- Centers for Disease Control and Prevention, Fort Collins, CO, United States
Despite the widespread recognition of the risks of disease transmission associated with international trade in domestic animals and movement of exotic animals, less is known about the role of rats in carrying pathogens between continents. The genus Bartonella, a highly prevalent and extremely diverse group of bacteria, includes species that are excellent sentinel organisms for evaluating the transoceanic and intra-continental movement of the pathogens carried by rats of the genus Rattus. The patterns of spatial distribution, occurrence, and genetic diversity of Bartonella species infecting rats and their arthropod ectoparasites depend on the geographic locations within metropolitan areas of the Americas, Africa, Asia, and Europe. One of the points addressed in this review is a comparison of the diversity of Bartonella species carried by rats in their original habitats in Southeast Asia and in the cities occupied by rats recently. The invasion of Rattus rats into new urban territories create significant risk for human health.
Introduction
Commensal Rats and Infectious Agents Carried by Rats in Urban Areas
People create dramatically new environments. This is especially evident in urban settings, which may favor some mammalian species that might become “synanthropic.” A number of factors contribute to the public health threat presented by synanthropic animals. According to the calculations provided by McFarlane et al. (2012), human cases of zoonotic infectious diseases caused by synanthropic animals happen 15 times more frequently compared to wild animals. Urban territories may provide suitable conditions for reproduction of arthropods serving as vectors for vector-borne diseases because of some specific environmental changes, such as attenuation of the temperature range and humidity (Shochat et al., 2006; Bradley and Altizer, 2007). The environments in cities have higher land surface temperature compared to surrounding natural habitats, so-called “urban heat islands” (Grimmond, 2007). The urban heat islands represent an example of the numerous potential environmental changes caused by urbanization that can affect distribution and prevalence of zoonotic and vector-borne diseases in cities and suburban areas.
Urban development changes rodent communities dramatically. Among mammalian species adapted to life in cities, certain species of the genus Rattus play especially evident and important roles because of their close association with human activities (Battersby et al., 2008). Living in close proximity to human houses and having high exploratory activity, rats frequently encounter people in cities and small towns. An extensive questionnaire conducted among residents of Baltimore, Maryland (N = 1,363) showed 64% of respondents seen rats on streets and alleys, 6% of residents noticed rats inside houses, and 1.2% of residents reported bites by rats (Childs et al., 1991). Microclimatic conditions in cities can affect development of the fleas found on rats (Krasnov et al., 2001) and, therefore, influence the transmission of some vector-borne bacteria. In addition, the urban environment affect availability of resources, specifically food, for rodents (Cevidanes et al., 2017), promoting higher concentration, and density of rats that in turn can facilitate density-dependent transmission of various zoonotic pathogens carried by these animals.
Rattus Rats as Invasive Species
Over 60 species belong to the genus Rattus (Musser and Carlton, 2005). Of these, only three species, the Norway rat (R. norvegicus), the black (roof) rat (R. rattus), and the Oriental house rat (R. tanezumi), have dispersed around the globe and colonized urban settings in most countries, and the range of the fourth invasive species, the Pacific rat (R. exulans), is limited to tropical Asia-Pacific areas (Kosoy et al., 2015). Rats of the genus Rattus became close neighbors to humans at a very early stage of human civilization. However, during the last century, distribution of rats has dramatically expanded to new geographic regions, and the rat invasion of the cities in different countries led to dramatic challenges (Khlyap et al., 2012). Importantly, the process of rat invasion from the region of their origin to other places continues. In the past, the distribution of rats depended mainly on cart traffic and ship routes, the latter being the leading means of crossing the oceans. In the modern world, the role of railroad, truck, and airplane in transporting rats is growing (Khlyap et al., 2016).
Application of molecular tools for barcoding animal species has challenged taxonomic identification of rats. Analysis of mitochondrial DNA of rats, previously identified as R. rattus, demonstrated the complex of separate species within the R. rattus-complex (Aplin et al., 2011). A recent survey of rats belonging to this complex collected across their global range and conducted by Aplin et al. (2011) allowed discriminating several genetic lineages within the black rat complex. Their investigations demonstrated that a diversification of these lineages happened in the early Middle Pleistocene within South Asia, southern and northern Indochina, and in the region close to Himalayas. These authors also identified two other currently recognized Rattus species as potential derivatives of a paraphyletic R. rattus (Aplin et al., 2011). Interestingly, their results demonstrated that three of four phylogenetic lineages within the R. rattus complex happened in prehistoric times. The distribution of particular genotypes matches historically documented patterns of human dispersal and trade. Based on this analysis, Aplin et al. (2011) concluded that commensalism arose multiple times in black rats and in widely separated geographic regions. Importantly, such a regionalism may account for spreading of pathogens associated with R. rattus (Aplin et al., 2011). Examining multiple samples of this species from different parts of the world, Aplin et al. (2011) reported ecologically and morphologically similar lineages of rats with distinct histories of invasion to other geographic regions. One of the lineages, designated as R. rattus I, has dispersed around the globe and exists alongside humans, while rats of another lineage (designated as R. rattus IV) have not spread beyond Bornean Malaysia, Indonesia, and the Philippines (Aplin et al., 2011; Lack et al., 2012).
Ecology of Rats in Urban Settings
Rodents in urban environments often show specific adaptations (Khlyap et al., 2012, 2016). Sometimes it may be challenging to define urban areas using ecological perspectives, especially when suburban areas are considered. It is also important to remember that ecological conditions in urban areas vary among countries. Specifically, the distribution of rats may greatly contrast between countries in the South and Southeast Asia where Rattus rats occupy practically all habitats and Europe and North America where rats are commonly restricted to city limits (Khlyap et al., 2012). Himsworth et al. (2014a) demonstrated that population density of urban rats varied significantly over short distances. Populations of rats are often largest in high-density residential areas (Khlyap et al., 2016).
Some big cities are particularly favorable places for urban rats because of their aging infrastructure, high moisture, and poverty rates. Easterbrook et al. (2005) estimated an outdoor Norway rat population in residential neighborhoods of Baltimore at around 48,000 individuals. Though these rats can disperse over long distances, investigations of marked animals demonstrated that they tend to live within small individual territories, sometimes not extending beyond a single building. Genetic analysis using microsatellite markers showed that rats demonstrate strong site fidelity (Gardner-Santana et al., 2009). However, there was evidence of infrequent, long-distance movements within the city indicated by capturing some rats 2–11.5 km away from the locations assigned based on the genetic analysis (Gardner-Santana et al., 2009). Among factors influencing rat presence and abundance, Himsworth et al. (2014b) suggested building condition and specific land use. In industrialized countries, rats commonly occupy sewer system of cities (Lund, 2015).
Pathogens and Movement Ecology of Animals
As invasive animal species rapidly become more prevalent in many parts of the world, relations between previously disconnected animal populations can promote the spread of pathogens carried by these animals (Crowl et al., 2008). On the other side of the coin, infectious agents detected in invasive animals may illustrate the “tracks” left during the spreading of their mammalian hosts. Genetic studies of animals provide irreplaceable tools for deciphering routes of invasion, but characterization of accompanying micro- and macro-parasites can provide additional support for such a goal. An assessment of risk of infectious diseases introduced by invasive animal hosts highlights importance of such information. Nevertheless, the selection of specific microbial species that can serve as markers for measuring the movement of animal hosts is not a trivial task. Clearly, the selected microorganisms should be prevalent in animal populations, but not too much so otherwise the omnipresent infections cannot be good indicators of animal movement. Secondly, these microbial species should be highly specific to their animal hosts to reflect the long history of coevolution between animals and microbes. Finally, their genetic variability should be high enough to reflect the routes of dispersion of the animals hosting these microbial agents.
Bartonella Species as Sentinel Organisms
Bartonella species comprise Gram-negative bacteria parasitizing mammalian erythrocytes and endothelial cells (Birtles, 2005). Infecting a wide variety of hosts, Bartonella may present in these animals as a subclinical and persistent bacteremia (Schülein et al., 2001). Bartonella species are an extremely diverse group of bacteria infecting various mammalian species, especially rodents (Kosoy et al., 2018), that also spans the symbiont-pathogen continuum (Segers et al., 2017). Moreover, these “vertebrate host-arthropod vector-bartonellae” tripartite systems appear to be globally distributed, phylogenetically complex, and provide a popular tool for ecological comparative analyses (Buffet et al., 2013; Brook et al., 2017; Kosoy et al., 2018). The rodent habitat represents an important factor for the transmission cycle of Bartonella in nature (Gutiérrez et al., 2015). Analyses of genetic diversity of Bartonella species based on sequencing approaches can be informative for comparing bacterial prevalence and diversity in rat populations across various spatial and temporal scales. However, the effect of urbanization on Bartonella prevalence and diversity in rat populations has never been sufficiently analyzed. The objectives of ecological studies determine the level of discrimination between compared strains or genotypes. In most situations, the investigators report discrimination of bacteria at the species level or compare sequence identity with a specific Bartonella type strain (Kosoy et al., 2018). In the absence of sequence data, reporting PCR-positive samples alone may overestimate bartonella prevalence in such ecological studies. Therefore, Kosoy et al. (2018) advocated that studies of prevalence of Bartonella should adhere to the standard of reporting only sequence-positive samples.
Methods
We thoroughly analyzed published literature concerning identification of Bartonella species in rats. For this review, we examine prevalence and diversity of Bartonella in rats belonging to the genus Rattus. Only in few instances, strictly for comparative purposes, we provided data on other mammalian species co-habiting with rats of Rattus. We conducted a literature search by various search engines, including PubMed, Scopus, BioOne, Medline, ScienceResearch, Google Scholar, OVID Medicine, and Web of Science. In the search, we used the following keywords: “Bartonella AND Rats,” “Bartonella AND Rodents,” “Bacteria AND Rattus,” “Rat-Borne Diseases,” “Rodent-Borne-Diseases,” “Urban Bacterial Diseases,” “Urban Rodents AND Pathogens,” and their combinations. We analyzed data from serological, molecular, and bacteriological detection of Bartonella in rats. Analyzing data obtained from different assays, we have given a priority to the results that included an identification of Bartonella species and genotypes.
Results and Discussion
Rattus Rat-Adapted Bartonella Species
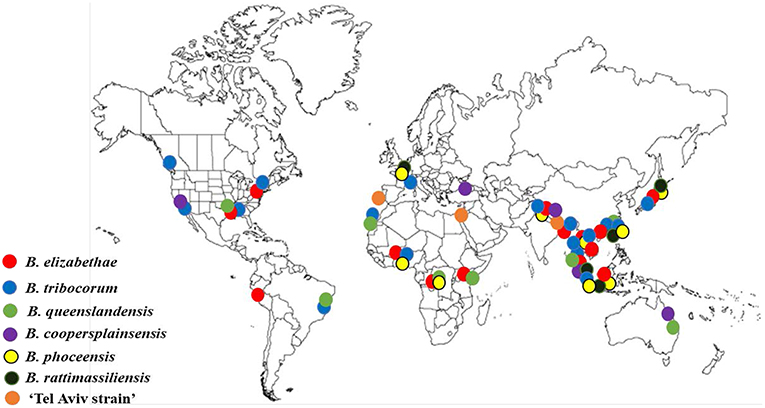
Overall, Bartonella species and genotypes found in Rattus rats are highly specific for rodents belonging to this genus (Kosoy et al., 2012; Buffet et al., 2013). Many rat-adapted Bartonella species have a worldwide distribution (Buffet et al., 2013; Hayman et al., 2013). The most prevalent Bartonella species, such as B. elizabethae, B. tribocorum, B. rattimassiliensis, and B. queenslandensis, are phylogenetically clustered in a well-demarcated lineage (Figure 1). Originally, all these species were described based on the sequence distances between the species for several housekeeping gene markers (Heller et al., 1998; Gundi et al., 2004, 2009). However, the question remains how to recognize the status of strains that occupy intermediate positions between the described Bartonella species. This question was not unique for Bartonella strains found in rats. To solve this problem, Kosoy et al. (2012) proposed to use a combination of genetic markers and ecological parameters for delineation of species complexes that include closely related genospecies, named strains, and unique genotypes carried by ecologically similar mammalian hosts. Following this proposal, the B. elizabethae complex sensu lato represents a large group of species and strains associated with the Old World rats (Kosoy et al., 2012; Buffet et al., 2013). A well-characterized, but unnamed strain “Tel-Aviv” also belongs to the B.elizabethae species complex (Harrus et al., 2009).
