- 1Department of Organismic and Evolutionary Biology, and Museum of Comparative Zoology, Harvard University, Cambridge, MA, United States
- 2Departments of Molecular, Cellular and Developmental Biology, and Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, MI, United States
Direct development is a reproductive mode in amphibians that has evolved independently from the ancestral biphasic life history in at least a dozen anuran lineages. Most direct-developing frogs, including the Puerto Rican coquí, Eleutherodactylus coqui, lack a free-living aquatic larva and instead hatch from terrestrial eggs as miniature adults. Their embryonic development includes the transient formation of many larval-specific features and the formation of adult-specific features that typically form postembryonically—during metamorphosis—in indirect-developing frogs. We found that pre-hatching developmental patterns of thyroid hormone receptors alpha (thra) and beta (thrb) and deiodinases type II (dio2) and type III (dio3) mRNAs in E. coqui limb and tail are conserved relative to those seen during metamorphosis in indirect-developing frogs. Additionally, thra, thrb, and dio2 mRNAs are expressed in the limb before formation of the embryonic thyroid gland. Liquid-chromatography mass-spectrometry revealed that maternally derived thyroid hormone is present throughout early embryogenesis, including stages of digit formation that occur prior to the increase in embryonically produced thyroid hormone. Eleutherodactylus coqui embryos take up much less 3,5,3′-triiodothyronine (T3) from the environment compared with X. tropicalis tadpoles. However, E. coqui tissue explants mount robust and direct gene expression responses to exogenous T3 similar to those seen in metamorphosing species. The presence of key components of the thyroid axis in the limb and the ability of limb tissue to respond to T3 suggest that thyroid hormone-mediated limb development may begin prior to thyroid gland formation. Thyroid hormone-dependent limb development and tail resorption characteristic of metamorphosis in indirect-developing anurans are evolutionarily conserved, but they occur instead in ovo in E. coqui.
Introduction
Direct development, a distinctive life-history mode in amphibians and other animals, has evolved in anurans multiple times from the ancestral biphasic life history; it characterizes many hundreds of living species (1). Even though direct-developing frogs typically lack both a free-living aquatic larval stage and a discrete, post-hatching metamorphosis, many species display a cryptic metamorphosis before hatching: adult-specific features, such as limbs, form precociously in the egg, and numerous tadpole-specific features are present initially but then are lost [Figure 1; (2, 3)]. Because such changes in frogs with indirect development are mediated by thyroid hormone (TH), the primary regulator of metamorphosis (4), evolutionary change in thyroid axis function and timing may underlie the numerous heterochronies observed between direct-developing and indirect-developing species (5–9). Yet, there have been few attempts to precisely delineate the role of this or other pertinent physiological mechanisms.
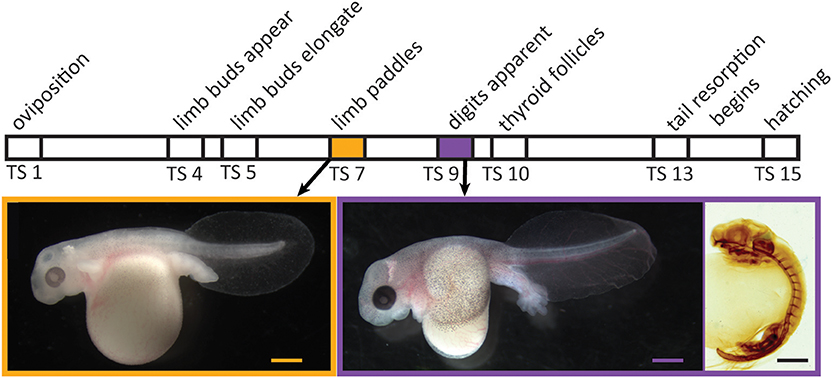
Figure 1. Relative timing of several developmental events during embryogenesis in Eleutherodactylus coqui. Images depict live TS stage 7 (Left) and stage 9 (Middle) embryos removed from overlying egg membranes, and a TS stage 9 embryo (Right) stained for type II collagen, which reveals the cartilaginous skeleton and notochord. Scale bars, 1 mm.
Embryonic development of direct-developing frogs, as seen in the Puerto Rican coquí, Eleutherodactylus coqui, appears to comprise a mosaic of TH-independent and TH-dependent features. We use the term “embryonic” to describe all in ovo development in E. coqui, although this period encompasses both the initial formation of major organ systems as well as the patterning, morphogenesis and growth that follows. Many of the latter events correspond to metamorphic changes in biphasic anurans. It was once thought that embryonic development in direct-developing species was primarily TH-independent (5). However, subsequent studies with exogenous T3 and with TH-synthesis inhibitors suggested at least a partial role for TH in terminal stages of limb development as well as tail resorption (6, 10). In E. coqui, for example, treatment with exogenous T3 causes precocious tail resorption but has little to no effect on limb elongation (11). Similarly, treatment with methimazole, a TH-synthesis inhibitor, inhibits only tail resorption and late stages of limb elongation but does not affect early limb differentiation or digit formation (8). The apparent TH-independence of early stages of limb development is correlated with the fact that limb bud, paddle and digit formation occur prior to formation of the embryonic thyroid gland [Figure 1; (12, 13)]. Thus, limb development in E. coqui comprises two periods: limb bud differentiation and paddle and digit morphogenesis, which precede formation of the thyroid gland and may be TH independent; and limb growth and elongation, which follow thyroid gland formation and are TH dependent. Experiments with TH-synthesis inhibitors, however, can only address the role of TH in the second period. The presumed TH independence of the first period remains to be verified experimentally.
All organs in the body are exposed to roughly the same concentration of circulating TH, primarily in the form of thyroxine (T4) and lower concentrations of 3,5,3′-triiodothyronine [T3; (14, 15)]. Hereafter, we use the term TH to refer to both T4 and T3. However, tissue-specific differences in uptake, metabolism, and action provide for diverse effects of TH in different tissues. Thus, tissue-specific changes in TH metabolism and action likely contribute to the heterochrony of developmental events observed in direct-developing anurans relative to biphasic species. Alternatively, the principal locus of change in hormonal control may involve a shift in the source of THs and when they are present in the embryo. Maternally derived TH is present at early developmental stages of all vertebrates examined so far. In most vertebrates, maternal TH is in the yolk; in most mammals, maternal TH can pass from mother to fetus via the placenta or milk. Yet, the role of maternally derived TH in amphibian embryos is poorly understood (16–18). If maternally derived THs are present in early embryos of E. coqui, they could influence limb development prior to formation of the embryonic thyroid gland. Finally, three different deiodinase enzymes control cellular metabolism of T4 in target tissues. In amphibians, two types of deiodinases play major roles during development. Deiodinase enzyme type II (Dio2) converts T4 into T3, which has at least 10 times greater affinity for TH receptors (TRs) than T4. Deiodinase type III (dio3) converts T4 to both T2 and reverse triiodothyronine (rT3), which are unable to bind TRs in most species. Thyroid hormones act by binding to two TR subtypes, designated alpha (α) and beta (β), to activate or repress transcription of TH target genes. Contrasting expression patterns of TRs and deiodinases may in part underlie the diverse, tissue-specific effects of TH in Xenopus species (19–26), and it is likely that changes in the temporal or spatial expression of deiodinases or TRs influence TH competence and action in target tissues in E. coqui.
Here we tested the hypothesis that developmental changes in TR and deiodinase mRNAs in developing E. coqui limb and tail, and in whole body TH content are conserved relative to those seen during metamorphosis in indirect-developing frogs. We also investigated whether E. coqui tissues are capable of responding directly to T3 action by mounting gene regulation responses similar to those seen in metamorphosing species. Taken together, our data support the hypothesis that limb development and tail resorption in E. coqui (8, 12) are mediated by conserved components of TH signaling. Additionally, our results suggest that maternal TH could facilitate limb development prior to formation of the embryonic thyroid gland.
Materials and Methods
Animal Care
Live adult Eleutherodactylus coqui were field-collected from introduced populations in Hilo, Hawaii, with the permission of the U.S. Fish and Wildlife Service (permits EX-14-06, EX-16-07, and EX-17-11). They were brought to Harvard University and maintained as a breeding colony in the Hanken laboratory (IACUC protocol #99-09-03); embryos were obtained following spontaneous matings. Following removal of the overlying chorion with watchmaker forceps in 2% cysteine (pH 8.5) in 10% Holtfreter solution, embryos were reared in 10% Holtfreter solution in Petri dishes at 22.5°C. Embryos were staged according to the normal table of Townsend & Stewart (TS; 1985), which defines 15 stages from fertilization (1) to hatching (15). Following internal fertilization, the adult female deposits embryos at TS stage 1.
Molecular Cloning and Sequence Validation
Partial cDNAs for dio2, dio3, thra, thrb, ribosomal protein L8 (rpL8), thyroid hormone induced bZip protein (thibz), and alpha-actinin 4 (actn4) (Genbank accession numbers MK784754, MK784753, MK784748, MK784749, MK784751, MK784750, MK784755) were isolated by PCR with exact primers (Table 1) using cDNA generated from RNA isolated from whole TS stage 13 embryos, and the resultant DNA fragments were subcloned into the pCR II plasmid. Exact primers for dio2, dio3, thra, thrb, rpL8, and thibz were designed from predicted full-length cDNA sequences provided by L. Sachs, N. Buisine, and G. Kerdivel (personal communication), while actn4 primers were designed from genomic sequences provided by A. Mudd, R. Harland, and D. Roksahr (personal communication). We also subcloned a partial cDNA for krüppel-like factor 9 (klf9) by degenerate PCR (oligonucleotide primers designed using CODEhop) using the same cDNA described above (Genbank accession number MK784752). The sequences of the subcloned partial cDNA fragments were confirmed by direct DNA sequencing and by comparing them against the full-length cDNAs provided by the investigators listed above.
