- 1Department of Physics and Astronomy, University of Southern California, Los Angeles, CA, United States
- 2Department of Chemistry, University of Southern California, Los Angeles, CA, United States
- 3Department of Biological Sciences, University of Southern California, Los Angeles, CA, United States
- 4Department of Earth Science, University of Southern California, Los Angeles, CA, United States
- 5Center for the Conservation of Biological Resources, Black Hills State University, Spearfish, SD, United States
- 6Department of Life Sciences, Texas A&M University, Corpus Christi, TX, United States
The terrestrial deep subsurface is host to significant and diverse microbial populations. However, these microbial populations remain poorly characterized, partially due to the inherent difficulty of sampling, in situ studies, and isolating of the in situ microbes. Motivated by the ability of microbes to gain energy from redox reactions at mineral interfaces, we here present in situ electrochemical colonization (ISEC) reactor as a method to directly study microbial electron transfer activity and to enable the capture and isolation of electrochemically active microbes. We installed a potentiostatically controlled ISEC reactor containing four working electrodes 1,500 m below the surface at the Sanford Underground Research Facility (SURF). The working electrodes were poised at different redox potentials to mimic energy-yielding mineral reducing and oxidizing reactions predicted to occur at this site. We present a 16S rRNA analysis of the in situ electrode-associated microbial communities, revealing the dominance of novel bacterial lineages under cathodic conditions. We also demonstrate that the in situ electrodes can be further used for downstream electrochemical laboratory enrichment and isolation of novel strains. Using this workflow, we isolated Bacillus, Anaerospora, Comamonas, Cupriavidus, and Azonexus strains from the electrode-attached biomass. Finally, the extracellular electron transfer (EET) activity of Comamonas strain (isolated at −0.19 V vs. SHE and designated WE1-1D1) and Bacillus strain (isolated at +0.53 V vs. SHE and designated WE4-1A1-BC) from and to a poised electrode, respectively, were confirmed in electrochemical reactors. Our study highlights the utility of in situ electrodes and electrochemical enrichment workflows to shed light on microbial activity in the deep terrestrial subsurface.
Introduction
Minerals that contain redox active elements (e.g., S, Fe, Mn) are abundant in subsurface environments, and can support the growth of microorganisms by acting as electron acceptors for heterotrophic respiration or electron donors for lithotrophic, and often autotrophic, metabolism (Nealson et al., 2002; Bach and Edwards, 2003; Edwards et al., 2005; Fredrickson and Zachara, 2008; Orcutt et al., 2011; Southam, 2012; Shi et al., 2016). This process of extracellular electron transfer (EET) to or from minerals is best characterized in a handful of Fe-reducing bacteria, especially Geobacter and Shewanella species (Lovley and Phillips, 1988; Myers and Nealson, 1988; Shi et al., 2016). To overcome the hurdle of electron transfer across the otherwise electrically-insulating cell envelope, these bacteria rely on a number of mechanisms: multiheme cytochrome complexes that bridge the periplasm and outer membrane (Myers and Myers, 1992; Hartshorne et al., 2009; White et al., 2013), microbial nanowires that reach out to distant electron acceptors (Reguera et al., 2005; Gorby et al., 2006), and soluble redox shuttles that diffusively link cells to external surfaces (Marsili et al., 2008; Von Canstein et al., 2008). EET can be electrochemically mimicked on electrode surfaces that function as surrogate electron acceptors (anodes) or donors (cathodes) to support microbial metabolism, depending on the poised potentials of these electrodes (Bond and Lovley, 2003; Gregory et al., 2004; Ross et al., 2011; Carmona-Martínez et al., 2013; Rowe et al., 2017a; Hirose et al., 2018). As a result, electrochemical enrichments have been applied to microbial samples from a variety of environments (Bond et al., 2002; Tender et al., 2002; Holmes et al., 2004; Kim et al., 2004; Rabaey and Verstraete, 2005; Reimers et al., 2006; White et al., 2009; Bond, 2010; Meyer et al., 2016). When combined with surveys of microbial community structure, these electrochemical techniques greatly expanded our understanding of the phylogenetic diversity of microbes capable of colonizing electrodes and led to the isolation of novel microorganisms capable of EET to anodes (Wrighton et al., 2008; Xing et al., 2008; Fedorovich et al., 2009; Badalamenti et al., 2016; Jangir et al., 2016; Kawaichi et al., 2018). While our mechanistic understanding of the molecular pathways that underlie inward EET (i.e., electron transfer from rather than to surfaces) lags behind metal reduction pathways, electrode-based techniques have highlighted the diversity of microbes capable of electron uptake from cathodes: acetogens, methanogens, as well as iron- and sulfur-oxidizers (Nevin et al., 2011; Summers et al., 2013; Bose et al., 2014; Beese-Vasbender et al., 2015; Deutzmann et al., 2015; Ishii et al., 2015; Rowe et al., 2015, 2017b).
The Sanford Underground Research Facility (SURF), located in the Black Hills of South Dakota, USA is the site of the former Homestake Gold Mine. It is hosted in quartz-veined, sulfide-rich segments of an Early Proterozoic, carbonate-facies iron-formation (Caddey et al., 1991; Hart et al., 2014; Lesko, 2015). The facility's tunnels provide access, maximally 4,850 ft (1,478 m) below ground surface (bgs), to deep subsurface fluids through a network of manifolds. The residence times of the accessible fluids range from very recent in the shallowest levels to much older (>10,000 years) fluids reaching the deeper levels (Murdoch et al., 2012; Osburn et al., 2014). The mine was converted to a state-run science facility in 2011, with emphasis on rare-process physics (Heise, 2015), and this accessible infrastructure provided a remarkable portal for in situ studies of the deep terrestrial biosphere. Osburn et al. (2014) combined geochemical data, energetic modeling, and 16S rRNA gene sequences from different boreholes to both assess the in situ microbial communities and predict energy-yielding metabolisms for unknown physiotypes. Notably, abundant energy was predicted for microorganisms from a variety of reactions including sulfur oxidation, iron oxidation, and manganese reduction, which motivated us to apply electrode-based techniques to assess and enrich for microbes capable of EET.
Here we report the deployment and operation of an in situ electrochemical colonization (ISEC) reactor fed by a manifold at the 4,850 ft (1,478 m) level of SURF. The reactor contained four working electrodes, poised at potentials spanning lower to higher, to act as an inexhaustible source or sink of electrons for capture of potential mineral oxidizing and reducing bacteria, with emphasis on mimicking sulfur oxidation, iron oxidation, and manganese reduction predicted as energy-yielding reactions in this manifold (Osburn et al., 2014). We hypothesized that physical electrodes, when deployed in situ, can serve as electron donors and/or acceptors to capture important bacterial lineages capable of EET from the deep subsurface. Further, we utilized these in situ electrodes for subsequent laboratory enrichment and isolation of novel strains, and directly tested the electron uptake by Comamonas isolate and electron donation by Bacillus isolate via a poised electrodes. To our knowledge, this effort represents the first potentiostatically-controlled multi-electrode reactor deployed in situ in a deep subsurface environment.
Materials and Methods
Field Measurements
The exploratory borehole (DUSEL 3A) studied is located at the 1,478 m (4,850 ft) below ground surface level close to the Yates shaft at SURF in Lead, South Dakota (USA). The 214 m-long borehole, drilled in 2009 intersects the Precambrian amphibolite metamorphic schists (Caddey et al., 1991; Hart et al., 2014) and has been capped with manifolds to prevent flooding. One year prior to this study, initial aqueous chemistry of DUSEL 3A fluid was obtained for thermodynamic modeling in February 2014 (Osburn et al., 2014). Further, detailed geochemical data were acquired during installation (December 2014) and conclusion (May 2015) of the ISEC reactor incubation. Aqueous chemistry (Table 1) was measured as described earlier (Osburn et al., 2014). Briefly, oxidation-reduction potential (ORP), conductivity, pH, temperature, and total dissolved solids (TDS) were measured in situ with an Ultrameter II 6PFCE (Myron L Company, Carlsbad, CA). The redox-sensitive species were measured using Hach DR/2400 portable field spectrophotometers and associated reaction kits (Hach Company, Loveland, CO). Major anions and cations were measured using ion chromatography with Metrohm column Metrosep A SUPP 150 and Metrosep C6−250/4.0, respectively (Metrohm, Fountain Valley, CA).
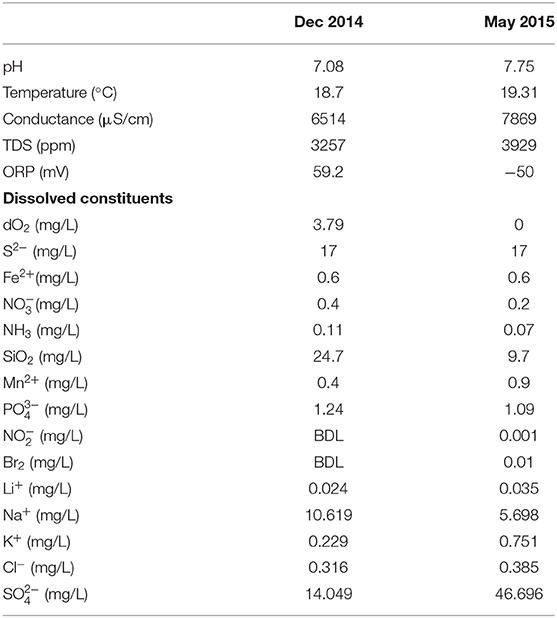
Table 1. Chemical composition of fluid from DUSEL 3A at the 4,850 ft (1,478 mbsl) level, Sanford Underground Research Facility (SD, USA), after commissioning and decommissioning of in situ electrochemical colonization (ISEC) reactor.
In situ Electrochemical Colonization Reactor
Fluid from DUSEL 3A was pumped through the ISEC reactor (Figure 1) for 5 months via peristaltic pumps (MasterFlex L/S Digital Drive, EW-07522-20, Cole-Parmer, USA) at a flow rate of 1 mL/min, providing a dilution rate of 1 day−1. Aqueous chemistry (ORP, conductivity, pH, and temperature) of the ISEC effluent was logged using a multiparameter HI9829 meter (Hanna Instruments, Woonsocket, RI). ISEC is a standard electrochemical half-cell design, custom fabricated from a 1 L spinner flask (CLS-1410, Chemglass Life Sciences, Vineland, NJ) to incorporate an Ag/AgCl reference electrode (3M NaCl, MF-2052, BASi, West Lafayette, IN) and a platinum counter electrode (CHI115, CH Instruments, Austin, TX). It holds a PTFE assembly of five threaded rods, each of which supported a working electrode (WE) composed of 6 × 7 cm carbon cloth (PW06, Zoltek, St. Louis, MO, USA). During enrichment, four working electrodes (WE1, WE2, WE3, and WE4) were poised at −0.19, +0.01, +0.26, and +0.53 V vs. SHE, respectively, using a four-channel potentiostat (EA164 Quadstat, EDaq, USA). Potentials were chosen to mimic conditions consistent with elemental sulfur oxidation (−0.19 V vs. SHE), iron oxidation (+0.01 V vs. SHE), and manganese reduction (+0.53 V vs. SHE), all predicted as putative energy-yielding metabolisms for the microbial community inhabiting the DUSEL 3A fluid (Osburn et al., 2014). Since the reduction potential of many cytochromes ranges from −0.5 to +0.3 V vs. SHE (Kracke et al., 2015) a fourth potential was also applied at +0.26 V vs. SHE. The reduction potentials were evaluated by: (1) converting the standard Gibbs free energy (G0) to standard reduction potential () of the reaction via (where: n refers to the number of electron transferred and F stands for the Faraday constant), and (2) using the geochemical data (specifically pH, temperature, and the concentration of SO, S2−, Mn2+, Fe2+) of DUSEL 3A liquid (from Dec 2014) to evaluate in situ reduction potential (Ered) via the Nernst Equation (where R refers to the gas constant, T denotes the temperature in kelvin, n refers to the number of electron transferred and F stands for the Faraday constant and Qr represents the reaction quotient of the cell reaction). All electrical connections were made using titanium insulated wire (KeegoTech, Menlo Park, CA, USA). The complete set-up was autoclaved along with the working and the counter electrodes. The reference electrode was ethanol-sterilized before it was inserted. Applied redox potentials and current production were controlled and recorded via the eCorder eCHART software (eDAQ Inc., Colorado Springs, CO). Before the deployment of the ISEC reactor in December 2014, the SURF facility made modifications to the manifold's design, which could potentially lead to backflow of fluid from a dehumidifier into the reactor. To avert this contamination, in December 2014, a check valve was installed upstream of the solenoid valve as shown in the schematic diagram of ISEC reactor (Figure 1). The reactor was incubated with a continuous flow of DUSEL 3A fluid for 5 months beginning December 2014, monitored monthly for proper operation, and finally retrieved in May 2015. During decommission, small sections of carbon cloth from each potential were collected for microscopy, active microbial community analysis, and further laboratory electrochemical enrichments as described below.