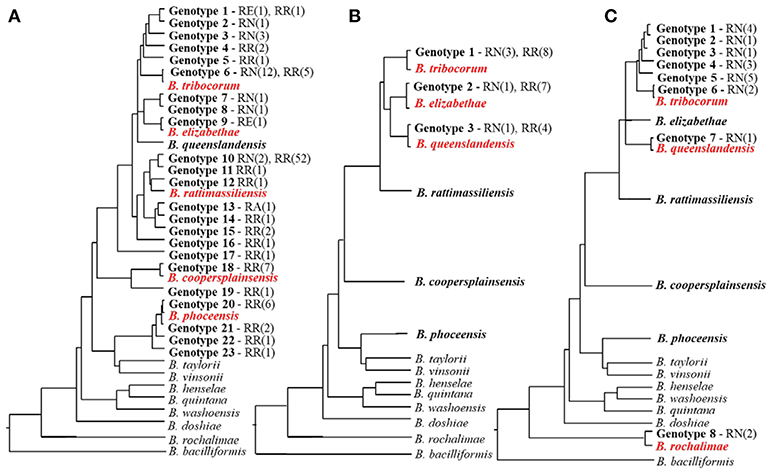
Figure 1. Diversity of Bartonella species and genotypes found in Rattus rats from rural Thailand (A), an urban slum in Nairobi, Kenya (B), and downtown Los-Angeles, California, U.S. (C) and their schematic phylogenetic relations. The names of Bartonella species found in a particular study are in red. The numbers of detected genotypes do not correspond to each other and are given solely for comparative purpose. The phylogenetic trees represent phylogenetic relationship of the gltA sequences of the named Bartonella species and unnamed genotypes obtained from three separate studies (Bai et al., 2009; Gundi et al., 2012; and Halliday et al., 2015) following the same methodology at one laboratory (CDC, Fort Collins, Colorado). The names of rat species are abbreviated as RN for R. norvegicus and RR for R. rattus; the number of isolates obtained from each species is in parentheses.
In addition, three more Bartonella species (B. phoceensis, B. coopersplainsensis, and B. rochalimae) infect Rattus rats worldwide (Buffet et al., 2013). Of these three species the first two (B. phoceensis and B. coopersplainsensis) are typical for Rattus rats, while B. rochalimae is an ubiquitous bacterial species detected in a wide range of mammals, and is especially common in wild carnivores and their fleas (Bai et al., 2016). B. phoceensis is a bacterium originally isolated from the blood of the rats of R. norvegicus from the city of Marseille, France (Gundi et al., 2004). Bartonella coopersplainsensis was isolated from the blood of a Cape York rat (Rattus leucopus) in Australia (Gundi et al., 2009).
Although stressing the high level of host-specificity for Rattus among B. elizabethae-like species, we have to admit that these species can also infect other mammals, e.g., Bandicota rats and Asian house shrews (Suncus murinus) from Bangladesh and Nepal (Bai et al., 2007; Gundi et al., 2010); Brush-furred rats (Lophuromys sp.) from Tanzania (Gundi et al., 2012); Namaqua rock rats (Aethomys namaquensis) and Bushveld gerbils (Tatera leucogaster) from South Africa (Pretorius et al., 2004); and Cairo spiny mice (Acomys cahirinus) from Israel (Morick et al., 2009). The unexpectedly broad host range of B. elizabethae-like species might be explained by the commonality of fleas that infest various rodent species. Thus, B. elizabethae, B. tribocorum, and B. queenslandensis DNA have been detected in Xenopsylla fleas collected not only from Rattus rats, but also from gerbils, Mus mice, and shrews worldwide (Tsai et al., 2010; Billeter et al., 2011; Bitam et al., 2012).
Bartonella Infections in Urban Populations of Rats
Asia
Bangladesh
Prevalence of Bartonella bacteria in R. rattus collected in Kamalapur, a low socioeconomic residential area of Dhaka, was 32.3% (32/99) (Bai et al., 2007). This rate was lower than that observed in two other co-habiting mammalian species: lesser bandicoot rats Bandicota bengalensis (63.2%) and house shrews Suncus murinus (42.9%). Bacteriological observations of small mammals captured in Dhaka indicated a diverse assemblage of genetic variants of Bartonella (Bai et al., 2007). The isolates obtained from R. rattus belonged to three groups, none of which aligned closely with previously described Bartonella species, whereas isolates obtained from lesser bandicoot rats Bandicota bengalensis were much closer or identical to B. elizabethae or B. tribocorum. Importantly, bartonellae isolated from two black rats captured in Dhaka were identical by the gltA gene to the isolates previously found in rats from Porto Santo Island, Portugal and New Orleans, Louisiana, USA. Later, multiple isolates similar to this type were found in R. rattus rats from Tel Aviv, Israel (Harrus et al., 2009).
China
The first study, which demonstrated a high prevalence of Bartonella in Rattus rats in Asian cities was conducted in Yunnan Province, the southwestern part of the mainland China (Ying et al., 2002). Culturing blood of rats from three cities (Yiliang, Baoshan, and Jianchuan) revealed 41.4% (24/58) Bartonella-positive animals among R. tanezumi subsp. flavipectus and 42.9% (3/7) among R. norvegicus. Rats in every investigated city along the coast of Fujian Province were infected by Bartonella species: 9.3% in Ningde, 9.5% in Fuzhou, 9.4% in Putian, 28.2% in Quanzhou, 17.4% in Xiamen, and 13.3% in Zhangzhou (Ye et al., 2009). Despite considerable heterogeneity and varying degrees of relatedness to B. elizabethae, all isolates from urban rats belonged to the same phylogenetic lineage. Isolates from rats from Fujian coastal regions belonged to three species: B. elizabethae, B. queenslandensis, and B. tribocorum (Ye et al., 2009).
Indonesia
Microscopic examination of blood smears of rodents from the Greater Jakarta area revealed 6.0% (13/218) Bartonella-positive rodents. Of 79 R. tanezumi, 49 R. norvegicus, and one R. exulans captured in three study sites (Bambu Apus, Penjaringan Harbor, and Ragunan Zoo), seven R. tanezumi rats and one R. norvegicus rat were positive for Bartonella species (Winoto et al., 2005). Three Bartonella species (B. phoceensis, B. rattimassiliensis, and B. elizabethae) were found in rats (R. tanezumi and R. norvegicus) from Jakarta (Winoto et al., 2005).
Japan
Interesting results were reported after investigation of rats collected in two cities and four suburban areas of Japan (Inoue et al., 2008). Bartonella isolates were obtained from R. rattus from suburban areas in Nakanoshima Island (2/4), Yoroshima Island (10/17), Tokunoshima Island (4/12), and Main Island (2/6). At the same time, all R. rattus rats captured in cities of Yokohama (n = 255) and Shimoda (n = 3) were free of Bartonella. All R. norvegicus from the cities (n = 85) and suburban areas (n = 20) were negative for Bartonella (Inoue et al., 2008). Investigating R. rattus rats captured from several places in Japan, Inoue et al. (2008) identified B. tribocorum, B. elizabethae, B. phoceensis, and B. rattimassiliensis based on two genetic markers.
Lao P.D.R
A molecular survey of rats was conducted in three urban areas of Laos (Angelakis et al., 2009). Bartonella DNA was found in 10.1% of R. rattus (n = 79) and 30.4% of R. exulans (n = 23) from Vientiane City, the largest city of Laos, which is located near its border with Thailand. Nine percent of R. rattus (n = 141) from the city of Luang Prabang in northern part of Laos were positive, as were 20.1% of R. rattus (n = 159) from a town in Luang Namtha Province near the border with Yunnan, China (Angelakis et al., 2009). Angelakis et al. (2009) identified three well-characterized Bartonella species (B. phoceensis, B. elizabethae, and B. tribocorum) and two additional genotypes (Lao/Nh1 and Lao/Nh2) related to B. tribocorum. In R. rattus rats, the authors found B. phoceensis (n = 1), B. elizabethae (n = 2), B. tribocorum (n = 2), and Lao/Nh2 (n = 3); while in R. exulans rats, the predominant species was B. tribocorum (n = 5) with single positive rats infected with either B. elizabethae or B. phoceensis (n = 1) (Angelakis et al., 2009).
Malaysia
Urban rats were captured as part of a pest management program in Kuala Lumpur (n = 59) and Pulau Pinang (n = 36). Of the 95 bacteriologically and molecularly tested rats, 13.5% of R. rattus (n = 58) and 13.8% of R. norvegicus (n = 37) were positive for Bartonella (Tay et al., 2014). Five species of Bartonella (B. tribocorum, B. rattimassiliensis, B. coopersplainsensis, B. elizabethae, and B. queenslandensis) were identified (Tay et al., 2014).
Nepal
Bartonella was detected in 39 (43.3%) of 90 R. rattus subsp. brunneusculus captured in three cities: Bhaktapur (33.3%; 12/26), Kathmandu (40.6%; 13/32), and Lalitpur (43.8%; 14/32) (Gundi et al., 2010). There were five species of Bartonella in R. rattus subsp. brunneusculus collected in three urban areas in Nepal, including B. elizabethae (n = 5), B. coopersplainsensis (n = 3), B. tribocorum (4), B. queenslandensis (n = 2), and B. phoceensis (n = 1). In addition, three genotypes were different from any described Bartonella species (Gundi et al., 2010).
Singapore
A molecular survey of commensal rodents in Singapore resulted in detection of Bartonella DNA in 75% (3/4) of R. norvegicus and 34.5% (10/29) of R. tanezumi (Neves et al., 2018). Annandale's rats (R. annandalei) captured outside the urban areas were negative for Bartonella. The only R. exulans captured in a city park was also free of Bartonella. Characterization of Bartonella genotypes circulating in rat populations within Singapore has revealed five species (Neves et al., 2018). Interestingly, the composition of Bartonella species differed depending on the rat species. B. coopersplainsensis, B. elizabethae, B. grahamii, and B. phoceensis were found in R. tanezumi; whereas B. tribocorum, B. rattimassiliensis, B. grahamii, and B. queenslandensis were found in R. norvegicus. A smaller number of Bartonella species was in R. exulans: B. tribocorum, B. rattimassiliensis, and B. phoceensis (Neves et al., 2018).
Taiwan
There were several studies of urban rats in Taiwan. The investigation of rodents captured in the Taichung area, including markets in the urban area, has found 52.7% of R. norvegicus (n = 169), 10% of R. rattus (n = 10), and 66.7% of R. losea (n = 3) (Hsieh et al., 2010) were positive for Bartonella. Among the 182 rats tested from Taichung, the city located in central Taiwan, the cultured organisms belonged to five species of Bartonella (Hsieh et al., 2010). Strains closely related to B. tribocorum and B. elizabethae were the most prevalent of them. In addition, B. rattimassilensis, B. grahamii, and B. phoceensis were isolated from R. norvegicus. Several R. norvegicus rats were co-infected with different Bartonella species: 10 rats with B. elizabethae-like and B. tribocorum-like organisms and one rat with B. phoceensis-like and B. tribocorum-like organisms (Hsieh et al., 2010).
Another study in Taichung resulted in detection of Bartonella in 9.4% (5/53) of R. norvegicus and 33.3% (1/3) of R. rattus (Lin et al., 2008). In another study in Taiwan, the same species of Bartonella (B. phoceensis, B. tribocorum, B. elizabethae, and B. rattimassiliensis) were detected in rats, but, in addition, B. queenslandensis was also reported (Tsai et al., 2010). The Bartonella detected in two R. norvegicus rats captured on the university campus was B. tribocorum. As we will show later, the diversity of Bartonella species in rats from natural habitats can be much higher. Interestingly, Lin et al. (2008) reported isolation of strains closely related to B. rochalimae from R. norvegicus captured Taichung, Taiwan.
Thailand
In spite of many investigations of Bartonella infections in rats from the fields and forest in different parts of Thailand, there were very limited efforts to investigate urban rats. There is only one report of investigation of urban rats captured in Bangkok in 2008: Bartonella bacteria were cultured from R. exulans (55.6%; n = 9) and R. rattus (67.6%; n = 34), and none from three R. norvegicus (Kim et al., 2016).
Middle East
Cyprus
Of 622 rats (402 R. norvegicus and 220 R. rattus subsp. frugivorus) collected in 51 different sites in Cyprus, 10.5% were found seropositive for Bartonella antibodies (Psaroulaki et al., 2010). The authors of this study used B. henselae antigens and, more likely, the reported antibodies were cross-reactive with other Bartonella species. The authors, however, claimed that the seropositivity rate significantly correlated with the presence of cat fleas, but not with the presence of rat fleas.
Israel
Bartonella DNA was detected in spleen samples of 19 out of 79 (24%) R. rattus captured in 19 suburban sites ranged from the kibbutz Dafna in the north of Israel to the City of Beersheba in the south (Morick et al., 2009). Bartonella strains obtained from R. rattus rats captured in Tel-Aviv, Israel were characterized by sequencing multiple genetic loci (gltA, ribC, rpoB, 16S RNA, groEL, and the intergenic spacer region ITS) (Harrus et al., 2009). These strains were identical among themselves, and sequences of each of the analyzed genes indicated a closeness to one of the two Bartonella species (B. tribocorum and B. elizabethae), while being different from both of these species (Harrus et al., 2009). The characterization of these strains supports a hypothesis that it could be a separate species of Bartonella.
Turkey
In a study conducted in an urban area of Zongulda in the western Black Sea Region of Turkey, only one rat out of 14 investigated, including eight R. rattus and six R. norvegicus was found to be Bartonella-positive (Çelebi et al., 2015). A Bartonella isolate obtained from R. rattus rats captured in Zongulda was identified as B. coopersplainsensis (Çelebi et al., 2015), the species which has been previously reported in Australia and Southeastern Asia (Gundi et al., 2009; Jiyipong et al., 2012).
