
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 04 August 2020
Sec. Systems Endocrinology
Volume 11 - 2020 | https://doi.org/10.3389/fendo.2020.00528
 Marta Greco1†
Marta Greco1† Eusebio Chiefari1†
Eusebio Chiefari1† Francesca Accattato1
Francesca Accattato1 Domenica M. Corigliano1
Domenica M. Corigliano1 Biagio Arcidiacono1
Biagio Arcidiacono1 Maria Mirabelli1
Maria Mirabelli1 Rossella Liguori1
Rossella Liguori1 Francesco S. Brunetti1
Francesco S. Brunetti1 Salvatore A. Pullano1
Salvatore A. Pullano1 Vincenzo Scorcia2
Vincenzo Scorcia2 Antonino S. Fiorillo1
Antonino S. Fiorillo1 Daniela P. Foti1
Daniela P. Foti1 Antonio Brunetti1*
Antonio Brunetti1*Objective: Recently, the role of circulating miRNAs as non-invasive biomarkers for the identification and monitoring of diabetes microvascular complications has emerged. Herein, we aimed to: identify circulating miRNAs differentially expressed in patients with and without diabetic retinopathy (DR); examine their predictive value; and understand their pathogenic impact.
Methods: Pooled serum samples from randomly selected matched patients with type 2 diabetes, either with or without DR, were used for initial serum miRNA profiling. Validation of the most relevant miRNAs was thereafter conducted by RT-qPCR in an extended sample of patients with DR and matched controls.
Results: Following miRNA profiling, 43 miRNAs were significantly up- or down-regulated in patients with DR compared with controls. After individual validation, 5 miRNAs were found significantly overexpressed in patients with DR. One of them, miR-1281, was the most up-regulated and appeared to be specifically related to DR. Furthermore, secreted levels of miR-1281 were increased in high glucose-cultured retinal cells, and there was evidence of a potential link between glucose-induced miR-1281 up-regulation and DR.
Conclusion: Our findings suggest miR-1281 as a circulating biomarker of DR. Also, they highlight the pathogenic significance of miR-1281, providing insights for a new potential target in treating DR.
Type 2 diabetes mellitus is an expanding global health problem, with an estimated prevalence of 8.8% in the general adult population (1), and a prediction of over 500 million affected people in the world by year 2030 (2). Chronic hyperglycemia in both type 1 and type 2 diabetes is associated with the development of long-term microvascular and macrovascular complications that may seriously compromise the patients' quality of life and life expectancy, and can contribute to high healthcare burden and costs (3).
Among diabetes-related microvascular complications (namely retinopathy, neuropathy and nephropathy), diabetic retinopathy (DR) is one of the most devastating and frequent injuries, affecting over 30% of patients with diabetes, and one of the leading causes of vision loss in the working-age population (4, 5). It is frequently the first microvascular complication to appear, and the risk of developing DR is directly related to the duration of diabetes and to the level of metabolic control (6).
In the recent years, a big effort has been made to identify novel biomarkers for both the prediction of diabetes (in particular type 2 diabetes) in individuals at risk of developing the disease, and for preventing the development of diabetes-related long-term complications. Several emerging lines of evidence in this field point to the relevance of circulating microRNAs (miRNAs) as disease markers (7, 8). MiRNAs are an evolutionarily conserved class of short non-coding single-stranded RNAs, which target over one-half of protein-coding transcripts (9, 10). They are involved in fundamental biological processes, such as cell differentiation and proliferation, apoptosis, tissue development, senescence and metabolism (9, 10). Recently, miRNAs have been implicated in the pathophysiology of many human diseases, including type 2 diabetes (10–12), as well as in the inflammatory and endothelial dysfunction that can trigger the development of vascular complications in diabetes (13–16). The application of circulating miRNAs as biomarkers for DR has gained much attention in recent years. In particular, at this level, miRNAs have been implicated in aberrant angiogenic growth of retinal endothelial cells, by adversely affecting the levels of vascular endothelial growth factor A (VEGFA) (17), a secreted mitogen which seems to play a role in DR, by inducing neovascularization and vascular permeability of retinal vessels (4, 18, 19). Research studies in this context have focused on a variety of in vivo and in vitro retinal models (20–22), as well as on human biological fluids from diabetic retinopathic patients (7, 23–26). However, little consistency emerges across these studies, such that solid conclusions cannot yet be made concerning the relationship between miRNAs and DR.
In the present work, we aimed at investigating miRNA expression patterns in serum samples of highly selected patients with type 2 diabetes, with and without DR. Among the newly identified miRNAs was miR-1281, which was found to be strongly associated with DR, and whose pathogenic role in this diabetic complication was also supported by a mechanistic explanation.
A case-control study for the identification of DR-associated plasma miRNA(s) was carried out in two steps. In the first step, an initial screening for serum miRNA profiling was performed. On a second step, we dealt with the individual validation of the most relevant miRNAs. The study was approved by the local ethics committee (Comitato etico regionale, Sezione Area Centro, Catanzaro, Italy, Protocol n. 116, May 14, 2015), and all participants gave written informed consent.
In the first step of the study, 5 unrelated men with type 2 diabetes and non-proliferative DR, in the absence of other diabetic complications, were enrolled at the Operative Unit of Endocrinology (University “Magna Græcia,” Catanzaro, Italy). A control group of 5 unrelated men with diabetes, but without DR, matched with the case group for clinical and biochemical features [age, BMI, duration of diabetes, blood pressure, glycated hemoglobin (HbA1c), lipid profile], in the absence of other disorders, and receiving the same medical treatment (metformin only), was recruited at the same time period from the same outpatient clinic (Table 1). At this step, only male patients were included to maximize matching between the two groups in consideration of the greater presence and severity of DR in men (27), and the known effects of sex hormones on miRNAs (28, 29). In the second step, 15 further unrelated patients with type 2 diabetes and DR, in the absence of other long-term complications, and 25 matched unrelated patients with type 2 diabetes, but without DR, were included (Table 1). In all participants, type 2 diabetes was diagnosed according to the American Diabetes Association criteria (30), whereas the diagnosis of non-proliferative DR was based on ophthalmological examination of the ocular fundus following dilation of the pupils, as reported before (19). Severity of DR was graded based on the worst eye, according to the International Clinical Diabetic Retinopathy Disease Severity Scale (31). Subjects affected by proliferative DR, or with pan-retinal photocoagulation, were excluded from the study. Diabetic nephropathy was excluded on the basis of an estimated glomerular filtration rate (eGFR) > 60 mL/min/1.73 m2, according to the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) formula (32), and on the absence of microalbuminuria (33). Neuropathy and macrovascular complications were excluded after appropriate clinical and instrumental examinations.
Obesity (BMI ≥ 30 kg/m2), ongoing inflammatory diseases [erythrocyte sedimentation rate (ESR) > 30 mm/h, and high-sensitivity C-reactive protein (hs-CRP) > 6 mg/L] and other intercurrent illnesses, poor glycemic control (HbA1c > 8%), smoking, insulin therapy and corticosteroid treatment were exclusion criteria.
All patients underwent anthropometric determinations and blood collection (after a 12 h overnight fasting) for the evaluation of fasting glycemia, HbA1c, lipid profile (total and HDL-cholesterol, and triglycerides), creatinine, and the inflammatory markers ESR and hs-CRP. Microalbuminuria was evaluated according to ADA recommendations (33).
