- 1Department of Marine Bioresources, Faculty of Bioresources, Mie University, Tsu, Japan
- 2Faculty of Science, Ushimado Marine Institute, Okayama University, Setouchi, Japan
- 3Department of Fisheries, Faculty of Agriculture, Kindai University, Nara, Japan
- 4Research Institute for Fisheries Science, Okayama Prefectural Technology Center for Agriculture, Forestry, and Fisheries, Setouchi, Japan
Crustacean reproduction has been hypothesized to be under complex endocrinological regulation by peptide hormones. To further improve our understanding of the mechanisms underlying this complex regulation, knowledge is needed regarding the hormones not only of the central nervous system (CNS) such as the X-organ/sinus gland (XOSG), brain, and thoracic ganglia, but also the peripheral gonadal tissues. For example, in vertebrates, some gonadal peptide hormones including activin, inhibin, follistatin, and relaxin are known to be involved in the reproductive physiology. Therefore, it is highly likely that some peptide factors from the ovary are serving as the signals among peripheral tissues and central nervous tissues in crustaceans. In this work, we sought to find gonadal peptide hormones and peptide hormone receptors by analyzing the transcriptome of the ovary of the kuruma prawn Marsupenaeus japonicus. The generated ovarian transcriptome data led to the identification of five possible peptide hormones, including bursicon-α and -β, the crustacean hyperglycemic hormone (CHH)-like peptide, insulin-like peptide (ILP), and neuroparsin-like peptide (NPLP). Dominant gene expressions for the bursicons were observed in the thoracic ganglia and the ovary, in the CNS for the CHH-like peptide, in the heart for NPLP, and in the ovary for ILP. Since the gene expressions of CHH-like peptide and NPLP were affected by a CHH (Penaeus japonicus sinus gland peptide-I) from XOSG, we produced recombinant peptides for CHH-like peptide and NPLP using Escherichia coli expression system to examine their possible peripheral regulation. As a result, we found that the recombinant NPLP increased vitellogenin gene expression in incubated ovarian tissue fragments. Moreover, contigs encoding putative receptors for insulin-like androgenic gland factor, insulin, neuroparsin, and neuropeptide Y/F, as well as several contigs encoding orphan G-protein coupled receptors and receptor-type guanylyl cyclases were also identified in the ovarian transcriptome. These results suggest that reproductive physiology in crustaceans is regulated by various gonadal peptide hormones, akin to vertebrates.
Introduction
As a one of the most important aquaculture target worldwide, the production of penaeid shrimps/prawns has been steadily increased for over the past 30 years. The species occupying the majority of current shrimp aquaculture is Litopenaeus vannamei and Penaeus monodon. Their production has increased ~430% from 1998 to 2008, and 190% from 2008 to 2018, achieving 5.7 million tons (Food and Agriculture Organization of the United Nations; www.fao.org/fishery/topic/16140/en). To enable sustainable penaeid shrimp production in future, efficient seed production technique is required. Information on the endocrine system governing reproductive physiology will help to have the similar efficient seed production technique as in other aquatic animals (1, 2).
A number of studies on endocrinological regulation of basic biological functions have so far focused on the central neurosecretory X-organ/sinus gland complex (XOSG) in the eyestalk among various crustacean species. It has been proved that various peptide hormones produced from the XOSG are regulating growth, metabolism, osmoregulation, and reproduction (3, 4), e.g., red pigment concentrating hormone (5), pigment dispersing hormone (6), crustacean hyperglycemic hormone (CHH) (7), molt-inhibiting hormone (MIH) (8), vitellogenesis- or gonad-inhibiting hormone (VIH/GIH) (9), mandibular organ-inhibiting hormone (MOIH) (10), and crustacean female sex hormone (CFSH) (11). Extensive works based on biological activity-oriented peptide purification and subsequent expansions of homologous cDNA cloning have been contributed to find the above peptide hormones. On the basis of these works, VIH is considered as a main regulator of reproductive process in terms of the inhibition of vitellogenin (VG, a major yolk protein precursor) synthesis. Furthermore, roles of peptide hormones from the central nervous system (CNS) other than XOSG, which includes the brain and thoracic ganglia, have been elucidated (12–14).
In Japan, the principal penaeid species is the kuruma prawn Marsupenaeus japonicus (former Penaeus japonicus), which is one of the most important aquatic resources. The reproductive processes of M. japonicus have also been extensively studied using vitellogenesis-related proteins and their genes (e.g., VG, cathepsin C, cortical rod protein, and thrombospondin) as indices of ovarian development (15–21). Among various peptide hormones which have been purified and characterized from the central XOSG (22–27), six type-I peptides of the crustacean hyperglycemic hormone (CHH) superfamily inhibit the expression of VG in the ovary (28, 29). Consequently, the six CHHs, called as P. japonicus sinus gland peptide-I (Pej-SGP-I), -II -III, -V, -VI, and –VII, have been hypothesized to be vitellogenesis-inhibiting hormones (VIHs), which explains why eyestalk-ablation acceralates ovarian development. In contrast to the hormones from central XOSG, only a few gender and reproductive organ-specific peripheral factors have been identified. An insulin-like androgenic gland factor of M. japonicus (Maj-IAG) is exclusively produced from the androgenic gland of the male gonad and supresses VG expression in the ovary (30, 31), which is presumed to control the development of male characteristics, like the orthologs in other decapods (32–34). An ovarian isoform of the crustacean female sex hormone of M. japonicus (Maj-CFSH-ov) is dominantly expressed in the ovary, but its function remains to be determined (35). Since some gonadal hormones, activin/inhibin (36–38), follistatin (39), and relaxin (40, 41), are known to be involved in the regulation of reproductive physiology in vertebrates, more attention should be paid for peptidergic factors from the gonad as well as the other peripheral tissues. Such factors may act as a feedback signal from the ovary to the CNS or as a signal that intermediate two VG synthetic site, the hepatopancreas and ovary, in female penaeid shrimps (42).
More recently, transcriptomic analysis has become an important tool for peptide/protein profiling in addition to the conventional approaches described above. The transcriptome data supports our comprehensive understanding of the mechanisms where target tissues are regulated. Indeed, many more transcripts encoding hormones homologous to those found in vertebrates or insects have been identified in various crustacean species (43–52). Some of the studies have shown the existence of transcripts for putative peptide hormones in the ovary: the red swamp crayfish Procambarus clarkii (47), the Australian red-craw crayfish Cherax quadricarinatus (49), and the giant freshwater prawn Macrobrachium rosenbergii (51). In addition to the peptide/protein hormone candidates, the transcriptomic data have been exploited for the identification and characterization of peptide hormone receptors (53, 54) and the elucidation of molecular pathways involved in the regulation of various biological functions (55, 56). Hence, transcriptomics is an essential tool in the quest to improve our understanding of reproductive biology underlying peptide hormones and their receptors in peripheral organs such as the gonads.
Herein, we performed next-generation RNA sequencing on M. japonicus ovary with the Illumina MiSeq system. The de novo assembled ovarian transcriptome data was searched for the putative peptide hormone precursors and peptide hormone receptors. Moreover, the effects of putative hormones on ovarian VG expression were examined using their recombinant peptides.
Materials and Methods
Total RNA Extraction
Adult M. japonicus were purchased from a local fish market in Okayama Prefecture, Japan. For tissue-specific gene expression analysis, the brain, eyestalk, thoracic ganglia, heart, hepatopancreas, intestine, and gonad were dissected from both three male (20.7 g average body weitht) and three female prawns (23.0 g average body weight; 1.0% average gonadosomatic index, GSI). The prawns are determined to be in intermolt (C0–C1) and early premolt (D0) stages thorugh the observation of the setal development of the pleopods using a method modified from the previous report (57). Ovarian developmental stages of the female prawns were determined as previtellogenic by histlogical analysis (20, 35). Tissues were stored in RNAlater solution (Thermo Fisher Scientific, MA, USA) at −20°C until further use. For RNA-sequencing, the ovary of two intermolt female prawns in previtellogenic stage (26.4 g body weight; 1.0% GSI) and early exogenous vitellogenic stage (56.3 g body weight; 2.5% GSI) were dissected out and stored as described above. Total RNA was isolated using the illustra RNAspin mini RNA isolation kit (GE Healthcare Bio-Sciences AB, Uppsala, Sweden).
Library Preparation and RNA-Sequencing
The concentration of the two total RNA samples isolated from the ovary was measured using the Qubit RNA BR assay kit (Thermo Fisher Scientific). The cDNA libraries were constructed with 1 μg of the total RNA using the NEBNext ultra directional RNA library prep kit for Illumina, NEBNext Poly(A) mRNA Magnetic Isolation Module, and NEBNext multiplex oligos for Illumina (index primers set 1; New England BioLabs, MA, USA). All protocols were performed according to the manufacturer's instructions with minor modifications, namely that the fragmentation of RNA was performed by 94°C followed by incubation for 7.5 min. The final library fragment size was estimated to be 200–870 bp (average of 520 bp) using the Agilent high sensitivity DNA kit (Agilent Technologies, CA, USA). The library was sequenced using the MiSeq with Reagent Kit v3 (Illumina, CA, USA) in the paired-end mode with a read length of 300 bases.
Data Processing and Bioinformatic Analyses
Bases with a quality score (QV < 20) were trimmed from the 5′ and 3′ ends of each read, and reads containing ≥ 30% of low quality bases (QV < 14) with <25 bp were removed before assembling. The preprocessed reads were assembled using the Trinity platform (58). The resultant contigs were analyzed using the Basic Local Alignment Search Tool + (BLAST+; version 2.3.0) to perform a homology search against the National Center for Biotechnology Information non-redundant (NCBI-nr) protein database (ver. 170504) with an E-value cutoff of 1 × 10−5. The above-mentioned operations were executed using the DNA Data Bank of Japan (DDBJ) Read Annotation Pipeline and the supercomputer at the Research Organization of Information and Systems (ROIS), National Institute of Genetics (NIG), Japan. The BLAST output contained a maximum of 30 hits for each sequence which were used to assign the functional Gene Ontology (GO) terms to the protein sequences and for further GO Slim analysis on Blast2GO (59, 60). The signal peptide was predicted using SignalP 4.1 Server (61).
