- 1Advanced DNA, Identification and Forensic Facility, The Environment Institute, Faculty of Sciences, University of Adelaide, Adelaide, SA, Australia
- 2School of Biological Sciences, University of Western Australia, Perth, WA, Australia
- 3CSIRO Land and Water, Floreat, WA, Australia
- 4Department of Environment and Water, State Herbarium of South Australia, Adelaide, SA, Australia
- 5Biodiversity and Conservation Science, Department of Biodiversity, Conservation and Attractions, Kensington, WA, Australia
- 6School of Life and Environmental Sciences, University of Sydney, Sydney, NSW, Australia
- 7Centre for Australian National Biodiversity Research (CSIRO), Canberra, ACT, Australia
- 8ARC Centre for Mine Site Restoration, School of Molecular and Life Sciences, Curtin University, Perth, WA, Australia
- 9Genecology Research Centre, University of the Sunshine Coast, Maroochydore, QLD, Australia
Building DNA barcode databases for plants has historically been ad hoc, and often with a relatively narrow taxonomic focus. To realize the full potential of DNA barcoding for plants, and particularly its application to metabarcoding for mixed-species environmental samples, systematic sequencing of reference collections is required using an augmented set of DNA barcode loci, applied according to agreed data generation and analysis standards. The largest and most complete reference collections of plants are held in herbaria. Australia has a globally significant flora that is well sampled and expertly curated by its herbaria, coordinated through the Council of Heads of Australasian Herbaria. There exists a tremendous opportunity to provide a comprehensive and taxonomically robust reference database for plant DNA barcoding applications by undertaking coordinated and systematic sequencing of the entire flora of Australia utilizing existing herbarium material. In this paper, we review the development of DNA barcoding and metabarcoding and consider the requirements for a robust and comprehensive system. We analyzed the current availability of DNA barcode reference data for Australian plants, recommend priority taxa for database inclusion, and highlight future applications of a comprehensive metabarcoding system. We urge that large-scale and coordinated analysis of herbarium collections be undertaken to realize the promise of DNA barcoding and metabarcoding, and propose that the generation and curation of reference data should become a national investment priority.
Introduction
The concept of utilizing DNA for large-scale identification purposes was first seriously proposed by Hebert et al. (2003), who considered DNA barcoding (hereafter “barcoding”) to be the only serious option for widespread taxonomic identification and description in the face of collapsing taxonomic expertise and the dramatic rate of biodiversity loss. Since then, barcoding has been a growing area of research (Figure 1) with over 6,000 papers published at the time of writing. In the years that followed Herbert et al.'s seminal paper, vigorous debate in the literature continued regarding the potential role of barcoding and its relationship with taxonomy. Serious concerns were raised that barcoding could be touted as a replacement for traditional taxonomic research, resulting in further erosion of funding and recognition of the intrinsic value of biological reference collections, their curation, and the taxonomic foundational nature of the studies upon which most other biological sciences are built (Ebach and Holdrege, 2005a,b; Gregory, 2005; Hebert and Gregory, 2005; Schindel and Miller, 2005; Will et al., 2005; de Carvalho et al., 2007; Miller, 2007). There has been greater recognition recently of the need for barcoding initiatives to be built on strong and dynamic taxonomic foundations in order to provide reliable identification capabilities (Collins and Cruickshank, 2013; Pečnikar and Buzan, 2014). Museums and herbaria are well placed to lead the development of such high quality barcoding reference libraries, utilizing the curated specimens they house (Puillandre et al., 2012). Indeed, the prevailing modern view amongst herbaria is that twentyfirst century collections institutions should be leading participants and collaborators in “big” data initiatives (Funk, 2018).
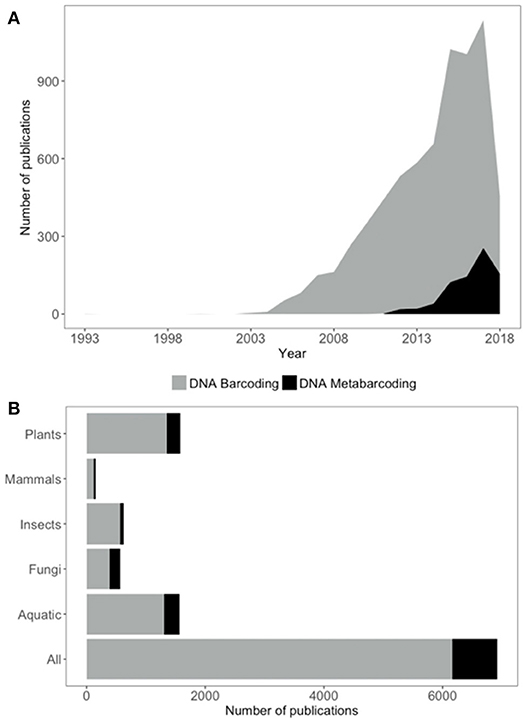
Figure 1. Publication trends in DNA barcoding and metabarcoding from ISI Web of Science. (A) Number of publications per year for DNA barcoding (gray) and DNA metabarcoding (black). (B) Number of DNA barcoding (gray) and metabarcoding (black) publications for different groups of organisms. Database searches undertaken 6th June 2018. See Supplementary Data for exact search terms used.
DNA metabarcoding (hereafter “metabarcoding”) is a special case of barcoding applied to samples that contain more than one organism. Metabarcoding utilizes the same reference databases as barcoding, but allows identification of taxa from mixed samples by using high throughput sequencing methods (Taberlet et al., 2012; Cristescu, 2014). Metabarcoding is an emerging area of research with over 600 papers at the time of writing (Figure 1). The potential of metabarcoding applications is expansive, with the prospect of rapidly determining the species composition of virtually any sample. From environmental monitoring (Kimmerling et al., 2018; Yan et al., 2018) to diet analysis (Xiong et al., 2017; Buglione et al., 2018; Robeson et al., 2018), enhanced biosecurity capabilities (Borrell et al., 2017) and detection of illegal trade (Arulandhu et al., 2017; de Boer et al., 2017), metabarcoding could provide cost-effective and reliable identification for a wide range of regulatory, conservation and commercial purposes. However, the practical applications of both barcoding and metabarcoding are often hampered by the absence of quality reference databases on which to base identifications.
Of all the taxonomic groups to which barcoding and metabarcoding methods are applied, identification of plants is the most common (Figure 1). Identification of aquatic organisms is also well represented in the literature, although the taxonomic breath of this group is obviously large and potentially overlapping with other groups. Identification of insects, fungi, and mammals are all less well represented with fewer than half the number of papers compared to those that address plants or aquatic organisms (Figure 1). Despite this appetite for genetic plant identification, systematic development of barcoding reference databases that comprise whole (or majority) flora of particular regions are sparse (but see de Vere et al., 2012; Saarela et al., 2013; Kuzmina et al., 2017). Instead, most studies develop partial reference databases and/or utilize genetic repositories such as GenBank to augment their resources which then requires post hoc data quality control (Leontidou et al., 2018; e.g., Kumar et al., 2018). The responsibility for accurate initial specimen identification and sequence data quality lies solely with the data generator for standard submissions to GenBank and there is often very little information upon which veracity can be assessed (e.g., voucher specimen meta-data, raw sequencing data files). Reliance on data that has not been derived from well curated and vouchered specimens poses risks of misidentification, an issue that is well recognized for GenBank data (Crocetta et al., 2015; Smith et al., 2016; Balakirev et al., 2017). A recent study of fish barcode sequences found evidence for potential errors in ~4% of sequences (Li et al., 2018). Other data repositories, such as the Barcode of Life Data system (BOLD) (Ratnasingham and Hebert, 2007), address this issue through the requirement for more complete meta-data and appropriately vouchered specimens before sequence data can be accepted. However, due to these stricter requirements, the number of species included in BOLD is far fewer than that represented in other repositories with less rigorous requirements. Recent estimates suggest that BOLD currently contains some barcoding information for ~20% of described land plants and, at the current rate of addition, it will take >64 years for the database to be complete (Wilkinson et al., 2017) without consideration of undescribed species.
The flora of Australia contains over 23,000 formally recognized species and is characterized by several biodiversity hotspots and high species and genus level endemism (Crisp et al., 1999; Costion et al., 2015; Keith and Tozer, 2017). It is well sampled and expertly curated across the nation's herbaria, coordinated through the Council of Heads of Australasian Herbaria (CHAH). To date, most DNA barcoding publications in Australia deal with a specific taxon of interest, e.g., Acacia (Nevill et al., 2013), Poaceae (Birch et al., 2017), with very few systematically approaching the task for specific regions in conjunction with herbaria (but see Costion et al., 2011; Shapcott et al., 2015, 2017; Howard et al., 2016). We posit that there now exists a tremendous opportunity to augment existing barcoding initiatives in Australia and undertake coordinated and systematic sequencing of the entire flora, harnessing the vast collections of herbarium specimens (including vouchered tissues in silica ready for DNA extraction) to provide a comprehensive and taxonomically robust reference database for plant barcoding and metabarcoding applications.
