
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Endocrinol., 13 August 2019
Sec. Experimental Endocrinology
Volume 10 - 2019 | https://doi.org/10.3389/fendo.2019.00517
This article is part of the Research TopicEpigenetic Control of Adaptive ThermogenesisView all articles
Homeothermic mammals including humans produce heat (also termed thermogenesis) inside their body to maintain constant body temperature. It has been shown that thermogenesis is elevated by several external factors; prominent being cold, diet, and physical exercise. This modulation ability is termed as “Adaptive Thermogenesis (AT).” Intake of high calorie diet was also shown to increase thermogenesis in laboratory animals, a phenomenon termed as diet-induced thermogenesis (DIT) (1). Research on finding out the mechanisms of DIT intensified in the recent years, as obesity and associated metabolic disorder increased rapidly all over the world. It is hoped that mechanisms of AT can be targeted to increase energy expenditure and provide protection against metabolic diseases including obesity. Brown adipose tissue (BAT) and skeletal muscle have emerged as the two major sites of AT. Major heat producer in BAT is a protein called uncoupling protein (UCP) 1 that dissipates proton gradient in mitochondria and thereby resulting in heat production (2). Few additional thermogenic mechanisms have also been reported in BAT such as futile TG lipolysis/esterification, creatine/phosphocreatine cycling, and ATP-dependent Ca2+-cycling (3–5). The understanding of mechanisms of AT in the skeletal muscle has been slow and forms the major focus of this review.
In the skeletal muscle, contributors to AT can be proteins of (i) Calcium (Ca2+)-handling, (ii) contractile apparatus, and (iii) mitochondrial metabolism. ATP utilization by these proteins can vary due to their relative expression and post-translational modification. This is additionally regulated by the abundance of signaling molecules (ions, lipids, etc.) in the micromilleu which in turn is determined by their transporter proteins. The term epigenetic has been classically applied to describe changes in protein expression via differential chromosomal compaction without genetic alteration. Herein, a more liberal interpretation of “epigenetics” is taken that encompasses all functional alterations irrespective of inheritance and genetic modifications. Such changes include transcriptional, post-transcriptional, translational, post-translational. We additionally discuss about the modulation of AT in the skeletal muscle by factors like microRNA, vitamins, and hormones.
Recent discoveries have made it clear that Ca2+-handling proteins are the key mediators of AT in the skeletal muscle (6). Epigenetic modifications of these proteins alter their physiological function thereby influence whole body metabolism. Ca2+ signaling proteins involved in different aspects of muscle function are: Ca2+-release from Sarcoplasmic reticulum SR (Ryanodine Receptor (RyR), IP3R); Ca2+-buffering inside SR (calsequestrin, calreticulin, parvalbumin, and sarcalumenin); Ca2+-uptake into SR (SERCA, SLN, PLB, and MLN); Ca2+-transport across the membrane (PMCA, MCU, MICU, TRP channels, STIM, Orai1). Function of these proteins can be regulated by epigenetic mechanisms and contribute to adaptive thermogenesis.
SERCA has emerged as the primary determinant of ATP utilization in muscle (7–9), therefore is a key player in adaptive thermogenesis. Mammals possess three distinct SERCA isoforms: SERCA 1 is predominantly expressed in fast twitch fibers; SERCA 2 is expressed in cardiac and slow twitch skeletal muscles; and SERCA 3 is found mostly in non-muscle cells (8, 10). SERCA function is chiefly regulated by micropeptides and post-translational modifications via factors including hormones (11).
Among the micropeptide regulators of SERCA, phospholamban (PLB) is the best characterized. PLB binds only to Ca2+ free state of SERCA and acts as an inhibitor. Once Ca2+ concentration in the microenvironment increases it gets dislodged from the SERCA allowing its unhindered activity (12). In contrast, SLN can interact with Ca2+ bound confirmations of SERCA promoting futile Ca2+ transport thus enhancing higher ATP utilization (10, 13, 14). This function of SLN leads to elevated energy expenditure resulting in heat production in the muscle and serves as a major regulator of non-shivering thermogenesis (NST). Interestingly, SLN is predominantly found in atria and skeletal muscle, while PLB expression is restricted to ventricle. PLB inhibition of SERCA is regulated by its phosphorylation at Ser-16 and Thr-17 residues by Ca2+/calmodulin- dependent protein kinase (CaM kinase) and protein kinase-A, respectively. On the other hand, SLN function is regulated by several factors; like its differential expression, its phosphorylation at Thr-5 residue, ATP concentration and membrane lipid composition (15–17). Recent studies have identified few more micropeptides like DWORF, MLN, and Ankyrin1 as regulators of SERCA (18–20). Although the exact function of these new regulators is not fully understood, their contribution to AT of muscle has been speculated. Interestingly, DWORF acts differently from all the other micropeptide regulators, even though is conserved and binds to the same SERCA groove as PLB and SLN. However, the expression of DWORF is limited to heart and slow twitch fibers like soleus, and it is proposed to be an activator of SERCA function (18). The fine tuning of SERCA activity by these micropeptide regulators might have a big impact on whole body energy homeostasis (shown in Figure 1), and an unrealized potential in adaptive thermogenesis.