For miRNA studies, approximately 10 mL of venous blood were collected from each patient after 10-12 h fasting in a serum separator tube (Vacutainer® SST™) and centrifuged within 1 h at 2,000 x g for 15 min at 4°C. Serum fractions were rapidly aliquoted into cryovial tubes and stored at −80°C for subsequent use. Prior to further processing, serum aliquots were thawed on ice, and subjected to a second centrifugation at 16,000 × g for 5 min at 4°C to discard remaining insoluble material. At this stage, samples were checked spectrophotometrically at 414 nm to exclude hemolysis.
Serum-circulating miRNAs were extracted from all patients enrolled in the study as enriched low molecular-weight RNA (LMW RNA), using a commercial column-based system (miRNeasy Serum/Plasma Kit, Qiagen). Briefly, for miRNA extraction, 200 μL of serum samples were used, and procedures were those recommended by the manufacturer, including the addition of synthetic exogenous C. elegans miR-39 for the evaluation of both miRNA extraction efficiency and data normalization. After miRNA quantification and purity control (A260/280 ratio = 1.9–2.1 and A260/230 ratio = 2.0–2.2) with a spectrometric method (Nanodrop 1000, Thermo Scientific), yielding between 14 and 30 ng RNA/μL (elution volume = 12 μL), equal amounts (100 ng) of RNA extracted from serum samples of well-selected, matched patients with type 2 diabetes, either with (n = 5) or without (n = 5) DR, were pooled and run at the same time. The LMW RNA fraction (a total of 500 ng for each RNA pool) was used as starting material for a subsequent one-step universal tailing reverse transcription reaction, carried out in a 100 μL reaction volume, using miScript II RT Kit (Qiagen) and a thermocycler system (GeneAmp 2700 Thermal Cycler, Applied Biosystems). MiRNA profiling of the two pools was performed by RT-qPCR on a 384 well-array, containing spotted forward primers for 372 miRNAs with detectable expression in serum (miScript miRNA PCR Array Human Serum & Plasma 384HC, Qiagen; Supplementary Table 1) and SYBR Green chemistry (miScript SYBR Green PCR Kit, Qiagen). Spike-in C. elegans miR-39, and 6 reference controls (SNORD61, SNORD68, SNORD72, SNORD95, SNORD96A, and RNU6B/RNU6-2) were used as normalizers. Reactions were carried out on QuantStudio™ 12K Flex Real-Time PCR System (Applied Biosystems), as indicated by the manufacturer's protocol. A free online data analysis software (https://geneglobe.qiagen.com/it/) was used for quality assessment and miRNA expression analysis. Fold-change values >2 and <0.5 were considered significant. MiRNAs with CT values > 30 were excluded.
LMW RNA was extracted from serum as described above. After reverse transcription reaction using 50 ng RNA as a template (miScript II RT Kit, Qiagen), individual validation of selected miRNAs was performed using RT-qPCR in all enrolled individuals (miScript SYBR Green PCR Kit, Qiagen; QuantStudio™ 12K Flex Real-Time PCR System, Applied Biosystems). Protocols were those indicated by the manufacturer. Normalization was obtained using the spike-in control C. elegans miR-39. All samples were tested in triplicates. A free online data analysis software (https://geneglobe.qiagen.com/it/) was used for quality assessment and miRNA expression analysis.
Human retinal pigment epithelial cells (ARPE-19) were cultured in Dulbecco's Modified Eagle Medium (DMEM)/F12 (Sigma Aldrich) supplemented with 10% fetal bovine serum (FBS) and 2 mM L-glutamine; human umbilical vein endothelial cells (HUVECs) were grown in ECM medium (ScienCell Research Laboratories), with the addition of 5% FBS. Penicillin (100 U/mL) and streptomycin (100 μg/mL) were added to each culture medium. All cultured cells were kept at 37°C in a humidified incubator with 5% CO2 atmosphere. For glucose treatment experiments, ARPE-19 and HUVEC cells were exposed to 5 or 25 mM glucose, and VEGFA expression was evaluated after 48 h incubation. Mycoplasma contamination was routinely evaluated, and only mycoplasma-free cells were used.
RNA extraction from ARPE-19 and HUVEC cells was performed using the Trizol reagent (Thermo Fisher Scientific) according to the manufacturer's protocol, followed by DNase treatment (Ambion) to eliminate DNA contaminations. cDNAs were obtained from 1 μg of total RNA, using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Sequence-specific primers for human VEGF and RPS9 were as already reported (19, 34). An Eppendorf Mastercycler ep realplex ES real-time thermocycler was used to perform RT-qPCR (19). SYBR Green fluorescence was measured and relative quantification was made against the RPS9 cDNA, used as an internal standard. All PCR reactions were performed in triplicate.
Extraction of the LMW RNA fraction containing miRNAs was obtained by a commercial column-based system (RNeasy MinElute Spin Columns, Qiagen), suitable for both cell supernatants and lysates. In our experimental conditions, 800 μL of supernatant or 700 μL of cell lysates (from 2 × 106 cells) were loaded per column. Fifty nanograms of RNA were used as template for reverse transcription using the miScript II RT Kit (Qiagen) and a thermocycler system (GeneAmp 2700 Thermal Cycler, Applied Biosystems). RT-qPCR was performed using miScript SYBR Green PCR Kit (Qiagen) and Eppendorf Mastercycler ep realplex ES real-time thermocycler (Eppendorf). Normalization was obtained using the spike-in control C. elegans miR-39.
Total cellular extracts were obtained as previously described (35), and Western blot analyses were performed to determine VEGFA protein expression in both ARPE-19 and HUVEC cells, using an anti-VEGFA specific antibody (sc-507, Santa Cruz Biotech).
The human VEGFA promoter construct, pGL3-2.6 (a kind gift from Dr. Sheehy, University College Dublin, Ireland) was transiently transfected into HUVEC cells, using LipofectAMINE 2000 reagent (Invitrogen Life Technology), and luciferase activity was assessed 48 h later using the dual-luciferase reporter assay system (Promega). Transient transfection of custom-synthesized miR-1281 mimic (miScript miRNA Mimics, Qiagen) was carried out using 5 nM mimic in the presence of HiPerFect Transfection Reagent (Qiagen), according to the manufacturer's protocol. VEGF gene expression was assessed 48 h post-transfection.
HUVEC and ARPE-19 cells were seeded in 12-well plates (0.2 × 106 cells/well, and 0.1 × 106 cells/well, respectively) to obtain confluency within 24 h, transfected with 150 ng (5 nM) miR-1281 mimic, and maintained in high glucose (25 mM) medium for 48 h, after which a wound was mechanically produced by scratching cell monolayers with a sterile 200-μL pipette tip. Cell migration was evaluated by quantitative image analysis of wound closure at 0, 4, 8, and 12 h for HUVEC cells, and up to 30 h for ARPE-19 cells after injury, using the ImageJ software (36). Percent of wound closure was calculated using the following equation:
Initially, each quantitative trait was tested for normality of distribution, using the Shapiro-Wilk normality test and, when required, it was log-transformed. Data were expressed as mean ± standard deviation (SD). The non-parametric Mann-Whitney U-test was used to compare intergroup differences in serum miRNA expression and other biochemical parameters. Spearman rank correlation analysis was used to explore the correlation between semiquantitative concentrations of screened miRNAs and clinical and biochemical parameters. Selected miRNAs were then forced in multivariable regression model adjusted for appropriate covariates. Receiver operating characteristic (ROC) analysis was performed to assess the discriminative capacity of selected miRNAs in identifying DR. A p < 0.05 was considered statistically significant. Data were analyzed using SPSS software version 20 (SPSS Inc.) and the miScript miRNA PCR array data analysis software (https://geneglobe.qiagen.com/it/).
