- Genomic and Applied Microbiology and Göttingen Genomics Laboratory, Institute of Microbiology and Genetics, Georg-August University of Goettingen, Göttingen, Germany
Nitriles are organic molecules with –C≡N as functional group and often toxic for living organisms. Detoxification can occur via nitrilases that degrade nitriles directly to carboxylic acids and ammonia, or with nitrile hydratases and amidases that convert nitriles to amides and subsequently to carboxylic acids and ammonia. Despite the knowledge of enzymatic degradation pathways, the influence of these compounds on the composition of bacterial communities is unknown. The tolerances of four phylogenetically different bacterial strains without known nitrile detoxification systems (Agrobacterium tumefaciens, Bacillus subtilis, Corynebacterium glutamicum, and Escherichia coli) to the toxic effects of nine nitriles and the corresponding carboxylic acids were determined. Based on these results, the effect of nitriles on diversity and composition of compost-derived bacterial communities was monitored over time by 16S rRNA gene amplicon-based and metagenome analyses. Acetone cyanohydrin, 2-phenylpropionitrile, and pyruvonitrile exhibited a lethal, phenylacetonitrile, 4-hydroxybenzonitrile, and cyclohexanecarbonitrile a growth-suppressing and succinonitrile, acetonitrile, and crotononitrile a growth-promoting effect on the studied communities. Furthermore, each nitrile had a specific community-shaping effect, e.g., communities showing growth-suppression exhibited high relative abundance of Paenibacillus. In general, analysis of all data indicated a higher resistance of Gram-positive than Gram-negative bacterial community members and test organisms to growth-suppressing nitriles. More than 70 putative nitrilase-encoding and over 20 potential nitrile hydratase-encoding genes were identified during analysis of metagenomes derived from nitrile-enrichments, underlining the high yet often unexplored abundance of nitrile-degrading enzymes.
Introduction
Nitriles are structurally diverse organic molecules with –C≡N as functional group. Naturally occurring nitriles are found in bacteria, fungi, plants, and animals as well as in marine and terrestrial habitats (Mahadevan, 1973; Legras et al., 1990; Scheuer, 1992; Fleming, 1999). The most widespread nitriles are cyanogenic glycosides, which often are present in plants (Conn, 1969) and other organisms like Myriopoda (Eyjólfsson, 1970). They can serve as storage form of reduced nitrogen (Poulton, 1990) or defense compound when degraded to highly toxic hydrogen cyanide (Fleming, 1999).
Toxicity of most nitriles originates not from the nitrile itself, but from cyanide release during degradation, e.g., by cytochrome P450 in eukaryotes (Tanii and Hashimoto, 1984; Grogan et al., 1992; Uetrecht and Trager, 2007). Subsequently, cyanide inhibits cytochrome c oxidases and in this way the electron transport chain (Willhite and Smith, 1981). Cytochrome P450 and cytochrome c oxidase are also present in many bacteria (Richter and Ludwig, 2003; Lewis and Wiseman, 2005) and therefore a similar mode of action as in eukaryotic organisms is indicated.
Detoxification and degradation are facilitated by two known enzymatic routes. The first involves nitrilases (EC 3.5.5.1), which belong to the carbon-nitrogen hydrolase superfamily and harbor a conserved catalytic triad consisting of glutamic acid, lysine, and cysteine (Pace and Brenner, 2001; Vergne-Vaxelaire et al., 2013). These enzymes degrade nitriles directly to carboxylic acids and ammonia. In the second route, nitriles are degraded to amides and subsequently to carboxylic acids and ammonia. The hydration of nitriles to amides is catalyzed by nitrile hydratases (NHases; EC 4.2.1.84), which consist of an α- and β-subunit and are classified as cobalt or ferric NHases. The subsequent hydrolysis of amides to carboxylic acids and ammonia is performed by amidases (EC 3.5.1.4; Ramteke et al., 2013).
Nitriles are also important industrial precursors for the production of bulk chemicals like acrylamide (Kurenkov, 1997) or part of various pharmaceuticals such as the anti-cancer drug letrozole or etravirine for HIV treatment (Fleming et al., 2010). Unintended release of man-made nitriles can lead to environmental accumulation and impact natural communities. In addition, some nitriles are used as herbicides, e.g., dichlobenil (Holtze et al., 2007) or bromoxynil (Baxter and Cummings, 2008). The effect of these synthetic compounds on bacterial communities, the key players in most nutrient cycles (Tanner, 1985; Alongi, 1994; Rashid et al., 2016), is almost unknown. While toxicity values and effects of many nitriles on mammals have been reported, comparable values are almost unavailable for microorganisms (Willhite and Smith, 1981; Ahmed and Farooqui, 1982; Tanii and Hashimoto, 1984; Wallig et al., 1988).
The aim of this work was to investigate the effect of nine nitriles and their corresponding degradation products (carboxylic acid and ammonia) on bacterial communities. The toxicity of these compounds was explored using four bacterial test strains containing no known nitrile detoxification system. Based on these results a community growth experiment was performed and revealed the specific impact of each nitrile on a bacterial community with respect to composition and diversity. Metagenome sequences provided insight into potential mechanisms of nitrile stress management, as quantity and distribution of nitrile-degrading enzymes was analyzed.
Results
Bacterial Life and Nitrile Toxicity
To establish stable nitrile-containing mixed cultures, it is necessary to determine the toxic effect of these compounds. Therefore, we analyzed the influence of a set of nine nitriles on the four model organisms Agrobacterium tumefaciens, Bacillus subtilis, Corynebacterium glutamicum, and Escherichia coli by using agar plates containing a rich complex or a minimal medium with different nitrile concentrations. The test organisms did not contain any known nitrile-degradation pathway. Additionally, the influence of corresponding carboxylic acids and ammonia was evaluated, as the products of a potential nitrile detoxification reaction also affect microbial growth.
Growth was monitored for 4 days on solid agar plates supplemented with a nitrile or the corresponding carboxylic acid and examined every 24 h (Supplementary Table 1). Compared to the Gram-negative strains (A. tumefaciens and E. coli), the Gram-positive strains (B. subtilis and C. glutamicum) showed better growth at high nitrile and carboxylic acid concentrations. Some of the tested strains revealed different behavior when comparing the influence of a nitrile and the corresponding carboxylic acid on their growth. For example, 4-hydroxybenzonitrile suppressed the growth of A. tumefaciens and E. coli even at 5 mM concentration but all organisms except A. tumefaciens were able to grow at 25 mM of 4-hydroxybenzoic acid (Figure 1). The higher susceptibility of the Gram-negative organisms was also observed for pyruvonitrile, as growth was inhibited at 15, 20, and 25 mM concentration, whereas Gram-positive strains grew at 15 mM. In addition, phenylacetic acid, 4-hydroxybenzoic acid, and pyruvic acid exhibited less growth-inhibiting effects than their nitrile counterparts. In contrast, cyclohexanecarbonitrile revealed no detectable effect on the growth of all tested organisms, but cyclohexanecarboxylic acid inhibited growth at 10 mM.
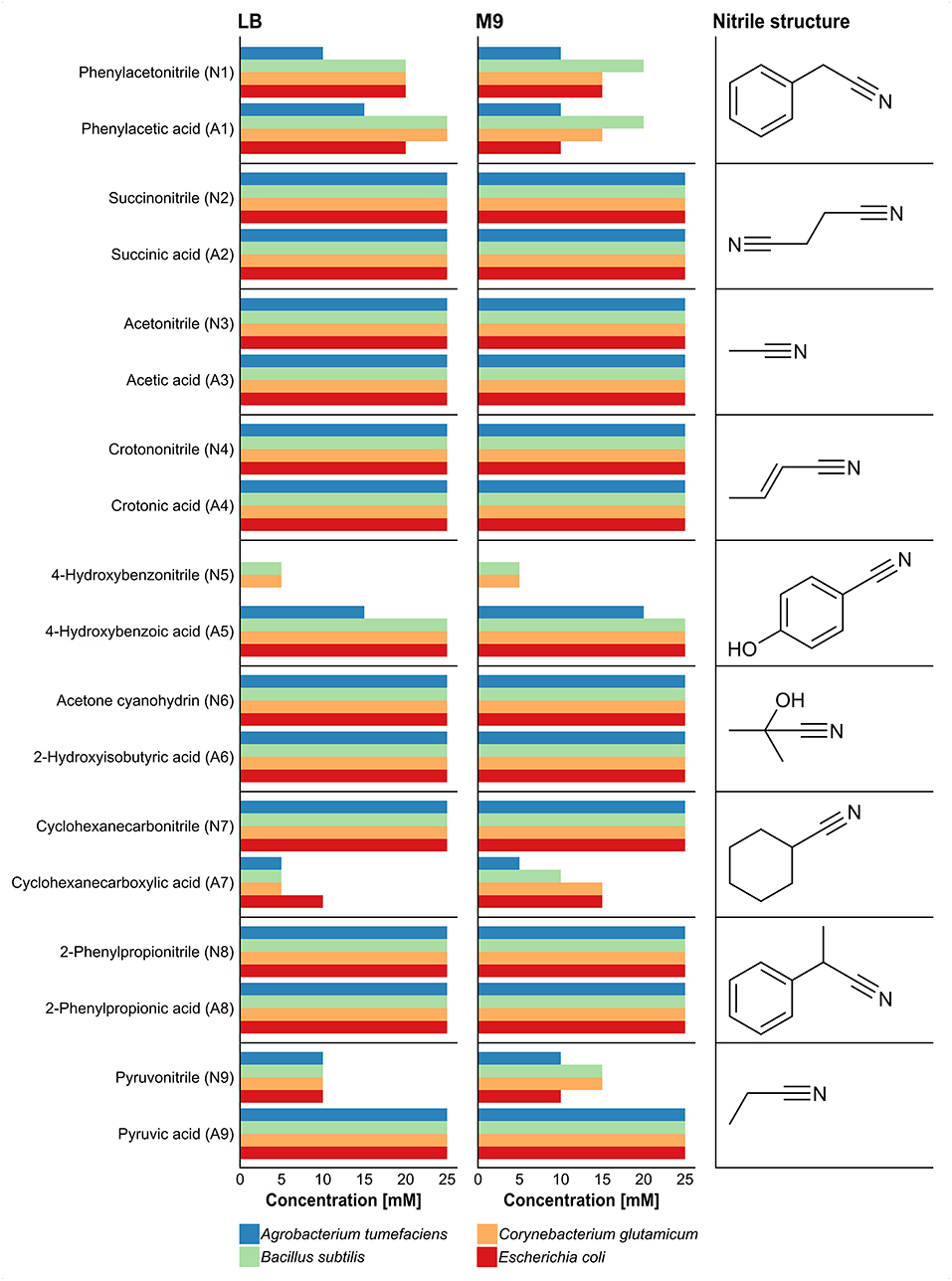
Figure 1. Effect of different nitriles and corresponding carboxylic acids on the growth of A. tumefaciens, B. subtilis, C. glutamicum, and E. coli after 4 days of incubation. Growth was performed in LB and M9-HEPES medium, and the tested concentrations of the nitriles and the corresponding carboxylic acids comprised 5, 10, 15, 20, and 25 mM. Length of bars represents maximal nitrile concentration for growth of each organism.
