
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 12 June 2020
Sec. Obesity
Volume 11 - 2020 | https://doi.org/10.3389/fendo.2020.00387
This article is part of the Research Topic Current Challenges for Targeting Brown Fat Thermogenesis to Combat Obesity View all 16 articles
 Sebastian Müller1,2,3
Sebastian Müller1,2,3 Aliki Perdikari1
Aliki Perdikari1 Dianne H. Dapito1
Dianne H. Dapito1 Wenfei Sun1
Wenfei Sun1 Bernd Wollscheid2
Bernd Wollscheid2 Miroslav Balaz1*
Miroslav Balaz1* Christian Wolfrum1*
Christian Wolfrum1*When exposed to cold temperatures, mice increase their thermogenic capacity by an expansion of brown adipose tissue mass and the formation of brite/beige adipocytes in white adipose tissue depots. However, the process of the transcriptional changes underlying the conversion of a phenotypic white to brite/beige adipocytes is only poorly understood. By analyzing transcriptome profiles of inguinal adipocytes during cold exposure and in mouse models with a different propensity to form brite/beige adipocytes, we identified ESRRG and PERM1 as modulators of this process. The production of heat by mitochondrial uncoupled respiration is a key feature of brite/beige compared to white adipocytes and we show here that both candidates are involved in PGC1α transcriptional network to positively regulate mitochondrial capacity. Moreover, we show that an increased expression of ESRRG or PERM1 supports the formation of brown or brite/beige adipocytes in vitro and in vivo. These results reveal that ESRRG and PERM1 are early induced in and important regulators of brite/beige adipocyte formation.
White and brown adipose tissue have divergent functions. White adipocytes store chemical energy within a single lipid droplet to release it back to the body when needed. In contrast, brown adipocytes primarily convert chemical energy into heat through the action of uncoupling protein 1 (UCP1) (1, 2). It is known that brown adipocytes do not only reside in a distinct fat pad in the interscapular region in rodents, but are also found interspersed between white adipocytes in other anatomical locations (3, 4). The latter have been termed brite (“brown-in-white”) or beige adipocytes and are identified by their multilocular appearance and the expression of UCP1 (5). Their formation can be induced through a plethora of stimuli, most prominently through exposure of the animals to a cold environment, by beta-adrenergic stimulation (6) and other endocrine factors (7, 8). Experimental evidence suggests that the arising brite/beige adipocytes can derive from de novo differentiation of stem cells (9, 10) as well as by direct interconversion of mature adipocytes (11–13).
The orphan nuclear estrogen related receptor gamma (ESRRG) is highly expressed in energy dependent tissues such as brain, heart, skeletal muscle, kidney and BAT and is responsible for cell-type specific function in the majority of those tissues (14). Studies have shown that ESRRG is not important for brown preadipocyte differentiation but for brown adipocyte function, acting in a complementary way together with other ESRR members in mitochondrial biogenesis and oxidative function (15). In fact, it was shown that ESRR members mediate the adaptive response of BAT to adrenergic stimulation (16). Moreover, ESRRG is necessary for the induction of UCP1 in in vitro differentiating cells (17) and for the maintenance of BAT thermogenic activity in vivo (18). ESRRG was shown to interact with GADD45γ to regulate UCP1 levels during cold adaption of brown adipose tissue (19). In regards to browning of white adipose tissue, the 1-benzyl-4-phenyl-1H-1,2,3-triazole derivative is shown to enhance the function of ESRRG and increase browning in vitro and in vivo (20). The potential transcription factor PGC1 and ESRR-induced regulator in muscle 1 (PERM1) has not been described in the context of adipose tissue so far, but was recently reported to regulate OXPHOS proteins in muscle in vitro and in vivo (21, 22). As ESRRG, PERM1 acts downstream of PGC1α, a master-regulator of mitochondrial capacity (23).
We aimed to study the latter process and showed that as early as 24 h after a cold stimulus, several known browning markers are significantly upregulated in the white adipocyte fraction. Importantly, we identified ESRRG and PERM1 as novel transcriptional regulators of brite/beige adipocyte formation.
To study the transcriptional events underlying the appearance of brite/beige adipocytes we housed 129/SV mice at 8°C for 24 h, 7 days or kept them at room temperature. In order to identify transcriptional changes induced by acute and prolonged cold exposure, we performed transcriptomic analysis of the mature inguinal white adipocyte (ingWA) fraction using RNA microarray. We excluded from further analysis two out of five individual mice housed at room temperature which already expressed high levels of Ucp1 (Figure S1), a phenomenon previously described in literature (24). Intriguingly, on the transcriptome level, the browning program was fully activated already after 24 h of cold exposure, as demonstrated by the upregulation of common browning makers Dio2, Fabp3, Cpt1b, Perilipin 5, Cox7a1, and Ucp1 (Figure 1A). Even though there is a clear difference in the multilocular adipocyte content (Figure S1B), the transcriptomic changes between 24 h and 7 days of cold exposure were only marginal (Figure 1B), indicating that the transcriptional program for brite/beige adipocyte formation is kept constant after the initial activation. A key feature in the appearance of brite/beige adipocytes is the increase in mitochondrial capacity (25, 26). We found specific nuclear encoded mitochondrial genes upregulated already after 24 h of cold exposure, indicating mitochondrial remodeling. On the other hand, mitochondrial encoded genes were only regulated after 7 days of cold exposure, which likely reflects the bulk increase in mitochondrial mass (Figure 1C). Analysis of early transcriptional changes revealed two genes, Esrrg and Perm1, to be significantly upregulated after 24 h of cold exposure (Figure 1A). We confirmed the expression of Esrrg and Perm1 by qRT-PCR in ingWA fraction 24 h after cold exposure in an independent cohort of mice (Figure 1D). We further validated the Esrrg and Perm1 expression in pure populations of mature adipocyte isolated by fluorescent activated cell sorting (FACS, Figure S1C). We used transgenic mice in the C57BL/6 background expressing green-fluorescent protein (GFP) under the control of the Ucp1 promoter housed at 8°C for 7 days. RNA expression analysis revealed Perm1 exclusive expression in the brite/beige adipocyte fraction, while Esrrg expression was enriched in the brite/beige compared to the white adipocytes (Figure 1E).
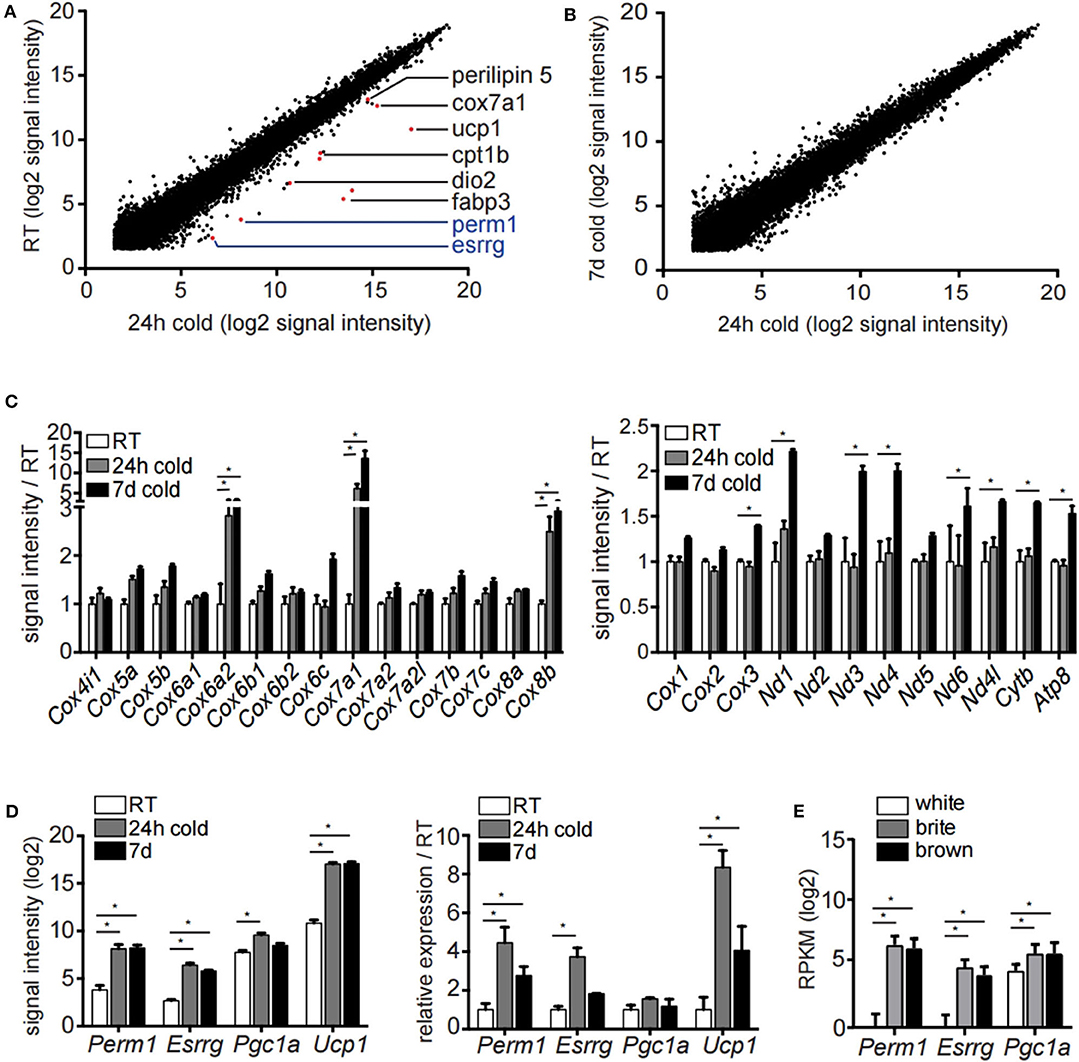
Figure 1. Microarray analysis of inguinal white adipocytes during cold exposure. (A,B) Signal intensities (log2) of all probes measured of the extracted RNA of the adipocyte fraction of ingWA comparing 24 h cold exposure and room temperature (A) or 24 h and 7 days cold exposure (B). Depicted in red are browning markers upregulated after cold treatment. (C) Signal intensity of specific nuclear encoded mitochondrial genes for OXPHOS complex IV (left) and all mitochondrial encoded genes (right) in room temperature, after 24 h and 7 days of cold exposure. Data are presented as microarray signal intensities relative to room temperature as mean with SEM (n = 3–5). (D) Expression of target genes Perm1, Esrrg, Pgc1α, and Ucp1 in room temperature, after 24 h and 7 days of cold exposure shown in the microarray (left) and in an independent cohort of mice via qRT-PCR (right). Data are presented as signal intensities (log2) or relative expression (ddCt) compared to room temperature, respectively, as mean with SEM (n = 3-5 per cohort). (E) Expression of target genes Perm1, Esrrg, and Pgc1α in FACS sorted mature adipocytes according to GFP expression under control of the Ucp1 promoter. Data are presented as mean RPKM (log2) with SD (n = 10 per cell type). Statistical testing (t-test) was performed between room temperature condition and cold housing for 24 h or 7 days, respectively, or between sorted white adipocytes and brite or brown adipocytes. A statistically significant result, meaning p-value < 0.05, is indicated with an asterisk (*).