We first performed miRNA profiling on pooled sera from a subgroup of 5 well-selected male patients with type 2 diabetes and non-proliferative DR (cases), and 5 matched patients with type 2 diabetes, but without DR (controls). Among the 372 miRNAs analyzed, 40 were up-regulated and 3 were down-regulated in pooled cases vs. pooled controls, following the threshold of ≤30 CT value and at least 2 average fold change (Table 2). In an attempt to perform a more stringent selection of differentially expressed miRNAs, only those with at least a 5-fold change in expression were chosen for the next step. Therefore, 23 potential miRNA candidates for DR were analyzed by RT-qPCR in an individual validation analysis, in which 15 further new cases and 25 new age- and BMI-matched controls were added to those analyzed in the first step of the study. As shown in Figure 1A, 5 of the 23 selected miRNAs (miR-1281, miR-4687-5p, miR-4688, miR-1260a, and miR-766-3p) showed a significant up-regulation in patients with DR compared to control patients. In accordance with the data retrieved from array-profiling, miR-1281 showed the largest fold change (over 9 average) (Figure 1A). On Spearman univariate correlation analysis, miR-4687-5p and miR-766-3p were correlated with female gender (ρ = 0.344, p = 0.018; ρ = 0.376, p = 0.009, respectively) and BMI (ρ = 0.400, p = 0.005; ρ = 0.322, p = 0.027, respectively), whereas miR-4688 and miR-1260a were correlated with BMI only (ρ = 0.426, p = 0.002; ρ = 0.298, p = 0.049, respectively). Interestingly, miR-1281 expression was significantly correlated solely with DR (ρ = 0.783), p < 0.001), thereby suggesting specificity for this diabetic complication. The association of miR-1281 with DR was further corroborated by multiple linear regression analysis (beta = 0.689, t = 6.632, p < 0.001), in which age, sex, BMI, and duration of diabetes were added as covariates.
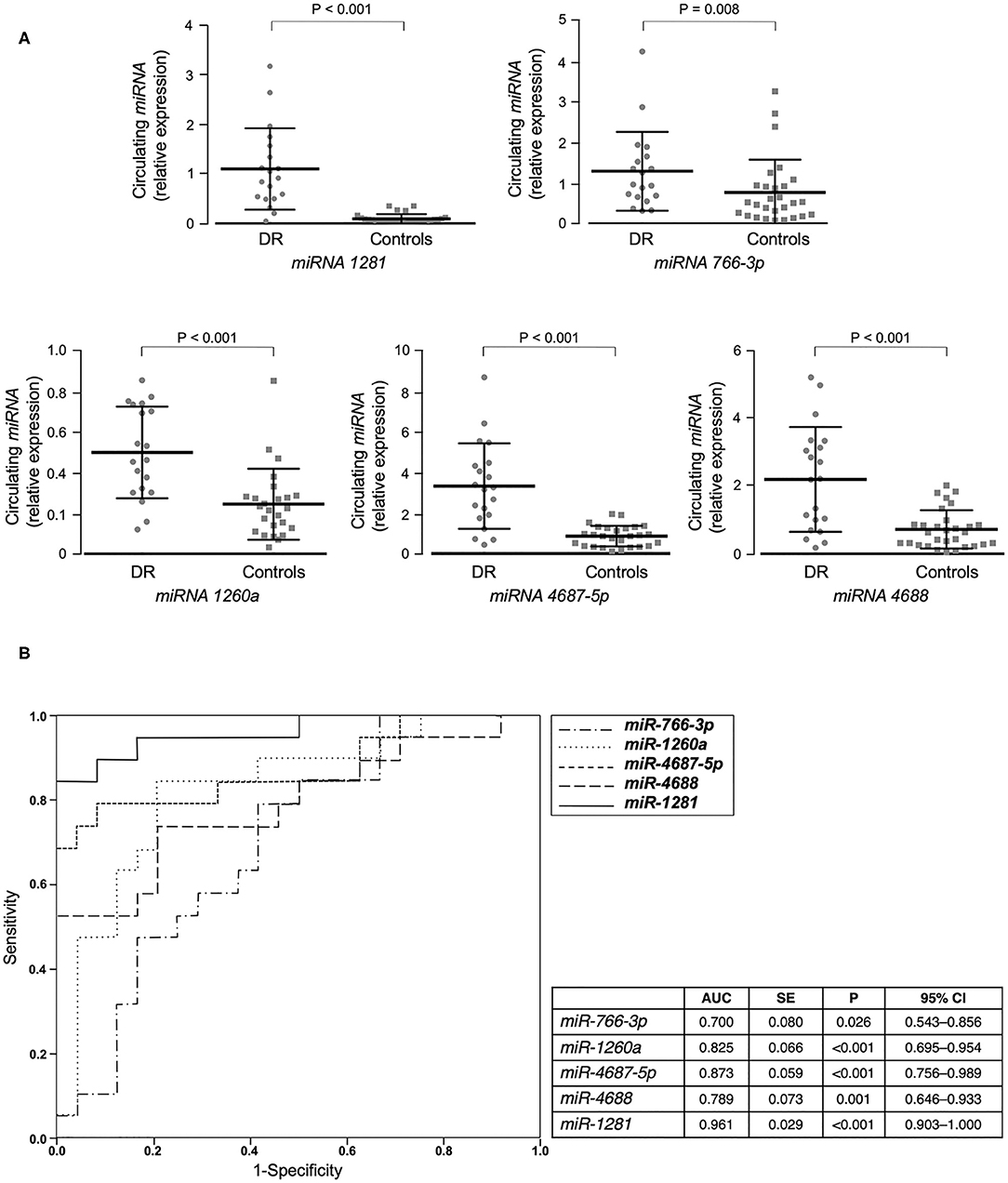
Figure 1. miRNAs validation and miRNA's accuracy. (A) Mean ± SD of validated circulating miRNAs in the serum of patients with type 2 diabetes, with (DR, n = 20) and without (controls, n = 25) DR. Expression levels of selected miRNAs were analyzed by RT-qPCR. The relative quantification measures the concentration of a specific miRNA and is calculated as 2^(−ΔCt) for each sample (37). ΔCt values are calculated as the raw Ct value—average of raw Ct values for the selected reference miRNA (cel-miR-39-3p) in each sample. Non-parametric Mann-Whitney test was used for comparisons among two groups. (B) ROC curves of validated miRNAs for the prediction of DR. Both sensitivity and specificity are indicated. AUC, area under curve; SE, standard error; CI, confidence interval.
Furthermore, ROC curve analyses were performed to assess sensitivity and specificity of selected circulating miRNAs in detecting DR. As shown in Figure 1B, all five selected miRNAs showed a significant area under the curve (AUC) and, among them, miR-1281 stood out as the best predictor of DR [AUC = 0.965 (0.902–1.000), p < 0.001], thus suggesting this miRNA as a potential novel biomarker for early diagnosis of DR.
