
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 17 September 2020
Sec. Neuroendocrine Science
Volume 11 - 2020 | https://doi.org/10.3389/fendo.2020.00640
Neuropeptides are the most abundant and diverse signal molecules in insects. They act as neurohormones and neuromodulators to regulate the physiology and behavior of insects. The majority of neuropeptides initiate downstream signaling pathways through binding to G protein-coupled receptors (GPCRs) on the cell surface. In this study, RNA-seq technology and bioinformatics were used to search for genes encoding neuropeptides and their GPCRs in the cowpea aphid Aphis craccivora. And the expression of these genes at different developmental stages of A. craccivora was analyzed by quantitative real-time PCR (qRT-PCR). A total of 40 candidate genes encoding neuropeptide precursors were identified from the transcriptome data, which is roughly equivalent to the number of neuropeptide genes that have been reported in other insects. On this basis, software analysis combined with homologous prediction estimated that there could be more than 60 mature neuropeptides with biological activity. In addition, 46 neuropeptide GPCRs were obtained, of which 40 belong to rhodopsin-like receptors (A-family GPCRs), including 21 families of neuropeptide receptors and 7 orphan receptors, and 6 belong to secretin-like receptors (B-family GPCRs), including receptors for diuretic hormone 31, diuretic hormone 44 and pigment-dispersing factor (PDF). Compared with holometabolous insects such as Drosophila melanogaster, the coding genes for sulfakinin, corazonin, arginine vasopressin-like peptide (AVLP), and trissin and the corresponding receptors were not found in A. craccivora. It is speculated that A. craccivora likely lacks the above neuropeptide signaling pathways, which is consistent with Acyrthosiphon pisum and that the loss of these pathways may be a common feature of aphids. In addition, expression profiling revealed neuropeptide genes and their GPCR genes that are differentially expressed at different developmental stages and in different wing morphs. This study will help to deepen our understanding of the neuropeptide signaling systems in aphids, thus laying the foundation for the development of new methods for aphid control targeting these signaling systems.
Insect neuropeptides are trace polypeptides produced by neurosecretory glands or neurosecretory cells in insects (1). They are the most abundant and diverse signal molecules and play important regulatory roles in the physiology and behavior in insects such as the growth, development, metabolism, reproduction, and feeding of insects (2). Most of the known neuropeptides are oligopeptides or small protein molecules composed of several to tens of amino acids. Biologically active mature neuropeptides are formed by complex post-translational processing of neuropeptide precursors. Neuropeptides act as signal molecules to exert their physiological functions as neuromodulators or neurohormones by binding to corresponding membrane receptors. Except for a few families of neuropeptides, such as prothoracicotropic hormone (PTTH), eclosion hormone (EH), and neuropeptide-like precursor 1 (NPLP1), the majority of neuropeptide receptors belong to the G protein-coupled receptors (GPCRs) superfamily (3). In recent years, insect GPCRs have been proposed to be used as potential targets to explore new insecticides or behavioral regulators, opening up new ways for the safe control of pests (4) By targeting neuropeptide GPCRs, future insecticides or behavioral regulators can prevent or over-stimulate the normal activity of these proteins, producing a lethal or deleterious effect on pests. Many species of aphids are important agricultural and forestry pests. At the same time, aphids are also model organisms for studying phenotypic plasticity, insect-plant interactions, insect-plant-virus interactions and endosymbionts and thus have attracted much attention from researchers (5).
In recent years, the continuous innovation of next-generation sequencing technology and bioinformatics technology has greatly promoted the prediction and identification of insect neuropeptides and their receptors (6–9). However, only Acyrthosiphon pisum has so far been comprehensively investigated for neuropeptides and their receptors in aphids (10–12). Aphis craccivora is a worldwide insect pest that causes harm to many crops such as cowpea, peanut and pea (13, 14). In addition to directly sucking plant sap, it can also cause fungal diseases and acts as a vector for various plant viruses such as cowpea mosaic virus (15) and peanut stripe virus (16). At present, A. craccivora is mainly controlled by chemical insecticides, resulting in increasingly severe insecticide resistance and ecological problems (17). So it is imperative to develop alternative new control strategy and identification of neuropeptides and their receptors of A. craccivora will be the first step toward achieving this goal.
In the present study, based on transcriptome sequencing and bioinformatics analysis, we screened the genes encoding neuropeptide precursors and their GPCRs in A. craccivora. We also conducted structural analysis and mature peptide prediction for the identified neuropeptide precursors and classified the neuropeptide receptors into subclasses based on phylogenetic analysis. The expression profiles of these neuropeptides and their GPCRs at different developmental stages were determined by quantitative real-time PCR (qRT-PCR). These identified gene sequences provide the basis for further investigation of the function of A. craccivora neuropeptides and their GPCRs.
Aphis craccivora was collected from peanut fields in Laixi, Shandong Province, China, in 2015. Then the collected aphids were reared for many generations with Vicia faba seedlings in our laboratory under the photoperiod of 14 L: 10 D, at 22 ± 1°C. For transcriptome sequencing in the next step, the heads of 1,000 wingless parthenogenetic adults were quickly removed into phosphate-buffered saline (PBS) buffer and immediately frozen with liquid nitrogen. To find as many target genes as possible, we also collected 50–60 whole-body aphids from each instar (including wingless and winged morphs) as a mixed sample to generate the second transcriptome. Total RNA extraction was performed from the above two samples using RNAiso Plus reagent (Takara, Dalian, China) following the manufacturer's instructions.
We used 2 μg total RNA to construct each library with the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (Illumina, San Diego, CA, USA). Paired-end sequencing was performed using an Illumina HiSeqTM 2,500 by Gene Denovo Biotechnology Co. (Guangzhou, China). After the raw sequencing reads were filtered to remove adaptors, low-quality sequences and reads with N content <5%, de novo assembly were performed using Trinity software with the default parameters (18). Finally, the annotation of unigene sequences were performed using a Blastx search against the NCBI non-redundant protein sequence (Nr), A manually annotated and reviewed protein sequence database (Swiss-Prot), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Clusters of Orthologous Groups (COG) databases with a cut-off E-value of 10−5.
tBlastn was utilized to search for unigenes encoding neuropeptides (including protein hormones) and their putative GPCRs from A. craccivora transcriptome using the homologs from A. pisum (10–12), D. melanogaster (19–21), Anopheles gambiae (22, 23), Apis mellifera (24, 25), Bombyx mori (26, 27), Tribolium castaneum (28, 29), Nilaparvata lugens (6), Rhodnius prolixus (30), and Chilo suppressalis (31) as the query sequences. The open reading frames (ORFs) of the candidate unigenes were predicted using the online tool ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and manually corrected based on homology to the published insect neuropeptides and their GPCRs. And then the encoded protein sequences were aligned to the Nr database using NCBI's Blastp program (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
For the neuropeptide precursors, SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) was employed to predict the signal peptides (32). The splice sites were predicted using the Known Motif and Insect Models methods of NeuroPred (http://stagbeetle.animal.uiuc.edu/cgi-bin/neuropred.py) (33) and were corrected based on the processing procedures of known insect neuropeptide precursors. Sulfinator (https://web.expasy.org/sulfinator/) was used to identify the sulfation of tyrosine residues (34). Cyclization of N-terminal glutamine/glutamic acid residue and amidation of the C-terminal glycine residue were predicted based on homology to the reported invertebrate neuropeptides. Multiple alignments of amino acid sequences were performed using Clustal X 2.0 and the results were visualized using GeneDoc software.
For the neuropeptide GPCRs, trans-membrane domains (TMDs) were predicted by TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM.NCBI) CD-Search tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was employed to search the conserved domain database (CDD) for the domain (35, 36).
Two unigene sequences in the transcriptome data were likely partial fragments from the same gene. RT-PCR was used to amplify and obtain longer sequences. Upstream and downstream primers were designed for each of the fragments using Primer Premier 5.0 software (Table S1). The cDNA template was synthesized using a PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China) The reaction procedure was as follows: pre-denaturation at 94°C for 3 min; 37 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and elongation at 72°C for 1 min; and a final elongation at 72°C for 5 min. The amplication products were sent to Invitrogen (Shanghai, China) for sequencing. The sequencing results were compared with the corresponding unigene sequences using Blastn to determine sequence identity.
Neuropeptide GPCRs of A. craccivora were compared with the reported homologs from A. pisum, N. lugens, D. melanogaster, B. mori, and T. castaneum. Amino acid sequences with a length shorter than 150 aa were removed from the data set for phylogenetic analysis. Multiple alignments were carried out using MAFFT (37). The maximum-likelihood (ML) phylogenetic trees were constructed using the FastTree software (version 2.1.7) under the default settings (38). Local support values were calculated based on the Shimodaira-Hasegawa (SH) test implemented within FastTree. Metabotropic glutamate receptor (mGluR) of D. melanogaster (encoded by CG11144) served as the outgroup for the phylogenetic tree of A-family GPCRs, and 5-hydroxytryptamine receptor (5-HTR) of D. melanogaster (encoded by CG1056) served as the outgroup for the phylogenetic tree of B-family GPCRs. The phylogenetic trees were finally edited and displayed using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree).