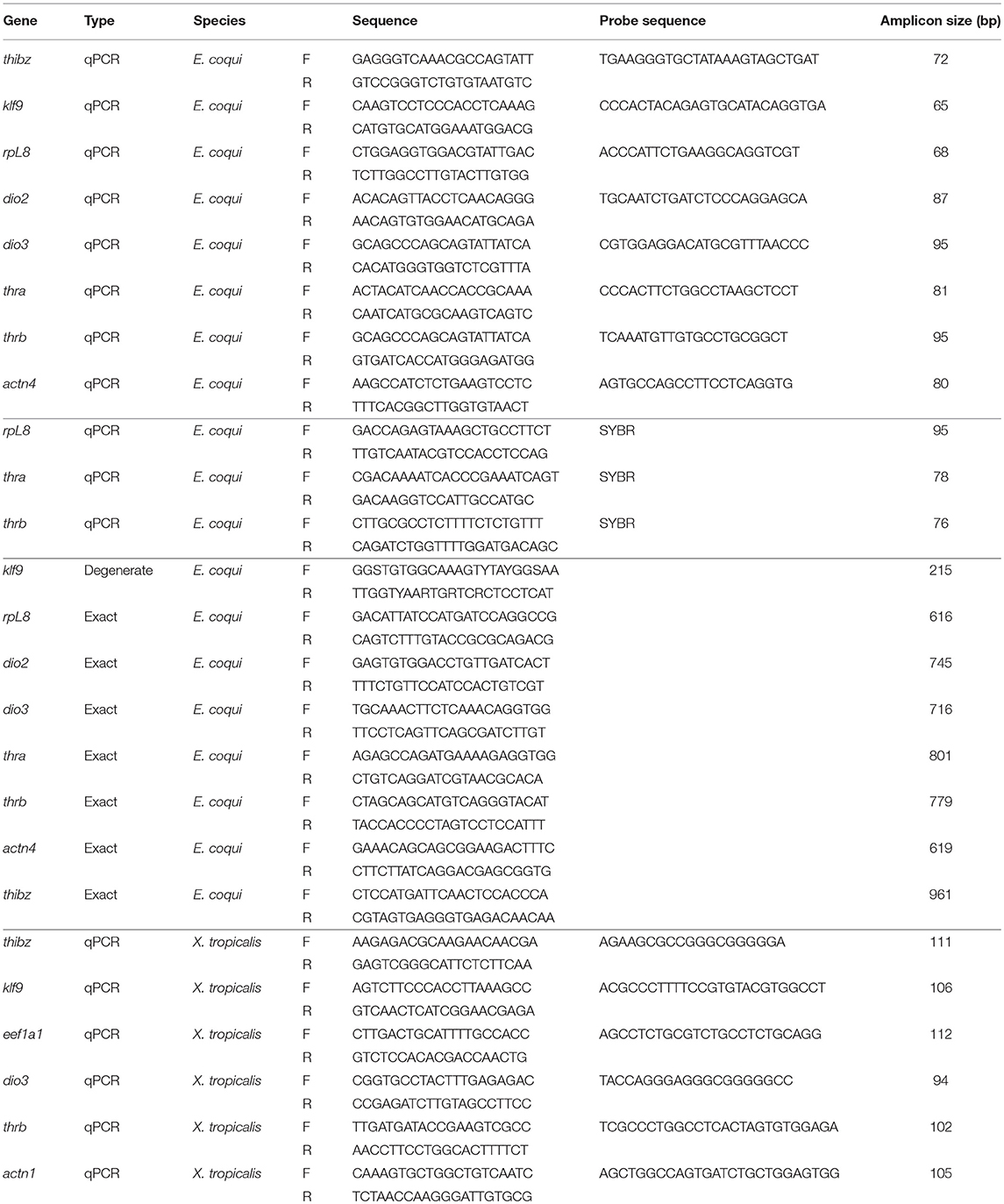
Table 1. Degenerate PCR, exact PCR, and qPCR primers and probes for Eleutherodactylus coqui and Xenopus tropicalis.
Prior to the full-length predicted cDNA sequences becoming available, oligonucleotide primers for SYBR-based reverse transcriptase quantitative PCR (RTqPCR) were designed based on the available mRNA sequences on Genbank for thra and thrb, and the previously cloned rpL8 [Genbank accession numbers AF201957.1 and AF201958.1; (8), Table 1]. For probe-based quantitative PCR (qPCR), primers and probes for actn4 were designed from the partial cloned cDNA sequence while dio2, dio3, thra, thrb, rpL8, thibz, and klf9 were designed based on the full-length sequences from other investigators listed above (Genbank accession numbers MK784763, MK784762, MK784757, MK784756, MK784760, MK784758, MK784759, MK784761).
Whole Body Extraction and Quantification of Iodothyronines Using LC-MS/MS
The iodothyronines T3, rT3, T4, and T2 were quantified from whole E. coqui embryos throughout development. Because embryos were not dissected from the yolk, all measurements include embryo and yolk TH content. Animals at different stages were anesthetized and snap frozen until extraction and LC-MS/MS analysis. Unfertilized oocytes were dissected from the ovaries of a newly sacrificed female and snap frozen. Between 15 and 20 embryos (~ 600 mg) were pooled to make one biological replicate. Three or four biological replicates were used for each developmental stage. Tissues were extracted for thyroid hormone analysis as described by Denver (27, 28) with the following modifications: stable isotope-labeled T3 and T4 (13C6 T3 and T4, Sigma) were used as an internal standard to correct for differences in extraction efficiency, and solid phase extraction with a Supel-Select SCX cartridge (60 mg 3 mL, Sigma) was used to further purify the extracted tissue. After conditioning the cartridge with 3 mL methanol (HPLC Grade, Sigma) and equilibrating it with 5 mL of 2% formic acid in water (HPLC Grade, Sigma), the sample was loaded, rinsed first with 3 mL 2% formic acid in water and then with 3 mL methanol, and finally eluted with 2 mL of freshly prepared 5% ammonium hydroxide in methanol. It was then evaporated to dryness under nitrogen flow and resuspended in 100 μl of 0.1% formic acid in methanol. Samples were measured at the Harvard Small Molecule Mass Spectrometry facility by using gradient liquid-chromatography mass-spectrometry (LC-MS/MS). Ten microliters of samples were injected on a C18 column (Kinetex 2.6 μm, 100 Å pore size, 150 × 2.1 mm, Phenomenex) in an Agilent 1290 HPLC coupled with an Agilent 6460 Triple Quad Mass Spectrometer. See Supplementary Information for the LC and MS parameters (Supplementary Tables 1, 2). Calibration curves were made in 0.1% formic acid in methanol with pure standards and the same amount of internal standard as the samples. Quantification results with a signal-to-noise (S/N) ratio >10 were used for the statistical analysis. Results with a ratio between 3 and 10 (purple type; Supplementary Table 3) were included in the graph (Figure 2) but not used in the statistical analysis; those with a ratio below 3 were not used (red type; Supplementary Table 3). We normalized iodothyronine content to the weight of the tissue extracted.
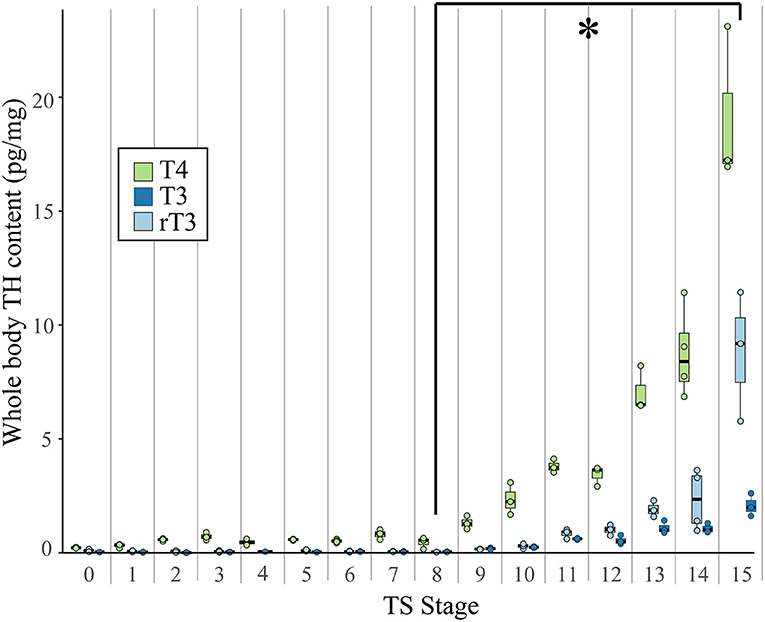
Figure 2. Whole body content of T4, T3, and rT3 in pooled E. coqui embryos at TS stages 1–15 and in unfertilized oocytes (TS 0) as quantified by LC-MS/MS. Whole body content of iodothyronines was normalized to sample weight; between 15 and 20 embryos were pooled to generate one biological replicate. Each value depicted in the graph is based on two-to-four replicates. Values based on fewer than three replicates are not included in the statistical analysis. All three iodothyronines increased significantly between TS stages 8 and 15 (post-hoc Dunn's test; p < 0.05), indicated by the asterisk (*). See Supplementary Data for a complete list of significant pairwise differences. Each boxplot represents median and range of the data.