The 129/SV mouse strain has a higher propensity of browning in the ingWA compared to C57BL/6 (27, 28). Therefore, we analyzed if this phenotypic difference can be recapitulated through whole transcriptome analysis. The adipocyte fractions of the ingWA, epididymal white (epiWA) and interscapular brown adipose tissue (iBA) were isolated from both strains housed at room temperature and analyzed by next-generation RNA sequencing. Classical browning markers Ucp1, Cox7a1, and Cidea were nearly absent in epiWA of C57BL/6 with very low expression in the 129/SV mouse model (Figure 2A), while they were expressed higher in the ingWA of 129/SV mice, indicating the presence of few brite/beige cells already at room temperature. Interestingly, we detected browning markers at higher levels in the iBA of C57BL/6 in comparison to 129/SV, potentially indicating a compensatory mechanism, to keep the total brown/brite/beige capacity constant. The expression levels of Esrrg and Perm1 accurately correlated with browning markers in the two mouse models (Figure 2B), suggesting a possible role in brite/beige and brown adipocytes. To validate that changes on the transcript level also result in increased protein abundance, we cold-exposed 129/SV mice for 3 and 7 days, respectively. We could show that PERM1 is not detectable at room temperature but is expressed after cold stimulation in ingWA (Figure 2C). Due to the unavailability of ESRRG specific antibodies, we could not confirm protein expression of ESRRG.
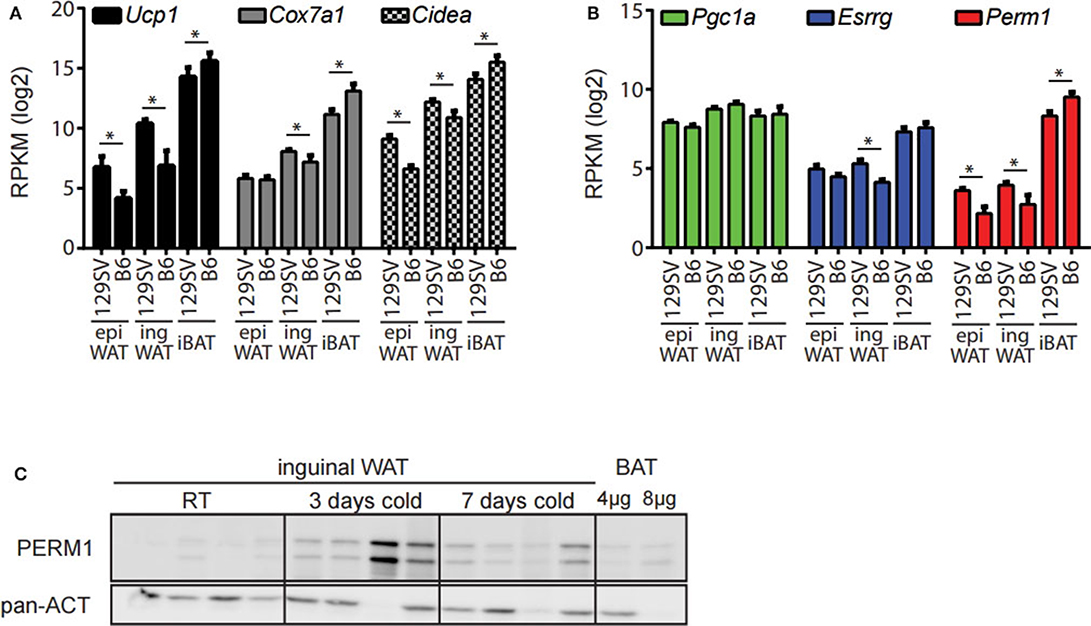
Figure 2. Esrrg and Perm1 mirror thermogenic capacity in adipocyte fractions of 129/SV and C57BL/6 mouse models. (A) Expression of common browning markers Ucp1, Cox7a1, and Cidea in the adipocyte fractions of epiWA, ingWA, and iBA in 129/SV or C57BL/6 mice housed at room temperature. Data are presented as mean RPKM with SEM (n = 4). (B) Expression of Pgc1α, Esrrg, and Perm1 in the adipocyte fractions of epiWA, ingWA and iBA in 129/SV or C57BL/6 mice housed at room temperature. Data are presented as mean RPKM with SEM (n = 4). (C) PERM1 and UCP1 protein levels in ingWAT of 129/SV mice at room temperature and after 3 and 7 days of cold exposure. BAT at 1/10 and 1/5 of protein loaded shown for comparison. Each lane represents a single mouse (n = 4). Statistical testing (t-test) was performed between the mouse strains (129SV & B6) in each of the different tissue derived adipocyte fractions. A statistically significant result, meaning p-value < 0.05, is indicated with an asterisk (*).
To characterize further the candidate genes Esrrg and Perm1, we analyzed the expression pattern in a tissue panel of C57BL/6 mice housed at room temperature. Interestingly, while we detected broad expression of Pgc1α and Esrrg, Perm1 expression was confined to tissues with a high energy demand, in particular iBAT, skeletal muscle and heart (Figure 3A), confirmed also on protein level (Figure 3B). The importance of ESRRG and PERM1 in energy metabolism is evident in global ESRRG knockout mice, which die shortly after birth, due to a heart-defect, as the organ cannot sustain its function despite an increase in mitochondrial number (29). In this study, Perm1 is considerably downregulated, in heart tissue of the Esrrg knockout mice (Figure 3C). Moreover, a recent study by Harms and colleagues describes a whitening of brown adipose tissue upon knockout of Prdm16, a master regulator of the brown adipocyte program (30), in mature brown adipocytes (31). Consistently, in the publicly available dataset, Perm1 is also among the highest downregulated proteins in the whitened BAT of these mice (Figure 3D).