The results provided a first insight into the possible inhibitory potential of the nitriles and carboxylic acids with respect to bacterial growth. The highest non-inhibitory concentration considering all four test strains was chosen for further experiments (see Material and Methods). In cases where the highest concentration of a nitrile and the corresponding carboxylic acid differed, the lower concentration was chosen for both substances. For substances inhibiting growth even at the lowest tested concentration, 5 mM were used subsequently.
Influence on Microbial Growth
To determine changes in bacterial communities due to the influence of nitriles and their corresponding carboxylic acids a growth experiment in liquid minimum medium was performed. A compost sample from the Experimental Botanical Garden Göttingen (Germany) served as inoculum.
To avoid nutrient depletion, cultures were transferred to fresh medium when growth curves started to flatten. A general reduction of the final OD with each cultivation passage was observed for the controls and the nitrile-based and carboxylic acid-based cultures (Figure 2). At the last measuring point of the carboxylic acid-supplemented cultures, seven of nine treatments revealed on average higher optical densities (OD between 0.5 and 0.9) than the control (OD of 0.4), indicating beneficial effects of the supplemented carboxylic acids. In case of the nitrile-supplemented cultures only three nitriles (succinonitrile, acetonitrile, and crotononitrile) exhibited a growth-supporting effect in comparison to the control without nitriles. The beneficial impact of acetonitrile was already obvious after 100 h incubation. Addition of cyclohexanecarbonitrile did not affect growth, whereas a negative effect on microbial growth was observed for phenylacetonitrile and 4-hydroxybenzonitrile. In contrast, cultures supplemented with acetone cyanohydrin, 2-phenylpropionitrile, or pyruvonitrile showed no growth at all after the first passage.
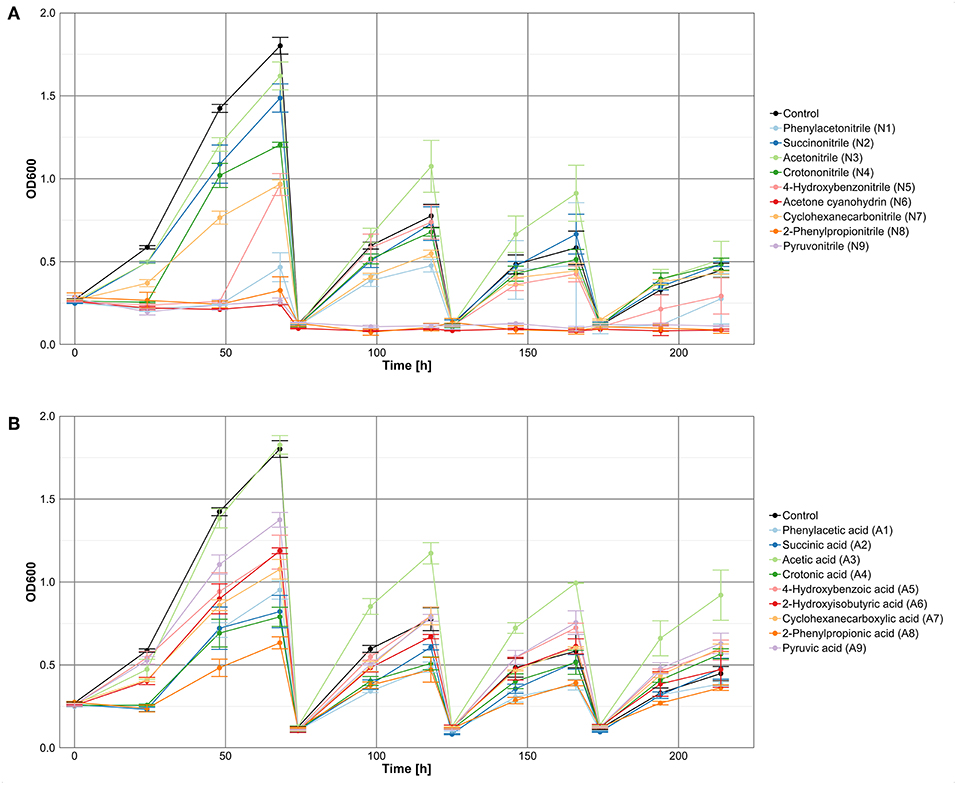
Figure 2. Growth of a microbial community in M9-HEPES treated with (A) nitriles or (B) the corresponding carboxylic acids.
Community Composition and Diversity
The bacterial community composition of the control cultures without addition of nitriles or the corresponding carboxylic acids was determined via amplicon-based 16S rRNA gene analysis (Figure 3, Supplementary Table 2). Analysis of Simpson indices of diversity (SID) at genus level revealed reduction of diversity from inoculum (SID 0.98) to the first sampling of the control after 68 h of incubation (SID 0.58). Diversity increased again after 118 h of incubation (SID 0.78) and stabilized between 166 and 214 h (SID 0.91 in both cases). Besides inoculum and control cultures, only communities showing growth after 214 h with the nitrile or the corresponding carboxylic acid were analyzed by 16S rRNA gene analysis (Figure 3). Cultures supplemented with acetone cyanohydrin, 2-phenylpropionitrile, and pyruvonitrile were excluded from this analysis due to absence of viability after first passage. Consequently, the respective carboxylic acid-supplemented cultures were also removed, as in these cases comparison between a nitrile and its carboxylic acid was not feasible.
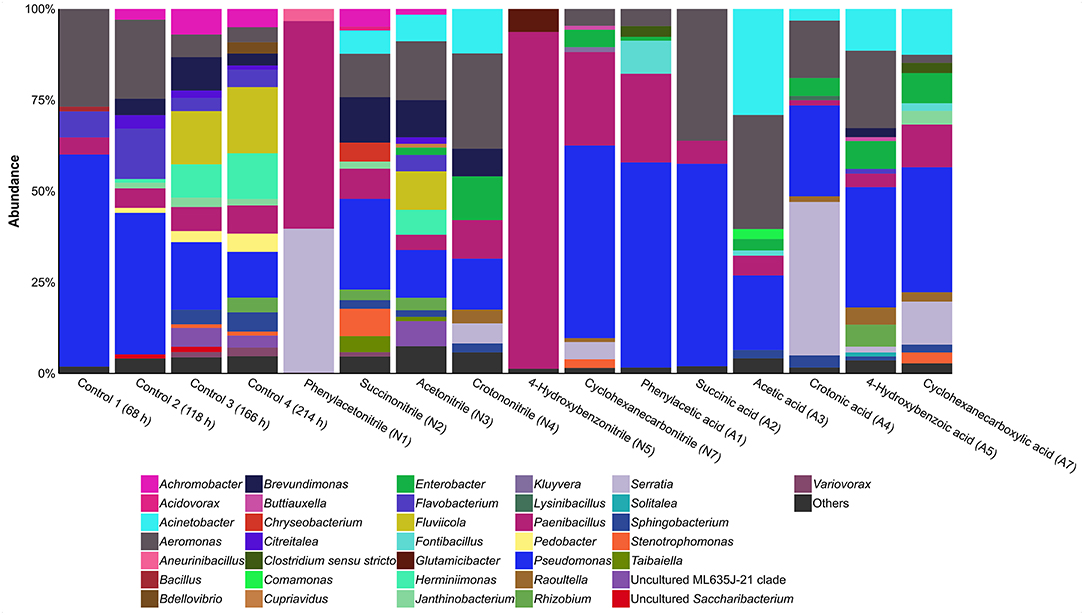
Figure 3. Relative abundance of bacterial genera based on 16S rRNA gene analysis in cultures containing different nitriles or carboxylic acids after 214 h of incubation. Others, genera with a relative abundance of >1%.