Molecular Cloning of Open Reading Frames of Hormonal Genes
Poly (A)+ RNA was prepared from 50 μg of the total RNA as described above using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England BioLabs). First-strand cDNA was synthesized from the purified Poly(A)+ RNA using the Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics, Mannheim, Germany) and an anchored-oligo(dT)18 primer. This first-strand cDNA was purified with AMPure XP (Beckman Coulter, IN, USA) and tailed with poly(A) using terminal transferase (Roche Diagnostics). The following PCRs were performed to obtain the correct open reading frame (ORF) of each target gene using the synthesized cDNA.
For 5′ -RACE, PrimeSTAR HS DNA polymerase or TaKaRa LA Taq DNA polymerase (Takara Bio, Shiga, Japan) were used with the cDNA on conventional PCR programs as per a previously described method (35). Adapter1 and adapter2, as forward primers, and bursA-R01 and -R02, as reverse primers, were used for the bursicon A subunit. Adapter1, adapter2, bursA-R01, and -R02 were used for the bursicon B subunit. Adapter1, adapter2, ilp-R01, -R07, -R10, R17, and R18 were used for the insulin-like peptide (Supplementary Table 1).
All PCR products were subcloned into the pGEM-T easy vector (Promega, WI, USA) after the addition of an adenine nucleotide at the 3′ ends. All plasmids were then sequenced on the 3730xl DNA analyzer (Applied Biosystems, CA, USA).
Construction of Plasmids for Recombinant Peptides
Expression plasmids for the M. japonicus CHH-like peptide and M. japonicus neuroparsin-like peptide (Maj-pCHH-B and Maj-NPLP, respectively), were prepared as per a previously described method (62). Both Maj-pCHH-B and Maj-NPLP cDNA fragments were amplified by PCR using the pchhbexF1/R1 and nplexF/R primer pairs, respectively (Supplementary Table 1). Each PCR product was mixed with the pET-44a(+) plasmid (Novagen, WI, USA), digested using Sma I and EcoR I (New England BioLabs), purified with AMPure XP, and then ligated. The thrombin protease cleavage site of the recombinant Maj-pCHH-B (rMaj-pCHH-B) expression plasmid was modified to a tobacco etch virus (TEV) protease cleavage site using the pchhbexF2/R2 primer pair (Supplementary Figure 1). Additionally, the thrombin protease cleavage site of the recombinant Maj-NPLP (rMaj-NPLP) expression plasmid was modified to a human rhinovirus 3C (HRV 3C) protease cleavage site using the nplexF2/R2 primer pair as per a previously described method (62). These modifications accompanied the substitution of N-terminal residues from Gln to Gly in rMaj-pCHH-B and from Ala to Gly in rMaj-NPLP (Supplementary Figure 2).
Expression and Purification of rMaj-pCHH-B and rMaj -NPLP
The transformation and culture of E. coli strain BL21(DE3) STAR (Thermo Fisher Scientific) with expression plasmids, induction of recombinant protein overexpression, and preparation of the soluble fraction of the cell lysate were performed as per previously described methods (62, 63).
The soluble fraction of the cell lysate containing the recombinant fusion protein, His-Nus-His-tagged rMaj-pCHH-B, was purified using the Ni Sepharose 6 Fast Flow resin (GE Healthcare). While the recombinant fusion protein was captured with the resin, the affinity tags were cleaved from rMaj-pCHH-B by protease digestion in a buffer containing 25 mM Tris-HCl (pH 7.4), 0.1 mM EDTA, 0.4 M urea, and ProTEV protease (Promega) at 20°C for 24 h. The untagged rMaj-pCHH-B was washed out from the resin and further purified by reverse phase high-performance liquid chromatography (RP-HPLC) on a Capcell Pak C18 SG300 column (150 × 6 mm; Shiseido, Tokyo, Japan) using the following program: a 2-min hold at 5% acetonitrile (MeCN) in 0.05% trifluoroacetic acid (TFA), a 5-min linear gradient of 5–25% MeCN in 0.05% TFA, a 15-min gradient of 29– 37% MeCN in 0.05% TFA, a 1.2-min gradient of 37– 85% MeCN in 0.05% TFA, and a 5-min hold at 85% MeCN in 0.05% TFA at a flow rate of 0.8 mL/min.
The soluble fraction containing recombinant His-Nus-His-tagged rMaj-NPLP was incubated with the Ni Sepharose 6 resin at 4°C for 20 h. The resin was then washed with washing buffer (20 mM phosphate buffer, 0.2 M NaCl, 50 mM imidazole, pH 7.4) and equilibrated with washing buffer without imidazole. For the cleavage of the tags, HRV 3C protease (Takara Bio) was added, and the resin slurry was incubated at 4°C for 3 days. Untagged rMaj-NPLP was eluted from the resin and further purified by RP-HPLC on the aforementioned column using the following program: a 1-min hold at 5% MeCN in 0.05% TFA, a 4-min linear gradient of 5–21% MeCN in 0.05% TFA, a 15-min gradient of 21– 29% MeCN in 0.05% TFA, a 3.25-min gradient of 29–85% MeCN in 0.05% TFA, and a 5-min hold at 85% MeCN in 0.05% TFA at a flow rate of 0.8 mL/min.
The mass spectra of the purified recombinant peptides were measured on an Agilent 6,520 Accurate-Mass Quadrupole-TOF mass spectrometer with an electrospray ionization (ESI) interface (Agilent Technologies) as we have previously described (62).
Ex-vivo Ovarian Incubation
The effect of one of the CHH family of peptides (Pej-SGP-I) on the mRNA expression of the putative hormones was assessed using our ex-vivo ovarian incubation system (28, 29). The same system was also used to assess the effects of rMaj-pCHH-B and rMaj-NPLP on vitellogenin (Maj-VG) expression. The recombinant Pej-SGP-I (rPej-SGP-I) was prepared as per previously described methods (62–64). Adult female prawns (22.5 g average body weight; 0.9% average gonadosomatic index, GSI) acted as donors of the ovary. As shown in Supplementary Figure 3, the abdominal part of the ovary, where left and right ovarian lobes were sticking to each other, was dissected out and divided into two lobes, and one lobe was incubated as control lobe (medium only), whereas the other received the hormone treatment (experimental). The adjacent part of ovary was kept as initial sample (without incubation) and as sample for histological analysis to determine the vitellogenic stage. Only a single sample set of the ovary (initial, control, and experimental) was prepared from one prawn and counted as n = 1; total 30 prawns were used for the experiment in Figure 5 (6 for each graph), 28 for Figure 6A, 24 for Figure 6B, and 12 for Figure 6C. All prawns used had immature ovary and were in intermolt to early premolt (C0, C1, D0, and D1) stages.
Following incubation, the ovarian tissue fragments were immersed in RNAlater solution and stored at −20°C. Total RNA extraction was then performed as described above.
Quantitative Real-Time Reverse Transcriptase PCR
Quantitative real-time reverse transcriptase PCR (qRT-PCR) was used for the quantification of the putative hormone genes, Maj-VG, and arginine kinase (Maj-AK). The sequences of primers and TaqMan probes used in this study have been listed in Supplementary Table 2. The concentrations of total RNA prepared from various tissues were quantified using the Qubit RNA BR assay kit, and for each sample 8 ng RNA was used for qRT-PCR. The qRT-PCR reactions were carried out using the iTaq universal probes one-step kit (Bio-Rad, CA, USA), and the same methods were used for the real-time monitoring of the fluorescence signal on the CFX96 real-time PCR detection system (Bio-Rad) as has been previously described (35). For the quantification of gene expression levels, relative standard curve method was used. DNA templates were amplified with respective gene-specific primer sets (Supplementary Table 2) so that they include qRT-PCR amplicon sequence of respective target genes. RNA standards were synthesized by in vitro transcription using in vitro Transcription T7 Kit (Takara Bio) with the DNA templates. The synthesized RNA standards were purified using the illustra RNAspin mini RNA isolation kit and quantified using the Qubit RNA BR assay kit as described above. The standard curves were generated using each RNA standard ranging from 40 ng to 0.4 pg prepared by serial 10-fold dilutions, and arbitrary values ranging from 40,000 to 0.4 were assigned correspondingly. Relative gene expression levels were determined based on the threshold cycles using the standard curves. Each standard had almost the same length (371–426 nt, Supplementary Table 2), and similar amplification efficiencies were achieved (95.9–99.8%) in the qRT-PCR.
For the incubated ovary samples, the relative expression of the target genes were standardized to those of Maj-AK, and expressed as a percent change from the initial (Figure 5) or 0 nM group (Figure 6) samples (35) (Supplementary Figure 3). In contrast, for tissue-specific gene expression analysis (Figure 4), the relative expression of the target genes were standardized to the total RNA input in the qRT-PCR to account for variations in the Maj-AK expression between tissues.
Statistical Analysis
Gene expression levels have been represented as the mean ± standard error mean (SEM). Statistical differences in gene expression levels were analyzed using the Wilcoxon signed rank test or one-way analysis of variance (ANOVA) followed by the Dunnett's post hoc test in the GraphPad Prism version 4.0 for Windows (GraphPad Software, CA, USA).
Results
RNA Sequencing and de novo Assembly
De novo assembly produced 98,509 contigs with a total size of ~91.8 Mbp. The N50 of the transcriptome was 1,676 bp long, and the longest contig was 15,684 bp long. Redundant contigs were eliminated based on a sequence similarity. Among the remaining 47,026 contigs, 24,033 (51.1%) had a BLAST hit, and 13,839 (29.4%) were assigned at least one GO term (Supplementary File). Following bioinformatics analysis on the ovarian transcriptome, several transcripts encoding putative hormones were identified as described below.
Bursicon
Two contigs putatively encoding bursicon, a heterodimer composed of burs-α and burs–β, were identified in the ovarian transcriptome. Additional cDNA cloning and homology analysis revealed that these were the α and β subunits; Maj-burs-α and Maj-burs-β, respectively. The Maj-burs-α precursor contains a signal peptide (19 amino acid residues) which, once cleaved, gives a mature peptide comprising of 121 amino acid residues. Additionally, the Maj-burs-β precursor also contains a signal peptide (30 amino acid residues) and once processed results in a mature peptide composed of 115 residues (Figure 1). Mature Maj-burs-α and Maj-burs-β both showed ≥ 50% amino acid sequence identity with the known bursicon subunits, and they both contain 11 Cys residues, 9 of which are conserved in the cystine knot-like domain (smart00041 in Conserved Protein Domain Family, NCBI) of the transforming growth factor-β superfamily (Supplementary Figure 4).

