- 1Institute of Environment and Sustainable Development, Banaras Hindu University, Varanasi, India
- 2Department of Biology, College of Science, Sultan Qaboos University, Muscat, Oman
In the present study, 11 cellulose degrading bacterial strains were isolated from water and soil samples of hot springs in the Chumathang village, Leh and Ladakh region, India. The isolated strains were identified as Bacillus subtilis, Bacillus aryabhattai, Bacillus stratosphericus, Bacillus altitudinis, and Brevibacterium frigoritolerans by biochemical and molecular approaches. All the strains were evaluated for the total cellulase, endoglucanase, exoglucanase, and β-glucosidase enzyme activities. On the basis of overall individual cellulose degrading enzyme activities, three strains were selected to develop consortium to enhance their cellulase enzyme activities. The potent cellulose degraders B. stratosphericus BHUJPV-H5, B. subtilis BHUJPV-H19, and B. subtilis BHUJPV-H12 were selected for the consortium development. The effect of cellulase activities of bacterial strains were evaluated ranged up to 6.06 and 0.72 mg ml−1 glucose by agro-residues of sugarcane bagasse and wheat straw, respectively, after 1 h of incubation. Total cellulase enzyme activity of consortia was recorded two times higher than the individual organisms. These strains can be used for enhancing bioethanol production from lignocellulosic biomass, which can consequently boost biofuel production industry.
Highlights
- Bacillus strains were isolated from hot spring's water and soil samples.
- Total cellulase enzyme activity was two times higher in the form of consortium.
- β-glucosidase activity was found to be high, with maximum 5.65 Uml−1.
- Use of wheat straw and sugarcane bagasse showed good amounts of glucose release on hydrolysis.
Introduction
Cellulose degrading bacteria have three classes of enzymes which act synergistically on the cellulosic matrix to degrade it to monomer sugars. These classes of enzymes are endoglucanases, exoglucanases, and β-glucosidases. In nature, cellulose degrading bacteria (e.g., Clostridium, Cellulomonas, Thermonospora, Bacillus, Bacteriodes, Ruminococcus, Erwinia, Acetovibrio, Microbispora, Streptomyces) have been isolated from different sources such as gut of termites, snail, bookworm, caterpillar (Gupta et al., 2011, 2012; Gupta and Verma, 2015), ruminants (bovine rum), soil samples (including deep subsurface soil), and human excreta (Robert and Bernalier-Donadille, 2003; Rastogi et al., 2009; Rawway et al., 2018). Lots of works in the past, since 1950s (Hungate, 1950), have been done to convert lignocellulosic biomass into bioethanol efficiently, but with little success (Gupta et al., 2012). Till date, no bacterial strain is reported with all three classes of cellulase enzymes needed for cellulose degradation. Hence, there is a need to explore novel bacterial strains for cellulose degradation into simple sugars which could be utilized as feedstock for bioethanol production.
Bacterial cellulases are preferred over fungal cellulases because of the following reasons. Their wide range of growth conditions and most importantly, thermostability. Growth rate of bacteria is much higher than fungal strains, which makes it easier for higher recombinant production of enzymes. Bacterial enzymes are also more complex in nature than their fungal counterparts, and hence provide improved function and synergy. Apart from this, the presence of bacterial strains in very harsh environmental conditions including extreme temperatures, pH, salt conditions, which often exist in the bioconversion chamber further adds an advantage. Fungal cellulases are extracted mainly from Trichoderma sp., Penicillium sp., Fusarium sp., Phanerochaete sp., Humicola sp., Schizophillum sp., and Aspergillus sp. (Gupta and Verma, 2015; Agrawal et al., 2016; Srivastava et al., 2018). These species also produce enzymes which have high thermal stability (Maki et al., 2009). Cellulase production reported from Paenibacillus and Bacillus strains have stability at high temperature and wide pH range, while Cellulomonas flavigena and Terendinibacter turnerae produce enzymes with multifunctionality and a broader substrate utilization (Maki et al., 2009). Peudomonas campinasensis BL11, a thermophilic bacterial isolate has growth temperatures over 25–60°C, wide pH range, and the ability to utilize a variety of saccharides and polysaccharides to produce xylanase, pectinase, cyclodextrin transferase, and two cellulases (Ko et al., 2007). Cellulosome production by Clostridium thermocellum and the ability to ferment sugars further offer prompts to use bacterial species for the bioconversion of biomass.
A novel thermophilic cellulose degrading bacterium (Herbinix hemicellulosilytica gen.) isolated from thermophilic biogas reactor (Koeck et al., 2015), and Herbivorax saccincola gen. nov., sp. nov., isolated by Koeck et al. (2016) from lab-scale biogas reactor, showed good total cellulase enzyme activities of 0.05 Umg−1 on crystalline cellulose (Koeck et al., 2016), which is comparable to the activity shown by C. thermocellum which is 0.09 Umg−1. Recent publications on thermophiles from Yellowstone national park suggest that thermophiles possess good cellulolytic activity (Hamilton-Brehm et al., 2010). Another study by Najar et al. (2018), reported the isolation of highly thermophilic bacteria from a north-east Indian hot spring. Hence, there is a need to isolate new thermophilic bacterial species in order to boost the biofuel production industry.
The cellulolytic enzymes work efficiently at temperatures around 50°C and in some studies, even at higher temperatures (Baharuddin et al., 2016). Verma et al. (2018) isolated the cellulase producing thermophilic Bacillus subtilis BHUJP-H1, Bacillus sp. BHUJP-H2, Bacillus licheniformis BHUJP-H3 from hot spring of Leh and Ladakh, indicates that these samples would provide bacteria inhabiting extreme environment, high temperature, and pressure tolerance (high altitude). This gives the hope to screen the bacterial strains with high cellulose degrading ability and help in enhanced bioethanol production. In this study, consortia of cellulose degrading microbes was developed to increase the cellulase activity of the bacterial strains (Agrawal et al., 2018a,b). Wheat straw and sugarcane bagasse, two major agro-wastes produced in the Northern part of India, were used as substrates for cellulase production. The higher activity of the cellulases on these substrates can lead to higher production of glucose and enhance bioethanol production.
The utilization of waste biomass to produce ethanol through cellulose degrading microbes not only reduce the emissions produced from the wastes, but also reduce the greenhouse gas emissions from fossil fuels usage, when used in a blended form. Moreover, bioethanol is a promising clean renewable biofuel, used mainly as a fuel additive which enhances the engine performance and simultaneously reduces the air pollution. With conventional fuels, such as gasoline and petroleum evolving huge amounts of greenhouse gases into the atmosphere, biofuels become the need of the hour. The higher production of glucose and bioethanol from the cellulosic biomass can boost up the biofuel production industry and this bioethanol can be amended in the gasoline by 20% to help reduce the fossil fuel consumption (Gupta and Verma, 2015; Agrawal et al., 2018c). The main aim of this study is to isolate thermophilic bacterial strains for developing efficient cellulose degrading consortium for glucose production from different agro-residues.
Materials and Methods
Isolation of Bacterial Strains
The water and soil samples were collected from the hot springs of Leh and Ladakh region of Jammu and Kashmir. The isolation was done through serial dilution and plating methods (Das et al., 2010). The plates were incubated at 45°C. The bacterial colonies were counted and picked the pure colonies on slant agar for further analysis. The pure cultures were also stored in 40% glycerol stock and kept at −80°C for further use. The bacterial strains were tested for the growth and morphology on nutrient agar plates at 48 h incubation. The strains were preliminarily tested for their cellulose degrading ability in carboxymethyl cellulose (CMC) plates. The plates were streaked with the bacterial strains on CMC agar media and incubated at 48–72 h for degradation of cellulose.
Biochemical Analysis
The selected strains were studied for their biochemical characterization. The biochemical tests performed were catalase test, Methyl Red Voges-Proskauer test, indole test, Gram's staining. Bacterial strains were grown in Mandel's medium and harvested at different intervals. CMC was used as the carbon source with 2% (w/v) mix in the Mandel's medium for determination of total cellulase and endoglucanase activity. Avicel (Sigmacell 20- Sigma Aldrich) with 2% (w/v) mix in the Mandel's media in place of CMC was used for the quantification of exoglucanase. For the estimation of β-glucosidase activity, p-nitrophenol-β-D-glucoside (p-NPG) with 400 μg/ml was used as a carbon source in Mandel's medium (Strahsburger et al., 2017). Total cellulase and endoglucanase activities were calculated at 72 h and exoglucanase and β-glucosidase activities were calculated at 24, 48, and 72 h (Zhang et al., 2009).