Nitrile-supplemented cultures indicated a general formation of two distinct bacterial community types. Cultures containing succinonitrile, acetonitrile, or crotononitrile showed a high diversity (SID 0.88, 0.91, and 0.86, respectively) when compared to cultures with phenylacetonitrile, 4-hydroxybenzonitrile or cyclohexanecarbonitrile (SID 0.52, 0.14, and 0.65, respectively). Cultures supplemented with the corresponding carboxylic acids (phenylacetic acid, 4-hydroxybenzoic acid, and cyclohexanecarboxylic acid) exhibited a similar pattern. Communities with acetic acid (SID 0.77), crotonic acid (SID 0.73), 4-hydroxybenzoic acid (SID 0.82), and cyclohexanecarboxylic acid (SID 0.83) exhibited a higher diversity compared to cultures supplemented with phenylacetic acid (SID 0.61) and succinic acid (SID 0.56). In addition, some carboxylic acid-containing cultures showed strong variations in diversity in relation to the corresponding nitrile-containing cultures. In case of succinonitrile and succinic acid, diversity decreased from SID 0.88 to 0.56. A similar trend was observed for acetonitrile (SID 0.91) and acetic acid (SID 0.77). In contrast, higher diversity was detected in 4-hydroxybenzoic acid-containing cultures (SID 0.82) compared to the corresponding nitrile (SID 0.14). These differences in diversity indicate a divergent effect of the nitrile group in comparison to the carboxylic acid group on microbial growth.
Nitrile-supplemented cultures showed distinct communities when compared with the carboxylic acid counterparts. Pseudomonas was present under most conditions, except in cultures containing phenylacetonitrile or 4-hydroxybenzonitrile. A similar occurrence was recorded for Aeromonas. Interestingly, Paenibacillus was the dominant genus in phenylacetonitrile- or 4-hydroxybenzonitrile-containing cultures with a relative abundance of 57 and 92%, respectively. Members of this genus are also present in all other cultures but in lower relative abundances. In addition, Brevundimonas was present (>1% relative abundance) in three of the six nitrile-treated cultures, but only in one containing a carboxylic acid. Furthermore, Acinetobacter and Aeromonas showed an on average higher relative abundance under acetic acid treatment compared to the nitrile-containing cultures. The relative abundances increased from 4 to 9% and 10 to 19%, respectively. In contrast, Pedobacter was only identified in the controls (>1% relative abundance), while Aneurinibacillus (3%), Chryseobacterium (5%), and Glutamicibacter (6%) were only present after treatment with phenylacetonitrile, succinonitrile or 4-hydroxybenzonitrile, respectively. Serratia showed high relative abundance in cultures containing phenylacetonitrile (40%) or crotonic acid (42%).
The combination the 16S rRNA gene analysis with the corresponding growth curves revealed further linkages between the density of a culture and its diversity. Cultures with phenylacetonitrile (SID 0.52) and 4-hydroxybenzonitrile (SID 0.14) did not only reveal reduced optical density, but a decreased diversity when compared to the cultures containing the corresponding carboxylic acids (SID 0.61 and 0.82, respectively). In contrast, succinic acid-containing cultures showed low diversity (SID 0.56), but exhibited higher density than the control (SID 0.91). Several cultures showed higher density and diversity than the control, indicating a growth-benefiting effect of succinonitrile, acetonitrile, 4-hydroxybenzoic acid, and cyclohexanecarboxylic acid.
How to Handle Nitriles
To assess in which way the different bacterial communities manage the supplemented nitriles, metagenomes of all cultures incubated for 214 h and showing growth were sequenced. Subsequently, the metagenomes were assembled, annotated, and analyzed. The samples were checked for presence of genes encoding nitrilases and nitrile hydratase subunits. Finally, the taxonomic assignment of the contigs harboring the targeted genes was used to infer the phylogenetic origin of the deduced enzymes (Supplementary Table 3).
In general, the metagenomes showed a similar taxonomic composition as previously determined via 16S rRNA gene analysis (Supplementary Table 4). Analysis of nitrile-degrading enzymes encoded by the metagenomes revealed a broad distribution and especially members of the genus Pseudomonas harbored several of the respective genes (Figure 4A). Furthermore, the untreated control cultures showed approx. a 2-fold higher abundance of nitrile-degrading enzymes when compared to other treatments except cyclohexanecarbonitril and succinic acid (Figure 4B).
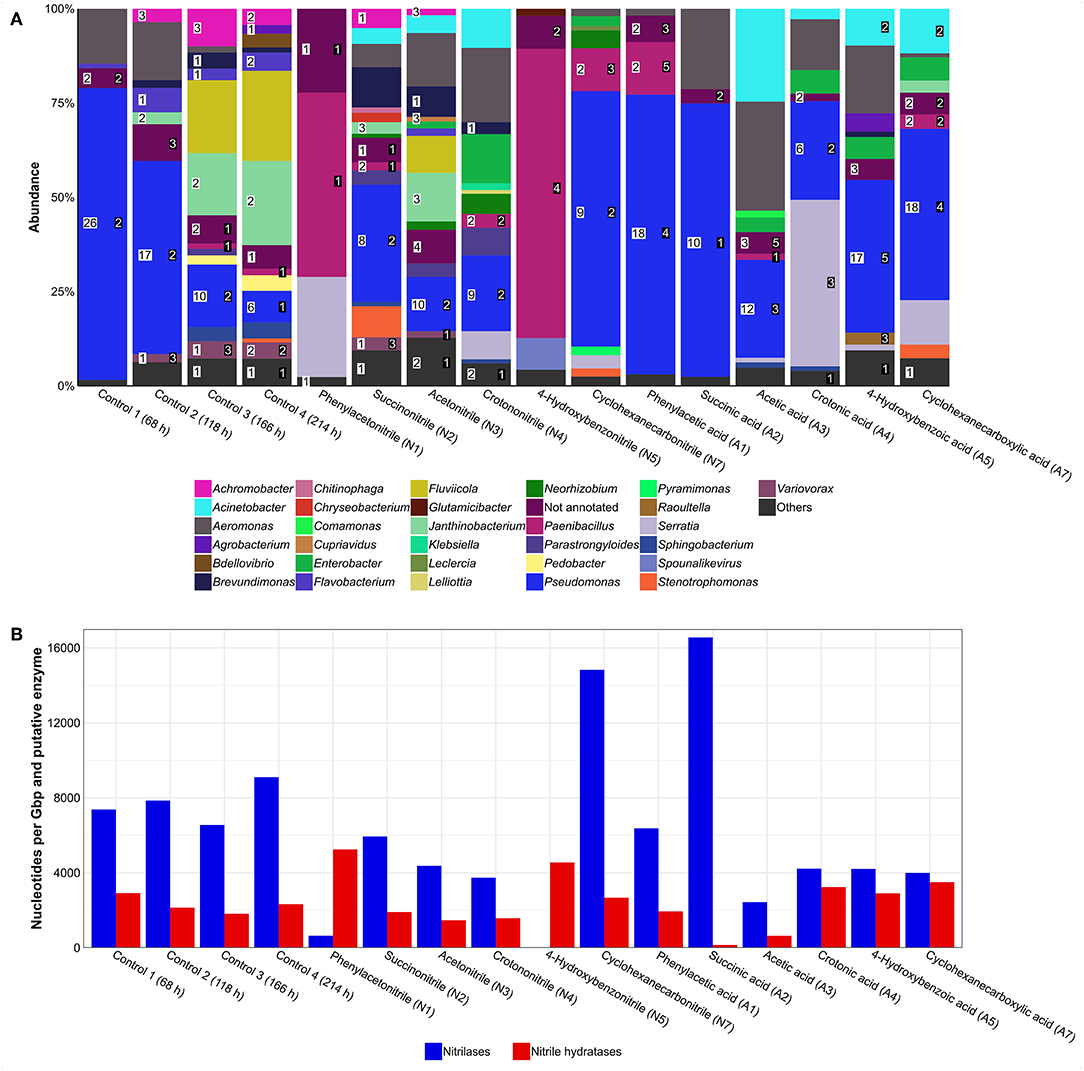
Figure 4. Putative nitrilases and nitrile hydratases found in metagenomes of enrichment cultures. (A) Distribution of enzymes. Numbers in white box represent number of nitrilases and numbers in black box represent number of nitrile hydratase subunits. Others, genera with an abundance of >1%. (B) Gene abundance based on coverage. Higher values represent an enrichment of nitrile-degrading genes and are potential indicators for nitrile degradation.
Regarding potential impacts of a nitrile on a bacterial community, several effects were observed. For example, the relative abundance of Pseudomonas increased in the cyclohexanecarbonitrile-containing culture compared to its carboxylic acid counterpart, while the diversity of putative nitrilases was reduced by 50% (Figure 4A). At the same time, abundance of nitrilase genes increased almost 4-fold, indicating their enrichment and correspondingly an increased potential for nitrile degradation in the community (Figure 4B).
A phylogenetic tree containing all identified and reference nitrilases (Figure 5) revealed no specific clustering based on the different treatments. Approximately 25% of all putative enzymes group together with β-cyano-L-alanine-degrading nitrilases. In addition, small clusters of arylacetonitrilases and cyanide dihydratases were found, containing 21 or 8 of the putative nitrilases, respectively. For the remaining 156 putative nitrilases, specific clustering was not recorded.
