- 1Instituto de Tecnologia Química e Biológica António Xavier, Universidade NOVA de Lisboa, Oeiras, Portugal
- 2Instituto de Biologia Experimental e Tecnológica, Oeiras, Portugal
Extracellular electron transfer pathways allow bacteria to transfer electrons from the cell metabolism to extracellular substrates, such as metal oxides in natural environments and electrodes in microbial electrochemical technologies (MET). Studies of electroactive microorganisms and mainly of Shewanella oneidensis MR-1 have demonstrated that extracellular electron transfer pathways relies on several multiheme c-type cytochromes. The small tetraheme cytochrome c (STC) is highly conserved among Shewanella species and is one of the most abundant cytochromes in the periplasmic space. It transfers electrons from the cell metabolism delivered by the inner-membrane tetraheme cytochrome CymA, to the porin-cytochrome complex MtrCAB in the outer-membrane, to reduce solid electron acceptors outside the cell, or electrodes in the case of MET. In this work knock-out strains of STC of S. oneidensis MR-1, expressing STC from distinct Shewanella species were tested for their ability to perform extracellular electron transfer, allowing to explore the effect of protein mutations in living organisms. These studies, complemented by a biochemical evaluation of the electron transfer properties of the individual proteins, revealed a considerable plasticity in the molecular components involved in extracellular electron transfer. The results of this work are pioneering and of significant relevance for future rational design of cytochromes in order to enhance extracellular electron transfer and thus contribute to the practical implementation of MET.
Introduction
The growing demand of renewable energy sources has increased the interest in microbial electrochemical technologies (MET), in particular of microbial fuel cells (MFC) (Logan and Regan, 2006; Arends and Verstraete, 2012; Ucar et al., 2017). In these devices, microorganisms are used as self-regenerating catalysts to convert chemical energy stored in biodegradable substances into electrical current. Accompanying the growth of this research field, MET have seen significant developments and nowadays, besides electrical power generation, they can also promote different sustainable technologies including wastewater treatment, water desalination, bioremediation of soils and aquifers and production of added value compounds (Logan and Rabaey, 2012; Bajracharya et al., 2016). It is the ability of electroactive organisms [i.e., organisms capable of interacting with an electrode (Koch and Harnisch, 2016)] to perform extracellular electron transfer that is at the heart of these systems, allowing them to exchange electrons with electrodes in MET (Richardson et al., 2012; White et al., 2016). Indeed, in their natural environment these organisms use insoluble compounds as terminal acceptors, allowing them to couple their metabolism with the reduction of insoluble electron acceptors outside of the cell, or vice-versa. Despite the great promise of these technologies for numerous applications, most of these systems are still restricted to lab-scale research projects, as the electron transfer rates between the organisms and the electrode are too low to design a viable scale-up process (Logan and Rabaey, 2012). This is a consequence of the fact that the organisms used in MET function as they would in their natural environment and did not evolved to grow and live in the conditions used in MET (Torres et al., 2010; Torres, 2014).
It is now well-known that electron transfer to electrodes is dependent on a chain of redox proteins, in particular multiheme c-type cytochromes, that conduct electrons from intracellular carriers through the periplasmic space and across the cell surface to reach the microbe-electrode interface (White et al., 2016). It has been demonstrated that the electron transfer performed by these proteins control the rate of electron transfer to electrodes (Torres et al., 2010; Strycharz-Glaven et al., 2011; Torres, 2014). Indeed, deletion of the genes and overexpression of these multiheme cytochromes were shown to modify current production in MET (Sydow et al., 2014; Teravest and Ajo-Franklin, 2015).
In recent years, significant efforts have been made to characterize the molecular details of the electron transfer processes performed by these proteins so rational manipulation can be used to enhance electron transfer rates (Fonseca et al., 2009; Paquete et al., 2014; Alves et al., 2017; Neto et al., 2017). However, the molecular factors that underpin the electron transfer rates in this redox chain are still unclear. In vitro studies of site directed mutants of these multiheme cytochromes have shown that electrostatic and thermodynamics are important factors in controlling electron transfer (Alves et al., 2017; Neto et al., 2017). However, it is still not known if changes in these parameters affect electron transfer in living cells, since cellular milieu influences protein folding, stability and the reactivity of the proteins (Kuznetsova et al., 2014; Gnutt and Ebbinghaus, 2016).
To explore this, in this study we investigated the ability of the electroactive bacterium Shewanella oneidensis MR-1 to perform extracellular electron transfer with STC orthologs. By relying on natural mutations, these STC proteins will mimic random mutations that are known to be functional, allowing to test the capacity of STC variants to perform electron transfer in living cells. STC is one of the most abundant proteins found in the periplasmic space of Shewanella (Tsapin et al., 2001; Meyer et al., 2004), and together with FccA is responsible for mediating electron transfer across the periplasmic space of Shewanella (Fonseca et al., 2013; Sturm et al., 2015). STC and FccA form two independent redox pathways between the tetraheme cytochrome CymA in the cytoplasmic membrane and the MtrCAB complex present at the cell surface (Fonseca et al., 2013) (Figure 1). In these pathways, CymA can receive electrons from the quinone pool, and then deliver them to the periplasmic proteins STC or FccA that then transfer the electrons to the MtrCAB complex at the outer membrane for the reduction of insoluble compounds or electrodes in the case of METs.
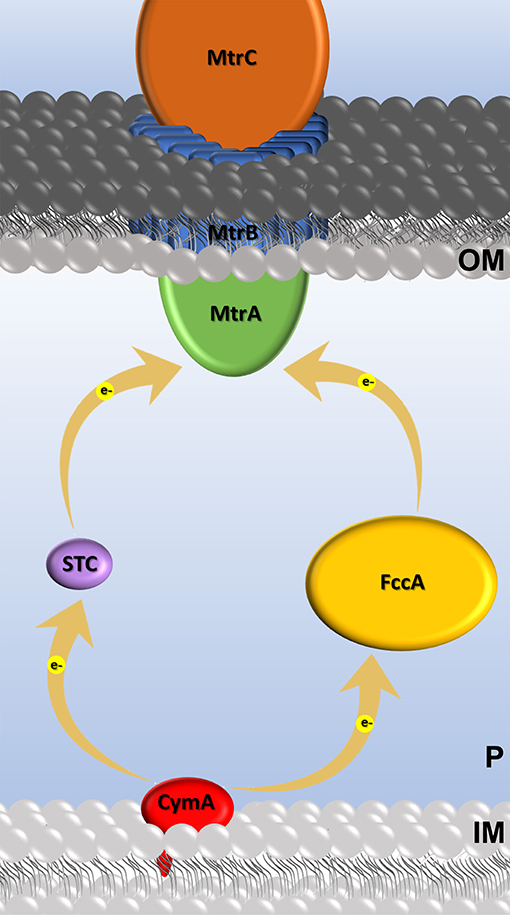
Figure 1. Cartoon of the extracellular electron transfer Mtr pathway in S. oneidensis and c-type cytochromes involved. Electron transfer events are represented by yellow arrows.
Until now, three-dimensional structures have only been obtained for STC from S. oneidensis and S. frigidimarina (Leys et al., 2002; Paixão et al., 2008), and structural analysis revealed that the structures are very similar. Despite the similar three-dimensional structure and aminoacid composition (>60 % identity), their redox properties are quite distinct (Pessanha et al., 2001; Fonseca et al., 2009). For example, heme I is the first heme to be oxidized in the STC from S. oneidensis, while in STC from S. frigidimarina it is heme IV (Pessanha et al., 2001; Fonseca et al., 2009).
In this work the structural and biochemical characterization of STC from S. algae is obtained for the first time, allowing to compare the different modes of action of three STC orthologous and unravel the molecular properties that are relevant for extracellular electron transfer in living cells. The S. oneidensis STC orthologous are from S. frigidimarina and S. algae, which reflect the different environments that these organisms can be found, since S. oneidensis is a fresh water organism (Myers and Nealson, 1988). While S. frigidimarina, is a marine organism, originally isolated from Antarctic sea ice being able to grow at lower temperatures (optimal growth temperature at ~20°C) and in high amounts of salt [up to 9% (wt/vol)] (Bowman et al., 1997), S. algae has been isolated from infected human samples (Vignier et al., 2013; Janda and Abbott, 2014). This organism is mostly associated with trauma and surgical manipulation that promote optimal conditions for these opportunistic pathogens to grow (Vignier et al., 2013).