In the remainder of this paper, we consider the technical advances that now make large-scale herbarium sampling the most cost-effective approach for database development and identify priorities for additional specimen and sequence collection to improve the taxonomic coverage and local completeness of reference databases. We also highlight future applications of a comprehensive barcoding and metabarcoding system for Australian plants.
Approaches for Barcoding and Metabarcoding in Plants
The issue of appropriate barcoding loci is a particular challenge for plants, where the mitochondrial cytochrome c oxidase 1 (CO1) locus used almost ubiquitously in animals with great success (but see Deagle et al., 2014), fails to provide useful resolution due to the generally slow rate of mitochondrial evolution in plants (Chase et al., 2007; Erickson et al., 2008). The challenge is threefold: (1) Selection of (an) appropriately variable marker(s) to allow a standardized barcoding approach that provide(s) sufficient resolution to distinguish species; (2) Selection of an appropriately short and variable marker to allow a standardized metabarcoding approach; and (3) Development of protocols for barcoding reference database building that are robust to advances in 1 and 2 above. Here we consider each of these challenges in turn.
Barcoding Approaches
A selection of standard barcoding loci for plants was made in 2009 by the Consortium for the Barcode of Life (CBOL) Plant Working Group which recommended that rbcL and matK be the loci of choice, plus other plastid or DNA markers as required. The decision was made with recognition that these two loci can only distinguish ~70% of species and represent a trade off in terms of universality, sequence quality, discriminatory power and cost of application (CBOL Plant Working Group, 2009). While rbcL and matK remain the standard, a number of other loci are now also routinely used in plant barcoding studies in order to improve resolution in the specific taxon of interest (Kress and Erickson, 2007; Fazekas et al., 2008; Vijayan and Tsou, 2010; Hollingsworth et al., 2011; Li et al., 2015), ITS is recommended as a third core locus for seed plants (China Plant Bol Group et al., 2011) and psbA-trnH is the most commonly used additional locus (Kress and Erickson, 2012). A number of cpDNA loci have been found to be variable within species in Australia (Byrne and Hankinson, 2012) and elsewhere (Dong et al., 2012; Shaw et al., 2014). These loci have also proved valuable in assessment of relationships at lower taxonomic levels and have been used to distinguish closely related species and intraspecific taxa (Shepherd et al., 2013; Anderson et al., 2016). Ultimately, as sequencing technology continues to advance, larger and more numerous genomic regions can be applied to barcoding to improve specimen identification capabilities for specific purposes (e.g., Tonti-Filippini et al., 2017). In particular, developments that facilitate the discovery and application of more informative nuclear loci will have a greater role to play, due to the limits of species resolution within the plastid genomes that currently dominate barcoding and metabarcoding loci thanks mainly to their relative abundance in specimen tissues (Hollingsworth et al., 2016).
Metabarcoding Approaches
Metabarcoding relies on determining unique taxa from mixed samples and so presents some specific requirements in terms of locus selection. To be truly universal, metabarcoding would require the inclusion of multiple loci, each optimized to capture species-level resolution in different taxa (e.g., CO1 for fauna, matK/rbcL for plants, ITS for fungi). However, whereas multiple loci sequence data can be combined for analysis of individual specimens in barcoding to improve taxonomic resolution (e.g., Li et al., 2017), the single bulk sample used in metabarcoding prohibits such treatment. Instead where multiple loci are utilized in metabarcoding, their results must be analyzed independently then combined to produce a list of likely taxa in the sample (Arulandhu et al., 2017). Further, variation in primer binding sites around metabarcoding loci often require the inclusion of multiple primer sets that are optimized to amplify various taxa, and mismatching of primer binding sites within a sample can lead to species detection failure (e.g., Miya et al., 2015).
Locus selection must also be mindful of the quality of DNA likely to be recovered from a sample. In samples where target organisms are likely to be alive, such as the microorganisms in soil, good quality DNA can be anticipated and hence longer loci employed. However, where remnants of organisms are the likely DNA source (such as trace plant or animal material in the environment), DNA is likely to be degraded and hence present challenges to the successful amplification of long loci. There are additional restrictions from the sequencing platform maximum read length, which is typically shorter than some of the longer barcoding loci (e.g., matK) meaning only partial sequences can be obtained. Therefore, short but highly informative loci are optimal for metabarcoding applications, with due consideration given to universality of priming sites within the target taxa (Coissac et al., 2012). Similarly to barcoding, the options for genomic regions to be applied to metabarcoding will expand as sequencing capabilities expand (Porter and Hajibabaei, 2018). There have been a number of modifications of the standard barcoding loci which target shorter, taxonomically informative parts of the sequence in metabarcoding studies especially when using degraded DNA (e.g., García-Robledo et al., 2013; de Vere et al., 2017; Erickson et al., 2017).
Metabarcoding of mixed samples has the potential for additional information to be derived from analyses, namely the quantification of DNA contributors as a proxy for abundance. However, while there can be a correlation between the proportions of sequencing reads and species in a mixture (Kraaijeveld et al., 2015; Evans et al., 2016) this is generally weak and not a reliable indicator of mixture proportions (Smart et al., 2017). There are multiple biases that can impact quantification accuracy; some specific to the loci being used (e.g., copy number in the genome, Angly et al., 2014; primer mismatches, Elbrecht and Leese, 2015; Piñol et al., 2015; Deiner et al., 2017) and some independent of loci choice (e.g., DNA isolation methods, Brooks et al., 2015; polymerase sequence affinity, Nichols et al., 2018; sampling approaches, Deiner et al., 2017). Various methods have been proposed to control for these biases to better facilitate accurate quantification of relative abundance from metabarcoding data, including the utilization of degenerate primers (Krehenwinkel et al., 2017a,b), incorporation of gene copy number information (Kembel et al., 2012) and the inclusion of control species to generate relative correction factors (Thomas et al., 2016). We anticipate that methods for more accurate abundance quantification through metabarcoding will be available in the coming years through further expansion of metabarcoding loci, improved understanding of biases, and the use of PCR-free sequencing approaches (Liu et al., 2016).
Requirements for Database Building
Given the variety of loci that have been targeted in barcoding and metabarcoding studies, along with the future capacity for further expansion in the breadth of genomic regions used, there is pressure for database-building approaches to maximize their future utility. Most of the effort in reliable database formation is in the collection and curation of taxonomically robust reference specimens, and their sampling for DNA analysis. Sequencing costs, although by no means trivial, are a minor component by comparison. Hence, there is great value in ensuring that the process of collection and sampling does not have to be repeated each time a new locus is optimized for barcoding or metabarcoding purposes. Both silica-dried DNA vouchers (Gaudeul and Rouhan, 2013) and extracted DNA banks (Hodkinson et al., 2007) are expected to become increasingly valued for this reason, thus having central locations for curation and storage of these resources is likely to facilitate more efficient database-building. There are well documented protocols available for how to create and curate DNA barcode reference libraries (Kress and Erickson, 2012). Future databases will ideally seek to employ sequencing methods that provide the most information (whilst maintaining cost efficiency and practicality), such as various whole-genome sequencing and target-enrichment approaches (Hollingsworth et al., 2016; Tonti-Filippini et al., 2017; Porter and Hajibabaei, 2018).
Herbaria as the Natural Home for Plant DNA Databases
We suggest that large-scale plant-barcoding reference-database building is highly relevant to the core business of all herbaria, due to their unique positioning as repositories of the world's flora. A recent review by a leading collections institution botanist states; “Collections are a gold mine of both information and tissue samples and more advances are on the way” (Funk, 2018). Effective plant barcoding and metabarcoding applications require the following with respect to reference databases: (1) Inclusion of multiple individuals from a broad suite of taxa to encompass a high proportion (if not all) of the possible species and to capture intraspecific diversity; (2) Accurate taxonomic identification of the included reference specimens; (3) Curation of reference specimens to retain as a resource for further study; (4) Availability of suitable material for destructive sampling for DNA extraction; (5) Extraction of DNA for sequencing and archiving; (6) Sequencing of a broad range of genomic regions according to standard protocols; (7) Data analysis according to standard protocols and meta-data sharing nomenclature; (8) Data archiving according to standard protocols; (9) Data fit for purpose, including ongoing data curation, and easily available for broad utility. We consider that points 1–4, which deal with the collection and curation of voucher material, are already fulfilled by herbaria. Point 5 considers the extraction and storage of DNA, which is also currently undertaken by many herbaria or in conjunction with research partners. Points 6–9, are concerned with data generation, analysis, storage and availability, which we contend should be further developed through consensus as part of large-scale and coordinated analyses of herbarium collections in conjunction with participating research organizations such as universities.