To investigate the potential role of miR-1281 in retinal damage, we first evaluated whether miR-1281 could be affected by glucose levels in vitro, in both ARPE-19 retinal epithelial cells and HUVEC endothelial cells exposed for 48 h to high (25 mM) glucose, a condition simulating that observed during sustained hyperglycemia. As shown in Figure 2A, miR-1281 was increased in the supernatant medium of cultured ARPE-19 cells, and decreased in the intracellular fraction in response to high glucose (p < 0.001), as measured by RT-qPCR. Under the same experimental conditions as those used for ARPE-19 cells, no differences in miR-1281 levels were observed in the extracellular fraction of HUVEC cells in relation to media glucose concentration, whereas miR-1281 was down-regulated in the intracellular location of HUVEC cells treated with high glucose (Figure 2B). In both cell lines, exposure to high glucose resulted in the up-regulation of VEGFA mRNA and protein expression compared to cells exposed to low (5 mM) glucose (p < 0.05 for ARPE cells; p < 0.01 for HUVEC cells) (Figure 2C), suggesting that a link exists between glucose levels, the release of miR-1281, and the expression of VEGFA.
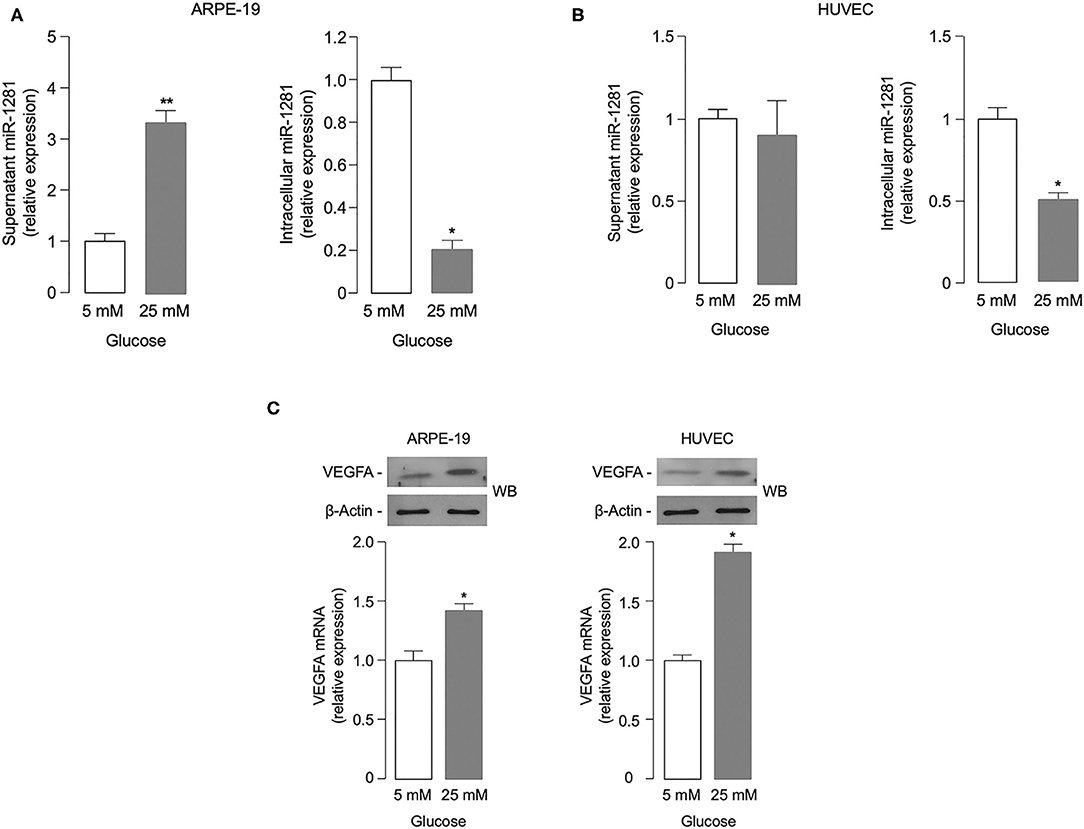
Figure 2. Effect of glucose on miR-1281 and VEGFA expression. (A,B) ARPE-19 epithelial cells and HUVEC endothelial cells were exposed to low (5 mM) or high (25 mM) glucose, and both extracellular (white bar) and intracellular (gray bar) levels of miR-1281 were obtained after 48 h incubation. Results are the mean ± SEM of three independent experiments, each in triplicate. *p < 0.05; **p < 0.001. (C) VEGFA mRNA and protein expression in ARPE-19 and HUVEC cells, as measured by RT-qPCR (bar graphs) and western blot (WB), respectively. mRNA levels are shown as means ± SEM of three independent experiments, each performed in triplicate. *p < 0.05. Representative WBs of VEGFA protein in both cell types are representative of at least 3 independent experiments. β-Actin, control of protein loading.
Based on the involvement of VEGFA in DR (4), it was intriguing to hypothesize that miR-1281 could be related to the microangiopathic damage of retinal vessels by up-regulating the expression of VEGFA, whose intimate role in retinal neovascularization and retinal hemorrhage is well-established (21). In an attempt to test this hypothesis, we examined the effect of miR-1281-enriched medium from in vitro cell cultures on VEGFA expression. As shown in Figure 3A, treatment with increasing amounts of conditioned medium from ARPE-19 epithelial cells exposed to high glucose caused a rise in both VEGFA mRNA and protein expression in HUVEC endothelial cells, thereby consistently supporting the hypothesis that the adverse effect of high glucose on retinal endothelial cell injury can be mediated, at least in part, by miR-1281 via VEGFA.
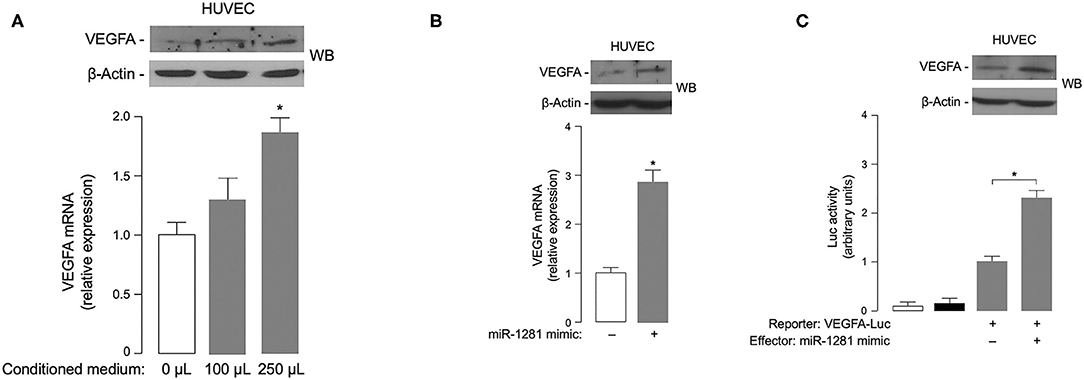
Figure 3. Effects of miR-1281 on VEGFA expression and cell migration. (A) VEGFA expression in HUVEC cells exposed to increasing amounts (0, 100, 250 μL) of conditioned medium from ARPE-19 cells, which underwent high glucose-induced miR-1281. VEGFA mRNA and protein levels were measured 48 h later by RT-qPCR and WB, respectively. VEGFA mRNA data are expressed as means ± SEM of three independent measurements, each performed in triplicate. *p < 0.05 vs. control (white bar). A representative WB of VEGFA out of three independent experiments is shown in the autoradiogram. β-actin, control of protein loading. (B) VEGFA mRNA and protein levels were measured as in (A), in HUVEC cells untreated or pretreated with 150 ng miR-1281 mimic for 48 h. Data for RT-qPCR and WB are representative of at least 3 independent experiments. *p < 0.05 vs. control (white bar). (C) Human VEGFA-Luc reporter vector (2 μg) was transfected into HUVEC cells, in the absence or presence of miR-1281 mimic (150 ng), and Luc activity was measured 48 h later. Data are means ± SEM for three separate transfections. Values are expressed as the factor of increase above the level of Luc activity obtained in cells transfected with VEGFA-Luc reporter vector in the absence of miR-1281 mimic (control), which is assigned an arbitrary value of 1. White bar, mock (no DNA); black bar, pGL3-basic (effector vector without an insert). *p < 0.05 vs. control. A representative WB of endogenous VEGFA in untreated and miR-1281 mimic-treated cells is shown in the autoradiogram.
