
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 18 October 2019
Sec. Experimental Endocrinology
Volume 10 - 2019 | https://doi.org/10.3389/fendo.2019.00705
Melanocortin-4 receptor (MC4R) plays important roles in regulation of multiple physiological processes including energy homeostasis, reproduction, sexual function, and other functions in mammals. Recent studies suggested that teleost MC4Rs have different physiological functions and pharmacological characteristics when compared to mammalian MC4Rs. In this study, we investigated spotted sea bass (Lateolabrax maculatus) MC4R (LmMC4R) physiology and pharmacology. Spotted sea bass mc4r consisted of a 984 bp open reading frame encoding a protein of 327 amino acids. LmMC4R was homologous to those of several teleost MC4Rs and human MC4R (hMC4R). qRT-PCR and in situ hybridization revealed that mc4r transcripts were highly expressed in the brain, followed by pituitary and liver. Brain mc4r transcripts were down-regulated in long-term and short-term fasting challenges. LmMC4R was a functional receptor with lower maximal binding and higher basal activity than hMC4R. THIQ was not able to displace 125I-NDP-MSH but could affect intracellular cAMP accumulation, suggesting that it was an allosteric ligand for LmMC4R. In vitro studies with spotted sea bass brain cells indicated that mRNA levels of neuropeptide Y and Agouti-related peptide were down-regulated by α-MSH. In summary, we cloned spotted sea bass MC4R, and showed that it had different pharmacological properties compared to hMC4R, and potentially different functions.
Melanocortin peptides are posttranslational products of proopiomelanocortin (POMC) that include α-, β-, and γ- melanocyte-stimulating hormones (α-, β-, and γ-MSH) and adrenocorticotropic hormone (ACTH) [reviewed in (1, 2)]. Melanocortin peptides exert their effects by activating melanocortin receptors (MCRs). Five MCRs have been cloned, named MC1R to MC5R based on the order in which they were first cloned [reviewed in (3, 4)]. MC4R belongs to Family A rhodopsin-like G protein-coupled receptors (GPCRs) and it primarily couples to the stimulatory G protein (Gs) to activate adenylyl cyclase, leading to increased level of intracellular cyclic adenosine monophosphate (cAMP) to activate downstream protein kinase A (PKA). α-MSH, ACTH and other POMC-derived peptides are endogenous agonists and Agouti-related peptide (AgRP) is endogenous antagonist of MC4R. In addition, analogs of α-MSH and some small molecules have also been identified as MC4R ligands. [Nle4, D-Phe7]-α-MSH (NDP-MSH) is a superpotent analog of α-MSH that is widely used in pharmacological studies of MCRs (5). THIQ, (N-[(3R)-1,2,3,4-tetrahydroisoquinolinium-3-ylcarbonyl]-(1R)-1-(4-chlorobenzyl)- 2-[4-cyclohexyl-4-(1H-1,2,4-triazol-1-ylmethyl)piperidin-1-yl]-2-oxoethylamine), is a small molecule agonist (6).
Activation of neurons expressing neuropeptide-Y (NPY) and AgRP increases food intake, while activation of neurons expressing POMC decreases food intake in human and mice [reviewed in (7)]. POMC-derived peptides, such as α-MSH and ACTH, are anorexigenic by activating MC4R. In human, two groups independently reported that MC4R frameshift mutations are associated with severe early-onset obesity in 1998 (8, 9). Since then, a total of at least 175 distinct MC4R mutations have been identified from patients associated with obesity and other diseases [reviewed in (10, 11)]. Mice lacking Mc4r expression have increased food intake and decreased energy expenditure, resulting in obesity and hyperinsulinemia (12). In addition to regulation of energy balance, recent studies reported that MC4R is also involved in reproductive functions via regulating hypothalamus-pituitary-gonad axis and prolactin secretion (13–15).
MC4R and other MCRs have also been identified in tetrapods and teleosts. In tetrapods, all MCRs (MC1R-MC5R) have been identified and higher MC4R expression was observed in central nervous system (3, 16). In teleosts, MC4R is expressed in both central and peripheral tissues (17–23). In cavefish (Astyanax mexicanus), non-synonymous mc4r mutations cause increased appetite and starvation resistance (24). In zebrafish, overexpression of AgRP leads to obesity phenotype (25). Intracerebroventricular (i.c.v) injection of MC4R agonist decreases food intake, while injection of MC4R antagonist increases food intake in goldfish and rainbow trout (Oncorhynchus mykiss) (26, 27). These results suggest that teleost MC4R also acts as a regulator in energy balance. Teleost MC4Rs are also associated with the onset of puberty, growth and body size, and sexual behaviors in a species-specific manner in different teleosts (28, 29). We showed that administration of MC4R ligands to spotted scat can change expression of genes related to reproduction (30).
Our previous studies showed that teleost MC4Rs have different pharmacological characteristics from mammalian MC4Rs. For example, compared to human MC4R (hMC4R), teleost MC4Rs display high basal activities (20–23). Moreover, THIQ acts as an orthosteric agonist to activate mammalian MC4Rs; however, it activates teleost MC4Rs allosterically (20, 22). Therefore, in the present study, we used spotted sea bass, Lateolabrax maculatus, as an animal model to systematically investigate LmMC4R physiology and pharmacology. We investigated mRNA expression and localization of mc4r in different tissues and changes in expression after fasting challenge. We also performed detailed pharmacological studies on LmMC4R including ligand binding and signaling. We included hMC4R in these experiments for comparison. We also isolated brain cells where mc4r was expressed most abundantly and stimulated these cells with α-MSH to evaluate the transcriptional changes of several genes associated with growth and energy balance.
All procedures involving fish followed the guidelines and were approved by the Animal Research and Ethics Committee of Ocean University of China (Permit Number: 20141201).
Total RNA was extracted from spotted sea bass brain using TRIzol (Invitrogen, Carlsbad, CA, USA). The concentration and integrity of total RNA were evaluated by the Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA). One microgram of RNA was used to synthesize first-strand cDNA using random primers and reverse transcriptase M-MLV with gDNA Eraser (TaKaRa, Japan). To amplify cDNA fragments of mc4r, PCR was performed and primers of mc4r were designed based on transcriptome databases (Table 1). PCRs were performed in a 25 μl mixture containing 1 μl cDNA, 0.5 μl of each primer, 2 μl dNTPs, 2.5 μl 10 × PCR buffer, 18.25 μl ddH2O, and 0.25 μl Taq DNA Polymerase (TaKaRa) with following program: initial denaturation at 94°C for 5 min, followed by 40 cycles at 94°C for 30 s, 60°C for 30 s and 72°C for 1 min. The reaction was terminated with a further extension of 5 min at 72°C. The amplification products were separated by 1.5% w/v agarose gel stained with ethidium bromide. Target fragment was purified by TIANgel Midi Purification kit (Tiangen, Beijing, China), and then subcloned into Trans1-T1 Escherichia coli (TransGen Biotech, Beijing, China). Two positive clones were sequenced on an ABI 3700 sequencer (Applied Biosystems, Foster City, CA, USA).
Multiple alignments of amino acid sequences of MC4Rs in different species were performed with DNAMAN 6.0 (Lynnon Biosoft, San Ramon, CA, USA). The percentage of similarity between amino acid sequences were calculated with DNAMAN 6.0. Phylogenetic tree based on amino acid sequences was constructed by Neighbor-joining and Maximum likelihood methods with Mega 6.0 software. The strength of branch relationships was assessed by bootstrap replication (N 1/4 1,000 replicates).