Collection and Curation of Physical Samples
Herbaria exist primarily to collect and help preserve the world's flora and interpret taxonomic relationships (point 1), but herbarium data also has a wide breadth of other important applications (Heberling and Isaac, 2017; James et al., 2018). It can be argued that no identification can be guaranteed to be 100% accurate (point 2) but that is the purpose of ongoing curation (point 3), to facilitate corrections and updates to taxonomic designations of specimens (Funk et al., 2005) and provide the means to avoid or correct the “error cascades” that can result from misidentification of reference materials (Bortolus, 2008).
One of the major reasons that herbaria have not historically been the automatic go-to place for barcoding reference specimens is the availability of material suitable for DNA extraction. The issue is twofold; firstly, herbaria are understandably reluctant to allow extensive destructive sampling of their often irreplaceable and finite resources (although it is usually permitted under controlled conditions, except in some special circumstances such as with type specimens or where destructive sampling would damage the future utility of the specimen); secondly, the methods of collection and preservation (plus the length of time since collection) of herbarium specimens generally leaves the DNA that can be extracted in various stages of degradation. Degraded DNA is usually difficult to barcode due to short fragment lengths, an issue of greatest concern with the longer DNA barcodes.
Several advances have now been made to largely overcome the above concerns. Firstly, it is now common practice for herbaria to include material suitable for DNA extraction in their field collection protocols; leaf or other suitable tissue can now be routinely used for DNA extraction without compromising the integrity of voucher specimens (Gaudeul and Rouhan, 2013; Funk et al., 2017). These tissues are also stored in a way that preserves the tissue for high quality DNA extraction, usually rapidly desiccated and stored on silica gel (point 4). In other special cases, extraction methods have been developed that avoid destructive sampling (Shepherd, 2017). Further, advances in sequencing approaches have reduced the requirements for long unfragmented sections of high molecular weight DNA (Hart et al., 2016; Hollingsworth et al., 2016) making herbarium material progressively more suitable for DNA extraction. However, some preservation methods used for herbarium specimens (particularly in the tropics where humidity often prevents efficient and continual desiccation) have a negative impact on plant DNA quality, such as the use of ethanol and formalin (Staats et al., 2011). These issues make historic collections preserved in this way more unsuitable for DNA extractions. Despite improvements in extraction and sequencing of DNA from herbarium specimens, there remain issues with potential sequence modification and sequencing errors that need to be understood and accounted for in downstream analyses (Staats et al., 2011; Weiß et al., 2016). Archiving of silica-dried leaf material for DNA extraction, as well as extracted DNA samples, are also increasingly routine procedures in herbaria and associated research organizations where appropriate long-term storage facilities are available (Seberg et al., 2016) (points 4 and 5).
Generation and Curation of Data
There is no global consensus on the best technical methods for generating and analyzing plant sequence data for building large-scale barcoding reference databases. Advances in technology will continue to reduce sequencing costs and increase the sequence coverage, hence adopting a single rigid sequencing approach is probably unwise. There is an opportunity for consortia of herbaria and other research institutions to develop and implement local standard protocols for sequencing and data analysis in the pursuit of complete regional flora databases (e.g., Kress and Erickson, 2012; Kuzmina et al., 2017) (points 6 and 7).
Appropriate archiving and publication to allow broad re-use of the data are important for effective plant DNA barcode databases (Wilkinson et al., 2016) (points 8 and 9). Currently, archiving of molecular data is routinely undertaken by partners of the International Nucleotide Sequence Database Collaboration (INSDC) (Karsch-Mizrachi et al., 2018), which have a variety of database products suitable for different data types. BOLD also accepts barcoding reference data and integrates with GenBank, with a particular identifier included to indicate that the sequence data meets the stricter barcode criteria with respect to reference specimen taxonomy and curation. With barcode data retrieved from existing herbarium collections, there is also the opportunity to link to the great wealth of information already collected and made available on specimens through national data portals such as the Australasian Virtual Herbarium (AVH; avh.chah.org.au), the Atlas of Living Australia (ALA; ala.org.au), and Canadensys (canadensys.net). Other international networks include the Global Genome Biodiversity Network (GGBN; ggbn.org) and the Global Biodiversity Information Facility (GBIF; gbif.org). The challenge lies in developing synergies between these offerings to ensure that data links remain dynamic and updates to taxonomy propagate throughout the system. DNA sequence data also requires curation, not just the specimens themselves. The release of the GGBN Data Standard specification (Droege et al., 2016) goes some way to supporting that end. In 2017, the GGBN conducted a public consultation on their proposed set of augmented terms dedicated to high throughput sequence library preparation, the results of which are yet to be released at the time of writing. International data standards for biodiversity information are developed and maintained by Biodiversity Information Standards, also known as the Taxonomic Databases Working Group, (TDWG; tdwg.org) which provides a technical framework for developing appropriate data sharing nomenclature. We anticipate that there will be further developments in public access portals over the coming years that will offer more fit-for-purpose solutions for archiving and accessing complex genomic data derived from reference specimens, the practical requirements of which will be best understood through collaborative efforts to undertake large-scale reference database generation.
Applications of Barcoding and Metabarcoding Based on Comprehensive Floral Reference Databases—an Australian Perspective
The practical applications of an accurate and affordable barcoding and metabarcoding system in Australia are substantial. Its development would provide many benefits through a concerted effort of national science infrastructure building. Here, we review the DNA reference data currently available through GenBank for Australian plant taxa and present a range of potential applications of a comprehensive floral DNA reference database, with both economic and environmental benefits to Australia. This provides a basis for the development of a comprehensive barcoding reference database for Australian plants.
DNA Reference Data for the Australian Flora
We used the Australian Plant Census (anbg.gov.au/chah/apc; accessed 21/3/2018 and 24/07/2018) to generate a list of accepted plant species names for Australian Embryophyta. We used the Matrix Maker script (Freyman and Thornhill, 2016) to search GenBank (searches conducted April and July 2018) against the resultant list of Australian plant names for seven commonly used barcoding loci (matK, rbcL, ITS, ndhF, trnL-trnF, atpB-rbcL, and psbA-trnH).
We found that 10,779 of 23,057 (47%) of Australian species listed in the Australian Plant Census (current taxa) had no DNA barcoding data available for any of the seven loci evaluated, and 44% have no data on GenBank of any kind. Only 14% of species had data for all three of the core loci (matK, rbcL, ITS). At the family level, 11 of 360 families (3%) had no data at all for any of the seven loci, with 130 families (36%) containing no data on the three core loci. For the largest 43 families, which each contain >100 species and together represent 78% of the flora of Australia, 50% of species had no data for the seven loci and only 12% of species had data for all three core loci. Results can be found in Supplementary Material. From our GenBank survey data we make recommendations for prioritizing taxa for future barcoding database-building initiatives in Australia to improve identification capacity (Table 1).

Table 1. Recommendations for priority taxa for barcode database building in Australia, based on a survey of data available in GenBank against the Australian Plant Census.
Application to Biodiversity Monitoring, Conservation, and Management
Biodiversity monitoring provides critical baseline data that enables change detection, and elucidation of temporal patterns in ecosystem dynamics (Lindenmayer and Gibbons, 2012; Schmeller et al., 2017). Australia has recognized the need for collection and curation of this science infrastructure, realized by the ongoing support of the Terrestrial Ecosystem Research Network (TERN; tern.org.au), which collects a wide variety of data and physical samples to enable characterization of Australian ecosystems and collaborates closely with CHAH to ensure taxonomic integrity of their data. Accurate and comprehensive monitoring programs are extremely resource intensive, as such there is an opportunity to augment these initiatives with more rapid methods for biodiversity assessment (Thomsen and Willerslev, 2015; Bonin et al., 2018), and indeed to validate the utility of these newer methods using comprehensive data collected from traditional monitoring approaches (Aylagas et al., 2016). In Australia, the availability of a comprehensive DNA barcoding reference database for plants would pave the way for more accurate and cost-efficient plant biodiversity monitoring tools to augment existing programs (Thompson and Newmaster, 2014).
Identification of plant species is also a fundamental component of conservation and management planning (Margules and Pressey, 2000) and of assessing potential impacts of development and changing land use (Glasson and Therivel, 2013). These activities have traditionally used standard morphological taxonomic approaches, although these can be problematic where taxonomic expertise is limited, for species that are difficult to distinguish using morphological characters, or where surveys need to be conducted at particular times of the year (e.g., when species are flowering and/or fruiting) (Hollingsworth et al., 2016). Barcoding of floras can establish a reference database that provides a basis for molecular identification of plant species at any time of the year and for small tissue samples. A project is underway to commence a reference database of species in the Pilbara region of Western Australia, an important area of economic development and assessment of flora for environmental impact assessment. Molecular identification of plants using barcoding has the potential to provide a timely and cost-effective approach for application in conservation planning and environmental impact assessment.