Africa
Benin
A molecular survey of Bartonella species in rats captured inside human residences and peridomestic areas was conducted in three zones of Cotonou where half of the population of the Republic of Benin resides (Martin-Alonso et al., 2016). Bartonella DNA was detected in 26.3% (5/19) of R. norvegicus captured in the zone 1 (20.8%), whereas none of the 110 R. rattus captured in the three zones were found positive for Bartonella. Three Bartonella species (B. elizabethae, B. tribocorum, and B. rochalimae) were found in spleens of R. norvegicus rats from an urban area near Nokoue Lake in Benin (Martin-Alonso et al., 2016).
DR Congo
In studies conducted in Djalusene, Kpandruma, Rethy, and Zaa, R. rattus was the only rat species captured inside houses and the domestic environment. Screening of spleen samples from the 25 captured R. rattus indicated the presence of Bartonella DNA in only one rat from Zaa (5.9%, 1/17) (Gundi et al., 2012). Out of 11 Bartonella sequences obtained from R. norvegicus rats, four were similar (96–100% homology) to the Bartonella strain 1-1C (GenBank EU551156), a B. rochalimae-like strain described from R. norvegicus in Taiwan. Interestingly, this strain was found only in R. norvegicus, not in R. rattus, although Bartonella prevalence was higher in the latter. Five sequences were similar with 97–100% homology to an uncultured Bartonella genotype previously detected in R. rattus in Nepal (Gundi et al., 2010). One sequence shared 100% homology with B. queenslandensis, a Bartonella species originally described in Australian rats (genera Melomys and Rattus) (Gundi et al., 2009). In another study conducted in Kisangani, the proportion of Bartonella-infected rats was significantly higher in R. rattus (25.0%, 5/20) than in R. norvegicus (15.1%; 16/106) (Laudisoit et al., 2014). Two R. rattus and two R. norvegicus captured during this study in a marketplace were co-infected with Bartonella and Rickettsia species (Laudisoit et al., 2014). A strain with 100% homology with the human strain of B. elizabethae was reported in one of the R. norvegicus rats (Laudisoit et al., 2014).
Ethiopia
Meheretu et al. (2013) reported only one rat PCR-positive for Bartonella among 19 R. rattus trapped from three localities in Tigray, the northernmost region of Ethiopia. In Aroresa, where R. rattus comprised 46.5% in the rodent community, none of 53 tested rats was positive. The only sequence obtained from a R. rattus rat from Golgolnaele, Ethiopia was different from all previously described Bartonella species, but clustered together with genotypes found exclusively in native rodent species (Stenocephalemys albipes and Arvicanthis dembeensis) from Ethiopia (Meheretu et al., 2013).
Kenya
In a cross-sectional rat survey conducted in Kibera, an urban slum in Nairobi City, 24 of the 220 (11%) trapped rats were Bartonella culture-positive, including R. norvegicus (50%; 5/10) and R. rattus (60%; 19/32). In contrast, in the rural area Asembo on the northern shore of Lake Victoria, where R. norvegicus were absent, prevalence of Bartonella in R. rattus was lower (13%; 2/16) compared to Nairobi (Halliday et al., 2015). The high infection prevalence observed in Rattus trapped at the Kibera site is more similar to prevalence ranges observed in studies of Asian Rattus populations than to other African populations. The Kibera study in Nairobi, the Kenyan capital, clearly has more intensive connection with the seaport in Mombasa (in terms of rodent movement from other seaports) than the Asembo site located inland near Lake Victoria. Therefore, the high prevalence of Bartonella-infected rats reported from the Kibera site could relate to repeated introductions of Rattus species to this site (Halliday et al., 2015). Three zoonotic Bartonella species were identified in rats captured in Kibera slum area in Nairobi. They were B. elizabethae (n = 7), B. tribocorum (n = 8), and B. queenslandensis (n = 4) among 19 infected R. rattus and B. elizabethae (n = 1), B. tribocorum (n = 3), and B. queenslandensis (n = 1) among five infected R. norvegicus (Halliday et al., 2015).
Madagascar
A study conducted in Central Madagascar showed that 58.9% (93/158) of R. rattus rats sampled in two human communities were positive for Bartonella (Brook et al., 2017). They found four species: B. elizabethae (28; 16.9%), B. phoceensis (40; 23.8%), B. rattimassiliensis (21; 12.5%), and B. tribocorum (1; 0.6%) (Brook et al., 2017).
Nigeria
A study to detect Bartonella species in commensal rodents and their ectoparasites was conducted in town of Vom, central Nigeria. Among rats trapped there in domestic and peridomestic habitats, 36 of 121 R. norvegicus (29.8%), and nine of 48 R. rattus (18.8%) were culture-positive for Bartonella (Kamani et al., 2013). The prevalence of Bartonella DNA found in that study was similar to the prevalence reported in rats from the Democratic Republic of Congo (Laudisoit et al., 2014). Bartonella strains detected in rats from Vom, a town in central Nigeria, were identical or closely related to B. elizabethae, B. tribocorum, and B. grahamii (Kamani et al., 2013). Of 36 sequences obtained from R. norvegicus, 26 had 98–100% similarity with B. elizabethae sequence. Nine of the sequences obtained from R. norvegicus had 97–98% similarity with B. tribocorum sequence, while one sequence had 98% similarity with B. grahamii. The Bartonella sequences obtained from R. rattus were identical to B. elizabethae.
South Africa
A significant difference of Bartonella prevalence was observed between two rat species (24% in R. norvegicus vs. 5% in R. rattus) in South Africa (Brettschneider et al., 2012). The authors of this study proposed a mathematical model explaining that the difference between these two co-occurring rat species might be due to the observed differences in flea infestation rates between these species. Trataris et al. (2012) reported Bartonella infections in 13% by culturing and in 25% by PCR among rats (R. norvegicus and R. rattus) sourced from a pest control company in the Ekurhuleni Metropolitan area, the East Rand region of Gauteng, South Africa. Some isolates from the rats were similar to B. elizabethae, while some were relatively similar to B. thailandensis originally described in Asian rats (Saisongkorh et al., 2009).
Uganda
A very low prevalence of Bartonella (1.3%; 3/228) was reported in R. rattus from villages of two districts of northwest Uganda, whereas a prevalence near 60% was in populations of local indigenous rodents (Billeter et al., 2014). The relatively low prevalence of infection may be due to the fact that Rattus rats were introduced into this area of the West Nile region relatively recently. Genotypes related to B. elizabethae were detected in three R. rattus rats (Billeter et al., 2014).
Europe
France
In contrast to numerous investigations of Bartonella infections in multiple sylvatic rodents in many European countries, reports of investigation of urban rats are very limited from this part of the world. Seventy-four rats (8 R. rattus and 66 R. norvegicus) were trapped in the center and suburb of the city of Marseille, France, and 20 of the R. norvegicus, but none of R. rattus were culture positive (Gundi et al., 2004). Three species of Bartonella were described based on characterization of the strains obtained from Norway rats from France. First, Heller et al. (1998) identified B. tribocorum from blood of two R. norvegicus rats captured near the Rhine River. Later, Gundi et al. (2004) isolated B. rattimassiliensis and B. phoceensis from rats R. norvegicus captured in the city of Marseille. A more recent search for Bartonella and various other pathogens and parasites was conducted in the urban park (Chanteraines) within Hauts-de-Seine, France (Desvars-Larrive et al., 2017). The prevalence of Bartonella culture-positive rats of R. norvegicus was very high (58.2%; 32/55). Surprisingly, the prevalence of Bartonella-DNA estimated by PCR was lower (31.4%; 27/86). The authors of this study were careful with identification of the Bartonella species detected in the rats, but short sequences of the obtained pap31 amplicons were identical to B. henselae sequences (Desvars-Larrive et al., 2017).
Spain
The only other reported surveys of urban rats in Europe were from Spain: one from Andalusia and another one from the Canary Islands. Bartonella was isolated from two of ten R. norvegicus collected in a suburban area of Seville, Andalusia (Márquez et al., 2008). The ITS sequence analysis from R. norvegicus from Andalusia showed two genetically different variants (Márquez et al., 2008). The first genotype belonged to B. tribocorum, closely related to the strain KM2519 detected in R. tanezumi in China (GenBank EF202169). The second genotype was detected in 17 individuals and was relatively close to, but different from B. elizabethae (Márquez et al., 2008).
In the Canary Islands, Bartonella was found in R. rattus from two islands, Tenerife and La Palma (Abreu-Yanes et al., 2018). Overall, the prevalence of Bartonella in the rats of the Canary Islands was 14.3%, with variations between 13.8% inside houses and 26.7% in peridomestic habitats. The only two Norway rats from Tenerife were tested Bartonella-free. Fifteen Bartonella sequences identified in rats of R. rattus from the Canary Islands belonged to B. tribocorum; B. queenslandensis was in four rats, and B. rochalimae was recovered from two rats. Interestingly, two different gltA haplotypes similar to B. elizabethae were detected only in house mice (Mus musculus), but not in any rat (Abreu-Yanes et al., 2018).
Portugal
A limited number of rats from Portugal (two R. norvegicus from Aguas de Moura and two R. rattus from Porto Santo Island in Madeira Archipelago) were culture-positive for Bartonella (Ellis et al., 1999). The strains found in Norway rats from Portugal were relatively close to B. tribocorum while a strain from R. rattus from an island of Madeira Archipelago was evidently different (Ellis et al., 1999). Interestingly, this strain from the Portuguese island was shown later to be identical to the strains described in some R. rattus in Dhaka, Bangladesh and in all infected black rats in Tel Aviv, Israel (Bai et al., 2007; Harrus et al., 2009).
North America
Canada
In Vancouver, Bartonella species were isolated from the blood of 25.2% (102/404) of R. norvegicus tested (Himsworth et al., 2015). All 102 Bartonella cultures isolated from R. rattus and R. norvegicus from Vancouver had identical gltA sequences to type strain of B. tribocorum (Himsworth et al., 2015).
United States
Ellis et al. (1999) identified Bartonella species in 19.4% of R. norvegicus and 12% R. rattus collected from multiple U.S. cities. Bartonella infection in R. norvegicus varied significantly between investigated cities. In 3/9 localities, a statistically significant higher-than-expected prevalence of Bartonella infection in R. norvegicus was observed in Los Angeles California (56%), and New Orleans, Louisiana (56.4%). Seven of 66 (10.6%) R. norvegicus were culture-positive in Baltimore, Maryland. Rats of R. rattus were infected with Bartonella species in five of seven cities with a range of prevalence from 9 to 60% (Ellis et al., 1999). No positive rats of R. norvegicus were found in four cities: Atlanta, Georgia; Rockport, Indiana; Reno, Nevada; and New York City. Surprisingly, no Bartonella-positive rats were found in New York City among 87 rats cultured during the survey conducted by Ellis et al. (1999). In two other studies conducted in five sites within New York City, 33% (25/133) R. norvegicus were Bartonella-positive by PCR (Firth et al., 2014) and 23% (30/133) by culturing (Peterson et al., 2017). Ellis et al. (1999) described seven genotypes of Bartonella among 63 isolates obtained from R. norvegicus. The most frequently identified genotype (28/74) was 99% similar to B. tribocorum. Two of the R. rattus isolates from Louisiana were identical to one from an R. norvegicus captured in the same locality. Surprisingly, the 11 other isolates obtained from R. rattus captured in the United States were distinct from those from R. norvegicus. Bartonella strains detected in six of 11 R. rattus rats were identical to the genotype common for cotton rats (Sigmodon hispidus) from Georgia. One variant matched another cotton rat genotype from Georgia. Several other studies of urban rats in the U.S. and Canada have followed the study conducted by Ellis et al. (1999). Most genotypes identified in R. norvegicus from downtown Los Angeles, California were B. tribocorum: 101 of 106 (95.3%) culture-positive rats were infected with this particular species. In addition to B. tribocorum, three rats were infected with B. queenslandensis and two rats with B. rochalimae (Gundi et al., 2012). Testing blood of the same animals by PCR demonstrated presence of B. rochalimae in 37 (18.5%) rats and B. queenslandensis in four rats (2.0%) (Gundi et al., 2012). In Baltimore, Maryland, antibodies against B. elizabethae were detected in 34.1% of rats of R. norvegicus (Easterbrook et al., 2007).
Bartonella bacteria were cultured from blood of 43.5% (87/200) of R. norvegicus trapped in 16 sites in downtown Los Angeles, California, while Bartonella DNA was detected in 67.5% (135/200) of the same rats (Gundi et al., 2012). Another investigation of Bartonella that targeted populations of rats in California was conducted in the San Francisco Bay Area, where morphologically identified black rats represent two genetically distinct lineages that have been equated to R. rattus and R. tanezumi (Conroy et al., 2013). Of 50 of these black rats from six locations within Alameda county, California, eight Bartonella-positive rats were found in two locations within the City of Oakland nearly three miles apart. Interestingly, four rats from one location carried B. tribocorum, the bacterium dominant in rats in Los Angeles; whereas four Bartonella-positive rats from another location carried another bacterium, B. coopersplainsensis, originally described from Rattus leucopus in Australia (Gundi et al., 2009) and the most prevalent species in rats from New Zealand (Helan et al., 2018). In the U.S., this bacterium was found only in R. rattus from New Orleans (Peterson et al., 2017).