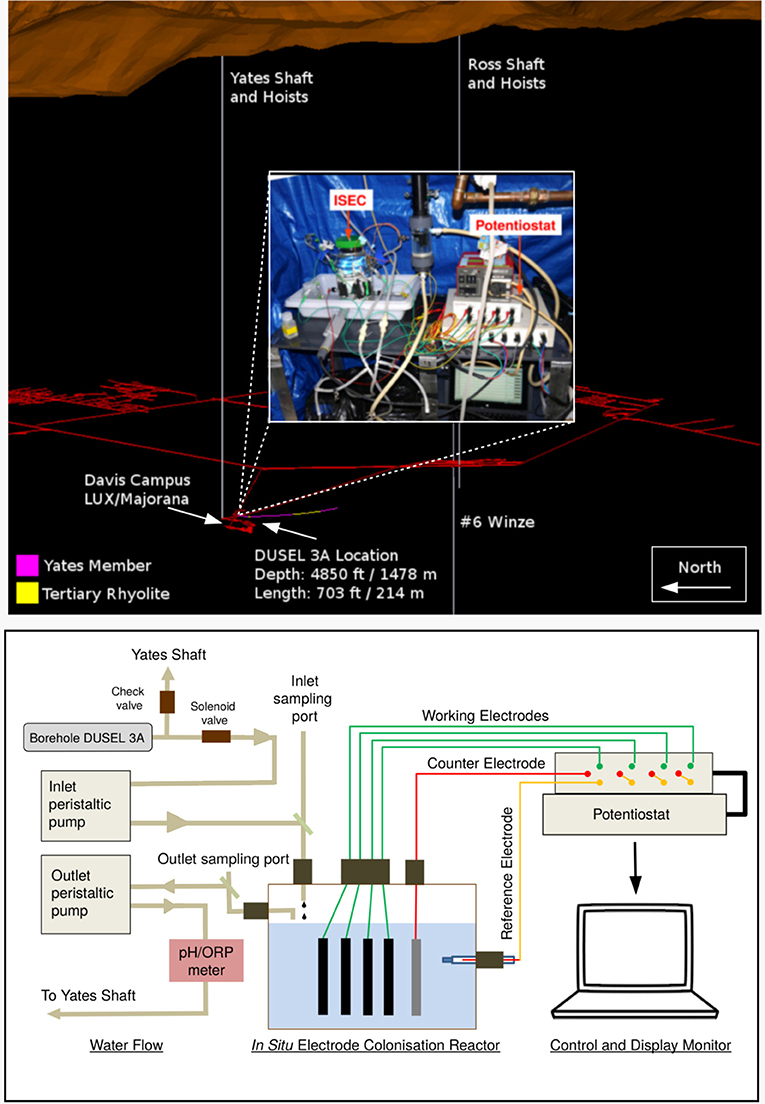
Figure 1. (Top) A 3-dimensional layout of Sanford Underground Research Facility (image to scale). DUSEL 3A, located at 4,850 ft level close to Davis Campus cutting the iron-rich Precambrian Yates member, was selected as the site for in situ electrochemical colonization (ISEC) reactor deployment. An image of the ISEC reactor after deployment (Dec 2014) is shown in the inset. (Bottom) Schematic diagram of ISEC reactor. The four carbon cloth working electrodes were poised at −0.19, +0.01, +0.26, and +0.53 V vs. SHE, respectively, using a 4-channel potentiostat acting as electron donors and acceptors in a single reactor. DUSEL 3A fluid was fed directly into the reactor and the effluent was pumped out to Yates shaft.
Secondary Laboratory Electrochemical Enrichment
At the conclusion of ISEC deployment, the carbon cloth working electrodes (3 × 6 cm) were placed into autoclaved 125 mL serum bottles containing filtered (0.22 μm) DUSEL 3A fluid filled to capacity and sealed with sterile butyl rubber stoppers and aluminum seals (Chemglass Life Sciences, USA). The serum bottles were stored in vacuum-sealed Mylar bags (ShieldPro, USA) with oxygen absorbers (OxyFree, USA) to minimize oxygen exposure during transportation. In the laboratory, secondary electrochemical enrichments were performed using the poised carbon cloth from the reactor in four separate three-electrode electrochemical cells (50 mL total volume). A standard three-electrode glass cell comprised of: (1) working electrode (WE): a carbon cloth with dimensions 1 × 1 cm; (2) counter electrode (CE): a platinum wire (CHI115, CH Instruments, USA), and (3) a reference electrode (RE): 1 M KCl Ag/AgCl reference electrode (CHI111, CH Instruments, USA). Specialty media was designed to mimic the in situ aqueous chemistry of DUSEL 3A at SURF, which included basal salts (L−1): NaCl (0.029 g), KH2PO4 (0.041 g), NH4Cl (0.160 g), FeSO4.7H2O (0.042 g), Na2SO4 (0.014 g), and amended with yeast extract (0.5 g), vitamins, and trace minerals (Kieft et al., 1999). Media was maintained at pH ~ 7.5 using phosphate buffer (10 mM). Enrichment of microbes capable of using electrode as electron donor was performed by augmenting SURF base media with sodium bicarbonate (10 mM), poising electrodes at −0.19 and +0.01 V vs. SHE, and continuous purging of a gas mix (CO2: air v:v 20:80) in the electrochemical cell. To enrich for microbes capable of using the electrode as the terminal electron acceptor, media was augmented with sodium acetate (5 mM), electrodes were poised at +0.26 and +0.53 V vs. SHE, and continuous N2 gas purging was used to maintain anoxic conditions. These secondary electrochemical enrichments were performed in batch mode (media changed every 7–10 days) for 2 months.
Active Microbial Community Analysis
The active microbial community was analyzed by extracting 16S ribosomal RNA (rRNA) transcripts from: (1) dehumidifier fluid (as a potential contaminant) (2 L), (2) DUSEL 3A fluid (2 L), (3) electrode-attached biofilms in ISEC (3 × 4 cm), (4) electrode-attached biofilms of the laboratory electrochemical enrichments (1 × 0.5 cm), and (5) Control. The DUSEL 3A and dehumidifier fluid were filtered through 0.22 μm sterivex filters (Millipore, USA) to collect microbiological samples. All microbiological samples were immediately placed on dry ice for transportation to the University of Southern California and kept in a −80°C freezer in the laboratory until extraction. Total RNA was extracted from the filters and electrode-attached biofilms (ISEC and secondary laboratory electrochemical enrichments) via physical (freeze, thaw, vortex), chemical (sodium dodecyl sulfate and EDTA), and biological (lysozyme) disruption of the cell wall prior to phenol-chloroform extraction as described previously (Reese et al., 2018). A control that included only reagents and no sample material was extracted alongside the samples to confirm lack of contamination. Any residual DNA was digested using TURBO DNA-free TM Kit (ThermoFisher Scientific, USA) and verified to be free of DNA by running a PCR of the RNA extraction. The samples were sent to Molecular Research DNA (Shallowater, TX, USA) for reverse transcription, library preparation, and high throughput sequencing. At the facility, sequencing library preparation included reverse transcribing to cDNA the hypervariable region V4 of the bacterial 16S rRNA transcript using 806R (Caporaso et al., 2011). Specifically, cDNA sequencing steps included purification and generation of blunt end cDNA followed by ligation to sequencing adapters, amplification using 515F and 806R priming set (Caporaso et al., 2011), denaturation, and sequencing. A total of 250 ng of double strand cDNA from each sample was used to prepare the libraries. All amplicons from different samples were pooled (10 pM each) and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, MA, USA). Deep sequencing was performed on Roche 454 FLX titanium platform using recommended reagents.
The sequences were processed using mothur (v 1.36.1) pipeline (Schloss et al., 2009). The barcodes and the primers were removed from each sequence, followed by trimming to remove any ambiguous base calls, average quality scores <20, and homopolymer runs more than 8 bp. A total of 422,490 high-quality unique sequences remained and the resulting average sequence length was 260 bp for all samples. The trimmed sequences were aligned with the SILVA-based reference alignment (Pruesse et al., 2007) using the Needleman-Wunsch pairwise alignment method (Needleman and Wunsch, 1970). Chimeras were removed using UCHIME (Edgar et al., 2011), and a distance matrix was created. The sequences were clustered to identify unique operational taxonomical units (OTUs) at the 97% level, followed by creating a shared OTU file amongst various samples, and the taxonomy was assigned using mothur formatted Ribosomal Database Project (RDP) training set version 16. The resultant microbiome was processed using “Phyloseq,” “Microbiome,” and “Vegan” package available in R (version 3.5.2). The dataset (OTU shared file, taxonomy file, and the metadata file) were introduced as a phyloseq objects in R. The microbiome data was pre-processed by removing contaminant OTUs from the extraction reagent. OTUs not represented in the DUSEL 3A (source fluid) were also removed from the dataset to account for any contamination from initial dehumidifier backflow. The resulting filtered microbiome data (including absolute abundance data from DUSEL 3A, ISEC, and Laboratory enrichments) was used to calculate the diversity indices, MDS plots, and statistics including Adonis and betadisper. The absolute abundance microbiome data of m phylotype/OTUs and n samples [, where cij are the number of reads for a phylotype/OTU (i) in the sample (j)] was transformed into the relative abundances by total sum scaling (). Next, to study the variation of relative abundance of a phylotype/OTU across sample the dataset was normalized to the maximum . The raw sequencing data have been uploaded to National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession number: SRR8537881-SRR8537882) under BioProject PRJNA262938. Custom scripts used for analysis the sff files via MOTHUR and further sequence processing via R can be found at https://github.com/yaminijangir/SURF_scripts.