To explore the possibility that miR-1281 independently directed VEGFA gene expression, transient transfection studies were undertaken using miR-mimic 1281 in HUVEC cells. As shown in Figure 3B, VEGFA mRNA expression was considerably increased in miR-mimic treated HUVEC cells as compared to miR-mimic negative control transfected cells. The increase in mRNA abundance paralleled the increase in VEGFA protein expression as detected by Western blot analysis of nuclear extracts from HUVEC cells transfected with miR-mimic 1281 (Figure 3B). Next, to test whether the VEGFA gene promoter could be transcriptionally regulated by miR-1281, HUVEC cells were transiently co-transfected with the luciferase (Luc) reporter plasmid VEGF 2.6-Luc, together with miR-1281 mimic. Under these experimental treatments, miR-1281 induced a significant increase in VEGF-Luc activity, which was paralleled by an increase in VEGFA protein abundance (Figure 3C), thereby indicating that miR-1281 positively regulates VEGFA protein expression through activation of VEGFA gene transcription.
To investigate the biological role of miR-1281 in relation to the molecular effects described on VEGFA expression, we finally performed wound healing assays in HUVEC cells to study cell migration and cell interactions in vitro. After cell transfection with miR-1281 mimic, we analyzed the closure of wounds mechanically created on the HUVEC cell monolayer as a surrogate of cell migration during wound healing in vivo. The images from cell scratch assay showed that directional cell migration was increased in miR-1281 transfected cells, in which wound closure was significantly enhanced compared with control cells at either 4, 8, or 12 h after scratch (Figure 4). In contrast, no differences in cell migration were found in ARPE-19 cells transfected or untransfected with miR-1281 mimic, in which wound closure following scratch occurred considerably later with respect to HUVEC cells (Figure 4). As proliferation and migration of retinal endothelial cells have been associated with the development of DR, these results sustain a role of miR-1281 in the regulation of factors known to be involved in this process, which include, among others, the VEGFA.
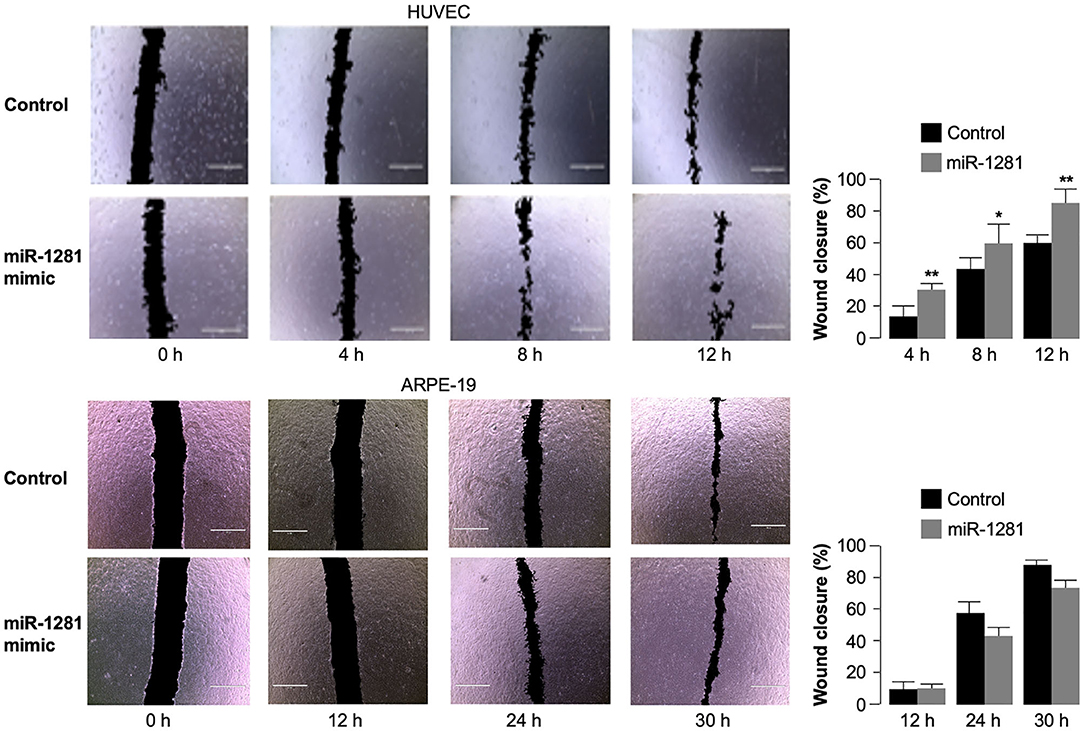
Figure 4. Effect of miR-1281 on cell migration. Wound healing assays were performed in HUVEC and ARPE-19 cells, either untransfected (Control), or transfected with miR-1281 mimic for 48 h. Wound closure was evaluated at the indicated time, after cell scratch, by phase contrast imaging (total magnification: 40 x), and analyzed with the ImageJ software. Quantitative data are expressed in bar graphs as mean ± SD of three independent assays, in which at least three images per condition were obtained. *p < 0.05; **p < 0.01 relative to untreated control cells at each time point.
In the present study, we investigated the differential expression of circulating miRNAs in a group of patients with type 2 diabetes, with and without non-proliferative DR. After comprehensive serum miRNA profiling and validation of individual miRNAs, 5 circulating miRNAs appeared to be highly up-regulated in patients with DR, compared to diabetic patients without DR. In particular, one of them, miR-1281, was the most up-regulated and seemed to be more specifically related to DR, displaying the strongest sensitivity and specificity in detecting this microvascular complication of diabetes.
In recent years, several works have investigated the association of miRNAs with DR (23–26, 37–52). In circulating miRNA profiling studies, miR-27b, miR-320a, and miR-126 were validated in patients with type 1 diabetes and DR (23, 42). In similar studies, miR-21, miR-181c, and miR-1179 were significantly upregulated in type 2 diabetic patients with proliferative DR vs. patients with non-proliferative DR (25), while miR-3197 and miR-2116-5p were higher in diabetic patients with DR, compared with patients without DR (37). In addtion, using an RNA-Seq approach, it was evidenced that miR-4448, miR-338-3p, miR-190a-5p, miR-485-5p, and miR-9-5p appeared differentially expressed in serum from retinopathic diabetic patients when compared to controls (49). However, while validation of miR-21 and miR-126 in DR was confirmed in other similar studies (39, 40, 44, 45), miR-320a, which was previously found to be upregulated in DR (23), was later reported to be significantly downregulated in another work (51), indicating, therefore, that discrepancies exist in this context. Several explanations can be provided for the lack of consensus in the literature in this regard. First, the selection of patients is very critical, as ethnicity, age, inclusion and exclusion criteria may substantially differ among studies. Second, differences in pre-analytical and analytical procedures may also account for discrepancies in the results. Third, there is no agreement on how miRNAs should be normalized, thus representing another issue in this debate. We believe that despite the remarkable number of studies on DR, all these observations indicate that additional miRNAs, including miR-1281, might still emerge as predictive indexes for DR.