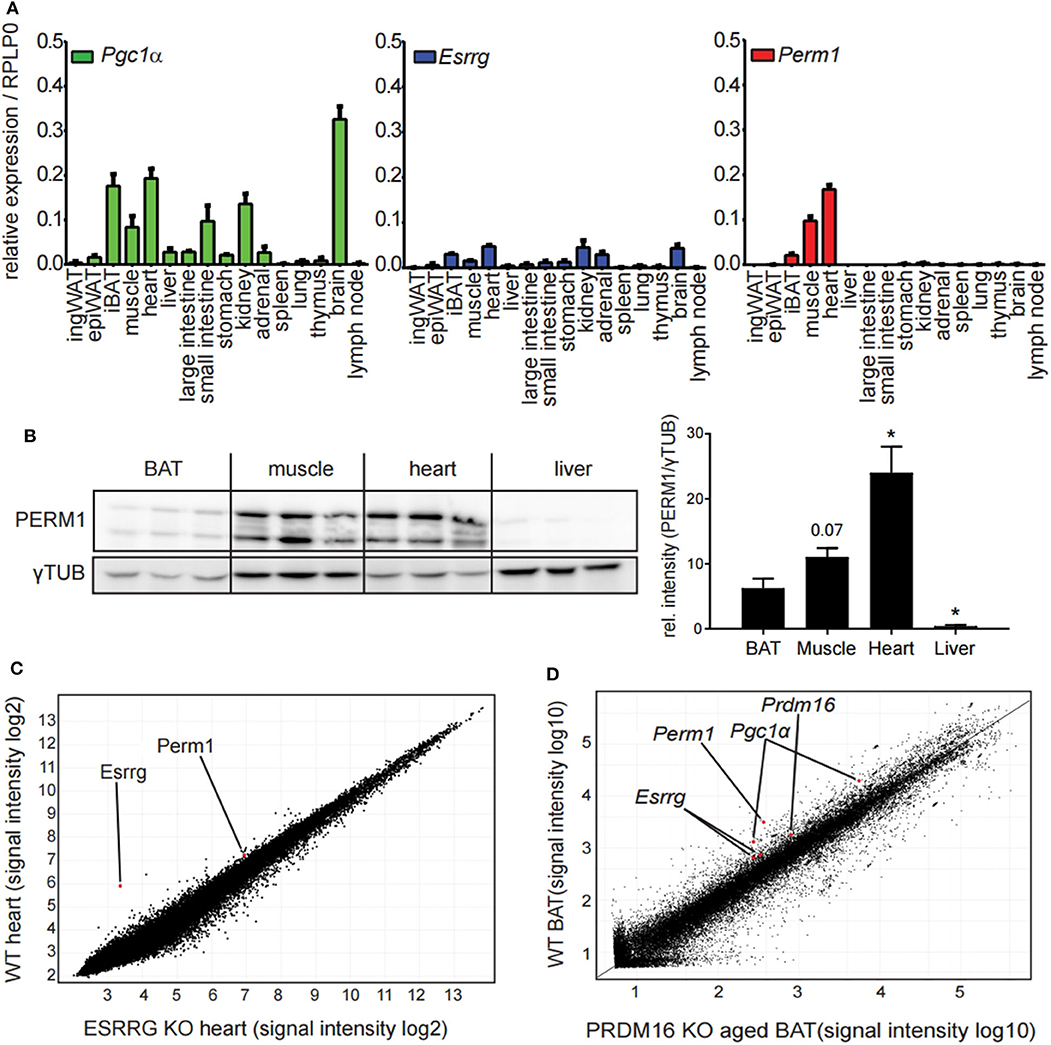
Figure 3. PERM1 expression is confined to tissues with a high energy demand and with ESRRG involved in energy metabolism. (A) Expression of Perm1, Esrrg, and Pgc1α in a panel of 16 tissues derived from C57BL/6 mice housed at room temperature. Data are shown as relative expression to reference gene RPLP0 with SEM (n = 5). (B) Representative western blot and quantification of PERM1 in BAT, muscle, heart, and liver. Each lane represents a single mouse (n = 3). (C) Microarray analysis of RNA extracted from heart tissue of ESRRG knockout mice as described in (29). Data were retrieved from GSE8199, plotted are all transcripts as log2 signal intensities (n = 3). (D) Representative microarray analysis of RNA extracted from BAT of aged mice, where PRDM16 is knocked out under the control of MYF5 promoter, leading to a whitening of BAT, as described in (31). Data were retrieved from GSE55080, depicted is one representative microarray with all probes as signal intensities (total n = 4). A statistically significant result, meaning p-value < 0.05, is indicated with an asterisk (*).
In order to establish an in vitro model to characterize the function and the role of PERM1 and ESRRG in brite/beige adipocyte formation, we isolated stromal-vascular fraction (SVF) of the epiWA, ingWA and iBA depots of 129/SV mice and subjected them to in vitro brite/beige adipocyte differentiation (10). To assess the dynamics of Perm1 and Esrrg expression during the differentiation of SVF cells into adipocytes, we harvested cells every 2 days starting from induction (day 0). Pparγ, as a general adipocyte marker, indicated adipocyte differentiation of cells derived from all three tissues, while browning markers Ucp1 and Cox7a1 were only detected in ingWA-derived SVF cells and were strongly induced in iBA-derived SVF cells during the differentiation time course (Figure 4A). While Pgc1α expression levels resembled these observations, Esrrg and Perm1 could not be detected in ingWA-derived differentiating cells and were induced at very low levels in the iBA derived differentiating SVF cells, indicating that these cells are not a suitable model to study PERM1 and ESRRG function. In conclusion, while Esrrg and Perm1 are strong indicators of browning capacity in vivo, their expression in vitro is limited.
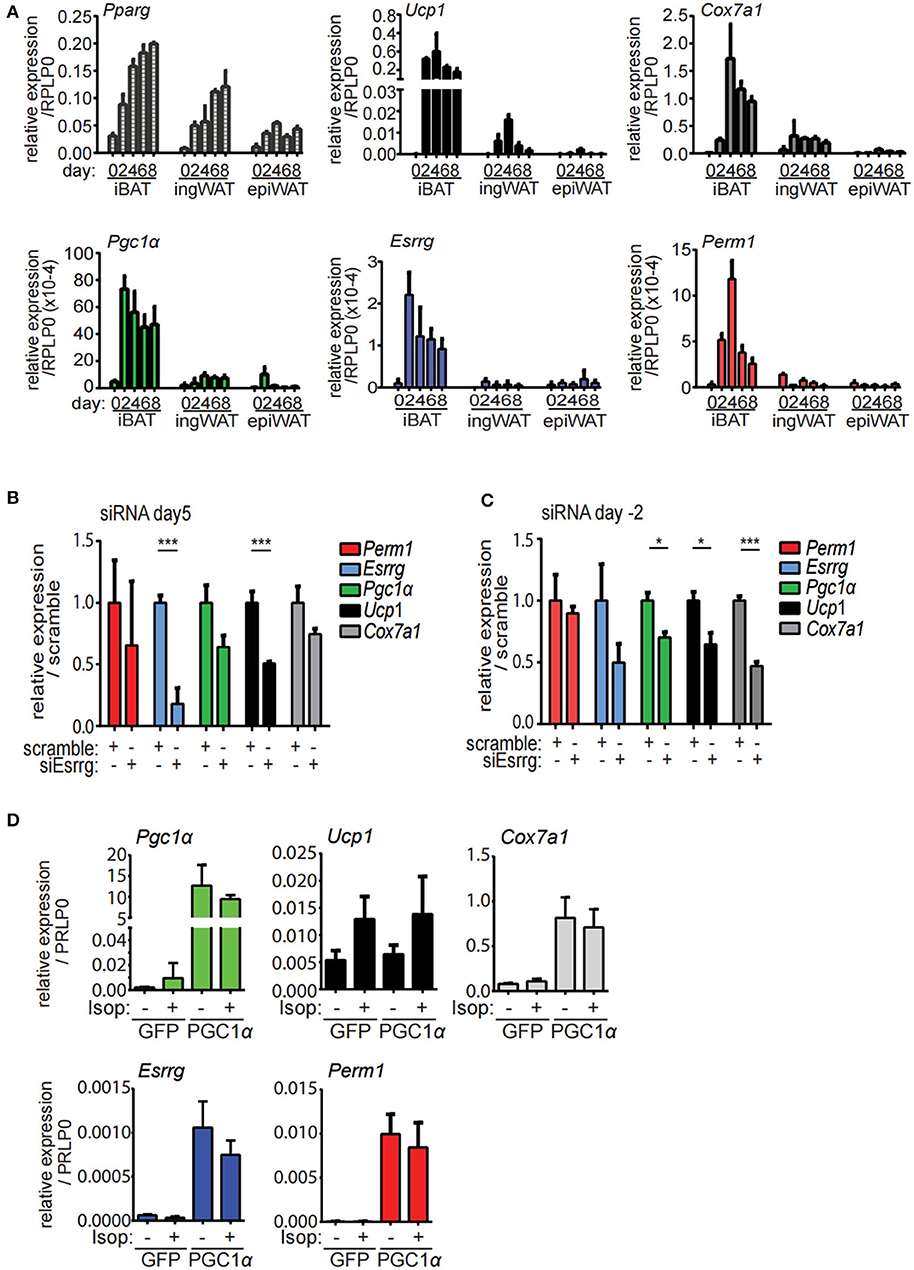
Figure 4. Esrrg and Perm1 have low expression levels in vitro but can be induced in a PGC1α dependent manner. (A) Time course of in vitro brite/beige adipocyte differentiation of SVF derived from iBA, ingWA or epiWA of 129/SV mice. Pparg (all isoforms) is shown as general adipocyte marker, Ucp1, and Cox7a1 as markers for brown adipocytes. Data are presented as relative expression to reference gene RPLP0 with SEM (n = 3). (B) Gene expression in immortalized brown mature adipocytes in vitro differentiated and treated with siRNAs targeting Esrrg at day 5 of differentiation. Data are presented as relative expression (ddCt) relative to non-targeting siRNA as mean with SD (n = 3). (C) Gene expression in immortalized brown pre-adipocytes treated with siRNAs targeting Esrrg 2 days prior to induction of differentiation. RNA was extracted on day 8 after induction and data are presented as expression (ddCt) relative to non-targeting siRNA as mean with SD (n = 3). (D) Gene expression in immortalized brown mature adipocytes in vitro differentiated and infected with adenovirus to overexpress PGC1α or GFP at day 5 after induction. Data are presented as relative expression to reference gene RPLP0 with SD (n = 3). A statistically significant result, meaning p-value < 0.05 and < 0.001, is indicated with an asterisk (*) and (***), respectively.