To determine the transcriptional expression levels of neuropeptides and their GPCRs at different life stages of A. craccivora, 20–60 aphids from each instar were collected separately for qRT-PCR analysis. From the third instar, we were able to distinguish winged from wingless aphids by wing buds of the thorax. For this reason, winged and wingless morphs of the aphids were collected separately from the 3rd, the 4th instar nymphs and adults. After total RNA was extracted as described above, cDNA was synthesized by reverse transcribing 1 μg of total RNA using a PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Dalian, China) according to the manufacturer's instructions. The gene-specific primers for qRT-PCR were designed using the NCBI online primer design tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Table S2). We select 40S ribosomal protein S8 gene (RPS8) as the reference gene to normalize the expression of the target genes (39).
The qRT-PCR reactions were run with SYBR Premix Ex Taq II (Tli RNaseH Plus) (Takara, Dalian, China) on an iQ™5 Multicolor Real-Time PCR Detection System (Bio-Rad Hercules, CA, USA). The qRT-PCR program was set as follows: 95°C for 3 min; 40 cycles of 95°C for 10 s and 58°C for 30 s; and followed by a melting cycle (from 60 to 95°C with an increase of 0.5°C per cycle). The experiment included 3 biological replicates and 3 technical replicates. The relative expression level of the target genes was calculated using the 2ΔΔCt method (40). Statistical analysis was performed using SPSS 20.0 (Systat Software Inc., London, UK). Statistical significance was evaluated using one-way ANOVA followed by the Tukey HSD test at the 0.05 level.
We sequenced the sample from the heads of A. craccivora wingless parthenogenetic adults and 56,634,657 clean reads (8.4 Gbp) were obtained. For the mixed sample, 70,096,246 clean reads (10.3 Gbp) were obtained. After assembly, 60,113 unigenes (N50 = 1,169) were obtained with an average length of 703 bp and a GC content of 38.08%. Among all the unigenes, a total of 45,150 were annotated in the Nr, Swissprot, KOG, and KEGG databases (Figure S1A), and the top 3 species with the most matches in Nr database were A. pisum, Diuraphis noxia, and Diachasma alloeum (Figure S1B). The transcriptome raw data have been submitted to the Sequence Read Archive (SRA) database at NCBI under the accession number SRP189414.
Based on the homology search and Nr-annotation, a total of 40 genes encoding neuropeptide precursors were identified, involving 32 neuropeptide families (Table 1; Supplementary Material 1) and covering most insect neuropeptide families (Table S3). However, a small number of family members were missing, including CNMamide (CNMa), corazonin (Crz), elevenin, neuroparsin, pigment-dispersing factor (PDF), RYamide (RYa), sulfakinin (SK), sex peptide (SP), trissin, arginine vasopressin-like peptide (AVLP), and adipokinetic hormone/corazonin-related peptide (ACP). Of the 40 neuropeptide genes obtained, 36 had intact ORFs, and the remaining four non-full-length sequences include three insulin-like peptide (ILP)-encoding genes and 1 PTTH -encoding gene (Table 1). The majority of A. craccivora neuropeptide precursors showed more than 90% similarity with their homologous sequences in other aphids, whereas a few had low similarity with homologous sequences in related species, such as proctolin (Proc), leucokinin (LK), and diuretic hormone 44 (DH44). Besides, orthologs of some A. pisum neuropeptides were not identified from A. craccivora transcriptome, including ion transport peptides (ITPs) and ILPs. However, the transcript encoding ITP-like was found in the A. craccivora transcriptome. Ten different genes coding for ILPs have been found in A. pisum genome but only six of these genes in A. craccivora, i.e., genes encoding ILP1, ILP4, ILP5, ILP7, ILP8, and ILP10. By contrast, the gene coding for NPLP3 possesses orthologs in A. craccivora but not in A. pisum.
During the process of neuropeptide production, a larger inactive neuropeptide precursor is first synthesized. One or several bioactive mature peptides are then formed through a series of enzymatic digestions and post-translational modifications, which are critical to ensure the biological activity and stability of mature neuropeptides (41). The signal peptide cleavage site, sulfation site, amidation site, and other modification sites of A. craccivora neuropeptide precursors were predicted, and the prediction results were revised based on the homology and conserved motif of the known invertebrate neuropeptides. A total of 164 mature peptides (including protein hormones) were predicted from 40 neuropeptide precursors in A. craccivora. Among them, more than 60 mature peptides could be biologically active. Meanwhile, a total of 52 mature peptides were predicted to be amidated at the C-terminus, and 11 sites of 6 mature peptides were predicted as potential sites of sulfation (Figure 1; Supplementary Material 2).
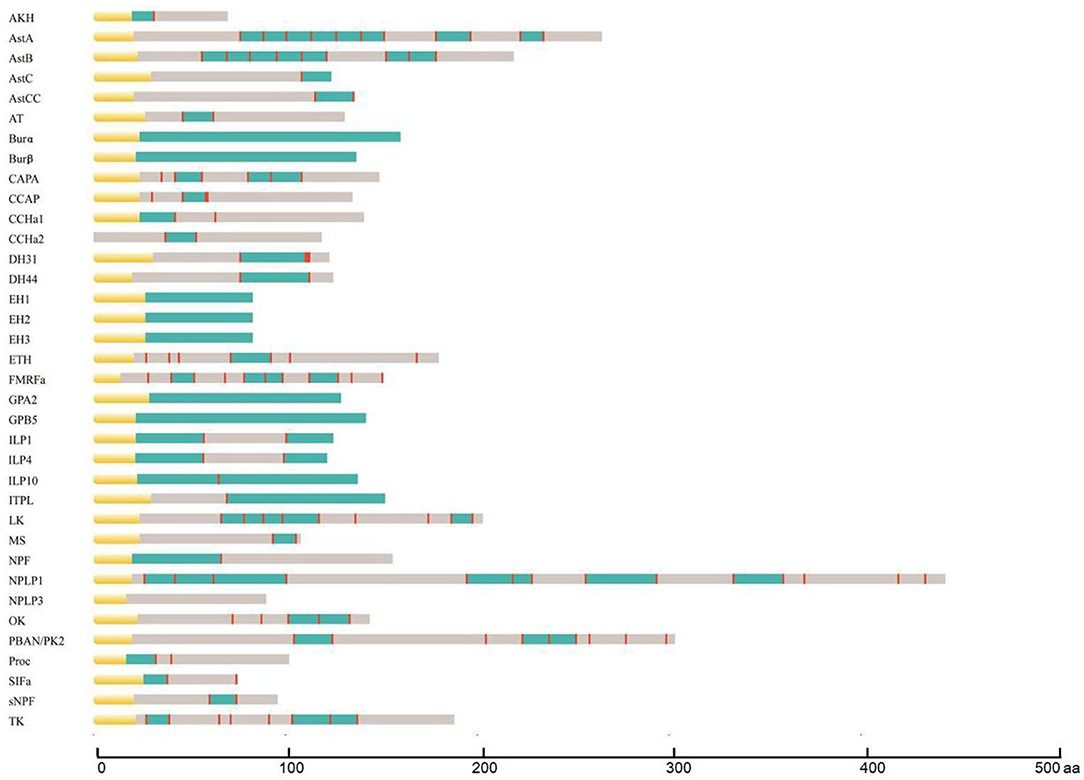
Figure 1. Schematic diagrams showing the organization of A. craccivora neuropeptide precursors. Yellow, signal peptide; blue, bioactive peptides; red, cleavage sites. Scale bars at bottom show length in amino acids (aa). Four incomplete sequences were not included; cleavage sites were not predicted in the NPLP3 precursor.
Most A. craccivora neuropeptides have highly conserved characteristic motifs within the same family. For example, the predicted 8 mature allatostatin A (AstA) peptides contain the C-terminal signature sequence of -Y/FXFGL/Iamide; adipokinetic hormone (AKH) has the sequence motif characterized by pQX6Wamide. Some mature neuropeptide sequences are identical in different aphids, such as short neuropeptide F (sNPF) and SIFamide (SIFa) of A. craccivora, A. pisum, and Myzus persicae. Even the sequences of mature peptides of DH31 and DH44 that are up to tens of amino acids are identical in A. craccivora and A. pisum (Supplementary Material 2).
