- 1Department of Biology, Miami University, Oxford, OH, United States
- 2Department of Psychology, University of Michigan, Ann Arbor, MI, United States
Background: We still do not understand the key drivers or prevalence of genetic monogamy in mammals despite the amount of attention that the evolution of mammalian monogamy has received. There have been numerous reviews of the hypotheses proposed to explain monogamy, some of which focused on animals in general, while others focused on particular classes like birds or mammals, or on specific orders within a class. Because monogamy is rare in mammals overall but relatively common in some of the orders in which it has been observed (e.g., Primates, Macroscelidea, and Carnivora), mammals provide a unique taxon in which to study the evolution and maintenance of monogamy However, the term “monogamy” encompasses related but separate phenomena; i.e., social monogamy (pair-living by opposite-sex conspecifics) and genetic monogamy or reproductive monogamy (mating exclusivity). A recent review of mammalian monogamy reported that 226 species (9%) in 9 orders (35%) were socially monogamous, although socially monogamous mammals are not necessarily genetically monogamous.
Methods: Since factors that predispose socially monogamous mammals to be genetically monogamous are still subject to debate, we conducted meta-analyses using model selection to determine the relative importance of several life history, demographic, and environmental factors in predicting genetic monogamy.
Results: We found sufficient data to include 41 species in our analysis, about 2x more than have been included in previous analyses of mammalian genetic monogamy. We found that living as part of a socially monogamous pair vs. in a group was the best predictor of genetic monogamy, either by itself or in combination with high levels of paternal care. A male-biased sex ratio and low population density were inversely related to the number of pairs that were genetically monogamous, but not to the production of intra-pair young or litters.
Conclusion: Our results agree with the results of some previous analyses but suggest that more than one factor may be important in driving genetic monogamy in mammals.
Introduction
The evolution of monogamy has long drawn attention from many scientists studying animal behavior, partially due to its rarity in some taxa and especially because monogamous behavior by males is puzzling because their reproductive success is typically driven by the number of females with which they are able to reproduce (Trivers, 1972). Although monogamy typically had been considered to encompass living with an opposite-sex conspecific, formation, and maintenance of a pair-bond, mating exclusivity, and biparental care (Kleiman, 1977; Wittenberger and Tilson, 1980; Mock and Fujioka, 1990), we now understand that “monogamous” species are quite variable in their social and reproductive behaviors (Westneat et al., 1990; Gowaty, 1996; Griffith et al., 2002) and that these social and reproductive aspects of monogamy may be under selection from different evolutionary pressures. This has resulted in the use of the terms social monogamy (who is living with whom) and reproductive or genetic monogamy (who is mating with whom; Wickler and Seibt, 1983; Fuentes, 1998, 2002; Reichard, 2003; Tecot et al., 2016). Although much research has been devoted to the evolution of social monogamy, genetic monogamy has received much less attention even though it may be more important from an evolutionary standpoint because it is linked directly to reproduction.
Current evidence suggests that female dispersion is the primary driver of social monogamy in mammals (Dobson et al., 2010; Lukas and Clutton-Brock, 2013). This conclusion is supported by phylogenetic analyses that show many independent transitions to social monogamy in mammals (Komers and Brotherton, 1997; Lukas and Clutton-Brock, 2013). In nearly all of these cases, social monogamy evolved where females lived solitarily in discrete home ranges and selection favored males that shared a home range with only one female (Brotherton et al., 1997; Lukas and Clutton-Brock, 2013). Other hypotheses argue that social monogamy provides protection from conspecific infanticide (Wolff and MacDonald, 2004; Opie et al., 2013), although this does not seem to be supported in mammals other than primates (Lukas and Clutton-Brock, 2013). Other traits associated with social monogamy such as paternal care, pair-bonding, group-living and cooperative breeding likely all evolved after the evolution of social monogamy (Komers and Brotherton, 1997; Lukas and Clutton-Brock, 2012, 2013; Opie et al., 2013). It is unclear, however, if genetic monogamy occurs as a consequence of the life history traits of socially monogamous species or is mainly driven by demographic or environmental factors.
Both males and females face trade-offs between seeking extra-pair mates or remaining genetically monogamous. Males face a trade-off between seeking extra-pair copulations (EPCs) and investing in their direct reproductive success by preventing cuckoldry of their mate, protecting their mate/offspring from predation/infanticide, or providing paternal care (Magrath and Komdeur, 2003; Shuster and Wade, 2003; Westneat and Stewart, 2003). Females might seek EPCs for benefits such as genetic variability in their offspring (Petrie and Kempenaers, 1998), mate confusion to prevent infanticide (Harcourt and Greenberg, 2001; Wolff and MacDonald, 2004) or seeking a higher quality mate (Kempenaers et al., 1992; Spencer et al., 1998; Griffith et al., 2002), but this may come at the loss of direct parental care or indirect parental care e.g., territory defense from their male social partner (Ihara, 2002; Westneat and Stewart, 2003).
Rationale
These trade-offs may be influenced by life history traits, demographic or environmental factors. Much of the previous research on genetic monogamy has focused on life history hypotheses, i.e., hypotheses that suggest that genetic monogamy is driven by traits such as group living or coloniality (Møller and Birkhead, 1993; Cohas and Allaine, 2009), breeding seasonality and synchrony (Stutchbury and Morton, 1995), or paternal care (Wade and Shuster, 2002; Magrath and Komdeur, 2003; Westneat and Stewart, 2003; Huck et al., 2014). Such evolved characteristics may determine the availability of EPCs and how likely an individual is to seek extra-pair mates. However, genetic monogamy may be driven more by demographic or environmental factors such as population density (Westneat and Sherman, 1997), a strongly male biased adult sex ratio (Fromhage et al., 2005), or environmental variability (Botero and Rubenstein, 2012). Despite previous attention from investigators, we still do not understand the most important drivers of mammalian genetic monogamy.
Objectives
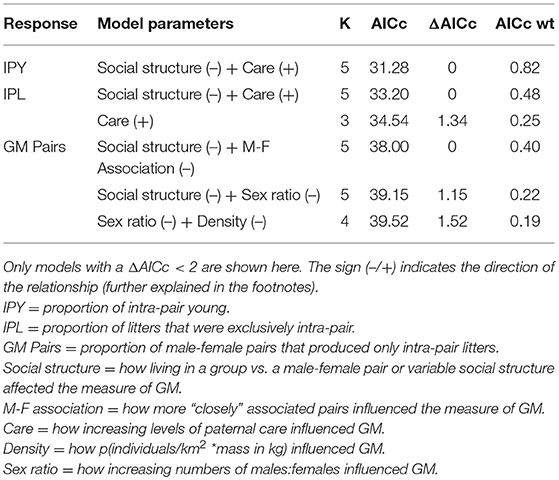
Prior analyses investigating the relative importance of different drivers of genetic monogamy have not reached consistent conclusions for several reasons. One reason is the difference in species included in the analyses. The first comparative studies examined mammals from various mating systems (Clutton-Brock and Isvaran, 2006; Isvaran and Clutton-Brock, 2007), while the more recent studies included only socially monogamous mammals (Cohas and Allaine, 2009; Huck et al., 2014). Even these latter studies defined social monogamy differently. In addition to including species from multiple mating systems vs. only socially monogamous mammals, each of these analyses assessed only a small subset of hypotheses proposed to explain genetic monogamy. In these various studies one or two of the following were found to be good predictors of genetic monogamy: the pattern of association between males and females (intermittent vs. continuous), the number of breeding females per social unit or group, the length of the mating season, social structure (single female, male-female pair, or group), and occurrence of paternal care. Although these studies found different explanations for genetic monogamy, the relative importance of these various hypotheses has not been directly compared. Therefore, our objective was to determine the relative importance of hypotheses proposed to explain genetic monogamy in socially monogamous mammals. We examined all hypotheses for which we could find sufficient data: four life-history, two demographic, and one environmental hypothesis (Table 1).