Total RNA was extracted from fresh tissues (pituitary, brain, liver, kidney, spleen, intestine, muscle, gonads, gill, heart) and treated with RNase-free DNase I (Thermo Scientific Corp, Waltham, MA, USA). M-MLV Reverse Transcriptase (Promega, Madison, WI, USA) was used for cDNA synthesis with oligo-dT (12-18) primers. The cDNA was subsequently used for amplification using specific primers based on mc4r sequence from transcriptome database. 18s mRNA expression was used as an internal reference for normalization (31). The quantitative reverse transcription PCR (qRT-PCR) reaction consisted of a total volume of 20 μl mixture containing 10 μl SYBR®FAST qPCR Master Mix (2X), 0.4 μl ROX reference dye, 2 ml template cDNA, 0.4 μl of each primer and 6.8 μl of nuclease-free water. PCR amplification was in a 96-well optical plate at 95°C for 5 s, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s, and finally followed by a dissociation curve to verify the specificity of amplified products. qRT-PCR was performed using the StepOne Plus Real-Time PCR system (Applied Biosystems) and the 2−ΔΔCT method was used to analyze the relative expression (32).
Brain, liver, and pituitary samples were fixed in buffered 4% paraformaldehyde for 24 h and then dehydrated with a graded series of ethanol solution (70–100%), cleared in xylene and embedded in paraffin. Seven-micron sections were cut for in situ hybridization. The primers for in situ hybridization of mc4r was listed in Table 1. Sense and antisense digoxigenin- (DIG)- labeled riboprobes were synthesized from the sequence (574–1,232 bp) of the mc4r using DIG RNA Labeling Kit (Roche Diagnostics, Mannheim, Germany). DIG in situ hybridization was performed as described previously (33). Briefly, the sections were rehydrated by a graded series of ethanol solution (100–70%) and then permeabilized with 0.1 M HCl for 8 min, followed by proteinase K (20 ng/μl) treatment for 20 min, prehybridized at 42°C for 1 h, and hybridized with DIG-labeled riboprobes (500 μg/ml) at 58°C overnight. After hybridization, the sections were washed and blocked with blocking reagent (Roche Diagnostics). DIG was detected with an alkaline phosphatase-conjugated anti-DIG antibody (Roche Diagnostics; diluted 1:1,000) and chromogenic development was conducted with NBT/BCIP (Roche Diagnostics). The samples were dehydrated by a series of graded ethanol, cleared in xylene, sealed with neutral resin and taken with a microscope (Olympus, Japan).
Approximately 40 basses per group (for short term fasting challenge, 100.00 ± 12.32 g) and 120 fish per group (for long term fasting challenge, 5.88 ± 0.26 g) were acclimatized in the aquaria at 25.2°C for at least 1 week. In short-term fasting, six basses per group (three duplicated groups were set) were sampled at 0, 1, 6, 12, and 24 h post fasting. In long-term fasting, 120 individuals of spotted sea bass were randomly divided into six 120-l-rectangular containers. Three to six fish per container were sampled after 8-week's fasting. All sampled fish were treated with eugenol and sampled immediately. Brain tissues were collected and temporarily placed into liquid nitrogen and finally stored at −80°C for mRNA extraction.
Spotted sea basses were anesthetized with MS-222 before decapitation. Brain tissue was removed and washed three times with phosphate buffered saline (PBS). Brain tissue was separated into small pieces in the tubes containing 500 μl trypsin (0.25%, Biological Industries, Kibbutz Beit Haemek, Israel) and then digested 5 min with 5 ml trypsin. After centrifugation for 5 min at 1,500 rpm, the supernatant was discarded and 50 mL medium containing 10 mL fetal bovine serum (FBS, Biological Industries), 500 μl antibiotics (Biological Industries) containing 100 U/mL penicillin and 100 mg/mL streptomycin, 39.5 mL M199 (Biosharp, Beijing, China) was added into each tube. The homogeneous cell suspensions were filtered into tubes by the screen mesh (40 μm Nylon, Corning, NY, USA). Finally, 1 mL cell suspension was added to each well of the 24-well plate and cells were incubated at 28°C. After 36 h incubation, α-MSH was added with fresh media with 20% FBS. Brain cells were collected after incubation and stored at −80°C for RNA extraction and qRT-PCR.
NDP-MSH, α-MSH, ACTH (1-24), and THIQ were used. NDP-MSH was purchased from Peptides International (Louisville, KY, USA), α-MSH was purchased from Pi Proteomics (Huntsville, AL, USA), ACTH (1-24) from Phoenix Pharmaceuticals (Burlingame, CA, USA), THIQ was purchased from Tocris Bioscience (Ellisville, MO, USA). 125I-NDP-MSH was iodinated as previously described (34) and 125I-cAMP was iodinated in our lab via chloramine T method (35, 36).
The N-terminal c-myc-tagged wild-type hMC4R subcloned into pcDNA3.1 was described previously (37). Open reading frame (ORF) of LmMC4R was identified by transcriptome data and the N-terminal c-myc-tagged ORF of LmMC4R was then subcloned into pcDNA3.1 vector by GenScript (Piscataway, NJ, USA) to obtain the plasmid expressing LmMC4R.
Three peptide ligands (α-MSH, ACTH (1-24) and NDP-MSH) and one small molecule ligand (THIQ) were used to evaluate ligand binding and signaling properties of LmMC4R. α-MSH and ACTH are endogenous agonists of all MCRs with the exception that α-MSH cannot activate MC2R. In this study, ACTH (1-24) instead of full-length ACTH (1-39) was used to investigate the pharmacological characteristics of LmMC4R as the first 24 amino acids of ACTH are highly conserved in different species ranging from human to teleosts and ACTH (1-24) is equipotent to the full-length ACTH (38, 39). THIQ is small molecule ligand of hMC4R and our previous studies revealed that they act as pharmacological chaperones, rescuing intracellularly retained hMC4R mutants (7, 40).
Human Embryonic Kidney (HEK) 293T cells (Manassas, VA, USA) were cultured and used for pharmacological assays. Plasmid transfection was performed as described before (41). At 48 h after transfection, binding, and signaling assays were performed.
HEK293T cells were transiently transfected with hMC4R or LmMC4R plasmid with N-terminal c-myc tag. Forty-eight hours after transfection, cells were incubated with mouse anti-myc 9E10 monoclonal antibody (Developmental Studies Hybridoma Bank, The University of Iowa, Iowa City, IA, USA) diluted 1:40 for 1 h. Cells were then washed and incubated with Alexa Fluor 488-labeled goat anti-mouse antibody (Invitrogen, Grand Island, NY, USA) diluted 1:2,000 for 1 h. The C6 Accuri Cytometer (Accuri Cytometers, Ann Arbor, MI, USA) was used for analysis. Fluorescence of cells expressing the empty vector (pcDNA3.1) was used for background staining. The expression of the LmMC4R was calculated as percentage of hMC4R expression using the following formula: [(LmMC4R – pcDNA3.1)/(hMC4R – pcDNA3.1) ×100%] (42).
For ligand binding, 48 h after transfection, cells were washed twice with warm DMEM containing 1 mg/mL bovine serum albumin (referred herein as DMEM/BSA). Subsequently, DMEM/BSA without or with different concentrations of unlabeled ligands and 80,000 cpm of [125I]-NDP-MSH were added to each well, with the total volume of 1 ml, and then the cells were incubated at 37°C for 1 h. The final concentration of various unlabeled ligands ranged from 10−11 to 10−6 M for NDP-MSH and THIQ, or from 10−10 to 10−5 M for α-MSH and ACTH (1-24). After incubation, the cells were washed twice with cold Hank's balanced salt solution containing 1 mg/mL BSA to terminate the reaction. Cells were then lysed by 0.5 M NaOH and collected for radioactive assays by gamma counter (Cobra II Auto-Gamma, Packard Bioscience, Frankfurt, Germany).