In New Orleans, Peterson et al. (2017) reported Bartonella infection by culture in 29 of 163 (17.8%) R. norvegicus and in five of 177 (2.8%) R. rattus. In Manhattan, New York City, 31 of 133 (23.3%) R. norvegicus rats tested were positive by PCR of spleen and heart tissues (Peterson et al., 2017). Peterson et al. (2017) reported significant differences in Bartonella diversity among rats between New Orleans and New York. In New Orleans, these authors detected B. coopersplanensis in five roof rats. They did not detect B. coopersplanensis in Norway rats in either New Orleans or New York City. However, they found four other Bartonella species from New Orleans, and those fell within the clades corresponding to B. rochalimae (13 positive by PCR only), B. elizabethae (10 positive by culture), B. tribocorum (19 by culture), and B. queenslandensis (five positive by culture). Among 29 Bartonella-positive Norway rats from New York City (Manhattan), 26 rats carried B. tribocorum, while three rats were infected with B. elizabethae (Peterson et al., 2017). Bartonella sequences recovered from these rats were located in the same clades as B. elizabethae, B. rochalimae, and B. tribocorum (Firth et al., 2014), while sequences obtained via tissue cultures were located in the same clade as B. elizabethae and B. tribocorum.
South America
Brazil
Bartonella were isolated from 5 of 26 (19%) R. norvegicus rats from two of five slum areas of Salvador, the third most populous city in Brazil (Costa et al., 2014). Conducting a wide survey of various rodent species in different parts of Brazil, Gonçalves et al. (2016) detected Bartonella species in two R. rattus from Mato Grosso (n = 3) and Goias (n = 4), while rats of the same species from Ceará (n = 12), and Pará (n = 8) were free of Bartonella. Of 14 strains of Bartonella isolated from R. norvegicus from Salvador, all but one were identical to B. queenslandensis, the remaining strain was close to B. tribocorum (Costa et al., 2014).
Peru
Analyzing Bartonella species in various animals inhabiting the villages in the Huaillacayan valley, Department of Ancash, Peru, Birtles et al. (1999) isolated two strains closely resembling B. elizabethae from rats collected in one village. This was the first reported identification of Bartonella species in a commensal rat from South America. Unfortunately, the authors were unable to identify the rat species. Investigating rodents in three villages in La Convencion Province of Peru for plague and Bartonella species, Martin-Alonso et al. (2014) tested 24 R. rattus from two residential areas. All 20 rats trapped in one area (Alto Ivochote) were free of Bartonella, whereas one of four rats captures in another area (Yoetoni) was PCR-positive for this bacterium. One sequence obtained by Martin-Alonso et al. (2014) from R. rattus from La Convencion Province, along with sequences obtained from two indigenous rodents species (H. perenensis and Oecomys sp.), had 98–99% sequence similarity to a genotypic variant obtained from Oryzomis palustris rats in the southeastern U. S. (Kosoy et al., 1997).
Oceania
Australia
Three novel Bartonella species were originally described from rats of the genus Rattus in Australia. Those are B. rattaustraliani found in rats of R. tunneyi, R. leucopus, and R. conatus; B. queenslandensis found in R. tunneyi, R. fuscipes, R. leucopus, and R. conatus; and B. coopersplainsensis found in R. leucopus (Gundi et al., 2009). The last two Bartonella species found in Australian rats were later discovered in rats from other continents. Two Bartonella species were detected in spleens of R. rattus from the Christmas Island, Australia (Dybing et al., 2016). Of 48 positive black rats found on this island, 28 rats carried B. phoceensis and eight rats carried a novel Bartonella genotype, potentially representing a new Bartonella species. Christmas Island is an Australian territory in the Indian Ocean lying in close proximity (360 km) to Jakarta, Indonesia. The authors considered the possibility that this Bartonella species had arrived in infected rats transported over the years on ships from nearby Indonesia (Dybing et al., 2016).
New Zealand
Using three molecular markers for identification of Bartonella DNA extracted from spleen samples of R. rattus collected in the Tongariro National Park, Helan et al. (2018) reported sequences matching B. coopersplainsensis and B. henselae from 15.4 (22/143) to 2.1% (3/143) of rats, respectively. Co-occurrence of B. coopersplainsensis and B. henselae sequences was observed in one rat. Even ff detection of prevalent B. coopersplainsensis was not surprising because this bacterium was described earlier in Australian rats (Gundi et al., 2009), the discovery of B. henselae in rats was very unexpected as this bacterium is typically found in domestic and wild cats and has never been reported previously in rats.
Distribution of Bartonella in Rat Populations Within Urban Territories
Ecological factors, including size and structure of rat populations and animal movement patterns, may determine prevalence and species diversity of Bartonella in rats within urban territories (Firth et al., 2014; Himsworth et al., 2015; Peterson et al., 2017). During the survey conducted by Halliday et al. (2015) in Kibera, an urban slum in Nairobi, Kenya, the proportion of infected Rattus overall varied from 0 to 60% by trapping zone (Figure 2). In three zones (A, B, and D), Mus musculus was the dominant species and only four Bartonella isolates were identified in rats (Figure 2). Concurrently, in two other zones, Rattus rats dominated in rodent populations presenting 51% in zone C and 40% in zone E. In these two zones (C and E), several species of Bartonella were reported (Figure 2; Halliday et al., 2015).
According to the investigation conducted in New Orleans, Louisiana by Peterson et al. (2017), prevalence of Bartonella infections ranged from 0 to 97% of rats. Most Bartonella-positive rats (85%) were found within a single housing block of New Orleans. All Bartonella-positive R. norvegicus rats were captured at two locations, where no roof rats were present. Bartonella-infected R. rattus were captured from five locations. Though two species of rats (R. norvegicus and R. rattus) were found in four of these five locations, no R. norvegicus rats were Bartonella-positive there. A single Bartonella-positive R. rattus was captured at a location where R. norvegicus were absent. Of five trapping locations in New York City, Bartonella-positive R. norvegicus rats were reported in each of them; however, prevalence of infected rats at these locations ranged from 10 to 85% (Peterson et al., 2017).
Himsworth et al. (2015) demonstrated significant geographic clustering of Bartonella-positive rats within Vancouver, Canada. The prevalence of B. tribocorum varied significantly by city block, from 0 to 60.5%. Analyzing various ecological factors affecting prevalence of B. tribocorum in R. norvegicus rats from Vancouver, Rothenburger et al. (2018) noticed that the infection was significantly lower within city blocks with one or more low-rise apartment buildings compared to blocks with none. There was no significant association of the infection prevalence with rat abundance, suggesting a lack of density-dependent pathogen transmission. According to this analysis, the infection rate positively correlated with high minimum temperatures and the authors suggested that a baseline minimum temperature could be important for survival of fleas that serve are vectors for transmission of B. tribocorum among rats (Rothenburger et al., 2018).
Abreu-Yanes et al. (2018) defined environmental parameters related to the presence of Bartonella DNA in rats in the Canary Islands. Specifically, their data suggest that occurrence of Bartonella on islands of this archipelago is influenced by biological and climatic conditions that vary among the islands. The probability of Bartonella infection in rodents in La Palma Island was four times higher compared to Tenerife Island and no Bartonella-positive rats were found in Lanzarote Island. A study carried out by Vicente and Gómez López (2012) showed that the flea Stenoponia tripectinata seem to have some preference for the conditions found on the four western islands, which include both Tenerife and La Palma, whereas this flea species was found in none of the 157 rodents from Lanzarote that belongs to the eastern group of islands. The results suggest that the ecological conditions on the Lanzarote Island are not suitable for the development of fleas S. tripectinata, while the ecological conditions on the La Palma Island (the most northwestern island of the archipelago) are more favorable for completing the life cycle of these fleas, which likely provide transmission of Bartonella infection between rats (Vicente and Gómez López, 2012).
Although Bartonella infections are prevalent in urban rat populations around the world and in many cities and prevalence of the infection can reach very high rates, we noted that rats in some cities and villages were not infected with Bartonella or the infection prevalence was quite low. At least, such situations were reported outside of Asia where we have not found reports of Bartonella-free populations of rats when a sufficient number of animals was tested. One of the first thoughts to explain the absence of Bartonella in some rat populations leads to representation of a kind of “island syndrome” wherein some parasites were absent or rare when a new rat population is established by invasion of a small number of individuals. In the regions where rats of the genus Rattus rarely occupy certain urban habitats, such as in areas of Europe and North America, populations of city rats are well-separated, in fact resembling “island populations.” This can explain the absence of Bartonella infections in rats from many big cities of the continental United States.
Noticeably, rats from the coastal cities on both Atlantic and Pacific sides (Los Angeles, Baltimore, New Orleans, New York, and Vancouver) carried Bartonella, while populations of rats in non-coastal cities were Bartonella-free (Ellis et al., 1999). In spite of a large number of rats investigated in Yokohama, Japan (255 R. rattus and 84 R. norvegicus), all tested animals were Bartonella-negative (Inoue et al., 2008). The rats of both R. rattus and R. norvegicus were Bartonella-free from some zones of the city of Cotonou, Benin (Martin-Alonso et al., 2016.
We admit that these observations are limited for supporting this statement. We can only speculate that continuous arrival of new rats through seaports is a crucially important factor for circulation of Bartonella among rats. Assuming an “island effect” on the formation of Bartonella communities in urban rat populations, we have to consider factors contributing to isolation of rat populations, such as time of establishing rat populations, distance and connectivity between continental rat populations and seaports where arrival of new rats is more likely to occur, rat population size, etc. (Papkou et al., 2016).
The “island effect” is not the only plausible explanation for the absence of Bartonella in rat populations within some urban areas and in some situations might not be the most important. When Bartonella bacteria are introduced into a new territory with invasive rats, the local conditions might not be favorable for the long-term circulation of the parasites within the newly established host population. Continuing an analogy borrowed from population genetics, a situation leads to the so-called “bottleneck effect” when bacteria are likely to be subjected to extreme changes in a host population. A critical issue is an availability of factors required for continuous transmission of Bartonella bacteria between rat hosts. A presence of appropriate flea species, particularly, the Oriental rat flea (X. cheopis) that can act as vectors, and the level of flea infestation, would be especially important factors for survival of Bartonella in rats. Unfortunately, data on the distribution of fleas in urban rat populations in the U.S. are quite sparse.
The absence of Bartonella infections in Rattus rats in some Ugandan villages and overall low prevalence in invasive rats in northwestern Uganda (Billeter et al., 2014) present another interesting situation that contrasts with numerous studies demonstrating a high prevalence of bartonellae among Rattus rats in Asia and in many places outside of Asia, e.g., Nairobi City. The relatively low prevalence of the infection in rats in this part of Africa may relate to relatively recent introduction of Rattus rats into the West Nile region. This fact is in contrast to the well-established rat populations in the cities on the coastal parts of Kenya and Tanzania, which probably have existed for millennia because of the historical dispersal of humans and their cargo via ships (Aplin et al., 2011). Despite intensive investigations, only a single rat was reported on a boat moored at Rhino Camp on the west side of the Albert Nile in the late 1930s (Hopkins, 1949; Amatre et al., 2009). Despite “fairly intensive” trapping efforts at 11 sites on land in the West Nile and West Madi regions along the west side of the Albert Nile, no R. rattus were captured during a survey undertaken in 1937 and 1938 by Hopkins (1949). Indeed, R. rattus might not have become established in the West Nile until the 1950s or even the 1960s as this rat was first identified in 1958 in the Ituri District of the DR Congo, which lies across the border from the West Nile Region (Borchert et al., 2007). This is perhaps not surprising as Hopkins (1949) suggested that R. rattus was first introduced to Uganda in the early Twentieth Century, a date that agrees with Delany's belief that this rat species first appeared in the country around 1910 (Delany, 1975). In this part of Africa, outsiders were restricted and movement of crops was limited until 1914 when the region became a British protectorate (Borchert et al., 2007). This likely could restrict the relocation of R. rattus and thereby limit the introduction of Bartonella infected rats. The extension of the “Kenya and Uganda Railways and Harbors” to the western Uganda in 1956 or the construction of the bridge across the Albert Nile at Pakwach in 1969 could have lead to gradual spread of Rattus rats to Arua near the border with DR Congo.
The Richness of Bartonella Bacterial Communities in Aboriginal Rats in Natural Habitats Compared to Urban Communities
Provided information suggests that rat-adapted Bartonella species originated from Asia. The first data supporting this claim came after collection of bacteria related to B. elizabethae in Rattus rats from three districts of Yunnan province of southwestern China (Ying et al., 2002). Following this report, multiple investigations of rats in Southeast Asia (Cambodia, Laos, Thailand, and Vietnam) have also confirmed ubiquitous distribution of this infection in native rat species of the genus Rattus (Bai et al., 2007, 2009; Angelakis et al., 2009; Jiyipong et al., 2012). In contrast to urban rats in many countries of the world where two rat species are observed, in Asia, various species of Rattus occupy natural habitats from tropical lowland to mountains. For instance, the ricefield rat (R. argentiventer) and the Malayan field rat (R. tiomanicus) are common in the rice fields and plantations and the Losea rat (R. losea) is more typical in gardens (Kosoy et al., 2015). At the same time, only few species of rats have evidently adapted to live closely to people; specifically, R. tanezumi rats are common in cities, small towns, and villages.