The mature neuropeptides of A. craccivora also exhibit some structural variability, which is mainly reflected by the expansion and truncation of the polypeptide sequences. The former mainly involves Proc, CCHamide (CCHa), and AstB, and the latter occurs in SIFa. The predicted Proc of A. craccivora is identical to that of M. persicae with an N-terminal extension compared to the pentapeptide proctolin (RYLPT) of most other insects (42); the effect of this expansion on Proc biological activity remains unknown. The N-terminus of A. craccivora CCHa1 has an addition of 4 amino acids (KQGA), and the N-terminus of CCHa2 has an additional Gly. There are two AstB peptides with a sequence motif of -WX7Wamide in A. craccivora, which has an extension of one amino acid residue in the middle of the polypeptide compared to the classical motif. This subtype of AstB has also been found in A. pisum, M. persicae, A. gossypii, N. lugens, and R. prolixus (6, 43). Therefore, it is speculated that -WX7Wamide may be a special AstB subtype that is prevalent in hemipteran insects. The predicted A. craccivora SIFa sequence (FRKPPFNGSIFamide) is consistent with that of A. pisum and M. persicae, but lacks an amino acid residue at the N-terminus compared to the classical motif.
In addition, variability is also reflected in the mutation of amino acid residues. The A. craccivora capability (CAPA) gene encodes a CAPA-pvk (periviscerokinin) peptide and a CAPA-pk (pyrokinin) peptide with the conserved characteristic motif, -PRVamide, and -PRLamide, respectively. This gene also encodes another polypeptide, EGLIAFPRIamide, which is identical to the CAPA-2 peptide of A. pisum and M. persicae (42, 44). This subtype of CAPA with -PRIamide is less common in insects. In addition to aphids, it is found only in a few insects such as T. castaneum, A. mellifera, and Camponotus floridanus (29, 45, 46).
The insect neuropeptide GPCRs belong to rhodopsin-like receptors (A-family GPCRs) or secretin-like receptors (B-family GPCRs). A total of 46 putative neuropeptide GPCRs were identified from the A. craccivora transcriptome data, among which 34 contained complete ORFs (Table 2; Supplementary Material 3). The encoded theoretical proteins had a length of 154–1184 amino acids. Forty of 46 GPCRs belong to the A-family (represented by 7tm_1 or PF0001 in the Pfam database), and six belong to the B-family (represented by 7tm_2 or PF0002 in the Pfam database) (Table 2). Phylogenetic analysis revealed that all A. craccivora neuropeptide GPCRs had unique orthologous proteins in A. pisum (Figures 2–4).
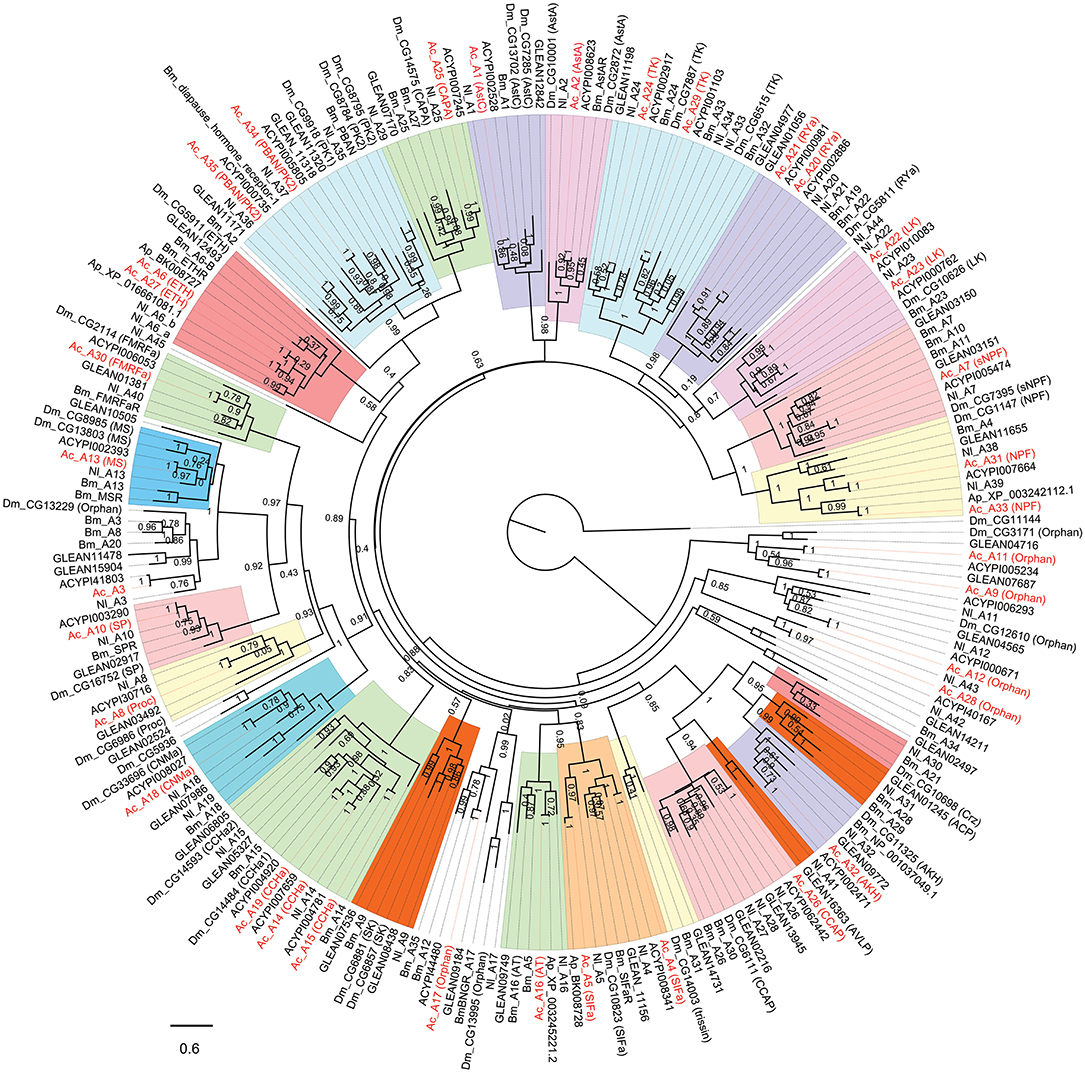
Figure 2. Phylogenetic tree of A-family neuropeptide GPCRs (LGRs are excluded). The maximum-likelihood tree was constructed using FastTree software with local support values calculated based on the Shimodaira-Hasegawa (SH) test. The tree is rooted by the metabotropic glutamate receptor CG11144 from D. melanogaster. Ac, A. craccivora; Dm, D. melanogaster; Ap, A. pisum; Bm, B. mori; Nl, N. lugens; ACYPIxxxxx, sequences from A. pisum; GLEANxxxxx, sequences from T. castaneum.
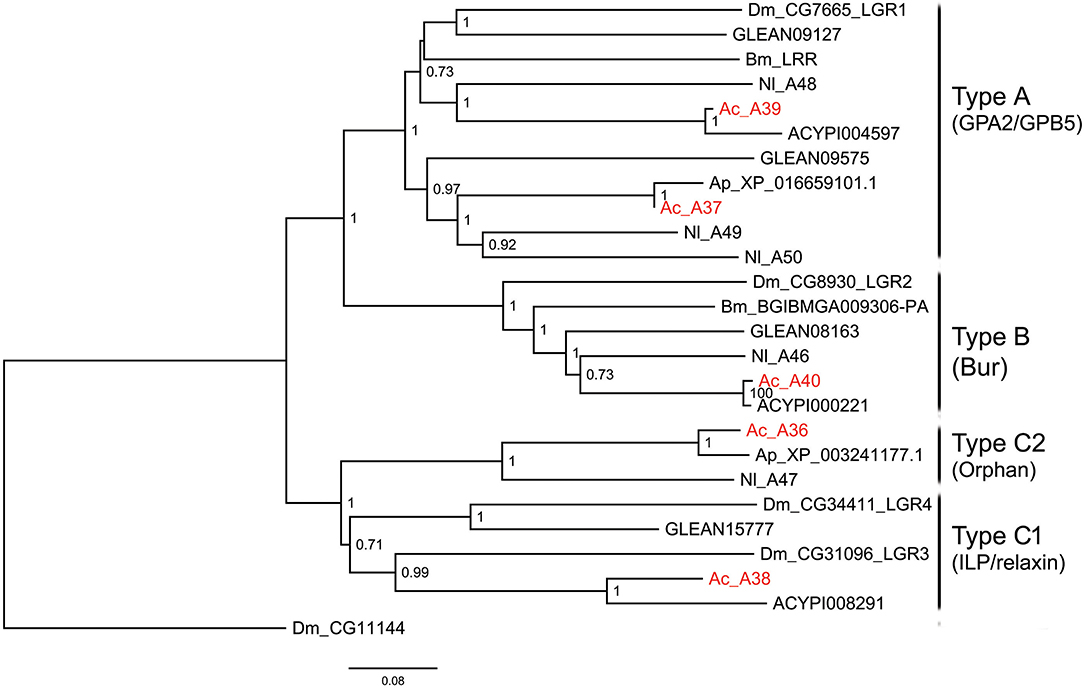
Figure 3. Phylogenetic tree of LGRs. The maximum-likelihood tree was constructed using FastTree software with local support values calculated based on the Shimodaira-Hasegawa (SH) test. The tree is rooted by the serotonin receptor CG1056 from D. melanogaster. Ac, A. craccivora; Dm, D. melanogaster; Ap, A. pisum; Bm, B. mori; Nl, N. lugens; ACYPIxxxxx, sequences from A. pisum; GLEANxxxxx, sequences from T. castaneum.