Application to Pollination Services
A variety of animals, predominantly insects, but also some birds, reptiles and mammals, provide pollination services for the majority of wild and cultivated plants (Potts et al., 2016; Hung et al., 2018). In Australia, some 65% of crops rely on the European honeybee (Apis melifera) for pollination (Keogh et al., 2010) and many likely also benefit from pollination from native pollinators (Garibaldi et al., 2013). Understanding pollination networks is key for optimizing the pollination services provided in both agricultural and natural settings and for examining the return of functionality to restored sites; by determining what floral resources are being utilized by pollinators, it is possible to develop strategies to maximize a desired outcome. For example, retaining or planting vegetation that can sustain pollinators between cropping seasons in order to maximize pollination services during crop flowering season (Asbjornsen et al., 2014).
Revealing pollination networks has traditionally relied on labor intensive observation studies, which record pollinator visitations to particular plant species. As pollination involves the physical movement of pollen from one plant to another via a pollinator, floral resource use can also be investigated by analyzing pollen collected by pollinators. This approach has been shown to better characterize pollination networks, revealing more interactions than visitation data alone (Bosch et al., 2009). Morphological identification of pollen through microscopy can be undertaken but requires significant time and skill, and taxonomic resolution is also generally limited to the genus or (more commonly) family level. Metabarcoding of collected pollen offers the opportunity to quickly determine the floral resources visited by pollinators and reveal pollination network associations (Bell et al., 2017). Metabarcoding has been shown to be capable of identifying more taxa than pollen morphological identification, particularly for infrequently detected species (Keller et al., 2015; Richardson et al., 2015b; Smart et al., 2017) as well as more connected plant-pollinator interaction networks (Pornon et al., 2017).
Identification to the species level relies on sufficiently complete barcode reference databases for plants (Cornman et al., 2015; Galliot et al., 2017; Pornon et al., 2017). Opportunities for quantifying pollen carriage using metabarcoding have also been investigated, with strong indications that the number of sequencing reads are correlated with taxon abundance (Keller et al., 2015; Richardson et al., 2015a; Pornon et al., 2016). These data are of great interest in pollination network research as they reveal the relative contributions of various pollen resources to pollinator diets and help quantify the importance of particular pollinators to plant species. However, further work is required to understand the limitations of the approach for accurate quantitative analysis of pollen loads (Bell et al., 2017). Accurate and comprehensive data on pollinator resource utilization in Australia would help in the design and implementation of strategies to secure pollination services for crops, and further incorporate the protection and promotion of pollination services in ecological restoration and conservation practices.
Application to Trade Monitoring and Enforcement
Understanding the biological components of traded materials is critical in order to uphold regulations regarding the legality, safety and legitimacy of products. The potential applications of barcoding and metabarcoding here are many, yet there has been very little utilization of them to date within Australia. This is in part due to the lack of comprehensive and reliable reference data. Plant barcoding and metabarcoding can be used to verify the identification of traded ornamental and agricultural plants, including timber (Dormontt et al., 2015; Ng et al., 2016; Staats et al., 2016), to detect the importation or movement of invasive species (Scriver et al., 2015; Xu et al., 2018), and determine the contents of botanical products such as traditional medicines (Newmaster et al., 2013; Seethapathy et al., 2015; Xin et al., 2015). Barcoding represents a potential additional tool for border security purposes as well as for businesses who could also utilize barcoding and metabarcoding services to internally verify compliance and demonstrate due diligence (Lowe et al., 2016).
Other Applications
In addition to the applications described above, this section briefly describes some additional applications that have been demonstrated in the international literature which could prove useful in the Australian context. Barcoding and metabarcoding of pollen and plant fragments have been utilized to assist in forensic applications where determining geographic origin is important (Ferri et al., 2009; Bell et al., 2016). Metabarcoding can be applied to the analysis of gut contents, enabling dietary analysis of animals. This application can assist in determining the threat posed to species from herbivory from certain animals (e.g., Nakahara et al., 2015; Nichols et al., 2016), as well as enabling the dietary requirements and food webs of threatened animal species and communities to be determined in order to facilitate both ex situ and in situ conservation efforts (De Barba et al., 2014; Srivathsan et al., 2015). The application of metabarcoding methods to ancient sedimentary DNA can also help elucidate historical ecosystem dynamics (Thomsen and Willerslev, 2015; Parducci et al., 2017) and has been shown to perform better than pollen analysis (Alsos et al., 2016; Niemeyer et al., 2017) for species richness estimation. Finally, metabarcoding of air samples has been demonstrated as effective in assessing pollen loads and therefore allergen risks, and has potential human health implications (Kraaijeveld et al., 2015). All of these broad applications of barcoding and metabarcoding would be facilitated by the availability of comprehensive barcode reference databases, particularly of plants.
Large-Scale and Coordinated Analyses of Herbarium Collections in Australia
DNA barcoding has developed at an accelerating pace over the last 15 years, with rapid improvements in information content thanks to high throughput sequencing approaches, determination of standard (core) loci, the building of reference databases, and the determination of community standards. However, plant barcoding in Australia is currently ad hoc, with no publicly available reference data for almost half of all Australian plant species. We recommend that a consortium of herbaria and other research institutions is formed with the explicit aim of developing a comprehensive DNA barcode reference database for the entire flora of Australia, starting with the plant families identified in Table 1. We further recommend the consortium pursue integrated funding opportunities that leverage existing resources, given the success of this consortium approach by Australian herbaria in other initiatives such as the AVH and ALA. Exemplar specimens from each species could be selected from existing collections or sourced through targeting sampling campaigns to capture both inter and intra specific species variation and geographic breadth. DNA extractions and sequencing could be undertaken in specialist partner laboratories in high volumes to maximize efficiencies and minimize methodological variation. Data analysis and curation could be centrally managed to ensure comprehensive and consistent data availability.
The applications for a robust and comprehensive plant barcoding system in Australia are diverse, and represent a range of potential benefits both economically and ecologically. From improved pollination services for agriculture, to more efficient monitoring of ecosystems, greater protection of threatened species, increased capacity for detection of invasive species and assurance of biological product authenticity, barcoding and metabarcoding can provide access to unprecedented amounts of biological information regarding the Australian environment and how we utilize and interact with it. In order to realize these benefits, we argue that large scale and coordinated analysis of herbarium collections are required to ensure the most cost-effective and taxonomically robust system be available to the community in perpetuity. The generation and curation of DNA reference data should become a national investment priority and recognized as essential science infrastructure.
Data Availability Statement
The dataset for this study can be found in the Supplementary Material.
Author Contributions
All authors were involved in manuscript inception. ED drafted the manuscript. ED, EB, and MW carried out data analysis. FE-V prepared the figure. All authors helped revise the manuscript and gave final approval for publication.
Funding
This work was supported by funding from the Australian Government Department of Agriculture and Water Resources as part of its Rural R&D for Profit program. PN was supported partly by the Australian Government through the Australian Research Council Industrial Transformation Training Centre for Mine Site Restoration (project number ICI150100041). FE-V was supported by CSIRO FSP Environomics.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The genesis of this paper came from discussions at the National Pollination DNA Barcoding Workshop held at the University of Adelaide in November 2017. The authors thank Ainsley Calladine of the State Herbarium of South Australia for assistance in comparison of GenBank data with the Australian Plant Census current taxonomy, and three reviewers whose constructive comments improved the manuscript. Finally, the authors would like to acknowledge the many and varied herbarium staff whose tireless work contributes to most botanical papers without sufficient recognition.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00134/full#supplementary-material
Supplementary Material. Excel file containing data for the paper. GenBank APC (first tab) is a table of the number of species within families of Australian plants (obtained from the Australian Plant Census) that have data on GenBank for seven barcoding loci (ITS, matK, rbcL, trnL-trnF, ndhF, atpB-rbcL, psbA-trnH). Data field descriptions (second tab) describes the field names used in GenBank APC in more detail. WoS Barcoding (third tab), WoS Metabarcoding (fourth tab) and WoS Groups (fifth tab) provide the data retrieved from Web of Science used to generate Figure 1 in the paper.