Overall, the prevalence of Bartonella infections was commonly high in rats in Asia, but not higher than in urban rats in Africa or Americas. For example, Jiyipong et al. (2012) reported Bartonella species in 9.6% of rats in Cambodia, 11.9% in Laos, and 11.0% in Thailand, all of which are lower than the rates reported in Los Angeles, U.S. (56%) or in Nairobi, Kenya (57%). While rats of various species of Rattus carried Bartonella in natural habitats in Asia, prevalence varied between species. For example, prevalence of infected individuals has significantly varied from 3.2% in R. exulans to 86.4% in R. norvegicus in Thailand (Bai et al., 2009) or from 10% of the R. rattus to 66.7% of the R. losea in Taiwan (Hsieh et al., 2010).
The most striking difference in Bartonella infection between aboriginal and invasive rats was in observed diversity of the bacteria. Bartonella bacteria found in urban rats in Africa, Europe, and North and South Americas belonged to one or a few species or genotypes; for example, all 102 infected rats from Vancouver, Canada carried bacteria identical to each other by the gltA, which is a quite sensitive genetic marker (Himsworth et al., 2015). The diversity of Bartonella strains found in rats inhabiting natural environments in Asia was very high. Investigating rats from 17 provinces in Thailand, Bai et al. (2009) identified 23 genetic variants, which clustered with not only B. coopersplainensis, B. elizabethae, B. phoceensis, B. rattimassiliensis, and B. tribocorum, but also with numerous Bartonella genotypes in intermediate positions between described species or were quite different from all known species. A novel genotype of Bartonella with the proposed name as “Candidatus Bartonella thailandensis” was identified in rats of Rattus surifer from Surin, a Thai province neighboring Cambodia (Saisongkorh et al., 2009). Klangthong et al. (2015) classified DNA sequences obtained from rats in Thailand into eight different cladograms. Bartonella sequences obtained from rats of several Rattus species from Southeast Asia represented over 40 different genetic variants and clustered into nine lineages (Jiyipong et al., 2012). All described rat-adapted Bartonella species were identified in rats (R. argentiventer, R. tanezumi, R. norvegicus, and Bandicota indica) from the Mekong Delta in Vietnam (Loan et al., 2015). The prevalence of Bartonella infection among rats trapped in farms, filed, and forest was 22.4%, much higher than the infection prevalence in rats that were purchased in city markets (8.7%). The highest prevalence was found in R. tanezumi (49.2%), followed by R. norvegicus (20.7%). No Bartonella was found in R. exulans. The species isolated from these rats were B. rattimassiliensis, B. tribocorum, B. elisabethae, B. coopersplainensis, and B. queenslandensis (corresponding to 43.8, 21.9, 18.8, 9.4, and 6.3% of 32 Bartonella-infected rats. Two species (B. rattimassiliensis and B. coopersplainensis) were identified in R. tanezumi only, while all other species of Bartonella were detected at least in two rat species (Loan et al., 2015). The prevalence of Bartonella species in rats from rural parts of Vietnam (Mekong Delta) was significantly lower than in Saigon Port, but the diversity of the species was evidently higher.
Phylogenetic Reconstruction of Global Dissemination of Bartonella by Rats
A comparison of Bartonella bacterial communities between aboriginal and invasive rats of the genus Rattus allows an investigation of the roles played by rats as carriers of these bacteria comparing the diversity of Bartonella genotypes in rats between Southeast Asia where presumably original diversification happened and other parts of the world where only two rat species were relatively recently introduced. Hayman et al. (2013) analyzed variations of one genetic locus (the gltA as the most widely used molecular marker for differentiation of Bartonella species) of 191 strains of rat-associated bartonellae from 17 countries. The phylogeographic analysis supported the hypotheses that Bartonella species likely originated in Southeast Asia. The analysis has also highlighted the role of R. rattus in disseminating Bartonella bacteria to other continents. Black rats have invaded most countries of the world with main introductions that happened through several commensalism events (Aplin et al., 2011). Furthermore, the phylogenetic analysis conducted by Hayman et al. (2013) demonstrated that diversification of species belonging to the B. elizabethae species complex occurred in Southeast Asia before some of the species belonging to this complex were transmitted to other geographic regions. Importantly, their analysis suggests that there were multiple disseminations of these bacteria within Asia and numerous introductions from Asia to other parts of the world. This conclusion is based on identification of several major clades of Bartonella strains of Southeast Asian origin that dispersed globally.
Specifically, Bartonella strains obtained from rats of the genus Rattus from different continents and countries were grouped into six major clades that presumably originated in Southeast Asia. Of those, one clade (A) is distributed globally with strains found in most regions, but not in Central Africa. Likely, R. norvegicus play the leading role in distributing Bartonella species belonging to this clade. The analysis also suggests that the process of bacterial dispersal in this species clade is still continuous. Strains belonging to another clade (B) were detected only in Asia and Western Europe. The strains belonging to the third clade (C) dispersed to countries of Africa, North and South America. Strains grouped into the D are found in Africa and North America, besides Asia. Clade E has limited geographic spread, with only a Eurasian distribution. Finally, the strains combined into the clade F are distributed across Pacific and were detected in East Asia, Australia, and western part of North America (Hayman et al., 2013).
Bartonella DNA in Rat Ectoparasites
Multiple field observations and limited experimental studies support the major role of rat fleas in transmission of bartonellae among rats. Gutiérrez et al. (2015) highlighted the importance of the level of the flea's host specificity, flea exchange between rodents, and flea abundance for success of transmission of Bartonella bacteria. The host specificity of some flea species, e.g., X. cheopis, may influence the acquisition or the restriction of specific Bartonella species and strains to rats. There are many reports of the presence of Bartonella DNA in ectoparasites collected from Rattus rats. Bartonella DNA was detected in 59.1% of 193 Xenopsylla cheopis fleas collected from 62 Rattus rats (R. exulans, R. norvegicus, and R. rattus) captured in Khon Kaen, the northeastern province of Thailand. Sequence analysis of DNA present in rat fleas from this area demonstrated the presence of Bartonella species similar to B. elizabethae, B. rattimassiliensis, B. rochalimae, and B. tribocorum (Billeter et al., 2013). Another study in Thailand targeting rats and their ectoparasites in villages from all regions of the country indicated that the prevalence of Bartonella DNA varied substantially depending on ectoparasite species (Klangthong et al., 2015). Of the multiple arthropods screened during this study, the highest prevalence of Bartonella DNA was in louse (Polyplax and Hoplopleura, 57.1%) and flea (X. cheopis, 25.8%) pools. Only few positive samples were found in pools of mites (Leptotrombidium and Ascoschoengastia, prevalence 1.7%) and ticks (Haemaphysalis species, prevalence 3.5%). Most identified sequences found in arthropods have been found in rats and belong to the B. elizabethae species complex. Eight sequences of B. tribocorum were detected from six flea pools. One genotype identified as B. queenslandensis (99.6% identity) was found in a flea pool. One flea pool was positive for Bartonella with genotype being not reported in rats, but still closely related to B. tribocorum (96.8 % identity). Some identified Bartonella sequences from tick and louse pools shared close similarity with B. coopersplainsensis. Interestingly, B. phoceensis DNA was detected solely from ectoparasites (louse, mite, and tick pools) (Klangthong et al., 2015). Fleas obtained from rats in Taiwan harbored DNA of several Bartonella species (Tsai et al., 2010). Bartonella DNA detected in eight lice (Polyplax) obtained from five Bartonella-positive R. norvegicus from Taiwan was identified as B. phoceensis. The authors noticed that fleas collected from adult rats (77.1%) more likely Bartonella-positive than fleas collected from juvenile rats (42.3%).
Laudisoit et al. (2014) reported a high prevalence of Bartonella DNA in rat fleas from Kisangani, D. R. Congo. Bartonella genotypes detected pools of fleas X. cheopis, ticks R. sanguineus, and mites Haemolaelaps from Nigeria were identical or similar B. elizabethae (97–100% similarity), but a genotype found in a flea Ctenophthalmus pool was B. tribocorum (97% similarity) (Kamani et al., 2013). Nine of 12 genetic variants detected in rat fleas in Uganda belonged to the B. elizabethae species complex (Bai et al., 2017). In Madagascar, B. elizabethae was found in fleas of Synopsyllus fonquerniei and X. cheopis, while B. phoceensis and B. rattimassiliensis were found in sucking lice of the genus Polyplax (Brook et al., 2017).
In New York City, Bartonella DNA recovered from Oriental rat fleas collected from Norway rats belonged to three Bartonella species. The most common sequences clustered with B. tribocorum, while sequences related to B. elizabethae and B. rochalimae were less common in fleas (Frye et al., 2015). There are more reports of Bartonella DNA detected in rat blood. The main message that we can deliver from these studies is that the range of Bartonella genotypes found in ectoparasites, especially in fleas matches very much the spectrum of bacterial species found in rats. This contrasts with some observations made during investigations of sylvatic rodent communities when a considerable discordance between genotypes of Bartonella obtained from several ‘rodent host-ectoparasite’ pairs was reported (Abbot et al., 2007). A recent experimental study conducted by McKee et al. (2018) supported vector capacity of Oriental rat fleas (X. cheopis) for transmission of a rat-adapted Bartonella species. Specifically, this experiment demonstrated acquisition of B. elizabethae by experimentally exposed rat fleas and excretion of Bartonella DNA in flea feces over several days (McKee et al., 2018).
Rat-Associated Bartonellosis in Humans
Pathogens carried by Norway and black rats can lead to significant morbidity and mortality in people around the world (Himsworth et al., 2013). The first species of rat-associated Bartonella proven to be a human pathogen was B. elizabethae (at that time described as Rochalimaea elizabethae) (Daly et al., 1993). This bacterium was isolated from blood of a 31-year old male patient with endocarditis admitted to the Saint Elizabeth Hospital in Massachusetts, U.S. The patient had no history of exposure to cats or other pets or intravenous drug use. A source of the infection remained mysterious until Ellis et al. (1999) found relatively similar bacteria in rats from the U.S. Ying et al. (2002) reported a variety of similar bacteria in rats in Southern China and, finally, an identical bacterium was found to be prevalent in rats from Vietnam (Loan et al., 2015). Later, an identical bacterium was identified in a febrile patient in Bangladesh (Faruque et al., 2017).
The strains identical or closely similar to rat-adapted Bartonella species, including B. elizabethae, B. tribocorum, and B. rattimassiliensis were identified in blood clots from eight febrile patients from two Thai provinces, Chiang Rai and Khon Kaen (Kosoy et al., 2010). These genotypes represented more than one-half of the Bartonella genotypes identified in human patients with fever of unknown etiology enrolled into this study. Importantly, some genotypes identified in rats from Los Angeles showed 98.8% similarity to the isolate obtained from a Thai patient (GenBank accession number GQ225706) (Gundi et al., 2012). Moreover, the strain of B. tribocorum identified in a Thai patient was identical to a Bartonella sequence detected in X. cheopis fleas collected from R. norvegicus rats in Los Angeles, California (Billeter et al., 2011). More recently, a strain of B. tribocorum was cultured with a bacteremia level of 60 colonies per 1ml from the blood of a 64-year old male patient with complaints of fatigue, muscle pain, and headache in France (Vayssier-Taussat et al., 2016).
Another species of Bartonella (B. rochalimae), though not specific for rats, was found in Rattus rats and their fleas from Asia and America (Billeter et al., 2011; Gundi et al., 2012). This bacterium was originally described as a human pathogen when it apparently caused fever and splenomegaly in a U.S. patient who became ill after traveling to Peru (Eremeeva et al., 2007). This bacterium was found in dogs and many wild mammalian species (foxes, rats, shrews, gerbils, and raccoons), suggesting that multiple reservoirs may be involved in its maintenance (Bai et al., 2016).
From our standpoint, the most intriguing and convincing case was reported from Tbilisi, the capital of the country of Georgia, where an 18-year old woman was admitted to hospital with a 2-week history of malaise, fever, and severe lymphadenopathy (Kandelaki et al., 2016). The patient lived in a residential area within Tbilisi and had not recently traveled outside the city. Based on lymphadenopathy and some other clinical manifestations, clinicians suspected cat scratch disease (CSD) although the patient denied any contact with cats. The clinical specimens were sent to the laboratory and results proved that indeed the bacterium found in samples from the patient belonged to the genus Bartonella. However, analyses that are more precise demonstrated that the strain was not B. henselae, the agent of CSD, but belonged to the B. elizabethae species complex. Thorough phylogenetic analysis involving seven molecular targets demonstrated that the bacterium had a divergence of 3.4% from B. elizabethae and 5.6% from B. tribocorum. Most importantly, this strain was identical to the Tel Aviv strain of Bartonella, which is prevalent and the only strain identified among R. rattus rats captured in Tel Aviv, Israel (Harrus et al., 2009).