To functionally characterize our candidate proteins, we used immortalized brown pre-adipocytes, which can be differentiated to mature adipocytes in vitro (32). Esrrg and Perm1 were silenced in proliferating preadipocytes (day−2) or in mature adipocytes (day 5), to study their role in regulation of brown/beige adipocyte formation and thermogenic activity, respectively. SiRNA mediated knockdown of Esrrg in mature brown adipocytes at day 5 of differentiation led to a significant reduction in Ucp1 and Cox7a1, while Pgc1α was not affected (Figure 4B), indicating that ESRRG drives brown adipocyte thermogenic activity. Moreover, siRNA mediated repression of Esrrg in preadipocytes prior to the induction of differentiation (siRNA treatment at day−2) resulted in significantly reduced expression of Ucp1, Cox7a1, and Pgc1α as well as Esrrg in mature adipocytes, indicating a role of Esrrg during the early phase of adipogenic differentiation (Figure 4C). Altogether, these data indicate that ESRRG controls both formation as well as the thermogenic activity of brown/beige adipocytes. We were unable to achieve knockdown of Perm1 with siRNA treatment; hence, no change in expression of other genes was observed (Figure S2A). As the expression levels of Esrrg and Perm1 in vitro are low, we aimed to increase their levels and analyze the concurrent functional implications. We employed an adenoviral system to overexpress either PGC1α or green fluorescent protein (GFP) as a control [kindly provided by Prof. Christoph Handschin (33)], as PGC1α was shown to increase Perm1 and Esrrg abundance in muscle studies (21). We demonstrated that Perm1 and Esrrg could be upregulated in a Pgc1α dependent manner in in vitro differentiated mature brown adipocytes (Figure 4D). Interestingly, Cox7a1 was co-regulated, while Ucp1 levels were unaffected. The latter can be explained by Ucp1 expression being already affected by control adenovirus treatment compared to untreated cells (Figure S2C). Moreover, the combination of adenoviral and siRNA treatment resulted in non-differentiating cells (data not shown). Notably, adenoviral overexpression of PGC1α in immortalized pre-adipocytes already lead to the expression of known target genes Esrra and Cox7a1 in a Pgc1α-dependent manner, but not Esrrg or Perm1 (Figure S2D).
Since endogenous expression of Esrrg and Perm1 is low in vitro, we set out to establish a model system, which does not rely on adenoviral transduction and allows for targeted expression of either Esrrg or Perm1. Therefore, we used the lentiviral pInducer21 system (34) to express Esrrg or Perm1 under the control of doxycycline, including GFP as a positive selection marker for transduced cells. Immortalized brown preadipocytes were infected and FACS sorted according to their GFP signals, to polyclonally generate six cell lines having negative, low or high GFP expression, corresponding to their ability to express either ESRRG or PERM1 upon doxycycline treatment. As a next step, we differentiated the pInducer_Esrrg cell lines (Figure 5A) in the presence of doxycycline during the last 3 days of differentiation. Indeed, we could show the effective induction of Esrrg, which inversely correlated with Ucp1 mRNA expression (Figure 5B). On the protein level, UCP1 was increased in an Esrrg-dependent manner, while the representative proteins of the oxidative phosphorylation chain (OXPHOS) were not affected (Figure 5C). Similar experiments with the pInducer_Perm1 cell lines demonstrated an efficient induction of PERM1 on transcript and protein level, while UCP1 remained unaffected (Figures 5D,E). Interestingly, components of the OXPHOS showed a trend to be positively affected by PERM1 expression. To corroborate this finding, we repeated the experiment with a prolonged doxycycline treatment (last 4 days of differentiation) of the pInducer_Perm1 cell lines. The omission of doxycycline during the 6-h isoproterenol stimulation led to a significant decrease in induced Perm1 mRNA (Figure S3A) and protein levels. Moreover we observed the recently described adverse effects of doxycycline on mitochondrial stability, by inhibiting the mitochondrial ribosome (35). Despite these confounding effects, we demonstrated an increase in representative OXPHOS proteins in a PERM1 dependent manner (Figures 5F,G). Moreover, these experiments did not reveal a direct regulation of Perm1 by Esrrg or vice versa (Figure S3B).
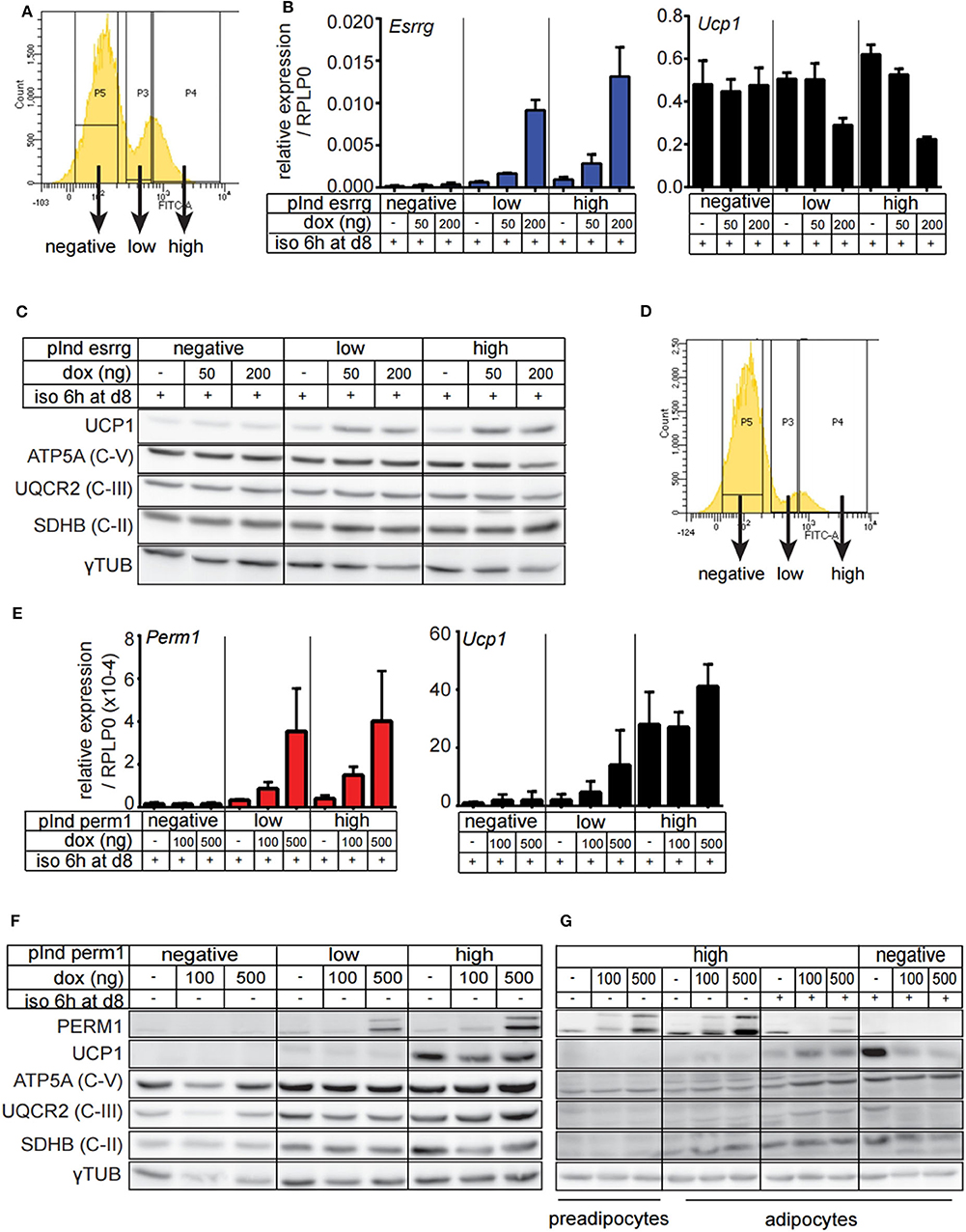
Figure 5. Generation and characterization of brown adipocyte cell lines expressing either ESRRG or PERM1 in a doxycycline dependent manner. (A) Representative FACS plots from immortalized brown pre-adipocytes transduced with lentiviral particles containing pInd21_ESRRG to polyclonally generate new cell lines where ESRRG is not expressed or is lowly or highly induced by doxycycline treatment, respectively. (B) Esrrg and Ucp1 expression in in vitro differentiated pInducer_ESRRG cell lines, treated with doxycycline from day 5 to 8. Data are presented as relative expression to reference gene RPLP0 with SD (n = 3). (C) UCP1, ATP5A, UQCR2, and SDHB expression in in vitro differentiated pInducer_ESRRG cell lines, treated with doxycycline from day 5 to 8. Gray dotted line indicates removal of the marker lane from the digital blot image. (D) Representative FACS plots from immortalized brown pre-adipocytes transduced with lentiviral particles containing pInd21_PERM2 to polyclonally generate new cell lines where PERM1 is not expressed or is lowly or highly induced by doxycycline treatment, respectively. (E) Perm1 and Ucp1 expression in in vitro differentiated pInducer_PERM1 cell lines treated with doxycycline from day 5 to 8. Data are presented as relative expression to reference gene RPLP0 with SD (n = 3). (F) UCP1, ATP5A, UQCR2, and SDHB protein levels in in vitro differentiated pInducer_PERM1 cell lines, treated with doxycycline from day 5 to 8. Gray dotted line indicates removal of the marker lane from the digital blot image. (G) UCP1, ATP5A, UQCR2, and SDHB expression in pInducer_PERM1 cell lines treated in preadipocyte state with doxycycline for 96 h or in vitro differentiated and treated with doxycycline from day 4 to 8. Gray dotted lines indicate removal of the marker lane from the digital blot image.