For intracellular cAMP evaluation, 48 h after transfection, HEK293T cells were washed twice with warm DMEM/BSA and then incubated with warm DMEM/BSA containing 0.5 mM isobutylmethylxanthine (Sigma-Aldrich) for 15 min. Subsequently, different concentrations of ligands were added to each well, with the total volume of 1 ml, to evaluate the ligand-stimulated intracellular cAMP levels. Final concentration of various unlabeled ligands ranged from 10−12 to 10−6 M. After 1 h incubation, the reaction was terminated on the ice and intracellular cAMP was collected by adding 0.5 M percholoric acid containing 180 μg/ml theophylline (Sigma-Aldrich) and 0.72 M KOH/0.6 M KHCO3 into each well. Intracellular cAMP levels were determined by radioimmunoassay as previously described (35) and 125I-cAMP was iodinated via chloramine T method (35, 36).
SPSS19.0 software was used to calculate the mean and standard error of the mean (S.E.M.) of gene expression results and results are presented as mean ± S.E.M. Significant differences of gene expression were determined by one-way ANOVA followed by Duncan's multiple range test with the significance level set at P < 0.05. GraphPad Prism 4.0 software (San Diego, CA, USA) was used to calculate the ligand binding and cAMP signaling parameters such as maximal binding (Bmax) and IC50 of ligand binding and maximal response (Rmax) and EC50 of cAMP signaling. The significance of differences in Bmax, IC50, Rmax and EC50 between LmMC4R and hMC4R were determined by Student's t-test by GraphPad Prism 4.0 software.
The bass mc4r gene sequence was identified from transcriptome databases (GenBank: SRR4409341/SRR4409397) (43). The total cDNA sequence of bass mc4r was 1,588 bp, containing an ORF of 984 bp that encoded a putative protein of 327 amino acids. We identified that the 5′ and 3′ untranslated region of bass mc4r was 493 and 114 bp, respectively. Like other GPCRs, LmMC4R had seven putative hydrophobic transmembrane domains (TMDs) with three extracellular loops (ECLs) and three intracellular loops (ICLs, Figure 1). The deduced amino acid sequence of TMDs, ECLs, and ICLs were significantly conserved to those of other species. The predicted amino acid sequence of LmMC4R was 94, 88, 83, 82, 70, 70, and 70% identical to European sea bass (Dicentrarchus labrax), fugu (Takifugu rubripes), common carp (Cyprinus carpio), zebrafish (Danio rerio), human (Homo sapiens), chicken (Gallus gallus) and mouse (Mus musculus) MC4Rs, respectively (Figure 2A). Phylogenetic tree analysis between LmMC4R and MC4Rs in other vertebrates revealed that LmMC4R was localized in a clade of teleost MC4Rs and was evolutionarily closer to European sea bass MC4R (Figure 2B).

Figure 1. Nucleotide and deduced amino acid sequence of LmMC4R. Positions of nucleotide and amino acid sequences are indicated on both sides. The seven TMDs are shaded in gray. The conserved motifs (PMY, DRY, and DPIIY) are highlighted in boxes. Underlines show initiation codon and most conserved residues in each TMD. Asterisk (*) denotes stop codon.
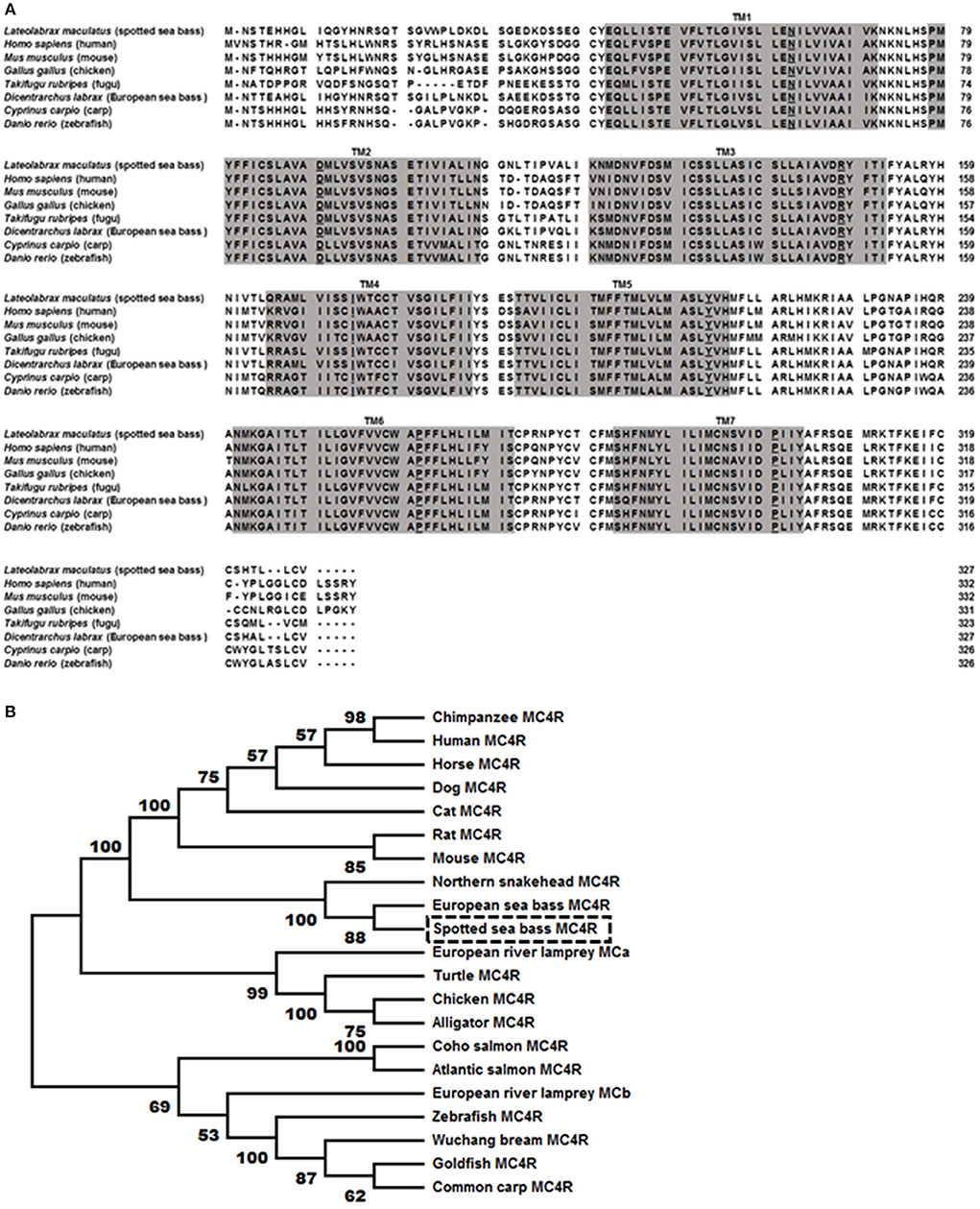
Figure 2. Comparison of amino acid sequences between LmMC4R and MC4Rs from other species (A) and phylogenetic tree of MC4R proteins (B). In (A), amino acids in shaded boxes indicate putative TMD 1-7, the most conserved residues in each TMD are underlined. In (B), trees were constructed using the neighbor-joining (NJ) method. Box shows LmMC4R. GenBank accession numbers: alligator (XP_006025279.1); Atlantic salmon (XP_014036044.1); cat (BBD19891.1); chicken (NP_001026685.1); chimpanzee (PNI69802.1); coho salmon (XP_020349696.1); common carp (CBX89936.1); dog (NP_001074193.1); European river lamprey MCRa (ABB36647.1); European river lamprey MCRb (ABB36648.1); European seabass (CBN82190.1); goldfish (CAD58853.1); horse (XP_001489706.1); human (NP_005903.2); mouse (NP_058673.2); Northern snakehead (AMM02541.1); rat (NP_037231.1); turtle (XP_024059247.1); Wuchang bream (AWA81516.1); zebrafish (NP_775385.1).