Table 1. Hypotheses* proposed to explain genetic monogamy in mammals.
The life history traits that we examined were paternal care, pair association, social structure, and breeding seasonality. The paternal care hypothesis predicts that care by the male parent, in addition to that provided by the mother, is critical for optimal offspring growth and survival. Due to this benefit, males seek paternity assurance to avoid caring for extra-pair offspring and females limit extra-pair matings to secure paternal care, both of which promote genetic monogamy (Birkhead and Møller, 1996; Gowaty, 1996). The pair association hypothesis predicts that the trade-off between mate guarding and pursuing EPCs determine levels of genetic monogamy (Clutton-Brock and Isvaran, 2006). Close association between a male and female makes it easier to guard ones' partner but harder for both individuals to obtain EPCs. The social structure hypothesis predicts that individuals living in groups would not be expected to be as genetically monogamous as those that live in male-female pairs (Møller and Birkhead, 1993) because there are more opportunities for EPCs in groups with multiple adults of one or both sexes. Finally, the breeding seasonality hypothesis makes two contrasting predictions. The first prediction is that when females are receptive at about the same time, males will benefit by seeking as many mating opportunities as possible. Thus, breeding synchrony provides more opportunities for mating and thus increases the opportunity for EPCs by males (Stutchbury and Morton, 1995; Stutchbury, 1998a,b). Consequently, there will also be more males nearby providing opportunities for females to engage in EPCs similar to what is observed in lek mating systems (Wagner, 1992). When reproduction occurs within a limited time, it may be too difficult to guard a mate and attempt to gain EPCs; therefore, those engaging in genetic monogamy may be making the best of the bad situation. But, if mate guarding occurs, then genetic monogamy would be favored; this is referred to as the “asynchrony hypotheses” by Neodorf (2004). Alternatively, the breeding seasonality hypothesis predicts that a long breeding season would constrain individuals to be genetically monogamous because females would be receptive asynchronously, thus opportunities for EPCs would be limited (Stutchbury and Morton, 1995; Westneat and Sherman, 1997) and competition for these matings would be high.
The demographic and environmental factors that we examined included population density, adult sex ratio, and climatic variability. These demographic and environmental factors could affect the occurrence of genetic monogamy by influencing the costs and benefits to males of pursuing EPCs or the potential costs of EPCs to females (Westneat and Stewart, 2003). For example, population density may be important because low densities would constrain individuals to be genetically monogamous because there would be limited opportunities to mate with other opposite-sex conspecifics, while genetic monogamy would be much less common at high population densities due to numerous opportunities to mate with multiple individuals (Westneat and Sherman, 1997). The adult sex ratio in the population may also influence genetic monogamy because male-biased sex ratios may result in a large number of extra-pair males seeking matings, resulting in increased opportunities for female extra-pair matings and increased costs associated with mate guarding (Fromhage et al., 2005). Alternatively, female-biased sex ratios could provide opportunities for males to mate with multiple females. Finally, climatic variability may be important because environments with low variation might reduce the genetic benefits of EPCs while highly variable climates favor EPCs that would increase genetic diversity in offspring (Botero and Rubenstein, 2012).
Methods
Search Strategy
To test functional hypotheses proposed to explain genetic monogamy in socially monogamous mammals, we first obtained a comprehensive list of mammals considered to be socially monogamous (229 species) from Lukas and Clutton-Brock (2013). For the purposes of our study, we define socially monogamous mammals as all pair-living mammals as well as group-living mammals that have a dominant breeding pair (sensu Cohas and Allaine, 2009; Lukas and Clutton-Brock, 2013). During our literature search and review of previous work on genetic monogamy, we also came across data from nine socially monogamous species not included in Lukas and Clutton-Brock (2013). We included them in our data set, bringing the total number of socially monogamous species to 238.
We then searched the primary literature for genetic parentage data on all these species using the ISI Web of Science database with combinations of the species' common or scientific name, along with the phrases “paternity” or “parentage analysis,” similar to Cohas and Allaine (2009). We first searched the species' common/scientific names and if this resulted in 20 or fewer results, we examined the titles of all these publications. In contrast, if there were more than 20 results, we combined additional search terms with the common/scientific names. So, for each species we performed 2 or 4 different database searches. We only included data from wild populations.
For each species with genetic parentage data, we calculated up to three different indices of genetic monogamy because genetic parentage data is not always presented in the same way in the literature. These three indices were: the proportion of a female's young sired by her social partner (referred to as intra-pair young, hereafter IPY); the proportion of all litters that were sired exclusively by a social pair (referred to as intra-pair litters, hereafter IPL), i.e., all offspring of a particular litter are assigned to one mother and one father (her social partner); and the proportion of pairs that had only within-pair offspring or litters (referred to as genetically monogamous pairs, hereafter GM pairs). Although we used three different indices, it is important to note that these are not three different outcomes but rather different attempts to quantify genetic monogamy. Furthermore, although many papers also included information which allowed us to determine which males were paired with which females, this was not always the case (e.g., Weston Glenn et al., 2009). For papers without this information, we calculated lower-end and upper-end estimates of our three indices of genetic monogamy based on the assumption that at least one of the sires of a litter was the male social partner when multiple-paternity was detected (although we understand it is possible that none of the sires were the male social partner). We then calculated one mean for each species from these lower and upper-end estimates and used these means in our models. We found two published papers with parentage data for three species and used the paper with the largest sample size and/or the paper from which we could calculate the specific index of genetic monogamy for our analyses.
For our list of socially monogamous mammalian species with genetic parentage data, we performed another set of literature searches to find information on variables that would allow us to examine functional hypotheses for genetic monogamy. For each variable of interest, we first searched the paper in which we had found the genetic parentage data for that species and the Mammalian Species account for that species when available. If we could not locate the information in these sources, we searched the ISI Web of Science database following procedures similar to those used in our initial literature search. We found enough data to test 7 hypotheses: 4 life history hypotheses, 2 demographic hypotheses, and 1 environmental hypothesis (Table 1; Supplementary Data Sheet 1). The variables we examined based on these hypotheses were: the type/amount of paternal care provided to young, frequency with which the male and female were closely associated, the type of social unit most common in the species, the seasonality of the species' breeding, population density (square root of the density multiplied by average mass of an adult individual), the population sex ratio, and climatic variability (as assessed by the Koppen-Geiger's climate classification, Peel et al., 2007) for the study site.