Estimation of Total Cellulase Activity
Crude enzyme (500 μl) was mixed with 50 mg of filter paper (Whatman No. 1) and 1.0 ml of 50 mM citrate buffer, then incubated for 1 h at 50°C. After that, 3 ml dinitro salicylic acid (DNS) reagent was added in reaction mixture to stop the reaction. The mixture was incubated in a water bath at 100°C for 10 min for the color development. The mixture was then allowed to cool and the optical density (OD) was measured at 540 nm (Miller et al., 1960). The OD of the enzyme assay was then used to calculate the concentration of the enzyme released against glucose standards. The FPase (Total cellulase) activity in U/ml was calculated using the following formula (Shareef et al., 2015):
Estimation of Endoglucanase Activity
Carboxymethyl cellulose sodium salt (Hi-Media) (2% w/v) solution was prepared in citrate buffer. Five hundred microliters of the crude enzyme was mixed with 500 μl of CMC (2% w/v) and incubated for 30 min at 50°C. After the incubation, the mixture was mixed with DNS reagent to arrest the reaction, then incubated in a water bath for 10 min at 100°C boiling water and later allowed to cool. Following that the OD of the mixture was taken at 540 nm. The CMCase activity in U/ml was calculated using following formula (Shareef et al., 2015):
Estimation of Exoglucanase Activity
Exoglucanase activity assay was performed using the protocol given by Zhang et al. (2009). Sigmacell 20 (Avicel) in the powder form (20 g) was mixed with 1,000 ml of acetate buffer. The bacterial cultures were grown in modified Mandel's medium with Avicel as a carbon source. Four hundred microliters of the crude enzymes were mixed with 1.6 ml of Avicel suspension (2% w/v). The mixtures were incubated for 2 h at 50°C. The reaction was stopped using 4 ml of glycine buffer. The hydrolysate produced was then assayed with the help of phenol sulfuric acid assay (Zhang et al., 2009). The hydrolysate (0.7 ml) was mixed with 0.7 ml of 5% phenol and 3.5 ml of concentrated sulfuric acid for color development. The mixture was then allowed to cool and OD were taken at 490 nm.
Estimation of β-Glucosidase Activity
p-Nitrophenol β-D-glucoside (5 mM) was used as substrate for the estimation of the β-glucosidase enzyme activity. The bacterial strains were grown in modified Mandel medium flooded with p-NPG as the carbon source. Crude enzyme (0.2 ml) was mixed with 1 ml of p-NPG suspension and 1.8 ml of acetate buffer. The mixtures were incubated at 50°C for 30 min. Four milliliters of glycine buffer was used to stop the reaction. The β-glucosidase activity in U/ml was calculated using this formula (Shareef et al., 2015):
Using Sugarcane Bagasse and Wheat Straw for Estimation of Total Cellulase Enzyme in Term of Glucose Production
Sugarcane and wheat straw were used to estimate the effect of crude enzymes on the agro-residues. Sugarcane bagasse, collected from a nearby cane juice store (25.284708°N, 83.000852°E, variety: CO-238) was washed, and oven dried at 50°C for 2 days. It was crushed and then autoclaved before its final use for the estimation. The wheat straw brought from a nearby store (25.264407°N, 82.9950523°E, variety: Malviya 510) was oven dried, crushed and autoclaved before use (Supplementary Figure 1). For the estimation of the degradation of the substrate by the enzyme, the estimation of the individual enzyme activity was measured (Zhang et al., 2009). The quantity of the substrates taken were 50 mg sugarcane bagasse and 50 mg wheat straw. The enzyme activity on sugarcane and wheat straw is calculated in U/ml by the following formula:
Total Cellulase, Endoglucanase, Exoglucanase and β-Glucosidase Activities Estimated in Different Combinations of Three Bacterial Strains
Total cellulase, endoglucanase, exoglucanase, and β-glucosidase enzyme activities were checked in presence of three consortia prepared from three bacterial strains (B. subtilis BHUJPV-H19, Bacillus stratosphericus BHUJPV-H5, and B. subtilis BHUJPV-H12). Initially the cultures were grown in nutrient broth for overnight (12 h). Consortia were prepared by inoculating equal volume of broth cultures into the Mandel's media with respective carbon sources for different enzyme activities to be quantified. The CMC was the carbon source used in Mandel's medium for total cellulase and endoglucanase activity estimation while avicel and p-nitrophenol-β-D-glucoside were used as carbon sources for the estimation of exoglucanase and β-glucosidase activities, respectively, by modified method of Zhang et al. (2009).
16 S rDNA Gene Amplification
The molecular identification of the bacterial strains was done through 16S rDNA sequencing. The isolation of genomic DNA was done by modified method of Russell and Sambrook (2001).
Amplification of the 16S rDNA
The universal primers were used for the amplification of 16S rDNA gene of each bacterial strains. The forward primer 27F (50-AGAGTTTGATCCTGGCTCAG-30) and reverse primers 939R (5′-CTTGTGCGGGCCCCCGTCAATTC-3′) and 1492R (5′-CAGGAAACAGCTATGAC-3′) were used [14]. The amplified product was purified using PCR purification kit (Invitrogen, PureLinkTM PCR purification kit, United States) for the sequencing of 16S rDNA gene. BLAST (basic local alignment sequence tool) was done of partial 16S rDNA gene sequence from NCBI data base. The obtained results with high similarity were used for identification of bacterial strains on the basis of query cover and identity percentage. Then partial 16S rDNA gene sequence submitted in the NCBI for getting accession number. The results of the 16S rDNA sequencing were used to establish the genetic relationship between the isolated bacterial strains. The software used for the purpose was Clustal W and Mega 4.0 with 500 bootstrap replications.
Statistical Analysis
All the enzymatic activities were carried out in triplicate and the results were expressed as mean ± standard error of independent replicates. Analysis of variance (ANOVA) with Duncan post hoc test was performed using SPSS (Statistical Package for the Social Sciences) software (version 21). The values of P ≤ 0.05 were considered statistically significant.
Results and Discussion
Morphology and Biochemical Characterization of the Bacterial Strains
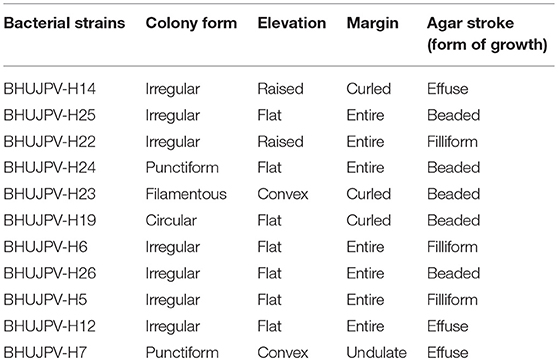
The isolated microbial strains showed different types of colony forms such as irregular flat type colony form with entire margins, punctiform, filamentous, and circular types. Also, strains such B. subtilis BHUJPV-H14 and B. subtilis BHUJPV-H22 showed raised elevation and B. subtilis BHUJPV-H23 and B. altitudinis BHUJPV-H7 showed convex elevation. The margins in the colony form of the bacterial cultures were mostly entire but some of them showed curled and undulate forms such as B. subtilis BHUJPV-H14, B. subtilis BHUJPV-H23, and B. subtilis BHUJPV-H19 with curled margins and B. altitudinis BHUJPV-H7 with undulate margins (Supplementary Figure 2; Table 1).
