- State Key Laboratory of Clean Energy Utilization, Zhejiang University, Hangzhou, China
Microalgae with a high growth rate and a carbon fixation rate present a promising potential to produce diverse renewable energy products and reduce greenhouse gas emissions. However, some microalgal species may have limited light-use efficiency and specific growth rate. To alleviate these issues, Nannochloropsis oceanica was subjected to 137Cs–γ radiation to obtain the desired mutant with enhanced light-use efficiency and increased biomass productivity. The N. oceanica mutant ZJU700 showed a 26.7% increase in biomass productivity after nuclear irradiation at a 700 GY dosage. It was found that the mutant had a 30.2% higher oxygen evolution rate than the wild type cells. High-throughput transcriptome sequencing showed that expression of photosynthetic related genes in the mutant were much higher than in wild type cells. Expression of the psbO gene increased by 455% in mutant ZJU700, contributing to an increased oxygen evolution rate by splitting water. Three up-regulated genes, namely petC, petF, and petH, resulted in enhanced electron transport during the photoreaction process. Up-regulation of many genes involved in the Calvin cycle indicated that CO2 fixation rate was likely to be increased to produce more carbohydrates in mutant, thereby contributing to the increased biomass productivity in mutant ZJU700.
Introduction
Human activities, such as combustion of fossil fuels have created a large amount of carbon dioxide (CO2), which can cause the greenhouse effect (Huang et al., 2015). Besides, the rapidly growing population of the world has increased the energy demand significantly (Khan et al., 2018). Among the numerous approaches to reduce the quantity of CO2 in the atmosphere, biotechnological methods employing microalgae have the exceptional advantages because of their natural negative carbon emission characters and the potential for producing highly valued byproducts (Sivakumar et al., 2014). Microalgae can be the feedstock for biofuels and value-added nutritional products with renewable, sustainable and economical characters (Khan et al., 2018). Microalgae may play a significant role in circular cascading biofuel systems (Wall et al., 2017). Among the microalgae strains, Nannochloropsis has been identified as a promising microalgal strain for its high lipid content and carbon fixation ability (Ma et al., 2013; Perin et al., 2015). The photosynthetic efficiency of microalgae converting light into biomass substantially influences the overall biomass productivity, which counts for the competitiveness of microalgae (Perin et al., 2015). Therefore, to make Nannochloropsis a competitive product on the market, biomass production needs to be optimized for its light use efficiency (Simionato et al., 2013; Moody et al., 2014). Among the optimization methods, microalgal strain mutation with nuclear radiation is a highly effective approach to improve their light-use efficiency.
Biological technologies have been widely used to improve targeted phenotypes of microalgae. At present, several studies have successfully improved Nannochloropsis characteristics through mutation. Ma et al. (2013) obtained a HP-1 mutant with increased biomass and enhanced light-use efficiency using heavy-ion radiation mutagenesis, however, the analytical methods were carried out at the macro level with no supporting information at the genetic level. Similarly, Perin et al. (2015) induced random mutagenesis using ethyl methane sulfonate, obtained a mutant with a higher biomass productivity by improving its photosynthetic ability, but again, no insights were given at the genetic level. Ermavitalini et al. (2017) studied the effects of 60Co-γ rays on Nannochloropsis and concluded that moderate dosage of γ rays could improve its biomass productivity and lipid content; however, the highest radiation dosage was 10 GY, which was too low to attain the best mutagenic results. Moreover, no further research was performed to determine the cause of this improvement. Liang et al. (2017) induced mutagenesis of Nannochloropsis by a chemical method and obtained fast-growing mutants. General gene expression analysis was carried out, but this research lacked photosynthetic analysis, such as oxygen evolution, which is the most relevant characterization of cell growth.
The researches above affirmed the feasibility of improving the light-use efficiency and biomass productivity of microalgae by mutagenesis. However, these researches lacked the interactive analysis between micro-level and macro-level. To improve light-use efficiency and biomass productivity, N. oceanica was mutated with 137Cs-γ radiation. The mutant strain ZJU700 showed a higher biomass productivity and oxygen evolution rate than the wild type. High-throughput transcriptome sequencing results showed that expression of photosynthesis-related genes were much higher in mutant ZJU700. Photosystem I (PSI), photosystem II (PSII) and electron transport processes were enhanced in the photoreaction. The Calvin cycle (dark reaction) was also enhanced and promoted CO2 fixation into carbohydrates. Moreover, the improved light-use efficiency induced by γ radiation contributed to higher biomass productivity in the mutant.
Materials and Methods
Microalgal Strain and Cultural Conditions
Nannochloropsis oceanica was obtained from Leadingtec biotechnology company, Shanghai, China. The F/2 seawater culture medium was used in the experiment as described elsewhere (Cheng et al., 2016), with the specific contents of NaNO3 and NaH2PO4 H2O adjusted to 2.64 and 0.20 mmol L−1, respectively.
The algal species were cultivated at 27°C in a 300 mL column glass bioreactor with 160 mm height, 56 mm inner diameter (ID) and 62 mm outer diameter (OD) in an artificial greenhouse, with the light intensity controlled at 7,000 Lux. A 1% CO2 mixture in nitrogen was bubbled into the bioreactor at a flow rate of 30 mL min−1. The bioreactor parameters and biomass measurements are described in our previous work (Cheng et al., 2016). All growth and oxygen evolution experiments were done in duplicate.