Data Analysis
In our analyses, we controlled for the phylogenetic relatedness across all of our species to account for the lack of independence among closely-related species (Garland et al., 1999; Freckleton et al., 2002). This type of phylogenetic regression is similar to analyses in previous comparative studies on genetic monogamy (Isvaran and Clutton-Brock, 2007; Huck et al., 2014). To accomplish this, we used a subset of a Mammalian supertree downloaded from TimeTree (on Dec 14, 2017), a publicly available phylogenetic tree synthesized from published phylogenies (Hedges et al., 2006, 2015; Kumar et al., 2017), that included our species of interest (Supplementary Image 1).
We analyzed our dataset using AICc model selection of phylogenetic least squares regression models that included different combinations of our predictor variables (Martins and Hansen, 1997; Freckleton et al., 2002). We created 12 a priori models that consisted of different combinations of the life history, demographic, and environmental hypotheses for which we had data from a sufficient number of species (Table 1), including models based on the results of previous comparative studies on mammalian extra-group paternity (see Supplementary Table 1; Clutton-Brock and Isvaran, 2006; Isvaran and Clutton-Brock, 2007; Cohas and Allaine, 2009; Lukas and Clutton-Brock, 2013). We ran 3 rounds of AICc model selection, one for each dependent variable, with the 12 a priori models using the means of IPY, IPL, and GM pairs. All of these variables were proportional data, therefore they were arcsine square-root transformed for the analyses. All presented coefficients are back-transformed. These models included a phylogenetic correction following the Brownian model of character evolution (Garland et al., 1999; Huck et al., 2014) and were weighted using to control for the wide range in sample sizes among studies (Gurevitch and Hedges, 1999; Griffith et al., 2002; Perry et al., 2002; Zaykin, 2011). For each model selection process, models were ranked from lowest- to highest-AICc and the ΔAICc and model Akaike weights were calculated. The weights are the probability that the model is the best model, given the data and other models in the candidate set (Wagenmakers and Farrell, 2004). All analyses were performed in R (R Core Team, 2018) using the ape (Paradis et al., 2004) and nlme packages (Pinheiro et al., 2018). All of our procedures followed the applicable meta-analysis standards set forward by the PRISMA statement (Moher et al., 2009).
Results
Study Selection and Characteristics
We found genetic data for 41 mammalian species (17% of the socially monogamous mammals on our list) in 53 different studies and used 46 of these studies for our analyses (Figure 1; Supplementary Data Sheet 1). We found the most species for the order Carnivora (14/41 = 34%), Primates (11/41 = 27%), and Rodentia (13/41 = 32%). Four other orders were represented by 1–3 species.
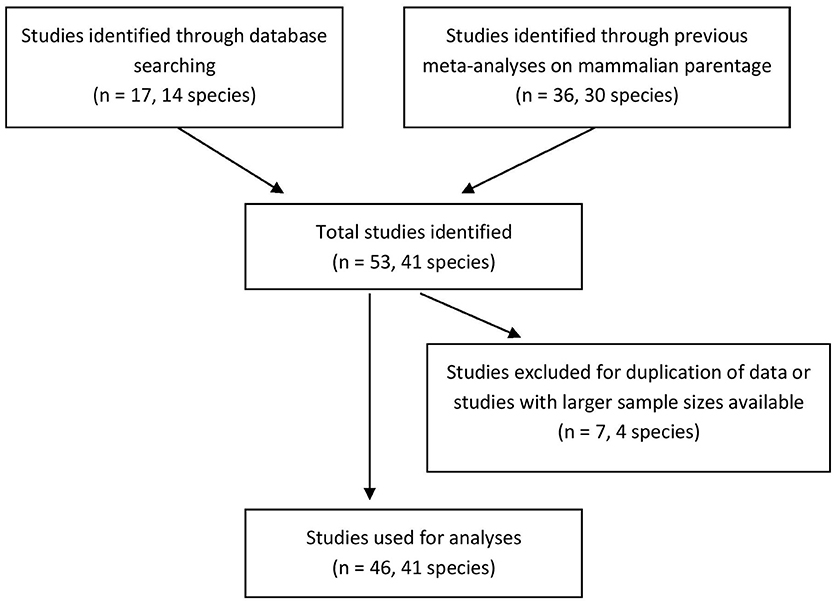
Figure 1. Flow chart for the literature search for studies of socially monogamous mammals with genetic parentage data. Sample sizes presented are the numbers of studies found.
In seven of our 41 species (17%), all pairs were genetically monogamous i.e., had no extra-pair young detected, although the sample size for the Bornean gibbon (Hylobates muelleri) was only 5 offspring. The mean ± SE proportion of IPY was 0.76 ± 0.03 (N = 39 species; range 0.17–1.00; median number of offspring/species = 40). The mean ± SE proportion of IPL was 0.73 ± 0.04 (N = 28 species; range: 0.34–1.00; median number of litters/species = 18). The mean ± SE proportion of GM pairs was 0.70 ± 0.04 (N = 34 species; range: 0.40–1.00; median number of pairs/species = 16).
Synthesized Findings
Each model competition procedure resulted in 1 to 3 models that were within 2 ΔAICc of the top model; these are shown in Table 2 (For all models, see Supplementary Table 1). The variable we refer to as social structure of the species was present in 4 of the 6 (66.7%) top models. Species that are group-living had lower levels of IPY, IPL, and GM pairs compared to species in which individuals lived in pairs or displayed intermediate social structure (Figure 2, Table 3). The level of paternal care appeared in all 3 of the top models for IPY and IPL, with an increasing level of paternal care always positively associated with more IPY/IPL (Figure 3, Table 3). The level of male-female association appeared in 1 of the top models for GM pairs; species where pairs were more closely associated had lower proportions of IPY than species with intermittent levels of association between the male and female. However, this outcome is likely to have been caused by multi-collinearity between pair association and social structure in the model, since this was the only model where pair association had this effect. Sex ratio appeared in two of the top models for GM pairs and population density appeared in one of them. Each of these variables had a negative effect on the measure of genetic monogamy. None of the other variables or combinations of variables appeared in a top model.
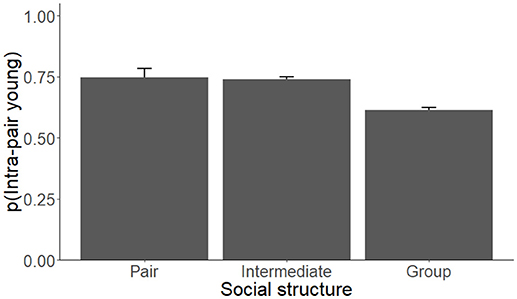
Figure 2. The proportion of intra-pair young (IPY) produced by species of socially monogamous mammals that live primarily in pairs, groups, or that are intermediate between these two types of social structure. The proportions presented are based on the coefficients from the top model for IPY (Tables 2, 3) and control for phylogenetic relatedness, differences in the level of paternal care, and are weighted by sample size.