This pioneering work is crucial to elucidate the properties that are relevant to advance the modification of multiheme proteins toward enhanced electron transfer rates necessary for the practical implementation of MET.
Materials and Methods
Bacterial Strains
The type strains S. oneidensis MR-1 (ATCC number 700550) was purchased from ATCC, while S. algae (DSM number 9167) and S. frigidimarina (DSM number 12253) were purchased from DSMZ. Gene deletion strains, S. oneidensis ΔSTC and S. oneidensis ΔSTC ΔFccA were kindly provided by Dr. Liang Shi and Dr. Johannes Gescher, respectively.
Production and Purification of STC From S. algae
Cells from S. algae were grown under aerobic conditions (180 rpm) in a batch culture at 37°C, using Terrific Broth (TB) medium supplemented with 10 g/L NaCl. Cells were allowed to grow for 24 h before harvesting by centrifugation (10,000 g, 15 min, 4°C). The cell pellet was resuspended in 20 mM Tris-HCl buffer, pH 7.6 containing a protease inhibitor cocktail and DNase. Cell disruption was obtained by two passages through a French Press at a pressure of 1,000 psi. Cell debris were removed by centrifugation at 10,000 × g for 15 min at 4°C, and the supernatant was ultracentrifuged at 200,000 × g for 1 h at 4°C. The supernatant containing the soluble protein fraction was loaded onto a diethylaminoethyl (DEAE) column (GE Healthcare) previously equilibrated with 20 mM Tris-HCl buffer (pH 7.6). A salt gradient from 0 to 1 M NaCl in the same buffer was applied and the fraction containing SaSTC was eluted at 300 mM NaCl. This fraction was dialyzed, concentrated and loaded onto a Q-Sepharose column (GE Healthcare), equilibrated previously with 20 mM Tris-HCl buffer (pH 7.6). The fraction containing SaSTC was eluted at 300 mM NaCl, using a salt gradient from 0 to 1 M NaCl in 20 mM Tris-HCl buffer (pH 7.6). As a final purification step the fraction containing SaSTC was dialyzed, concentrated and loaded onto a hydroxylapatite (HTP) column (Bio-Rad Laboratories), pre-equilibrated with 10 mM potassium phosphate buffer (pH 7.6). The fraction containing SaSTC did not bind to the column and was eluted in the washout volume.
The chromatographic fractions were routinely analyzed by SDS-PAGE and UV–visible spectroscopy to select those containing the protein of interest. The purity of the protein was revealed as a single band in the gel. Pure cytochrome samples had a typical absorbance ratio, ASoretPeak/A280nm, larger than 7.0.
NMR Experiments
SaSTC protein for NMR experiments was lyophilized and dissolved using 2H2O (99.9 atom %) to a final concentration of approximately 2 mM. The ionic strength was adjusted to 100 mM by the addition of KCl. The NMR spectra obtained before and after the lyophilization were identical, indicating that the protein structure of SaSTC was not affected by this procedure. NMR spectra were acquired on Bruker spectrometers operating at 500 MHz and 25°C using oxidized and reduced samples. Protein reduction was achieved with a concentrated solution of sodium dithionite prepared in the same buffer as the protein.
The fully reduced 1H heme signals were assigned following routine strategies (Silva and Louro, 2017), using several 1H-NOESY spectra with different mixing times with the aid of a TOCSY spectrum. Assignment of the heme methyl signals in the fully oxidized protein was done using a combination of 13C-1H-HMQC experiments, NOESY and TOCSY experiments as previously performed for the homologous proteins (Pessanha et al., 2001; Fonseca et al., 2009). Furthermore, 1H-NOESY spectra collected using partially oxidized samples were used to cross-assign the methyl signals, from the reduced to the oxidized state following previous described procedures (Salgueiro et al., 1992). Partially oxidized protein was prepared by adding with a gastight syringe controlled amounts of air to the NMR tube containing reduced protein.
The Bruker TopSpin program was used to visualize the NMR spectra and the CARA program was used for the assignment work. The proton spectra were calibrated using the water signal as an internal reference.
Cyclic Voltammetry
For protein film voltammetry (PFV) experiments pure SaSTC (in 20 mM phosphate buffer with 100 mM KCl at pH 7.6) was mixed with poly-L-lysine solution (0.1% in H20 from SIGMA) in a mixture of 1:1 to facilitate the adsorption of the protein to the electrode (Alves et al., 2017). A small volume of this mixture was deposited onto pyrolytic graphite edge (PGE) electrode and was left to dry. The PGE electrode was polished with aqueous Al2O3 slurry (1.0 μm), rinsed with H2O and dried with a tissue before exposure to STC. Experiments were performed at 25°C using a three-electrode cell configuration consisting of the PGE electrode, the Ag/AgCl (3M KCl) reference electrode and a platinum wire counter electrode, that was housed in a Coy anaerobic chamber. Cyclic voltammetry was performed in 20 mM phosphate buffer with 100 mM KCl at different pH values using a CHI electrochemical analyzer from CHI Instruments controlled by the manufactures' software (version 10.12). The acquisition of voltammograms at different pH values was achieved by replacing the existing buffer with the buffer of interest. After each experiment the pH was measured and used as the actual pH of the experiment. Reduction potentials are reported with respect to the SHE by addition of 0.207 V to those measured. The currents from the electrochemical data were normalized using QSOAS (Fourmond et al., 2009).
Thermodynamic Characterization
The thermodynamic characterization of SaSTC was achieved using a thermodynamic model that considers four redox centers and one ionizable center (Fonseca et al., 2009). Briefly, the detailed thermodynamics of STC can be described with a total of 32 microstates (Paquete and Louro, 2014). The relative population of these microstates given by the Boltzmann distribution is defined by 15 thermodynamic parameters: four reduction potentials, one for each individual heme, the pKa of the ionizable center, six redox interaction energies between each pair of heme and six redox-Bohr interaction energies between the hemes and the ionizable center (Fonseca et al., 2009). When working in conditions of fast intramolecular electron exchange and slow intermolecular electron exchange in the NMR timescale, it is possible to define five oxidation states and discriminate a set of NMR resonances for the protons of the hemes in each of the five oxidation states (Louro, 2006). The resonance positions and linewidths of the heme methyl groups of SaSTC were followed for the redox titrations monitored by NMR spectroscopy. These chemical shifts are dependent on the ratio between the molar fraction of the microstates in which a particular heme is oxidized and the states in which the same heme is reduced, allowing to determine the relative microscopic redox potentials of the hemes in any particular stage of oxidation (Paquete and Louro, 2014). To calibrate the relative reduction potentials, the electrochemical data obtained by PFV were used. The thermodynamic model was simultaneously fitted to the electrochemical and NMR data with the Nelder-Mead Simplex algorithm implemented in MATLAB (MathWorks Inc) (Lagarias et al., 1998), to define the 15 thermodynamic parameters that describe SaSTC.
Crystallization and Structure Determination of STC From S. algae
SaSTC was crystallized by the hanging drop vapor diffusion method where 1 μL precipitant was mixed with 1 μL of a 40 mg/mL protein solution and equilibrated against a 500 μL reservoir at 21°C. The reservoir solution contained 0.1 M Bicine pH 8.5 and 3.7 M Ammonium Sulfate as precipitant, similar to the previously crystallized STC from S. oneidensis (Leys et al., 2002). Crystals were harvested and flash-cooled in liquid nitrogen directly from the crystallization drop, and diffraction data was collected at 100 K to a resolution of 1.15 Å at the Diamond Light Source (DLS) beamline I04 (Didcot, UK). The images were processed with autoPROC (Vonrhein et al., 2011), which makes use of XDS (Kabsch, 2010) and the CCP4 suite (Winn et al., 2011) for integration and conversion of integrated intensities to structure factors. Due to the crystal diffraction quality it was possible to extend data resolution to 1.04 Å, by integrating spots toward the corners of the detector at the expense of some completeness. The structure was solved by molecular replacement (MR) using PHASER in the CCP4 suite (McCoy et al., 2007) and STC from S. oneidensis (Leys et al., 2002) as phasing model. The model was corrected and completed with COOT (Emsley et al., 2010) after an initial refinement using REFMAC5 in the CCP4 suite (Murshudov et al., 1997). Structure refinement was performed using PHENIX (Afonine et al., 2012). Throughout the refinement, the model was periodically checked and corrected with COOT against σA-weighted 2|Fo| – |Fc| and |Fo| – |Fc| electron-density maps. Solvent molecules were added automatically by PHENIX and validated by inspection of electron-density maps in COOT. In the final refinement cycles, hydrogen atoms were included in calculated positions with the PHENIX READYSET tool and anisotropic displacement parameters (ADPs) were refined for all non-solvent non-hydrogen atoms. The final values of R and R-free (Brünger, 1992) were 0.151 and 0.178, respectively and the maximum likelihood estimate of the overall coordinate error was 0.11 Å. The refinement statistics are presented in Table SM5. The stereochemical quality of the model was analyzed with MOLPROBITY (Chen et al., 2010) and there are no outliers in the Ramachandran (Ramachandran and Sasisekharan, 1968) ϕ, φ plot. The coordinates and structure factors have been submitted to the Protein Data Bank in Europe (Velankar et al., 2010) with accession codes PDB ID 6HR0. Pictures were produced using Pymol (http://www.pymol.org).