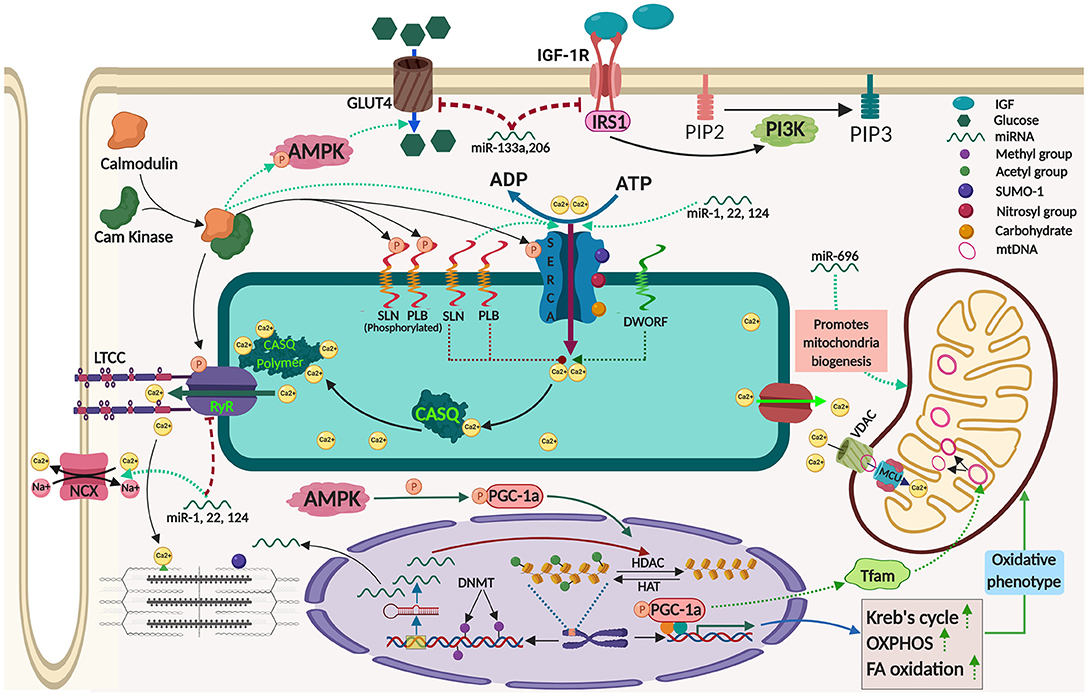
Figure 1. Mechanisms contributing to adaptive thermogenesis in the skeletal muscle and its regulation by epigenetic processes. The image was created by Biorender.com.
Several post-translational modifications including glutathionylation, SUMOylation, O-GlcNAcylation, glycosylation, nitration, and acetylation have been proposed to modulate SERCA activity. Glutathionylation of SERCA mainly occurs at Cys674 residue and has isoform specific effects (11, 21, 22). Peroxinitrite, one critical molecule in glutathionylation has different effects on distinct SERCA isoforms. Peroxynitrite promotes SERCA2a glutathionylation increasing Ca2+-transport during artherosclerosis whereas, it inhibits Ca2+-uptake of SERCA1a by oxidizing it at lower levels of glutathione (21, 22). Several investigations addressed SUMOylation of SERCA, although SERCA2a is better studied. In this type of modification small ubiquitin like modifier (SUMO) 1 binds to SERCA at Lys 480 & 585 residues (11, 23). SUMOylation upregulates SERCA2a activity and is cardioprotective, while its effects on SERCA1a are still unknown. Literatures suggested SUMOylation-mediated increased SERCA function is due to blocking of acetylation of SERCA (11). Acetylation is another post-translational modification that acts antagonistic to that of SUMOylation (24). Glycosylation involves addition of carbohydrate molecules to SERCA at elevated glucose levels in the muscle hindering SERCA2a activity and therefore has been considered as a pharmacological target in diabetes and cardiovascular diseases (25). O-GlcNAcylation is one of the specific modification among glycosylation in which N-acetylglucosamine is added to SERCA2a at serine and threonine residues. Similar to other glycosylation. O-GlcNAcylation reduces SERCA2a activity directly as well as indirectly by increasing PLB expression level during hyperglycemia (25). Another post-translational modification of SERCA is nitration, where a nitro group is added to tyrosine residues. Level of nitration in SERCA2a increases during hyperglycemic condition as a result of higher oxidative stress (26), hence might be critical in metabolic diseases like diabetes. Although regulation of SERCA activity by various epigenetic and post-translational modifications are important in energy homeostasis but their individual roles in AT are yet to be deciphered.