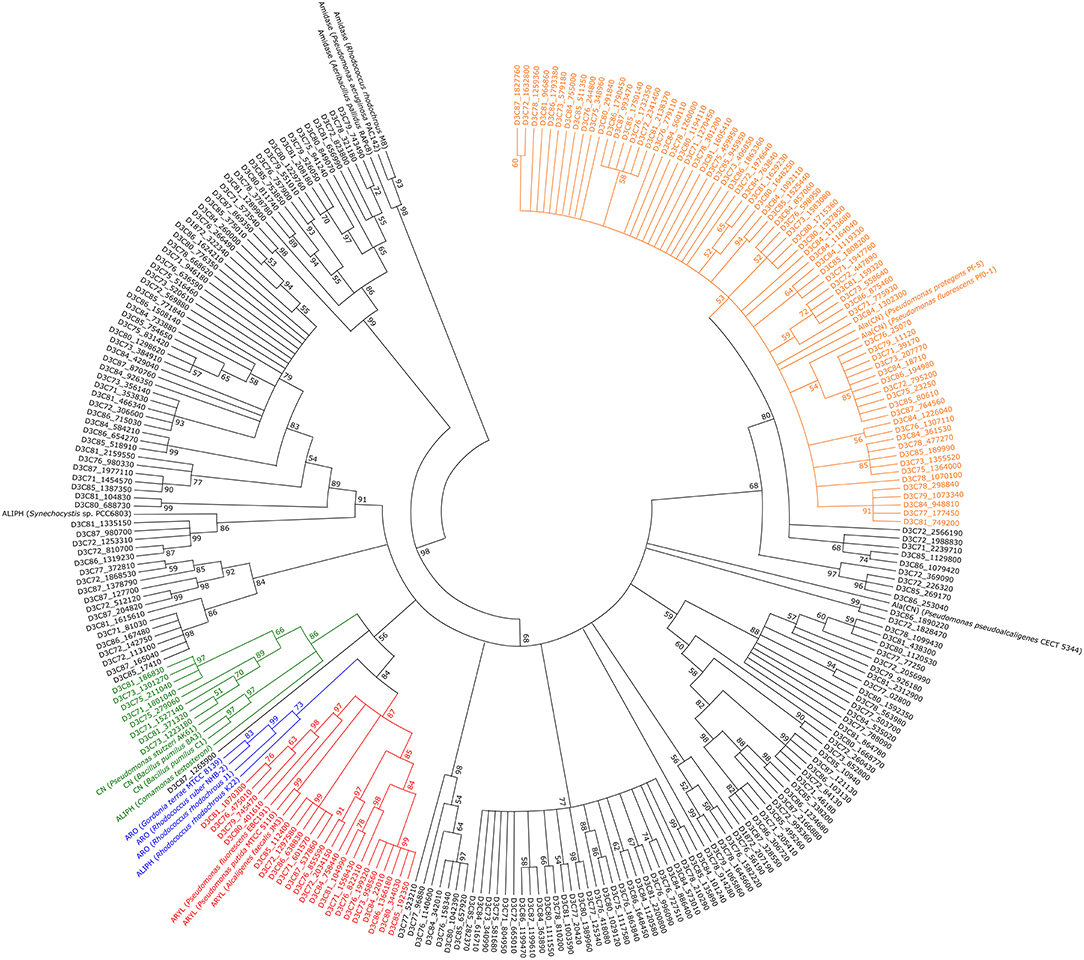
Figure 5. Phylogenetic tree containing all putative metagenome-derived nitrilases. 500 bootstrap replicates were calculated and tree nodes were condensed at values below 50 %. ALIPH, aliphatic nitrilase; ARO, aromatic nitrilase; ARY, arylacetonitrilase; ALA(CN), β-cyano-L-alanine-nitrilase; CN, cyanide dihydratase. Green, putative cluster of cyanide dihydratases; blue, putative cluster of aromatic nitrilases; red, putative cluster of arylacetonitrilases; orange, putative cluster of β-cyano-L-alanine-nitrilases.
Discussion
Studies on the susceptibility of prokaryotes to several bioactive compounds such as antibiotics (Jorgensen and Ferraro, 2009), cytostatics (Marhan, 1995), fungicides (Bending et al., 2007), insecticides (Malathi et al., 2017), and nematicides (Abo-El-Dahab et al., 1977) are performed frequently. To our knowledge literature systematically addressing bacterial sensitivity toward nitriles and their degradation products is rare. Results of the plate-based toxicity assay, determining growth of A. tumefaciens, B. subtilis, C. glutamicum, and E. coli, indicate several differences in their susceptibility to the tested chemicals. Both Gram-positive strains revealed increased tolerance to the used nitriles as more biomass was observed compared to the Gram-negative strains.
Among the tested compounds, 4-hydroxybenzonitrile was the most toxic. The closely related compounds 3,5-diiodo-4-hydroxybenzonitrile (ioxynil) and 3,5-dibromo-4-hydroxybenzonitrile (bromoxynil) are common herbicides (Holly, 1964; Holtze et al., 2008) and bromoxynil has a documented negative impact on the soil microbial biomass (Abbas et al., 2014), indicating general toxicity of this group of chemicals. The second most toxic chemical was pyruvonitrile, which has structural similarities to the key metabolite pyruvate. This similarity leads to interference with central metabolism and deleterious effects. The toxicity of several carboxylic acids was reduced compared to the corresponding nitriles, except for cyclohexanecarboxylic acid. Its toxicity was between 2- and 5-fold higher for the tested prokaryotes than that of its nitrile counterpart. The respective mechanism remains unknown.
The toxicity assay served mainly as an initial test to define working concentrations for the investigation of bacterial communities. Optical density data, representative for the viability of a community, provided a first insight into the influence of the tested compounds on bacterial communities. In case of the nitriles, only succinonitrile, acetonitrile, and crotononitrile (Figure 2A) exhibited a positive effect on microbial growth, indicating nitrile-degrading abilities of the predominant bacterial community members. For acetonitrile, this effect was already experimentally confirmed through the successful isolation of nitrile-degrading strains from similar cultures (Egelkamp et al., 2017). All other nitrile-containing cultures revealed reduced density indicating weak or no degradation ability or tolerance levels with respect to the tested compounds.
For the differences between the solid and liquid cultures with the substrates acetone cyanohydrin, 2-phenylpropionitrile, and pyruvonitrile, two explanations are likely. First, in case of cyanohydrins, spontaneous decomposition has been described (Tanii, 2017; Yerabolu et al., 2017), and probably similar mechanisms also apply for other nitriles. Consequently, different stabilities of the nitriles under the here applied conditions could have influenced the observed results. Second, the compost sample did not contain any acetone cyanohydrin, 2-phenylpropionitrile, or pyruvonitrile resistant or degrading microorganisms. However, 16S rRNA gene analysis of the inoculum revealed the presence of A. tumefaciens, B. subtilis, C. glutamicum, and E. coli. As these species successfully grew during the initial plate test, the first assumption seems to us more likely. Furthermore, degrading organisms or at least nitrilases for acetone cyanohydrin and 2-phenylpropionitrile have already been identified (Nagasawa et al., 1990; Gilligan et al., 1993; Layh et al., 1997, 1998; Bauer et al., 1998; Gerasimova et al., 2004; Schreiner et al., 2010; Bhatia et al., 2013), indicating the existing potential to handle these chemicals.
Regardless of specific impacts of the used chemicals, diversity of the enrichments generally decreased compared to the inoculum. This is in accordance with other studies, as often more than 99% of all environmental microorganisms cannot be cultivated under laboratory conditions (Torsvik et al., 1996; Davis et al., 2005; Solden et al., 2016). However, re-diversification of the controls over the course of the experiment was observed and is explained by the prolonged incubation time. Extended incubation is a well-known factor for increasing the number of isolates on solid media (Davis et al., 2005). Apparently, regular transfer to fresh medium and therefore prolonged incubation without nutrient deficiency leads also to an increase of proliferating and fast-growing bacteria in liquid medium. Slow-growing organisms get the possibility to overcome their lag period and proliferate during longer incubation times. This re-diversification has a severe impact on results using liquid cultures, as several genera would be missed in case of earlier sampling and analysis. In this experiment, diversity stabilized between the third and fourth sampling time. The 16S rRNA gene data after 214 h of incubation revealed diverse individual cultures, stating that nitriles and their degradation products impact bacterial community composition and diversity. An increased relative abundance of Gram-positive genera in cultures containing growth-suppressing nitriles was observed. This is in accordance with the initial results showing better growth of Gram-positive bacteria on nitrile-supplemented agar plates than Gram-negative bacteria. It appears that Gram-positive bacteria are more resistant to the negative influence of these specific compounds. Still, the overall dominance of Paenibacillus in the 4-hydroxybenzonitrile-containing sample was unexpected. We did not find any experimentally confirmed nitrile-degrading enzyme for this genus in literature. However, predictions for putative nitrilase-encoding genes and closely related cyanide dihydratases exist (Benedik and Sewell, 2018). This would explain the observed phenomenon. On the contrary, neither database entries nor literature on nitrile-degrading enzymes were found for Brevundimonas, which showed higher abundances in the nitrile-containing cultures compared to the corresponding carboxylic acid-containing cultures. For Serratia nitrile degradation has been described (Jin et al., 2010; Santoshkumar et al., 2010), but respective genes were not identified in the metagenome derived from the phenylacetonitrile-treated culture. Nevertheless, with an abundance of 40% Serratia seems to be well-adapted to the applied nitrile and probably has found, like Brevundimonas, a new way to handle this type of compounds. Thus, further investigation of all three genera are potentially of great value for the discovery of novel nitrile-degrading enzymes and resistance mechanisms.
A few reports mentioned nitrile-degrading activity of Acinetobacter (Yamamoto et al., 1990; Finnegan et al., 1991), but not for Aeromonas. The absence of putative nitrilases and nitrile hydratases for Aeromonas and only few nitrile hydratase subunits assigned to Acinetobacter indicate the rarity of respective genes in both genera. The reduced average abundance of both genera in nitrile-treated cultures might result from a higher susceptibility to possible toxic effects of the nitriles and subsequent displacement of these genera.