Cloning of STC
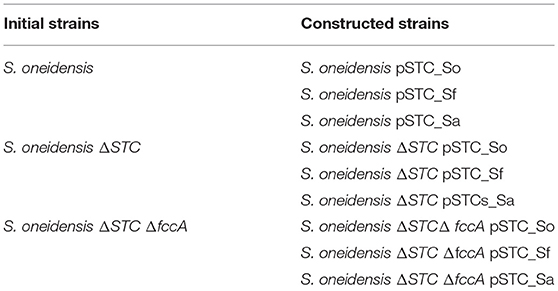
DNA fragments containing the gene that express STC (cctA gene) from S. oneidensis, S. algae and S. frigidimarina were amplified via polymerase chain reaction (PCR) from the genomic DNA of each bacteria using the primers listed in Table SM1. To include a ribosome binding site (RBS), the genes were first amplified with primers containing the RBS sequence (Table SM1). The PCR products were ligated into the pBBR1MCS-2 vector using the restriction enzymes EcoRV and BamHI originating three vectors (pSTC_So; pSTC_Sf; pSTC_Sa), each containing the STC gene from the respective Shewanella species (S. oneidensis; S. algae; S. frigidimarina). These vectors were inserted into the strains, S. oneidensis, S. oneidensis ΔSTC, and S. oneidensis ΔSTC ΔFccA, creating new strains (Table 1). The vector pBBR1MCS-2 was a gift from Kenneth Peterson (Addgene plasmid # 85168) (Kovach et al., 1995). All strains were routinely cultivated in LB medium at 30°C and 180 rpm. Strains containing the pBBR1MCS-2 vector were grown in the presence of kanamycin (50 μg/mL).
Decolorization of Methyl Orange
Biodecolorization experiments were performed at 30°C and 180 rpm using Shewanella basal medium (SBM) (Marsili et al., 2008) containing 20 mM lactate as the electron donor and sole carbon source and methyl orange (MO) to obtain the final concentration of 50 μM. SBM minimal medium consists of (per liter) 0.225 g K2HPO4, 0.225 g KH2PO4, 0.46 g NaCl, 0.225 g (NH4)2SO4, 0.117 g MgSO4·7H2O, and 10 mM HEPES, adjusted to pH 7.0. In addition, 5 ml·l−1 of 10 % (w/v) casamino acids and 10 ml·l−1 trace minerals solution (per liter: 1.5 g NTA, 0.1 g MnCl2·4H2O, 0.3 g FeSO4·7H2O, 0.17 g CoCl2·6H2O, 0.1 g ZnCl2, 0.04 g CuSO4·5H2O, 0.005 g AlK(SO4)2·12H2O, 0.005 g H3BO3, 0.09 g Na2MoO4, 0.12 g NiCl2, 0.02 g NaWO4·2H2O, and 0.10 g Na2SeO4), were added (Marsili et al., 2008). Hungate tubes were filled with 10 ml of SBM medium supplemented with lactate and MO, sparged with N2 for 15 min to ensure anaerobic conditions, and then sealed with rubber stoppers.
Bacterial cells were grown overnight in LB medium at 30°C and 180 rpm. They were subsequently washed with SBM minimal medium and used to inoculate the anaerobic dye containing medium to have approximately 2 × 108 cells.ml−1. Decolorization was followed over time at 460 nm using a colorimeter WPA Model C07500 (Biochrom). Controls without inoculation or inoculated with inactivated bacteria (heated to 90°C for 15 min) were used as control. All experiments were carried out in triplicate and repeated three times independently.
Binding Experiments
For the binding experiments the cytochromes (SaSTC; SfSTC; SoCymA; SoMtrA) were lyophilized and dissolved using 2H2O (99.9 atom %) to a final concentration of approximately 0.5 mM. Pure proteins of SfSTC, SoSTC, SoCymA, and SoMtrA were obtained as previously described (Paquete et al., 2010; Fonseca et al., 2013). The ionic strength of the samples was adjusted to 100 mM by the addition of KCl. 1H-1D-NMR data were obtained on a Bruker Avance II 500 MHz NMR spectrometer equipped with a QXI probe at 25°C. Samples containing 50 μM of one cytochrome were titrated against increasing amounts of another cytochrome. NMR spectra were recorded for each addition. All cytochromes were used in the fully oxidized state.
To determine binding affinities, the chemical shift perturbations (Δδbind) of the NMR signals from a cytochrome (CytA) resulting from the complex formation with another cytochrome (CytB) were plotted against the molar ratio (R) of [CytB]/[CytA]. The 1H-NMR spectra were processed and analyzed using the Bruker TopSpin 2.1 software. Chemical shifts are reported in parts per million (ppm) and the spectra were calibrated using the water signal as internal reference. The data were fitted using least squares minimization to a 1:1 binding model using equations (1) and (2) (Worrall et al., 2003):
where is the maximal chemical shift perturbation of the NMR signals resulting from the complex formation between CytA and CytB, Kd is the dissociation constant, [CytA]0 is the initial concentration of CytA, and [CytB]0 is the stock concentration of CytB. Binding affinities between the membrane multiheme cytochromes CymA and MtrA from S. oneidensis with the periplasmic redox partner STC belonging to different Shewanella species were determined. When several methyl signals belonging to an individual heme were clearly visible, the data obtained for all methyl signals were used to define the dissociation constant. Experimental uncertainty was estimated from the spectral resolution of the NMR acquisition. Only chemical shift perturbations larger than 0.025 ppm were considered significant (Díaz-Moreno et al., 2005).
Results
Thermodynamic Characterization of SaSTC
The purity of SaSTC was confirmed by SDS-PAGE gel stained with Coomassie (Figure SM1). The assignment in the oxidized and reduced state of the heme methyl signals in the NMR spectra of SaSTC was achieved (Tables SM2, SM3). The values are comparable to those obtained for the homologous SoSTC and SfSTC (Pessanha et al., 2001; Fonseca et al., 2009). Given the sensitivity of the NMR chemical shift to the environment this suggests that the heme core architecture of this protein is similar to that of the other two. 1H-NOESY spectra collected using partially oxidized samples were used to cross-assign the methyl signals from reduced to the oxidized state of SaSTC (Figure 2) (Salgueiro et al., 1992), and measure the fraction of oxidation of the individual hemes, which is crucial for the detailed microscopic thermodynamic characterization of this protein.