RyR is the primary Ca2+-release channel of SR; while RyR1 is dominant in skeletal muscle, RyR2 predominates in cardiac. Genetic alterations of RyR that make the channel leaky are the basis of diseases like malignant hyperthermia and central core disease. Some studies have demonstrated epigenetic regulation of RyR gene expression in health and disease, suggesting that same regulations might play important role in adaptive thermogenesis. Rokach et al. showed that in recessive RYR1-linked myopathies, Histone deacetylase (HDAC4 and HDAC5), and DNA methyltransferases (DNMT1 and DNMT2) are upregulated leading to chromatin condensation and reduction in RyR1 transcript level. Increased HDACs also sequester mef2, a major muscle transcriptional factor that might affect expression of many genes including miRNAs. They have proposed that these epigenetic regulators can be targeted pharmacologically to treat several inherited muscular disorders (27). Thiol rich RyR protein is responsive toward phosphorylation, oxidation, nitrosylation, and glutathionylation (28–30). RyR1 phosphorylation at serine and threonine residues by CamKII and PKA, respectively, has been suggested to make the channel unstable and cause leak of Ca2+ from the SR having severe consequences on Ca2+-signaling in the cytosol. RyR1 phosphorylation plays a key role in adaptation to cold (31). Nitrosylation of RyR at thiol groups is facilitated by O2 level in the muscle and increases Ca2+-release from the channel (29). Thus, RyR might play an important role in AT either directly by Ca2+-induced bioenergetics or indirectly via Ca2+-mediated signaling.
AT of skeletal muscle is highly dependent on ATP utilization by the contractile apparatus. This can be regulated by relative myosin isoform expression, their post-translational modifications and bioenergetics of contractile apparatus. Interestingly, all are affected during physical activity for AT recruitment in the muscle. Exercise mediates conversion of slow twitch-to-fast twitch fiber in soleus by myosin heavy chain (MHC) gene expression via chromatin remodeling. During this transition, histone deacetylation down-regulates the MHC-I gene, whereas histone acetylation upregulate fast type IIx and IIb MHC genes. Alteration in relative expression of myosin isoforms has also been seen in condition that influence AT such as aging, high fat/sugar diet; whereas in cold adaptation it is still less explored. Interestingly, a newly characterized confirmation of myosin called super relaxed state (SRX) by Stewart et al. throws light on its potential role in cold adaptation (32). In normal condition ATP turnover by myosin is <0.1 s, but in SRX, myosin can slowly split ATP with turnover time (~230 s at 24°C) inside the core myosin filament in skeletal muscle. Further, activity of SRX myosin can be enhanced in cold through phosphorylation by Myosin light chain kinase (MLCK) or by substitution of GTP for ATP.
Energy used by myosin is also influenced by epigenetic modifications of other partner proteins of contractile apparatus. Troponin-T binds to tropomyosin and control acto-myosin cross bridge formation thereby determining ATP usage by myosin. Sumoylation of actin by SUMO-1 has been reported in the skeletal muscle in response to exercise training indicating its potential in adaptive thermogenesis (33). Resistance exercise training in older adults showed marked increase in the expression of slow troponin-T isoforms by alternative splicing of the pre-mRNA. Parallely differential expression of several tropomyosin isoforms (Tpm1.6, Tpm1.7, Tpm2.1, etc.) distinctly regulate myosin ATPase thus affecting energy expenditure and thermogenesis by the muscle (34). Hence, epigenetic regulation of contractile machinery proteins may serve as a mechanism to influence energy usage in the muscle having an effect on adaptive thermogenesis.
The SR Ca2+-handling machinery and contractile apparatus determine the cytosolic Ca2+-level that can regulate the Ca2+-signaling into the nucleus and mitochondria. Thus, all the physiological functions of the skeletal muscle is intimately coupled to mitochondrial metabolism, which in turn depend on modulation of: (1) mitochondrial transcriptional machinery, (2) nuclear gene expression, and (3) Ca2+-influx into the mitochondrial lumen. Each of these components can be affected by epigenetic mechanisms mediating the adaptive changes in the thermogenic state of the skeletal muscle. Mitochondrial metabolism is critically governed by PGC-1α, a master transcriptional regulator encoded by a nuclear gene (PPARGC1A). The activity of PGC-1α in skeletal muscle regulates chromatin remodeling via by acetylation/deacetylation (SIRT1, GCN5) and methylation (PRMT1) in response to energetically drained energy/substrate conditions like fasting, exercise, calorie restriction (35, 36). The PGC-1α function is fine-tuned by post-translational modifications, including phosphorylation (by AKT, GSK-3, CLK2, CAMKKB, PKA, S6K, P38, and AMPK), dephosphorylation (calcineurin), ubiquitination (SCF), and O-GlcNAc modification. Level of PGC-1α protein and its activity is closely intertwined with muscle metabolic status via epigenetic mechanisms that respond to number of physiological cues like hypoxia, exercise, calorie restriction, and aging.
AMPK serves as another key energy sensor and its activation enhances glucose uptake (by increasing GLUT4 translocation), fatty acid oxidation, and mitochondrial biogenesis in the skeletal muscle. In fact, AMPK can also regulate PGC-1α by direct phosphorylation at Thr177 and Ser538 residues thereby increasing the transcription of PGC-1α-target genes (37). Interestingly, AMPK is activated by high AMP/ATP ratio which is usually associated with elevated cytosolic Ca2+-level in the muscle, where Ca2+-dependent signaling is already induced. Therefore, AMPK serves as a key integrator of Ca2+-dependent and ATP-dependent pathways.