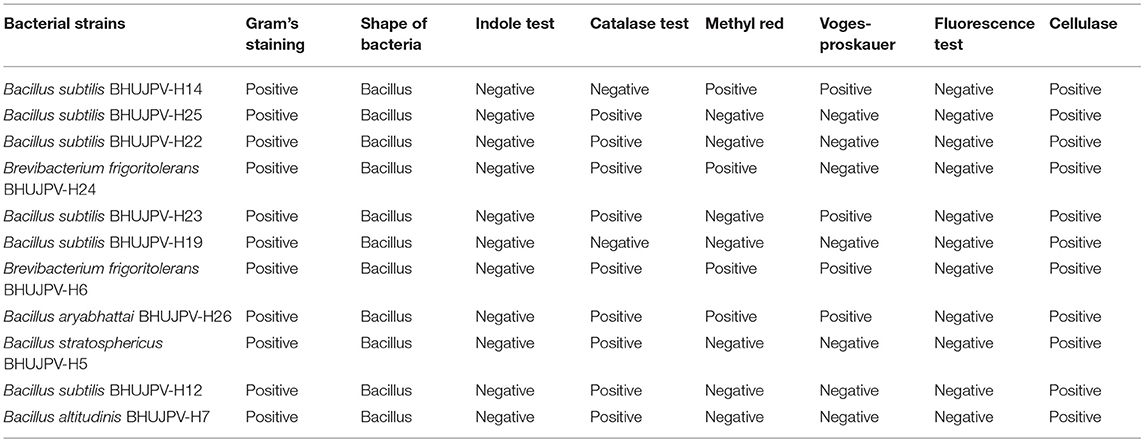
All strains showed positive test for Gram's staining and found rod shape cells. All strains showed negative results for Indole test. B. subtilis BHUJPV-H14 and B. subtilis BHUJPV-H19 showed negative results for catalase test while all others showed positive results for catalase test. In methyl-red test the B. subtilis BHUJPV-H14, B. frigoritolerans BHUJPV-H24, B. frigoritolerans BHUJPV-H6, and B. aryabhattai BHUJPV-H26 were found positive while others showed negative results for the same. The Voges-Proskauer test showed positive results in B. subtilis BHUJPV-H14, B. subtilis BHUJPV-H23, B. frigoritolerans BHUJPV-H6, and B. aryabhattai BHUJPV-H26, and the remaining others showed negative results for the test. All the strains showed positive results for cellulase test qualitatively with good zone of clearance and hence were taken further for the quantification of the enzyme activities, along with their molecular identification (Supplementary Figures 3, 4; Table 2).
Molecular Identification of Bacteria Strains
16S rDNA gene sequencing was done and submitted in NCBI followed by BLAST result. The sequence for these strains were submitted in NCBI to obtain the accession number. Most of the identified bacterial strains were showed maximum similarity with Bacillus strains with BLAST results. After BLAST result, the bacterial strains were identified as B. subtilis BHUJPV-H14 (Accession number: MG550890), B. subtilis BHUJPV-H25 (Accession number: MG596982), B. subtilis BHUJPV-H22 (Accession number: MG596980), B. frigoritolerans BHUJPV-H24 (Accession number: MG596981), B. subtilis BHUJPV-H23 (Accession number: MG596987), B. subtilis BHUJPV-H19 (Accession number: MG596986), B. frigoritolerans BHUJPV-H6 (Accession number: MF664678), B. aryabhattai BHUJPV-H26 (Accession number: MG596983), B. stratosphericus BHUJPV-H5 (Accession number: MF664677), B. subtilis BHUJPV-H12 (Accession number: MF776630), and B. altitudinis BHUJPV-H7 (Accession number: MF664679). Many Bacillus strains have been reported in the past that have been isolated from samples of high temperatures. Yadav et al. (2015), reported nine genera that belong to Bacillaceae which are, Bacillus, Halobacillus, Lysinibacillus, Oceanobacillus, Paenibacillus, Salinibacillus, Sediminibacillus, Thallasobacillus, and Virgibacillus. Another study by Acharya and Chaudhary (2012), reported the isolation of B. licheniformis WBS1 and Bacillus sp. WBS from a hot spring West Bengal. Mohammad et al. (2017), identified isolated B. licheniformis and Thermomonas hydrothermalis from Jordan hot springs.
In the phylogenetic tree analysis, bacterial isolates B. altitudinis BHUJPV-H14 and BHUJPV-H19 were closely related. Strains B. frigoritolerans BHUJPV-H24 and B. aryabhattai BHUJPV-H26 to B. stratospericus BHUJPV-H5 to B. altitudinis BHUJPV-H7 showed very close similarity in the phylogenetic tree analysis. B. subtilis BHUJPV-H22 to B. subtilis BHUJPV-H25, B. subtilis BHUJPV-H14 to B. subtilis BHUJPV-H19 were found to be very closely related. B. aryabhattai BHUJPV-H26 and B. frigoritolerans IND-13 showed close similarity. These two strains are also closely related to B. altitudinis HEM05. B. stratospehricus BHUJPV-H5 is also closely related to B. subtilis KRI, B. subtilis BHUJPV-H14 and B. subtilis BHUJPV-H19. B. frigoritolerans BHUJPV-H6 is closely related to B. subtilis BHUJPV-H12, B. aryabhattai CN13-5, B. subtilis GX S18, B. subtilis GX S-5 (Supplementary Figure 5).
Enzyme Quantification
Estimation of Total Cellulase Activity in Bacterial Strains
The total cellulase activity of the identified bacterial strains ranged as high as 0.14 Uml−1. Bacterial strains B. stratosphericus BHUJPV-H5, B. subtilis BHUJPV-H12, B. subtilis BHUJPV-H19, and B. subtilis BHUJPV-H23 showed 0.14, 0.13, 0.12, and 0.14 Uml−1 total cellulase enzyme activity, respectively (Figure 1). Bacterial strains such as B. subtilis BHUJPV-H14 and B. frigoritolerans BHUJPV-H24 showed total cellulase activities of 0.03 and 0.04 Uml−1. All other bacterial strains showed low total cellulase activity when quantified. In a similar study, by Dipasquale et al. (2014), maximum cellulase activity was found to be 10.3 Uml−1 (purified crude extract) in Thermosipho sp. strain 3. In a study by Al Azkawi et al. (2018) B. subtilis S1 was found to be producing 2.963 U/ml of cellulase enzyme.
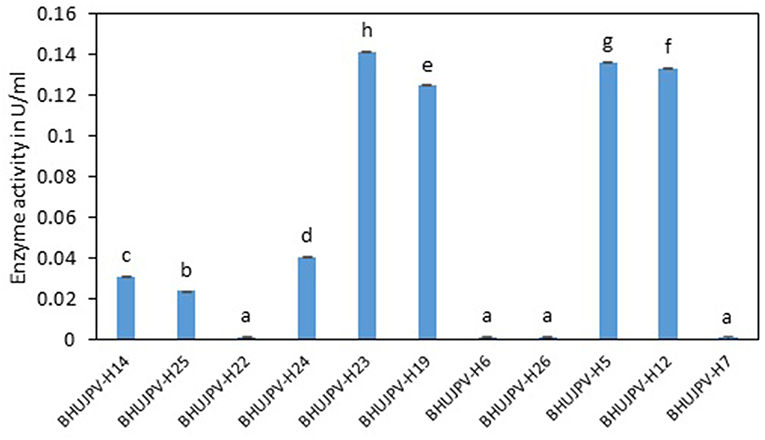
Figure 1. Total cellulase activity of the isolated bacterial strains at 72 h of enzyme harvest. Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
Estimation of Endoglucanase Activity in Bacterial Strains
Similarly, high endoglucanase enzyme activities were observed in B. stratosphericus BHUJPV-H5, B. subtilis BHUJPV-H12, B. subtilis BHUJPV-H19, and B. subtilis BHUJPV-H23 with activities 0.35, 0.21, 0.23, and 0.26 Uml−1, respectively, at 72 h. Other strains such as B. subtilis BHUJPV-H22 showed moderate amount of endoglucanase activity (0.08 Uml−1). All other bacterial strains produced enzyme at a much lower activity rate with 0.03 Uml−1 by B. subtilis BHUJPV-H14, 0.02 Uml−1 by B. subtilis BHUJPV-H25, B. frigoritolerans BHUJPV-H24, B. frigoritolerans BHUJPV-H6, and B. aryabhattai BHUJPV-H26 while 0.04 Uml−1 endoglucanase enzyme activity was observed in B. altitudinis BHUJPV-H7 at 72 h incubation (Figure 2). Similarly, Ladeira et al. (2015) observed the endoglucanase activity 0.29 Uml−1 by Bacillus sp. at 50°C incubation.