Pedobacter is known for broad antibiotic resistance potential and heavy metal tolerance (Romaniuk et al., 2018; Tipayno et al., 2018; Viana et al., 2018), but apparently these mechanisms are not beneficial to handle problematic nitriles or carboxylic acids. Additionally, genes for nitrile degradation were not predicted for members of this genus. The same applies for Aneurinibacillus, Chryseobacterium, and Glutamicibacter. However, every genus showed growth in at least one nitrile-containing culture. Thus, the underlying mechanism remains unclear.
Previous assumptions that nitriles and their handling are a common feature in nature (Conn, 1969; Eyjólfsson, 1970; Mahadevan, 1973; Legras et al., 1990; Poulton, 1990; Scheuer, 1992; Fleming, 1999) were confirmed as many nitrile-degrading enzymes were identified in different genera, even in the control culture. The high abundance of nitrilase-encoding genes in the cyclohexanecarbonitrile-containing treatment demonstrates enrichment of respective genes.
Phylogenetic analysis revealed a high diversity of the encountered putative nitrilases as they were assigned to several different groups. As previous studies have shown, nitrilases often cluster based on their substrate specificity (Howden and Preston, 2009). Thus, clustering of aromatic nitrilases and arylacetonitrilases is explained by the similar substrates. The R. rhodochrous K22 nitrilase preferably acts on aliphatic nitriles but is similar to benzonitrile-degrading nitrilases with respect to sequence (Kobayashi et al., 1990), explaining its clustering with other aromatic nitrilases.
Conclusion
The here employed experimental setup for the investigation of the impact of nitriles and their degradation products on bacterial communities showed that these compounds significantly influence the composition of bacterial communities and partially lead to a strong reduction of diversity. In addition, it revealed strains with potential for remediation purposes such as Paenibacillus in case of 4-hydroxybenzonitrile.
In conclusion, this work provides insight on the influence of different nitriles and their corresponding carboxylic acids on bacterial growth. A higher susceptibility of Gram-negative strains (A. tumefaciens and E. coli) to growth-suppressing effects of nitriles compared to the Gram-positive strains (B. subtilis and C. glutamicum) was observed during toxicity tests. Accordingly, subsequent experiments with liquid cultures and a soil-derived microbial community exhibited higher abundance of Gram-positive organisms in samples containing growth-suppressing nitriles. Nevertheless, the here presented results are just a first observation of this phenomenon and the number of tested organisms and chemicals should be increased to provide insights in the underlying mechanisms.
The liquid culture experiment with a compost community as inoculum highlighted problems when transferring complex communities for several times to fresh medium, as the maximal optical densities were reduced each passage. Nevertheless, 16S rRNA gene analysis of the control cultures after 68, 118, 166, and 214 h of incubation revealed a re-diversification and stabilization of the bacterial community composition, indicating that the time point for analysis of the chemically-treated cultures after 214 h was appropriate. In addition, a lethal effect of acetone cyanohydrin, 2-phenylpropionitrile, and pyruvonitrile on microbial communities was documented. Consequently, environmental contamination with these compounds should be prevented, as bioremediation is most likely problematic. In general, most of the nitriles were more toxic than the corresponding carboxylic acids, indicating a specific influence of the nitrile group. Nevertheless, in some cases the carboxylic acids exhibited strong negative effects on culture growth or diversity. This demonstrates the importance of testing the influence of educts and products of an enzymatic reaction before starting with screening experiments to avoid unintended lethal effects.
Materials and Methods
Media and Stock Solutions
For growth of microorganisms, LB-Miller medium (10 g tryptone, 10 g NaCl, and 5 g yeast extract per liter; Miller, 1972) or M9-HEPES was used. M9-HEPES was prepared using 15.5 g HEPES, 1 g NH4Cl, and 0.5 g NaCl per liter. The pH was adjusted to 7.0 before autoclaving. Ten mL ATCC vitamin supplement (LGC Standards, Teddington, UK), 10 mL ATCC trace mineral supplement, 10 mL glucose solution (20 %), 1 mL MgSO4 · 7 H2O solution (1 M), 1 mL CaCl2 solution (14.7 g/l), and 1 mL thiamine-HCl solution (1 mg/mL) were added afterwards. For toxicity determination, following chemicals were solved in H2O, DMSO, or ethanol, subsequently sterile-filtered to ensure sterility and added to the medium after autoclaving: phenylacetonitrile (N1), acetonitrile (N3), 2-hydroxyisobutyric acid (A6), 2-phenylpropionic acid (A8) (all from TCI Chemicals, Eschborn, Germany), succinonitrile (N2), crotononitrile (N4), 4-hydroxybenzonitrile (N5), acetone cyanohydrin (N6), cyclohexanecarbonitrile (N7), 2-phenylpropionitrile (N8), pyruvonitrile (N9), phenylacetic acid (A1), succinic acid (A2), acetic acid (A3), crotonic acid (A4), 4-hydroxybenzoic acid (A5), cyclohexanecarboxylic acid (A7), and pyruvic acid (A9) (all from Sigma-Aldrich, Taufkirchen, Germany). In case of carboxylic acids, an equimolar amount of ammonia was added in form of ammonia water (Carl Roth, Karlsruhe, Germany) to resemble the degradation of nitriles leading to the equimolar production of carboxylic acids and ammonia. For agar plates, 15 g agar per liter was added before autoclaving.
Nitrile Toxicity on Selected Strains
Agrobacterium tumefaciens NTL4(pCF372)(pCF218) (Swiderska et al., 2001), Bacillus subtilis NCIB 3610 (Cohn, 1872), Corynebacterium glutamicum ATCC 13032 (Kinoshita et al., 1958), and Escherichia coli W3110 were used as model organisms. Pre-cultures of these strains were grown in LB medium at 30°C and 180 rpm (Multitron shaker, Infors, Einsbach, Germany) for 24 h. Afterwards, each strain was plated on a quadrant of a LB or M9-HEPES plate containing one of the nine nitriles or the respective carboxylic acid in a concentration of 5, 10, 15, 20, and 25 mM. As control, LB or M9-HEPES agar plates without nitriles and carboxylic acids were used. Every 24 h, the growth on the plates was visually analyzed and categorized on a scale of 0 to 4 by comparison with the control. In case of no growth a 0 was given, whereas similar growth compared to the control was treated as 4. The remaining values were given for ~25% (= 1), 50% (= 2), and 75% (= 3) of colony size and opaqueness when compared to the control.
Nitrile Toxicity on a Microbial Community
Compost (100 g, pH 7.5) of the Experimental Botanical Garden Göttingen, Germany (51°33′22.6′′N 9°57′16.2′′E) was solved in 500 mL ddH2O and subsequently filtered with a 2.7 μm GF/D glass fiber filter (Whatman, Little Chalfont, UK). Baffled flasks (total volume 100 mL) were filled with 35 mL M9-HEPES containing one of the nitriles or carboxylic acids at following concentrations: phenylacetonitrile (N1) 10 mM, succinonitrile (N2) 25 mM, acetonitrile (N3) 25 mM, crotononitrile (N4) 25 mM, 4-hydroxybenzonitrile (N5) 5 mM, acetone cyanohydrin (N6) 25 mM, cyclohexanecarbonitrile (N7) 5 mM, 2-phenylpropionitrile (N8) 25 mM, and pyruvonitrile (N9) 10 mM. Concentrations of carboxylic acids were identical to that of the corresponding nitriles. Flasks filled only with M9-HEPES served as control. All treatments were prepared in triplicates. The solved and filtered compost (5 mL) was added to each baffled flask and the cultures were incubated at 25°C and 75 rpm (Multitron shaker). To monitor the growth of the cultures, the optical density at 600 nm (OD600) was measured. To prevent starvation of the microorganisms, cultures were transferred after 3 days to fresh medium containing the same nitrile or carboxylic acid at the concentrations used before. The new cultures were adjusted to an OD600 of 0.1 and incubated again at 25°C and 75 rpm. In case that cultures showed no growth, the cultures were not transferred to fresh medium but further incubated. The procedure was repeated two additional times after 2 days of incubation per growth period. The final cultures were again incubated for 2 days.
DNA Extraction
DNA of the compost sample solved in water was isolated before and after filtering using the PowerMax soil DNA isolation kit (MOBIO Laboratories, Carlsbad, CA, USA) according to the manufacturer's protocol. DNA of the M9-HEPES-containing enrichment cultures was isolated using the MasterPure Complete DNA and RNA purification kit as recommended by the manufacturer (Epicentre, Madison, WI, USA). Concentration of isolated DNA was measured using a NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA) or with a Quant-iT dsDNA HS assay kit and a Qubit fluorometer as recommended by the manufacturer (Invitrogen, Carlsbad, CA, USA).