Activation of mitochondrial function in the skeletal muscle is also influenced by Ca2+-regulated transcription factors like (CaMK) isoforms and calcineurin/NFAT. Role of calcineurin/NFAT in muscle fiber type determination is well-known (38), but recent studies have shown that they decode the local Ca2+-signal and mediate that to nucleus for expression of mitochondrial metabolic genes (39). Another important mitochondrial transcriptional regulator in the muscle is myocyte enhancer factor (MEF) 2. It is suggested that MEF-2 strikes a balance between HDACs-mediated transcriptional repression and activation by factors like NFAT, PGC-1α, and MAPK.
The acute increase in oxidative metabolism in muscle is achieved by Ca2+-influx into the mitochondria and the same process can also be tapped for adaptive thermogenesis. Mitochondrial Ca2+-influx is governed by two channels: voltage-dependent anion channel (VDAC), located on the outer mitochondrial membrane and mitochondrial calcium uniporter (MCU) on the inner mitochondrial membrane. It has been shown that MCU overexpression results in increased protein synthesis leads to hypertrophic phenotype and protects mice from denervation-induced atrophy in muscles via IGF1/AKT and PGC-1α4 pathways. Important role of MCU in skeletal muscle metabolism is further highlighted by the identification of mutations in MICU1, an MCU regulator, in patients with proximal muscle weakness (40). On the other hand, VDAC1 expression was significantly up-regulated in the skeletal muscle upon cold adaptation of mice which suggest role for VDAC in adaptive thermogenesis (41). Recent studies has also thrown light on Mitochondrial Ca2+-influx due to SR-Ca2+-handling by the action of a small protein called SLN that regulate SERCA-mediated Ca2+-uptake. Evidences shows that mitochondrial metabolism is intertwined with muscle thermogenic genes speculating its potential role in AT.
AT can further be modulated by additional factors such as miRNA, vitamins, and hormones. We chose to discuss them separately as the role of these factors is broad and their role in skeletal muscle AT is being updated every day.
The miRNAs are short non-coding RNA of 20-24 nucleotides that modify target gene expression post-transcriptionally. Several miRNAs have been discovered and characterized in the skeletal muscle termed as myo-miRs (Table 1). Research has evidenced that physiological responses like lipid utilization, endothelial function and mitochondrial activity are modulated by miRNAs in skeletal muscle. Expression of NCX (sodium/calcium exchanger 1), inositol-1,4,5-trisphosphate receptor 1 (IP3R1), and SERCA-2a (sarcoplasmic reticulum Ca+2 ATPase-2a) are targeted by miR-1, miR-25, and miR-214, respectively. A recent study showed that miR-1a, miR-22, and miR-124 cause RyR1 down-regulation in the skeletal muscles of multi-minicore disease. Interestingly, miRNA has been shown to have additional role other than mRNA regulation by directly interfering with protein-protein interaction. Soller et al. have shown that miR-1 and miR-21 binds to PLB with low dissociation constant reversing its inhibition on SERCA (43). High-fat diet (HFD) induced modifications of gene expression is in part mediated by miRNAs such as miR-1a, miR-133a, and miR-206. Both miR-133a and miR-206 upregulated after HFD intake targeting insulin-growth factor (IGF)-1 and IGF-1R mRNA leading to drop in the IGF-1 signaling pathway (59).
Role of miRNA in various types of exercise training has been studied in details (Table 1). Exercise training (both acute and endurance) down-regulate and upregulate different sets of miRNA that positively co-regulate the expression of HDAC4 and NRF-1 leading to muscle regeneration, and mitochondrial biogenesis (60). Angiogenesis via VEGF-A expression are regulated by opposing action of miR-206 and miR-1 (61). The miR-1 suppress HDAC4, in turn, upregulates follistatin (FLN), a fusion promoting factor and antagonize the myogenic inhibitor myostatin (MSN) and SARS (seryl-tRNA synthetase) contrary to the action of miR-206 (62). It has been studied that muscle hypertrophy was enhanced by expression of c-Met, HGF, IGF-1, SRF, and LIF genes by down-regulation of miR-1 and miR-133a via functional overload in plantaris muscle (49). Exercise upregulated PGC-1α mediates its effects by downregulating miR-696 and upregulating miR-23 expression leading to mitochondrial biogenesis in skeletal muscle (52, 63). Two miRNAs miR-499 and miR-208b coexpressed with Myh7 and Myh7b genes in slow myofiber genes and regulated by miR-208a (expressed in Myh6 gene) for slow to fast twitch fiber type switching (64). Thyroid hormone upregulated miR-133a1 in a Thyroid receptor (TR)-dependent manner and promotes slow-to-fast muscle switch by repressing TEA domain family member 1 (TEAD1) (65). Expression of the phosphatase and tensin homolog (PTEN) and forkhead box O1 (FOXO1) is suppressed by miR-486 to promote muscle hypertrophy (66).