Mutation and Screening of Microalgae
Solutions of N. oceanica with initial OD750 of 1.5 were placed in fifteen 50 mL triangle bottles (three bottles for each radiation dosage) and irradiated with 137Cs-γ rays at dosages of 200, 300, 500, 700, and 900 Gy and a dose rate of 1 Gy min−1. Experiments were performed at the Academy of Agricultural Sciences, Zhejiang Province, China. The nuclear radiation generator was the same as described in the previous work (Cheng et al., 2016). After radiation, microalgal cells were placed in an incubator for 1 month for self-healing at 20°C and under an illumination of 1,000 Lux. The survival recovery percentage at each irradiation dosage was calculated as follows:
The survival recovery percentage was defined as 100% for the wild type, because no cells were died when there was no radiation.
Afterwards, 0.2 μL of microalgae from each triangle bottle treatment was injected into agar plates made of F/2 seawater medium described above with 1.5% agar, which was irradiated at different dosages, for purification of a single strain. After purifying thousands of mutants on agar plates, single colonies were picked into 96-well plate with F/2 seawater medium and cultivating in 96-well plate for 7 days, three strains exhibiting the highest biomass concentration at each radiation dosage [reflected by the optical density value from microplate reader (Tecan Infinite 200, Switzerland), OD405 was used as instructed] were screened and incubated for 8 days in 20 mL test tubes containing 5 mL of culture medium. During the large-scale cultivation, cell concentrations were measured by the spectrophotometer with a wavelength at 750 nm (Griffiths et al., 2011) and the strains with the fastest growth rate were further screened.
The relationship between dried weight of microalgal biomass and optical density of 750 nm is as follows:
Instrumental Analysis of N. oceanica and the Mutant
Oxygen evolution was measured in duplicate by Oxygraph (Hansatech, UK). For each treatment, a 1 mL culture sample was taken into the center column for the oxygen evolution test. The center column was illuminated at 350 μmol (m2·s)−1 using a red-light source. The oxygen evolution rate was calculated using the accumulated oxygen concentration and the time of the steady increase phase as following:
where Ct1 is the accumulated O2 concentration at time t1 and Ct0 is the accumulated O2 concentration at time t0, t1 = t0 + 60 s. The oxygen concentration was measured every second, while the data points shown in the figures represent every 5 s.
Main components (protein, carbohydrates, and lipids) in N. oceanica and the mutant were measured on the 4th day of 1% CO2 bubbling cultivation. The measurement methods of total carbohydrates content in N. oceanica biomass were determined by modified Anthrone method (Carrieri et al., 2010). The measurement methods of proteins and lipids contents in N. oceanica biomass were same as described in a previous work (Su et al., 2010).
Gene Expression Analysis of Mutant ZJU700
On the 4th day of cultivation, two duplicate samples of wild type and mutant ZJU700 were filtered, quick-frozen in liquid nitrogen, collected and stored at −80 °C, which were sent to Shanghai OE Biotech Co., Ltd for further analysis. High-throughput transcriptome sequencing was conducted on both the wild type and mutant ZJU700 samples. The manufacturer's protocols were followed when using the mirVana miRNA kit (Thermofisher) for RNA extraction; the Truseq stranded mRNA LT sample prep kit (Illumina) for sequencing library preparation; and the Bioanalyzer's High Sensitivity DNA kit (Agilent) for quality control of the libraries. Gene expression levels were compared between the wild type and mutant ZJU700.
Significance Test
The P-value analysis was conducted by the software “IBM SPSS Statistics 20,” which could give significance test result (P-value) when there were two replicates or more. This test could tell if the difference between two samples was significant.
(1) Input the two FPKM values of the first sample (two replicates) in “Column VAR00001,” input the two FPKM values of the second sample (two replicates) in “Column VAR00002.”
(2) Click “Analyze-Compare Means-Paired-Samples T Test.”
(3) Choose “VAR00001” in “Variable1” and choose “VAR00002” in “Variable2.” Click “OK” for analyze.
(4) Read the number in “Sig. (2-tailed)” of “Paired Samples Test” as the result of P-value for the significance test.
(5) P-value < 0.05 means the difference between the two samples was significant.
Results and Discussion
Mutating and Screening Nannochloropsis With Nuclear Radiation
Figure 1A shows the survival recovery percentage of N. oceanica after self-healed for 30 days from the irradiation with various dosages of γ rays. The survival recovery percentage decreased with the increasing γ radiation dosages, indicating that microalgae had a higher mortality rate under higher radiation intensity. This was because γ rays from nuclear radiation generate oxidative stress on thylakoid structures in microalgal cells, thereby impacting the PSII and Calvin cycle, or even causing death (Fuma et al., 2009).
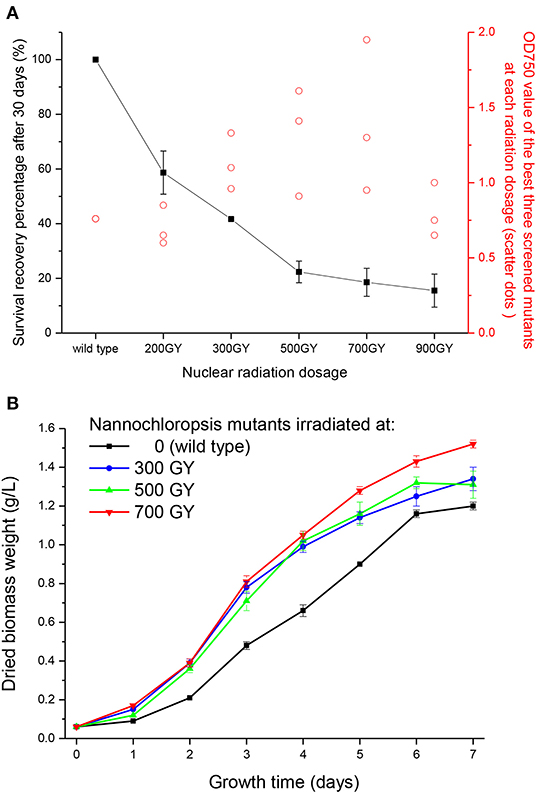
Figure 1. Survival recovery percentage and growth curves of Nannochloropsis mutants after various nuclear radiation dosages. (A) Survival recovery percentage on the 30th day after nuclear radiation and OD 750 value of the best three screened mutants at each radiation dosage (scatter dots). (B) Growth curves of screened Nannochloropsis mutants.