The results of several serological surveys supported a potential exposure of people to rat-adapted Bartonella species. A survey of 630 drug users conducted in Baltimore, Maryland, reported seroprevalence of antibodies to rat-specific B. elizabethae (33%), 3-fold higher than prevalence to the cat-specific B. henselae (11%) or louse-transmitted B. quintana (10%) (Comer et al., 1996). A similar survey conducted in Central and East Harlem in New York City showed an even higher prevalence of seroreactivity to B. elizabethae (46%) compared to antibody positivity observed to B. henselae (10%) and B. quintana (2%) (Comer et al., 2001). A study of homeless people in Stockholm, Sweden reported high seroprevalence (52%) to B. elizabethae (Ehrenborg et al., 2008). McVea et al. (2018) reported exposure to rat-associated Bartonella species among intravenous drug users in an impoverished neighborhood of Vancouver, Canada. A retrospective serological survey of archived specimens indicated that Bartonella antibodies are prevalent among febrile patients in the Kathmandu Valley of Nepal (Myint et al., 2011). When 11 cases with high titers were compared to eight different Bartonella antigens, the highest titers (ranged from 1:256 to 1:2,048) reported in three patients were against the antigen of B. elizabethae.
The Movement of Bartonella by Rats From Southeast Asia to Urban Centers in Other Parts of the World
A high diversity of Bartonella species and strains on the one side and association of specific Bartonella species with mammalian hosts on the other, in our case with rats of the genus Rattus, provide an opportunity for reconstructing the movement of these bacteria from the jungles of Southeast Asia to cities on all continents except for Antarctica. The studies of Bartonella strains associated with the rats of genera Rattus and Bandicota demonstrated that these bacteria cluster into a separate phylogenetic lineage (Heller et al., 1998; Ellis et al., 1999; Ying et al., 2002; Castle et al., 2004; Gundi et al., 2004, 2009). These Bartonella species likely originated in Southeast Asia and subsequently dispersed from Asia with Rattus rats because of human activity. Later these bacteria became common and widespread in urban and peridomestic environments around the world (Childs et al., 1999; Ellis et al., 1999).
We have to distinguish roles played by rats as hosts of Bartonella within the lands where they had originally diversified from rats that have been translocated in recent human history. Data supporting the hypothesis of the Old World origin of Rattus rat-associated Bartonella species include the widespread occurrence of genetically related isolates of Bartonella species in R. norvegicus from Portugal, the United States, and South America (Buffet et al., 2013). On the other hand, there is an evident difference between the Bartonella isolates obtained from rats and from indigenous rodents of America (Ellis et al., 1999). The first evidence Bartonella genotypes from Southeastern Asia being related to B. elizabethae came from identification of the high diversity of Bartonella in R. tanezumi rats in several cities of southern China and in lesser bandicoot rats (Bandicota bengalensis) and black rats (R. rattus) in Dhaka, Bangladesh (Ying et al., 2002; Bai et al., 2007). A few of the multitude of genotypes found in these rats were identical to sequences of cultures from Rattus rats found in France, Portugal, and the United States.
We propose to consider a “source-sink” ecological model developed in the field of population ecology of animals and plants (Pulliam, 1988) for comprehending the differences described earlier in this article. According to the original scheme, rat populations distributed across source habitats within the native range in Asia (“source”) are self-sustaining; while the rat populations introduced to other continents (“sink”) can be maintained continuously only by immigration of rats from natural habitats. Assuming the role of aboriginal rat populations as “sources” and the role of commensal rats in the continents where rats were introduced relatively recently as “sinks,” we can propose one more component for inclusion into this conceptual model. The assemblage of Bartonella strains in rats inhabiting big cities in Asia is commonly less diverse compared to populations of rats in native habitats and rural areas within the range of natural origin, but more diverse than in cities of other continents. These communities represent intermediate positions in the “source-sink” model. For example, the number of rat species in Bangkok, Ho Chi Minh City, Yangon, and others is restricted to only two commensal rat species common to urban areas in Africa, Americas, and Europe; but have a reduced diversity compared to communities observed in the forests or fields in Southeast Asia outside cities. The reduced number of rats in big Asian cities can explain the intermediate rate of Bartonella circulated within these rat populations. For example, the diversity of Bartonella species observed in rats from Dhaka, Bangladesh (Bai et al., 2007) was higher than in rats from cities in Americas and Europe (Gundi et al., 2012), but lower than in natural and agricultural settings in Thailand (Bai et al., 2009). Although all described Rattus rat-associated species of Bartonella have a worldwide distribution, the diversity of Bartonella genotypes in rats from natural habitats of southeastern Asia is much higher compared to a number of strains reported in all cities of the world outside Asia.
In a number of studies, Bartonella infection prevalence was higher in R. norvegicus compared to R. rattus (Ellis et al., 1999; Hsieh et al., 2010; Martin-Alonso et al., 2016). In some situations, this difference can be explained by the load of ectoparasites carried by these rats, but likely this is not the sole explanation. Brettschneider et al. (2012) noticed a similar effect and argued that more detailed biological research on Bartonella infections is needed to explain such observations. Based on comparative phylogeography of invasive rats in the United States, Lack et al. (2013) came to conclusion that rats of R. norvegicus may contribute to a greater diversity of pathogens from various international sources and spread them across the U.S. compared to R. rattus. Their premise is based the data suggesting that gene flow among populations was higher for the Norway rats compared to R. rattus (Lack et al., 2013). In addition, their analyses support their hypothesis that R. norvegicus rats invade both Atlantic and Pacific coasts of the U.S. and likely from different points of their origin (Lack et al., 2013). A comparison of Bartonella observed in both R. norvegicus and R. rattus in the US cities (Ellis et al., 1999) and phylogenetic analysis of Bartonella isolates conducted by Hayman et al. (2013) support this supposition.
Conclusions
Bartonella species, being a highly prevalent and extremely diverse group of bacteria, are excellent sentinel organisms for evaluating the transoceanic and intra-continental movement of the pathogens by rats of the genus Rattus. The analyses described in this article confirmed the role of human-mediated distribution of invasive rat species in dissemination of rat-adapted parasites. Intensive collections and characterization of the Bartonella strains recovered from Rattus rats allowed the demonstration of the global dissemination of such strains from Asia to Africa, Australia, Europe, and finally to the Americas. Phylogenetic analyses of rat-adapted strains represent an interesting model for investigating pathogen-host coevolution. The interesting question remains how introduction of specialized parasites introduced via invasive rodent hosts can alter parasite community dynamics. Finally, the accumulation of reports of human cases associated with rat-borne Bartonella species has increased concern about public health consequences of the global distribution of these bacteria and their introduction to urban centers.
Author Contributions
MK and YB contributed conception and design of the study, organized the database, wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kenneth Gage, Irina Goodrich, and three reviewers for their careful reading of the manuscript and constructive comments.
References
Abbot, P., Aviles, A. E., Eller, L., and Durden, L. A. (2007). Mixed infections, cryptic diversity, and vector-borne pathogens: evidence from Polygenis fleas and Bartonella species. Appl. Environ. Microbiol. 73, 6045–6052. doi: 10.1128/AEM.00228-07
Abreu-Yanes, E., Martin-Alonso, A., Martin-Carrillo, N., Livia, K. G., Marrero-Gagliardi, A., Valladares, B., et al. (2018). Bartonella in Rodents and ectoparasites in the Canary Islands, Spain: new insights into host-vector-pathogen relationships. Microb. Ecol. 75, 264–273. doi: 10.1007/s00248-017-1022-y
Amatre, G., Babi, N., Enscore, R. E., Ogen-Odoi, A., Atiku, L. A., Akol, A., et al. (2009). Flea diversity and infestation prevalence on rodents in a plague-endemic region of Uganda. Am. J. Trop. Med. Hyg. 81, 718–724. doi: 10.4269/ajtmh.2009.09-0104
Angelakis, E., Khamphoukeo, K., Grice, D., Newton, P. N., Roux, V., Aplin, K., et al. (2009). Molecular detection of Bartonella species in rodents from the Lao PDR. Clin. Microbiol. Infect. 2, 95–97. doi: 10.1111/j.1469-0691.2008.02177.x
Aplin, K. P., Suzuki, H., Chinen, A. A., Chesser, R. T., Ten Have, J., Donnellan, S. C., et al. (2011). Multiple geographic origins of commensalism and complex dispersal history of black rats. PLoS ONE 6:e26357. doi: 10.1371/journal.pone.0026357
Bai, Y., Gilbert, A., Fox, K., Osikowicz, L., and Kosoy, M. (2016). Bartonella rochalimae and B. vinsonii subsp. berkhoffii in wild carnivores from Colorado, USA. J Wildl Dis. 52, 844–849. doi: 10.7589/2016-01-015
Bai, Y., Kosoy, M. Y., Lerdthusnee, K., Peruski, L. F., and Richardson, J. H. (2009). Prevalence and genetic heterogeneity of Bartonella strains cultured from rodents from 17 provinces in Thailand. Am. J. Trop. Med. Hyg. 81, 811–816. doi: 10.4269/ajtmh.2009.09-0294
Bai, Y., Montgomery, S. P., Sheff, K. W., Chowdhury, M. A., Breiman, R. F., Kabeya, H., et al. (2007). Bartonella strains in small mammals from Dhaka, Bangladesh, related to Bartonella in America and Europe. Am. J. Trop. Med. Hyg. 77, 567–570. doi: 10.4269/ajtmh.2007.77.567
Bai, Y., Osikowicz, L. M., Kosoy, M. Y., Eisen, R. J., Atiku, L. A., and Mpanga, J. T. (2017). Comparison of zoonotic bacterial agents in fleas collected from small mammals or host-seeking fleas from a Ugandan region where plague is endemic. mSphere. 2:e00402–17. doi: 10.1128/mSphere.00402-17
Battersby, S., Hirschhorn, R. B., and Amman, B. R. (2008). “Commensal rodents,” in Public Health Significance of Urban Pests, eds. X. Bonnefoy, H. Kampen, K. Sweeney, and World Health Organization (Copenhagen: World Health Organization), 387–419.
Billeter, S. A., Borchert, J. N., Atiku, L. A., Mpanga, J. T., Gage, K. L., and Kosoy, M. Y. (2014). Bartonella species in invasive rats and indigenous rodents from Uganda. Vector Borne Zoonotic Dis. 14, 182–188. doi: 10.1089/vbz.2013.1375
Billeter, S. A., Colton, L., Sangmaneedet, S., Suksawat, F., Evans, B. P., and Kosoy, M. Y. (2013). Molecular detection and identification of Bartonella species in rat fleas from northeastern Thailand. Am. J. Trop. Med. Hyg. 89, 462–465. doi: 10.4269/ajtmh.12-0483
Billeter, S. A., Gundi, V. A., Rood, M. P., and Kosoy, M. Y. (2011). Molecular detection and identification of Bartonella species in Xenopsylla cheopis fleas (Siphonaptera: Pulicidae) collected from Rattus norvegicus rats in Los Angeles, California. Appl. Environ. Microbiol. 77, 7850–7852. doi: 10.1128/AEM.06012-11
Birtles, R. J. (2005). Bartonellae as elegant hemotropic parasites. Ann. N. Y. Acad. Sci. 1063, 270–279. doi: 10.1196/annals.1355.044
Birtles, R. J., Canales, J., Ventosilla, P., Alvarez, E., Guerra, H., Llanos-Cuentas, A., et al. (1999). Survey of Bartonella species infecting intradomicillary animals in the Huayllacallán Valley, Ancash, Peru, a region endemic for human bartonellosis. Am. J. Trop. Med. Hyg. 60, 799–805. doi: 10.4269/ajtmh.1999.60.799
Bitam, I., Rolain, J. M., Nicolas, V., Tsai, Y. L., Parola, P., Gundi, V. A., et al. (2012). A multi-gene analysis of diversity of Bartonella detected in fleas from Algeria. Comp. Immunol. Microbiol. Infect. Dis. 35, 71–76. doi: 10.1016/j.cimid.2011.11.002
Borchert, J. N., Mach, J. J., and Linder, T. J. (2007). “Invasive Rats and Bubonic Plague in Northwest Uganda,” in Managing Vertebrate Invasive Species: Proceedings of an International Symposium, eds G. W. Witmer, W. C. Pitt, and K. A. Fagerstone (Fort Collins: USDA/APHIS Wildlife Services, National Wildlife Research Center), 283–293.
Bradley, C. A., and Altizer, S. (2007). Urbanization and the ecology of wildlife diseases. Trends Ecol. Evol. 22, 95–102. doi: 10.1016/j.tree.2006.11.001
Brettschneider, H., Anguelov, R., Chimimba, C. T., and Bastos, A. D. (2012). A mathematical epidemiological model of gram-negative Bartonella bacteria: does differential ectoparasite load fully explain the differences in infection prevalence of Rattus rattus and Rattus norvegicus? J. Biol. Dyn. 6, 763–781. doi: 10.1080/17513758.2012.705906
Brook, C. E., Bai, Y., Yu, E. O., Ranaivoson, H. C., Shin, H., Dobson, A. P., et al. (2017). Elucidating transmission dynamics and host-parasite-vector relationships for rodent-borne Bartonella spp. in Madagascar. Epidemics 20, 56–66. doi: 10.1016/j.epidem.2017.03.004
Buffet, J. P., Kosoy, M., and Vayssier-Taussat, M. (2013). Natural history of Bartonella-infecting rodents in light of new knowledge on genomics, diversity and evolution. Future Microbiol. 8, 1117–1128. doi: 10.2217/fmb.13.77
Castle, K. T., Kosoy, M., Lerdthusnee, K., Phelan, L., Bai, Y., Gage, K. L., et al. (2004). Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am. J. Trop. Med. Hyg. 70, 429–433.