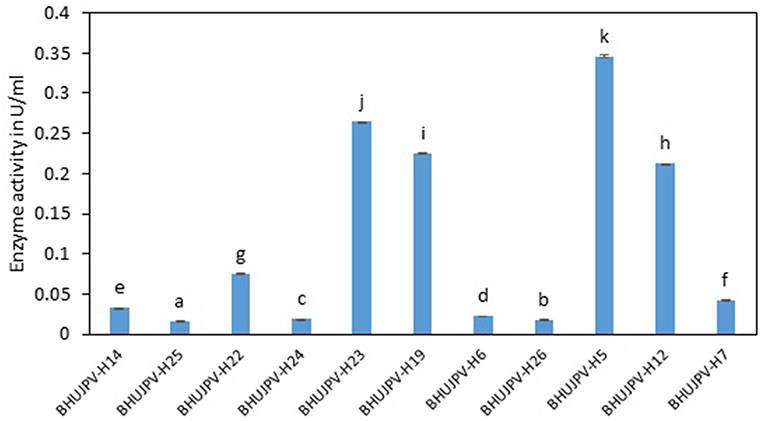
Figure 2. Endoglucanase activity of the isolated bacterial strains at 72 h of enzyme harvest. Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
Estimation of Exoglucanase Activity in Bacterial Strains
The exoglucanase activity of the bacterial strains ranged as high as 0.06 Uml−1 at 24 h of enzyme harvest and its assay in B. aryabhattai BHUJPV-H26. The B. altitudinis BHUJPV-H7 showed enzyme activity of 0.03 Uml−1, while B. subtilis BHUJPV-H22, B. subtilis BHUJPV-H23, B. frigoritolerans BHUJPV-H6, and B. stratosphericus BHUJPV-H5 showing exoglucanase activity of 0.02 Uml−1 at 24 h incubation (Figure 3). At 48 h incubation, no efficient exoglucanase activity was observed in the bacterial cultures except B. subtilis BHUJPV-H12 which showed an activity of 0.03 Uml−1. Other strains such as B. subtilis BHUJPV-H14, B. frigoritolerans BHUJPV-H6, B. stratosphericus BHUJPV-H5, and B. altitudinis BHUJPV-H7 showed anactivity of 0.02 Uml−1. Remaining strains showed very low exoglucanse activity at 48 h of enzyme harvest. The exoglucanase activity of the bacterial strains with enzymes harvested at 72 h showed somewhat similar results to 24 and 48 h enzyme harvest. The exoglucanse activity of the isolated bacterial strains at 72 h of enzyme harvest ranged upto 0.02 Uml−1. B. subtilis BHUJPV-H14 and B. subtilis BHUJPV-H12 showed 0.02 Uml−1 of enzyme activity while other strains such as B. subtilis BHUJPV-H25, B. subtilis BHUJPV-H22, B. frigoritolerans BHUJPV-H24, B. subtilis BHUJPV-H19, B. frigoritolerans BHUJPV-H6, B. aryabhattai BHUJPV-H26, B. altitudinis BHUJPV-H7 showed enzyme activities of 0.01 Uml−1. Ladeira et al. (2015) also studied the exoglucanase activity in Bacillus sp. and found it to be 0.83 Uml−1.
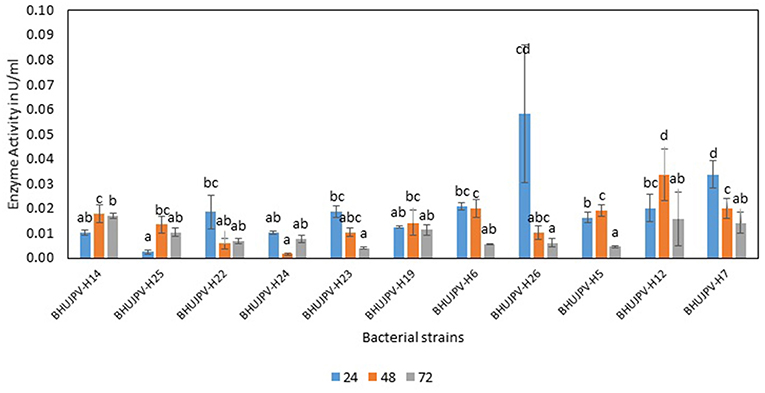
Figure 3. Exoglucanase enzyme activity of the bacterial strains in the modified Mandel media at different time intervals. Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
β-Glucosidase Activity
The β-glucosidase activity of the isolated bacterial cultures at 24 h of harvest ranged as high as 5.03 Uml−1 in B. altitudinis BHUJPV-H7. B. subtilis BHUJPV-H19, B. frigoritolerans BHUJPV-H6, and B. subtilis BHUJPV-H12 showed enzyme activity of 2.55, 2.63, and 1.24 Uml−1, respectively. Other strains such as B. subtilis BHUJPV-H25, B. subtilis BHUJPV-H23, and B. stratosphericus BHUJPV-H5 showed enzyme activities of 1.30, 1.87, and 1.33 Uml−1, respectively. B. subtilis BHUJPV-H22, B. frigoritolerans BHUJPV-H24, and B. aryabhattai BHUJPV-H26 showed activities of 0.43, 0.50, and 0.40 Uml−1. The enzyme activities harvested at 48 h from B. subtilis BHUJPV-H23 was 4.14 Uml−1. B. subtilis BHUJPV-H25, B. stratosphericus BHUJPV-H5, and B. subtilis BHUJPV-H12 showed enzyme activities of 3.53, 1.37, and 0.43 Uml−1, respectively. The other strains such as B. subtilis BHUJPV-H14, B. subtilis BHUJPV-H22, B. frigoritolerans BHUJPV-H24, B. subtilis BHUJPV-H19, B. frigoritolerans BHUJPV-H6, B. aryabhattai BHUJPV-H26, and B. altitudinis BHUJPV-H7 showed enzyme activities of 0.79, 0.36, 0.83, 0.54, 0.22, 0.65, and 0.79 Uml−1, respectively, at 48 h of enzyme harvest from the bacterial cultures. The enzyme activities of the isolated bacterial strains at 72 h of incubation ranged as high as 5.65 Uml−1 in B. subtilis BHUJPV-H19. The enzyme activities of other bacterial strains at 72 h of incubation include B. subtilis BHUJPV-H23, and B. aryabhattai BHUJPV-H26 with enzyme activities of 4.21, and 4.14 Uml−1. B. subtilis BHUJPV-H14, B. subtilis BHUJPV-H22, B. frigoritolerans BHUJPV-H24, B. frigoritolerans BHUJPV-H6, B. stratosphericus BHUJPV-H5, and B. altitudinis BHUJPV-H7 were found to possess enzyme activities of 0.43, 0.43, 0.54, 0.86, 0.50, and 0.43 Uml−1, respectively, at 72 h incubation (Figure 4). Sharma et al. (2015) observed β-glucosidase activity of 3.1 Uml−1 by P. janthinellum EMS-UV-8. High β-glucosidase activity can improve the overall rate of reaction by reducing end product inhibition (Agrawal et al., 2016, 2017).
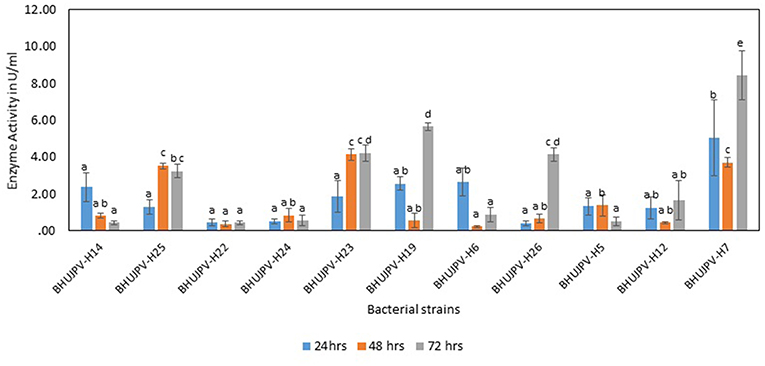
Figure 4. β-glucosidase activity of different bacterial strains. Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
Effect of Cellulase Enzyme Activity on Different Agro-Residues
Enzyme Cellulase Activity on Sugarcane Bagasse
A very high activity was shown by B. subtilis BHUJPV-H19 with 6.06 mg/ml of glucose released in the hydrolysate with 60 min exposure to the sugarcane bagasse. B. subtilis BHUJPV-H23 released 4.69 mg/ml of glucose. Similarly, B. stratosphericus BHUJPV-H5 and B. subtilis BHUJPV-H12 allowing the release of 4.62 and 4.42 mg/ml of glucose in the hydrolysate. The strains B. subtilis BHUJPV-H14, B. subtilis BHUJPV-H25, B. subtilis BHUJPV-H22, B. frigoritolerans BHUJPV-H24, BHUJPV-H26, and B. altitudinis BHUJPV-H7 released low amounts of glucose in the hydrolysate, with 0.10, 0.02, 0.11, 0.10, 0.30, 0.11, and 0.14 mg/ml of glucose, respectively, at 72 h of enzyme harvest (Figure 5).