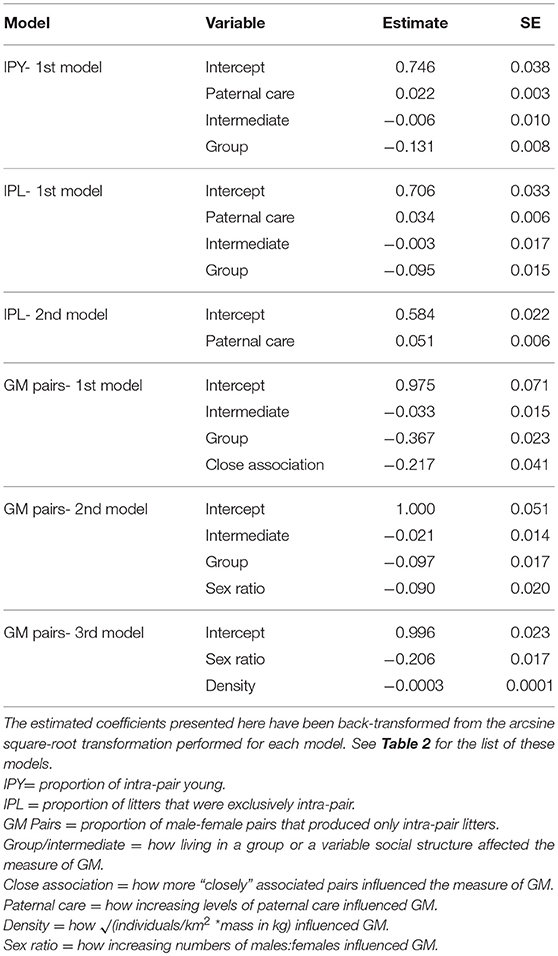
Table 3. Model coefficients and standard errors from the top phylogenetic regression models explaining three indices of genetic monogamy (GM).
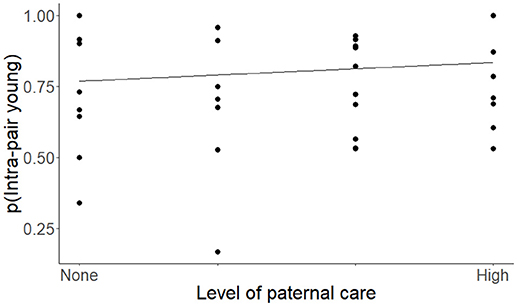
Figure 3. The proportion of intra-pair young (IPY) produced by species of socially monogamous mammals with differing levels of paternal care, ranked from species that provide no paternal care to those that provide intensive paternal care. The regression is based on the coefficients from the top model for IPY (Tables 2, 3) and controls for phylogenetic relatedness, differences in social structure, and is weighted by sample size.
Discussion
Summary of Main Findings
Our results show that no one model was consistently the best for all our indices of genetic monogamy. Life history variables, specifically social structure and paternal care, were the only type of predictor variables for IPY and IPL and the life history variable referred to as social structure was in two of the top models for GM pairs. Demographic variables, specifically sex ratio and population density, only appeared in two of the top models for GM pairs.
The only predictive variable found for all three indices of genetic monogamy was social structure (Table 2), specifically living in a socially monogamous pair or intermediately (i.e., sometimes as a monogamous pair and sometimes in a group) as opposed to living primarily in a group. Individuals that lived as a member of a socially monogamous pair showed higher levels of genetic monogamy compared to individuals found primarily in a group with other sexually mature conspecifics. Living as a member of a pair was an important predictor of genetic monogamy either by itself or in combination with other life history variables such as paternal care, the amount of male-female association or sex ratio. Paternal care was important in explaining IPY and IPL, especially in combination with pair living (Table 2). Sex ratio and population density were the only demographic variables in any top model. They were found in the top models for GM pairs but not in models for any other index of genetic monogamy. No other demographic or environmental variables were in any of our top models. These results suggest that the levels of genetic monogamy across socially monogamous mammals are likely not driven by merely one variable but a combination of variables, and different variables may be more important for different species (see also Klug, 2018).
Our study, including twice as many species as prior studies, is consistent with results from the largest previous comparative study of socially monogamous mammals (Cohas and Allaine, 2009) that showed that group-living species had higher levels of EPY than pair-living species. Although there are numerous possible benefits to group-living, including increased group vigilance resulting in increased protection from predation, foraging benefits, increased production, or survival of offspring due to helping by other group members (Krause and Ruxton, 2002; Ward and Webster, 2016), it appears that a major cost to group living is being unable to prevent one's mate from engaging in EPCs (Suter et al., 2009; Nichols et al., 2015).
A high level of paternal care was also associated with increased levels of genetic monogamy for all models in which it was present (Table 3). The positive relationship between levels of paternal care and IPY is consistent with the results of Huck et al. (2014), even when we include group-living species in the data set (see also Kvarnemo, 2006). Previous studies show that paternal care likely evolved after transitions to social monogamy (Komers and Brotherton, 1997; Lukas and Clutton-Brock, 2013). For this reason, we suggest that genetic monogamy may have co-evolved with paternal behavior when selection initially favored males that were more affiliative toward females and offspring and, subsequently these affiliative behaviors became modified into paternal care (Komers and Brotherton, 1997; Dillard and Westneat, 2016). Selection might then have favored males that provided parental care only when paternity was certain to allow males to limit investing in offspring sired by other males. If females engaged in EPCs and males were able to detect this, then these females could face high costs from losing male investment and may then have decreased or refrained from engaging in extra-pair mating to ensure male care. Over evolutionary time, females might have produced offspring that were increasingly reliant on male care or male care might have resulted in greater fitness benefits through the quality or number of offspring. Either of these scenarios could result in stabilizing selection for male care and genetic monogamy (Smith, 1977; Dunbar, 1995; Wade and Shuster, 2002; Stockley and Hobson, 2016). This may be the explanation for the high level of genetic monogamy in the California mouse (Peromyscus californicus), for example, where providing paternal care and remaining genetically monogamous results in greater reproductive success for a male than the alternative (Gubernick and Teferi, 2000; Ribble, 2003).
Demographic variables were not good predictors of IPY or IPL but the combination of sex ratio and either social structure or population density were two of the top models for GM pairs, with increases in population density or the proportion of adult males in the population having a negative effect on GM pairs. This was consistent with our predictions and also has been found in previous avian studies (Westneat and Sherman, 1997). Higher population densities may provide greater opportunities for EPCs since there would be a greater likelihood of encountering opposite-sex conspecifics, thus lower costs of pursuing EPCs while resulting in increased difficulty in guarding ones' own mate to prevent EPCs. A male-biased sex ratio likely has similar effects because unpaired or subdominant males may pursue matings with paired females.