Figure 1. Amino acid sequences of putative hormones. Sequences were deduced from transcriptome analysis of M. japonicus ovary and additional cDNA cloning. Sequences shown in magenta and green represent the predicted signal sequences and cleavage sites, respectively. Underlined sequences represent CPRP in Maj-pCHH-B and C-peptide in Maj-ILP1, respectively.
Neuroparsin-Like Peptide
The ovarian transcriptome contained one contig encoding a precursor of Maj-NPLP which contains a 24 amino acid-long signal peptide and 78 amino acid-long mature peptide (Figure 1). We identified 12 conserved Cys residues, which were a characteristic of the crustacean neuroparsin family, in the mature Maj-NPLP peptide. Maj-NPLP shares 46% amino acid sequence identity with an neuroparsin in the greasyback shrimp Metapenaeus ensis (MeNPLP) which has been hypothesized to regulate ovarian maturation (65). Additionally, Maj-NPLP shares 32% amino acid sequence identity with the ovary ecdysteroidogenic hormone (OEH) in Aedes aegypti, the yellow fever mosquito (66) (Supplementary Figure 5).
CHH-Family Peptide
We found one contig encoding the precursor of a CHH-like peptide in the ovarian transcriptome. Additional cDNA cloning revealed that this precursor consisted of a 25 amino acid signal peptide, a 22 amino acid CHH precursor-related peptide (CPRP), an RXRR cleavage signal, and a 74 amino acid mature hormone (Figure 1). Considering the existence of CPRP and the absence of a single Gly residue 5 amino acids downstream of the first Cys residue, we considered this peptide to belong to the type-I subfamily of the CHH superfamily. On the other hand, there was no C-terminal amidation signal, a characteristic feature of type-I subfamily precursors. Additionally, there was a single-residue insertion seven amino acids downstream of the third Cys residue. Based on the primary structure nomenclature of the M. japonicus CHH-family peptides (25, 67), we categorized this peptide as a putative CHH-B (Maj-pCHH-B). Its mature peptide sequence showed 37–46% amino acid identity with the Marsupenaeus CHH-family peptides (Supplementary Figure 6). Phylogenetic analyses of the CHH superfamily characterized in the XOSG of M. japonicus so far (22, 23, 25, 26, 67) suggests that Maj-pCHH-B is diverse and most likely from the typical type-I and type-II subfamilies (Figure 2).
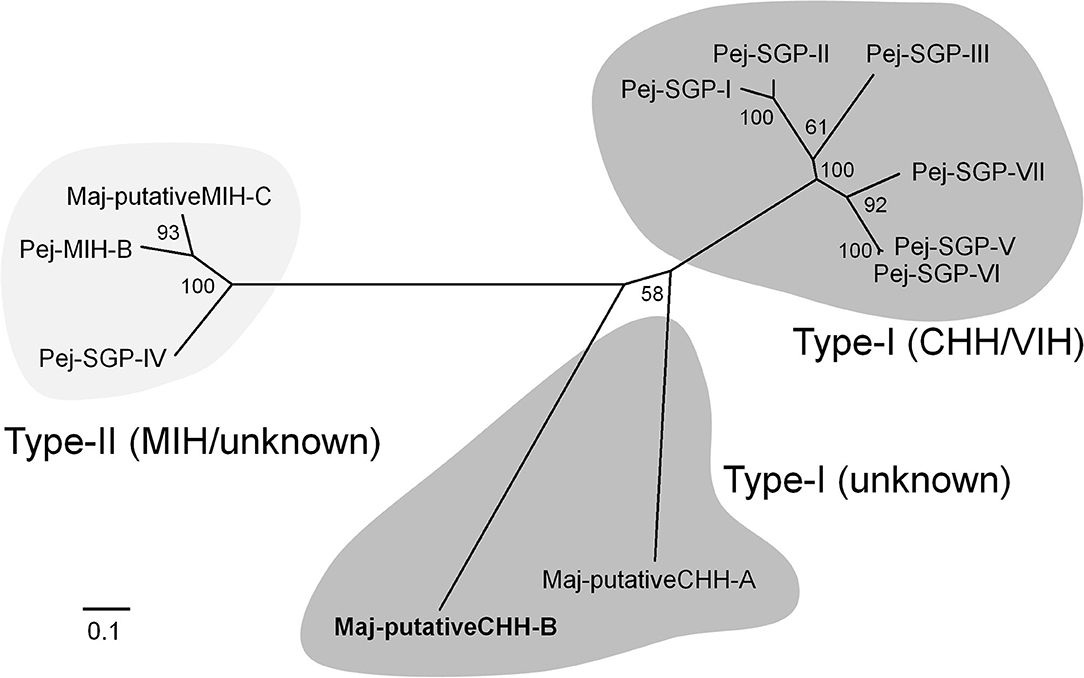
Figure 2. Molecular phylogenetic tree of M. japonicus CHH-family peptides. This phylogenetic tree was constructed by the neighbor-joining method with computation of the evolutionary distances using the JTT matrix-based method. Values at the nodes represent the percentage of 1,000 bootstrap replicates. The scale bar shows the number of substitutions per site. Amino acid sequences have been cited from the previous reports (25, 26, 67).
Insulin-Like Peptide
A putative insulin-like peptide precursor (Maj-ILP1) was identified in the ovarian transcriptome of M. japonicas. The full-length ORF of Maj-ILP1 was obtained by several rounds of 5′ -RACE. The precursor comprised a signal peptide composed of 20 residues, a B-chain consisting of 30 amino acid residues, an RXRR cleavage signal, a C-peptide composed of 131 amino acid residues, the other RXRR cleavage signal, and an A-chain composed of 24 amino acid residues (Figure 1). The primary structure of Maj-ILP1 was distinct from IAG in the same species (Maj-IAG); mature peptide (deduced B- and A- chains) of Maj-ILP1 shared only 28.6% amino acid sequence identity with that of Maj-IAG (Supplementary Figure 7). Phylogenetic analysis of known insulin/relaxin family shows that Maj-ILP1 is part of the insulin/insulin-related peptide group and not the IAG or relaxin groups (Figure 3).
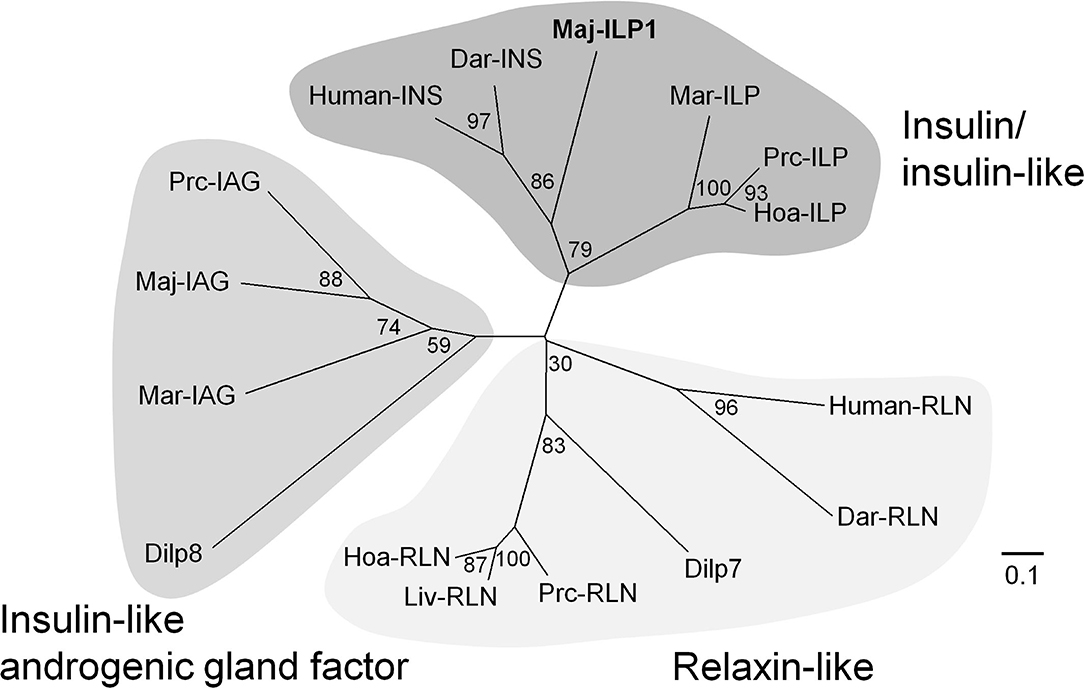
Figure 3. Molecular phylogenetic tree of insulin-family peptides. This phylogenetic tree was constructed by the neighbor-joining method with computation of the evolutionary distances using Poisson correction. Values at the nodes represent the percentage of 1,000 bootstrap replicates. The scale bar shows the number of substitutions per site. Accession numbers of cDNA sequences used are as follows: human-INS, BC005255; human-RLN, A06846; Dar-INS, Dar-RLN, JN215212; Dilp7, NP_570070; Dilp8, NP_648949; Maj-IAG, BAK20460. The other sequences have been cited from a recent report (48).
Putative Hormone Receptors
Based on BLAST analysis, several contigs encoding the G-protein coupled receptor family, the receptor guanylyl cyclase family, and the insulin receptor family were found in the present ovarian transcriptome (Table 1). A full-length ORF of contig N13763 was obtained by additional cDNA cloning, and its seven-transmembrane domain showed 62% amino acid sequence identity to the neuropeptide Y (NPY) receptor in the Nevada dampwood termite Zootermopsis nevadensis. Additional cDNA cloning also revealed that the ORF of contig N28645 was composed of an N-terminal leucine-rich repeat domain and a seven-transmembrane domain that was highly similar to the transmembrane domain of the relaxin-family peptide receptors (cd15137 in Conserved Protein Domain Family). Furthermore, contigs N06137 and N35023 were the most similar to the receptor for OEH in A. aegypti (66) and to IAGR in the Chinese white shrimp Fenneropenaeus chinensis (53), respectively.