Meanwhile, photosynthesis of the remaining cells was more likely to be affected under higher radiation intensity, thus generating more phenotypes. After 8 days of large-scale cultivation in test tubes, the average cell concentration of the three best screened strains (reflected by OD750 value) were shown in Figure 1A. These three strains showed various growth phenotypes, which might have led to a large error bar. Little effect was observed on the screened mutant under the lowest irradiation (200 GY) as the cell concentration was nearly identical to the wild type. The biomass concentration of screened mutants increased with increasing radiation dosages, peaking at 700 GY, and then decreased at the irradiation of 900 GY. This might because genes in Nannochloropsis mutant cells were destructured and recombined to a greater extent under a higher radiation intensity, with the survived cells undergoing greater changes via acute evolutionary processes. After 700 GY radiation dosage, the average biomass concentration of the screened mutants increased by 84.2% compared with the wild type; however, the biomass concentration (OD750 value) decreased to 0.80 when the radiation dosage further increased to 900 GY, which might due to the extensive damage to the mutant cells caused by the excessive radiation.
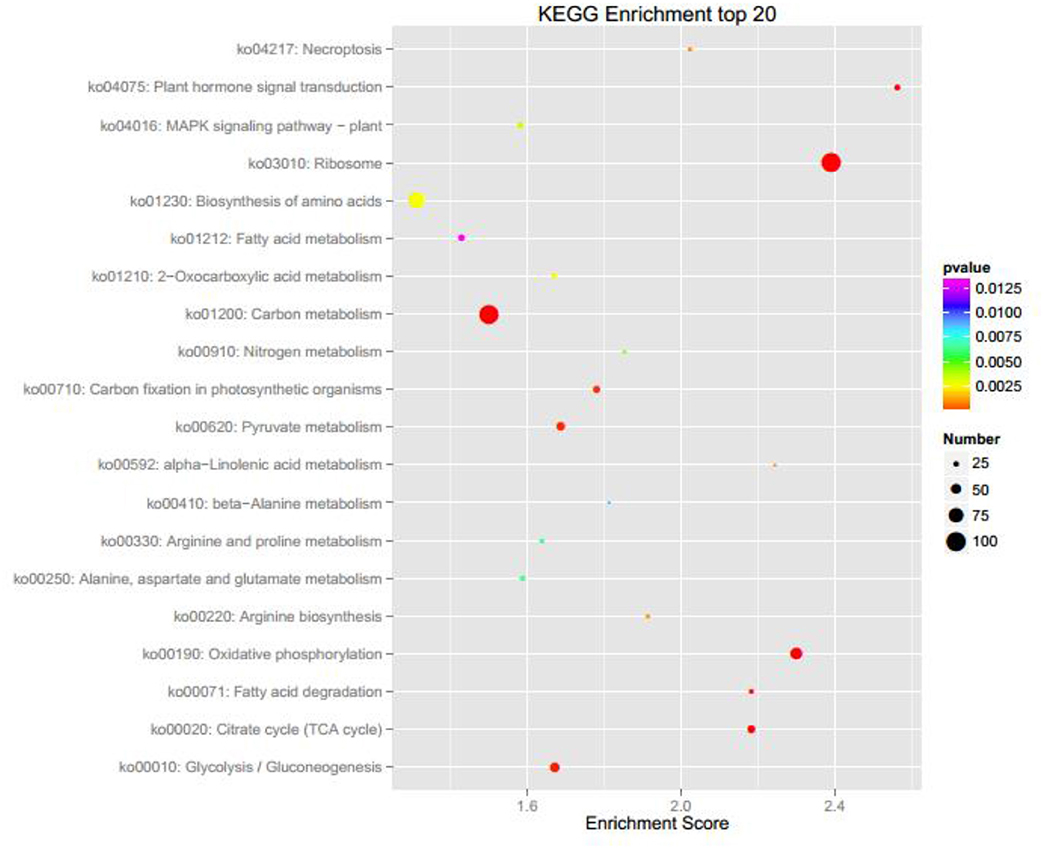
Wild type Nannochloropsis and mutant strains with the highest concentrations at radiation dosages of 300, 500, and 700 GY were then cultivated in the column photobioreactors. Growth curves were shown in Figure 1B. The dried biomass weight of all mutants were higher than the wild type. The mutant strain irradiated at 700 GY showed the highest biomass productivity, which was 26.7% higher than that of the wild type. This mutant strain showed the best growth phenotype after several rounds of screening; thus, it was designated as Nannochloropsis ZJU700 and used for the subsequent experiments. The mutant strain ZJU700 had been cultivated for tens of generations in the lab and showed similar growth phenotype among each generation, which proved the stability of this mutant. On the 4th day of the 1% CO2 bubbling cultivation, the carbohydrates content of the mutant (30%) was higher than the wild type N. oceanica (23%), whereas proteins and lipids content of the mutant (27 and 32%, respectively) were a little lower than the wild type (30 and 35%, respectively). The following two pathways were among the KEGG enrichment top 20 of wild type N. oceanica and the mutant: carbon fixation in photosynthetic organisms and citrate cycle, which were the most important pathways for carbon and biomass accumulation (Figure 2). This result indicated that the carbon fixation process changed greatly in the mutant ZJU700 cells, and finally led to its higher biomass productivity.