Until now, only few studies have been published in which authors had identified miR-1281 as a potential biomarker in human diseases. Among these disease states were abdominal aortic aneurysm (53), coronary heart disease and pulmonary arterial hypertension (54), chronic nephropathy (55), immune thrombocytopenic purpura (56), and malignant pleural mesothelioma (57). Our findings in the present work are consistent with these studies, implicating miR-1281 as a potential biomarker to track disease progression or aid in disease diagnosis. Furthermore, our results in the present work are in agreement with other studies indicating that miR-1281 is up-regulated in the central cornea of patients with diabetes (58), as well as in plasma and urine samples of patients with chronic kidney disease, a condition that is often secondary to poorly controlled diabetes (55).
Apart from the histone deacetylase 4 (HDAC4) gene, that has been recently shown as a direct target of miR-1281 in pulmonary artery smooth muscle cells (54), no other targets of miR-1281 have been identified yet. Our in vitro results from human ARPE-19 retinal cells show that the extracellular release of miR-1281 following high glucose treatment significantly exceeded the release under low glucose conditions, and this behavior only partially reflected that of endothelial HUVEC cells, in which miR-1281 was also significantly reduced in the intracellular environment, although to a lesser extent than in ARPE-19 cells, but was unchanged in the extracellular medium. Based on these data, and on the observation that treatment with miR-1281 resulted in the increase in both VEGFA mRNA abundance and VEGFA promoter-driven Luc activity in HUVEC cells, it can be speculated that increased release of miR-1281 by epithelial retinal cells in instances of chronic glucose excess, such as in diabetes, might play a pathogenetic role in DR through the activation of VEGFA protein production from adjacent endothelial cells, thereby inducing VEGFA-mediated neoangiogenesis and vascular damage of retinal vessels. How miR-1281 upregulates VEGFA gene transcription is not clear and requires further studies. TargetScan prediction analysis suggests that this miRNA could potentially target mRNA sequences of the hypoxia-inducible factor-1α inhibitor (HIF1AN). The accumulation of HIF-1α and the impairment of Von Hippel-Lindau functional activity, due to miR-1281-induced HIF1AN instability/degradation, would both increase HIF-1—a master regulator of hypoxia—and, as a consequence, the enhanced transcription of hypoxia-responsive genes, including VEGFA. Whether miR-1281 has a functional role in glucose-induced retinal damage by VEGFA via HIF-1 has to be proved. If this interpretation is correct, it seems tempting to imply miR-1281 as a novel pathogenetic marker of retinal microangiopathic damage in patients with diabetes, which may explain, at least in part, how chronic sustained hyperglycemia could abnormally up-regulate VEGFA in retinal cells. Furthermore, the stimulatory effect of miR-1281 on HUVEC cell migration observed in this study indeed supports the biological role of this miRNA in DR. In this context, VEGFA definitely plays a role, although the potential link of miR-1281 to other factors involved in the directional migration of retinal endothelial cells, such as adhesion molecules (59), cannot be excluded.
On the other hand, the involvement of selected miRNAs in a growing number of pathophysiological conditions, including diabetes and its microvascular complications, has also been reported by others, in both clinical and non-clinical settings (14, 17). As reported above, the attempt to identify a sensitive and specific biomarker related to DR among circulating miRNAs has produced, so far, poor consistency between results (21, 60–62). Although other miRNAs may also contribute to the development of DR (17, 63–65), our results herein indicate, for the first time, that miR-1281 could indeed be a potential novel biomarker of DR. Compared to other studies, the main strengths of our work are the enrollment of highly selected patients with type 2 diabetes and their genetic homogeneous makeup with no genetic admixture (66). Also, of particular interest is the simultaneous demonstration as to how overproduction of miR-1281 in retinal cells may pathophysiologically contribute to retinal vascular damage, providing a mechanistic explanation for the adverse effect of this miRNA on DR. This should contribute to exceed the main limit of our study, mainly represented by its small sample size, in addition to differences in triglyceride levels in DR patients vs. controls as possible confounder to the results of this work.
Our findings identify miR-1281 as a novel sensitive and specific non-invasive biomarker for the early detection of DR. Furthermore, they suggest that, by positively regulating the expression of the neoangiogenic factor VEGFA in HUVEC endothelial cells under glucose excess conditions, miR-1281 can play a pathogenic role in the development of DR. Further studies with larger numbers of patients are required in order to fully validate these results in a clinical setting of patients with diabetes.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by Comitato etico regionale, Sezione Area Centro, Catanzaro, Italy, Protocol no. 116, May 14, 2015. The patients/participants provided their written informed consent to participate in this study.
MG, FA, DC, and BA performed experiments in vitro and validation of selected miRNAs in individual samples. MG and EC analyzed data, performed statistical analysis and contributed to manuscript draft. MM and RL helped collecting blood samples and clinical data from ambulatory patients. FB provided valuable suggestions and drafted figures. SP and AF contributed reagents, materials. VS performed ophthalmological examinations for diabetic retinopathy. DF conceived and helped supervise the project and contributed to data interpretation. AB conceived and supervised the study and wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Article Processing Charge was funded by the Department of Health Sciences, University “Magna Graecia” of Catanzaro, Italy.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2020.00528/full#supplementary-material
1. International Diabetes Federation. IDF Atlas. 9th ed. Available online at: http://www.diabetesatlas.org (accessed March 27, 2020).
2. Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diab Res Clin Pract. (2010) 87:4–14. doi: 10.1016/j.diabres.2009.10.007
3. Li R, Zhang P, Barker LE, Chowdhury FM, Zhang X. Cost-effectiveness of interventions to prevent and control diabetes mellitus: a systematic review. Diabetes Care. (2010) 33:1872–94. doi: 10.2337/dc10-0843
4. Antonetti DA, Klein R, Gardne TW. Diabetic retinopathy. N Engl J Med. (2012) 366:1227–39. doi: 10.1056/NEJMra1005073
5. Yau JW, Rogers SL, Kawasaki R, Lamoureux EL, Kowalski JW, Bek T, et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care. (2012) 35:556–64. doi: 10.2337/dc11-1909
6. Wang PH, Lau J, Chalmers TC. Meta-analysis of effects of intensive blood-glucose control on late complications of type I diabetes. Lancet. (1993) 34:1306–9. doi: 10.1016/0140-6736(93)90816-Y
7. Raffort J, Hinault C, Dumortier O, Van Obberghen E. Circulating micro- RNAs and diabetes: potential applications in medical practice. Diabetologia. (2015) 58:1978–92. doi: 10.1007/s00125-015-3680-y
8. Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. (2014) 37:1375–83. doi: 10.2337/dc13-1847
9. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
11. Guay C, Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol. (2013) 9:513–21. doi: 10.1038/nrendo.2013.86
12. Tang X, Tang G, Ozcan S. Role of microRNAs in diabetes. Biochim Biophys Acta. (2008) 1779:697–701. doi: 10.1016/j.bbagrm.2008.06.010
13. Kantharidis P, Wang B, Carew RM, Lan HY. Diabetes complications: the microRNA perspective. Diabetes. (2011) 60:1832–7. doi: 10.2337/db11-0082
14. Zhang Y, Sun X, Icli B, Feinberg MW. Emerging roles for microRNAs in diabetic microvascular disease: novel targets for therapy. Endocr Rev. (2017) 38:145–68. doi: 10.1210/er.2016-1122
15. De Rosa S, Arcidiacono B, Chiefari E, Brunetti A, Indolfi C, Foti DP. Type 2 diabetes mellitus and cardiovascular disease: genetic and epigenetic links. Front Endocrinol. (2018) 9:2. doi: 10.3389/fendo.2018.00002
16. Beltrami C, Angelini TG, Emanueli C. Noncoding RNAs in diabetes vascular complications. J Mol Cell Cardiol. (2014) 89:42–50. doi: 10.1016/j.yjmcc.2014.12.014
17. Barutta F, Bellini S, Mastrocola R, Bruno G, Gruden G. MicroRNA and microvascular complications of diabetes. Int J Endocrinol. (2018) 2018:6890501. doi: 10.1155/2018/6890501
18. Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. (2003) 9:669–76. doi: 10.1038/nm0603-669
19. Chiefari E, Ventura V, Capula C, Randazzo G, Scorcia V, Fedele M, et al. A polymorphism of HMGA1 protects against proliferative diabetic retinopathy by impairing HMGA1-induced VEGFA expression. Sci Rep. (2016) 6:39429. doi: 10.1038/srep39429
20. Mortuza R, Feng B, Chakrabarti S. miR-195 regulates SIRT1-mediated changes in diabetic retinopathy. Diabetologia. (2014) 57:1037–46. doi: 10.1007/s00125-014-3197-9
21. McArthur K, Feng B, Wu Y, Chen S, Chakrabarti S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes. (2011) 60:1314–23. doi: 10.2337/db10-1557
22. Xiong F, Du X, Hu J, Li T, Du S, Wu, et al. Altered retinal microRNA expression profiles in early diabetic retinopathy: an in silico analysis. Curr Eye Res. (2014) 39:720–9. doi: 10.3109/02713683.2013.872280
23. Zampetaki A, Willeit P, Burr S, Yin X, Langley SR, Kiechl S, et al. Angiogenic microRNAs linked to incidence and progression of diabetic retinopathy in type 1 diabetes. Diabetes. (2016) 65:216–27. doi: 10.2337/db15-0389
24. Wang C, Wan S, Yang T, Niu D, Zhang A, Yang C, et al. Increased serum micro- RNAs are closely associated with the presence of microvascular complications in type 2 diabetes mellitus. Sci Rep. (2016) 6:20032. doi: 10.1038/srep20032
25. Qing S, Yuan S, Yun C, Hui H, Mao P, Wen F, et al. Serum miRNA biomarkers serve as a fingerprint for proliferative diabetic retinopathy. Cell Physiol Biochem. (2014) 34:1733–40. doi: 10.1159/000366374
26. He M, Wang W, Yu H, Wang D, Cao D, Zeng Y, et al. Comparison of expression profiling of circular RNAs in vitreous humour between diabetic retinopathy and non-diabetes mellitus patients. Acta Diabetol. (2020) 57:479–89. doi: 10.1007/s00592-019-01448-w
27. Ozawa GY, Bearse MA Jr., Adams AJ. Male-female differences in diabetic retinopathy? Curr Eye Res. (2015) 40:234–9. doi: 10.3109/02713683.2014.958500
28. Sharma S, Eghbali M. Influence of sex differences on microRNA gene regulation in disease. Biol Sex Differ. (2014) 5:3. doi: 10.1186/2042-6410-5-3
29. Klinge CM. miRNAs and estrogen action. Trends Endocrinol Metab. (2012) 5:223–33. doi: 10.1016/j.tem.2012.03.002
30. American Diabetes Association. Classification and diagnosis of diabetes: standards of medical care in diabetes−2020. Diabetes Care. (2020) 43(Suppl. 1):S14–31. doi: 10.2337/dc20-S002
31. Wilkinson CP, Ferris FL, Klein RE, Lee PP, Agardh CD, Davis M, et al. Global diabetic retinopathy project group. proposed international clinical diabetic retinopathy and diabetic macular edema disease severity scales. Ophthalmology. (2003) 110:1677–82. doi: 10.1016/S0161-6420(03)00475-5
32. Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, Feldman HI, et al. CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration). a new equation to estimate glomerular filtration rate. Ann Intern Med. (2009) 150:604–12. doi: 10.7326/0003-4819-150-9-200905050-00006
33. American Diabetes Association. Diabetic nephropathy. Diabetes Care. (2002) 25(Suppl. 1):S85–9. doi: 10.2337/diacare.25.2007.S85
34. Chiefari E, Paonessa F, Iiritano S, Le Pera I, Palmieri D, Brunetti G, et al. The cAMP-HMGA1-RBP4 system: a novel biochemical pathway for modulating glucose homeostasis. BMC Biol. (2009) 7:24. doi: 10.1186/1741-7007-7-24
35. Brunetti A, Foti D, Goldfine ID. Identification of unique nuclear regulatory proteins for the insulin receptor gene, which appear during myocyte and adipocyte differentiation. J Clin Invest. (1993) 92:1288–95. doi: 10.1172/JCI116702
36. Corigliano DM, Syed R, Messineo S, Lupia A, Patel R, Reddy CVR, et al. Indole and 2,4-thiazolidinedione conjugates as potential anticancer modulators. PeerJ. (2018) 6:e5386. doi: 10.7717/peerj.5386
37. Ji H, Yi Q, Chen L, Wong L, Liu Y, Xu G, et al. Circulating miR-3197 and miR-2116-5pas novel biomarkers for diabetic retinopathy. Clin Chim Acta. (2020) 501:147–53. doi: 10.1016/j.cca.2019.10.036
38. García de la Torre N, Fernández-Durango R, Gómez R, Fuentes M, Roldán-Pallarés M, Donate J, et al. Expression of angiogenic microRNA in endothelial progenitor cells from type 1 diabetic patients with and without diabetic retinopathy. Invest Ophthalmol Vis Sci. (2015) 56:4090–8. doi: 10.1167/iovs.15-16498
39. Qin LL, An MX, Liu YL, Xu HC, Lu ZQ. MicroRNA-126: a promising novel biomarker in peripheral blood for diabetic retinopathy. Int J Ophthalmol. (2016) 10:530–4. doi: 10.18240/ijo.2017.04.05