Bass mc4r mRNA expression in brain and peripheral tissues (pituitary, intestine, muscle, skin, spleen, liver, gill, kidney, stomach, heart and gonad) was analyzed by qRT-PCR (Figure 3A). The expression of 18s mRNA, a stable reference gene, was used as an internal control for normalization. Bass mc4r mRNA was highly expressed in the brain, followed by pituitary, liver and other peripheral tissues.
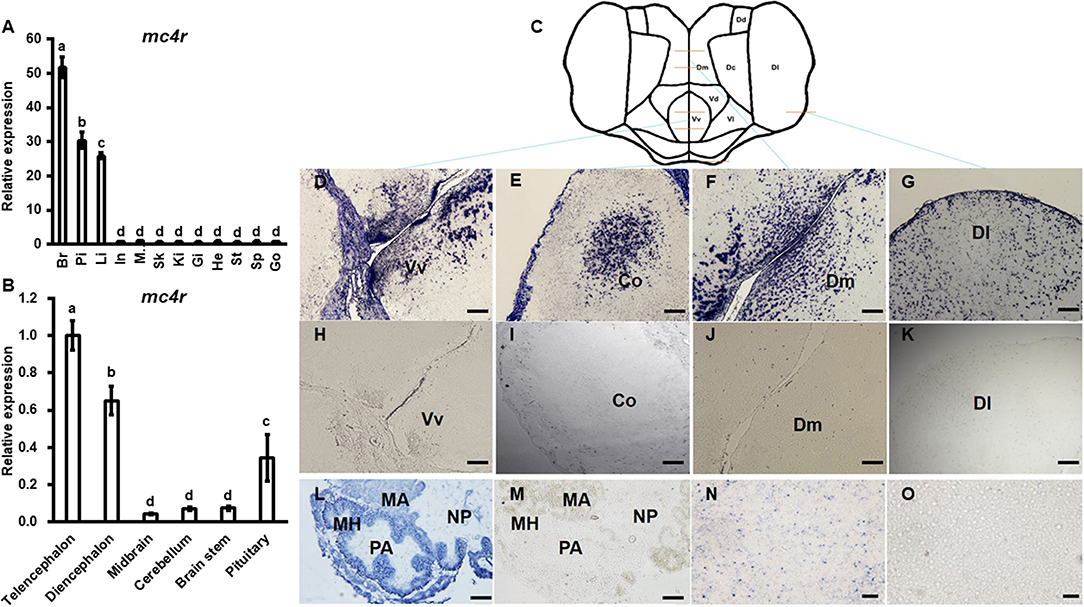
Figure 3. Expression of bass mc4r mRNA in various tissues (A) and brain regions (B) and localization in brain (D–K), pituitary (L,M) and liver (N,O). Data are presented as means ± S.E., n = 6. Different letters indicate significant difference (P < 0.05, one-way ANOVA followed by Duncan's multiple range test). In, intestines; Pi, pituitary; Mu, muscle; Br, brain; Sk, skin; Sp, spleen; Li, liver; Gi, gill; Ki, kidney; St, stomach; He, heart; Go, gonad. The structure and cut regions of telencephalon is shown in (C). (D–O) show the bass mc4r mRNA localization in brain, pituitary and liver, respectively. Hybridization of sense bass mc4r-cRNA probe did not display specific signals in the brain samples (H–K,M,O), showing specificity of the signal. Vv, ventral telencephalon; Co, central region of the olfactory bundle; DI, lateral part of the dorsal telencephalon area; Dm, medial part of the dorsal telencephalon part. PA, pro-adenohypophysis; MH, meso-adeno hypophysis; MA, meta adenohypophysis; NP, neurohypophysis. Scale bars of figs. (D–K) indicate 50 μm and figs. (L–O) indicate 20 μm.
The relative mRNA expression of bass mc4r was evaluated in different brain regions (Figure 3B). Higher mc4r mRNA expression was observed in telencephalon, diencephalon and pituitary gland (Figure 3B). In situ hybridization showed that mc4r was localized in the ventral part of the ventral telencephalon (Vv) (Figures 3D,H). The mc4r-expressing cells were observed in the brain regions of the central region of the olfactory bundle (Co) (Figures 3E,I), the lateral part of the dorsal telencephalon area (Dl) (Figures 3G,K), and the medial part of the dorsal telencephalon part (Dm) (Figures 3F,J). We also observed the localization of bass mc4r mRNA in cells of pituitary (Figures 3L,M) and liver (Figures 3N,O).
To evaluate the potential physiological functions of bass mc4r in regulating energy homeostasis, we analyzed brain mc4r mRNA expressions in short-term and long-term fasting experiments. Short-term fasting did not change body weight (data not shown). Although there was no change at 1 h after fasting, there was significant decrease in mc4r expression at 6, 12, and 24 h (Figure 4A). Expression of agrp was significantly decreased at 1, 12, and 24 h (Figure 4B), and expression of npy was significantly decreased at 6 and 12 h (Figure 4C).
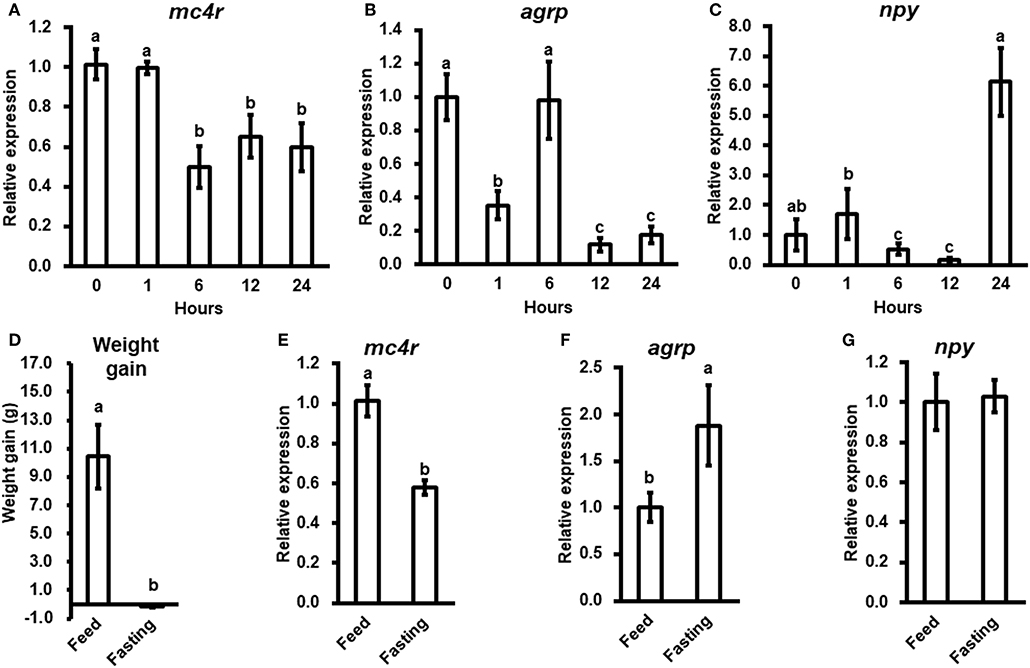
Figure 4. The relative mRNA expression of brain mc4r, agrp and npy during short-term (A–C) and long-term (E–G) fasting challenge and body weight of spotted sea bass in long-term fasting challenge (D). Data are presented as means ± S.E., n = 6. Different letters indicate significant difference (P < 0.05, one-way ANOVA followed by Duncan's multiple range test).