Since ESRRG and PERM1 can positively regulate brown adipocyte function in vitro, we aimed to investigate whether their overexpression also supports browning of white adipose tissue in vivo. Therefore, we directly injected adenoviruses into the right ingWA fat-pad of 129/SV mice, which led to the overexpression of either Esrrg or Perm1, and control virus for LacZ expression into the left ingWA fat pad of the same mouse (Figure 6A). Two days after injection, our target proteins were only expressed locally, and we could not detect a spread to liver on transcript (Figure S4) and protein level (Figure 6B). Two days after the injections, we exposed the animals to 8°C for 7 days to induce browning of ingWA. Intriguingly, despite varying degrees of browning in the individual animals, we observed an asymmetric browning between the fat pads injected with virus overexpressing Esrrg and the control virus, supporting that Esrrg overexpression lead to increased levels of UCP1 in vivo (Figure 6C). Perm1 overexpression on the other hand, led to a selective increase in Complex 1, 3, and 5 of the OXPHOS, while UCP1 levels remained rather constant (Figures 6D,E). In summary, based on our data we propose that ESRRG and PERM1 support browning in vitro and in vivo. While ESRRG regulates UCP1 abundance, PERM1 controls the amount of the OXPHOS components.
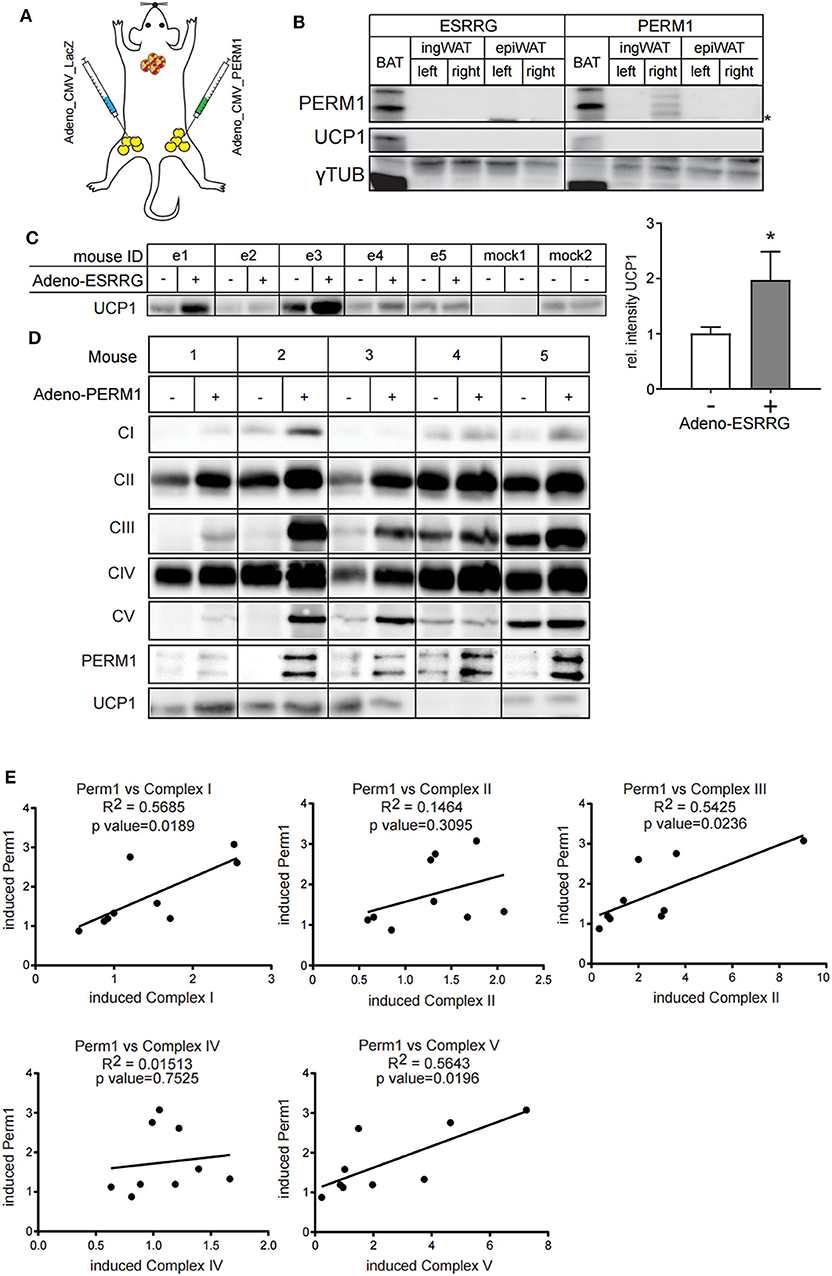
Figure 6. ESRRG and PERM1 positively regulate browning of ingWAT in vivo. (A) Schematic illustration of the direct injection of adenoviral particles leading to the overexpression of LacZ (left ingWA fat pad) and PERM1 (right ingWA fat pad) in the same mouse. (B) PERM1 detection in the ingWA fat pad injected with the adenoviral particles for PERM1 overexpression. (C) Representative western blot and quantification of UCP1 in the ingWA fat pad injected with adenoviral particles for ESRRG overexpression (+). (D) Representative blots for OXPHOS complex subunits, UCP1 and PERM1 expression in ingWA fat pad injected with the adenoviral particles of PERM1 overexpression. (E) Correlation of PERM1 expression with different OXPHOS subunits as shown by protein expression in nine mice injected in one ingWA fat pad with adenoviral particles for PERM1 overexpression. A statistically significant result, meaning p-value < 0.05, is indicated with an asterisk (*).
During cold exposure, mice must increase their thermogenic capacity to maintain their body temperature by increasing brown adipose tissue mass and forming brite/beige adipocytes within white adipose tissue depots (36, 37). Experimental evidence demonstrates that brite/beige adipocytes can arise from de novo differentiated stem cells (9, 10) or by conversion of mature adipocytes in the ingWA (12, 13). In this study, we solely analyzed the floating adipocyte fraction after cold exposure, thereby excluding cold induced transcriptional changes in stromal cells, as is described in whole tissue analyses (38). We could show that common browning factors are already fully upregulated on a transcriptional level after 24 h of cold stimulation, while morphological and UCP1 protein expressing brite/beige adipocytes are only detectable after several days of cold exposure, depending on the mouse model used (9, 12, 25). Mitochondria are the main functional organelles for non-shivering thermogenesis (26). Based on our transcriptomic profile, we detected a selective upregulation of specific OXPHOS components after 24 h of cold exposure, indicating remodeling concurrent with expansion of mitochondria, during the conversion of a phenotypic white to a phenotypic brite/beige cell. Similar observations have been made in ultra-structural analyses of mitochondria during cold adaption in inguinal white adipocytes (39). On a proteomic level, it has been shown, that brown fat mitochondria rather resemble those of muscle than of white adipose tissue (40). Intriguingly, ESRRG and PERM1, the candidates we propose to govern this phenotypic conversion, are important regulators of mitochondrial capacity in muscle tissues (22, 41).
The orphan nuclear receptor ESRRG has already been shown to be necessary for the induction of UCP1 in in vitro differentiating cells (17) as well as for maintaining BAT thermogenic activity in vivo (16, 18). We corroborated these results by showing that knockdown of Esrrg in brown preadipocytes or in mature brown adipocyte in vitro leads to a decrease in UCP1 expression. This data is consistent with the phenotype of adipocyte-specific Esrrg knockout mice, which display marked downregulation of BAT-selective genes, pronounced whitening of BAT and cold intolerance (18). Moreover, it has recently been shown that ESRRG interacts with GADD45γ to regulate UCP1 levels during cold adaption of brown adipose tissue (19). According to our data, ESRRG exerts a similar function in regulating UCP1 induction during the formation of brite/beige adipocytes. Cold exposure of 129/SV mice for 24 h showed increased levels of Esrrg and Ucp1 in the mature adipocyte fraction of the ingWAT.
The potential transcription factor PERM1 has not been described in the context of adipose tissue so far, but was recently reported to regulate OXPHOS proteins in muscle in vitro and in vivo (21, 22). We observed very low expression levels of PERM1 in brown adipocytes in vitro similar to a study comparing in vitro differentiated C2C12 cells to primary murine muscle, where PERM1 was more than 100-fold less abundant in C2C12 cells (42).
ESRRG and PERM1 act downstream of PGC1α, a master-regulator of mitochondrial capacity (23). Since PGC1α is highly regulated post-transcriptionally (43, 44), the transcript level alone is of limited informational value. When we analyzed the transcriptional profiles of the adipocyte fractions of different adipose tissue depots in 129/SV and C57BL/6 mice, we found that although Pgc1a expression is similar between the two strains and different depots, Esrrg and Perm1 expression accurately mirrors the increased thermogenic capacity of 129/SV mice. Moreover, we gathered further evidence that total thermogenic capacity is kept rather constant (45, 46) by demonstrating that C57BL/6 mice have higher expression of classical browning marker genes in interscapular brown adipocytes, while in 129/SV mice, expression of browning markers is distributed between interscapular brown and inguinal white adipocyte fractions. In vivo, we could show that overexpression of PERM1, although does not increase UCP1 expression, correlates with OXPHOS components regulation, suggesting an important role of PERM1 in mitochondrial function. Multiple reports indicate that regulating brite adipocyte formation is sufficient to elicit a metabolic phenotype (24, 47–49). Therefore, molecules inducing brown/beige adipocyte formation and activity such as irisin, BMP7, FGF21, natriuretic peptides and mineralocorticoid receptor antagonists have attracted a lot of attention in recent years (44, 50–54). Here, we show that ESRRG and PERM1 govern the phenotypic conversion of white to brite/beige adipocytes. However, future studies are needed to validate the therapeutic potential of these two transcriptional regulators in animal models of obesity.