40. Jiang Q, Lyu XM, Yuan Y, Wang L. Plasma miR-21 expression: an indicator for the severity of type 2 diabetes with diabetic retinopathy. Biosci Rep. (2017) 37:BSR20160589. doi: 10.1042/BSR20160589
41. Kamalden TA, Macgregor-Das AM, Kannan SM, Dunkerly-Eyring B, Khaliddin N, Xu Z, et al. Exosomal MicroRNA-15a transfer from the pancreas augment diabetic complications by inducing oxidative stress. Antioxid Redox Signal. (2017) 27:913–30. doi: 10.1089/ars.2016.6844
42. Barutta F, Bruno G, Matullo G, Chaturvedi N, Grimaldi S, Schalkwijk C, et al. MicroRNA-126 and micro-/macrovascular complications of type 1 diabetes in the EURODIAB prospective complication study. Acta Diabetol. (2017) 54:133–9. doi: 10.1007/s00592-016-0915-4
43. Zou HL, Wang Y, Gang Q, Zhang Y, Sun Y. Plasma level of miR-93 is associated with higher risk to develop type 2 diabetic retinopathy. Graefes Arch Clin Exp Ophthalmol. (2017) 255:1159–66. doi: 10.1007/s00417-017-3638-5
44. Mazzeo A, Beltramo E, Lopatina T, Gai C, Trento M, Porta M. Molecular and functional characterization of circulating vesicles from diabetic patients with and without retinopathyand healthy subjects. Exp Eye Res. (2018) 176:69–77. doi: 10.1016/j.exer.2018.07.003
45. Mazzeo A, Lopatina T, Gai C, Trento M, Porta M, Beltramo E. Functional analysis of miR-21-3p, miR-30b-5p and miR-150-5p shuttled by extracellular vesicles from diabetic subjects reveal their association with diabetic retinopathy. Exp Eye Res. (2019) 184:56–63. doi: 10.1016/j.exer.2019.04.015
46. Dantas da Costa e Silva ME, Polina ER, Crispim D, Sbruzzi RC, Lavinsky D, Mallmann F, et al. Plasma levels of miR-29b and miR-200bin type 2 diabetic retinopathy. J Cell Mol Med. (2019) 23:1280–7. doi: 10.1111/jcmm.14030
47. Blum A, Meerson A, Rohana H, Jabaly H, Nahul N, Celesh D, et al. MicroRNA-423 may regulate diabetic vasculopathy. Clin Exp Med. (2019) 19:469–77. doi: 10.1007/s10238-019-00573-8
48. Pastukh N, Meerson A, Kalish D, Jabaly H, Blum A. Serum miR-122 levels correlate with diabetic retinopathy. Clin Exp Med. (2019) 19:255–60. doi: 10.1007/s10238-019-00546-x
49. Li Z, Dong Y, He C, Pan X, Liu D, Yang J, et al. RNA-Seq revealed novel non-proliferative retinopathy specific circulating miRNAs in T2DM patients. Front Genet. (2019) 10:531. doi: 10.3389/fgene.2019.00531
50. Polina ER, Oliveira FM, Sbruzzi RC, Crispim D, Canani LH, Santos KG. Gene polymorphism and plasma levels of miR-155 in diabetic retinopathy. Endocr Connect. (2020) 8:1591–9. doi: 10.1530/EC-19-0446
51. Prado MSG, Lemos de Jesus M, Cunha de Goes T, Oliveira Mendonça LS, Martins Kaneto C. Downregulation of circulating miR-320a and target gene prediction in patients with diabetic retinopathy. BMC Res Notes. (2020) 13:155. doi: 10.1186/s13104-020-05001-9
52. Liu X, Li J, Li X. miR-142-5p regulates the progression of diabetic retinopathy by targeting IGF1. Int J Immunopathol Pharmacol. (2020) 34:2058738420909041. doi: 10.1177/2058738420909041
53. Zhang W, Shang T, Huang C, Yu T, Liu C, Qiao T, et al. Plasma microRNAs serve as potential biomarkers for abdominal aortic aneurysm. Clin Biochem. (2015) 48:988–92. doi: 10.1016/j.clinbiochem.2015.04.016
54. Li Y, Li L, Qian Z, Lin B, Chen J, Luo Y, et al. Phosphatidylinositol 3-kinase-DNA methyltransferase 1-miR-1281-histone deacetylase 4 regulatory axis mediates platelet-derived growth factor-induced proliferation and migration of pulmonary artery smooth muscle cells. J Am Heart Assoc. (2018) 7:e007572. doi: 10.1161/JAHA.117.007572
55. Muralidharan J, Ramezani A, Hubal M, Knoblach S, Shrivastav S, Karandish S, et al. Extracellular microRNA signature in chronic kidney disease. Am J Physiol Renal Physiol. (2017) 312:F982–91. doi: 10.1152/ajprenal.00569.2016
56. Zuo B, Zhai J, You L, Zhao Y, Yang J, Weng Z, et al. Plasma microRNAs characterising patients with immune thrombocytopenic purpura. Thromb Haemost. (2017) 117:1420–31. doi: 10.1160/TH-16-06-0481
57. Bononi I, Comar M, Puozzo A, Stendardo M, Boschetto P, Orecchia S, et al. Circulating microRNAs found dysregulated in ex-exposed asbestos workers and pleural mesothelioma patients as potential new biomarkers. Oncotarget. (2016) 7:82700–11. doi: 10.18632/oncotarget.12408
58. Funari VA, Winkler M, Brown J, Dimitrijevich SD, Ljubimov AV, Saghizadeh M. Differentially expressed wound healing-related microRNAs in the human diabetic cornea. PLoS ONE. (2013) 8:e84425. doi: 10.1371/journal.pone.0084425
59. Olson JA, Whitelaw CM, McHardy KC, Pearson DW, Forrester JV. Soluble leucocyte adhesion molecules in diabetic retinopathy stimulate retinal capillary endothelial cell migration. Diabetologia. (1997) 40:1166–71. doi: 10.1007/s001250050802
60. Ye P, Liu J, He F, Xu W, Yao K. Hypoxia-induced deregulation of miR-126 and its regulative effect on VEGF and MMP-9 expression. Int J Med Sci. (2013) 11:17–23. doi: 10.7150/ijms.7329
61. Meng S, Cao I, Zhang X, Fan Y, Fang L, Wang C, et al. Downregulation of microRNA-130a contributes to endothelial progenitor cell dysfunction in diabetic patients via its target Runx3. PLoS ONE. (2013) 8:e68611. doi: 10.1371/journal.pone.0068611
62. Martinez B, Peplow PV. MicroRNAs as biomarkers of diabetic retinopathy and disease progression. Neural Regen Res. (2019) 14:1858–69. doi: 10.4103/1673-5374.259602
63. Shao J, Fan G, Yin X, Gu Y, Wang X, Xin Y, et al. A novel transthyretin/STAT4/miR-223-3p/FBXW7 signaling pathway affects neovascularization in diabetic retinopathy. Mol Cell Endocrinol. (2019) 498:110541. doi: 10.1016/j.mce.2019.110541
64. Fu X, Ou B. miR-152/LIN28B axis modulates high-glucose-induced angiogenesis in human retinal endothelial cells via VEGF signaling. J Cell Biochem. (2020) 121:954–62. doi: 10.1002/jcb.28978
65. Chiefari E, Tanyolaç S, Iiritano S, Sciacqua A, Capula C, Arcidiacono B, et al. A polymorphism of HMGA1 is associated with increased risk of metabolic syndrome and related components. Sci Rep. (2013) 3:1491. doi: 10.1038/srep01491
Keywords: diabetic retinopathy, type 2 diabetes, miRNA profiling, serum biomarkers, VEGFA, HUVEC endothelial cells, ARPE-19 epithelial cells
Citation: Greco M, Chiefari E, Accattato F, Corigliano DM, Arcidiacono B, Mirabelli M, Liguori R, Brunetti FS, Pullano SA, Scorcia V, Fiorillo AS, Foti DP and Brunetti A (2020) MicroRNA-1281 as a Novel Circulating Biomarker in Patients With Diabetic Retinopathy. Front. Endocrinol. 11:528. doi: 10.3389/fendo.2020.00528
Received: 06 May 2020; Accepted: 29 June 2020;
Published: 04 August 2020.
Edited by:
Marcus M. Seldin, University of California, Irvine, United StatesReviewed by:
Guido Sebastiani, University of Siena, ItalyCopyright © 2020 Greco, Chiefari, Accattato, Corigliano, Arcidiacono, Mirabelli, Liguori, Brunetti, Pullano, Scorcia, Fiorillo, Foti and Brunetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio Brunetti, YnJ1bmV0dGlAdW5pY3ouaXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.