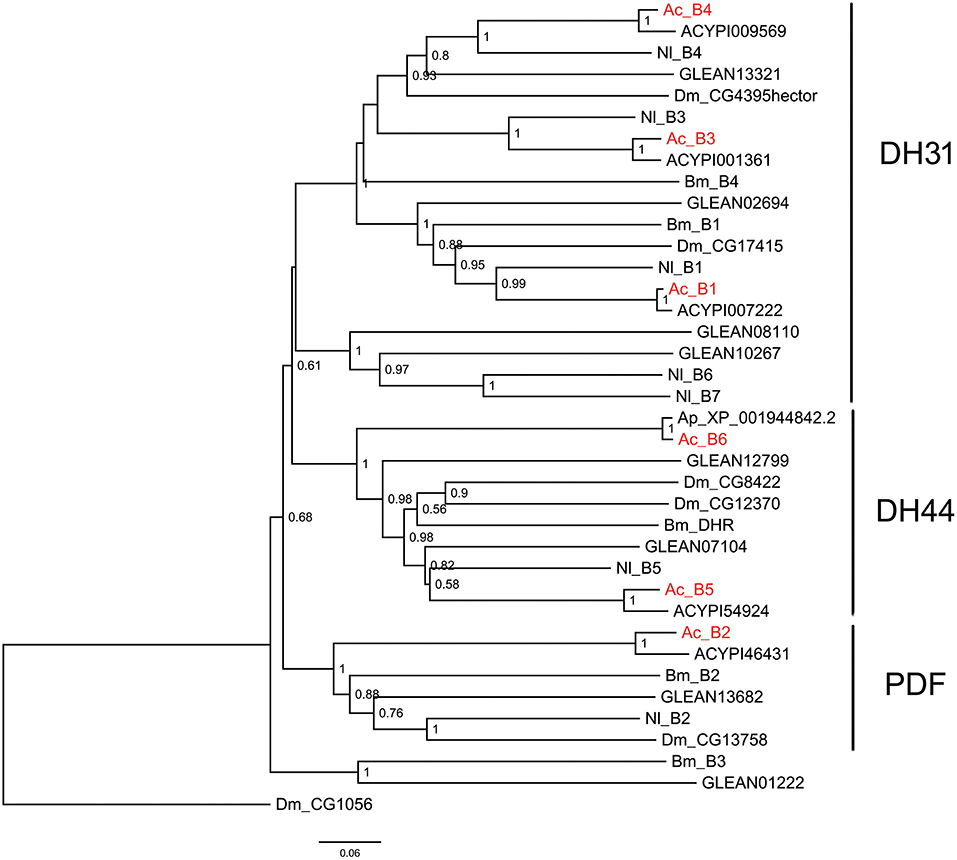
Figure 4. Phylogenetic tree of B-family neuropeptide GPCRs. The maximum-likelihood tree was constructed using FastTree software with local support values calculated based on the Shimodaira-Hasegawa (SH) test. The tree is rooted by the serotonin receptor CG1056 from D. melanogaster. Ac, A. craccivora; Dm, D. melanogaster; Ap, A. pisum; Bm, B. mori; Nl, N. lugens; ACYPIxxxxx, sequences from A. pisum; and GLEANxxxxx, sequences from T. castaneum.
In insects, neuropeptide (including protein hormones) receptors account for more than two-thirds of A-family GPCRs (12). According to the binding ligands, A-family neuropeptide GPCRs of A. craccivora were divided into 21 groups: receptors for RYa, LK, tachykinin (TK), SIFa, ecdysis-triggering hormone (ETH), CAPA, PK, Proc, FMRFamide (FMRFa), SP, mysupression (MS), AstA/AstC, CCHa, CNMa, allatotropin (AT), sNPF, NPF, crustacean cardioactive peptide (CCAP), AKH, Bursicon (Bur), and GPA2/GPB5 (Figures 2, 3). In addition to these GPCRs, 7 orphan receptors, A3, A9, A11, A12, A17, A28, and A36, were clustered into six orphan receptor branches (Figures 2, 3). No orthologous genes encoding the receptors for SK, Crz, AVLP, and trissin were found in A. craccivora. A recent study found that elevenin in N. lugens regulates body color via the receptor Nl_42 (47), but no ortholog of this receptor was found in A. craccivora (Figure 2).
Leucine-rich repeat-containing GPCRs (LGRs) are a special class of neuropeptide/protein hormone receptors in A-family GPCRs. They act as receptors for glycoprotein hormone or insulin/relaxin-related proteins and play important regulatory roles in development and reproduction of insects (48, 49). According to the structure characteristics, LGRs are divided into type A, type B, and type C. The main differences among the 3 types are the number of leucine-rich repeats (LRRs), the presence or absence of a low density lipoprotein receptor-like cysteine-rich (LDLa) motif, and type-specific hinge region (48, 49). In this study, a total of 5 LGRs (A36-A40) were identified from A. craccivora (Figure 3). Among them, only A40 has a complete ORF. CDD analysis indicated that A40 has 15 LRR motifs and no LDLa motif, showing typical structural characteristics of a type B LGR. A36 has an N-terminus containing 7 LDLa repeats and 8 LRR motifs, consistent with the typical characteristics of a type C LGR. The presence of conserved domains was not detected in other LGRs due to incomplete sequences. Phylogenetic analysis revealed that A. craccivora A37 and A39 were clustered into the branch of GPA2/GPB5 receptor. A40 is the ortholog of D. melanogaster LGR2, which has been confirmed to be a receptor for Bursicon and is also the first deorphanized LGR in invertebrates (50, 51). It has been confirmed that dILP8 is a ligand for LGR3 in D. melanogaster (52), and the A38 receptor of A. craccivora is clustered into this branch. Thus, it is speculated that A38 may be an insulin-like receptor. In addition, there is an orphan receptor, A36, clustered into the C2 LGR subfamily (Figure 3).
B-family GPCRs are a small class of receptors that differ structurally and functionally from other families. Their remarkable structural feature is a long N-terminal domain. The B-family can be further subdivided into three subfamilies: subfamily B1, B2, and B3 (53). Neuropeptide GPCRs belong to the B1 subfamily and contain three types of neuropeptide/protein hormone receptors: calcitonin receptor (also known as DH31 receptor), corticotropin releasing factor (CRF)-like diuretic hormone receptor (also known as DH44 receptor), and PDF receptor (19). All these three types of receptors were identified in A. craccivora and involved a total of 6 sequences (B1–B6) (Figure 4). Phylogenetic analysis revealed that A. craccivora B1, B3, and B4 were clustered in the branch of DH31 receptor; B5 and B6 were clustered in the branch of DH44 receptor; and B2 was clustered in the branch of PDF receptor (Figure 4).
The expression profiles of A. craccivora neuropeptides and their GPCRs at different developmental stages were determined by qRT-PCR. For most of the measured neuropeptide-encoding genes, the expression abundance showed a trend of decreasing first and then increasing during development, i.e., the expression level was significantly downregulated at the 2nd and 3rd instar stages (Figure 5). For the neuropeptide GPCR genes, most were differentially expressed at different developmental stages and in different wing morphs, and moreover, most were more abundant in the adult stage than in other stages (Figure 6). The two TK receptors, A24 and A29, had basically the same developmental-stage expression pattern, while the expression patterns of the two members of NPF receptor family, A31 (NPFR ortholog) and A33 (NPY2R ortholog), were different (Figure 6). Overall, the gene expression patterns of most neuropeptide-encoding genes and their presumed GPCR-encoding genes were not consistent (Figures 5, 6).