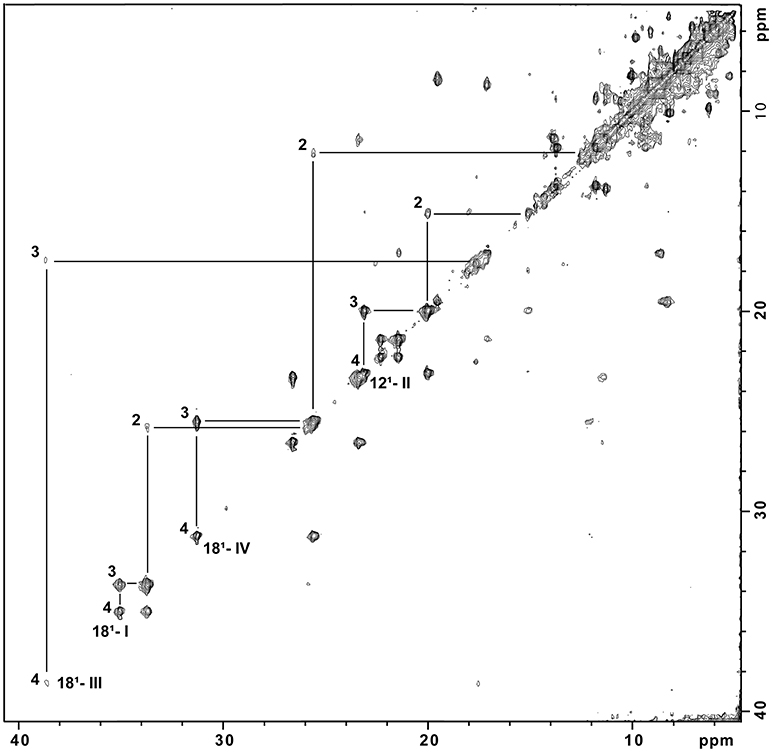
Figure 2. 1H-NOESY spectrum of partially reduced STC from S. algae at pH 7.0. Cross peaks resulting from intermolecular electron exchange between different redox stages are indicated for one methyl group from each heme, identified using the IUPAC nomenclature (M181I; M121II; M181III; M181IV) in the different stages of oxidation (1, 2, 3, and 4). The Roman numerals correspond to the four hemes, respectively.
The NMR spectra of partially oxidized samples of SaSTC showed that each heme substituent displays five discrete NMR signals that correspond to each of the five possible macroscopic stages of oxidation that this protein can assume from fully reduced to fully oxidized (Paquete and Louro, 2014).
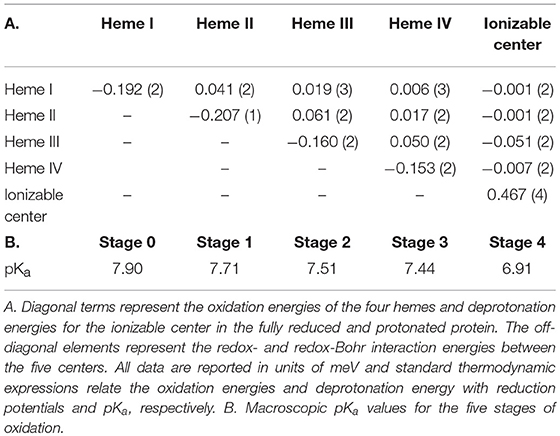
The fitting of the thermodynamic model to the data obtained from the heme methyl groups 181, 121, 121, and 181 to monitor the degree of oxidation of the individual hemes at different pH values, together with the data obtained by PFV allowed the determination of the thermodynamic parameters of SaSTC (Table 2).
The thermodynamic model shows a good agreement with the experimental data and therefore is appropriate to describe the redox- and redox-Bohr properties of the STC in the physiological range (Figure SM2). The detailed thermodynamic characterization of SaSTC shows that the reduction potentials of the hemes are negative, which are expected for bis-histidinyl-ligated heme groups with substantial exposure to the solvent (Dolla et al., 1994). The positive values for the interaction between pairs of hemes, indicates negative homotropic cooperativity with the oxidation of a particular heme rendering the oxidation of its neighbors more difficult. The interaction energies between the hemes and the ionizable center are negative, a sign of positive heterotropic cooperativity, whereby the oxidation of the hemes facilitates the deprotonation of the ionizable center, and vice-versa.
The relative order of oxidation of the hemes from SaSTC differs from SoSTC and SfSTC (Table 3). However, it has in common the fact that heme III has always the highest potential, and therefore it is always the last heme to be oxidized (Pessanha et al., 2003; Fonseca et al., 2009). Heme III is also the heme with the strongest redox-Bohr interaction which has a value similar to those reported for protonation of heme propionates (Pessanha et al., 2003; Fonseca et al., 2009). The order of oxidation for the other three hemes is different for the three proteins and this must arise from changes in the aminoacid composition or the structure.
Structural Determination and Analysis of SaSTC
The crystal structure of STC was determined to 1.04 Å resolution and deposited in PDB with accession number 6HR0 (Tables SM4, SM5). The electron density is well defined from residue 1 to 90, which represents the mature protein after removal of the signal sequence, containing the four hemes, a phosphate ion and 171 water molecules.
The structure is very similar to the previously determined crystal structure of SoSTC (Leys et al., 2002) and the NMR structure of SfSTC (Paixão et al., 2008), showing an oval shape consisting of three α-helices and five helical turns which are connected by extended loops. Embedded in the structure are the four hemes, which are well exposed to the surroundings and arranged in a linear manner with heme I and II, and heme III and IV oriented perpendicular toward each other. Heme II and III are in parallel planes (Figure 3). The length of the α-helices and the turns are highly conserved between the two crystallized STCs, however with some differences in the loop region in the C-terminal part from the conserved His62 until Ser85. SoSTC possesses no secondary structural elements in this loop, while SaSTC possesses an α-helical turn from Cys75 until Ser77. Also, SaSTC possesses a longer α-helix in the C-terminus despite being one amino acid shorter than SoSTC. This results in conformational changes between the C-terminal residues thus the packing of the amino acids around heme IV.
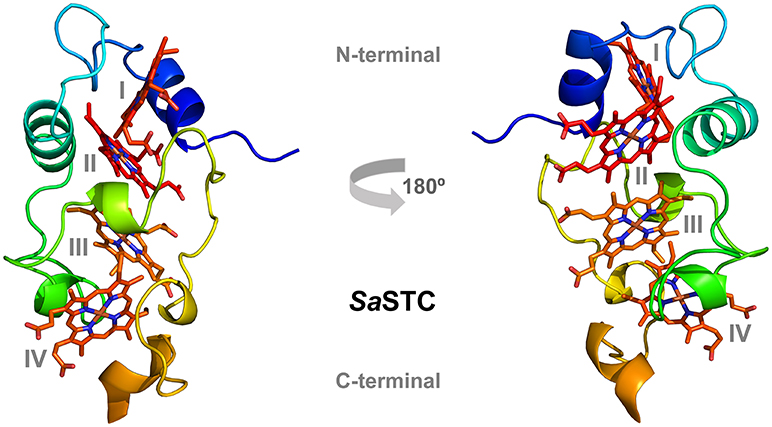
Figure 3. Three-dimensional structure of SaSTC (PDB ID: 6HR0). The hemes are represented in red and orange, with heme I and II in the N-terminal part of the structure and heme III and IV in the C-terminal part.
Decolorization of Methyl Orange
Decolorization of MO provides a convenient signal at 465 nm to monitor the ability of Shewanella to perform extracellular electron transfer (Cai et al., 2012; Cao et al., 2013). This is a consequence of the fact that MO does not cross the outer membrane and therefore mimetizes the behavior of any extracellular electron acceptor of similar or more positive potential that interacts with the extracellular electron transfer pathway of Shewanella (Cai et al., 2012). The disappearance of the peak at 465 nm reflects the reductive cleavage of the conjugated azo-bond in the MO (Cai et al., 2012; Liu et al., 2013), and all the Shewanella strains used in this work have the ability to decolorize MO (Figure 4).
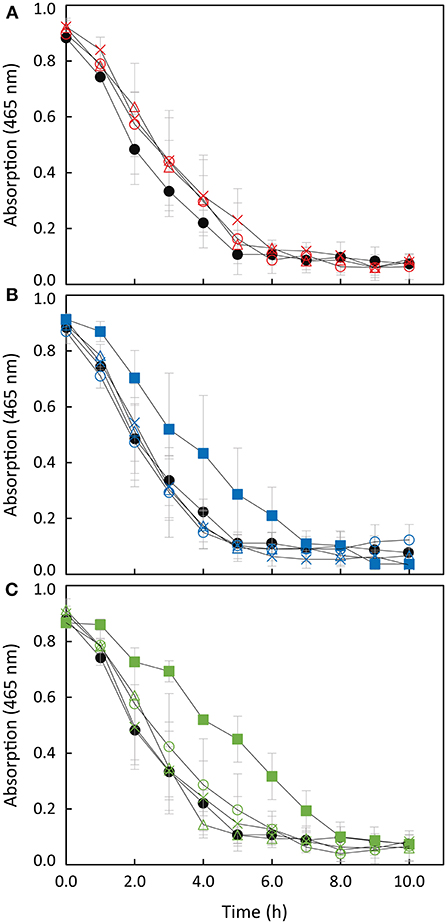
Figure 4. Anaerobic reduction of MO by S. oneidensis wild-type (filled circles) and (A) strains overexpressing SoSTC (open triangles), SfSTC (open circles) and SaSTC (crosses); (B) S. oneidensis ΔSTC (filled squares) and complemented strains [ΔSTC with SoSTC (open triangles), SfSTC (open circles) and SaSTC (crosses)]; (C) S. oneidensis ΔSTCΔFccA (filled squares) and complemented strains [ΔSTCΔFccA with SoSTC (open triangles) and SfSTC (open circles) and SaSTC (crosses)]. The error bars represent standard deviations (SD) of the measurements.