In long-term fasting experiment, when compared to the initial body weight (5.88 ± 0.26 g), spotted sea bass in feeding group increased body weight (16.32 ± 2.23 g) by 10.44 g, while spotted sea bass in fasting group decreased body weight (5.73 ± 0.07 g) by 0.15 g (Figure 4D). We observed that mc4r expression was significantly down-regulated (Figure 4E), agrp mRNA expression was significantly up-regulated (Figure 4F) while there was no significant change in npy expression (Figure 4G).
We stimulated isolated brain cells (Figure S1) with 10−7 or 10−6 M α-MSH for 3 h. We showed that mc4r mRNA expression was not altered (Figure 5A), while mRNA expression of agrp and npy was significantly down-regulated (Figures 5B,C).
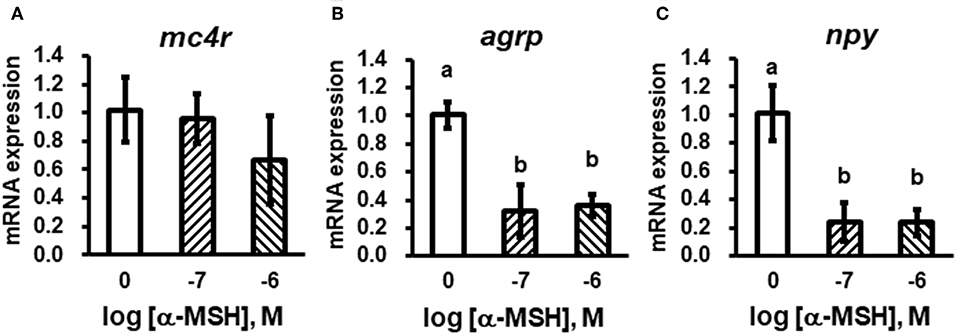
Figure 5. In vitro evaluation of bass mc4r (A), agrp (B), and npy (C) mRNA expression in isolated brain cells. Data are presented as means ± S.E., n = 6. Different letters indicate significant differences compared to control (P < 0.05, student t-test).
The cell surface and total expressions of LmMC4R were only ~2.1% to those of hMC4R, showing significant difference (Figure S2). Competitive ligand binding assays were performed to investigate the binding property of LmMC4R. Different concentrations of unlabeled ligands including α-MSH, ACTH (1-24), NDP-MSH, and THIQ, were used to compete with a fixed amount of 125I-NDP-MSH. Maximal binding value of the LmMC4R was around 20% of that of the hMC4R (Figure 6 and Table 2). Both hMC4R and LmMC4R bound to NDP-MSH with the highest affinity. When unlabeled α-MSH, NDP-MSH or ACTH (1-24) was used as the ligand, LmMC4R showed significantly lower IC50 values compared to those of hMC4R (Table 2). When THIQ was used as the unlabeled competitor, LmMC4R was not able to displace 125I-NDP-MSH, whereas dose-dependent displacement of 125I-NDP-MSH binding to the hMC4R was observed (Figure 6D and Table 2).
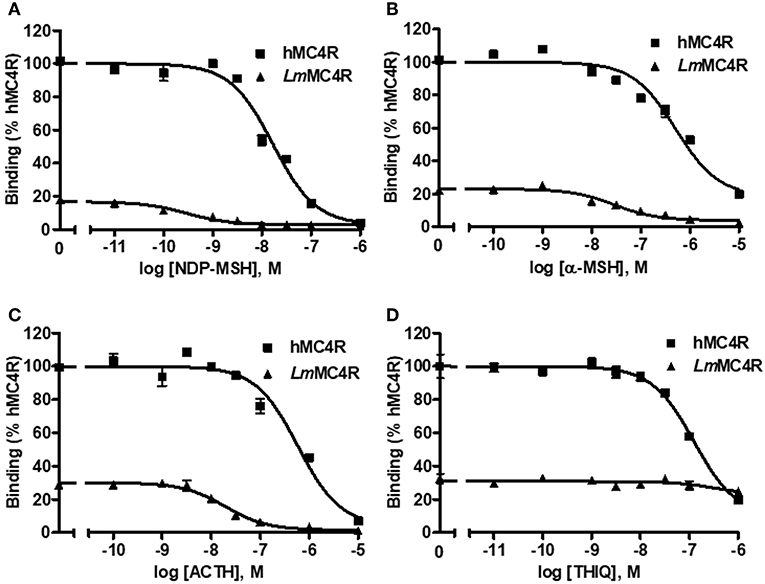
Figure 6. Ligand binding properties of LmMC4R. HEK293T cells were transiently transfected with hMC4R or LmMC4R plasmids. Forty-eight hours after transfection, different concentrations of unlabeled NDP-MSH (A), α-MSH (B), ACTH (1-24) (C), and THIQ (D) were used to displace the binding of 125I-NDP-MSH, rescpectively. Data are expressed as % of hMC4R binding ± range from duplicate measurements within one experiment. The curves are representative of 3 independent experiments.
Dose-dependent increase of intracellular cAMP was observed when LmMC4R was stimulated by NDP-MSH, α-MSH, ACTH (1-24) and THIQ (Figure 7). The maximal responses of LmMC4R in response to NDP-MSH, α-MSH, and ACTH were 144.57 ± 8.65%, 216.86 ± 16.88%, and 125.56 ± 5.45%, respectively, of those of hMC4R, whereas the maximal response of LmMC4R in response to THIQ was 57.78 ± 5.32% of that of hMC4R (Figure 7 and Table 3). EC50s of NDP-MSH and THIQ for LmMC4R were significantly higher than those for hMC4R, and EC50s of α-MSH and ACTH (1-24) for LmMC4R and hMC4R were not significantly different (Figure 7 and Table 3).
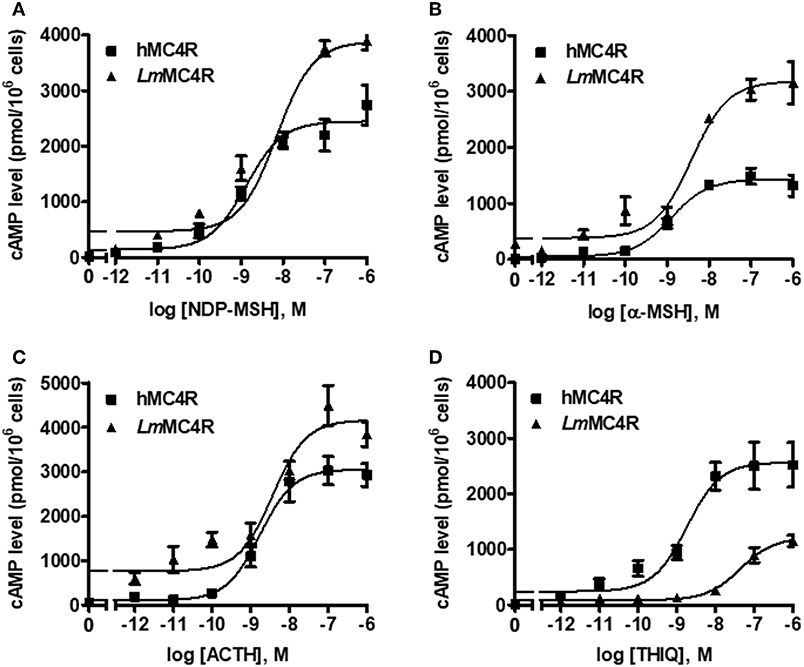
Figure 7. Signaling properties of LmMC4R. HEK293T cells were transiently transfected with hMC4R or LmMC4R plasmids. Forty-eight hours after transfection, different concentrations of NDP-MSH (A), α-MSH (B), ACTH (1-24) (C), and THIQ (D) was used to stimulate the cells transfected with hMC4R or LmMC4R. Data are expressed as mean ± SEM from triplicate measurements within one experiment. The curves are representative of three independent experiments.