There are a number of possible reasons that we found different top models depending on the index of genetic monogamy that we examined. Variation in the number of offspring produced per reproductive bout could influence the outcome of model selection. Approximately 24% of species in our data set give birth to singletons, e.g., many primates, while others such as canids have larger litters. The difference in the number of offspring per reproductive bout could result in different associations between IPY, IPL, and GM pairs. For example, EPCs may result in a small proportion of EPY for species with litter sizes much greater than one. If 5 pairs each have 5 offspring with 1 EPY each, 100% of the pairs would be considered non-GM but only 20% of the total number of offspring would by IPY. Additionally, the length of the studies and of the pair-bonds differ greatly among species in the data set. Although some studies provided data from multiple breeding seasons or years (e.g., Alpine marmots, Ferrandiz-Rovira et al., 2016), others either did not follow pairs across multiple seasons or only sampled within one breeding season (e.g., Bornean gibbon, Oka and Takenaka, 2001). Finally, although our three indices of genetic monogamy are each different ways to quantify genetic monogamy, they are not necessarily of equal biological importance. Previous studies have focused on EPY (or IPY for our study), and this may be the best index from an evolutionary perspective because it is the closest to reproductive success, which is often used to index fitness (Gimenez and Gaillard, 2018).
Limitations
We had to exclude several potentially biologically important variables from our analyses due to the lack of field data from a sufficient number of species. These variables include the role of female spacing, sexually transmitted disease, relatedness between members of a pair, potential genetic benefits of EPC, and other environmental variables that may influence the interactions between unpaired individuals e.g., habitat structure (Biagolini et al., 2017). Female spacing has been proposed to be a very important driver of social monogamy (Komers and Brotherton, 1997; but see Dobson et al., 2010) and may also influence genetic monogamy due to the inability of males to control access to more than one female (see also Isvaran and Clutton-Brock, 2007). Furthermore, under certain circumstances the presence of sexually transmitted diseases may selectively favor genetic monogamy because mating with one only one opposite-sex conspecific allows individuals to decrease the probability of being infected (Loehle, 1995; Thrall et al., 1997; Kokko et al., 2002; McLeod and Day, 2014). Additionally, relatedness between members of a pair and the genetic benefits of EPY can influence the levels of EPCs in a variety of socially monogamous species (Blomqvist et al., 2002; Varian-Ramos and Webster, 2012; Leclaire et al., 2013; Arct et al., 2015). If we were able to include additional variables in our analysis, we might have found different variables in our top models. Future studies focusing on these potentially important variables may increase our understanding of mammalian genetic monogamy or increase our confidence that the variables in our top models are the most important drivers of genetic monogamy.
Furthermore, some of these variables might interact, be highly related to each other, or be bidirectional, making it challenging to unravel the most important predictors of genetic monogamy. For example, estrous synchrony and spatial distribution of females could interact to affect the probability of males obtaining EPCs. If all females are fertile or in estrus at the same time but are dispersed as opposed to being clustered, the prospects for a male seeking EPCs would differ. Additionally, variables such as population density and seasonality may be highly related, as seen in vole populations where population density typically is lower in winter and spring than in the fall (Getz et al., 1993). Finally, some variables may be bidirectional i.e., they may influence genetic monogamy as well as being influenced by the level of genetic monogamy (Andersson, 1994; Alonzo, 2010). For example, if females engage in EPCs we would predict the level of paternal care to decrease. Conversely, the level of paternal care could also influence the likelihood of females seeking EPCs (Westneat et al., 1990; Birkhead and Møller, 1992; Andersson, 1994; Alonzo, 2010). These examples highlight some of the challenges in trying to encapsulate relevant factors into a model that predicts genetic monogamy across numerous species.
Although a comparative approach can allow us to determine predictors of genetic monogamy across mammals, we realize that all mammalian species do not fit the patterns found. For example, some group-living species in our study had high levels of genetic monogamy despite the overall finding that group-living species had lower levels (Patzenhauerová et al., 2013; Ferrandiz-Rovira et al., 2016). Tight synchronous breeding (Platner, 2005; Hilgartner et al., 2012) and male territory defense (Sommer and Tichy, 1999; Sommer, 2003) are hypothesized to drive high levels of genetic monogamy in two of our species despite low degrees of pair association and no paternal care in both species. Although comparative studies may detect overall evolutionary or environmental trends, the results are not expected to adequately explain the complex processes that result in genetic monogamy in every species.
Another important consideration is how genetic monogamy might differentially benefit conspecifics within or between populations. Differential benefits among individuals may complicate the interpretation of data analyzed at the species-level. Some hypotheses proposed to explain genetic monogamy (e.g., the paternal care hypothesis) predict that individuals will display uniform mating behaviors within a population, other hypotheses predict a lack of uniformity in genetic monogamy. For example, selection for genetic compatibility between members of a pair may result in pairs with low genetic compatibility being less likely to be genetically monogamous than more genetically compatible pairs within the same population (Griffith et al., 2002). Furthermore, populations living in highly variable environments may face different selection pressures and evolutionary trade-offs resulting in intraspecific differences in levels of genetic monogamy (Bishop et al., 2004; Streatfeild et al., 2011). We did not attempt to account for this intraspecific variation in our models because we only found a few species with genetic parentage data from more than one population.
Conclusions
Our study provides the most comprehensive comparative examination of genetic monogamy in mammals to date. Previous studies have primarily focused on life history traits that have been proposed to explain levels of genetic monogamy, and our integration of demographic and environmental variables with these life history variables allowed us to better test additional hypotheses. Although we found no single model that best explained all our indices of genetic monogamy, our results strongly demonstrate that social structure and paternal care are important in explaining variation in genetic monogamy of mammalian species, with some evidence for a couple demographic variables. Data from more species would allow us to determine if any additional variables are important drivers of genetic monogamy. Furthermore, data on variables not yet examined in many socially monogamous species e.g., relatedness between members of the breeding pair, would allow us to test additional hypotheses proposed to explain genetic monogamy. Based on the available data, our results suggest that genetic monogamy is likely to be a consequence of multiple factors in mammals, but that social structure and paternal care appear to be especially important.
Author Contributions
CL, AS, and NS all participated in the literature search and in compiling data. CL organized the dataset, conducted the data analyses, and wrote most of the first draft of the manuscript. AS and NS contributed to and revised the manuscript. All authors read and revised numerous versions of the manuscript. All authors approved of the final version of the submitted manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Brian Keane and members of the Solomon-Keane lab group for helpful suggestions on a previous version of this manuscript. The work on this manuscript was supported by National Institutes of Health (1R15HD075222-01A1) awarded to NS, Brian Keane and Bruce Cushing (a colleague at UT El Paso).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00139/full#supplementary-material
Supplementary Table 1. Model selection for the level of intra-pair young across 39 mammalian species. See Table 1 for more information on the variables included in model selection.