Isolation and Electrochemical Measurements of Pure Isolates
Electrode-attached biomass from each of WE1 and WE4 was streaked on R2A agar plates (Reasoner and Geldreich, 1985) and incubated at 30°C to obtain single colonies. Morphologically distinct colonies were re-streaked on fresh R2A agar plates, resulting in multiple isolates. For taxonomic classification, isolates were grown to late exponential phase in liquid R2A at 30°C and DNA extraction performed using the UltraClean® Microbial DNA Isolation kit (Mo Bio Laboratories, Carlsbad, CA, USA). The bacterial 16S rRNA gene was PCR-amplified from the extracted DNA using primers 8F and 1492R (Integrated DNA Technologies, USA). The purified PCR product (PureLink PCR Purification Kit, Life Technologies, CA, USA) was sent for sanger sequencing to Genewiz (South Plainfield, NJ, USA) from the 1492R primer. The 600 bp length sequences of the isolated strains have been deposited to NCBI SRA (accession numbers: MK483257, MK483258). The chronoamperometry (i.e., current vs. time at a fixed potential) measurement of each isolate was performed in a standard three-electrode glass electrochemical cell (50 mL volume) in triplicates.
Comamonas sp. (designated strain WE1-1D1) was grown aerobically to stationary phase from frozen stocks (−80°C in 20% glycerol) in R2A media and the culture was used to inoculate 750 mL of SURF base medium at 1% (v:v). After reaching the mid-exponential phase, the culture was pelleted by centrifugation at 5,369 × g for 10 min, washed three times and re-suspended in 15 mL fresh SURF base medium without sodium bicarbonate, yeast extract, and trace minerals. Five milliliters of this suspension was introduced into three separate electrochemical cells, already containing 50 mL SURF base medium (excluding sodium bicarbonate, yeast extract, and trace minerals), after the abiotic current was stabilized to a steady baseline. The electrochemical cell was continuously purged with 100% purified compressed air, and chronoamperometry was performed with the working electrode poised at −0.012 V vs. SHE. After cyclic voltammetry was completed at a scan rate of 1 mV/s, chronoamperometric measurement was resumed at −0.012 V vs. SHE, and ~10.7 mM potassium cyanide was added after ~1.75 h. Cell densities were determined by plate counts.
Bacillus sp. (designated strain WE4-1A1-BC) was grown aerobically to stationary phase from frozen stocks (−80°C in 20% glycerol) in R2A media and the culture was used to inoculate 500 mL of SURF base medium at 1% (v:v). After reaching mid-exponential phase, the culture was pelleted by centrifugation at 7,000 × g for 10 min, washed two times and resuspended in 10 mL fresh SURF base medium without sodium acetate. A total of 5 mL of the suspension was introduced into the electrochemical cell, containing 50 mL SURF base medium augmented with sodium acetate (5 mM), after the abiotic current was stabilized to a steady baseline. The electrochemical cell was continuously purged with N2 to maintain anaerobic conditions, and chronoamperometry was performed with the working electrode poised at +0.53 V vs. SHE. Cell densities were determined by plate counts. The reproducibility of the experiment was tested in triplicates.
Microscopy
For scanning electron microscopy (SEM), electrode samples were fixed overnight in 2.5% glutaraldehyde. Samples were then subjected to an ethanol dehydration series (25, 50, 75, 90, and 100% v/v ethanol for 15 min each treatment) and critical point drying (Autosamdri 815 critical point drier, Tousimis Inc., Rockville, MD, USA). The samples were then mounted on aluminum stubs, coated with gold (Sputter coater 108, Cressington Scientific), and imaged at 5 keV using a JEOL JSM 7001F low vacuum field emission SEM.
Results and Discussion
In situ Current Response and Active Microbial Community Composition
Large negative currents were detected on both WE1 and WE2, −276.68 ± 43.80 μA and −168.76 ± 26.82 μA, respectively, consistent with the expected cathode oxidation conditions. While significantly smaller in magnitude, negative currents were also detected on WE3 and WE4 at higher (nominally anodic) potentials (Table 2). It is important to note that the currents observed in the ISEC reactor are a combination of both biotic and abiotic redox reactions occurring on electrode surfaces. Dissolved oxygen of the SURF 3A liquid varied decreased from 3.79 mg/L to zero during the course of ISEC deployment causing oxygen leakage into the reactor and associated abiotic oxygen reduction currents. In May 2015 (during decommissioning of the ISEC reactor), ORP values of DUSEL 3A fluid and the effluent of the ISEC reactor were −50 mV (Table 1) and 200 mV (Table 2), respectively; confirming oxygen leakage inside the ISEC reactor. The dissolved organic carbon measured at SURF 3A hole during installation of ISEC was 239.04 ± 1.96 μM.
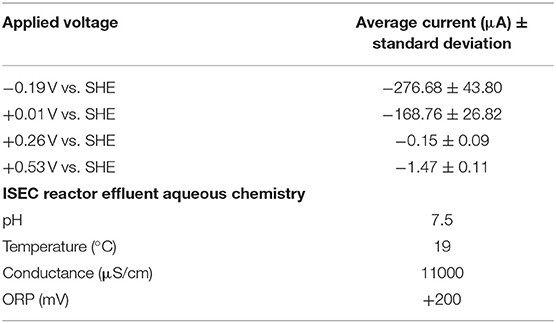
Table 2. Mean current (last 40 days) observed at various redox potentials in the in situ electrochemical colonization (ISEC) reactor and aqueous water chemistry of ISEC reactor effluent.
Nucleic-acid analysis has proven to be effective for characterizing the phylogenetic, taxonomic, and functional structure of microbial assemblages. Since extracted DNA may originate from extracellular DNA pools, dead, and/or dormant cells, sequenced 16S rRNA genes represent the total microbial community rather than its active fraction. On the other hand, ribosomal RNA transcripts describe the active portion of the microbial community (Jansson et al., 2012) and hence, an RNA-based molecular approach was chosen for this study. Herein we present the taxonomic assignments in terms of relative percent of classified sequences with respect to the total number of sequences.
The in situ microbial community analyses determined by pyrosequencing the 16S rRNA transcripts, revealed that bacteria dominated DUSEL 3A fluid (99.65%). The bacterial sequences most closely matched the phyla Proteobacteria (83.23%), Bacteroidetes (10.14%), Firmicutes (0.50%), Chloroflexi (0.13%), Planctomycetes (0.003%). Some sequences (5.82%) did not match to any phyla and have been labeled as unclassified. Within Proteobacteria, the majority of sequences were identified as Gammaproteobacteria (26.03%), followed by unclassified Proteobacteria class (24.45%), Deltaproteobacteria (17.78%), Alphaproteobacteria (10.30%), and Betaproteobacteria (4.66%) (Figure 2). The archaeal community in DUSEL 3A fluid was dominated by sequences related to Crenarchaeota (0.19%), Euryarchaeota (0.13%), and Thaumarchaeota (0.03%). In a DNA-based study, Osburn et al. (2014) also observed a predominance of Bacteria over Archaea in the in situ fluid at the 4,850-ft level of SURF. They further observed a higher abundance of Firmicutes, which we attribute to the DNA- vs. RNA-based microbial community analysis because this phylum has been shown to experience dormancy through sporulation. Additionally, this difference can also arise from the Firmicutes exhibiting large 16S rRNA gene copy number (Větrovsk and Baldrian, 2013).
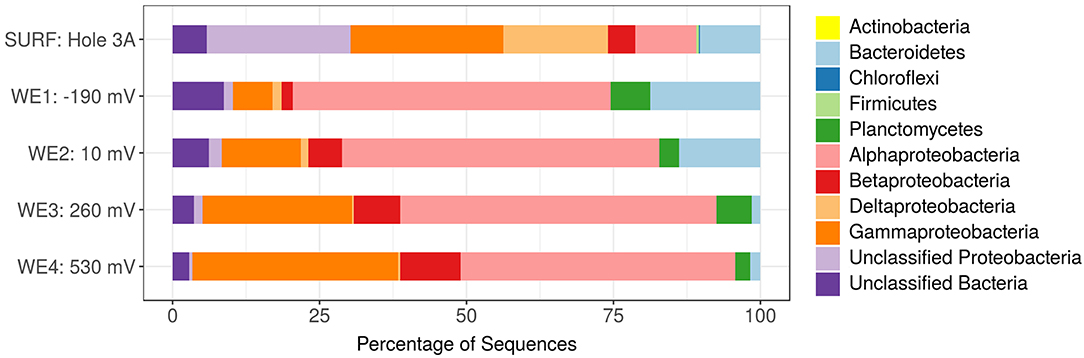
Figure 2. Phylum classification of the active in situ bacterial community of DUSEL 3A fluid (labeled SURF: Hole 3A) and the four separate working electrodes in the in situ electrochemical colonization (ISEC) reactor (labeled WE1-WE4). The unclassified (e.g., putatively novel microbiota) were relatively more abundant at the reducing potential (−0.19 V vs. SHE) where bacteria use the electrode as the electron donor.
Importantly, the active microbial community structure revealed some shifts between the DUSEL 3A in situ fluid and the electrode-attached biofilms of the ISEC reactor. The majority of sequences obtained from electrode-attached biomass of ISEC working electrodes were most closely related to the Proteobacteria, specifically the Alphaproteobacteria (46.75–54.03%), Gammaproteobacteria (6.68–35.96%), Betaproteobacteria (1.98–10.24%), unclassified (0.57–2.17%), and Deltaproteobacteria (0.29–1.54%). The increased relative abundance of sequences identified as Gammaproteobacteria and Betaproteobacteria was observed with increasing electrode potential. The overall relative abundance of sequences related to Planctomycetes phylum substantially increased from the DUSEL 3A fluid (0.003%) to ISEC working electrodes (3–6%). On the other hand, the relative abundance of sequences identified as Chloroflexi and Firmicutes phyla decreased from 0.13 and 0.49% to 0.001 and 0.002%, respectively. The relative abundance of sequences related to Bacteroidetes from the electrode-attached biofilm (across different working electrodes) of the ISEC reactor ranged from 1.45 to 18.76% and unclassified OTUs at the phyla level ranged from 2.83 to 8.72%. The ISEC reactor captured the majority of the bacterial community, but some members, including Methanobacteria, unclassified Clostridia, Methylophilaceae, and Moraxellaceae, were not represented. These populations were originally present in low abundances (<0.13%) in DUSEL 3A fluid. Suspected oxygen leakage into the ISEC reactor potentially hindered the survival of obligate anaerobes belonging to the class Methanobacteria and Clostridia (Dworkin and Falkow, 2006). Many OTUs which were unclassified (at the phylum level) along with Bacteroidetes and Alphaproteobacteria favored the lower potential electrodes; this observation highlights the potential for largely uncharacterized microbial lineages to perform extracellular electron uptake in subsurface environments. Within the sequences classified as Archaea, the phylum Thaumarchaeota (ammonia-oxidizing Candidatus Nitrosoarchaeum) was identified on all ISEC working electrodes. However, Crenarchaeota sequences (0.002%) were only present at an electrode potential of −0.19 V. It is important to note the universal primers (515F and 806R) employed in this study may underestimate the diversity of archaea present in the environment and primers specific for archaeal domain should be employed (Bahram et al., 2019).
Based on the 16S rRNA sequencing data, the bacterial communities of the electrode attached biofilms demonstrated potential-dependent distribution patterns (Figure 3). Below we discuss these patterns in light of previously reported physiotypes. However, we stress that the presence of specific lineages on electrodes is not in itself evidence of EET, since poised electrodes may support microbial consortia capable of diverse metabolisms, including syntrophy, heterotrophy or fermentation, depending on the microenvironment formed at the electrodes. This makes isolation and electrochemical characterization fundamental to confirming and investigating EET processes from the electrode-attached biofilm community.