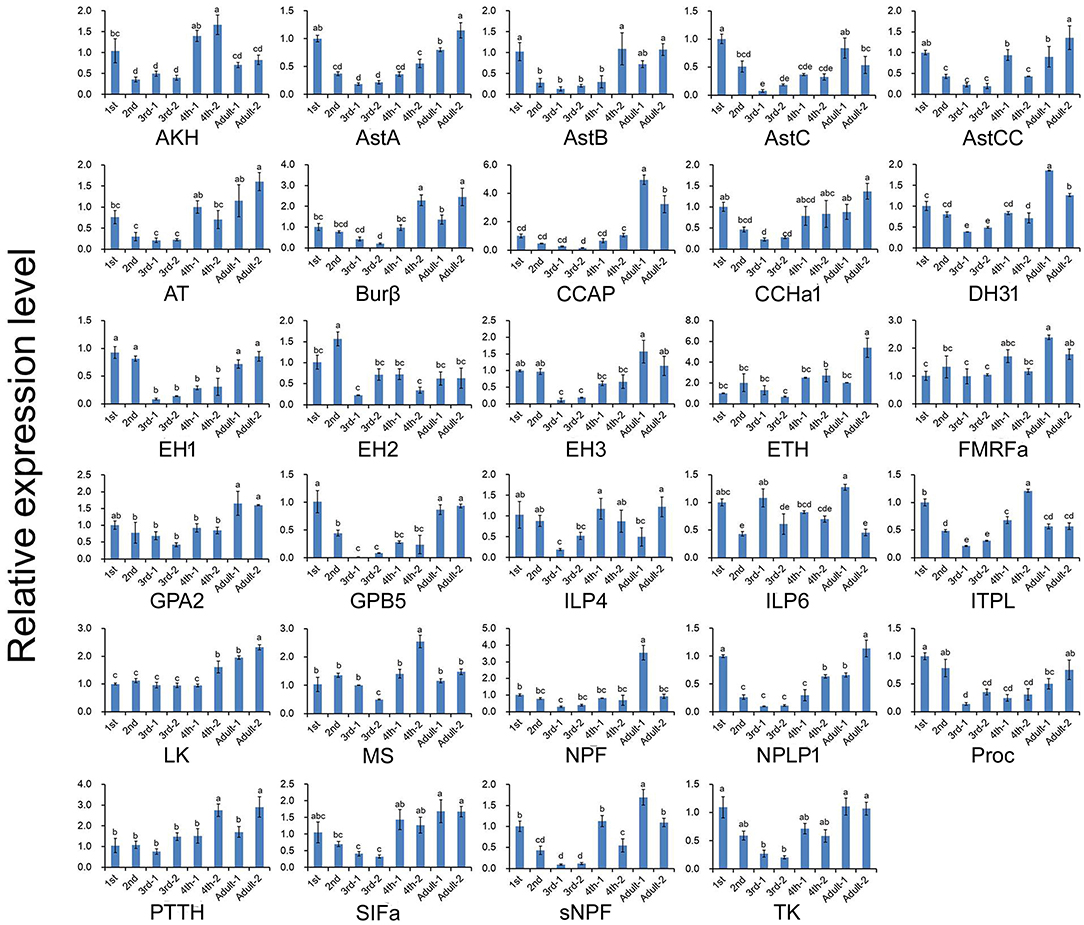
Figure 5. Relative expression levels of neuropeptide-encoding genes at different developmental stages of A. craccivora. The transcript level was measured via qRT-PCR and normalized against RPS8. Data are presented as the mean ± SD. The data were statistically analyzed by one-way ANOVA followed by Tukey's HSD test. Different lowercase letters indicate significant differences at the 0.05 level. “−1” indicates wingless aphids; “−2” indicates winged aphids.
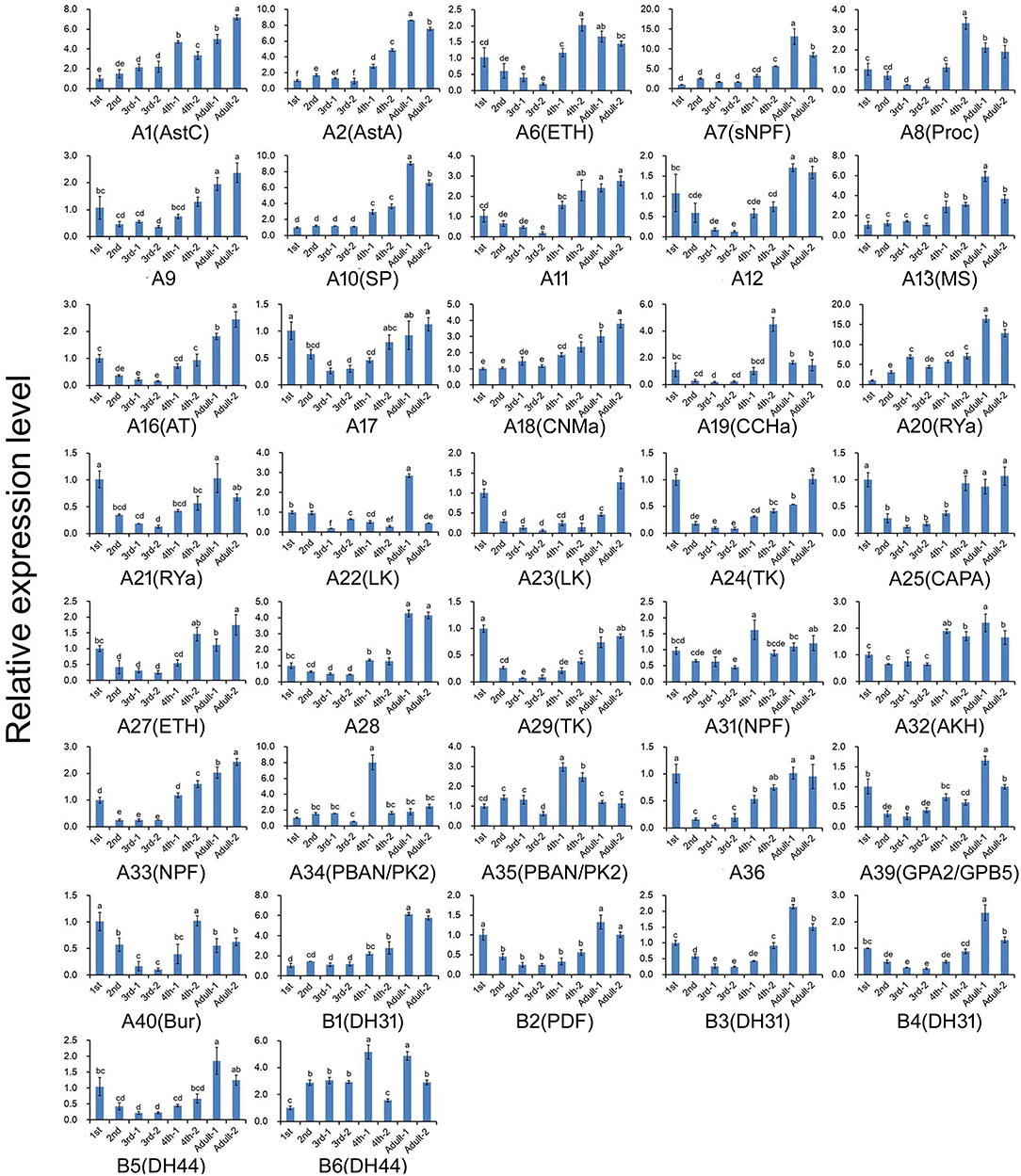
Figure 6. Relative expression levels of the genes encoding G protein-coupled receptors for the neuropeptides at different developmental stages of A. craccivora. The transcript level was measured via qRT-PCR and normalized against RPS8. Data are presented as the mean ± SD. The data were statistically analyzed by one-way ANOVA followed by Tukey's HSD test. Different lowercase letters indicate significant differences at the 0.05 level. Neuropeptide receptors A1–A40 are abbreviated as A1–A40, respectively; Neuropeptide receptors B1–B6 are abbreviated as B1–B6, respectively. And the receptors are marked with their predicted neuropeptide ligand partners in brackets; orphan receptors are not marked. “−1” indicates wingless aphids; “−2” indicates winged aphids.
Based on RNA-seq technology, we identified a large number of genes encoding neuropeptides and their GPCRs from A. craccivora in this study. The types and numbers of these genes are basically similar to those found in other insects, indicating that RNA-seq can be used as a powerful tool for the excavation of these genes from insects in the absence of whole-genome data. For some neuropeptide signaling systems, A. craccivora seems to lack both receptor and neuropeptide precursor (SK, Crz, AVLP, trissin, and elevenin). For others, the neuropeptide was not found or is lacking, but its presumed GPCR is present (e.g., PDF, RYa, CNMa, etc.). Why were they not found? There may be three main reasons. Firstly, the sequenced samples probably did not cover all stages of the A. craccivora life cycle, and some genes may not have been expressed in these measured samples; in other cases, some genes are expressed at low abundance in certain tissues or at certain developmental stages and the transcripts would be further diluted in the mixed sample. Secondly, due to the lack of strong sequence conservation among some neuropeptide precursors or neuropeptide GPCRs, their clear orthologs could not be found in A. craccivora based on homology searches. Thirdly, the neuropeptides may truly be absent in this species, a phenomenon that is common in insects. For example, sex peptide was reported only in the Drosophila genus; while neuroparsin is missing in D. melanogaster and other Drosophila species (54). Compared to D. melanogaster, B. mori, and T. castaneum, we found neither the genes coding for SK, Crz, AVLP, trissin, and elevenin in A. craccivora nor the genes coding for their receptors. If both neuropeptide and its receptor are truly absent, it is more likely that this signaling system is lacking as a whole based on neuropeptide-receptor co-evolution in this aphid species. Likewise, the former four signaling systems are not found in A. pisum (10, 12). These absent pathways are mainly involved in feeding, ecdysis and osmotic homeostasis in other insects (55–58), suggesting the absense is the result of constant adaptation to the environment during evolution of aphids. SK, Crz, and AVLP neuropeptide signaling systems were found in another hemipteran insect, N. lugens, but the trissin signaling system was absent in this species (6). For the neuropeptide GPCRs, two receptor genes for LK, SIFa and NPF have been identified from A. craccivora, A. pisum, and N. lugens (Figure 2), suggesting that each of these receptor genes duplicated in the evolution. These 3 types of receptors are mainly functionally involved in water balance, sexual behavior, and feeding behavior (59–62). However, we cannot conclude from these findings that the replication of these genes is a common feature of hemipterans, because they are a highly diverse group of insects.