By analyzing the transcriptome of adipocytes of different mouse models and inguinal adipocytes during cold exposure, we identified ESRRG and PERM1 as regulators of brite/beige adipocyte formation. Both ESRRG and PERM1 are under the control of the PGC1α transcriptional network and upon cold exposure, their expression is increased in ingWA. While ESRRG controls UCP1 levels, PERM1 is regulating the components of the oxidative phosphorylation chain and both processes together are key events in the phenotypic conversion of a white to a brite/beige adipocyte.
Our paper lacks the exact molecular mechanism through which PERM1 and ESRRG modulate the browning process. We show descriptive evidence from several angles, but in the scope of this study lack the appropriate in vivo models, to show a mechanistic link to certain physiological functions. Future studies are needed to validate the metabolic impact of PERM1 and ESRRG on mitochondrial function and brown/beige adipocyte thermogenic activity.
All chemicals and reagents were obtained from Sigma-Aldrich, unless specified otherwise.
Coding transcript sequences for Esrrg and Perm1 were retrieved from Ensembl and synthesized (GenScript) in sequence verified puc57 carrier plasmids flanked by NruI and XhoI restriction sites (Perm1: 2455 residues; Esrrg: 1408 residues). Both constructs were cloned into pENTR1A_dual_selection (Invitrogen) entry vectors for further use.
Adenoviral particles carrying overexpression constructs under control of the CMV promotor to express either GFP or PGC1α were a kind gift by Prof. Handschin and described elsewhere (55). Plasmids to produce adenoviral particles to overexpress ESRRG or PERM1 were generated with the pAd/CMV/V5-DEST Gateway Vector Kit (Invitrogen) by gateway cloning through a one-step LR-clonase (Invitrogen) reaction. Directing of insertion and nucleotide sequence of the insert was verified by sequencing analysis (Microsynth). Viruses were produced and passaged in HEK 293A cells (Invitrogen), purified with the Adenovirus Standard Purification ViraKit (Virapur) according to manufacturer‘s instructions and titrated by FACS analysis (GFP, PGC1α) or plague assays (ESRRG, PERM1, LacZ).
The destination vector pINDUCER21 (ORF-EG) for lentivirus production was a gift from Stephen Elledge & Thomas Westbrook (Addgene plasmid # 46948) (34). ESRRG and PERM1 sequences were inserted into the pINDUCER21 plasmid by gateway cloning through a one-step LR-clonase (Invitrogen) reaction. Directing of insertion and nucleotide sequence of the insert was verified by sequencing analysis (Microsynth). Lentiviral particles were produced in HEK 293T cells (Thermo Fischer) by co-transfection of psPAX2 (a gift from Didier Trono; Addgene plasmid # 12260), pMD2.g (a gift from Didier Trono; Addgene plasmid # 12259) and pINDUCER21 plasmid in a 9:1:10 ratio using Lipofectamine 2000 (Thermo Fisher). Cells were incubated with DMEM supplemented with 10 % FBS, 1.1 % BSA and 5 mM sodium butyrate for 36 h. Supernatant was harvested, centrifuged for 5 min at 200 g to pellet cellular debris and filtered (0.45 μm). Before freezing the crude virus preparation, 5 μg/ml polybrene was added.
129S2/SvPasCrl wild-type mice for all experiments were acquired from Charles-River at 4 weeks of age. C57BL/6N wild-type mice were bred in-house. Ucp1-GFP mice were previously described (12). All experiments were performed with young adult (12 weeks old) male mice kept on an inverted 12 h dark/light cycle and fed chow diet ad libitum. Cold stimulation was performed in temperature and humidity-controlled climate chambers (Memmert) with 4-5 animals housed in type II cages at an air-temperature of 8° C. All animal procedures were approved by the Veterinary office of the Canton of Zürich.
Purified adenoviral particles were pre-incubated for 2 h at room temperature with 1.2 % poly-lysine. Animals were anesthetized by intra-peritoneal injection of 50 μl of a 1:1 mixture of Xylosine (1:5 in PBS) and Ketamine (1:2.5 in PBS). Adenoviruses (50 μl, 5.0 x 109 PFU/ml per fat pad) were directly injected into the ingWAT fat pad through the skin.
For tissue sampling, animals were euthanized in carbon dioxide atmosphere. Popliteal lymph nodes were carefully removed, and adipose tissue depots were dissected and snap frozen in liquid nitrogen. For tissue sampling after adenoviral injections, euthanized animals were perfused utilizing a peristaltic pump with PBS containing 5 mM EDTA.
Dissected adipose tissues were minced and incubated in 5 ml collagenase buffer (25 mM KHCO3, 12 mM KH2PO4, 1.2 mM MgSO4, 4.8 mM KCl, 120 mM NaCl, 1.2 mM CaCl2, 5 mM glucose, 2.5% BSA, 2 mg/ml collagenase type II (clostridium histolyticum); sterile filtered (0.2 μm) for 60 min at 37 °C on a shaker and repeatedly re-suspended. The reaction was stopped by addition of 5 ml DMEM (Gibco) supplemented with 10 % FBS (Gibco) and 1 % Pen/Strep (Gibco) and centrifugation for 5 min at 200 g. The floating adipocyte fraction was carefully removed and filtered through 100 μm cell strainer (BD bioscience). Before further processing, the adipocyte fraction was washed three times by careful resuspension in separate tubes with 1 ml PBS (Gibco) followed by centrifugation for 2 min at 200 g. The SVF pellet was filtered through a 40 μm cell strainer (BD bioscience) and incubated in 1 ml erythrocyte lysis buffer (154 mM NH4lC, 10 mM KHCO3 and 0.1 m M EDTA) for 5 min at 24°C. After centrifugation for 5 min at 200 g, the SVF cells were re-suspended in DMEM supplemented with 10 % FBS and 1 % Pen/Strep.
FACS sorting of adipocytes according to their GFP expression was described previously (56). Briefly, cell sorting of adipocytes was performed using an Aria III high-speed sorter (BD Bioscience). A nozzle of 130 mm diameter, sheath pressure of 10 psi and a standard 4-way purity mask as described in the sorter manual was used during all sorts. Transgenic Ucp1-eGFP mice, expressing the eGFP protein under the control of Ucp1 promoter, were cold acclimated at 8°C for 7 days and the mature adipocyte fractions from iBAT and iWAT were separated by FACS using eGFP. The adipocyte population was first defined in the forward and side scatter by size and internal complexity characteristics. The GFP+ population was defined in the respective gate and the mature brown adipocytes were isolated from the mature fraction of iBAT. The same strategy was applied for the mature adipocyte fraction of iWAT to isolate the GFP+ population, which constitutes brite adipocytes, and the adjacent GFP− population that constitutes white adipocytes. For each sample, 3 or 6 same gender mice were pooled per sample and 500–3,000 cells were collected directly in RNA Lysis Buffer (Qiagen) and kept on ice until frozen and stored at −80°C.
All cells were routinely passaged in complete DMEM (Gibco) supplemented with 10 % FBS (Gibco) and 1 % Pen/Strep (Gibco), unless stated otherwise. Isolated SVF cells were plated confluent at a density of 100 000 per well in a 96-well plate (day−2). Brown adipogenesis was induced (day 0) by addition of 5 mM dexamethasone, 0.5 mg/ml insulin, 0.5 mM isobutyl methylxanthine, 1 μM rosiglitazone and 1 nM T3 in complete DMEM. Two days after induction the medium was switched to complete DMEM supplemented with 0.5 mg/ml insulin and 1 nM T3, the medium was refreshed every other day until the cells were harvested (day 8). Immortalized brown pre-adipocytes as well as derived pINDUCER21_ESRRG and pINDUCER21_PERM1 cell lines were grown to confluence. Differentiation was induced the next day by addition of complete DMEM supplemented with 20 nM insulin, 0.5 mM isobutyl methylxanthine, 125 μM indomethacin, 1 μM dexamethasone, 1 nM T3 and 1 μM rosiglitazone. Two days after induction the medium was switched to complete DMEM supplemented with 20 nM insulin and 1 nM T3, consecutively the medium was refreshed every other day until the cells were harvested (day 8). Isoproterenol (1 μM) and doxycycline treatments of the pINDUCER cell lines were performed in addition, at the indicated time-points. To knockdown candidate proteins, 15 nmol siRNA smart-pools (4 siRNAs per gene, Dharmacon) were reverse-transfected into pre-adipocytes (day−2) or mature adipocytes (day 5) using Lipofectamine RNAiMAX (Invitrogen).
Adenoviral infections were performed at day 5 of differentiation. Purified viral particles were pre-incubated in complete DMEM supplemented with 1.2 % poly-lysine (vol/vol) for 2 h at room temperature, before being added to the culture at MOI of 10, if not indicated otherwise.
For lentiviral infection of immortalized brown pre-adipocytes, crude virus preparation was added to sub-confluent cells and the culture plate centrifuged at 200 g for 10 min. After 1-h incubation at 37°C the crude virus was diluted 1:1 with complete DMEM. After 24 h the medium was changed to complete DMEM. The infected cells were passaged for 2 weeks and the integration of the pINDUCER construct verified by visible GFP expression through microscopy. Polyclonal FACS sorting was performed at the ETHZ Flow Cytometry Core Facility on a FACS Aria III (BD Biosciences) flow cytometer.