Interestingly, N. oceanica reached its highest biomass productivity at a radiation dosage of 700 GY, which was the same dosage at which Chlorella sp. peaked in our previous study (Cheng et al., 2016). Conversely, Spirulina (a prokaryotic organism) grew fastest after irradiation at 9,000 GY (Cheng et al., 2017). Therefore, the “best intensity” of Spirulina was 10 times higher than that of Nannochloropsis and Chlorella (eukaryotic microalgal strains). These results indicated that eukaryotic strains were more sensitive to γ rays from nuclear radiation and were more likely to attain the best following irradiation at around 700 GY, whereas prokaryotic strains, such as Spirulina would achieve the best phenotype at a much higher radiation intensity.
Higher Oxygen Evolution Rate in Mutant ZJU700
To further explore the reason for the improved growth phenotype of Nannochloropsis mutant ZJU700, oxygen evolution rates of mutant ZJU700 and the wild type were measured (Figure 3). On the 4th day, the oxygen evolution rate of mutant ZJU700 was 44.63 nmol/mg/min, which was 2.7% higher than the wild type (Figure 3B). When taking the biomass concentration into account, the oxygen evolution rate of mutant ZJU700 was 41.95 nmol/mL/min, which was 30.4% higher than the wild type (Figure 3b). This result shows that mutant ZJU700 produces oxygen much faster than the wild type.
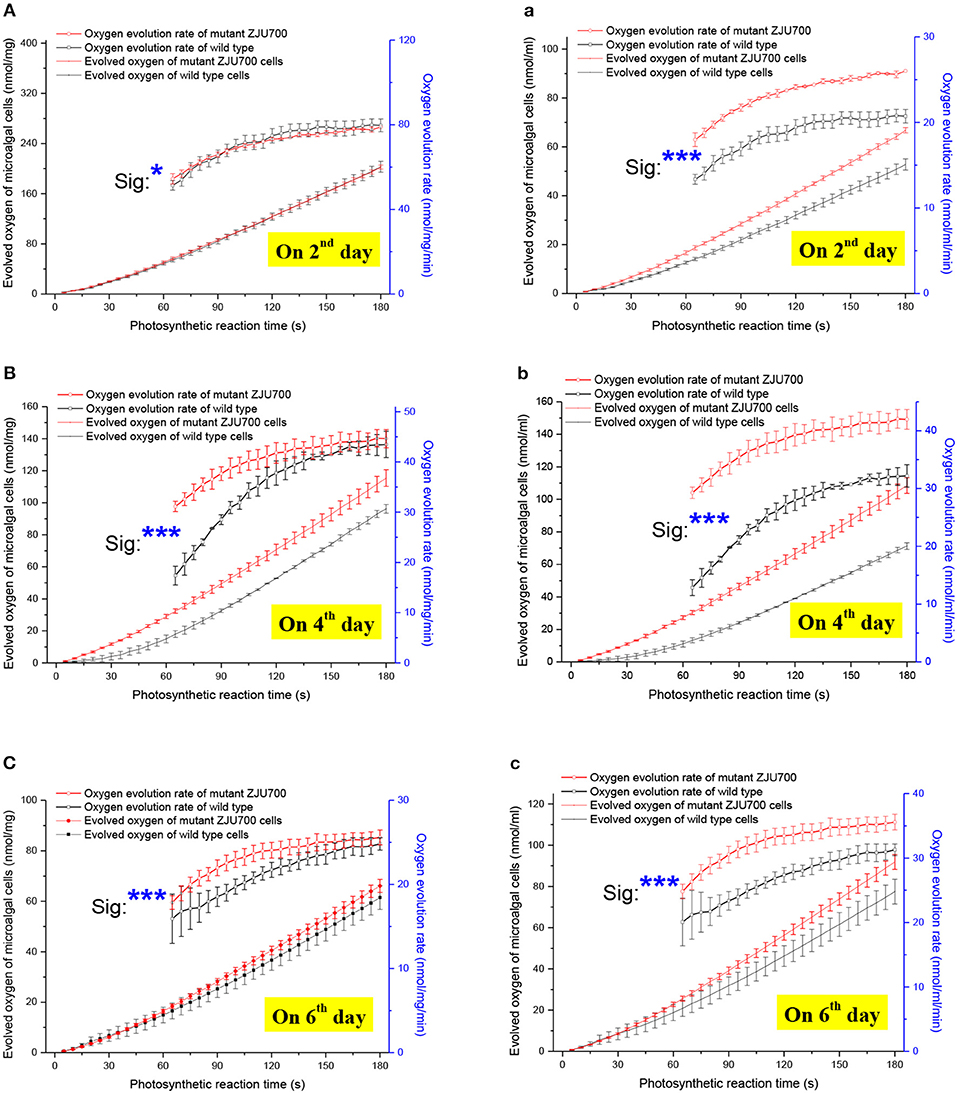
Figure 3. Oxygen evolution rate of microalgal cells of Nannochloropsis mutant ZJU700. Normalized to dried biomass weight: (A) Day 2, (B) Day 4, (C) Day 6. Normalized to culture volume: (a) Day 2, (b) Day 4, (c) Day 6.