The ΔSTC Shewanella strain decolorizes MO at a significant lower rate than wild-type, while the decolorization by the double knock-out Shewanella strain ΔSTC ΔFccA is even lower (see Figure SM3). This is in agreement with the fact that both STC and FccA act as the main electron transfer shuttles in the periplasmic space of this electroactive organism, transferring electrons from SoCymA to SoMtrA (Fonseca et al., 2013) (see Figure 1). The residual decolorization activity found in the double mutant can derive from the fact that a fraction of MtrA is known to exist free in the periplasm and this fraction can receive electrons from CymA (Schuetz et al., 2009).
When STC is expressed using a complementing plasmid, the ability to decolorize MO is restored to the same rate as the wild-type, indicating the importance of STC in the extracellular electron transfer pathway of Shewanella. Interestingly, strains complemented with SfSTC and SaSTC decolorize MO at similar rate to that of the strain complemented with the native SoSTC. This indicates that SfSTC and SaSTC can also interact and exchange electrons efficiently with SoCymA and SoMtrA. It is known from the literature that CymA, in contrast with other members of the NapC/NirT family of proteins, is promiscuous with respect to redox partners (Schwalb et al., 2003).
Binding Experiments of SfSTC and SaSTC With CymA and MtrA
NMR spectroscopy is exquisitely sensitive to changes in the chemical environment, which makes it the ideal technique to study protein-protein interactions (Ubbink, 2009). For electron transfer to occur between cytochromes, the hemes in both proteins must be in close proximity. Therefore, when changes in the chemical shifts of the signals from the hemes of one cytochrome occur in the presence of another, this indicates an interaction between the proteins that likely leads to electron transfer (Fonseca et al., 2013). This type of experiment was performed for SfSTC and SaSTC with MtrA and CymA from S. oneidensis individually. These experiments indicated that an interaction occurs between SaSTC and both SoMtrA and SoCymA and also between SfSTC and both SoMtrA and SoCymA (Figure SM4). Fitting of a 1:1 binding model to the data (Fonseca et al., 2013) allowed the determination of the dissociation constants of the various STCs with SoMtrA, and with SoCymA (Table 4). In all three cytochromes heme IV is always the heme that is most affected, and therefore the one near the binding region.
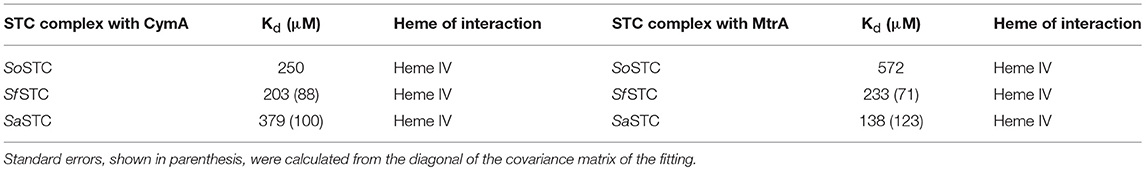
Table 4. Pairwise interactions tested for SoSTC (Fonseca et al., 2013), SaSTC and SfSTC (this work) with SoMtrA and SoCymA.
While for CymA the binding constants obtained for the different STCs are highly similar to the ones obtained with SoSTC, the Kd values obtained for MtrA binding are quite distinct. The binding of SfSTC and SaSTC to MtrA is stronger than of SoSTC.
Discussion
The elucidation of the molecular basis of the electron transfer processes performed by multiheme c-type cytochromes is crucial for the efficient rational manipulation of electroactive organisms toward specific properties and toward their practical implementation in MET (Fonseca et al., 2009; Paquete et al., 2014; Alves et al., 2017; Neto et al., 2017). As one of the most abundant protein in the periplasmic space of Shewanella, STC plays an important role in extracellular electron transfer (Gordon et al., 2000; Sturm et al., 2015), being crucial together with FccA for the electron transfer across the periplasm (Fonseca et al., 2013; Alves et al., 2015; Sturm et al., 2015). This is clearly observed in the decolorization assay with MO, where the double knock-out mutant of these two proteins presents half of the decolorization rate when compared with the wild-type strain. Moreover, the expression of STC from a plasmid restores the MO decolorization rate observed in the wild-type organism, showing the importance of this protein in the extracellular electron transfer pathway of Shewanella.
Interestingly, all the STCs tested in this work restore the MO decolorization rate to that of the wild-type organism. However, despite their high sequence identity (Figure 5), the three cytochromes are different in several aspects, including their redox properties (Table 3), their surface shape and electrostatics (Figure 6).
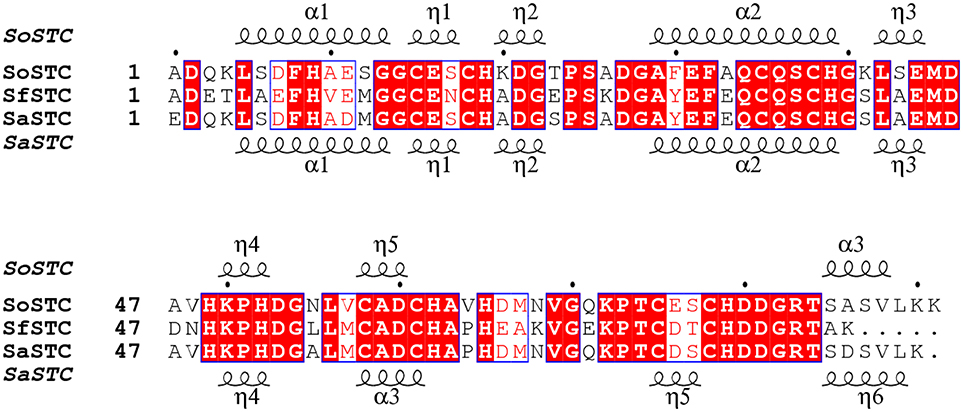
Figure 5. Aminoacid sequence comparison of STC from S. oneidensis, S. frigidimarina, and S. algae. The α and η symbols represent long and short α-helices regions in the three-dimensional structure, respectively.
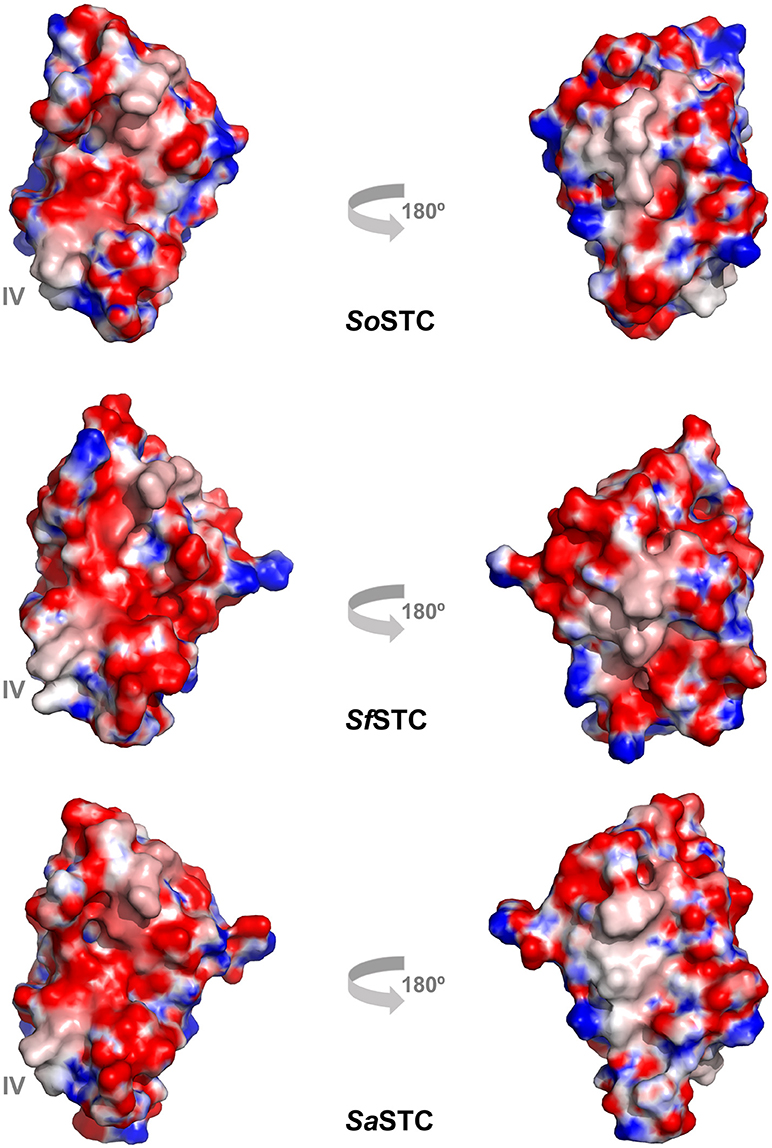
Figure 6. Electrostatic surface potential (−10 to +10 kT/e) of SoSTC (PDB ID 1M1P), SfSTC (PDB ID 2K3V) and of SaSTC (PDB ID 6HR0). The surfaces are shown in the same orientation as the ribbon representation of SaSTC in Figure 4, and were calculated using the APBS plugin in Pymol (Baker et al., 2001).