Quantitative PCR
Dechorionated embryos were anesthetized by immersion in 10% Holtfreter solution with drops of 2% neutral-buffered MS-222 added until the embryos no longer responded to toe pinches (between 30 and 60 s). Limbs and tails were dissected and homogenized in TriZol reagent (Invitrogen) and kept at −20°C until RNA isolation. Total RNA was isolated following the manufacturer's protocol within 3 weeks of homogenization. Because qPCR primers did not span exon-exon boundaries, genomic DNA was removed with an Ambion DNA-free kit (cat. #AM1906). Controls with no reverse-transcriptase verified that removal of genomic DNA was complete. Total RNA was quantified with a Qubit Fluorometer 3.0 and checked for purity on a Nanodrop spectrophotometer. For SYBR Green RTqPCR assays, 200 ng of total RNA was used for input for each reaction. For probe-based qPCR, 660 ng of total RNA for each sample was synthesized into cDNA with iScript Reverse Transcriptase Supermix for RT-qPCR (BioRad). Complementary DNA was kept at −20°C until the qPCR assay was performed. mRNA levels were analyzed with either Ssoadvanced Universal Probes Supermix (BioRad) or an iTaq Universal SYBR Green One-Step kit (BioRad) on a CFX384 machine. See Supplementary Data for qPCR cycling conditions. Optimal qPCR conditions were determined with temperature gradient and cDNA dilutions for dynamic range of input. Standard curves showed high efficiency of reaction (90–105%), and R2 was equal to or >0.98 for all primer sets. No template controls showed no amplification. All oligonucleotides are listed in Table 1. All SYBR and probe-based qPCR experiments were done in simplex. The relative mRNA levels were determined as described by Schmittgen and Livak (29). For the developmental expression studies, target-gene expression was normalized to the reference gene rpL8, which did not show significant variation across development [rpL8 mRNA values are given in Supplementary Table 5; see also (8)]. In the in vivo and the tissue explant T3 response experiments, E. coqui target gene mRNA levels were normalized to the reference genes rpL8 and actn4, which was unaffected by T3 treatment. Small, statistically insignificant changes in reference gene mRNAs could have led to a small underestimation of the effect of T3 in these experiments.
For Xenopus tropicalis, qPCR primers and probes for thrb, klf9, thibz, dio3, elongation factor 1 alpha (eef1a1) and alpha-actinin 1 (actn1) were designed from publicly available sequences (Genbank accession numbers XM_012964865.2, NM_001113674.1, XM_018092557.1, NM_001113667.2, NM_001016692.2, and NM_001079198.1). For tissue explant experiments, X. tropicalis target gene expression was normalized to eef1a1 and actn1.
Treatment of E. coqui in vivo
Eleutherodactylus coqui embryos were dechorionated into 10% Holtfreter solution at least 24 h prior to immersion in T3. One mM stock T3 in DMSO or 0.01 N NaOH was diluted to make 50 nM T3 in 10% Holtfreter solution. We chose 50 nM T3 because it has been shown to induce tail resorption in E. coqui (8), and a 46-h timepoint to allow enough time for induction of T3 response genes. We chose TS stage 9 embryos because the last third of limb development is TH-dependent (8), but TS stage 9 is still prior to thyroid gland activation. T3 treatment solutions were refreshed every 8–12 h. After 46 h (n = 12–14 TS-9 embryos), dechorionated embryos were anesthetized as described above and limbs and tails were dissected, from which total RNA was extracted using TriZol reagent.
Measurement of Environmental T3 Uptake in X. tropicalis and E. coqui
To determine if E. coqui embryos are capable of taking up TH from their surrounding environment, we immersed dechorionated TS stage 9 E. coqui embryos or NF 51–55 X. tropicalis tadpoles in 30 mL (E. coqui) or at least 500 mL (X. tropicalis) 10% Holtfreter solution with either 1 nM (n = 4–6 biological replicates/treatment) or 50 nM (n = 3–4 biological replicates/treatment) stable isotope-labeled T3. We chose TS stage 9 E. coqui embryos to match the in vivo T3 treatment experiments and selected X. tropicalis tadpoles with developing limbs with similar morphology to E. coqui TS stage 9. Approximately twenty E. coqui individuals (600 mg tissue) or two tadpoles were pooled were pooled to make one biological replicate. Tadpoles were either ordered from Xenopus1 (Ann Arbor, Michigan, U.S.A.) or derived from the Hanken lab colony. Stock 100 μg/mL stable isotope-labeled T3 was diluted to either 1 or 50 nM T3. After either 8 or 24 h in 1 nM labeled T3 solution or 46 h in 50 nM T3 solution, X. tropicalis tadpoles and E. coqui embryos (with yolk removed) were anesthetized with neutral-buffered 2% MS-222, rinsed three times in PBS and snap frozen until extraction. On average, E. coqui embryos were more densely packed in T3 solution (5.9 mg tissue per mL media) than X. tropicalis tadpoles (2.0 mg tissue per mL media); however, E. coqui embryos are routinely cultured in these conditions with no ill effects. Tissue was extracted as described above. Because we measured whole body content of stable isotope-labeled T3 as a proxy for T3 uptake, we used 25 ng of stable isotope-labeled rT3 as an internal standard to correct for extraction efficiency.
Tissue Explant Culture and T3 Treatments
To further investigate if thyroid axis components in the E. coqui limb and tail are functional, we cultured E. coqui and X. tropicalis limb and tail explants (30, 31), treated them with T3, and assayed gene expression. We treated NF stage 52–54 (32) X. tropicalis tadpoles and TS stage 9 E. coqui embryos with 50 U/mL of penicillin-streptomycin added to aquarium or Petri dish solution for 24 h prior to dissection. Tadpoles and embryos were terminally anesthetized and dipped into 70% ethanol to sterilize the epidermis before dissection. Four X. tropicalis and two E. coqui individuals were pooled to make a single biological replicate of each species. Tissues were dissected into ice-cold 1:1.5-diluted Leibowitz-15 media (Gibco) containing 50 U/mL penicillin-streptomycin, 50 mg/mL gentamicin and 10 mM HEPES. Prior to T3 treatment tissues were cultured overnight in media supplemented with insulin (500 ug/mL) on a laboratory bench at room temperature (21°C) with gentle shaking (50 rpm). The next morning, stock T3 was diluted in 0.01 N NaOH and added to the media to a final concentration of 50 nM. Media and T3 were changed every 8–12 h. After treatment for 8 or 46 h, limb and tail explants were rinsed three times in phosphate-buffered saline (PBS) and homogenized in TriZol. RNA was isolated according to the manufacturer's protocol.
Statistical Analysis
Statistical analyses of qPCR data were done with RStudio version 1.0.136 and visualized with ggplot2 (https://ggplot2.tidyverse.org/). Developmental timeline qPCR and iodothyronine content data followed a non-normal distribution as determined by Q-Q plots and the Shapiro-Wilk test; Levene's test determined that TH content data additionally had unequal variance. Log10-transformed data were not normally distributed. Therefore, a Kruskal-Wallis test was used to determine if there were significant differences among groups, and a post-hoc Dunn's test with the Benjamini and Hochberg (BH) correction was used to identify stages that differ from each other while adjusting for multiple comparisons. We performed a least squares regression on T4, T3, and rT3 data sets to investigate possible differences in iodothyronines kinetics during development. For the developmental timeline qPCR data, statistical tests were performed on data pooled from two independent experiments (see Supplementary Data for data from each experiment). For in vivo and in vitro T3 treatment experiments, Student's t-test was used to identify significant differences between T3-treated groups and controls.
Results
Predicted Proteins of Isolated E. coqui cDNAs Contain Conserved Domains
Most isolated cDNAs contained functional domains of orthologous proteins. The predicted E. coqui TRα and TRβ sequences cover amino acids 11–281 (65%), and amino acids 9–273 (69%) of the orthologous X. tropicalis proteins, respectively. Both predicted TR protein sequences contain the DNA-binding domain and most of the ligand-binding domain. Alignments show that the predicted protein sequence of the E. coqui TRα DNA-binding domain has 97% identity to the X. tropicalis DNA-binding domain, while the TRα ligand-binding domain shared between the predicted E. coqui and X. tropicalis sequences are 98% identical. The DNA-binding domain of the predicted E. coqui TRβ sequence is 100% identical to the DNA-binding domain in X. tropicalis TRβ, and the ligand-binding domain is 95% identical. The predicted partial E. coqui Dio2 sequence covers amino acids 2–254 (98%) of X. tropicalis Dio2 and the partial E. coqui Dio3 sequence covers amino acids 7–252 (90%) of X. tropicalis Dio3. Additionally, the predicted protein sequence of both dio2 and dio3 isolated cDNAs contain the selenocysteine site and the thioredoxin domain. Both thioredoxin domains share 86% identity with the orthologous X. tropicalis thioredoxin domain. The partial predicted amino acid sequence of E. coqui Klf9 covers amino acids 194–264 (25%) of X. tropicalis Klf9 and contains the three characteristic zinc-finger domains (100% identity) in the C-terminus of X. tropicalis Klf9. The isolated E. coqui thibz sequence covers amino acids 159–335 (53%) of X. tropicalis NFIL3-like (synonym for thbzip) and lacks the highly conserved basic leucine zipper domain. Even without the highly conserved basic leucine zipper domain, the predicted E. coqui protein sequence still clusters with other orthologous NFIL3-like proteins, rather than with other proteins with the basic leucine zipper domain (NFIL3 and CREB1) in maximum likelihood trees of these three orthologous vertebrate proteins (data not shown). Similarly, the other partial predicted E. coqui sequences cluster with other orthologous genes rather than with other closely related proteins containing similar domains (data not shown). We confirmed all isolated E. coqui cDNAs against the full-length transcript provided by investigators listed in the methods. Finally, we also performed BLASTx and BLASTn searches with the isolated E. coqui cDNA sequences. All cloned sequences have high similarity to predicted orthologous genes in frog species and other vertebrates (Supplementary Table 4).