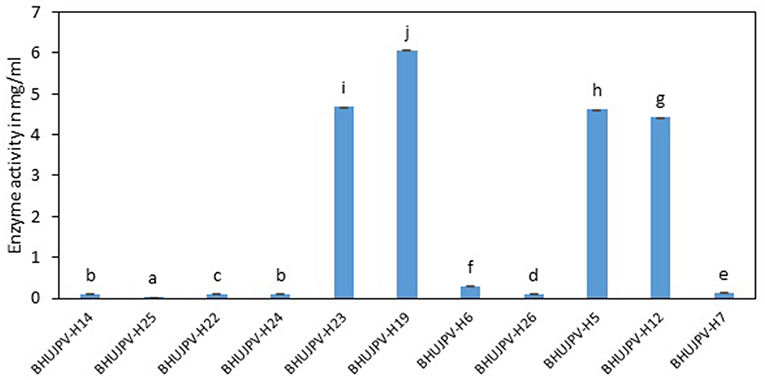
Figure 5. Enzyme activity on wheat straw (Sugars released from sugarcane bagasse). Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
Enzyme Cellulase Activity on Wheat Straw
Comparatively lower activity was shown on wheat straw than the sugarcane bagasse by the isolated cellulolytic bacterial strains. The strains B. subtilis BHUJPV-H12, B. subtilis BHUJPV-H23, B. subtilis BHUJPV-H19, B. frigoritolerans BHUJPV-H6, and B. altitudinis BHUJPV-H7 showed higher glucose release of 0.72, 0.48, 0.39, 0.22, and 0.24 mg/ml, respectively. Other strains B. subtilis BHUJPV-H14, B. subtilis BHUJPV-H25, B. subtilis BHUJPV-H22, B. frigoritolerans BHUJPV-H24, and B. aryabhattai BHUJPV-H26 released 0.03, 0.23, 0.22, 0.03, and 0.04 mg/ml of glucose, respectively, at 72 h of enzyme harvest (Figure 6).
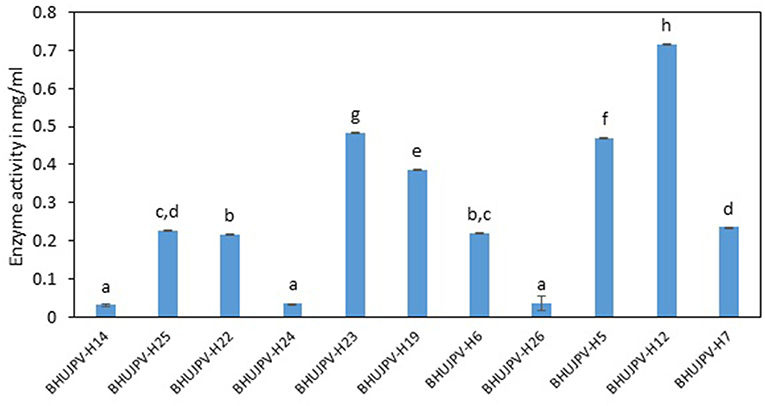
Figure 6. Enzyme activity on sugarcane bagasse (Sugars released on degrading wheat straw). Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
A study on raw poplar biomass by Tabassum et al. (2018) observed the release of 2.30 mg/ml of reducing sugars after 6 h of incubation, whilw the commercial cellulase released 3.85 mg/ml reducing sugars. The bacterial strains in the form of consortia in general showed the increase in the enzyme activity in comparison to the enzyme harvest of a single bacterial strain. The enzyme activities of all four enzymes were quantified at different time intervals, i.e., 24, 48, and 72 h. Kato et al. (2005), studied the coexistence of five bacterial isolates namely Clostridium straminisolvens CSK1, Clostridium sp. strain FG4, Pseudoxanthomonas sp. strain M1-3, Brevibacillus sp. strain M1-5, and Bordetella sp. strain M1-6, in the form of mixed cultures to study the coexistence among themselves, which exhibited functional and structural stability. Here in this study, we have focused on the higher enzyme cultivation study, rather than going for the mechanism involved behind the same. A study by Nair et al. (2018) successfully optimized the cellulase activity to 2.42 U/ml in B. velezensis ASN1. A similar kind of study was done by Sivakumar et al. (2016) to isolate and characterize B. licheniformis from compost.
Consortium A
The total cellulase activity was found to be high in consortium A with 0.15 Uml−1 at 24 h, 0.24 Uml−1 at 48 h, and 0.30 Uml−1 at 72 h of enzyme harvest. When compared with the total cellulase activities of the individual bacterial strains, it was found that the total cellulase activity at 72 h was on lower side. The endoglucanase activity of the consortium exhibited 0.02, and 0.21 Uml−1 of enzyme activity at 24, and 72 h of incubation and subsequent enzyme harvest, respectively. Endoglucanase activity of the isolated bacterial strains did not show higher enzyme activity in the form of consortium than the individual bacterial strains. The β-glucosidase activity of prepared Consortium A showed 8.15 Uml−1 at 72 h of enzyme harvest (Figures 7–10).
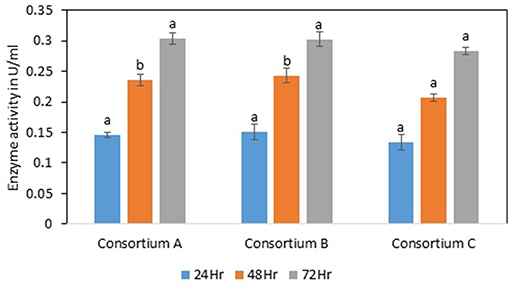
Figure 7. Total cellulase activity of different consortia. Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
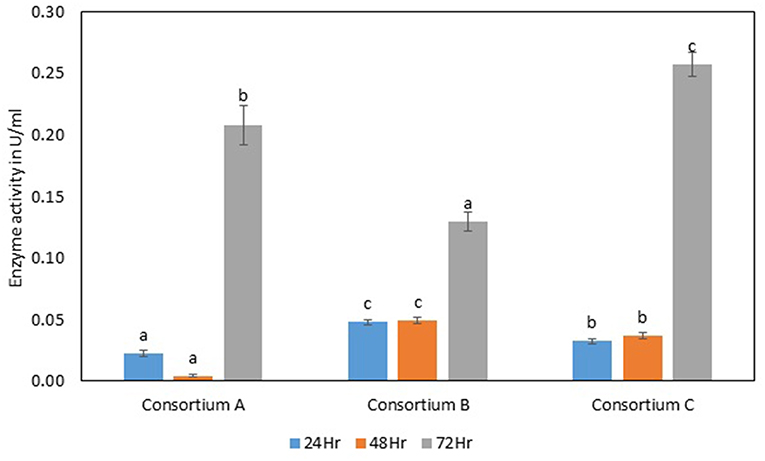
Figure 8. Endoglucanase activity of different consortia. Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
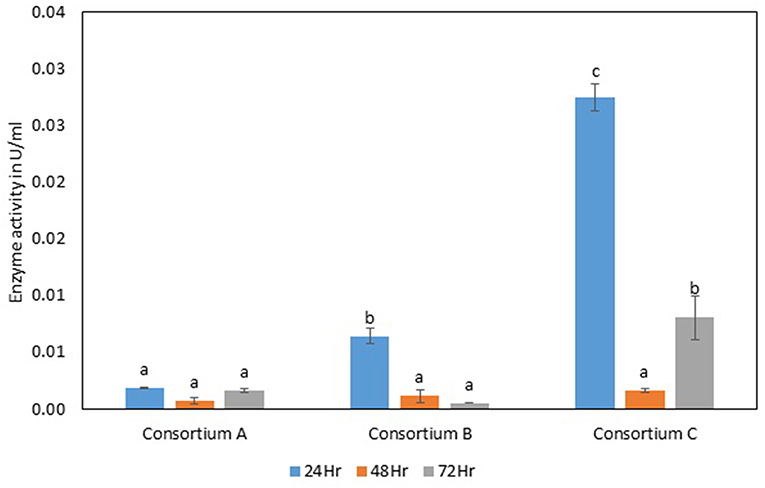
Figure 9. Exoglucanase activity of different strains. Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
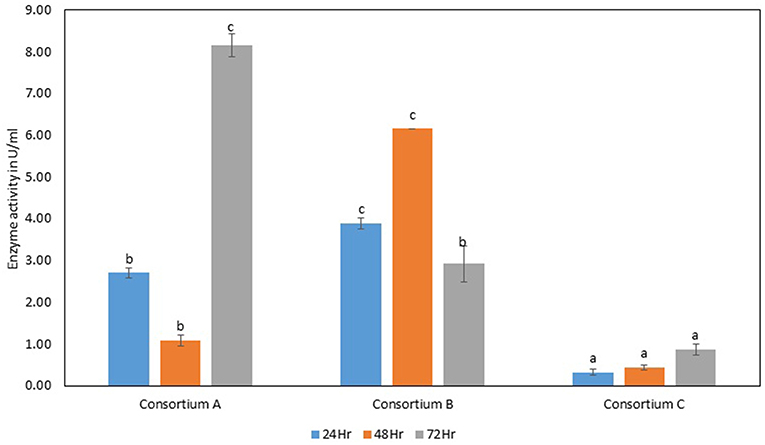
Figure 10. β-glucosidase activity of different strains. Values are the mean ± SE (standard error). Mean values in each bar with the same alphabet superscript(s) do not differ significantly, but different alphabet superscript is showed significantly different between each treatment by Duncan post hoc test (p ≤ 0.05).