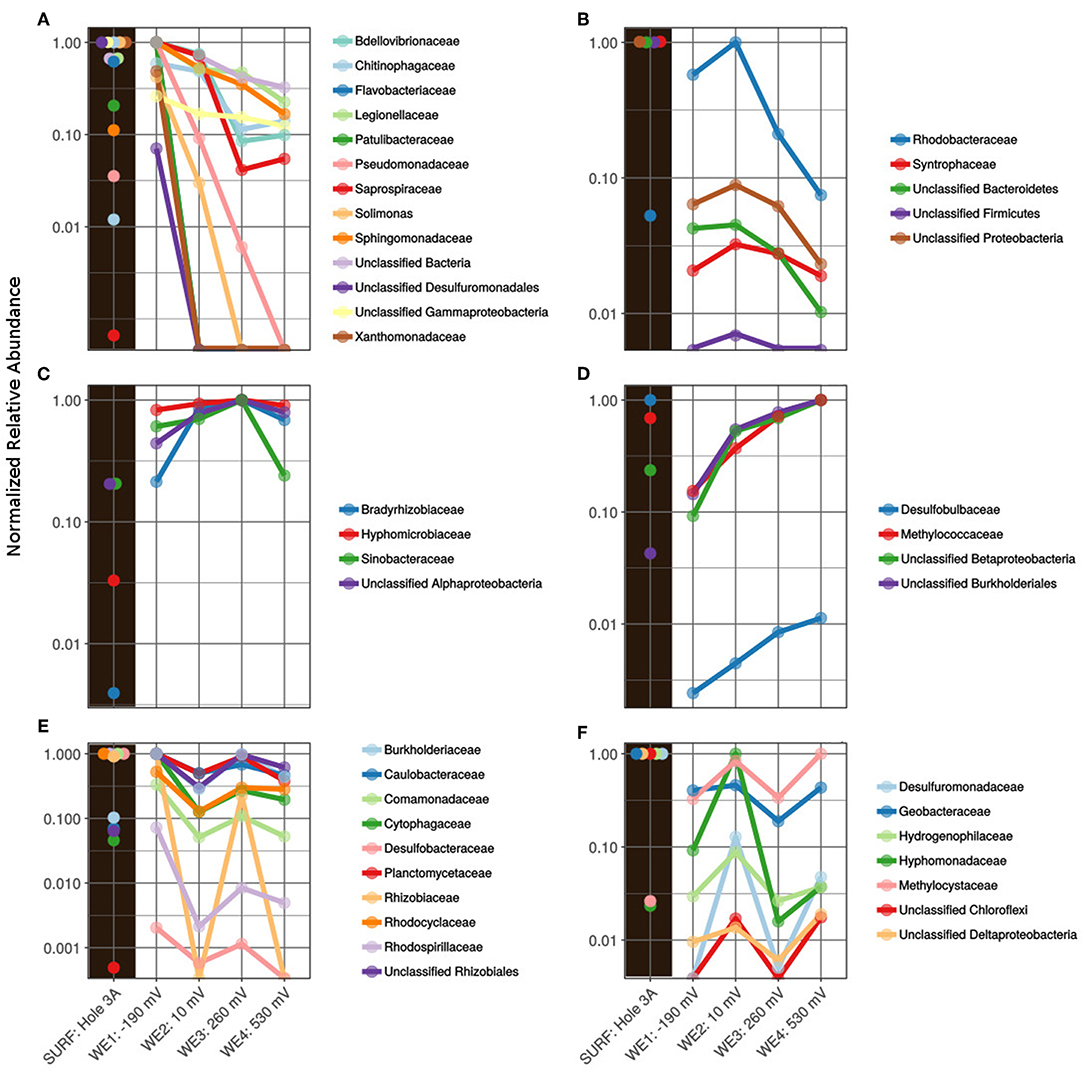
Figure 3. Active family-level normalized relative abundance of bacterial community present in the in situ electrochemical colonization (ISEC) reactor sorted by poised electrodes: (A) maximum at WE1: −0.19 V vs. SHE, (B) maximum at WE2: +0.01 V vs. SHE, (C) maximum at WE3: + 0.26 V vs. SHE, (D) maximum at WE4: +0.53 V vs. SHE, (E) maximum at WE1 and WE3, and (F) maximum at WE2 and WE4, with reference to the inoculum-fluid of SURF DUSEL 3A (labeled as SURF: Hole 3A).
At a potential of −0.19 V (WE1), chosen to mimic sulfur oxidation, the electrode was dominated by sequences closely related to the families Bdellovibrionaceae, Chitinophagaceae, Flavobacteraceae, Legionellaceae, Patulibacteraceae, Psuedomonadaceae, Saprospiraceae, Solimonas, Sphingomonadaceae, Xanthomonadaceae, and unclassified Desulfuromonadales (Figure 3A). Consistent with this, members of Flavobacterium (within the Flavobacteraceae family) have been shown to attach to and metabolize sulfur minerals, and this has been postulated to create micro-oxic environments for sulfur oxidizers (Wright et al., 2013). Some Flavobacteria genera have also co-existed with mat-forming neutrophilic iron oxidizers (Fleming et al., 2014), and some Mn-oxidizing strains belonging to Flavobacterium have been isolated from caves of upper Tennessee river basin (Carmichael et al., 2013). A Pseudomonas/Pseudoalteromonas-like Gammaproteobacteria was previously shown to putatively catalyze iron(II) oxidation under micro-aerobic conditions, isolated from volcanic seamount (Sudek et al., 2009), which may contribute to the formation of iron mats in the deep marine subsurface. Other lineages captured at this potential included Patulibacteraceae, Saprospiraceae, Chitinophagaceae, Sphingomonadaceae, and Solimonas, each putatively containing aerobic respiratory metabolism among their members (Saddler and Bradbury, 2005; Takahashi et al., 2006; Glaeser and Kämpfer, 2014; Kim et al., 2014; McIlroy and Nielsen, 2014; Rosenberg, 2014). The potential +0.01 V vs. SHE (WE2), chosen to mimic iron oxidation, favored the colonization of members from bacterial families Rhodobacteraceae, Syntrophaceae, and many OTUs were identified as unclassified at the phyla level (Figure 3B). The Rhodobacteraceae family has been widely studied to understand banded iron formation in early Earth (Kappler et al., 2005; Hegler et al., 2008). The abundance of many unclassified from the Desulfuromonadales, Gammaproteobacteria, Bacteroidetes, Firmicutes, and Proteobacteria at the lower electrode potentials highlights the importance of alternate sources of reducing power available in the subsurface. The families Bradyrhizobiaceae and Hyphomicrobiaceae, were predominantly abundant on WE3 poised at +0.26 V vs. SHE (Figure 3C). Sequences related to mesophilic sulfate-reducing Desulfobulbaceae (Kuever, 2014) and type 1 methanotrophic Methylococcaceae (Bowman, 2014) were relatively dominant on WE4 poised at +0.53 V vs. SHE, which was chosen to mimic manganese reduction (Figure 3D).
Some microbes adapt to broad ranges of environments (“habitat generalists”) as their key survival strategy, while others specialize in certain niches (“habitat specialists”) (Tienderen and Van, 1991). In this study, we observed that some bacterial populations colonized working electrodes specific to a particular habitat—low and high electrode potential (Figures 3E,F, respectively). Among these, members of the bacterial families Comamonadaceae, Rhodocyclaceae, Burkholderiaceae, Caulobacteraceae, Hyphomonadaceae, Hydrogenophilaceae, Desulfobacteraceae, Desulfuromonadaceae, Geobacteraceae, have been shown to interact with minerals (Lovley, 1991). Rhodoferax ferrireducens sp. Strain T118T, a member of the Comamonadaceae family, can support growth by oxidizing acetate with iron(III) reduction (Finneran et al., 2003). Also, novel strains from the Delftia genus (Comamonadaceae) and Azonexus (Rhodocyclaceae) were demonstrated to donate electrons to poised electrodes (+0.522 V vs. SHE) in anaerobic conditions (Jangir et al., 2016). A member of the Burkholderiaceae family, metallophilic gram-negative Cupriavidus metallidurans, has demonstrated to actively biomineralized particulate Au (Reith et al., 2009). Desulfuromonadaceae family contains members are capable of sulfur and Fe(III) reduction (Greene, 2014), and more recently Desulfuromonas soudanensis WTL was isolated using electrochemical enrichments from anoxic deep subsurface brine (Badalamenti et al., 2016) with 38 multiheme cytochromes. In this study, the relative predominance of Desulfuromonadaceae at lower and higher potentials indicates their ability to survive/grow in niches (Figure 3F). Finally, Geobacteraceae, consisting of the widely studied genus Geobacter, has been documented to putatively reduce insoluble Fe(III) and Mn(IV) (Röling, 2014) and mimicked on electrodes for anode reduction (Bond and Lovley, 2003). This group has also been shown to uptake electron from graphite electrodes coupled with anaerobic respiration (Gregory et al., 2004).
To summarize, the ISEC reactor captured a wide range of metabolically active bacteria, including (but not exclusively) well-studied groups, such as Geobacteraceae, shown to be capable of microbe-mineral interactions in earlier studies.
Current Production and Microbial Community Composition in Laboratory Electrochemical Enrichment
After 5 months of in situ colonization at SURF, sections of biomass-containing electrodes from ISEC reactor were transferred to laboratory electrochemical reactors. Within 8 days, currents developed on both cathodic (−0.19 and +0.01 V vs. SHE) and anodic potentials (+0.26 and +0.53 V vs. SHE), which will be henceforth named as cathodic current (transfer of electrons from electrode to microbe) and anodic current (transfer of electrons from microbe to electrode). Since the WE3 (+0.26 V vs. SHE) was lost due to an electrical short between the counter and the working electrode on the 42nd day of laboratory incubation, further analysis was performed only on WE1, WE2, and WE4 of the laboratory electrochemical enrichments. The average currents (for the last media change) corresponding to WE1 and WE4 were −43.46 ± 7.76 μA, −0.83 ± 0.06 μA, and 582.56 ± 63.14 μA, respectively (Figure 4). Separate incubation with sterile media under identical operating conditions (abiotic control) resulted in an average current of −24.976 ± 0.74 μA at WE1, −1.705 ± 0.128 μA at WE2, and 0.925 ± 0.23 μA at WE4. The observed cathodic or anodic currents from laboratory electrochemical enrichments at WE1 and WE4 were very large compared to the sterile controls demonstrating electron uptake and donation to poised electrodes via enriched microbes. However, the sterile (control) cathodic current at WE2 was higher than laboratory enrichments indicating that at this potential it is difficult to interpret the difference between the abiotic oxygen reduction with microbially mediated oxygen reduction at the electrode. Hence, cyclic voltammetry (CV) was performed to diagnose the reaction with a scan rate of at 10 mV/s on the biofilms of colonized electrodes of WE1, WE2, and WE4 and compared to the abiotic control (Figure 4). CV of electrode-attached biofilm at WE1 and WE2 revealed presence of reversible diffusion controlled peaks indicating presence of redox active mediators shown to enhance EET (Marsili et al., 2008; Von Canstein et al., 2008). CV of electrode-attached biofilm at WE4 revealed the classic sigmoidal shape (Torres et al., 2008) previously observed for pure EET-capable strains under turnover conditions, suggesting enrichment for biofilm containing electrode-bound, non-diffusing, redox enzymes capable of coupling oxidation of the electron donor (in this case acetate) to reduction of the electrode. Interestingly, the formal potential (as defined in Richter et al., 2009) of this biofilm as +0.2 V vs. SHE lies between the formal potential of G. sulfurreducens WT as −0.15 V vs. SHE (Richter et al., 2009) and the peak potential of outer-membrane cytochromes of S. oneidensis MR-1 as +0.21 V vs. SHE (Xu et al., 2016). The overall increase in the potentiostatic current, the difference in the biotic cyclic voltammogram (compared to abiotic control) and cell biomass on the electrodes observed through SEM image (Figure 5), point to the electrochemical enrichment of bacterial communities capable of electron transfer to/from the electrode.