Çelebi, C., Karagöz, A., Öktem, M. A., Carhan, A., Ozkazanc, N. K., Babur, C., et al. (2015). Bartonella species in wild small mammals in Western Black Sea Region of Turkey. Vet. Fakültesi dergisi 62, 183–187. doi: 10.1501/Vetfak_0000002678
Cevidanes, A., Altet, L., Chirife, A. D., Proboste, T., and Millán, J. (2017). Drivers of Bartonella infection in micromammals and their fleas in a Mediterranean peri-urban area. Vet. Microbiol. 203, 181–188. doi: 10.1016/j.vetmic.2017.03.023
Childs, J. E., Ellis, B. A., Nicholson, W. L., Kosoy, M., and Sumner, J. W. (1999). Shared vector-borne zoonoses of the Old World and New World: home grown or translocated? Schweiz. Med. Wochenschr. 129, 1099–1105.
Childs, J. E., Glass, G. E., Ksiazek, T. G., Rossi, C. A., Oro, J. G., and Leduc, J. W. (1991). Human-rodent contact and infection with lymphocytic choriomeningitis and Seoul viruses in an inner-city population. Am. J. Trop. Med. Hyg. 44, 117–121. doi: 10.4269/ajtmh.1991.44.117
Comer, J. A., Diaz, T., Vlahov, D., Monterroso, E., and Childs, J. E. (2001). Evidence of rodent-associated Bartonella and Rickettsia infections among intravenous drug users from Central and East Harlem, New York City. Am. J. Trop. Med. Hyg. 65, 855–860. doi: 10.4269/ajtmh.2001.65.855
Comer, J. A., Flynn, C., Regnery, R. L., Vlahov, D., and Childs, J. E. (1996). Antibodies to Bartonella species in inner-city intravenous drug users in Baltimore, Md. Arch. Intern. Med. 156, 2491–2495. doi: 10.1001/archinte.1996.00440200111014
Conroy, C. J., Rowe, K. C., Rowe, K. M., Kamath, P. L., Aplin, K. P., Lucia Hui, L., et al. (2013). Cryptic genetic diversity in Rattus of the San Francisco Bay region, California. Biol. Invasions 15, 741–758. doi: 10.1007/s10530-012-0323-9
Costa, F., Porter, F. H., Rodrigues, G., Farias, H., de Faria, M. T., Wunder, E. A., et al. (2014). Infections by Leptospira interrogans, Seoul virus, and Bartonella spp. among Norway rats (Rattus norvegicus) from the urban slum environment in Brazil. Vector Borne Zoonotic Dis. 14, 33–40. doi: 10.1089/vbz.2013.1378
Crowl, T. A., Crist, T. O., Parmenter, R. R., Belovsky, G., and Lugo, A. E. (2008). The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 6, 238–246. doi: 10.1890/070151
Daly, J. S., Worthington, M. G., Brenner, D. J., Moss, C. W., Hollis, D. G., Weyant, R. S., et al. (1993). Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 31, 872–881.
Desvars-Larrive, A., Pascal, M., Gasqui, P., Cosson, J. F., Benoît, E., Lattard, V., et al. (2017). Population genetics, community of parasites, and resistance to rodenticides in an urban brown rat (Rattus norvegicus) population. PLoS ONE 12:e0184015. doi: 10.1371/journal.pone.0184015
Dybing, N. A., Jacobson, C., Irwin, P., Algar, D., and Adams, P. J. (2016). Bartonella species identified in rodent and feline hosts from island and Mainland Western Australia. Vector Borne Zoonotic Dis. 16, 238–244. doi: 10.1089/vbz.2015.1902
Easterbrook, J. D., Kaplan, J. B., Vanasco, N. B., Reeves, W. K., Purcell, R. H., Kosoy, M. Y., et al. (2007). A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. Epidemiol. Infect. 135, 1192–1199. doi: 10.1017/S0950268806007746
Easterbrook, J. D., Shields, T., Klein, S. L., and Glass, G. E. (2005). Norway rat population in Baltimore, Maryland, 2004. Vector Borne Zoonotic Dis. 5, 296–299. doi: 10.1089/vbz.2005.5.296
Ehrenborg, C., Byström, R., Hjelm, E., Friman, G., and Holmberg, M. (2008). High Bartonella spp. seroprevalence in a Swedish homeless population but no evidence of trench fever. Scand. J. Infect. Dis. 40, 208–215. doi: 10.1080/00365540701632972
Ellis, B. A., Regnery, R. L., Beati, L., Bacellar, F., Rood, M., Glass, G. G., et al. (1999). Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J. Infect. Dis. 180, 220–224.
Eremeeva, M. E., Gerns, H. L., Lydy, S. L., Goo, J. S., Ryan, E. T., Mathew, S. S., et al. (2007). Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N. Engl. J. Med. 356, 2381–2387. doi: 10.1056/NEJMoa065987
Faruque, L. I., Zaman, R. U., Gurley, E. S., Massung, R. F., Alamgir, A. S., Galloway, R. L., et al. (2017). Prevalence and clinical presentation of Rickettsia, Coxiella, Leptospira, Bartonella and chikungunya virus infections among hospital-based febrile patients from December 2008 to November 2009 in Bangladesh. BMC Infect. Dis. 17:141. doi: 10.1186/s12879-017-2239-6
Firth, C., Bhat, M., Firth, M. A., Williams, S. H., Frye, M. J., Simmonds, P., et al. (2014). Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. MBio 5, 1–16. doi: 10.1128/mBio.01933-14
Frye, M. J., Firth, C., Bhat, M., Firth, M. A., Che, X., and Lee, D. (2015). Preliminary survey of ectoparasites and associated pathogens from Norway rats in New York City. J. Med. Entomol. 52, 253–259. doi: 10.1093/jme/tjv014
Gardner-Santana, L. C., Norris, D. E., Fornadel, C. M., Hinson, C. M., Klein, S. L., and Glass, G. E. (2009). Commensal ecology, urban landscapes, and their influence on the genetic characteristics of city-dwelling Norway rats (Rattus norvegicus). Mol. Ecol. 8, 2766–2778. doi: 10.1111/j.1365-294X.2009.04232.x
Gonçalves, L. R., Favacho, A. R., Roque, A. L., Mendes, N. S., Fidelis, O. L., Benevenute, J. L., et al. (2016). Association of Bartonella species with wild and synanthropic rodents in different Brazilian biomes. Appl. Environ. Microbiol. 82, 7154–7164. doi: 10.1128/AEM.02447-16
Grimmond, S. U. E. (2007). Urbanization and global environmental change: local effects of urban warming. Geograph. J. 173, 83–88. doi: 10.1111/j.1475-4959.2007.232_3.x
Gundi, V. A., Billeter, S. A., Rood, M. P., and Kosoy, M. Y. (2012). Bartonella spp. in rats and zoonoses, Los Angeles, California, USA. Emerg. Infect. Dis. 18, 631–633. doi: 10.3201/eid1804.110816
Gundi, V. A., Kosoy, M. Y., Myint, K. S., Shrestha, S. K., Shrestha, M. P., Pavlin, J. A., et al. (2010). Prevalence and genetic diversity of Bartonella species detected in different tissues of small mammals in Nepal. Appl. Environ. Microbiol. 76, 8247–8254. doi: 10.1128/AEM.01180-10
Gundi, V. A. K. B., Davoust, B., Khamis, A., Boni, M., Raoult, D., and La Scola, B. (2004). Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol. 42: 3816–3818. doi: 10.1128/JCM.42.8.3816-3818.2004
Gundi, V. A. K. B., Taylor, C., Raoult, D., and La Scola, B. (2009). Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov. and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int. J. Syst. Evol. Microbiol. 59, 2956–2961. doi: 10.1099/ijs.0.002865-0
Gutiérrez, R., Krasnov, B., Morick, D., Gottlieb, Y., Khokhlova, I. S., and Harrus, S. (2015). Bartonella infection in rodents and their flea ectoparasites: an overview. Vector Borne Zoonotic Dis. 15, 27–39. doi: 10.1089/vbz.2014.1606
Halliday, J. E., Knobel, D. L., Agwanda, B., Bai, Y., Breiman, R. F., Cleaveland, S., et al. (2015). Prevalence and diversity of small mammal-associated Bartonella species in rural and urban Kenya. PLoS Negl. Trop. Dis. 9:e0003608. doi: 10.1371/journal.pntd.0003608
Harrus, S., Bar-Gal, G. K., Golan, A., Elazari-Volcani, R., Kosoy, M. Y., Morick, D., et al. (2009). Isolation and genetic characterization of a Bartonella strain closely related to Bartonella tribocorum and Bartonella elizabethae in Israeli commensal rats. Am. J. Trop. Med. Hyg. 81, 55–58. doi: 10.4269/ajtmh.2009.81.55
Hayman, D. T., McDonald, K. D., and Kosoy, M. Y. (2013). Evolutionary history of rat-borne Bartonella: the importance of commensal rats in the dissemination of bacterial infections globally. Ecol. Evol. 3, 3195–3203. doi: 10.1002/ece3.702
Helan, J. V. G., Grinberg, A., Gedye, K., Potter, M. A., and Harrus, S. (2018). Molecular detection of Bartonella coopersplainsensis and B. henselae in rats from New Zealand. N. Z. Vet. J. 66, 257–260. doi: 10.1080/00480169.2018.1483781
Heller, R., Riegel, P., Hansmann, Y., Delacour, G., Bermond, D., Dehio, C., et al. (1998). Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int. J. Syst. Bacteriol. 48 (Pt 4), 1333–1339. doi: 10.1099/00207713-48-4-1333
Himsworth, C. G., Bai, Y., Kosoy, M. Y., Wood, H., DiBernardo, A., Lindsay, R., et al. (2015). An investigation of Bartonella spp., Rickettsia typhi, and Seoul hantavirus in rats (Rattus spp.) from an inner-city neighborhood of Vancouver, Canada: is pathogen presence a reflection of global and local rat population structure? Vector Borne Zoonotic Dis. 15, 21–26. doi: 10.1089/vbz.2014.1657
Himsworth, C. G., Jardine, C. M., Parsons, K. L., Feng, A. Y., and Patrick, D. M. (2014a). The characteristics of wild rat (Rattus spp.) populations from an inner-city neighborhood with a focus on factors critical to the understanding of rat-associated zoonoses. PLoS ONE 9:e91654. doi: 10.1371/journal.pone.0091654
Himsworth, C. G., Parsons, K. L., Feng, A. Y., Kerr, T., Jardine, C. M., and Patrick, D. M. (2014b). A mixed methods approach to exploring the relationship between Norway rat (Rattus norvegicus) abundance and features of the urban environment in an inner-city neighborhood of Vancouver, Canada. PLoS ONE 9:e97776. doi: 10.1371/journal.pone.0097776
Himsworth, C. G., Parsons, K. L., Jardine, C., and Patrick, D. M. (2013). Rats, cities, people, and pathogens: a systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector Borne Zoonotic Dis. 13, 349–359. doi: 10.1089/vbz.2012.1195
Hopkins, G. H. E. (1949). Report on Rats, Fleas and Plague in Uganda. East African Standard Ltd. For the Government Printer of Uganda. 52.
Hsieh, J. W., Tung, K. C., Chen, W. C., Lin, J. W., Chien, L. J., Hsu, Y. M., et al. (2010). Epidemiology of Bartonella infection in rodents and shrews in Taiwan. Zoonoses Public Health 57, 439–446. doi: 10.1111/j.1863-2378.2009.01234.x
Inoue, K., Maruyama, S., Kabeya, H., Yamada, N., Ohashi, N., Sato, Y., et al. (2008). Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl. Environ. Microbiol. 74, 5086–5092. doi: 10.1128/AEM.00071-08
Jiyipong, T., Jittapalapong, S., Morand, S., Raoult, D., and Rolain, J. M. (2012). Prevalence and genetic diversity of Bartonella spp. in small mammals from Southeastern Asia. Appl. Environ. Microbiol. 78, 8463–8466. doi: 10.1128/AEM.02008-12
Kamani, J., Morick, D., Mumcuoglu, K. Y., and Harrus, S. (2013). Prevalence and diversity of Bartonella species in commensal rodents and ectoparasites from Nigeria, West Africa. PLoS Negl. Trop. Dis. 7:e2246. doi: 10.1371/journal.pntd.0002246
Kandelaki, G., Malania, L., Bai, Y., Chakvetadze, N., Katsitadze, G., Imnadze, P., et al. (2016). Human Lymphadenopathy caused by ratborne Bartonella, Tbilisi, Georgia. Emerg. Infect. Dis. 22, 544–546. doi: 10.3201/eid2203.151823
Khlyap, L., Glass, G., and Kosoy, M. (2012). “Rodents in urban ecosystems of Russia and the USA,” in Rodents: Habitat, Pathology and Environmental Impact. eds A. Triunveri and D. Scalise (Nova Scientific Publishers), 1–22.
Khlyap, L. A., Kosoy, M., Popov, V. P., Cosson, J. F., and Morand, S. (2016). Rats of the genus Rattus as hosts for natural focal infectious agents. Med. Parazitol (Mosk). 47–52.