Given the steady increase of accumulated oxygen (over 3 min) during the oxygen evolution measurement, constant oxygen evolution rates were obtained in both the wild type and mutant ZJU700, indicating steady downstream carbon assimilation during the Calvin cycle to pull the photosynthetic oxygen evolution by consuming its products. A higher carbon fixation rate could be inferred from the higher oxygen evolution rate, thereby contributing to the higher growth rate in the mutant. On the 6th day, the oxygen evolution rates of mutant ZJU700 and the wild type both decreased when compared with those on the 4th day, indicating that the carbon fixation rates decreased and the growth of the microalgae shifted from the logarithmic to the stationary phase.
Enhanced Photoreaction in Mutant ZJU700
The O2 evolution was significantly affected by photosynthetic and respiration processes. Gene expression levels between the wild type Nannochloropsis and mutant ZJU700 were analyzed, expression levels of photosynthesis related genes were much higher in the mutant than the wild type, while the respiration related genes were not found differently expressed.
A photoreaction splits water into oxygen, synthesizes NADPH and ATP, and provides energy and materials for carbon fixation in cells. After nuclear radiation mutagenesis, mutant ZJU700 showed not only a faster oxygen evolution rate at the macro level, but also a higher expression of photoreaction-related genes at the genetic level (Figure 4).
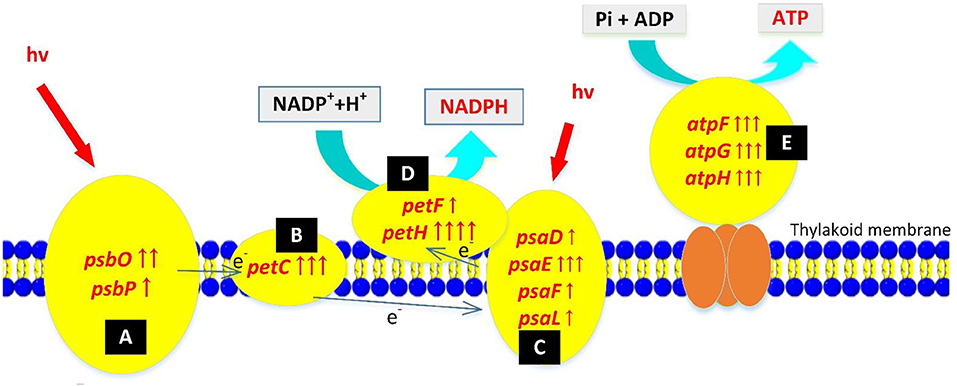
Figure 4. Up-regulation of photoreaction-related genes in the Nannochloropsis mutant ZJU700 after nuclear radiation. Yellow ovals represent protein complexes with up-regulated genes. One red upward arrow means that log2 (fold change of up-regulated gene expression) is higher than 1, two red upward arrows means that log2 (fold change of up-regulated gene expression) is higher than 2, etc. (A) Photosystem II complex; (B) Cytochrome b6/f complex; (C) Photosystem I complex; (D)ferredoxin and ferredoxin-NADP+reductase; (E) ATP synthase complex.
Up-Regulation of Genes Involved in Photosystems II and I
Photosystem II is a key system found in the thylakoid membranes of oxygenic photosynthetic organisms (Cady et al., 2008). In mutant ZJU700, genes of two PSII subunits genes, psbO and psbP, were expressed 4.55 and 2.02 times higher than the wild type (Table 1). The psbO gene is the most important enzyme complex in PSII, which catalyses oxygen evolution by splitting water (De Las Rivas and Barber, 2004; Rumpho et al., 2008). The psbP gene is required for PSII stability. Higher psbP expression has been attributed to a marked increase in the fluorescence quantum yield (Fv/Fm) (Ifuku et al., 2005; Yi et al., 2007). The up-regulation of PSII-related genes might well enhance the water splitting process and produced oxygen faster in mutant ZJU700.
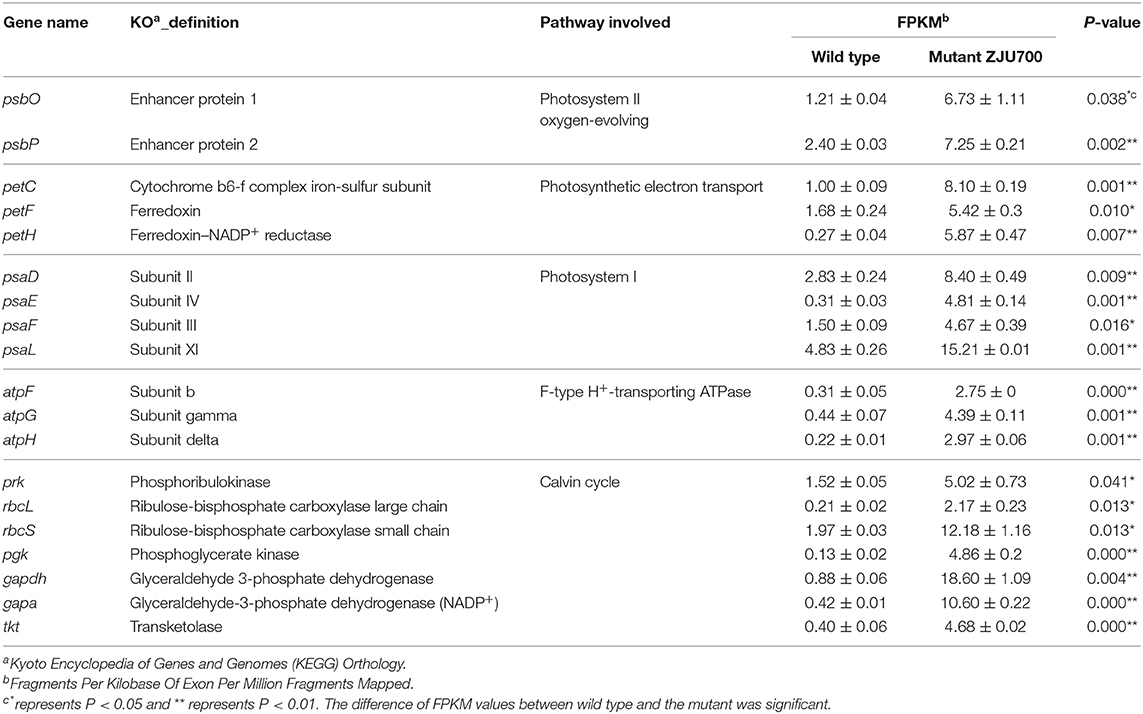
Table 1. Annotation and regulation of differentially expressed genes related to the photoreaction and Calvin cycle in the Nannochlorpsis mutant ZJU700.