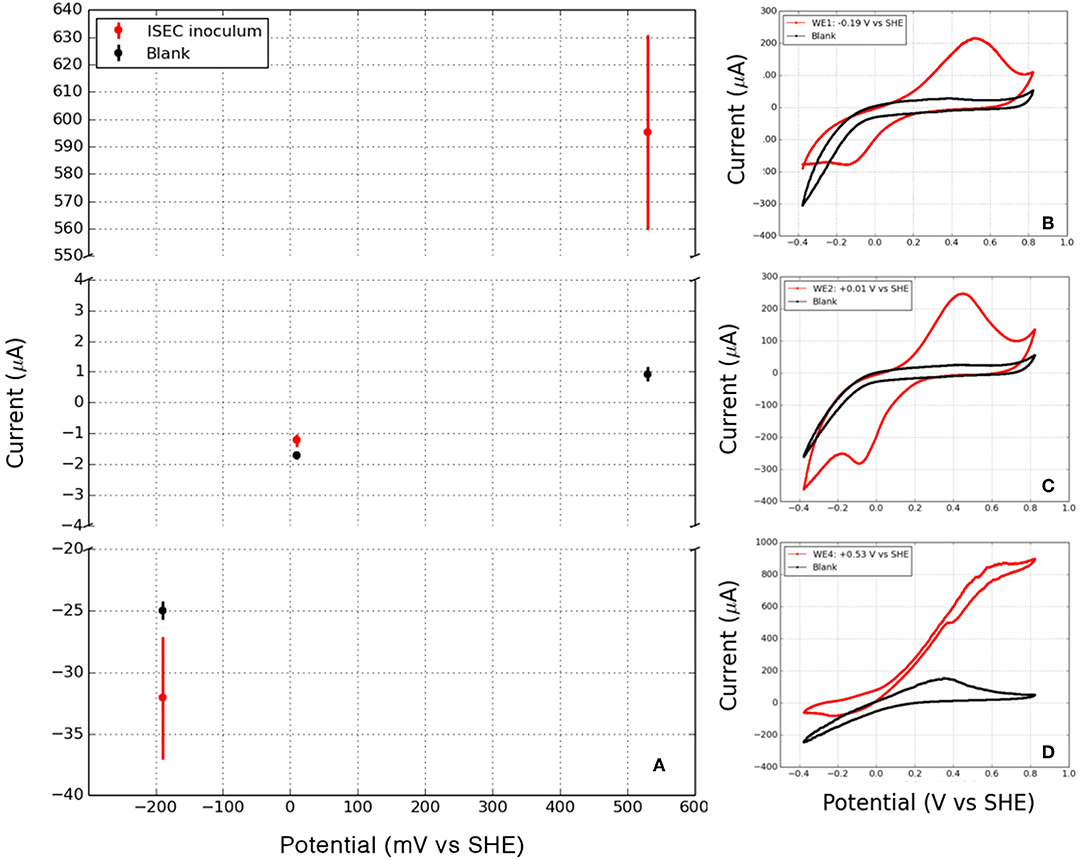
Figure 4. (A) Mean cathodic and anodic currents observed in laboratory enrichments at WE1 (−0.19 V vs. SHE), WE2 (+0.01 V vs. SHE) and WE4 (+0.53 V vs. SHE). Cyclic voltammetry of the working electrodes (including attached biofilms) and the corresponding abiotic controls (blank) at various potentials: (B) WE1: −0.19 V vs. SHE, (C) WE2: +0.010 V vs. SHE, and (D) WE4: +0.53 V vs. SHE, points to redox activity mediated by the in situ microorganisms.

Figure 5. Scanning electron micrograph of the enrichment electrode-associated biomass at WE1 at −0.19 V vs. SHE (left), WE2 at +0.01 V vs. SHE (center), and WE4 at 0.53 V vs. SHE (right) (scale = 1 μm).
The active bacterial community analysis of the laboratory electrochemical enrichments indicates growth of multiple unclassified bacterial phyla on reducing (cathodic) potentials, similar to the ISEC reactor (Figure 6). The dominance of Desulfuromonadaceae and Geobacteraceae observed at the most oxidizing (anodic) potential of +0.53 V vs. SHE (mimicking manganese reduction) (Figure 6) explains the high anodic current and the classic catalytic sigmoidal CV signature (Torres et al., 2008). Members of these families are capable of anaerobic respiration utilizing a variety of compounds as electron acceptors, including sulfur, Mn(IV), Fe(III), and poised electrodes (Lovley, 2006). Interestingly, Comamonadaceae and Rhodocyclaceae were present in all the poised electrodes suggesting an ability to transfer electrons to/from the poised electrodes. Many diverse bacterial families were enriched at the lower/reducing potentials −0.19 and +0.01 V vs. SHE mimicking sulfur and iron oxidation) including the Plactomycetaceae, Caulobacteraceae, Bradyrhizobiaceae, Hyphomicrobiaceae, Sphingomonadaceae, and Hydrogenophilaceae. The higher diversity of subsurface organisms at lower potential electrodes, including those that could not be unclassified at the phyla level, stress the need for further experiments to investigate extracellular electron uptake pathways.

Figure 6. Active family-level normalized relative abundance of bacterial community present in laboratory electrochemical enrichment. The size of the symbol represents the normalized relative abundance of that group across various habitat types (depicted by different colors).
Microbial Community Structure Diversity Measurements
The observed richness (number of OTUs) decreased from the source with various electrochemical enrichments (Figure 7). Nevertheless, ISEC was able to capture at least 50% of the observed OTUs at DUSEL 3A. Based on Shannon's diversity index, there was only weak statistical evidence (Welch Two Sample T-test) of similarities in microbial community structures between DUSEL 3A and ISEC (t = 3.2394, p = 0.04235) or between DUSEL 3A and laboratory enrichment (t = 3.2527, p = 0.07365). It is interesting to note that the bacterial community attached to electrode poised at +0.01 V vs. SHE (mimicking iron oxidation) has consistent higher Shannon diversity index than the communities on differently poised electrodes. OTU evenness remains constant from DUSEL 3A to ISEC and laboratory enrichment (except for the laboratory electrode poised at +0.53 V vs. SHE). The consistent lower alpha diversity of biofilm enriched on the electrode poised at +0.53 V vs. SHE (laboratory) is likely due to the fact that acetate was the only energy source provided. Multidimensional scaling (via PCoA analysis), evaluated using various methods, points to clustering based on enrichment (Figure 7). Further Adonis tests performed on Bray-Curtis points to variation with each enrichment step [R2 = 0.54918, Pr(>F) = 0.001].
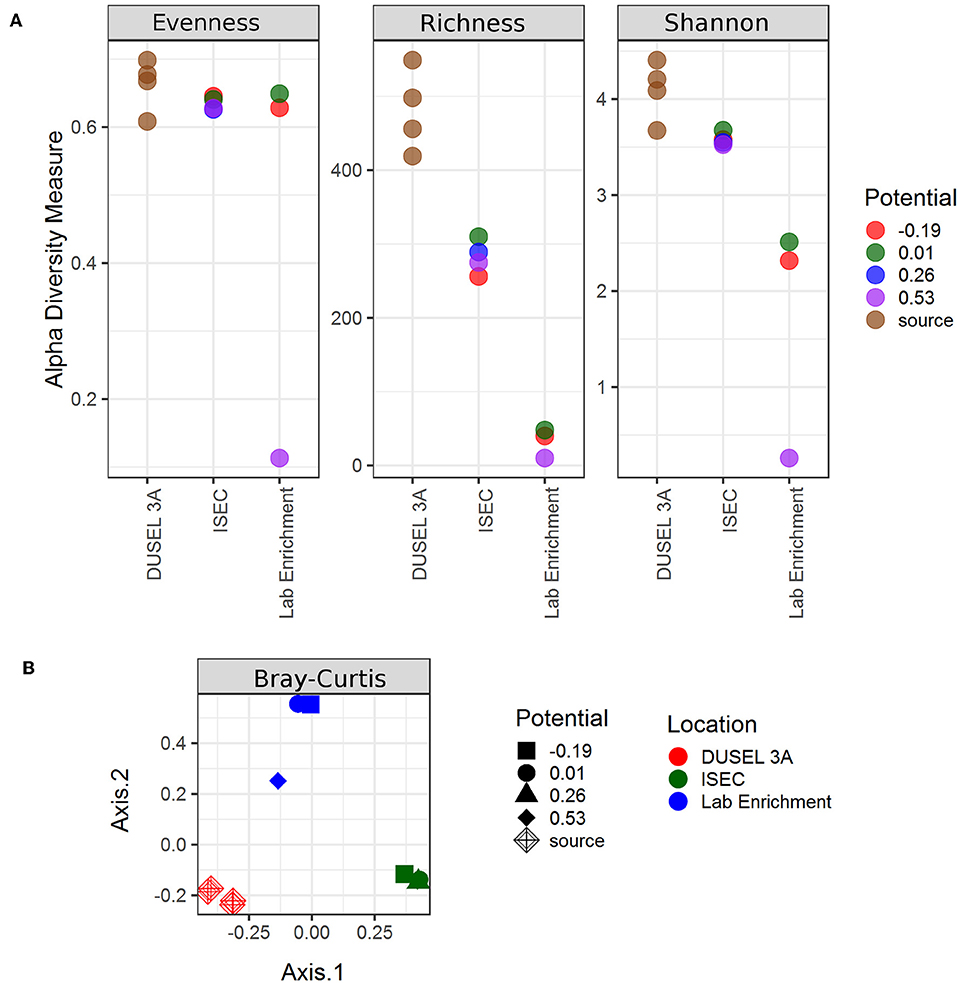
Figure 7. Alpha and beta diversity indices evaluated for the samples. Both the indices point to variation in the microbial community structure from ISEC to lab electrochemical enrichment. (A) Estimates of evenness, richness, and Shannon diversity index of the microbial community as a function of location. (B) Multidimensional scaling plot evaluated using Bray–Curtis method to the clustering of the microbial community based on location.
Isolation and Electrochemical Characterization of Isolated Strains
Bacterial strains were isolated as pure cultures originating from the biomass associated with WE1 (0.19 V vs. SHE) and WE4 (+0.53 V vs. SHE). Phylogenetic analyses of the 16S rRNA gene sequences demonstrated that these strains belonged to the genera Bacillus, Anaerospora, Cupriavidus, Azonexus, and Comamonas. While Bacillus, Anaerospora, Cupriavidus, and Azonexus were isolated from WE4 (+0.53 V vs. SHE), two strains of Comamonas were isolated from both WE1 (−0.19 V vs. SHE) and WE4 (+0.53 V vs. SHE). Bacillus, Anaerospora, and Comamonas were isolated on heterotrophic media R2A. Whereas, Cupriavidus and Azonexus were enriched as iron-reducers on Fe(III)-NTA and poorly crystalline iron oxide, respectively (Lovley, 1991). Further electrochemical studies were performed on isolated Comamonas sp. (strain WE1-1D1) from WE1 (−0.19 V vs. SHE) and Bacillus sp. (strain WE4-1A1-BC) from WE4 (+0.53 V vs. SHE).