RNA extraction of adipocyte fractions from single mice for microarray analysis was performed using the RNeasy Mini Kit (Qiagen). For NGS analysis, adipocytes fractions of three mice were pooled and lysed in Trizol reagent (Life Technologies). Samples were centrifuged at 14,000 x g for 15 min and the interphase transferred to a new tube. After addition of chloroform, RNA in the colorless upper phase was purified with the RNeasy Mini Kit (Qiagen). From whole tissues and in vitro differentiated adipocytes, RNA was extracted with the Trizol reagent according to the manufacturer‘s instructions followed by DNase digestion (Invitrogen) and purification through NaOAc precipitation. RNA quantity and purity were controlled with a Nanodrop-2000 spectrophotometer. Of in vitro differentiated SVF cells and pInducer cell lines RNA extraction, DNase digestion and direct on-column cDNA synthesis was performed using the MultiMACS system (Miltenyi Biotec) according to the manufacturer's instructions.
Reverse transcription was performed using the High Capacity cDNA Reverse transcription kit (Applied Biosystems) with 1 μg RNA per reaction or the MultiMACS system (Miltenyi Biotec). QRT-PCR was performed with Fast Sybr Green Master Mix (Life Technologies) in 10 μl reactions in 96-well format on StepOnePlus (Applied Biosystems) or in 384-well on ViiA7 (Applied Biosystems) systems. All primers used are listed in Supplemental Table 1.
RNA probe labeling, purification, hybridization to the microarray and scanning was performed by the Functional Genomics Center Zurich (FGCZ) utilizing the Mouse Gene Expression v2 4 × 44K, G4846A Microarray Kit (Agilent). Raw data extraction was performed by the FGCZ, in brief, the raw probe intensities were extracted with Agilent's Feature Extraction software (v9.5.3.1). Log2-transformation and normalization using the quantile normalization method were performed with R (v3.1). To find genes with significant expression changes between groups, empirical Bayes statistics were applied to the data by moderating the standard errors of the estimated values using the limma-package (v3.18) (57). P-values were obtained from the moderated t-statistic and corrected for multiple testing with the Benjamini–Hochberg method.
RNA sequencing was performed utilizing the HiSeq 2000 (Illumina) platform as previously described (56). Raw data was analyzed at the FGCZ. In brief, sequences were aligned to the Mus musculus reference genome (build GRCm38) and quantification of gene level expression was carried out using RSEM (Version 1.2.12) (58, 59). RNAseq data of pure murine white, brite/beige and brown adipocytes was previously published (56) and is available in the European Nucleotide Archive under accession number PRJEB20634.
In vitro differentiated adipocytes were scraped off the culture plates in RIPA buffer [50 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 % NP40, protease inhibitor cocktail (Roche)]. Muscle and heart tissues were powdered on dry ice using a mortar and pestle prior to re-suspension in RIPA. Lysates were further processed with a TissueLyser LT (Qiagen; 5 min at 50 Hz) and sonicated in an ice-cooled branson-type sonication bath for 30 min before being cleared of debris by centrifugation at 14,000 g for 15 min at 4°C. For adipose tissues, RIPA buffer was supplemented with 0.5 % sodium deoxycholate, 1 % Triton-X and 10 % glycerol and additionally centrifuged at 14,000 g for 15 min at 23°C to remove the liquid fat layer prior to clearing of debris. Protein concentration of the supernatants was determined either by DC Protein Assay (Bio-Rad) or Pierce BCA assay (Thermo Scientific). Equal amounts of proteins were separated on SDS-polyacrylamide gels in a Mini-Protean tetra (Bio-Rad) apparatus, before transfer to a nitrocellulose membrane (PerkinElmer). Antibodies used for probing were UCP1 (1:1000, ab10983, Abcam), PERM1 (C1orf170, 1:1000, HPA031711, Atlas Antibodies), total OXPHOS (1:500, ab110413, Abcam), pan-Actin (D18C11, 1:1000, CS#8456, Cell Signaling) and γ-tubulin (1:6000, T6557, Sigma-Aldrich). Chemiluminescent signals of the HRP-conjugated secondary antibodies (Calbiochem) were visualized by a LAS 4000 mini ImageQuant system (GE Healthcare Life Sciences).
For in vivo studies, littermates were randomly assigned to the groups. Sample sizes were determined on the basis of previous experiments using similar methodologies. The animal numbers used for all experiments are indicated in the corresponding figure legends. All cell culture experiments were performed with 2–3 technical replicates for RNA and protein analysis, and independently reproduced 2–4 times. Results are reported as mean ± SEM or SD, as indicated in the figure legends. Two-tailed unpaired Student's T-test was applied on comparison of two groups. In case of non-normal data distribution, a non-parametric Wilcoxon test was performed. ANOVA was applied on comparisons of multiple groups. Pearson's correlation coefficient was calculated, and all statistical analyses were performed using GraphPad Prism 6 and R version 3.4.4. Statistical differences are indicated as *for P < 0.05, **for P < 0.01 and ***for P < 0.001.
All datasets generated for this study are included in the article/Supplementary Material.
The animal study was reviewed and approved by Kantonales Vetamt Zürich.
SM and CW designed the study. BW and CW supervised the experiments. SM, AP, DD, and WS performed the experiments. SM and BW performed the proteome analysis. SM, AP, WS, and MB performed the transcriptome analysis. SM, MB, and CW wrote the paper. All authors reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2020.00387/full#supplementary-material
Figure S1. Ucp1 expression in ingWA of individual mice during cold exposure. (A) Ucp1 expression levels in the adipocyte fraction of ingWA of individual mice house at room temperature or cold exposed for 24 h or 7 days, respectively. Dotted line separates groups at room temperature with low or high Ucp1 expression. Data is presented as log2 microarray signal intensity. (B) Representative immunohistochemical images of inguinal WAT (H&E stained) from a different cohort of mice housed at room temperature (RT), 24 h or 7 days at 8°C. Major morphological changes are visible after 7 days of cold exposure, only minor changes after 24 h. Micrographs were taken at 10x magnification. (C) Representative FACS plots from interscapular BAT (iBAT) and inguinal WAT (iWAT), and gating strategy for GFP+ brown adipocytes, GFP+ brite and GFP− white adipocytes.
Figure S2. In vitro characterization of ESRRG and PERM1. (A) Gene expression in immortalized brown mature adipocytes in vitro differentiated and treated with siRNAs targeting Perm1 at day 5 of differentiation. Data are presented as relative expression (ddCt) relative to non-targeting siRNA as mean with SD (n = 3). (B) Gene expression in immortalized brown mature adipocytes in vitro differentiated and treated with siRNAs targeting Perm1 2 days prior to differentiation. Data are presented as relative expression (ddCt) relative to non-targeting siRNA as mean with SD (n = 3). (C) Ucp1 expression in immortalized brown mature adipocytes in vitro differentiated and infected either with mock or with adenovirus to overexpress PGC1α or GFP at day 5 after induction. Data are presented as relative expression to reference gene RPLP0 with SD (n = 3). (D) Gene expression in immortalized brown mature adipocytes infected with different concentrations of adenoviral particles for PGC1α overexpression. Data are presented as relative expression to the lowest virus concentration (1/32 dilution; n = 1 per condition).
Figure S3. Esrrg and Perm1 do not induce each other in in vitro differentiated brown adipocytes (A) Perm1 expression after doxycycline is omitted during 6-h isoproterenol stimulation. Data are presented as relative expression to reference gene RPLP0 as mean with SD (n = 3). (B) Perm1 and Esrrg expression in in in vitro differentiated pInducer_ESRRG and pInducer_PERM1 cell lines, respectively, treated with doxycycline from day 5 to 8. Data are presented as relative expression to reference gene RPLP0 as mean with SD (n = 3).
Figure S4. Overexpression of PERM1 or ESRRG by direct adenoviral injection is confined to the fat pad injected. (A) Esrrg or Perm1 expression in the fat pad injected with the respective adenovirus. Data are presented as relative expression to reference gene RPLP0 (n = 1 mouse per virus).
1. Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. (2004) 84:277–359. doi: 10.1152/physrev.00015.2003
2. Cinti S. The adipose organ. Prostaglandins Leukot Essent Fatty Acids. (2005) 73:9–15. doi: 10.1016/j.plefa.2005.04.010
3. Young P, Arch JR, Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Lett. (1984) 167:10–4. doi: 10.1016/0014-57938480822-4
4. Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Penicaud L, et al. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci. (1992) 103(Pt 4):931–42.