16S rRNA Gene Sequencing
For amplification of the V3-V4 region of bacterial 16S rRNA genes, the primers S-D-Bact-0341-b-S-17 and S-D-Bact-0785-a-A-21 (Klindworth et al., 2013) were used. The primers contained adapters for Illumina MiSeq sequencing (Ilumina, San Diego, CA, USA). The DNA isolated from solved compost or M9-HEPES-containing enrichment cultures was used as template. PCR reaction mixtures (50 μl final volume) contained 10 μl 5-fold Phusion GC buffer, 0.2 μM of each of the primers, 0.2 μM MgCl2, 2.5 μl DMSO, 200 μM of each of the four dNTPs, 1 U of Phusion DNA polymerase (Thermo Fisher Scientific), and 25 ng template DNA. For amplification, the following protocol was used: initial denaturation at 98°C for 1 min, 25 cycles of denaturation at 98°C for 45 s, annealing at 60°C for 45 s, and elongation at 72°C for 30 s, followed by final elongation at 72°C for 5 min. PCR reactions for each culture were performed in triplicate, pooled in equal amounts, and purified with the NucleoMag 96 PCR kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's protocol. For quantification of the PCR products, a Qubit fluorometer was used in combination with the Quant-iT dsDNA HS assay kit as recommended by the manufacturer (Invitrogen). The purified amplicons were indexed using the Nextera XT DNA library prep kit according to the instructions of the manufacturer (Illumina). Dual index paired-end sequencing (2 × 300 bp) with v3 chemistry was done on an Illumina MiSeq platform as recommended by the manufacturer (Illumina).
16S rRNA Gene Sequence Analysis
CASAVA data analysis software (Illumina) was used for demultiplexing and clipping of sequence adapters from raw sequences. Before removing sequences with an average quality score below 20 and unresolved bases with split_libraries_fastq.py from QIIME 1.9.1 (Caporaso et al., 2010), paired-end sequences were merged using PEAR v0.9.10 with default parameters (Zhang et al., 2014). Default settings of cutadapt 1.14 (Martin, 2011) were used for removal of non-clipped reverse and forward primer sequences. Subsequently, the UNOISE2 pipeline of USEARCH 9.2.64 (Edgar, 2010) was used to remove, dereplicate, and denoise reads shorter than 380 bp, leading to zero-radius operational taxonomic units (zOTUs). Furthermore, chimeric sequences were removed. For this purpose, UCHIME2 was used in reference mode against the SILVA SSU database version 128 (Yilmaz et al., 2014). To create a zOTU table, the quality-filtered sequences were mapped to chimera-free OTUs using USEARCH. With parallel_assign_taxonomy_blast.py taxonomic classification of the zOTU sequences against the SILVA database was done. Filter_otu_table.py was used for removal of chloroplasts, unclassified OTUs, and extrinsic domain OTUs. Finally, the lowest number of sequences by random subsampling (12,100 reads per sample) was used for sample comparison at the same surveying effort.
Metagenome Sequencing and Analysis
DNA isolated from each culture triplicate was pooled in equal amounts. Paired-end metagenome sequencing was performed with an Illumina HiSeq 4000 (2 × 150 bp) using the Nextera XT DNA library prep kit. For quality-trimming and verification of the paired-end reads, Trimmomatic version 0.36 (Bolger et al., 2014) and FastQC version 0.11.5 (Andrews, 2010) were used. Assembly and coverage calculation were performed with SPAdes version 3.10 (Bankevich et al., 2012). Contigs >200 bp were annotated with Prokka 1.11 (Seemann, 2014). For taxonomic assignment of nitrilases, a BLASTn search (Altschul et al., 1990) was performed against the NCBI non-redundant database. Abundance of nitrile-degrading genes was calculated by mapping quality-filtered reads against the sequence-based identified putative nitrile-converting genes with Bowtie2 version 2.3.4.2 (Langmead and Salzberg, 2012). Normalization was done by dividing the number of mapped nucleotides by the total number of nucleotides. Subsequently, these values were normalized against the number of putative target genes.
Phylogenetic Tree
Evolutionary analyses were conducted in MEGA X version 10.0.5 (Kumar et al., 2018). Alignment of the amino acid sequences was done with MUSCLE (Edgar, 2004) and standard settings. The evolutionary history was inferred by using the Maximum Likelihood method and LG model (Le and Gascuel, 2008) as recommended by MEGA X's build-in model test tool. The tree with the highest log likelihood (−31012.19) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 1.1643)]. This analysis involved 276 amino acid sequences. A total of 491 positions were in the final dataset. Extensive subtree pruning regrafting and a very weak branch swap filter were used. Phylogeny was tested by calculating 500 bootstrap replicates and nodes with values below 50% were condensed for the final image. Reference sequences were recovered from the NCBI database (accession numbers in parentheses): For aromatic nitrilases, Rhodococcus rhodochrous J1 (BAA01994) (Kobayashi et al., 1992a), R. ruber NHB-2 (CCN27135) (Kaplan et al., 2013), and Gordonia terrae MTCC 8139 (AGR86048) (Kumar et al., 2015) were used as references. Aliphatic nitrilases were derived from Comamonas testosteroni (AAA82085) (Lévy-Schil et al., 1995), Synechocystis sp. PCC6803 (BAA10717) (Heinemann et al., 2003), and R. rhodochrous K22 (BAA02127) (Kobayashi et al., 1992b), while arylacetonitrilases were obtained from Alcaligenes faecalis JM3 (BAA02684) (Kobayashi et al., 1993), Pseudomonas fluorescens EBC191 (AAW79573) (Kiziak et al., 2005), and Pseudomonas putida MTCC 5110 (ABV21758) (Banerjee et al., 2009). β-Cyano-L-alanine nitrilases were derived from Pseudomonas fluorescens Pf0-1 (ABA74312) (Howden et al., 2009; Silby et al., 2009), Pseudomonas protegens Pf-5 (AAY92181) (Paulsen et al., 2005; Howden et al., 2009), and Pseudomonas pseudoalcaligenes CECT 5344 (CDM40486) (Wibberg et al., 2014; Acera et al., 2017). Cyanide dihydratases were retrieved from Pseudomonas stutzeri AK61 (AIY29195) (Crum et al., 2015), Bacillus pumilus C1 (AF392815) (Jandhyala et al., 2003), and B. pumilus 8A3 (AAN77003) (Jandhyala et al., 2003). Furthermore, three amidases were included, originating from Pseudomonas aeruginosa PAC142 (P11436) (Ambler et al., 1987), R. rhodochrous M8 (AAX83004) (Riabchenko et al., 2006), and Geobacillus pallidus RAPc8 (AAO23013) (Pereira et al., 1998).
Author Contributions
RH and RD conceived the study. RE and TZ performed the experiments. RE, DS, and RH analyzed and interpreted the data. RE, RH, and RD wrote the manuscript. All authors interpreted the results and reviewed the final version of the manuscript.
Funding
The work of RE was supported by the Fonds der Chemischen Industrie im Verband der Chemischen Industrie e.V. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mechthild Bömeke and Melanie Heinemann for technical support. We acknowledge support by the Open Access Publication Funds of the University of Göttingen.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2019.00103/full#supplementary-material
References
Abbas, Z., Akmal, M., Khan, K. S., and Fayyaz-ul-Hassan (2014). Effect of buctril super (bromoxynil) herbicide on soil microbial biomass and bacterial population. Brazilian Arch. Biol. Technol. 57, 9–14. doi: 10.1590/S1516-89132014000100002
Abo-El-Dahab, M., El-Goorani, M., and El-Wakil, M. (1977). Effect of certain chemicals on the vitro growth of Agrobacterium tumefaciens and on the gall formation on artificially infected plants. Egypt. J. Phytophatol. 9, 43–57.
Acera, F., Carmona, M. I., Castillo, F., Quesada, A., and Blasco, R. (2017). A cyanide-induced 3-cyanoalanine nitrilase in the cyanide-assimilating bacterium Pseudomonas pseudoalcaligenes strain CECT 5344. Appl. Environ. Microbiol. 83, 1–14. doi: 10.1128/AEM.00089-17
Ahmed, A. E., and Farooqui, M. Y. (1982). Comparative toxicities of aliphatic nitriles. Toxicol. Lett. 12, 157–163. doi: 10.1016/0378-4274(82)90179-5
Alongi, D. M. (1994). The role of bacteria in nutrient recycling in tropical mangrove and other coastal benthic ecosystems. Hydrobiologia 285, 19–32. doi: 10.1007/BF00005650
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ambler, R. P., Auffret, A. D., and Clarke, P. H. (1987). The amino acid sequence of the aliphatic amidase from Pseudomonas aeruginosa. FEBS Lett. 215, 285–290. doi: 10.1016/0014-5793(87)80163-1
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Banerjee, A., Dubey, S., Kaul, P., Barse, B., Piotrowski, M., and Banerjee, U. C. (2009). Enantioselective nitrilase from Pseudomonas putida: cloning, heterologous expression, and bioreactor studies. Mol. Biotechnol. 41, 35–41. doi: 10.1007/s12033-008-9094-z
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bauer, R., Knackmuss, H.-J., and Stolz, A. (1998). Enantioselective hydration of 2-arylpropionitriles by a nitrile hydratase from Agrobacterium tumefaciens strain d3. Appl. Microbiol. Biotechnol. 49, 89–95. doi: 10.1007/s002530051142
Baxter, J., and Cummings, S. P. (2008). The degradation of the herbicide bromoxynil and its impact on bacterial diversity in a top soil. J. Appl. Microbiol. 104, 1605–1616. doi: 10.1111/j.1365-2672.2007.03709.x
Bending, G. D., Rodríguez-Cruz, M. S., and Lincoln, S. D. (2007). Fungicide impacts on microbial communities in soils with contrasting management histories. Chemosphere 69, 82–88. doi: 10.1016/j.chemosphere.2007.04.042
Benedik, M. J., and Sewell, B. T. (2018). Cyanide-degrading nitrilases in nature. J. Gen. Appl. Microbiol. 64, 90–93. doi: 10.2323/jgam.2017.06.002
Bhatia, S. K., Mehta, P. K., Bhatia, R. K., and Bhalla, T. C. (2013). An isobutyronitrile-induced bienzymatic system of Alcaligenes sp. MTCC 10674 and its application in the synthesis of α-hydroxyisobutyric acid. Bioprocess Biosyst. Eng. 36, 613–625. doi: 10.1007/s00449-012-0817-y
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cohn, F. (ed.). (1872). “Untersuchungen über Bacterien,” in Beiträge zur Biologie der Pflanzen (Breslau: J. U. Kern's Verlag), 127–224.