In this study, we demonstrated that spotted sea bass mc4r encoded a protein of 327 amino acids with seven transmembrane domains and conversed motifs such as PMY, DRY, and DPIIY (DPxxY) (Figure 1). Compared to transmembrane domains, extracellular N-terminal domain and ECLs were less conservative (Figure 2). Similar results have also been reported in other teleosts (20–22, 44, 45). Cys residues have been shown to be critical for MC4R integrity possibly by forming disulfide bonds (46). We identified 15 Cys residues in LmMC4R, as in other teleost MC4Rs (19, 21, 45), suggesting that the number of Cys residues was highly conserved in MC4Rs during teleost evolution. Amino acid sequence of LmMC4R was ~94% identical to European sea bass MC4R, ~80% identical to several other teleost MC4Rs including zebrafish, fugu and carp, and was ~70% identical to mammalian MC4Rs (Figure 2).
We observed the highest mc4r expression in brain (Figure 3). This is consistent with previous studies that non-mammalian MC4Rs are also abundantly expressed in brain (36). Expression patterns of the teleost mc4r are much wider when compared to those in mammals. In addition to brain, teleost mc4r are also expressed in pituitary and certain peripheral tissues including eyes, liver, gonads, spleen, and gastrointestinal tract (36). The spotted sea bass mc4r was also highly expressed in pituitary, similar to findings in barfin flounder, goldfish, zebrafish, and European sea bass (36). Recent studies showed teleost MC4Rs might play important role in regulating gonadal development (21, 22). In this study, we observed that mc4r expression in spotted sea bass gonad was low (Figure 3). Taken together, these results suggested that wider expression of teleost mc4r might be associated with roles in regulating multiple physiological functions.
In mice, changes in food intake represent 60% of the total effect of the MC4R in regulating energy homeostasis (47). In Mexican cavefish (Astyanax mexicanus), mc4r mutations associated with signaling efficiency contribute to physiological adaptations to nutrient-poor conditions by increasing appetite, growth, and starvation resistance (24). We observed short-term fasting led to down-regulation of mc4r with fluctuating changes in agrp and npy expression while long-term fasting resulted in down regulated mc4r with up-regulated agrp (Figure 4). We hypothesize that MC4R might be more important in regulating long-term energy balance. Moreover, in vitro studies showed incubation of isolated brain cells with α-MSH could decrease npy and agrp mRNA expressions, although it did not change mc4r expression (Figure 5). All these results showed the conserved function of MC4R in regulating food intake (47). Further studies need to investigate the distinct functions of AGRP or NPY neurons in regulating food intake with MC4R due to the fact they showed different transcriptional patterns during fasting (Figure 4).
Detailed pharmacological studies were further performed on LmMC4R. We observed that the cell surface expression of LmMC4R was significantly lower than that of hMC4R, which might explain the differences of total binding between LmMC4R and hMC4R. Ligand binding experiments also showed that LmMC4R bound to α-MSH and ACTH (1-24) with similar affinities (Figure 6). Compared with hMC4R, LmMC4R showed significantly higher binding affinity to NDP-MSH (~60-fold higher), α-MSH (~20-fold higher), and ACTH (1-24, ~20-fold higher) (Figure 6), consistent with previous studies of swamp eel, spotted scat, orange-spotted grouper, fugu and rainbow trout MC4Rs (21–23, 48, 49).
In cAMP signaling assays, α-MSH and ACTH (1-24) stimulated LmMC4R and hMC4R with similar potencies (Figure 7). THIQ could bind to hMC4R and displace the 125I-NDP-MSH in a dose-dependent manner, suggesting that binding sites of THIQ and NDP-MSH were overlapping. THIQ could not displace 125I-NDP-MSH binding at LmMC4R; however, THIQ stimulated intracellular cAMP accumulation at LmMC4R with an EC50 of 63.88 nM, which was significantly higher than that of hMC4R. We propose that THIQ might act as an allosteric agonist at LmMC4R, similar to our previous studies in grass carp and swamp eel (21, 22).
In agreement with previous studies that teleost MC4Rs showed high constitutive activity in cAMP pathway (20–23), this study observed that LmMC4R had ~9-fold higher constitutive activity than that of hMC4R (Figure 7 and Table 3). N-termini act as an important modulator in regulating constitutive activities in GPCRs (50–52). Although amino acid sequences of MC4Rs are conserved from mammals to teleosts, N-termini of LmMC4R and other teleost MC4Rs were less conserved to those of hMC4R, raising the possibility that variations of residues in N-termini might lead to high constitutive activities in teleost. Indeed, hMC4R has also been shown to have constitutive activity (53) and mutations leading to decreased constitutive activity or other loss-of-functions are believed to be associated with obesity pathogenesis (16, 54, 55). However, in agriculture (aquaculture), the farmed animals with lower MC4R constitutive activity may show a higher food efficiency, lower basal metabolism and faster weight gain, increasing the economic benefits of agriculture (aquaculture). Inverse agonists that decrease fish MC4R constitutive activity might be used in aquaculture.
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
YL, Y-XT, H-SW, and XQ: conceptualization. K-QZ, Z-SH, and W-JL: project administration. YL and Y-XT: supervision. YL, Y-XT, and XQ: methodology. Z-SH and K-QZ: writing—original draft. YL, Y-XT, H-SW, and XQ: writing—review and editing.
Research in the authors' laboratories were supported by grants from China Agriculture Research System (CARS-47), Ocean University of China-Auburn University (OUC-AU) Joint Center Grants Program, Fundamental Research Funds for the Central Universities in China (841712004), and China Scholarship Council (201606330081).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2019.00705/full#supplementary-material
ACTH, adrenocorticotropic hormone; AgRP, Agouti-related peptide; ECL, extracellular loop; FBS, fetal bovine serum; GH, growth hormone; GHRH, growth hormone-releasing hormone; GPCR, G protein-coupled receptor; HEK 293T cells, human embryonic kidney 293T cells; hMC4R, human MC4R; ICL, intracellular loop; i.c.v., intracerebroventricular; IGF, insulin-like growth factor; MCR, melanocortin receptor; MSH, melanocyte-stimulating hormone; NDP-MSH, [Nle4, D-Phe7]-α-MSH; NPY, neuropeptide-Y; ORF, open reading frame; PBS, phosphate buffered saline; PKA, protein kinase A; SST, somatostatins; THIQ, N-[(3R)-1, 2, 3, 4-tetrahydroisoquinolinium-3-ylcarbonyl]-(1R)-1-(4-chlorobenzyl)-2-[4-cyclohexyl-4-(1H-1, 2, 4-triazol-1-ylmethyl)piperidin-1-yl]-2-oxoethylamine; TMD, transmembrane domain.