Several hormones influencing muscle metabolism (like insulin, leptin, GLP-1, thyroid) undergo drastic changes in their circulating levels in conditions that can have impact on AT (67). Steroids and thyroids are the best studied hormones to regulate protein expression in muscles and catecholamines are known to enhance muscle energy expenditure (68). Similarly, many vitamins (e.g., Vitamin D) directly or indirectly mediate their effects by altering intracellular calcium dynamics. Their conspicuous pleiotropic gene expression are known to modulate energy status and also affect protein expression in different muscle types (69). However, their specific role in regulating AT in the skeletal muscle is still poorly defined. It is interesting to note that few cytokines that were previously thought not to regulate energy homeostasis have been demonstrated to do so and are now reclassified as hormones. Defining their function in regulation of muscle energy utilization will clarify their role in AT.
Epigenetic mechanisms significantly control AT in skeletal muscle. Skeletal muscle constitute more than 40% of the body weight and its energy status serves as a major determinant of metabolic rate. Even a minute alteration in muscle metabolic state can have remarkable change in the whole body energy expenditure. So, epigenetic mechanisms regulating skeletal muscle AT can hugely control energy homeostasis. Some of these mechanisms can serve as good targets for manipulation of energy expenditure to counter metabolic disorders such as obesity and type 2 diabetes. Researchers have tried to develop pharmacological agents to tap muscle metabolism for treatment of obesity. However, these efforts have not resulted in any product yet as epigenetic control of muscle AT is still not well-defined. Another major direction that future studies should address is whether epigenetic regulation of muscle AT affects the functioning of other organs. Secretion of myokines and their effect in modulation of many organs including white fat depot, liver has recently been illustrated. Another poorly defined area is role of vitamins in regulation of muscle metabolism and NST capacity that can play a key role in adaptive thermogenesis. Future studies in these directions will unravel mechanisms of epigenetic regulation that will help in designing strategies to counter metabolic disorders by activation of AT.
SP prepared the figure and BS prepared the table. All authors drafted the manuscript together, critically revised the work, and approved the final version.
NB was funded by the Science and Engineering Research Board (SERB), DST, India (ECR/2016/001247) and Ramalingaswamy Fellowship, Department of Biotechnology (DBT), India (BT/RLF/Re-entry/41/2014). BS was supported by Junior Research Fellowship from Council of Scientific and Industrial Research (CSIR), India.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Muthu Periasamy, Dr. Meghna Pant, Ojas Tikoo, Andrea Bergano, Surat Giri, and Debasa Mukherjee for manuscript editing and valuable comments.
1. Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced thermogenesis. Nature. (1979) 281:31–5. doi: 10.1038/281031a0
2. Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. (2004) 84:277–359. doi: 10.1152/physrev.00015.2003
3. Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell. (2015) 163:643–55. doi: 10.1016/j.cell.2015.09.035
4. Ikeda K, Kang Q, Yoneshiro T, Camporez JP, Maki H, Homma M, et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med. (2017) 23:1454–65. doi: 10.1038/nm.4429
5. Blondin DP, Frisch F, Phoenix S, Guérin B, Turcotte ÉE, Haman F, et al. Inhibition of intracellular triglyceride lipolysis suppresses cold-induced brown adipose tissue metabolism and increases shivering in humans. Cell Metab. (2017) 25:438–7. doi: 10.1016/j.cmet.2016.12.005
6. Pant M, Bal NC, Periasamy M. Sarcolipin: a key thermogenic and metabolic regulator in skeletal muscle. Trends Endocrinol Metab. (2016) 27:881–92. doi: 10.1016/j.tem.2016.08.006
7. Smith IC, Bombardier E, Vigna C, Tupling AR. ATP consumption by sarcoplasmic reticulum Ca(2)(+) pumps accounts for 40-50% of resting metabolic rate in mouse fast and slow twitch skeletal muscle. PLoS ONE. (2013) 8:e68924. doi: 10.1371/journal.pone.0068924
8. Periasamy M, Maurya SK, Sahoo SK, Singh S, Sahoo SK, Reis FCG, et al. Role of SERCA pump in muscle thermogenesis and metabolism. Compr Physiol. (2017) 7:879–90. doi: 10.1002/cphy.c160030
9. Fajardo VA, Bombardier E, Vigna C, Devji T, Bloemberg D, Gamu D, et al. Co-expression of SERCA isoforms, phospholamban and sarcolipin in human skeletal muscle fibers. PLoS ONE. (2013) 8:e84304. doi: 10.1371/journal.pone.0084304
10. Periasamy M, Kalyanasundaram A. SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve. (2007) 35:430–2. doi: 10.1002/mus.20745
11. Stammers AN, Susser SE, Hamm NC, Hlynsky MW, Kimber DE, Kehler DS, et al. The regulation of sarco(endo)plasmic reticulum calcium-ATPases (SERCA). Can J Physiol Pharmacol. (2015) 93:843–54. doi: 10.1139/cjpp-2014-0463
12. Kirchberber MA, Tada M, Katz AM. Phospholamban: a regulatory protein of the cardiac sarcoplasmic reticulum. Recent Adv Stud Cardiac Struct Metab. (1975) 5:103–5.