Kim, K. S., Inoue, K., Kabeya, H., Sato, S., Takada, T., Pangjai, D., et al. (2016). Prevalence and diversity of Bartonella species in wild small mammals in Asia. J. Wildl. Dis. 52, 10–21. doi: 10.7589/2015-01-015
Klangthong, K., Promsthaporn, S., Leepitakrat, S., Schuster, A. L., McCardle, P. W., Kosoy, M., et al. (2015). The distribution and diversity of Bartonella species in rodents and their ectoparasites across Thailand. PLoS ONE 10:e0140856. doi: 10.1371/journal.pone.0140856
Kosoy, M., Bai, Y., Sheff, K., Morway, C., Baggett, H., Maloney, S. A., et al. (2010). Identification of Bartonella infections in febrile human patients from Thailand and their potential animal reservoirs. Am. J. Trop. Med. Hyg. 82, 1140–1145. doi: 10.4269/ajtmh.2010.09-0778
Kosoy, M., Hayman, D. T., and Chan, K. S. (2012). Bartonella bacteria in nature: where does population variability end and a species start? Infect. Genet. Evol. 12, 894–904. doi: 10.1016/j.meegid.2012.03.005
Kosoy, M., Khlyap, L., Cosson, J. F., and Morand, S. (2015). Aboriginal and invasive rats of genus Rattus as hosts of infectious agents. Vector Borne Zoonotic Dis. 15, 3–12. doi: 10.1089/vbz.2014.1629
Kosoy, M., McKee, C., Albayrak, L., and Fofanov, Y. (2018). Genotyping of Bartonella bacteria and their animal hosts: current status and perspectives. Parasitology 145, 543–562. doi: 10.1017/S0031182017001263
Kosoy, M. Y., Regnery, R. L., Tzianabos, T., Marston, E. L., Jones, D. C., Green, D., et al. (1997). Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am. J. Trop. Med. Hyg. 57, 578–588. doi: 10.4269/ajtmh.1997.57.578
Krasnov, B. R., Khokhlova, I. S., Fielden, L. J., and Burdelova, N. V. (2001). Effect of air temperature and humidity on the survival of pre-imaginal stages of two flea species (Siphonaptera: Pulicidae). J. Med. Entomol. 38, 629–637. doi: 10.1603/0022-2585-38.5.629
Lack, J. B., Greene, D. U., Conroy, C. J., Hamilton, M. J., Braun, J. K., Mares, M. A., et al. (2012). Invasion facilitates hybridization with introgression in the Rattus rattus species complex. Mol. Ecol. 21, 3545–3561. doi: 10.1111/j.1365-294X.2012.05620.x
Lack, J. B., Hamilton, J., Braun, J. K., Mares, M. A., and Van Den Bussche, R. A. (2013). Comparative phylogeography of invasive Rattus rattus and Rattus norvegicus in the U.S. reveals distinct colonization histories and dispersal. Biol. Invasions 15, 1067–1087. doi: 10.1007/s10530-012-0351-5
Laudisoit, A., Falay, D., Amundala, N., Akaibe, D., de Bellocq, J. G., Van Houtte, N., et al. (2014). High prevalence of Rickettsia typhi and Bartonella species in rats and fleas, Kisangani, Democratic Republic of the Congo. Am. J. Trop. Med. Hyg. 90, 463–468. doi: 10.4269/ajtmh.13-0216
Lin, J. W., Chen, C. Y., Chen, W. C., Chomel, B. B., and Chang, C.C. (2008). Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J. Med. Microbiol. 57(Pt 12), 1496–1501. doi: 10.1099/jmm.0.2008/004671-0
Loan, H. K., Cuong, N. V., Takhampunya, R., Klangthong, K., Osikowicz, L., Kiet, B. T., et al. (2015). Bartonella species and trombiculid mites of rats from the Mekong Delta of Vietnam. Vector Borne Zoonotic Dis. 15, 40–47. doi: 10.1089/vbz.2014.1604
Lund, M. (2015). “Commensal rodents,” in Rodent pests and their control. CAB International, 2nd Edn, eds. A. P. Buckle and R. H. Smith, (Wallingford Oxon: University of Huddersfield), 23–43.
Márquez, F. J., Rodríguez-Liébana, J. J., Pachón-Ibáñez, M. E., Docobo-Pérez, F., Hidalgo-Fontiveros, A., Bernabeu-Wittel, M., et al. (2008). Molecular screening of Bartonella species in rodents from south western Spain. Vector Borne Zoonotic Dis. 8, 695–700. doi: 10.1089/vbz.2007.0257
Martin-Alonso, A., Houemenou, G., Abreu-Yanes, E., Valladares, B., Feliu, C., and Foronda, P. (2016). Bartonella spp. in Small Mammals, Benin. Vector Borne Zoonotic Dis. 16, 229–237. doi: 10.1089/vbz.2015.1838
Martin-Alonso, A., Soto, M., Foronda, P., Aguilar, E., Bonnet, G., Pacheco, R., et al. (2014). Bartonella spp. and Yersinia pestis reservoirs, Cusco, Peru. Emerg. Infect. Dis. 20, 1069–1070. doi: 10.3201/eid2006.131194
McFarlane, R., Sleigh, A., and McMichael, T. (2012). Synanthropy of wild mammals as a determinant of emerging infectious diseases in the Asian-Australasian region. Ecohealth 9, 24–35. doi: 10.1007/s10393-012-0763-9
McKee, C. D., Osikowicz, L. M., Schwedhelm, T. R., Maes, S. E., Enscore, R. E., Gage, K. L., et al. (2018). Acquisition of Bartonella elizabethae by experimentally exposed Oriental rat fleas (Xenopsylla cheopis; Siphonaptera, Pulicidae) and excretion of Bartonella DNA in flea feces. J. Med. Entomol. 55, 1292–1298. doi: 10.1093/jme/tjy085
McVea, D. A., Himsworth, C. G., Patrick, D. M., Lindsay, L. R., Kosoy, M., and Kerr, T. (2018). Exposure to rats and rat-associated Leptospira and Bartonella species among people who use drugs in an impoverished, inner-city neighborhood of Vancouver, Canada. Vector Borne Zoonotic Dis. 18, 82–88. doi: 10.1089/vbz.2017.2179
Meheretu, Y., Leirs, H., Welegerima, K., Breno, M., Tomas, Z., Kidane, D., et al. (2013). Bartonella prevalence and genetic diversity in small mammals from Ethiopia. Vector Borne Zoonotic Dis. 13, 164–175. doi: 10.1089/vbz.2012.1004
Morick, D., Baneth, G., Avidor, B., Kosoy, M. Y., Mumcuoglu, K. Y., and Mintz, D. (2009). Detection of Bartonella spp. in wild rodents in Israel using HRM real-time PCR. Vet. Microbiol. 139, 293–297. doi: 10.1016/j.vetmic.2009.06.019
Musser, G. G., and Carlton, M. D. (2005). “Superfamily Muroidea,” in Mammal Species of the World, 3rd Edn, eds D. E. Wilson and D. M. Reeder (The Johns Hopkins University Press), 894–1531.
Myint, K. S., Gibbons, R. V., Iverson, J., Shrestha, S. K., Pavlin, J. A., Mongkolsirichaikul, D., et al. (2011). Serological response to Bartonella species in febrile patients from Nepal. Trans. R. Soc. Trop. Med. Hyg. 105, 740–742. doi: 10.1016/j.trstmh.2011.08.002
Neves, E. S., Mendenhall, I. H., Borthwick, S. A., Su, Y. C. F., and Smith, G. J. D. (2018). Detection and genetic characterization of diverse Bartonella genotypes in the small mammals of Singapore. Zoonoses Public Health 65, e207–e215. doi: 10.1111/zph.12430
Papkou, A., Gokhale, C. S., Traulsen, A., and Schulenburg, H. (2016). Host-parasite coevolution: why changing population size matters. Zoology 119, 330–338. doi: 10.1016/j.zool.2016.02.001
Peterson, A. C., Ghersi, B. M., Alda, F., Firth, C., Frye, M. J., Bai, Y., et al. (2017). Rodent-borne Bartonella infection varies according to host species within and among cities. Ecohealth 14, 771–782. doi: 10.1007/s10393-017-1291-4
Pretorius, A. M., Beati, L., and Birtles, R. J. (2004). Diversity of Bartonellae associated with small mammals inhabiting Free State province, South Africa. Int. J. Syst. Evol. Microbiol. 54, 1959–1967. doi: 10.1099/ijs.0.03033-0
Psaroulaki, A., Antoniou, M., Toumazos, P., Mazeris, A., Ioannou, I., Chochlakis, D., et al. (2010). Rats as indicators of the presence and dispersal of six zoonotic microbial agents in Cyprus, an island ecosystem: a seroepidemiological study. Trans. R. Soc. Trop. Med. Hyg. 104, 733–739. doi: 10.1016/j.trstmh.2010.08.005
Pulliam, H. R. (1988). Sources, sinks, and population regulation. Am. Naturalist 132, 652–661. doi: 10.1086/284880
Rothenburger, J. L., Himsworth, C. G., Nemeth, N. M., Pearl, D. L., and Jardine, C. M. (2018). Beyond abundance: how microenvironmental features and weather influence Bartonella tribocorum infection in wild Norway rats (Rattus norvegicus). Zoonoses Public Health 65, 339–351. doi: 10.1111/zph.12440
Saisongkorh, W., Wootta, W., Sawanpanyalert, P., Raoult, D., and Rolain, J. M. (2009). “Candidatus Bartonella thailandensis”: a new genotype of Bartonella identified from rodents. Vet. Microbiol. 139, 197–201. doi: 10.1016/j.vetmic.2009.05.011
Schülein, R., Seubert, A., Gille, C., Lanz, C., Hansmann, Y., Piémont, Y., et al. (2001). Invasion and persistent intracellular colonization of erythrocytes. A unique parasitic strategy of the emerging pathogen Bartonella. J. Exp. Med. 193, 1077–1086.
Segers, F. H., Kešnerová, L., Kosoy, M., and Engel, P. (2017). Genomic changes associated with the evolutionary transition of an insect gut symbiont into a blood-borne pathogen. ISME J. 11, 1232–1244. doi: 10.1038/ismej.2016.201
Shochat, E., Warren, P. S., Faeth, S. H., McIntyre, N. E., and Hope, D. (2006). From patterns to emerging processes in mechanistic urban ecology. Trends Ecol. Evol. 21, 186–191. doi: 10.1016/j.tree.2005.11.019
Tay, S. T., Mokhtar, A. S., Zain, S. N., and Low, K. C. (2014). Isolation and molecular identification of Bartonella from wild rats (Rattus species) in Malaysia. Am. J. Trop. Med. Hyg. 90, 1039–1042. doi: 10.4269/ajtmh.13-0273
Trataris, A. N., Rossouw, J., Arntzen, L., Karstaedt, A., and Frean, J. (2012). Bartonella spp. in human and animal populations in Gauteng, South Africa, from 2007 to 2009. Onderstepoort J. Vet. Res. 79, 452–460. doi: 10.4102/ojvr.v79i2.452
Tsai, Y. L., Chuang, S. T., Chang, C. C., Kass, P. H., and Chomel, B. B. (2010). Bartonella species in small mammals and their ectoparasites in Taiwan. Am. J. Trop. Med. Hyg. 83, 917–923. doi: 10.4269/ajtmh.2010.10-0083
Vayssier-Taussat, M., Moutailler, S., Féménia, F., Raymond, P., Croce, O., La Scola, B., et al. (2016). Identification of novel zoonotic activity of Bartonella spp., France. Emerg. Infect. Dis. 22, 457–462. doi: 10.3201/eid2203.150269
Vicente, S., and Gómez López, M. (2012). Presence of Stenoponia tripectinata (Tiraboschi, 1902) (Siphonaptera, Ctenophtalmidae) in murine (Rodentia) from the Canary Islands. Acta Parasitol. 57, 190–193. doi: 10.2478/s11686-012-0024-7
Winoto, I. L., Goethert, H., Ibrahim, I. N., Yuniherlina, I., Stoops, C., Susanti, I., et al. (2005). Bartonella species in rodents and shrews in the greater Jakarta area. Southeast Asian J. Trop. Med. Public Health 36, 1523–1529.
Ye, X., Li, G. W., Yao, M. L., Luo, W., and Su, L. Q. (2009). Study on the prevalence and genotypes of Bartonella species in rodent hosts from Fujian coastal regions. Zhonghua Liu Xing Bing Xue Za Zhi. 30, 989–992.
Keywords: bacteria, Bartonella, invasive species, movement ecology, rats, Rattus, urban zoonoses
Citation: Kosoy M and Bai Y (2019) Bartonella Bacteria in Urban Rats: A Movement From the Jungles of Southeast Asia to Metropoles Around the Globe. Front. Ecol. Evol. 7:88. doi: 10.3389/fevo.2019.00088
Received: 03 October 2018; Accepted: 07 March 2019;
Published: 28 March 2019.
Edited by:
Claire M. Jardine, University of Guelph, CanadaReviewed by:
Soichi Maruyama, Nihon University, JapanArmanda Bastos, University of Pretoria, South Africa
Dean E. Biggins, United States Geological Survey, United States
Copyright © 2019 Kosoy and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Kosoy, a29zb3ltaWNoYWVsQGdtYWlsLmNvbQ==
 Michael Kosoy
Michael Kosoy Ying Bai
Ying Bai