1. Smith AI, Funder JW. Proopiomelanocortin processing in the pituitary, central nervous system, and peripheral tissues. Endocr Rev. (1988) 9:159–79. doi: 10.1210/edrv-9-1-159
2. Dores RM, Lecaude S. Trends in the evolution of the proopiomelanocortin gene. Gen Comp Endocrinol. (2005) 142:81–93. doi: 10.1016/j.ygcen.2005.02.003
3. Cone RD. Studies on the physiological functions of the melanocortin system. Endocr Rev. (2006) 27:736–49. doi: 10.1210/er.2006-0034
4. Tao YX. Melanocortin receptors. Biochim Biophys Acta. (2017) 1863:2411–3. doi: 10.1016/j.bbadis.2017.08.001
5. Sawyer TK, Sanfilippo PJ, Hruby VJ, Engel MH, Heward CB, Burnett JB, et al. 4-Norleucine, 7-D-phenylalanine-α-melanocyte-stimulating hormone: a highly potent α-melanotropin with ultralong biological activity. Proc Natl Acad Sci USA. (1980) 77:5754–8. doi: 10.1073/pnas.77.10.5754
6. Sebhat IK, Martin WJ, Ye Z, Barakat K, Mosley RT, Johnston DB, et al. Design and pharmacology of N-[(3R)-1,2,3,4-tetrahydroisoquinolinium- 3-ylcarbonyl]-(1R)-1-(4-chlorobenzyl)- 2-[4-cyclohexyl-4-(1H-1,2,4-triazol- 1-ylmethyl)piperidin-1-yl]-2-oxoethylamine (1), a potent, selective, melanocortin subtype-4 receptor agonist. J Med Chem. (2002) 45:4589–93. doi: 10.1021/jm025539h
7. Tao YX. The melanocortin-4 receptor: physiology, pharmacology, and pathophysiology. Endocr Rev. (2010) 31:506–43. doi: 10.1210/er.2009-0037
8. Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. (1998) 20:113–4. doi: 10.1038/2407
9. Yeo GS, Farooqi IS, Aminian S, Halsall DJ, Stanhope RG, O'rahilly S. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat Genet. (1998) 20:111–2. doi: 10.1038/2404
10. Tao YX. Mutations in melanocortin-4 receptor and human obesity. Prog Mol Biol Transl Sci. (2009) 88:173–204. doi: 10.1016/S1877-1173(09)88006-X
11. Hinney A, Volckmar AL, Knoll N. Melanocortin-4 receptor in energy homeostasis and obesity pathogenesis. Prog Mol Biol Transl Sci. (2013) 114:147–91. doi: 10.1016/B978-0-12-386933-3.00005-4
12. Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. (1997) 88:131–41. doi: 10.1016/S0092-8674(00)81865-6
13. Khong K, Kurtz SE, Sykes RL, Cone RD. Expression of functional melanocortin-4 receptor in the hypothalamic GT1-1 cell line. Neuroendocrinology. (2001) 74:193–201. doi: 10.1159/000054686
14. Watanobe H, Schioth HB, Izumi J. Pivotal roles of α-melanocyte-stimulating hormone and the melanocortin 4 receptor in leptin stimulation of prolactin secretion in rats. J Neurochem. (2003) 85:338–47. doi: 10.1046/j.1471-4159.2003.01683.x
15. Roa J, Herbison AE. Direct regulation of GnRH neuron excitability by arcuate nucleus POMC and NPY neuron neuropeptides in female mice. Endocrinology. (2012) 153:5587–99. doi: 10.1210/en.2012-1470
16. Tao YX. Molecular mechanisms of the neural melanocortin receptor dysfunction in severe early onset obesity. Mol Cell Endocrinol. (2005) 239:1–14. doi: 10.1016/j.mce.2005.04.012
17. Ringholm A, Fredriksson R, Poliakova N, Yan YL, Postlethwait JH, Larhammar D, et al. One melanocortin 4 and two melanocortin 5 receptors from zebrafish show remarkable conservation in structure and pharmacology. J Neurochem. (2002) 82:6–18. doi: 10.1046/j.1471-4159.2002.00934.x
18. Cerda-Reverter JM, Ringholm A, Schioth HB, Peter RE. Molecular cloning, pharmacological characterization, and brain mapping of the melanocortin 4 receptor in the goldfish: involvement in the control of food intake. Endocrinology. (2003) 144:2336–49. doi: 10.1210/en.2002-0213
19. Kobayashi Y, Tsuchiya K, Yamanome T, Schioth HB, Kawauchi H, Takahashi A. Food deprivation increases the expression of melanocortin-4 receptor in the liver of barfin flounder, Verasper moseri. Gen Comp Endocrinol. (2008) 155:280–7. doi: 10.1016/j.ygcen.2007.05.010
20. Li JT, Yang Z, Chen HP, Zhu CH, Deng SP, Li GL, et al. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in spotted scat, Scatophagus argus. Gen Comp Endocrinol. (2016) 230–1:143–52. doi: 10.1016/j.ygcen.2016.04.010
21. Li L, Yang Z, Zhang YP, He S, Liang XF, Tao YX. Molecular cloning, tissue distribution, and pharmacological characterization of melanocortin-4 receptor in grass carp (Ctenopharyngodon idella). Domest Anim Endocrinol. (2017) 59:140–51. doi: 10.1016/j.domaniend.2016.11.004
22. Yi TL, Yang LK, Ruan GL, Yang DQ, Tao YX. Melanocortin-4 receptor in swamp eel (Monopterus albus): cloning, tissue distribution, and pharmacology. Gene. (2018) 678:79–89. doi: 10.1016/j.gene.2018.07.056
23. Rao YZ, Chen R, Zhang Y, Tao YX. Orange-spotted grouper melanocortin-4 receptor: modulation of signaling by MRAP2. Gen Comp Endocrinol. (2019) 284:113234. doi: 10.1016/j.ygcen.2019.113234
24. Aspiras AC, Rohner N, Martineau B, Borowsky RL, Tabin CJ. Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc Natl Acad Sci USA. (2015) 112:9668–73. doi: 10.1073/pnas.1510802112
25. Song Y, Cone RD. Creation of a genetic model of obesity in a teleost. FASEB J. (2007) 21:2042–9. doi: 10.1096/fj.06-7503com
26. Cerda-Reverter JM, Schioth HB, Peter RE. The central melanocortin system regulates food intake in goldfish. Regul Pept. (2003) 115:101–13. doi: 10.1016/S0167-0115(03)00144-7
27. Schjolden J, Schioth HB, Larhammar D, Winberg S, Larson ET. Melanocortin peptides affect the motivation to feed in rainbow trout (Oncorhynchus mykiss). Gen Comp Endocrinol. (2009) 160:134–8. doi: 10.1016/j.ygcen.2008.11.003
28. Lampert KP, Schmidt C, Fischer P, Volff JN, Hoffmann C, Muck J, et al. Determination of onset of sexual maturation and mating behavior by melanocortin receptor 4 polymorphisms. Curr Biol. (2010) 20:1729–34. doi: 10.1016/j.cub.2010.08.029
29. Smith CC, Harris RM, Lampert KP, Schartl M, Hofmann HA, Ryan MJ. Copy number variation in the melanocortin 4 receptor gene and alternative reproductive tactics the swordtail Xiphophorus multilineatus. Environ Biol Fishes. (2015) 98:23–33. doi: 10.1007/s10641-014-0234-y
30. Jiang DN, Li JT, Tao YX, Chen HP, Deng SP, Zhu CH, et al. Effects of melanocortin-4 receptor agonists and antagonists on expression of genes related to reproduction in spotted scat, Scatophagus argus. J Comp Physiol B. (2017) 187:603–12. doi: 10.1007/s00360-017-1062-0