13. Traaseth NJ, Ha KN, Verardi R, Shi L, Buffy JJ, Masterson LR, et al. Structural and dynamic basis of phospholamban and sarcolipin inhibition of Ca(2+)-ATPase. Biochemistry. (2008) 47:3–13. doi: 10.1021/bi701668v
14. Bal NC, Maurya SK, Sopariwala DH, Sahoo SK, Gupta SC, Shaikh SA, et al. Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nat Med. (2012) 18:1575–9. doi: 10.1038/nm.2897
15. Shaikh SA, Sahoo SK, Periasamy M. Phospholamban and sarcolipin: are they functionally redundant or distinct regulators of the Sarco(Endo)Plasmic Reticulum Calcium ATPase? J Mol Cell Cardiol. (2016) 91:81–91. doi: 10.1016/j.yjmcc.2015.12.030
16. Bal NC, Singh S, Reis FCG, Maurya SK, Pani S, Rowland LA, et al. Both brown adipose tissue and skeletal muscle thermogenesis processes are activated during mild to severe cold adaptation in mice. J Biol Chem. (2017) 292:16616–25. doi: 10.1074/jbc.M117.790451
17. Paran CW, Verkerke AR, Heden TD, Park S, Zou K, Lawson HA, et al. Reduced efficiency of sarcolipin-dependent respiration in myocytes from humans with severe obesity. Obesity. (2015) 23:1440–9. doi: 10.1002/oby.21123
18. Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. (2016) 351:271–5. doi: 10.1126/science.aad4076
19. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. (2015) 160:595–606. doi: 10.1016/j.cell.2015.01.009
20. Desmond PF, Labuza A, Muriel J, Markwardt ML, Mancini AE, Rizzo MA, et al. Interactions between small ankyrin 1 and sarcolipin coordinately regulate activity of the sarco(endo)plasmic reticulum Ca(2+)-ATPase (SERCA1). J Biol Chem. (2017) 292:10961–72. doi: 10.1074/jbc.M117.783613
21. Viner RI, Huhmer AF, Bigelow DJ, Schoneich C. The oxidative inactivation of sarcoplasmic reticulum Ca(2+)-ATPase by peroxynitrite. Free Radic Res. (1996) 24:243–59. doi: 10.3109/10715769609088022
22. Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, et al. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat Med. (2004) 10:1200–7. doi: 10.1038/nm1119
23. Kho C, Lee A, Jeong D, Oh JG, Chaanine AH, Kizana E, et al. SUMO1-dependent modulation of SERCA2a in heart failure. Nature. (2011) 477:601–5. doi: 10.1038/nature10407
24. Foster DB, Liu T, Rucker J, O'Meally RN, Devine LR, Cole RN, et al. The cardiac acetyl-lysine proteome. PLoS ONE. (2013) 8:e67513. doi: 10.1371/journal.pone.0067513
25. Clark RJ, McDonough PM, Swanson E, Trost SU, Suzuki M, Fukuda M, et al. Diabetes and the accompanying hyperglycemia impairs cardiomyocyte calcium cycling through increased nuclear O-GlcNAcylation. J Biol Chem. (2003) 278:44230–7. doi: 10.1074/jbc.M303810200
26. Tang WH, Cheng WT, Kravtsov GM, Tong XY, Hou XY, Chung SK, et al. Cardiac contractile dysfunction during acute hyperglycemia due to impairment of SERCA by polyol pathway-mediated oxidative stress. Am J Physiol Cell Physiol. (2010) 299:C643–53. doi: 10.1152/ajpcell.00137.2010
27. Rokach O, Sekulic-Jablanovic M, Voermans N, Wilmshurst J, Pillay K, Heytens L, et al. Epigenetic changes as a common trigger of muscle weakness in congenital myopathies. Hum Mol Genet. (2015) 24:4636–7. doi: 10.1093/hmg/ddv195
28. Sun J, Xu L, Eu JP, Stamler JS, Meissner G. Nitric oxide, NOC-12, and S-nitrosoglutathione modulate the skeletal muscle calcium release channel/ryanodine receptor by different mechanisms. An allosteric function for O2 in S-nitrosylation of the channel. J Biol Chem. (2003) 278:8184–9. doi: 10.1074/jbc.M211940200
29. Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci USA. (2001) 98:11158–62. doi: 10.1073/pnas.201289098
30. Marks AR. Calcium cycling proteins and heart failure: mechanisms and therapeutics. J Clin Invest. (2013) 123:46–52. doi: 10.1172/JCI62834
31. Danila CI, Hamilton SL. Phosphorylation of ryanodine receptors. Biol Res. (2004) 37:521–5. doi: 10.4067/S0716-97602004000400005
32. Stewart MA, Franks-Skiba K, Chen S, Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci USA. (2010) 107:430–5. doi: 10.1073/pnas.0909468107
33. Gehlert S, Klinz FJ, Willkomm L, Schiffer T, Suhr F, Bloch W. Intense resistance exercise promotes the acute and transient nuclear translocation of small ubiquitin-related modifier (SUMO)-1 in human myofibres. Int J Mol Sci. (2016) 17:E646. doi: 10.3390/ijms17050646