Both gram-negative Comamonas and gram-positive Bacillus have previously been reported in microbial fuel cell communities. While the genus Comamonas has been previously enriched on both cathodic and anodic communities of microbial fuel cells (Xing et al., 2010; Ishii et al., 2012; Sun et al., 2012), the genus Bacillus has only been reported from anodic communities (Nimje et al., 2009; Ismail and Jaeel, 2013; Tang et al., 2014). Some species of these genera are capable of interacting with minerals. The Comamonas sp. strain IST-3 oxidizes Fe(II) for energy gain (Blöthe and Roden, 2009) and Bacillus can reduce Fe(III) but without conserving energy through this interaction (de Castro and Ehrlich, 1970).
Comamonas sp. (designated strain WE1-1D1) was studied in a half-cell reactor for electron uptake at working potential of −0.012 V vs. SHE. The media was constantly purged with 100% purified compressed air to supply oxygen as the electron acceptor. Upon inoculation of Comamonas sp. (designated strain WE1-1D1) to the reactors, the cathodic current (electron uptake from electrodes) increased from −0.51 to −1.67 μA (Figure 8), the planktonic cell density increased from 7 ± 1 × 108 to 2 ± 1 × 109 CFU/mL and SEM images confirmed cellular attachment with the formation of a monolayer biofilm on the WE (Figure 9). To further validate electron uptake by the strain, potassium cyanide (an electron transport chain inhibitor) was added resulting in the collapse of the cathodic current (Figure 8). Cyclic voltammetry shows a catalytic wave (reflecting cathode oxidation) with an onset potential of 50–100 mV (Figure 8). Taken together, the rise in the planktonic cell density, increase in the cathodic current, and presence of cells on the WE (Figure 9), point to electron uptake from the poised electrode by Comamonas sp. (strain WE1-1D1). Previously, electrochemical studies performed on the mineral oxidizing bacteria show a diversity of EET mechanisms (Summers et al., 2013; Bose et al., 2014; Deutzmann et al., 2015; Ishii et al., 2015; Rowe et al., 2015). Further characterization of the isolate is essential in understanding its potential for EET and pinpoint the mechanisms.
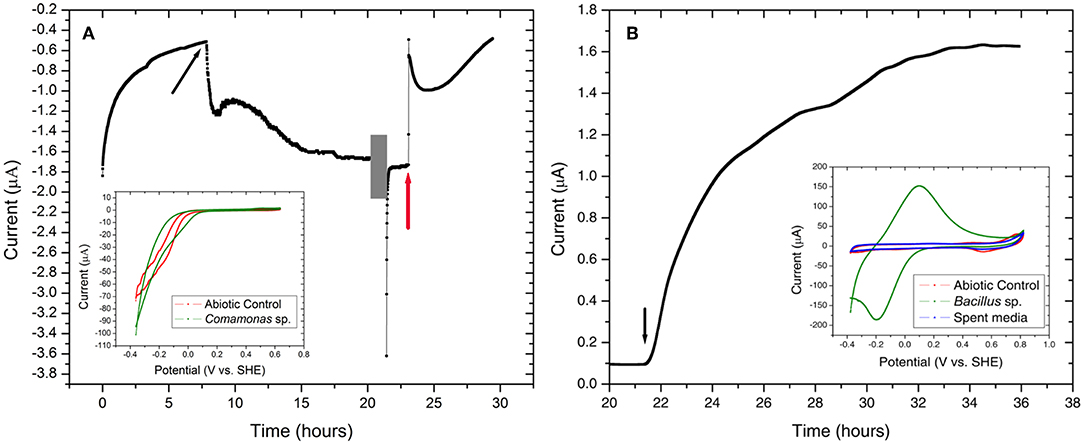
Figure 8. Chronoamperometry of the isolated strains (A) Comamonas sp. (strain WE1-1D1) and (B) Bacillus sp. (strain WE4-1A1-BC) grown at −0.012 V vs. SHE and +0.53 V vs. SHE, respectively. (A) An increase in cathodic current was observed with the addition of Comamonas sp. (strain WE1-1D1) (black arrow) to the electrochemical cell. At 20 h (gray box), CV (inset) was performed on the electrode-attached biofilm and potassium cyanide was injected at 23 h (red arrow). (B) Bacillus sp. (strain WE4-1A1-BC) was enriched on working electrode poised at +0.53 V vs. SHE with acetate as the electron donor and carbon source. Black arrow indicates the time point when the bioreactor was inoculated with the cells. Cyclic voltammogram of the abiotic control, electrode-attached biofilm and the spent media is shown in the inset.
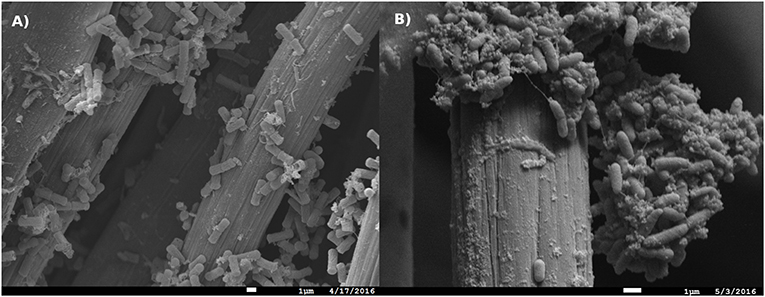
Figure 9. Scanning electron micrograph of (A) Bacillus sp. (strain WE4-1A1-BC) and (B) Comamonas sp. (strain WE1-1D1) biomass associated with the working electrodes (Scale = 1 μm).
Bacillus sp. (strain WE4-1A1-BC) was studied in a half-cell reactor with working electrode poised at +0.53 V vs. SHE. Upon inoculation of Bacillus sp. (strain WE4-1A1-BC) to the electrochemical reactors, the anodic current rose from 0.1 to 1.6 μA (Figure 8) and SEM images confirmed cellular attachment with the formation of a monolayer biofilm (Figure 9). The total planktonic cell density decreased from 2 ± 1 × 108 to 4 ± 1 × 106 CFU/mL indicating possible cell lysis as free planktonic cells. Cyclic voltammetry shows a reversible peak centered at 0 V vs. SHE in the biofilm-associated working electrode. To study whether this peak arose from any soluble redox mediators released by the organism, cyclic voltammetry was performed on the spent media (planktonic cells were spun down 15 × at 7,000 × g). Cyclic voltammogram of the spent media also lacked the reversible peak indicating that redox components bound to the gram-positive bacteria (Figure 8) were responsible for EET. Furthermore, HPLC measurements, as described previously (Bretschger et al., 2010), confirmed a decrease in acetate concentration by 0.8 mM (12%) in 13 h. Based on these measurements, the coulombic efficiency was calculated as 1.81% using the ratio of the total coulombs produced during the experiment to the theoretical amount of coulombs assuming oxidation of acetate to CO2. In comparison, coulombic efficiency of a well-known electrode reducer S. oneidensis MR-1 for electricity generation from lactate (to acetate) is ~20% (Watson and Logan, 2010). Taken collectively, the overall increase in anodic current, reduction in acetate concentration, the shift in biotic cyclic voltammogram relative to abiotic CV, and presence of cells on the WEs (Figure 9), suggest that Bacillus sp. (strain WE4-1A1-BC) is capable of electron transfer to the poised electrode. Additional genetic and physical studies are required to investigate the precise EET mechanism from this strain.
Conclusions
Our results highlight the potential for poised electrodes to serve as in situ observatories to capture for further downstream studies electrochemically active microbes in the deep terrestrial subsurface. The in situ electrodes deployed at the SURF captured many families represented in the borehole fluid, while also showing potential-dependent clear shifts in the relative abundance of specific lineages. Although the ISEC reactor electrodes were poised to mimic sulfur oxidation, iron oxidation and manganese reduction, the microbial community structure pointed to sequences matching families capable of diverse metabolisms, likely reflecting interactions within and across the microbial communities colonizing separate electrochemical niches in the single cultivation vessel. Since downstream laboratory incubations were performed in separate electrochemical cells with different environmental conditions, the microbial structure of the anode-attached biofilms differed significantly from the cathode-attached biofilm. Using this workflow we were able to separately enrich and ultimately isolate cathode-oxidizing and anode-reducing microbes originating from the same environment. Further genetic and physical studies are needed to elucidate the mechanisms responsible for EET in the isolated strains.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Accession numbers: SRR8537881, SRR8537882, MK483257, and MK483258.
Author Contributions
ME-N, JA, and YJ: conception or design of the work. YJ, BR, GW, and CA: field work. YJ, AK, NB, and LZ: data collection. YJ and ME-N: data analysis and interpretation. YJ and ME-N: drafting the article. ME-N, JA, and BR: critical revision of the article.
Funding
This work was supported by the NASA Astrobiology Institute (Life Underground project) under cooperative agreement NNA13AA92A. AK and NB were supported by the National Science Foundation DIMENSIONS grant DEB-1542527 for electrochemical characterization of the isolated strains.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the science and support staff at SURF, including Jaret Heise, Tom F. Regan, Kathy Hart, and Oren T. Loken, for mine access, reactor deployment, and facilitating sample collection. We are also grateful to Oxana Gorbantenko and Forrest Cain, of Black Hills State University, for monitoring the ISEC reactor during the extended deployment. In addition, we acknowledge Duane Moser, Ken Nealson, Magdalena Osburn, Douglas LaRowe, Shuai Xu, Pratixaben Savalia, Lina Bird, and Annette Rowe for providing their valuable input. Scanning Electron Microscopy was performed at the University of Southern California's Core Center of Excellence in Nanoscale Imaging. This manuscript has been released as a pre-print at bioRxiv (Jangir et al., 2019).
References
Bach, W., and Edwards, K. J. (2003). Iron and sulfide oxidation within basalt ocean crust: implications for chemolithoautotrophic microbial biomass production. Geochim. Cosmochim. Acta 67, 3871–3887. doi: 10.1016/S0016-7037(03)00304-1
Badalamenti, J. P., Summers, Z. M., Chan, C. H., Gralnick, J. A., and Bond, D. R. (2016). Isolation and genomic characterization of ‘Desulfuromonas soudanensis WTL', a metal- and electrode-respiring bacterium from anoxic deep subsurface brine. Front. Microbiol. 7:913. doi: 10.3389/fmicb.2016.00913
Bahram, M., Anslan, S., Hildebrand, F., Bork, P., and Tedersoo, L. (2019). Newly designed 16S rRNA metabarcoding primers amplify diverse and novel archaeal taxa from the environment. Environ. Microbiol. Rep. 11, 487–494. doi: 10.1111/1758-2229.12684
Beese-Vasbender, P. F., Nayak, S., Erbe, A., Stratmann, M., and Mayrhofer, K. J. J. (2015). Electrochemical characterization of direct electron uptake in electrical microbially influenced corrosion of iron by the lithoautotrophic SRB Desulfopila corrodens strain IS4. Electrochim. Acta 167, 321–329. doi: 10.1016/j.electacta.2015.03.184
Blöthe, M., and Roden, E. E. (2009). Microbial iron redox cycling in a circumneutral-pH groundwater seep. Appl. Environ. Microbiol. 75, 468–473. doi: 10.1128/AEM.01817-08
Bond, D. R. (2010). “Electrodes as electron acceptors, and the bacteria who love them,” in Geomicrobiology: Molecular and Environmental Perspective, eds L. Barton, M. Mandl, and A. Loy (Dordrecht: Springer), 385–399. doi: 10.1007/978-90-481-9204-5_18
Bond, D. R., Holmes, D. E., Tender, L. M., and Lovley, D. R. (2002). Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295, 483–485. doi: 10.1126/science.1066771
Bond, D. R., and Lovley, D. R. (2003). Electricity production by geobacter sulfurreducens attached to electrodes electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 69, 1548–1555. doi: 10.1128/AEM.69.3.1548-1555.2003
Bose, A., Gardel, E. J., Vidoudez, C., Parra, E. A., and Girguis, P. R. (2014). Electron uptake by iron-oxidizing phototrophic bacteria. Nat. Commun. 5:3391. doi: 10.1038/ncomms4391
Bowman, J. P. (2014). “The family Methylococcaceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer), 411–440. doi: 10.1007/978-3-642-38922-1_237
Bretschger, O., Cheung, A. C. M., Mansfeld, F., and Nealson, K. H. (2010). Comparative microbial fuel cell evaluations of Shewanella spp. Electroanalysis 22, 883–894. doi: 10.1002/elan.200800016
Caddey, S. W., Bachman, R. L., Campbell, T. J., Reid, R. R., and Otto, R. P. (1991). The Homestake Gold Mine, an Early Proterozoic Iron-Formation-Hosted Gold Deposit. Lawrence County, SD: Bulletin. Available online at: https://pubs.er.usgs.gov/publication/b1857J (accessed April 19, 2018).