Tissue-Specific Expression of the Putative Hormones
Tissue-specific expression levels of the putative hormone genes were examined by qRT-PCR analysis (Figure 4). We found that Maj-pCHH-B was mainly expressed in the nervous system and in the intestine of both male and female prawns. Expression in the ovary was low compared to the nervous system, and we did not detect any significant sexual dimorphism in the expression pattern. Additionally, Maj-NPLP was expressed primarily in the heart. Apparent expression was also observed in the thoracic ganglia and in the intestine. Maj-burs-α and Maj-burs-β are primarily expressed in the thoracic ganglia of both sexes and in the ovary. The levels of Maj-burs-α and -β expression in the ovary were significantly higher compared to those in the testis. Maj-ILP1 displayed a gender-specific expression pattern as it was expressed primarily in the ovary. Clear expression was also observed in the nervous system, while no expression was observed in the hepatopancreas of both sexes.
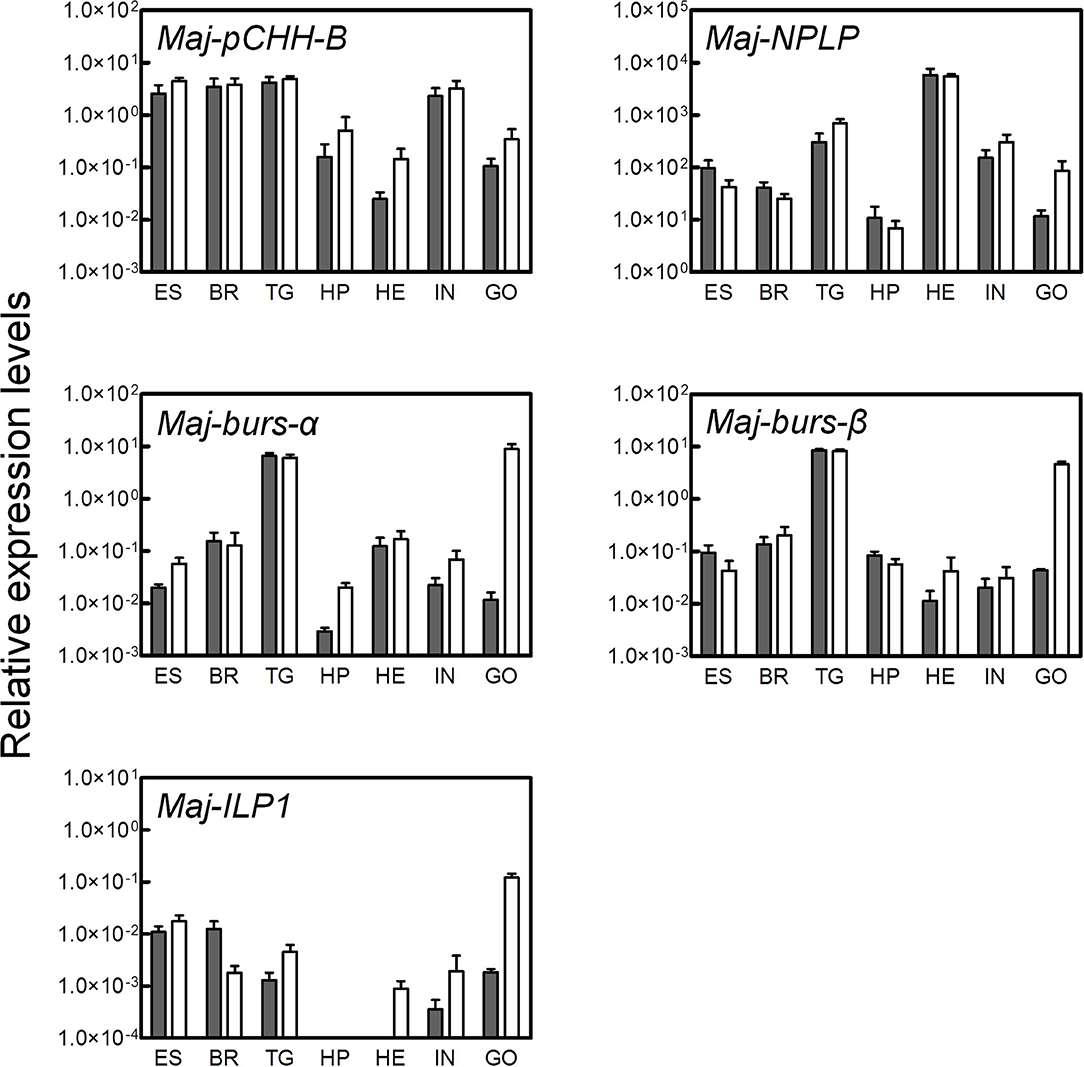
Figure 4. Tissue-specific expression analysis of the putative hormones. Gene expression of the putative hormones were examined by qRT-PCR in various tissues (ES, eyestalk; BR, brain; TG, thoracic ganglia; HP, hepatopancreas; HE, heart; IN, intestine; GO, gonad). Relative expression levels per 8 ng of the total RNA have been represented as mean ± SEM (n = 3). Open and solid bars represent female and male prawns, respectively.
Effect of Eyestalk VIH on the Expression of the Putative Hormones
To investigate the potential involvement of the putative hormones in vitellogenesis, the effects of Pej-SGP-I, a VIH of M. japonicus (28), on hormone gene expression were first examined (Figure 5). In an ex-vivo ovarian incubation system, we found that Maj-pCHH-B mRNA level was significantly reduced by 50 nM rPej-SGP-I. Maj-NPLP expression was also reduced by rPej-SGP-I, but not changed (p = 0.063). Maj-burs-α, Maj-burs-β, and Maj-ILP1 mRNA levels were not affected by rPej-SGP-I.
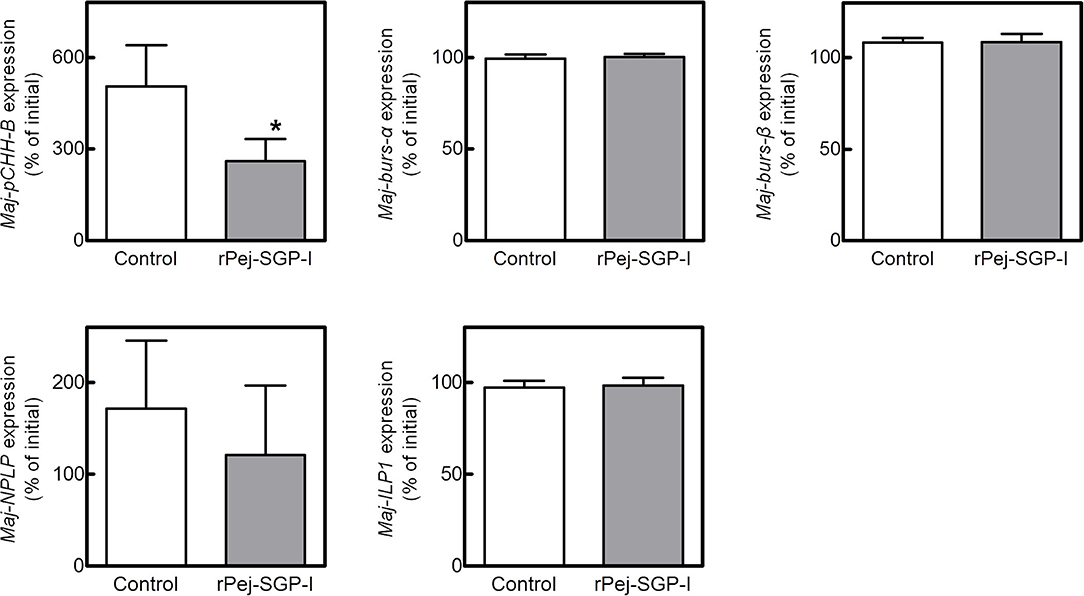
Figure 5. Effect of rPej-SGP-I on mRNA levels of the putative hormones in the ovary. Relative expression levels have been shown as the percentage change relative to that of the initial expression (mean ± SEM, n = 6). Statistical differences between control and rPej-SGP-I-treated groups were analyzed by the Wilcoxon signed rank test (*P < 0.05).
Preparation of rMaj-NPLP and rMaj-pCHH-B
Based on the above results, we further investigated potential involvement of Maj-NPLP and Maj-pCHH-B in vitellogenesis using recombinant peptides. rMaj-NPLP and rMaj-pCHH-B were expressed both as Nus-tagged fusion proteins; they were mostly recovered in the soluble fraction of cell lysates and successfully purified using a Ni-sepharose resin and subsequent HPLC. The deconvoluted mass spectra of untagged and HPLC-purified recombinant peptides have been shown in Supplementary Figure 8. Electrospray ionization (ESI) mass spectrum of rMaj-NPLP revealed a molecular mass of 8,357.1 which agreed with the calculated value (8,368.7) minus 12 Da, suggesting the presence of 6 disulfide bonds in the structure. Similarly, the observed molecular mass of rMaj-pCHH-B was 8,391.7 which was similar to the calculated value (8,397.6) minus 6 Da, indicating the presence of 3 disulfide bonds in the structure.
Effect of rMaj-NPLP and rMaj-pCHH-B on Ovarian VG Expression
The effects of the recombinant peptides on Maj-VG expression were assessed in the ovarian incubation system. Although rMaj-NPLP did not affect Maj-VG expression at lower doses, Maj-VG expression increased at higher doses with significant changes observed in the ovary fragments treated with 20 nM rMaj-NPLP (Figure 6A). In comparison, rMaj-pCHH-B treatment did not affect the Maj-VG expression up to 200 nM (Figure 6B). When 200 nM rMaj-pCHH-B was co-incubated with 50 nM rPej-SGP-I, rMaj-pCHH-B acted neither cooperatively nor antagonistically on the vitellogenesis-inhibiting activity of rPej-SGP-I (Figure 6C).