34. Gordon AM, Regnier M, Homsher E. Skeletal and cardiac muscle contractile activation: tropomyosin “rocks and rolls”. News Physiol Sci. (2001) 16:49–55. doi: 10.1152/physiologyonline.2001.16.2.49
35. Cheng Z, Almeida FA. Mitochondrial alteration in type 2 diabetes and obesity: an epigenetic link. Cell Cycle. (2014) 13:890–7. doi: 10.4161/cc.28189
36. Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. (2008) 582:46–53. doi: 10.1016/j.febslet.2007.11.034
37. Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci USA. (2007) 104:12017–22. doi: 10.1073/pnas.0705070104
38. McCullagh KJ, Calabria E, Pallafacchina G, Ciciliot S, Serrano AL, Argentini C, et al. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc Natl Acad Sci USA. (2004) 101:10590–5. doi: 10.1073/pnas.0308035101
39. Maurya SK, Herrera JL, Sahoo SK, Reis FCG, Vega RB, Kelly DP, et al. Sarcolipin signaling promotes mitochondrial biogenesis and oxidative metabolism in skeletal muscle. Cell Rep. (2018) 24:2919–31. doi: 10.1016/j.celrep.2018.08.036
40. Logan CV, Szabadkai G, Sharpe JA, Parry DA, Torelli S, Childs AM, et al. Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat Genet. (2014) 46:188–93. doi: 10.1038/ng.2851
41. Bal NC, Maurya SK, Singh S, Wehrens XH, Periasamy M. Increased reliance on muscle-based thermogenesis upon acute minimization of brown adipose tissue function. J Biol Chem. (2016) 291:17247–57. doi: 10.1074/jbc.M116.728188
42. Poddar S, Kesharwani D, Datta M. Histone deacetylase inhibition regulates miR-449a levels in skeletal muscle cells. Epigenetics. (2016) 11:579–87. doi: 10.1080/15592294.2016.1188247
43. Soller KJ, Yang J, Veglia G, Bowser MT. Reversal of Phospholamban inhibition of the Sarco(endo)plasmic Reticulum Ca2+-ATPase (SERCA) using short, protein-interacting RNAs and oligonucleotide analogs. J Biol Chem. (2016) 291:21510–8. doi: 10.1074/jbc.M116.738807
44. Gurha P, Abreu-Goodger C, Wang T, Ramirez MO, Drumond AL, van Dongen S, et al. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation. (2012) 125:2751–61. doi: 10.1161/CIRCULATIONAHA.111.044354
45. Li X, Guo L, Zhou X, Gao X, Liang P. miRNAs regulated overexpression of ryanodine receptor is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Sci Rep. (2015) 5:14095. doi: 10.1038/srep14095
46. Ringholm S, Biensø RS, Kiilerich K, Guadalupe-Grau A, Aachmann-Andersen NJ, Saltin B, et al. Bed rest reduces metabolic protein content and abolishes exercise-induced mRNA responses in human skeletal muscle. Am J Physiol Endocrinol Metab. (2011) 301:E649–E658. doi: 10.1152/ajpendo.00230.2011
47. Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab. (2008) 295:E1333–E1340. doi: 10.1152/ajpendo.90562.2008
48. Mueller M, Breil FA, Lurman G, Klossner S, Flück M, Billeter R, et al. Different molecular and structural adaptations with eccentric and conventional strength training in elderly men and women. Gerontology. (2011) 57:528–38. doi: 10.1159/000323267
49. McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. (2007) 102:306–13. doi: 10.1152/japplphysiol.00932.2006
50. Keller P, Vollaard NBJ, Gustafsson T, Gallagher IJ, Sundberg CJ, Rankinen T, et al. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol. (2011) 110:46–59. doi: 10.1152/japplphysiol.00634.2010
51. Latouche C, Natoli A, Reddy-Luthmoodoo M, Heywood SE, Armitage JA, Kingwell BA. MicroRNA-194 modulates glucose metabolism and its skeletal muscle expression is reduced in diabetes. PLoS ONE. (2016) 11:e0155108. doi: 10.1371/journal.pone.0155108
52. Aoi W, Naito Y, Mizushima K, Takanami Y, Kawai Y, Ichikawa H, et al. The microRNA miR-696 regulates PGC-1{alpha} in mouse skeletal muscle in response to physical activity. Am J Physiol Endocrinol Metab. (2010) 298:E799–806. doi: 10.1152/ajpendo.00448.2009
53. Xu Y, Zhao C, Sun X, Liu Z, Zhang J. MicroRNA-761 regulates mitochondrial biogenesis in mouse skeletal muscle in response to exercise. Biochem Biophys Res Commun. (2015) 467:103–8. doi: 10.1016/j.bbrc.2015.09.113