References
Alsos, I. G., Sjögren, P., Edwards, M. E., Landvik, J. Y., Gielly, L., Forwick, M., et al. (2016). Sedimentary ancient DNA from Lake Skartjørna, Svalbard: assessing the resilience of arctic flora to Holocene climate change. Holocene 26, 627–642. doi: 10.1177/0959683615612563
Anderson, B. M., Barrett, M. D., Krauss, S. L., and Thiele, K. (2016). Untangling a species complex of arid zone grasses (Triodia) reveals patterns congruent with co-occurring animals. Mol. Phylogenet. Evol. 101, 142–162. doi: 10.1016/j.ympev.2016.05.014
Angly, F. E., Dennis, P. G., Skarshewski, A., Vanwonterghem, I., Hugenholtz, P., and Tyson, G. W. (2014). CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2:11. doi: 10.1186/2049-2618-2-11
Arulandhu, A. J., Staats, M., Hagelaar, R., Voorhuijzen, M. M., Prins, T. W., Scholtens, I., et al. (2017). Development and validation of a multi-locus DNA metabarcoding method to identify endangered species in complex samples. Gigascience 6:18. doi: 10.1093/gigascience/gix080
Asbjornsen, H., Hernandez-Santana, V., Liebman, M., Bayala, J., Chen, J., Helmers, M., et al. (2014). Targeting perennial vegetation in agricultural landscapes for enhancing ecosystem services. Renew. Agric. Food Syst. 29, 101–125. doi: 10.1017/S1742170512000385
Aylagas, E., Borja, Á., Irigoien, X., and Rodríguez-Ezpeleta, N. (2016). Benchmarking DNA metabarcoding for biodiversity-based monitoring and assessment. Front. Mar. Sci. 3:e00096. doi: 10.3389/fmars.2016.00096
Balakirev, E. S., Saveliev, P. A., and Ayala, F. J. (2017). Complete mitochondrial genomes of the Cherskii's sculpin Cottus czerskii and Siberian taimen Hucho taimen reveal GenBank entry errors: incorrect species identification and recombinant mitochondrial genome. Evol. Bioinformatics 13:7. doi: 10.1177/1176934317726783
Bell, K. L., Burgess, K. S., Okamoto, K. C., Aranda, R., and Brosi, B. J. (2016). Review and future prospects for DNA barcoding methods in forensic palynology. Forensic Sci. Int. Genet. 21, 110–116. doi: 10.1016/j.fsigen.2015.12.010
Bell, K. L., Fowler, J., Burgess, K. S., Dobbs, E. K., Gruenewald, D., Lawley, B., et al. (2017). Applying pollen DNA metabarcoding to the study of plant–pollinator interactions. Appl. Plant Sci. 5:1600124. doi: 10.3732/apps.1600124
Birch, J. L., Walsh, N. G., Cantrill, D. J., Holmes, G. D., and Murphy, D. J. (2017). Testing efficacy of distance and tree-based methods for DNA barcoding of grasses (Poaceae tribe Poeae) in Australia. PLoS ONE 12:22. doi: 10.1371/journal.pone.0186259
Bonin, A., Taberlet, P., Zinger, L., and Coissac, E. (2018). Environmental DNA: For Biodiversity Research and Monitoring. Oxford, UK: Oxford University Press.
Borrell, Y. J., Miralles, L., Do Huu, H., Mohammed-Geba, K., and Garcia-Vazquez, E. (2017). DNA in a bottle-Rapid metabarcoding survey for early alerts of invasive species in ports. PLoS ONE 12:17. doi: 10.1371/journal.pone.0183347
Bortolus, A. (2008). Error cascades in the biological sciences: the unwanted consequences of using bad taxonomy in ecology. Ambio 37, 114–118. doi: 10.1579/0044-7447(2008)37[114:ECITBS]2.0.CO;2
Bosch, J., Martín González, A. M., Rodrigo, A., and Navarro, D. (2009). Plant–pollinator networks: adding the pollinator's perspective. Ecol. Lett. 12, 409–419. doi: 10.1111/j.1461-0248.2009.01296.x
Brooks, J. P., Edwards, D. J., Harwich, M. D., Rivera, M. C., Fettweis, J. M., Serrano, M. G., et al. (2015). The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 15:66. doi: 10.1186/s12866-015-0351-6
Buglione, M., Maselli, V., Rippa, D., De Filippo, G., Trapanese, M., and Fulgione, D. (2018). A pilot study on the application of DNA metabarcoding for non-invasive diet analysis in the Italian hare. Mammalian Biol. 88, 31–42. doi: 10.1016/j.mambio.2017.10.010
Byrne, M., and Hankinson, M. (2012). Testing the variability of chloroplast sequences for plant phylogeography. Austr. J. Bot. 60, 569–574. doi: 10.1071/BT12146
CBOL Plant Working Group (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. 106, 12794–12797. doi: 10.1073/pnas.0905845106
Chase, M. W., Cowan, R. S., Hollingsworth, P. M., Van Den Berg, C., Madriñán, S., Petersen, G, Wilkinson, M., et al. (2007). A proposal for a standardised protocol to barcode all land plants. Taxon 56, 295–299.
Li, D. Z., Gao, L. M., Li, H. T., Wang, H., Ge, X. J., et al. (2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19641–19646. doi: 10.1073/pnas.1104551108
Coissac, E., Riaz, T., and Puillandre, N. (2012). Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol. Ecol. 21, 1834–1847. doi: 10.1111/j.1365-294XX.2012.05550.x
Collins, R. A., and Cruickshank, R. H. (2013). The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 13, 969–975. doi: 10.1111/1755-0998.12046
Cornman, R. S., Otto, C. R., Iwanowicz, D., and Pettis, J. S. (2015). Taxonomic characterization of honey bee (Apis mellifera) pollen foraging based on non-overlapping paired-end sequencing of nuclear ribosomal loci. PLoS ONE 10:e0145365. doi: 10.1371/journal.pone.0145365
Costion, C., Ford, A., Cross, H., Crayn, D., Harrington, M., and Lowe, A. (2011). Plant DNA barcodes can accurately estimate species richness in poorly known floras. PLoS ONE 6:e26841. doi: 10.1371/journal.pone.0026841
Costion, C. M., Edwards, W., Ford, A. J., Metcalfe, D. J., Cross, H. B., Harrington, M. G., et al. (2015). Using phylogenetic diversity to identify ancient rain forest refugia and diversification zones in a biodiversity hotspot. Divers. Distrib. 21, 279–289. doi: 10.1111/ddi.12266
Crisp, M., West, J., and Linder, H. (1999). “Biogeography of the terrestrial flora (section 3),” in Flora of Australia Volume 1 Introduction, eds. A. Orchard and H. Thompson. (Melbourne, VIC: Australian Biological Resources Study/CSIRO Publishing), 321–367.
Cristescu, M. E. (2014). From barcoding single individuals to metabarcoding biological communities: towards an integrative approach to the study of global biodiversity. Trends Ecol. Evol. 29, 566–571. doi: 10.1016/j.tree.2014.08.001
Crocetta, F., Mariottini, P., Salvi, D., and Oliverio, M. (2015). Does GenBank provide a reliable DNA barcode reference to identify small alien oysters invading the Mediterranean Sea? J. Mar. Biol. Assoc. U. K. 95, 111–122. doi: 10.1017/S0025315414001027
de Barba, M., Miquel, C., Boyer, F., Mercier, C., Rioux, D., Coissac, E., et al. (2014). DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Mol. Ecol. Resour. 14, 306–323. doi: 10.1111/1755-0998.12188
de Boer, H. J., Ghorbani, A., Manzanilla, V., Raclariu, A. C., Kreziou, A., Ounjai, S., et al. (2017). DNA metabarcoding of orchid-derived products reveals widespread illegal orchid trade. Proc. R. Soc. B-Biol. Sci. 284:9. doi: 10.1098/rspb.2017.1182
de Carvalho, M. R., Bockmann, F. A., Amorim, D. S., Brandão, C. R. F., De Vivo, M., De Figueiredo, J. L., et al. (2007). Taxonomic impediment or impediment to taxonomy? A commentary on systematics and the cybertaxonomic-automation paradigm. Evol. Biol. 34, 140–143. doi: 10.1007/s11692-007-9011-6
de Vere, N., Jones, L. E., Gilmore, T., Moscrop, J., Lowe, A., Smith, D., et al. (2017). Using DNA metabarcoding to investigate honey bee foraging reveals limited flower use despite high floral availability. Sci. Rep. 7:42838. doi: 10.1038/srep42838
de Vere, N., Rich, T. C., Ford, C. R., Trinder, S. A., Long, C., Moore, C. W., et al. (2012). DNA barcoding the native flowering plants and conifers of Wales. PLoS ONE 7:e37945. doi: 10.1371/journal.pone.0037945
Deagle, B. E., Jarman, S. N., Coissac, E., Pompanon, F., and Taberlet, P. (2014). DNA metabarcoding and the cytochrome c oxidase subunit I marker: not a perfect match. Biol. Lett. 10:20140562. doi: 10.1098/rsbl.2014.0562
Deiner, K., Bik, H. M., Machler, E., Seymour, M., Lacoursiere-Roussel, A., Altermatt, F., et al. (2017). Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895. doi: 10.1111/mec.14350
Dong, W., Liu, J., Yu, J., Wang, L., and Zhou, S. (2012). Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 7:e35071. doi: 10.1371/journal.pone.0035071
Dormontt, E. E., Boner, M., Braun, B., Breulmann, G., Degen, B., Espinoza, E., et al. (2015). Forensic timber identification: it's time to integrate disciplines to combat illegal logging. Biol. Conserv. 191, 790–798. doi: 10.1016/j.biocon.2015.06.038
Droege, G., Barker, K., Seberg, O., Coddington, J., Benson, E., Berendsohn, W. G., et al. (2016). The global genome biodiversity network (GGBN) data standard specification. Database 2016:baw125. doi: 10.1093/database/baw125
Ebach, M. C., and Holdrege, C. (2005a). DNA barcoding is no substitute for taxonomy. Nature 434, 697–697. doi: 10.1038/434697b
Ebach, M. C., and Holdrege, C. (2005b). More taxonomy, not DNA barcoding. BioScience 55, 822–824. doi: 10.1641/0006-3568(2005)055[0823:MTNDB]2.0.CO;2
Elbrecht, V., and Leese, F. (2015). Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass—sequence relationships with an innovative metabarcoding protocol. PLoS ONE 10:e0130324. doi: 10.1371/journal.pone.0130324
Erickson, D. L., Reed, E., Ramachandran, P., Bourg, N. A., Mcshea, W. J., and Ottesen, A. (2017). Reconstructing a herbivore's diet using a novel rbcL DNA mini-barcode for plants. AoB Plants 9:plx015-plx015. doi: 10.1093/aobpla/plx015
Erickson, D. L., Spouge, J., Resch, A., Weigt, L. A., and Kress, J. W. (2008). DNA barcoding in land plants: developing standards to quantify and maximize success. Taxon 57, 1304–1316.