Changes in Whole Body Iodothyronine Content During Embryonic E. coqui Development
Using LC-MS/MS, we detected the iodothyronines T4, T3, and rT3 in unfertilized oocytes and at every stage of development (Figure 2). Thyroxine content (pg/mg body weight) was highest, followed by rT3 and then T3. We detected T2 only at TS stages 14 and 15, when hatching occurs, and at this point, T2 content was less than all other iodothyronine content and ranged between 0.04 and 0.78 pg/mg body weight (Supplementary Table 3). The three quantifiable iodothyronines were low and relatively constant up to TS stage 8, after which stage they showed statistically significant increases [T3: Kruskal-Wallis rank sum test, X2 = 43.2 (df = 15), p < 0.001; T4: Kruskal-Wallis rank sum test, X2 = 43.7 (df = 15), p < 0.001; rT3: Kruskal-Wallis rank sum test, X2 = 39.7 (df = 14), p < 0.001]. Whole body content of all three iodothyronines showed statistically significant increases between stages 8 and 13 (post-hoc Dunn's test; p = 0.048, 0.033, 0.035 for T3, T4, and rT3, respectively). The velocity of change was slower for rT3 and T3 compared with T4. Stage was a significant predictor for all three iodothyronines [T3: F = 70.8 (df = 45), p < 0.001; T4: F = 54.2 (df = 46) p < 0.001; rT3: F = 23.7 (df = 43), p < 0.001]. Although all iodothyronines are positively correlated with stage, the velocity of change was slower for rT3 and T3 (slope of least squares regression (LSR) line, b = 0.2964 and 0.1009, respectively) compared to T4 (LSR, b = 0.7984). Tissue content of all three iodothyronines was highest at TS 15. Note also that oocytes and early embryos (TS 0–5) of E. coqui have large yolk deposits, which may increase the S/N ratio and cause an underestimation of iodothyronine content in the embryo and yolk at these stages.
Changes in Thyroid Hormone Receptor and Deiodinase mRNA Levels in the Embryonic Tail
Both thra and thrb mRNAs in the E. coqui tail showed statistically significant changes during development [Figure 3A; thra: Kruskal-Wallis rank sum test, X2 = 20.18 (df = 4), p < 0.001; thrb: Kruskal-Wallis rank sum test, X2 = 26.78 (df = 4), p < 0.001]. Thyroid hormone receptor α and thrb mRNA in the tail bud are approximately equal at TS stage 5 (Figure 3A). Thyroid hormone receptor α mRNA in the tail at hatching is between 2.1- and 4-fold higher than the early tail (TS stages 5 and 7, post-hoc Dunn's test, p = 0.002 and 0.03, respectively). Thyroid hormone receptor β mRNA follows a similar pattern—it increased 4-fold between the onset of tail resorption (TS 13) and hatching (TS 15)—although thra increased only 1.8-fold over the same interval (Figure 3A). Thyroid hormone receptor β mRNA at hatching (TS 15) is between 18- and 47-fold higher than in the early tail (TS stage 5 and TS stage 7, post-hoc Dunn's test, p < 0.001 and p = 0.002, respectively).
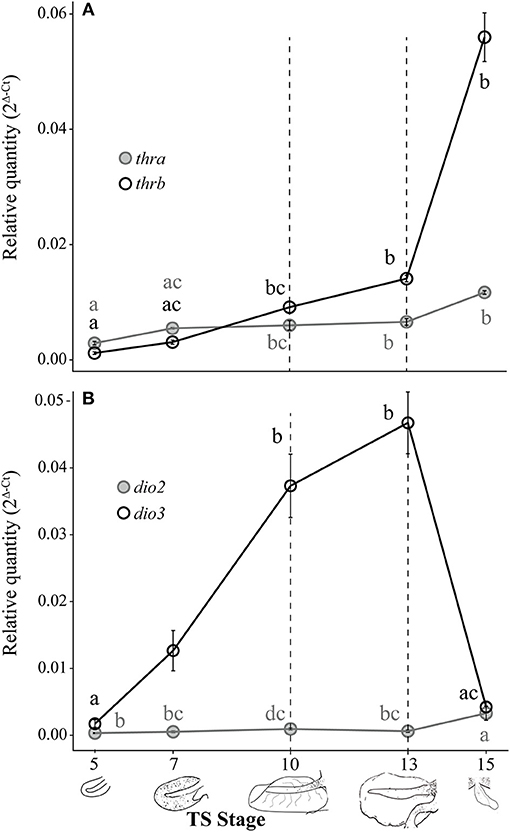
Figure 3. Relative thra and thrb mRNA levels (A) and dio2 and dio3 mRNA levels (B) in the pre-hatching tail of E. coqui. Dashed vertical line at TS stage 10 marks when thyroid follicles are first visible in histological sections; the line at TS 13 indicates the onset of tail resorption. Drawings on the x-axis depict tail growth and resorption before hatching. Each expression value is represented as a circle centered on the mean of 3–7 individuals ± SE. Lower-case letters in gray (thra and dio2) and black (thrb and dio3) indicate significant pairwise differences between groups (post-hoc Dunn's test, p < 0.05). See Supplementary Data for a complete list of pairwise comparisons.
Deiodinase type II and dio3 mRNAs significantly changed during tail development (Figure 3B; dio2: Kruskal-Wallis rank sum test, X2 = 17.37 (df = 4), p = 0.002; dio3: Kruskal-Wallis rank sum test, X2 = 26.11 (df = 4), p < 0.001). Patterns of deiodinase mRNA in the developing tail were essentially the opposite of those seen in the limb. Deiodinase type II mRNA was low throughout tail development and resorption but rose almost 10-fold as hatching neared (TS 15; Figure 3B). At hatching (TS 15), dio2 mRNA was higher than at TS 5, 7 and 13 (post-hoc Dunn's test, p = 0.001, 0.029, and 0.031, respectively). Deiodinase type III mRNA increased 27-fold between TS 5 and 13 (post-hoc Dunn's test, p < 0.001) and then decreased steeply (11-fold) between the onset of tail resorption and hatching (post-hoc Dunn's test, p = 0.007). Repeated experiments demonstrate the similar patterns of thra, thrb, dio2, and dio3 expression (Supplementary Figure 1).
Changes in Thyroid Hormone Receptor and Deiodinase mRNA Levels in the Embryonic Hind Limb
Both thra and thrb mRNAs in the E. coqui hind limb showed statistically significant changes during development (Figure 4A; thra: Kruskal-Wallis rank sum test, X2 = 20.66 (df = 4), p < 0.001; thrb: Kruskal-Wallis rank sum test, X2 = 25.36 (df = 4), p < 0.001). The level of thra mRNA was greater than thrb mRNA in the limb bud until TS 10, when the thra mRNA level began to decrease and continued to decline through hatching (Figure 4A). The peak thra mRNA level at TS 10 coincides with the appearance of thyroid follicles (13); thra mRNA in the hind limb at this stage was significantly higher than in the limb bud at TS 5 (post-hoc Dunn's test, p = 0.001), in the limb paddle at TS stage 7 (p = 0.009) and in the fully formed froglet limb at TS 15 (post-hoc Dunn's test, p = 0.002). At hatching, thra mRNA level was lower than thrb mRNA levels. Between paddle (TS 7) and toepad formation (TS 13), thrb mRNA rose ~21-fold to a peak at TS 13. At TS 13, thrb expression was significantly higher than in the limb bud and paddle (Figure 4A; TS 5 and 7; post-hoc Dunn's test, p < 0.001 and p = 0.001, respectively). Thyroid hormone receptor β mRNA drops almost 1.5-fold between TS 13 and hatching.
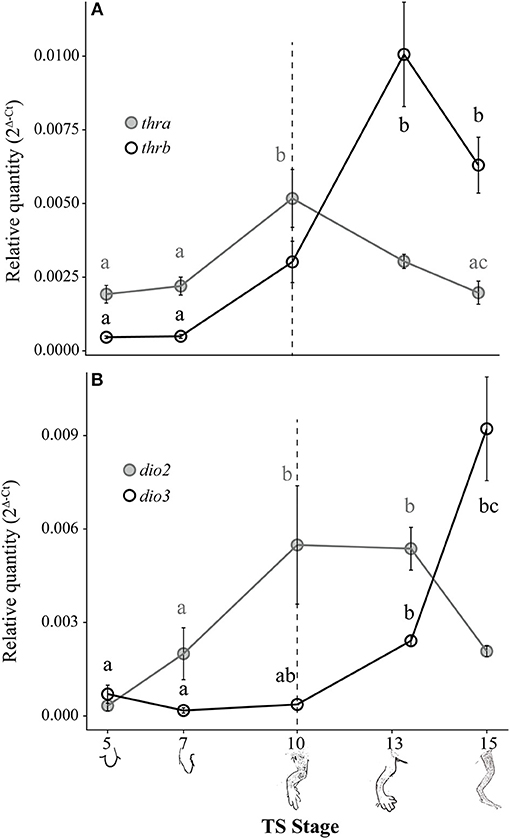
Figure 4. Relative thra and thrb mRNA levels (A) and dio2 and dio3 mRNA levels (B) in the pre-hatching hind limb of E. coqui. Dashed vertical line at TS stage 10 marks when thyroid follicles are first visible in histological sections. Drawings on the x-axis depict sequential formation of the limb. Each expression value is represented as a circle centered on the mean of 5–7 hind limb pairs ± SE. Lower-case letters in gray (thra and dio2) and black (thrb and dio3) indicate significant pairwise differences between groups (post-hoc Dunn's test, p < 0.05). See Supplementary Data for a complete list of pairwise comparisons.