Supplementary Table 2. Model selection for the level of intra-pair litters across 28 mammalian species. See Table 1 for more information on the variables used in model selection.
Supplementary Table 3. Model selection for the level of genetically monogamous pairs across 34 mammalian species. See Table 1 for more information on the variables included in model selection.
Supplementary Data Sheet 1. This data sheet contains a list of all the species used in our analysis, the indices of genetic monogamy calculated from parentage data for each species, and the seven variables used in our analyses for each species, along with all the references for these.
Supplementary Image 1. The phylogeny of the 41 Mammalian species used for the phylogenetic regressions. Phylogeny was created by sub-setting the Mammalian supertree accessed from TimeTree (http://www.timetree.org/) on Dec 14, 2017 to the species used in each analysis.
References
Alonzo, S. H. (2010). Social and coevolutionary feedbacks between mating and parental investment. Trends Ecol. Evol. 25, 99–108. doi: 10.1016/j.tree.2009.07.012
Arct, A., Drobniak, S. M., and Cichon, M. (2015). Genetic similarity between mates predicts extrapair paternity-a meta-analysis of bird studies. Behav. Ecol. 26, 959–968. doi: 10.1093/beheco/arv004
Biagolini, C. Jr, Westneat, D. F., and Francisco, M. R. (2017). Does habitat structural complexity influence the frequency of extra-pair paternity in birds? Behav. Ecol. Sociobiol. 71:101. doi: 10.1007/s00265-017-2329-x
Birkhead, T. R., and Møller, A. P. (1992). Sperm Competition in Birds: Evolutionary Causes and Consequences. San Diego, CA: Academic Press.
Birkhead, T. R., and Møller, A. P. (1996). “Monogamy and sperm competition in birds,” in Partnerships in Birds: The Study of Monogamy, ed J. M. Black (New York, NY: Oxford University Press), 323–343.
Bishop, J. M., Jarvis, J. U. M., Spinks, A. C., Bennett, N. C., and O'Ryan, C. (2004). Molecular insight into patterns of colony composition and paternity in the common mole-rat Cryptomys hottentotus hottentotus. Mol. Ecol. 13, 1217–1229. doi: 10.1111/j.1365-294X.2004.02131.x
Blomqvist, D., Andersson, M., Küpper, C., Cuthill, I. C., Kis, J., Lanctot, R. B., et al. (2002). Genetic similarity between mates and extra-pair parentage in three species of shorebirds. Nature 419, 613–615. doi: 10.1038/nature01104
Botero, C. A., and Rubenstein, D. R. (2012). Fluctuating environments, sexual selection and the evolution of flexible mate choice in birds. PLoS ONE 7:e32311. doi: 10.1371/journal.pone.0032311
Brotherton, P. N., Pemberton, J. M., Komers, P. E., and Malarky, G. (1997). Genetic and behavioural evidence of monogamy in a mammal, Kirk's dik-dik (Madoqua kirkii). Proc. R. Soc. B Biol. Sci. 264, 675–681.
Clutton-Brock, T. H., and Isvaran, K. (2006). Paternity loss in contrasting mammalian societies. Biol. Lett. 2, 513–516. doi: 10.1098/rsbl.2006.0531
Cohas, A., and Allaine, D. (2009). Social structure influences extra-pair paternity in socially monogamous mammals. Biol. Lett. 5, 313–316. doi: 10.1098/rsbl.2008.0760
Dillard, J. R., and Westneat, D. F. (2016). Disentangling the correlated evolution of monogamy and cooperation. Trends Ecol. Evol. 31, 503–513. doi: 10.1016/j.tree.2016.03.009
Dobson, F. S., Way, B. M., and Baudoin, C. (2010). Spatial dynamics and the evolution of social monogamy in mammals. Behav. Ecol. 21, 747–752. doi: 10.1093/beheco/arq048
Dunbar, R. I. M. (1995). The mating system of callitrichid primates: I. Conditions for the coevolution of pair bonding and twinning. Anim. Behav. 50, 1057–1070. doi: 10.1016/0003-3472(95)80106-5
Ferrandiz-Rovira, M., Allainé, D., Callait-Cardinal, M. P., and Cohas, A. (2016). Mate choice for neutral and MHC genetic characteristics in Alpine marmots: different targets in different contexts? Ecol. Evol. 6, 4243–4257. doi: 10.1002/ece3.2189
Freckleton, R. P., Harvey, P. H., and Pagel, M. (2002). Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 160, 712–726. doi: 10.1086/343873
Fromhage, L., Elgar, M. A., and Schneider, J. M. (2005). Faithful without care: the evolution of monogyny. Evolution 59, 1400–1405. doi: 10.1111/j.0014-3820.2005.tb01790.x
Fuentes, A. (1998). Re-evaluating primate monogamy. Am. Anthropol. 100, 890–907. doi: 10.1525/aa.1998.100.4.890
Fuentes, A. (2002). Patterns and trends in primate pair bonds. Int. J. Primatol. 23, 953–978. doi: 10.1023/A:1019647514080
Garland, T., Midford, P. E., and Ives, A. R. (1999). An introduction to phylogenetically based statistical methods, with a new method for confidence intervals on ancestral value. Integr. Comp. Biol. 39, 374–388. doi: 10.1093/icb/39.2.374
Getz, L. L., McGuire, B., and Pizzuto, T. (1993). Social organization of the prairie vole (Microtus ochrogaster). J. Mammal. 74, 44–58. doi: 10.2307/1381904
Gimenez, O., and Gaillard, J.-M. (2018). Estimating individual fitness in the wild using capture-recapture data. Popul. Ecol. 60, 101–109. doi: 10.1007/s10144-017-0598-x
Gowaty, P. A. (1996). “Battles of the sexes and orgins of monogamy,” in Partnerships in Birds: The Study of Monogamy, ed J. M. Black (New York, NY: Oxford University Press), 21–52.