Evans, N. T., Olds, B. P., Renshaw, M. A., Turner, C. R., Li, Y., Jerde, C. L., et al. (2016). Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding. Mol. Ecol. Resour. 16, 29–41. doi: 10.1111/1755-0998.12433
Fazekas, A. J., Burgess, K. S., Kesanakurti, P. R., Graham, S. W., Newmaster, S. G., Husband, B. C., et al. (2008). Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 3:e2802. doi: 10.1371/journal.pone.0002802
Ferri, G., Alù, M., Corradini, B., and Beduschi, G. (2009). Forensic botany: species identification of botanical trace evidence using a multigene barcoding approach. Int. J. Legal Med. 123, 395–401. doi: 10.1007/s00414-009-0356-5
Freyman, W. A., and Thornhill, A. H. (2016). Matrix Maker [Computer software]. Available online at: https://github.com/wf8/matrixmaker
Funk, V. A. (2018). Collections-based science in the 21st Century. J. Syst. Evol. 56, 175–193. doi: 10.1111/jse.12315
Funk, V. A., Gostel, M., Devine, A., Kelloff, C. L., Wurdack, K., Tuccinardi, C., et al. (2017). Guidelines for collecting vouchers and tissues intended for genomic work (Smithsonian Institution): Botany Best Practices. Biodiversity Data J. 2017:e11625. doi: 10.3897/BDJ.5.e11625
Funk, V. A., Hoch, P. C., Prather, L. A., and Wagner, W. L. (2005). The importance of vouchers. Taxon 54, 127–129. doi: 10.2307/25065309
Galliot, J. N., Brunel, D., Berard, A., Chauveau, A., Blanchetete, A., Lanore, L., et al. (2017). Investigating a flower-insect forager network in a mountain grassland community using pollen DNA barcoding. J. Insect Conserv. 21, 827–837. doi: 10.1007/s10841-017-0022-z
García-Robledo, C., Erickson, D. L., Staines, C. L., Erwin, T. L., and Kress, W. J. (2013). Tropical plant–herbivore networks: Reconstructing species interactions using DNA barcodes. PLOS ONE 8:e52967. doi: 10.1371/journal.pone.0052967
Garibaldi, L. A., Steffan-Dewenter, I., Winfree, R., Aizen, M. A., Bommarco, R., Cunningham, S. A., et al. (2013). Wild pollinators enhance fruit set of crops regardless of honey bee abundance. Science 339, 1608–1611. doi: 10.1126/science.1230200
Gaudeul, M., and Rouhan, G. (2013). A plea for modern botanical collections to include DNA-friendly material. Trends Plant Sci. 18, 184–185. doi: 10.1016/j.tplants.2012.12.006
Glasson, J., and Therivel, R. (2013). Introduction to environmental impact assessment. London: Routledge.
Gregory, T. R. (2005). DNA barcoding does not compete with taxonomy. Nature 434, 1067–1067. doi: 10.1038/4341067b
Hart, M. L., Forrest, L. L., Nicholls, J. A., and Kidner, C. A. (2016). Retrieval of hundreds of nuclear loci from herbarium specimens. Taxon 65, 1081–1092. doi: 10.12705/655.9
Heberling, J. M., and Isaac, B. L. (2017). Herbarium specimens as exaptations: New uses for old collections. Am. J. Bot. 104, 963–965. doi: 10.3732/ajb.1700125
Hebert, P. D., Cywinska, A., and Ball, S. L. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Hebert, P. D., and Gregory, T. R. (2005). The promise of DNA barcoding for taxonomy. Syst. Biol. 54, 852–859. doi: 10.1080/10635150500354886
Hodkinson, T. R., Waldren, S., Parnell, J. A.N., Kelleher, C. T., Salamin, K., and Salamin, N. (2007). DNA banking for plant breeding, biotechnology and biodiversity evaluation. J. Plant Res. 120, 17–29. doi: 10.1007/s10265-006-0059-7
Hollingsworth, P. M., Graham, S. W., and Little, D. P. (2011). Choosing and using a plant DNA barcode. PLOS ONE 6:e19254. doi: 10.1371/journal.pone.0019254
Hollingsworth, P. M., Li, D.-Z., Van Der Bank, M., and Twyford, A. D. (2016). Telling plant species apart with DNA: from barcodes to genomes. Phil. Trans. R. Soc. B 371:20150338. doi: 10.1098/rstb.2015.0338
Howard, M. G., Mcdonald, W. J., Forster, P. I., Kress, W. J., Erickson, D., Faith, D. P., et al. (2016). Patterns of phylogenetic diversity of subtropical rainforest of the Great Sandy Region, Australia indicate long term climatic refugia. PLoS ONE 11:e0153565. doi: 10.1371/journal.pone.0153565
Hung, K. L. J., Kingston, J. M., Albrecht, M., Holway, D. A., and Kohn, J. R. (2018). The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B Biol. Sci. 285:8. doi: 10.1098/rspb.2017.2140
James, S. A., Soltis, P. S., Belbin, L., Chapman, A. D., Nelson, G., Paul, D. L., et al. (2018). Herbarium data: Global biodiversity and societal botanical needs for novel research. Appl. Plant Sci. 6:8. doi: 10.1002/aps3.1024
Karsch-Mizrachi, I., Takagi, T., and Cochrane, G. (2018). The international nucleotide sequence database collaboration. Nucleic Acids Res. 46, D48–D51. doi: 10.1093/nar/gkx1097
Keith, D. A., and Tozer, M. G. (2017). “Girt: A continental synthesis of Australian vegetation,” in Australian Vegetation, 3rd revised edn, ed D. A. Keith (Cambridge, UK: Cambridge University Press), 3–39.
Keller, A., Danner, N., Grimmer, G., Ankenbrand, M., Von Der Ohe, K., et al. (2015). Evaluating multiplexed next-generation sequencing as a method in palynology for mixed pollen samples. Plant Biol. 17, 558–566. doi: 10.1111/plb.12251
Kembel, S. W., Wu, M., Eisen, J. A., and Green, J. L. (2012). Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput. Biol. 8:e1002743. doi: 10.1371/journal.pcbi.1002743
Keogh, R., Robinson, A., and Mullins, I. J. (2010). Pollination Aware: The real value of pollination in Australia. Barton: RIRDC.