A total of 40 genes enconding neuropeptides were identified from A. craccivora. Compared with another aphid species A. pisum (10), ITP and several ILPs were not detected from A. craccivora. ITP is a member of the crustacean hyperglycaemic hormone/ITP (CHH/ITP) superfamily, which stimulates the ileum to transport Cl− ion and may act as an antidiuretic hormone in insects (63, 64). In addition to ITP peptide, the ITP gene also encodes an alternatively spliced variant without C-terminal amidation (known as ITP-like or ITPL) (64). Only the transcript encoding ITPL was found in the A. craccivora transcriptome, whereas the ITP-encoding transcript was not. Generally, invertebrate genomes contain multiple genes encoding ILPs including 10 ILP precursor-encoding genes in the A. pisum genome (10), while only 6 of this family were found in A. craccivora. In contrast to ITPs and ILPs, an NPLP3 ortholog is present in A. craccivora but not in A. pisum. There are 4 NPLP-encoding genes, NPLP1-4, in D. melanogaster (65); NPLP2 and NPLP3 were found in A. mellifera (25). However, only one NPLP-encoding gene, NPLP1, was found in A. pisum (10). We found NPLP1 and NPLP3 in A. craccivora, which is consistent with that previously reported in a hemipteran insect, Diaphorina citri (7), while NPLP1, NPLP3, and NPLP4 are present in another hemipteran insect, N. lugens (6). Interestingly, no genes encoding PTTH orthologs were originally identified in hemipteran insects such as A. pisum and R. prolixus, probably due to insufficient conservation of PTTH sequences (6, 10). However, we used the sequences of PTTH precursors reported in other insects as a query to conduct a homology search and found a unigene in the A. craccivora. Moreover, a homologous sequence (ARM65502.1) was also found in A. pisum by the Blast program. PTTH is a polypeptide hormone that stimulates prothoracic glands to synthesize and release ecdysone and it was first cloned and characterized in B. mori (66). Mature B. mori PTTH is a homodimer. Each monomer contains 7 conserved Cys residues, of which 6 Cys form 3 internal disulfide bonds and the 7th Cys forms a disulfide bond between two monomers (66, 67). PTTH was later identified in several lepidopterans (68) and D. melanogaster (69). Compared with other neuropeptides, the sequence similarity of PTTH between different insects was relatively lower. However, the position of Cys residues and the folding structure of dimers were highly conservative (69–72). Tanaka et al. (6) used B. mori PTTH as a query to search for the analog in the N. lugens transcriptome and finally found a unigene whose theoretical protein has conserved structural features of PTTH. This is the first PTTH-like gene identified from hemimetabolous insects. We also found a unigene from the A. craccivora transcriptome that lacks the 5′-end of the ORF and may encode a partial sequence of the PTTH precursor. Sequence alignment revealed that the PTTH-like precursor of A. craccivora lacks the Cys residue at position 4 (Figure S2). The PTTH ortholog in A. pisum (ARM65502.1) also lacks the Cys residue at this position. Notably, all the 7 Cys residues of PTTH monomer are involved in the formation of disulfide bonds, which is required for PTTH to achieve biological activity (70). The effect of the absence of the fourth Cys residue on the formation of the PTTH advanced structure and whether it affects its normal biological function in aphids deserves further investigation.
We also carried out the expression profiling of genes encoding neuropeptides and their receptors in different developmental stages. Unfortunately, the low expression level of some measured genes reduced the accuracy of their qRT-PCR data. Thus, such genes were not included in the results (Figures 5, 6). The expression and functions of insect neuropeptides may change with development (73). In the present study, the tested neuropeptide genes also showed different levels of expression during development of A. craccivora. However, whether the differential temporal expressions of these genes have a biological role on development needs further investigation. It is worth noting that NPF was expressed in A. craccivora wingless adults at a significantly higher level compared with that in winged adults. NPF is homologous to the vertebrate neuropeptide Y (NPY) and its functional roles are well-known in regulation of feeding behavior in a wide range of insects (60). This neuropeptide was also suggested to play a positive role in the regulation of food intake in A. pisum using RNAi knockdown (74). In view of this, whether the differential expression of NPF between morphs has an impact on feeding behavior of different wing-morph adults or not, worths further research.
Nutritional conditions are key environmental factors that affect the growth and development of insects. The ILPs play a main role in systematically regulating the growth of the body in response to nutritional conditions (75). A prominent feature of insect ILPs is that multiple genes encode different ILPs. For example, 8 ILPs have been identified in D. melanogaster (76, 77), while 39 ILPs have been identified in B. mori (78). The synthesis and secretion of different ILPs in the same insect species exhibits different temporal and spatial pattern, and their functions are also different (79). In A. pisum, ILP5 has attracted the attention of researchers due to its abundant expression. It is speculated that this gene may be involved in the rapid development of aphids (10). It has also been reported that ILP5 is involved in embryonic development during wing differentiation in A. pisum (79). From the RPKM values, the expression abundance of ILP5 gene in A. craccivora is much higher than other ILP-encoding genes, which is consistent with that reported in A. pisum (80). The ILP5 transcript was detected from both the head as well as the mixed whole-body samples of A. craccivora. Besides, it was abundantly expressed in the head of wingless adult aphids. A. pisum ILP5 gene shows the similar expression pattern: it is expressed systemically in A. pisum adults and third instar nymphs with the highest expression level in the head of wingless adults (including the antennae) (81). This expression characteristic of ILP5 in aphids suggests the importance of its biological function and deserves further in-depth investigation.
In this study, the sequence similarity between neuropeptide precursors is very high among different species of aphids. Several neuropeptide genes, such as sNPF, SIFa, AstC, AstCC, and Proc, even encode identical mature active peptides in different species of aphids. For neuropeptide GPCRs, besides high sequence similarity, all of the identified A. craccivora neuropeptide GPCRs have a one-to-one orthologous relationship with the homologs of A. pisum. This result indicates that the aphid neuropeptide GPCRs are also highly conserved reflected by their types and numbers. In summary, the neuropeptide signaling systems in aphids are highly conserved and provide potential insecticide targets for the control of aphids. In addition, most neuropeptide signalings are conserved in both holometabolous and hemimetabolous insects, suggesting that they play key roles in the physiological processes of insects (3). Some neuropeptide signaling systems are lost in specific species, possibly due to functional redundancy that occurs during adaptation and evolution or other neuropeptide signalings taking over their functions (3).
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
T-XL and M-JQ supervised the whole project. XL and LD performed the experiments and wrote the manuscript with equal contributions. X-JJ, QJ, and C-JQ provided technical support. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2018YFD0201000), the China Agriculture Research System (Grant No. CARS-13), the Major Scientific and Technological Innovation Projects in Shandong Province (2019JZZY010702), and the Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2018E21).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2020.00640/full#supplementary-material
1. Nässel DR, Winther AME. Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol. (2010) 92:42–104. doi: 10.1016/j.pneurobio.2010.04.010
2. Altstein M, Nässel DR. Neuropeptide signaling in insects. In: Geary TG, Maule AG, editor. Neuropeptide Systems as Targets for Parasite and Pest Control. Boston, MA: Springer (2010). p. 155–65.