Griffith, S. C., Owens, I. P. F., and Thuman, K. A. (2002). Extra pair paternity in birds: a review of interspecific variation and adaptive function. Mol. Ecol. 11, 2195–2212. doi: 10.1046/j.1365-294X.2002.01613.x
Gubernick, D. J., and Teferi, T. (2000). Adaptive significance of male parental care in a monogamous mammal. Proc. R. Soc. Lond. B 267, 147–150. doi: 10.1098/rspb.2000.0979
Gurevitch, J., and Hedges, L. V. (1999). Statistical issues in ecological meta-analysis. Ecology 80, 1142–1149. doi: 10.1890/0012-9658(1999)080[1142:SIIEMA]2.0.CO;2
Harcourt, A. H., and Greenberg, J. (2001). Do gorilla females join males to avoid infanticide? A quantitative model. Anim. Behav. 62, 905–915. doi: 10.1006/anbe.2001.1835
Hedges, S. B., Dudley, J., and Kumar, S. (2006). TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972. doi: 10.1093/bioinformatics/btl505
Hedges, S. B., Marin, J., Suleski, M., Paymer, M., and Kumar, S. (2015). Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845. doi: 10.1093/molbev/msv037
Hilgartner, R., Fichtel, C., Kappeler, P. M., and Zinner, D. (2012). Determinants of pair-living in red-tailed sportive lemurs (Lepilemur ruficaudatus). Ethology 118, 466–479. doi: 10.1111/j.1439-0310.2012.02033.x
Huck, M., Fernandez-Duque, E., Babb, P., and Schurr, T. (2014). Correlates of genetic monogamy in socially monogamous mammals: insights from Azara's owl monkeys. Proc. R. Soc. B Biol. Sci. 281:20140195. doi: 10.1098/rspb.2014.0195
Ihara, Y. (2002). A model for evolution of male parental care and female multiple mating. Am. Nat. 160, 235–244. doi: 10.1086/341019
Isvaran, K., and Clutton-Brock, T. (2007). Ecological correlates of extra-group paternity in mammals. Proc. R. Soc. B Biol. Sci. 274, 219–224. doi: 10.1098/rspb.2006.3723
Kempenaers, B., Verheyen, G. R., Van den Broeck, M., Burke, T., Van Broeckhoven, C., and Dhondt, A. (1992). Extra-pair paternity results from female preference for high-quality males in the blue tit. Nature 357:494. doi: 10.1038/357494a0
Klug, H. (2018). Why Monogamy? A review of potential ultimate drivers. Front. Ecol. Evol. 6:30. doi: 10.3389/fevo.2018.00030
Kokko, H., Ranta, E., Ruxton, G., and Lundberg, P. (2002). Sexually transmitted disease and the evolution of mating systems. Evolution 56, 1091–1100. doi: 10.1111/j.0014-3820.2002.tb01423.x
Komers, P. E., and Brotherton, P. N. M. (1997). Female space use is the best predictor of monogamy in mammals. Proc. R. Soc. London. Ser. B Biol. Sci. 264, 1261–1270. doi: 10.1098/rspb.1997.0174
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Kvarnemo, C. (2006). Evolution and maintenance of male care: is increased paternity a neglected benefit of care? Behav. Ecol. 17, 144–148. doi: 10.1093/beheco/ari097
Leclaire, S., Nielsen, J. F., Sharp, S. P., and Clutton-Brock, T. H. (2013). Mating strategies in dominant meerkats: evidence for extra-pair paternity in relation to genetic relatedness between pair mates. J. Evol. Biol. 26, 1499–1507. doi: 10.1111/jeb.12151
Loehle, C. (1995). Social barriers to pathogen transmission in wild animal populations. Ecology 76, 326–335. doi: 10.2307/1941192
Lukas, D., and Clutton-Brock, T. (2012). Cooperative breeding and monogamy in mammalian societies. Proc. R. Soc. B Biol. Sci. 279, 2151–2156. doi: 10.1098/rspb.2011.2468
Lukas, D., and Clutton-Brock, T. H. (2013). The evolution of social monogamy in mammals. Science 341, 526–530. doi: 10.1126/science.1238677
Magrath, M. J. L., and Komdeur, J. (2003). Is male care compromised by additional mating opportunity? Trends Ecol. Evol. 18, 424–430. doi: 10.1016/S0169-5347(03)00124-1
Martins, E. P., and Hansen, T. F. (1997). Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am. Nat. 149, 646–667. doi: 10.1086/286013
McLeod, D. V., and Day, T. (2014). Sexually transmitted infection and the evolution of serial monogamy. Proc. R. Soc. B Biol. Sci. 281:20141726. doi: 10.1098/rspb.2014.1726
Mock, D. W., and Fujioka, M. (1990). Monogamy and long-term pair bonding in vertebrates. Trends Ecol. Evol. 5, 39–43. doi: 10.1016/0169-5347(90)90045-F
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G., Altman, D., Antes, G., et al. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6:e1000097. doi: 10.1371/journal.pmed.1000097
Møller, A. P., and Birkhead, T. R. (1993). Certainty of paternity covaries with paternal care in birds. Behav. Ecol. Sociobiol. 33, 261–268. doi: 10.1007/BF02027123
Neodorf, D. (2004). Extrapair paternity in birds: understanding variation among species. Auk 121, 302–307. doi: 10.1642/0004-8038(2004)121[0302:EPIBUV]2.0.CO;2
Nichols, H. J., Cant, M. A., and Sanderson, J. L. (2015). Adjustment of costly extra-group paternity according to inbreeding risk in a cooperative mammal. Behav. Ecol. 26, 1486–1494. doi: 10.1093/beheco/arv095
Oka, T., and Takenaka, O. (2001). Wild gibbons' parentage tested by non-invasive DNA sampling and PCR-amplified polymorphic microsatellites. Primates 42, 67–73. doi: 10.1007/BF02640690
Opie, C., Atkinson, Q. D., Dunbar, R. I. M., and Shultz, S. (2013). Male infanticide leads to social monogamy in primates. Proc. Natl. Acad. Sci. U.S.A. 110, 13328–13332. doi: 10.1073/pnas.1307903110
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Patzenhauerová, H., Šklíba, J., Bryja, J., and Šumbera, R. (2013). Parentage analysis of Ansell's mole-rat family groups indicates a high reproductive skew despite relatively relaxed ecological constraints on dispersal. Mol. Ecol. 22, 4988–5000. doi: 10.1111/mec.12434
Peel, M. C., Finlayson, B. L., and McMahon, T. A. (2007). Updated world map of the Köppen-Geiger climate classification. Hydrol. Earth Syst. Sci. 11, 1633–1644. doi: 10.5194/hess-11-1633-2007
Perry, G., Garland, T., and Garland Jr, T. (2002). Lizard home ranges revisited: effects of sex, body size, diet habitat, and phylogeny. Ecology 83, 1870–1885. doi: 10.2307/3071771
Petrie, M., and Kempenaers, B. (1998). Extra-pair paternity in birds: explaining variation between species and populations. Trends Ecol. Evol. 13, 52–57. doi: 10.1016/S0169-5347(97)01232-9
Pinheiro, J., Bates, D., DebRoy, S., and Sarkar, D. (2018). nlme: Linear and Nonlinear Mixed Effects Models. Available online at: https://cran.r-project.org/web/packages/nlme/citation.html (Accessed March 28, 2018).
Platner, K. (2005). Das genetische Paarungssystem von Lepilemur ruficaudatus. [Diploma]. Göttingen: Georg-August-Universität zu Göttingen.
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Reichard, U. H. (2003). “Monogamy: past and present,” in Monogamy: Mating Strategies and Partnerships in Birds, Humans and Other Mammals, eds U. H. Reichard and C. Boesch (Cambridge: Cambridge University Press), 3–25.
Ribble, D. O. (2003). “The evolution of social and reproductive monogamy in Peromyscus, evidence from Peromyscus californicus (the California mouse),” in Monogamy: Mating Strategies and Partnerships in Birds, Humans and Other Mammals, eds U. H. Reichard and C. Boesch (Cambridge: Cambridge University Press), 81–92.