The sequence identity between the three STC is very high, with SoSTC possessing 64.8 and 83.5% identity to SfSTC and SaSTC, respectively, providing an explanation for the similarity between the structures that are observed. This is further well reflected by a visualization of the electrostatic surface potential of the three proteins, which shows a similar relative negative charge distribution on the side where heme I and IV are exposed, and presence of some hydrophobic patches on the side exposing heme II and III (Figure 6).
However, it is possible to observe small differences around heme IV, the heme that interacts with both redox partners MtrA and CymA. A closer look into this region it is possible to observe that SaSTC is slightly more positively charged than SoSTC, while SfSTC is more hydrophobic. Moreover, there are some conformational differences in the C-terminal loop region, which may alter the packing of the amino acids surrounding heme IV, and thus the exposure of the heme to the surface. These structural differences may justify the different dissociation constants observed for the different STC orthologs and SoMtrA (Table 4), allowing us to suggest that the interacting site in MtrA is a negatively charged region. This would explain the lower dissociation constant obtained for SaSTC that is more positively charged around heme IV and therefore more attractive to the negative region of MtrA, and the higher dissociation constant of SoSTC that contains a more negatively charged region around this heme thus being less attractive to MtrA.
The lower dissociation constant observed between SaSTC and SoMtrA indicates that the equilibrium is shifted toward a greater residence time of the bound form between these two proteins (Corzo, 2006). If we consider that the binding of the STC and MtrA within the periplasmic space of Shewanella is diffusion limited, the mean life-time of the SaSTC complex with SoMtrA is in the order of 7 microseconds, while for the native SoSTC-SoMtrA complex is ~2 μs (Corzo, 2006). This however is not reflected in the decolorization assay, where the three proteins decolorize MO at the same rate. This can be a consequence of the already fast association/dissociation process that occurs between STC and MtrA. Therefore any difference in affinity in the μM range would not be sufficient to reflect a difference in overall electron transfer rate. This suggest, that, at least in the periplasmic space, mutations that affect binding affinities in this order of magnitude in vitro are not sufficient to modify the electron transfer rates of the overall process in vivo.
Besides complex formation, biological electron transfer also depends on the driving force of the reaction (Marcus, 1982). The thermodynamic properties obtained for the three STC orthologous show that the order of oxidation of the hemes are different. Interestingly, in all the proteins, heme III is the first heme to be reduced (Table 3). This can be explained by the fact that heme III is the less exposed heme, presenting a higher reduction potential in all three proteins. With different order of oxidation of the hemes for the various STCs, the way their electrostatic properties change from the fully reduced to the fully oxidized state is different, and do not match the optimized sequence of electron egress from the hemes of the native SoSTC. However, this distinct mode of action of the different proteins is not reflected into changes in the overall electron transfer properties, since the three proteins lead to similar rates of MO decolorization by S. oneidensis. This difference may be however relevant for the distinct conditions in which their organisms of origin live, since slight differences in their biochemical properties may provide advantages when colonizing different environments.
The stronger interaction of SfSTC and SaSTC with SoMtrA in comparison with that of SoSTC indicates that MtrA has a higher discrimination capacity of its redox partners than CymA, where the dissociation constants were very similar with all STC orthologs tested. This similar behavior suggests that electrostatics do not play a significant role in the recognition process of CymA. This can derived from the fact that CymA evolved to interact with several proteins in the periplasmic space (Schwalb et al., 2003), and therefore the recognition site is likely flexible to accommodate the different redox partners. The lack of the three-dimensional structure of this protein prevent us to elucidate how CymA recognizes and binds to its redox partners, including STC.
The results obtained in this work show that the evolutionary pressure in these organisms tuned the properties of these proteins to efficiently transfer electrons between CymA and MtrA in the different environmental conditions, and not in the conditions relevant for the operation of MET. This may be a consequence of the fact that electroactive organisms in their natural environment, often encounter limitations in nutrients, carbon and electron source, and even in electron acceptors, which limits their biomass production and lower their metabolic rate. In contrast, in MET there is an excess of nutrients, electron donor and acceptor, and the systems are always working at full speed.
This work has demonstrated that natural selection has operated in these proteins in a way that it preserves the aminoacids that are crucial to maintain the three-dimensional structure and preserve functionality even while modifying those aminoacids that alter their biochemical properties, and are advantageous when colonizing different environments. This guarantees that the proteins are still able to interact with each other and transfer electrons, preventing metabolic arrest. It also shows that we still have ample room to modify these proteins and optimize them toward enhanced electron transfer properties that are adequate to advance the practical implementation of MET.
Conclusion
The decolorization assays together with the detailed structural and functional properties of STC demonstrated that there is still ample room to modify the extracellular electron transfer chain of Shewanella without disturbing its overall performance. Given the complementary roles of STC and FccA in the periplasmic space of Shewanella, we expect that mutations in FccA will reveal the same plasticity. These results are of utmost importance for the ultimate goal of enhancing EET for biotechnological applications since it suggests that mutations designed to enhance the electron transfer across the microbe-electrode interface are unlikely to adversely affect the electron-transfer performance of the remainder trans-periplasmatic redox chain.
Author Contributions
BF and CP constructed the plasmids and performed the decolorization assays. BF, CP, RL, and LS were responsible for the thermodynamic characterization of SaSTC. The crystallization and structure determination of SaSTC were performed by IT, EM, and PM. The work was planned and organized by CP and RL. The results obtained were discussed with all authors. The paper was written by CP, BF, EM, and RL.
Funding
This work was supported by Fundação para a Ciência e a Tecnologia (FCT) Portugal [SFRH/BPD/96952/2013 to CP, SFRH/BPD/93164/2013 to BF, SFRH/BPD/93164/2013 to EM, PD/BD/135187/2017 to IT, PTDC/BBB-BQB/4178/2014, PTDC/BBB-BQB/3554/2014 and ERA-MBT/0003/2014 Grants], by Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER funds through COMPETE2020—Programa Operacional Competitividade e Internacionalização (POCI), by ITQB research unit GREEN-it Bioresources for sustainability (UID/Multi/04551/2013), and by the European Union's Horizon 2020 research and innovation programme under grant agreement No 810856.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The NMR spectrometers at CERMAX are integrated in the National NMR Network (PTNMR) and are partially supported by Infrastructure Project No 022161 (co-financed by FEDER through COMPETE 2020, POCI and PORL and FCT through PIDDAC). Beamtime at I04 at Diamond Light Source (DLS) and assistance from the beamline staff during the synchrotron data collections are gratefully acknowledged (http://www.diamond.ac.uk/Beamlines/Mx/I04.html).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2019.00002/full#supplementary-material
References
Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., et al. (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367. doi: 10.1107/S0907444912001308
Alves, A. S., Costa, N. L., Tien, M., Louro, R. O., and Paquete, C. M. (2017). Modulation of the reactivity of multiheme cytochromes by site-directed mutagenesis: moving towards the optimization of microbial electrochemical technologies. J. Biol. Inorg. Chem. 22, 87–97. doi: 10.1007/s00775-016-1409-0
Alves, M. N., Neto, S. E., Alves, A. S., Fonseca, B. M., Carrêlo, A., Pacheco, I., et al. (2015). Characterization of the periplasmic redox network that sustains the versatile anaerobic metabolism of Shewanella oneidensis MR-1. Front. Microbiol. 6, 1–10. doi: 10.3389/fmicb.2015.00665
Arends, J. B., and Verstraete, W. (2012). 100 years of microbial electricity production: three concepts for the future. Microb. Biotechnol. 5, 333–346. doi: 10.1111/j.1751-7915.2011.00302.x
Bajracharya, S., Sharma, M., Mohanakrishna, G., Dominguez Benneton, X., Strik, D. P. B. T. B., Sarma, P. M., et al. (2016). An overview on emerging bioelectrochemical systems (BESs): Technology for sustainable electricity, waste remediation, resource recovery, chemical production and beyond. Renew. Energy 98, 153–170. doi: 10.1016/j.renene.2016.03.002
Baker, N. A., Sept, D., Joseph, S., Holst, M. J., and McCammon, J. A. (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98, 10037–10041. doi: 10.1073/pnas.181342398
Bowman, J. P., McCammon, S. A., Brown, M. V., Nichols, D. S., and McMeekin, T. A. (1997). Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63, 3068–3078.