Kimmerling, N., Zuqert, O., Amitai, G., Gurevich, T., Armoza-Zvuloni, R., Kolesnikov, I., et al. (2018). Quantitative species-level ecology of reef fish larvae via metabarcoding. Nat. Ecol. Evol. 2:306. doi: 10.1038/s41559-017-0413-2
Kraaijeveld, K., Weger, L. A., Ventayol García, M., Buermans, H., Frank, J., Hiemstra, P. S., et al. (2015). Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Mol. Ecol. Resour. 15, 8–16. doi: 10.1111/1755-0998.12288
Krehenwinkel, H., Kennedy, S., Pekár, S., Gillespie, R. G., and Johnston, S. (2017a). A cost-efficient and simple protocol to enrich prey DNA from extractions of predatory arthropods for large-scale gut content analysis by Illumina sequencing. Methods Ecol. Evol. 8, 126–134. doi: 10.1111/2041-210X.12647
Krehenwinkel, H., Wolf, M., Lim, J. Y., Rominger, A. J., Simison, W. B., and Gillespie, R. G. (2017b). Estimating and mitigating amplification bias in qualitative and quantitative arthropod metabarcoding. Sci. Rep. 7:17668. doi: 10.1038/s41598-017-17333-x
Kress, W. J., and Erickson, D. L. (2007). A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2:e508. doi: 10.1371/journal.pone.0000508
Kress, W. J., and Erickson, D. L. (eds.). (2012). “DNA barcodes: methods and protocols,” in DNA Barcodes (New York, NY: Springer), 3–8. doi: 10.1007/978-1-61779-591-6
Kumar, J. U. S., Krishna, V., Seethapathy, G. S., Ganesan, R., Ravikanth, G., and Shaanker, R. U. (2018). Assessment of adulteration in raw herbal trade of important medicinal plants of India using DNA barcoding. 3 Biotech 8:135. doi: 10.1007/s13205-018-1169-3
Kuzmina, M. L., Braukmann, T. W. A., Fazekas, A. J., Graham, S. W., Dewaard, S. L., Rodrigues, A., et al. (2017). Using herbarium-derived DNAs to assemble a large-scale DNA barcode library for the vascular plants of Canada. Appl. Plant Sci. 5:11. doi: 10.3732/apps.1700079
Leontidou, K., Vernesi, C., De Groeve, J., Cristofolini, F., Vokou, D., and Cristofori, A. (2018). DNA metabarcoding of airborne pollen: new protocols for improved taxonomic identification of environmental samples. Aerobiologia 34, 63–74. doi: 10.1007/s10453-017-9497-z
Li, Q. W., Wu, J. H., Wang, Y. S., Lian, X. M., Wu, F. L., Zhou, L., et al. (2017). The phylogenetic analysis of Dalbergia (Fabaceae: Papilionaceae) based on different DNA barcodes. Holzforschung 71, 939–949. doi: 10.1515/hf-2017-0052
Li, X. B., Shen, X. J., Chen, X., Xiang, D., Murphy, R. W., and Shen, Y. Y. (2018). Detection of potential problematic Cytb gene sequences of fishes in GenBank. Front. Genet. 9:5. doi: 10.3389/fgene.2018.00030
Li, X. W., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y. T., and Chen, S. L. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. 90, 157–166. doi: 10.1111/brv.12104
Lindenmayer, D., and Gibbons, P. (2012). Biodiversity Monitoring in Australia. Collingwood: CSIRO Publishing.
Liu, S., Wang, X., Xie, L., Tan, M., Li, Z., Su, X., et al. (2016). Mitochondrial capture enriches mito-DNA 100 fold, enabling PCR-free mitogenomics biodiversity analysis. Mol. Ecol. Resour. 16, 470–479. doi: 10.1111/1755-0998.12472
Lowe, A. J., Dormontt, E. E., Bowie, M. J., Degen, B., Gardner, S., Thomas, D., et al. (2016). Opportunities for improved transparency in the timber trade through scientific verification. BioScience 66, 990–998. doi: 10.1093/biosci/biw129
Margules, C. R., and Pressey, R. L. (2000). Systematic conservation planning. Nature 405:243. doi: 10.1038/35012251
Miller, S. E. (2007). DNA barcoding and the renaissance of taxonomy. Proc. Natl. Acad. Sci. 104, 4775–4776. doi: 10.1073/pnas.0700466104
Miya, M., Sato, Y., Fukunaga, T., Sado, T., Poulsen, J. Y., Sato, K., et al. (2015). MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2:150088. doi: 10.1098/rsos.150088
Nakahara, F., Ando, H., Ito, H., Murakami, A., Morimoto, N., Yamasaki, M., et al. (2015). The applicability of DNA barcoding for dietary analysis of sika deer. DNA Barcodes 3, 200–206. doi: 10.1515/dna-2015-0021
Nevill, P. G., Wallace, M. J., Miller, J. T., and Krauss, S. L. (2013). DNA barcoding for conservation, seed banking and ecological restoration of Acacia in the Midwest of Western Australia. Mol. Ecol. Resour. 13, 1033–1042. doi: 10.1111/1755-0998.12060
Newmaster, S. G., Grguric, M., Shanmughanandhan, D., Ramalingam, S., and Ragupathy, S. (2013). DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 11:222. doi: 10.1186/1741-7015-11-222
Ng, K. K. S., Lee, S. L., Tnah, L. H., Nurul-Farhanah, Z., Ng, C. H., Lee, C. T., et al. (2016). Forensic timber identification: a case study of a CITES listed species, Gonystylus bancanus (Thymelaeaceae). Forensic Sci. Int. 23, 197–209. doi: 10.1016/j.fsigen.2016.05.002
Nichols, R. V., Åkesson, M., and Kjellander, P. (2016). Diet assessment based on rumen contents: a comparison between DNA metabarcoding and macroscopy. PLOS ONE 11:e0157977. doi: 10.1371/journal.pone.0157977
Nichols, R. V., Vollmers, C., Newsom, L. A., Wang, Y., Heintzman, P. D., Leighton, M., et al. (2018). Minimizing polymerase biases in metabarcoding. Mol. Ecol. Resour. 18, 927–939. doi: 10.1111/1755-0998.12895
Niemeyer, B., Epp, L. S., Stoof-Leichsenring, K. R., Pestryakova, L. A., and Herzschuh, U. (2017). A comparison of sedimentary DNA and pollen from lake sediments in recording vegetation composition at the Siberian treeline. Mol. Ecol. Resour. 17, e46–e62. doi: 10.1111/1755-0998.12689
Parducci, L., Bennett, K. D., Ficetola, G. F., Alsos, I. G., Suyama, Y., Wood, J. R., et al. (2017). Ancient plant DNA in lake sediments. New Phytol. 214, 924–942. doi: 10.1111/nph.14470
Pečnikar, Ž.F., and Buzan, E. V. (2014). 20 years since the introduction of DNA barcoding: from theory to application. J. Appl. Genet. 55, 43–52. doi: 10.1007/s13353-013-0180-y
Piñol, J., Mir, G., Gomez-Polo, P., and Agustí, N. (2015). Universal and blocking primer mismatches limit the use of high-throughput DNA sequencing for the quantitative metabarcoding of arthropods. Mol. Ecol. Resour. 15, 819–830. doi: 10.1111/1755-0998.12355
Pornon, A., Andalo, C., Burrus, M., and Escaravage, N. (2017). DNA metabarcoding data unveils invisible pollination networks. Sci. Rep. 7:5. doi: 10.1038/s41598-017-16785-5
Pornon, A., Escaravage, N., Burrus, M., Holota, H., Khimoun, A., Mariette, J., et al. (2016). Using metabarcoding to reveal and quantify plant-pollinator interactions. Sci. Rep. 6:27282. doi: 10.1038/srep27282
Porter, T. M., and Hajibabaei, M. (2018). Scaling up: A guide to high-throughput genomic approaches for biodiversity analysis. Mol. Ecol. 27, 313–338. doi: 10.1111/mec.14478
Potts, S. G., Imperatriz-Fonseca, V., Ngo, H. T., Aizen, M. A., Biesmeijer, J. C., Breeze, T. D., et al. (2016). Safeguarding pollinators and their values to human well-being. Nature 540, 220–229. doi: 10.1038/nature20588
Puillandre, N., Bouchet, P., Boisselier-Dubayle, M. C., Brisset, J., Buge, B., Castelin, M., et al. (2012). New taxonomy and old collections: integrating DNA barcoding into the collection curation process. Mol. Ecol. Resour. 12, 396–402. doi: 10.1111/j.1755-0998.2011.03105.x
Ratnasingham, S., and Hebert, P. D. N. (2007). BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Richardson, R. T., Lin, C.-H., Quijia, J. O., Riusech, N. S., Goodell, K., and Johnson, R. M. (2015a). Rank-based characterization of pollen assemblages collected by honey bees using a multi-locus metabarcoding approach. Appl. Plant Sci. 3:1500043. doi: 10.3732/apps.1500043
Richardson, R. T., Lin, C.-H., Sponsler, D. B., Quijia, J. O., Goodell, K., and Johnson, R. M. (2015b). Application of ITS2 metabarcoding to determine the provenance of pollen collected by honey bees in an agroecosystem. Appl. Plant Sci. 3:1400066. doi: 10.3732/apps.1400066