Consortium B
The total cellulase activity of consortium B was also higher than the individual strains. The total enzyme activity of consortium B was 0.15, 0.24, and 0.30 Uml−1 at 24, 48, and 72 h of enzyme harvest, respectively. The endoglucanase activity in consortium B also showed similar results, with lower endoglucanase activity, 0.05, 0.05, and 0.13 Uml−1 of enzyme activity at 24, 48, and 72 h of enzyme cultivation, respectively, when compared with enzyme activity of individual strains with an activity of either 0.23 Uml−1 or 0.21 Uml−1. The exoglucanase activities of the prepared consortium was found to be 0.01, 0.00, and 0.00 Uml−1 at 24, 48, and 72 h of enzyme harvest, respectively, which is again almost negligible (Figures 7–10). The β-glucosidase activity of the prepared Consortium B showed some good results. Enzyme activity was found to be 3.89, 6.16, and 2.91 Uml−1 at 24, 48, and 72 h of enzyme harvest, respectively.
Consortium C
The total cellulase activity of the prepared consortia yet again showed good results in consortium C. Total cellulase activity by consortium C showed 0.13, 0.21, and 0.28 Uml−1 at 24, 48, and 72 h, respectively, which was higher in comparison to individual total cellulase activities of the bacterial strains. The endoglucanase activity of the consortium C showed low activity in comparison to individual bacterial strains with 0.03, 0.04, and 0.26 Uml−1 at 24, 48, and 72 h, respectively. Exoglucanase activity in the consortium C was also found to be on lower side in comparison to enzyme activity of individual bacterial strains. The activity was found to be 0.02, 0.00, and 0.01 Uml−1 at 24, 48, and 72 h, respectively. β-glucosidase activity of consortium C was found to be 0.32, 0.43, and 0.86 Uml−1 at 24, 48, and 72 h, respectively (Figures 7–10). This enzyme activity in consortium C was found to be very low in comparison to enzyme activity of individual bacterial strains.
In a study by Dabhi et al. (2014), it was observed that a highest of 0.178 Uml−1 of total cellulase activity, 1.716 Uml−1 of endoglucanase activity, and 0.602 Uml−1 of β-glucosidase activity through the bacterial isolates in the form of consortia. Another study by Park et al. (2012), observed the release of 0.466 Uml−1 of endoglucanase, 0.014 Uml−1 of exoglucanase, and 0.041 Uml−1 of β-glucosidase, when the cultures were used in the form of consortia. The enzyme activities in the consortia showed two-fold increase in total cellulase activity, but the endoglucanase activity decreased significantly. The results obtained in the quantification of the total cellulase enzyme revealed that the total production was much higher than other B. subtilis strains or the work carried out in the literature. The total cellulase activity in a similar work done Gupta et al. (2012), yielded 0.012–0.196 U/ml. It was reported that B. subtilis showed 1.08 U/mg total cellulase activity (Heck et al., 2002). In a recent study by Ladeira et al. (2015), the avicelase or exoglucanase activity was estimated to be 0.83 Uml−1 and endoglucanase activity 0.29 Uml−1 at cultivation time of 120 and 168 h, respectively. A study by Dipasquale et al. (2014) was carried out on a thermophilic microbe showing cellulose degradation as well as detergent compatibility. Another study with different lignocellulolytic enzyme activity in the consortium yielded results of CMCase 3.8 Umg−1, xylanase 8.9 Umg−1 at pH 5 on day two, and β-glucosidase activity of 5.0 Umg−1 on day five. A comparative study of different microbes isolated and their maximum enzyme activities is mentioned in the Table 3. In this study, better results were found in terms of enzyme activity and might prove beneficial to the bio-based product industries. Further investigation of the consortia on different agro-residues can yield fruitfully significant results.
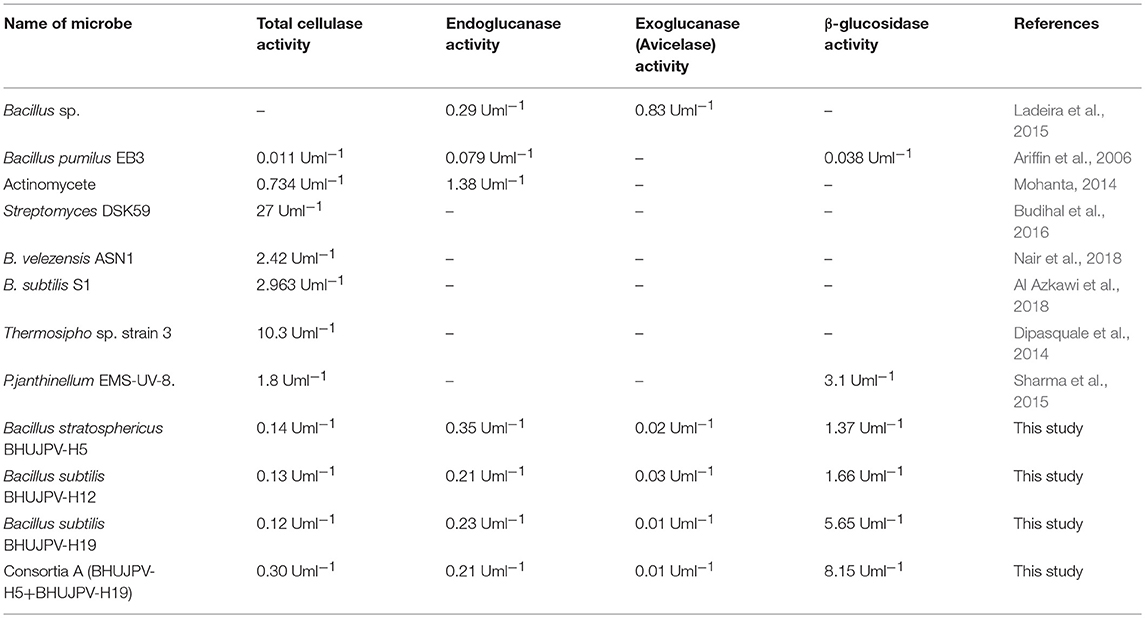
Table 3. A comparative study of enzyme activities of different microbes in literature and present study.
Future Perspectives
The thermophilic bacterial strains can be effectively used for the consolidated bioprocessing approach and also favors the rate of reaction in terms of hydrolysis product. These strains can also be tested for their activity on different agro-residues such as rice husk, cotton dust. The thermophiles can also be used effectively if the substrate is pre-treated with the help of ionic liquids and eutectic solvents (Satlewal et al., 2018a,b). Ionic liquids are known to effectively loosen up the lignin-hemicellulose-cellulose complex (Satlewal et al., 2018b). Another major insight that can be looked into is the production of ligninolytic enzymes and other hemicellulose degrading enzymes by the identified bacterial strains. This may help in combined biomass processing of the agro-wastes. The bioprocessing approach together with the use of thermophiles could lead to the production of highly cost-effective bioethanol.