Brünger, A. T. (1992). Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475. doi: 10.1038/355472a0
Cai, P.-J., Xiao, X., He, Y.-R., Li, W.-W., Chu, J., Wu, C., et al. (2012). Anaerobic biodecolorization mechanism of methyl orange by Shewanella oneidensis MR-1. Appl. Microbiol. Biotechnol. 93, 1769–1776. doi: 10.1007/s00253-011-3508-8
Cao, D. M., Xiao, X., Wu, Y. M. Y. Y., Ma, X. B., Wang, M. N., Wu, Y. M. Y. Y., et al. (2013). Role of electricity production in the anaerobic decolorization of dye mixture by exoelectrogenic bacterium Shewanella oneidensis MR-1. Bioresour. Technol. 136, 176–181. doi: 10.1016/j.biortech.2013.02.083
Chen, V. B., Arendall, W. B. III., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., et al. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21. doi: 10.1107/S0907444909042073
Corzo, J. (2006). Time, the forgotten dimension of ligand binding teaching. Biochem. Mol. Biol. Educ. 34, 413–416. doi: 10.1002/bmb.2006.494034062678
Díaz-Moreno, I., Díaz-Quintana, A., Molina-Heredia, F. P., Nieto, P. M., Hansson, Ö., De la Rosa, M. A., et al. (2005). NMR analysis of the transient complex between membrane photosystem I and soluble cytochrome c 6. J. Biol. Chem. 280, 7925–7931. doi: 10.1074/jbc.M412422200
Dolla, A., Blanchard, L., Guerlesquin, F., and Bruschi, M. (1994). The protein moiety modulates the redox potential in cytochromes c. Biochimie 76, 471–479. doi: 10.1016/0300-9084(94)90171-6
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. doi: 10.1107/S0907444910007493
Fonseca, B. M., Paquete, C. M., Neto, S. E., Pacheco, I., Soares, C. M., and Louro, R. O. (2013). Mind the gap: cytochrome interactions reveal electron pathways across the periplasm of Shewanella oneidensis MR-1. Biochem. J. 449, 101–108. doi: 10.1042/BJ20121467
Fonseca, B. M., Saraiva, I. H., Paquete, C. M., Soares, C. M., Pacheco, I., Salgueiro, C. A., et al. (2009). The tetraheme cytochrome from Shewanella oneidensis MR-1 shows thermodynamic bias for functional specificity of the hemes. J. Biol. Inorg. Chem. 14, 375–385. doi: 10.1007/s00775-008-0455-7
Fourmond, V., Hoke, K., Heering, H. A., Baffert, C., Leroux, F., Bertrand, P., et al. (2009). SOAS: a free program to analyze electrochemical data and other one-dimensional signals. Bioelectrochemistry 76, 141–147. doi: 10.1016/j.bioelechem.2009.02.010
Gnutt, D., and Ebbinghaus, S. (2016). The macromolecular crowding effect – from in vitro into the cell. Biol. Chem. 397, 37–44. doi: 10.1515/hsz-2015-0161
Gordon, E. H. J., Pike, A. D., Hill, A. E., Cuthbertson, P. M., Chapman, S. K., and Reid, G. A. (2000). Identification and characterization of a novel cytochrome c(3) from Shewanella frigidimarina that is involved in Fe(III) respiration. Biochem J. 349(Pt 1), 153–158. doi: 10.1042/0264-6021:3490153
Janda, J. M., and Abbott, S. L. (2014). The genus Shewanella : from the briny depths below to human pathogen. Crit. Rev. Microbiol. 40, 293–312. doi: 10.3109/1040841X.2012.726209
Kabsch, W. (2010). XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132. doi: 10.1107/S0907444909047337
Koch, C., and Harnisch, F. (2016). Is there a specific ecological niche for electroactive microorganisms? ChemElectroChem 3, 1282–1295. doi: 10.1002/celc.201600079
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kuznetsova, I. M., Turoverov, K. K., and Uversky, V. N. (2014). What macromolecular crowding can do to a protein. Int. J. Mol. Sci. 15, 23090–23140. doi: 10.3390/ijms151223090
Lagarias, J. C., Reeds, J. A., Wright, M. H., and Wright, P. E. (1998). Convergence properties of the nelder–mead simplex method in low dimensions. SIAM J. Optim. 9, 112–147. doi: 10.1137/S1052623496303470
Leys, D., Meyer, T. E., Tsapin, A. S., Nealson, K. H., Cusanovich, M. A., and Van Beeumen, J. J. (2002). Crystal structures at atomic resolution reveal the novel concept of “electron-harvesting” as a role for the small tetraheme cytochrome c. J. Biol. Chem. 277, 35703–35711. doi: 10.1074/jbc.M203866200
Liu, G., Zhou, J., Meng, X., Fu, S. Q., Wang, J., Jin, R., et al. (2013). Decolorization of azo dyes by marine Shewanella strains under saline conditions. Appl. Microbiol. Biotechnol. 97, 4187–4197. doi: 10.1007/s00253-012-4216-8
Logan, B. E., and Rabaey, K. (2012). Conversion of wastes into bioelectricity and chemicals by using microbial electrochemical technologies. Science 337, 686–690. doi: 10.1126/science.1217412
Logan, B. E., and Regan, J. M. (2006). Electricity-producing bacterial communities in microbial fuel cells. Trends Microbiol. 14, 512–518. doi: 10.1016/j.tim.2006.10.003
Louro, R. O. (2006). Proton thrusters: overview of the structural and functional features of soluble tetrahaem cytochromes c3. JBIC J. Biol. Inorg. Chem. 12, 1–10. doi: 10.1007/s00775-006-0165-y
Marcus, R. A. (1982). Electron transfer reactions. J. Mol. Struct. 81:296. doi: 10.1016/0022-2860(82)85348-9
Marsili, E., Rollefson, J. B., Baron, D. B., Hozalski, R. M., and Bond, D. R. (2008). Microbial biofilm voltammetry: direct electrochemical characterization of catalytic electrode-attached biofilms. Appl. Environ. Microbiol. 74, 7329–7337. doi: 10.1128/AEM.00177-08
McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007). Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. doi: 10.1107/S0021889807021206
Meyer, T. E., Tsapin, A. I., Vandenberghe, I., De Smet, L., Frishman, D., Nealson, K. H., et al. (2004). Identification of 42 possible cytochrome c genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. Omi. A J. Integr. Biol. 8, 57–77. doi: 10.1089/153623104773547499
Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255. doi: 10.1107/S0907444996012255
Myers, C. R., and Nealson, K. H. (1988). Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240, 1319–1321. doi: 10.1126/science.240.4857.1319
Neto, S. E., de Melo-Diogo, D., Correia, I. J., Paquete, C. M., and Louro, R. O. (2017). Characterization of OmcA mutants from Shewanella oneidensis MR-1 to investigate the molecular mechanisms underpinning electron transfer across the microbe-electrode interface. Fuel Cells 17, 1–11. doi: 10.1002/fuce.201700023
Paixão, V. B., Salgueiro, C. A., Brennan, L., Reid, G. A., Chapman, S. K., and Turner, D. L. (2008). The solution structure of a tetraheme cytochrome from Shewanella frigidimarina reveals a novel family structural motif. Biochemistry 47, 11973–11980. doi: 10.1021/bi801326j