Shuster, S. M., and Wade, M. J. (2003). Mating Systems and Strategies. Princeton University Press. Available online at: http://books.google.com/books?hl=en&lr=&id=bD4FV4uHtlsC&pgis=1 (Accessed November 1, 2014).
Smith, J. M. (1977). Parental investment: a prospective analysis. Anim. Behav. 25, 1–9. doi: 10.1016/0003-3472(77)90062-8
Sommer, S. (2003). Effects of habitat fragmentation and changes of dispersal behaviour after a recent population decline on the genetic variability of noncoding and coding DNA of a monogamous Malagasy rodent. Mol. Ecol. 12, 2845–2851. doi: 10.1046/j.1365-294X.2003.01906.x
Sommer, S., and Tichy, H. (1999). Major histocompatibility complex (MHC) class II polymorphism and paternity in the monogamous Hypogeomys antimena, the endangered, largest endemic Malagasy rodent. Mol. Ecol. 8, 1259–1272. doi: 10.1046/j.1365-294X.1999.00687.x
Spencer, P. B. S., Horsup, A. B., and Marsh, H. D. (1998). Enhancement of reproductive success through mate choice in a social rock-wallaby, Petrogale assimilis (Macropododae) as revealed by microsatellite markers. Behav. Ecol. Sociobiol. 43, 1–9. doi: 10.1007/s002650050460
Stockley, P., and Hobson, L. (2016). Paternal care and litter size coevolution in mammals. Proc. R. Soc. B Biol. Sci. 283:20160140. doi: 10.1098/rspb.2016.0140
Streatfeild, C. A., Mabry, K. E., Keane, B., Crist, T. O., and Solomon, N. G. (2011). Intraspecific variability in the social and genetic mating systems of prairie voles, Microtus ochrogaster. Anim. Behav. 82, 1387–1398. doi: 10.1016/j.anbehav.2011.09.023
Stutchbury, B. J., and Morton, E. S. (1995). The effect of breeding synchrony on extra-pair mating systems in songbirds. Behaviour 132, 675–690. doi: 10.1163/156853995X00081
Stutchbury, B. J. M. (1998a). Breeding synchrony best explains variation in extra-pair mating system among avian species. Behav. Ecol. Sociobiol. 43, 221–222. doi: 10.1007/s002650050485
Stutchbury, B. J. M. (1998b). Female mate choice of extra pair males breeding synchrony is important. Behav. Ecol. Sociobiol. 43, 213–215. doi: 10.1007/s002650050483
Suter, S. M., Bielanska, J., Röthlin-Spillmann, S., Strambini, L., and Meyer, D. R. (2009). The cost of infidelity to female reed buntings. Behav. Ecol. 20, 601–608. doi: 10.1093/beheco/arp037
Tecot, S. R., Singletary, B., and Eadie, E. (2016). Why “monogamy” isn't good enough. Am. J. Primatol. 78, 340–354. doi: 10.1002/ajp.22412
Thrall, P. H., Antonovics, J., and Bever, J. D. (1997). Sexual transmission of disease and host mating dystems: within-season reproductive success. Am. Nat. 149, 485–506. doi: 10.1086/286001
Trivers, R. L. (1972). “Parental investment and sexual selection,” in Sexual Selection and the Descent of Man, ed B. Campbell (London: Heinemann), 136–179.
Varian-Ramos, C. W., and Webster, M. S. (2012). Extrapair copulations reduce inbreeding for female red-backed fairy-wrens, Malurus melanocephalus. Anim. Behav. 83, 857–864. doi: 10.1016/j.anbehav.2012.01.010
Wade, M. J., and Shuster, S. M. (2002). The evolution of parental care in the context of sexual selection: a critical reassessment of parental investment theory. Am. Nat. 160, 285–292. doi: 10.1086/341520
Wagenmakers, E. J., and Farrell, S. (2004). AIC model selection using Akaike weights. Psychon. Bull. Rev. 11, 192–196. doi: 10.3758/BF03206482
Wagner, R. H. (1992). Extra-pair copulations in a lek: the secondary mating system of monogamous razorbills. Behav. Ecol. Sociobiol. 31, 63–71. doi: 10.1007/BF00167817
Ward, A., and Webster, M. (2016). Sociality: The Behaviour of Group-Living Animals. Basel: Springer International Publishing.
Westneat, D. F., and Sherman, P. W. (1997). Density and extra-pair fertilizations in birds: a comparative analysis. Behav. Ecol. Sociobiol. 41, 205–215. doi: 10.1007/s002650050381
Westneat, D. F., Sherman, P. W., and Morton, M. L. (1990). The ecology and evolution of extra-pair copulations in birds. Curr. Ornithol. 7, 331–369.
Westneat, D. F., and Stewart, I. R. K. (2003). Extra-pair paternity in birds: causes, correlates, and conflict. Annu. Rev. Ecol. Evol. Syst. 34, 365–396. doi: 10.1146/annurev.ecolsys.34.011802.132439
Weston Glenn, J. L., Civitello, D. J., and Lance, S. L. (2009). Multiple paternity and kinship in the gray fox (Urocyon cinereoargenteus). Mamm. Biol. 74, 394–402. doi: 10.1016/j.mambio.2008.10.003
Wickler, W., and Seibt, U. (1983). “Monogamy: an ambiguous concept,” in Mate Choice, ed P. Bateson (New York, NY: Cambridge University Press), 33–50.
Wittenberger, J. F., and Tilson, R. L. (1980). The evolution of monogamy: hypotheses and evidence. Annu. Rev. Ecol. Syst. 11, 197–232. doi: 10.1146/annurev.es.11.110180.001213
Wolff, J. O., and MacDonald, D. W. (2004). Promiscuous females protect their offspring. Trends Ecol. Evol. 19, 127–134. doi: 10.1016/j.tree.2003.12.009
Keywords: genetic monogamy, social monogamy, extra-pair paternity, mammals, paternal care, social structure
Citation: Lambert CT, Sabol AC and Solomon NG (2018) Genetic Monogamy in Socially Monogamous Mammals Is Primarily Predicted by Multiple Life History Factors: A Meta-Analysis. Front. Ecol. Evol. 6:139. doi: 10.3389/fevo.2018.00139
Received: 14 May 2018; Accepted: 24 August 2018;
Published: 19 September 2018.
Edited by:
Matjaz Kuntner, National Institute of Biology (NIB), SloveniaReviewed by:
Peter L. Hurd, University of Alberta, CanadaJames Dale, Massey University, New Zealand
Copyright © 2018 Lambert, Sabol and Solomon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nancy G. Solomon, c29sb21vbmdAbWlhbWlvaC5lZHU=
 Connor T. Lambert
Connor T. Lambert Anne C. Sabol
Anne C. Sabol Nancy G. Solomon
Nancy G. Solomon