Conn, E. E. (1969). Cyanogenic glycosides. J. Agric. Food Chem. 17, 519–526. doi: 10.1021/jf60163a014
Crum, M. A., Park, J. M., Mulelu, A. E., Sewell, B. T., and Benedik, M. J. (2015). Probing C-terminal interactions of the Pseudomonas stutzeri cyanide-degrading CynD protein. Appl. Microbiol. Biotechnol. 99, 3093–3102. doi: 10.1007/s00253-014-6335-x
Davis, K. E., Joseph, S. J., and Janssen, P. H. (2005). Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71, 826–834. doi: 10.1128/AEM.71.2.826-834.2005
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Egelkamp, R., Schneider, D., Hertel, R., and Daniel, R. (2017). Nitrile-degrading bacteria isolated from compost. Front. Environ. Sci. 5:56. doi: 10.3389/fenvs.2017.00056
Eyjólfsson, R. (1970). “Recent advances in the chemistry of cyanogenic glycosides,” in Fortschritte der Chemie organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products, eds W. Herz, H. Grisebach, and A. I. Scott (Vienna: Springer Vienna), 74–108. doi: 10.1007/978-3-7091-7123-3_2
Finnegan, I., Toerien, S., Abbot, L., Smit, F., and Raubenheimer, H. G. (1991). Identification and characterisation of an Acinetobacter sp. capable of assimilation of a range of cyano-metal complexes, free cyanide ions and simple organic nitriles. Appl. Microbiol. Biotechnol. 36, 142–144. doi: 10.1007/BF00164715
Fleming, F. F. (1999). Nitrile-containing natural products. Nat. Prod. Rep. 16, 597–606. doi: 10.1039/a804370a
Fleming, F. F., Yao, L., Ravikumar, P. C., Funk, L., and Shook, B. C. (2010). Nitrile-containing pharmaceuticals: efficacious roles of the nitrile pharmacophore. J. Med. Chem. 53, 7902–7917. doi: 10.1021/jm100762r
Gerasimova, T., Novikov, A., Osswald, S., and Yanenko, A. (2004). Screening, characterization and application of cyanide-resistant nitrile hydratases. Eng. Life Sci. 4, 543–546. doi: 10.1002/elsc.200402160
Gilligan, T., Yamada, H., and Nagasawa, T. (1993). Production of S-(+)-2-phenylpropionic acid from (R,S)-2-phenylpropionitrile by the combination of nitrile hydratase and stereoselective amidase in Rhodococcus equi TG328. Appl. Microbiol. Biotechnol. 39, 720–725. doi: 10.1007/BF00164456
Grogan, J., DeVito, S. C., Pearlman, R. S., and Korzekwa, K. R. (1992). Modeling cyanide release from nitriles: prediction of cytochrome P450 mediated acute nitrile toxicity. Chem. Res. Toxicol. 5, 548–552. doi: 10.1021/tx00028a014
Heinemann, U., Engels, D., Bürger, S., Kiziak, C., Mattes, R., and Stolz, A. (2003). Cloning of a nitrilase gene from the cyanobacterium Synechocystis sp. strain PCC6803 and heterologous expression and characterization of the encoded protein. Appl. Environ. Microbiol. 69, 4359–4366. doi: 10.1128/AEM.69.8.4359-4366.2003
Holly, K. (1964). Ioxynil and bromoxynil as herbicides. Int. J. Pest Manag. Part C 10, 256–258. doi: 10.1080/05331856409432879
Holtze, M. S., Hansen, H. C. B., Juhler, R. K., Sørensen, J., and Aamand, J. (2007). Microbial degradation pathways of the herbicide dichlobenil in soils with different history of dichlobenil-exposure. Environ. Pollut. 148, 343–351. doi: 10.1016/j.envpol.2006.10.028
Holtze, M. S., Sørensen, S. R., Sørensen, J., and Aamand, J. (2008). Microbial degradation of the benzonitrile herbicides dichlobenil, bromoxynil and ioxynil in soil and subsurface environments - insights into degradation pathways, persistent metabolites and involved degrader organisms. Environ. Pollut. 154, 155–168. doi: 10.1016/j.envpol.2007.09.020
Howden, A. J., Harrison, C. J., and Preston, G. M. (2009). A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J. 57, 243–253. doi: 10.1111/j.1365-313X.2008.03682.x
Howden, A. J., and Preston, G. M. (2009). Nitrilase enzymes and their role in plant-microbe interactions. Microb. Biotechnol. 2, 441–451. doi: 10.1111/j.1751-7915.2009.00111.x
Jandhyala, D., Berman, M., Meyers, P. R., Sewell, B. T., Willson, R. C., and Benedik, M. J. (2003). CynD, the cyanide dihydratase from Bacillus pumilus: gene cloning and structural studies. Appl. Environ. Microbiol. 69, 4794–4805. doi: 10.1128/AEM.69.8.4794-4805.2003
Jin, L.-Q., Liu, Z.-Q., Zheng, Y.-G., and Shen, Y.-C. (2010). Identification and characterization of Serratia marcescens ZJB-09104, a nitrile-converting bacterium. World J. Microbiol. Biotechnol. 26, 817–823. doi: 10.1007/s11274-009-0238-5
Jorgensen, J. H., and Ferraro, M. J. (2009). Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 49, 1749–1755. doi: 10.1086/647952
Kaplan, O., Veselá, A. B., Petríčková, A., Pasquarelli, F., Pičmanová, M., Rinágelová, A., et al. (2013). A comparative study of nitrilases identified by genome mining. Mol. Biotechnol. 54, 996–1003. doi: 10.1007/s12033-013-9656-6
Kinoshita, S., Nakayama, K., and Akita, S. (1958). Taxonomical study of glutamic acid accumulating bacteria, Micrococcus glutamicus nov. sp. Bull. Agric. Chem. Soc. Japan 22, 176–185. doi: 10.1080/03758397.1958.10857463
Kiziak, C., Conradt, D., Stolz, A., Mattes, R., and Klein, J. (2005). Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology 151, 3639–3648. doi: 10.1099/mic.0.28246-0
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Kobayashi, M., Izui, H., Nagasawa, T., and Yamada, H. (1993). Nitrilase in biosynthesis of the plant hormone indole-3-acetic acid from indole-3-acetonitrile: cloning of the Alcaligenes gene and site-directed mutagenesis of cysteine residues. Proc. Natl. Acad. Sci. U.S.A. 90, 247–251. doi: 10.1073/pnas.90.1.247
Kobayashi, M., Komeda, H., Yanaka, N., Nagasawa, T., and Yamada, H. (1992a). Nitrilase from Rhodococcus rhodochrous J1: sequencing and overexpression of the gene and identification of an essential cysteine residue. J. Biol. Chem. 267, 20746–20751.
Kobayashi, M., Yanaka, N., Nagasawa, T., and Yamada, H. (1990). Purification and characterization of a novel nitrilase of Rhodococcus rhodochrous K22 that acts on aliphatic nitriles. J. Bacteriol. 172, 4807–4815. doi: 10.1128/jb.172.9.4807-4815.1990
Kobayashi, M., Yanaka, N., Nagasawa, T., and Yamada, H. (1992b). Primary structure of an aliphatic nitrile-degrading enzyme, aliphatic nitrilase, from Rhodococcus rhodochrous K22 and expression of its gene and identification of its active site residue. Biochemistry 31, 9000–9007. doi: 10.1021/bi00152a042
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, V., Seth, A., Kumari, V., Kumar, V., and Bhalla, T. C. (2015). Purification, characterization and in-silico analysis of nitrilase from Gordonia terrae. Protein Pept. Lett. 22, 52–62. doi: 10.2174/0929866521666140909154537
Kurenkov, V. F. (1997). “Acrylamide polymers,” in Handbook of Engineering Polymeric Materials, ed N. P. Cheremisinoff (New York, NY: Marcel Dekker, 61–72.