31. Wang HL, Wen HS, Li Y, Zhang KQ, Liu Y. Evaluation of potential reference genes for quantitative RT-PCR analysis in spotted sea bass (Lateolabrax maculatus) under normal and salinity stress conditions. PeerJ. (2018) 6:e5631. doi: 10.7717/peerj.5631
32. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
33. Qi X, Zhou W, Lu D, Wang Q, Zhang H, Li S, et al. Sexual dimorphism of steroidogenesis regulated by GnIH in the goldfish, Carassius auratus. Biol Reprod. (2013) 88:89. doi: 10.1095/biolreprod.112.105114
34. Mo XL, Yang R, Tao YX. Functions of transmembrane domain 3 of human melanocortin-4 receptor. J Mol Endocrinol. (2012) 49:221–35. doi: 10.1530/JME-12-0162
35. Steiner AL, Kipnis DM, Utiger R, Parker C. Radioimmunoassay for the measurement of adenosine 3',5'-cyclic phosphate. Proc Natl Acad Sci USA. (1969) 64:367–73. doi: 10.1073/pnas.64.1.367
36. Tao YX, Huang H, Wang ZQ, Yang F, Williams JN, Nikiforovich GV. Constitutive activity of neural melanocortin receptors. Meth Enzymol. (2010) 484:267–79. doi: 10.1016/B978-0-12-381298-8.00014-9
37. Tao YX, Segaloff DL. Functional characterization of melanocortin-4 receptor mutations associated with childhood obesity. Endocrinology. (2003) 144:4544–51. doi: 10.1210/en.2003-0524
38. Schwyzer R. ACTH: a short introductory review. Ann NY Acad Sci. (1977) 297:3–26. doi: 10.1111/j.1749-6632.1977.tb41843.x
39. Dores RM, Londraville RL, Prokop J, Davis P, Dewey N, Lesinski N. Molecular evolution of GPCRs: melanocortin/melanocortin receptors. J Mol Endocrinol. (2014) 52:T29–42. doi: 10.1530/JME-14-0050
40. Huang H, Wang W, Tao YX. Pharmacological chaperones for the misfolded melanocortin-4 receptor associated with human obesity. Biochim Biophys Acta. (2017) 1863:2496–507. doi: 10.1016/j.bbadis.2017.03.001
41. Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. (1987) 7:2745–52. doi: 10.1128/MCB.7.8.2745
42. Wang SX, Fan ZC, Tao YX. Functions of acidic transmembrane residues in human melanocortin-3 receptor binding and activation. Biochem Pharmacol. (2008) 76:520–30. doi: 10.1016/j.bcp.2008.05.026
43. Zhang XY, Wen HS, Wang HL, Ren YY, Zhao J, Li Y. RNA-Seq analysis of salinity stress-responsive transcriptome in the liver of spotted sea bass (Lateolabrax maculatus). PLoS ONE. (2017) 12:1–18. doi: 10.1371/journal.pone.0173238
44. Jangprai A, Boonanuntanasarn S, Yoshizaki G. Characterization of melanocortin 4 receptor in Snakeskin Gourami and its expression in relation to daily feed intake and short-term fasting. Gen Comp Endocrinol. (2011) 173:27–37. doi: 10.1016/j.ygcen.2011.04.021
45. Wei R, Yuan D, Zhou C, Wang T, Lin F, Chen H, et al. Cloning, distribution and effects of fasting status of melanocortin 4 receptor (MC4R) in Schizothorax prenanti. Gene. (2013) 532:100–7. doi: 10.1016/j.gene.2013.09.068
46. Tarnow P, Schoneberg T, Krude H, Gruters A, Biebermann H. Mutationally induced disulfide bond formation within the third extracellular loop causes melanocortin 4 receptor inactivation in patients with obesity. J Biol Chem. (2003) 278:48666–73. doi: 10.1074/jbc.M309941200
47. Balthasar N, Dalgaard LT, Lee CE, Yu J, Funahashi H, Williams T, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. (2005) 123:493–505. doi: 10.1016/j.cell.2005.08.035
48. Haitina T, Klovins J, Andersson J, Fredriksson R, Lagerstrom MC, Larhammar D, et al. Cloning, tissue distribution, pharmacology and three-dimensional modelling of melanocortin receptors 4 and 5 in rainbow trout suggest close evolutionary relationship of these subtypes. Biochem J. (2004) 380:475–86. doi: 10.1042/bj20031934
49. Klovins J, Haitina T, Fridmanis D, Kilianova Z, Kapa I, Fredriksson R, et al. The melanocortin system in Fugu: determination of POMC/AGRP/MCR gene repertoire and synteny, as well as pharmacology and anatomical distribution of the MCRs. Mol Biol Evol. (2004) 21:563–79. doi: 10.1093/molbev/msh050
50. Zhang M, Tong KP, Fremont V, Chen J, Narayan P, Puett D, et al. The extracellular domain suppresses constitutive activity of the transmembrane domain of the human TSH receptor: implications for hormone-receptor interaction and antagonist design. Endocrinology. (2000) 141:3514–7. doi: 10.1210/endo.141.9.7790
51. Nishi S, Nakabayashi K, Kobilka B, Hsueh AJ. The ectodomain of the luteinizing hormone receptor interacts with exoloop 2 to constrain the transmembrane region. Studies using chimeric human and fly receptors. J Biol Chem. (2002) 277:3958–64. doi: 10.1074/jbc.M109617200
52. Paavola KJ, Stephenson JR, Ritter SL, Alter SP, Hall RA. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J Biol Chem. (2011) 286:28914–21. doi: 10.1074/jbc.M111.247973
53. Tao YX. Constitutive activity in melanocortin-4 receptor: biased signaling of inverse agonists. Adv Pharmacol. (2014) 70:135–54. doi: 10.1016/B978-0-12-417197-8.00005-5
54. Srinivasan S, Lubrano-Berthelier C, Govaerts C, Picard F, Santiago P, Conklin BR, et al. Constitutive activity of the melanocortin-4 receptor is maintained by its N-terminal domain and plays a role in energy homeostasis in humans. J Clin Investig. (2004) 114:1158–64. doi: 10.1172/JCI200421927
Keywords: spotted sea bass, melanocortin-4 receptor, signaling, allosteric modulator, constitutive activity
Citation: Zhang K-Q, Hou Z-S, Wen H-S, Li Y, Qi X, Li W-J and Tao Y-X (2019) Melanocortin-4 Receptor in Spotted Sea Bass, Lateolabrax maculatus: Cloning, Tissue Distribution, Physiology, and Pharmacology. Front. Endocrinol. 10:705. doi: 10.3389/fendo.2019.00705
Received: 07 May 2019; Accepted: 30 September 2019;
Published: 18 October 2019.
Edited by:
Yong Zhu, East Carolina University, United StatesReviewed by:
Giulia Baldini, University of Arkansas for Medical Sciences, United StatesCopyright © 2019 Zhang, Hou, Wen, Li, Qi, Li and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Li, eXVubGkwMTE2QG91Yy5lZHUuY24=; Ya-Xiong Tao, dGFveWF4aUBhdWJ1cm4uZWR1
†These authors have contributed equally to this work as co-first authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.