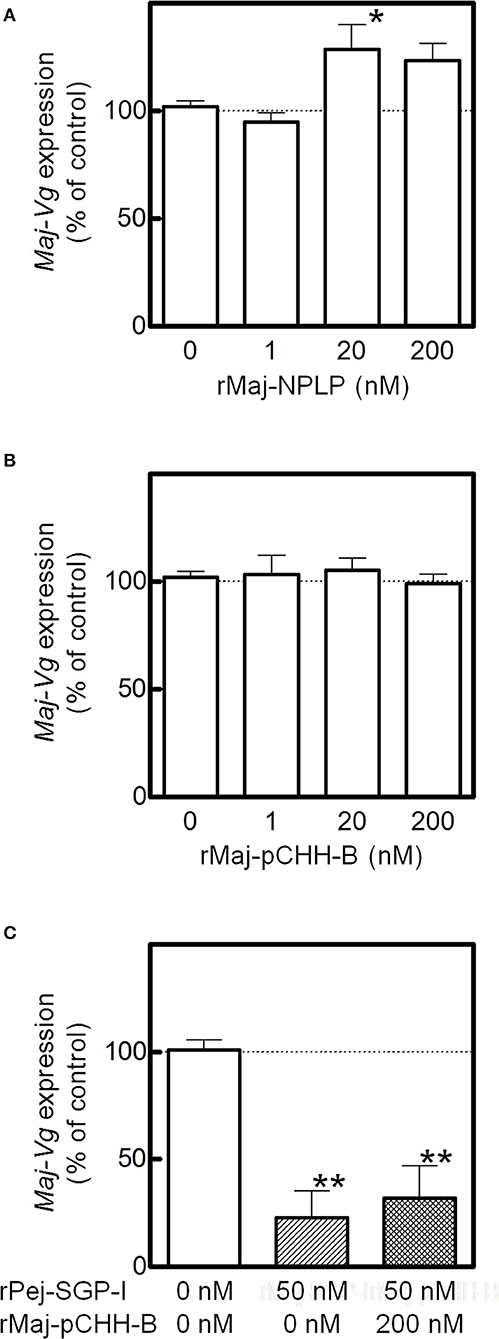
Figure 6. Analyses of rMaj-NPLP and rMaj-pCHH-B functions. Maj-VG expressions in ex-vivo ovarian fragments incubated with (A) rMaj-NPLP and (B) rMaj-pCHH-B are examined. Expression levels have been represented as the percentage change relative to those of 0 nM control groups. The differences between controls and the other groups are tested for significance using a one-way ANOVA followed by the Dunnett's post test (*P < 0.05; n = 4–8 for rMaj-pCHH-B and 6–8 for rMaj-NPLP). (C) Maj-VG expression was also examined following incubation without hormones, with rPej-SGP-I alone, and with both rPej-SGP-I and rMaj-pCHH-B. The differences between controls and the other groups are tested for significance using a one-way ANOVA followed by the Dunnett's post test (**P < 0.01; n = 4).
Discussion
The hypothalamus-pituitary-gonad is the main axis controlling vertebrate reproductive processes. In crustaceans, the XOSG and brain ganglia appear to correspond to the axis. Peptide hormones secreted from these niches, especially from the XOSG, have been well-characterized, and their involvements in reproductive processes have been studied. Conversely, there is little information on gonadal hormones. Most penaeid shrimps synthesize VG in the hepatopancreas and in the ovary (42). For example, in M. japonicus, the same VG transcript is present in both tissues, but their expression dynamics differ slightly during vitellogenesis (17, 20, 21). Therefore, some peptide factors from the ovary is thought to be serving as feedback signals among peripheral tissues and CNS. Additional studies on gonadal hormones will improve our understanding of the mechanisms behind crustacean reproduction. Thus, in this study, we analyzed the ovarian transcriptome of M. japonicus and identified Maj-burs-α, Maj-burs-β, Maj-NPLP, Maj-pCHH-B, and Maj-ILP1 (Figure 1) as some possible peptide hormones produced in M. japonicus ovary. Moreover, we examined functions of Maj-NPLP and Maj-pCHH-B. These data, in combination with previous work (35), suggest a peripheral regulation of the ovary as shown in Figure 7 and below.
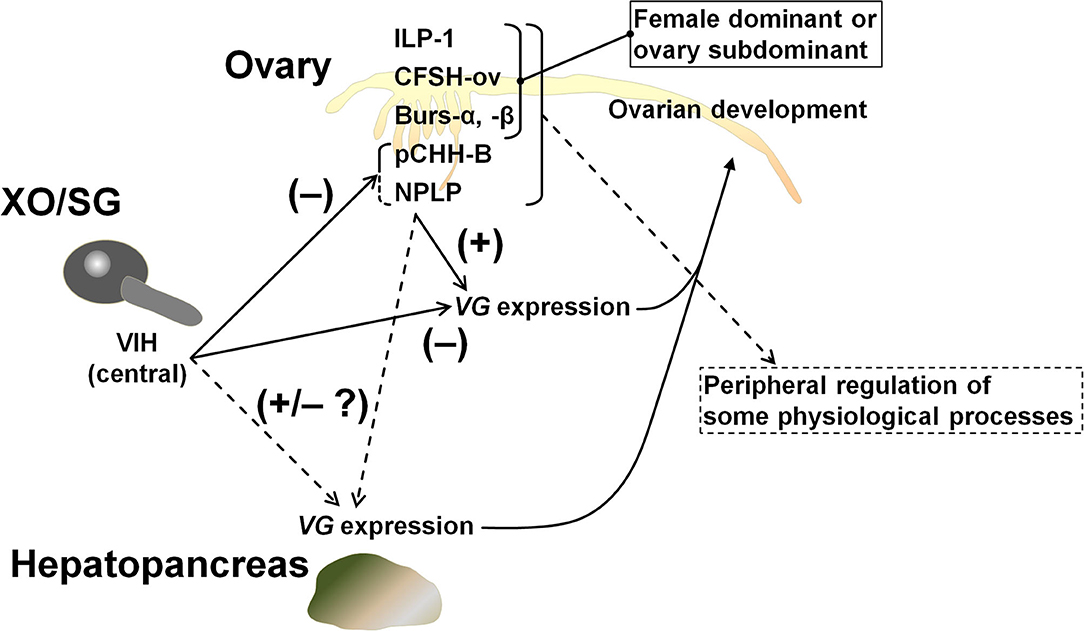
Figure 7. Schematic model of peripheral regulation of the ovary. It is highly likely that Maj-NPLP is a vitellogenesis-regulating factor in M. japonicus considering its activity and the existence of a putative receptor in the ovary. The other female dominant or ovary subdominant hormones may play a role in some physiological processes. Maj-CFSH-ov has been previously characterized (35).
Regarding neuroparsin, its vitellogenesis-inhibiting activity in terms of the inhibition of the juvenile hormone system has been reported in the migratory locust Locusta migratoria (68). In contrast, OEH of the neuroparsin family has a gonadotrophic effect in A. aegypti (66, 69); the OEH as well as several ILPs stimulate the ovarian ecdysteroid production, which induces VG synthesis in the fat body. In crustacean species, a neuroparsin-like peptide (MeNPLP) which is produced by the hepatopancreas and has been reported to induce VG expression in M. ensis (65). Transcriptomic analysis of Fenneropenaeus merguiensis (the banana shrimp) ovary shows that the expression of a neuroparsin precursor is higher in the vitellogenic stage compared to that in the non-vitellogenic stage (70). In the present study, we showed that Maj-NPLP has a stimulatory effect on VG synthesis in the ovary (Figure 6A). Although inhibitory effect of Pej-SGP-I on Maj-NPLP expression is not clear (Figure 5), it is likely that other five VIHs (Pej-SGP-II, -III, -V. -VI, and -VII) (28) regulate Maj-NPLP expression in the ovary. Taken together, we concluded that the NPLP is a regulator of reproductive process in arthropods. Multiple isoforms of NPLP are often found in a single crustacean species (47, 49, 65) with differing tissue-specific expression. The expression pattern of Maj-NPLP is similar to that of Mar-NP-2 in M. rosenbergii (51) which is expressed predominantly in the thoracic ganglia, heart, and gonads. Regarding the Maj-NPLP receptor, contig N06137 (Table 1) has significant sequence similarity to the OEH receptor (66) as well as a similar domain structure (i.e., extracellular venus fly trap domain, a single transmembrane domain, and intracellular tyrosine kinase domain), thereby suggesting that Maj-NPLP may act on the ovary through endocrine or autocrine/paracrine modes of action via this receptor. Such information may be used for further characterization of the role of NPLPs in M. japonicus vitellogenesis.
Bursicon, a heterodimer composed of burs-α (burs) and burs-β (pburs), regulates cuticle tanning and wing expansion after ecdysis in insects. Additionally, bursicon of the blue crab Callinectes sapidus (CasBurs) appears to be involved in the deposition and thickening of new cuticle as well as granulation of hemocytes (71). Reflecting the constitutive increased expression of the β subunit compared to the α subunit, the ββ homodimer as well as the αβ heterodimer are found in the pericardial organ of C. sapidus, but their intrinsic functioning is unknown. However, characteristics of bursicon, a member of the TGF-β superfamily (Supplementary Figure 4) and the dimerization patterns of the subunits, are analogous to those of the activin/inhibin family of proteins which participate in the regulation of reproductive physiology in mammals. Consequently, these data support our hypothesis of the role of bursicon in crustacean reproductive processes. In fact, bursicon has been reported to stimulate VG expression in the ovary of Penaeus monodon, also known as the black tiger shrimp (13). However, only the heterodimer (i.e., Pmbursα and Pmbursβ subunits) exhibit such a stimulatory effect, whereas the αα and ββ homodimers do not. In contrast, the bursicon receptor ortholog DLGR2, which is encoded in the rickets gene in the fruit fly Drosophila melanogaster (72), is not found in our M. japonicus ovarian transcriptome. Although contig N28645 (Table 1) has a similar domain organization to DLGR2 in terms of the N-terminal leucine-rich repeat domain and the seven-transmembrane receptor domain, their overall sequence similarity is low.