Deiodinase type II and dio3 mRNAs both showed statistically significant but contrasting patterns throughout limb development [Figure 4B; dio2: Kruskal-Wallis rank sum test, X2 = 18.65 (df = 4), p < 0.001; dio3: Kruskal-Wallis rank sum test, X2 = 25.76 (df = 4), p < 0.001]. Deiodinase type II mRNA increased 16-fold between limb bud (TS 5) and digit formation (TS 10) and remained at this level through subsequent limb growth (TS 13; post-hoc Dunn's test, p = 0.007 and p < 0.001, respectively). Deiodinase type II mRNA decreased 2.6-fold between TS 13 and hatching to the level originally present in the newly formed limb bud (e.g., TS 5). Deiodinase type III mRNA remained low throughout most of limb development, but it increased 25-fold between the initial formation of thyroid follicles (TS 10) and hatching (TS 15; post-hoc Dunn's test, p = 0.001). Repeated experiments show the same general contrasting mRNA expression patterns for dio2, dio3, thra, and thrb (Supplementary Figure 2).
Exogenous T3 Induced Gene Expression Responses in the TS 9 E. coqui Tail, but Not the Limb
To determine if E. coqui tissues are capable of mounting a gene regulation response to exogenous T3, we performed in vivo T3 treatments (Figure 5A). Immersion of TS 9 E. coqui embryos in 50 nM T3 for 8 h caused a significant induction of klf9 (Student's t-test, t = 5.61 (df = 21.74), p < 0.001) and thibz (Student's t-test, t = 6.20 (df = 12.42), p < 0.001) in the tail. Immersion in 50 nM T3 for 46 h additionally significantly induced thrb mRNA (Supplementary Figure 4). In contrast, the identical treatment significantly increased only thibz expression (Figure 5B; Student's t-test, t = 3.11 (df = 18.92), p = 0.006) in the limb.
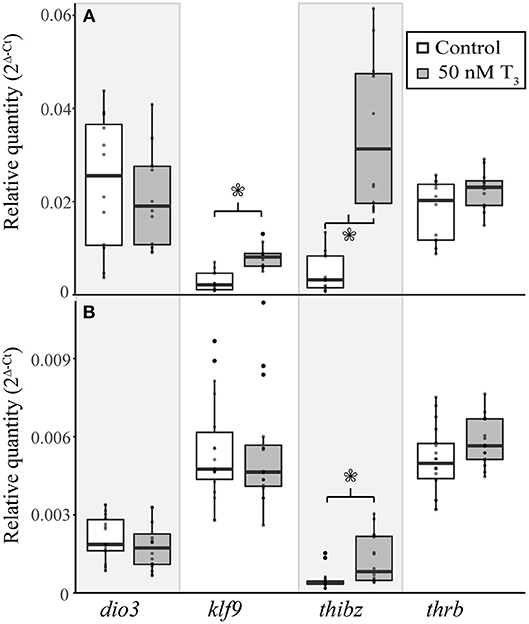
Figure 5. Exogenous treatment with 50 nM T3 for 8 h induces gene expression in the tail of E. coqui embryos at TS stage 9 (A), but not in limbs at the same stage (B). Boxes and whiskers depict the median and range of 12–16 individuals from two independent experiments. Asterisks indicate a significant change in expression (Student's t-test, p < 0.05).
E. coqui Embryos Took up Significantly Less T3 From the Environment Than Did X. tropicalis Tadpoles
Because previous studies suggested that E. coqui limbs are insensitive to TH, and because we observed a weak TH response in our in vivo experiments, we wanted to confirm that immersion in T3 increased tissue content of T3. We quantified stable isotope-labeled T3 tissue content after immersing X. tropicalis tadpoles or E. coqui embryos in stable isotope-labeled T3 solution under three conditions. We chose 50 nM T3 and 46 h treatment to match the E. coqui in vivo T3 response experiments. We also chose two conditions that represent relevant time points from previous studies of larval Xenopus species: (1) treatment with 1 nM T3 for 8 h is sufficient for X. tropicalis' whole body T3 content to surpass the T3 concentration in the surrounding media (33), and (2) treatment with 1 nM T3 for 24 h is sufficient to induce gene expression responses in X. tropicalis (31, 34). After immersing E. coqui in 1 nM labeled T3 for 8 and 24 h, we detected endogenous T3 but not labeled T3. However, we detected labeled T3 in X. tropicalis tissue at both 8 and 24 h (Table 2). We detected stable isotope-labeled T3 in both E. coqui and X. tropicalis tissue following 46-h treatment with 50 nM T3. Total content of labeled T3 in X. tropicalis tissue was ~63 times that found in E. coqui tissues [Table 2, Student's t-test, t = −3.20 (df = 2.00), p = 0.085]. Additionally, X. tropicalis has ~ 875 times more stable isotope-labeled T3 than endogenous T3 content. In contrast, stable isotope-labeled T3 in E. coqui is approximately equal to endogenous T3 content.
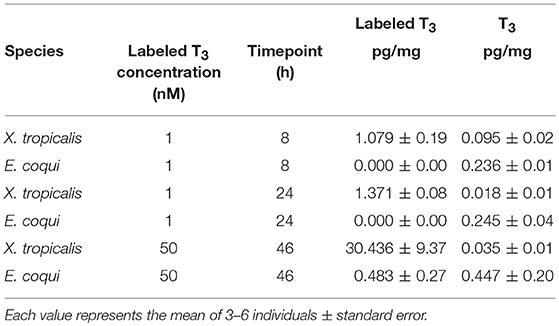
Table 2. Nieuwkoop and Faber stage 51–55 Xenopus tropicalis tadpoles have more labeled T3 tissue content than do TS stage 9 E. coqui embryos after immersion in labeled T3 for 8, 24, or 46 h.
Exogenous T3 Strongly Induced T3 Response Genes in TS Stage 9 E. coqui Limb Explants
Treatment with 50 nM T3 for 8 h significantly increased dio3 [Student's t-test, t = 8.40 (df = 4.00), p = 0.001), klf9 (Student's t-test, t = 14.41 (df = 4.18), p < 0.001], thibz [Student's t-test, t = 9.64 (df = 4.01), p < 0.001], and thrb [Student's t-test, t = 8.26 (df = 4.39), p < 0.001] mRNAs in explants of X. tropicalis tail (Figure 6A). The same treatment caused a significant increase in dio3 [Student's t-test, t = 3.49 (df = 3.00), p = 0.040], klf9 [Student's t-test, t = 13.66 (df = 3.08), p < 0.001], and thibz [Student's t-test, t = 21.50 (df = 2.07), p = 0.002] mRNAs in X. tropicalis limb explants (Figure 6B). Similarly, exogenous T3 increased dio3 [Student's t-test, t = 2.56 (df = 6.47), p = 0.040], klf9 [Student's t-test, t = 2.67 (df = 8.22), p = 0.028], thibz [Student's t-test, t = 3.54 (df = 8.00), p = 0.008], and thrb [Student's t-test, t = 5.38 (df = 8.62), p < 0.001] mRNAs in explants of E. coqui tail (Figure 6C). Deiodinase type III [Student's t-test, t = 2.61 (df = 9.80), p = 0.027], klf9 [Student's t-test, t = 6.11 (df = 8.05), p < 0.001], thibz [Student's t-test, t = 6.49 (df = 8.00), p < 0.001], and thrb [Student's t-test, t = 7.70 (df = 8.80), p < 0.001] increased after the same treatment in E. coqui limb explants (Figure 6D).
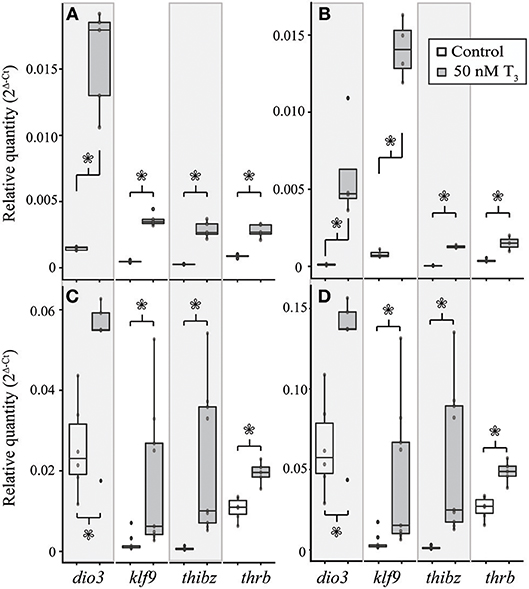
Figure 6. Treatment with 50 nM exogenous T3 for 8 h induces deiodinase type III (dio3), krüppel-like factor 9 (klf9), thyroid hormone induced bZip protein (thibz), and thrb expression in NF stage 52–54 X. tropicalis tail (A) and limb (B) explants and in TS stage 9 E. coqui tail (C) and limb explants (D). Asterisks indicate a significant increase in mRNA levels (Student's t-test, p < 0.05).
