, A. J. E. (2006). Physical and chemical interactions between aphids and plants. J. Exp. Bot. 57, 729–737. doi: 10.1093/jxb/erj089
Xue, C., Liu, Z., Dai, L., Bu, J., Liu, M., Zhao, Z., et al. (2018). Changing host photosynthetic, carbohydrate, and energy metabolisms play important roles in phytoplasma infection. Phytopathology 108, 1067–1077. doi: 10.1094/PHYTO-02-18-0058-R
Yi, J.-C., Lim, T.-H. L., and Cha, B. (2001). Changes in phytoplasma densities in witches' broom-infected jujube trees over seasons. Plant Pathol. J. 17, 295–299.
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., et al. (2005). Constans activates suppressor of overexpression of constans 1 flowering locus T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778. doi: 10.1104/pp.105.066928
Keywords: false blossom, Vaccinium macrocarpon, transcriptome analysis, RNASeq, RT-qPCR, host manipulation hypothesis
Citation: Pradit N, Rodriguez-Saona C, Kawash J and Polashock J (2019) Phytoplasma Infection Influences Gene Expression in American Cranberry. Front. Ecol. Evol. 7:178. doi: 10.3389/fevo.2019.00178
Received: 15 February 2019; Accepted: 03 May 2019;
Published: 24 May 2019.
Edited by:
David Georges Biron, Centre National de la Recherche Scientifique (CNRS), FranceCopyright © 2019 Pradit, Rodriguez-Saona, Kawash and Polashock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James Polashock, amFtZXMucG9sYXNob2NrQGFycy51c2RhLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.