Maj-pCHH-B expression was suppressed by a central VIH, suggesting its possible involvement in the regulation of vitellogenesis, but we were unable to show this experimentally using the recombinant peptide (Figure 6B). And Maj-pCHH-B acted neither antagonistically nor cooperatively on the vitellogenesis-inhibiting activity of Pej-SGP-I (Figure 6C). Considering the dominant gene expression pattern in the CNS including the eyestalk, Maj-pCHH-B may act as a neurotransmitter, much like the ion transport peptides (ITP and ITP-L) (73, 74), the ortholog of the CHH superfamily in insects (4, 75). Further functional analysis of Maj-pCHH-B, Mj-putativeCHH, and Mj-putativeMIH-C (25) are required to elucidate the diverse biological functions of the CHH superfamily in M. japonicus. As for the primary structure, the single-residue insertion seven amino acids downstream of the third Cys residue (Supplementary Figure 6) is also reported in a CHH from the Pacific white shrimp Litopenaeus vannamei (76). Although some venom peptides from spiders and centipedes, which are members of the CHH superfamily, have an unusual number of amino acid residues between the third and fourth or fourth and fifth Cys residues, they share the common tertiary CHH superfamily scaffold (62, 77). Thus, this suggests that Maj-pCHH-B also possess a similar backbone fold.
As shown by our previous studies, six CHH-family peptides from M. japonicus XOSG inhibit the ovarian Maj-VG expression (28, 29). Hence, the receptor for the CHH-family of peptides is most likely found in the ovary. However, the report of functional CHH-family receptor molecule, in which specific ligand-receptor interaction is proved, is currently very limited (78, 79). Contig N13763 (Table 1) shares 37% amino acid sequence identity with the ITPL receptor in the silkworm Bombyx mori (BNGR-A24). Since the ITPL receptor has lower but definite affinity to ITP, N13763 should be investigated as a potential CHH-family receptor in future. In contrast, the contig N13763 shows higher similarity with the NPY receptor. Interestingly, the existence of NPY/F in crustacean species has been reported in some transcriptome analyses (43–49, 51, 52), and NPF has been suggested to stimulate ovarian development in M. rosenbergii (80). Therefore, NPF may have a similar function in M. japonicus. In L. vannamei, it is suggested that a receptor guanylyl cyclase (LvGC) is CHH receptor and is involved in the regulation of IAG expression (78). Two contigs encoding receptor guanylyl cyclase family found in the ovarian transcriptome in this study (N05324 and N33402, Table 1) do not contain the extracellular ligand domain, and their sequence similarities cannot be examined.
Recent advances in transcriptomic analysis have revealed multiple molecular species of an insulin family in a single crustacean species. For example, three ILPs (i.e., IAG, ILP, and relaxin-like) have been reported in L. vannamei, M. rosenbergii, and P. clarkii, respectively (48). Considering the present discovery of Maj-ILP1 and the result of phylogenetic analyses (Figure 3), there may be the third member of the relaxin-like molecular species in M. japonicus. Multiple ILPs have also been identified in insects such as B. mori (81), D. melanogaster (82), and the red flour beetle Tribolium castaneum (83). In T. castaneum ILP2 and ILP3 regulate VG expression in downstream juvenile hormone signaling (84), and in A. aegypti ILP3 controls egg production through the stimulation of yolk uptake and ecdysteroid production in the ovary (85). Arthropod ILP has been suggested to be a factor which links nutritional status and reproductive status (86, 87). Taken together, the female-specific and gonad-dominant Maj-ILP1 may also have such functions. Furthermore, our M. japonicus ovarian transcriptome contained a putative insulin receptor/IAGR (Table 1). Although Fenneropenaeus IAGR is not detected in any female tissues (53), the presence of IAGR homolog in M. japonicus ovary can account for the inhibitory effect of Maj-IAG on Maj-VG expression (31). The possibly important relationship between the insulin signaling pathway and reproduction should be studied in terms of the ligand as well as the receptor and downstream factors, such as the components of the signaling pathway which have been revealed to be conserved in M. japonicus by transcriptome analysis (Supplementary Figure 9).
In summary, we reported putative peptide hormones and receptors obtained through mRNA-sequencing analysis of the ovary of M. japonicus. Results of this study suggest a possible peripheral regulation by these hormones in the crustacean reproductive physiology (Figure 7). Factors involved in vitellogenesis regulation, ovary-specific expression patterns, and putative receptors for neuroparsin, ILP, and other peptide hormones are fascinating starting points for further detailed characterization. Above all, the effects of the ovarian hormones as well as central VIH on Maj-VG expression in the hepatopancreas should be addressed to outline the endocrine regulation of vitellogenesis. In addition, the transcriptome data generated in this work can also be utilized to further study hormonal functions. For example, target genes with expression patterns that are affected by VIH and other putative hormones can be efficiently searched using the combination with ex-vivo culture system and transcriptome analysis. Effective use of transcriptomic data from central and peripheral tissues will allow the comprehensive understanding of the regulatory mechanism of reproduction and other physiological processes in crustaceans.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, Data for DRA010103 is available in DDBJ Sequence Read Archive.
Author Contributions
NT conceived the idea for the project, conducted most of the experiments, and analyzed the results. YK and KI analyzed the transcriptomic data. NT and TS prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the Grants-in-Aid for Scientific Research (Nos. 15K07576 and 18K05819) from the Ministry of Education, Culture, Sport, Science, and Technology (MEXT) of Japan and by the SUNBOR GRANT from the Suntory Institute for Bioorganic Research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mrs. T. Shiokawa of the Division of Instrumental Analysis, Okayama University for her technical assistance in ESI mass spectrometry. We thank Dr. T. Okumura of the National Research Institute of Aquaculture, Japan Fisheries Research and Education Agency for his helpful suggestions in this work. We are grateful to Dr. Yasuhisa Kayano, former Director of the Research Institute for Fisheries Science, Okayama Prefectural Technology Center for Agriculture, Forestry, and Fisheries, for providing facilities to perform the MiSeq sequencing. Computations were partially performed on the NIG supercomputer at the ROIS National Institute of Genetics of Japan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2020.00541/full#supplementary-material
References
1. Zohar Y, Muñoz-Cueto JA, Elizur A, Kah O. Neuroendocrinology of reproduction in teleost fish. Gen Comp Endocrinol. (2010) 165:438–55. doi: 10.1016/j.ygcen.2009.04.017
2. Yaron Z, Levavi-Sivan B. Hormonal control of reproduction and growth | Endocrine Regulation of Fish Reproduction, In: Farrell AP, editor. Encyclopedia of Fish Physiology. Cambridge, MA: Academic Press. (2020). pp. 1500–8. doi: 10.1016/B978-0-12-374553-8.00058-7
3. Keller R. Crustacean neuropeptides: structures, functions and comparative aspects. Experientia. (1992) 48:439–48. doi: 10.1007/BF01928162
4. Webster SG, Keller R, Dircksen H. The CHH-superfamily of multifunctional peptide hormones controlling crustacean metabolism, osmoregulation, moulting, and reproduction. Gen Comp Endocrinol. (2012) 175:217–33. doi: 10.1016/j.ygcen.2011.11.035
5. Fernlund P, Josefsson L. Crustacean color-change hormone: amino acid sequence and chemical synthesis. Science. (1972) 177:173–5. doi: 10.1126/science.177.4044.173
6. Fernlund P. Structure of a light-adapting hormone from the shrimp, pandalus borealis. Biochim Biophys Acta. (1976) 439:17–25. doi: 10.1016/0005-2795(76)90155-0
7. Keller R, Wunderer G. Purification and amino acid composition of the neurosecretory hyperglycemic hormone from the sinus gland of the shore crab, Carcinus maenas. Gen Comp Endocrinol. (1978) 34:328–35. doi: 10.1016/0016-6480(78)90256-3
8. Webster SG. Amino acid sequence of putative moult-inhibiting hormone from the crab Carcinus maenas. Proc R Soc London Ser B Biol Sci. (1991) 244:247–52. doi: 10.1098/rspb.1991.0078
9. Soyez D, van Deijnen JE, Martin M. Isolation and characterization of a vitellogenesis-inhibiting factor from sinus glands of the lobster, Homarus americanus. J Exp Zool. (1987) 244:479–84. doi: 10.1002/jez.1402440314
10. Wainwright G, Webster SG, Wilkinson MC, Chung JS, Rees HH. Structure and significance of mandibular organ-inhibiting hormone in the crab, Cancer pagurus. involvement in multihormonal regulation of growth and reproduction. J Biol Chem. (1996) 271:12749–54. doi: 10.1074/jbc.271.22.12749
11. Zmora N, Chung JS. A novel hormone is required for the development of reproductive phenotypes in adult female crabs. Endocrinology. (2014) 155:230–9. doi: 10.1210/en.2013-1603