PSI is a pigment-protein complex embedded in the thylakoid membrane and catalyses the photoreduction of ferredoxin and transfers electrons (Reilly and Nelson, 1988). Similar to PSII, four nuclear-encoded subunits of PSI were up-regulated in mutant ZJU700 (Table 1), thus promoting electron transport processes in PSI. The up-regulation of PSI-related genes was likely to contribute to the enhanced light absorption and electron transportation abilities.
Up-Regulation of Genes Involved in Photosynthetic Electron Transport
The petC, petF, and petH genes, which transport electrons at different sites during the photoreaction, were expressed 709, 223, and 2,109% times higher in mutant ZJU700 (Table 1). The petC gene encodes an iron-sulfur protein, which is an electron carrier and plays important roles in vital activities (Zusman et al., 2004). Overexpression of petF could distribute photosynthetic electrons to a variety of essential metabolic pathways and also increase the efficiency of scavenging reactive oxygen species, thereby alleviating damage to organelles (Floß et al., 1997; Lin et al., 2013). The petH gene is a flavoenzyme that catalyses the terminal step in PSI-associated electron transport, converting NADP+ and H+ into NADPH (Martínez-Júlvez et al., 1996). γ radiation led to the up-regulation of the above genes, thereby enhancing photosynthetic electron transportation in the PSI and provided more NADPH substrate for the dark reaction.
Up-Regulation of Genes Involved in ATP Synthase
ATP synthase is the most important enzyme that catalyses the formation of ATP during the photoreaction, and is driven by the energy provided from downstream proton transport along the proton gradient across membranes (Yoshida et al., 2001) (Equation 2).
Three subunit genes (atpF, atpG, and atpH) in this large protein complex were up-regulated after γ radiation (Table 1), which more efficiently catalyzed the conversion from ADP to ATP, thus providing more energy (ATP) for the dark reaction.
Enhanced Calvin Cycle in Mutant ZJU700
The Calvin cycle, an energy-consuming process that converts CO2 into organic carbon, is an open system connected to light photosynthetic reactions and plays a central role in algal metabolism (Jablonsky et al., 2011; Yang et al., 2017).
Results of transcriptome sequencing showed that most Calvin cycle-related genes were up-regulated in mutant ZJU700 (Figure 5). The rbcL and rbcS genes, which catalyse the primary carboxylation of ribulose 1,5-bisphosphate to yield two 3-phosphoglycerate molecules (Hügler and Sievert, 2011), were expressed 9.38 and 5.18 times higher, respectively, than the wild type (Table 1). The expression of gapdh, which catalyses the conversion of 1,3-bisphosphoglycerate into glyceraldehyde-3-phosphate (Michelet et al., 2013), increased by 2,024%. The prk gene catalyses phosphorylation of ribulose-5-phosphate to ribulose-1,5-bisphophate using ATP generated by thylakoid ATP synthase (Michelet et al., 2013). In mutant ZJU700, prk gene was expressed 2.30 times higher than in the wild type.
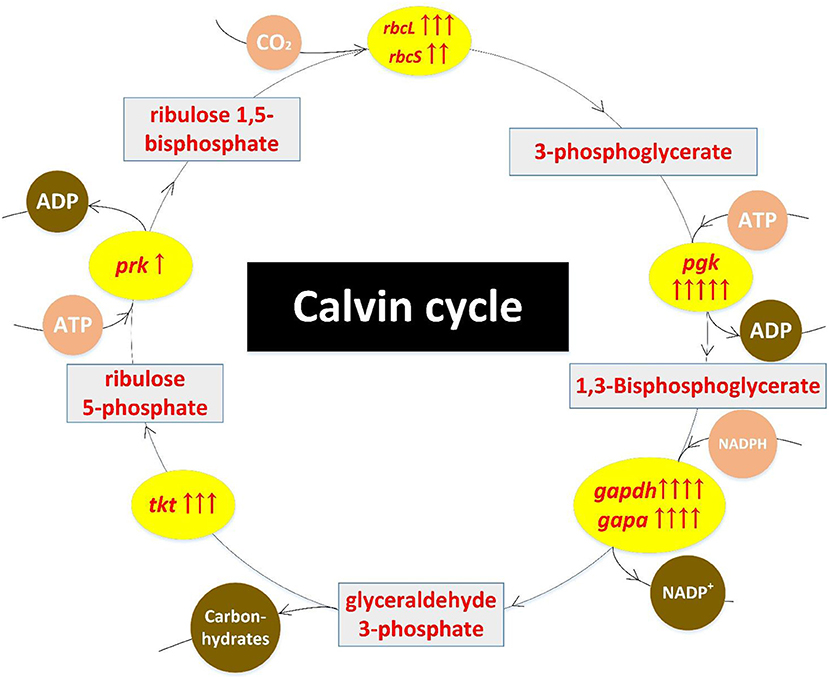
Figure 5. Up-regulation of Calvin cycle-related genes in the Nannochloropsis mutant ZJU700 after nuclear radiation. Yellow ovals represent enhanced enzymes with up-regulated genes. One red upward arrow means that log2 (fold change of up-regulated gene expression) is higher than 1, two red upward arrows means that log2 (fold change of up-regulated gene expression) is higher than 2, etc.