Robeson, M. S., Khanipov, K., Golovko, G., Wisely, S. M., White, M. D., Bodenchuck, M., et al. (2018). Assessing the utility of metabarcoding for diet analyses of the omnivorous wild pig (Sus scrofa). Ecol. Evol. 8, 185–196. doi: 10.1002/ece3.3638
Saarela, J. M., Sokoloff, P. C., Gillespie, L. J., Consaul, L. L., and Bull, R. D. (2013). DNA Barcoding the Canadian Arctic Flora: Core Plastid Barcodes (rbcL plus matK) for 490 Vascular Plant Species. PLoS ONE 8:36. doi: 10.1371/journal.pone.0077982
Schindel, D. E., and Miller, S. E. (2005). DNA barcoding a useful tool for taxonomists. Nature 435, 17–17. doi: 10.1038/435017b
Schmeller, D., Bohm, M., Arvanitidis, C., Barber-Meyer, S., Brummitt, N., Chandler, M., et al. (2017). Building capacity in biodiversity monitoring at the global scale. Biodiversity Conserv. 26, 2765–2790. doi: 10.1007/s10531-017-1388-7
Scriver, M., Marinich, A., Wilson, C., and Freeland, J. (2015). Development of species-specific environmental DNA (eDNA) markers for invasive aquatic plants. Aquat. Bot. 122, 27–31. doi: 10.1016/j.aquabot.2015.01.003
Seberg, O., Droege, G., Barker, K., Coddington, J. A., Funk, V., Gostel, M., et al. (2016). Global Genome Biodiversity Network: saving a blueprint of the Tree of Life – a botanical perspective. Ann. Bot. 118, 393–399. doi: 10.1093/aob/mcw121
Seethapathy, G. S., Ganesh, D., Santhosh Kumar, J. U., Senthilkumar, U., Newmaster, S. G., Ragupathy, S., et al. (2015). Assessing product adulteration in natural health products for laxative yielding plants, Cassia, Senna, and Chamaecrista, in Southern India using DNA barcoding. Int. J. Legal Med. 129, 693–700. doi: 10.1007/s00414-014-1120-z
Shapcott, A., Forster, P. I., Guymer, G. P., Mcdonald, W. J. F., Faith, D. P., Erickson, D., et al. (2015). Mapping biodiversity and setting conservation priorities for SE Queensland's rainforests using DNA barcoding. PLoS ONE 10:e0122164. doi: 10.1371/journal.pone.0122164
Shapcott, A., Liu, Y., Howard, M., Forster, P. I., Kress, W. J., Erickson, D. L., et al. (2017). Comparing floristic diversity and conservation priorities across South East Queensland regional rain forest ecosystems using phylodiversity indexes. Int. J. Plant Sci. 178, 211–229. doi: 10.1086/690022
Shaw, J., Shafer, H. L., Leonard, O. R., Kovach, M. J., Schorr, M., and Morris, A. B. (2014). Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am. J. Bot. 101, 1987–2004. doi: 10.3732/ajb.1400398
Shepherd, K. A., Perkins, A., Collins, J., Byrne, M., and Thiele, K. R. (2013). Morphological and molecular evidence supports the recognition of a new subspecies of the critically endangered Pityrodia scabra (Lamiaceae). Invert. Syst. 26, 1–12. doi: 10.1071/SB12009
Shepherd, L. D. (2017). A non-destructive DNA sampling technique for herbarium specimens. PLoS ONE 12:e0183555. doi: 10.1371/journal.pone.0183555
Smart, M., Cornman, R. S., Iwanowicz, D., Mcdermott-Kubeczko, M., Pettis, J. S., Spivak, M. S., et al. (2017). A comparison of honey bee-collected pollen from working agricultural lands using light microscopy and ITS metabarcoding. Environ. Entomol. 46, 38–49. doi: 10.1093/ee/nvw159
Smith, B. E., Johnston, M. K., and Lucking, R. (2016). From GenBank to GBIF: phylogeny-based predictive niche modeling tests accuracy of taxonomic identifications in large occurrence data repositories. PLoS ONE 11:15. doi: 10.1371/journal.pone.0151232
Srivathsan, A., Sha, J., Vogler, A. P., and Meier, R. (2015). Comparing the effectiveness of metagenomics and metabarcoding for diet analysis of a leaf-feeding monkey (Pygathrix nemaeus). Mol. Ecol. Resour. 15, 250–261. doi: 10.1111/1755-0998.12302
Staats, M., Arulandhu, A. J., Gravendeel, B., Holst-Jensen, A., Scholtens, I., Peelen, T., et al. (2016). Advances in DNA metabarcoding for food and wildlife forensic species identification. Anal. Bioanal. Chem. 408, 4615–4630. doi: 10.1007/s00216-016-9595-8
Staats, M., Cuenca, A., Richardson, J. E., Vrielink-Van Ginkel, R., Petersen, G., Seberg, O., et al. (2011). DNA damage in plant herbarium tissue. PLoS ONE 6:e28448. doi: 10.1371/journal.pone.0028448
Taberlet, P., Coissac, E., Pompanon, F., Brochmann, C., and Willerslev, E. (2012). Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. doi: 10.1111/j.1365-294XX.2012.05470.x
Thomas, A. C., Deagle, B. E., Eveson, J. P., Harsch, C. H., and Trites, A. W. (2016). Quantitative DNA metabarcoding: improved estimates of species proportional biomass using correction factors derived from control material. Mol. Ecol. Resour. 16, 714–726. doi: 10.1111/1755-0998.12490
Thompson, K. A., and Newmaster, S. G. (2014). Molecular taxonomic tools provide more accurate estimates of species richness at less cost than traditional morphology-based taxonomic practices in a vegetation survey. Biodiversity Conserv. 23, 1411–1424. doi: 10.1007/s10531-014-0672-z
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Tonti-Filippini, J., Nevill, P. G., Dixon, K., and Small, I. (2017). What can we do with 1000 plastid genomes? Plant J. 90, 808–818. doi: 10.1111/tpj.13491
Vijayan, K., and Tsou, C. H. (2010). DNA barcoding in plants: taxonomy in a new perspective. Curr. Sci. 99, 1530–1541.
Weiß, C. L., Schuenemann, V. J., Devos, J., Shirsekar, G., Reiter, E., Gould, B. A., et al. (2016). Temporal patterns of damage and decay kinetics of DNA retrieved from plant herbarium specimens. R. Soc. Open Sci. 3:160239. doi: 10.1098/rsos.160239
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3:18. doi: 10.1038/sdata.2016.18
Wilkinson, M. J., Szabo, C., Ford, C. S., Yarom, Y., Croxford, A. E., Camp, A., et al. (2017). Replacing Sanger with Next Generation Sequencing to improve coverage and quality of reference DNA barcodes for plants. Sci. Rep. 7:46040. doi: 10.1038/srep46040
Will, K. W., Mishler, B. D., and Wheeler, Q. D. (2005). The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 54, 844–851. doi: 10.1080/10635150500354878
Xin, T., Li, X., Yao, H., Lin, Y., Ma, X., Cheng, R., et al. (2015). Survey of commercial Rhodiola products revealed species diversity and potential safety issues. Sci. Rep. 5:8337. doi: 10.1038/srep08337
Xiong, M. Y., Wang, D. J., Bu, H. L., Shao, X. N., Zhang, D., Li, S., et al. (2017). Molecular dietary analysis of two sympatric felids in the Mountains of Southwest China biodiversity hotspot and conservation implications. Sci. Rep. 7:12. doi: 10.1038/srep41909
Xu, S. Z., Li, Z. Y., and Jin, X. H. (2018). DNA barcoding of invasive plants in China: A resource for identifying invasive plants. Mol. Ecol. Resour. 18, 128–136. doi: 10.1111/1755-0998.12715
Keywords: natural history collections, barcoding reference database, conservation, science infrastructure, taxonomy
Citation: Dormontt EE, van Dijk K, Bell KL, Biffin E, Breed MF, Byrne M, Caddy-Retalic S, Encinas-Viso F, Nevill PG, Shapcott A, Young JM, Waycott M and Lowe AJ (2018) Advancing DNA Barcoding and Metabarcoding Applications for Plants Requires Systematic Analysis of Herbarium Collections—An Australian Perspective. Front. Ecol. Evol. 6:134. doi: 10.3389/fevo.2018.00134
Received: 08 June 2018; Accepted: 21 August 2018;
Published: 13 September 2018.
Edited by:
Michael David Martin, Norwegian University of Science and Technology, NorwayReviewed by:
David Vieites, Consejo Superior de Investigaciones Científicas (CSIC), SpainWei Lun Ng, Sun Yat-sen University, China
Steven G. Newmaster, University of Guelph, Canada
Copyright © 2018 Dormontt, van Dijk, Bell, Biffin, Breed, Byrne, Caddy-Retalic, Encinas-Viso, Nevill, Shapcott, Young, Waycott and Lowe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eleanor E. Dormontt, ZWxlYW5vci5kb3Jtb250dEBhZGVsYWlkZS5lZHUuYXU=
 Eleanor E. Dormontt
Eleanor E. Dormontt Kor-jent van Dijk
Kor-jent van Dijk Karen L. Bell2,3
Karen L. Bell2,3 Margaret Byrne
Margaret Byrne Francisco Encinas-Viso
Francisco Encinas-Viso Paul G. Nevill
Paul G. Nevill Michelle Waycott
Michelle Waycott Andrew J. Lowe
Andrew J. Lowe