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 (Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Carmichael, M. J., Carmichael, S. K., Santelli, C. M., Strom, A., and Auer, S. L. B. (2013). Mn(II)-oxidizing bacteria are abundant and environmentally relevant members of ferromanganese deposits in caves of the upper tennessee river basin. Geomicrobiol. J. 30, 779–800. doi: 10.1080/01490451.2013.769651
Carmona-Martínez, A. A., Harnisch, F., Kuhlicke, U., Neu, T. R., and Schröder, U. (2013). Electron transfer and biofilm formation of Shewanella putrefaciens as function of anode potential. Bioelectrochemistry 93, 23–29. doi: 10.1016/j.bioelechem.2012.05.002
de Castro, A. F., and Ehrlich, H. L. (1970). Reduction of iron oxide minerals by a marine Bacillus. Antonie Van Leeuwenhoek 36, 317–327. doi: 10.1007/BF02069033
Deutzmann, J. S., Sahin, M., and Spormann, A. M. (2015). Extracellular enzymes facilitate electron uptake in biocorrosion and bioelectrosynthesis. MBio 6, e00496–e00415. doi: 10.1128/mBio.00496-15
Dworkin, M., and Falkow, S. (2006). The Prokaryotes: A Handbook on the Biology of Bacteria. New York, NY: Springer. doi: 10.1007/0-387-30743-5
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Edwards, K. J., Bach, W., and McCollom, T. M. (2005). Geomicrobiology in oceanography: microbe-mineral interactions at and below the seafloor. Trends Microbiol. 13, 449–456. doi: 10.1016/j.tim.2005.07.005
Fedorovich, V., Knighton, M. C., Pagaling, E., Ward, F. B., Free, A., and Goryanin, I. (2009). Novel electrochemically active bacterium phylogenetically related to Arcobacter butzleri, isolated from a microbial fuel cell. Appl. Environ. Microbiol. 75, 7326–7334. doi: 10.1128/AEM.01345-09
Finneran, K. T., Johnsen, C. V., and Lovley, D. R. (2003). Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int. J. Syst. Evol. Microbiol. 53, 669–673. doi: 10.1099/ijs.0.02298-0
Fleming, E. J., Cetinić, I., Chan, C. S., Whitney King, D., and Emerson, D. (2014). Ecological succession among iron-oxidizing bacteria. ISME J. 8, 804–815. doi: 10.1038/ismej.2013.197
Fredrickson, J. K., and Zachara, J. M. (2008). Electron transfer at the microbe-mineral interface: a grand challenge in biogeochemistry. Geobiology 6, 245–253. doi: 10.1111/j.1472-4669.2008.00146.x
Glaeser S. P Kämpfer P. (2014). “The family Sphingomonadaceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer).
Gorby, Y. A., Yanina, S., McLean, J. S., Rosso, K. M., Moyles, D., Dohnalkova, A., et al. (2006). Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. U.S.A. 103, 11358–11363. doi: 10.1073/pnas.0604517103
Greene, A. C. (2014). “The family Desulfuromonadaceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer).
Gregory, K. B., Bond, D. R., and Lovley, D. R. (2004). Graphite electrodes as electron donors for anaerobic respiration. Environ. Microbiol. 6, 596–604. doi: 10.1111/j.1462-2920.2004.00593.x
Hart, K., Trancynger, T. C., Roggenthen, W., and Heise, J. (2014). “Topographic, geologic, and density distribution modeling in support of physics experiments at the Sanford Underground Research Facility (SURF),” in Proceedings of the South Dakota Academy of Science, Vol. 93, 33–41.
Hartshorne, R. S., Reardon, C. L., Ross, D., Nuester, J., Clarke, T. A., Gates, A. J., et al. (2009). Extracellular electron transfer via microbial nanowires. Proc. Natl. Acad. Sci. 106, 3968–3973. doi: 10.1073/pnas.0900086106
Hegler, F., Posth, N. R., Jiang, J., and Kappler, A. (2008). Physiology of phototrophic iron(II)-oxidizing bacteria: implications for modern and ancient environments. FEMS Microbiol. Ecol. 66, 250–260. doi: 10.1111/j.1574-6941.2008.00592.x
Heise, J. (2015). The Sanford underground research facility at homestake. J. Phys. Conf. Ser. 606:012015. doi: 10.1088/1742-6596/606/1/012015
Hirose, A., Kasai, T., Aoki, M., Umemura, T., Watanabe, K., and Kouzuma, A. (2018). Electrochemically active bacteria sense electrode potentials for regulating catabolic pathways. Nat. Commun. 9:1083. doi: 10.1038/s41467-018-03416-4
Holmes, D. E., Bond, D. R., O'Neil, R. A., Reimers, C. E., Tender, L. R., and Lovley, D. R. (2004). Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb. Ecol. 48, 178–190. doi: 10.1007/s00248-003-0004-4
Ishii, S., Logan, B. E., and Sekiguchi, Y. (2012). Enhanced electrode-reducing rate during the enrichment process in an air-cathode microbial fuel cell. Appl. Microbiol. Biotechnol. 94, 1087–1094. doi: 10.1007/s00253-011-3844-8
Ishii, T., Kawaichi, S., Nakagawa, H., Hashimoto, K., and Nakamura, R. (2015). From chemolithoautotrophs to electrolithoautotrophs: CO2 fixation by Fe(II)-oxidizing bacteria coupled with direct uptake of electrons from solid electron sources. Front. Microbiol. 6:994. doi: 10.3389/fmicb.2015.00994
Ismail, Z. Z., and Jaeel, A. J. (2013). Sustainable power generation in continuous flow microbial fuel cell treating actual wastewater: influence of biocatalyst type on electricity production. ScientificWorldJournal 2013:713515. doi: 10.1155/2013/713515
Jangir, Y., French, S., Momper, L. M., Moser, D. P., Amend, J. P., and El-Naggar, M. Y. (2016). Isolation and characterization of electrochemically active subsurface Delftia and Azonexus species. Front. Microbiol. 7:756. doi: 10.3389/fmicb.2016.00756
Jangir, Y., Karbelkar, A. A., Beedle, N. M., Zinke, L. A., Wanger, G., Anderson, C. M., et al. (2019). In situ electrochemical studies of the terrestrial deep subsurface biosphere at the sanford underground research facility, South Dakota, USA. bioRxiv [Preprint]. doi: 10.1101/555474
Jansson, J. K., Neufeld, J. D., Moran, M. A., and Gilbert, J. A. (2012). Omics for understanding microbial functional dynamics. Environ. Microbiol. 14, 1–3. doi: 10.1111/j.1462-2920.2011.02518.x
Kappler, A., Pasquero, C., Konhauser, K. O., and Newman, D. K. (2005). Deposition of banded iron formations by anoxygenic phototrophic Fe(II)-oxidizing bacteria. Geology 33:865. doi: 10.1130/G21658.1
Kawaichi, S., Yamada, T., Umezawa, A., McGlynn, S. E., Suzuki, T., Dohmae, N., et al. (2018). Anodic and cathodic extracellular electron transfer by the filamentous bacterium Ardenticatena maritima 110S. Front. Microbiol. 9:68. doi: 10.3389/fmicb.2018.00068
Kieft, T. L., Fredrickson, J. K., Onstott, T. C., Gorby, Y. A., Kostandarithes, H. M., Bailey, T. J., et al. (1999). Dissimilatory reduction of Fe(III) and other electron acceptors by a Thermus isolate. Appl. Environ. Microbiol. 65, 1214–21.
Kim, B. H., Park, H. S., Kim, H. J., Kim, G. T., Chang, I. S., Lee, J., et al. (2004). Enrichment of microbial community generating electricity using a fuel-cell-type electrochemical cell. Appl. Microbiol. Biotechnol. 63, 672–681. doi: 10.1007/s00253-003-1412-6
Kim, S.-J., Moon, J.-Y., Weon, H.-Y., Ahn, J.-H., Chen, W.-M., and Kwon, S.-W. (2014). Solimonas terrae sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 64, 1218–1222. doi: 10.1099/ijs.0.055574-0
Kracke, F., Vassilev, I., and Krömer, J. O. (2015). Microbial electron transport and energy conservation - The foundation for optimizing bioelectrochemical systems. Front. Microbiol. 6:575. doi: 10.3389/fmicb.2015.00575
Kuever, J. (2014). “The family Desulfobulbaceae,” in The Prokaryotes (Berlin; Heidelberg: Springer), 75–86. doi: 10.1007/978-3-642-39044-9_267
Lesko, K. T. (2015). The Sanford underground research facility at homestake (SURF). Phys. Proc. 61, 542–551. doi: 10.1016/j.phpro.2014.12.001
Lovley, D. R. (1991). Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55, 259−287. doi: 10.1016/S0065-2911(04)49005-5
Lovley, D. R. (2006). Bug juice: harvesting electricity with microorganisms. Nat. Rev. Microbiol. 4, 497–508. doi: 10.1038/nrmicro1442
Lovley, D. R., and Phillips, E. J. P. (1988). Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54, 1472–1480.
Marsili, E., Baron, D. B., Shikhare, I. D., Coursolle, D., Gralnick, J. A., and Bond, D. R. (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105, 3968–3973. doi: 10.1073/pnas.0710525105
McIlroy, S. J, and Nielsen, P. H. (2014). “The family Saprospiraceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer).