3. Caers J, Verlinden H, Zels S, Vandersmissen HP, Vuerinckx K, Schoofs L. More than two decades of research on insect neuropeptide GPCRs: an overview. Front Endocrinol. (2012) 3:151. doi: 10.3389/fendo.2012.00151
4. Audsley N, Down RE. G protein coupled receptors as targets for next generation pesticides. Insect Biochem Mol Biol. (2015) 67:27–37. doi: 10.1016/j.ibmb.2015.07.014
5. The International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. (2010) 8:e1000313. doi: 10.1371/journal.pbio.1000313
6. Tanaka Y, Suetsugu Y, Yamamoto K, Noda H, Shinoda T. Transcriptome analysis of neuropeptides and G-protein coupled receptors (GPCRs) for neuropeptides in the brown planthopper Nilaparvata lugens. Peptides. (2014) 53:125–33. doi: 10.1016/j.peptides.2013.07.027
7. Wang Z, Zhou W, Hameed MS, Liu J, Zeng X. Characterization and expression profiling of neuropeptides and G-protein-coupled receptors (GPCRs) for neuropeptides in the Asian citrus psyllid, Diaphorina citri (Hemiptera: Psyllidae). Int J Mol Sci. (2018) 19:3912. doi: 10.3390/ijms19123912
8. Li F, Zhao X, Zhu S, Wang T, Li T, Woolfley T, et al. Identification and expression profiling of neuropeptides and neuropeptide receptor genes in Atrijuglans hetaohei. Gene. (2020) 743:144605. doi: 10.1016/j.gene.2020.144605
9. Zhang H, Bai J, Huang S, Liu H, Lin J, Hou Y. Neuropeptides and G-protein coupled receptors (GPCRs) in the red palm weevil Rhynchophorus ferrugineus olivier (Coleoptera: Dryophthoridae). Fron Physiol. (2020) 11:159. doi: 10.3389/fphys.2020.00159
10. Huybrechts J, Bonhomme J, Minoli S, Prunier-Leterme N, Dombrovsky A, Abdel-Latief M, et al. Neuropeptide and neurohormone precursors in the pea aphid, Acyrthosiphon pisum. Insect Mol Biol. (2010) 19:87–95. doi: 10.1111/j.1365-2583.2009.00951.x
11. Li C, Chen M, Sang M, Liu X, Wu W, Li B. Comparative genomic analysis and evolution of family-B G protein-coupled receptors from six model insect species. Gene. (2013) 519:1–12. doi: 10.1016/j.gene.2013.01.061
12. Li C, Yun X, Hu X, Zhang Y, Sang M, Liu X, et al. Identification of G protein-coupled receptors in the pea aphid, Acyrthosiphon pisum. Genomics. (2013) 102:345–54. doi: 10.1016/j.ygeno.2013.06.003
13. Brady CM, White JA. Cowpea aphid (Aphis craccivora) associated with different host plants has different facultative endosymbionts. Ecol Entomol. (2013) 38:433–7. doi: 10.1111/een.12020
14. Brady CM, Asplen MK, Desneux N, Heimpel GE, Hopper KR, Linnen CR, et al. Worldwide populations of the aphid Aphis craccivora are infected with diverse facultative bacterial symbionts. Microb Ecol. (2014) 67:195–204. doi: 10.1007/s00248-013-0314-0
15. Coceano PG, Peressini S, Bianchi GL. The role of winged aphid species in the natural transmission of soybean mosaic potyvirus to soybean in North-east Italy. Phytopathol Mediterr. (1998) 37:111–8.
16. Chen K, Xu Z, Zhang Z, Fang X, Yan L, Higgins MC, et al. Biological characters and sequence analysis of coat protein gene of Chinese strains of peanut stripe virus (PStV). Chin J Oil Crop Sci. (1999) 21:55–9, 64.
17. Fouad EA, Abou-Yousef HM, Abdallah IS, Kandil MA. Resistance monitoring and enzyme activity in three field populations of cowpea aphid (Aphis craccivora) from Egypt. Crop Prot. (2016) 81:163–7. doi: 10.1016/j.cropro.2015.12.015
18. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. (2011) 29:644–52. doi: 10.1038/nbt.1883
19. Brody T, Cravchik A. Drosophila melanogaster G protein-coupled receptors. J Cell Biol. (2000) 150:83–8. doi: 10.1083/jcb.150.2.F83
20. Broeck JV. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides. (2001) 22:241–54. doi: 10.1016/S0196-9781(00)00376-4
21. Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. (2001) 11:1126–42. doi: 10.1101/gr.169901
22. Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, et al. G protein-coupled receptors in Anopheles gambiae. Science. (2002) 298:176–8. doi: 10.1126/science.1076196
23. Riehle MA, Garczynski SF, Crim JW, Hill CA, Brown MR. Neuropeptides and peptide hormones in Anopheles gambiae. Science. (2002) 298:172–5. doi: 10.1126/science.1076827
24. Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJP. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog Neurobiol. (2006) 80:1–19. doi: 10.1016/j.pneurobio.2006.07.005
25. Hummon AB, Richmond TA, Verleyen P, Baggerman G, Huybrechts J, Ewing MA, et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science. (2006) 314:647–9. doi: 10.1126/science.1124128
26. Fan Y, Sun P, Wang Y, He X, Deng X, Chen X, et al. The G protein-coupled receptors in the silkworm, Bombyx mori. Insect Biochem Mol Biol. (2010) 40:581–91. doi: 10.1016/j.ibmb.2010.05.005
27. Roller L, Yamanaka N, Watanabe K, Daubnerová I, Žitnan D, Kataoka H, et al. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem Mol Biol. (2008) 38:1147–57. doi: 10.1016/j.ibmb.2008.04.009
28. Hauser F, Cazzamali G, Williamson M, Park Y, Li B, Tanaka Y, et al. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front Neuroendocrinol. (2008) 29:142–65. doi: 10.1016/j.yfrne.2007.10.003
29. Li B, Predel R, Neupert S, Hauser F, Tanaka Y, Cazzamali G, et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. (2008) 18:113–22. doi: 10.1101/gr.6714008
30. Ons S, Sterkel M, Diambra L, Urlaub H, Rivera-Pomar R. Neuropeptide precursor gene discovery in the Chagas disease vector Rhodnius prolixus. Insect Mol Biol. (2011) 20:29–44. doi: 10.1111/j.1365-2583.2010.01050.x
31. Xu G, Gu GX, Teng ZW, Wu SF, Huang J, Song QS, et al. Identification and expression profiles of neuropeptides and their G protein-coupled receptors in the rice stem borer Chilo suppressalis. Sci Rep. (2016) 6:28976. doi: 10.1038/srep28976
32. Petersen TN, Brunak S, von Heijne G, Nielsen H. Signalp 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. (2011) 8:785–6. doi: 10.1038/nmeth.1701
33. Southey BR, Amare A, Zimmerman TA, Rodriguez-Zas SL, Sweedler JV. NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. (2006) 34:W267–72. doi: 10.1093/nar/gkl161
34. Monigatti F, Gasteiger E, Bairoch A, Jung E. The sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics. (2002) 18:769–70. doi: 10.1093/bioinformatics/18.5.769
35. Marchler-Bauer A, Bryant SH. CD-search: protein domain annotations on the fly. Nucleic Acids Res. (2004) 32:W327–31. doi: 10.1093/nar/gkh454
36. Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. (2015) 43:D222–6. doi: 10.1093/nar/gku1221
37. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. (2013) 30:772–80. doi: 10.1093/molbev/mst010
38. Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE. (2010) 5:e9490. doi: 10.1371/journal.pone.0009490
39. Yang C, Pan H, Liu Y, Zhou X. Temperature and development impacts on housekeeping gene expression in cowpea aphid, Aphis craccivora (Hemiptera: Aphidiae). PLoS ONE. (2015) 10:e0130593. doi: 10.1371/journal.pone.0130593
40. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
41. Nässel DR, Larhammar D. Neuropeptides and peptide hormones. In: Galizia C, Lledo PM, editor. Neurosciences-From Molecule to Behavior: A University Textbook. Berlin: Springer Spektrum (2013). p. 213–37. doi: 10.1007/978-3-642-10769-6_11
42. Christie AE. In silico analyses of peptide paracrines/hormones in Aphidoidea. Gen Comp Endocrinol. (2008) 159:67–79. doi: 10.1016/j.ygcen.2008.07.022
43. Sterkel M, Urlaub H, Rivera-Pomar R, Ons S. Functional proteomics of neuropeptidome dynamics during the feeding process of Rhodnius prolixus. J Proteome Res. (2011) 10:3363–71. doi: 10.1021/pr2001012
44. Predel R, Wegener C. Biology of the CAPA peptides in insects. Cell Mol Life Sci. (2006) 63:2477–90. doi: 10.1007/s00018-006-6187-3
45. Boerjan B, Cardoen D, Bogaerts A, Landuyt B, Schoofs L, Verleyen P. Mass spectrometric profiling of (neuro)-peptides in the worker honeybee, Apis mellifera. Neuropharmacology. (2010) 58:248–58. doi: 10.1016/j.neuropharm.2009.06.026
46. Schmitt F, Vanselow JT, Schlosser A, Kahnt J, Rössler W, Wegener C. Neuropeptidomics of the carpenter ant Camponotus floridanus. J Proteome Res. (2015) 14:1504–14. doi: 10.1021/pr5011636