54. Zhou T, Meng X, Che H, Shen N, Xiao D, Song X, et al. Regulation of insulin resistance by multiple MiRNAs via targeting the GLUT4 signalling pathway. Cell Physiol Biochem. (2016) 38:2063–78. doi: 10.1159/000445565
55. Yamamoto H, Morino K, Nishio Y, Ugi S, Yoshizaki T, Kashiwagi A, et al. MicroRNA-494 regulates mitochondrial biogenesis in skeletal muscle through mitochondrial transcription factor A and Forkhead box j3. Am J Physiol Endocrinol Metab. (2012) 303:E1419–E1427. doi: 10.1152/ajpendo.00097.2012
56. Sun Y, Cui D, Zhang Z, Zhang Q, Ji L, Ding S. Voluntary wheel exercise alters the levels of miR-494 and miR-696 in the skeletal muscle of C57BL/6 mice. Comp Biochem Physiol B Biochem Mol Biol. (2016) 202:16–22. doi: 10.1016/j.cbpb.2016.07.002
57. Fernandes T, Hashimoto Nara Y, Magalhães Flávio C, Fernandes Fernanda B, Casarini Dulce E, Carmona Adriana K, et al. Aerobic exercise training–induced left ventricular hypertrophy involves regulatory microRNAs, decreased angiotensin-converting enzyme-angiotensin II, and synergistic regulation of angiotensin-converting enzyme 2-angiotensin (1-7). Hypertension. (2011) 58:182–9. doi: 10.1161/HYPERTENSIONAHA.110.168252
58. Fernandes T, Magalhaes FC, Roque FR, Phillips MI, Oliveira EM. Exercise training prevents the microvascular rarefaction in hypertension balancing angiogenic and apoptotic factors: role of microRNAs-16,−21, and−126. Hypertension. (2012) 59:513–20. doi: 10.1161/HYPERTENSIONAHA.111.185801
59. Frias Fde T, de Mendonca M, Martins AR, Gindro AF, Cogliati B, Curi R, et al. MyomiRs as markers of insulin resistance and decreased myogenesis in skeletal muscle of diet-induced obese mice. Front Endocrinol. (2016) 7:76. doi: 10.3389/fendo.2016.00076
60. Russell AP, Lamon S, Boon H, Wada S, Guller I, Brown EL, et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J Physiol. (2013) 591:4637–53. doi: 10.1113/jphysiol.2013.255695
61. Stahlhut C, Suarez Y, Lu J, Mishima Y, Giraldez AJ. miR-1 and miR-206 regulate angiogenesis by modulating VegfA expression in zebrafish. Development. (2012) 139:4356–64. doi: 10.1242/dev.083774
62. Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol. (2010) 189:1157–69. doi: 10.1083/jcb.200912093
63. Safdar A, Abadi A, Akhtar M, Hettinga BP, Tarnopolsky MA. miRNA in the regulation of skeletal muscle adaptation to acute endurance exercise in C57Bl/6J male mice. PLoS ONE. (2009) 4:e5610. doi: 10.1371/journal.pone.0005610
64. van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. (2009) 17:662–73. doi: 10.1016/j.devcel.2009.10.013
65. Zhang D, Wang X, Li Y, Zhao L, Lu M, Yao X, et al. Thyroid hormone regulates muscle fiber type conversion via miR-133a1. J Cell Biol. (2014) 207:753–66. doi: 10.1083/jcb.201406068
66. Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, et al. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA. (2010) 107:4218–23. doi: 10.1073/pnas.1000300107
67. Liu Z, Long W, Fryburg DA, Barrett EJ. The regulation of body and skeletal muscle protein metabolism by hormones and amino acids. J Nutr. (2006) 136:212S−7S. doi: 10.1093/jn/136.1.212S
68. Børsheim E, Knardahl S, Høstmark AT, Bahr R. Adrenergic control of post-exercise metabolism. Acta Physiol Scand. (1998) 162:313–23. doi: 10.1046/j.1365-201X.1998.0321e.x
Keywords: skeletal muscle, muscle mitochondria, calcium handling mechanisms, SERCA (sarco(endo)plasmic reticulum Calcium ATPase), energy homeostasis, thermogenesis
Citation: Sahu B, Pani S, Swalsingh G and Bal NC (2019) Non and Epigenetic Mechanisms in Regulation of Adaptive Thermogenesis in Skeletal Muscle. Front. Endocrinol. 10:517. doi: 10.3389/fendo.2019.00517
Received: 11 May 2019; Accepted: 15 July 2019;
Published: 13 August 2019.
Edited by:
Feng Xu, Institute of Molecular and Cell Biology (A*STAR), SingaporeReviewed by:
Lei Sun, National University of Singapore, SingaporeCopyright © 2019 Sahu, Pani, Swalsingh and Bal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naresh C. Bal, bmFyZXNoLmJhbEBraWl0YmlvdGVjaC5hYy5pbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.