In both species, the magnitude of increase for all genes was greater in the limb than in the tail (Table 3). The same trends were observed after treating tissue explants with 50 nM T3 for 46 h (Supplementary Figure 5). In tail explants, T3 induced fold changes of a similar order of magnitude for thibz (between 42- and 44-fold) and thrb (between 1.8- and 3.8-fold), but not for dio3 and klf9; in E. coqui, dio3, and klf9 mRNAs increased 2- and 9.9-fold, respectively, while dio3 and klf9 mRNAs increased 11.9- and 12.5-fold in X. tropicalis. In limb explants, dio3 and thibz mRNA differed by an order of magnitude between species. Deiodinase type III mRNA increased 58.8-fold in X. tropicalis limb explants, while dio3 mRNA increased 3.7-fold in E. coqui limb tissue. Thyroid hormone induced bZip protein mRNA increased only 37-fold in X. tropicalis limb explants, while dio3 mRNA increased 180-fold in E. coqui limb explants.
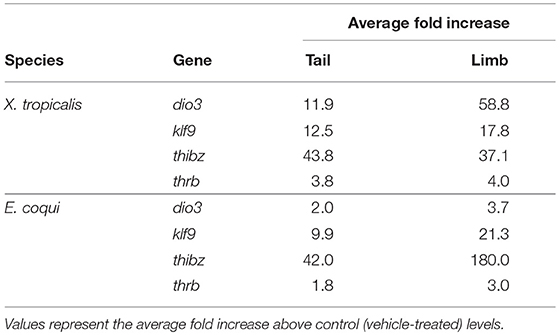
Table 3. Induction of deiodinase type III (dio3), krüppel-like factor 9 (klf9), thyroid hormone induced bZip protein (thibz), and thyroid hormone receptor β (thrb) in tail and limb explants of NF stages 52–54 Xenopus tropicalis and TS stage 9 Eleutherodactylus coqui after treatment with 50 nM T3 for 8 h.
Discussion
In this study we show that the core TH signaling components are evolutionarily conserved in Eleutherodactylus coqui limb and tail tissue. We also show that developmental patterns of thra, thrb, dio2, and dio3 mRNAs, and whole-body TH content in E. coqui closely match those reported during metamorphosis of Xenopus species. We also find maternal T4, T3, and rT3 in unfertilized eggs and early embryos of E. coqui, which may mediate TR signaling prior to embryonic thyroid gland formation. This is the first published report of TH metabolites and maternally derived TH in a direct-developing frog. Additionally, we demonstrate that E. coqui tissues show robust gene expression responses to exogenous T3 similar to those seen in metamorphosing species. Eleutherodactylus coqui embryos take up much less T3 from the environment compared with X. tropicalis. This difference likely explains the relatively weak and variable gene expression responses seen in vivo in E. coqui, and was likely a significant confounding factor for previously published results.
Developmental Profiles of Whole Body Iodothyronine Content
Temporal dynamics of whole-body iodothyronine content in direct-developing E. coqui mirror those described for indirect-developing frogs, which retain the ancestral biphasic life history: Scaphiopus hammondii (28), Rana catesbeiana (35), Bufo marinus (36), Bufo japonicus (37), and Xenopus laevis (33). Anuran metamorphosis comprises three successive stages: premetamorphosis, when little to no TH is present; prometamorphosis, when TH concentrations slowly rise; and a rapid metamorphic climax characterized by a peak in TH concentrations. The temporal profile of TH content in embryonic E. coqui similarly defines three successive periods: (1) Low TH content characterizes the first half of development, prior to thyroid follicle formation (TS 1–8). (2) After thyroid follicles appear, TH content gradually rises until tail resorption began (TS 9–12). (3) TH content dramatically increases, with a peak in TH at or just prior to hatching (TS 13–15). In addition to amphibians, many other vertebrates experience peak concentrations of TH at life history transitions—at hatching in precocial birds (38), at the larval-to-juvenile transition in several fish species (39–41), at ~14 days post-partum in rats and mice (42, 43), and at birth in humans (44).
Thyroid hormones are present throughout early embryogenesis and the subsequent period of pre-hatching development in E. coqui (TS 1–9), beginning up to eight days before thyroid follicles can be detected histologically (13). These hormones are almost certainly maternal in origin. Similarly, T4 and T3 have been detected in yolk and gastrulating embryos of four other anuran species—Bufo marinus (36), Rana catesbeiana (35), Bombina orientalis (45), and Xenopus laevis (16). Early Xenopus tropicalis embryos express key TH signaling components (46). Indeed, TH signaling is also functional in the Xenopus tadpole central nervous system (CNS) before thyroid gland formation (16, 18). Maternally derived TH has a conserved role in vertebrate CNS development (47) and embryogenesis (17, 48, 49). Therefore, it seems likely that direct-developing frogs require maternal TH for normal neural development, as do most vertebrate species, although we do not evaluate that hypothesis here.
Maternal TH may regulate limb development occurring before the differentiation of the embryonic thyroid gland in direct-developing frogs. In metamorphosing anurans, TH signaling is required for terminal limb differentiation (22), but the initial stages of limb development are TH-independent. For example, tadpoles immersed in methimazole, a TH-synthesis inhibitor, develop a long limb-bud-like structure (24), and thyroidectomized tadpoles develop calcification centers in the hind limb (50, 51). In E. coqui, the limb bud proliferates and digits develop prior to the appearance of embryonic thyroid follicles (TS stages 9–10) [Figure 3; (8, 13)]. Two hypotheses could account for this observation: (1) E. coqui relies on maternal TH, rather than embryonically produced TH, to regulate early stages of digit patterning and growth (TS 6–9); or (2) paddle and digit formation in E. coqui proceed independently of TH. Our data show that requisite components of TH signaling are present at this time. Future investigation should evaluate the functional role of TH during this critical developmental period. A switch from embryonic to maternally synthesized TH for the regulation of early limb development, if it occurred, could explain the heterochronic shift in limb development and would represent an evolutionary novelty in direct-developing species.
Thyroid Hormone Receptor α, thrb, Dio2, and Dio3 mRNA Expression Patterns During Development and T3 Response in the Embryonic Tail
Tail resorption in Xenopus tropicalis occurs late in metamorphosis and is mediated by TRβ (52). Because tail resorption in E. coqui occurs late in embryogenesis and requires T3 (8), we expected that thra, thrb, dio2, and dio3 mRNA dynamics in the E. coqui tail would mirror those described in Xenopus. Our results support this hypothesis: in the E. coqui tail, a rise in thrb expression coincides with the rise in embryonic TH content, consistent with a role for thrb in mediating tail resorption.
Deiodinase type II and dio3 mRNA expression patterns in the developing E. coqui tail are also similar to those described in indirect-developing species in which these deiodinase enzymes are critical for coordinating metamorphosis (20). Elevated dio3 expression protects the tail from an early apoptotic response to T3 until metamorphic climax in Xenopus (26); E. coqui tail resorption also begins at TS 13, when dio3 expression significantly decreases. Although they serve different functions, the tail serves a critical role in both species: the larval Xenopus tail is a critical locomotor organ, whereas the embryonic E. coqui tail functions in respiration. In both species, maintenance of the tail is accomplished in part by dio3 inactivation of T4 and T3.
Given the conservation of mRNA dynamics in the E. coqui tail, we wanted to determine whether the tissue could respond to exogenous T3. In Xenopus species, treatment with exogenous T3 induces transcription of direct T3 response genes dio3, klf9, thibz, and thrb (19, 53–56). Exogenous T3 induces significant increases in the mRNA of three of these T3 response genes, klf9, thibz, and thrb, supporting the hypothesis that TH signaling components are conserved and mediate tail resorption in E. coqui.
Thyroid Hormone Receptor α, thrb, Dio2, and Dio3 mRNA Expression Patterns During Development and T3 Response in the Embryonic Hind Limb
Thyroid hormone receptor α, thrb, dio2, and dio3 mRNA expression patterns parallel those described in Xenopus species in the period leading up to and during metamorphosis (33, 57). In indirect-developing frogs, TRα has a critical role in controlling post-embryonic developmental timing (58–60) and in promoting proliferation in the hind limb during metamorphosis (61–63). Constitutive thra expression supports a proliferative and competence-establishing role for TRα in E. coqui. In the E. coqui limb, a rise in thrb expression coincides with the rise in embryonic TH content, consistent with TRβ autoinduction and tissue sensitization to TH described in Xenopus (64).
The tissue-specific patterns of dio2 and dio3 underlie the differential sensitivity of limb and tail tissue in metamorphosing frogs. Deiodinase type II expression is constitutive in the developing limb of Xenopus laevis, causing the limb to be sensitive to small amounts of T3 produced during premetamorphosis (23). Similarly, elevated dio2 expression in E. coqui limbs throughout most of limb development, including several days prior to formation of the embryonic thyroid gland, supports a role for TH-mediated limb development and growth.