12. Chen T, Zhang L-P, Wong N-K, Zhong M, Ren C-H, Hu C-Q. Pacific white shrimp (Litopenaeus vannamei) vitellogenesis-inhibiting hormone (VIH) is predominantly expressed in the brain and negatively regulates hepatopancreatic vitellogenin (VTG) gene expression. Biol Reprod. (2014) 90:69–76. doi: 10.1095/biolreprod.113.115030
13. Sathapondecha P, Panyim S, Udomkit A. A novel function of bursicon in stimulation of vitellogenin expression in black tiger shrimp, Penaeus monodon. Aquaculture. (2015) 446:80–7. doi: 10.1016/j.aquaculture.2015.04.027
14. Tiu SH-K, Chan S-M. The use of recombinant protein and RNA interference approaches to study the reproductive functions of a gonad-stimulating hormone from the shrimp Metapenaeus ensis. FEBS J. (2007) 274:4385–95. doi: 10.1111/j.1742-4658.2007.05968.x
15. Yamano K, Qiu G-F, Unuma T. Molecular cloning and ovarian expression profiles of thrombospondin, a major component of cortical rods in mature oocytes of Penaeid shrimp, Marsupenaeus japonicas 1. Biol Reprod. (2004) 70:1670–8. doi: 10.1095/biolreprod.103.025379
16. Tsutsui N, Kim YK, Jasmani S, Ohira T, Wilder MN, Aida K. The dynamics of vitellogenin gene expression differs between intact and eyestalk ablated kuruma prawn Penaeus (Marsupenaeus) japonicus. Fish Sci. (2005) 71:249–56. doi: 10.1111/j.1444-2906.2005.00957.x
17. Tsutsui N, Kawazoe I, Ohira T, Jasmani S, Yang WJ, Wilder MN, et al. Molecular characterization of a cDNA encoding vitellogenin and its expression in the hepatopancreas and ovary during vitellogenesis in the kuruma prawn, Penaeus japonicus. Zoolog Sci. (2000) 17:651–60. doi: 10.2108/zsj.17.651
18. Qiu GF, Yamano K, Unuma T. Cathepsin C transcripts are differentially expressed in the final stages of oocyte maturation in kuruma prawn Marsupenaeus japonicus. Comp Biochem Physiol B Biochem Mol Biol. (2005) 140:171–81. doi: 10.1016/j.cbpc.2004.09.027
19. Okumura T, Kim YK, Kawazoe I, Yamano K, Tsutsui N, Aida K. Expression of vitellogenin and cortical rod proteins during induced ovarian development by eyestalk ablation in the kuruma prawn, Marsupenaeus japonicus. Comp Biochem Physiol A Mol Integr Physiol. (2006) 143:246–53. doi: 10.1016/j.cbpa.2005.12.002
20. Okumura T, Yamano K, Sakiyama K. Vitellogenin gene expression and hemolymph vitellogenin during vitellogenesis, final maturation, and oviposition in female kuruma prawn, Marsupenaeus japonicus. Comp Biochem Physiol A Mol Integr Physiol. (2007) 147:1028–37. doi: 10.1016/j.cbpa.2007.03.011
21. Kim YK, Tsutsui N, Kawazoe I, Okumura T, Kaneko T, Aida K. Localization and developmental expression of mRNA for cortical rod protein in kuruma prawn Marsupenaeus japonicus. Zool Sci. (2005) 22:675–80. doi: 10.2108/zsj.22.675
22. Yang W-J, Aida K, Nagasawa H. Amino acid sequences and activities of multiple hyperglycemic hormones from the kuruma prawn, Penaeus japonicus. Peptides. (1997) 18:479–85. doi: 10.1016/S0196-9781(96)00332-4
23. Yang W-J, Aida K, Terauchi A, Sonobe H, Nagasawa H. Amino acid sequence of a peptide with molt-inhibiting activity from the kuruma prawn Penaeus japonicus. Peptides. (1996) 17:197–202. doi: 10.1016/0196-9781(95)02122-1
24. Yang W-J, Aida K, Nagasawa H. Characterization of chromatophorotropic neuropeptides from the kuruma prawn Penaeus japonicus. Gen Comp Endocrinol. (1999) 114:415–24. doi: 10.1006/gcen.1999.7266
25. Yamano K, Unuma T. Expressed sequence tags from eyestalk of kuruma prawn, Marsupenaeus japonicus. Comp Biochem Physiol A Mol Integr Physiol. (2006) 143:155–61. doi: 10.1016/j.cbpa.2005.11.005
26. Ohira T, Katayama H, Tominaga S, Takasuka T, Nakatsuji T, Sonobe H, et al. Cloning and characterization of a molt-inhibiting hormone-like peptide from the prawn Marsupenaeus japonicus. Peptides. (2005) 26:259–68. doi: 10.1016/j.peptides.2004.09.003
27. Kotaka S, Ohira T. cDNA cloning and in situ localization of a crustacean female sex hormone-like molecule in the kuruma prawn Marsupenaeus japonicus. Fish Sci. (2018) 84:53–60. doi: 10.1007/s12562-017-1152-7
28. Tsutsui N, Nagakura-Nakamura A, Nagai C, Ohira T, Wilder MN, Nagasawa H. The ex vivo effects of eyestalk peptides on ovarian vitellogenin gene expression in the kuruma prawn Marsupenaeus japonicus. Fish Sci. (2013) 79:33–8. doi: 10.1007/s12562-012-0566-5
29. Tsutsui N, Katayama H, Ohira T, Nagasawa H, Wilder MN, Aida K. The effects of crustacean hyperglycemic hormone-family peptides on vitellogenin gene expression in the kuruma prawn, Marsupenaeus japonicus. Gen Comp Endocrinol. (2005) 144:232–9. doi: 10.1016/j.ygcen.2005.06.001
30. Banzai K, Ishizaka N, Asahina K, Suitoh K, Izumi S, Ohira T. Molecular cloning of a cDNA encoding insulin-like androgenic gland factor from the kuruma prawn Marsupenaeus japonicus and analysis of its expression. Fish Sci. (2011) 77:329–35. doi: 10.1007/s12562-011-0337-8
31. Katayama H, Kubota N, Hojo H, Okada A, Kotaka S, Tsutsui N, et al. Direct evidence for the function of crustacean insulin-like androgenic gland factor (IAG): Total chemical synthesis of IAG. Bioorg Med Chem. (2014) 22:5783–9. doi: 10.1016/j.bmc.2014.09.031
32. Rosen O, Manor R, Weil S, Gafni O, Linial A, Aflalo ED, et al. A sexual shift induced by silencing of a single insulin-like gene in crayfish: ovarian upregulation and testicular degeneration. PLoS ONE. (2010) 5:e15281. doi: 10.1371/journal.pone.0015281
33. Ventura T, Manor R, Aflalo ED, Weil S, Rosen O, Sagi A. Timing sexual differentiation: full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol Reprod. (2012) 86:1510–9. doi: 10.1095/biolreprod.111.097261
34. Ventura T, Manor R, Aflalo ED, Weil S, Raviv S, Glazer L, et al. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology. (2009) 150:1278–86. doi: 10.1210/en.2008-0906
35. Tsutsui N, Kotaka S, Ohira T, Sakamoto T. Characterization of distinct ovarian isoform of crustacean female sex hormone in the kuruma prawn Marsupenaeus japonicus. Comp Biochem Physiol A Mol Integr Physiol. (2018) 217:7–16. doi: 10.1016/j.cbpa.2017.12.009
36. Ling N, Ying S-Y, Ueno N, Shimasaki S, Esch F, Hotta M, et al. Pituitary FSH is released by a heterodimer of the β-subunits from the two forms of inhibin. Nature. (1986) 321:779–82. doi: 10.1038/321779a0
37. Robertson DM, Klein R, de Vos FL, McLachlan RI, Wettenhall REH, Hearn MTW, et al. The isolation of polypeptides with FSH suppressing activity from bovine follicular fluid which are structurally different to inhibin. Biochem Biophys Res Commun. (1987) 149:744–9. doi: 10.1016/0006-291X(87)90430-X
38. Vale W, Rivier J, Vaughan J, McClintock R, Corrigan A, Woo W, et al. Purification and characterization of an FSH releasing protein from porcine ovarian follicular fluid. Nature. (1986) 321:776–9. doi: 10.1038/321776a0
39. Ueno N, Ling N, Ying S-Y, Esch F, Shimasaki S, Guillemin R. Isolation and partial characterization of follistatin: a single-chain Mr 35,000 monomeric protein that inhibits the release of follicle-stimulating hormone. Proc Natl Acad Sci USA. (1987) 84:8282–6. doi: 10.1073/pnas.84.23.8282
40. Bathgate RAD, Samuel CS, Burazin TCD, Layfield S, Claasz AA, Reytomas IGT, et al. Human relaxin gene 3 (H3) and the equivalent mouse relaxin (M3) gene. novel members of the relaxin peptide family. J Biol Chem. (2002) 277:1148–57. doi: 10.1074/jbc.M107882200
41. Hisaw FL. Experimental relaxation of the pubic ligament of the guinea pig. Exp Biol Med. (1926) 23:661–3. doi: 10.3181/00379727-23-3107
42. Wilder M, Okumura T, Tsutsui N. Reproductive mechanisms in crustacea focusing on selected prawn species: vitellogenin structure, processing and synthetic control. Aqua-BioSci Monogr. (2010) 3:73–110. doi: 10.5047/absm.2010.00303.0073
43. Christie AE. Expansion of the Litopenaeus vannamei and Penaeus monodon peptidomes using transcriptome shotgun assembly sequence data. Gen Comp Endocrinol. (2014) 206:235–54. doi: 10.1016/j.ygcen.2014.04.015
44. Christie AE. Prediction of Scylla olivacea (Crustacea; Brachyura) peptide hormones using publicly accessible transcriptome shotgun assembly (TSA) sequences. Gen Comp Endocrinol. (2016) 230–231:1–16. doi: 10.1016/j.ygcen.2016.03.008
45. Christie AE, Chi M. Prediction of the neuropeptidomes of members of the Astacidea (Crustacea, Decapoda) using publicly accessible transcriptome shotgun assembly (TSA) sequence data. Gen Comp Endocrinol. (2015) 224:38–60. doi: 10.1016/j.ygcen.2015.06.001
46. Ventura T, Cummins SF, Fitzgibbon Q, Battaglene S, Elizur A. Analysis of the central nervous system transcriptome of the Eastern rock lobster Sagmariasus verreauxi reveals its putative neuropeptidome. PLoS ONE. (2014) 9:e97323. doi: 10.1371/journal.pone.0097323
47. Veenstra JA. The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen Comp Endocrinol. (2015) 224:84–95. doi: 10.1016/j.ygcen.2015.06.013
48. Veenstra JA. Similarities between decapod and insect neuropeptidomes. PeerJ. (2016) 4:e2043. doi: 10.7717/peerj.2043
49. Nguyen TV, Cummins SF, Elizur A, Ventura T. Transcriptomic characterization and curation of candidate neuropeptides regulating reproduction in the eyestalk ganglia of the Australian crayfish, Cherax quadricarinatus. Sci Rep. (2016) 6:38658. doi: 10.1038/srep38658