Paquete, C. M., Fonseca, B. M., Cruz, D. R., Pereira, T. M., Pacheco, I., Soares, C. M., et al. (2014). Exploring the molecular mechanisms of electron shuttling across the microbe/metal space. Front. Microbiol. 5:318. doi: 10.3389/fmicb.2014.00318
Paquete, C. M., and Louro, R. O. (2014). Unveiling the details of electron transfer in multicenter redox proteins. Acc. Chem. Res. 47, 56–65. doi: 10.1021/ar4000696
Paquete, C. M., Saraiva, I. H., Calçada, E., and Louro, R. O. (2010). Molecular basis for directional electron transfer. J. Biol. Chem. 285, 10370–10375. doi: 10.1074/jbc.M109.078337
Pessanha, M., Brennan, L., Xavier, A. V., Cuthbertson, P. M., Reid, G. A., Chapman, S. K., et al. (2001). NMR structure of the haem core of a novel tetrahaem cytochrome isolated from Shewanella frigidimarina: Identification of the haem-specific axial ligands and order of oxidation. FEBS Lett. 489, 8–13. doi: 10.1016/S0014-5793(00)02383-8
Pessanha, M., Louro, R. O., Correia, I. J. I. J., Rothery, E. L., Pankhurst, K. L. K. L., Reid, G. A., et al. (2003). Thermodynamic characterization of a tetrahaem cytochrome isolated from a facultative aerobic bacterium, Shewanella frigidimarina : a putative redox model for flavocytochrome c 3. Biochem. J. 495, 489–495. doi: 10.1042/bj20021408
Ramachandran, G. N., and Sasisekharan, V. (1968). Conformation of polypeptides and proteins. Adv. Protein Chem. 23, 283–437. doi: 10.1016/S0065-3233(08)60402-7
Richardson, D. J., Fredrickson, J. K., and Zachara, J. M. (2012). Electron transport at the microbe-mineral interface: a synthesis of current research challenges. Biochem. Soc. Trans. 40, 1163–1166. doi: 10.1042/BST20120242
Salgueiro, C. A., Turner, D. L., Santos, H., LeGall, J., and Xavier, A. V. (1992). Assignment of the redox potentials to the four haems in Desulfovibrio vulgaris cytochrome c3 by 2D-NMR. FEBS Lett. 314, 155–158. doi: 10.1016/0014-5793(92)80963-H
Schuetz, B., Schicklberger, M., Kuermann, J., Spormann, A. M., Gescher, J., Kuermann, J., et al. (2009). Periplasmic electron transfer via the c-type cytochromes Mtra and Fcca of Shewanella oneidensis Mr-1. Appl. Environ. Microbiol. 75, 7789–7796. doi: 10.1128/AEM.01834-09
Schwalb, C., Chapman, S. K., and Reid, G. A. (2003). The tetraheme cytochrome cyma is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis. Biochemistry 42, 9491–9497. doi: 10.1021/bi034456f
Silva, A. V., and Louro, R. O. (2017). Biomolecular NMR assignment: illustration using the heme signals in horse cytochrome c. J. Chem. Educ. 94, 1280–1284. doi: 10.1021/acs.jchemed.7b00123
Strycharz-Glaven, S. M., Snider, R. M., Guiseppi-Elie, A., and Tender, L. M. (2011). On the electrical conductivity of microbial nanowires and biofilms. Energy Environ. Sci. 4:4366. doi: 10.1039/c1ee01753e
Sturm, G., Richter, K., Doetsch, A., Heide, H., Louro, R. O., and Gescher, J. (2015). A dynamic periplasmic electron transfer network enables respiratory flexibility beyond a thermodynamic regulatory regime. ISME J. 9, 1802–1811. doi: 10.1038/ismej.2014.264
Sydow, A., Krieg, T., Mayer, F., Schrader, J., and Holtmann, D. (2014). Electroactive bacteria: molecular mechanisms and genetic tools. Appl. Microbiol. Biotechnol. 98, 8481–8495. doi: 10.1007/s00253-014-6005-z
Teravest, M. A., and Ajo-Franklin, C. M. (2015). Transforming exoelectrogens for biotechnology using synthetic biology. Biotechnol. Bioeng. 113, 687–697. doi: 10.1002/bit.25723
Torres, C. I. (2014). On the importance of identifying, characterizing, and predicting fundamental phenomena towards microbial electrochemistry applications. Curr. Opin. Biotechnol. 27, 107–114. doi: 10.1016/j.copbio.2013.12.008
Torres, C. I., Marcus, A. K., Lee, H. S., Parameswaran, P., Krajmalnik-Brown, R., and Rittmann, B. E. (2010). A kinetic perspective on extracellular electron transfer by anode-respiring bacteria. FEMS Microbiol. Rev. 34, 3–17. doi: 10.1111/j.1574-6976.2009.00191.x
Tsapin, A. I., Vandenberghe, I., Nealson, K. H., Scott, J. H., Meyer, T. E., Cusanovich, M. A., et al. (2001). Identification of a small tetraheme cytochrome c and a flavocytochrome c as two of the principal soluble cytochromes c in Shewanella oneidensis strain MR1. Appl. Environ. Microbiol. 67, 3236–3244. doi: 10.1128/AEM.67.7.3236-3244.2001
Ubbink, M. (2009). The courtship of proteins: understanding the encounter complex. FEBS Lett. 583, 1060–1066. doi: 10.1016/j.febslet.2009.02.046
Ucar, D., Zhang, Y., and Angelidaki, I. (2017). An overview of electron acceptors in microbial fuel cells. Front. Microbiol. 8:643. doi: 10.3389/fmicb.2017.00643
Velankar, S., Best, C., and Beuth, B. (2010). PDBe: protein data bank in Europe. Nucleic Acids Res. 38, D308–D317. doi: 10.1093/nar/gkp916
Vignier, N., Théodose, R., Barreau, M., Baubion, E., Olive, C., Cabié, A., et al. (2013). Human infection with Shewanella putrefaciens and S. algae: report of 16 cases in martinique and review of the literature. Am. J. Trop. Med. Hyg. 89, 151–156. doi: 10.4269/ajtmh.13-0055
Vonrhein, C., Flensburg, C., Keller, P., Sharff, A., Smart, O., Paciorek, W., et al. (2011). Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 67, 293–302. doi: 10.1107/S0907444911007773
White, G. F., Edwards, M. J., Gomez-Perez, L., Richardson, D. J., Butt, J. N., and Clarke, T. A. (2016). Mechanisms of bacterial extracellular electron exchange. Adv. Microb. Physiol. 68, 87–138. doi: 10.1016/bs.ampbs.2016.02.002
Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242. doi: 10.1107/S0907444910045749
Keywords: extracellular electron transfer, Shewanella, small tetraheme cytochrome, microbial fuel cells, methyl orange, orthologous proteins, microbial electrochemical technologies
Citation: Fonseca BM, Silva L, Trindade IB, Moe E, Matias PM, Louro RO and Paquete CM (2019) Optimizing Electroactive Organisms: The Effect of Orthologous Proteins. Front. Energy Res. 7:2. doi: 10.3389/fenrg.2019.00002
Received: 01 October 2018; Accepted: 11 January 2019;
Published: 29 January 2019.
Edited by:
Adrián Escapa, Universidad de León, SpainReviewed by:
Sarah Glaven, United States Naval Research Laboratory, United StatesG. Velvizhi, Indian Institute of Chemical Technology (CSIR), India
Copyright © 2019 Fonseca, Silva, Trindade, Moe, Matias, Louro and Paquete. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ricardo O. Louro, bG91cm9AaXRxYi51bmwucHQ=
Catarina M. Paquete, Y3BhcXVldGVAaXRxYi51bmwucHQ=
 Bruno M. Fonseca
Bruno M. Fonseca Luís Silva
Luís Silva Inês B. Trindade1
Inês B. Trindade1 Elin Moe
Elin Moe Pedro M. Matias
Pedro M. Matias Ricardo O. Louro
Ricardo O. Louro Catarina M. Paquete
Catarina M. Paquete