In indirect-developing species, including Xenopus and spadefoot toads (Scaphiopus), concentrations between 1 and 10 nM T3 are sufficient to promote precocious metamorphosis, tail resorption, and gene expression responses in limbs and tail (31, 65, 66). However, previous studies report that the E. coqui limb has no morphological response to high doses of exogenous T3 (11). Our study is the first to characterize mRNA expression changes in a direct-developing frog species in response to exogenous T3. Treatment of E. coqui embryos with exogenous T3 prior to formation of the thyroid follicles increases expression of four direct T3 response genes in the tail, consistent with studies in Xenopus species (19, 53–55). However, limbs of the same embryos do not respond to T3, despite the high dose administered (50 nM T3). The lack of response previously observed in direct-developing species may be confounded by an inability of T3 to reach the limb tissue. We observe a weak induction of T3 response genes in TS stage 7 limbs, a full two days before E. coqui begins to produce TH (Supplementary Figure 3). It is possible that this response occurs because the adult epidermis is not yet fully formed and T3 is better able to penetrate into the tissue, or because there is less endogenous T3 present at TS 7 than at TS 9. In either case, the ability to respond to T3 prior to thyroid gland formation is similar to biphasic species; tadpoles are also TH competent as soon as they hatch. Finally, the similar robust gene regulation response induced in E. coqui and X. tropicalis limb explants suggests that the limb tissue itself is similarly competent in both species. Overall, these data support the hypothesis that TH plays a role in E. coqui limb development and may do so prior to formation of the embryonic thyroid gland.
Here we support previous claims that later stages of limb development in E. coqui are TH-dependent but we additionally show that TH-signaling components are present during earlier stages, and that E. coqui limb tissue is sensitive to T3. Eleutherodactylus coqui eggs are provisioned with maternally derived TH, which may mediate organogenesis before differentiation and activity of the embryo's own thyroid gland. Altogether, our data suggest that the TH-mediated molecular module active during post-hatching metamorphosis in indirect-developing frogs has been shifted prior to hatching in direct-developing species.
Ethics Statement
This study was carried out in accordance with the recommendations of the Harvard Faculty of Arts and Sciences Institutional Animal Care and Use Committee. The protocol was approved by the Harvard Faculty of Arts and Sciences Institutional Animal Care and Use Committee.
Author Contributions
ML designed experiments and performed experiments, interpreted data, and wrote the manuscript. RD and JH contributed to experimental design, edited the manuscript, and discussed data interpretation.
Funding
This study was supported by a Graduate Women in Science Fellowship and NSF grant IOS 11456115 to RD, an NSF Doctoral Dissertation Improvement Grant #1701591, and Miyata and Goelet Grants from the Museum of Comparative Zoology. Published by a grant from the Wetmore Colles fund of the Museum of Comparative Zoology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Laurent Sachs, Nicolas Buisine, Gwenneg Kerdivel, Austin Mudd, Richard Harland, and Dan Rokhsar for providing sequences that made this work possible. We also thank Charles Vidoudez for technical help in tissue extraction and TH quantification.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2019.00307/full#supplementary-material
References
1. Pough FH, Andrews R, Crump M, Savitzky AH, Wells KD, Brandley MC. Herpetology. 4th ed. Sunderland, MA: Sinauer Associates, Inc. (2015).
2. Elinson RP, del Pino EM. Developmental diversity of amphibians. WIREs Dev Biol. (2012) 1:345–69. doi: 10.1002/wdev.23
3. Hanken J, Klymkowsky MW, Summers CH, Seufert DW, Ingebrigsten N. Cranial ontogeny in the direct-developing frog, Eleutherodactylus coqui (Anura: Leptodactylidae), analyzed using whole-mount immunohistochemistry. J Morphol. (1992) 211:95–118. doi: 10.1002/jmor.1052110111
4. Denver RJ. Neuroendocrinology of amphibian metamorphosis. In: Shi Y-B, editor. Current Topics in Developmental Biology. Burlington: Elsevier Inc. (2013), 195–227.
5. Lynn WG. A Study of the Thyroid in Embryos of Eleutherodactylus nubicola. The Anatomical Record, 64, p.525-535, 2 plates.
6. Lynn WG, Peadon AM. The role of the thyroid gland in direct development in the Anuran, Eleutherodactylus martinicensis. Growth. (1955) 19:263–86.
7. Schlosser G, Roth G. Development of the retina is altered in the directly developing frog Eleutherodactylus coqui (Leptodactylidae). Neurosci Lett. (1997) 224:153–6. doi: 10.1016/S0304-3940(97)00174-2
8. Callery EM, Elinson RP. Thyroid hormone-dependent metamorphosis in a direct developing frog. Proc Natl Acad Sci USA. (2000) 97:2615–20. doi: 10.1073/pnas.050501097
9. Elinson RP. Metamorphosis in a frog that does not have a tadpole. In: Shi Y-B, editor. Current Topics in Developmental Biology. Vol. 103. Burlington: Elsevier Inc. (2013), 259–76. doi: 10.1016/B978-0-12-385979-2.00009-5
10. Lynn WG. The effects of thiourea and phenylthiourea upon the development of Eleutherodactylus ricordii. Biol Bull. (1948) 94:1–15. doi: 10.2307/1538202
11. Elinson RP. Leg development in a frog without a tadpole (Eleutherodactylus coqui). J Exp Zool. (1994) 270:202–10. doi: 10.1002/jez.1402700209
12. Townsend D, Stewart M. Direct development in Eleutherodactylus coqui (Anura: Leptodactylidae): a staging table. Copeia. (1985) 1985:423–36. doi: 10.2307/1444854
13. Jennings DH, Hanken J. Mechanistic basis of life history evolution in anuran amphibians: thyroid gland development in the direct-developing frog, Eleutherodactylus coqui. Gen Comp Endocrinol. (1998) 111:225–32. doi: 10.1006/gcen.1998.7111
14. Rosenkilde P. Thyroid hormone synthesis in metamorphosing and adult Xenopus laevis. Gen Comp Endocrinol. (1978) 34:95–6.
15. Buscaglia M, Leloup J, De Luze A. The role and regulation of monodeiodination of thyroxine to 3,5,3'-triidothyronine during amphibian metamorphosis. In: Balles M, Bownes M, editors. Metamorphosis. Oxford: Clarendon Press. (1985), 273–293.
16. Fini JB, Le Mével S, Palmier K, Darras VM, Punzon I, Richardson SJ, et al. Thyroid hormone signaling in the Xenopus laevis embryo is functional and susceptible to endocrine disruption. Endocrinology. (2012) 153:5068–81. doi: 10.1210/en.2012-1463
17. Morvan-Dubois G, Fini JB, Demeneix B. A. Is thyroid hormone signaling relevant for vertebrate embryogenesis? In: Shi Y-B, editor. Current Topics in Developmental Biology. Vol 103. Burlington: Elsevier Inc (2013), 365–96. doi: 10.1016/B978-0-12-385979-2.00013-7
18. Le Blay K, Préau L, Morvan-Dubois G, Demeneix B. Expression of the inactivating deiodinase, Deiodinase 3, in the pre-metamorphic tadpole retina. PLoS ONE. (2018) 13:e0195374. doi: 10.1371/journal.pone.0195374
19. Wang Z, Brown DD. Thyroid hormone-induced gene expression program for amphibian tail resorption. J Biol Chem. (1993) 268:16270–8.
20. Becker K, Stephens K. The type 2 and type 3 iodothyronine deiodinases play important roles in coordinating development in Rana catesbeiana tadpoles. Endocrinology. (1997) 138:2989–97. doi: 10.1210/endo.138.7.5272
21. Galton VA. Iodothyronine 5′-deiodinase activity in the amphibian Rana catesbeiana at different stages of the life cycle. Endocrinology. (1988) 122:1746–50.
22. Schreiber AM, Das B, Huang H, Marsh-Armstrong N, Brown DD. Diverse developmental programs of Xenopus laevis metamorphosis are inhibited by a dominant negative thyroid hormone receptor. Proc Natl Acad Sci USA. (2001) 98:10739–44. doi: 10.1073/pnas.191361698
23. Cai L, Brown DD. Expression of type II iodothyronine deiodinase marks the time that a tissue responds to thyroid hormone-induced metamorphosis in Xenopus laevis. Dev Biol. (2004) 266:87–95. doi: 10.1016/j.ydbio.2003.10.005
24. Brown DD, Cai L, Das B, Marsh-Armstrong N, Schreiber A, Juste R. Thyroid hormone controls multiple independent programs required for limb development in Xenopus laevis metamorphosis. Proc Natl Acad Sci USA. (2005) 102:12455–8. doi: 10.1073/pnas.0505989102
25. Buchholz DR, Paul BD, Fu L, Shi Y-B. Molecular and developmental analyses of thyroid hormone receptor function in Xenopus laevis, the African clawed frog. Gen Comp Endocrinol. (2006) 145:1–19. doi: 10.1016/j.ygcen.2005.07.009
26. Nakajima K, Fujimoto K, Yaoita Y. Regulation of thyroid hormone sensitivity by differential expression of the thyroid hormone receptor during Xenopus metamorphosis. Genes Cells. (2012) 17:645–59. doi: 10.1111/j.1365-2443.2012.01614.x
27. Denver RJ. Acceleration of anuran amphibian metamorphosis by corticotropin-releasing hormone-like peptides. Gen Comp Endocrinol. (1993) 91:38–51. doi: 10.1006/gcen.1993.1102
28. Denver RJ. Hormonal correlates of environmentally induced metamorphosis in the western spadefoot toad, Scaphiopus hammondii. Gen Comp Endocrinol. (1998) 110:326–36. doi: 10.1006/gcen.1998.7082
29. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. (2008) 3:1101–8. doi: 10.1038/nprot.2008.73
30. Tata JR, Kawahara A, Baker BS. Prolactin inhibits both thyroid hormone-induced morphogenesis and cell death in cultured amphibian larval tissues. Dev Biol. (1991) 146:72–80. doi: 10.1016/0012-1606(91)90447-B
31. Bonett RM, Hoopfer ED, Denver RJ. Molecular mechanisms of corticosteroid synergy with thyroid hormone during tadpole metamorphosis. Gen Comp Endocrinol. (2010) 168:209–19. doi: 10.1016/j.ygcen.2010.03.014
32. Nieuwkoop P, Faber J. Normal Table of Xenopus laevis (Daudin). New York, NY: Garland Publishing Inc. (1994). Available online at: http://www.xenbase.org/anatomy/alldev.do.