47. Uchiyama H, Maehara S, Ohta H, Seki T, Tanaka Y. Elevenin regulates the body color through a G protein-coupled receptor NlA42 in the brown planthopper Nilaparvata lugens. Gen Comp Endocrinol. (2018) 258:33–8. doi: 10.1016/j.ygcen.2017.07.017
48. Van Hiel MB, Vandersmissen HP, Van Loy T, Broeck JV. An evolutionary comparison of leucine-rich repeat containing G protein-coupled receptors reveals a novel LGR subtype. Peptides. (2012) 34:193–200. doi: 10.1016/j.peptides.2011.11.004
49. Van Loy T, Vandersmissen HP, Van Hiel MB, Poels J, Verlinden H, Badisco L, et al. Comparative genomics of leucine-rich repeats containing G protein-coupled receptors and their ligands. Gen Comp Endocrinol. (2008) 155:14–21. doi: 10.1016/j.ygcen.2007.06.022
50. Luo CW, Dewey EM, Sudo S, Ewer J, Hsu SY, Honegger HW, et al. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc Natl Acad Sci USA. (2005) 102:2820–5. doi: 10.1073/pnas.0409916102
51. Mendive FM, Van Loy T, Claeysen S, Poels J, Williamson M, Hauser F, et al. Drosophila molting neurohormone bursicon is a heterodimer and the natural agonist of the orphan receptor DLGR2. FEBS Lett. (2005) 579:2171–6. doi: 10.1016/j.febslet.2005.03.006
52. Vallejo DM, Juarez-Carreño S, Bolivar J, Morante J, Dominguez M. A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science. (2015) 350:aac6767. doi: 10.1126/science.aac6767
53. Harmar AJ. Family-B G-protein-coupled receptors. Genome Biol. (2001) 2:reviews3013–1. doi: 10.1186/gb-2001-2-12-reviews3013
54. Veenstra JA. What the loss of the hormone neuroparsin in the melanogaster subgroup of Drosophila can tell us about its function. Insect Biochem Mol Biol. (2010) 40:354–61. doi: 10.1016/j.ibmb.2010.03.001
55. Zels S, Dillen S, Crabbé K, Spit J, Nachman RJ, Broeck JV. Sulfakinin is an important regulator of digestive processes in the migratory locust, Locusta migratoria. Insect Biochem Mol Biol. (2015) 61:8–16. doi: 10.1016/j.ibmb.2015.03.008
56. Kim YJ, Spalovská-Valachová I, Cho KH, Zitnanova I, Park Y, Adams ME, et al. Corazonin receptor signaling in ecdysis initiation. Proc Natl Acad Sci USA. (2004) 101:6704–9. doi: 10.1073/pnas.0305291101
57. Aikins MJ, Schooley DA, Begum K, Detheux M, Beeman RW, Park Y. Vasopressin-like peptide and its receptor function in an indirect diuretic signaling pathway in the red flour beetle. Insect Biochem Mol Biol. (2008) 38:740–8. doi: 10.1016/j.ibmb.2008.04.006
58. Roller L, CiŽmár D, Gáliková Z, Bednár B, Daubnerová I, Žitnan D. Molecular cloning, expression and identification of the promoter regulatory region for the neuropeptide trissin in the nervous system of the silkmoth Bombyx mori. Cell Tissue Res. (2016) 364:499–512. doi: 10.1007/s00441-015-2352-z
59. Al-Anzi B, Armand E, Nagamei P, Olszewski M, Sapin V, Waters C, et al. The leucokinin pathway and its neurons regulate meal size in Drosophila. Curr Biol. (2010) 20:969–78. doi: 10.1016/j.cub.2010.04.039
60. Nassel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: any similarities to vertebrate neuropeptide Y signaling? Peptides. (2011) 32:1335–55. doi: 10.1016/j.peptides.2011.03.013
61. Terhzaz S, Rosay P, Goodwin SF, Veenstra JA. The neuropeptide SIFamide modulates sexual behavior in Drosophila. Biochem Bioph Res Commun. (2007) 352:305–10. doi: 10.1016/j.bbrc.2006.11.030
62. Dow JA. Insights into the malpighian tubule from functional genomics. J Exp Biol. (2009) 212:435–45. doi: 10.1242/jeb.024224
63. Audsley N, McIntosh C, Phillips JE. Isolation of a neuropeptide from locust corpus cardiacum which influences ileal transport. J Exp Biol. (1992) 173:261–74. doi: 10.1016/S0079-6123(08)61172-3
64. Begum K, Li B, Beeman RW, Parka Y. Functions of ion transport peptide and ion transport peptide-like in the red flour beetle Tribolium castaneum. Insect Biochem Mol Biol. (2009) 39:717–25. doi: 10.1016/j.ibmb.2009.08.005
65. Baggerman G, Cerstiaens A, De Loof A, Schoofs L. Peptidomics of the larval Drosophila melanogaster central nervous system. J Biol Chem. (2002) 277:40368–74. doi: 10.1074/jbc.M206257200
66. Kawakami A, Kataoka H, Oka T, Mizoguchi A, Kimura-Kawakami M, Adachi T, et al. Molecular cloning of the Bombyx mori prothoracicotropic hormone. Science. (1990) 247:1333–5. doi: 10.1126/science.2315701
67. Kataoka H, Nagasawa H, Isogai A, Ishizaki H, Suzuki A. Prothoracicotropic hormone of the silkworm, Bombyx mori: amino acid sequence and dimeric structure. Agric Biol Chem. (1991) 55:73–86. doi: 10.1080/00021369.1991.10870535
68. Marchal E, Vandersmissen HP, Badisco L, Van De Velde S, Verlinden H, Iga M, et al. Control of ecdysteroidogenesis in prothoracic glands of insects: a review. Peptides. (2010) 31:506–19. doi: 10.1016/j.peptides.2009.08.020
69. McBrayer Z, Ono H, Shimell MJ, Parvy JP, Beckstead RB, Warren JT, et al. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell. (2007) 13:857–71. doi: 10.1016/j.devcel.2007.11.003
70. Ishibashi J, Kataoka H, Isogai A, Kawakami A, Saegusa H, Yagi Y, et al. Assignment of disulfide bond location in prothoracicotropic hormone of the silkworm, Bombyx mori: a homodimeric peptide. Biochemistry. (1994) 33:5912–9. doi: 10.1021/bi00185a031
71. Shionoya M, Matsubayashi H, Asahina M, Kuniyoshi H, Nagata S, Riddiford LM, et al. Molecular cloning of the prothoracicotropic hormone from the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol. (2003) 33:795–801. doi: 10.1016/S0965-1748(03)00078-X
72. Wei ZJ, Zhang QR, Kang L, Xu WH, Denlinger DL. Molecular characterization and expression of prothoracicotropic hormone during development and pupal diapause in the cotton bollworm, Helicoverpa armigera. J Insect Physiol. (2005) 51:691–700. doi: 10.1016/j.jinsphys.2005.03.005
73. Nässel DR, Zandawala M. Recent advances in neuropeptide signaling in Drosophila, from genes to physiology and behavior. Prog Neurobiol. (2019) 179:101607. doi: 10.1016/j.pneurobio.2019.02.003
74. Li X, Qu MJ, Zhang Y, Li JW, Liu TX. Expression of neuropeptide F gene and its regulation of feeding behavior in the pea aphid, Acyrthosiphon pisum. Fron physiol. (2018) 9:87. doi: 10.3389/fphys.2018.00087
75. Okamoto N, Yamanaka N. Nutrition-dependent control of insect development by insulin-like peptides. Curr Opin Insect Sci. (2015) 11:21–30. doi: 10.1016/j.cois.2015.08.001
76. Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. (2001) 11:213–21. doi: 10.1016/S0960-9822(01)00068-9
77. Colombani J, Andersen DS, Léopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. (2012) 336:582–5. doi: 10.1126/science.1216689
78. Antonova Y, Arik AJ, Moore W, Riehle MA, Brown MR. Insulin-like peptides: structure, signaling, and function. In: Gilbert LI, editor. Insect Endocrinology. San Diego, CA: Academic Press (2012). p. 63–92. doi: 10.1016/B978-0-12-384749-2.10002-0
79. Guo SS, Zhang M, Liu TX. Insulin-related peptide 5 is involved in regulating embryo development and biochemical composition in pea aphid with wing polyphenism. Front Physiol. (2016) 7:31. doi: 10.3389/fphys.2016.00031
80. Sabater-Muñoz B, Legeai F, Rispe C, Bonhomme J, Dearden P, Dossat C, et al. Large-scale gene discovery in the pea aphid Acyrthosiphon pisum (Hemiptera). Genome Biol. (2006) 7:R21. doi: 10.1186/gb-2006-7-3-r21
Keywords: Aphis craccivora, neuropeptides, G protein-coupled receptors (GPCRs), expression profiling, transcriptome
Citation: Li X, Du L, Jiang X-J, Ju Q, Qu C-J, Qu M-J and Liu T-X (2020) Identification and Characterization of Neuropeptides and Their G Protein-Coupled Receptors (GPCRs) in the Cowpea Aphid Aphis craccivora. Front. Endocrinol. 11:640. doi: 10.3389/fendo.2020.00640
Received: 01 May 2020; Accepted: 06 August 2020;
Published: 17 September 2020.
Edited by:
Leo T. O. Lee, University of Macau, ChinaReviewed by:
Kevin Qu, The Chinese University of Hong Kong, ChinaCopyright © 2020 Li, Du, Jiang, Ju, Qu, Qu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming-Jing Qu, MTM0NTUyNzc1ODBAMTYzLmNvbQ==; Tong-Xian Liu, dHhsaXVAbndzdWFmLmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.