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Layh, N., Hirrlinger, B., Stolz, A., and Knackmuss, H.-J. (1997). Enrichment strategies for nitrile-hydrolysing bacteria. Appl. Microbiol. Biotechnol. 47, 668–674. doi: 10.1007/s002530050993
Layh, N., Parratt, J., and Willetts, A. (1998). Characterization and partial purification of an enantioselective arylacetonitrilase from Pseudomonas fluorescens DSM 7155. J. Mol. Catal. B Enzym. 5, 467–474. doi: 10.1016/S1381-1177(98)00075-7
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Legras, J. L., Chuzel, G., Arnaud, A., and Galzy, P. (1990). Natural nitriles and their metabolism. World J. Microbiol. Biotechnol. 6, 83–108. doi: 10.1007/BF01200927
Lévy-Schil, S., Soubrier, F., Crutz-Le Coq, A. M., Faucher, D., Crouzet, J., and Pétré, D. (1995). Aliphatic nitrilase from a soil-isolated Comamonas testosteroni sp.: gene cloning and overexpression, purification and primary structure. Gene 161, 15–20. doi: 10.1016/0378-1119(95)00242-X
Lewis, D. F. V., and Wiseman, A. (2005). A selective review of bacterial forms of cytochrome P450 enzymes. Enzyme Microb. Technol. 36, 377–384. doi: 10.1016/j.enzmictec.2004.07.018
Mahadevan, S. (1973). Role of oximes in nitrogen metabolism in plants. Annu. Rev. Plant Physiol. 24, 69–88. doi: 10.1146/annurev.pp.24.060173.000441
Malathi, V. M., Jalali, S. K., Lyju, V. J., Gracy, R. G., More, R. P., Anandham, R., et al. (2017). Associated bacterial diversity of insecticide-susceptible and -resistant brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae) analyzed by culture-dependent and -independent methods. Phytoparasitica 45, 683–693. doi: 10.1007/s12600-017-0629-3
Marhan, J. (1995). Mutagenicity of cytostatic drugs in a bacterial system. I. Ames test. Folia Microbiol. 40, 457–461. doi: 10.1007/BF02814722
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17:10. doi: 10.14806/ej.17.1.200
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Nagasawa, T., Mauger, J., and Yamada, H. (1990). A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3. Purification and characterization. Eur. J. Biochem. 194, 765–772. doi: 10.1111/j.1432-1033.1990.tb19467.x
Pace, H. C., and Brenner, C. (2001). The nitrilase superfamily: classification, structure and function. Genome Biol. 2:reviews0001.1. doi: 10.1186/gb-2001-2-1-reviews0001
Paulsen, I. T., Press, C. M., Ravel, J., Kobayashi, D. Y., Myers, G. S., Mavrodi, D. V., et al. (2005). Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23, 873–878. doi: 10.1038/nbt1110
Pereira, R. A., Graham, D., Rainey, F. A., and Cowan, D. A. (1998). A novel thermostable nitrile hydratase. Extremophiles 2, 347–357. doi: 10.1007/s007920050078
Ramteke, P. W., Maurice, N. G., Joseph, B., and Wadher, B. J. (2013). Nitrile-converting enzymes: an eco-friendly tool for industrial biocatalysis. Biotechnol. Appl. Biochem. 60, 459–481. doi: 10.1002/bab.1139
Rashid, M. I., Mujawar, L. H., Shahzad, T., Almeelbi, T., Ismail, I. M. I., and Oves, M. (2016). Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol. Res. 183, 26–41. doi: 10.1016/j.micres.2015.11.007
Riabchenko, L., Podcherniaev, D., Kotlova, E., and Yanenko, A. (2006). Cloning the amidase gene from Rhodococcus rhodochrous M8 and its expression in Escherichia coli. Genetika 42, 1075–1082. doi: 10.1134/S1022795406080060
Richter, O.-M. H., and Ludwig, B. (2003). “Cytochrome c oxidase - Structure, function, and physiology of a redox-driven molecular machine,” in Reviews of Physiology, Biochemistry and Pharmacology, eds S. Amara, E. Bamberg, M. Blaustein, H. Grunicke, R. Jahn, W. Lederer, et al. (Berlin: Springer Berlin Heidelberg), 47–74. doi: 10.1007/s10254-003-0006-0
Romaniuk, K., Ciok, A., Decewicz, P., Uhrynowski, W., Budzik, K., Nieckarz, M., et al. (2018). Insight into heavy metal resistome of soil psychrotolerant bacteria originating from King George Island (Antarctica). Polar Biol. 41, 1319–1333. doi: 10.1007/s00300-018-2287-4
Santoshkumar, M., Nayak, A. S., Anjaneya, O., and Karegoudar, T. B. (2010). A plate method for screening of bacteria capable of degrading aliphatic nitriles. J. Ind. Microbiol. Biotechnol. 37, 111–115. doi: 10.1007/s10295-009-0663-3
Scheuer, P. J. (1992). Isocyanides and cyanides as natural products. Acc. Chem. Res. 25, 433–439. doi: 10.1021/ar00022a001
Schreiner, U., Hecher, B., Obrowsky, S., Waich, K., Klempier, N., Steinkellner, G., et al. (2010). Directed evolution of Alcaligenes faecalis nitrilase. Enzyme Microb. Technol. 47, 140–146. doi: 10.1016/j.enzmictec.2010.05.012
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Silby, M. W., Cerdeño-Tárraga, A. M., Vernikos, G. S., Giddens, S. R., Jackson, R. W., Preston, G. M., et al. (2009). Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 10:R51. doi: 10.1186/gb-2009-10-5-r51
Solden, L., Lloyd, K., and Wrighton, K. (2016). The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 31, 217–226. doi: 10.1016/j.mib.2016.04.020
Swiderska, A., Berndtson, A. K., Cha, M. R., Li, L., Beaudoin, G. M., Zhu, J., et al. (2001). Inhibition of the Agrobacterium tumefaciens TraR quorum-sensing regulator. Interactions with the TraM anti-activator. J. Biol. Chem. 276, 49449–49458. doi: 10.1074/jbc.M107881200
Tanii, H. (2017). Allyl nitrile: toxicity and health effects. J. Occup. Health 59, 104–111. doi: 10.1539/joh.16-0147-RA
Tanii, H., and Hashimoto, K. (1984). Studies on the mechanism of acute toxicity of nitriles in mice. Arch. Toxicol. 55, 47–54. doi: 10.1007/BF00316585
Tanner, A. C. (1985). “The role of bacteria in the cycling of nutrients within the maritime antarctic environment,” in Antarctic Nutrient Cycles and Food Webs, eds W. Siegfried, P. Condy, and R. Laws (Berlin: Springer Berlin Heidelberg), 123–127. doi: 10.1007/978-3-642-82275-9_18
Tipayno, S. C., Truu, J., Samaddar, S., Truu, M., Preem, J.-K., Oopkaup, K., et al. (2018). The bacterial community structure and functional profile in the heavy metal contaminated paddy soils, surrounding a nonferrous smelter in South Korea. Ecol. Evol. 8, 6157–6168. doi: 10.1002/ece3.4170
Torsvik, V., Sørheim, R., and Goksøyr, J. (1996). Total bacterial diversity in soil and sediment communities - a review. J. Ind. Microbiol. Biotechnol. 17, 170–178. doi: 10.1007/BF01574690
Uetrecht, J. P., and Trager, W. (2007). “Oxidation pathways and the enzymes that mediate them,” in Drug Metabolism: Chemical and Enzymatic Aspects, eds J. P. Uetrecht and W. Trager (New York, NY: Informa Healthcare), 33–108. doi: 10.1201/b14488
Vergne-Vaxelaire, C., Bordier, F., Fossey, A., Besnard-Gonnet, M., Debard, A., Mariage, A., et al. (2013). Nitrilase activity screening on structurally diverse substrates: providing biocatalytic tools for organic synthesis. Adv. Synth. Catal. 355, 1763–1779. doi: 10.1002/adsc.201201098
Viana, A. T., Caetano, T., Covas, C., Santos, T., and Mendo, S. (2018). Environmental superbugs: the case study of Pedobacter spp. Environ. Pollut. 241, 1048–1055. doi: 10.1016/j.envpol.2018.06.047
Wallig, M. A., Gould, D. H., Fettman, M. J., and Willhite, C. C. (1988). Comparative toxicities of the naturally occurring nitrile 1-cyano-3,4-epithiobutane and the synthetic nitrile n-valeronitrile in rats: differences in target organs, metabolism and toxic mechanisms. Food Chem. Toxicol. 26, 149–157. doi: 10.1016/0278-6915(88)90111-1
Wibberg, D., Luque-Almagro, V. M., Igeño, M. I., Bremges, A., Roldán, M. D., Merchán, F., et al. (2014). Complete genome sequence of the cyanide-degrading bacterium Pseudomonas pseudoalcaligenes CECT5344. J. Biotechnol. 175, 67–68. doi: 10.1016/j.jbiotec.2014.02.004
Willhite, C. C., and Smith, R. P. (1981). The role of cyanide liberation in the acute toxicity of aliphatic nitriles. Toxicol. Appl. Pharmacol. 59, 589–602. doi: 10.1016/0041-008X(81)90314-8
Yamamoto, K., Ueno, Y., Otsubo, K., Kawakami, K., and Komatsu, K. (1990). Production of S-(+)-ibuprofen from a nitrile compound by Acinetobacter sp. strain AK226. Appl. Environ. Microbiol. 56, 3125–3129.
Yerabolu, J. R., Liotta, C. L., and Krishnamurthy, R. (2017). Anchimeric-assisted spontaneous hydrolysis of cyanohydrins under ambient conditions: implications for cyanide-initiated selective transformations. Chem. A Eur. J. 23, 8756–8765. doi: 10.1002/chem.201701497
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: nitriles, nitrilases, nitrile hydratases, nitrile toxicity, nitrile degradation, metagenomics
Citation: Egelkamp R, Zimmermann T, Schneider D, Hertel R and Daniel R (2019) Impact of Nitriles on Bacterial Communities. Front. Environ. Sci. 7:103. doi: 10.3389/fenvs.2019.00103
Received: 09 February 2019; Accepted: 18 June 2019;
Published: 03 July 2019.
Edited by:
Rajeshwar P. Sinha, Banaras Hindu University, IndiaReviewed by:
Olimpio Montero, Spanish National Research Council (CSIC), SpainSamuel Ita Eduok, University of Uyo, Nigeria
Copyright © 2019 Egelkamp, Zimmermann, Schneider, Hertel and Daniel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rolf Daniel, rdaniel@gwdg.de
 Richard Egelkamp
Richard Egelkamp Till Zimmermann
Till Zimmermann Dominik Schneider
Dominik Schneider Robert Hertel
Robert Hertel Rolf Daniel
Rolf Daniel