Conclusions
The bacterial strains isolated from hot water spring samples have been identified as mostly Bacillus species and some Brevibacterium species. Bacterial strain B. stratosphericus BHUJPV-H5 showed endoglucanase activity of 0.35 Uml−1 at 72 h of enzyme harvest. B. subtilis BHUJPV-H12 showed β-glucosidase enzyme activity of 3.78 Uml−1 with high sugar release of 4.42 and 0.72 mgml−1 in sugarcane bagasse and wheat straw, respectively. B. subtilis BHUJPV-H19 showed β-glucosidase enzyme activity of 5.65 Uml−1. Consortium A showed total cellulase enzyme activity of 0.30 Uml−1, endoglucanase activity of 0.21 Uml−1, and high β-glucosidase activity of 8.15 Uml−1 at 72 h. Hence, this consortium A can be used for further modifications and also higher production of glucose from the agro-residues.
Data Availability
The datasets for this manuscript are not publicly available because we have submitted this research manuscript as original and novel research for making microbial consortia for cellulose degradation. All data is original not published anywhere, only molecular characterized data 16SrRNA sequences have been submitted in NCBI-DATA for getting accession numbers. Requests to access the datasets should be directed to JV, dmVybWFfYmh1QHlhaG9vLmNvLmlu.
Author Contributions
This research was conducted and written by SS. Experiment designed and edited by JV and NS. DJ as co-author did isolation and characterization of all microbial strains which has been used in this study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors are highly grateful to Council of Scientific and Industrial Research [Grant no. 38(1414)/16/EMR-II] for providing the funds to carry out the research work. Authors thank DST and DBT for providing funds for research equipment's. Authors are also grateful to the Director of the Institute of Environment and Sustainable Development for providing the infrastructure facilities. Authors thank Prof. Janardan Yadav, and Mr. Satya Prakash for their help in this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2019.00061/full#supplementary-material
Supplementary Figure 1. Pre-treated wheat straw and sugarcane bagasse used as substrate.
Supplementary Figure 2. External morphology of the identified bacterial strains.
Supplementary Figure 3. Biochemical analysis of the bacterial strains.
Supplementary Figure 4. CMC hydrolysis of the bacterial strains of CMC-congo red plates.
Supplementary Figure 5. Phylogenetic tree of the molecularly identified bacterial strains. The phylogenetic tree between 39 taxa was generated using the UPGMA method.
Abbreviations
CMC, Carboxymethyl Cellulose; NA, Nutrient agar; Uml−1, International units per milliliter; ml, Milliliter; w/v, Weight/Volume; EDTA, Ethylene diamine tetra acetic acid; μg, Microgram; FPase, Enzyme activity on filter paper; mg, Milligram; °C, Degree Celsius; CO2, Carbon Dioxide; N2O, Dinitrogen oxide; CH4, Methane; MR, Methyl Red; VP, Voges-Proskauer; OD, Optical density; nm, Nanometre; DNS, Dinitrosalicylic acid; h, Hours.
References
Acharya, S., and Chaudhary, A. (2012). Alkaline cellulase produced by a newly isolated thermophilic Aneurinibacillus thermoaerophilus WBS2 from hot spring, India. Afr. J. Microbiol. Res. 6, 5453–545S8. doi: 10.5897/AJMR11.1491
Agrawal, R., Bhadana, B., Mathur, A. S., Kumar, R., Gupta, R. P., and Satlewal, A. (2018a). Improved enzymatic hydrolysis of pilot scale pretreated rice straw at high total solids loading. Front. Energy Res. 6:115. doi: 10.3389/fenrg.2018.00115
Agrawal, R., Satlewal, A., Mathur, A. S., Gupta, R. P., Raj, T., Kumar, R., et al. (2018b). Kinetic and enzyme recycling studies of immobilized β-glucosidase for lignocellulosic biomass hydrolysis. Environ. Eng. Manag. J. 17, 1385–1398. doi: 10.30638/eemj.2018.137
Agrawal, R., Satlewal, A., Sharma, B., Mathur, A., Gupta, R., Tuli, D., et al. (2017). Induction of cellulases by disaccharides or their derivatives in Penicillium janthinellum EMS-UV-8 mutant. Biofuels 8, 615–622. doi: 10.1080/17597269.2016.1242692
Agrawal, R., Semwal, S., Kumar, R., Mathur, A., Gupta, R. P., Tuli, D. K., et al. (2018c). Synergistic enzyme cocktail to enhance hydrolysis of steam exploded wheat straw at pilot scale. Front. Energy Res. 6:122. doi: 10.3389/fenrg.2018.00122
Agrawal, R., Srivastava, A., and Verma, A. K. (2016). Immobilization of β-glucosidase onto silicon oxide nanoparticles and augment of phenolics in sugarcane juice. J. Food Sci. Technol. 53, 3002–3012. doi: 10.1007/s13197-016-2269-x
Al Azkawi, A. S., Sivakumar, N., and Al Bahry, S. (2018). Bioprocessing of cardboard waste for cellulase production. Biomass Convers. Bior. 8, 597–606. doi: 10.1007/s13399-018-0309-7
Ariffin, H., Abdullah, N., Umi Kalsom, M. S., Shirai, Y., and Hassan, M. A. (2006). Production and characterization of cellulase by Bacillus pumilus EB3. Int. J. Eng. Technol. 3, 47–53.
Baharuddin, M., Ahmad, A., La Nafie, N., and Zenta, F. (2016). Cellulase enzyme activity of bacillus circulans from larvae Cossus cossus in lignocellulosic substrate. Am. J. Biomed. Life Sci. 4, 21–25. doi: 10.11648/j.ajbls.20160402.13
Budihal, S. R., Agsar, D., and Patil, S. R. (2016). Enhanced production and application of acidothermophilic Streptomyces cellulase. Bioresour. Technol. 200, 706–712. doi: 10.1016/j.biortech.2015.10.098
Dabhi, B. K., Vyas, R. V., and Shelat, H. N. (2014). Use of banana waste for the production of cellulolytic enzymes under solid substrate fermentation using bacterial consortium. Int. J. Curr. Microbiol. App. Sci. 3, 337–346.
Das, A., Bhattacharya, S., and Murali, L. (2010). Production of cellulase from a thermophilic Bacillus sp. isolated from cow dung. Am. Eurasian J. Agric. Environ. Sci. 8, 685–691.