γ radiation led to the up-regulation of Calvin cycle-related genes and was likely to accelerate biomass accumulation while consuming more ATP generated from upstream photoreactions. This was echoed by the enhanced upstream processes, such as the higher oxygen evolution rate.
Conclusion
Nannochloropsis oceanica was mutated with 137Cs-γ radiation. The mutant strain ZJU700 showed a 26.7% increase in biomass productivity. Macro level results indicated that the oxygen evolution rate of the mutant was 30.2% higher than the wild type. Gene analysis via high-throughput transcriptome sequencing showed that expression of photosynthesis-related genes in both the photoreaction and Calvin cycle (dark reaction) of mutant ZJU700 were much higher than that of the wild type. Results at both the macro and genetic levels consistently suggest that mutant ZJU700 might have an improved light-use efficiency induced by γ radiation, which contributed to its higher biomass productivity.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
HL, JC, and JZ designed the study. HL, ZW, and XZ performed the experiment. HL and SC analyzed the data. HL and JC drafted the article. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by National Key R&D program-China (2017YFE0122800).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Prof. Dismukes at Rutgers University for his support and help in the oxygen evolution test.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2020.00143/full#supplementary-material
References
Cady, C. W., Crabtree, R. H., and Brudvig, G. W. (2008). Functional models for the oxygen-evolving complex of photosystem II. Coord. Chem. Rev. 252, 444–455. doi: 10.1016/j.ccr.2007.06.002
Carrieri, D., Momot, D., Brasg, I. A., Ananyev, G., Lenz, O., Bryant, D. A., et al. (2010). Boosting autofermentation rates and product yields with sodium stress cycling: application to production of renewable fuels by cyanobacteria. Appl. Environ. Microbiol. 76, 6455–6462. doi: 10.1128/AEM.00975-10
Cheng, J., Lu, H., He, X., Yang, W., Zhou, J., and Cen, K. (2017). Mutation of Spirulina sp. by nuclear irradiation to improve growth rate under 15% carbon dioxide in flue gas. Bioresour. Technol. 238, 650–656. doi: 10.1016/j.biortech.2017.04.107
Cheng, J., Lu, H., Huang, Y., Li, K., Huang, R., Zhou, J., et al. (2016). Enhancing growth rate and lipid yield of Chlorella with nuclear irradiation under high salt and CO2 stress. Bioresour. Technol. 203, 220–227. doi: 10.1016/j.biortech.2015.12.032
De Las Rivas, J., and Barber, J. (2004). Analysis of the structure of the PsbO protein and its implications. Photosyn. Res. 81, 329–343. doi: 10.1023/B:PRES.0000036889.44048.e4
Ermavitalini, D., Sari, I.P., Prasetyo, E.N., Abdulgani, N., and Saputro, T.B. (2017). Effect of gamma 60Co irradiation on the lipid content and fatty acid composition of Nannochloropsis sp. microalgae. AIP Conf. Proc. 1854:020009. doi: 10.1063/1.4985400
Floß, B., Igloi, G. L., Cassier-Chauvat, C., and Mühlenhoff, U. (1997). Molecular characterization and overexpression of the petF gene from Synechococcus elongatus: evidence for a second site of electrostatic interaction between ferredoxin and the PS I-D subunit. Photosyn. Res. 54, 63–71. doi: 10.1023/A:1005823620291
Fuma, S., Ishii, N., Takeda, H., Miyamoto, K., Yanagisawa, K., Doi, K., et al. (2009). Effects of acute γ-irradiation on the aquatic microbial microcosm in comparison with chemicals. J. Environ. Radioact. 100, 1027–1033. doi: 10.1016/j.jenvrad.2009.06.007
Griffiths, M. J., Garcin, C., van Hille, R. P., and Harrison, S. T. L. (2011). Interference by pigment in the estimation of microalgal biomass concentration by optical density. J. Microbiol. Methods 85, 119–123. doi: 10.1016/j.mimet.2011.02.005
Huang, Y., Cheng, J., Lu, H., Huang, R., Zhou, J., and Cen, K. (2015). Simultaneous enhancement of microalgae biomass growth and lipid accumulation under continuous aeration with 15% CO2. RSC Adv. 5, 50851–50858. doi: 10.1039/C5RA08401F
Hügler, M., and Sievert, S. M. (2011). Beyond the calvin cycle: autotrophic carbon fixation in the Ocean. Ann. Rev. Mar. Sci. 3, 261–289. doi: 10.1146/annurev-marine-120709-142712
Ifuku, K., Yamamoto, Y., Ono, T.-A., Ishihara, S., and Sato, F. (2005). PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol. 139, 1175–1184. doi: 10.1104/pp.105.068643
Jablonsky, J., Bauwe, H., and Wolkenhauer, O. (2011). Modeling the Calvin-Benson cycle. BMC Syst. Biol. 5, 1–13. doi: 10.1186/1752-0509-5-185