33. Krain LP, Denver RJ. Developmental expression and hormonal regulation of glucocorticoid and thyroid hormone receptors during metamorphosis in Xenopus laevis. J Endocrinol. (2004) 181:91–104. doi: 10.1677/joe.0.1810091
34. Bagamasbad PD, Bonett RM, Sachs L, Buisine N, Raj S, Knoedler JR, et al. Deciphering the regulatory logic of an ancient, ultraconserved nuclear receptor enhancer module. Molecul Endocrinol. (2015) 29:856–72. doi: 10.1210/me.2014-1349
35. Fujikara K, Suzuki S. Thyroxine and thyroglobulin in eggs and embryos of bullfrog. Zool Sci. (1991) 8:1166.
36. Weber GM, Farrar ES, Tom CKF, Grau EG. Changes in whole-body thyroxine and triiodothyronine concentrations and total content during early development and metamorphosis of the toad Bufo marinus. Gen Comp Endocrinol. (1994) 94:62–71. doi: 10.1006/gcen.1994.1060
37. Niinuma TM, Hirano T, Kikuyama S. Changes in tissue concentrations of thyroid hormones in metamorphosing toad larvae. Zool Sci. (1991) 8:345–50.
38. De Groef B, Grommen SVH, Darras VM. Hatching the cleidoic egg: the role of thyroid hormones. Front Endocrinol. (2013) 4:1–10. doi: 10.3389/fendo.2013.00063
39. Brown D. The role of thyroid hormone in zebrafish and axolotl development. Proc Natl Acad Sci USA. (1997) 94:13011–6. doi: 10.1073/pnas.94.24.13011
40. De Jesus EGT, Toledo JD, Simpas MS. Thyroid hormones promote early metamorphosis in grouper (Epinephelus coioides) larvae. Gen Comp Endocrinol. (1998) 112:10–6. doi: 10.1006/gcen.1998.7103
41. Reddy PK, Lam TJ. Role of thyroid hormones in tilapia larvae (Oreochromis mossambicus): 1. Effects of the hormones and an antithyroid drug on yolk absorption, growth and development. Fish Physiol Biochem. (1992) 9:473–85. doi: 10.1007/BF02274228
42. Babu S, Sinha RA, Mohan V, Rao G, Pal A, Pathak A, et al. Effect of hypothyroxinemia on thyroid hormone responsiveness and action during rat postnatal neocortical development. Exp Neurol. (2011) 228:91–8. doi: 10.1016/j.expneurol.2010.12.012
43. Hadj-Sahraoui N, Seugnet I, Ghorbel MT, Demeneix B. Hypothyroidism prolongs mitotic activity in the post-natal mouse brain. Neurosci Lett. (2000) 280:79–82. doi: 10.1016/S0304-3940(00)00768-0
44. Buchholz DR. More similar than you think: Frog metamorphosis as a model of human perinatal endocrinology. Dev Biol. (2015) 408:188–95. doi: 10.1016/j.ydbio.2015.02.018
45. Jennings D.H. (1997). Evolution of Endocrine Control of Development in Direct-Developing Amphibians. Unpublished Ph.D. dissertation. University of Colorado, Boulder.
46. Duarte-Guterman P, Langlois VS, Pauli BD, Trudeau VL. Expression and T3 regulation of thyroid hormone- and sex steroid-related genes during Silurana (Xenopus) tropicalis early development. Gen Comp Endocrinol. (2010) 166:429–35. doi: 10.1016/j.ygcen.2009.12.008
47. Bernal J. Thyroid hormone receptors in brain development and function. Nat Clin Pract Endocrinol Metab. (2007) 3:249–59. doi: 10.1038/ncpendmet0424
48. Havis E, Le Mével S, Morvan Dubois G, Shi D, Scanlan TS, Demeneix B, et al. Unliganded thyroid hormone receptor is essential for Xenopus laevis eye development. EMBO J. (2006) 25:4943–51. doi: 10.1038/sj.emboj.7601356
49. Morvan Dubois G, Sebillot A, Kuiper GJM, Verhoelst CHJ, Darras VM, Visser TH, et al. Deiodinase activity is present in Xenopus laevis during early embryogenesis. Endocrinology. (2006) 147:4941–9. doi: 10.1210/en.2006-0609
50. Terry GS. Effects of the extirpation of the thyroid gland upon ossification in Rana pipiens. J Exp Zool. (1918) 24:567–87. doi: 10.1002/jez.1400240306
51. Allen BM. The results of thyroid removal in the larvae of Rana pipiens. J Exp Zool. (1983) 24:499–519. doi: 10.1002/jez.1400240303
52. Nakajima K, Tazawa I, Yaoita Y. Thyroid hormone receptor α - and β -knockout Xenopus tropicalis tadpoles reveal subtype-specific roles during development. Endocrinology. (2017) 159:733–43. doi: 10.1210/en.2017-00601
53. Ranjan M, Wong J, Shi YB. Transcriptional repression of Xenopus TRβ gene is mediated by a thyroid hormone response element located near the start site. J Biol Chem. (1994) 269:24699–705.
54. Furlow DJ, Kanamori A. The transcription factor basic transcription element-binding protein 1 is a direct thyroid hormone response gene in the frog Xenopus laevis. Endocrinology. (2002) 143:3295–305. doi: 10.1210/en.2002-220126
55. Das B, Heimeier RA, Buchholz DR, Shi Y-B. Identification of direct thyroid hormone response genes reveals the earliest gene regulation programs during frog metamorphosis. J Biol Chem. (2009) 284:34167–78. doi: 10.1074/jbc.M109.066084
56. Helbing CC, Werry K, Crump D, Domanski D, Veldhoen N, Bailey CM. Expression profiles of novel thyroid hormone-responsive genes and proteins in the tail of Xenopus laevis tadpoles undergoing precocious metamorphosis. Molecul Endocrinol. (2003) 17:1395–409. doi: 10.1210/me.2002-0274
57. Yaoita Y, Brown DD. A correlation of thyroid hormone receptor gene expression with amphibian metamorphosis. Genes Dev. (1990) 4:1917–24. doi: 10.1101/gad.4.11.1917
58. Wen L, Shi YB. Unliganded thyroid hormone receptor α controls developmental timing in Xenopus tropicalis. Endocrinology. (2015) 156:721–34. doi: 10.1210/en.2014-1439
59. Choi J, Suzuki KT, Sakuma T, Shewade L, Yamamoto T, Buchholz D. Unliganded thyroid hormone receptor α regulates developmental timing via gene repression in Xenopus tropicalis. Endocrinology. (2015) 156:735–44. doi: 10.1210/en.2014-1554
60. Buchholz DR, Shi Y. Dual function model revised by thyroid hormone receptor alpha knockout frogs. Gen Comp Endocrinol. (2018) 265:214–8. doi: 10.1016/j.ygcen.2018.04.020
61. Denver RJ. The molecular basis of thyroid hormone-dependent central nervous system remodeling during amphibian metamorphosis. Compar Biochem Physiol Part C, Pharmacol Toxicol Endocrinol. (1998) 119:219–28. doi: 10.1016/S0742-8413(98)00011-5
62. Denver RJ, Hu F, Scanlan TS, Furlow DJ. Thyroid hormone receptor subtype specificity for hormone-dependent neurogenesis in Xenopus laevis. Dev Biol. (2009) 326:155–68. doi: 10.1016/j.ydbio.2008.11.005
63. Choi J, Ishizuya-Oka A, Buchholz DR. Growth, development, and intestinal remodeling occurs in the absence of thyroid hormone receptor a in tadpoles of Xenopus tropicalis. Endocrinology. (2017) 158:1623–33. doi: 10.1210/en.2016-1955
64. Tata JR. Autoinduction of nuclear hormone receptors during metamorphosis and its significance. Insect Biochem Mol Biol. (2000) 30:645–51. doi: 10.1016/S0965-1748(00)00035-7
65. Buckbinder L, Brown DD. Thyroid hormone-induced gene expression changes in the developing frog limb. J Biol Chem. (1992) 267:25786–91.
Keywords: embryo, direct development, thyroid hormone, amphibians, evolution, metamorphosis, maternal effects, life history
Citation: Laslo M, Denver RJ and Hanken J (2019) Evolutionary Conservation of Thyroid Hormone Receptor and Deiodinase Expression Dynamics in ovo in a Direct-Developing Frog, Eleutherodactylus coqui. Front. Endocrinol. 10:307. doi: 10.3389/fendo.2019.00307
Received: 05 February 2019; Accepted: 29 April 2019;
Published: 24 May 2019.
Edited by:
Marco António Campinho, Center of Marine Sciences (CCMAR), PortugalReviewed by:
Paula Duarte-Guterman, University of British Columbia, CanadaSalvatore Benvenga, University of Messina, Italy
Copyright © 2019 Laslo, Denver and Hanken. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mara Laslo, mlaslo@g.harvard.edu
 Mara Laslo
Mara Laslo Robert J. Denver
Robert J. Denver James Hanken
James Hanken