Dipasquale, L., Romano, I., Picariello, G., Calandrelli, V., and Lama, L. (2014). Characterization of a native cellulase activity from an anaerobic thermophilic hydrogen-producing bacterium Thermosipho sp. strain 3. Ann. Microbio. 64, 1493–1503. doi: 10.1007/s13213-013-0792-9
Gupta, A., and Verma, J. P. (2015). Sustainable bio-ethanol production from agro-residues: a review. Renew. Sust. Energ. Rev. 41, 550–567. doi: 10.1016/j.rser.2014.08.032
Gupta, P., Samant, K., and Sahu, A. (2012). Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. Microbiol. 2012:578925. doi: 10.1155/2012/578925
Gupta, R., Khasa, Y. P., and Kuhad, R. C. (2011). Evaluation of pretreatment methods in improving the enzymatic saccharification of cellulosic materials. Carbohyd. Polym. 84, 1103–1109. doi: 10.1016/j.carbpol.2010.12.074
Hamilton-Brehm, S. D., Mosher, J. J., Vishnivetskaya, T., Podar, M., Carroll, S., Allman, S., et al. (2010). Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl. Environ. Microbiol. 76, 1014–1020. doi: 10.1128/AEM.01903-09
Heck, J. X., Hertz, P. F., and Ayub, M. A. (2002). Cellulase and xylanase productions by isolated Amazon Bacillus strains using soybean industrial residue based solid-state cultivation. Braz. J. Microbiol. 33, 213–218. doi: 10.1590/S1517-83822002000300005
Kato, S., Haruta, S., Cui, Z. J., Ishii, M., and Igarashi, Y. (2005). Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl. Environ. Microbiol. 71, 7099–7106. doi: 10.1128/AEM.71.11.7099-7106.2005
Ko, C. H., Chen, W. L., Tsai, C. H., Jane, W. N., Liu, C. C., and Tu, J. (2007). Paenibacillus campinasensis BL11: a wood material-utilizing bacterial strain isolated from black liquor. Bior. Technol. 98, 2727–2733. doi: 10.1016/j.biortech.2006.09.034
Koeck, D. E., Ludwig, W., Wanner, G., Zverlov, V. V., Liebl, W., and Schwarz, W. H. (2015). Herbinix hemicellulosilytica gen. nov., sp. nov., a thermophilic cellulose-degrading bacterium isolated from a thermophilic biogas reacto. Int. J. Syst. Evol. Microbiol. 65, 2365–2371. doi: 10.1099/ijs.0.000264
Koeck, D. E., Mechelke, M., Zverlov, V. V., Liebl, W., and Schwarz, W. H. (2016). Herbivorax saccincola gen. nov., sp. nov., a cellulolytic, anaerobic, thermophilic bacterium isolated via in sacco enrichments from a lab-scale biogas reactor. Int. J. Syst. Evol. Microbiol. 66, 4458–4463. doi: 10.1099/ijsem.0.001374
Ladeira, S. A., Cruz, E., Delatorre, A. B., Barbosa, J. B., and Leal Martins, M. L. (2015). Cellulase production by thermophilic Bacillus sp: SMIA-2 and its detergent compatibility. Electron J. Biotech. 18, 110–115. doi: 10.1016/j.ejbt.2014.12.008
Maki, M., Leung, K. T., and Qin, W. (2009). The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 5:500. doi: 10.7150/ijbs.5.500
Miller, G. L., Blum, R., Glennon, W. E., and Burton, A. L. (1960). Measurement of carboxymethylcellulase activity. Anal. Biochem. 1, 127–132. doi: 10.1016/0003-2697(60)90004-X
Mohammad, B. T., Al Daghistani, H. I., Jaouani, A., Abdel-Latif, S., and Kennes, C. (2017). Isolation and characterization of thermophilic bacteria from jordanian hot springs: Bacillus licheniformis and Thermomonas hydrothermalis isolates as potential producers of thermostable enzymes. Int. J. Microbiol. 2017:6943952. doi: 10.1155/2017/6943952
Mohanta, Y. K. (2014). Isolation of cellulose-degrading actinomycetes and evaluation of their cellulolytic potential. J. Biosci. Bioeng. 2, 1–5. doi: 10.13189/bb.2014.020101
Nair, A. S., Al-Battashi, H., Al-Akzawi, A., Annamalai, N., Gujarathi, A., Al-Bahry, S., et al. (2018). Waste office paper: a potential feedstock for cellulase production by a novel strain Bacillus velezensis ASN1. Waste Manag. 79, 491–500. doi: 10.1016/j.wasman.2018.08.014
Najar, I. N., Sherpa, M. T., Das, S., Verma, K., Dubey, V. K., and Thakur, N. (2018). Geobacillus yumthangensis sp. nov., a thermophilic bacterium isolated from a north-east Indian hot spring. Int. J. Syst. Evol. Microbiol. 68, 3430–3434. doi: 10.1099/ijsem.0.003002
Park, J. I., Steen, E. J., Burd, H., Evans, S. S., Redding-Johnson, A. M., and Batth, T. (2012). A thermophilic ionic liquid-tolerant cellulase cocktail for the production of cellulosic biofuels. PLoS ONE 7:e37010. doi: 10.1371/journal.pone.0037010
Rastogi, G., Muppidi, G. L., Gurram, R. N., Adhikari, A., Bischoff, K. M., Hughes, S. R., et al. (2009). Isolation and characterization of cellulose-degrading bacteria from the deep subsurface of the Homestake gold mine, Lead, South Dakota, USA. J. Ind. Microbiol. Biotechnol. 36, 585–598. doi: 10.1007/s10295-009-0528-9
Rawway, M., Ali, S. G., and Badawy, A. S. (2018). Isolation and identification of cellulose degrading bacteria from different sources at assiut governorate (Upper Egypt). J. Ecol. Heal. Environ. Int. J. 6, 15–24. doi: 10.18576/jehe/060103
Robert, C., and Bernalier-Donadille, A. (2003). The cellulolytic microflora of the human colon: evidence of microcrystalline cellulose-degrading bacteria in methane-excreting subjects. FEMS Microbiol. Ecol. 46, 81–89. doi: 10.1016/S0168-6496(03)00207-1
Russell, D. W., and Sambrook, J. (2001). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory.
Satlewal, A., Agrawal, R., Bhagia, S., Sangoro, J., and Ragauskas, A. J. (2018a). Natural deep eutectic solvents for lignocellulosic biomass pretreatment: recent developments, challenges and novel opportunities. Biotechnol. Adv. 36, 2032–2050. doi: 10.1016/j.biotechadv.2018.08.009
Satlewal, A., Agrawal, R., Das, P., Bhagia, S., Pu, Y., Puri, S. K., et al. (2018b). Assessing the facile pretreatments of bagasse for efficient enzymatic conversion and their impacts on structural and chemical properties. ACS Sustain. Chem. Eng. 7, 1095–1104. doi: 10.1021/acssuschemeng.8b04773
Shareef, I., Satheesh, M., and Christopher, S. X. (2015). Isolation and identification of cellulose degrading microbes. Int. J. Innov. Res. Sci. Eng. 8, 6788–6793. doi: 10.15680/IJIRSET.2015.0408012
Sharma, B., Agrawal, R., Singhania, R. R., Satlewal, A., Mathur, A., Tuli, D., et al. (2015). Untreated wheat straw: potential source for diverse cellulolytic enzyme secretion by Penicillium janthinellum EMS-UV-8 mutant. Bioresour. Technol. 196, 518–524. doi: 10.1016/j.biortech.2015.08.012
Sivakumar, N., Al Zadjali, A., Al Bahry, S., Elshafie, A., and Eltayeb, E. A. (2016). Isolation and characterization of cellulolytic Bacillus licheniformis from compost. Afr. J. Biotechnol. 15, 2434–2446. doi: 10.5897/AJB2016.15641
Srivastava, N., Srivastava, M., Mishra, P. K., Gupta, V. K., Molina, G., Rodriguez-Couto, S., et al. (2018). Applications of fungal cellulases in biofuel production: advances and limitations. Renew. Sust. Energ. Rev. 82, 2379–2386. doi: 10.1016/j.rser.2017.08.074
Strahsburger, E., de Lacey, A. M. L., Marotti, I., DiGioia, D., Biavati, B., and Dinelli, G. (2017). In vivo assay to identify bacteria with β-glucosidase activity. Electron J. Biotechnol. 30, 83–87. doi: 10.1016/j.ejbt.2017.08.010
Tabassum, F., Irfan, M., Shakir, H. A., and Qazi, J. I. (2018). RSM based optimization of nutritional conditions for cellulase mediated Saccharification by Bacillus cereus. J. Biol. Eng. 12:7. doi: 10.1186/s13036-018-0097-4
Verma, J. P., Jaiswal, D. K., Krishna, R., Prakash, S., Yadav, J., and Singh, V. (2018). Characterization and screening of thermophilic Bacillus strains for developing plant growth promoting consortium from hot spring of Leh and Ladakh region of India. Front. Microbiol. 9:1293. doi: 10.3389/fmicb.2018.01293
Keywords: cellulase, endoglucanase, exoglucanase, β-glucosidase, sugarcane bagasse, wheat straw, glucose, consortium
Citation: Singh S, Jaiswal DK, Sivakumar N and Verma JP (2019) Developing Efficient Thermophilic Cellulose Degrading Consortium for Glucose Production From Different Agro-Residues. Front. Energy Res. 7:61. doi: 10.3389/fenrg.2019.00061
Received: 24 February 2019; Accepted: 07 June 2019;
Published: 02 July 2019.
Edited by:
Anli Geng, Ngee Ann Polytechnic, SingaporeReviewed by:
Ruchi Agrawal, Indian Oil Corporation, IndiaAlok Satlewal, Indian Oil Corporation, India
Copyright © 2019 Singh, Jaiswal, Sivakumar and Verma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jay Prakash Verma, dmVybWFfYmh1QHlhaG9vLmNvLmlu; anB2Lmllc2RAYmh1LmFjLmlu
 Saurabh Singh
Saurabh Singh Durgesh Kumar Jaiswal1
Durgesh Kumar Jaiswal1 Jay Prakash Verma
Jay Prakash Verma