5. Rosenwald M, Wolfrum C. The origin and definition of brite versus white and classical brown adipocytes. Adipocyte. (2014) 3:4–9. doi: 10.4161/adip.26232
6. Himms-Hagen J, Cui J, Danforth E Jr., Taatjes DJ, Lang SS, Waters BL, et al. Effect of CL-316,243, a thermogenic beta 3-agonist, on energy balance and brown and white adipose tissues in rats. Am J Physiol. (1994) 266:R1371–82. doi: 10.1152/ajpregu.1994.266.4.R1371
7. Bartelt A, Heeren J. Adipose tissue browning and metabolic health. Nat Rev Endocrinol. (2014) 10:24–36. doi: 10.1038/nrendo.2013.204
8. Frontini A, Vitali A, Perugini J, Murano I, Romiti C, Ricquier D, et al. White-to-brown transdifferentiation of omental adipocytes in patients affected by pheochromocytoma. Biochim Biophys Acta. (2013) 1831:950–9. doi: 10.1016/j.bbalip.2013.02.005
9. Wang QA, Tao C, Gupta RK, Scherer PE. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat Med. (2013) 19:1338–44. doi: 10.1038/nm.3324
10. Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. (2012) 150:366–76. doi: 10.1016/j.cell.2012.05.016
11. Cinti S. Transdifferentiation properties of adipocytes in the adipose organ. Am J Physiol Endocrinol Metab. (2009) 297:E977–86. doi: 10.1152/ajpendo.00183.2009
12. Rosenwald M, Perdikari A, Rulicke T, Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nat Cell Biol. (2013) 15:659–67. doi: 10.1038/ncb2740
13. Lee YH, Petkova AP, Konkar AA, Granneman JG. Cellular origins of cold-induced brown adipocytes in adult mice. FASEB J. (2015) 29:286–99. doi: 10.1096/fj.14-263038
14. Alaynick WA, Way JM, Wilson SA, Benson WG, Pei L, Downes M, et al. ERRγ regulates cardiac, gastric, and renal potassium homeostasis. Mol. Endocrinol. (2010) 24:299–309. doi: 10.1210/me.2009-0114
15. Gantner ML, Hazen BC, Eury E, Brown EL, Kralli A. Complementary roles of estrogen-related receptors in brown adipocyte thermogenic function. Endocrinology. (2016) 157:4770–81. doi: 10.1210/en.2016-1767
16. Brown EL, Hazen BC, Eury E, Wattez JS, Gantner ML, Albert V, et al. Estrogen-related receptors mediate the adaptive response of brown adipose tissue to adrenergic stimulation. iScience. (2018) 2:221–37. doi: 10.1016/j.isci.2018.03.005
17. Dixen K, Basse AL, Murholm M, Isidor MS, Hansen LH, Petersen MC, et al. ERRgamma enhances UCP1 expression and fatty acid oxidation in brown adipocytes. Obesity. (2013) 21:516–24. doi: 10.1002/oby.20067
18. Ahmadian M, Liu S, Reilly SM, Hah N, Fan W, Yoshihara E, et al. ERRgamma preserves brown fat innate thermogenic activity. Cell Rep. (2018) 22:2849–59. doi: 10.1016/j.celrep.2018.02.061
19. Gantner ML, Hazen BC, Conkright J, Kralli A. GADD45γ regulates the thermogenic capacity of brown adipose tissue. Proc Natl Acad Sci USA. (2014) 111:11870–5. doi: 10.1073/pnas.1406638111
20. Xu S, Mao L, Ding P, Zhuang X, Zhou Y, Yu L, et al. 1-Benzyl-4-phenyl-1H-1,2,3-triazoles improve the transcriptional functions of estrogen-related receptor γ and promote the browning of white adipose. Bioorg Med Chem. (2015) 23:3751–60. doi: 10.1016/j.bmc.2015.03.082
21. Cho Y, Hazen BC, Russell AP, Kralli A. Peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. J Biol Chem. (2013) 288:25207–18. doi: 10.1074/jbc.M113.489674
22. Cho Y, Hazen BC, Gandra PG, Ward SR, Schenk S, Russell AP, et al. Perm1 enhances mitochondrial biogenesis, oxidative capacity, and fatigue resistance in adult skeletal muscle. FASEB J. (2016) 30:674–87. doi: 10.1096/fj.15-276360
23. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. (1998) 92:829–39. doi: 10.1016/S0092-8674(00)81410-5
24. Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. (2014) 156:304–16. doi: 10.1016/j.cell.2013.12.021
25. Gospodarska E, Nowialis P, Kozak LP. Mitochondrial turnover: a phenotype distinguishing brown adipocytes from interscapular brown adipose tissue and white adipose tissue. J Biol Chem. (2015) 290:8243–55. doi: 10.1074/jbc.M115.637785
26. Shabalina IG, Petrovic N, de Jong JM, Kalinovich AV, Cannon B, Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Rep. (2013) 5:1196–203. doi: 10.1016/j.celrep.2013.10.044
27. Kozak LP. The genetics of brown adipocyte induction in white fat depots. Front Endocrinol (Lausanne). (2011) 2:64. doi: 10.3389/fendo.2011.00064
28. Li Y, Bolze F, Fromme T, Klingenspor M. Intrinsic differences in BRITE adipogenesis of primary adipocytes from two different mouse strains. Biochim Biophys Acta. (2014) 1841:1345–52. doi: 10.1016/j.bbalip.2014.06.003
29. Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. (2007) 6:13–24. doi: 10.1016/j.cmet.2007.06.007
30. Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. (2007) 6:38–54. doi: 10.1016/j.cmet.2007.06.001
31. Harms MJ, Ishibashi J, Wang W, Lim HW, Goyama S, Sato T, et al. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. (2014) 19:593–604. doi: 10.1016/j.cmet.2014.03.007
32. Klein J, Fasshauer M, Klein HH, Benito M, Kahn CR. Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action. Bioessays. (2002) 24:382–8. doi: 10.1002/bies.10058
33. Handschin C, Lin J, Rhee J, Peyer AK, Chin S, Wu PH, et al. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell. (2005) 122:505–15. doi: 10.1016/j.cell.2005.06.040
34. Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA. (2011) 108:3665–70. doi: 10.1073/pnas.1019736108
35. Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, et al. Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. (2015) 10:1681–91. doi: 10.1016/j.celrep.2015.02.034
36. Lowell BB, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. (1993) 366:740–2. doi: 10.1038/366740a0
37. Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. (1997) 387:90–4. doi: 10.1038/387090a0
38. Rosell M, Kaforou M, Frontini A, Okolo A, Chan YW, Nikolopoulou E, et al. Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am J Physiol Endocrinol Metab. 306:E945–64. doi: 10.1152/ajpendo.00473.2013
39. Loncar D. Convertible adipose tissue in mice. Cell Tissue Res. (1991) 266:149–61. doi: 10.1007/BF00678721
40. Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metab. (2009) 10:324–35. doi: 10.1016/j.cmet.2009.08.014
41. Rangwala SM, Wang X, Calvo JA, Lindsley L, Zhang Y, Deyneko G, et al. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. J Biol Chem. (2010) 285:22619–29. doi: 10.1074/jbc.M110.125401
42. Deshmukh AS, Murgia M, Nagaraj N, Treebak JT, Cox J, Mann M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol Cell Proteomics. (2015) 14:841–53. doi: 10.1074/mcp.M114.044222
43. Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. (2011) 93:884S−90. doi: 10.3945/ajcn.110.001917
44. Fisher FM, Kleiner S, Douris N, Fox EC, Mepani RJ, Verdeguer F, et al. FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. (2012) 26:271–81. doi: 10.1101/gad.177857.111
45. Schulz TJ, Huang P, Huang TL, Xue R, McDougall LE, Townsend KL, et al. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. (2013) 495:379–83. doi: 10.1038/nature11943
46. Nedergaard J, Cannon B. The browning of white adipose tissue: some burning issues. Cell Metab. (2014) 20:396–407. doi: 10.1016/j.cmet.2014.07.005
47. Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell. (2015) 163:643–55. doi: 10.1016/j.cell.2015.09.035
48. Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. (2012) 150:620–32. doi: 10.1016/j.cell.2012.06.027
49. Shinoda K, Ohyama K, Hasegawa Y, Chang HY, Ogura M, Sato A, et al. Phosphoproteomics identifies CK2 as a negative regulator of beige adipocyte thermogenesis and energy expenditure. Cell Metab. (2015) 22:997–1008. doi: 10.1016/j.cmet.2015.09.029
50. Armani A, Cinti F, Marzolla V, Morgan J, Cranston GA, Antelmi A, et al. Mineralocorticoid receptor antagonism induces browning of white adipose tissue through impairment of autophagy and prevents adipocyte dysfunction in high-fat-diet-fed mice. FASEB J. (2014) 28:3745–57. doi: 10.1096/fj.13-245415
51. Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. (2012) 81:463–8. doi: 10.1038/nature10777
52. Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. (2008) 454:1000–4. doi: 10.1038/nature07221
53. Liu D, Ceddia RP, Collins S. Cardiac natriuretic peptides promote adipose 'browning' through mTOR complex-1. Mol Metab. (2018) 9:192–8. doi: 10.1016/j.molmet.2017.12.017
54. Bordicchia M, Liu D, Amri EZ, Ailhaud G, Dessi-Fulgheri P, Zhang C, et al. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J Clin Invest. (2012) 122:1022–36. doi: 10.1172/JCI59701
55. Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. (2000) 106:847–56. doi: 10.1172/JCI10268
56. Perdikari A, Leparc GG, Balaz M, Pires ND, Lidell ME, Sun W, et al. BATLAS: deconvoluting brown adipose tissue. Cell Rep. (2018) 25:784–97. doi: 10.1016/j.celrep.2018.09.044
57. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
58. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
Keywords: beige adipocytes, brown adipocyte, adipogenesis, estrogen receptor, adipose tissue
Citation: Müller S, Perdikari A, Dapito DH, Sun W, Wollscheid B, Balaz M and Wolfrum C (2020) ESRRG and PERM1 Govern Mitochondrial Conversion in Brite/Beige Adipocyte Formation. Front. Endocrinol. 11:387. doi: 10.3389/fendo.2020.00387
Received: 14 February 2020; Accepted: 15 May 2020;
Published: 12 June 2020.
Edited by:
Patrick C. N. Rensen, Leiden University Medical Center, NetherlandsReviewed by:
Didier F. Pisani, Centre National de la Recherche Scientifique (CNRS), FranceCopyright © 2020 Müller, Perdikari, Dapito, Sun, Wollscheid, Balaz and Wolfrum. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Wolfrum, Y2hyaXN0aWFuLXdvbGZydW1AZXRoei5jaA==; Miroslav Balaz, bWlyb3NsYXYtYmFsYXpAZXRoei5jaA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.