Khan, M. I., Shin, J. H., and Kim, J. D. (2018). The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Fact. 17, 1–21. doi: 10.1186/s12934-018-0879-x
Liang, S., Guo, L., Lin, G., Zhang, Z., Ding, H., Wang, Y., et al. (2017). Improvement of Nannochloropsis oceanica growth performance through chemical mutation and characterization of fast growth physiology by transcriptome profiling. Chinese J. Oceanol. Limnol. 35, 792–802. doi: 10.1007/s00343-017-6023-7
Lin, Y. H., Pan, K. Y., Hung, C. H., Huang, H. E., Chen, C. L., Feng, T. Y., et al. (2013). Overexpression of ferredoxin, PETF, enhances tolerance to heat stress in Chlamydomonas reinhardtii. Int. J. Mol. Sci. 14, 20913–20929. doi: 10.3390/ijms141020913
Ma, Y., Wang, Z., Zhu, M., Yu, C., Cao, Y., Zhang, D., et al. (2013). Increased lipid productivity and TAG content in Nannochloropsis by heavy-ion irradiation mutagenesis. Bioresour. Technol. 136, 360–367. doi: 10.1016/j.biortech.2013.03.020
Martínez-Júlvez, M., Hurley, J. K., Tollin, G., Gómez-Moreno, C., and Fillat, M. F. (1996). Overexpression in E. coli of the complete petH gene product from Anabaena: purification and properties of a 49 kDa ferredoxin-NADP+ reductase. Biochim. Biophys. Acta 1297, 200–206. doi: 10.1016/S0167-4838(96)00024-6
Michelet, L., Zaffagnini, M., Morisse, S., Sparla, F., Pérez-Pérez, M. E., Francia, F., et al. (2013). Redox regulation of the Calvin–Benson cycle: something old, something new. Front. Plant Sci. 4:470. doi: 10.3389/fpls.2013.00470
Moody, J. W., McGinty, C. M., and Quinn, J. C. (2014). Global evaluation of biofuel potential from microalgae. Proc. Natl. Acad. Sci. U.S.A. 111, 8691–8696. doi: 10.1073/pnas.1321652111
Perin, G., Bellan, A., Segalla, A., Meneghesso, A., Alboresi, A., and Morosinotto, T. (2015). Generation of random mutants to improve light-use efficiency of Nannochloropsis gaditana cultures for biofuel production. Biotechnol. Biofuels 8, 1–13. doi: 10.1186/s13068-015-0337-5
Reilly, P., and Nelson, N. (1988). Photosystem I complex. Mol. Biol. Photosynth. 1, 485–496. doi: 10.1007/978-94-009-2269-3_22
Rumpho, M. E., Worful, J. M., Lee, J., Kannan, K., Tyler, M. S., Bhattacharya, D., et al. (2008). Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc. Natl. Acad. Sci. U.S.A. 105, 17867–17871. doi: 10.1073/pnas.0804968105
Simionato, D., Basso, S., Giacometti, G. M., and Morosinotto, T. (2013). Optimization of light use efficiency for biofuel production in algae. Biophys. Chem. 182, 71–78. doi: 10.1016/j.bpc.2013.06.017
Sivakumar, M., Ranjith Kumar, R., Shashirekha, V., and Seshadri, S. (2014). Influence of carbon-dioxide on the growth of Spirulina sp. (MCRC-A0003) isolated from Muttukadu backwaters, South India. World J. Microbiol. Biotechnol. 30, 2775–2781. doi: 10.1007/s11274-014-1688-y
Su, H., Cheng, J., Zhou, J., Song, W., and Cen, K. (2010). Hydrogen production from water hyacinth through dark- and photo-fermentation. Int. J. Hydrogen Energy 35, 8929–8937. doi: 10.1016/j.ijhydene.2010.06.035
Wall, D. M., McDonagh, S., and Murphy, J. D. (2017). Cascading biomethane energy systems for sustainable green gas production in a circular economy. Bioresour. Technol. 243, 1207–1215. doi: 10.1016/j.biortech.2017.07.115
Yang, B., Liu, J., Ma, X., Guo, B., Liu, B., Wu, T., et al. (2017). Genetic engineering of the Calvin cycle toward enhanced photosynthetic CO2 fixation in microalgae. Biotechnol. Biofuels 10:299. doi: 10.1186/s13068-017-0916-8
Yi, X., Hargett, S. R., Liu, H., Frankel, L. K., and Bricker, T. M. (2007). The PsbP protein is required for photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J. Biol. Chem. 282, 24833–24841. doi: 10.1074/jbc.M705011200
Yoshida, M., Muneyuki, E., and Hisabori, T. (2001). ATP synthase–a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677. doi: 10.1038/35089509
Keywords: microalgae, radiation mutagenesis, biomass productivity, oxygen evolution, photosynthesis
Citation: Lu H, Cheng J, Wang Z, Zhang X, Chen S and Zhou J (2020) Enhancing Photosynthetic Characterization and Biomass Productivity of Nannochloropsis Oceanica by Nuclear Radiation. Front. Energy Res. 8:143. doi: 10.3389/fenrg.2020.00143
Received: 04 November 2019; Accepted: 10 June 2020;
Published: 28 July 2020.
Edited by:
David M. Wall, University College Cork, IrelandReviewed by:
Changhong Yao, Sichuan University, ChinaBaohua Zhu, Ocean University of China, China
Yandu Lu, Hainan University, China
Copyright © 2020 Lu, Cheng, Wang, Zhang, Chen and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Cheng, juncheng@zju.edu.cn
 Hongxiang Lu
Hongxiang Lu Jun Cheng
Jun Cheng Zhenyi Wang
Zhenyi Wang