Meyer, J. L., Jaekel, U., Tully, B. J., Glazer, B. T., Wheat, C. G., Lin, H. T., et al. (2016). A distinct and active bacterial community in cold oxygenated fluids circulating beneath the western flank of the Mid-Atlantic ridge. Sci. Rep. 6, 1–14. doi: 10.1038/srep22541
Murdoch, L. C., Germanovich, L. N., Wang, H., Onstott, T. C., Elsworth, D., Stetler, L., et al. (2012). Hydrogeology of the vicinity of Homestake mine, South Dakota, USA. Hydrogeol. J. 20, 27–43. doi: 10.1007/s10040-011-0773-7
Myers, C. R., and Myers, J. M. (1992). Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174, 3429–3438. doi: 10.1128/jb.174.11.3429-3438.1992
Myers, C. R., and Nealson, K. H. (1988). Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240, 1319–1321. doi: 10.1126/science.240.4857.1319
Nealson, K. H., Belz, A., and McKee, B. (2002). Breathing metals as a way of life: geobiology in action. Antonie van Leeuwenhoek. 81, 215–222. doi: 10.1023/A:1020518818647
Needleman, S. B., and Wunsch, C. D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48, 443–453. doi: 10.1016/0022-2836(70)90057-4
Nevin, K. P., Hensley, S. A., Franks, A. E., Summers, Z. M., Ou, J., Woodard, T. L., et al. (2011). Electrosynthesis of organic compounds from carbon dioxide is catalyzed by a diversity of acetogenic microorganisms. Appl. Environ. Microbiol. 77, 2882–2886. doi: 10.1128/AEM.02642-10
Nimje, V. R., Chen, C.-Y., Chen, C.-C., Jean, J.-S., Reddy, A. S., Fan, C.-W., et al. (2009). Stable and high energy generation by a strain of Bacillus subtilis in a microbial fuel cell. J. Power Sources 190, 258–263. doi: 10.1016/j.jpowsour.2009.01.019
Orcutt, B. N., Sylvan, J. B., Knab, N. J., and Edwards, K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. doi: 10.1128/MMBR.00039-10
Osburn, M. R., LaRowe, D. E., Momper, L. M., and Amend, J. P. (2014). Chemolithotrophy in the continental deep subsurface: Sanford Underground Research Facility (SURF), USA. Front. Microbiol. 5:610. doi: 10.3389/fmicb.2014.00610
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Rabaey, K., and Verstraete, W. (2005). Microbial fuel cells: novel biotechnology for energy generation. Trends Biotechnol. 23, 291–298. doi: 10.1016/j.tibtech.2005.04.008
Reasoner, D. J., and Geldreich, E. E. (1985). A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49, 1–7.
Reese, B. K., Zinke, L. A., Sobol, M. S., LaRowe, D. E., Orcutt, B. N., Zhang, X., et al. (2018). Nitrogen cycling of active bacteria within oligotrophic sediment of the Mid-Atlantic Ridge Flank. Geomicrobiol. J. 35, 468–483. doi: 10.1080/01490451.2017.1392649
Reguera, G., McCarthy, K. D., Mehta, T., Nicoll, J. S., Tuominen, M. T., and Lovley, D. R. (2005). Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101. doi: 10.1038/nature03661
Reimers, C. E., Girguis, P., Stecher, H. A., Tender, L. M., Ryckelynck, N., and Whaling, P. (2006). Microbial fuel cell energy from an ocean cold seep. Geobiology 4, 123–136. doi: 10.1111/j.1472-4669.2006.00071.x
Reith, F., Etschmann, B., Grosse, C., Moors, H., Benotmane, M. A., Monsieurs, P., et al. (2009). Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. Proc. Natl. Acad. Sci. U.S.A. 106, 17757–17762. doi: 10.1073/pnas.0904583106
Richter, H., Nevin, K. P., Jia, H., Lowy, D. A., Lovley, D. R., and Tender, L. M. (2009). Cyclic voltammetry of biofilms of wild type and mutant Geobacter sulfurreducens on fuel cell anodes indicates possible roles of OmcB, OmcZ, type IV pili, and protons in extracellular electron transfer. Energy Environ. Sci. 2:506. doi: 10.1039/b816647a
Röling W. F. M. (2014). “The family Geobacteraceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer).
Rosenberg, E. (2014). “The family Chitinophagaceae,” in The Prokaryotes, eds E. Rosenberg, E.F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin; Heidelberg: Springer).
Ross, D. E., Flynn, J. M., Baron, D. B., Gralnick, J. A., and Bond, D. R. (2011). Towards electrosynthesis in Shewanella: energetics of reversing the Mtr pathway for reductive metabolism. PLoS ONE 6:e16649. doi: 10.1371/journal.pone.0016649
Rowe, A. R., Chellamuthu, P., Lam, B., Okamoto, A., and Nealson, K. H. (2015). Marine sediments microbes capable of electrode oxidation as a surrogate for lithotrophic insoluble substrate metabolism. Front. Microbiol. 6:784. doi: 10.3389/fmicb.2014.00784
Rowe, A. R., Rajeev, P., Jain, A., Pirbadian, S., Okamotao, A., Gralnick, J. A., et al. (2017a). Tracking electron uptake from a cathode into Shewanella cells: implications for generating maintenance energy from solid substrates. bioRxiv [Preprint]. doi: 10.1128/mBio.02203-17
Rowe, A. R., Yoshimura, M., LaRowe, D. E., Bird, L. J., Amend, J. P., Hashimoto, K., et al. (2017b). In situ electrochemical enrichment and isolation of a magnetite-reducing bacterium from a high pH serpentinizing spring. Environ. Microbiol. 19, 2272–2285. doi: 10.1111/1462-2920.13723
Saddler, G. S, and Bradbury, J. F. (2005). “Xanthomonadales ord. nov.,” in Bergey's Manual® of Systematic Bacteriology, eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (Boston, MA: Springer).
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shi, L., Dong, H., Reguera, G., Beyenal, H., Lu, A., Liu, J., et al. (2016). Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 14, 651–662. doi: 10.1038/nrmicro.2016.93
Southam, G. (2012). Minerals as substrates for life: the prokaryotic view. Elements 8, 101–106. doi: 10.2113/gselements.8.2.101
Sudek, L. A., Templeton, A. S., Tebo, B. M., and Staudigel, H. (2009). Microbial ecology of Fe (hydr)oxide mats and basaltic rock from Vailulu'u Seamount, American Samoa. Geomicrobiol. J. 26, 581–596. doi: 10.1080/01490450903263400
Summers, Z. M., Gralnick, J. A., and Bond, D. R. (2013). Cultivation of an obligate Fe(II)-oxidizing lithoautotrophic bacterium using electrodes. MBio 4:e00420–12. doi: 10.1128/mBio.00420-12
Sun, Y., Wei, J., Liang, P., and Huang, X. (2012). Microbial community analysis in biocathode microbial fuel cells packed with different materials. AMB Express 2:21. doi: 10.1186/2191-0855-2-21
Takahashi, Y., Matsumoto, A., Morisaki, K., and Omura, S. (2006). Patulibacter minatonensis gen. nov., sp. nov., a novel actinobacterium isolated using an agar medium supplemented with superoxide dismutase, and proposal of Patulibacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 56, 401–406. doi: 10.1099/ijs.0.63796-0
Tang, J., Yang, G., Wen, J., Yu, Z., Zhou, S., and Liu, Z. (2014). Bacillus thermophilum sp. nov., isolated from a microbial fuel cell. Arch. Microbiol. 196, 629–634. doi: 10.1007/s00203-014-1001-3
Tender, L. M., Reimers, C. E., Stecher, H. A., Holmes, D. E., Bond, D. R., Lowy, D. A., et al. (2002). Harnessing microbially generated power on the seafloor. Nat. Biotechnol. 20, 821–825. doi: 10.1038/nbt716
Tienderen, P. H., and Van (1991). Evolution of generalists and specialist in spatially heterogeneous environments. Evolution 45:1317. doi: 10.2307/2409882
Torres, C. I., Marcus, A. K., Parameswaran, P., and Rittmann, B. E. (2008). Kinetic experiments for evaluating the nernst–monod model for anode-respiring bacteria (ARB) in a biofilm anode. Environ. Sci. Technol. 42, 6593–6597. doi: 10.1021/es800970w
Větrovsk,ý, T., and Baldrian, P. (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE 8:e57923. doi: 10.1371/journal.pone.0057923
Von Canstein, H., Ogawa, J., Shimizu, S., and Lloyd, J. R. (2008). Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74, 615–623. doi: 10.1128/AEM.01387-07
Watson, V. J., and Logan, B. E. (2010). Power production in MFCs inoculated with Shewanella oneidensis MR-1 or mixed cultures. Biotechnol. Bioeng. 105, 489–498. doi: 10.1002/bit.22556
White, G. F., Shi, Z., Shi, L., Wang, Z., Dohnalkova, A. C., Marshall, M. J., et al. (2013). Rapid electron exchange between surface-exposed bacterial cytochromes and Fe(III) minerals. Proc. Natl. Acad. Sci. U.S.A. 110, 6346–6351. doi: 10.1073/pnas.1220074110
White, H. K., Reimers, C. E., Cordes, E. E., Dilly, G. F., and Girguis, P. R. (2009). Quantitative population dynamics of microbial communities in plankton-fed microbial fuel cells. ISME J. 3, 635–646. doi: 10.1038/ismej.2009.12
Wright, K. E., Williamson, C., Grasby, S. E., Spear, J. R., and Templeton, A. S. (2013). Metagenomic evidence for sulfur lithotrophy by Epsilonproteobacteria as the major energy source for primary productivity in a sub-aerial arctic glacial deposit, Borup Fiord Pass. Front. Microbiol. 4:63. doi: 10.3389/fmicb.2013.00063
Wrighton, K. C., Agbo, P., Warnecke, F., Weber, K. A., Brodie, E. L., DeSantis, T. Z., et al. (2008). A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2, 1146–1156. doi: 10.1038/ismej.2008.48
Xing, D., Cheng, S., Logan, B. E., and Regan, J. M. (2010). Isolation of the exoelectrogenic denitrifying bacterium Comamonas denitrificans based on dilution to extinction. Appl. Microbiol. Biotechnol. 85, 1575–1587. doi: 10.1007/s00253-009-2240-0
Xing, D., Zuo, Y., Cheng, S., Regan, J. M., and Logan, B. E. (2008). Electricity generation by Rhodopseudomonas palustris DX-1. Environ. Sci. Technol. 42, 4146–4151. doi: 10.1021/es800312v
Keywords: microbial electrochemical cell, Sanford Underground Research Facility, Bacillus, Comamonas, in situ studies, extracellular electron transfer, electromicrobiology
Citation: Jangir Y, Karbelkar AA, Beedle NM, Zinke LA, Wanger G, Anderson CM, Reese BK, Amend JP and El-Naggar MY (2019) In situ Electrochemical Studies of the Terrestrial Deep Subsurface Biosphere at the Sanford Underground Research Facility, South Dakota, USA. Front. Energy Res. 7:121. doi: 10.3389/fenrg.2019.00121
Received: 22 June 2019; Accepted: 17 October 2019;
Published: 01 November 2019.
Edited by:
Chi Ho Chan, University of Minnesota Twin Cities, United StatesReviewed by:
Fabrizio Colosimo, University of New Hampshire, United StatesCraig Lee Moyer, Western Washington University, United States
Jonathan P. Badalamenti, University of Minnesota Twin Cities, United States
Copyright © 2019 Jangir, Karbelkar, Beedle, Zinke, Wanger, Anderson, Reese, Amend and El-Naggar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamed Y. El-Naggar, bW5hZ2dhciYjeDAwMDQwO3VzYy5lZHU=
 Yamini Jangir
Yamini Jangir Amruta A. Karbelkar
Amruta A. Karbelkar Nicole M. Beedle3
Nicole M. Beedle3 Laura A. Zinke
Laura A. Zinke Greg Wanger
Greg Wanger Cynthia M. Anderson
Cynthia M. Anderson Brandi Kiel Reese
Brandi Kiel Reese Jan P. Amend
Jan P. Amend Mohamed Y. El-Naggar
Mohamed Y. El-Naggar