50. Powell D, Knibb W, Remilton C, Elizur A. De-novo transcriptome analysis of the banana shrimp (Fenneropenaeus merguiensis) and identification of genes associated with reproduction and development. Mar Genomics. (2015) 22:71–8. doi: 10.1016/j.margen.2015.04.006
51. Suwansa-ard S, Thongbuakaew T, Wang T, Zhao M, Elizur A, Hanna PJ, et al. In silico neuropeptidome of female Macrobrachium rosenbergii based on transcriptome and peptide mining of eyestalk, central nervous system and ovary. PLoS ONE. (2015) 10:e0123848. doi: 10.1371/journal.pone.0123848
52. Toullec J-Y, Corre E, Mandon P, Gonzalez-Aravena M, Ollivaux C, Lee C-Y. Characterization of the neuropeptidome of a Southern Ocean decapod, the antarctic shrimp Chorismus antarcticus: focusing on a new decapod ITP-like peptide belonging to the CHH peptide family. Gen Comp Endocrinol. (2017) 252:60–78. doi: 10.1016/j.ygcen.2017.07.015
53. Guo Q, Li S, Lv X, Xiang J, Sagi A, Manor R, et al. A putative insulin-like androgenic gland hormone receptor gene specifically expressed in male Chinese shrimp. Endocrinology. (2018) 159:2173–85. doi: 10.1210/en.2017-03253
54. Sharabi O, Manor R, Weil S, Aflalo ED, Lezer Y, Levy T, et al. Identification and characterization of an insulin-like receptor involved in crustacean reproduction. Endocrinology. (2016) 157:928–41. doi: 10.1210/en.2015-1391
55. Uengwetwanit T, Ponza P, Sangsrakru D, Wichadakul D, Ingsriswang S, Leelatanawit R, et al. Transcriptome-based discovery of pathways and genes related to reproduction of the black tiger shrimp (Penaeus monodon). Mar Genomics. (2018) 37:69–73. doi: 10.1016/j.margen.2017.08.007
56. Chen K, Li E, Li T, Xu C, Wang X, Lin H, et al. Transcriptome and molecular pathway analysis of the hepatopancreas in the pacific white shrimp litopenaeus vannamei under chronic low-salinity stress. PLoS ONE. (2015) 10:e0131503. doi: 10.1371/journal.pone.0131503
57. Chan S-M, Rankin SM, Keeley LL. Characterization of the molt stages in penaeus vannamei: setogenesis and hemolymph levels of total protein, ecdysteroids, and glucose. Biol Bull. (1988) 175:185–92. doi: 10.2307/1541558
58. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. (2013) 8:1494–512. doi: 10.1038/nprot.2013.084
59. Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. (2008) 36:3420–35. doi: 10.1093/nar/gkn176
60. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. (2005) 21:3674–6. doi: 10.1093/bioinformatics/bti610
61. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. (2011) 8:785–6. doi: 10.1038/nmeth.1701
62. Tsutsui N, Sakamoto T, Arisaka F, Tanokura M, Nagasawa H, Nagata K. Crystal structure of a crustacean hyperglycemic hormone (CHH) precursor suggests structural variety in the C-terminal regions of CHH superfamily members. FEBS J. (2016) 283:4325–39. doi: 10.1111/febs.13926
63. Inoue H, Tsutsui N, Nagai C, Nagata K, Tanokura M, Nagasawa H. Crystallization and preliminary X-ray analysis of crustacean hyperglycaemic hormone from the kuruma prawn Marsupenaeus japonicus in its weakly active precursor form. Acta Crystallogr Sect F Struct Biol Cryst Commun. (2011) 67:1586–9. doi: 10.1107/S1744309111040140
64. Katayama H, Ohira T, Aida K, Nagasawa H. Significance of a carboxyl-terminal amide moiety in the folding and biological activity of crustacean hyperglycemic hormone. Peptides. (2002) 23:1537–46. doi: 10.1016/S0196-9781(02)00094-3
65. Yang SP, He J-G, Sun CB, Chan SF. Characterization of the shrimp neuroparsin (MeNPLP): RNAi silencing resulted in inhibition of vitellogenesis. FEBS Open Bio. (2014) 4:976–86. doi: 10.1016/j.fob.2014.09.005
66. Vogel KJ, Brown MR, Strand MR. Ovary ecdysteroidogenic hormone requires a receptor tyrosine kinase to activate egg formation in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. (2015) 112:5057–62. doi: 10.1073/pnas.1501814112
67. Katayama H, Ohira T, Nagasawa H. Crustacean peptide hormones: structure, gene expression and function. Aqua-BioScience Monogr. (2013) 6:49–90. doi: 10.5047/absm.2013.00602.0049
68. Girardie J, Boureme D, Couillaud F, Tamarelle M, Girardie A. Anti-juvenile effect of neuroparsin A, a neuroprotein isolated from locust corpora cardiaca. Insect Biochem. (1987) 17:977–83. doi: 10.1016/0020-1790(87)90106-5
69. Brown MR, Graf R, Swiderek KM, Fendley D, Stracker TH, Champagne DE, et al. Identification of a steroidogenic neurohormone in female mosquitoes. J Biol Chem. (1998) 273:3967–71. doi: 10.1074/jbc.273.7.3967
70. Saetan U, Sangket U, Deachamag P, Chotigeat W. Ovarian transcriptome analysis of vitellogenic and non-vitellogenic female banana shrimp (Fenneropenaeus merguiensis). PLoS ONE. (2016) 11:e0164724. doi: 10.1371/journal.pone.0164724
71. Chung JS, Katayama H, Dircksen H. New functions of arthropod bursicon: inducing deposition and thickening of new cuticle and hemocyte granulation in the blue crab, callinectes sapidus. PLoS ONE. (2012) 7:e46299. doi: 10.1371/journal.pone.0046299
72. Luo C-W, Dewey EM, Sudo S, Ewer J, Hsu SY, Honegger H-W, et al. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc Natl Acad Sci USA. (2005) 102:2820–5. doi: 10.1073/pnas.0409916102
73. Johard HAD, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Förster C, et al. Peptidergic clock neurons in Drosophila : ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol. (2009) 516:59–73. doi: 10.1002/cne.22099
74. Dai L, Zitnan D, Adams ME. Strategic expression of ion transport peptide gene products in central and peripheral neurons of insects. J Comp Neurol. (2007) 500:353–67. doi: 10.1002/cne.21192
75. Montagné N, Desdevises Y, Soyez D, Toullec J-Y. Molecular evolution of the crustacean hyperglycemic hormone family in ecdysozoans. BMC Evol Biol. (2010) 10:62. doi: 10.1186/1471-2148-10-62
76. Ventura-López C, Gómez-Anduro G, Arcos FG, Llera-Herrera R, Racotta IS, Ibarra AM. A novel CHH gene from the Pacific white shrimp Litopenaeus vannamei was characterized and found highly expressed in gut and less in eyestalk and other extra-eyestalk tissues. Gene. (2016) 582:148–60. doi: 10.1016/j.gene.2016.02.011
77. Undheim EAB, Grimm LL, Low C-F, Morgenstern D, Herzig V, Zobel-Thropp P, et al. Weaponization of a hormone: convergent recruitment of hyperglycemic hormone into the venom of arthropod predators. Structure. (2015) 23:1283–92. doi: 10.1016/j.str.2015.05.003
78. Guo Q, Li S, Lv X, Xiang J, Manor R, Sagi A, et al. Sex-biased CHHs and their putative receptor regulate the expression of IAG gene in the shrimp Litopenaeus vannamei. Front Physiol. (2019) 10:1525. doi: 10.3389/fphys.2019.01525
79. Nagai C, Mabashi-Asazuma H, Nagasawa H, Nagata S. Identification and characterization of receptors for ion transport peptide (ITP) and ITP-like (ITPL) in the silkworm Bombyx mori. J Biol Chem. (2014) 289:32166–77. doi: 10.1074/jbc.M114.590646
80. Tinikul Y, Engsusophon A, Kruangkum T, Thongrod S, Tinikul R, Sobhon P. Neuropeptide F stimulates ovarian development and spawning in the female giant freshwater prawn, Macrobrachium rosenbergii, and its expression in the ovary during ovarian maturation cycle. Aquaculture. (2017) 469:128–36. doi: 10.1016/j.aquaculture.2016.11.026
81. Mizoguchi A, Okamoto N. Insulin-like and IGF-like peptides in the silkmoth Bombyx mori: discovery, structure, secretion, and function. Front Physiol. (2013) 4:217. doi: 10.3389/fphys.2013.00217
82. Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. (2006) 51:1–24. doi: 10.1146/annurev.ento.51.110104.151011
83. Li B, Predel R, Neupert S, Hauser F, Tanaka Y, Cazzamali G, et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. (2008) 18:113–22. doi: 10.1101/gr.6714008
84. Sheng Z, Xu J, Bai H, Zhu F, Palli SR. Juvenile hormone regulates vitellogenin gene expression through insulin-like peptide signaling pathway in the red flour beetle, Tribolium castaneum. J Biol Chem. (2011) 286:41924–36. doi: 10.1074/jbc.M111.269845
85. Brown MR, Clark KD, Gulia M, Zhao Z, Garczynski SF, Crim JW, et al. An insulin-like peptide regulates egg maturation and metabolism in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. (2008) 105:5716–21. doi: 10.1073/pnas.0800478105
86. Badisco L, van Wielendaele P, Vanden Broeck J. Eat to reproduce: a key role for the insulin signaling pathway in adult insects. Front Physiol. (2013) 4:202. doi: 10.3389/fphys.2013.00202
Keywords: peptide hormone, Marsupenaeus japonicus, ovary, reproduction, transcriptome, vitellogenesis
Citation: Tsutsui N, Kobayashi Y, Izumikawa K and Sakamoto T (2020) Transcriptomic Analysis of the Kuruma Prawn Marsupenaeus japonicus Reveals Possible Peripheral Regulation of the Ovary. Front. Endocrinol. 11:541. doi: 10.3389/fendo.2020.00541
Received: 24 April 2020; Accepted: 06 July 2020;
Published: 19 August 2020.
Edited by:
Haihui Ye, Xiamen University, ChinaReviewed by:
Shihao Li, Institute of Oceanology, Chinese Academy of Sciences, ChinaShogo Haraguchi, Showa University, Japan
Copyright © 2020 Tsutsui, Kobayashi, Izumikawa and Sakamoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naoaki Tsutsui, dHN1MkBiaW8ubWllLXUuYWMuanA=
 Naoaki Tsutsui
Naoaki Tsutsui Yasuhisa Kobayashi
Yasuhisa Kobayashi Kouichi Izumikawa4
Kouichi Izumikawa4 Tatsuya Sakamoto
Tatsuya Sakamoto